Abstract
Background & Aims
Cholangiocarcinoma (CCA), heterogeneous biliary tumours with dismal prognosis, lacks accurate early diagnostic methods especially important for individuals at high-risk (i.e. those with primary sclerosing cholangitis [PSC]). Here, we searched for protein biomarkers in serum extracellular vesicles (EVs).
Methods
EVs from patients with isolated PSC (n = 45), concomitant PSC-CCA (n = 44), PSC who developed CCA during follow-up (PSC to CCA; n = 25), CCAs from non-PSC aetiology (n = 56), and hepatocellular carcinoma (n = 34) and healthy individuals (n = 56) were characterised by mass spectrometry. Diagnostic biomarkers for PSC-CCA, non-PSC CCA, or CCAs regardless of aetiology (Pan-CCAs) were defined and validated by ELISA. Their expression was evaluated in CCA tumours at a single-cell level. Prognostic EV biomarkers for CCA were investigated.
Results
High-throughput proteomics of EVs identified diagnostic biomarkers for PSC-CCA, non-PSC CCA, or Pan-CCA, and for the differential diagnosis of intrahepatic CCA and hepatocellular carcinoma, which were cross-validated by ELISA using total serum. Machine learning-based algorithms disclosed CRP/FIBRINOGEN/FRIL for the diagnosis of PSC-CCA (local disease [LD]) vs. isolated PSC (AUC = 0.947; odds ratio [OR] =36.9) and, combined with carbohydrate antigen 19-9, overpowers carbohydrate antigen 19-9 alone. CRP/PIGR/VWF allowed the diagnosis of LD non-PSC CCAs vs. healthy individuals (AUC = 0.992; OR = 387.5). It is noteworthy that CRP/FRIL accurately diagnosed LD Pan-CCA (AUC = 0.941; OR = 89.4). Levels of CRP/FIBRINOGEN/FRIL/PIGR showed predictive capacity for CCA development in PSC before clinical evidence of malignancy. Multi-organ transcriptomic analysis revealed that serum EV biomarkers were mostly expressed in hepatobiliary tissues, and single-cell RNA sequencing and immunofluorescence analysis of CCA tumours showed their presence mainly in malignant cholangiocytes. Multivariable analysis unveiled EV prognostic biomarkers, with COMP/GNAI2/CFAI and ACTN1/MYCT1/PF4V associated negatively and positively with patients’ survival, respectively.
Conclusions
Serum EVs contain protein biomarkers for the prediction, early diagnosis, and prognostication of CCA that are detectable using total serum, representing a tumour cell-derived liquid biopsy tool for personalised medicine.
Impact and implications
The accuracy of current imaging tests and circulating tumour biomarkers for cholangiocarcinoma (CCA) diagnosis is far from satisfactory. Most CCAs are considered sporadic, although up to 20% of patients with primary sclerosing cholangitis (PSC) develop CCA during their lifetime, constituting a major cause of PSC-related death. This international study has proposed protein-based and aetiology-related logistic models with predictive, diagnostic, or prognostic capacities by combining two to four circulating protein biomarkers, moving a step forward into personalised medicine. These novel liquid biopsy tools may allow the (i) easy and non-invasive diagnosis of sporadic CCAs, (ii) identification of patients with PSC with higher risk for CCA development, (iii) establishment of cost-effective surveillance programmes for the early detection of CCA in high-risk populations (e.g. PSC), and (iv) prognostic stratification of patients with CCA, which, altogether, may increase the number of cases eligible for potentially curative options or to receive more successful treatments, decreasing CCA-related mortality.
Keywords: Cholangiocarcinoma, Primary sclerosing cholangitis, Protein biomarkers, Extracellular vesicles, Liquid biopsy, Mass spectrometry, Single-cell RNA-sequencing
Graphical abstract

Highlights
-
•
Circulating EV-proteins allow for CCA risk prediction, early and differential (vs. HCC) diagnosis, and prognostication.
-
•
Serum CRP/FIBRINOGEN/FRIL levels discriminated patients with early-stage PSC-CCA from those with PSC.
-
•
Most pan-CCA biomarkers are mainly expressed in malignant cholangiocytes within human CCA tumours.
-
•
Serum CRP/FIBRINOGEN/FRIL/PIGR levels predict CCA development in patients with PSC before radiological tumour evidence.
-
•
The abundance of EV-proteins COMP/GNAI2/CFAI and ACTN1/MYCT1/PF4V independently predict patients’ overall survival.
Introduction
Cholangiocarcinoma (CCA) includes a heterogeneous group of malignant tumours that can emerge at any location of the biliary system. According to the anatomical origin, they are classified as intrahepatic (iCCA), perihilar (pCCA), or distal (dCCA).1,2 Although CCA is still considered a rare type of cancer, its global incidence and related mortality rates have been alarmingly increasing in the last decades.1 Moreover, the silent growth of these tumours strongly compromises their early detection, limiting patients’ accessibility to potentially curative options (i.e. tumour resection).1,3
CCA diagnosis requires imaging methods and further cytological/histological confirmation.3,4 The cancer biomarker carbohydrate antigen 19-9 (CA19-9) is the only liquid-biopsy tool currently used in clinics to help in CCA diagnosis, but its diagnostic power is low, specially at early CCA stages. The suboptimal accuracy of current non-invasive diagnostic approaches reflects the need for cytological/histological confirmation. Nevertheless, tumour biopsy or brushing is sometimes discouraged owing to patients’ fragility and advanced disease stages, risk of bleeding and peritoneal seeding, and/or the low amount of tissue collected that may not be sufficient for cytological/histological confirmation, especially in pCCA and dCCA cases.5
Most CCAs are considered sporadic and lack clear aetiology, although some well-established conditions significantly increase the odds of CCA development, including the presence of choledochal cysts, biliary stones, cirrhosis, certain viruses, or biliary diseases (e.g. primary sclerosing cholangitis [PSC]).1,6 In particular, PSC is a chronic, cholestatic, and immune-mediated liver disease of unknown aetiology, characterised by liver cell death, fibrosis, and hepatic failure. PSC confers a substantial risk for CCA development (up to 20% lifetime risk), resulting in premature death.1,2,7 Early diagnosis of CCA in patients with PSC using non-invasive methods is challenging, as there are overlapping radiological features between benign and malignant biliary strictures. All these pieces of evidence highlight the need of accurate non-invasive biomarkers for CCAs as a way to establish surveillance programmes for their early detection in high-risk populations and also to provide a faster diagnosis of sporadic CCAs, ultimately decreasing cancer-related mortality.
Extracellular vesicles (EVs), which are nanometre lipid-bilayered spheres, have arisen as a promising source of biomarkers for human diseases.8 These vesicles, released from cells and found in biofluids, contain distinct types of biomolecules (e.g. proteins, nucleic acids, lipids, and metabolites) and participate in cell-to-cell communication, being useful tools for biomarker discovery.9 In this study, we aimed to characterise the proteomic profile of serum EVs from individuals with PSC-associated CCA (PSC-CCA), CCAs from non-PSC aetiology, PSC who developed CCA during follow-up (PSC to CCA), isolated PSC, and hepatocellular carcinoma (HCC) and healthy individuals as a way to identify accurate biomarkers to predict CCA development, to early diagnose CCA, and to estimate prognosis of patients with CCA.
