Abstract
Microbial lipids are considered promising and environmentally friendly substitutes for fossil fuels and plant-derived oils. They alleviate the depletion of limited petroleum storage and the decrement of arable lands resulting from the greenhouse effect. Microbial lipids derived from oleaginous yeasts provide fatty acid profiles similar to plant-derived oils, which are considered as sustainable and alternative feedstocks for use in the biofuel, cosmetics, and food industries. Rhodotorula toruloides is an intriguing oleaginous yeast strain that can accumulate more than 70% of its dry biomass as lipid content. It can utilize a wide range of substrates, including low-cost sugars and industrial waste. It is also robust against various industrial inhibitors. However, precise control of the fatty acid profile of the lipids produced by R. toruloides is essential for broadening its biotechnological applications. This mini-review describes recent progress in identifying fatty synthesis pathways and consolidated strategies used for specific fatty acid-rich lipid production via metabolic engineering, strain domestication. In addition, this mini-review summarized the effects of culture conditions on fatty acid profiles in R. toruloides. The perspectives and constraints of harnessing R. toruloides for tailored lipid production are also discussed in this mini-review.
Graphical abstract
Keywords: Microbial lipid, Fatty acid alteration, Metabolic engineering, Rhodotorula toruloides
Introduction
Owing to the depletion of petroleum storage and the growing world population, lipids derived from plants and animals have been exploited as an alternative to petroleum for producing biofuel and oleochemicals (Chemat et al. 2021; Ndiaye et al. 2019; Riazi et al. 2020; Shaah et al. 2021). However, production limitations (long harvesting time and unstable production affected by geographical and climate factors) and environmental issues (environmental pollution and competence of arable land for food crops) of plant- and animal-based lipids have forced scientists to seek new alternatives (Singh et al. 2020a; Meijaard et al. 2020; Toldrá-Reig et al. 2020). To date, much attention has been paid to the development of microbial lipids because of their short harvesting time and fewer environmental concerns (Bao et al. 2021; Liu et al. 2021c; Aamer Mehmood et al. 2021; Uthandi et al. 2021).
Rhodotorula toruloides is a non-conventional red yeast, also known as Rhodosporidium toruloides, R. rubescens, R. glutinis, or R. gracilis, that belongs to the subphylum Pucciniomycotina within the division Basidiomycota (Abeln and Chuck 2021; Koh et al. 2014). This strain is an excellent microbial lipid producer and has received great attention owing to its outstanding lipogenic ability. R. toruloides can accumulate up to 70% of its dry cellular weight as lipid content under nutrient-limiting conditions (nitrogen, phosphate, and sulfate) (Wang et al. 2018; Wu et al. 2010, 2011). It is also well known for its robustness, as it can produce lipids by utilizing a wide range of substrates from low-cost industrial by-products (Zhao et al. 2022; Wen et al. 2020; Saini et al. 2020; Park et al. 2018b). Numerous studies have described ways to improve lipid production in R. toruloides, including robust mutant selection, bioprocess optimization, and metabolic engineering (Zhao et al. 2022; Wen et al. 2020; Saini et al. 2020; Park et al. 2018b). In addition, available genome sequence (Dinh et al. 2019; Martín-Hernández et al. 2021; Zhu et al. 2012), characterized lipid metabolism (Jagtap et al. 2021), and accessible genetic tools (Bonturi et al. 2022; Jiao et al. 2019; Otoupal et al. 2019) for R. toruloides have also been established and developed. Two efficient and time-saving transformation methods by using Lithium-acetate (Tsai et al. 2017) and electroporation (Liu et al. 2017) were established to shorten the experimental time of the traditional method via Agrobacterium tumefaciens-mediated transformation (ATMT) (Lin et al. 2014). In addition, RNAi machinery was also proven to be functional for gene downregulation in R. toruloides (Liu et al. 2019). Genome editing toolboxes such as CRISPR-Cas9 system were also reported to achieve single or multi-genes editing in R. toruloides (Jiao et al. 2019; Otoupal et al. 2019; Schultz et al. 2019). All these advanced genetic tools and knowledge accelerated the improvement of the control of lipid production in R. toruloides and broaden its applications.
Microbial lipids rich in triacylglycerols are promising and attractive alternatives to existing petroleum-, plant-, and animal-based lipids. A triacylglycerol molecule consists of three fatty acid (acyl) chains attached to an alcohol glycerol backbone (Yoshinaga 2021). Carbon length, desaturation level, and position determine the physicochemical and biological properties of fatty acid (FA) chains attached to triacylglycerols (Cook and McMaster 2002; Temkov and Muresan 2021; Lee et al. 2022; Saini et al. 2021; Falomir-Lockhart et al. 2019). The lipids from R. toruloides have FA compositions that are similar to those of plant-derived oils. Despite the variation of fatty acid profiles existing in different R. toruloides strains, lipids produced in R. toruloides mainly consist of C16-C18 long-chain FAs (palmitic acid [PA], C16:0, 15–40% of total fatty acid (TFA); palmitoleic acid [POA], C16:1, 1–2% of TFA; stearic acid [SA], C18:0, 10–20% of TFA; oleic acid [OA], C18:1, 40–60% of TFA; linoleic acid [LA], C18:2, 1–10% of TFA; α-linolenic acid [ALA], C18:3, 1–2% of TFA). In addition, a small proportion of FAs, such as myristic acid (MA, C14:0, 2% of TFA), arachidic acid (AA, C20:0, less than 0.5% of TFA), docosanoic acid (DA, C22:0, less than 0.5% of TFA), and tetracosanoic acid (TA, C24:0, less than 1% of TFA), have also been reported in R. toruloides (Krikigianni et al. 2022; Zhang et al. 2022a; Liu et al. 2021a). Therefore, R. toruloides-derived lipids could potentially be exploited as feedstock for plant oil substitutes in different applications, such as biodiesel, biolubricants, cosmetics, nutritional supplements, plastics, and coating materials (Carmona-Cabello et al. 2021; Papadaki et al. 2018; Adrio 2017; Lopez-Huertas 2010; Yu et al. 2014). For example, lipids rich in OA (over 70% of TFA) are preferred for biodiesel production due to their appropriate fluidity and stability during storage (Graef et al. 2009). Lipids with high saturated FA profiles are desired as equivalents to coconut (PA, 23–30%; SA, 32–37%; OA, 30–37%; LA, 2–4% of TFA) and cocoa (PA, 26%; SA, 35%; OA, 33%; LA, 3% of TFA) butter in food industries (Lipp et al. 2001; Papanikolaou and Aggelis 2010). Moreover, lipids rich in polyunsaturated fatty acids (PUFA) such as docosahexaenoic acid (C22:6, DHA) and eicosapentaenoic acid (C20:5, EPA) (16% and 17% of TFA) are considered as substitutes to fish oils (Lee et al. 2019). Lipids containing conjugated linoleic acids (CLAs, a group of isomers of LA), γ-linolenic acid (GLA, C18:3), and nervonic acid (NA, C24:1), which are relatively low in their sources, are considered as valuable lipids because of their clinical benefits (Szczepańska et al. 2022). However, commercially desired lipids are often produced at low concentrations or with inappropriate FA compositions in microbial lipids, which may lead to complex downstream processes and result in high production costs (Ochsenreither et al. 2016; Barta et al. 2021). Therefore, harnessing R. toruloudes for specific FA-rich lipid production is a vital issue for industrialization. Most studies have focused on maximizing the lipid-producing ability of R. toruloides (Zhao et al. 2022; Wen et al. 2020). Hence, few review papers have summarized and discussed strategies for controlling FA composition in R. toruloides. Further, a thorough discussion of the culture conditions that affect FA profiles in R. toruloides has not yet been done. In this mini-review, we summarize the advances in the control and production of tailored lipids and the effects of culture conditions on FA profiles in R. toruloides (Table 1).
Table 1.
