Abstract
Methanol has recently gained significant attention as a potential carbon substrate for the production of fuels and chemicals, owing to its high degree of reduction, abundance, and low price. Native methylotrophic yeasts and bacteria have been investigated for the production of fuels and chemicals. Alternatively, synthetic methylotrophic strains are also being developed by reconstructing methanol utilization pathways in model microorganisms, such as Escherichia coli. Owing to the complex metabolic pathways, limited availability of genetic tools, and methanol/formaldehyde toxicity, the high-level production of target products for industrial applications are still under development to satisfy commercial feasibility. This article reviews the production of biofuels and chemicals by native and synthetic methylotrophic microorganisms. It also highlights the advantages and limitations of both types of methylotrophs and provides an overview of ways to improve their efficiency for the production of fuels and chemicals from methanol.
Keywords: Native methylotrophs, Synthetic methylotrophs, Pichia pastoris, Methylobacterium extorquens, Bacillus methanolicus, Escherichia coli
1. Introduction
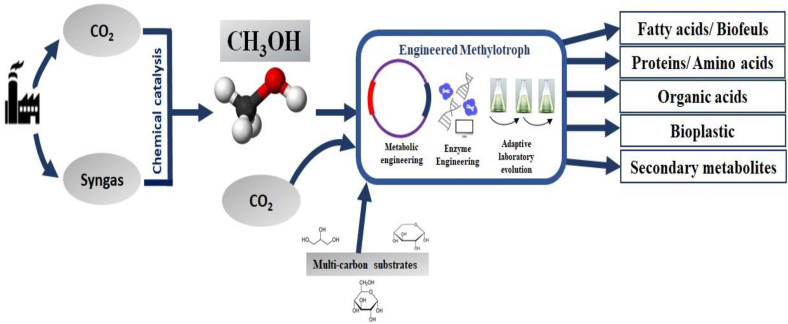
Increasing environmental pollution and the depletion of food resources have inspired industrial biotechnologists to focus on developing environmentally friendly and renewable carbon substrates for producing fuels and chemicals using microbial cell factories [1]. Methanol (CH3OH), a non-food carbon feedstock, is an attractive substrate for microbial production of fuels and chemicals for several reasons [2]. First, it can be readily produced from syngas components using catalytic methods such as the photocatalytic conversion of CO2, electrocatalytic reduction of CO2, hydrogenation of CO or CO2 [3], and catalytic conversion of methane (CH4) [4]. Methanol accounts for 11% of all chemicals derived from syngas [2]. Furthermore, it can be produced from CO/CO2 or methane using anaerobic acetogens or aerobic methanotrophs, respectively [5,6]. Additionally, the degree of reduction for methanol and glucose is 6 and 4, respectively, which makes methanol a higher energy-content compound. The high degree of reduction shows that methanol contains 50% more electrons per carbon atom than glucose and these extra electrons can be used to produce reduced products such as alcohols, fatty acids, and carboxylic acids in larger quantities from methanol using microbial chassis [7]. Furthermore, the price of methanol is comparable to that of glucose, and the annual methanol production capacity for 2021 has increased to 110 million metric tons [8]. Therefore, the development of a methanol-based biorefinery for producing fuels and chemicals is in demand [5].
Although, syngas or C1 gases can be directly used for the microbial production of fuels and chemicals, owing to the low gas-liquid mass transfer rate, the reduced growth of syngas-utilizing microbes and varying/toxic components of syngas limits its commercial application for various target products. In contrast, methanol is completely miscible in water and can overcome the mass transfer barrier, resulting in a higher titer of the target products [6]. Therefore, there is a need to develop an integrated platform involving the chemical production of methanol from syngas and its subsequent fermentation into the desired product using microbial platforms (Fig. 1).
Fig. 1.
Overview of integrated bio-refinery for the production of fuels and chemicals from methanol. CO2 or syngas produced from fossil fuels can be converted to methanol by chemical conversion which can be subsequently utilized by engineered methylotrophs for the production of desired chemicals. In addition, the methylotrophs can be converted to auto-methylotroph or hetero-methylotroph by metabolic engineering strategies to enhance the biomass or productivity of the microorganism.
In nature, native aerobic methylotrophs can convert methanol to formaldehyde by methanol oxidoreductase and subsequently assimilate formaldehyde into cell biomass. Although methanol oxidation can generate more electrons than glucose oxidation, however, the electron transport chain of native methylotrophs favors the oxidation of reduced electron carriers such as NAD(P)H to support cell growth, rather than allowing their intracellular accumulation, thus reducing the availability of such reduced electron carriers for the production of target bioproducts [9]. In addition, the limited genetic tools and toxicity of either methanol or formaldehyde must be overcome to produce fuels and chemicals at the industrial level using native methylotrophs [3]. To achieve methanol-based bio-manufacturing on an industrial scale, the development of microbial platforms that can efficiently convert methanol into valuable chemicals is required. In recent years, synthetic methylotrophs have gained much attention for the conversion of model microorganisms such as E. coli, Corynebacterium glutamicum, and Saccharomyces cerevisiae into methanol-utilizing microbes. Although the growth rates and product yields of synthetic methylotrophs are much lower than those of native methylotrophs, their potential is sufficient to be investigated for the production of fuels and chemicals [10]. In this review, we comprehensively discuss the potential and limitations of various native and synthetic methylotrophs for the production of fuels and chemicals.
2. Methanol as a carbon substrate for native methylotrophs
Methylotrophs are a group of organisms that can utilize C1 compounds such as methane or methanol as carbon and energy sources. Native methylotrophs grow relatively fast on methanol, with a doubling time of approximately 0.3 h−1, and can assimilate methanol at higher rates into cellular biomass (∼16 mmol/g DCW/h) [5]. In nature, methylotrophs include both methylotrophic bacteria and yeasts, which can oxidize methanol to formaldehyde, a toxic intermediate. Formaldehyde is then either converted to cell biomass using various pathways or further oxidized to CO2 to generate energy in the form of NADH [11]. In this section, we review and compare the natural metabolic pathways and enzymes involved in methanol and formaldehyde assimilation in native methylotrophic bacteria and yeasts. We also review metabolic engineering strategies for producing different chemicals using native methylotrophs and discuss the limitations of chemical production and methanol utilization.
2.1. Oxidation of methanol to formaldehyde
In all methanol-utilizing methylotrophs, methanol is first oxidized to formaldehyde, either by methanol dehydrogenases (MDH) in methylotrophic bacteria or by alcohol oxidases (AODs) in methylotrophic yeasts. Depending on the electron acceptor, various types of methanol-oxidizing enzymes have been reported in methylotrophic bacteria and yeasts, including pyrroloquinoline quinone (PQQ)-dependent MDHs or NAD-dependent MDHs in methylotrophic bacteria and O2-dependent AODs in methylotrophic yeasts [11].
MDHs, in gram-negative methylotrophic bacteria, contain a periplasmic PQQ-dependent prosthetic group that captures electrons from methanol oxidation, passes them to cytochrome C, and is thus processed via the electron transport chain to reach the terminal electron acceptor. This electron transfer drives a proton gradient force that is sufficient to produce less than one ATP molecule, at the expense of one oxidized methanol molecule [12]. In the model gram-negative methylotrophic bacteria Methylobacterium extorquens AM1, the MxaF1 MDH is considered the main catalyst for the oxidation of methanol to formaldehyde. However, the oxidation of methanol to formaldehyde involves at least 15 genes, of which 14 genes are co-transcribed to oxidize methanol to formaldehyde [13]. However, the disadvantage of PQQ-dependent MDHs is their dependence on molecular O2; for chemical production, some metabolites must be produced under anaerobic conditions [14].
O2-dependent AODs are found in the peroxisomes of eukaryotic yeasts such as Pichia pastoris and use FAD as a cofactor to regenerate energy. Methanol oxidation by AODs produces formaldehyde and hydrogen peroxide (H2O2) in peroxisomes, which can be further metabolized by the central metabolism. O2-dependent AODs act under aerobic conditions, similar to PQQ-dependent MDHs; however, electron conservation is poor for O2-dependent AODs compared to PQQ-dependent MDHs because AOD electrons are directly transferred to molecular O2 with a high Gibbs free change (Table 1) [11,15].
Table 1.
Comparison of different methanol oxidation enzymes under physiological conditions calculated by Whitaker et al. [9], using the indicated temperature (T oC) for growth.
Nicotinamide adenine dinucleotide (NAD)-dependent MDHs are found in the cytoplasm of gram-positive methylotrophs such as Bacillus methanolicus. NAD-dependent MDH utilizes NAD as a cofactor and electron acceptor, can act under aerobic and anaerobic conditions, and produces NADH in the form of reducing equivalents directly from the electrons obtained via methanol oxidation. Unlike PQQ-dependent MDHs, NAD-dependent MDHs require a single gene to oxidize methanol to formaldehyde with the help of an activator protein [11].
2.2. Assimilation of formaldehyde into cell biomass in native methylotrophs
Formaldehyde (HCOH) can be assimilated into cell biomass using various metabolic pathways or can be dissimilated into CO2. Four distinct formaldehyde assimilation pathways have been identified. The serine, ribulose monophosphate (RuMP), and ribulose bisphosphate (RuBP) pathways have been identified in bacteria, whereas yeasts typically utilize the xylulose monophosphate pathway (XuMP) to assimilate formaldehyde into cell biomass [11]; however, all of these pathways result in the same product (three carbon compounds from C1 formaldehyde), which is further metabolized to carry important cellular functions.
2.2.1. Formaldehyde fixation through the serine cycle
M. extorquens is a well-studied example of a serine pathway in methylotrophic bacteria. Following its formation in the periplasmic space, formaldehyde crosses the cytoplasmic membrane and reacts with C1 carrier molecules such as tetrahydrofolate (H4F) or tetrahydromethanopterin (H4MPT). In the H4F pathway, formaldehyde reacts with H4F and is converted to methylene-H4F, whereas in the H4MPT pathway, formaldehyde is first formed by the reaction of tetrahydromethanopterin enzymes, which are either subsequently converted to methylene-H4F by H4F dependent enzymes or dissipated to CO2 to produce reducing equivalents. It has been suggested that simultaneous condensation reactions occur for both pathways during the serine cycle, which helps to convert the overproduced formaldehyde into non-toxic compounds for cell biomass [16]. The methylene-H4F enters the serine cycle via condensation with glycine. Serine is converted to oxaloacetate by a series of reactions in the serine cycle, which is further converted to malyl-CoA and subsequently cleaved to form acetyl-CoA along with the regeneration of glyoxylate. Acetyl-CoA enters the ethylmalonyl-CoA (EMC) and polyhydroxybutyrate (PHB) pathways and a portion of the tricarboxylic acid cycle (TCA cycle) [17]. During growth on methanol, the expression of genes involved in the EMC pathway in M. extorquens was higher than those grown on multi-carbon substrates. The overall stoichiometric reaction for the production of pyruvate (an important precursor of major metabolites) via formaldehyde in the serine cycle is given by Equation (1) [9].
| CO2 + 2HCHO + 2NADH + 2ATP = pyruvate + 2NAD+ + 2ADP + FPH2 | (Equation 1) |
2.2.2. Formaldehyde fixation via the RuMP pathway
The RuMP pathway, in which three formaldehyde molecules are used to form a 3-carbon compound, can be divided into three parts: formaldehyde fixation, cleavage, and rearrangement. During fixation, formaldehyde is fixed with ribulose-5-phosphate (Ru5P) to form hexulose 6-phosphate (H6P) by 3-hexulose-6-phosphate synthase (HPS), which is subsequently converted to fructose 6-phosphate (F6P) by 6-phospho-3-hexuloisomerase (PHI). Three main metabolic pathways in RuMP-utilizing methylotrophic bacteria, namely Embden–Meyerhof–Parnas (EMP), pentose phosphate (PPP), and Entner–Doudoroff (ED) pathways, are used to utilize F6P. B. methanolicus is a primary example of RuMP-pathway-utilizing bacteria [18]. During cleavage, F6P is either converted to fructose 1, 6-bisphosphatase (FBP) by the FBP aldolase variant and subsequently to glyceraldehyde 3-phosphate (GAP) and dihydroxyacetone phosphate (DHAP), or 2-keto-3-deoxy-6- phosphogluconate (KDGP) by the KDGP aldolase variant and subsequently converted to pyruvate.
The FBP aldolase variant is superior to the KDGP variant as it regenerates one NADPH molecule and two ATPs, whereas the KDGP variant only regenerates one NADPH molecule. The rearrangement of the RuMP pathway is utilized to regenerate Ru5P via xylulose 5-phosphate (Xu5P) and ribose 5-phosphate (R5P). Two variants, transaldolase (TA) and sedoheptulose 1, 7-bisphosphatase (SBPase), were identified in the rearrangement region. The overall stoichiometric reaction of pyruvate formation through the most efficient transaldolase variant is given by Equation (2) [9].
| 3HCHO + NAD+ + ADP = pyruvate + NADH + ATP | (Equation 2) |
2.2.3. Formaldehyde assimilation via the XuMP pathway
The XuMP pathway is shared by methylotrophic yeasts, in which formaldehyde is condensed with Xu5P in the peroxisome by Dihydroxyacetone Synthase (DAS) to form dihydroxyacetone (DHA) and GAP. DHA is further phosphorylated by Dihydroxyacetone Kinase (DAK) to form DHAP, which is subsequently condensed to form FBP. FBP is converted to F6P, which is used for the regeneration of Xu5P and the synthesis of cellular metabolites via the formation of pyruvate through the GAP intermediate. Pyruvate then enters the TCA cycle to produce cellular biomass [11]. The RuMP and XuMP pathways are quite similar in the fixation, cleavage, and rearrangement of formaldehyde; however, they differ in the channeling of formaldehyde to PPP. In the RuMP pathway, formaldehyde reacts with Ru5P and then forms F6P via isomerization. However, in the XuMP pathway, formaldehyde reacts with the glycolaldehyde group of Xu5P to produce DHA and GAP which are then converted to F6P through a series of reactions [19]. The overall stoichiometry for pyruvate formation through the XuMP pathway is given by Equation (3) [11].
| 3HCHO + NAD+ + 2ADP = Pyruvate + NADH +2ATP | (Equation 3) |
2.2.4. Formaldehyde assimilation via the RuBP pathway
Some bacteria, such as Beijerinckia mobilis, contain another pathway for the fixation of formaldehyde into cell biomass, in which formaldehyde is first converted to CO2 by two formaldehyde dehydrogenases, NAD(P)-dependent glutathione (GSH) formaldehyde dehydrogenase, and phenazine methosulfate (PMS). B. mobilis also contains formate dehydrogenase, which indicates the formation of CO2 via formate. The presence of ribulose-1, 5-bisphosphate carboxylase (Rubisco) in B. mobilis confirms that the CO2 formed from formaldehyde is fixed in the cell biomass, making this strain a chemoautotroph. CO2 enters the RuBP cycle by reacting with Ribulose 1, 5-bisphosphate (RuBP). Similar to the RuMP pathway, the Ru5P pathway uses transketolase and transaldolase rearrangements to convert G3P into Xu5P to regenerate RuBP [20]. The stoichiometric equation for pyruvate formation from formaldehyde is given in Equation (4).
| 3HCHO + 7ATP + NAD+ = pyruvate + NADH + 7ADP | (Equation 4) |
2.2.5. Formaldehyde dissimilation to CO2
Formaldehyde dissimilation into CO2 is crucial for generating energy and for avoiding the accumulation of formaldehyde at toxic levels. Dissimilation proceeds via the conversion of formaldehyde to formate followed by several steps to CO2. Cofactors involved in the dissimilation of formaldehyde into CO2 include H4F, GSH, H4MPT, and mycothiol (MSH). In the aerobic methylotrophs H4F and H4MPT, both pathways are necessary to sustain microbial growth in methanol. The simultaneous activation of the H4F and H4MPT pathways plays two key roles: (1) detoxification of formaldehyde and (2) generation of energy to drive methanol oxidation [9,11]. It has been reported that in M. extorquens, the formation of CO2 begins before the assimilation pathway. This indicates that the carbon flux goes only to the assimilation branch when the dissimilation branch increases to the poised level. Such a strategy is important for preventing the accumulation of toxic molecules such as formaldehyde, glycine, and glyoxylate [16].
2.3. Biotechnological applications of native methylotrophs
The microbial production of chemicals usually depends on the amount of carbon flux towards the target products, which can theoretically be improved via metabolic engineering and culture conditions. The yield and productivity of microorganisms for the production of chemicals can be evaluated based on biomass production and their ability to utilize carbon substrates for biomass production. A basic stoichiometric equation was developed for the conversion of methanol to biomass as given in Equation (5) [21].
| 1.63CH3OH + 1.39O2 + 0.23NH3 + nutrients = CH1.69O0.38N0.24 (biomass) + 0.63CO2 + 2.76H2O+733 kJ | (Equation 5) |
Owing to their complex metabolism, native methylotrophs can be used for the production of various chemicals ranging from organic acids to proteins and alcohols, with the help of metabolic engineering (Fig. 2, Fig. 3). Natural products from native methylotrophic bacteria include poly (3-hydroxybutyrate) (PHB) and amino acids, whereas methylotrophic yeasts are the most widely used for recombinant protein production [6]. In this section, different chemicals produced by native or engineered methylotrophs are discussed in detail, focusing on the limitations of methanol utilization for chemical production.