Materials and methods
Study population
Serum from individuals with (i) isolated PSC (n = 45), (ii) concomitant PSC and CCA (PSC-CCA; n = 44), (iii) PSC who developed CCA during follow-up (PSC to CCA; n = 25), (iv) CCAs from non-PSC aetiology (n = 56), and (v) HCC (n = 34) and (vi) healthy individuals (n = 56) were obtained from Donostia University Hospital (San Sebastian, Spain), Oslo University Hospital Rikshospitalet (Oslo, Norway), Medical University Hospital of Warsaw (Warsaw, Poland), University Medical Center Hamburg-Eppendorf (Hamburg, Germany), Heidelberg University Hospital (Heidelberg, Germany), The Christie NHS Foundation Trust (Manchester, UK), and Salamanca University Hospital (Salamanca, Spain). A detailed description of the study population is provided in the Supplementary information.
Isolation and characterisation of EVs
Serum EV isolation and characterisation are described in the Supplementary information.
High-throughput proteomics
Proteomic analysis of serum EVs was performed on a nanoACQUITY UPLC System connected to an LTQ Orbitrap XL for the ‘Orbitrap cohort’ and on a timsTOF Pro with PASEF for the ‘timsTOF cohort’. Peptide quantity normalisation and batch effect correction were accomplished with the proBatch R package. Details are outlined in the Supplementary information.
Diagnostic biomarker selection
A schematic flowchart of the strategy followed for diagnostic biomarker identification is displayed in Fig. S1. Serum EV protein biomarkers were classified into whether they are specific for PSC-CCA, non-PSC CCA, or Pan-CCA, according to the criteria detailed in Fig. S2 and in Supplementary information.
Expression analysis of protein biomarkers in human tissues
To evaluate the potential origin of serum EV protein biomarkers, their expression was analysed in human tissues bulk RNA datasets, in a healthy liver single-cell RNA-sequencing (scRNA-seq) dataset, in scRNA-seq data from patients with CCA, and in formalin-fixed, paraffin-embedded (FFPE) human tumour samples as described in the Supplementary information.
Statistical analysis
Statistical analyses were performed as indicated in the Supplementary information.
Results
Differential abundance of EV proteins in patients with PSC-CCA, patients with PSC, and healthy individuals revealed candidate biomarkers to diagnose CCA
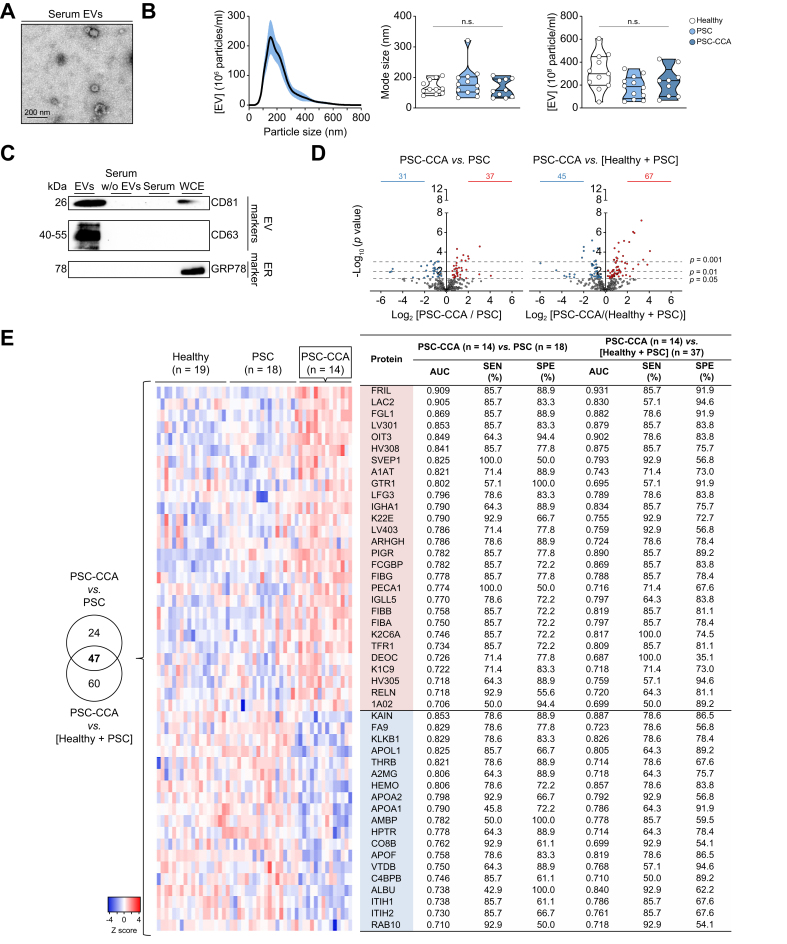
Serum EVs isolated from patients with PSC-CCA, patients with isolated PSC, or healthy individuals were characterised by transmission electron microscopy (TEM), nanoparticle tracking analysis (NTA), and immunoblotting. TEM images confirmed the prevalence of classic cup-shaped, rounded morphology vesicles with an average size smaller than 200 nm (Fig. 1A). Particles analysed by NTA showed similar size and concentration in all groups, being particularly enriched in 170 nm in diameter (mode size), revealing exosomes and/or small and medium-size microvesicles as the principal components of the isolated EV fraction (Fig. 1B). By immunoblotting, EV protein markers tetraspanins CD63 and CD81 were highly enriched in the isolated EV fraction when compared with total serum, EV-depleted serum (serum without EVs), and whole-cell extract (WCE) from normal human cholangiocytes, whereas the endoplasmic reticulum marker GRP78 was absent in isolated EVs, substantiating the proper isolation and high purity of the obtained EV fraction (Fig. 1C).
Fig. 1.
Isolated serum EV fractions are enriched in exosomes and microvesicles, and contain proteins with diagnostic capacity for PSC-CCA.
Characterisation of serum EVs by (A) TEM, (B) NTA, and (C) immunoblotting. (D) Volcano plot of identified proteins in PSC-CCA vs. PSC and in PSC-CCA vs a ‘non-malignant control group’ including healthy individuals and patients with PSC. Significantly enriched proteins are coloured in red and proteins with lower abundance in blue (parametric Student's t test). (E) Venn diagram with the number of proteins with significant AUC values in both comparatives. Heatmap and diagnostic values of EV proteins altered in patients with concomitant PSC-CCA compared with PSC and to ‘the non-malignant control group’. Enriched proteins are coloured in red, and proteins with lower abundance in blue. n.s. (Kruskal–Wallis test). CCA, cholangiocarcinoma; ER, endoplasmic reticulum; EV, extracellular vesicle; NTA, nanoparticle tracking analysis; PSC-CCA, concomitant PSC and CCA; PSC, primary sclerosing cholangitis; SEN, sensitivity; SPE, specificity; TEM, transmission electron microscopy; WCE, whole cell extract.
The proteomic profile of EVs was then characterised by mass spectrometry (MS) (‘Orbitrap MS cohort’). Univariable analysis revealed distinct EV protein profiles when comparing individuals with PSC-CCA, individuals with isolated PSC, or healthy individuals. Compared with PSC, the abundance of 68 proteins was altered in PSC-CCA (37 up, 31 down). When comparing patients with PSC-CCA with a group composed of both healthy individuals and patients with PSC (‘non-malignant control group’), the abundance of 67 proteins was found increased and 45 decreased in PSC-CCA (Fig. 1D). Next, the diagnostic capacity of single candidate biomarkers was investigated. Forty-seven EV proteins showed significant AUC values for CCA diagnosis, with FRIL providing the highest score when compared with that in individuals with isolated PSC (AUC = 0.909) or with that in the non-malignant control group (AUC = 0.931) (Fig. 1E).
