To the Editor,
Understanding early tumorigenic events have obvious diagnostic and prognostic implications, particularly in the skin since this is the site of more US cancer diagnoses than any other organ (Rogers et al., 2015). Actinic keratosis (AK) are dysplastic keratinocyte lesions characterized by irregularly shaped scaly papules frequently manifesting on sun-damaged skin, and are considered precursor lesions which can develop into cutaneous squamous cell carcinoma (cSCC), the second most common keratinocyte cancer (Siegel et al., 2017). Delineating the molecular events that characterize transition from normal skin to AK and on to established skin cancer has been the focus of a growing number of studies employing various technologies to identify markers of both initiation and progression of AK and cSCC (Chitsazzadeh et al., 2016, Hameetman et al., 2013, Kim et al., 2021, Lambert et al., 2014, Padilla et al., 2010, Ra et al., 2011, Thomson et al., 2021, Zheng et al., 2021).
Traditionally, cancer biology focusses on change within the tumor compartment for diagnosis and staging. Recent genomic profiling of AK has been similar as multiple studies have focused on DNA analysis to identify clonal chromosomal abnormalities and somatic mutations (Kim et al., 2021, Thomson et al., 2021), and transcriptional analysis has been limited to whole skin (Chitsazzadeh et al., 2016, Hameetman et al., 2013, Padilla et al., 2010) or the epidermal compartment alone, isolated using laser capture microdissection (Lambert et al., 2014, Zheng et al., 2021). This approach is in line with a mutation-centric view of cancer supported by the clear requirement for a tumor to harbor multiple somatic DNA changes, without consideration of the dermal compartment, populated predominantly by fibroblasts and, in the case of AK and cSCC, infiltrating immune cells. Since sun-damaged skin can harbor multiple so called tumor driver-gene mutations caused by prolonged exposure to UV radiation (Martincorena et al., 2015, South et al., 2014) it becomes challenging to differentiate tumor from non-tumor and tumor from pre-cancerous lesions based on mutation burden alone.
As part of a pilot study to evaluate current technologies available to assess a large cohort of archival matched patient samples, we employed the Nanostring Digital Spatial Profiler (DSP) to compare transcriptional change in sun-protected skin with different regions of matched AK from the six individuals. The Nanostring DSP allows for the selection of tissue compartments of interest for RNA analysis using photocleavable RNA probes isolated on the basis of antibody staining. Here we used a pan-cytokeratin antibody to differentiate the epidermal from dermal compartment and assayed over 18,000 RNA probes from multiple regions of interest (ROI) comparing sun protected skin (n=5–6 from 6 individuals for a total of 34 ROI), the center of an AK lesion (n=3 for a total of 18 ROI) and edge of the AK lesion (n=2–3 for a total of 17 ROI) representing sun-damaged skin, from six unrelated individuals (Figure 1A).
Figure 1: Spatial Profiling identifies the greatest transcriptional change in the cytokeratin negative compartment comparing sun-protected skin with actinic keratosis.