Approaches used to alter FA composition in R. toruloides
| Approach | Strategy | Parental strain | Carbon source | Major FA profile | References |
|---|---|---|---|---|---|
|
Metabolic engineering |
Overexpression of RtELO1 | CECT13085 | Glycerol | Increased OA from 50% to 60–70% of TFA | Fillet et al. (2016) |
| Overexpression of RtELO1 | ATCC 10657 | Glucose | Increased OA from 50% to 60–70% of TFA | Liu et al. (2018) | |
| Gene disruption of RtELO2 | ATCC 10657 | Glucose | No change in FA profile | Liu et al. (2018) | |
| Overexpression of genomic RtOLE1 | TK16-DMKU3 | Glucose | Increased OA from 50 to 62% of TFA | Tsai et al. (2019) | |
| Gene disruption of RtFAD2 | ATCC 10657 (Δku70e) | Glucose | Increased OA from 30 to 60% of TFA | Liu et al. (2021b) | |
| Overexpression of genomic RtFAD2 | TK16-DMKU3 | Glucose | Increased LA from 14 to 28% and ALA from 1.2% to 3.9% of TFA | Wu et al. (2021) | |
| Co-overexpression of codon-optimized MaFAD2 and FvFAD2 | AS 2.1389 | Glucose | Increased LA from 5 to 27% of TFA | Wang et al. (2016) | |
| Disruption of RtALD1 and overexpression of codon-optimized MaFAD2 | ATCC 10657 | Glucose | Increased LA | Liu et al. (2018) | |
| Disruption of RtFAD2 and co-overexpression of codon-optimized MaFAD2 and MaFAD6 | ATCC 10657 (Δku70e) | Glucose | Increased OA to 60.1% of TFA and produced 27.3% GLA of TFA | Liu et al. (2021b) | |
| Co-overexpression of plant derived KCSs and OLE1 | CECT 13085 | Glucose | Produced EA and NA (20% of TFA) | Fillet et al. (2017) | |
|
Mutant isolation |
ALE at 37°C | TK16-DMKU3 | Glucose | Mutant L1-1 produced 86% OA of TFA at 37°C | Wu et al. (2018) |
| ALE with wheat straw hydrolysate | NRRL Y-1091 | Glucose, xylose | Mutant CH4 and CH5 produced 40–47% OA and 3.5% LA of TFA | Liu et al. (2021d) | |
| UV mutagenesis and selected mutant under ethanol and H2O2 or cerulenin | NBRC 8766 | Glucose | Increased PA (5.6–6.5%) and SA (12.7–13.5%) of TFA | Yamada et al. (2017) | |
| UV mutagenesis and selected mutant under ethanol and H2O2 or LiCl | 2.1389 | Glucose | Increased PA (23–24%) and SA (10–12%) of TFA | Guo et al. (2019) | |
| ARTP and NTG mutagenesis and selected mutant by color | NP11 | Glucose | No change in FA profile | Zhang et al. (2016a) | |
| ARTP followed by ALE with tea waste hydrolysate | ACCC 20341 | Tea waste hydrolysate | Increased 5.5% ALA of TFA | Qi et al. (2020) | |
|
Optimization of bioprocess |
Optimized C/N ratio | R-ZL mutant (from AS 2.1389) | Sucrose | Increased PA (21–26% of TFA) and ALA (6–15% of TFA) | Ye et al. (2021) |
| Optimized C/P ratio | Y4 mutant (from AS 2.1389) | Glucose | Increased OA from 50 to 67% of TFA | Wu et al. (2010) | |
| Optimized C/S ratio | Y4 mutant (from AS 2.1389) | Glucose | Increased saturated FA (MA, PA and SA) up to 63% of TFA | Wu et al. (2011) | |
| Used C5 sugars (arabinose and xylose) | CBS14 (from CBS) | Arabinose or/and xylose | Slightly increased PA (25–30% of TFA) | Wiebe et al. (2012) | |
| Used crude glycerol as a carbon source | ATCC 10788 | Crude glycerol | Increased 40% saturated FAs (PA and SA) of TFA | Uperty et al. (2017, 2018) | |
| Used acetic acid as a carbon source and optimized C/N ratio | AS 2.13 | Acetic acid | Slightly increased OA to 50% of TFA from 42% of TFA while using glucose | Huang et al. (2016) | |
| Used propionic acid (C3) as a carbon source | NCYC 1576 | Propionic acid | Produced 31% of odd-chain fatty acids (C17:0 and C17:1) of TFA | Krikigianni et al. (2022) | |
| Applied essential oils from plants | ATCC 10788 | Glycerol and limonene | EOs from clove, cinnamon pine, and orange increase SA or PA contents | Uprety and Rakshit (2018) | |
| Supplemented with Mg2+ | 1588 | Wood hydrolysate | Produced ETAs (0.03% of TFA) | Saini et al. (2022a) | |
| Supplemented with Cu2+ | 1588 | Wood hydrolysate | Produced DHA (0.05% of TFA) and GLA (0.12% of TFA) | Saini et al. (2022a) | |
| Supplemented with Zn2+or Fe2+ | 1588 | Wood hydrolysate | Produced GLA (0.2% of TFA) | Saini et al. (2022a) | |
| Optimized oxygen availability | NCYC 921 | Carob pulp syrup | Content of PUFAs elevated as oxygen availability increased | Parreira et al. (2015) | |
| Increased cultivation temperature to 37°C | TK16-DMKU3 | Glucose | Increased the content of saturated FAs (PA and SA) content (30% from 17%) | Wu et al. (2020) | |
| Decreased cultivation temperature to 15°C | YM25079 | Glucose | The content of LA and ALA increased from 22 and 8% to 35% and 21% of TFA | He et al. (2015) | |
| Assessed the pH effects on FA profile | NCYC 921 | Glucose | FA profiles slightly changed at various pH | Dias et al. (2016) | |
| Exposed light during cultivation | NBRC 10032 | Glucose | Increased POA and ALA to 3.6% and 4.5% of TFA from 1.6% and 1.1% of TFA | Pham et al. (2020) | |
| Electro-fermentation with redox mediator Neutral Red | DSM 4444 | Glucose | Increased the content of saturated FAs (from 37 to 50% of TFA) | Arbter et al. (2019) |
PA palmitic acid, POA palmitoleic acid, SA stearic acid, OA oleic acid, LA linoleic acid, ALA α-linolenic acid, GLA γ-linolenic acid, DHA docosahexaenoic acid, EA erucic acid, ETA eicosatrienoic acid, NA nervonic acid, PUFA polyunsaturated fatty acid, TFA total fatty acid
Harnessing de novo FA synthesis pathways in R. toruloides
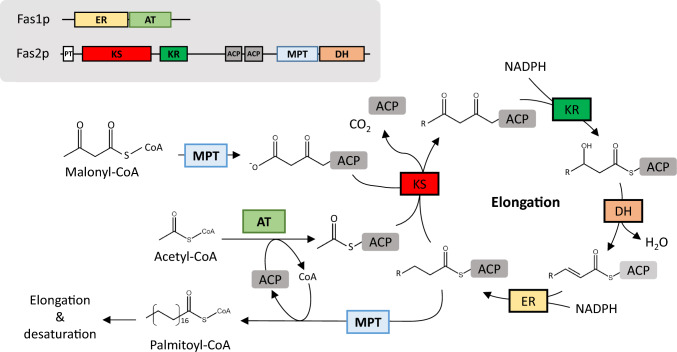
The FA composition of yeasts is precisely synthesized and regulated by FA synthases (Fass), elongases (Elos), and desaturases (Fads) (Heil et al. 2019; Singh et al. 2020b; Takaku et al. 2020; Matsuzawa et al. 2020). FA synthase, an essential enzyme in FA synthesis, catalyzes the reaction between acetyl-CoA and malonyl-CoA to produce C16-C18 FAs (Heil et al. 2019). R. toruloides Fas (RtFas) consists of two subunits (α- and β-subunits), which are encoded by FAS1 (RtFAS1) and FAS2 (RtFAS2), to form a multifunctional enzyme (Fischer et al. 2015; Zhu et al. 2012) (Fig. 1). RtFas1 contains acyltransferase and enoyl reductase domains. RtFas2 contains phosphopantetheinyl transferase, ketoacyl synthase, ketoacyl reductase, two acyl carrier proteins, malonyl/palmitoyl transferase, and dehydratase domains (Fig. 1). Liu et al. (2019) demonstrated that knocking down RtFAS1 and RtFAS2 using RNA interference decreased lipid content, but did not change the FA profile in R. toruloides NP11. Heterologous expression of RtFas in Saccharomyces cerevisiae also showed no effect on the FA composition (Zhou et al. 2016). These studies imply that FAS may not be a good candidate for manipulating FA composition in R. toruloides. However, enzyme engineering of the KS domain of Fas in the oleaginous yeast Yarrowia lipolytica enhanced the content of medium chain FAs (Rigouin et al. 2017), which suggests that engineering Fas may be applied in R. toruloides to modify the FA composition.
Fig. 1.
Schematic diagram of fatty acid synthase and fatty acid synthesis in R. toruloides cells. Abbreviations indicate components of the fatty acid synthase system. ACP acyl carrier protein, AT acyltransferase, DH dehydratase, ER enoyl reductase, KR ketoacyl reductase, KS ketoacyl synthase, MPT malonyl/palmitoyl transferase, PT phosphopantetheine transferase
Once malonyl-CoA is elongated to the carbon length of the acyl chain to C16, C16 acyl-CoA is released from the barrel-shaped Fas in R. toruloides. The released C16 acyl-CoA is further modified by FA Elos and Fads in R. toruloides (Zhu et al. 2012) (Fig. 2). Fads catalyze the desaturation of FAs by introducing double bonds into C–C bonds, which generate monounsaturated FAs (MUFAs) and polyunsaturated FAs (PUFAs) (Takaku et al. 2020; Wang et al. 2022a). Elos mediate the elongation of FAs by adding extra carbon atoms to the acyl chains. All desaturases and elongases exhibit different substrate specificities and spatial and temporal characteristics (Cerone and Smith 2022; Szczepańska et al. 2022). Therefore, elucidation of the functions of Fads and Elos could assist in the control of FA composition and production of customized lipids in R. toruloides.
Fig. 2.
Fatty acid elongation and desaturation pathways in R. toruloides. Genes encoding FA synthase, elongases, and desaturases are in red
The functions of Elos have been well studied in the model yeast S. cerevisiae and other yeasts used in the industry (Kihara 2012; Uemura 2012; Rigouin et al. 2018; Matsuzawa et al. 2020); however, few studies have elucidated the functions of Elos in R. toruloides. A patent (Fillet et al. 2016) reported that overexpression of RtELO1 significantly enhanced the OA content to 70% of the TFA in R. toruloides. In another patent (Liu et al. 2018), overexpression of RtELO1 led to an increase in OA. In addition, disruption of RtELO2 did not affect FA profiles (Liu et al. 2018). Conversely, studies on RtFads have also been conducted. R. toruloides Δ9-desaturase (RtOle1) catalyzes the conversion of SA to OA by introducing a cis-double bond at the Δ9 position of acyl-CoA. OA, one of the most essential and abundant FA in R. toruloides, provides appropriate fluidity and excellent thermal and oxidative stability for oleochemical applications such as biodiesel, biolubricants, and hydraulic fluids (Wang et al. 2022b). OA production is determined by RtOle1, which is a rate-limiting enzyme in LA and ALA biosynthesis (Nagao et al. 2019). Hence, RtOle1 plays a vital role in controlling the FA profiles. Overexpression of RtOle1 in R. toruloides have been shown to effectively increase OA content up to 70% and only slightly increase POA content (Tsai et al. 2019). Liu et al. (2021b) suggested that RtOle1 has a strong substrate preference for stearoyl-CoA over palmitoyl-CoA. In addition, blocking the downstream desaturation of OA into LA by disrupting RtFAD2 could also increase the OA content from 30 to 60% TFA. LA and ALA are essential FAs that humans cannot synthesize. Hence, lipids with high LA and ALA contents have high value as nutraceuticals in the human diet (Chen and Liu 2020). Liu et al. (2021b) also demonstrated that R. toruloides FAD2 and FAD4 encode Δ12/Δ15 desaturase and ∆9/Δ12/Δ15 desaturase, respectively, which produce LA and ALA from OA by introducing C–C double bonds at the Δ12 and Δ15 positions of FAs. Additionally, the co-overexpression of RtOLE1 and RtFAD2 has been applied to produce LA-rich lipids (Wu et al. 2021). Nevertheless, downregulation of RtOLE1 expression by elevated LA and ALA was observed in R. toruloides (Wu et al. 2021). Furthermore, two regulatory elements, ORE1 and ORE2, upstream of RtOLE1 have been reported (Liu et al. 2021b). The authors suggested that ORE1 positively regulates R. toruloides FAD gene transcription. Therefore, elucidating the transcription factors associated with the regulatory elements of RtOLE1 is necessary to manipulate the FA desaturation pathway. For example, S. cerevisiae transcriptional factors Spt2 and Mga2 upregulate essential genes involved in unsaturated FA biosynthesis (Zhang et al. 2022b; Sinha et al. 2022). The oleaginous Y. lipolytica Mga2 protein also regulates desaturase gene expression (Liu et al. 2015). Taken together, these studies show that manipulating the endogenous FA synthesis pathway in R. toruloides is an effective strategy for tailoring lipid production. Furthermore, the regulation of the native FA synthesis pathway should also be considered for a rational design to enhance specific FA production.