Serum EVs contain protein biomarkers for the early, accurate, and aetiology-based diagnosis of CCAs
To test whether these candidate CCA biomarkers are specific for patients with PSC or have a common diagnostic ability also for CCAs of non-PSC aetiology, previous samples and new additional ones from individuals with PSC, PSC-CCA, or non-PSC CCA or healthy individuals were analysed in a newer and more powerful mass spectrometer (‘timsTOF cohort’). All EV proteins commonly identified in timsTOF analysis, in Orbitrap MS, and/or in a non-PSC CCA dataset previously published by our group10 were selected for subsequent analysis and identification of aetiology-related CCA protein biomarkers (Fig. S2). In particular, five proteins specifically allowed the differential diagnosis of patients with PSC-CCA vs. patients with isolated PSC, harbouring LG3BP, IGLL5, and IGKC the highest diagnostic capacity (AUC = 0.710, 0.687, and 0.654, respectively) (Fig. 2A). By contrast, the abundance of seven proteins allowed the specific diagnosis of non-PSC CCAs, providing AUC values up to 0.854 (for TSN9) when compared with that in healthy controls (Fig. 2B). Considering patients with CCA regardless of aetiology (Pan-CCA), the abundance of 63 EV proteins was altered compared with that in the non-malignant control group, observing that most biomarkers were general for all CCAs (Fig. 2C). Among them, FIBG, HEMO, and PON1 stood out as the best individual diagnostic biomarkers, displaying AUC values of 0.823, 0.795, and 0.777, respectively. Importantly, most candidate biomarkers retained their diagnostic accuracy when considering only patients with local disease (LD) (Fig. 2A–C). Notably, a subanalysis including only samples not previously analysed by Orbitrap MS revealed similar sensitivity and specificity values (Fig. S3).
Fig. 2.
Serum EV protein biomarkers for the diagnosis of CCA according to tumour aetiology.
Biomarkers for the specific diagnosis of (A) CCA in patients with PSC (specific PSC-CCA biomarkers), (B) CCA in patients without PSC (non-PSC CCA biomarkers), and (C) CCA regardless of aetiology (Pan-CCA biomarkers; the best 53 biomarkers are displayed). Enriched proteins are coloured in red, and proteins with lower abundance in blue. ‘–’, n.s. AUC. CCA, cholangiocarcinoma; EV, extracellular vesicle; LD, local disease; PSC-CCA, concomitant PSC-CCA; PSC, primary sclerosing cholangitis; SEN, sensitivity; SPE, specificity.
Pan-CCA EV biomarkers are detected in total serum by immunoblotting and ELISA, aiding diagnosis
We next evaluated whether the Pan-CCA EV biomarkers are merely detected within serum EVs or whether they are amenable to be measured using total serum. By immunoblotting, a selection of Pan-CCA biomarkers (e.g. VWF, PIGR, FIBG, FIBB, FGL1, CRP, FRIL, and OIT3) was analysed in isolated EVs, EV-depleted serum (serum without EVs), and total serum from patients with CCA and healthy individuals. Almost all analysed proteins were enriched in isolated EVs, but, importantly, they were detectable using whole serum (Fig. 3A and B). To ascertain the potential translation of these biomarkers into clinics, total serum from previous patients and additional ones was used to measure the levels of these proteins by ELISA, which is a technique already implemented in daily clinical practice. Of note, the levels of most candidate biomarkers measured in total serum by ELISA correlated with the ones measured in serum EVs by MS (Fig. S4), reinforcing the usefulness of this technique as a translational approach. Although no differences were observed between patients with PSC and healthy individuals, total serum levels of FRIL, CRP, FGL1, VWF, FIBRINOGEN, and OIT3 were increased in patients with PSC-CCA and non-PSC CCA when compared with the non-malignant control group, displaying great accuracy for Pan-CCA diagnosis (AUC values up to 0.886; Fig. 3C). PIGR serum levels were found augmented in patients with isolated PSC, when compared with healthy individuals, but it also allowed the diagnosis of PSC-CCA and non-PSC CCA when compared with their clinically relevant controls (patients with PSC and healthy individuals, respectively). Notably, increased CRP and FRIL levels were linked to the greatest risk of CCA occurrence in patients with PSC, with odds ratio (OR) values of 18.1 (95% CI 4.9–67.5) and 17.3 (95% CI 4.7–64.2), respectively. Similarly, increased PIGR and CRP levels provided the highest CCA diagnostic accuracies when the absence of PSC is clinically confirmed, with AUC values of 0.953 and 0.934, respectively (Fig. 3C). A subanalysis considering only samples that were not previously analysed by MS confirmed the diagnostic powerfulness of these biomarkers, validating the results (Fig. S5). The high diagnostic capacity of these proteins was also observed when considering only patients with LD, thus confirming their value for early CCA diagnosis (Fig. S6).
Fig. 3.
Candidate EV protein biomarkers are detected using total serum and aid the diagnosis of CCA.
(A) Representative immunoblots of selected biomarkers in serum EVs, serum without EVs, and total serum subfractions (10 μg protein loaded per column) of patients with CCA and healthy individuals. (B) Representative immunoblots of selected biomarkers in total serum (50 μg protein loaded per column) of patients with iCCA, pCCA, or dCCA, and healthy individuals. (C) Levels of CRP, FRIL, FGL1, VWF, PIGR, FIBRINOGEN, and OIT3 measured by ELISA in serum samples from patients with PSC, PSC-CCA, and non-PSC CCA and healthy individuals and diagnostic values for Pan-CCA vs. the non-malignant group (healthy + PSC), PSC-CCA vs. PSC, and CCA vs. healthy individuals. n.s.; ∗p <0.05; ∗∗p <0.01; ∗∗∗∗p <0.0001 (Kruskal–Wallis test). AI, accuracy index; CCA, cholangiocarcinoma; dCCA, distal CCA; EV, extracellular vesicle; iCCA, intrahepatic CCA; NPV, negative predictive value; OR, odds ratio; Pan-CCA, CCA regardless of aetiology; pCCA, perihiliar CCA; PPV, positive predictive value; PSC-CCA, concomitant PSC and CCA; PSC, primary sclerosing cholangitis; ROC, receiver operating characteristic; SEN, sensitivity; SPE, specificity.
We next explored potential associations of these candidate biomarkers with CCA subtypes, disease status, and serum CA19-9 levels. The levels of most individual candidate biomarkers were independent of CCA subtype and disease status. Still, PIGR serum levels were higher in patients with pCCA or dCCA than in those with iCCA, although patients with iCCA had increased levels in comparison with their respective controls (Fig. S7). Considering disease status, patients with metastatic disease (MD) displayed increased FGL1, when compared with patients with LD or locally advanced disease (LAD), although elevated FGL1 serum levels were still observed in patients with LD and LAD compared with both healthy individuals or patients with PSC (Fig. S8). Serum levels of VWF, FIBRINOGEN, and OIT3 positively correlated with serum CA19-9 levels (Fig. S9A), whereas the presence of cirrhosis did not alter biomarker levels (Fig. S9B).
