Abstract
Infections caused by multi-drug resistant Enterobacterales (MDR-E) are difficult to treat and cause significant mortality, especially in developing countries. This study characterized the phenotypic and genotypic profiles of 49 randomly selected beta-lactam resistant MDR-E previously isolated from patients being managed in hospitals in Nigeria using whole genome sequencing. The study isolates exhibited 85.5% resistance to 3rd generation cephalosporins and 65.3% resistance to carbapenems. The blaTEM-1B (29, 59.2%), blaCTX-M-15 (38, 77.6%), and blaNDM-1 (17, 51.5%) were the most common penicillinase, ESBL, and carbapenem resistant genes across isolates, respectively. Seventeen (45%) of blaCTX-M-15 was carried on the insertion sequence ISEc9 while blaNDM-1 (11, 64.7%) were associated with ISEc33. None of the 21 plasmids detected were associated with β-lactamase genes. Higher resistance rates were found in E. coli ST-88 (n = 2) and the high-risk ST-692 (n = 2). For Klebsiella species, the high-risk clones ST-476 (n = 8) and ST-147 (n = 3) predominated and had higher phenotypic resistance rates and higher number of AMR genes. The mechanisms and pattern of antibiotic resistance differ from patterns previously described with isolates harbouring a wide range of AMRGs. The detection of several chromosomally mediated carbapenemases in our study also represents a significant finding that warrants further investigation to better understand its’ implications for clinical practice and public health. The selected MDR-Es were found to be pan-susceptible to tigecycline and had very low resistance to fosfomycin, suggesting a potential for these as empiric treatments. A surveillance approach incorporating both conventional laboratory techniques and modern molecular techniques is essential for the comprehensive characterization of the emergence and dissemination of antimicrobial resistance in Enterobacterales infections within Nigeria.
Subject terms: Infectious diseases, Phylogeny, Evolution, Molecular biology, Diseases, Medical research, Molecular medicine
Introduction
Infections with multi-drug resistant Enterobacterales (MDR-E) are a growing global threat. The threat of MDR-E infections, such as extended-spectrum beta lactamase producing Enterobacterales (ESBL-E) and carbapenem-resistant Enterobacterales (CRE), is of great concern in low- and middle-income African countries. These countries have the highest global burden of antimicrobial-resistant (AMR) infections with 114.8 deaths per 100,000 population. Despite this burden, these countries have a striking dearth of medical literature addressing this critical problem in these regions1. The resistance profiles of CRE often extends beyond beta-lactam agents to include, aminoglycosides, tetracyclines and chloramphenicol as reported in recent scientific studies2–4.
There are several mechanisms of resistance mediating MDR-E, including the production of carbapenemases, AmpC enzymes, and ESBLs5. Published data indicate significant differences in the mortality rates of patients with MDR-E infections, such as those caused by CRE versus those resistant by other mechanisms6–8. The infections caused by MDR-E are a particular global concern because the encoding genes are often located on mobile genetic elements, such as highly transmissible plasmids and integrons, that may spread among different bacterial species and potentially cause epidemics9.
Despite its high burden, there is a paucity of studies characterizing MDR-E strains specifically from Nigeria—the most populous country in Africa10,11, which hampers our ability to implement targeted antimicrobial stewardship and epidemiologic control measures. To address this critical gap in knowledge, our study undertakes a comprehensive phenotypic and molecular characterization of beta-lactam resistant MDR-E strains isolated from patients attending six hospitals in Northern Nigeria. By unraveling the mechanisms underlying resistance and understanding the current strain landscape, our research aims to provide essential insights necessary for developing effective interventions, optimizing treatment strategies, and guiding infection control practices in this LMIC setting.
This study characterized contemporary strains of MDR-E to define their phenotypic and genotypic antibiotic resistance profiles. This included elucidating the mechanisms of resistance to cephalosporins and carbapenems, carriage of AMR genes on mobile genetic elements, and correlating sequence types with antimicrobial resistance profiles. We also aimed to determine genetic relatedness of these MDR-E and suggest ways to combat infections caused by these pathogens.
Results
Bacterial strains and sample source
We studied 49 non-duplicate Enterobacterales isolates as shown in Table 1. Of these, 83.7% were recovered from blood (n = 41), 12.2% from urine (n = 6), and 4.1% from wound swabs (n = 2). Over half (57.1%, n = 28) of the isolates were Klebsiella spp.
Table 1.
Distribution of Enterobacterales characterized in study.
| Genus | Enterobacterales species | n (%) | Isolate source |
|---|---|---|---|
| Citrobacter spp | Citrobacter freundii | 2 (4.1) | BC, U |
| Enterobacter spp | Enterobacter cloacae subsp. cloacae | 9 (18.4) | BC |
| Enterobacter hormaechei | 1 (2.0) | WS | |
| Escherichia spp | Escherichia coli | 6 (12.2) | BC, U |
| Klebsiella spp | Klebsiella pneumoniae | 17 (34.7) | BC, U |
| Klebsiella quasipneumoniae | 10 (20.4) | BC | |
| Serratia spp | Serratia ureilytica | 4 (8.16) | BC, U |
| Total | 49 (100.0) |
Isolate source key: BC Blood culture, U Urine, WS Wound swab.
Antimicrobial susceptibility testing results
Antimicrobial susceptibility testing was performed for the 49 MDR-E against 17 different antibiotics and also to determine ESBL production. Results are shown in Table 2 and detailed results are presented in Table S1. The prevalence of resistance for third generation cephalosporins among the study isolates was 85.5% to cefotaxime and 80.0% to ceftriaxone. The overall ESBL production among study isolates was 30.3%. Resistance rate to meropenem was 65.3% and carbapenemase production as identified by the mCIM method detected in 55.1% of the isolates.
