Abstract
cis-Aconitate decarboxylase (ACOD1, IRG1) converts cis-aconitate to the immunomodulatory and antibacterial metabolite itaconate. Although the active site residues of human and mouse ACOD1 are identical, the mouse enzyme is about fivefold more active. Aiming to identify the cause of this difference, we mutated positions near the active site in human ACOD1 to the corresponding residues of mouse ACOD1 and measured resulting activities in vitro and in transfected cells. Interestingly, Homo sapiens is the only species with methionine instead of isoleucine at residue 154 and introduction of isoleucine at this position increased the activity of human ACOD1 1.5-fold in transfected cells and 3.5-fold in vitro. Enzyme activity of gorilla ACOD1, which is almost identical to the human enzyme but has isoleucine at residue 154, was similar to the mouse enzyme in vitro. Met154 in human ACOD1 forms a sulfur-π bond to Phe381, which is positioned to impede access of the substrate to the active site. It appears that the ACOD1 sequence has changed at position 154 during human evolution, resulting in a pronounced decrease in activity. This change might have offered a selective advantage in diseases such as cancer.
Subject terms: Enzyme mechanisms, Enzymes, Molecular evolution, Evolutionary genetics, Dendritic cells, Monocytes and macrophages, Antimicrobial responses, Innate immunity
Introduction
cis-Aconitate decarboxylase (ACOD1) catalyses the decarboxylation of cis-aconitate to produce itaconic acid, which is a key immunomodulatory metabolite of activated macrophages. It regulates cytokine production and inhibits viral replication1,2 and also plays roles in immune defences against bacterial pathogens3.
Published data suggest that mouse macrophages produce higher levels of itaconic acid than human cells, even though no direct comparison has been reported. Concentrations of 1.5–8 mM have been found in the mouse macrophage cell line RAW264.7 and in primary mouse bone-marrow-derived macrophages (BMDM) upon activation with LPS4–6. Levels in the human macrophage-like cell line dTHP1 were low upon LPS activation and simultaneous treatment with LPS and IFNγ was required to reach an intracellular itaconate concentration of 460 µM7. Primary human macrophages contained only approx. 60 µM itaconate upon LPS activation5.
Correspondingly, the mouse ACOD1 enzyme is more active with a significantly higher catalytic rate constant kcat of 4.9 s−1 in comparison to the human enzyme with 0.9 s−18. The Michaelis-constants KM were similar. The active site residues of both enzymes are completely conserved8, suggesting that the higher activity of the murine orthologue is determined by residues outside the active site. The mechanism of ACOD1 catalysis includes an opening of the active site by tilting the smaller domain relative to the larger domain (Fig. 1)8,9. This opening is required for the substrate to get to the active site. It is expected that the enzyme’s activity is determined not only by the conserved active site residues interacting with the substrate, but also by residues involved in the opening and closing movements. To determine the structural basis for the difference in ACOD1 activity between mice and human, we investigated residues near the active site and at the domain interface that might be relevant for the higher activity of mouse ACOD1 (mACOD1)8,9.
Figure 1.

Crystal structure of a subunit of the human ACOD1 homodimer (PDB ID code 6R6U)8. Mutated residues differing between human and mouse ACOD1 are shown as sticks in cyan and conserved active site residues as magenta sticks. All these residues are close to the interface between the smaller (green) and larger (yellow) domain.