To enhance the diagnostic capacity, machine learning-based binary logistic models (LMs) were performed with 70% of the samples (training), and the generated model was further evaluated in the resulting 30% (testing 30%) and in a subset including only samples with LD-CCA (testing LD). A model combining CRP/FRIL allowed the identification of CCA independently of disease aetiology with high accuracy (AUC = 0.923, 0.905, and 0.941 in the training, testing 30%, and testing LD cohorts, respectively; Fig. 4A). If PSC was present, the combination of CRP/FIBRINOGEN/FRIL (LM1) or CRP/FRIL (LM2) allowed the diagnosis of PSC-CCA with great accuracy (AUC = 0.856 [training] and 0.907 [testing 30%] for LM1 and AUC = 0.847 [training] and 0.901 [testing 30%] for LM2), particularly in LD-CCA (AUC = 0.947 for LM1 and 0.925 for LM2) (Fig. 4B). It is noteworthy that although most of the single biomarkers were not significantly superior to CA19-9 (Fig. S9C), the combination of the LMs with CA19-9 levels significantly overpowered the diagnostic capacity of CA19-9 alone, particularly at LD-CCA (AUC = 0.960 for LM1 and 0.965 for LM2 vs. 0.735 for CA19-9; Fig. S10). Finally, when the absence of PSC is clinically confirmed, an LM combining CRP/PIGR/VWF provided almost the maximum diagnostic capacity (AUC = 0.994 and 1.000 in the training and testing 30% cohorts, respectively) for CCA compared with that for healthy individuals, also at early tumour stages (AUC = 0.992 for LD-CCA). A model combining only CRP/VWF might also help in the diagnosis of CCAs arising from non-PSC aetiologies, with AUC values of 0.979 and 0.960 in the training and testing 30% cohorts, respectively, retaining its diagnostic capacity for the early diagnosis of CCA (AUC = 0.968; Fig. 4C).
Fig. 4.
LMs combining ELISA-validated serum protein biomarkers enable the accurate diagnosis of CCA in patients with or without PSC.
Binary logistic regression models in the training (70%) cohort, as well as in the testing 30% and LD cohorts for CCA diagnosis (A) regardless of disease aetiology, (B) in patients with PSC, and (C) in patients without PSC. AI, accuracy index; CCA, cholangiocarcinoma; LD, local disease; LM, logistic model; OR, odds ratio; Pan-CCA, CCA regardless of aetiology; PSC, primary sclerosing cholangitis; ROC, receiver operating characteristic; SEN, sensitivity; SPE, specificity.
Serum Pan-CCA biomarkers aid the differential diagnosis of iCCA and HCC
Serum levels of FGL1, CRP, PIGR, FIBRINOGEN, VWF, FRIL, or OIT3 measured by ELISA were found increased in patients with iCCA compared with patients with HCC, with accuracy indexes up to 86.2%. It is noteworthy that increased serum levels of FGL1 and CRP were linked to the greatest risk of iCCA presence, with ORs of 67.7 (95% CI 3.3–1,397.0) and 42.6 (95% CI 11.8–154), respectively, in comparison with HCC, and the combination of these two biomarkers increased the odds to 144 (95% CI 8–2,590) (Fig. 5A).
Fig. 5.
Serum EV protein biomarkers for the differential diagnosis of iCCA vs. HCC.
(A) Levels and diagnostic values of FGL1, CRP, PIGR, FIBRINOGEN, VWF, FRIL, and OIT3 measured by ELISA in serum samples from patients with iCCA and HCC. Heatmaps, Venn diagrams, and diagnostic values of specific EV proteins for the diagnosis of (B) iCCA and (C) HCC. Enriched proteins are coloured in red, and proteins with lower abundance in blue. ∗p <0.05; ∗∗p <0.01; ∗∗∗p <0.001; ∗∗∗∗p <0.0001 (Mann–Whitney test). AI, accuracy index; EV, extracellular vesicle; HCC, hepatocellular carcinoma; iCCA, intrahepatic cholangiocarcinoma; NPV, negative predictive value; PPV, positive predictive value; PSC, primary sclerosing cholangitis; ROC, receiver operating characteristic; SEN, sensitivity; SPE, specificity.
To identify additional biomarkers for the differential diagnosis of iCCA and HCC, the proteomic profile of serum EVs from patients with HCC, iCCA, and PSC and healthy individuals was evaluated. Thirteen specific iCCA candidate biomarkers (six up and seven down) were obtained, with APOC3, ALBU, and AMPN providing the highest AUC values (0.736, 0.732, and 0.726, respectively; Fig. 5B). In resemblance, 58 HCC-specific EV protein biomarkers were identified, with 30 exhibiting an increased abundance in serum EVs from patients with HCC, compared with patients with iCCA and healthy individuals, whereas 28 of them displayed decreased levels (Fig. 5C). Importantly, ANGP1 provided the highest diagnostic capacity in discriminating patients with HCC and iCCA, with AUC values of 0.850.
EV protein biomarkers are mainly expressed in liver and differ across specific cell populations
To decipher the potential origin of candidate serum EV protein biomarkers for CCA, the expression of the ELISA-validated biomarkers was first analysed in a human multi-organ (n = 61 tissues/organs) transcriptomic dataset (Fig. 6A and Fig. S11A). Data revealed that all these EV protein biomarkers are expressed in hepatobiliary tissues. Interestingly, some EV biomarkers –including FGA, FGG, FGB, CRP, FGL1, and OIT3 – were almost exclusively expressed in hepatobiliary tissues, representing more than 70% of the total expression detected in human organs.
Fig. 6.
Human multi-organ transcriptome and scRNA-seq reveal the potential origin of serum EV protein biomarkers.
(A) Normalised expression of candidate serum biomarkers in 61 human tissues/organs from the Consensus dataset of the Human Protein Atlas. (B) tSNE plot and normalised expression of candidate biomarkers in each liver cell type from normal liver scRNA-seq (GSE115469). (C) tSNE plot and cell type proportion from 12 iCCA tumours (GSE151530). Biomarker expressing-positive cells and relative biomarker expression within the cell types of iCCA tumours. (D) Representative IF images of PIGR, FIBG, CRP, and FGL1 and colocalisation with CK19+-positive cells. Scale bars = 50 μm ∗∗∗∗p <0.0001 (Kruskal–Wallis test). CCA, cholangiocarcinoma; CK19, cytokeratin 19; EV, extracellular vesicle; HCC, hepatocellular carcinoma; iCCA, intrahepatic CCA; IF, immunofluorescence; NC, normalised counts; NK, natural killer; PSC-CCA, concomitant PSC and CCA; PSC, primary sclerosing cholangitis; RNA-seq, RNA sequencing; ROC, receiver operating characteristic; sCRNA-seq, single-cell RNA sequencing; SL, surrounding liver; tSNE, t-distributed stochastic neighbour embedding.
Human liver scRNA-seq showed that the expression of some candidate biomarkers was cell type-specific, whereas others were indiscriminately expressed along different liver cell types (Fig. 6B). For instance, PIGR was almost exclusively expressed in cholangiocytes, whereas endothelial cells had the highest expression of OIT3 and VWF. In addition, the main source of FGA, FGG, FGB, CRP, and FGL1 was hepatocytes, whereas the expression of FTL was more diverse, being detected in all liver cell types.