Table 2.
Antimicrobial resistance profiles of Enterobacterales by species.
| Citrobacter spp (2) | E. coli (6) | Enterobacter spp (10) | Klebsiella spp (28) | S. ureilytica (3) | Total resistance n (%) | |
|---|---|---|---|---|---|---|
| Amikacin | 0 (0.0) | 0 (0.0) | 7 (50.0) | 7 (50.0) | 0 (0.0) | 14 (28.6) |
| Gentamicin | 2 (100.0) | 6 (100.0) | 8 (80.0) | 26 (92.9) | 0 (0.0) | 42 (85.7) |
| Aztreonam | 0 (0.0) | 6 (100.0) | 9 (90.0) | 25 (89.3) | 0 (0.0) | 40 (81.6) |
| Ampicillin Sulbactam | 2 (100.0) | 6 (100.0) | 10 (100.0) | 28 (100.0) | 3 (100.0) | 48 (100.0) |
| Piperacillin Tazobactam | 2 (100.0) | 6 (100.0) | 8 (80.0) | 23 (82.1) | 0 (0.0) | 39 (79.6) |
| Cefotaxime | 2 (100.0) | 6 (100.0) | 9 (90.0) | 27 (96.4) | 3 (100.0) | 47 (95.9) |
| Cefepime# | 2 (100.0) | 6 (100.0) | 9 (90.0) | 27 (96.4) | 0 (0.0) | 44 (89.8) |
| Ceftriaxone | 2 (100.0) | 6 (100.0) | 9 (90.0) | 27 (96.4) | 1 (33.3) | 45 (91.8) |
| Ceftazidime | 2 (100.0) | 6 (100.0) | 9 (90.0) | 27 (96.4) | 0 (0.0) | 44 (89.8) |
| Ceftazidime Avibactam | 2 (100.0) | 4 (66.7) | 8 (80.0) | 15 (53.6) | 0 (0.0) | 29 (59.2) |
| Ceftolozane Tazobactam | N/R | 6 (100.0) | 9 (90.0) | 23 (82.1) | 0 (0.0) | 38 (80.9) |
| Meropenem | 2 (100.0) | 4 (66.7) | 8 (80.0) | 18 (64.3) | 0 (0.0) | 32 (65.3) |
| Colistin* | 0 (0.0) | 0 (0.0) | 9 (90.0) | 2 (7.1) | 3 (100.0) | 14 (28.6) |
| Tigecycline | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Fosfomycin** | 0 (0.0) | 0 (0.0) | 0 (0.0) | 7 (25.0) | 0 (0.0) | 7 (14.3) |
| Trimethoprim Sulphamethoxazole | 2 (100.0) | 6 (100.0) | 9 (90.0) | 27 (96.4) | 0 (0.0) | 44 (89.8) |
| ESBL*** | 0 (0.0) | 2 (33.3) | 1 (10.0) | 12 (44.9) | 0 (0.0) | 15 (30.6) |
| mCIM | 1 (50.0) | 4 (66.7) | 8 (80.0) | 14 (50.0) | 0 (0.0) | 27 (55.1) |
*Except for colistin, all I breakpoints represented as resistant. #SDD cefepime E. coli is reported as R.
**For fosfomycin, NS represented as R.
***ESBL is ≥ 3 times decrease in MIC when cephalosporin is tested alone compared with the addition of an inhibitor.
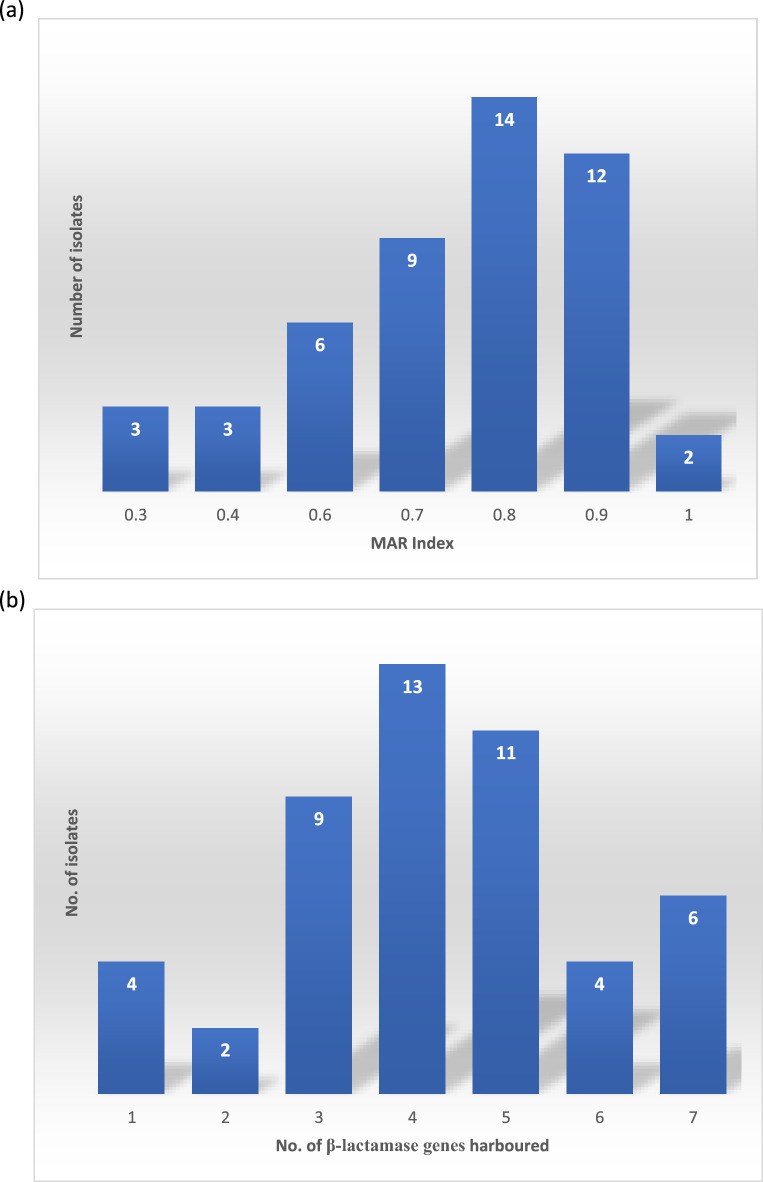
In accordance with the selection criteria, all isolates included in the study exhibited resistance to at least one antibiotic within three distinct classes, indicating multidrug resistance (MDR). A total of 14 (34%) of the isolates were resistant to antibiotic agents in at least eight of nine antibiotic classes tested. There was a total of 30 different beta-lactam resistance genes found in this study with the isolates harbouring a minimum of one and a maximum of seven (average of 4) beta-lactamase genes. The multiple antimicrobial resistance (MAR) index and number of β-lactam resistance associated antimicrobial resistance genes (ARGs) harboured by the isolates are shown in Fig. 1 below.