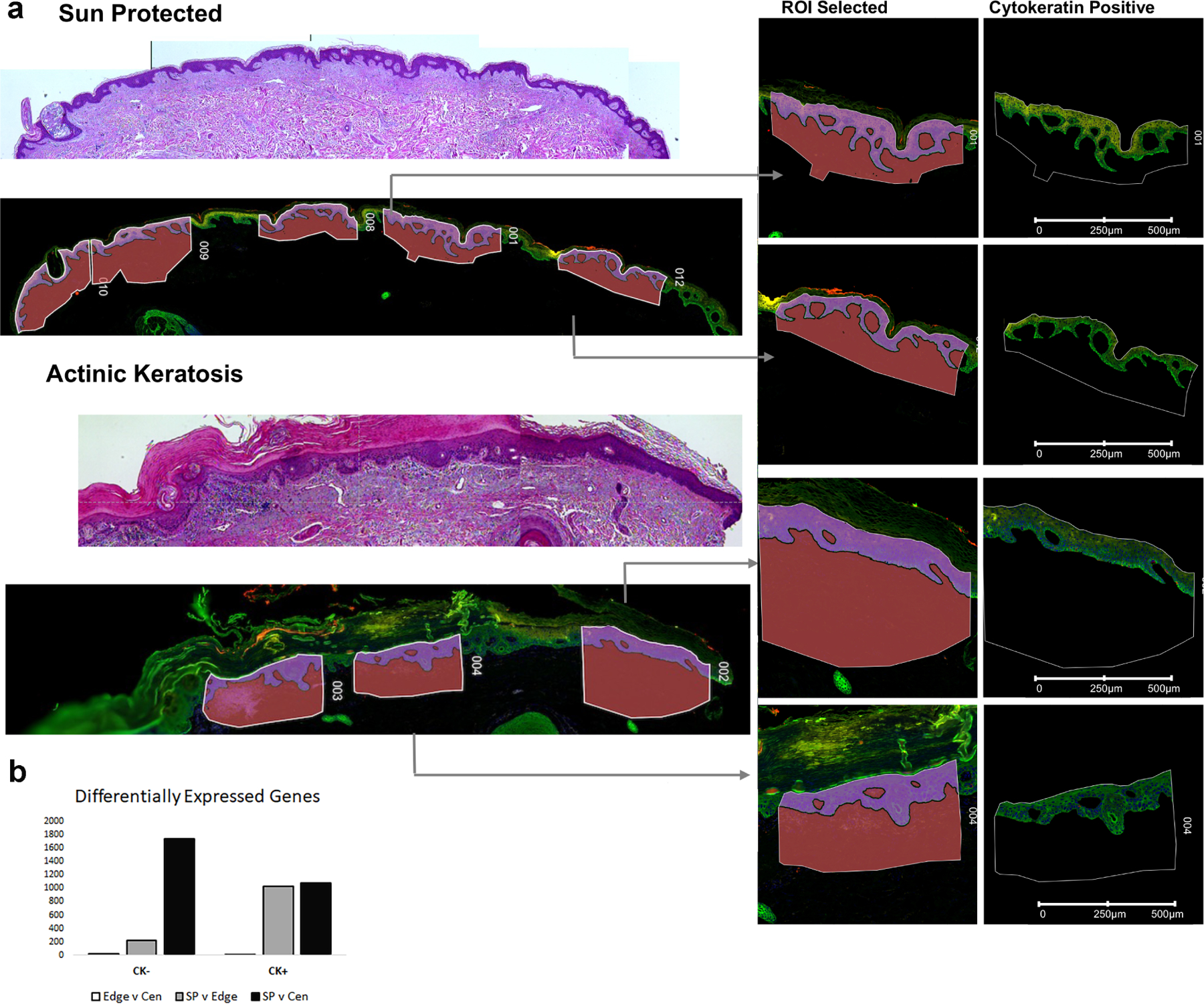
a: Digital Spatial Profiler identified regions of interest (ROI) highlights examples of areas of patient tissue sampled for transcriptional profiling. b: Differentially expressed genes identified using a pairwise FDR adjusted p-value of less than 0.05 and an overall t-test statistic of 0.05 highlights greater number DEGs (y-axis) in the cytokeratin negative compartment (CK-) comparing sun protected normal skin (SP) with actinic keratosis center (Cen). CK+ = cytokeratin positive compartment, Edge = actinic keratosis edge.