ScRNA-seq data reveals specific cell types within human CCA tumours expressing EV protein biomarkers
We next investigated the expression of the aforementioned Pan-CCA biomarkers in the cell populations within human CCA tumours (Fig. 6C and Fig. S12). When analysing the single-cell transcriptome of the GSE151530 and GSE125449 datasets, composed of CCA tumour samples from 12 and 10 patients, respectively, the highest percentage of cells expressing FGA, FGG, FGB, CRP, FGL1, and PIGR transcripts were malignant cholangiocytes, whereas VWF- and OIT3-expressing cells were principally endothelial cells. Meanwhile, the expression of FTL was more widespread, being detected in all the main tumour cells, with a predominant increase in T cells and tumour cholangiocytes. We then validated these findings by immunofluorescence and found that PIGR, FIBG, CRP, and FGL1 were strongly detected in both human PSC-CCA and non-PSC CCA tissues, mostly co-localising with cytokeratin 19 (CK19)+ cells, thus confirming their expression specifically in malignant cholangiocytes within tumours, regardless of disease aetiology. In contrast, tissue specimens from patients with PSC or surrounding livers displayed almost negligible signals (Fig. 6D).
Serum EV proteins allowed the prediction of CCA development in patients with PSC before clinical evidence of malignancy
The abundance of several proteins (18 up and 14 down) was altered in serum EVs from patients with PSC without clinical evidence of malignancy at sampling but who progressed to CCA during follow-up (PSC to CCA) compared with patients with PSC who did not develop CCA in more than 5 years after PSC diagnosis, providing AUC values up to 0.868. These values remained almost unaltered when considering only patients with samples obtained more than 6 months before CCA diagnosis (Fig. 7A). In fact, when stratifying patients with PSC to CCA according to the period between sampling and CCA diagnosis (<6 months, 6–12 months, and >12 months), no differences were observed in their abundance, indicating that these alterations might be detected in patients with PSC more than 1 year before clinical evidence of the tumour (Fig. S13).
Fig. 7.
Serum proteins allow the prediction of CCA development in patients with PSC.
(A) Heatmap and diagnostic values of specific EV proteins for the differential identification of patients with PSC who progressed to CCA over time (PSC to CCA) and non-malignant PSC. Enriched proteins in red, and proteins with lower abundance in blue. (B) Levels and diagnostic values of FIBRINOGEN, CRP, PIGR, and FRIL in total serum from patients with PSC to CCA, PSC-CCA, and non-malignant PSC. (C) Binary logistic regression models for the prediction of CCA development in patients with PSC. ‘–’, n.s. AUC; ∗p <0.05; ∗∗∗p <0.001; ∗∗∗∗p <0.0001 (Kruskal–Wallis test). m, months from sampling to CCA diagnosis; AI, accuracy index; CCA, cholangiocarcinoma; EV, extracellular vesicle; iCCA, intrahepatic CCA; LM, logistic model; NPV, negative predictive value; OR, odds ratio; PPV, positive predictive value; PSC-CCA, concomitant PSC and CCA; PSC, primary sclerosing cholangitis; ROC, receiver operating characteristic; SEN, sensitivity; SPE, specificity; CA19-9, carbohydrate antigen 19-9.
Considering that OIT3, PIGR, FRIL, CRP, and FIBG may also have predictive value for CCA development in patients with PSC, their serum levels were next analysed by ELISA. Although no differences were observed for OIT3 (data not shown), the levels of FIBRINOGEN, CRP, PIGR, and FRIL were already found increased in serum from the PSC to CCA group, when compared with patients with non-malignant PSC, providing differential AUC values up to 0.828 (Fig. 7B). In addition, the serum levels of these biomarkers were similar to the ones detected in patients with confirmed PSC-CCA. At the individual level, the comparison of the predictive capacity of the four ELISA-validated serum candidate biomarkers with CA19-9, total bilirubin, aspartate transaminase (AST), alanine transaminase (ALT), gamma glutamyltransferase (GGT), and alkaline phosphatase (ALP) for the development of CCA in patients with PSC revealed that FIBRINOGEN was significantly superior to almost all the above-mentioned parameters (apart from bilirubin). CRP, FRIL, and PIGR significantly outperformed AST, ALT, or GGT (Fig. S14A). Importantly, all patients with PSC showcasing higher levels in these four predictive biomarkers developed CCA during follow-up, whereas patients without elevation in any of these biomarkers did not progress to CCA. A logistic regression model including CRP/FIBRINOGEN/FRIL showed that patients with PSC displaying increased levels in these three proteins have 44-fold augmented risk of developing CCA during follow-up, when compared with patients with non-malignant PSC, providing positive and negative predictive values of 91.7% and 80%, respectively. In contrast, increased serum CA19-9 levels in patients with PSC were not indicative of a greater risk for CCA development (OR 2.2, 95% CI 0.7–7.1) (Fig. 7C). Of note, CRP/FIBRINOGEN/FRIL and CRP/FIBRINOGEN/FRIL/PIGR LMs significantly overpowered not only the common markers of cholestatic liver injury (ALT, AST, total bilirubin, GGT, and ALP) but also CA19-9 (Fig. S14B).
Serum EVs hold proteins capable of estimating the prognosis of patients with CCA
Univariable analysis revealed that 108 EV proteins may also estimate survival in patients with CCA, regardless of disease aetiology (Fig. 8A). Multivariable analysis of prognostic biomarker candidates that were independent of clinical and demographic variables (Fig. S15A and B) revealed that high levels of COMP, GNAI2, A1AG1, CFAI, and LRP1 are independent predictors of worse overall survival (OS), whereas increased abundance of MYCT1, ACTN1, KPYM, and PF4V predict better prognosis (Fig. 8B).
Fig. 8.
Association of serum EV protein levels with patients’ outcome.
(A) Schematic representation of the strategy used to define prognostic biomarkers. (B) Multivariable analysis of serum EV proteins with prognostic value independent of the clinical/demographic variables sex, age, PSC, cirrhosis, CCA subtype, disease status, CA19-9, and surgical intervention. Kaplan–Meier curves of OS for each prognostic biomarker in 10 years of follow-up. (C) Kaplan–Meier curve, Cox regression analysis, and log-rank test of patients with CCA according to the ‘bad prognostic’ (COMP/GNAI2/CFAI) and ‘good prognostic’ (ACTN1/MYCT1/PF4V) panels. AI, accuracy index; CA19-9, carbohydrate antigen 19-9; CCA, cholangiocarcinoma; dCCA, distal CCA; EV, extracellular vesicle; HR, hazard ratio; iCCA, intrahepatic CCA; LAD, locally advanced disease; LD, local disease; LM, logistic model; MD, metastatic disease; mOS, median overall survival; NPV, negative predictive value; OR, odds ratio; OS, overall survival; pCCA, perihiliar CCA; PPV, positive predictive value; PSC-CCA, concomitant PSC and CCA; PSC, primary sclerosing cholangitis; ROC, receiver operating characteristic; SEN, sensitivity; SPE, specificity.
In the multi-organ RNA-sequencing (RNA-seq) data, all the nine genes were found to be expressed in hepatobiliary tissues, with ORM1 (coding for A1AG1) being almost exclusively present in the liver (Fig. S16A). At a cellular level, in healthy livers, some genes were predominantly detected in a cell type-specific manner (i.e. PF4V1 in cholangiocytes and MYCT1 in endothelial cells; Fig. S16B). Similar findings were observed in human CCA tumours, with malignant cholangiocytes being the main cell type expressing not only PF4V1 but also ORM1 and CFI. However, although MYCT1 was almost exclusively found in endothelial cells, the expression of COMP, GNAI2, LRP1, ACTN1, and PKM (coding for KPYM) was ubiquitous in all cell types within human CCA tumours (Fig. S17).