Figure 1.
(a) Multiple Antibiotic Resistance Index of Enterobacterales in study. (b) Isolates with number of β-lactam resistance genes harboured.
Antimicrobial resistance genes and the beta-lactamase mobilome
In silico identification of ARGs showed that isolates harboured a wide range of plasmid-mediated and chromosomally encoded resistance genes. Genes encoding resistance to beta-lactam drugs, which include carbapenemases, ESBLs, and AmpCs, are shown in the Table 3 below, along with frequency of detection in the Enterobacterales isolates studied.
Table 3.
Beta-lactam resistance genes detected among studied Enterobacterales.
| Antimicrobial resistance gene | Frequency n (%) | Antimicrobial resistance gene | Frequency n (%) | Antimicrobial resistance gene | Frequency n (%) |
|---|---|---|---|---|---|
| Penicillinase genes (n = 93) | AmpC and ESBL genes (n = 84) | Carbapenemase genes (n = 33) | |||
| blaTEM-1B | 29 (59.2) | blaCTX-M-15 | 38 (77.6) | blaNDM-1 | 17 (51.5) |
| blaSHV-187 | 22 (44.9) | blaOXA-1 | 27 (55.1) | blaNDM-5 | 12 (36.4) |
| blaOKP-B-15 | 9 (18.4) | blaACT-2 | 5 (1.22) | blaOXA-48 | 3 (9.1) |
| blaCMH-3 | 8 (16.3) | blaSST-1 | 3 (6.12) | blaOXA-181 | 1 (3.0) |
| blaOKP-B-2 | 7 (14.3) | blaCMY-2 | 2 (4.08) | ||
| blaSHV-67 | 3 (6.12) | blaCMY-48 | 2 (4.08) | ||
| blaTEM-1A | 3 (6.12) | blaSHV-106/blaSHV-28 | 2 (4.08) | ||
| blaOXA-2 | 2 (4.08) | blaCTX-M-14 | 1 (2.04) | ||
| blaSHV-11 | 2 (4.08) | blaDHA-1 | 1 (2.04) | ||
| blaTEM-1D | 2 (4.08) | blaDHA-22 | 1 (2.04) | ||
| blaCMH-6 | 1 (2.04) | blaSHV-12 | 1 (2.04) | ||
| blaMAL-2 | 1 (2.04) | blaACT-5 | 1 (2.04) | ||
| blaOKP-A-9 | 1 (2.04) | ||||
| blaOXA-320 | 1 (2.04) | ||||
| blaOXA-534 | 1 (2.04) | ||||
| blaOXA-9 | 1 (2.04) | ||||
The most common AMR gene detected was the blaCTX-M-15 with 38 (77.6%) isolates harbouring the gene. Of these blaCTX-M-15, 19 (50.0%) of them did not appear on mobile genetic elements (MGEs) and notably, there were no other AMR genes in close proximity to them. In contrast, the other 50% of blaCTX-M-15 carried on MGEs were rarely carried alone and were associated with several combinations of blaTEM-1A, blaTEM-1b, aph(3'')-Ib, aac(3)-Iia, sul2, and tet(A). (See Supplementary Table 1) The MGEs carrying the blaCTX-M-15 were primarily insertion sequences with the most common association being ISEc9 (17), IS5075 (1), and ISKpn43 (1).
The blaOXA-1 was the next most common ESBL gene being present in 27 (55.1%) isolates. Only two of these were mapped to IS26 insertion sequence while the others 25 (81.5%) were not mapped to MGEs. All blaOXA-1 genes were located in close association with blaOXA-320, blaOXA-534, catB3 and aac(6')-Ib-cr.