Metabolic engineering by introduction of exogenous genes for specific high-value lipid production
In addition to manipulating the native FA synthesis pathway, overexpression of genes from plants or other fungi was attempted in R. toruloides to produce specific high-value lipids. Disrupting native aldehyde dehydrogenase (ALD1) and overexpressing FAD2 from Mortierella alpina and FAD3 from Aleurites fordiiek led to a total ALA content of up to 49% of TFA (Liu et al. 2018). Tsai et al. (2019) enhanced the OA content from 50 to 70% of TFA by introducing S. cerevisiae OLE, which encodes a Δ9-desaturase. Wang et al. (2016) successfully enhanced LA content five-fold and achieved final LA titers of up to 1.3 g/L under flask culture conditions by introducing FAD2 from M. alpina and Fusarium verticillioides (Wang et al. 2016). GLA is a valuable omega-6 FA predominantly present in plants in relatively low amounts. GLA has anti-inflammatory properties and has applications in the treatment of several diseases (Liu et al. 2021b). Furthermore, Liu et al. (2021b) succeeded in producing GLA by disrupting native RtFAD2 and co-overexpressing M. alpine FAD2 and FAD6. The OA and GLA contents were further increased to 60.1% and 27.3% of TFA, respectively, in 2-L bioreactors.
FAs with carbon lengths of 20 or more are known as very-long-chain FAs (VLCFAs). VLCFAs, such as erucic acid (EA, C22:1) and NA, are potentially renewable feedstocks for the production of plastics, cosmetics, resins, nylon, and lubricants (Fillet et al. 2017). Additionally, NA has the potential to be used in the treatment of several neurological diseases (Liu et al. 2021a). Hence, EA and NA are high-value FAs. However, R. toruloides cannot naturally produce either EA or NA. To produce VLCFA-rich lipids, Fillet et al. (2017) introduced various plant-derived 3-ketoacyl-CoA synthases (KCSs) into R. toruloides and successfully produced EA and NA. By increasing the KCS copy number and co-overexpressing plant-derived ELOs, the engineered strain produced more EA and NA, which accounted for 27% of TFA in 7-L bioreactors.
Domestication of R. toruloides strains for tailored lipid production
Although metabolic engineering is a powerful strategy for harnessing R. toruloides for tailored lipid production, lack of information of the genetic background is the main limitation for strain engineering. Therefore, other strategies, such as adaptive laboratory evolution (ALE) and mutagenesis by physical/chemical methods, are also popular for isolating mutants with the desired traits for tailored lipid production in R. toruloides (Wen et al. 2020). The mutation rate is one of the main differences between ALE and mutagenesis using physicochemical methods. ALE domesticates parental strains with simultaneous mutation rates by growing several generations under selective pressure conditions (Phaneuf et al. 2020). Mutagenesis by physical and chemical methods commonly introduces a high mutation rate by random DNA double-strand breakage with UV irradiation and chemical agents. Although mutagenesis by physical and chemical methods is an effective way to introduce mutations into the genome, isolated mutants usually possess undesired defects (Arora et al. 2020). Owing to the advantages and disadvantages of the two methods, researchers have employed ALE and mutagenesis by physical and chemical methods in R. toruloides.
Wu et al. (2018) attempted to overcome the high temperatures that occur during fermentation by isolating thermotolerant R. toruloides DMKU3-TK16. The thermotolerant strain L1-1 was isolated under heat stress at 37°C using the ALE method. The isolated strain exhibited improved growth and lipid productivity at 37°C and a high OA content (90%) when cultured at 37°C. Liu et al. (2021d) attempted to improve the tolerance of R. toruloides to toxic lignocellulosic hydrolysates for lipid production. The isolated strains showed improved growth in media containing lignocellulosic biomass hydrolysate. According to their data, the evolved strains CH4 and CH5 showed improved OA and LA content (calculated). However, most evolved strains did not show significant FA profile alterations compared to the parental strain (R. toruloides NRRL Y-1091). Mutagenesis by UV has also been used for strain improvement in R. toruloides. Yamada et al. (2017) attempted to improve the lipid-producing ability of R. toruloides NBRC 8766 by UV irradiation and mutant selection under ethanol and H2O2 or cerulenin stress. Two strains with improved lipid production (8766 2-31 M and 8766 3-11C) were isolated. The FA profiles of these two strains showed considerably increased PA content (12.7% and 13.5%) with decreased MA and LA contents. Another study adopted a similar strategy of UV mutagenesis followed by mutant selection under ethanol and H2O2 or LiCl stress (Guo et al. 2019). The research team obtained two strains (R-ZL2 and R-ZL13) with improved lipid production from R. toruloides strain 2.1389. The R-ZL2 and R-ZL13 strains exhibited higher saturated FA profiles with increased PA (23 and 24%) and SA (12 and 10%) content, respectively. These two studies demonstrated that UV mutagenesis combined with stress selection could effectively domesticate R. toruloides to produce FA-rich lipids. Zhang et al. (2016a) obtained mutants of R. toruloides NP11 using atmospheric and room temperature plasma (ARTP) and nitrosoguanidine (NTG) methods. However, they did not apply stress selection after mutagenesis. According to their results, the isolated mutant XR-2 did not show a significant change in the FA profile compared with the parental strain R. toruloides NP11. In contrast, Qi et al. (2020) applied stress selection using inhibitory lignocellulosic hydrolysates after ARTP. Mutant RM18 exhibited improved ALA content (5.5%, 1.6 times more than the parental strain). These studies (Yamada et al. 2017; Guo et al. 2019; Zhang et al. 2016a; Qi et al. 2020) imply that stress selection after physical or chemical mutagenesis is necessary for screening mutants for desired FA production.
Culture conditions affect FA profiles in R. toruloides
Numerous strategies, such as optimizing culture conditions and waste utilization, have been applied to enhance lipid production in R. toruloides to produce economically competitive lipids (Zhao et al. 2022). However, the effects of each strategy on the FA profiles of lipids varied. Therefore, we have consolidated the progress obtained and provided a wide scope in the following sections. The information can help design rational bioprocesses to enhance specific FA-rich lipid production and assist in tailored lipid production.
Nitrogen, phosphate, and sulfate starvation
Nitrogen (N), phosphate (P), and sulfate (S) starvation can promote lipid accumulation in R. toruloides (Wang et al. 2018; Wu et al. 2010, 2011), and nitrogen limitation is the most adapted method. Different strategies affect lipid synthesis via different pathways, resulting in different metabolic fluxes. Therefore, the N, P, and S concentrations also affected the FA profiles. Ye et al. (2021) found that, when using sucrose as the sole carbon source and adjusting the C/N ratio to over 80 with ammonium sulfate or ammonium nitrate in the culture media, the FA contents of PA and ALA significantly increased in cells. Increasing the C/P ratio with glucose as a carbon source led to an increase in the OA content from 50 to 67% (Wu et al. 2010). In a study of S-limitation (Wu et al. 2011), lipids produced under increased C/S conditions favored saturated FAs (MA, PA, and SA) up to 60%. According to these studies, the strategies used to trigger lipid production should focus on enhancing the specific FA content.
Carbohydrates
Carbohydrates significantly affect FA profiles by activating key metabolic genes involved in gluconeogenesis, the glyoxylate cycle, and the tricarboxylic acid cycle (Sun et al. 2021; Patel et al. 2015). Most microbes possess a unique carbon catabolite mechanism. Microbes metabolize sugars sequentially because glucose represses the utilization of other sugars (Sun et al. 2021; Patel et al. 2015). Therefore, strategies using a primary carbon substrate or a defined ratio of carbon substrates have been reported for R. toruloides. Wiebe et al. (2012) demonstrated that R. toruloides could utilize C5 (arabinose and xylose) and C6 (glucose) sugars for lipid production. The lipids produced using arabinose and xylose as the sole carbon sources showed increased PA and LA contents compared to glucose, indicating that C5 (xylose and arabinose) sugars significantly changed the FA profiles. However, the FA profiles of the lipids produced using mixed sugars of glucose, arabinose, and xylose did not change significantly compared to those produced using glucose alone. Lignocellulosic biomass hydrolysate is a promising sustainable feedstock for microbial lipid production. After pretreatment and saccharification of lignocellulosic biomass, the complex links among lignin, cellulose, and hemicellulose are broken down, thus releasing fermentable sugars, such as glucose, xylose, or arabinose (Saini et al. 2022b). Osorio-González et al. (2019) demonstrated that R. toruloides strains had higher PA and SA contents when using C5 hydrolysate than C6 hydrolysate. Therefore, selecting carbohydrates and monitoring the carbohydrate content in the media during fermentation is vital for designing a rational fermentation process for desired FA production.
Crude substrates
In addition to using a single carbon source or defined carbon source, non-monomeric substrates containing abundant carbon sources from industrial, agricultural, municipal solid, and biomass wastes have been extensively studied and employed for lipid production in R. toruloides owing to their low cost (Abeln and Chuck 2021; Zhao et al. 2022).
Crude glycerol (CG) is a widely available product in the biodiesel industry. Approximately 10% (w/w) of glycerol is generated in every batch of biodiesel and is considered a waste (Lee et al. 2014; Uprety et al. 2016). Uprety et al. (2017, 2018) indicated that R. toruloides cells grown in media containing CG contained nearly 40% saturated FA (PA and SA), and a reduction in OA (from 60 to 47% of TFA) was observed compared to cells grown with pure glycerol. CG usually consists of glycerol and other impurities, such as glycerol, soap, methanol, ash, FA methyl esters (FAME), and salt. Uprety et al. (2018) reported that the decrease in OA in cells resulted from various impurities, particularly soap. Therefore, impurities in CG should be noted for tailored lipid production.