Lastly, a panel comprising three ‘bad prognostic biomarkers’ (COMP/GNAI2/CFAI) showed the finest OS predictive capacity (Fig. 8C). The median OS (mOS) of patients without elevations on any of these biomarkers was 102 months. In contrast, higher levels of two to three of these biomarkers were associated with shorter mOS (5 months). Remarkably, the risk of death of these patients was increased by 30 times (hazard ratio 30.4, 95% CI 4.0–230.0) when compared with patients with low levels of these three proteins. Similarly, a panel including three ‘good prognostic biomarkers’ (ACTN1/MYCT1/PF4V) showed the finest OS prediction in patients with CCA. Patients with increased serum levels of these three biomarkers showed an mOS of 102 months, compared with the 9 months observed when lower levels are detected in at least one of these serum EV proteins, with a 6-fold increased risk of death. Of note, these panels retained their prognostic capacity when considering only patients who underwent potentially curative surgery (Fig. S15C).
Discussion
The precise, non-invasive, and early diagnosis of CCA remains a major challenge, particularly in patients at high risk, such as those with PSC. Here, we conducted an international multicentric study in which accurate EV protein biomarkers for the diagnosis of CCAs were identified. By high-throughput proteomics, novel biomarkers for the prediction of CCA development in patients with PSC; for the early and accurate diagnosis of PSC-related CCAs, non-PSC CCAs, and Pan-CCAs; and for the differential diagnosis of iCCA and HCC were disclosed. Importantly, we cross-validated these results by measuring the levels of candidate biomarkers in total serum by ELISA, confirming that these biomarkers are amenable to be detected in raw biological fluids, ensuring their potential translation into clinics. In addition, proteins present in circulating EVs not only allowed the precise diagnosis but also estimated the survival in patients with CCA. Human multi-organ transcriptomic analysis revealed that most diagnostic and prognostic biomarkers were expressed in hepatobiliary tissues. scRNA-seq analysis of normal liver and CCA tumours pointed out that the expression of candidate biomarkers was cell-specific, being particularly detected in malignant cholangiocytes within CCA tumours, thus reinforcing this novel tumour cell-derived liquid biopsy strategy.
Although the majority of CCAs emerge without a clear aetiology, PSC is a well-established risk factor. Up to 20% of patients with PSC might develop CCA during their lifetime, which commonly appears within the first year after PSC diagnosis and in younger people (40–50 years), when compared with CCAs from other aetiologies (∼65 years).11,12 In fact, CCA is currently responsible for more than 30% of PSC-associated premature deaths,13 constituting a substantial health and social problem. Current screening strategies for CCA diagnosis in patients with PSC have scant clinical value owing to their low accuracy. Serum CA19-9 levels are generally not elevated in early CCA stages and are also increased in ∼30% of patients with isolated PSC, and up to 7% of the general population are unable to express CA19-9 because of FUT3 activity deficiency,14 strongly limiting the usefulness of CA19-9 as a routine diagnostic screening tool. At the radiological level, benign biliary strictures closely resemble the initial malignant changes, which makes the early and appropriate diagnosis of CCA in patients with PSC extremely challenging. All these limitations markedly impact on CCA detection, which is an accidental event in up to 40% of PSC-CCA cases, when liver transplantation was required or even at autopsy.15 Previous studies have proposed several serum biomarkers for the differential diagnosis of CCA and PSC, although most of them did not include patients with concomitant PSC-CCA,14 raising major concerns when translating these findings into clinics.
Our study has contributed with novel protein biomarkers for CCAs. It is important to highlight that we have included only strictly selected patients, with biopsy-proven CCA confirmation, increasing the robustness of this analysis. It is noteworthy that the combination of protein-based models, including two to three biomarkers, with CA19-9 significantly improved the diagnostic capacity of CA19-9 alone. Most of these novel biomarkers and LMs are independent of anatomical CCA subtype and revealed high diagnostic accuracy also at early tumour stages. Similarly, the elevation of proposed biomarkers was not altered by the presence/absence of cirrhosis. In fact, >90% of patients with CCA included in our cohort did not present liver cirrhosis at sampling, thus suggesting that the levels of these biomarkers are not affected by cirrhosis. Our data indicate that the combination of serum CRP/FIBRINOGEN/FRIL levels might be particularly useful for the early diagnosis of PSC-CCA, whereas combining CRP/PIGR/VWF might aid the diagnosis of CCA arising in patients without PSC. In addition, the identified serum protein biomarkers also aid in the differential diagnosis of iCCA and HCC, which is currently an unmet need in clinical practice. Furthermore, by using serum samples from patients with PSC who had no clinical evidence of malignant masses at the time of sampling but who developed CCA during follow-up, we proposed FIBRINOGEN, CRP, PIGR, and FRIL not only as candidate diagnostic biomarkers but also as novel predictive biomarkers of CCA development in patients with PSC. Although no clinical features of tumour development were observed in these patients, we may not discount the fact that small undetectable lesions might be already present at sampling. Consequently, these predictive biomarkers should be prospectively validated in the near future.
EVs were shown to recapitulate features of their cells of origin.8 Considering that CCAs are highly heterogeneous, desmoplastic, and stroma-enriched tumours,1,2 scRNA-seq analysis might provide a better characterisation of biomarker-expressing cell populations. The majority of the most promising biomarkers for CCA, which displayed an increased abundance in serum EVs when compared with controls, were found chiefly expressed in malignant cholangiocytes. We further evaluated the protein levels of PIGR, FIBG, CRP, and FGL1 in human FFPE CCA tumours by immunofluorescence, confirming their marked expression in malignant cholangiocytes. It is worth noting that unravelling the type of cells that might be actively secreting biomarker-containing EVs could also postulate them as novel potential cell-specific therapeutic targets for CCA. For instance, PIGR, FIBG, and FGL1 have been studied in other gastrointestinal cancers and characterised as key contributors of tumorigenesis.[16], [17], [18] Consequently, their role in cholangiocarcinogenesis deserves future analysis.
Apart from harbouring a predictive and diagnostic capacity in CCA, we were also able to identify novel EV protein prognostic biomarkers that may aid to predict OS in patients regardless of disease status, serum CA19-9 levels, and CCA subtype. We herein propose two prognostic panels, one comprising ‘bad prognostic biomarkers’ (COMP/GNAI2/CFAI) and another one including ‘good prognostic biomarkers’ (ACTN1/MYCT1/PF4V), that allowed predicting OS, even in patients undergoing surgery with curative intent. Despite reports stating the capacity of these biomarkers to predict cancer prognosis is scarce, higher levels of COMP were recently associated with increased metastasis and circulating tumour cell count in patients with breast cancer.19 Similarly, decreased serum levels of PF4V were indicative of tumour progression and predicted worse OS and recurrence in patients with prostate cancer.20 Future studies should be conducted to confirm the prognostic value of these biomarkers and their role in CCA pathobiology.