Thirty-three isolates harboured carbapenemase genes, which were primarily blaNDM-1 (35.0%) and blaNDM-5 (25.0%) of which three were susceptible to meropenem (details are shown in the Supplementary Table 1). For the rest of the 46 isolates, the genotypic resistance profiles matched the phenotypic resistance profiles. In addition, the resistance genes identified by Xpert Carba-R testing and WGS were in agreement (the identification of blaOXA-48 by GeneXpert is inclusive of the blaOXA-48 variant blaOXA-181). Most blaNDM-1 (11, 64.7%) were associated with the insertion sequence ISEc33 and the others not on any MGE. In contrast, blaNDM-5 were mostly not located on MGEs (10, 83.3%) with the remaining two associated with IS5. The one blaOXA-181 was associated with the rarely reported insertion sequence ISKpn19 of ISKra4 family.
While no beta-lactamase gene was mapped to plasmids, a total of 21 plasmids in 10 classes were detected in this study for E. coli including Col (BS512) (3), ColKP3 (1), IncB/O/K/Z (1), IncFIA (5), IncFIB (AP001918) (5), IncFII (5), IncFII(pAMA1167-NDM-5) (2), IncI1-I(Alpha) (3), IncQ1 (1), IncX3 (3). A total of 18 plasmids were detected among all isolates of Klebsiella species, including IncFIB (AP001918) (5), IncFII (5), IncFII(pAMA1167-NDM-5) (2), IncI1-I(Alpha) (3), IncQ1 (1), IncX3 (3).
Sequence types of Enterobacterales in study
The ST-88 (n = 2) E. coli had high MAR index of 0.7 and harboured relatively higher number of AMR genes (average of 21 genes) than other STs. The two ST-692 had lower average MAR index (average = 0.55), and lower total number of AMR genes (n = 17) than ST-88.
Of the 28 Klebsiella spp strains, the most common STs were ST-476 (n = 8), followed by ST-147 (n = 3). There were two each of ST-14, ST-1788, ST-25, and ST-429 (Supplementary Table 1). There was one each of ST-101, ST-1224, ST-13, ST-17, ST-336, ST-353, and ST-978. For analysis. the STs were grouped as the commonest ST-476, ST-147, and ‘others’. All ST-147 and ST-476 isolates had at least 17 AMR genes, harboured carbapenemase genes, and were carbapenem resistant unlike the ‘other’ group of STs. The ST-147 group had the highest MAR index average of 7.7, followed by the ST-476 group which had an average MAR index of 7.2 compared to 6.2 average MAR index of the ‘other’ group.
The most frequent Enterobacter cloacae ST was ST-1107 (7) with an average MAR index of 6.4, and with one ST-167 that had lower MAR index of 3. There were two strains of Citrobacter freundii, (ST-22), both with an average 19 AMR genes.
Phylogenetic relatedness of Enterobacterales in study
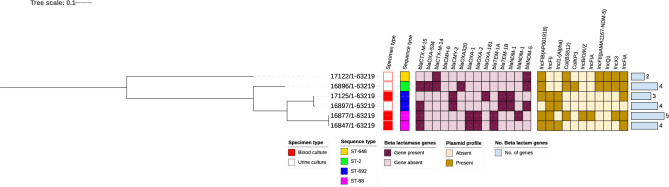
The SNP -based phylogenetic relatedness of E. coli is captured in the Fig. 2 below indicating the STs, carbapenemase genes, other significant antimicrobial resistance genes, and total number of antimicrobial resistance genes detected. Pairwise SNP difference for all isolates was > 20 implying there were no outbreak strains among the tested strains.
Figure 2.
SNP-based phylogenetic relatedness of E. coli STs, plasmids, and resistance genes. SNP-based phylogenetic tree of MDR E. coli isolates visualized in Interactive Tree of Life tool (iTOL). SNP-based phylogenetic tree of MDR E. coli isolates visualized in Interactive Tree of Life tool (iTOL). Shown for each isolate from left to right are colour strips for specimen source, and sequence type, heat map for beta-lactamase gens, and bar chart of number of beta-lactamase genes harboured. Tree was midpoint rooted.
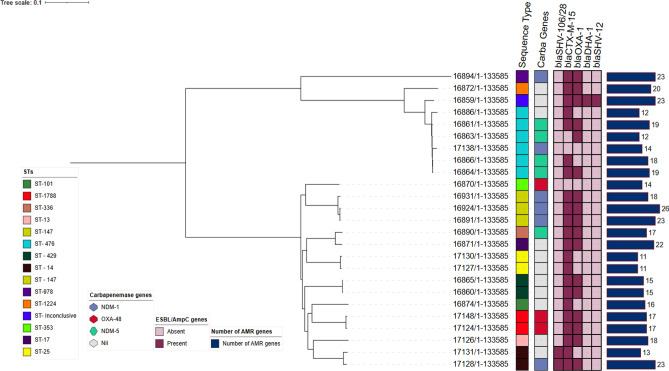
None of the Klebsiella spp strains in this collection appeared to be outbreak-related as SNP distances were > 20 for all isolates. The phylogenetic tree shown in Fig. 3 depicts the STs, carbapenemase genes, other significant AMR genes, and total number of AMR genes detected. See Supplementary Table 1 for pairwise SNP differences.
Figure 3.
SNP-based phylogenetic relatedness of Klebsiella species STs, plasmids, and resistance genes. SNP-based phylogenetic tree of MDR E. coli isolates visualized in Interactive Tree of Life tool (iTOL). Shown for each isolate from left to right are colour strips for specimen source, and sequence type, heat map for beta-lactamase gens, and bar chart of number of beta-lactamase genes harboured. Tree was midpoint rooted.