Results
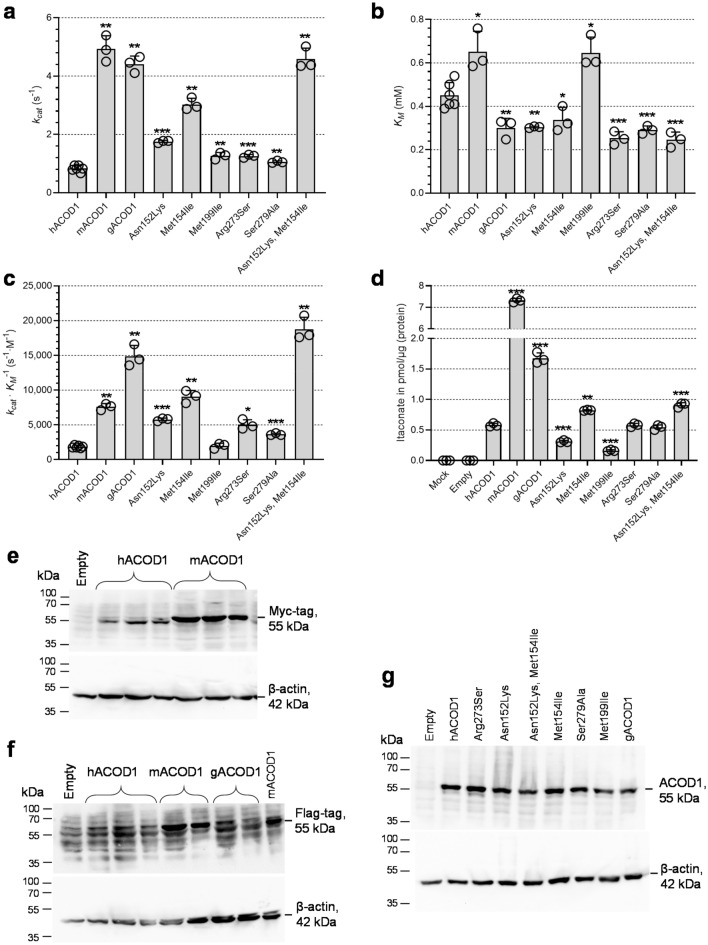
We selected five amino acid positions close to the active site and to the interface of the protein’s two domains, which differ between human and mouse ACOD1 (Fig. 1). We individually mutated the corresponding amino acids of the human sequence to their mouse counterparts. The mutations were Asn152Lys, Met154Ile, Met199Ile, Arg273Ser and Ser279Ala (Fig. 2). Recombinant proteins were produced in E. coli and their activity was tested in vitro (Fig. 3a–c). In parallel, A549 cells (which do not express the endogenous ACOD1 gene) were transfected with wild-type and mutated plasmids, followed by measurement of intracellular itaconic acid concentrations (Fig. 3d). All mutations had a positive effect on the catalytic rate constant kcat in vitro (Fig. 3a). Especially for the Met154Ile mutant, kcat and itaconic acid production in cells increased strongly in comparison to wild-type human ACOD1 (hACOD1). The mutations also slightly improved substrate affinity and resulted in decreased Michaelis-constants KM for all mutants except Met199Ile (Fig. 3b). A double mutant combining the mutations with the highest effect on kcat, Asn152Lys and Met154Ile, had an even higher kcat value. Notably, this double mutant had the highest catalytic efficiency (kcat/KM ratio) of all tested enzymes (Fig. 3c). The results of the in vitro assay and the transfection experiments corresponded well. However, while the Asn152Lys and Met154Ile double mutant had a similar activity as mouse ACOD1 in vitro, the transfection experiments with mouse ACOD1 resulted in much greater itaconic acid accumulation as compared to transfection with the double mutant of hACOD1 (Fig. 3d). This discrepancy can be explained by higher expression of mACOD1, in comparison to the human enzyme and the mutants. In order to measure expression levels of the expressed proteins, we made use of their C-terminal Flag and Myc tags. The mouse protein was readily detected in immunoblots with antibodies either against the Myc-tag or Flag-tag of the recombinant proteins, and the signal was considerably stronger in comparison to human or gorilla ACOD1 (Fig. 3e,f). Human wild type and mutant ACOD1 and gorilla ACOD1 (gACOD1) were all expressed at a similar level (Fig. 3g). It, therefore, appears that mACOD1 is expressed more efficiently than the human or gorilla sequences in our transfection system.
