Abstract
The floral nectar of angiosperms harbors a variety of microorganisms that depend predominantly on animal visitors for their dispersal. Although some members of the genus Acinetobacter and all currently known species of Rosenbergiella are thought to be adapted to thrive in nectar, there is limited information about the response of these bacteria to variation in the chemical characteristics of floral nectar. We investigated the growth performance of a diverse collection of Acinetobacter (n = 43) and Rosenbergiella (n = 45) isolates obtained from floral nectar and the digestive tract of flower-visiting bees in a set of 12 artificial nectars differing in sugar content (15% w/v or 50% w/v), nitrogen content (3.48/1.67 ppm or 348/167 ppm of total nitrogen/amino nitrogen), and sugar composition (only sucrose, 1/3 sucrose + 1/3 glucose + 1/3 fructose, or 1/2 glucose + 1/2 fructose). Growth was only observed in four of the 12 artificial nectars. Those containing elevated sugar concentration (50% w/v) and low nitrogen content (3.48/1.67 ppm) were limiting for bacterial growth. Furthermore, phylogenetic analyses revealed that the ability of the bacteria to grow in different types of nectar is highly conserved between closely related isolates and genotypes, but this conservatism rapidly vanishes deeper in phylogeny. Overall, these results demonstrate that the ability of Acinetobacter spp. and Rosenbergiella spp. to grow in floral nectar largely depends on nectar chemistry and bacterial phylogeny.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00248-022-02088-4.
Keywords: Acinetobacter, Floral nectar, Bee, Phylogeny, Rosenbergiella
Introduction
Microbial growth in natural habitats largely depends on local environmental conditions, including diverse physico-chemical factors and the availability of key nutrients [1]. In host-associated microorganisms, such local environmental conditions can vary across potential hosts or even among different parts of the same host, so that each microhabitat may select for microbial populations displaying specific phenotypes and/or genotypes depending on the prevailing growth constraints and associated selective regime [2, 3]. Additionally, many microbial traits seem to be phylogenetically conserved, so that closely related taxa display more similar trait values than distant relatives do [4].
Land plants are hosts of diverse microbial communities, and specifically, the flowers of angiosperms provide diverse ephemeral habitats for microbial growth [5], of which floral nectar has received much attention in recent years. While floral nectar is assumed to be initially sterile, it often becomes rapidly colonized after anthesis by microorganisms (particularly yeasts and bacteria) associated with pollinators or other flower-visiting animals [6–8]. However, the typical chemical properties of floral nectar impose strong constraints for microbial life in this habitat [8–13]. In general, floral nectar is characterized by moderate to high sugar concentrations, which may exert high osmotic pressure, and low concentrations of other substances essential for microbial growth, such as amino acids and other nitrogen sources [11, 14–17].
Previous research has documented the presence of phylogenetically diverse bacteria in the floral nectar of diverse plant species worldwide [6, 18–25]. Members of the genera Acinetobacter and Rosenbergiella (Gammaproteobacteria) rank among the most frequent nectar inhabitants [6, 8, 9, 18, 26]. Furthermore, these two genera have been found in the mouthparts and digestive tract of diverse pollinators and other flower visitors, including insects (e.g., honeybees, bumblebees, and beetles) and hummingbirds [25, 27–29]. Although floral nectar is a complex solution of several nutrients, most of the research on microbial growth in nectar has focused on individual components (e.g., only sugars or only nitrogen sources). In this regard, it has been demonstrated that some Acinetobacter species and all Rosenbergiella species can tolerate high sugar concentrations (up to 60% w/v) and feed on different carbon and nitrogen sources [27, 30–36]. However, the combined effect of different chemical characteristics of nectar on the growth of nectar-inhabiting bacteria remains largely unexplored. Moreover, although phylogenetic affiliation seems to account for some variability in the growth on different nutrient sources of Acinetobacter species, so that closely related lineages (species or isolates) perform more similarly than do distantly related lineages [32, 36], it has not been determined yet if this factor has some influence on the growth of Acinetobacter and other bacterial taxa in floral nectar.
In this study, we investigated the growth performance of a diverse collection of Acinetobacter and Rosenbergiella isolates obtained from floral nectar and bees from different geographical origins in a set of twelve artificial nectars differing in their basic properties, namely, overall sugar and nitrogen content and sugar composition. Growth profiles were compared under the hypotheses that (i) under the combination of elevated sugar and low nitrogen content, the growth of Acinetobacter and Rosenbergiella isolates in nectar is limited, and (ii) the growth ability of these bacteria in different nectar types is determined by their phylogenetic affiliation.
Materials and Methods
Isolates
Forty-three Acinetobacter isolates and 45 Rosenbergiella isolates were used in this study (Tables S1 and S2, respectively). Studied isolates had been obtained between 2011 and 2018 from flowers and bees collected in different locations of Europe (Belgium, France, and Spain), the USA (California and Hawaii Island), South Africa, and Japan. Each of the isolates came from separate flowers or bees. All bacterial isolates analyzed in this study were grown on trypticase soy agar (TSA; Merck Life Science, Overijse, Belgium) at 25 °C and stored at − 80 °C in brain heart infusion (BHI) broth (Becton Dickinson, Erembodegem, Belgium) containing 25% glycerol (Merck Life Science) until further use.
Species-level classification of isolates was achieved by analyzing the sequence of a number of housekeeping genes commonly used as taxonomic markers of the Gammaproteobacteria, including atpD, which encodes the ATP synthase β-chain; gyrB, encoding the DNA gyrase subunit B; and/or rpoB, encoding the β subunit of RNA polymerase (see Supplementary Methods). Previous work has revealed that these housekeeping genes offer greater resolution than the 16S ribosomal RNA (rRNA) gene for discriminating between the Acinetobacter lineages (rpoB) and the Rosenbergiella lineages (atpD, gyrB, and rpoB) typically found in floral nectar and insects [30, 31, 33, 35].
Acinetobacter isolates included representatives of A. nectaris (n = 35; 81.4% of the total number of isolates of this genus) and A. boissieri (n = 8; 18.6%), which earlier studies have identified as the most prevalent species of the genus in floral nectar [18, 20, 23, 29] (Table S1). Ten A. nectaris isolates were obtained from the mouthparts, honey crop, or gut of honeybees (Apis mellifera) collected in Stanford campus (California, USA), whereas the remaining 25 A. nectaris isolates and all A. boissieri isolates had been retrieved from floral nectar of 11 plant species from nine families collected in Belgium (13 A. nectaris isolates from 2 plant species), Spain (5 A. nectaris isolates from 4 plant species and 8 A. boissieri isolates from 6 plant species), and the USA (7 A. nectaris isolates from Epilobium canum (Onagraceae)) (Table S1).
Most Rosenbergiella isolates (n = 41, 91.1% of the total number of isolates of this genus) had been obtained from floral nectar of diverse plant species (15 plant species from 12 families) collected in different countries, but four isolates (8.9%) had been retrieved from bees collected in Stanford Campus (California, USA) (Table S2). Rosenbergiella isolates included all validated species of the genus, namely R. epipactidis (n = 16 isolates (35.6%): 1 from France, 1 from Japan, 5 from Spain, and 9 from the USA, all of them from floral nectar), R. nectarea (n = 16 isolates (35.6%): 6 nectar isolates from Belgium, 2 nectar isolates from France, and 4 nectar isolates from the USA, plus 4 isolates from the mouthparts of honeybees (3 isolates) or the gut of a bumblebee (Bombus sp., 1 isolate) collected in the USA), R. collisarenosi (n = 6 isolates (13.3%): 2 from Belgium, 3 from Spain, and 1 from the USA, all of them from floral nectar), and R. australiborealis (n = 3 isolates (6.7%) found in nectar samples from South Africa) (Table S2). Additionally, 3 nectar isolates from Spain (6.7%) and one nectar isolate from Hawaii (USA, 2.2%) belonged to “R. gaditana” and “R. metrosideri,” respectively, two new Rosenbergiella species which are pending formal recognition [30].
Growth in Artificial Nectars
All isolates were tested for their ability to grow in twelve types of artificial nectar. These nectars varied in total sugar concentration (15% or 50% w/v, coded as “s” and “S,” respectively; i.e., lowercase for low sugar level and uppercase for high sugar level), the availability of nitrogen sources (3.48/1.67 ppm and 348/167 ppm of total nitrogen/amino nitrogen; “n” and “N,” respectively; i.e., lowercase for low nitrogen level and uppercase for high nitrogen level), and their sugar composition (only sucrose, 1/3 sucrose + 1/3 glucose + 1/3 fructose, or 1/2 glucose + 1/2 fructose; coded as “S” [only sucrose], “M” [mix of sucrose and hexoses], and “H” [only hexoses], respectively). Values of total sugar concentration used in this study (15% or 50% w/v) resemble those commonly found in nectar from different plant species, which typically range from 15 to 40% w/v but may reach > 50% (e.g., under warm conditions and low relative humidity) [37–39]. Moreover, the sugar composition and ratios considered in this study (only sucrose, 1/3 sucrose + 1/3 glucose + 1/3 fructose, or 1/2 glucose + 1/2 fructose) correspond to the sucrose dominant nectar, balanced nectar, and fructose-glucose dominant nectar categories (S, SFG, and FG types, respectively) established by Percival [40] for natural nectars. Additionally, the amino acid concentration of nectar has been observed in ranges from 0.3 to 12.5 µmol/mL in naturally pollinated plants, depending on the pollinator type [41, 42]. Assuming an average molecular weight of 136.9 g/mol and an average nitrogen content of 14.7% for proteinogenic amino acids, those previously reported amino acid values correspond to 6 ppm and 251.6 ppm of amino nitrogen, respectively (but note that floral nectar may contain non-proteinogenic amino acids and other nitrogen sources). Therefore, the nitrogen content of the artificial nectars used in this study (3.48/1.67 ppm and 348/167 ppm of total nitrogen/amino nitrogen in the “n” and “N” treatments, respectively) is also similar to the content found in natural nectars. A three-letter code was used to name each artificial nectar depending on these three basic properties (Table 1).