Analysis of these DSP data identified the expected changes in the epidermal compartment comparing sun-protected skin to AK demonstrating an upregulation of gene expression associated with P38 MAPK, TP53, ERK and DNA damage associated pathways and in line with previous studies (Chitsazzadeh et al., 2016, Zheng et al., 2021) (Figure 2A and Supplemental Tables 1&2). Comparison of the dermal compartment revealed up-regulation of pathways associated with matrix metalloproteinases, cell adhesion and extracellular matrix (ECM) remodeling as well as multiple immune related pathways including innate immune system, MHC class II antigen presentation and toll-like receptor signaling (Supplemental Tables 3&4). Differential expression analysis of batch corrected, normalized read counts using a pairwise FDR adjusted p-value of less than 0.05, for those genes with an overall t-test statistic of 0.05 revealed the greatest change in transcriptional activity in the dermal compartment when comparing sun protected skin with the center of the AK lesion. The fewest transcriptional changes in our matched samples were evident from comparing AK edge with AK center, again with the greatest change being in the dermal compartment (Figure 1B and Supplemental Table 5). Interestingly, comparing sun protected skin with AK edge revealed more change in the epidermal compartment suggesting a progressive model where early transcriptional change from sun protected skin to AK edge (assumed to be sun damaged normal skin) is evident in the epidermal compartment which leads to activation of multiple signaling cascades within the dermal compartment and significant changes to ECM remodeling, presumably with a major contribution from infiltration of immune cells (Figure 2B).
Figure 2: Pathway changes in the progression from sun-protected skin to actinic keratosis.
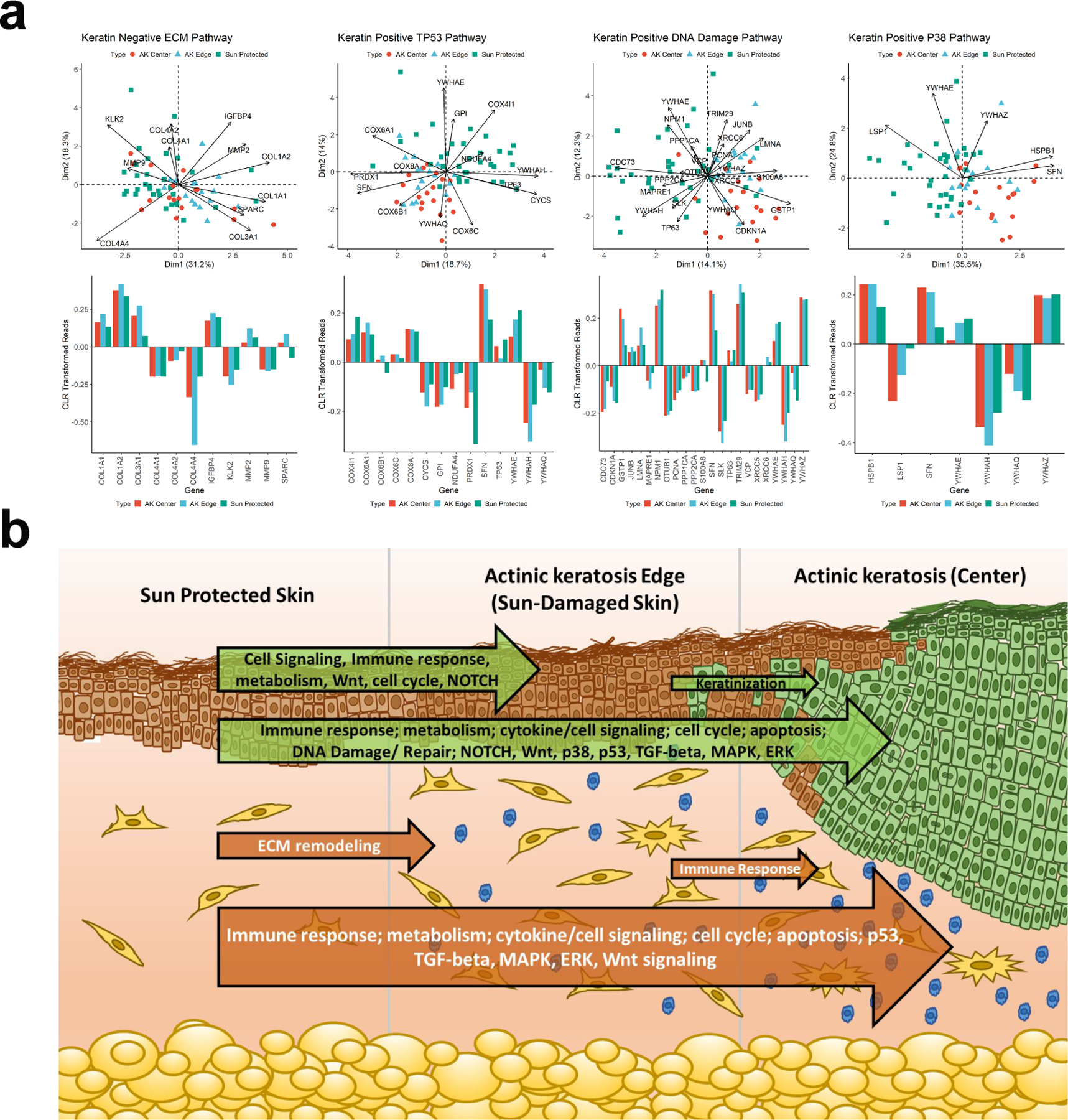
a: Biplots showing centered log-ratio (CLR) transformed reads comparing sun-protected and actinic keratosis samples. Individual samples are shown with three color and symbol combinations and the mean values are larger symbols on the PCA plane with the CLR standardized gene expression plotted. For CK-negative samples the SP and AK edge cluster together, and for CK-positive samples AK edge and center cluster together relative to the SP samples. The bar charts help interpret the directionality of the projections which can be inferred from the PCA plot but is simplified by the bar graphs.
b: Graphical representation of transcriptional change moving through sun protected skin to actinic keratosis. Major pathways (identified using methodology described in Ben-Ari Fuchs et al., 2016) changing comparing each of the three compartments are indicated within the arrows representing the overall extent of transcriptional change, green arrows represent transcriptional change within the cytokeratin positive compartment while orange arrows represent the dermal (cytokeratin negative) compartment.