Figure 2.
Sequence alignment of ACOD1 from human (IRG1_HUMAN), gorilla (G3RFY3_GORGO), chimpanzee (H2Q7N6_PANTR) and mouse (IRG1_MOUSE). Positions that were mutated are displayed with a pink background and non-conserved residues are shown in black.
Figure 3.
Activities of ACOD1 mutants. (a–c) kcat, KM and kcat/KM values of wild-type human, mouse and gorilla ACOD1 and human ACOD1 mutants, resulting from in vitro enzyme assays using purified enzymes and HPLC/UV quantification of itaconate production. (d) Proteins corresponding to (a) were expressed in transfected A549 cells and intracellular itaconic acid was measured by LC/MS 24 h post transfection. Individual measurements are represented by open circles and error bars represent standard deviations. Significant P-values calculated with Welch's unequal variances t-test for comparison with hACOD1 values are indicated by asterisks: *P < 0.05, **P < 0.01, ***P < 0.001 (Supplementary Table 1). (e–g) Immunoblots of ACOD1 proteins overexpressed in transfected A549 cells. (e) Overexpressed hACOD1 and mACOD1 plasmids were detected by Myc-tag antibody (three independent transfections). (f) Overexpressed hACOD1, mACOD1 and gACOD1 detected by Flag-tag antibody. (g) Overexpressed wild-type and mutant hACOD1 and gACOD1 detected by anti-human ACOD1 antibody. Loading internal control is β-actin. Empty, transfection with the empty pCMV6-Entry vector. Original blots are presented in Supplementary Figs. 1–3.
Interestingly, the methionine at position 154 is specific to humans, and sequences of Neandertals and Denisovans that we analysed also have the Met154 codon. Essentially all other known mammalian ACOD1 sequences, including those of other hominids, have an isoleucine at position 154 (Fig. 2). This indicates that Met154 was specifically acquired during human evolution before the population split between humans, Neandertals and Denisovans. Because Met154 is human-specific, hACOD1 was expected to have a lower activity than ACOD1 from our closest relatives, chimpanzee and gorilla. Gorilla and human ACOD1 differ at 5 positions (Fig. 2). Recombinant gorilla ACOD1 was highly active both in the in vitro assay and in the transfected cells and was more active than the Met154Ile mutant of hACOD1 (Fig. 3a,d). Chimpanzee ACOD1 (cACOD1) could not be produced in our E. coli system and it was expressed only weakly in the transfected cells, without detectable itaconic acid production.
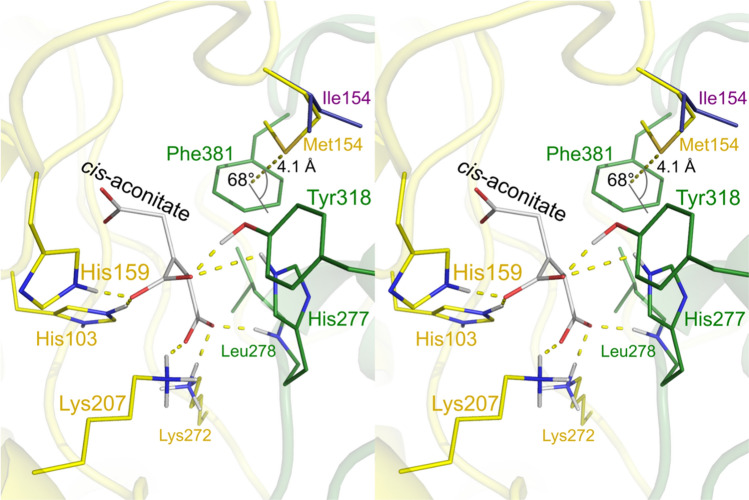
A sulfur-π (Met-aromatic) bond10 between Met154 and Phe381 was identified by the computer program MetAromatic11,12 (Fig. 4, Supplementary Table 2). This human-specific interaction connects the larger and the smaller domain, suggesting that it stabilizes the enzyme’s closed conformation.