In conclusion, we here demonstrated that serum EVs contain protein biomarkers for the prediction of CCA development in PSC, as well as for the early tumour detection in individuals with PSC, in individuals without PSC, and in individuals with CCAs regardless of disease aetiology, which are amenable to be detected using total serum. This has significant clinical relevance, as early CCA detection in asymptomatic patients with PSC was associated with improved OS and recurrence-free survival following liver transplantation21,22 and also because patients with ‘very early’ iCCA undergoing surgery or liver transplantation display better prognosis, when compared with patients with more advanced CCA.23 This study also reinforces the idea that patients with CCAs arising from different aetiologies (e.g. PSC vs. non-PSC) may have common and different serum EV proteins, and therefore, there is a need to use proper, well-defined biomarkers to identify CCA in specific patient subgroups, moving a step forward into the personalised diagnosis of CCA. This, together with the fact that most of these candidate biomarkers are preferentially expressed in malignant cholangiocytes within CCA tumours, reinforces our approach as an innovative tumour cell-derived liquid biopsy strategy. To confirm the translational capacity of these novel predictive, diagnostic, and prognostic biomarkers, a next prospective validation phase using larger patient cohorts should be conducted. This would open a new avenue for the early non-invasive diagnosis of CCA, consequently enabling a prompt therapeutic intervention and improving patients’ welfare and outcome.
Abbreviations
A1AG1, alpha-1-acid glycoprotein 1; ACTN1, alpha-actinin-1; AI, accuracy index; ALBU, albumin protein; ALP, alkaline phosphatase; ALT, alanine transaminase; AST, aspartate transaminase; CA19-9, carbohydrate antigen 19-9; CCA, cholangiocarcinoma; CFAI, complement factor I; CK19, cytokeratin 19; COMP, cartilage oligomeric matrix protein; CRP, C reactive protein; dCCA, distal CCA; ELISA, enzyme-linked immunosorbent assay; ER, endoplasmic reticulum; EV, extracellular vesicle; FFPE, formalin-fixed, paraffin-embedded; FGB, fibrinogen beta chain; FGG, fibrinogen gamma chain; FGL1, fibrinogen-like protein 1; FIBA, fibrinogen alpha chain; FIBB, fibrinogen beta chain; FIBG, fibrinogen gamma chain; FRIL, ferritin light chain protein; FTL, ferritin light chain; FUT3, fucosyltransferase 3 (Lewis blood group); GGT, gamma glutamyltransferase; GNAI2, guanine nucleotide-binding protein G(i) subunit alpha-2; GRP78, endoplasmic reticulum chaperone BiP; HCC, hepatocellular carcinoma; HR, hazard ratio; iCCA, intrahepatic CCA; KPYM, pyruvate kinase; LAD, locally advanced disease; LD, local disease; LM, logistic model; LRP1, LDL receptor-related protein 1; MD, metastatic disease; mOS, median overall survival; MS, mass spectrometry; MYCT1, MYC Target 1; NC, normalised count; NK, natural killer; NPV, negative predictive value; NTA, nanoparticle tracking analysis; OIT3, oncoprotein-induced transcript 3; OR, odds ratio; OS, overall survival; ORM1, orosomucoid 1; Pan-CCA, CCA regardless of aetiology; pCCA, perihiliar CCA; PF4V, platelet factor 4 variant; PIGR, polymeric immunoglobulin receptor; PKM, Pyruvate Kinase M1/2; PPV, positive predictive value; PSC, primary sclerosing cholangitis; PSC-CCA, concomitant PSC and CCA; RNA-seq, RNA sequencing; ROC, receiver operating characteristic; SL, surrounding liver; scRNA-seq, single-cell RNA-sequencing; SEN, sensitivity; SPE, specificity; TEM, transmission electron microscopy; VWF, von Willebrand factor protein/gene; WCE, whole-cell extract.
Financial support
This work was supported by the following: Spanish Carlos III Health Institute (ISCIII) (JMB [FIS PI18/01075, PI21/00922 and Miguel Servet Program CON14/00129 and CPII19/00008], MJP [FIS PI17/00022, PI20/00186], RIRM [FIS PI20/00189], and PMR [Sara Borrell CD19/00254 and Miguel Servet Program CP22/00073]) cofinanced by ‘Fondo Europeo de Desarrollo Regional’ (FEDER); CIBERehd (ISCIII): JMB, PMR, MJP, and LB, Spain; IKERBASQUE, Basque foundation for Science (to JMB and PMR); Diputación Foral Gipuzkoa’ (2020-CIEN-000067-01 and 2021-CIEN-000029-04-01 to PMR); Department of Health of the Basque Country (2019111024 to MJP; 2022111070 to PMR; and 2017111010 and 2020111077 to JMB), ‘Euskadi RIS3’ (2019222054, 2020333010, and 2022333032 to JMB; and 2022333041 to MJP), and BIOEF (Basque Foundation for Innovation and Health Research: EiTB Maratoia BIO15/CA/016/BD to JMB). The following also provided support: La Caixa Scientific Foundation (HR17-00601 to JMB), ‘Fundación Científica de la Asociación Española Contra el Cáncer’ (AECC Scientific Foundation, to JMB), PSC Partners US (to JMB) and PSC Supports UK (to JMB: 06119JB), European Union’s Horizon 2020 Research and Innovation Program (grant number 825510, ESCALON: to JMB), and AMMF – The Cholangiocarcinoma Charity (EU/2019/AMMFt/001, to JMB and PMR). MJP was funded by the Spanish Ministry of Economy and Competitiveness (MINECO: ‘Ramón y Cajal’ Program RYC-2015-17755), and AL by the Basque Government (PRE_2017_1_0345). The funding sources had no involvement in study design, data collection and analysis, decision to publish, or preparation of the article.
Conflicts of interest
The authors disclose no conflict of interest for this study.
Please refer to the accompanying ICMJE disclosure forms for further details.
Authors’ contributions
Study concept and design: AL, TF, PMR, JMB; sample collection and data acquisition: AL, MA, PM, PO, EZ, MMG, CS, AA, CJO, ALC, MM, TP, MV, RJA, MTD, AL, JWV, RIRM, LIS, YPC, FJCC, IR, MK, CI, JB, LMNC, JMFP, FE, MJP, JBA, LB,THK, TF, PMR, JMB; data analysis and interpretation: AL, MA, PO, EZ, CJO, PMR, JMB; statistical analysis: AL, MA, EZ, CJO; drafting of the manuscript: AL, PMR, JMB; funding: LB, PMR, JMB; read and critically revised the manuscript and agreed to the published version: all authors.
Data availability
EV isolation and characterisation protocols have been submitted to EV-TRACK knowledgebase (EV-TRACK ID: EV210077), and EV-proteomic results have been uploaded to PRIDE – Proteomics Identification Database – EMBL-EBI (Orbitrap cohort: PXD026197; timsTOF cohort: PXD036943 and PXD037007).
Acknowledgements
We would like to express their gratitude to Dr Ana Martinez Amesti and Maite Miranda, from the Microscopy Unit at the University of the Basque Country (UPV/EHU) for their help and support in transmission electron microscopy (TEM) experiments. We would also like to thank Dr Ibon Martinez for his advice and tips regarding statistical analysis and to Dr Beatriz Nafria, Dr Marie Ines Hitler, Dr Benjamin Goeppert, and Dr Stephanie Rossler for all their help in collecting human samples. This article is based upon work from COST Action CA18122 European Cholangiocarcinoma Network (Euro-Cholangio-Net) supported by COST (European Cooperation in Science and Technology: www.cost.eu), in collaboration with the European Network for the Study of Cholangiocarcinoma (ENSCCA: http://www.enscca.org/), the International Primary Sclerosing Cholangitis Study Group (iPSCSG: https://www.ipscsg.org/), the European Reference Network on Rare Liver Diseases (ERN-Rare Liver: https://rare-liver.eu/), and the European Reference Network on Rare Adult Cancers (solid tumours; EURACAN: https://euracan.eu/).
Footnotes
Author names in bold designate shared co-first authorship.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhep.2023.02.027.