These data are intriguing since previous studies of AK have focused on the cell of origin, the keratinocyte, and while clear differences in transcription have been identified comparing AK with sun-protected or UV-exposed normal skin, fewer changes are observed comparing AK with cSCC (Chitsazzadeh et al., 2016, Padilla et al., 2010, Zheng et al., 2021). Indeed, AK and cSCC keratinocytes are characterized by a high tumor mutation burden and gross chromosomal changes (Chitsazzadeh et al., 2016, Ra et al., 2011, Thomson et al., 2021), and a compelling hypothesis supported by the data presented here is that key molecular changes which influence progression of AK are in fact found in the dermal compartment. Further work with a wider sample set and inclusion of matched cSCC samples will be needed to test this hypothesis and the DSP platform offers the ability to do so in archival, formalin fixed samples. A recent study combining single cell and spatial transcriptomics to compare 10 SCC samples with patient matched normal skin also included separation of the dermal compartment and highlighted intercellular communication between the stroma and the leading edge of invasive SCC (Ji et al., 2020). This observation was also noted in an earlier single cell study of head and neck SCC, where the leading edge of the tumor was highlighted as the most prognostic region (Puram et al., 2017). These studies support the idea that the dermal compartment may well hold key prognosticating information in the context of AK and cSCC.
Collectively, these data suggest that while the epidermal compartment harbors the cell of origin for SCC and AK, the greatest change in transcriptional homeostasis in the progression from sun-protected skin to AK is observed in the dermal compartment and may well be a fruitful avenue of investigation for delineating molecular markers of initiation and progression.
Supplementary Material
Acknowledgments
This research was funded by the National Cancer Institute, grant numbers P01CA229112 and T32CA078447. This project utilized the Genomics and Flow Cytometry Shared Resources at the Sidney Kimmel Cancer Center, supported by the NCI, grant 5P30CA056036–17.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Human Subjects
All participants provided written, informed consent and this protocol was approved by the Institutional Review Board at the University of Arizona (Protocol Title: Skin Cancer Prevention Program Biorepository; Protocol Number: 1200000229).