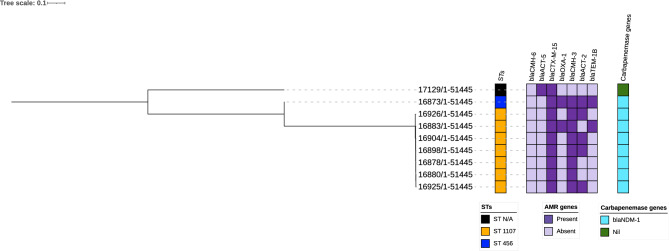
None of the Enterobacter cloacae strains in this collection appeared to be outbreak-related as SNP distances were > 20 for all isolates. The phylogenetic tree shown in Fig. 4 depicts the STs, carbapenemase genes, other significant AMR genes, and total number of AMR genes detected (See Supplementary Table 1) for pairwise SNP differences.
Figure 4.
SNP-based phylogenetic relatedness of Enterobacter cloacae STs, plasmids, and resistance genes. SNP-based phylogenetic tree of MDR E. coli isolates visualized in Interactive Tree of Life tool (iTOL). Shown for each isolate from left to right are colour strips for specimen source, and sequence type, heat map for beta-lactamase gens, and bar chart of number of beta-lactamase genes harboured. Tree was midpoint rooted.
Discussion
Antimicrobial resistance, particularly to broad-spectrum carbapenems, is a significant global challenge in medical and public health. Of great concern is the increasing prevalence of antibiotic resistance to beta-lactam antibiotics, which are commonly used as a first-line treatment for infections, particularly in developing nations where they are more cost-effective and easily accessible than other types of antibiotics. This poses a critical threat to infection care and highlights the urgent need for interventions to combat antimicrobial resistance.
Resistance to tigecycline among E. coli isolates was not observed in our study, including those strains that were carbapenem resistant. This is contrary to the 23% tigecycline resistance for carbapenem-resistant isolates reported in a surveillance study in the USA12. It is worth noting that tigecycline is not typically available or utilized in clinical settings in Nigeria. Except for K. pneumoniae which were 75% susceptible to fosfomycin, all other MDR-E studied here were susceptible. While fosfomycin is typically reserved for treatment of uncomplicated UTI, there is an unharnessed potential for use as empiric therapy for infections such as sepsis13. Tapping into the potential of tigecycline and fosfomycin especially for settings with high carbapenem resistance is worth investigating further especially because it is cheaper and potentially carbapenem sparing.
A wide array of resistance genes (AMRGs) was found among the Enterobacterales in this study with most of the phenotypic resistance to beta-lactam antibiotics observed, corresponding with expected AMRGs except for the three isolates that harboured carbapenemase genes which were not resistant to carbapenems. This is likely because the genes were not expressed14. Genes encoding resistance to penicillins, cephalosporins, and carbapenems were detected both on mobile genetic elements and on the chromosome. The blaTEM-1B and blaSHV-187 were the most common penicillinase genes detected and always occurred with other ESBL or carbapenemase genes. The finding of blaCTX-M-15 and blaOXA-1 genes as the more common ESBL genes are in keeping with other studies from the African continent4,15,16. The insertion sequence located upstream of the gene and reported as responsible for its mobilization is however different as this study had almost complete ISEc9 unlike previously reported ISEc117. This study findings also contrasts with other studies which have found the blaCTX-M-15 to be primarily carried on plasmids15,18.
In concordance with other studies conducted in similar settings, the blaNDM-1 and blaNDM-5 were the most common carbapenemase genes detected11,19,20. The strains of Enterobacterales in this study were highly diverse with a significant number of potentially unreported/inconclusive STs that need further analysis. The E. coli ST-648, which is considered a high-risk clone mostly reported in hospital settings, and was identified in this setting. Consistent with previous studies, it was recovered from the hospital, was resistant to most antibiotics tested, and harboured a large number of AMR genes21. Newly emerging high risk sequence types of K. pneumoniae in this study include the ST-101 and ST-147 which had previously been reported as circulating outside the African continent suggests a more global dissemination of this clone22,23. The predominant Enterobacter cloacae, ST-1107, is a high-risk clone associated with the blaOXA-48 carbapenemase gene and has been implicated in hospital outbreaks24. Similarly, C. freundii ST-22 had previously been characterized as being associated with MDR infections in hospital setting25,26. While many of the isolates were collected from the same hospitals around the same time period, there was no suggestion of an outbreak strain across all isolates based on ST analyses of individual species although the purposive sampling from a large collection of strains might hinder accurate determination of outbreaks.
While carbapenemases not located on mobile genetic elements are relatively uncommon27,28, our study detected these in high numbers outside of MGEs. These chromosomal carbapenemases conferring intrinsic and stable resistance is a concerning development as can lead to more propagation and transmission of these MDR bacteria.
In conclusion, our study has identified high risk Enterobacterales strains that exhibit both phenotypic and genotypic mechanisms of antimicrobial resistance, posing a serious threat to public health in Nigeria. The emergence and dissemination of these multidrug and extensively drug-resistant strains is a major concern, highlighting the urgent need for comprehensive surveillance and monitoring programs which include molecular methods. Our findings emphasize the importance of incorporating both conventional laboratory techniques and modern molecular techniques to provide a comprehensive characterization of antimicrobial resistance in Enterobacterales infections within Nigeria. This approach can facilitate the early detection and prompt response to emerging resistance patterns, allowing for the implementation of effective infection control measures and the development of new therapeutic options.