Table 1.
Characteristics of the artificial nectars used in this study for testing the growth performance of Acinetobacter and Rosenbergiella isolates
| Artificial nectar code | Sugar levela | Nitrogen levelb | Sugar compositionc |
|---|---|---|---|
| snS | Low (15%) | Low (3.48/1.67) | S |
| sNS | Low (15%) | High (348/167) | S |
| SnS | High (50%) | Low (3.48/1.67) | S |
| SNS | High (50%) | High (348/167) | S |
| snM | Low (15%) | Low (3.48/1.67) | 1/3 S, 1/3 G, 1/3 F |
| sNM | Low (15%) | High (348/167) | 1/3 S, 1/3 G, 1/3 F |
| SnM | High (50%) | Low (3.48/1.67) | 1/3 S, 1/3 G, 1/3 F |
| SNM | High (50%) | High (348/167) | 1/3 S, 1/3 G, 1/3 F |
| snH | Low (15%) | Low (3.48/1.67) | 1/2 G, 1/2 F |
| sNH | Low (15%) | High (348/167) | 1/2 G, 1/2 F |
| SnH | High (50%) | Low (3.48/1.67) | 1/2 G, 1/2 F |
| SNH | High (50%) | High (348/167) | 1/2 G, 1/2 F |
aTotal percentage of sugars (w/v)
bConcentration of total nitrogen/amino nitrogen (all values are in ppm)
cS: sucrose; G: glucose; F: fructose
Artificial nectars were prepared as indicated in the Supplementary Methods and then added to different rows of sterile 96-well plates (180 μL per well) (BRAND GmbH + Co KG, Wertheim, Germany). A positive control (1/10 × tryptic soy broth, TSB; Merck Life Science) and a negative control (filter-sterilized distilled water) were included in all plates, and the order of the artificial nectars and controls in the rows of the plates (six for artificial nectars + two for the controls) was randomized in each replicate of the assay. Assay plates were kept in refrigeration (4 °C) overnight and then left at room temperature for 20 min until inoculation with bacterial cells. Eleven columns of the assay plates were then inoculated with 20 μL per well of a different cell suspension (c.107 colony forming units per mL) in saline solution prepared and starved as indicated in the Supplementary Methods (i.e., eleven isolates were tested per plate), whereas the wells of the twelfth column were inoculated with 20 μL of sterile saline solution to serve as microbe-free controls. The order of isolates and the microbe-free control in the columns of the plates was randomized in all assay plates. All assays (growth of each Acinetobacter or Rosenbergiella isolate in each artificial nectar) were repeated at least three times, and some randomly chosen isolates were inoculated in two different columns of the same plate to test for intraplate reproducibility of the assays. Inoculated plates were covered with a breathable membrane (Breath-Easy; Diversified Biotech, Boston, MA, USA) and incubated with no agitation for 7 days at 25 °C. Optical density (OD) values at a wavelength of 600 nm were determined for each isolate and test condition by putting the assay plates into a benchtop spectrophotometer (Multiskan GO; Thermo Fisher, Merelbeke, Belgium) just after inoculation (day 0) and after 3 and 7 days of incubation; a brief shaking (30 s) was applied to each assay plate just before the OD determinations. Normalized OD readings were calculated by subtracting the OD values of the microbe-free control wells from the OD values of the test wells.
Phylogenetic Reconstruction
Maximum likelihood (ML) phylogenetic trees were generated for all Acinetobacter and Rosenbergiella isolates and sequence types (or genotypes; STs), defined as sequences (rpoB for Acinetobacter spp. and concatenation of atpD, gyrB, and rpoB sequences for Rosenbergiella spp.) differing in at least one nucleotide, as indicated in Supplementary Methods. Briefly, nucleotide sequences obtained for the studied isolates and some reference strains (e.g., Acinetobacter calcoaceticus NIPH 2245 T and Phaseolibacter flectens ATCC 12775 T, used as outgroups in the trees built for Acinetobacter and Rosenbergiella, respectively) were included in multiple alignments generated by MUSCLE [43]. ML trees of isolates and STs were built for the rpoB gene sequences of Acinetobacter (861 bp) and a concatenation of atpD + gyrB + rpoB sequences of Rosenbergiella (1863 bp) using PhyML v.3.0 [44] with smart model selection based on the Akaike information criterion (AIC) [45] (see details in Figs. S1 and S2), and the resulting trees were visualized and edited with the Molecular Evolutionary Genetics Analysis v.11 (MEGA11) software [46].
Additionally, to assess the phylogenetic conservatism of measured traits at the species level (see “Data Analysis”), a ML genome-based tree was generated for the two Acinetobacter and the six Rosenbergiella species included in this study using the up-to-date bacterial core gene set (UBCG2) pipeline [47] (Supplementary Methods).
Data Analysis
Unless otherwise indicated, all data analyses described in the following sections were performed using R version 4.1.0 [48] run on RStudio v.1.4.1717 [49]. Required R libraries are indicated below.
Exploratory Data Analysis and Data Normalization
The growth performance (GP) of each isolate in each artificial nectar after 3 and 7 days of incubation was evaluated by subtracting to the increase in the normalized OD value (see above) obtained in such condition (ΔOD) the increase in the normalized OD value obtained in the negative control that did not contain any sugar or nitrogen source ((ΔODctrl) which accounted for possible growth due to, for example, nutrient reserves remaining after the starvation step [32]); i.e., GP = ΔOD − ΔODctrl = (ODf − OD0) − (ODf-ctrl − OD0-ctrl), where OD0 and ODf are the OD values on day 0 and after incubation (3 or 7 days), respectively, and OD0-ctrl and ODf-ctrl are the equivalent values obtained for the negative control.
Intra-plate reproducibility of the assay testing for growth in artificial nectars (i.e., agreement of the GP values obtained for replicates ran in the same plate) was evaluated by calculating Lin’s concordance correlation coefficient (CCC) for agreement on continuous measures, which quantifies the agreement between two measures of the same variable and ranges from − 1 (strong discordance) to 1 (perfect agreement) [50, 51]. For each combination of artificial nectar and incubation time, Lin’s CCC was calculated using the epi.ccc() function of the R library “epiR” v. 2.0.33 [52]. Scatter plots of the replicate data and the corresponding linear regression line were generated using “ggplot2” v.3.3.5 [53].
All trait values were further processed by calculating the average of the GP values obtained for the three replicates of each isolate and test condition (GPavg), and the distributions of GPavg values obtained in each test condition for Acinetobacter vs. Rosenbergiella and the different species tested were visualized by violin plots using the R library “vioplot” v.0.3.7 [54]. To allow a fairer comparison of growth data across genera and species, GPavg values were converted into Z-scores (i.e., GPavg of a particular isolate minus average GPavg of all 88 isolates (43 Acinetobacter + 45 Rosenbergiella isolates) for the same test, divided by the corresponding standard deviation) using Microsoft Excel 2016 (Redmond, WA, USA). For a given test condition (artificial nectar and incubation time), isolates with positive Z-scores had a growth performance above the mean of the studied set of isolates, whereas those with negative Z-scores had a growth performance below the mean. Finally, the mean Z-score of the isolates belonging to each ST and species of Acinetobacter and Rosenbergiella was calculated.
Nonparametric Factorial Analysis of Variance
The effect of different factors — namely, taxonomic affiliation (at the genus or species level), incubation time (3 or 7 days), and the type of artificial nectar (Table 1) — and their interactions on the growth results of all the isolates in the different types of artificial nectar (expressed as a vector of Z-scores) was analyzed by repeated-measures factorial analysis of variance (ANOVA) of aligned rank transformed (ART) data, as implemented in the R library “ARTool” [55, 56]. ART-based ANOVA is a non-parametric statistical test that relies on a preprocessing step that “aligns” the data for each possible main effect or interaction (i.e., effects are estimated as marginal means and then stripped from the response variable so that all effects but one are removed) before assigning averaged ranks, after which conventional ANOVA procedures are used [55, 56]. P values < 0.05 were considered significant.
Phylogenetically Informed Analyses
Further analysis of growth performance data was achieved by applying different methods which account for the non-independence of data due to shared ancestry. Such phylogenetically informed methods were applied at the isolate, genotype (ST), and species level to evaluate the magnitude of phylogenetic correlations in studied traits. The inputs of the analyses were (i) an R data frame with the Z-scores (or average Z-scores) obtained for Acinetobacter and Rosenbergiella isolates, STs, or species, and (ii) the ML phylogenetic trees built from housekeeping gene sequences or genome assemblies, and converted into ultrametric trees (i.e., trees where all tips are at equal distance from the root) by the force.ultrametric() function of “phytools” v.0.7–80 [57]. When necessary, phylogenetically independent contrasts (PICs) [58] were calculated by applying the pic() function of the R library “ape” v.5.5 [59] to the Z-scores obtained for each trait (growth in different artificial nectars after 3 and 7 days of incubation). Holm’s correction was applied to P values whenever multiple comparisons were made, and corrected P values < 0.05 were considered significant.
Pairwise correlations between PICs obtained for different traits for Acinetobacter and Rosenbergiella isolates were assessed by the non-parametric Spearman rank test, as implemented in the R library “Hmisc” v.4.5–0 [60]. Correlation matrices were visualized using the R library “corrplot” v.0.90 [61].
Visualization of the trait data obtained for obtained for Acinetobacter and Rosenbergiella isolates, STs, and species on the corresponding ML trees was achieved by generating phylogenetic heatmaps using the phylo.heatmap() function of “phytools.” In addition, we used the fitContinuous() function of the R library “geiger” v.2.0.7 [62] to determine which model of trait evolution provided the best fit to the phenotypic data. Nine different models were tested (Brownian motion, Ornstein–Uhlenbeck, early-burst, trend, Pagel’s λ, Pagel’s κ, Pagel’s δ, drift, and white noise model; see Supplementary Methods), and their relative likelihood was assessed by calculating their AIC values and Akaike weights [63]. When no evolutionary model yielded an Akaike weight ≥ 0.5, it was concluded that none of them fit the data substantially better than the others [32, 36, 64, 65].