Fig. 2.
Production of fuels and chemicals from methylotrophic yeasts. The native key enzymes of methanol assimilation in methylotrophic yeasts are shown in purple. The overexpressed endogenous and heterologously expressed key enzymes for the target product are shown in red and blue, respectively. The solid arrow shows a single reaction while the dashed arrow shows multiple reactions. Enzyme abbreviations:AOX, alcohol oxidase; FDH, Formate dehydrogenase; DAS, Dihydroxyacetone synthase; DAK, dihydroxyacetone kinase; FBA, Fructose-1,6-bisphosphate aldolase; FBPase, Fructose 1,6-bisphosphatase; CrtB, phytoene synthase; CrtI, phytoene desaturase; ValS, valencene synthase; ADH, alcohol dehydrogenase; ERG9, squalene synthase; ERG1, squalene epoxidase; PgDDS, dammarenediol synthase gene; FAS, Fatty acid synthase; TesB, thioesteras; ScADH5, alcohol dehydrogenase; CpFAH, Fatty acid hydroxylase; DGAT, acyl-CoA: diacylglycerol acyltransferase; npgA, phosphopantetheinyl transferase; 6-MSAS, 6-methylsalicylic acid synthase, citABCDE, gene cluster for citrinin synthesis; undB, desaturase-like decarboxylase; D-LDH, d-lactate dehydrogenase; ASPDH, Aspartate dehydrogenase; ADC, l-Aspartate-α-decarboxylases. Metabolite abbreviations: CO2, Carbon dioxide; DHA, Dihydroxyacetone; GAP, glyceraldehyde 3-phosphate; DHAP, Dihydroxyacetone phosphate, F1,6BP, Fructose 1,6 bisphosphate; Xu5P, Xylulose 5-phosphate; GAP, Glyceraldehyde 3-phosphate.
Fig. 3.
Production of fuels and chemicals from native methylotrophic bacteria. Methylotrophic bacteria can either use the RuMP pathway or the serine cycle for methanol assimilation. Methylotrophic bacteria utilizing the serine cycle contain the EMC pathway and TCA cycle for biomass generation while methylotrophic bacteria utilizing the RuMP pathway mainly use the TCA cycle for biomass generation. The RuMP pathway is shown in blue while the serine cycle is shown in purple. The native key enzymes of methanol assimilation in methylotrophic bacteria are shown in purple. The overexpressed endogenous and heterologously expressed key enzymes for the target product are shown in red and blue, respectively. The solid arrow shows a single reaction while the dashed arrow shows multiple reactions. Enzyme abbreviations:PQQ-MDH, PQQ-dependent methanol dehydrogenase; NAD-MDH, NAD-dependent methanol dehydrogenase; HPS, 3-hexulose-6-phosphate synthase; PHI, 6-phospho-3-hexuloisomerase; PFK, 6-phosphofructokinase; FBA, fructose-bisphosphate aldolase; FDH, Formate dehydrogenase; RPE, Ribulose-5-phosphate 3-epimerase; RPI, Ribose-5-phosphate isomerase; SHMT, Serine hydroxymethyltransferase; Mtd, Methylene-tetrahydromethanopterin; lysA, mesodiaminopimelate decarboxylase; cadA, lysine decarboxylase; patA, putrescine aminase; patD, 5-aminopentanal dehydrogenase; DavB, lysine 2-monooxygenase; DavA, 5-aminovaleramidase; HDI, homoserine dehydrogenase; HDII, homoserine dehydrogenase; thrC, threonine synthase; cad, cis-aconitic acid decarboxylase; gabA, glutamate decarboxylase; phaCAC, PHA synthase from A. caviae; phaC, PHA synthase; alsSD, acetolactate synthase; budAB, acetolactate decarboxylase; mcr, malonyl-CoA reductase; Ter, trans-2-enoyl-CoA reductase; Adh2, bifunctional aldehyde/alcohol dehydrogenase; RCM, (R)-3-hydroxybutyryl coenzyme A (CoA)-specific coenzyme B12-dependent mutases; ccr, crotonyl-CoA carboxylase; zssl, α-humulene synthase; vioABCD, gene cluster for violacein synthesis. Metabolite abbreviations: Ru5P, Ribulose 5-Phosphate; CH2 H4F, N5,N10-methylenetetrahydromethanopterin; H4MPT, Tetrahydromethanopterin; H4F, tetrahydrofolate; PEP, Phosphoenolpyruvate; H6P, Hexulose-6-phosphate; F6P, Fructose-6-phosphate; FBP, Fructose 1,6-bisphophate; DHAP, Dihydroxyacetone phosphate; GAP, Glyceraldehyde 3-phosphate; Xu5P, Xylulose 5-phosphate; E4P, Erythrose 4-phosphate; S7P, sedoheptulose-7-phosphate; R5P, ribose-5-phosphate; Ru5P, Ribulose-5-phosphate; CO2, Carbon dioxide; 3-HP, 3- Hydroxy propionic acid; PHB, Poly (3-Hydroxybutyrate); 2-HIBA, 2-Hydroxyisobutyric acid; FPP, Farnesyl diphosphate; P (3HB-co-3HV-co-3HHX), Poly (3-hydroxybutyrate-co-3-hydroxyvalerate-co-3-hydroxyhexanoate); GABA, γ-aminobutyric acid.
2.3.1. Production of fuels and chemicals from methylotrophic yeasts
Yeast has several advantages as a production host, such as a high tolerance to toxic solvents, pH, and temperature changes. In addition, the organelle-based yeast structures provide options for targeted metabolic engineering to enhance the desired carbon flux. In methylotrophic yeasts, enzymes involved in methanol metabolism are localized in peroxisomes which provide better tolerance to methanol or formaldehyde [19]. Furthermore, methylotrophic yeasts can utilize a wide range of sugar substrates along with methanol, which can increase biomass and product titer. Due to these characteristics, methylotrophic yeasts such as P. pastoris and Hansenula polymorpha (H. polymorpha) have been used for a wide range of products (Table 2).
Table 2.
Production of fuels and chemicals using native methylotrophic yeasts.
| Product | Strain | Metabolic engineering strategy | Substrate | Culture condition | Product titer/yield | Ref. |
|---|---|---|---|---|---|---|
| Free fatty acids (FFA) | P. pastoris | Blocking FFA consumption (ΔFAA1, ΔFAA2, and ΔPOX1), enhancing acetyl-CoA (Overexpression of mmACL, bbXFPK, ckPTA) and NADPH (overexpression scIDP2), improving methanol assimilation (overexpression of DAS2) | Methanol | Fed-batch using methanol as the sole carbon source in minimal media (MM), methanol concentration was kept below 20 g/l | 23 g/l | [30] |
| Fatty alcohol | P. pastoris | Heterologous expression of mmCAR, npgA, scADH5 Overexpression of FaCoAR in FFA overproducing strain |
Methanol | Fed-batch using methanol as the sole carbon source in MM, methanol concentration was kept below 20 g/l | 2 g/l | [30] |
| FFA | O. polymorpha | ΔFAA | Methanol | Shake flask using 10 g/l methanol in MM | Growth defects | [31] |
| Methanol, xylose, and glucose | Shake flask using MM with 20 g/l glucose, 10 g/l xylose and 10 g/l methanol | 1.2 g/l | ||||
| FFA | O. polymorpha | ΔFAA; enhancing methanol assimilation (overexpression of FBP1and RPE); enhancing acetyl-coA (overexpression of mmACL) enhancing NADPH (overexpression of ZWF1and ScIDP2); ALE-derived activation of LPL1and IZH3 | Methanol | Fed-batch fermenter with initial culture using 10 g/l methanol in MM, continuous feeding of methanol after 24 h | 15.9 g/l | [32] |
| Methanol and glucose | Fed-batch fermenter with initial culture using 20 g/l glucose in MM followed by continuous feeding of methanol after 15 h | 9.1 g/l | ||||
| Methanol and glycerol | Fed-batch fermenter with initial culture using 20 g/l glycerol in MM followed by continuous feeding of methanol after 24 h | 13.2 g/l | ||||
| Ricinoleic acid | P. pastoris | Δ12 desaturase, Heterologous expression of cpFAH and cpDGAT1 | Methanol andglycerol | Fed-batch fermenter, Initial growth on glycerol followed by methanol induction and addition of 0.5% methanol every 24 h |
495 μg/ml | [33] |
| γ- Linoleic acid (GLA) | P. pastoris | Heterologous expression of Δ6-Desaturase under methanol oxidation promoter (MOX) | Methanol | Fed-batch fermenter using 2% methanol in MM | 11.9% GLA of Total fatty acids | [35] |
| 6-methylsalicylic acid | P. pastoris | Heterologous expression of npgA, 6-MSAS | Methanol and glycerol | Fed-batch fermenter with initial growth on glycerol in MM followed by methanol induction for 20 h | 2.2 g/l | [27] |
| Citrinin | P. pastoris | Heterologous expression of pksCT, npgA; mpl6; mpl7 overexpression of mpl1, mpl2 and mpl4 | Methanol | Fed-batch fermenter with 0.5% methanol added after every 24 h | 0.6 mg/l | [28] |
| D-lactic acid | P. pastoris | Heterologous expression of d-LDH | Methanol | Test tube fermentation using methanol as the sole carbon source in YPM (Yeast extract-peptone-Mannitol medium) for 96 h | 3.48 g/l | [38] |
| β-alanine | P. pastoris | Heterologous expression of multiple copies of ADCs; overexpression of aspartate dehydrogenase | Methanol and glycerol | Fed-batch with two-stage fermentation using 40 g/l of glycerol for the growth phase followed by the production phase maintaining 3 g/l of methanol | 5.6 g/l | [39] |
| α-alkenes | P. pastoris |
ΔFAA; Heterologous expression of UndB |
Methanol | Shake flask containing 20 g/l methanol in MM | 1.6 mg/l | [36] |
| Monacolin J | P. pastoris | Expression of monacolin J and lovastatin biosynthesis pathway; distribution of the pathway in two strains to use as co-cultures | Methanol and glycerol | Fed-batch with initial growth using glycerol followed by methanol induction | 593.9 mg/l | [29] |
| Lovastatin | P. pastoris | Expression of lovastatin biosynthesis pathway; distribution of the pathway in two strains to use as co-cultures | Methanol and glycerol | Fed-batch with initial growth using glycerol followed by methanol induction | 250.8 mg/l | [29] |
| Malic acid | P. pastoris | Introduction of malic acid producing genes | Methanol | Fed-batch fermentation using methanol and yeast extract in MM | 2.79 g/l | [41] |
| Glutathione | H. polymorpha DL-1 | Overexpression of GSH2 and MET4 | Methanol | Fed-Batch using methanol in MM | 250 mg/l | [134] |
| Lycopene | P. pastoris | Heterologous expression of genes encoding crtB, crtE, and crtI; Overexpression of HMG1 |
Glycerol and methanol | Fed-batch fermenter using two-stage cultivation with initial growth on glycerol in BSM (Basal salt medium) followed by methanol induction | 714 mg/l | [24] |
| Nootkatone | P. pastoris | Heterologous expression of HPO, CPR, and valence synthase; Overexpression of ADH, HMG1 |
Glucose and methanol | Fed-batch fermenter, biphasic cultivation using glucose and methanol in BSM, the addition of dodecane overlay to prevent evaporation | 208 mg/l | [25] |
| Dammarenediol-II | P. pastoris | Heterologous expression of PgDDS; overexpression of ERG1; repression of ERG7 | Methanol and squalene | BMMY media with 0.5% methanol supplementation every 24 h, Exogenous addition of squalene | 1.073 mg/g DCW | [26] |
2.3.1.1. Methylotrophic yeasts for secondary metabolite production
Yeast-derived secondary metabolites represent a vast variety of natural products with applications in nutraceuticals, pharmaceuticals, and agrochemicals. Among the secondary metabolites, isoprenoids, especially terpenoids and polyketides, have gained considerable attention for their production by methylotrophic yeasts. In yeast, isoprenoids are produced via the mevalonate pathway (MVA), which uses acetyl-CoA as a precursor. As acetyl-CoA is an essential precursor of many cellular metabolites involved in biomass production, extensive metabolic engineering to increase the flux towards the MVA pathway and cofactor regeneration is required to produce secondary metabolites from yeasts [19].
The production of terpenoids in P. pastoris proceeds via farnesyl diphosphate (FPP), the central metabolite in the production of terpenoids. In P. pastoris, FPP is produced in peroxisomes, which contain enzymes crucial for methanol and formaldehyde oxidation. Additionally, it has been shown that peroxisomes of P. pastoris can proliferate upon the addition of methanol, which can in turn increase the storage space and pool of intermediates for terpenoid production. These properties make P. pastoris a suitable host for the production of peroxisomal terpenoids [22]. In the first case of terpenoid production, only 1.96 mg/L of lycopene was produced from methanol in engineered P. pastoris via the lycopene biosynthesis pathway. A comparison of the different carbon sources showed that glucose was the most favorable for the production of lycopene from P. pastoris. Using glucose in minimal media along with methanol, 4.6 mg lycopene/g DCW was obtained during batch fermentation [23]. Later, lycopene production was enhanced to 9.319 mg/g DCW by increasing the flux of geranylgeranyl diphosphate (GGPP) from FPP and using optimized culture conditions with glucose and methanol as co-substrates [24]. Wriessnegger et al. showed that the co-expression of premnaspirodiene oxygenase, cytochrome P450 reductase, and valence synthase allows (+)-valence to be produced in glucose/methanol biphasic fermentation from P. pastoris, which can then be converted to trans-nootkatol and subsequently oxidized to (+)-nootkatone by intrinsic alcohol dehydrogenase. Additionally, enhancing the expression of hydroxyl-methylglutaryl-CoA reductase and alcohol dehydrogenase produced 208 mg/L of (+)-nootkatone during biphasic cultivation with glucose and methanol as substrates [25]. To produce fuels and chemicals, downregulation and upregulation of competitive pathways and metabolite precursors, respectively, are necessary to obtain a high target production titer. Liu et al. enhanced the dammarenediol-II titer to 0.736 mg/g DCW by reducing the competitive consumption and increasing the supply of 2,3-oxidosqualene [26]. Polyketide production from methylotrophic yeasts has also been demonstrated through heterologous expression of the desired pathways. For example, 6-methylsalicylic acid was produced at a final titer of 2.2 g/L in P. pastoris [27]. Similarly, complex polyketides such as citrinin were produced by expression of the citrinin synthesis pathway, and it was shown that P. pastoris can be used as a model organism for the synthesis of complex polyketides [28].
One strategy to enhance product formation is to split the pathway into two strains and then use a co-culture system to produce the desired metabolite. Such a strategy could reduce the metabolic burden in a single-strain process, which could be caused by different metabolites during the growth and production phases. Liu et al. used this strategy to produce monacolin J and lovastatin by splitting the upstream and downstream modules into different cells of P. pastoris. Using P. pastoris co-cultures, the amounts of monacolin J and lovastatin increased by 55% and 71%, respectively, compared with a single strain containing the complete pathway. In the fed-batch fermenter, 593.9 and 250.8 mg/L of monacolin J and lovastatin, respectively, were produced by the co-cultures system using methanol as a sole carbon substrate [29].
2.3.1.2. Production of fatty acids and fatty acid-derived products from methylotrophic yeasts
Fatty acids are essential cellular components that can be used for the production of biofuels and chemicals. The main hindrance to the production of fatty acids or fatty acid-derived products from methanol is the toxicity of formaldehyde accumulated during growth in methanol. Additionally, fatty acids production requires a large supply of NADPH as an energy source. A comparison of P. pastoris and S. cerevisiae showed that NADPH/NADP+ levels were much higher in P. pastoris than in S. cerevisiae, making it an ideal host for the production of free fatty acids (FFA) and fatty acid derivatives [30].
Cai et al. showed that fatty acids can be overproduced from P. pastoris (up to 23.4 g/L) by increasing the flux to acetyl-CoA, diverting the flux towards fatty acid synthesis, and decreasing the accumulation of toxic formaldehyde. As a result, 23.4 g/L of FFA was produced from methanol in fed-batch fermentation using P. pastoris with a yield of 0.078 g FFA/g methanol. The FFA-overproducing strain was further engineered to produce fatty alcohols; however, the titer of fatty alcohols was lower (2 g/L of methanol) than that of FFA. The lower production of fatty alcohols was attributed to the excretion of FFA, as approximately 41% of FFA was transported outside the cells and recovered from the aqueous phase [30]. Similarly, disruption of the fatty acyl-CoA synthase gene (faa1) in O. polymorpha can produce 674 mg/L of FFA from glucose as the sole carbon source. However, this engineered strain was unable to grow on methanol as the sole carbon source, and the co-substrate of xylose and methanol enhanced methanol utilization, with a final titer of 1.2 g/L of FFA in fed-batch fermentation [31]. Later, it was found that growth inhibition in methanol for Δfaa1 in O. polymorpha was associated with the enhanced toxicity of methanol and formaldehyde. By rescuing O. polymorpha from formaldehyde toxicity, the overexpressing PPP and gluconeogenesis pathway produced 15.5 g/L of FFA from methanol as a carbon source in two-stage fermentation with glycerol using O. polymorpha [32].