Contributor Information
Pedro M. Rodrigues, Email: pedro.rodrigues@biodonostia.org.
Jesus M. Banales, Email: jesus.banales@biodonostia.org.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Banales J.M., Marin J.J.G., Lamarca A., Rodrigues P.M., Khan S.A., Roberts L.R., et al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol. 2020;17:557–588. doi: 10.1038/s41575-020-0310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodrigues P.M., Olaizola P., Paiva N.A., Olaizola I., Agirre-Lizaso A., Landa A., et al. Pathogenesis of cholangiocarcinoma. Annu Rev Pathol. 2021;16:433–463. doi: 10.1146/annurev-pathol-030220-020455. [DOI] [PubMed] [Google Scholar]
- 3.Izquierdo-Sanchez L., Lamarca A., La Casta A., Buettner S., Utpatel K., Klumpen H.J., et al. Cholangiocarcinoma landscape in Europe: diagnostic, prognostic and therapeutic insights from the ENSCCA Registry. J Hepatol. 2022;76:1109–1121. doi: 10.1016/j.jhep.2021.12.010. [DOI] [PubMed] [Google Scholar]
- 4.Macias R.I.R., Cardinale V., Kendall T.J., Avila M.A., Guido M., Coulouarn C., et al. Clinical relevance of biomarkers in cholangiocarcinoma: critical revision and future directions. Gut. 2022;71:1669–1683. doi: 10.1136/gutjnl-2022-327099. [DOI] [PubMed] [Google Scholar]
- 5.Valle J.W., Borbath I., Khan S.A., Huguet F., Gruenberger T., Arnold D., et al. Biliary cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27:v28–v37. doi: 10.1093/annonc/mdw324. [DOI] [PubMed] [Google Scholar]
- 6.Clements O., Eliahoo J., Kim J.U., Taylor-Robinson S.D., Khan S.A. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma: a systematic review and meta-analysis. J Hepatol. 2020;72:95–103. doi: 10.1016/j.jhep.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Karlsen T.H., Folseraas T., Thorburn D., Vesterhus M. Primary sclerosing cholangitis – a comprehensive review. J Hepatol. 2017;67:1298–1323. doi: 10.1016/j.jhep.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 8.Lapitz A., Arbelaiz A., Olaizola P., Aranburu A., Bujanda L., Perugorria M.J., et al. Extracellular vesicles in hepatobiliary malignancies. Front Immunol. 2018;9:2270. doi: 10.3389/fimmu.2018.02270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsova P., Ibrahim S.H., Verma V.K., Morton L.A., Shah V.H., LaRusso N.F., et al. Extracellular vesicles in liver pathobiology: small particles with big impact. Hepatology. 2016;64:2219–2233. doi: 10.1002/hep.28814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arbelaiz A., Azkargorta M., Krawczyk M., Santos-Laso A., Lapitz A., Perugorria M.J., et al. Serum extracellular vesicles contain protein biomarkers for primary sclerosing cholangitis and cholangiocarcinoma. Hepatology. 2017;66:1125–1143. doi: 10.1002/hep.29291. [DOI] [PubMed] [Google Scholar]
- 11.Chapman M.H., Webster G.J., Bannoo S., Johnson G.J., Wittmann J., Pereira S.P. Cholangiocarcinoma and dominant strictures in patients with primary sclerosing cholangitis: a 25-year single-centre experience. Eur J Gastroenterol Hepatol. 2012;24:1051–1058. doi: 10.1097/MEG.0b013e3283554bbf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weismuller T.J., Trivedi P.J., Bergquist A., Imam M., Lenzen H., Ponsioen C.Y., et al. Patient age, sex, and inflammatory bowel disease phenotype associate with course of primary sclerosing cholangitis. Gastroenterology. 2017;152:1975–1984 e1978. doi: 10.1053/j.gastro.2017.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boonstra K., Weersma R.K., van Erpecum K.J., Rauws E.A., Spanier B.W., Poen A.C., et al. Population-based epidemiology, malignancy risk, and outcome of primary sclerosing cholangitis. Hepatology. 2013;58:2045–2055. doi: 10.1002/hep.26565. [DOI] [PubMed] [Google Scholar]
- 14.Vedeld H.M., Folseraas T., Lind G.E. Detecting cholangiocarcinoma in patients with primary sclerosing cholangitis – the promise of DNA methylation and molecular biomarkers. JHEP Rep. 2020;2 doi: 10.1016/j.jhepr.2020.100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boberg K.M., Bergquist A., Mitchell S., Pares A., Rosina F., Broomé U., et al. Cholangiocarcinoma in primary sclerosing cholangitis: risk factors and clinical presentation. Scand J Gastroenterol. 2002;37:1205–1211. doi: 10.1080/003655202760373434. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X., Wang F., Huang Y., Ke K., Zhao B., Chen L., et al. FGG promotes migration and invasion in hepatocellular carcinoma cells through activating epithelial to mesenchymal transition. Cancer Manag Res. 2019;11:1653–1665. doi: 10.2147/CMAR.S188248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y., Qiao H.X., Zhou Y.T., Hong L., Chen J.H. Fibrinogen-like protein 1 promotes the invasion and metastasis of gastric cancer and is associated with poor prognosis. Mol Med Rep. 2018;18:1465–1472. doi: 10.3892/mmr.2018.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tey S.K., Wong S.W.K., Chan J.Y.T., Mao X., Ng T.H., Yeung C.L.S., et al. Patient pIgR-enriched extracellular vesicles drive cancer stemness, tumorigenesis and metastasis in hepatocellular carcinoma. J Hepatol. 2022;76:883–895. doi: 10.1016/j.jhep.2021.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Papadakos K.S., Hagerling C., Ryden L., Larsson A.M., Blom A.M. High levels of expression of cartilage oligomeric matrix protein in lymph node metastases in breast cancer are associated with reduced survival. Cancers. 2021;13:5876. doi: 10.3390/cancers13235876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang M., Guan J., Huo Y.L., Song Y.S., Chen L.Z. Downregulation of serum CXCL4L1 predicts progression and poor prognosis in prostate cancer patients treated by radical prostatectomy. Asian J Androl. 2019;21:387–392. doi: 10.4103/aja.aja_117_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eaton J.E., Welle C.L., Bakhshi Z., Sheedy S.P., Idilman I.S., Gores G.J., et al. Early cholangiocarcinoma detection with magnetic resonance imaging versus ultrasound in primary sclerosing cholangitis. Hepatology. 2021;73:1868–1881. doi: 10.1002/hep.31575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azad A.I., Rosen C.B., Taner T., Heimbach J.K., Gores G.J. Selected patients with unresectable perihilar cholangiocarcinoma (pCCA) derive long-term benefit from liver transplantation. Cancers. 2020;12:3157. doi: 10.3390/cancers12113157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sapisochin G., Facciuto M., Rubbia-Brandt L., Marti J., Mehta N., Yao F.Y., et al. Liver transplantation for “very early” intrahepatic cholangiocarcinoma: international retrospective study supporting a prospective assessment. Hepatology. 2016;64:1178–1188. doi: 10.1002/hep.28744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
EV isolation and characterisation protocols have been submitted to EV-TRACK knowledgebase (EV-TRACK ID: EV210077), and EV-proteomic results have been uploaded to PRIDE – Proteomics Identification Database – EMBL-EBI (Orbitrap cohort: PXD026197; timsTOF cohort: PXD036943 and PXD037007).