Finally, the statistical dependence among the Z-scores obtained for isolates, STs, and species due to their phylogenetic relationships was evaluated by calculating four different phylogenetic signal metrics commonly used in comparative biology, namely, Blomberg’s K, Pagel’s λ, Moran’s I, and Abouheif’s Cmean [36, 65–73]. K and λ assume a Brownian motion model of trait evolution (i.e., random walk with constant trait variance over time [58]), and the closer their values are to zero, the more phylogenetically independent a trait is, while values of 1 for these metrics correspond to the Brownian motion expectation, and values > 1 mean that close relatives are more similar than expected under Brownian motion [69, 70]. Therefore, K and λ can be used to assess the strength (or “effect size”) of phylogenetic structuring [67, 72]. In contrast, I and Cmean are autocorrelation indices that depend on the structure and size of the phylogeny, but which are not based on any evolutionary model and are unable to provide information on the strength of the phylogenetic signal [67, 72]. Therefore, the values of I and Cmean cannot be quantitatively compared [72]. Computation of all these metrics was performed using the multiPhylosignal() function of R library “picante” v.1.8.2 [74] for calculation of K, the phylosig() function of “phytools” for λ, and the abouheif.moran() function of “adephylo” v.1.1–11 [75] for I and Cmean (method = “Abouheif” and “oriAbouheif” for computation of phylogenetic proximity between the tips of trees, respectively). Statistical significance was tested in all cases by randomization with 1000 repetitions.
Results
Identification of Sequence Types and Phylogenetic Reconstruction
Analysis of rpoB sequences identified a total of 18 STs (ST01A to ST18A) among the Acinetobacter isolates included in this study, whereas 24 STs (ST01R to ST24R) were identified for the concatenation of atpD + gyrB + rpoB sequences among the Rosenbergiella isolates (Tables S1 and S2). STs represented between one and ten conspecific isolates of nectar or bee origin, but never from both habitats. However, two Acinetobacter and five Rosenbergiella STs included isolates from different plant species, and one Rosenbergiella ST grouped two isolates retrieved from honeybees with one obtained from a bumblebee (Tables S1 and S2).
In general, isolates or STs belonging to the same species, as determined by phylogenetic proximity to the corresponding type strain or a reference strain, formed well-supported clades (> 90% bootstrap) in the ML trees built for Acinetobacter or Rosenbergiella (Figs. S1 and S2). However, the combined analysis of atpD, gyrB, and rpoB sequences did not provide enough resolution for differentiating between the single isolate of “R. metrosideri” (JB07) and R. epipactidis isolates (Fig. S2). Furthermore, two major subclades were detected within R. nectaris, one of which grouped three out of the four Rosenbergiella isolates of bee origin analyzed in the present study (namely, B1A, B4A, and B5A) and was phylogenetically close to the fourth one (B3A) (Fig. S2). Nevertheless, the branching of such R. nectarea subclades was only moderately supported (77.5% bootstrap). Examination of the ML genome tree generated from the UBCG2 set confirmed the robustness of the clades grouping the type strains of the two Acinetobacter species and the six Rosenbergiella species analyzed in this study, as well as the phylogenetic relatedness between R. epipactidis and ‘R. metrosideri’ (Fig. S3).
Growth in Artificial Nectars
All Acinetobacter and Rosenbergiella isolates showed very little or null growth in the nutrient-free negative control (ΔODctrl values after 7 days of incubation were in the range of 0.000–0.045 (mean ± S.D. = 0.001 ± 0.004) for Acinetobacter isolates and of 0.000–0.017 (mean ± S.D. = 0.001 ± 0.002) for Rosenbergiella isolates). Scarce or null growth was also observed in eight of the twelve artificial nectars studied, including most of the nectars with a high sugar content and all the nectars containing the lowest nitrogen content (ΔODavg values after 7 days of incubation [ΔODavg7] were < 0.05 in the following artificial nectars: snS, SnS, snM, SnM, SNM, snH, SnH, and SNH; Tables 1 and S3). Therefore, only the growth data obtained for the following four artificial nectars was considered in subsequent analyses: sNS (low sugar/high nitrogen/only sucrose; ΔODavg7 = 0.005–0.489), sNM (low sugar/high nitrogen/mixture of sucrose, glucose, and fructose; ΔODavg7 = 0.067–0.401), sNH (low sugar/high nitrogen/only hexoses; ΔODavg7 = 0.049–0.393), and SNS (high sugar/high nitrogen/only sucrose; ΔODavg7 = − 0.009–0.071) (Table S3). Intra-plate reproducibility of the assays testing for growth in these four artificial nectars on day 3 and 7 was generally good to excellent, as the CCC 95% confidence intervals included in all cases values > 0.8 (Table S4) and the regression lines between repeated measures were close to the line of perfect concordance (i.e., y = x; Fig. S4).
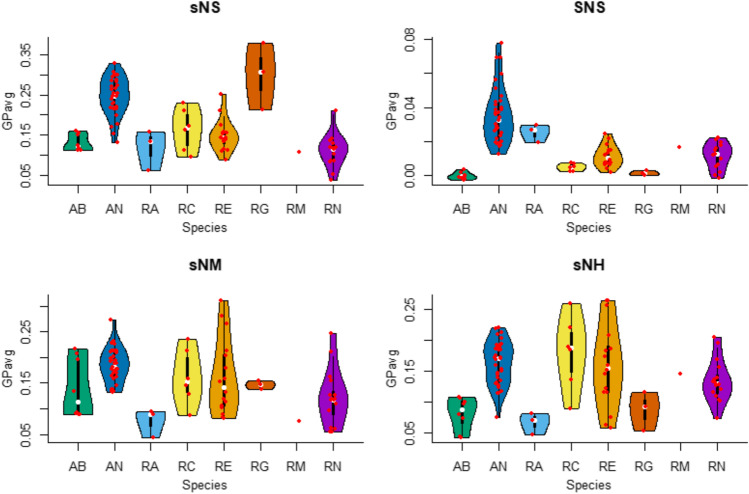
In general, the average growth performance (i.e., GPavg) values obtained in the different artificial nectars for the isolates of each Acinetobacter and Rosenbergiella species were broadly distributed and showed several outliers (Fig. 1). ART-based ANOVA revealed that the growth of isolates in artificial nectars significantly depended on their taxonomic affiliation (at the genus and species level) and the two-way interactions between the taxonomic affiliation and the type of nectar, and between the taxonomic affiliation and the incubation time, whereas the type of nectar and incubation time had by themselves a lower and non-significant influence on growth differences (Table 2).
Fig. 1.
Violin plots showing the growth results, expressed as average growth performance (GPavg), in different artificial nectars of the Acinetobacter and Rosenbergiella species tested in this study. Note that, in general, the GPavg values obtained for most species in each artificial nectar were broadly distributed around the median value (white dots contained within the thick black bars representing the interquartile range [IQR] that are shown inside the violin plots). Outlier values (red dots located beyond the boundaries of the thin black lines representing 1.5 × IQR) are evident in some plots. Abbreviations: AB, Acinetobacter boissieri (n = 8); AN, Acinetobacter nectaris (n = 35); RA, Rosenbergiella australiborealis (n = 3); RC, Rosenbergiella collisarenosi (n = 6); RE, Rosenbergiella epipactidis (n = 16); RG, ‘Rosenbergiella gaditana’ (n = 3); RM, ‘Rosenbergiella metrosideri’ (n = 1); RN, Rosenbergiella nectarea (n = 16). Artificial nectar codes are as in Table 1
Table 2.
Results obtained in the non-parametric factorial analysis of variance (ANOVA) of the growth performance of Acinetobacter and Rosenbergiella isolates in artificial nectars using aligned rank transformed data
| Factors | Species-level analysisa | Genus-level analysisa | ||||||
|---|---|---|---|---|---|---|---|---|
| df | df res | F | P | df | df res | F | P | |
| Artificial nectar (AN)b | 3 | 560 | 0.38 | 0.768 | 3 | 602 | 0.50 | 0.685 |
| Incubation time (IT)c | 1 | 560 | < 0.01 | 0.954 | 1 | 602 | 0.16 | 0.688 |
| Taxonomic affiliation (TA)d | 7 | 80 | 26.78 | < 0.001* | 1 | 86 | 52.43 | < 0.001* |
| AN × IT | 3 | 560 | 0.22 | 0.884 | 3 | 602 | 0.31 | 0.817 |
| AN × TA | 21 | 560 | 11.94 | < 0.001* | 3 | 602 | 13.82 | < 0.001* |
| IT × TA | 7 | 560 | 6.93 | < 0.001* | 1 | 602 | 22.54 | < 0.001* |
| AN × IT × TA | 21 | 560 | 1.23 | 0.223 | 3 | 602 | 0.84 | 0.474 |
adf: degrees of freedom; df res: degrees of freedom of residuals, F: F statistics; P: P values (statistically significant values are denoted by an asterisk)
bArtificial nectar types: low sugar/high nitrogen/only sucrose (sNS); low sugar/high nitrogen/mixture of sucrose, glucose, and fructose (sNM); low sugar/high nitrogen/only hexoses (sNH); and high sugar/high nitrogen/only sucrose (SNS) (see details in Table 1)
cIncubation time of assay plates (3 days vs. 7 days)
dTaxonomic affiliation of studied isolates at the species level (A. boissieri [n = 8], A. nectaris [n = 35], R. australiborealis [n = 3], R. collisarenosi [n = 6], R. epipactidis [n = 16], ‘R. gaditana’ [n = 3], ‘R. metrosideri’ [n = 1], or R. nectarea [n = 16]; species names pending of validation are indicated between quotation marks) or genus level (Acinetobacter [n = 43] vs. Rosenbergiella [n = 45])
Phylogenetic-Based Analysis of Trait Variation
Correlation analysis of the PICs obtained from Z-scores data revealed 11 and 8 significant correlations between the assimilation assays for Acinetobacter and Rosenbergiella, respectively (39.3% and 28.6%, respectively, of the total number of pairwise comparisons, n = 28) (Fig. S5). All significant correlations obtained for Acinetobacter and Rosenbergiella were positive (Spearman’s ρ = 0.513–0.817 and 0.695–0.868, respectively). Moreover, the PICs obtained for growth in a same artificial nectar after 3 and 7 days of incubation were highly correlated (Fig. S5), so subsequent phylogenetic-based analyses were only performed using the results obtained on day 3.