Methylotrophic yeasts have been widely used to produce fatty acid derivatives. Co-expression of genes is a useful strategy for expressing multiple genes involved in chemical biosynthesis. Ricinoleic acid is a long-chain hydroxyl fatty acid that has applications in the manufacturing industry. Heterologous co-expression of fatty acid hydroxylase and diacylglycerol acyl transferase has been used to produce 495 μg/mL of ricinoleic acid in P. pastoris using methanol induction in two-stage fermentation [33]. Additionally, the co-expression of enzymes involved in the elongation of polyunsaturated fatty acids, such as Δ6-desaturase and Δ5-desaturase, produced C22 polyunsaturated fatty acids in P. pastoris [34]. Similarly, 697 mg/L γ-linoleic acid was produced under optimized fermentation conditions using H. polymorpha [35]. Long-chain α-alkenes are important chemicals that can be used in the production of lubricants and surfactants. In P. pastoris, the decarboxylation of fatty acids by the expression of UndB from Pseudomonas putida produced 1.6 mg/L of α-alkenes using methanol as the sole carbon substrate [36]. However, the titer of α-alkenes was lower than that of other fatty acid-derived products, probably because of the low specificity of the decarboxylase enzyme UndB [36].
2.3.1.3. Methylotrophic yeasts for pyruvate and TCA-derived products
To produce pyruvate or TCA cycle-derived products in methylotrophic yeasts, the expression of heterologous genes should be sufficiently high to compete with the natural pathways involved in biomass formation. Additionally, push and pull-up strategies must be employed to drive the flux toward the target product. However, the production of chemicals in methylotrophic yeasts usually suffers from a low copy number of heterologously expressed genes because the integration of these genes is usually employed to obtain the target recombinant strain [37]. By integrating the d-LDH encoding d-lactate dehydrogenase gene at the ribosomal DNA locus, the expression of d-LDH in P. pastoris was enhanced to produce d-lactic acid from pyruvate. Additionally, by adjusting the copy number of d-LDH and culture optimization for the recombinant strain produced 3.48 g/L d-lactic acid was produced using methanol as the carbon substrate in 96 h [38]. Similarly, P. pastoris was engineered to produce β-alanine via the expression of l-aspartate-α-decarboxylases (ADCs) from B. subtilis, and its copy number was optimized to enhance the expression of ADCs. Additionally, enhancing the precursor supply from aspartate produced 5.6 g/L β-alanine using methanol as the carbon substrate [39]. Similarly, Zhang et al. used multicopy integration to obtain a high expression of genes encoding pyruvate decarboxylase, fumarase, and malate dehydrogenase in P. pastoris. Using glycerol as the initial substrate and adding 0.5% methanol every 12 h produced 0.47, 9.42, and 42.28 g/L of Fumaric, succinic, and malic acid, respectively [40]. Later, Guo et al. showed that redistributing the flux towards malic acid and blocking the key module of the XuMP pathway in P. pastoris can produce 2.79 g/L of malic acid when methanol is the sole carbon substrate [41,42].
2.3.2. Production of fuels and chemicals using methylotrophic bacteria
Methylotrophic bacteria usually have a higher growth rate on methanol as the sole carbon source than methylotrophic yeasts. Many products can be produced through the metabolic engineering of different methylotrophic bacteria by targeting the main formaldehyde assimilation pathways (serine and RuMP pathways). Additionally, the downstream pathways for formaldehyde assimilation vary among different methylotrophic bacteria, which enhances the product range compared with methylotrophic yeasts. However, extensive metabolic engineering of methylotrophic bacteria is required to produce high titers of the target products (Table 3). In this section, we review metabolic engineering strategies for the production of valuable chemicals using natural methylotrophic bacteria.
Table 3.
Production of fuels and chemicals using native or synthetic methylotrophic bacteria.
| Product |
Strain |
Methanol assimilation pathway |
Metabolic engineering strategy |
Substrate |
Culture condition |
Product titer/yield |
Ref. |
|---|---|---|---|---|---|---|---|
| Native methylotrophic bacteria | |||||||
| L-glutamate | B. methanolicus | RuMP | Δhom-1 | Methanol | Fed-batch fermentation with automatic control for methanol feeding | 32 g/l | [42] |
| Methanol and methionine | Fed-batch fermentation with automatic control for methanol along with methionine feeding | 69 g/l | |||||
| L-lysine | B. methanolicus | RuMP | NOA2 mutant strain | Methanol and amino acids | Fed-batch fermentation with automatic control for methanol supply; supplementation of threonine, lysine, and methionine | 65 g/l | [42] |
| Cadaverine | B. methanolicus | RuMP | Heterologous expression of cadA and ldcC; overexpression of l-lysine synthesis pathway | Methanol | Fed-batch fermentation using methanol as the sole carbon source | 11.3 g/l | [44] |
| 5-AVA | B. methanolicus | RuMP | Expression of 5-AVA biosynthesis pathway from cadaverine and l-lysine | Methanol | Shake flask cultures using methanol as the sole carbon source | 23.7 mg/l | [45] |
| Methanol andcadaverine | Shake flask cultures using methanol along with supplementation of cadaverine | 77 mg/l | |||||
| GABA | B. methanolicus | RuMP | Heterologous expression of gadSt and gadB | Methanol | Fed-batch fermentation employing a pH shift from 6.5 to 4.6 using methanol as the sole carbon source | 9 g/l | [46] |
| Acetoin | B. methanolicus | RuMP | Heterologous expression of alsSD operon, overexpression of malic enzyme, and isocitrate lyase | Methanol | Shake flask using methanol as the carbon source | 0.42 g/l | [57] |
| 2-HIBA | M. extorquens AM1 | Serine cycle | Expression of B12-dependent mutases | Methanol | Fed-batch fermentation using methanol as the sole carbon source under nitrogen-limiting conditions | 2.1 g/l | [53] |
| α-humulene | M. extorquens AM1 | Serine cycle | Heterologous expression of α-humulene synthase and FFP; heterologous expression of the mevalonate pathway | Methanol | Fed-batch fermentation using methanol as the sole carbon source, cumate as inducer, and dodecane as an organic overlay | 1.65 g/l | [64] |
| Itaconic acid | M. extorquens AM1 | Serine cycle | Heterologous expression of cis-aconitic acid decarboxylase; repression of phaR | Methanol | Fed-batch fermentation using methanol as the sole carbon source | 31.6 mg/l | [56] |
| Violacein | M. extorquens AM1 | Serine cycle | Heterologous expression of operon VioABCDE followed by random mutagenesis | Methanol | Shake flask cultures using methanol as the sole carbon source | 67.8 mg/l | [65] |
| Methanol and acetate | Shake flask cultures using methanol and acetate | 118 mg/l | |||||
| Mesaconic acid | M. extorquens AM1 | Serine cycle | Expression of yciA (thioestrase) | Methanol | Shake flask using methanol as the sole carbon source | 70 mg/l | [54] |
| 3-Hydroxypropionic acid | M. extorquens AM1 | Serine cycle | Heterologous expression of mcr; enhancing the expression by promoter optimization and multicopy expression of mcr | Methanol | Shake flask using methanol as the sole carbon source | 69.8 mg/l | [58] |
| Mevalonate | M. extorquens AM1 | Serine cycle | Construction of mevalonate biosensor along with mevalonate biosynthesis pathway | Methanol | Fed-batch fermentation using methanol as the sole carbon source | 2.67 g/l | [61] |
| Crotyl diphosphate | M. extorquens AM1 | Serine cycle | Expression of THKM82V, IPK and ADH2 | Methanol and crotonol | Shake flask using methanol along with supplementation of exogenous crotonal | 0.60 μg/mL | [135] |
| Crotonic acid | M. extorquens AM1 | Serine cycle | Heterologous expression of isocitrate lyase and malate synthase for activation of the glyoxylate shunt, deletion of EMC pathway genes | Methanol | Shake flask using methanol as the sole carbon source | Growth defects on sole methanol | [55] |
| Acetate | Shake flasks using initial growth on acetate; supplementation of 3-nitropropionate to repress glyoxylate shunt during production phase on acetate | 0.2 mg/l | |||||
| PHB | M. extorquens sp. K. | Serine cycle | N/A | Methanol | Fed-batch fermentation using methanol as the sole carbon source under nitrogen and mineral salt-deficient conditions | 136 g/l; 66% of total DCW | [49] |
| P(3HB-co-3HV-co-3HHX) | M. extorquens AM1 | Serine cycle | Deletion of native phaC, heterologous expression of phaC from Aeromonas cavaie, overexpression of β-ketothiolase and acetoacetyl-CoA reductase | Methanol | Fed-batch fermentation using methanol as the sole carbon source under cobalt-deficient conditions | 43.6% of DCW | [136] |
| PHB | M. extorquens AM1 | Serine cycle | N/A | Methanol | Fed-batch fermentation with controlled methanol supplementation (0.01 g/l) in minimal media | 46% of DCW | [48] |
| P(3HB-co-3HV) | Methylobacterium sp. GW2 | Serine cycle | N/A | Methanol and valerate | Fed-batch fermentation using 0.5% methanol as carbon source; supplementation of valerate | 30% of DCW | [50] |
| Synthetic methylotrophic bacteria | |||||||
| Succinic Acid | E. coli | RuMP | Heterologous expression of NAD-dependent MDH; heterologous expression of RuMP pathway | Glucose and methanol | Fed-batch fermentation using 100 g/l glucose and 6.4 g/l methanol. Methanol was used as an auxiliary substrate | 68.75 g/l, 1.45% carbon derived from methanol | [113] |
| Ethanol | E. coli | Modified Serine cycle | Deletion of aceB, gcvP, glcB, frdB, ldhA, gcl, expression of the modified serine cycle, expression of the ethanol production pathway | LB, methanol, and xylose | Shake flask using LB medium supplemented with 30 mM xylose and 200 mM methanol | 36.3 mM, 33.8% of carbon derived from methanol | [117] |
| Acetate | E. coli | Modified Serine cycle | ΔaceB, ΔgcvP, ΔglcB, ΔfrdB, ΔldhA, Δgcl, expression of modified serine cycle, expression of the ethanol production pathway | LB, methanol, and xylose | Shake flask using LB medium supplemented with 30 mM xylose and 200 mM methanol | 37 mM, 27.2% of carbon derived from methanol | [117] |
| 1-butanol | E. coli | RuMP | Δrpe, ΔrpiAB, expression of NAD-dependent MDH, Hps, and Phi followed by adaptive evolution, expression of the 1-butanol production pathway | Xylose and methanol | Fed-batch fermentation using 87 mM of methanol and 100 mM of xylose | 2.0 g/L, 71% of carbon derived from methanol | [137] |
| Acetone | E. coli | RuMP | Δpgi, ΔfrmA, Expression of MDH, Hps, and Phi, Expression of non-oxidative pentose phosphate pathway; expression of acetone formation pathway |
Glucose and methanol | Fed-batch fermentation using 260 mM glucose and 38.3 mM methanol | 45.0 mM, 3.6% of carbon derived from methanol | [75] |
| E. coli | Δpgi, ΔrpiAB, Δedd, ΔfrmA, expression of RuMP pathway followed by ALE | Glucose and methanol | Fed-batch fermentation using 36 g/l glucose and 500 mM methanol | 1 g/L, 22% of carbon derived from methanol | [138] | ||
| Cadaverine | C. glutamicum | RuMP | Δald, Δfdh; heterologous expression of NAD-dependent MDH, Hps, and Phi | Ribose and methanol | Shake flask using 20 mM ribose and 200 mM | 1.5 g/l, 15% of carbon derived from methanol | [119] |
| Naringenin | E. coli | RuMP | Expression of NAD-dependent Mdh fromBacillus stearothermophilus, Expression of Hps and Phi from B. methanolicus, Expression of Coumaroyl CoA ligase and Chalcone synthase | Co-utilization of methanol and yeast extract | 3.5 mg/L 18% of carbon derived from methanol | [139] | |
| D-allulose | E. coli | RuMP | Coupling of allulose monophosphate, RuMP, and PPP | Xylose and methanol | Fed-batch fermenter using 102 mM xylose and 97 mM methanol in the presence of yeast extract and tryptone | 0.512 mM d-allulose/mM methanol | [116] |
| Succinic Acid | E. coli | RuMP | Expression of methanol dissimilation pathway along with RuMP pathway | Glucose, methanol, and formate | Fed-batch using 50 g/l glucose, 2 g/l formate, and 200 mM methanol. Methanol and formate were used as auxiliary substrates | 63.42 g/l | [114] |
2.3.2.1. Production of amino acids and amino acids-derived products
Among methylotrophic bacteria, B. methanolicus has been widely used for the production of amino acids such as lysine, threonine, glutamate, and serine because of its higher growth rate on methanol as the sole carbon source. In B. methanolicus, genes for methanol assimilation are present on the plasmid and can be upregulated to enhance the growth rate. Brautaset et al. showed that by overexpressing pyruvate carboxylase and repressing the competing pathway for glutamate synthesis, l-lysine could be overproduced (65 g/L titer) by B. methanolicus when methanol-fed batch fermentation was used. Additionally, in the same study, repression of homoserine dehydrogenase genes caused significantly elevated glutamate production (69 g/L titer) from methanol [42]. M. extorquens can naturally produce serine from glycine; thus, this methylotrophic bacterium is a suitable host for serine production. However, metabolic engineering for the high production of serine from the living cells of M. extorquens would decrease the growth of this methylotrophic bacterium, because serine is a crucial component for formaldehyde assimilation. Therefore, resting cells of M. extorquens were used by Siriote et al. to produce 54.5 g/L of l-serine from glycine and methanol. However, the yield of serine from methanol was low (8.3%), probably because of the higher activity of MDH compared to transhydroxymethylase in resting cells [43].
Amino acids produced by methylotrophic bacteria can be further utilized to produce different products using heterologous pathways. For example, Naerdal et al. produced 11.3 g/L of cadaverine from a lysine-overproducing strain of B. methanolicus by expressing the lysine decarboxylase gene along with media optimization using methanol as the sole carbon substrate [44]. Brito et al. screened different pathways for the production of 5-Aminovelerate (5-AVA) using a recombinant strain of B. methanolicus and showed that 5-AVA can be produced either by the expression of 2-monooxygenase and 5-aminovaleramidase from l-lysine or by the expression of putrescine and 5-aminopentanal dehydrogenase from cadaverine. 5-AVA production was enhanced to 0.02 g/L by utilizing the two above-mentioned pathways from the recombinant strain of B. methanolicus using methanol as the sole carbon source [45]. However, the production of amino acid-derived chemicals is far below that of the precursor amino acid itself, because of the high flux of competing pathways. Irla et al. took advantage of glutamate production from methanol to produce 9 g/L γ-aminobutyric acid by expressing glutamate decarboxylase genes in B. methanolicus using two-stage fed-batch methanol fermentation [46].
2.3.2.2. Methylotrophic bacteria for the production of EMC-derived products
PHB can naturally accumulate in M. extorquens AM1 strains as a storage material using EMC pathway intermediates, and its production is enhanced under nitrogen-limiting conditions [47]. Although M. extorquens AM1 has been used for the production of PHB [48], the highest recorded production of PHB was from the M. extorquens K strain, which produced 160 g/L of PHB (66% of dry cell weight) in an automated fed-batch fermenter under nitrogen-limiting conditions [49]. Similarly, using methanol as the sole carbon source under cobalt-deficient conditions, the accumulation of p (3-hydroxybutyrate-co-3-hydroxy valerate) P (3HBco3HV) was observed in M. extorquens AM1 [47]. Furthermore, Methylobacterium sp. GW2 strain accumulated 40% of the PHB when methanol was the sole carbon source and was able to accumulate P (3HBco3HV) upon the addition of valeric acid to the culture media [50]. However, the titer of PHB and co-polymers is even lower than that of industrial horse E. coli, which cannot naturally produce PHB. Recombinant E. coli strains have been shown to produce up to 80% PHB using glucose as the carbon and energy source [51]. The lower production of PHB in methylotrophic bacteria is associated with the fact that the EMC pathway plays a major role in the production of cell biomass from methanol, and the production of competing metabolites must be repressed to further enhance PHB production.