Figure 4.
Stereo view of the hACOD1 active site and the sulfur-π bond between Met154 and Phe381. The active site of the hACOD1 structure (PDB ID code 6R6U) is shown with the putative conformation of the substrate cis-aconitate described in Chen et al.8. In this model, the substrate is bound by residues of the larger domain (yellow) and the smaller domain (green). The sulfur atom of Met154 is located at 4.1 Å distance from the Phe381 ring centre (dashed yellow line) at an angle of 68° to the plane of the aromatic ring. The Ile154 residue of the aligned mACOD1 structure of Chun et al.9 (PDB ID code 7BR9) is shown in purple.
Discussion
A cause for the different activities of mouse and human ACOD1 was identified. It was found that a human-specific residue, Met154, markedly reduces the activity of ACOD1. All other hominid ACOD1 sequences have an isoleucine at this position. The gorilla enzyme was found to be even more active than the mouse enzyme. This was unexpected, as the sequences of gorilla and human ACOD1 are 99% identical (Fig. 2). It suggests that during the evolution of Homo sapiens from the last common primate ancestor, a mutation at position 154 to methionine was acquired, leading to several-fold lower activity of the enzyme. Thus, it is expected that itaconate levels are lower in human ACOD1-expressing cells in comparison to other hominids.
Changing Asn152 to Lys also increased activity in the present study, especially in the double mutant with Met154Ile. We have previously identified a variant allele of human ACOD1 in African ethnicity that changes Asn152 to Ser, leading to a 50% increase in enzyme activity8, underlining the importance of this position for enzyme activity. Asn152 is conserved in primates, but is not well conserved throughout mammals.
Human evolution from a common primate ancestor was driven by large changes in genome structure, by gene duplications and changes in non-coding sequences that regulate gene activity13. In addition, non-synonymous point-mutations in protein-encoding sequences resulted in gene inactivation. Only a small number of human-specific amino acid substitutions were identified that resulted in changes in protein function and biochemical properties. The brain-related proteins FOXP214,15, MCPH116 and ASPM17 and the male reproduction-associated protamines PRM1 and PRM218 contain amino acid substitutions that arguably were targets of positive selection, indicating that the substitutions have an effect on protein function19. The Met154 in hACOD1 represents a single amino acid change during human evolution for which the biochemical consequences could be clarified.
The mechanism of ACOD1 catalysis includes an opening of the active site by tilting the smaller domain relative to the larger domain (Fig. 1)8,9. The side chains of residues 152 and 154 are located at the interface of the smaller and larger domain, and residue 154 also lines a hydrophobic pocket in the active centre. A sulfur-π bond was identified between Met154 and Phe381 of hACOD1, which links the two domains and stabilizes the enzyme’s closed conformation (Fig. 4). Sulfur-π bonds yield significant additional stabilization in comparison to purely hydrophobic interactions20,21. A higher stability of the closed conformation of hACOD1 would reduce substrate access to the active site and could thereby decrease overall enzyme activity.
The catalytic rate constant kcat of mACOD1 was 5.9 fold higher than of hACOD1 in the present study. The catalytic efficiency kcat/KM, which is a measure of enzyme catalysis at low substrate concentrations, differed by a factor of 4.1. It should be noted that these in vitro enzyme parameters are not the only predictors of itaconate levels upon macrophage activation. Rates of transcription and translation and transcript as well as protein stability are also important, as are the rate of itaconate catabolism, secretion into the extracellular environment, and substrate availability. The higher activity of mouse ACOD1 may therefore not be the only reason for the much higher levels of itaconate in mouse cells. Further research is needed in order to identify the relative contributions of all parameters that ultimately determine intracellular itaconate accumulation.