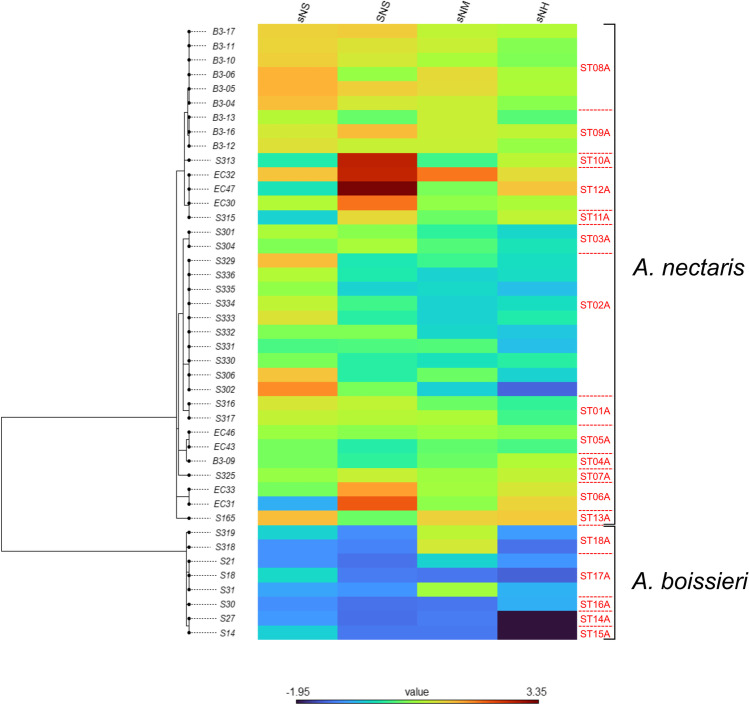
Mapping of the trait data to the ML trees generated for Acinetobacter suggested some clade dependence of the trait values. In particular, A. boissieri isolates and STs displayed lower growth values in most test conditions than A. nectaris (see the phylogenetic heatmaps in Figs. 2 and S6). Furthermore, some intraspecies differentiation was found within A. nectaris and A. boissieri at the isolate and ST level (Figs. 2 and S6). Evolutionary model fitting of studied traits indicated that Pagel’s κ and λ models were the most supported for Acinetobacter isolates and STs (each model fit the Z-scores obtained for two artificial nectars) (Table S5). Phylogenetic signal analysis of the Z-scores obtained for Acinetobacter isolates and STs yielded significant results for all artificial nectars when tested by Moran’s and Abouheif’s methods, and by Pagel’s method (100% of significant tests, in all cases; Table 3). Blomberg’s K values were low (1.12·10−6–5.67 × 10−6) but significant for all traits when analyzed for Acinetobacter isolates, and non-significant for all traits except growth in the sNH nectar after 3 days of incubation (K = 1.35 × 10−4) when analyzed for STs (Table 3).
Fig. 2.
Phylogenetic heatmap of the trait values obtained for the different Acinetobacter isolates (shown in rows) in the growth assays performed in this study. Artificial nectar codes (shown in columns) are as in Table 1. The phylogenetic tree shown on the left corresponds to the maximum likelihood (ML) tree depicted in Fig. S1. The sequence type (ST; shown in red characters) and species affiliation of isolates are indicated at the right side of the heatmap (see also Table S1). Note that some clade dependence of the trait values (growth in the four artificial nectars under analysis) is observed, with A. boissieri isolates displaying in most cases lower trait values than A. nectaris
Table 3.
Phylogenetic signal of the growth of Acinetobacter and Rosenbergiella isolates, sequence types (STs), and species in different artificial nectars
| Phylogenetic tree (N)a | Phylogenetic signal metricb | Artificial nectarsc,d | |||
|---|---|---|---|---|---|
| sNS | SNS | sNM | sNH | ||
| Acinetobacter | |||||
| Isolates (43) | K | 2.23 × 10−6 (0.004*) | 4.52 × 10−6 (0.004*) | 1.12 × 10−6 (0.004*) | 5.67 × 10−6 (0.004*) |
| λ | 0.901 (< 0.001*) | 0.980 (< 0.001*) | 0.870 (0.001*) | 0.983 (< 0.001*) | |
| I | 0.573 (0.004*) | 0.760 (0.004*) | 0.499 (0.004*) | 0.723 (0.004*) | |
| Cmean | 0.595 (0.004*) | 0.769 (0.004*) | 0.520 (0.004*) | 0.742 (0.004*) | |
| STs (18) | K | 3.21 × 10−6 (0.541) | 5.38 × 10−6 (0.541) | 8.76 × 10−6 (0.527) | 1.35 × 10−4 (0.024*) |
| λ | 0.642 (0.004*) | 0.945 (< 0.001*) | 0.433 (0.033*) | 0.998 (< 0.001*) | |
| I | 0.427 (0.004*) | 0.594 (0.004*) | 0.501 (0.004*) | 0.654 (0.004*) | |
| Cmean | 0.491 (0.004*) | 0.613 (0.004*) | 0.557 (0.004*) | 0.691 (0.004*) | |
| Rosenbergiella | |||||
| Isolates (45) | K | 1.05 × 10−6 (0.026*) | 1 × 10−6 (0.004*) | 2 × 10−6 (0.004*) | 1.84 × 10−6 (0.004*) |
| λ | 0.834 (< 0.001*) | 0.797 (0.002*) | 0.982 (0.025*) | 0.911 (0.022*) | |
| I | 0.439 (0.004*) | 0.446 (0.004*) | 0.406 (0.004*) | 0.452 (0.004*) | |
| Cmean | 0.480 (0.004*) | 0.460 (0.004*) | 0.417 (0.004*) | 0.467 (0.004*) | |
| STs (24) | K | 0.042 (0.086) | 0.105 (0.004*) | 0.036 (0.086) | 0.041 (0.075) |
| Λ | 0.564 (1) | 0.973 (0.006*) | 6.62 × 10−5 (1) | 6.62 × 10−5 (1) | |
| I | − 0.078 (0.572) | 0.344 (0.020*) | 0.033 (0.5) | 0.187 (0.152) | |
| Cmean | 0.064 (0.551) | 0.394 (0.020*) | 0.061 (0.551) | 0.224 (0.120) | |
| Species (8) | K | 0.099 (0.888) | 0.154 (0.444) | 0.044 (0.888) | 0.131 (0.483) |
| λ | 6.68 × 10−5 (1) | 6.68 × 10−5 (1) | 6.68 × 10−5 (1) | 6.68 × 10−5 (1) | |
| I | − 0.257 (1) | − 0.414 (1) | − 0.278 (1) | − 0.328 (1) | |
| Cmean | − 0.103 (1) | − 0.099 (1) | − 0.099 (1) | − 0.084 (1) | |
aPhylogenetic tree used for computation of phylogenetic signal indices (see details in the main text). N: number of isolates, STs, or genomes available for analysis
bK: Bloomberg’s K; λ: Pagel’s λ; I: Moran’s I; Cmean: Abouheif’s Cmean
cArtificial nectar codes are as in Table 1
dPhylogenetic signal metric estimates and Holm-corrected P-values. Significant P values (< 0.05) are denoted by an asterisk
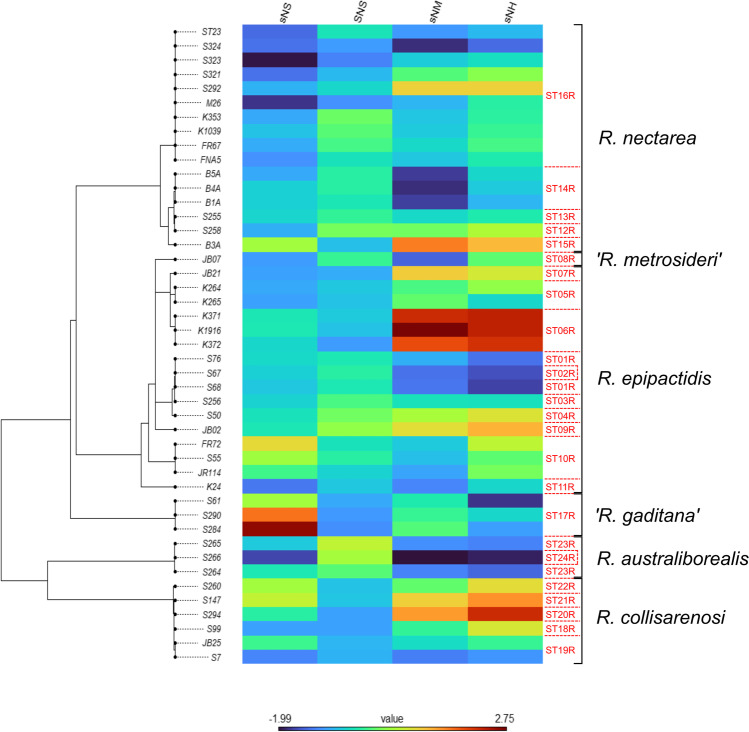
Trait differentiation between clades at the isolate and ST level was less evident in the phylogenetic heatmaps generated for Rosenbergiella ML (Figs. 3 and S7). Pagel’s κ was the most supported evolutionary model for Rosenbergiella isolates (Z-scores obtained in 3 out of 4 artificial nectars), followed by Pagel’s λ model (Z-scores obtained in the sNS nectar). In contrast, the Z-scores obtained for Rosenbergiella STs fit in most cases a white noise model of evolution, whereas Pagel’s κ was only supported for growth in the SNS nectar (Table S5). Significant results were obtained for all artificial nectars and all phylogenetic signal metrics at the isolate level (Table 3). Blomberg’s K values were low (< 2 × 10−6) in all cases, whereas high λ values were obtained for growth in the sNM and the sNH nectars (λ > 0.91). ST-based analysis yielded significant results for all phylogenetic signal metrics for growth in the SNS nectar, but not in the other nectars containing a lower sugar concentration (sNS, sNM, and sNH; Table 3).