Since PHB production in M. extorquens AM1 proceeds via the EMC pathway, the production of EMC-derived chemicals from M. extorquens AM1 is also a suitable option because of the overflow of the EMC pathway, especially under nutrient-deficient conditions. As many genes involved in methanol metabolism are upregulated during the accumulation of PHB in M. extorquens AM1, the production of EMC-derived chemicals other than PHB requires strong expression of the target genes to outperform competing PHB production, as downregulation of the EMC pathway or PHB production can affect methanol metabolism [52]. Rohde et al. expressed a mesophilic B12-dependent acyl-CoA mutase enzyme in M. extorquens AM1 and channeled 3-hydroxybutyryl-CoA to 2-hydroxybutyric acid (2-HIBA). The resulting strain produced 2.1 g/L of 2-HIBA in a fed-batch fermenter with methanol as the sole carbon source [53]. In addition, EMC pathway-derived carboxylic acids were produced by the expression of different thioesterases in M. extorquens AM1. Sonntag et al. produced 70 mg/L mesaconic acid and 60 mg/L methylsuccinic acid from methanol using a recombinant strain of M. extorquens AM1 [54]. Another strategy for producing EMC-derived products from M. extorquens using methanol is to use a downstream formaldehyde assimilation pathway other than EMC. To enhance the titer of EMC-derived products, an alternative glyoxylate shunt pathway was constructed as the primary pathway for cell growth in M. extorquens AM1. The strain was able to grow on acetate as a carbon source using an alternative glyoxylate shunt pathway instead of the EMC pathway; severe growth defects were observed in methanol due to low expression of MDH in the recombinant strain, indicating the importance of the EMC pathway for methanol metabolism in M. extorquens AM1 [55].
2.3.2.3. Methylotrophic bacteria for pyruvate and TCA-derived products
The presence of a downstream TCA cycle in methylotrophic bacteria provides an opportunity for the production of TCA-derived chemicals using methanol as the sole carbon source. TCA cycle in M. extorquens AM1 was exploited for itaconic acid production by expressing a codon-optimized cis-aconitic acid decarboxylase gene. However, only 31.6 mg/L of itaconic acid, with productivity of 0.056 mg/L/h, was produced using the engineered strain from methanol as the sole carbon source. Further attempts to increase the metabolic flux towards the TCA cycle by obstructing PHB production failed to enhance itaconic acid production. Transcriptomic analysis revealed that many genes involved in methanol metabolism are derived from the PHB accumulation pathway, and disruption of the PHB pathway leads to the rewiring of methanol metabolism in M. extorquens AM1 [56]. Drejer et al. produced 0.26 g/L of acetoin from methanol by integrating acetoin synthesis genes into B. methanolicus. The titer of acetoin was enhanced to 0.42 g/L by enhancing the flux towards pyruvate with the overexpression of malic enzyme and isocitrate lyase with a yield of 0.07 g acetoin/g methanol [57]. To demonstrate the feasibility of M. extorquens AM1 for the production of acetyl-CoA-derived products, the malonyl-CoA reductase gene was integrated into M. extorquens AM1 to produce 3-hydroxypropionic acid. However, because of the conversion of 3-HP to propionyl-CoA (an important metabolite of the EMC pathway), only 69.8 mg/L of 3-HP was produced by the recombinant strain of M. extorquens AM1 [58]. Zhu et al. introduced the mevalonate synthesis pathway, wherein optimized expression of acetoacetyl-CoA thiolase (phaA) produced 215 mg/L and 2.22 g/L of mevalonate in shake flasks and fed-batch fermenters, respectively, from M. extorquens AM1 [59]. In another study, genes encoding trans-enoyl-CoA reductase and alcohol dehydrogenase were heterologously expressed along with EMC crotonase to produce 15.1 mg/L of 1-butanol using ethylamine as a substrate [60]. However, the low amount of acetyl-CoA is a bottleneck in the production of acetyl-CoA-derived products. The acetyl-CoA amount was enhanced by 7% with the help of a transcriptional regulator, which led to the production of 2.67 g/L mevalonate from M. extorquens using methanol as the sole carbon source; however, this titer is still far below the industrial requirement [61].
2.3.2.4. Other chemicals produced by methylotrophic bacteria
M. extorquens AM1 has a natural methylerythritol-4-phosphate (MEP) pathway for the production of secondary metabolites, such as carotenoids [62]; however, the production of secondary metabolites in the MVA pathway is superior [63]. The EMC pathway of M. extorquens AM1 was exploited for the production of sesquiterpenoids by Sonntag et al. using heterologous expression of the MVA pathway, starting from the acetoacetyl-CoA of the EMC pathway. Heterologous expression of codon-optimized α-humulene synthase, farnesyl pyrophosphate, and the MVA pathway produced 1.65 g/L of α-humulene from the carotenoid-deficient strain of M. extorquens AM1 when methanol was the sole carbon source during fed-batch fermentation [64]. Hoa et al. demonstrated violacein production in the shikimate pathway of M. extorquens AM1 by expressing the violacein synthesis genes. Random mutagenesis of the recombinant strain increased flux from phosphoenolpyruvate (PEP) to the shikimate pathway by condensing PEP with erythrose-4-phosphate. It was shown that acetate can be a suitable substrate along with methanol to increase the shikimate pathway-derived products. Moreover, 118 mg/L violacein was produced from a recombinant strain of M. extorquens AM1 using acetate and methanol as carbon substrates [65].
2.3.3. Recombinant protein production using methylotrophs
Owing to the availability of a strong methanol-inducible promoter (AOX) of the alcohol oxidase (AOD) gene, P. pastoris has been exploited for the expression and production of foreign proteins [66] (Table 4). In addition, using an AOX promoter, a large number of proteins can be secreted extracellularly, which simplifies the purification step. The AOX promoter can be repressed by glucose, but its expression is not repressed in the presence of glycerol [67]. Additionally, the AOX promoter showed higher expression in the presence of methanol; however, above a certain limit (3.65 g/dm3), methanol was toxic to cells, mainly because of the formation of formaldehyde, which inhibits growth and lowers the product titer. Although the specific production rate should increase at higher concentrations of methanol, owing to the toxic nature of methanol-derived metabolites, the highest production rate was achieved at much lower concentrations. Therefore, the strategy for the production of proteins from methylotrophic yeasts usually includes initial growth on glycerol as a carbon source to produce higher biomass, followed by the use of methanol to express or induce the recombinant protein via the AOX promoter. This two-stage fermentation strategy can produce up to 400 g/L of dry cell weight (DCW), which can ultimately increase productivity based on higher biomass [68].
Table 4.
Recombinant Protein production using methylotrophs.
| Protein |
Strain |
Methanol assimilation pathway |
Metabolic engineering strategy |
Substrate |
Culture condition |
Product titer |
Ref. |
|---|---|---|---|---|---|---|---|
| Methylotrophic Yeasts | |||||||
| Hepatitis B surface antigen (HBsAg) | P. pastoris | XuMP | Expression of HBsAg under AOX1 promoter | Methanol and glycerol | Chemostat fermentation using glycerol to obtain high cell density followed by methanol induction | 187.71 mg/l | [69] |
| Trypsinogen | P. pastoris | XuMP | Expression of porcine trypsinogen using AOX1 promoter | Methanol and glucose | Continuous chemostat fermentation using glucose and methanol | 210 mg/l | [67] |
| Cellulase | P. pastoris | XuMP | TrCBH2 encoding cellulase under DES promoter | Methanol and glycerol | Two-stage Fed-batch fermentation using glycerol to obtain high cell density followed by methanol induction | 18 g/l | [140] |
| Human serum albumin (HSA) | P. pastoris | XuMP | Expression of Hsa gene under AOX1 promoter | Methanol and glycerol | Two-stage fed-batch fermentation using glycerol for growth followed by methanol induction for 96 h | 8.86 g/l | [141] |
| P. pastoris (ALE-based methanol tolerant strain) | XuMP | Expression of Has gene under AOX1 promoter in Adapted strain | Methanol and glucose | Two-stage fed-batch fermentation using glucose for growth followed by methanol induction for 48 h | 350 mg/l | [142] | |
| Plectasin | P. pastoris | XuMP | Expression of gene encoding plectasin | Methanol and glycerol | Two-stage fed-batch fermentation using glycerol for growth followed by methanol induction for 120 h | 748.6 μg/ml | [143] |
| Xylanase | P. pastoris | XuMP | Heterologous expression of xyn11A under AOX1 promoter | Methanol | Fed-batch using methanol as the sole carbon source | 1.16 g/l | [71] |
| Methylotrophic bacteria | |||||||
| Green fluorescent protein (GFP) | M. extorquens AM1 | Serine cycle | Heterologous expression of GFP under mxaF promoter | Methanol | Fed-batch using methanol as sole carbon source | 4 g/L | [72] |
| Enterocin P | M. extorquens AM1 | Serine cycle | Heterologous expression of EntP under mxaF promoter | Methanol | Shake flask using CHOI-medium containing methanol as sole carbon source | 155 ng/ml | [73] |
The hepatitis B vaccine, based on the hepatitis B surface antigen (HbsAg), is produced from P. pastoris on an industrial scale [66]. Traditional fermentation for HbsAg includes high initial biomass fermentation using glycerol in batch or fed-batch mode with subsequent induction with methanol. Heat dissipation and O2 transfer are major challenges faced in industrial fermentation for HbsAg production. To overcome these issues, Rahimi et al. developed a continuous chemostat-based fermentation process to produce HbsAg from P. pastoris and enhanced the volumetric productivity of HbsAg to 1.699 mg HbsAg g/L/h, which is higher than the 1.38 mg HbsAg g/L/h in the traditional fermentation [69]. P. pastoris can use different substrates for growth, such as glucose, mannitol, glycerol, or methanol. Although glucose is thought to repress the expression of AOX1, it has been shown that co-utilizing glucose and methanol can produce 1.4 times more biomass than using methanol alone. This strategy was used to produce trypsinogen from a recombinant strain of P. pastoris [67]. However, using methanol and sorbitol as co-substrates, the product formation rate was higher, but a lower cell density was achieved [70]. Therefore, to achieve the production of recombinant proteins from methylotrophic yeasts, process optimization needs to be considered in two-stage fermentation, and should be balanced in such a way that higher biomass and product can be produced simultaneously.
Although two-stage fermentation is usually used for the production of recombinant proteins from P. pastoris, the processing time for two-stage fermentation is between 100 h and 150 h, which increases the production cost at the industrial level. To reduce production time, a single-phase xylanase production process was conducted using methanol as the only substrate. Xylanase production was 1.16 g/L after 48 h using the single-phase process, which was higher than that of traditional two-phase fermentation. Additionally, most of the xylanase was excreted into the supernatant; however, using methanol as the sole carbon source produced less biomass (up to 34 g/L of total DCW) than two-stage fermentation [71].
Similarly, methylotrophic bacteria, such as M. extorquens, have also been used to produce recombinant proteins in fed-batch fermenters. Green fluorescent protein (GFP) production was achieved with a yield of 0.33 g/g methanol using M. extorquens containing the heterologous expression of GFP [72]. Similarly, Enterocin P (EntP), a bacteriocin, was produced from M. extorquens by heterologous expression of the EntP gene from Enterococcus faecium P13 using methanol as the sole carbon source [73]. However, the production of recombinant proteins from M. extorquens is much lower than that of its methylotrophic yeast counterpart. The lower production of proteins from M. extorquens can be attributed to either a decrease in cell growth due to the lower cell metabolism capability during the production of recombinant proteins or the accumulation of storage compounds such as PHB using the carbon derived from methanol oxidation [72].
2.4. Limitations of native methylotrophs and their solutions for the production of fuels and chemicals
Although methanol is a cheap substrate compared to other sugar-based substrates, a methanol-based biorefinery can only be achieved if the product titer or productivity from the native methylotrophic strains meets industrial requirements. The assimilation of methanol to biomass at the formaldehyde oxidation level in the RuMP pathway proceeds at rates equal to biomass generation by sugar substrates such as glucose. However, the complex and interdependent pathways in native methylotrophs make it difficult to divert the flux toward product formation [7]. In this section, strategies for overcoming the limitations of native methylotrophic strains in methanol assimilation and metabolism are discussed.
2.4.1. The balance between the pathways during methanol assimilation must be tightly regulated
Methanol assimilation in native methylotrophs requires a balance between methanol oxidation, formaldehyde assimilation, and the conversion of formaldehyde to biomass precursors, such as acetyl-CoA or pyruvate. In native methylotrophs, formaldehyde assimilation, and methanol oxidation are tightly regulated along with other metabolic pathways, such as glycolysis, PPP, and the ED pathway [4]. An imbalance among these pathways can result in a shortage of precursors for the serine, RuMP, or XuMP pathways, causing lower formaldehyde assimilation. This imbalance can also lead to a shortage of biomass precursors such as pyruvate or acetyl-CoA and energy precursors such as NADH. A similar balance is required for formaldehyde assimilation and dissimilation pathways to generate energy for methanol oxidation and to avoid the accumulation of toxic formaldehyde. During the EMC cycle of M. extorquens AM1, it has been shown that the growth of methanol and balance between the growth and oxidation of methanol is maintained at the EMC level by the regeneration of glyoxylate from acetyl-CoA [74]. Therefore, metabolic engineering strategies for native methylotrophic organisms must be finely tuned to avoid growth defects in methanol.
One strategy to enhance methanol assimilation via formaldehyde fixation is to increase the precursors of formaldehyde assimilation in the respective metabolic pathways. For example, the expression of non-oxidative PPP has been proven useful in enhancing the regeneration of precursors in the RuMP pathway [75]. Because more precursors are available to fix formaldehyde via the RuMP pathway, the flux towards the assimilation of formaldehyde would be greater than that towards the dissimilation pathway. Alternatively, formaldehyde-inducible promoters or biosensors can be used to enhance precursor regeneration [76]. Substrate channeling is another strategy for enhancing the flux towards the required metabolites to increase the target product or methanol metabolism [77]. The scaffold-based assembly of key enzymes involved in methanol assimilation, such as MDH, FDH, 6-phosphofructokinase, transketolase, and FBP aldolase, can be useful for obtaining higher biomass and enhancing methanol utilization in native methylotrophs [78].
2.4.2. Overcoming the carbon and energy loss during methanol assimilation
The major methanol assimilation pathways described above have both advantages and disadvantages. For example, the production of pyruvate through the serine cycle is higher (0.5 mol/mol methanol); however, this pathway consumes 1 mol ATP/mol methanol. In contrast, in the RuMP and XuMP pathways, one-carbon loss occurs during the formation of acetyl-CoA using DHAP as an intermediate. As a result, these pathways produce less pyruvate than the serine cycle. However, RuMP and XuMP are known to be energy-conservative pathways that can generate ATP (0.33 mol ATP/mol methanol in RuMP; 0.66 mol ATP/mol methanol in XuMP) [6]. NADH molecules produced during the dissimilation pathway of methanol metabolism drive energy by converting it to ATP via oxidative phosphorylation. Although the dissimilation pathway of methanol oxidation supplies energy, carbon loss in the form of CO2 is a major hurdle in the development of native methylotrophs for fuel and chemical production. A recent study showed that CO2 generated by the dissimilation pathway in M. extorquens AM1 is fixed by carboxylases present in the EMC cycle. 13C labeling confirmed that approximately 50% of the carbon in PHB originated from CO2 [74]. According to stoichiometric Equation (5), approximately 38% of the carbon in PHB is lost in the form of CO2 whereas 62% of the carbon is used for cell biomass. Additionally, approximately 0.85 g of O2 is required to oxidize 1 g of methanol [21]. However, it has been reported that approximately 50% of carbon is lost during the dissimilation pathway for growth on methanol [79]. In addition, the dissimilation of formaldehyde has been shown to increase in fermenter studies, which poses another limitation for upgrading the process. For example, stoichiometric model-based analysis of P. pastoris showed that only 18% of the methanol used was dissimilated by yeast cells in shake flask cultures, whereas this value could reach 70–80% in fed-batch fermentation [80,81].
Therefore, theoretically, the flux towards the assimilation of formaldehyde needs to be enhanced to efficiently utilize methanol for cell biomass. However, the dissimilation pathway in native methylotrophs is considered the main source of energy in the form of NADH, and its knockout has been associated with impaired growth in native methylotrophic organisms [82]. This is because the genes involved in the TCA cycle are downregulated in native methylotrophs when methanol is the sole carbon source, and depending on the microorganism, EMC, ED, or EMP are the major pathways for biomass generation [83]. Therefore, a suitable option to prevent carbon loss in the form of CO2 is to construct CO2 fixation pathways, such as CBB, to produce cell biomass from the released CO2 by native methylotrophs (Fig. 4C). Recently, Gassler et al. constructed a Calvin Benson Bassham cycle (CBB) in P. pastoris that enabled the utilization of CO2 in the recombinant strain. However, at least eight genes need to be expressed in P. pastoris to create a functional CBB cycle, followed by the deletion of AOX1, DAS1, and DAS2 along with adaptive laboratory evolution (ALE). The resulting P. pastoris strain was able to grow on CO2 as the sole carbon source with a maximum growth rate of 0.018 h−1; however, methanol growth was hampered by the deletion of key enzymes involved in methanol metabolism [84]. In another attempt, the CBB cycle was constructed in M. extorquens AM1 to produce biomass using CO2 as the sole carbon source. However, a fully autotrophic strain was not obtained, and regeneration of energy from methanol was used to produce higher cell biomass in CO2 and methanol co-feeding [85]. These results show that autotrophic growth can be achieved in native methylotrophic strains, which can be exploited as functional auto-methylotrophic microorganisms capable of producing biomass from methanol and CO2 simultaneously (Fig. 4D). However, it is important to consider that the fixation of CO2 into the CBB cycle can deprive cells of the energy required to maintain methanol fixation in formaldehyde. To overcome this energy requirement, the co-utilization of substrates (Fig. 4B) such as xylose and methanol can be a fruitful strategy for the development of hetero-auto-methylotrophs. In addition, because sugar carbon sources use the TCA cycle as the main cycle for biomass formation, the utilization of sugar substrates in engineered cells with an enhanced TCA cycle can be applied. Thus, the TCA cycle produces biomass, whereas the other cycles of methanol/formaldehyde assimilation can be exploited for the target products.