R. toruloides can utilize volatile FAs (VFAs), such as acetic acid (C2), propionic acid (C3), and butyric acid (C4) (Gao et al. 2017) for lipid production. VFAs can be obtained through the degradation of organic waste biomass by anaerobic digestion (Park et al. 2018a; Llamas et al. 2020), and thus VFAs can be exploited as cheap carbon sources for lipid production in yeasts (Park et al. 2018a, b; Llamas et al. 2020). However, different VFAs caused different FA changes in R. toruloides. R. toruloides grown in media with acetic acid (C2) as the sole carbon source generates high OA-lipids (nearly 50% of TFA) (Huang et al. 2016; Krikigianni et al. 2022). When propionic acid (C3) was used as the sole carbon source, the lipids had a high content of saturated FAs (65.9% of TFA) and a low MUFA content of 16% (Krikigianni et al. 2022). Notably, when propionic acid C3 was used, odd-chain FAs (margaric acid, C17:0, and heptadecenoic acid, C17:1) were generated, accounting for 30.8% of TFA. When fermented brewer’s spent grain contained mixed VFAs, the production of odd-chain FAs (ODFAs) was not observed, indicating that VFA composition influenced the FA profiles in R. toruloides. ODFAs are unusual and rare FAs that are found in natural sources. They are valuable FAs because they are associated with several health benefits, such as regulating allergies, psoriasis, and autoimmune disorders, and reducing the risk of metabolic disorders (Dąbrowski and Konopka 2022). Therefore, using VFAs as a carbon source for lipid production in R. toruloides could alter the native FA profile and produce unusual FA.
Plant-derived essential oils (EOs) can also alter the FA profiles of oleaginous microbes (Uprety and Rakshit 2018; Uprety et al. 2022). EOs are volatile and hydrophobic compounds mainly composed of terpenic hydrocarbons and oxygenated derivatives. EOs can impact the carbon flux toward a specific type of FAs inside microbes. Hence, EOs have been applied to alter the FA profile of R. toruloides (Uprety and Rakshit 2018). The EOs from clove-, cinnamon-, pine-, and orange-supplemented growth media for R. toruloides significantly increased the saturated FA content to 29%, 15%, 14%, and 36% of TFA, respectively. Furthermore, the EOs from orange also elevated PA content to 41% of TFA (Uprety and Rakshit 2018). Therefore, EOs are good inducers of FA production in R. toruloides.
Trace metal salts
Trace metal salts, such as zinc sulfate, copper sulfate, ferric chloride, manganese sulfate, and magnesium sulfate have also been commonly used as micronutrients in growth media to promote microbial growth (Saini et al. 2022a, b). These micronutrients serve as co-factors for several enzymes involved in lipid synthesis and desaturation pathways, and affect their catalytic activities (Singh et al. 2016; Ma et al. 2009; Romero et al. 2018). Furthermore, these metals may alter the catalytic activities of desaturase enzymes, resulting in alterations in the FA composition of lipids (Cai et al. 2020). Metal supplementation with wood hydrolysate as a carbon source causes the production of unusual PUFAs in R. toruloides (Saini et al. 2022a, b). When cells were grown in media supplemented with Mg2+, R. toruloides cells produced eicosatrienoic acid (ETA, C20:3) (0.03% of TFA). Similarly, an unusual DHA (0.05% of TFA) was produced by R. toruloides in media supplemented with Cu2+. In addition, adding Zn2+, Fe2+, and Cu2+ increased the content of GLA, which is also an unusual FA in R. toruloides, from 0.07% to 0.22%, 0.2%, and 0.12% of TFA, respectively. EA, DHA, and GLA are essential FA in humans; hence, they are high-value compounds. These results indicated that the addition of specific trace metals induced the production of high-value PUFAs in R. toruloides.
Oxygen
Oxygen availability is a critical parameter, especially during high cell density and large-scale fermentation, because of the unequal oxygen distribution in the microenvironment of the vessels, which affects cell growth. Oxygen is also a key molecule involved in FA desaturation. Fads introduce C–C double bonds into acyl chains by reducing oxygen (Sperling et al. 2003). Because these are oxygen-dependent reactions, low oxygen availability results in a higher degree of lipid saturation (Abeln and Chuck 2021). In addition, oxygen levels affect the activation of crucial genes, such as S. cerevisiae OLE1 in the FA saturation pathway through transcriptional factors (Romero et al. 2018; Burr et al. 2016). Therefore, oxygen is essential for the desaturation of FA by Fas. Choi et al. (1982) observed that the degree of unsaturation was significantly affected by the specific oxygen uptake rate of R. toruloides. Parreira et al. (2015) also confirmed that the PUFAs fraction increased as oxygen availability increased. These results indicated that oxygen availability is associated with PUFA synthesis in R. toruloides. Furthermore, the oxygen-activation pathways of Fas should be confirmed in R. toruloides.
Temperature
The growth temperature greatly influences various life processes in organisms, including gene transcription, metabolic activity, cell growth, nutrient absorption, cell survival, energy production, and membrane fluidity (Price and Sowers 2004; Fonseca et al. 2019). Microbial cells can modulate membrane fluidity to adapt to environmental temperature. Temperature influences membrane fluidity; decreasing temperature results in reduced viscosity and a more rigid membrane, whereas increasing temperature leads to increased viscosity and a looser membrane (Fonseca et al. 2019; Renne and Kroon 2018). Hence, microbial cells change their FA composition in the membrane lipids to maintain membrane fluidity. An increase in unsaturated FAs content reduces the melting point and increases membrane fluidity (Chen et al. 2022). On the other hand, an increase in saturated FAs content elevates their melting point and decreases membrane fluidity (Mejía-Barajas et al. 2018).
A growth temperature of 30°C is typically adopted for lipid production in R. toruloides. However, higher and lower temperatures significantly influence the FA profile of R. toruloides. The FA profile of the lipids in R. toruloides cells grown at 37°C increased the content of saturated FAs (PA and SA) (30% from 17%), which led to a higher saturated level than that at 30°C (Wu et al. 2020). Notably, the increase in saturated FA at 37°C may result from reduced RtFad2 desaturase activity (Wu et al. 2021). These reports suggest that temperature affects Fad activity and alters membrane FA composition, which may help cells maintain membrane fluidity. In contrast, when R. toruloides cells were grown at 15°C, the content of PUFAs, including LA and ALA, increased significantly compared to that at 25°C. The LA and ALA contents increased from 22 to 35% and from 8 to 21%, respectively (He et al. 2015). He et al. (2015) also demonstrated that the mRNA transcription level of a putative FA desaturase was five-fold higher in cells grown at 15°C than in cells grown at 25°C. In Y. lipolytica, the transcriptional factor Mga2, encoded by YALI0B12342g, may be associated with the upregulation of FAD2 at low temperatures (Tezaki et al. 2017). Therefore, elucidating the mechanism of low-temperature-induced FAD2 expression is necessary for R. toruloides to produce PUFAs.
pH
The commonly set pH for lipid production in R. toruloides is 5–6 (Li et al. 2006; Yang et al. 2014; Zhang et al. 2016b; Zeng et al. 2017). However, different medium pH values influence the FA profiles at different levels (Hall and Ratledge 1977). Similar scenarios were first observed in R. toruloides IIP-30 (formerly known as R. glutinis IIP-30) in the pH range of 3–6 (Johnson et al. 1992). Dias et al. (2016) also reported that medium pH values slightly altered FA profiles in R. toruloides. A slight MUFA percentage (OA) increased in the pH range 4.0–5.5. The PUFA percentages (LA and ALA) decreased at pH 4.5–5.5. Although the alteration in FA profiles did not show strong pH dependency, a specific pH range could assist in specific FA-rich lipid production based on their studies.
Light
Rhodotorula toruloides is a promising strain for carotenoid production. The carotenoid and lipid biosynthesis pathways are related because acetyl-CoA molecules are shared as precursors (Bruder et al. 2019). As a critical photo-inducer, light can promote carotenoid production by increasing microbial growth and activity of enzymes essential for carotenoid biosynthesis (Frengova and Beshkova 2009). In addition, photo-induced reactive oxygen species (ROS) may be associated with the activation of carotenoid synthesis pathways through the transcription factor bZIP in R. toruloides (Lin et al. 2017). Pham et al. (2020) demonstrated that R. toruloides cells grown under light conditions showed higher POA (two-fold) and ALA (four-fold) contents of TFA compared to those grown under dark conditions. Their results indicate that light could be a helpful factor for controlling FA profiles in R. toruloides.
Electro-fermentation
Electro-fermentation (EF) is a promising technique for improving the performance of bioprocesses that has been applied to increase lipid production in R. toruloides (Arbter et al. 2019). EF with the redox mediator Neutral Red (NR) caused a significant shift in the FA profile from OA to PA, resulting in a highly saturated FA composition (from 37 to 50%) in R. toruloides (Arbter et al. 2019). EF with NR is an excellent technique for producing highly saturated lipids as an alternative source of plant lipids such as coconut oil. In addition, EF with different media and strains may be applicable to R. toruloides.
Conclusion
Numerous review articles have summarized the different strategies for maximizing lipid production in R. toruloides. However, few have summarized and compared the impact of each method on FA composition. Thus, this mini-review describes strategies and advanced progress for controlling the FA composition in R. toruloudes through metabolic engineering. In addition, this mini-review also summarized the parameters of culture conditions such as substrate utilization and bioprocess control that have impacts on FA profile in R. toruloides. R. toruloides exhibits flexibility in producing lipids that are rich in saturated FA, MUFA, PUFA, and even unusual FAs. Therefore, R. toruloides is a promising platform for producing specific FA lipids. Further studies are necessary to reveal the mechanisms underlying the described strategies and findings to precisely control FA synthesis and bioprocess in R. toruloides for specific FA lipid production.
