Abstract
The Catabolism of tryptophan modulates the immunosuppressive microenvironment in tumors. KYNU (Kynureninase) served as an enzyme involved in amino acid tryptophan catabolism through the kynurenine pathway. The molecular and clinical characteristics of KYNU remain unclear, and the impact of KYNU on the immune response has not been reported until now. We analyzed large-scale transcriptome data and related clinical information on 2994 breast cancer patients to characterize KYNU's role in breast cancer. There was a strong correlation between KYNU expression and major molecular and clinical characteristics, and it was more likely to be overexpressed in patients with higher malignancy subtypes. Inflammatory and immune responses were strongly correlated with KYNU. KYNU was also associated with immune modulators at the pan-cancer level, particularly its potential synergistic role with other immune checkpoints in breast cancer. KYNU expression was linked to the malignancy grade of breast cancer and predicted poorer outcomes. Tryptophan catabolism might play an important role in modulating the tumor immune microenvironment through KYNU. More significantly, KYNU might synergize with CTLA4, PDL2, IDO1, and other immune checkpoints, contributing to the development of combination cancer immunotherapy targeting KYNU and other checkpoints. As far as we are aware, this is the biggest and most thorough study describing KYNU's role in breast cancer.
Keywords: Cancer immunotherapy, KYNU, Immune response, Inflammatory activity, Breast cancer
1. Introduction
Globally, breast cancer (BC) is the most common cancer type among women [1,2]. In the past few decades, BC treatment strategies evolved rapidly, and including mature systemic therapies, surgery, chemotherapy, radiotherapy, endocrine therapy, and targeted therapy, have improved survival [3]. In spite of variable therapeutic methods, BC remains the leading cause of cancer-related mortality among females [1]. Many studies reported that catabolism of the amino acid tryptophan (Trp) was involved in regulating anti-tumor immune response through the expression of the rate-limiting enzymes indoleamine 2,3-dioxygenase 1 (IDO1), IDO2, and tryptophan 2,3-dioxygenase (TDO2) through Kynurenine Pathway (KP) [4,5,6,7]. The oxidation of Trp could be catalyzed by these enzymes and converted by formamidases to Kynurenine (Kyn). Kynureninase (KYNU) is located on 2q22.2, encoding Kynurenine enzyme, which is a pyridoxal-5′-phosphate (PLP)3-dependent enzyme [8]. KYNU could catalyze hydrolytic cleavage of l-kynurenine to l-alanine and anthranilic acid [9]. As reported, Plasma Kyn concentrations and Kyn/Trp ratios are often elevated in advanced-stage cancer patients and are associated with poor outcomes [10]. Furthermore, Tumor microenvironment expression of IDO1 and TDO is associated with immunosuppression [11,12]. As well as IDO1 and TDO2, KYNU is a key enzyme in the metabolism of Trp and Kyn and might be an important cooperator of immune suppression.
It has been reported that KYNU is related to multiple diseases, including immune system diseases [13,14,15], central nervous system diseases [16], psychiatric diseases [17], adrenal diseases [18], and variable types of cancers. There are several studies exploring the relationship between KYNU expression and cancers. Some studies suggested KYNU is a cancer suppression gene. It was indicated that KYNU expression was down-regulated in high aggressive osteosarcoma cell lines [19] and was negatively associated with survival of lung adenocarcinoma [20]. However, some studies implied the opposite results in colon cancer and cutaneous squamous cell carcinoma [21,22]. In breast cancer, recent studies reported that in AhR depletion BC cells, whose tumorigenic properties were decreased, KYNU expression was downregulated [23]. Another study indicated KYNU could suppress BC cell proliferation, tumor growth and development [24]. Another one indicated KYNU could upderpin CD44-promoted breast tumor cell invasion [25]. KYNU's specific expression pattern and potential impact on other immune cell populations, as well as immune modulators in BC, is unknown.