Itaconate is an immune-regulatory compound with beneficial or detrimental effects in different kinds of diseases. It is an anti-infective and anti-inflammatory immunometabolite22. On the other hand, ACOD1 has also been reported to enhance tumour growth and reduce T-cell activity23–26 and aggravate sepsis in a mouse model27. One may hypothesize that the change to Met154 in human ACOD1, reducing itaconic acid synthesis, could contribute to a higher resistance against cancer, whereas the frequent Asn152Lys variant counteracts this reduction of enzyme activity, potentially improving host defence against infections. However, it is difficult to infer from the above reports about the role, if any, of ACOD1 activity in human evolution because most functional studies on ACOD1 (and itaconate) have been conducted in mouse models. Nonetheless, the differences between murine and human ACOD1 reported herein can serve as stepping stones for further studies on human ACOD1 function.
Materials and methods
Plasmids
Plasmids are listed in Supplementary Table 3. Plasmids pCAD29 and pCAD39 were used for E. coli expression of residues 4–461 of hACOD1 (GenBank NM_001258406) and residues 4–462 of mACOD1 (GenBank NM_008392) with N-terminal StrepTagII and TEV protease cleavage site8. Transfections were done with plasmids pCMV6Entry-hIrg1 and pCMV6Entry-mIrg1 for expression of full-length hACOD1 and mACOD1 with C-terminal Myc-tag and Flag-tag. Single point mutations were introduced into the hACOD1 expression vectors pCAD29 and pCMV6Entry-hIrg18 by QuikChange mutagenesis (see Supplementary Table 4 for QuikChange primers).
The cACOD1 and gACOD1 plasmids for expression in E. coli were cloned with the Golden Mutagenesis method, using pCAD29_hIRG1_4-461_pvp008 as the template28. Primers were designed with the GoldenMutagenesis R library (see Supplementary Tables 5 and 6 for primers and PCRs). All primers had the tail GCGGGTACCGGTCTC, including a KpnI site. PCR products were cloned into the KpnI site of the vector pUC19exBsaI. pUC19exBsaI, a gift of Martin Bommer (Max Delbrück Center, Berlin, Germany), was derived from pUC19 by removing its BsaI site by introducing a silent point mutation. The resulting clones were used in Golden Gate reactions29 with the backbone vector pET-T7pro-ter, also a gift of Martin Bommer. pET-T7pro-ter was derived from pET-28a(+) by replacing the sequence from the NcoI site to the NotI site (CCATGG…GCGGCCGC) with the sequence CAATGAGAGACCGGT ACCGGTCTCAAGGTT AGTAAGCGGCCGC for Golden Gate cloning with BsaI (underlined). For Golden Gate cloning, the pET-T7pro-ter vector was diluted to 50 ng/µl (14.6 pM). The pU19exBsaI-derived plasmids were diluted to twice the molar concentration of pET-T7pro-ter (29.2 pM). The reactions were set up on ice with 1 µl of each plasmid, 2 µl T4 ligase buffer (NEB), 1 µl BsaI-Hfv2 (NEB) and 0.5 µl T4 DNA ligase (400 U/µl, NEB) and water to 10 µl and were incubated for 30 cycles of 5 min at 37 °C and 5 min at 16 °C, followed by incubation at 16 °C overnight. The reactions were heated for 5 min at 60 °C and then 5 µl were used for transformation of 50 µl OneShot OmniMAX 2 T1 chemically competent cells. This resulted in the clones pCAD161 (chimpanzee ACOD1, cACOD1) and pCAD183 (gACOD1). cACOD1 and gACOD1 cDNA sequences were cloned into pCMV6Entry by SLIC cloning30. The vector backbone fragment was generated by EcoRI and NotI digestion of pCMV6Entry-hIrg1. The full open reading frames of cACOD1 and gACOD1, including the missing terminal codons, were obtained by PCR from E. coli expression plasmids with long primers (Supplementary Tables 5, 6). For cACOD1, a full-length ORF (cACOD1.1, pCAD181, primers cCAD-pCMV6-LF and cCAD_1-480_pCMV6_R) and an N-terminally truncated ORF, starting at position 11 (cACOD1.11, pCAD180, primers cCAD-pCMV6-SF and cCAD_1-480_pCMV6_R), were cloned with the Zero blunt TOPO cloning kit (Thermo Fisher Scientific), generating clones TOPO4 and TOPO5. The N-terminus of the truncated cACOD1.11 ORF is the same as in hACOD1 (MMLKSITES…). The inserts of TOPO4 and 5 clones were SLIC cloned into the pCMV6Entry vector by using a QuickFusion kit (Absource Diagnostics), generating clones cACOD1.11 (pCAD180) and cACOD1.1 (pCAD181). gACOD1 was SLIC cloned into pCMV6Entry directly.