Abbreviations
- AA
Arachidic acid
- ACP
Acyl carrier protein
- ALA
α-Linolenic acid
- ALD
Aldehyde dehydrogenase
- ALE
Adaptive laboratory evolution
- ARTP
Atmospheric and room temperature plasma
- AT
Acyltransferase
- ATMT
Agrobacterium tumefaciens-mediated transformation
- C
Carbon
- CG
Crude glycerol
- DA
Docosanoic acid
- DH
Dehydratase
- DHA
Docosahexaenoic acid
- EA
Erucic acid
- EF
Electro-fermentation
- ELO
Elongase
- EO
Essential oil
- ER
Enoyl reductase
- ETA
Eicosatrienoic acid
- FA
Fatty acid
- FAD
Fatty acid desaturase
- FAME
Fatty acid methyl ester
- FAS
Fatty acid synthase
- GLA
γ-Linolenic acid
- KCS
Ketoacyl-CoA synthase
- KR
Ketoacyl reductase
- KS
Ketoacyl synthase
- LA
Linoleic acid
- MA
Myristic acid
- MPT
Malonyl/palmitoyl transferase
- MUFA
Monounsaturated fatty acid
- N
Nitrogen
- NA
Nervonic acid
- NR
Neutral red
- NTG
Nitrosoguanidine
- OA
Oleic acid
- ODFA
Odd-chain fatty acid
- P
Phosphate
- PA
Palmitic acid
- POA
Palmitoleic acid
- PT
Phosphopantetheine transferase
- PUFA
Polyunsaturated fatty acid
- ROS
Reactive oxygen species
- Rt
Rhodotorula toruloides
- S
Sulfate
- SA
Stearic acid
- TA
Tetracosanoic acid
- TFA
Total fatty acid
- UV
Ultraviolet
- VFA
Volatile fatty acid
- VLCFA
Very-long-chain fatty acid
Author contributions
WCC, HK and KF discussed the content and direction. WCC wrote the main manuscript text and prepared figures and table.
Funding
Open access funding provided by Osaka University. This study was supported in part by the NEDO Project (Project code: P20011) of the Ministry of Economy, Trade, and Industry (METI), Japan, and by the COI-NEXT Program (Grant number: JPMJPF2106) of Japan Science and Technology (JST).
Data availability
The data supporting the findings of this mini-review are available in the references cited within this article.
Declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aamer Mehmood M, Shahid A, Malik S, Wang N, Rizwan Javed M, Nabeel Haider M, Verma P, Umer Farooq Ashraf M, Habib N, Syafiuddin A, Boopathy R. Advances in developing metabolically engineered microbial platforms to produce fourth-generation biofuels and high-value biochemicals. Bioresour Technol. 2021;337:125510. doi: 10.1016/j.biortech.2021.125510. [DOI] [PubMed] [Google Scholar]
- Abeln F, Chuck CJ. The history, state of the art and future prospects for oleaginous yeast research. Microb Cell Fact. 2021;20:221. doi: 10.1186/s12934-021-01712-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrio JL. Oleaginous yeasts: Promising platforms for the production of oleochemicals and biofuels. Biotechnol Bioeng. 2017;114:1915–1920. doi: 10.1002/bit.26337. [DOI] [PubMed] [Google Scholar]
- Arbter P, Sinha A, Troesch J, Utesch T, Zeng AP. Redox governed electro-fermentation improves lipid production by the oleaginous yeast Rhodosporidium toruloides. Bioresour Technol. 2019;294:122122. doi: 10.1016/j.biortech.2019.122122. [DOI] [PubMed] [Google Scholar]
- Arora N, Yen H-W, Philippidis GP. Harnessing the power of mutagenesis and adaptive laboratory evolution for high lipid production by oleaginous microalgae and yeasts. Sustainability. 2020;12:5125. doi: 10.3390/su12125125. [DOI] [Google Scholar]
- Bao W, Li Z, Wang X, Gao R, Zhou X, Cheng S, Men Y, Zheng L. Approaches to improve the lipid synthesis of oleaginous yeast Yarrowia lipolytica: a review. Renew Sustain Energy Rev. 2021;149:111386. doi: 10.1016/j.rser.2021.111386. [DOI] [Google Scholar]
- Barta DG, Coman V, Vodnar DC. Microalgae as sources of omega-3 polyunsaturated fatty acids: Biotechnological aspects. Algal Res. 2021;58:102410. doi: 10.1016/j.algal.2021.102410. [DOI] [Google Scholar]
- Bonturi N, Pinheiro MJ, de Oliveira PM, Rusadze E, Eichinger T, Liudžiūtė G, De Biaggi JS, Brauer A, Remm M, Miranda EA, Ledesma-Amaro R, Lahtvee PJ. Development of a dedicated Golden Gate Assembly Platform (RtGGA) for Rhodotorula toruloides. Metab Eng Commun. 2022;15:e00200. doi: 10.1016/j.mec.2022.e00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr R, Stewart EV, Shao W, Zhao S, Hannibal-Bach HK, Ejsing CS, Espenshade PJ. Mga2 transcription factor regulates an oxygen-responsive lipid homeostasis pathway in fission yeast. J Biol Chem. 2016;291:12171–12183. doi: 10.1074/jbc.M116.723650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Yu XH, Chai J, Liu CJ, Shanklin J. A conserved evolutionary mechanism permits Δ9 desaturation of very-long-chain fatty acyl lipids. J Biol Chem. 2020;295:11337–11345. doi: 10.1074/jbc.RA120.014258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Cabello M, García IL, Papadaki A, Tsouko E, Koutinas A, Dorado MP. Biodiesel production using microbial lipids derived from food waste discarded by catering services. Bioresour Technol. 2021;323:124597. doi: 10.1016/j.biortech.2020.124597. [DOI] [PubMed] [Google Scholar]
- Cerone M, Smith TK. Desaturases: Structural and mechanistic insights into the biosynthesis of unsaturated fatty acids. IUBMB Life. 2022;74:1036–1051. doi: 10.1002/iub.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemat F, Vian MA, Ravi HK. Toward petroleum-free with plant-based chemistry. Curr Opin Green Sustain Chem. 2021;28:100450. doi: 10.1016/j.cogsc.2021.100450. [DOI] [Google Scholar]
- Chen J, Liu H. Nutritional indices for assessing fatty acids: a mini-review. Int J Mol Sci. 2020;21:5695. doi: 10.3390/ijms21165695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zhang X, Li S, Cui J, Yang X, Zhang Q. The MAP-kinase HOG1 controls cold adaptation in Rhodosporidium kratochvilovae by promoting biosynthesis of polyunsaturated fatty acids and glycerol. Curr Microbiol. 2022;79:253. doi: 10.1007/s00284-022-02957-8. [DOI] [PubMed] [Google Scholar]
- Choi SY, Ryu DD, Rhee JS. Production of microbial lipid: Effects of growth rate and oxygen on lipid synthesis and fatty acid composition of Rhodotorula gracilis. Biotechnol Bioeng. 1982;24:1165–1172. doi: 10.1002/bit.260240513. [DOI] [PubMed] [Google Scholar]
- Cook HW, McMaster CR (2002) Chapter 7 fatty acid desaturation and chain elongation in eukaryotes. New Compr Biochem 36:181–204. 10.1016/S0167-7306(02)36009-5
- Dąbrowski G, Konopka I. Update on food sources and biological activity of odd-chain, branched and cyclic fatty acids—a review. Trends Food Sci Technol. 2022;119:514–529. doi: 10.1016/j.tifs.2021.12.019. [DOI] [Google Scholar]
- Dias C, Silva C, Freitas C, Reis A, da Silva TL. Effect of medium pH on Rhodosporidium toruloides NCYC 921 carotenoid and lipid production evaluated by flow cytometry. Appl Biochem Biotechnol. 2016;179:776–787. doi: 10.1007/s12010-016-2030-y. [DOI] [PubMed] [Google Scholar]
- Dinh HV, Suthers PF, Chan SHJ, Shen Y, Xiao T, Deewan A, Jagtap SS, Zhao H, Rao CV, Rabinowitz JF, Maranas CD. A comprehensive genome-scale model for Rhodosporidium toruloides IFO0880 accounting for functional genomics and phenotypic data. Metab Eng Commun. 2019;9:e00101. doi: 10.1016/j.mec.2019.e00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falomir-Lockhart LJ, Cavazzutti GF, Giménez E, Toscani AM. Fatty acid signaling mechanisms in neural cells: fatty acid receptors. Front Cell Neurosci. 2019;13:162. doi: 10.3389/fncel.2019.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillet S, Ronchel C, Callejo C, Fajardo MJ, Moralejo H, Adrio JL. Engineering Rhodosporidium toruloides for the production of very long-chain monounsaturated fatty acid-rich oils. Appl Microbiol Biotechnol. 2017;101:7271–7280. doi: 10.1007/s00253-017-8461-8. [DOI] [PubMed] [Google Scholar]
- Fillet SC, Suárez González B, Ronchel Barreno MC, Velasco Álvarez J, Fondevila JLA (2016) Production of microbial oils with an elevated oleic acid content, WO 2016185073. Google Patent. https://patents.google.com/patent/WO2016185073A1/en. Accessed 27 October 2022.