Fig. 4.
Strategies to enhance the methanol assimilation or biomass in native methylotrophic microorganisms. (A) Construction of RuMP pathway in serine cycle utilizing bacteria. The enzymes shown in red were heterologously expressed or overexpressed to make the functional RuMP pathway in M. extorquens AM1. The Solid arrow shows a single reaction while the dashed arrow shows multiple reactions. (B) Strategies for Co-utilization of sugar substrates such as glucose (gold color), glycerol (green color), or xylose (blue color) to convert RuMP utilizing methylotroph to hetero-methylotroph. The solid arrow shows a single reaction while the dashed arrow shows multiple reactions. (C) Strategy to convert methylotroph into autotroph by constructing CBB cycle in native methylotrophs. The methanol conversion to biomass can be blocked at formaldehyde or formate level in RuMP/XuMP or serine cycle utilizing microbe, respectively. In this way, methanol will be used to generate the energy while CO2 produced by the dissimilation pathway will be used to produce biomass. (D) Strategy to convert methylotroph into auto-methylotroph by constructing CBB cycle in native methylotrophs either using RuMP, XuMP, or serine cycle. The CO2 produced by the dissimilation pathway can be converted to biomass. The methanol will be converted to biomass and also will be used to generate energy. (E) Enzyme engineering of MDH to increase the formaldehyde formation from methanol. (F) Adaptive laboratory evolution of native methylotrophic microorganisms to increase methanol/formaldehyde tolerance and enhanced methanol assimilation. Enzymes abbreviations:Mdh, Methanol dehydrogenase; Hps, 3-hexulose-6-phosphate synthase; Phi, 6-phospho-3-hexuloisomerase; Pfk, 6-phosphofructokinase; G6dph, Glucose-6-phosphate dehydrogenase; GlpF, glycerol transporter; GlpK, glycerol kinase; GlpD, glycerol 3-phosphate dehydrogenase; GspA, glycerol 3-phosphate dehydrogenase; XylA, xylose isomerase; XylB, xylulose kinase; HK, hexokinase. Metabolite abbreviations: CH2 H4F, N5,N10-methylenetetrahydromethanopterin; CO2, Carbon dioxide; H6P, Hexulose-6-phosphate; F6P, Fructose-6-phosphate; FBP, Fructose 1,6-bisphosphate; DHAP, Dihydroxyacetone phosphate; GAP, Glyceraldehyde 3-phosphate; Ru5P, Ribulose 5-Phosphate; G6P, glucose-6-phosphate; 6-PG, 6-phosphogluconate; G3P, glyceraldehyde-3-phosphate; 2-PG, 2-phosphoglycerate; H4MPT, Tetrahydromethanopterin; H4F, tetrahydrofolate; Xu5P, Xylulose 5-phosphate; Gly3P, glycerol 3-phosphate; ADP, Adenosine diphosphate; ATP, Adenosine triphosphate, NAD(P)+, Nicotinamide adenine dinucleotide phosphate; NAD(P)H, reduced form of NAD(P)+; NAD+, Nicotinamide adenine dinucleotide; NADH, Reduced form of NAD+; 3PGA, 3-phosphoglycerate; RuBP, Ribulose 1,5-bisphosphate.
Nonetheless, NADH regeneration during the dissimilation pathway can be used to enhance the product titer. Schroer et al. showed that regenerating NADH by overexpressing FDH led to high production of butanediol in the whole cell system of P. pastoris [86]. Another option is to develop the RuMP pathway in M. extorquens AM1 to produce higher cell biomass along the serine cycle (Fig. 4A). It has been predicted that an approximately 33% biomass enhancement can be observed if both pathways are functional in M. extorquens; however, experimental data have led to an enhancement of 16% of the biomass [87].
2.4.3. Improving the catalytic properties of MDH to enhance the methanol oxidation
One of the key restraints in the development of native methylotrophs for industrial-based production is the poor catalytic and thermodynamic characteristics of MDH. MDH is a reversible enzyme that converts methanol to formaldehyde and vice versa. However, the catalytic efficiency of MDH for the conversion of formaldehyde to methanol is 1000-fold more than the conversion of methanol to formaldehyde [88]. In addition, the Gibbs free energy of NAD-dependent MDH for methanol oxidation is lower at higher temperatures, which is the probable reason for using PQQ-dependent MDH from mesophilic bacteria and AOD from methylotrophic yeasts [89]. To utilize methanol efficiently, traditional enzyme engineering of MDH is required to enhance the catalytic efficiency of methanol-to-formaldehyde conversion (Fig. 4 E) [90].
Recently, an NAD-dependent variant was isolated from the Cupriavidus necator N-1 strain, which showed higher activity towards methanol than the MDH of B. methanolicus. The activity of this enzyme was further improved by directed evolution, which increased the Kcat/Km value 6-fold compared with the MDH of B. methanolicus [91]. Engineering native methylotrophs with catalytically improved MDH should be considered to enhance the biomass and production of methanol. Enhancing the activity of MDH using activator proteins can also enhance methanol oxidation to formaldehyde [92]. However, the balance between formaldehyde assimilation and dissimilation must be considered to drive methanol oxidation in the cell biomass. One strategy involves the use of engineered MDH in native methylotrophs along with formaldehyde biosensors to drive formaldehyde assimilation.
2.4.4. Low availability of electrons to drive metabolites during growth on methanol
Native methylotrophs also do not produce metabolites at high levels, because of the unavailability of electrons to drive metabolite production. In methylotrophic bacteria, the low availability of electrons arises because the electron transport chain tends to oxidize the reduced form of energy, such as NADH, for use in methanol oxidation, rather than permitting their accumulation to drive the production of metabolites. In contrast, the electrons produced in methylotrophic yeasts during the oxidation of methanol are directly transferred to O2 rather than passing through the electron chain, which results in reduced ATP production in methylotrophic yeasts due to methanol oxidation [7]. Additionally, as these electrons are readily used for the oxidation of methanol, a strategy needs to be developed that can readily divert these electrons toward product formation to increase the titer of energy-dependent products, such as fatty acids or higher alcohols. Recently, electron channeling has gained interest in the scientific community for increasing the titer of products [93]. This strategy can be employed in native methylotrophs to improve the electron availability for the target product.
2.4.5. Formaldehyde toxicity needs to be overcome to enhance the methanol assimilation
Methanol assimilation in native methylotrophs proceeds by formaldehyde as an intermediate, and the accumulation of formaldehyde negatively affects the cell structure; therefore, its assimilation is tightly regulated in native methylotrophs. Thus, the dissimilation of formaldehyde into CO2 may be a more favorable reaction than its assimilation, which generates energy. However, CO2 is not further used by the above-mentioned pathways to increase the biomass metabolites; rather, it is used only to generate energy for methanol oxidation. One suitable option is to screen for methylotrophs that are highly tolerant to methanol and formaldehyde. Guo et al. isolated a methylotrophic strain of Methylomonas sp. ZR1 can use methane or methanol as the sole carbon source and can utilize up to 35–40 g/L methanol. In addition, this strain is a native producer of C30 carotenoids, making it a suitable host for the production of isoprenoids from methanol [94]. Using formaldehyde transcriptional factors, B. methanolicus cells were made more tolerant to higher amounts of methanol, and the cell growth rate was improved by adding additional copies of the hps and phi genes [95]. Co-utilization of different substrates can also be employed to overcome formaldehyde toxicity in native methylotrophic strains. For example, it has been shown that methanol toxicity in Pichia methanolica arises because of formaldehyde rather than methanol toxicity, and formaldehyde toxicity can be overcome using glucose and methanol as co-substrates [96].
3. Construction of synthetic methylotrophs for methanol utilization
Although native methylotrophs grow faster on methanol, owing to the underdeveloped genetic tools, the complex metabolic pathways and difficulties in controlling the metabolic flux towards the target product have led to the scientific community's interest in synthetic methylotrophs for the production of biofuels and chemicals [97]. Synthetic methylotrophy involves the expression of methanol assimilation pathways/genes in industrial non-methylotrophic workhorses, such as E. coli and S. cerevisiae, to utilize these microbes as methanol-fixing microorganisms. In this section, we review current progress in the development of synthetic methylotrophs.
3.1. Heterologous expression of MDH for synthetic methylotrophs
The most important enzyme for methanol assimilation in non-native methylotrophs is MDH, the heterologous expression of which has proven difficult owing to its lower kinetic properties. The choice of MDH for constructing synthetic methylotrophs varies from organism to organism; however, NAD-dependent MDHs have mostly been considered for the development of synthetic methylotrophs. For example, in the case of E. coli (a gram-negative bacterium), MDH from gram-negative M. extorquens AM1 initially seemed to be a desirable choice owing to its higher activity than other MDHs. However, MDH from M. extorquens is PQQ-dependent, and E. coli does not naturally produce the PQQ co-factor. Therefore, supplementation with PQQ cofactor is required, which is economically unsuitable. Additionally, approximately 15 genes, including cytochrome CL or cytochrome C oxidase, are involved in methanol oxidation by PQQ-dependent MDH. However, in the case of NAD-dependent MDH from B. methanolicus, only one gene is required for MDH expression. Additionally, electrons can be directly used by E. coli without the requirement of any foreign electron transport chain [9,11]. However, the key issue in the expression of NAD-dependent MDH is the irreversible action of the enzyme, which favors the reduction of formaldehyde to methanol over its oxidation of methanol to formaldehyde. Although irreversible fixation of formaldehyde to H6P or F6P via the RuMP pathway provides the driving force for formaldehyde fixation, these downstream enzymes must be expressed at higher ratios (1:10 MDH to HPS) to produce sufficient amounts of H6P. Substrate channeling is the key strategy for pulling formaldehyde formation towards biomass by expressing the required enzymes in supramolecular assemblies or cascades [5].
3.2. Heterologous expression of formaldehyde assimilation pathway in synthetic methylotrophs
For formaldehyde assimilation into biomass using synthetic methylotrophs, the RuMP pathway is a suitable option, because most genes involved in formaldehyde assimilation by the RuMP pathway are present in E. coli. These genes participate in glycolysis, ED, and PPP pathways; therefore, theoretically, to create a functional RuMP pathway in E. coli, only the expression of hexulose-6-phosphate synthase (Hps) and 6-phospho-3-hexuloisomerase (Phi) is required. Additionally, the RuMP pathway appears energetically favorable over the serine cycle for its expression in non-methylotrophic bacteria, because formaldehyde assimilation in the serene cycle proceeds in the form of methylene-THF. In the RuMP cycle, one molecule of NADPH and ATP is generated for the formation of each pyruvate molecule, whereas in the serine cycle, NADPH and ATP are consumed for pyruvate production [5,9,11].
3.3. Formaldehyde toxicity in synthetic methylotrophs
In native methylotrophs, the genes involved in formaldehyde metabolism are highly upregulated upon the addition of methanol to accommodate fast methanol oxidation; however, in synthetic methylotrophs, formaldehyde toxicity can be a major problem for cell survival. Formaldehyde biosensors [76], formaldehyde inducible promoters [98], and adaptive laboratory evolution (ALE) (Fig. 4F) [99,100] have been exploited to overcome formaldehyde toxicity and to obtain a balance between MDH activity and formaldehyde assimilation [76].
3.4. Synthetic methylotrophic strain development
Non-native methylotrophic bacteria, such as Corynebacterium glutamicum [101], Bacillus subtilis [102], and E. coli [75,99,103], have been developed to use methanol as a carbon substrate by expressing NAD-dependent MDH, HPS, and HPI to create the functional RuMP pathway. Although 13C labeling showed the utilization of methanol as a central metabolite, the above-mentioned strains were not able to grow solely in methanol, and very low conversion of methanol to central metabolites was achieved [99,101]. To enhance methanol utilization by synthetic E. coli, Price et al. expressed MDH, HPS, and HPI in a self-assembled scaffold using SH-3 ligand interactions. Additionally, to slow down the reduction of formaldehyde to CO2, an NADH sink was constructed using lactate dehydrogenase to convert pyruvate into lactate, which uses NADH for enzymatic reactions [78]. The enzyme assembly of MDH, HPS, and HPI along with NADH sink increased methanol utilization up to 1.7 mM/h using resting cells of E. coli; however, the resulting strain was still unable to grow on methanol as the sole carbon source [78].
A key limitation of synthetic methylotrophs is the downstream conversion of formaldehyde to F6P or H6P. Maintaining a low formaldehyde concentration is important to drive methanol oxidation. Only 50 μM μM formaldehyde is required for 250 mM methanol to reach the reaction equilibrium of NAD-dependent MDH in E. coli [104,105]. Therefore, driving formaldehyde toward the RuMP pathway is a major step in obtaining a synthetic methylotrophic E.coli strain. Woolston et al. improved the regeneration of Ru5P by activating the sedoheptulose bisphosphatase variant of the RuMP pathway to drive formaldehyde assimilation into biomass but argued that the low and reversible activity of MDH is a major bottleneck in the development of synthetic methylotrophy [105]. To avoid formaldehyde toxicity, NAD-dependent MDH and RuMP cycles were constructed in an engineered E. coli strain capable of growing on gluconate. Furthermore, adaptive laboratory evolution increased methanol consumption and growth rate in methanol-essential E. coli strains in the presence of gluconate and methanol [106]. Similarly, Cruz et al. demonstrated that the reductive glycine cycle, which can produce glycine from formate, can be enabled in S. cerevisiae. The utilization of formate to enhance precursors for formaldehyde assimilation can be applied as a co-substrate strategy. Additionally, He et al. proposed a homoserine cycle for the autotrophic growth of E. coli on methanol and showed that this pathway can outperform the naturally existing RuMP pathway or serine cycle for biomass production [107].
To avoid plasmid loss, S. cerevisiae was constructed for methanol utilization by integrating the AOD and XuMP pathways from P. pastoris wherein 1.4 g/L of methanol was consumed, which increased the cell growth by 3.31% in glucose/methanol cultures. Furthermore, methanol consumption was increased to 2.35 g/L using yeast extract in the culture media, and an 11% increase in growth was observed in the engineered strain when methanol was used as an auxiliary substrate [108]. A reductive glycine pathway was recently constructed, which showed methanol-dependent growth in E. coli [109]. ALE, rather than rational site-directed enzyme engineering, has been proven to be more effective for the construction of synthetic methylotrophic strains. Using ALE, a synthetic methylotrophic strain of E. coli was constructed that could grow on methanol as the sole carbon source. Although the growth rate of the ALE-developed strain was comparable to that of native methylotrophs, with a doubling time of 8 h, a maximum OD600 of 2 was achieved [110]. Similarly, a strain was constructed that could grow on CO2 as the sole carbon source [111], and on CO2 and formate [112]. In the future, a combination of CO2 or formate fixation with methanol utilization could be used to construct strains capable of growing on waste C1 carbon substrates.
3.5. Production of fuels and chemicals using synthetic methylotrophs
To date, no synthetic methylotrophs that can grow solely on methanol and simultaneously produce the target chemical have been developed. To date, most strategies have focused on the utilization of methanol as an auxiliary substrate, primarily to drive energy for the production of chemicals in co-substrate cultivation. As the energy generated by methanol oxidation in synthetic methylotrophs can be used to drive the production of chemicals, Zhang et al. constructed a methanol assimilation pathway in a succinic acid-producing strain of E. coli. The NADH produced by the methanol oxidation in glucose-methanol fermentation was used by the recombinant strain of E. coli and the production of succinic acid enhanced anaerobic fermentation with a yield of 0.98 g succinic acid/g methanol [113]. Recently, Guo et al. combined formaldehyde dissimilation and assimilation pathways in an E. coli strain to exploit energy generation for succinic acid production. Additionally, they enhanced the CO2 fixation ability of this strain by introducing pyruvate decarboxylase. Using resting glycolated E. coli cells, the toxic effects of methanol metabolism can be minimized in synthetic methylotrophs [114]. Similarly, a synthetic methylotrophic strain of E. coli was used to produce lysine via NADPH regeneration and methanol metabolism [115]. A recombinant E. coli strain was used to produce d-allulose by coupling methanol reduction, PPP, allulose, and RuMP pathways. The engineered strain produced 70.7 mM d-allulose with productivity of 0.512 mM/mM in methanol [116]. A modified serine cycle was constructed in E. coli that could utilize CO2 and methanol and was used to produce ethanol from methanol [117]. Glutamate [118] and cadaverine [119] production has also been demonstrated by synthetic methylotrophic strains of C. glutamicum by co-utilizing methanol and sugar substrates.