Enzyme assays
ACOD1 proteins were produced in E.coli and purified as described8, using plasmids pCAD29 and pCAD39 for wild-type hACOD1 (aa 4–461) and mACOD1 (aa 4–461). A detailed protocol is available at protocols.io (10.17504/protocols.io.14egn2npyg5d/v2). Purified proteins were stored in GF buffer (10 mM HEPES, pH 7.4, 10% [v/v] glycerol, 150 mM NaCl, 1 mM TCEP) at −80 °C. Enzyme assays were performed in triplicates as described7. Briefly, 125 μL 0.2 M sodium phosphate buffer, pH 6.5, 15 μL enzyme and 10 µL cis-aconitate (neutralized, in water) were incubated at 37 °C for 10 min, followed by heat inactivation and HLPC quantification of itaconic acid. Assays were performed with seven different substrate concentrations. Curves of enzyme rate v over substrate concentration [S] were fitted using GraphPad Prism with the Michaelis–Menten equation v = kcat[S]/(KM + [S]) to determine kcat and KM.
Transfections
Human epithelial A549 adenocarcinoma cells (DSMZ no. ACC107) were transfected using Lipofectamine LTX and PLUS Reagent (ThermoFisher #15338100) for 24 or 48 h and itaconate production was measured as described8 according to our validated LC–MS/MS assay31.
Western blots
After aspirating the supernatant, the transfected A549 cells were washed once in ice-cold PBS and lysed in ice-cold RIPA buffer (containing protease inhibitor). Upon quantification of protein by Pierce BCA Protein Assay (ThermoFisher, #23225), lysates were diluted in 4× Laemmli sample buffer, then heat-denatured at 95 °C for 10 min. Equal amounts of protein extracts and 5 µl marker (PageRuler Plus Prestained Protein Ladder, #26620, ThermoFisher) were separated by 10% SDS-PAGE and transferred to a 0.45 µm nitrocellulose membrane (Amersham Protran Premium 0.45 µm NC Nitrocellulose Blotting Membrane, GE Healthcare #10600013). The membrane was blocked with 5% non-fat dry milk for 1 h at room temperature. After 3 × 10 min TBST (0.1% Tween 20 in TBS) washing, the membrane were incubated over night at 4 °C with primary antibodies specific to human ACOD1 (D6H2Y, Cell Signalling #77510; diluted 1:1000 in TBST), DYKDDDDK flag-tag (D6W5B, Cell Signaling, #14793S, diluted 1:1000 in 5% BSA), or Myc-tag (71D10, Cell Signalling #2278, diluted 1:1000 in 5% BSA in TBST), followed by incubation with the second antibody, goat anti-rabbit IgG-HRP (Southern Biotech #4030-05, diluted 1:5000 in TBST) for 1 h at room temperature. The bands were visualized by enhanced chemiluminescence via Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare #RPN2232). After stripping the membrane for 30 min at 60 °C in 62.5 mM Tris–HCl, 2% SDS, pH 6.8, β-actin bands were visualized using HRP-conjugated anti-β-Actin antibody (Abcam #ab49900, diluted 1:20,000 in TBST for 30 min at room temperature) via the ECL reagent.