Methods
Study design, sample collection, isolate primary processing and final selection
A total of 49 MDR-E isolates were selected from our archives and processed for this study. These were selected randomly from the archive of over 200 MDR-E isolated from the parent studies of Community-Acquired Bacteremic Syndromes in Young Nigerian Children (CABSYNC) and Community-acquired Pneumonia and Invasive Bacterial Diseases (CAPIBD). These 49 were chosen as a convenience number. All 49 non-duplicate isolates were obtained from clinical specimens collected from the cities of Kano and Abuja. Clinical isolates were collected from patients with suspected blood stream infections, urinary tract infections (UTIs), and wound infections. In Kano, the clinical isolates were obtained from Aminu Kano Teaching Hospital (AKTH), Hasiya Bayero Children’s Hospital (HBCH), and Murtala Mohammed Specialist Hospital (MMSH) while in Abuja, isolates were obtained from the University of Abuja teaching hospital (UATH) and National Hospital Abuja (NHA). Ethical approval was obtained from all the respective hospitals for the primary studies with these isolates collected from 2016 to 2021. Informed consent was collected from all participants and/or their legal guardians. The isolates have been stored in − 80 °C freezers since isolation from original samples.
Isolation, identification, and antibiotic susceptibility testing of Enterobacterales isolates
Primary processing was dependent on the specimen following standard laboratory procedures with cysteine lactose electrolyte deficient agar-CLED (Oxoid, Basingstoke, UK) for urine, 5% Sheep blood agar (SBA), chocolate agar, and MacConkey agar for positive automated blood culture vials, SBA, and MacConkey agar (Oxoid, Basingstoke, UK) for other specimen types. MacConkey and SBA were incubated in ambient air and Chocolate agar in 5% CO2 for 18–24 h at 35–37 °C. The identification and antimicrobial susceptibility testing (AST) of suspected Enterobacterales bacterial colonies were conducted using the Phoenix system (BD Diagnostic Systems, Sparks, MD, US) and interpreted according to Clinical and Laboratory Standards Institute (CLSI) M100 30th Edition guidelines29. The control strains E. coli ATCC 25922, and K. pneumoniae ATCC 700600 were used for quality control. At both IFAIN and Cepheid sites, Xpert® Carba-R (Cepheid, Sunnyvale, CA, USA) was used for identification of five carbapenem resistance genes (blaKPC, blaVIM, blaOXA-48, blaIMP, and blaNDM).
Isolate secondary processing
A total of 49 MDR-Es identified as above were randomly selected from our archives and shipped to Cepheid (Sunnyvale, CA, USA), for whole genome sequencing. Inclusion criteria was isolates identified as MDR-Es based on predefined resistance patterns and retrieved only from the six hospitals in the primary studies. We excluded duplicate isolates, isolates that were not MDR-Es, isolates with incomplete data, and those collected from hospitals other than the six mentioned here.
Isolates were re-identified using Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry MALDI-TOF MS (Bruker Daltonics, Germany) and their AST profiles re-confirmed with MicroScan Detect Neg MIC 2 on the WalkAway 40 SI system (Beckman Coulter, Inc., USA). ESBL confirmation was done according to the MicroScan software rules. Whole genome sequencing (WGS) was conducted and in silico prediction of antimicrobial susceptibility phenotype performed from WGS output. Carbapenem resistance was defined as phenotypic resistance to ertapenem, imipenem, or meropenem (CLSI)29. Multi-drug resistance was defined as resistance to at least one agent in three or more antimicrobial classes. The multiple antibiotic resistance (MAR) index for each isolate was calculated using the formula MAR = a/b, where 'a' represents the number of antibiotics the test isolate exhibited resistance to, and 'b' represents the total number of antibiotics to which the test isolates were exposed.
Whole-genome sequencing for detection and characterization of antimicrobial resistance genes, plasmids, and insertion sequences
Pure cultures of each organism were grown overnight in trypticase soy broth (Hardy Diagnostics, USA) and genomic DNA were extracted using the Qiagen DNeasy Blood and Tissue kit (Qiagen, USA). Sequencing libraries were prepared from extracted DNA using Nextera XT DNA Library Kit (Illumina, USA) and were sequenced using the Miseq instrument with Reagent Kit v3 (Illumina, USA) chemistry. Raw reads were trimmed for quality and de novo assembled using CLC Genomics Workbench v. 21.0.4 (Qiagen Digital Insights, Denmark). Sequence analysis of bacterial isolates included identification of best matching reference using K-mer spectra, multilocus sequence typing (MLST), and drug resistance analysis using CLC Microbial Genomics Module 21.1 (Qiagen) with ResFinder database (downloaded on 2021-05-30, Center for Genomic Epidemiology). All procedures were done in accordance with manufacturer protocols. Prediction of AMR was conducted by using Mobile Element Finder v1.0.3 (2020-10-09) and selecting Acquired Antimicrobial Resistance genes (ResFinder)30. We located the mobile genetic elements (MGEs) associated with resistance genes by using Mobile Element Finder with database v1.0.2 (2020-06-09)30. Each resistance gene was classified as being carried by a plasmid, other MGE, or as not associated with an MGE. Plasmids were detected using PlasmidFinder-2.0 with threshold for minimum at 95% identity and minimum 60% coverage using draft genome assemblies31. The AMR gene substrates were crosschecked on the CARD database website32. The high-quality Illumina paired-ends reads generated were assembled de novo into the draft genome sequence for every isolate using the CLC genomic workbench. Quality assessment for genome assemblies was also carried out using the CLC genomic workbench.