3.6. Limitations of synthetic methylotrophs for the production of fuels and chemicals
The main hurdle in the development of synthetic methylotrophs is the condensation of C1 molecules to produce C3 or C4 compounds, which can be used for cell biomass formation. For example, the oxidation of methanol to formaldehyde requires a sufficient amount of energy and flux balance between the upstream and downstream pathways to produce cellular biomass owing to formaldehyde toxicity [7,9]. Therefore, for the development of synthetic methylotrophs and their subsequent use in industrial production, efficient production of energy and biomass should be achieved. Formaldehyde is toxic to cellular processes; thus, E. coli naturally has a formaldehyde detoxification pathway linked to glutathione (GSH) by GSH-dependent formaldehyde dehydrogenase [120]. To utilize formaldehyde for cell biomass, the formaldehyde detoxification pathway must be repressed; however, efficient expression of formaldehyde assimilation genes is required to rescue cells from formaldehyde toxicity.
The development of enzyme cascades using engineered or improved enzymes for methanol assimilation in cell-free systems could be another strategy for efficiently converting methanol into the target product. However, such a strategy has limited use in industrial-based applications because the cofactor requirement differs for each enzyme. Additionally, it is difficult to maintain balance and reaction conditions to achieve favorable productivity. Furthermore, the balance between formaldehyde and NADPH needs to be maintained to direct the reaction toward formaldehyde assimilation. A methanol condensation cycle combining the RuMP pathway with non-oxidative glycolysis was constructed and used to produce ethanol from methanol. However, the in vivo construction of this pathway has not yet been reported [121]. Recently, a pathway for the production of ethylene glycol, glycolate, ethanol, and glycerate from formaldehyde, methanol, and formate was demonstrated. The utilization of such pathways can be integrated with the natural metabolism of target microorganisms and exploited for the production of desired chemicals from methanol [86].
4. Discussion and future perspectives
In recent years, methanol has gained considerable attention as a C1 carbon substrate for the bioproduction of fuels and chemicals. Methanol possesses more energy than glucose [122]; therefore, it is an attractive substrate for the bio-production of reduced fuels and chemicals. Furthermore, the fact that gas fermentation is limited by gas-liquid mass transfer [6] highlights methanol as a preferable carbon substrate for industrial biotechnology. This emphasizes the need to develop an integrated methanol-based biorefinery.
Native methylotrophs can be engineered to produce various chemicals. Owing to different methanol assimilation pathways, carbon and energy efficiencies vary among different methylotrophic strains. Therefore, the choice of methylotrophs must be carefully considered for specific chemical production. For instance, during methanol assimilation, the RuMP and XuMP pathways utilize one formaldehyde molecule and one Ru5P or Xu5P molecule, respectively, to generate two G3P molecules. During the conversion of G3P to acetyl-CoA, one carbon is lost in the form of CO2, which can reduce the carbon yield during the production of acetyl-CoA-derived products [123]. In comparison, serine cycle-utilizing microorganisms can convert formaldehyde to acetyl-CoA without any carbon loss [123,124], which can enhance the carbon yield of acetyl-CoA-derived products such as PHB [49], 2-HIBA [53], mevalonate [61] and α-humulene [64]. Additionally, serine cycle-utilizing microorganisms contain the EMC pathway, which plays a major role in the regeneration of glyoxylate and possesses many CoA-esters such as propionyl-CoA, ethylmalonyl-CoA, methylsuccinyl-CoA, and mesaconyl-CoA. The production of C5 di-carboxylic acids derived from the CoA-esters of the EMC pathway is a promising option [125], as most of these carboxylic acids are commercially unavailable. Moreover, the EMC pathway is known to be upregulated during growth on methanol compared to the growth on multi-carbon compounds and can fix half of the carbon lost during the dissimilation of formaldehyde to CO2 [17]. The PHB and EMC pathways share the first two steps that convert acetyl-CoA to 3HB-CoA; therefore, enhancing the flux towards the EMC pathway can be an appropriate solution for the production of EMC pathway-derived products [21,125].
The RuMP pathway is known to be more energy-conservative than the serine cycle [123,124]. For instance, for the generation of one mol of acetyl-CoA, the serine cycle utilizes two mol of NADH and two mol of ATP, whereas the RuMP pathway generates one mol of ATP and 2 mol of NADH [123]. NADH can be further converted to NADPH by overexpressing NADH kinase, which can be exploited for the production of NADPH-dependent chemicals, such as l-lysine [115]. Additionally, RuMP pathway-utilizing bacteria, such as B. methanolicus encode genes for the complete TCA cycle, and a significant amount of acetyl-CoA enters the first reaction of the TCA cycle during growth on methanol [126,127]. Therefore, by controlling the carbon flux from pyruvate to the TCA cycle, B. methanolicus can be exploited for the production of amino acids, such as l-glutamate [118], l-lysine [42], and amino acid-derived chemicals, such as cadaverine [44]. On the other hand, methylotrophic yeasts, such as P. pastoris, are Crabtree-negative microorganisms that tend to distribute carbon flux towards energy and biomass formation rather than surplus metabolites. Consequently, 50% of the carbon is lost in the form of CO2 the formaldehyde dissimilation pathway. This results in a lower accumulation of metabolites that can be used for the production of chemicals using methanol as a substrate [128]. Nonetheless, during growth on methanol, P. pastoris has high NADPH/NADP+ levels, making it a suitable host for the production of reduced chemicals such as fatty acids [30,32] and terpenoids [[24], [25], [26]]. In addition, P. pastoris has a relatively higher flux towards the pentose phosphate pathway (PPP) than towards the TCA cycle or glycolysis [128,129]. A higher carbon flux towards PPP allows the generation of E4P, which can be exploited for the production of aromatic compounds such as tyrosine, resveratrol, and naringenin [130].
To enhance the economic feasibility, the product titer and productivity of native methylotrophs should be enhanced. The low rate of methanol oxidation by native methylotrophs, low formaldehyde assimilation by native pathways, higher rate of formaldehyde dissimilation to CO2, the toxicity of formaldehyde, low availability of electrons for product formation, and interconnected methanol assimilation pathways need to be addressed. More efficient genetic tools should be developed for the metabolic engineering of native methylotrophs. The gas-liquid mass transfer limitations must be overcome to meet the high requirements of O2 for the oxidation of methanol [[9], [10], [11]]. Although proof-of-concept production of different chemicals has been demonstrated using engineered native methylotrophs, recombinant protein production from methylotrophic yeasts and amino acid production from methylotrophic bacteria are high enough to compete with sugar-based bioprocesses. All natural formaldehyde assimilation pathways are cyclic, which makes it difficult to drive the carbon flux toward the desired product. Therefore, the construction of linear pathways in native methylotrophs can be considered to enhance C1 metabolism [[131], [132], [133]]. Substrate and electron channeling can also be applied to enhance the product titer in native methylotrophs [93]; however, such a strategy must be designed in such a way that it does not affect the native metabolism of methylotrophs. Additionally, the expression of target enzymes in native methylotrophs can be enhanced by screening for strong promoters, site-directed enzyme engineering, and ALE. To overcome methanol toxicity, a co-substrate strategy can be applied to methylotrophic bacteria, which can also increase biomass. Screening methylotrophic strains that are more resistant to methanol and the development of efficient formaldehyde biosensors are promising options.
Recently, synthetic methylotrophs have been considered an attractive substitute for native methylotrophic strains based on their well-studied metabolism and the availability of developed genetic resources. Unfortunately, efficient methanol utilization in non-natural methylotrophs has not yet been achieved. The methanol utilization rate of synthetic methylotrophs is far below that of native methylotrophs [79,110] even when methanol is used as an auxiliary substrate [80]. To achieve an efficient synthetic methylotroph for the production of fuels and chemicals, enzyme engineering, systems biology, and synthetic biology must be integrated to develop novel and efficient pathways for methanol or formaldehyde assimilation. Additionally, ALE could be a fruitful strategy coupled with rational enzyme engineering to obtain a synthetic methylotroph strain capable of growing efficiently on methanol as the sole carbon source. In conclusion, efficient methanol-based microbial chassis could be developed based on strain engineering of both native and synthetic methylotrophs for the methanol-based bioproduction of fuels and chemicals as a suitable alternative to sugar-based bioproduction.
CRediT authorship contribution statement
Arslan Sarwar: Conceptualization, Investigation, Writing - original draft. Eun Yeol Lee: Writing - Review & Editing, Supervision, Funding acquisition, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the C1 Gas Refinery Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science and ICT (2015M3D3A1A01064882). This research was also supported by the Korea Institute of Marine Science & Technology Promotion (KIMST), funded by the Ministry of Oceans and Fisheries, Korea (20220532).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Ali A., Shen P.K. Recent advances in graphene-based platinum and palladium electrocatalysts for the methanol oxidation reaction. J Mater Chem. 2019;7(39):22189–22217. doi: 10.1039/c9ta06088j. [DOI] [Google Scholar]
- 2.Stacheter A., Noll M., Lee C.K., Selzer M., Glowik B., Ebertsch L., et al. Methanol oxidation by temperate soils and environmental determinants of associated methylotrophs. ISME J. 2013;7(5):1051–1064. doi: 10.1038/ismej.2012.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabarius J.T., Wegat V., Roth A., Sieber V. Synthetic methylotrophy in yeasts: towards a circular bioeconomy. Trends Biotechnol. 2021;39(4):348–358. doi: 10.1016/j.tibtech.2020.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Khirsariya P., Mewada R.K. Single step oxidation of methane to methanol–towards better understanding. Procedia Eng. 2013;51:409–415. doi: 10.1016/j.proeng.2013.01.057. [DOI] [Google Scholar]
- 5.Antoniewicz M.R. Synthetic methylotrophy: strategies to assimilate methanol for growth and chemicals production. Curr Opin Biotechnol. 2019;59:165–174. doi: 10.1016/j.copbio.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Frazão C.J.R., Walther T. Syngas and methanol‐based biorefinery concepts. Chem Ing Tech. 2020;92(11):1680–1699. doi: 10.1002/cite.202000108. [DOI] [Google Scholar]
- 7.Dalena F., Senatore A., Marino A., Gordano A., Basile M., Basile A. In: Chapter 1 - methanol production and applications: an overview. Basile A., Dalena F., editors. Elsevier; Methanol: 2018. pp. 3–28. [Google Scholar]
- 8.The Methanol industry USA. 2022. https://www.methanol.org/the-methanol-industry/ Available from. : (accessed on 9th January 2023) [Google Scholar]
- 9.Whitaker W.B., Sandoval N.R., Bennett R.K., Fast A.G., Papoutsakis E.T. Synthetic methylotrophy: engineering the production of biofuels and chemicals based on the biology of aerobic methanol utilization. Curr Opin Biotechnol. 2015;33:165–175. doi: 10.1016/j.copbio.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Wang J., Qin R., Guo Y., Ma C., Wang X., Chen K., et al. Engineering the native methylotrophs for the bioconversion of methanol to value-added chemicals: current status and future perspectives. Green Chem. Eng. 2022 doi: 10.1016/j.gce.2022.10.005. [DOI] [Google Scholar]
- 11.Zhang W., Zhang T., Wu S., Wu M., Xin F., Dong W., et al. Guidance for engineering of synthetic methylotrophy based on methanol metabolism in methylotrophy. RSC Adv. 2017;7(7):4083–4091. doi: 10.1039/c6ra27038g. [DOI] [Google Scholar]
- 12.Williams P.A., Coates L., Mohammed F., Gill R., Erskine P.T., Coker A., et al. The atomic resolution structure of methanol dehydrogenase from Methylobacterium extorquens. Acta Crystallogr. 2005;61(Pt 1):75–79. doi: 10.1107/S0907444904026964. [DOI] [PubMed] [Google Scholar]
- 13.Good N.M., Moore R.S., Suriano C.J., Martinez-Gomez N.C. Contrasting in vitro and in vivo methanol oxidation activities of lanthanide-dependent alcohol dehydrogenases XoxF1 and ExaF from Methylobacterium extorquens AM1. Sci Rep. 2019;9(1):4248. doi: 10.1038/s41598-019-41043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le T.K., Lee Y.J., Han G.H., Yeom S.J. Methanol dehydrogenases as a key biocatalysts for synthetic methylotrophy. Front Bioeng Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.787791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernauer L., Radkohl A., Lehmayer L.G.K., Emmerstorfer-Augustin A. Komagataella phaffii as emerging model organism in fundamental research. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.607028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vorholt J.A., Marx C.J., Lidstorm M.E., Thauer R.K. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J Bacteriol. 2000;182(23):6645–6650. doi: 10.1128/JB.182.23.6645-6650.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smejkalova H., Erb T.J., Fuchs G. Methanol assimilation in Methylobacterium extorquens AM1: demonstration of all enzymes and their regulation. PLoS One. 2010;5(10) doi: 10.1371/journal.pone.0013001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yurimoto H., Kato N., Sakai Y. Assimilation, dissimilation, and detoxification of formaldehyde, a central metabolic intermediate of methylotrophic metabolism. Chem Rec. 2005;5(6):367–375. doi: 10.1002/tcr.20056. [DOI] [PubMed] [Google Scholar]
- 19.Hartner F.S., Glieder A. Regulation of methanol utilisation pathway genes in yeasts. Microb Cell Factories. 2006;5:39. doi: 10.1186/1475-2859-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dedysh S.N., Smirnova K.V., Khmelenina V.N., Suzina N.E., Liesack W., Trotsenko Y.A. Methylotrophic autotrophy in Beijerinckia mobilis. J Bacteriol. 2005;187(11):3884–3888. doi: 10.1128/JB.187.11.3884-3888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schrader J., Schilling M., Holtmann D., Sell D., Filho M.V., Marx A., et al. Methanol-based industrial biotechnology: current status and future perspectives of methylotrophic bacteria. Trends Biotechnol. 2009;27(2):107–115. doi: 10.1016/j.tibtech.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 22.Gould S.J., McCollum D., Spong A.P., Heyman J.A., Subramani S. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992;8(8):613–628. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
- 23.Bhataya A., Schmidt-Dannert C., Lee P.C. Metabolic engineering of Pichia pastoris X-33 for lycopene production. Process Biochem. 2009;44(10):1095–1102. doi: 10.1016/j.procbio.2009.05.012. [DOI] [Google Scholar]
- 24.Zhang X., Wang D., Duan Y., Zheng X., Lin Y., Liang S. Production of lycopene by metabolically engineered Pichia pastoris. Biosci Biotechnol Biochem. 2020;84(3):463–470. doi: 10.1080/09168451.2019.1693250. [DOI] [PubMed] [Google Scholar]