- Fischer M, Rhinow D, Zhu Z, Mills DJ, Zhao ZK, Vonck J, Grininger M. Cryo-EM structure of fatty acid synthase (FAS) from Rhodosporidium toruloides provides insights into the evolutionary development of fungal FAS. Protein Sci. 2015;24:987–95. doi: 10.1002/pro.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca F, Pénicaud C, Tymczyszyn EE, Gómez-Zavaglia A, Passot S. Factors influencing the membrane fluidity and the impact on production of lactic acid bacteria starters. Appl Microbiol Biotechnol. 2019;103:6867–6883. doi: 10.1007/s00253-019-10002-1. [DOI] [PubMed] [Google Scholar]
- Frengova GI, Beshkova DM. Carotenoids from Rhodotorula and Phaffia: Yeasts of biotechnological importance. J Ind Microbiol Biotechnol. 2009;36:163. doi: 10.1007/s10295-008-0492-9. [DOI] [PubMed] [Google Scholar]
- Gao R, Li Z, Zhou X, Cheng S, Zheng L. Oleaginous yeast Yarrowia lipolytica culture with synthetic and food waste-derived volatile fatty acids for lipid production. Biotechnol Biofuels. 2017;10:247. doi: 10.1186/s13068-017-0942-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef G, LaVallee BJ, Tenopir P, Tat M, Schweiger B, Kinney AJ, Van Gerpen JH, Clemente TE. A high-oleic-acid and low-palmitic-acid soybean: agronomic performance and evaluation as a feedstock for biodiesel. Plant Biotechnol J. 2009;7:411–421. doi: 10.1111/j.1467-7652.2009.00408.x. [DOI] [PubMed] [Google Scholar]
- Guo M, Cheng S, Chen G, Chen J. Improvement of lipid production in oleaginous yeast Rhodosporidium toruloides by ultraviolet mutagenesis. Eng Life Sci. 2019;19:548–556. doi: 10.1002/elsc.201800203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MJ, Ratledge C. Lipid accumulation in an oleaginous yeast (Candida 107) growing on glucose under various conditions in a one- and two-stage continuous culture. Appl Environ Microbiol. 1977;33:577–584. doi: 10.1128/aem.33.3.577-584.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Yang Z, Hu B, Ji X, Wei Y, Lin L, Zhang Q. Correlation of polyunsaturated fatty acids with the cold adaptation of Rhodotorula glutinis. Yeast. 2015;32:683–690. doi: 10.1002/yea.3095. [DOI] [PubMed] [Google Scholar]
- Heil CS, Wehrheim SS, Paithankar KS, Grininger M. Fatty acid biosynthesis: chain length regulation and control. Chembiochem. 2019;20:2298–2321. doi: 10.1002/cbic.201800809. [DOI] [PubMed] [Google Scholar]
- Huang XF, Liu JN, Lu LJ, Peng KM, Yang GX, Liu J. Culture strategies for lipid production using acetic acid as sole carbon source by Rhodosporidium toruloides. Bioresour Technol. 2016;206:141–149. doi: 10.1016/j.biortech.2016.01.073. [DOI] [PubMed] [Google Scholar]
- Jagtap SS, Deewan A, Liu JJ, Walukiewicz HE, Yun EJ, Jin YS, Rao CV. Integrating transcriptomic and metabolomic analysis of the oleaginous yeast Rhodosporidium toruloides IFO0880 during growth under different carbon sources. Appl Microbiol Biotechnol. 2021;105:7411–7425. doi: 10.1007/s00253-021-11549-8. [DOI] [PubMed] [Google Scholar]
- Jiao X, Zhang Y, Liu X, Zhang Q, Zhang S, Zhao ZK. Developing a CRISPR/Cas9 system for genome editing in the Basidiomycetous yeast Rhodosporidium toruloides. Biotechnol J. 2019;14:e1900036. doi: 10.1002/biot.201900036. [DOI] [PubMed] [Google Scholar]
- Johnson V, Singh M, Saini VS, Sista VR, Yadav NK. Effect of pH on lipid accumulation by an oleaginous yeast: Rhodotorula glutinis IIP-30. World J Microbiol Biotechnol. 1992;8:382–384. doi: 10.1007/BF01198749. [DOI] [PubMed] [Google Scholar]
- Kihara A. Very long-chain fatty acids: elongation, physiology and related disorders. J Biochem. 2012;152:387–395. doi: 10.1093/jb/mvs105. [DOI] [PubMed] [Google Scholar]
- Koh CMJ, Liu Y, Moehninsi LL. Molecular characterization of KU70 and KU80 homologues and exploitation of a KU70-deficient mutant for improving gene deletion frequency in Rhodosporidium toruloides. BMC Microbiol. 2014;14:50. doi: 10.1186/1471-2180-14-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikigianni E, Matsakas L, Rova U, Christakopoulos P, Patel A. Investigating the bioconversion potential of volatile fatty acids: Use of oleaginous yeasts Rhodosporidium toruloides and Cryptococcus curvatus towards the sustainable production of biodiesel and odd-chain fatty acids. Appl Sci. 2022;12:6541. doi: 10.3390/app12136541. [DOI] [Google Scholar]
- Lee JJ, Chen L, Shi J, Trzcinski A, Chen WN. Metabolomic profiling of Rhodosporidium toruloides grown on glycerol for carotenoid production during different growth phases. J Agric Food Chem. 2014;62:10203–10209. doi: 10.1021/jf502987q. [DOI] [PubMed] [Google Scholar]
- Lee AV, You L, Oh SY, Li Z, Code A, Zhu C, Fisher-Heffernan RE, Regnault TRH, De Lange CFM, Huber LA, Karrow NA. Health benefits of supplementing nursery pig diets with microalgae or fish oil. Animals. 2019;9:80. doi: 10.3390/ani9030080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YY, Tang TK, Chan ES, Phuah ET, Lai OM, Tan CP, Wang Y, Ab Karim NA, Mat Dian NH, Tan JS. Medium chain triglyceride and medium-and long chain triglyceride: Metabolism, production, health impacts and its applications—a review. Crit Rev Food Sci Nutr. 2022;62:4169–4185. doi: 10.1080/10408398.2021.1873729. [DOI] [PubMed] [Google Scholar]
- Li YH, Liu B, Zhao ZB, Bai FW. Optimization of culture conditions for lipid production by Rhodosporidium toruloides. Chin J Biotechnol. 2006;22:650–656. doi: 10.1016/S1872-2075(06)60050-2. [DOI] [PubMed] [Google Scholar]
- Lin X, Wang Y, Zhang S, Zhu Z, Zhou YJ, Yang F, Sun W, Wang X, Zhao ZK. Functional integration of multiple genes into the genome of the oleaginous yeast Rhodosporidium toruloides. FEMS Yeast Res. 2014;14:547–555. doi: 10.1111/1567-1364.12140. [DOI] [PubMed] [Google Scholar]
- Lipp M, Simoneau C, Ulberth F, Anklam E, Crews C, Brereton P, de Greyt W, Schwack W, Wiedmaier C. Composition of genuine cocoa butter and cocoa butter equivalents. J Food Compost Anal. 2001;14:399–408. doi: 10.1006/jfca.2000.0984. [DOI] [Google Scholar]
- Liu L, Markham K, Blazeck J, Zhou N, Leon D, Otoupal P, Alper HS. Surveying the lipogenesis landscape in Yarrowia lipolytica through understanding the function of a Mga2p regulatory protein mutant. Metab Eng. 2015;31:102–111. doi: 10.1016/j.ymben.2015.07.004. [DOI] [PubMed] [Google Scholar]
- Liu H, Jiao X, Wang Y, Yang X, Sun W, Wang J, Zhang S, Zhao ZK. Fast and efficient genetic transformation of oleaginous yeast Rhodosporidium toruloides by using electroporation. FEMS Yeast Res. 2017 doi: 10.1093/femsyr/fox017. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang Y, Liu H, Jiao X, Zhang Q, Zhang S, Zhao ZK. RNA interference in the oleaginous yeast Rhodosporidium toruloides. FEMS Yeast Res. 2019;19:031. doi: 10.1093/femsyr/foz031. [DOI] [PubMed] [Google Scholar]
- Liu F, Wang P, Xiong X, Zeng X, Zhang X, Wu G. A review of nervonic acid production in plants: Prospects for the genetic engineering of high nervonic acid cultivars plants. Front Plant Sci. 2021;12:626625. doi: 10.3389/fpls.2021.626625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Koh CMJ, Yap SA, Cai L, Ji L. Understanding and exploiting the fatty acid desaturation system in Rhodotorula toruloides. Biotechnol Biofuels. 2021;14:73. doi: 10.1186/s13068-021-01924-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Moradi H, Shi S, Darvishi F. Yeasts as microbial cell factories for sustainable production of biofuels. Renewab Sustain Energy Rev. 2021;143:110907. doi: 10.1016/j.rser.2021.110907. [DOI] [Google Scholar]
- Liu Z, Radi M, Mohamed ETT, Feist AM, Dragone G, Mussatto SI. Adaptive laboratory evolution of Rhodosporidium toruloides to inhibitors derived from lignocellulosic biomass and genetic variations behind evolution. Bioresour Technol. 2021;333:125171. doi: 10.1016/j.biortech.2021.125171. [DOI] [PubMed] [Google Scholar]
- Liu Y, Koh CM, Ji L (2018) Methods for efficient production of polyunsaturated fatty acids (PUFA) in Rhodosporidium and Rhodotorula species. Google patent. https://patents.google.com/patent/US10081821B2/en. Accessed 27 October 2022.