Neandertal and Denisovan ACOD1 sequence analysis
The ACOD1 coding sequences of Neandertals and Denisovans were analysed with the UCSC Genome Browser (https://genome.ucsc.edu) and with support by Janet Kelso (Max Planck Institute for Evolutionary Anthropology, Leipzig, Germany). The codon of Met154 was covered by one Neandertal sequence read and more than 20 reads for Denisovans.
Supplementary Information
Acknowledgements
We would like to thank Claudia Wylegalla for expert technical assistance.
Author contributions
K.B. designed the study, performed experiments, analysed data and wrote the first draft of the manuscript. F.F.C. performed experiments, analysed data and edited the manuscript. M.Z., I.Y. and C.C. performed experiments and analysed data. W.B. and F.P. provided resources and edited the manuscript. All authors have read and approved the final version of the manuscript.
Funding
The study was funded by the China Scholarship Council (CSC) and by internal funds of the Helmholtz Centre for Infection Research. Open Access funding enabled and organized by Projekt DEAL.
Data availability
The raw data and materials that support the findings of this study are available from the corresponding authors upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fangfang Chen and Israfil Yalcin.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-37373-w.
References
- 1.O'Neill LAJ, Artyomov MN. Itaconate: the poster child of metabolic reprogramming in macrophage function. Nat. Rev. Immunol. 2019;19:273–281. doi: 10.1038/s41577-019-0128-5. [DOI] [PubMed] [Google Scholar]
- 2.Sohail A, et al. Itaconate and derivatives reduce interferon responses and inflammation in influenza A virus infection. PLoS Pathog. 2022;18:e1010219. doi: 10.1371/journal.ppat.1010219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peace CG, O'Neill LA. The role of itaconate in host defense and inflammation. J. Clin. Invest. 2022;132:e148548. doi: 10.1172/JCI148548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meiser J, et al. Itaconic acid indicates cellular but not systemic immune system activation. Oncotarget. 2018;9:32098–32107. doi: 10.18632/oncotarget.25956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michelucci A, et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. USA. 2013;110:7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mills EL, et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen F, et al. Citraconate inhibits ACOD1 (IRG1) catalysis, reduces interferon responses and oxidative stress, and modulates inflammation and cell metabolism. Nat. Metab. 2022;4:534–546. doi: 10.1038/s42255-022-00577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen F, et al. Crystal structure of cis-aconitate decarboxylase reveals the impact of naturally occurring human mutations on itaconate synthesis. Proc. Natl. Acad. Sci. USA. 2019;116:20644–20654. doi: 10.1073/pnas.1908770116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun HL, Lee SY, Kim K-H, Lee CS, Oh T-J, Park HH. The crystal structure of mouse IRG1 suggests that cis-aconitate decarboxylase has an open and closed conformation. PLoS ONE. 2020;15:e0242383. doi: 10.1371/journal.pone.0242383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reid KSC, Lindley PF, Thornton JM. Sulphur-aromatic interactions in proteins. FEBS Lett. 1985;190:209–213. doi: 10.1016/0014-5793(85)81285-0. [DOI] [Google Scholar]
- 11.Weber DS, Warren JJ. A survey of methionine-aromatic interaction geometries in the oxidoreductase class of enzymes: What could Met-aromatic interactions be doing near metal sites? J. Inorg. Biochem. 2018;186:34–41. doi: 10.1016/j.jinorgbio.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Weber DS, Warren JJ. The interaction between methionine and two aromatic amino acids is an abundant and multifunctional motif in proteins. Arch. Biochem. Biophys. 2019;672:108053. doi: 10.1016/j.abb.2019.07.018. [DOI] [PubMed] [Google Scholar]