- 25.Wriessnegger T., Augustin P., Engleder M., Leitner E., Muller M., Kaluzna I., et al. Production of the sesquiterpenoid (+)-nootkatone by metabolic engineering of Pichia pastoris. Metab Eng. 2014;24:18–29. doi: 10.1016/j.ymben.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Liu X.B., Liu M., Tao X.Y., Zhang Z.X., Wang F.Q., Wei D.Z. Metabolic engineering of Pichia pastoris for the production of dammarenediol-II. J Biotechnol. 2015;216:47–55. doi: 10.1016/j.jbiotec.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Gao L., Cai M., Shen W., Xiao S., Zhou X., Zhang Y. Engineered fungal polyketide biosynthesis in Pichia pastoris: a potential excellent host for polyketide production. Microb Cell Factories. 2013;12(1):77. doi: 10.1186/1475-2859-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue Y., Kong C., Shen W., Bai C., Ren Y., Zhou X., et al. Methylotrophic yeast Pichia pastoris as a chassis organism for polyketide synthesis via the full citrinin biosynthetic pathway. J Biotechnol. 2017;242:64–72. doi: 10.1016/j.jbiotec.2016.11.031. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y., Tu X., Xu Q., Bai C., Kong C., Liu Q., et al. Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol. Metab Eng. 2018;45:189–199. doi: 10.1016/j.ymben.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 30.Cai P., Wu X., Deng J., Gao L., Shen Y., Yao L., et al. Methanol biotransformation toward high-level production of fatty acid derivatives by engineering the industrial yeast Pichia pastoris. Proc Natl Acad Sci USA. 2022;119(29) doi: 10.1073/pnas.2201711119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y., Zhai X., Yu W., Feng D., Shah A.A., Gao J., et al. Production of free fatty acids from various carbon sources by Ogataea polymorpha. Bioresour Bioprocess. 2022;9(1) doi: 10.1186/s40643-022-00566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao J., Li Y., Yu W., Zhou Y.A.-O. Rescuing yeast from cell death enables overproduction of fatty acids from sole methanol. Nat Metab. 2022;4(7):932–943. doi: 10.1038/s42255-022-00601-0. https://doi: 10.1038/s42255-022-00601-0 [DOI] [PubMed] [Google Scholar]
- 33.Meesapyodsuk D., Chen Y., Ng S.H., Chen J., Qiu X. Metabolic engineering of Pichia pastoris to produce ricinoleic acid, a hydroxy fatty acid of industrial importance. J Lipid Res. 2015;56(11):2102–2109. doi: 10.1194/jlr.M060954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim S.H., Roh K.H., Kim K.S., Kim H.U., Lee K.R., Kang H.C., et al. Coexpression of multiple genes reconstitutes two pathways of very long-chain polyunsaturated fatty acid biosynthesis in Pichia pastoris. Biotechnol Lett. 2014;36(9):1843–1851. doi: 10.1007/s10529-014-1550-1. [DOI] [PubMed] [Google Scholar]
- 35.Khongto B., Laoteng K., Tongta A. Fermentation process development of recombinant Hansenula polymorpha for gamma-linolenic acid production. J Microbiol Biotechnol. 2010;20(11):1555–1562. doi: 10.4014/jmb.1003.03004. [DOI] [PubMed] [Google Scholar]
- 36.Cai P., Li Y., Zhai X., Yao L., Ma X., Jia L., et al. Microbial synthesis of long-chain α-alkenes from methanol by engineering Pichia pastoris. Bioresour Bioprocess. 2022;9(1) doi: 10.1186/s40643-022-00551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marx H., Mecklenbrauker A., Gasser B., Sauer M., Mattanovich D. Directed gene copy number amplification in Pichia pastoris by vector integration into the ribosomal DNA locus. FEMS Yeast Res. 2009;9(8):1260–1270. doi: 10.1111/j.1567-1364.2009.00561.x. [DOI] [PubMed] [Google Scholar]
- 38.Yamada R., Ogura K., Kimoto Y., Ogino H. Toward the construction of a technology platform for chemicals production from methanol: D-lactic acid production from methanol by an engineered yeast Pichia pastoris. World J Microbiol Biotechnol. 2019;35(2):37. doi: 10.1007/s11274-019-2610-4. [DOI] [PubMed] [Google Scholar]
- 39.Miao L., Li Y., Zhu T. Metabolic engineering of methylotrophic Pichia pastoris for the production of β-alanine. Bioresour Bioprocess. 2021;8(1) doi: 10.1186/s40643-021-00444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang T., Ge C., Deng L., Tan T., Wang F. C4-dicarboxylic acid production by overexpressing the reductive TCA pathway. FEMS Microbiol Lett. 2015;362(9) doi: 10.1093/femsle/fnv052. [DOI] [PubMed] [Google Scholar]
- 41.Guo F., Dai Z., Peng W., Zhang S., Zhou J., Ma J., et al. Metabolic engineering of Pichia pastoris for malic acid production from methanol. Biotechnol Bioeng. 2021;118(1):357–371. doi: 10.1002/bit.27575. [DOI] [PubMed] [Google Scholar]
- 42.Brautaset T., Jakobsen O.M., Degnes K.F., Netzer R., Naerdal I., Krog A., et al. Bacillus methanolicus pyruvate carboxylase and homoserine dehydrogenase I and II and their roles for L-lysine production from methanol at 50 degrees C. Appl Microbiol Biotechnol. 2010;87(3):951–964. doi: 10.1007/s00253-010-2559-6. [DOI] [PubMed] [Google Scholar]
- 43.Sirirote P., Yamane T., Shimizu S. Production of L-serine from methanol and Glycine by resting cells of a methylotroph under automatically controlled conditions. J Ferment Technol. 1986;64(5):389–396. doi: 10.1016/0385-6380(86)90025-7. [DOI] [Google Scholar]
- 44.Naerdal I., Pfeifenschneider J., Brautaset T., Wendisch V.F. Methanol-based cadaverine production by genetically engineered Bacillus methanolicus strains. Microb Biotechnol. 2015;8(2):342–350. doi: 10.1111/1751-7915.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brito L.F., Irla M., Naerdal I., Le S.B., Delepine B., Heux S., et al. Evaluation of heterologous biosynthetic pathways for methanol-based 5-aminovalerate production by thermophilic Bacillus methanolicus. Front Bioeng Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.686319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irla M., Nærdal I., Brautaset T., Wendisch V.F. Methanol-based γ-aminobutyric acid (GABA) production by genetically engineered Bacillus methanolicus strains. Ind Crop Prod. 2017;106:12–20. doi: 10.1016/j.indcrop.2016.11.050. [DOI] [Google Scholar]
- 47.Suzuki T., Yamane T., Shimizu S. Kinetics and effect of nitrogen source feeding on production of poly-β-hydroxybutyric acid by fed-batch culture. Appl Microbiol Biotechnol. 1986;24(5):366–369. doi: 10.1007/BF00294591. [DOI] [Google Scholar]
- 48.Bourque D., Pomerleau Y., Groleau D. High-cell-density production of poly-β-hydroxybutyrate (PHB) from methanol by Methylobacterium extorquens: production of high-molecular-mass PHB. Appl Microbiol Biotechnol. 1995;44(3):367–376. doi: 10.1007/BF00169931. [DOI] [Google Scholar]
- 49.Suzuki T., Yamane T., Shimizu S. Mass production of poly-β-hydroxybutyric acid by fully automatic fed-batch culture of methylotroph. Appl Microbiol Biotechnol. 1986;23(5):322–329. doi: 10.1007/BF00257027. [DOI] [Google Scholar]
- 50.Yezza A., Fournier D., Halasz A., Hawari J. Production of polyhydroxyalkanoates from methanol by a new methylotrophic bacterium Methylobacterium sp. GW2. Appl Microbiol Biotechnol. 2006;73(1):211–218. doi: 10.1007/s00253-006-0458-7. [DOI] [PubMed] [Google Scholar]
- 51.Wu H., Fan Z., Jiang X., Chen J., Chen G.Q. Enhanced production of polyhydroxybutyrate by multiple dividing. E. coli. Microb Cell Fact. 2016;15(1):128. doi: 10.1186/s12934-016-0531-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korotkova N., Lidstrom M.E. Connection between poly-beta-hydroxybutyrate biosynthesis and growth on C(1) and C(2) compounds in the methylotroph Methylobacterium extorquens AM1. J Bacteriol. 2001;183(3):1038–1046. doi: 10.1128/JB.183.3.1038-1046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rohde M.-T., Tischer S., Harms H., Rohwerder T. Production of 2-hydroxyisobutyric acid from methanol by Methylobacterium extorquens AM1 expressing (R)-3-Hydroxybutyryl coenzyme A-isomerizing enzymes. Appl Environ Microbiol. 2017;83(3) doi: 10.1128/AEM.02622-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonntag F., Buchhaupt M., Schrader J. Thioesterases for ethylmalonyl-CoA pathway derived dicarboxylic acid production in Methylobacterium extorquens AM1. Appl Microbiol Biotechnol. 2014;98(10):4533–4544. doi: 10.1007/s00253-013-5456-y. [DOI] [PubMed] [Google Scholar]
- 55.Schada von Borzyskowski L., Sonntag F., Poschel L., Vorholt J.A., Schrader J., Erb T.J., et al. Replacing the ethylmalonyl-CoA pathway with the glyoxylate shunt provides metabolic flexibility in the central carbon metabolism of Methylobacterium extorquens AM1. ACS Synth Biol. 2018;7(1):86–97. doi: 10.1021/acssynbio.7b00229. [DOI] [PubMed] [Google Scholar]
- 56.Lim C.K., Villada J.C., Chalifour A., Duran M.F., Lu H., Lee P.K.H. Designing and engineering Methylorubrum extorquens AM1 for itaconic acid production. Front Microbiol. 2019;10:1027. doi: 10.3389/fmicb.2019.01027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drejer E.B., Chan D.T.C., Haupka C., Wendisch V.F., Brautaset T., Irla M. Methanol-based acetoin production by genetically engineered Bacillus methanolicus. Green Chem. 2020;22(3):788–802. doi: 10.1039/c9gc03950c. [DOI] [Google Scholar]
- 58.Yang Y.M., Chen W.J., Yang J., Zhou Y.M., Hu B., Zhang M., et al. Production of 3-hydroxypropionic acid in engineered Methylobacterium extorquens AM1 and its reassimilation through a reductive route. Microb Cell Factories. 2017;16(1):179. doi: 10.1186/s12934-017-0798-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu W.L., Cui J.Y., Cui L.Y., Liang W.F., Yang S., Zhang C., et al. Bioconversion of methanol to value-added mevalonate by engineered Methylobacterium extorquens AM1 containing an optimized mevalonate pathway. Appl Microbiol Biotechnol. 2016;100(5):2171–2182. doi: 10.1007/s00253-015-7078-z. [DOI] [PubMed] [Google Scholar]
- 60.Hu B., Lidstrom M.E. Metabolic engineering of Methylobacterium extorquens AM1 for 1-butanol production. Biotechnol Biofuels. 2014;7(1):156. doi: 10.1186/s13068-014-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang W.F., Cui L.Y., Cui J.Y., Yu K.W., Yang S., Wang T.M., et al. Biosensor-assisted transcriptional regulator engineering for Methylobacterium extorquens AM1 to improve mevalonate synthesis by increasing the acetyl-CoA supply. Metab Eng. 2017;39:159–168. doi: 10.1016/j.ymben.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 62.Vuilleumier S., Chistoserdova L., Lee M.C., Bringel F., Lajus A., Zhou Y., et al. Methylobacterium genome sequences: a reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources. PLoS One. 2009;4(5) doi: 10.1371/journal.pone.0005584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin V.J.J., Pitera D.J., Withers S.T., Newman J.D., Keasling J.D. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol. 2003;21(7):796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 64.Sonntag F., Kroner C., Lubuta P., Peyraud R., Horst A., Buchhaupt M., et al. Engineering Methylobacterium extorquens for de novo synthesis of the sesquiterpenoid alpha-humulene from methanol. Metab Eng. 2015;32:82–94. doi: 10.1016/j.ymben.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 65.Quynh Le H.T., Anh Mai D.H., Na J.G., Lee E.Y. Development of Methylorubrum extorquens AM1 as a promising platform strain for enhanced violacein production from co-utilization of methanol and acetate. Metab Eng. 2022;72:150–160. doi: 10.1016/j.ymben.2022.03.008. [DOI] [PubMed] [Google Scholar]
- 66.Manfrao-Netto J.H.C., Gomes A.M.V., Parachin N.S. Advances in using Hansenula polymorpha as chassis for recombinant protein production. Front Bioeng Biotechnol. 2019;7:94. doi: 10.3389/fbioe.2019.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paulová L., Hyka P., Branská B., Melzoch K., Kovar K. Use of a mixture of glucose and methanol as substrates for the production of recombinant trypsinogen in continuous cultures with Pichia pastoris Mut+ J Biotechnol. 2012;157(1):180–188. doi: 10.1016/j.jbiotec.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 68.Cereghino G., Cereghino J., Ilgen C., Cregg J. Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr Opin Biotechnol. 2002;13(4):329–332. doi: 10.1016/S0958166902003300. [DOI] [PubMed] [Google Scholar]
- 69.Rahimi A., Hosseini S.N., Karimi A., Aghdasinia H., Arabi Mianroodi R. Enhancing the efficiency of recombinant hepatitis B surface antigen production in Pichia pastoris by employing continuous fermentation. Biochem Eng J. 2019;141:112–119. doi: 10.1016/j.bej.2018.10.019. [DOI] [Google Scholar]
- 70.Ramon R., Ferrer P., Valero F. Sorbitol co-feeding reduces metabolic burden caused by the overexpression of a Rhizopus oryzae lipase in Pichia pastoris. J Biotechnol. 2007;130(1):39–46. doi: 10.1016/j.jbiotec.2007.02.025. [DOI] [PubMed] [Google Scholar]
- 71.Cayetano-Cruz M., Pérez de los Santos A.I., García-Huante Y., Santiago-Hernández A., Pavón-Orozco P., López y López V.E., et al. High level expression of a recombinant xylanase by Pichia pastoris cultured in a bioreactor with methanol as the sole carbon source: purification and biochemical characterization of the enzyme. Biochem Eng J. 2016;112:161–169. doi: 10.1016/j.bej.2016.04.014. [DOI] [Google Scholar]
- 72.Belanger L., Figueira M.M., Bourque D., Morel L., Beland M., Laramee L., et al. Production of heterologous protein by Methylobacterium extorquens in high cell density fermentation. FEMS Microbiol Lett. 2004;231(2):197–204. doi: 10.1016/S0378-1097(03)00956-X. [DOI] [PubMed] [Google Scholar]
- 73.Gutierrez J., Bourque D., Criado R., Choi Y.J., Cintas L.M., Hernandez P.E., et al. Heterologous extracellular production of enterocin P from Enterococcus faecium P13 in the methylotrophic bacterium Methylobacterium extorquens. FEMS Microbiol Lett. 2005;248(1):125–131. doi: 10.1016/j.femsle.2005.05.029. [DOI] [PubMed] [Google Scholar]
- 74.Peyraud R., Kiefer P., Christen P., Massou S., Portais J.-C., Vorholt J.A. Demonstration of the ethylmalonyl-CoA pathway by using 13C metabolomics. Proc Natl Acad Sci U S A. 2009;106(12):4846–4851. doi: 10.1073/pnas.0810932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bennett R.K., Gonzalez J.E., Whitaker W.B., Antoniewicz M.R., Papoutsakis E.T. Expression of heterologous non-oxidative pentose phosphate pathway from Bacillus methanolicus and phosphoglucose isomerase deletion improves methanol assimilation and metabolite production by a synthetic Escherichia coli methylotroph. Metab Eng. 2018;45:75–85. doi: 10.1016/j.ymben.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 76.Woolston B.M., Roth T., Kohale I., Liu D.R., Stephanopoulos G. Development of a formaldehyde biosensor with application to synthetic methylotrophy. Biotechnol Bioeng. 2018;115(1):206–215. doi: 10.1002/bit.26455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y.H. Substrate channeling and enzyme complexes for biotechnological applications. Biotechnol Adv. 2011;29(6):715–725. doi: 10.1016/j.biotechadv.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 78.Price J.V., Chen L., Whitaker W.B., Papoutsakis E., Chen W. Scaffoldless engineered enzyme assembly for enhanced methanol utilization. Proc Natl Acad Sci U S A. 2016;113(45):12691–12696. doi: 10.1073/pnas.1601797113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu T., Zhao T., Bankefa O.E., Li Y. Engineering unnatural methylotrophic cell factories for methanol-based biomanufacturing: challenges and opportunities. Biotechnol Adv. 2020;39 doi: 10.1016/j.biotechadv.2019.107467. [DOI] [PubMed] [Google Scholar]
- 80.Jorda J., de Jesus S.S., Peltier S., Ferrer P., Albiol J. Metabolic flux analysis of recombinant Pichia pastoris growing on different glycerol/methanol mixtures by iterative fitting of NMR-derived (13)C-labelling data from proteinogenic amino acids. Nat Biotechnol. 2014;31(1):120–132. doi: 10.1016/j.nbt.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 81.Vanz A., Lünsdorf H., Adnan A., Nimtz M., Gurramkonda C., Khanna N., et al. Physiological response of Pichia pastoris GS115 to methanol-induced high level production of the Hepatitis B surface antigen: catabolic adaptation, stress responses, and autophagic processes. Microb Cell Factories. 2012;11(1):103. doi: 10.1186/1475-2859-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chistoserdova L., Crowther G.J., Vorholt J.A., Skovran E., Portais J.C., Lidstrom M.E. Identification of a fourth formate dehydrogenase in Methylobacterium extorquens AM1 and confirmation of the essential role of formate oxidation in methylotrophy. J Bacteriol. 2007;189(24):9076–9081. doi: 10.1128/JB.01229-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Muller J.E., Litsanov B., Bortfeld-Miller M., Trachsel C., Grossmann J., Brautaset T., et al. Proteomic analysis of the thermophilic methylotroph Bacillus methanolicus MGA3. Proteomics. 2014;14(6):725–737. doi: 10.1002/pmic.201300515. [DOI] [PubMed] [Google Scholar]