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Author Contributions Statement (CRediT-compliant)
Conceptualization: all authors; data curation: all authors; formal analysis: all authors; funding acquisition: all authors; investigation: all authors; methodology: all authors; project administration: all authors; resources: all authors; software: all authors; supervision: all authors; validation: all authors; visualization: all authors; writing – original draft: all authors; and writing – review and editing: all authors.
Data Availability Statement
All data are available on request. Datasets related to this article can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE221001, hosted by the Gene Expression Omnibus (accession number GSE221001).
References
- Ben-Ari Fuchs S, Lieder I, Stelzer G, Mazor Y, Buzhor E, Kaplan S, et al. GeneAnalytics: An Integrative Gene Set Analysis Tool for Next Generation Sequencing, RNAseq and Microarray Data. OMICS 2016;20(3):139–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitsazzadeh V, Coarfa C, Drummond JA, Nguyen T, Joseph A, Chilukuri S, et al. Cross-species identification of genomic drivers of squamous cell carcinoma development across preneoplastic intermediates. Nat Commun 2016;7:12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hameetman L, Commandeur S, Bavinck JN, Wisgerhof HC, de Gruijl FR, Willemze R, et al. Molecular profiling of cutaneous squamous cell carcinomas and actinic keratoses from organ transplant recipients. BMC Cancer 2013;13:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji AL, Rubin AJ, Thrane K, Jiang S, Reynolds DL, Meyers RM, et al. Multimodal Analysis of Composition and Spatial Architecture in Human Squamous Cell Carcinoma. Cell 2020;182(2):497–514 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, Shin S, Jung SH, Park YM, Park GS, Lee SH, et al. Genomic Progression of Precancerous Actinic Keratosis to Squamous Cell Carcinoma. J Invest Dermatol 2021. [DOI] [PubMed]
- Lambert SR, Mladkova N, Gulati A, Hamoudi R, Purdie K, Cerio R, et al. Key differences identified between actinic keratosis and cutaneous squamous cell carcinoma by transcriptome profiling. Br J Cancer 2014;110(2):520–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015;348(6237):880–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla RS, Sebastian S, Jiang Z, Nindl I, Larson R. Gene expression patterns of normal human skin, actinic keratosis, and squamous cell carcinoma: a spectrum of disease progression. Arch Dermatol 2010;146(3):288–93. [DOI] [PubMed] [Google Scholar]
- Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017;171(7):1611–24 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ra SH, Li X, Binder S. Molecular discrimination of cutaneous squamous cell carcinoma from actinic keratosis and normal skin. Mod Pathol 2011;24(7):963–73. [DOI] [PubMed] [Google Scholar]
- Rogers HW, Weinstock MA, Feldman SR, Coldiron BM. Incidence Estimate of Nonmelanoma Skin Cancer (Keratinocyte Carcinomas) in the U.S. Population, 2012. JAMA Dermatol 2015;151(10):1081–6. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- South AP, Purdie KJ, Watt SA, Haldenby S, den Breems N, Dimon M, et al. NOTCH1 mutations occur early during cutaneous squamous cell carcinogenesis. J Invest Dermatol 2014;134(10):2630–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J, Bewicke-Copley F, Anene CA, Gulati A, Nagano A, Purdie K, et al. The Genomic Landscape of Actinic Keratosis. J Invest Dermatol 2021;141(7):1664–74 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Capell BC, Parekh V, O’Day C, Atillasoy C, Bashir HM, et al. Whole-Exome and Transcriptome Analysis of UV-Exposed Epidermis and Carcinoma In Situ Reveals Early Drivers of Carcinogenesis. J Invest Dermatol 2021;141(2):295–307 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available on request. Datasets related to this article can be found at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE221001, hosted by the Gene Expression Omnibus (accession number GSE221001).