Fig. 3.
Phylogenetic heatmap of the trait values obtained for the different Rosenbergiella isolates (shown in rows) in the growth assays performed in this study. Artificial nectar codes (shown in columns) are as in Table 1. The phylogenetic tree shown on the left corresponds to the maximum likelihood (ML) tree depicted in Fig. S2. The sequence type (ST; shown in red characters) and species affiliation of isolates is indicated at the right side of the heatmap (see also Table S2). Species names pending of validation are indicated between quotation marks. Note that trait differentiation between clades is less evident than for Acinetobacter isolates (see Fig. 2), as closely related isolates displayed in some cases divergent trait values
Mapping of the trait data to the ML tree generated from genome sequences showed no clear pattern of trait variation across Rosenbergiella species but confirmed the phenotypic differences between Acinetobacter species (A. nectaris displayed higher average growth values in all test conditions than A. boissieri; Fig. S8). The Z-scores obtained for Acinetobacter and Rosenbergiella species usually best fit a white noise model, whereas no evolutionary model of trait evolution performed substantially better that the others in the analysis of the Z-scores obtained for the SNS nectar (Table S5). No phylogenetic signal metric yielded significant results for any artificial nectar in the species-level analysis (Table 3).
Discussion
The high osmotic pressure and highly unbalanced carbon to nitrogen ratio often found in floral nectar are major hurdles to microbial life that determine the assembly of microbial communities in this microhabitat [9–12, 14, 17, 32]. However, there is still limited information on how different microorganisms respond to different types of nectar. The phylogenetic determination of growth variation in floral nectar was addressed in the present study by analyzing the growth performance of a diverse collection of Acinetobacter and Rosenbergiella isolates in artificial nectars differing in three basic chemical properties, namely, overall sugar content, overall nitrogen content, and sugar composition.
A first important result of this study was that all the Acinetobacter and Rosenbergiella isolates tested did not grow or only displayed very scarce growth in the artificial nectars containing the lowest amount of nitrogen (3.48 ppm), suggesting that these bacteria may preferentially grow in nitrogen-rich nectars. Unfortunately, to our knowledge, no published report on bacterial presence in floral nectar includes information about the nitrogen content of nectar samples, so the hypothetical preference of Acinetobacter spp. and Rosenbergiella spp. for nitrogen-rich nectars cannot be confirmed or refuted with field data. Additionally, it should be noted that even though the nectar-inhabiting representatives of these bacterial genera seem unable to grow in low-nitrogen media, they might be adapted to survive nitrogen limitation in a quiescent state and resume growth upon arrival of nitrogen inputs from external sources (e.g., pollen grains that fall into nectar or the frass of insects that visit flowers), the metabolic by-products of other nectar microbes such as yeasts, and/or cell debris [17, 32, 76]. For example, it has been recently demonstrated that the Acinetobacter species typically found in floral nectar can induce pollen germination and bursting, and protein release into solution, which might benefit bacterial growth [76].
Furthermore, our results confirmed that the high osmotic pressure resulting from elevated sugar concentrations is an efficient filter of microbial growth in floral nectar [10], as this factor hindered the growth of most isolates that were analyzed. In fact, negligible growth of all Acinetobacter and Rosenbergiella isolates was obtained in the SNM and SNH nectars, which contained the highest sugar level (50% w/v), a high nitrogen content (348 ppm in total), and either a mixture of sucrose and hexoses (SNM) or only hexoses (SNH). Nevertheless, some isolates grew in the SNS nectar that had the same overall sugar and nitrogen content as SNM and SNH but included sucrose as the only sugar source. Nectar sugar concentration varies significantly in natural plant populations, both within and between species, and may be affected by microclimatic conditions [77, 78]. Most plant species have nectar sugar concentrations ranging from 15 to 40% w/v, whereas concentrations over 50% are only frequent under warm conditions and low relative humidity [37–39]. Results of the present study thus prompt the testable prediction that densities of Acinetobacter and Rosenbergiella in nectar, and perhaps other nectar bacteria too (see Pusey [79]), should decline in seasons, habitats, or regions characterized by consistently warm and dry conditions. However, Von Arx et al. [80] observed that the abundance of nectar bacteria in two night-blooming plants of the Sonoran Desert reached in some cases 106 CFU/mL, which is within the range of values reported in studies performed in temperate regions [6, 24]. Moreover, drought often does not result in more concentrated nectar [81], and it has been observed that some bacteria such as Fructobacillus (phylum Bacillota) grow better in synthetic nectar at warmer temperatures [82]. It should also be noted that the molar concentration of sugar solutions with the same percentage of sugar (% w/v) differs depending on the monosaccharide content: the higher the monosaccharide content, the higher the molarity. Since water activity (aw) depends on molarity, the monosaccharide solutions will have a lower aw, which may explain why amongst the high sugar nectars, only the one with only sucrose (i.e., SNS) was able to support some growth. To our knowledge, most previous osmotolerance tests carried out on the Acinetobacter and Rosenbergiella species analyzed in this study have incorporated sucrose as the only sugar source [27, 30, 31, 33, 35]. Specific solute effects of sucrose, glucose, and fructose have been reported for other bacterial species and could lead to differential responses to osmotic stress [83]. Additionally, the lack of growth in the SNM and SNH nectars might not only be due to the high osmotic pressure resulting from elevated monosaccharide concentrations, but also to other factors such as the differences in viscosity and/or dissociation constants (which result in pH changes) of the different sugar types making up the solution mixture [84].
A common expectation in trait biology in scenarios where evolution mainly proceeds by vertical gene inheritance is that closely related organisms resemble each other more than compared to distant relatives [65, 69]. However, events such as gene loss, horizontal gene transfer — which is particularly common among the Pseudomonadota [85]—, and convergent evolution may reduce the phylogenetic signal of functional traits [86]. In this regard, the ANOVA results obtained in this study indicated that growth variation in artificial nectars mostly depended on the genus and species identity of tested isolates, whereas the nectar type and the incubation time of test plates explained a limited amount of the total variation in the data, mostly in interaction with the taxonomy of isolates. Within genus Acinetobacter, phenotypic differentiation between A. nectaris and A. boissieri was evident when the trait data obtained in the present study was mapped onto the ML trees built from rpoB sequences and the UBCG2 set. Furthermore, significant phylogenetic signal was detected for growth in the sNS, sNM, sNH, and SNS nectars by most methods of analysis, thus suggesting that the distribution of trait values among Acinetobacter isolates and STs was not random but significantly correlated with phylogeny. Nevertheless, evolutionary model fitting and the low Blomberg’s K values obtained for all traits under study for Acinetobacter isolates and STs suggest that the evolution of such traits departed from a pure Brownian motion. The evolutionary models most supported for traits with significant phylogenetic signal were Pagel’s κ and λ, both of which are transformations of the Brownian motion model that raise all branch lengths in the phylogenetic tree to a power κ or multiply internal branches by a factor λ, respectively (but note that κ = 1 and λ = 1 correspond to pure Brownian motion) [73]. Similar phenotypic differentiation between A. nectaris and A. boissieri and departure from a pure Brownian motion model has been recently reported for assimilation of diverse nitrogen sources [32], but the physiological mechanisms responsible for the growth differences between these and other recently described nectar-inhabiting species of Acinetobacter (A. pollinis, which seems to be the closest relative to A. nectaris, and A. baretiae and A. rathckeae, which belong to the same clade as A. boissieri [27]) remain to be elucidated.
Significant phylogenetic signal was also obtained for growth of Rosenbergiella in all artificial nectars at the isolate level and for growth in the SNS nectar at the ST level, even when no clear phenotypic differentiation was found between the representatives of this genus in the phylogenetic heatmaps generated from the concatenation of housekeeping gene sequences or the UBCG2 set. In contrast, no significant phylogenetic signal was detected for any trait in the species-level analysis. Moreover, the growth data obtained for Rosenbergiella STs and Acinetobacter and Rosenbergiella species in the sNS, sNM, and SNH nectars best fit the white noise model of trait evolution, which assumes that trait values follow a random normal distribution and species have no significant trait covariance [65]. Altogether, these results suggest that the ability to grow in floral nectar is highly conserved between closely related isolates and genotypes of Acinetobacter and Rosenbergiella, but this pattern vanishes deeper in the phylogeny (i.e., between congeneric species or between the genera Acinetobacter and Rosenbergiella). It remains unclear how this hypothesis fits within the framework for predicting the phylogenetic conservatism of functional traits proposed by Martini et al. [87], by which the dispersion and the depth of clades that contain a given trait are correlated with its complexity, so that complex traits encoded by many genes are highly phylogenetically conserved and found in a few deep clades, whereas simpler traits (e.g., the ability to use simple nutrients) are phylogenetically dispersed and found in small clades.
Habitat differences are often linked to trait variation, as different environmental conditions can select for different phenotypes. For example, Pozo et al. [13] found that nectar-inhabiting isolates of the yeasts Metschnikowia reukaufii and Metschnikowia gruessii that originated from a same plant host (at the family and/or species level) tended to show more similar overall growth profiles in media containing different nutrient sources or growth inhibitors than did isolates from other hosts. Moreover, we recently reported that, in general, Acinetobacter isolates of bee origin showed higher growth than nectar isolates in media containing as the only nitrogen source L-cysteine or L-tryptophan [32]. Given the different microenvironmental conditions of floral nectar and the digestive tract of bees (e.g., sugar-dominated, nitrogen-poor chemical composition vs. complex chemical composition, determined by the presence of dietary and excretion waste, respectively), it may be expected that bacterial isolates from these habitats display some phenotypic differences. However, as our collection of isolates was biased towards isolates from a limited selection of plant families and bee species collected in a few countries, the habitat and geographic origin of isolates were not taken into account in our data analysis. A detailed characterization of additional isolates from different animal visitors of flowers (including different insect orders and non-insect vectors of microbes) and more diverse plant hosts (e.g., including plant species from tropical regions, which seem to be important reservoirs of nectar microbes [88]) is required for a fair habitat-based comparison of bacterial phenotypes.