- 13.Hollox EJ, Zuccherato LW, Tucci S. Genome structural variation in human evolution. Trends Genet. 2022;38:45–58. doi: 10.1016/j.tig.2021.06.015. [DOI] [PubMed] [Google Scholar]
- 14.Enard W, et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J, Webb DM, Podlaha O. Accelerated protein evolution and origins of human-specific features: Foxp2 as an example. Genetics. 2002;162:1825–1835. doi: 10.1093/genetics/162.4.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evans PD, et al. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science. 2005;309:1717–1720. doi: 10.1126/science.1113722. [DOI] [PubMed] [Google Scholar]
- 17.Evans PD, et al. Adaptive evolution of ASPM, a major determinant of cerebral cortical size in humans. Hum. Mol. Genet. 2004;13:489–494. doi: 10.1093/hmg/ddh055. [DOI] [PubMed] [Google Scholar]
- 18.Wyckoff GJ, Wang W, Wu C-I. Rapid evolution of male reproductive genes in the descent of man. Nature. 2000;403:304–309. doi: 10.1038/35002070. [DOI] [PubMed] [Google Scholar]
- 19.Suntsova MV, Buzdin AA. Differences between human and chimpanzee genomes and their implications in gene expression, protein functions and biochemical properties of the two species. BMC Genomics. 2020;21:535. doi: 10.1186/s12864-020-06962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ringer AL, Senenko A, Sherrill CD. Models of S/π interactions in protein structures: Comparison of the H2S–benzene complex with PDB data. Protein Sci. 2007;16:2216–2223. doi: 10.1110/ps.073002307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valley CC, et al. The methionine-aromatic motif plays a unique role in stabilizing protein structure. J. Biol. Chem. 2012;287:34979–34991. doi: 10.1074/jbc.M112.374504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGettrick AF, O'Neill LA. Two for the price of one: itaconate and its derivatives as an anti-infective and anti-inflammatory immunometabolite. Curr. Opin. Immunol. 2022;80:102268. doi: 10.1016/j.coi.2022.102268. [DOI] [PubMed] [Google Scholar]
- 23.Roberts L, Leighton I, Schwarz B, Wehrly T, Evans T, Bosio C. Itaconate indirectly influences expansion of effector T cells following vaccination with Francisella tularensis live vaccine strain. Cell. Immunol. 2022;373:104485. doi: 10.1016/j.cellimm.2022.104485. [DOI] [PubMed] [Google Scholar]
- 24.Zhao H, et al. Myeloid-derived itaconate suppresses cytotoxic CD8(+) T cells and promotes tumour growth. Nat. Metab. 2022;4:1660–1673. doi: 10.1038/s42255-022-00676-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss JM, et al. Itaconic acid mediates crosstalk between macrophage metabolism and peritoneal tumors. J. Clin. Invest. 2018;128:3794–3805. doi: 10.1172/JCI99169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pires-Afonso Y, et al. Elucidating tumour-associated microglia/macrophage diversity along glioblastoma progression and under ACOD1 deficiency. Mol. Oncol. 2022;16:3167–3191. doi: 10.1002/1878-0261.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu R, et al. Aconitate decarboxylase 1 is a mediator of polymicrobial sepsis. Sci. Transl. Med. 2022;14:eabo2028. doi: 10.1126/scitranslmed.abo2028. [DOI] [PubMed] [Google Scholar]
- 28.Püllmann P, Ulpinnis C, Marillonnet S, Gruetzner R, Neumann S, Weissenborn MJ. Golden mutagenesis: An efficient multi-site-saturation mutagenesis approach by golden gate cloning with automated primer design. Sci. Rep. 2019;9:10932. doi: 10.1038/s41598-019-47376-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PLoS ONE. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 31.Winterhoff M, et al. Establishment, validation, and initial application of a sensitive LC–MS/MS assay for quantification of the naturally occurring isomers itaconate, mesaconate, and citraconate. Metabolites. 2021;11:270. doi: 10.3390/metabo11050270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data and materials that support the findings of this study are available from the corresponding authors upon reasonable request.