- Llamas M, Dourou M, González-Fernández C, Aggelis G, Tomás-Pejó E. Screening of oleaginous yeasts for lipid production using volatile fatty acids as substrate. Biomass Bioenergy. 2020;138:105553. doi: 10.1016/j.biombioe.2020.105553. [DOI] [Google Scholar]
- Lopez-Huertas E. Health effects of oleic acid and long chain omega-3 fatty acids (EPA and DHA) enriched milks. A review of intervention studies. Pharmacol Res. 2010;61:200–207. doi: 10.1016/j.phrs.2009.10.007. [DOI] [PubMed] [Google Scholar]
- Ma Z, Chu CH, Cheng D. A novel direct homogeneous assay for ATP citrate lyase. J Lipid Res. 2009;50:2131–2135. doi: 10.1194/jlr.D900008-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Hernández GC, Müller B, Chmielarz M, Brandt C, Hölzer M, Viehweger A, Passoth V. Chromosome-level genome assembly and transcriptome-based annotation of the oleaginous yeast Rhodotorula toruloides CBS 14. Genomics. 2021;113:4022–4027. doi: 10.1016/j.ygeno.2021.10.006. [DOI] [PubMed] [Google Scholar]
- Matsuzawa T, Kamisaka Y, Maehara T, Takaku H, Yaoi K. Identification and characterization of two fatty acid elongases in Lipomyces starkeyi. Appl Microbiol Biotechnol. 2020;104:2537–2544. doi: 10.1007/s00253-020-10401-9. [DOI] [PubMed] [Google Scholar]
- Meijaard E, Brooks TM, Carlson KM, Slade EM, Garcia-Ulloa J, Gaveau DLA, Lee JSH, Santika T, Juffe-Bignoli D, Struebig MJ, Wich SA, Ancrenaz M, Koh LP, Zamira N, Abrams JF, Prins HHT, Sendashonga CN, Murdiyarso D, Furumo PR, Macfarlane N, Hoffmann R, Persio M, Descals A, Szantoi Z, Sheil D. The environmental impacts of palm oil in context. Nat Plants. 2020;6:1418–1426. doi: 10.1038/s41477-020-00813-w. [DOI] [PubMed] [Google Scholar]
- Mejía-Barajas J, Montoya-Pérez R, Manzo-Avalos S, Cortés-Rojo C, Riveros-Rosas H, Cervantes C, Saavedra-Molina A (2018) Fatty acid addition and thermotolerance of Kluyveromyces marxianus. FEMS Microbiol Lett 365:fny043. https://doi:doi:10.1093/femsle/fny043 [DOI] [PubMed]
- Nagao K, Murakami A, Umeda M. Recent progress in biophysical research of biological membrane systems. Chem Pharm Bull. 2019;67:327–332. doi: 10.1248/cpb.c18-01001. [DOI] [PubMed] [Google Scholar]
- Ndiaye M, Arhaliass A, Legrand J, Roelens G, Kerihuel A. Reuse of waste animal fat in biodiesel: Biorefining heavily-degraded contaminant-rich waste animal fat and formulation as diesel fuel additive. Renew Energy. 2019;145:1073–1079. doi: 10.1016/j.renene.2019.06.030. [DOI] [Google Scholar]
- Ochsenreither K, Glück C, Stressler T, Fischer L, Syldatk C. Production strategies and applications of microbial single cell oils. Front Microbiol. 2016;7:1539. doi: 10.3389/fmicb.2016.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otoupal PB, Ito M, Arkin AP, Magnuson JK, Gladden JM, Skerker JM. Multiplexed CRISPR-Cas9-based genome editing of Rhodosporidium toruloides. mSphere. 2019 doi: 10.1128/mSphere.00099-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadaki A, Fernandes KV, Chatzifragkou A, Aguieiras ECG, da Silva JAC, Fernandez-Lafuente R, Papanikolaou S, Koutinas A, Freire DMG. Bioprocess development for biolubricant production using microbial oil derived via fermentation from confectionery industry wastes. Bioresour Technol. 2018;267:311–318. doi: 10.1016/j.biortech.2018.07.016. [DOI] [PubMed] [Google Scholar]
- Papanikolaou S, Aggelis George. Yarrowia lipolytica: a model microorganism used for the production of tailor-made lipids. Eur J Lipid Sci Technol. 2010;112:639–654. doi: 10.1002/EJLT.200900197. [DOI] [Google Scholar]
- Park YK, Dulermo T, Ledesma-Amaro R, Nicaud JM. Optimization of odd chain fatty acid production by Yarrowia lipolytica. Biotechnol Biofuels. 2018;11:158. doi: 10.1186/s13068-018-1154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YK, Nicaud JM, Ledesma-Amaro R. The engineering potential of Rhodosporidium toruloides as a workhorse for biotechnological applications. Trends Biotechnol. 2018;36:304–317. doi: 10.1016/j.tibtech.2017.10.013. [DOI] [PubMed] [Google Scholar]
- Parreira TM, Freitas C, Reis A, Roseiro J, da Silva TL. Carbon concentration and oxygen availability affect lipid and carotenoid production by carob pulp syrup-grown Rhodosporidium toruloides NCYC 921. Eng Life Sci. 2015;15:815–823. doi: 10.1002/elsc.201500002. [DOI] [Google Scholar]
- Patel A, Pruthi V, Singh RP, Pruthi PA. Synergistic effect of fermentable and non-fermentable carbon sources enhances TAG accumulation in oleaginous yeast Rhodosporidium kratochvilovae HIMPA1. Bioresour Technol. 2015;188:136–144. doi: 10.1016/j.biortech.2015.02.062. [DOI] [PubMed] [Google Scholar]
- Pham KD, Shida Y, Miyata A, Takamizawa T, Suzuki Y, Ara S, Yamazaki H, Masaki K, Mori K, Aburatani S, Hirakawa H, Tashiro K, Kuhara S, Takaku H, Ogasawara W. Effect of light on carotenoid and lipid production in the oleaginous yeast Rhodosporidium toruloides. Biosci Biotechnol Biochem. 2020;84:1501–1512. doi: 10.1080/09168451.2020.1740581. [DOI] [PubMed] [Google Scholar]
- Phaneuf PV, Yurkovich JT, Heckmann D, Wu M, Sandberg TE, King ZA, Tan J, Palsson BO, Feist AM. Causal mutations from adaptive laboratory evolution are outlined by multiple scales of genome annotations and condition-specificity. BMC Genom. 2020;21:514. doi: 10.1186/s12864-020-06920-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price PB, Sowers T. Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc Natl Acad Sci USA. 2004;101:4631–4636. doi: 10.1073/pnas.0400522101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Shen P, Hu R, Xue T, Jiang X, Qin L, Chen Y, Huang J. Carotenoids and lipid production from Rhodosporidium toruloides cultured in tea waste hydrolysate. Biotechnol Biofuels. 2020;13:74. doi: 10.1186/s13068-020-01712-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne MF, de Kroon AIPM. The role of phospholipid molecular species in determining the physical properties of yeast membranes. FEBS Lett. 2018;592:1330–1345. doi: 10.1002/1873-3468.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazi B, Mosby JM, Millet B, Spatari S. Renewable diesel from oils and animal fat waste: implications of feedstock, technology, co-products and ILUC on life cycle GWP. Resour Conserv Recycl. 2020;161:104944. doi: 10.1016/j.resconrec.2020.104944. [DOI] [Google Scholar]
- Rigouin C, Gueroult M, Croux C, Dubois G, Borsenberger V, Barbe S, Marty A, Daboussi F, André I, Bordes F. Production of medium chain fatty acids by Yarrowia lipolytica: Combining molecular design and TALEN to engineer the fatty acid synthase. ACS Synth Biol. 2017;6:1870–1879. doi: 10.1021/acssynbio.7b00034. [DOI] [PubMed] [Google Scholar]
- Rigouin C, Croux C, Borsenberger V, Ben Khaled M, Chardot T, Marty A, Bordes F. Increasing medium chain fatty acids production in Yarrowia lipolytica by metabolic engineering. Microb Cell Fact. 2018;17:142. doi: 10.1186/s12934-018-0989-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero AM, Jordá T, Rozès N, Martínez-Pastor MT, Puig S. Regulation of yeast fatty acid desaturase in response to iron deficiency. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863:657–668. doi: 10.1016/j.bbalip.2018.03.008. [DOI] [PubMed] [Google Scholar]
- Saini R, Hegde J, Brar SK, Vezina P. Advanced biofuel production and road to commercialization: An insight into bioconversion potential of Rhodosporidium sp. Biomass Bioenergy. 2020;132:105439. doi: 10.1016/j.biombioe.2019.105439. [DOI] [Google Scholar]
- Saini RK, Prasad P, Sreedhar RV, Akhilender Naidu K, Shang X, Keum YS. Omega-3 polyunsaturated fatty acids (PUFAs): Emerging plant and microbial sources, oxidative stability, bioavailability, and health benefits—a review. Antioxidants (Basel) 2021;10:1627. doi: 10.3390/antiox10101627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini R, Gonzalez CSO, Hegde K, Brar SK, Vezina P. Evaluating the Effect of Trace metal salts on lipid accumulation ability of Rhodosporidium toruloides-1588 using wood hydrolysate as a carbon source. Bioenerg Res. 2022 doi: 10.1007/s12155-022-10521-2. [DOI] [Google Scholar]
- Saini R, Osorio-Gonzalez CS, Hegde K, Brar SK, Vezina P. Investigating the ability of Rhodosporidium toruloides-1588 to use furfural as a carbon source and its degradation: an enzyme identification study. Sustain Energy Fuels. 2022;6:4331–4337. doi: 10.1039/D2SE00772J. [DOI] [Google Scholar]
- Schultz JC, Cao M, Zhao H. Development of a CRISPR/Cas9 system for high efficiency multiplexed gene deletion in Rhodosporidium toruloides. Biotechnol Bioeng. 2019;116:2103–2109. doi: 10.1002/bit.27001. [DOI] [PubMed] [Google Scholar]
- Shaah MAH, Hossain MS, Allafi FAS, Alsaedi A, Ismail N, Ab Kadir MO, Ahmad MI. A review on non-edible oil as a potential feedstock for biodiesel: Physicochemical properties and production technologies. RSC Adv. 2021;11:25018–25037. doi: 10.1039/D1RA04311K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Barrow CJ, Puri M, Tuli DK, Mathur AS. Combination of calcium and magnesium ions prevents substrate inhibition and promotes biomass and lipid production in thraustochytrids under higher glycerol concentration. Algal Res. 2016;15:202–209. doi: 10.1016/j.algal.2016.02.024. [DOI] [Google Scholar]
- Singh D, Sharma D, Soni SL, Sharma S, Sharma PK, Jhalani A. A review on feedstocks, production processes, and yield for different generations of biodiesel. Fuel. 2020;262:116553. doi: 10.1016/j.fuel.2019.116553. [DOI] [Google Scholar]
- Singh K, Graf B, Linden A, Sautner V, Urlaub H, Tittmann K, Stark H, Chari A. Discovery of a regulatory subunit of the yeast fatty acid synthase. Cell. 2020;180:1130–1143. doi: 10.1016/j.cell.2020.02.034. [DOI] [PubMed] [Google Scholar]
- Sperling P, Ternes P, Zank TK, Heinz E. The evolution of desaturases. Prostaglandins Leukot Essent Fatty Acids. 2003;68:73–95. doi: 10.1016/s0952-3278(02)00258-2. [DOI] [PubMed] [Google Scholar]
- Sun C, Shah AM, Yang J, Wang Z, Zhu L, Song Y. Transcriptome analysis of oleaginous fungus Mucor circinelloides WJ11 in response to exogenous soybean oil as carbon source. Nat Prod Commun. 2021 doi: 10.1177/1934578X211023366. [DOI] [Google Scholar]