Previous research has revealed that trait variation in Metschnikowia yeasts is to some extent determined by the total sugar concentration and the relative fructose content in the floral nectar of the host plant [12]. In the present study, we did not analyze the chemical composition of the nectar samples from which the studied isolates were obtained and, therefore, we could not determine the impact of the host’s nectar chemistry on the growth performance of Acinetobacter and Rosenbergiella. Moreover, the floral nectar of many plant species contains antimicrobial proteins and secondary compounds putatively protecting nectar from microbial invasion [12, 89–93], a factor which was not considered in the preparation of the artificial nectars used in this study. However, the chemical composition of floral nectar varies widely at the interspecies and intraspecies level (and even between flowers of a same plant or the nectaries of a same flower), and can depend on climatic conditions [15, 94, 95]. Additionally, nectar microbes can modify nectar chemistry, either by consuming sugars or other nutrients, by modifying its pH, by releasing diverse metabolites, and/or by reducing the concentration of some toxins of plant origin [93, 96, 97]. Therefore, determining if trait variation in nectar microbes is linked to nectar chemistry might be challenging. Finally, for the sake of simplification, the growth experiments carried out in this study included cells of a single isolate of Acinetobacter or Rosenbergiella. However, different genotypes and/or species of these genera frequently co-occur in floral nectar, and they can also coexist with other bacteria and yeasts [18, 26]. Therefore, our experiments did not account for possible mechanisms of microbe-microbe interaction resulting in growth facilitation or inhibition that may take place in nature [9]. In any case, to our knowledge, this is the first study demonstrating that phylogenetic factors determine the ability of nectar bacteria to grow in artificial media mimicking the conditions of osmotic pressure and nitrogen limitation found in natural nectars. As many relevant traits of nectar bacteria, such as their ability to alter nectar’s chemistry and interact with floral visitors (e.g., through the production of volatile organic compounds), depend on their osmotolerance and nitrogen scavenging ability, we consider that bacterial phylogeny should be incorporated as a variable in future studies analyzing the ecological relevance of these microorganisms.
Conclusion
In conclusion, the results of this study demonstrate that elevated sugar concentration and low nitrogen content are major hurdles for the growth of Acinetobacter and Rosenbergiella in nectar. Furthermore, our results reveal that the ability of these microorganisms to grow in different types of nectar is highly conserved across closely related isolates and genotypes (STs), but this conservatism rapidly vanishes deeper in phylogeny. The importance of other factors, such as the habitat and geographical origin of isolates, in determining the phenotypic differentiation between isolates, STs, and species of nectar- and/or bee-associated bacteria remains to be evaluated in future studies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Prof. Tadashi Fukami (Stanford University), Dr. Rachel Vannette (UC Davis), and Dr. Megan Morris (Lawrence Livermore National Lab) for providing some of the strains used in this study and useful feedback on previous drafts of this manuscript, Christel Verreth for providing excellent technical assistance, and the US Department of the Interior (National Park Service) and Rhonda Loh for the scientific research and collecting permit HAVO-2013-SCI-0003. The constructive comments from the journal’s editor and four anonymous reviewers are gratefully acknowledged.
Author Contribution
Conceptualization and resources: all authors. Investigation, formal analysis, and data curation: JRM-P, SA-P. Writing — original draft preparation: JRM-P, SA-P. Writing — review and editing: all authors. Supervision: SA-P, BL. Funding: SA-P, BL, CdV, RRJ.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This work was supported by funding from the European Union’s Horizon 2020 research and innovation program (to SA-P and BL; Marie Skłodowska-Curie Grant Agreement No. 742964), the Spanish Ministry of Science and Innovation (to SA-P; “Ramón y Cajal” contract, RYC2018-023847-I), Fundación BBVA (to CdV; Red Leonardo, I Convocatoria de Ayudas a Investigadores, Innovadores y Creadores Culturales), and the Deutsche Forschungsgemeinschaft (to RRJ; DFG, JU2856/2–2).
Data Availability
All relevant results are included in this paper or available as supplementary materials at the journal’s website. The sequence data used in this study is available at the GenBank/ENA/DDBJ databases under the accession numbers shown in the main text, supplementary methods, and/or Tables S1 and S2.
Declarations
Disclaimer
The funders had no role in the preparation of the manuscript or in the decision to publish.
Ethics Approval
Ethics approval was not required. All bacterial isolates used in this study were collected following local regulations and, when required, the specifications of the Nagoya Protocol on Access and Benefit Sharing (ABS).
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
References
- 1.Atlas RM, Bartha R. Microbial ecology: fundamentals and applications. 4. Menlo Park: Benjamin-Cummings Publishing Company; 1998. [Google Scholar]
- 2.Herrera CM, Pozo MI, Bazaga P. Clonality, genetic diversity and support for the diversifying selection hypothesis in natural populations of a flower-living yeast. Mol Ecol. 2011;20(21):4395–4407. doi: 10.1111/j.1365-294X.2011.05217.x. [DOI] [PubMed] [Google Scholar]
- 3.Read DS, Woodcock DJ, Strachan NJ, Forbes KJ, Colles FM, Maiden MC, Clifton-Hadley F, Ridley A, Vidal A, Rodgers J, Whiteley AS, Sheppard SK (2013) Evidence for phenotypic plasticity among multihost Campylobacter jejuni and C. coli lineages, obtained using ribosomal multilocus sequence typing and Raman spectroscopy. Appl Environ Microbiol 79(3):965–973. 10.1128/AEM.02521-12 [DOI] [PMC free article] [PubMed]
- 4.Martiny JB, Jones SE, Lennon JT, Martiny AC (2015) Microbiomes in light of traits: a phylogenetic perspective. Science 350(6261):aac9323. 10.1126/science.aac9323 [DOI] [PubMed]
- 5.Aleklett K, Hart M, Shade A. The microbial ecology of flowers: an emerging frontier in phyllosphere research. Botany. 2014;92(4):253–266. doi: 10.1139/cjb-2013-0166. [DOI] [Google Scholar]
- 6.Fridman S, Izhaki I, Gerchman Y, Halpern M. Bacterial communities in floral nectar. Environ Microbiol Rep. 2012;4(1):97–104. doi: 10.1111/j.1758-2229.2011.00309.x. [DOI] [PubMed] [Google Scholar]
- 7.Herrera CM, de Vega C, Canto A, Pozo MI. Yeasts in floral nectar: a quantitative survey. Ann Bot. 2009;103(9):1415–1423. doi: 10.1093/aob/mcp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pozo MI, Lievens B, Jacquemyn H. Impact of microorganisms on nectar chemistry, pollinator attraction and plant fitness. In: Peck RL, editor. Nectar: production, chemical composition and benefits to animals and plants. New York: Nova Science Publishers Inc.; 2015. pp. 1–40. [Google Scholar]
- 9.Álvarez-Pérez S, Lievens B, Fukami T. Yeast-bacterium interactions: the next frontier in nectar research. Trends Plant Sci. 2019;24(5):393–401. doi: 10.1016/j.tplants.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Herrera CM, Canto A, Pozo MI, Bazaga P. Inhospitable sweetness: nectar filtering of pollinator-borne inocula leads to impoverished, phylogenetically clustered yeast communities. Proc Biol Sci. 2010;277(1682):747–754. doi: 10.1098/rspb.2009.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lievens B, Hallsworth JE, Pozo MI, Ben Belgacem Z, Stevenson A, Willems KA, Jacquemyn H. Microbiology of sugar-rich environments: diversity, ecology and system constraints. Environ Microbiol. 2015;17(2):278–298. doi: 10.1111/1462-2920.12570. [DOI] [PubMed] [Google Scholar]
- 12.Pozo MI, Lachance MA, Herrera CM. Nectar yeasts of two southern Spanish plants: the roles of immigration and physiological traits in community assembly. FEMS Microbiol Ecol. 2012;80(2):281–293. doi: 10.1111/j.1574-6941.2011.01286.x. [DOI] [PubMed] [Google Scholar]
- 13.Pozo MI, Herrera CM, Van den Ende W, Verstrepen K, Lievens B, Jacquemyn H (2015) The impact of nectar chemical features on phenotypic variation in two related nectar yeasts. FEMS Microbiol Ecol 91(6):fiv055. 10.1093/femsec/fiv055 [DOI] [PubMed]
- 14.Herrera CM. Scavengers that fit beneath a microscope lens. Ecology. 2017;98(10):2725–2726. doi: 10.1002/ecy.1874. [DOI] [PubMed] [Google Scholar]
- 15.Nicolson SW, Thornburg RW. Nectar chemistry. In: Nicolson SW, Nepi M, Pacini E, editors. Nectaries and Nectar. Dordrecht: Springer-Verlag; 2007. pp. 215–264. [Google Scholar]
- 16.Palmer-Young EC, Farrell IW, Adler LS, Milano NJ, Egan PA, Junker RR, Irwin RE, Stevenson PC. Chemistry of floral rewards: intra- and interspecific variability of nectar and pollen secondary metabolites across taxa. Ecol Monogr. 2019;89(1):e01335. doi: 10.1002/ecm.1335. [DOI] [Google Scholar]