Multi-Locus sequence typing (MLST) of E. coli
The PubMLST—Achtman scheme was performed to identify the sequence types (STs) and clonal complexes (CCs) of the isolates33. Isolates with 100% match against known MLST alleles were assigned STs and CCs. Those without perfectly matching alleles were identified as unknown or inconclusive STs34. In silico MLST-analyses were performed using previously described seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA)35.
Calling SNPs and inferring phylogeny
The FASTA files generated from WGS were uploaded unto the CSI Phylogeny 4.1 service of Centre for Genomic Epidemiology (https://cge.cbs.dtu.dk/services/CSIPhylogeny/). CSI Phylogeny outputs were generated based on a selected reference sequence for the different Enterobacterales and downloaded as Newick and text files. Thresholds for SNP calling were for depth = 10 ×, for SNP quality − 30, for map quality − 25, and 1.96 for minimum Z score36. Visualization annotation, and management of tree files were performed using the interactive Tree of Life tool—iTOL v6 (http://itol.embl.de/itol.cgi). Pairwise SNP differences between genomes were computed to determine if isolates of different origins were related with SNP distances < 21 indicating close relatedness and 21–50 indicating more distant relatedness37.
Ethics statement
The ethics review boards of National Hospital Abuja (NHA) reviewed and gave approval for the study (Approval number NHA/EC/033/2018). The ethics review boards of Hasiya Bayero Children’s hospital, Murtala Mohammed specialist hospital (Approval numbers NHREC/21/08/2008/AKTH/EC/872 and AKTH/MAC/SUB/12A/P-3/VI/972) and Gwagwalada specialist hospital (Approval number FCTA/HHSS/NH/GEN/54/II/128) gave approval for the parent CAPIBD and CABSYNC studies. All bacteria isolates used here were recovered from submitted clinical specimens at the selected hospitals. All procedures we performed were in accordance with the guidelines and regulations of the ethics review board.
Data collection and analyses
The data collected and recorded in Microsoft Excel was processed and analysed using STATA software (StataCorp. 2019. TX: StataCorp LLC). The distribution of phenotypic and genotypic features was calculated and determined as frequencies and ratios. The relevant data for this study is included within the research article and can also be accessed as supplementary information.
Supplementary Information
Acknowledgements
Isolates used in this study were from the primary studies of Community-Acquired Bacteremic Syndromes in Young Nigerian Children (CABSYNC) and Community-acquired Pneumonia and Invasive Bacterial Diseases (CAPIBD) funded by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (award number R01AI097493) and the Bill & Melinda Gates Foundation (grant number OPP1034619). The resources for isolate transportation, MALDITOF, Xpert Carba-R and WGS of isolates were provided by Cepheid, Sunnyvale, CA. IFAIN supported other components of isolate characterization such as identification, susceptibility testing with Phoenix, mCIM and other phenotypic characterization tests. We recognize the contribution of IFAIN and Cepheid laboratory staff towards the processing the bacteria.
Author contributions
S.K.O. and F.C.T. conceived the study. N.M., I.A.T., C.D., R.E. and G.O. designed and conducted the test protocols. NM wrote the first draft manuscript. S.K.O., C.D., G.O., I.A.T., V.O., A.J., F.H., B.J., E.O. and F.C.T. revised the manuscript. All authors read and approved the final manuscript.
Data availability
The datasets used and analyzed during the current study are available from the corresponding author upon request. All data generated or analyzed during this study are also included in this published article and its supplementary information files. For whole genome sequencing, raw reads were uploaded to the National Center for Biotechnology (NCBI) database and can be accessed using BioProject ID: PRJNA952997.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-37621-z.
References
- 1.Murray CJ, et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet. 2022;399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford PA. Extended-spectrum β-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 2001;14:933. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myat TO, et al. ESBL- and carbapenemase-producing enterobacteriaceae in patients with bacteremia, Yangon, Myanmar, 2014. Emerg. Infect. Dis. 2017;23:857. doi: 10.3201/eid2305.161100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medugu N, et al. Molecular characterization of multi drug resistant Escherichia coli isolates at a tertiary hospital in Abuja, Nigeria. Sci. Rep. 2022;12:1–10. doi: 10.1038/s41598-022-19289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Queenan AM, Bush K. Carbapenemases: The versatile β-lactamases. Clin. Microbiol. Rev. 2007;20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiotos K, et al. Increased 30-day mortality associated with carbapenem-resistant enterobacteriaceae in children. Open Forum Infect. Dis. 2018 doi: 10.1093/ofid/ofy222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamma PD, et al. The likelihood of developing a carbapenem-resistant Enterobacteriaceae infection during a hospital stay. Antimicrob. Agents Chemother. 2019 doi: 10.1128/AAC.00757-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tamma PD, et al. Comparing the outcomes of patients with carbapenemase-producing and non-carbapenemase-producing carbapenem-resistant enterobacteriaceae bacteremia. Clin. Infect. Dis. 2017;64:257–264. doi: 10.1093/cid/ciw741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant enterobacteriaceae: The impact and evolution of a global menace. J. Infect. Dis. 2017;215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shettima SA, Tickler IA, dela Cruz, C. M. & Tenover, F. C. Characterisation of carbapenem-resistant Gram-negative organisms from clinical specimens in Yola, Nigeria. J. Glob. Antimicrob. Resist. 2020;21:42–45. doi: 10.1016/j.jgar.2019.08.017. [DOI] [PubMed] [Google Scholar]