- Szczepańska P, Hapeta P, Lazar Z. Advances in production of high-value lipids by oleaginous yeasts. Crit Rev Biotechnol. 2022;42:1–22. doi: 10.1080/07388551.2021.1922353. [DOI] [PubMed] [Google Scholar]
- Takaku H, Matsuzawa T, Yaoi K, Yamazaki H. Lipid metabolism of the oleaginous yeast Lipomyces starkeyi. Appl Microbiol Biotechnol. 2020;104:6141–6148. doi: 10.1007/s00253-020-10695-9. [DOI] [PubMed] [Google Scholar]
- Temkov M, Mureșan V. Tailoring the structure of lipids, oleogels and fat replacers by different approaches for solving the trans-fat issue—a review. Foods. 2021;10:1376. doi: 10.3390/foods10061376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tezaki S, Iwama R, Kobayashi S, Shiwa Y, Yoshikawa H, Ohta A, Horiuchi H, Fukuda R. Δ12-fatty acid desaturase is involved in growth at low temperature in yeast Yarrowia lipolytica. Biochem Biophys Res Commun. 2017;488:165–170. doi: 10.1016/j.bbrc.2017.05.028. [DOI] [PubMed] [Google Scholar]
- Toldrá-Reig F, Mora L, Toldrá F. Trends in biodiesel production from animal fat waste. Appl Sci. 2020;10:3644. doi: 10.3390/app10103644. [DOI] [Google Scholar]
- Tsai YY, Ohashi T, Kanazawa T, Polburee P, Misaki R, Limtong S, Fujiyama K. Development of a sufficient and effective procedure for transformation of an oleaginous yeast, Rhodosporidium toruloides DMKU3-TK16. Curr Genet. 2017;63:359–371. doi: 10.1007/s00294-016-0629-8. [DOI] [PubMed] [Google Scholar]
- Tsai YY, Ohashi T, Wu CC, Bataa D, Misaki R, Limtong S, Fujiyama K. Delta-9 fatty acid desaturase overexpression enhanced lipid production and oleic acid content in Rhodosporidium toruloides for preferable yeast lipid production. J Biosci Bioeng. 2019;127:430–440. doi: 10.1016/j.jbiosc.2018.09.005. [DOI] [PubMed] [Google Scholar]
- Uemura H. Synthesis and production of unsaturated and polyunsaturated fatty acids in yeast: current state and perspectives. Appl Microbiol Biotechnol. 2012;95:1–12. doi: 10.1007/s00253-012-4105-1. [DOI] [PubMed] [Google Scholar]
- Uprety BK, Rakshit SK. Use of essential oils from various plants to change the fatty acids profiles of lipids obtained from oleaginous yeasts. J Am Oil Chem Soc. 2018;95:135–148. doi: 10.1002/aocs.12006. [DOI] [Google Scholar]
- Uprety BK, Chaiwong W, Ewelike C, Rakshit SK. Biodiesel production using heterogeneous catalysts including wood ash and the importance of enhancing byproduct glycerol purity. Energy Convers Manag. 2016;115:191–199. doi: 10.1016/j.enconman.2016.02.032. [DOI] [Google Scholar]
- Uprety BK, Dalli SS, Rakshit SK. Bioconversion of crude glycerol to microbial lipid using a robust oleaginous yeast Rhodosporidium toruloides ATCC 10788 capable of growing in the presence of impurities. Energy Convers Manag. 2017;135:117–128. doi: 10.1016/j.enconman.2016.12.071. [DOI] [Google Scholar]
- Uprety BK, Samavi M, Rakshit SK. Contribution of specific impurities in crude glycerol towards improved lipid production by Rhodosporidium toruloides ATCC 10788. Bioresour Technol Rep. 2018;3:27–34. doi: 10.1016/j.biteb.2018.05.011. [DOI] [Google Scholar]
- Uprety BK, Morrison EN, Neil Emery RJ, Farrow SC. Customizing lipids from oleaginous microbes: Leveraging exogenous and endogenous approaches. Trends Biotechnol. 2022;40:482–508. doi: 10.1016/j.tibtech.2021.09.004. [DOI] [PubMed] [Google Scholar]
- Uthandi S, Kaliyaperumal A, Srinivasan N, Thangavelu K, Muniraj IK, Zhan X, Gathergood N, Gupta VK. Microbial biodiesel production from lignocellulosic biomass: new insights and future challenges. Crit Rev Environ Sci Technol. 2021;52:2197–2225. doi: 10.1080/10643389.2021.1877045. [DOI] [Google Scholar]
- Wang Y, Zhang S, Pötter M, Sun W, Li L, Yang X, Jiao X, Zhao ZK. Overexpression of Δ12-fatty acid desaturase in the oleaginous yeast Rhodosporidium toruloides for production of linoleic acid-rich lipids. Appl Biochem Biotechnol. 2016;180:1497–1507. doi: 10.1007/s12010-016-2182-9. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang S, Zhu Z, Shen H, Lin X, Jin X, Jiao X, Zhao ZK. Systems analysis of phosphate-limitation-induced lipid accumulation by the oleaginous yeast Rhodosporidium toruloides. Biotechnol Biofuels. 2018;11:148. doi: 10.1186/s13068-018-1134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Shi TQ, Lin L, Wei P, Ledesma-Amaro R, Ji XJ. Engineering Yarrowia lipolytica to produce tailored chain-length fatty acids and their derivatives. ACS Synth Biol. 2022;11:2564–2577. doi: 10.1021/acssynbio.2c00305. [DOI] [PubMed] [Google Scholar]
- Wang K, Shi TQ, Wang J, Wei P, Ledesma-Amaro R, Ji XJ. Engineering the lipid and fatty acid metabolism in Yarrowia lipolytica for sustainable production of high oleic oils. ACS Synth Biol. 2022;11:1542–1554. doi: 10.1021/acssynbio.1c00613. [DOI] [PubMed] [Google Scholar]
- Wen Z, Zhang S, Odoh CK, Jin M, Zhao ZK. Rhodosporidium toruloides—a potential red yeast chassis for lipids and beyond. FEMS Yeast Res. 2020 doi: 10.1093/femsyr/foaa038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiebe MG, Koivuranta K, Penttilä M, Ruohonen L. Lipid production in batch and fed-batch cultures of Rhodosporidium toruloides from 5 and 6 carbon carbohydrates. BMC Biotechnol. 2012;12:26. doi: 10.1186/1472-6750-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Hu C, Jin G, Zhao X, Zhao ZK. Phosphate-limitation mediated lipid production by Rhodosporidium toruloides. Bioresour Technol. 2010;101:6124–6129. doi: 10.1016/j.biortech.2010.02.111. [DOI] [PubMed] [Google Scholar]
- Wu S, Zhao X, Shen H, Wang Q, Zhao ZK. Microbial lipid production by Rhodosporidium toruloides under sulfate-limited conditions. Bioresour Technol. 2011;102:1803–1807. doi: 10.1016/j.biortech.2010.09.033. [DOI] [PubMed] [Google Scholar]
- Wu Chih-Chan, Tsai Yung-Yu, Ohashi Takao, Misaki Ryo, Limtong Savitree, Fujiyama Kazuhito. Isolation of a thermotolerant Rhodosporidium toruloides DMKU3-TK16 mutant and its fatty acid profile at high temperature. FEMS Microbiol Lett. . 2018 doi: 10.1093/femsle/fny203. [DOI] [PubMed] [Google Scholar]
- Wu CC, Ohashi T, Misaki R, Limtong S, Fujiyama K. Ethanol and H2O2 stresses enhance lipid production in an oleaginous Rhodotorula toruloides thermotolerant mutant L1-1. FEMS Yeast Res. 2020 doi: 10.1093/femsyr/foaa030. [DOI] [PubMed] [Google Scholar]
- Wu CC, Ohashi T, Kajiura H, Sato Y, Misaki R, Honda K, Limtong S, Fujiyama K. Functional characterization and overexpression of Δ12-desaturase in the oleaginous yeast Rhodotorula toruloides for production of linoleic acid-rich lipids. J Biosci Bioeng. 2021;131:631–639. doi: 10.1016/j.jbiosc.2021.02.002. [DOI] [PubMed] [Google Scholar]
- Yamada R, Kashihara T, Ogino H. Improvement of lipid production by the oleaginous yeast Rhodosporidium toruloides through UV mutagenesis. World J Microbiol Biotechnol. 2017;33:99. doi: 10.1007/s11274-017-2269-7. [DOI] [PubMed] [Google Scholar]
- Yang X, Jin G, Gong Z, Shen H, Bai F, Zhao ZK. Recycling biodiesel-derived glycerol by the oleaginous yeast Rhodosporidium toruloides Y4 through the two-stage lipid production process. Biochem Eng J. 2014;91:86–91. doi: 10.1016/j.bej.2014.07.015. [DOI] [Google Scholar]
- Ye Z, Sun T, Hao H, He Y, Liu X, Guo M, Chen G. Optimising nutrients in the culture medium of Rhodosporidium toruloides enhances lipids production. AMB Express. 2021;11:149. doi: 10.1186/s13568-021-01313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga K. Development of analytical methods and nutritional studies using synthetic fatty acids and triacylglycerols. J Oleo Sci. 2021;70:1–9. doi: 10.5650/jos.ess20196. [DOI] [PubMed] [Google Scholar]
- Yu AQ, Pratomo Juwono NK, Leong SSJ, Chang MW. Production of fatty acid-derived valuable chemicals in synthetic microbes. Front Bioeng Biotechnol. 2014;2:78. doi: 10.3389/fbioe.2014.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Bian D, Xie Y, Jiang X, Li X, Li P, Xie T. Utilization of food waste hydrolysate for microbial lipid and protein production by Rhodosporidium toruloides Y2. J Chem Technol Biotechnol. 2017;92:666–673. doi: 10.1002/jctb.5049. [DOI] [Google Scholar]
- Zhang C, Shen H, Zhang X, Yu X, Wang H, Xiao S, Wang J, Zhao ZK. Combined mutagenesis of Rhodosporidium toruloides for improved production of carotenoids and lipids. Biotechnol Lett. 2016;38:1733–1738. doi: 10.1007/s10529-016-2148-6. [DOI] [PubMed] [Google Scholar]
- Zhang S, Skerker JM, Rutter CD, Maurer MJ, Arkin AP, Rao CV. Engineering Rhodosporidium toruloides for increased lipid production. Biotechnol Bioeng. 2016;113:1056–1066. doi: 10.1002/bit.25864. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kamal R, Li Q, Yu X, Wang Q, Zhao ZK. Comparative fatty acid compositional profiles of Rhodotorula toruloides haploid and diploid strains under various storage conditions. Fermentation. 2022;8:467. doi: 10.3390/fermentation8090467. [DOI] [Google Scholar]
- Zhang Y, Pang J, Liu S, Nie K, Deng L, Wang F, Liu J. Harnessing transcription factor Mga2 and fatty acid elongases to overproduce palmitoleic acid in Saccharomyces cerevisiae. Biochem Eng J. 2022;181:108402. doi: 10.1016/j.bej.2022.108402. [DOI] [Google Scholar]
- Zhao Y, Song B, Li J, Zhang J. Rhodotorula toruloides: an ideal microbial cell factory to produce oleochemicals, carotenoids, and other products. World J Microbiol Biotechnol. 2022;38:13. doi: 10.1007/s11274-021-03201-4. [DOI] [PubMed] [Google Scholar]
- Zhou YJ, Buijs NA, Zhu Z, Qin J, Siewers V, Nielsen J. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat Commun. 2016;7:11709. doi: 10.1038/ncomms11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Zhang S, Liu H, Shen H, Lin X, Yang F, Zhou YJ, Jin G, Ye M, Zou H, Zhao ZK. A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat Commun. 2012;3:1112. doi: 10.1038/ncomms2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the findings of this mini-review are available in the references cited within this article.