- 17.Pozo MI, Jacquemyn H (2019) Addition of pollen increases growth of nectar-living yeasts. FEMS Microbiol Lett 366(15):fnz191. 10.1093/femsle/fnz191 [DOI] [PubMed]
- 18.Álvarez-Pérez S, Herrera CM. Composition, richness and nonrandom assembly of culturable bacterial-microfungal communities in floral nectar of Mediterranean plants. FEMS Microbiol Ecol. 2013;83(3):685–699. doi: 10.1111/1574-6941.12027. [DOI] [PubMed] [Google Scholar]
- 19.Álvarez-Pérez S, Herrera CM, de Vega C. Zooming-in on floral nectar: a first exploration of nectar-associated bacteria in wild plant communities. FEMS Microbiol Ecol. 2012;80(3):591–602. doi: 10.1111/j.1574-6941.2012.01329.x. [DOI] [PubMed] [Google Scholar]
- 20.Bartlewicz J, Lievens B, Honnay O, Jacquemyn H. Microbial diversity in the floral nectar of Linaria vulgaris along an urbanization gradient. BMC Ecol. 2016;16:18. doi: 10.1186/s12898-016-0072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Vega C, Álvarez-Pérez S, Albaladejo RG, Steenhuisen S-L, Lachance M-A, Johnson SD, Herrera Carlos M. The role of plant–pollinator interactions in structuring nectar microbial communities. J Ecol. 2021;109(9):3379–3395. doi: 10.1111/1365-2745.13726. [DOI] [Google Scholar]
- 22.Jacquemyn H, Lenaerts M, Brys R, Willems K, Honnay O, Lievens B. Among-population variation in microbial community structure in the floral nectar of the bee-pollinated forest herb Pulmonaria officinalis L. PLoS ONE. 2013;8(3):e56917. doi: 10.1371/journal.pone.0056917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacquemyn H, Lenaerts M, Tyteca D, Lievens B. Microbial diversity in the floral nectar of seven Epipactis (Orchidaceae) species. Microbiologyopen. 2013;2(4):644–658. doi: 10.1002/mbo3.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsuji K, Fukami T. Community-wide consequences of sexual dimorphism: evidence from nectar microbes in dioecious plants. Ecology. 2018;99(11):2476–2484. doi: 10.1002/ecy.2494. [DOI] [PubMed] [Google Scholar]
- 25.Vannette RL, Fukami T. Dispersal enhances beta diversity in nectar microbes. Ecol Lett. 2017;20(7):901–910. doi: 10.1111/ele.12787. [DOI] [PubMed] [Google Scholar]
- 26.Sharaby Y, Rodríguez-Martínez S, Lalzar M, Halpern M, Izhaki I. Geographic partitioning or environmental selection: what governs the global distribution of bacterial communities inhabiting floral nectar? Sci Total Environ. 2020;749:142305. doi: 10.1016/j.scitotenv.2020.142305. [DOI] [PubMed] [Google Scholar]
- 27.Álvarez-Pérez S, Baker LJ, Morris MM, Tsuji K, Sanchez VA, Fukami T, Vannette RL, Lievens B, Hendry TA (2021) Acinetobacter pollinis sp. nov., Acinetobacter baretiae sp. nov. and Acinetobacter rathckeae sp. nov., isolated from floral nectar and honey bees. Int J Syst Evol Microbiol 71(5). 10.1099/ijsem.0.004783. [DOI] [PubMed]
- 28.Bosmans L, Pozo MI, Verreth C, Crauwels S, Wilberts L, Sobhy IS, Wäckers F, Jacquemyn H, Lievens B. Habitat-specific variation in gut microbial communities and pathogen prevalence in bumblebee queens (Bombus terrestris) PLoS ONE. 2018;13(10):e0204612. doi: 10.1371/journal.pone.0204612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samuni-Blank M, Izhaki I, Laviad S, Bar-Massada A, Gerchman Y, Halpern M. The role of abiotic environmental conditions and herbivory in shaping bacterial community composition in floral nectar. PLoS ONE. 2014;9(6):e99107. doi: 10.1371/journal.pone.0099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Álvarez-Pérez S, de Vega C, Vanoirbeek K, Tsuji K, Herrera CM, Jacquemyn H, Junker RR, Fukami T, Michiels C, Lievens B. Phylogenomic analysis of the genus Rosenbergiella and description of Rosenbergiella gaditana sp. nov., and Rosenbergiella metrosideri sp. nov., isolated from floral nectar. Int J Syst Evol Microbiol, submitted. [DOI] [PubMed]
- 31.Álvarez-Pérez S, Lievens B, Jacquemyn H, Herrera CM. Acinetobacter nectaris sp. nov. and Acinetobacter boissieri sp. nov. isolated from floral nectar of wild mediterranean insect-pollinated plants. Int J Syst Evol Microbiol. 2013;63(4):1532–1539. doi: 10.1099/ijs.0.043489-0. [DOI] [PubMed] [Google Scholar]
- 32.Álvarez-Pérez S, Tsuji K, Donald M, Van Assche A, Vannette RL, Herrera CM, Jacquemyn H, Fukami T, Lievens B. Nitrogen assimilation varies among clades of nectar- and insect-associated acinetobacters. Microb Ecol. 2021;81(4):990–1003. doi: 10.1007/s00248-020-01671-x. [DOI] [PubMed] [Google Scholar]
- 33.Halpern M, Fridman S, Atamna-Ismaeel N, Izhaki I. Rosenbergiella nectarea gen. nov., sp. nov., in the family Enterobacteriaceae, isolated from floral nectar. Int J Syst Evol Microbiol. 2013;63(11):4259–4265. doi: 10.1099/ijs.0.052217-0. [DOI] [PubMed] [Google Scholar]
- 34.Kim PS, Shin NR, Kim JY, Yun JH, Hyun DW, Bae JW. Acinetobacter apis sp. nov., isolated from the intestinal tract of a honey bee Apis mellifera. J Microbiol. 2014;52(8):639–645. doi: 10.1007/s12275-014-4078-0. [DOI] [PubMed] [Google Scholar]
- 35.Lenaerts M, Alvarez-Pérez S, de Vega C, Van Assche A, Johnson SD, Willems KA, Herrera CM, Jacquemyn H, Lievens B. Rosenbergiella australoborealis sp. nov., Rosenbergiella collisarenosi sp. nov. and Rosenbergiella epipactidis sp. nov., three novel bacterial species isolated from floral nectar. Syst Appl Microbiol. 2014;37(6):402–411. doi: 10.1016/j.syapm.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Van Assche A, Álvarez-Pérez S, de Breij A, De Brabanter J, Willems KA, Dijkshoorn L, Lievens B. Phylogenetic signal in phenotypic traits related to carbon source assimilation and chemical sensitivity in Acinetobacter species. Appl Microbiol Biotechnol. 2017;101(1):367–379. doi: 10.1007/s00253-016-7866-0. [DOI] [PubMed] [Google Scholar]
- 37.Baker HG. Sugar concentrations in nectars from hummingbird flowers. Biotropica. 1975;7(1):37–41. doi: 10.2307/2989798. [DOI] [Google Scholar]
- 38.Chalcoff VR, Aizen MA, Galetto L. Nectar concentration and composition of 26 species from the temperate forest of South America. Ann Bot. 2006;97(3):413–421. doi: 10.1093/aob/mcj043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willmer P. Pollination and floral ecology. Princeton: Princeton University Press; 2011. [Google Scholar]
- 40.Percival MS. Types of nectar in angiosperms. New Phytol. 1961;60(3):235–281. doi: 10.1111/j.1469-8137.1961.tb06255.x. [DOI] [Google Scholar]
- 41.Baker HG, Baker I. Amino acids in nectar and their evolutionary significance. Nature. 1973;241:543–545. doi: 10.1038/241543b0. [DOI] [Google Scholar]
- 42.Baker HG, Baker I. The occurrence and significance of amino acids in floral nectar. Plant Syst Evol. 1986;151:175–186. doi: 10.1007/BF02430273. [DOI] [Google Scholar]
- 43.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 45.Lefort V, Longueville JE, Gascuel O. SMS: Smart Model Selection in PhyML. Mol Biol Evol. 2017;34(9):2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamura K, Stecher G, Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim J, Na SI, Kim D, Chun J. UBCG2: Up-to-date bacterial core genes and pipeline for phylogenomic analysis. J Microbiol. 2021;59(6):609–615. doi: 10.1007/s12275-021-1231-4. [DOI] [PubMed] [Google Scholar]
- 48.R Core Team (2021) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.r-project.org/. Accessed 23 May 2022.
- 49.RStudio Team (2021) RStudio: Integrated Development Environment for R. RStudio, PBC, Boston, MA. http://www.rstudio.com/. Accessed 23 May 2022.
- 50.Lin L. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255–268. doi: 10.2307/2532051. [DOI] [PubMed] [Google Scholar]
- 51.Lin LIK. A note on the concordance correlation coefficient. Biometrics. 2000;56(1):324–325. [Google Scholar]
- 52.Stevenson M, Sergeant E, Nunes T, Heuer C, Marshall J, Sanchez J, Thornton R, Reiczigel J, Robison-Cox J, Sebastiani P, Solymos P, Yoshida K, Jones G, Pirikahu S, Firestone S, Kyle R, Popp J, Jay M, Reynard C, Cheung A, Singanallur N (2021). epiR: tools for the analysis of epidemiological data. R package version 2.0.33. https://CRAN.R-project.org/package=epiR. Accessed 23 May 2022.
- 53.Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2016. [Google Scholar]
- 54.Adler D, Kelly ST (2021) vioplot: violin plot. R package version 0.3.7. https://github.com/TomKellyGenetics/vioplot. Accessed 23 May 2022
- 55.Kay M, Elkin L, Higgins J, Wobbrock J (2021) ARTool: aligned rank transform for nonparametric factorial ANOVAs. R package version 0.11.1. 10.5281/zenodo.594511. Accessed 1 July 2022
- 56.Wobbrock J, Findlater L, Gergle D, Higgins J (2011) The aligned rank transform for nonparametric factorial analyses using only ANOVA procedures. In: Proceedings of the SIGCHI Conference on Human Factors in Computing Systems. Association for Computing Machinery, New York, pp 143–146. 10.1145/1978942.1978963
- 57.Revell LJ. Phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 58.Felsenstein J. Phylogenies and the comparative method. Am Nat. 1985;125(1):1–15. doi: 10.1086/284325. [DOI] [PubMed] [Google Scholar]
- 59.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35(3):526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 60.Harrell FE, Dupont C, et al. (2021). Hmisc: Harrell miscellaneous. R package version 4.5–0. https://CRAN.R-project.org/package=Hmisc. Accessed 23 May 2022.