- 11.Medugu N, et al. Molecular characterization of multi drug resistant Escherichia coli isolates at a tertiary hospital in Abuja, Nigeria. Sci. Rep. 2022 doi: 10.1038/s41598-022-19289-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulens MPH, S. N. et al. Carbapenem-Resistant enterobacterales in individuals with and without health care risk factors—Emerging infections program, United States, 2012–2015. Am J Infect Control0, 1–8 (2022). [DOI] [PMC free article] [PubMed]
- 13.Williams PCM, et al. The potential of fosfomycin for multi-drug resistant sepsis: An analysis of in vitro activity against invasive paediatric Gram- negative bacteria. J. Med. Microbiol. 2019;68:711. doi: 10.1099/jmm.0.000973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodford N, Sundsfjord A. Molecular detection of antibiotic resistance: When and where? J. Antimicrob. Chemother. 2005;56:259–261. doi: 10.1093/jac/dki195. [DOI] [PubMed] [Google Scholar]
- 15.Agyekum A, et al. blaCTX-M-15 carried by IncF-type plasmids is the dominant ESBL gene in Escherichia coli and Klebsiella pneumoniae at a hospital in Ghana. Diagn. Microbiol. Infect. Dis. 2016;84:328–333. doi: 10.1016/j.diagmicrobio.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 16.Duru C, et al. Molecular characterization of invasive Enterobacteriaceae from pediatric patients in Central and Northwestern Nigeria. PLoS ONE. 2020;15:165. doi: 10.1371/journal.pone.0230037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirel L, Naas T, Nordmann P. Genetic support of extended-spectrum β-lactamases. Clin. Microbiol. Infect. 2008;14:75–81. doi: 10.1111/j.1469-0691.2007.01865.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhuo C, Li XQ, Zong ZY, Zhong NS. Epidemic plasmid carrying bla CTX-M-15 in Klebsiella penumoniae in China. PLoS ONE. 2013;8:52222. doi: 10.1371/journal.pone.0052222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spread of Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates Producing NDM-Type Metallo-β-Lactamase in Myanmar - PMC. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9431462/. [DOI] [PMC free article] [PubMed]
- 20.Sugawara Y, et al. Characterization of blaNDM-5-harbouring Klebsiella pneumoniae sequence type 11 international high-risk clones isolated from clinical samples in Yangon General Hospital, a tertiary-care hospital in Myanmar. J. Med. Microbiol. 2021 doi: 10.1099/jmm.0.001348. [DOI] [PubMed] [Google Scholar]
- 21.Eger E, et al. Circulation of extended-spectrum beta-lactamase-producing Escherichia coli of pandemic sequence types 131, 648, and 410 among hospitalized patients, caregivers, and the community in Rwanda. Front. Microbiol. 2021 doi: 10.3389/fmicb.2021.662575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonnin RA, et al. Emergence of new non-clonal group 258 high-risk clones among Klebsiella pneumoniae Carbapenemase-Producing K. pneumoniae Isolates, France. Emerg. Infect. Dis. 2020;26:1212–1220. doi: 10.3201/eid2606.191517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaidullina ER, et al. Genomic analysis of the international high-risk clonal lineage Klebsiella pneumoniae sequence type 395. Genome Med. 2023;15:9. doi: 10.1186/s13073-023-01159-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lpez-Hernndez I, et al. Carbapenemase-producing gram-negative bacteria in Andalusia, Spain, 2014–2018. Emerg. Infect. Dis. 2020;26:2218–2222. doi: 10.3201/eid2609.191772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruegsegger L, et al. Multidrug-resistant gram-negative bacteria in burn patients. Antimicrob. Agents Chemother. 2022 doi: 10.1128/aac.00688-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hadjadj L, et al. Outbreak of carbapenem-resistant enterobacteria in a thoracic-oncology unit through clonal and plasmid-mediated transmission of the bla OXA-48 gene in Southern France. Front. Cell. Infect. Microbiol. 2022 doi: 10.3389/fcimb.2022.1048516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petit A, Ben-yaghlane-bouslama H, Sofer L, Labia R. Characterization of chromosomally encoded penicillinases in clinical isolates of Klebsiella pneumoniae. J. Antimicrob. Chemother. 1992;29:629–638. doi: 10.1093/jac/29.6.629. [DOI] [PubMed] [Google Scholar]
- 28.Nordmann P, Mariotte S, Naas T, Labia R, Nicolas MH. Biochemical properties of a carbapenem-hydrolyzing beta-lactamase from Enterobacter cloacae and cloning of the gene into Escherichia coli. Antimicrob. Agents Chemother. 1993;37:939. doi: 10.1128/AAC.37.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clinical and Laboratory Standards Institute. CLSI M100 30th Edition. Journal of Services Marketing vol. 30th (2020).
- 30.Johansson MHK, et al. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021;76:101–109. doi: 10.1093/jac/dkaa390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carattoli A, et al. In Silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Comprehensive Antibiotic Resistance Database. https://card.mcmaster.ca/.
- 33.Jolley, K. A., Bray, J. E. & Maiden, M. C. J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications [version 1; referees: 2 approved]. Wellcome Open Res3, (2018). [DOI] [PMC free article] [PubMed]
- 34.Larsen MV, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wirth T, et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE. 2014;9:e104984. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pightling AW, et al. Interpreting whole-genome sequence analyses of foodborne bacteria for regulatory applications and outbreak investigations. Front. Microbiol. 2018;9:1482. doi: 10.3389/fmicb.2018.01482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author upon request. All data generated or analyzed during this study are also included in this published article and its supplementary information files. For whole genome sequencing, raw reads were uploaded to the National Center for Biotechnology (NCBI) database and can be accessed using BioProject ID: PRJNA952997.