- 84.Gassler T., Sauer M., Gasser B., Egermeier M., Troyer C., Causon T., et al. The industrial yeast Pichia pastoris is converted from a heterotroph into an autotroph capable of growth on CO(2) Nat Biotechnol. 2020;38(2):210–216. doi: 10.1038/s41587-019-0363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schada von Borzyskowski L., Carrillo M., Leupold S., Glatter T., Kiefer P., Weishaupt R., et al. An engineered Calvin-Benson-Bassham cycle for carbon dioxide fixation in Methylobacterium extorquens AM1. Metab Eng. 2018;47:423–433. doi: 10.1016/j.ymben.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 86.Schroer K., Peter Luef K., Stefan Hartner F., Glieder A., Pscheidt B. Engineering the Pichia pastoris methanol oxidation pathway for improved NADH regeneration during whole-cell biotransformation. Metab Eng. 2010;12(1):8–17. doi: 10.1016/j.ymben.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 87.Yuan X.J., Chen W.J., Ma Z.X., Yuan Q.Q., Zhang M., He L., et al. Rewiring the native methanol assimilation metabolism by incorporating the heterologous ribulose monophosphate cycle into Methylorubrum extorquens. Metab Eng. 2021;64:95–110. doi: 10.1016/j.ymben.2021.01.009. [DOI] [PubMed] [Google Scholar]
- 88.Krog A., Heggeset T.M., Muller J.E., Kupper C.E., Schneider O., Vorholt J.A., et al. Methylotrophic Bacillus methanolicus encodes two chromosomal and one plasmid born NAD+ dependent methanol dehydrogenase paralogs with different catalytic and biochemical properties. PLoS One. 2013;8(3) doi: 10.1371/journal.pone.0059188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zavec D., Troyer C., Maresch D., Altmann F., Hann S., Gasser B., et al. Beyond alcohol oxidase: the methylotrophic yeast Komagataella phaffii utilizes methanol also with its native alcohol dehydrogenase Adh2. FEMS Yeast Res. 2021;21(2) doi: 10.1093/femsyr/foab009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yi J., Lee J., Sung B.H., Kang D.K., Lim G., Bae J.H., et al. Development of Bacillus methanolicus methanol dehydrogenase with improved formaldehyde reduction activity. Sci Rep. 2018;8(1) doi: 10.1038/s41598-018-31001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu T.Y., Chen C.T., Liu J.T., Bogorad I.W., Damoiseaux R., Liao J.C. Characterization and evolution of an activator-independent methanol dehydrogenase from Cupriavidus necator N-1. Appl Microbiol Biotechnol. 2016;100(11):4969–4983. doi: 10.1007/s00253-016-7320-3. [DOI] [PubMed] [Google Scholar]
- 92.Ochsner A.M., Muller J.E., Mora C.A., Vorholt J.A. In vitro activation of NAD-dependent alcohol dehydrogenases by Nudix hydrolases is more widespread than assumed. FEBS Lett. 2014;588(17):2993–2999. doi: 10.1016/j.febslet.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 93.Park S.Y., Eun H., Lee M.H., Lee S.Y. Metabolic engineering of Escherichia coli with electron channelling for the production of natural products. Nat Catal. 2022;5(8):726–737. doi: 10.1038/s41929-022-00820-4. [DOI] [Google Scholar]
- 94.Guo W., Li D., He R., Wu M., Chen W., Gao F., et al. Synthesizing value-added products from methane by a new Methylomonas. J Appl Microbiol. 2017;123(5):1214–1227. doi: 10.1111/jam.13581. [DOI] [PubMed] [Google Scholar]
- 95.Jakobsen O.M., Benichou A., Flickinger M.C., Valla S., Ellingsen T.E., Brautaset T. Upregulated transcription of plasmid and chromosomal ribulose monophosphate pathway genes is critical for methanol assimilation rate and methanol tolerance in the methylotrophic bacterium Bacillus methanolicus. J Bacteriol. 2006;188(8):3063–3072. doi: 10.1128/JB.188.8.3063-3072.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wakayama K., Yamaguchi S., Takeuchi A., Mizumura T., Ozawa S., Tomizuka N., et al. Regulation of intracellular formaldehyde toxicity during methanol metabolism of the methylotrophic yeast Pichia methanolica. J Biosci Bioeng. 2016;122(5):545–549. doi: 10.1016/j.jbiosc.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 97.Nguyen A.D., Hwang I.Y., Chan J.Y., Lee E.Y. Reconstruction of methanol and formate metabolic pathway in non-native host for biosynthesis of chemicals and biofuels. Biotechnol Bioproc Eng. 2016;21(4):477–482. doi: 10.1007/s12257-016-0301-7. [DOI] [Google Scholar]
- 98.Rohlhill J., Sandoval N.R., Papoutsakis E.T. Sort-seq approach to engineering a formaldehyde-inducible promoter for dynamically regulated Escherichia coli growth on methanol. ACS Synth Biol. 2017;6(8):1584–1595. doi: 10.1021/acssynbio.7b00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bennett R.K., Gregory G.J., Gonzalez J.E., Har J.R.G., Antoniewicz M.R., Papoutsakis E.T. Improving the methanol tolerance of an Escherichia coli methylotroph via adaptive laboratory evolution enhances synthetic methanol utilization. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.638426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hennig G., Haupka C., Brito L.F., Ruckert C., Cahoreau E., Heux S., et al. Methanol-essential growth of Corynebacterium glutamicum: adaptive laboratory evolution overcomes limitation due to methanethiol assimilation pathway. Int J Mol Sci. 2020;21(10) doi: 10.3390/ijms21103617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Witthoff S., Schmitz K., Niedenfuhr S., Noh K., Noack S., Bott M., et al. Metabolic engineering of Corynebacterium glutamicum for methanol metabolism. Appl Environ Microbiol. 2015;81(6):2215–2225. doi: 10.1128/AEM.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gao B., Zhao N., Deng J., Gu Y., Jia S., Hou Y., et al. Constructing a methanol-dependent Bacillus subtilis by engineering the methanol metabolism. J Biotechnol. 2022;343:128–137. doi: 10.1016/j.jbiotec.2021.12.005. [DOI] [PubMed] [Google Scholar]
- 103.Muller J.E.N., Meyer F., Litsanov B., Kiefer P., Potthoff E., Heux S., et al. Engineering Escherichia coli for methanol conversion. Metab Eng. 2015;28:190–201. doi: 10.1016/j.ymben.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 104.Bennett B.D., Kimball E.H., Gao M., Osterhout R., Van Dien S.J., Rabinowitz J.D. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5(8):593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Woolston B.M., King J.R., Reiter M., Van Hove B., Stephanopoulos G. Improving formaldehyde consumption drives methanol assimilation in engineered E. coli. Nat Commun. 2018;9(1):2387. doi: 10.1038/s41467-018-04795-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meyer F., Keller P., Hartl J., Groninger O.G., Kiefer P., Vorholt J.A. Methanol-essential growth of Escherichia coli. Nat Commun. 2018;9(1):1508. doi: 10.1038/s41467-018-03937-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.He H., Hoper R., Dodenhoft M., Marliere P., Bar-Even A. An optimized methanol assimilation pathway relying on promiscuous formaldehyde-condensing aldolases in E. coli. Metab Eng. 2020;60:1–13. doi: 10.1016/j.ymben.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 108.Dai Z., Gu H., Zhang S., Xin F., Zhang W., Dong W., et al. Metabolic construction strategies for direct methanol utilization in Saccharomyces cerevisiae. Bioresour Technol. 2017;245(Pt B):1407–1412. doi: 10.1016/j.biortech.2017.05.100. [DOI] [PubMed] [Google Scholar]
- 109.Kim S., Lindner S.N., Aslan S., Yishai O., Wenk S., Schann K., et al. Growth of E. coli on formate and methanol via the reductive glycine pathway. Nat Chem Biol. 2020;16(5):538–545. doi: 10.1038/s41589-020-0473-5. [DOI] [PubMed] [Google Scholar]
- 110.Chen F.Y., Jung H.W., Tsuei C.Y., Liao J.C. Converting Escherichia coli to a synthetic methylotroph growing solely on methanol. Cell. 2020;182(4):933–946 e14. doi: 10.1016/j.cell.2020.07.010. [DOI] [PubMed] [Google Scholar]
- 111.Gleizer S., Ben-Nissan R., Bar-On Y.M., Antonovsky N., Noor E., Zohar Y., et al. Conversion of Escherichia coli to generate all biomass carbon from CO(2) Cell. 2019;179(6):1255–1263 e12. doi: 10.1016/j.cell.2019.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bang J., Hwang C.H., Ahn J.H., Lee J.A., Lee S.Y. Escherichia coli is engineered to grow on CO(2) and formic acid. Nat Microbiol. 2020;5(12):1459–1463. doi: 10.1038/s41564-020-00793-9. [DOI] [PubMed] [Google Scholar]
- 113.Zhang W., Zhang T., Song M., Dai Z., Zhang S., Xin F., et al. Metabolic engineering of Escherichia coli for high yield production of succinic acid driven by methanol. ACS Synth Biol. 2018;7(12):2803–2811. doi: 10.1021/acssynbio.8b00109. [DOI] [PubMed] [Google Scholar]
- 114.Guo F., Wu M., Zhang S., Feng Y., Jiang Y., Jiang W., et al. Improved succinic acid production through the reconstruction of methanol dissimilation in Escherichia coli. Bioresour Bioprocess. 2022;9(1) doi: 10.1186/s40643-022-00547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang X., Wang X., Lu X., Ma C., Chen K., Ouyang P. Methanol fermentation increases the production of NAD(P)H-dependent chemicals in synthetic methylotrophic Escherichia coli. Biotechnol Biofuels. 2019;12:17. doi: 10.1186/s13068-019-1356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Guo Q., Liu M.M., Zheng S.H., Zheng L.J., Ma Q., Cheng Y.K., et al. Methanol-dependent carbon fixation for irreversible synthesis of d-allulose from d-xylose by engineered Escherichia coli. J Agric Food Chem. 2022;70(44):14255–14263. doi: 10.1021/acs.jafc.2c06616. [DOI] [PubMed] [Google Scholar]
- 117.Yu H., Liao J.C. A modified serine cycle in Escherichia coli coverts methanol and CO(2) to two-carbon compounds. Nat Commun. 2018;9(1):3992. doi: 10.1038/s41467-018-06496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tuyishime P., Wang Y., Fan L., Zhang Q., Li Q., Zheng P., et al. Engineering Corynebacterium glutamicum for methanol-dependent growth and glutamate production. Metab Eng. 2018;49:220–231. doi: 10.1016/j.ymben.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 119.Lessmeier L., Pfeifenschneider J., Carnicer M., Heux S., Portais J.C., Wendisch V.F. Production of carbon-13-labeled cadaverine by engineered Corynebacterium glutamicum using carbon-13-labeled methanol as co-substrate. Appl Microbiol Biotechnol. 2015;99(23):10163–10176. doi: 10.1007/s00253-015-6906-5. [DOI] [PubMed] [Google Scholar]
- 120.Gonzalez C.F., Proudfoot M., Brown G., Korniyenko Y., Mori H., Savchenko A.V., et al. Molecular basis of formaldehyde detoxification. Characterization of two S-formylglutathione hydrolases from Escherichia coli, FrmB and YeiG. J Biol Chem. 2006;281(20):14514–14522. doi: 10.1074/jbc.M600996200. [DOI] [PubMed] [Google Scholar]
- 121.Bogorad I.W., Chen C.T., Theisen M.K., Wu T.Y., Schlenz A.R., Lam A.T., et al. Building carbon-carbon bonds using a biocatalytic methanol condensation cycle. Proc Natl Acad Sci U S A. 2014;111(45):15928–15933. doi: 10.1073/pnas.1413470111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wegat V., Fabarius J.T., Sieber V. Synthetic methylotrophic yeasts for the sustainable fuel and chemical production. Biotechnol Biofuels Bioprod. 2022;15(1):113. doi: 10.1186/s13068-022-02210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang X., Yuan Q., Luo H., Li F., Mao Y., Zhao X., et al. Systematic design and in vitro validation of novel one-carbon assimilation pathways. Metab Eng. 2019;56:142–153. doi: 10.1016/j.ymben.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 124.Zhan C., Li X., Yang Y., Nielsen J., Bai Z., Chen Y. Strategies and challenges with the microbial conversion of methanol to high-value chemicals. Biotechnol Bioeng. 2021;118(10):3655–3668. doi: 10.1002/bit.27862. [DOI] [PubMed] [Google Scholar]
- 125.Alber B.E. Biotechnological potential of the ethylmalonyl-CoA pathway. Appl Microbiol Biotechnol. 2011;89(1):17–25. doi: 10.1007/s00253-010-2873-z. [DOI] [PubMed] [Google Scholar]
- 126.Irla M., Wendisch V.F. Efficient cell factories for the production of N-methylated amino acids and for methanol-based amino acid production. Microb Biotechnol. 2022;15(8):2145–2159. doi: 10.1111/1751-7915.14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Muller J.E., Heggeset T.M., Wendisch V.F., Vorholt J.A., Brautaset T. Methylotrophy in the thermophilic Bacillus methanolicus, basic insights and application for commodity production from methanol. Appl Microbiol Biotechnol. 2015;99(2):535–551. doi: 10.1007/s00253-014-6224-3. [DOI] [PubMed] [Google Scholar]
- 128.Guo F., Qiao Y., Xin F., Zhang W., Jiang M. Bioconversion of C1 feedstocks for chemical production using Pichia pastoris. Trends Biotechnol. 2023 doi: 10.1016/j.tibtech.2023.03.006. [DOI] [PubMed] [Google Scholar]
- 129.Nie Y., Huang M., Lu J., Qian J., Lin W., Chu J., et al. Impacts of high beta-galactosidase expression on central metabolism of recombinant Pichia pastoris GS115 using glucose as sole carbon source via (13)C metabolic flux analysis. J Biotechnol. 2014;187:124–134. doi: 10.1016/j.jbiotec.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 130.Kumokita R., Bamba T., Inokuma K., Yoshida T., Ito Y., Kondo A., et al. Construction of an l-tyrosine chassis in Pichia pastoris enhances aromatic secondary metabolite production from glycerol. ACS Synth Biol. 2022;11(6):2098–2107. doi: 10.1021/acssynbio.2c00047. [DOI] [PubMed] [Google Scholar]
- 131.Chou A., Lee S.H., Zhu F., Clomburg J.M., Gonzalez R. An orthogonal metabolic framework for one-carbon utilization. Nat Metab. 2021;3(10):1385–1399. doi: 10.1038/s42255-021-00453-0. [DOI] [PubMed] [Google Scholar]
- 132.He H., Edlich-Muth C., Lindner S.N., Bar-Even A. Ribulose monophosphate shunt provides nearly all biomass and energy required for growth of E. coli. ACS Synth Biol. 2018;7(6):1601–1611. doi: 10.1021/acssynbio.8b00093. [DOI] [PubMed] [Google Scholar]
- 133.Wang C., Ren J., Zhou L., Li Z., Chen L., Zeng A.P. An aldolase-catalyzed new metabolic pathway for the assimilation of formaldehyde and methanol to synthesize 2-Keto-4-hydroxybutyrate and 1,3-propanediol in Escherichia coli. ACS Synth Biol. 2019;8(11):2483–2493. doi: 10.1021/acssynbio.9b00102. [DOI] [PubMed] [Google Scholar]
- 134.Ubiyvovk V.M., Ananin V.M., Malyshev A.Y., Kang H.A., Sibirny A.A. Optimization of glutathione production in batch and fed-batch cultures by the wild-type and recombinant strains of the methylotrophic yeast Hansenula polymorpha DL-1. BMC Biotechnol. 2011;11(1):8. doi: 10.1186/1472-6750-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang J., Zhang C.T., Yuan X.J., Zhang M., Mo X.H., Tan L.L., et al. Metabolic engineering of Methylobacterium extorquens AM1 for the production of butadiene precursor. Microb Cell Factories. 2018;17(1):194. doi: 10.1186/s12934-018-1042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Orita I., Nishikawa K., Nakamura S., Fukui T. Biosynthesis of polyhydroxyalkanoate copolymers from methanol by Methylobacterium extorquens AM1 and the engineered strains under cobalt-deficient conditions. Appl Microbiol Biotechnol. 2014;98(8):3715–3725. doi: 10.1007/s00253-013-5490-9. [DOI] [PubMed] [Google Scholar]
- 137.Chen C.T., Chen F.Y., Bogorad I.W., Wu T.Y., Zhang R., Lee A.S., et al. Synthetic methanol auxotrophy of Escherichia coli for methanol-dependent growth and production. Metab Eng. 2018;49:257–266. doi: 10.1016/j.ymben.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 138.Bennett R.K., Dillon M., Gerald Har J.R., Agee A., von Hagel B., Rohlhill J., et al. Engineering Escherichia coli for methanol-dependent growth on glucose for metabolite production. Metab Eng. 2020;60:45–55. doi: 10.1016/j.ymben.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 139.Whitaker W.B., Jones J.A., Bennett R.K., Gonzalez J.E., Vernacchio V.R., Collins S.M., et al. Engineering the biological conversion of methanol to specialty chemicals in Escherichia coli. Metab Eng. 2017;39:49–59. doi: 10.1016/j.ymben.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 140.Mellitzer A., Ruth C., Gustafsson C., Welch M., Birner-Grunberger R., Weis R., et al. Synergistic modular promoter and gene optimization to push cellulase secretion by Pichia pastoris beyond existing benchmarks. J Biotechnol. 2014;191:187–195. doi: 10.1016/j.jbiotec.2014.08.035. [DOI] [PubMed] [Google Scholar]
- 141.Zhu W., Gong G., Pan J., Han S., Zhang W., Hu Y., et al. High level expression and purification of recombinant human serum albumin in Pichia pastoris. Protein Expr Purif. 2018;147:61–68. doi: 10.1016/j.pep.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 142.Moser J.W., Prielhofer R., Gerner S.M., Graf A.B., Wilson I.B., Mattanovich D., et al. Implications of evolutionary engineering for growth and recombinant protein production in methanol-based growth media in the yeast Pichia pastoris. Microb Cell Factories. 2017;16(1):49. doi: 10.1186/s12934-017-0661-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang J., Yang Y., Teng D., Tian Z., Wang S., Wang J. Expression of plectasin in Pichia pastoris and its characterization as a new antimicrobial peptide against Staphyloccocus and Streptococcus. Protein Expr Purif. 2011;78(2):189–196. doi: 10.1016/j.pep.2011.04.014. [DOI] [PubMed] [Google Scholar]