- 61.Wei T, Simko V (2021) R package ‘corrplot’: visualization of a correlation matrix (Version 0.90). https://github.com/taiyun/corrplot. Accessed 23 May 2022.
- 62.Pennell MW, Eastman JM, Slater GJ, Brown JW, Uyeda JC, FitzJohn RG, Alfaro ME, Harmon LJ. geiger v2.0: an expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics. 2014;30(15):2216–2218. doi: 10.1093/bioinformatics/btu181. [DOI] [PubMed] [Google Scholar]
- 63.Akaike H. A new look at the statistical model identification. IEEE Trans Autom Control. 1974;19:716–723. doi: 10.1109/TAC.1974.1100705. [DOI] [Google Scholar]
- 64.Álvarez-Pérez S, Dhami MK, Pozo MI, Crauwels S, Verstrepen KJ, Herrera CM, Lievens B, Jacquemyn H. Genetic admixture increases phenotypic diversity in the nectar yeast Metschnikowia reukaufii. Fungal Ecol. 2021;49:101016. doi: 10.1016/j.funeco.2020.101016. [DOI] [Google Scholar]
- 65.Narwani A, Alexandrou MA, Herrin J, Vouaux A, Zhou C, Oakley TH, Cardinale BJ. Common ancestry is a poor predictor of competitive traits in freshwater green algae. PLoS ONE. 2015;10(9):e0137085. doi: 10.1371/journal.pone.0137085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abouheif E (1999) A method for testing the assumption of phylogenetic independence in comparative data. Evol Ecol Res 1:895–909. http://www.evolutionary-ecology.com/abstracts/v01/1152.html. Accessed 23 May 2022.
- 67.Alonso C, Pérez R, Bazaga P, Herrera CM. Global DNA cytosine methylation as an evolving trait: phylogenetic signal and correlated evolution with genome size in angiosperms. Front Genet. 2015;6:4. doi: 10.3389/fgene.2015.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blomberg SP, Garland T, Jr, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 69.Kamilar JM, Cooper N. Phylogenetic signal in primate behaviour, ecology and life history. Phil Trans R Soc B. 2013;368:20120341. doi: 10.1098/rstb.2012.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Molina-Venegas R, Rodríguez MA. Revisiting phylogenetic signal; strong or negligible impacts of polytomies and branch length information? BMC Evol Biol. 2017;17(1):53. doi: 10.1186/s12862-017-0898-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moran PAP. Notes on continuous stochastic phenomena. Biometrika. 1950;37:17–23. doi: 10.2307/2332142. [DOI] [PubMed] [Google Scholar]
- 72.Münkemüller T, Lavergne S, Bzeznik B, Dray S, Jombart T, Schiffers K, Thuiller W. How to measure and test phylogenetic signal. Methods Ecol Evol. 2012;3:743–756. doi: 10.1111/j.2041-210X.2012.00196.x. [DOI] [Google Scholar]
- 73.Pagel M. Inferring the historical patterns of biological evolution. Nature. 1999;401:877–884. doi: 10.1038/44766. [DOI] [PubMed] [Google Scholar]
- 74.Kembel SW, Cowan PD, HelmusMR CornwellWK, Morlon H, Ackerly DD, Blomberg SP, Webb CO. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 75.Jombart T, Balloux F, Dray S. adephylo: new tools for investigating the phylogenetic signal in biological traits. Bioinformatics. 2010;26(15):1907–1909. doi: 10.1093/bioinformatics/btq292. [DOI] [PubMed] [Google Scholar]
- 76.Christensen SM, Munkres I, Vannette RL. Nectar bacteria stimulate pollen germination and bursting to enhance microbial fitness. Curr Biol. 2021;31(19):4373–4380.e6. doi: 10.1016/j.cub.2021.07.016. [DOI] [PubMed] [Google Scholar]
- 77.Corbet SA, Unwin DM, Prys-Jones OE. Humidity, nectar and insect visits to flowers, with special reference to Crataegus, Tilia and Echium. Ecol Entomol. 1979;4(1):9–22. doi: 10.1111/j.1365-2311.1979.tb00557.x. [DOI] [Google Scholar]
- 78.Corbet SA, Willmer PG, Beament JWL, Unwin DM, Prys-Jones OE. Post-secretory determinants of sugar concentration in nectar. Plant Cell Environ. 1979;2(4):293–308. doi: 10.1111/j.1365-3040.1979.tb00084.x. [DOI] [Google Scholar]
- 79.Pusey PL. The role of water in epiphytic colonization and infection of pomaceous flowers by Erwinia amylovora. Phytopathology. 2000;90(12):1352–1357. doi: 10.1094/PHYTO.2000.90.12.1352. [DOI] [PubMed] [Google Scholar]
- 80.von Arx M, Moore A, Davidowitz G, Arnold AE. Diversity and distribution of microbial communities in floral nectar of two night-blooming plants of the Sonoran Desert. PLoS ONE. 2019;14(12):e0225309. doi: 10.1371/journal.pone.0225309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Descamps C, Quinet M, Jacquemart A-L. The effects of drought on plant-pollinator interactions: what to expect? Environ Exp Bot. 2021;182:104297. doi: 10.1016/j.envexpbot.2020.104297. [DOI] [Google Scholar]
- 82.Russell KA, McFrederick QS (2021) Elevated temperature may affect nectar microbes, nectar sugars, and bumble bee foraging preference. Microb Ecol. 10.1007/s00248-021-01881-x [DOI] [PMC free article] [PubMed]
- 83.Fredsgaard C, Moore DB, Al Soudi AF, Crisler JD, Chen F, Clark BC, Schneegurt MA. Relationships between sucretolerance and salinotolerance in bacteria from hypersaline environments and their implications for the exploration of Mars and the icy worlds. Int J Astrobiol. 2017;16(2):156–162. doi: 10.1017/S1473550416000240. [DOI] [Google Scholar]
- 84.Mizzi L, Maniscalco D, Gaspari S, Chatzitzika C, Gatt R, Valdramidis VP. Assessing the individual microbial inhibitory capacity of different sugars against pathogens commonly found in food systems. Lett Appl Microbiol. 2020;71(3):251–258. doi: 10.1111/lam.13306. [DOI] [PubMed] [Google Scholar]
- 85.Abby SS, Tannier E, Gouy M, Daubin V. Lateral gene transfer as a support for the tree of life. Proc Natl Acad Sci USA. 2012;109(13):4962–4967. doi: 10.1073/pnas.1116871109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boucher Y, Douady CJ, Thane Papke R, Walsh DA, Boudreau MR, Nesbø CL, Case RJ, Ford Doolittle W. Lateral gene transfer and the origins of prokaryotic groups. Annul Rev Genet. 2003;37:283–328. doi: 10.1146/annurev.genet.37.050503.084247. [DOI] [PubMed] [Google Scholar]
- 87.Martiny AC, Treseder K, Pusch G. Phylogenetic conservatism of functional traits in microorganisms. ISME J. 2013;7(4):830–838. doi: 10.1038/ismej.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Canto A, Herrera CM, Rodriguez R. Nectar-living yeasts of a tropical host plant community: diversity and effects on community-wide floral nectar traits. PeerJ. 2017;5:e3517. doi: 10.7717/peerj.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aizenberg-Gershtein Y, Izhaki I, Santhanam R, Kumar P, Baldwin IT, Halpern M. Pyridine-type alkaloid composition affects bacterial community composition of floral nectar. Sci Rep. 2015;5:11536. doi: 10.1038/srep11536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carter C, Healy R, O'Tool NM, Naqvi SM, Ren G, Park S, Beattie GA, Horner HT, Thornburg RW. Tobacco nectaries express a novel NADPH oxidase implicated in the defense of floral reproductive tissues against microorganisms. Plant Physiol. 2007;143(1):389–399. doi: 10.1104/pp.106.089326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heil M. Nectar: generation, regulation and ecological functions. Trends Plant Sci. 2011;16(4):191–200. doi: 10.1016/j.tplants.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 92.Park S, Thornburg RW. Biochemistry of nectar proteins. J Plant Biol. 2009;52:27–34. doi: 10.1007/s12374-008-9007-5. [DOI] [Google Scholar]
- 93.Vannette RL, Fukami T. Nectar microbes can reduce secondary metabolites in nectar and alter effects on nectar consumption by pollinators. Ecology. 2016;97(6):1410–1419. doi: 10.1890/15-0858.1. [DOI] [PubMed] [Google Scholar]
- 94.Canto A, Pérez R, Medrano M, Castellanos MC, Herrera CM. Intraplant variation in nectar sugar composition in two Aquilegia species (Ranunculaceae): contrasting patterns under field and greenhouse conditions. Ann Bot. 2007;99(4):653–660. doi: 10.1093/aob/mcl291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Herrera CM, Pérez R, Alonso C. Extreme intraplant variation in nectar sugar composition in an insect-pollinated perennial herb. Am J Bot. 2006;93(4):575–581. doi: 10.3732/ajb.93.4.575. [DOI] [PubMed] [Google Scholar]
- 96.Tucker CM, Fukami T. Environmental variability counteracts priority effects to facilitate species coexistence: evidence from nectar microbes. Proc Biol Sci. 2014;281(1778):20132637. doi: 10.1098/rspb.2013.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vannette RL, Fukami T. Contrasting effects of yeasts and bacteria on floral nectar traits. Ann Bot. 2018;121(7):1343–1349. doi: 10.1093/aob/mcy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant results are included in this paper or available as supplementary materials at the journal’s website. The sequence data used in this study is available at the GenBank/ENA/DDBJ databases under the accession numbers shown in the main text, supplementary methods, and/or Tables S1 and S2.