The present study systematically examined the potential role of KYNU in inducing immune responses and inflammatory activities, as well as its possible relationship to immune modulators. Molecularly and clinically, this is the first integrative study to characterize KYNU expression in breast cancer.
2. Materials and methods
2.1. Data
The TCGA dataset was downloaded using GDCRNATools (access date: January 16, 2022) [26]. After normalizing counts data using TMM method in edgeR [27], the voom method in limma package was used to transform them [28]. In more than half of the samples, genes with a cpm greater than 1 were retained. We collected standardized survival data from the TCGA Pan-Cancer Clinical Data Resource (TCGA-CDR) [29]. An online database known as cBioPortal was used to download the METABRIC dataset, which contains a total of 1904 cases [30] (access date: January 20, 2022).
2.2. Bioinformatics analysis
The clusterProfiler package was used to analyze the Gene Ontology (GO) functional enrichment of genes associated with KYNU [31]. Significant GO terms were those with an adjusted P-value lower than 0.05. The Immunology Database and Analysis Portal (ImmPort) database (https://www.immport.org/home) was used to download genes related to immunity [32]. To estimate the abundance of GO gene sets associated with immune functions and inflammation, gene set variation analysis (GSVA) was performed [33,34]. TISIDB data (http://cis.hku.hk/TISIDB/) were used to investigate the correlation between KYNU and immune modulators at pan-cancer levels [35], which is an integrated resource for interactions between tumors and the immune system. An analysis of correlations between KYNU expression, metagenes, and T cell immune functions was conducted using Spearman coefficients.
2.3. Statistical analysis
In order to calculate the correlations between continuous variables, Spearman correlation analyses were performed. Pearson's Chi-squared test, Student t-test, or one-way ANOVA were used to assess differences between groups. We used the statistical software R (version 3.6.0; http://www.r-project.org/) for all statistical tests. Other statistical calculations and graphs were carried out with the help of several packages, including ggplot2 [36], pheatmap, pROC [37], circlize [38] and corrgram [39]. Statistics tests were conducted with a two-sided P-value of less than 0.05 to indicate that the results were statistically significant.
3. Results
3.1. Breast cancer clinical and molecular characteristics associated with KYNU expression
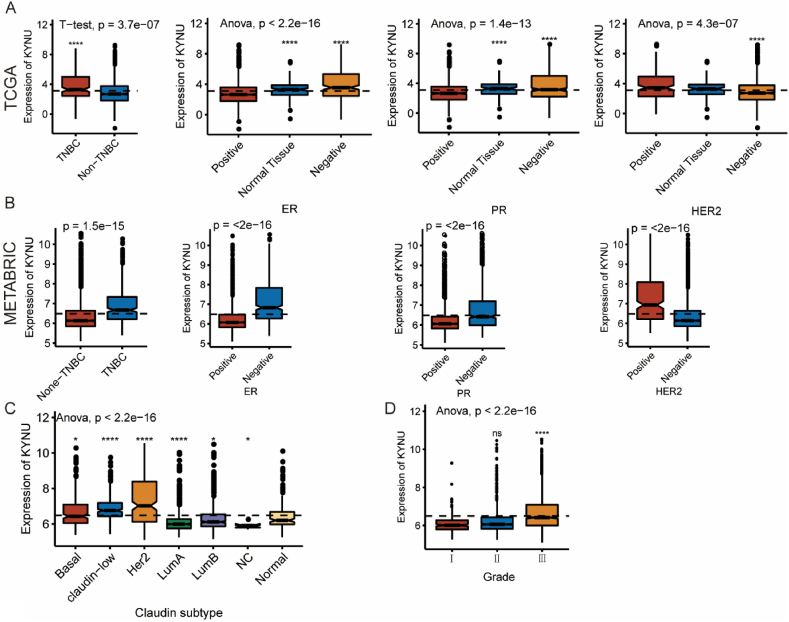
To investigate the association between KYNU expression and clinical characteristics of breast cancer patients. Patients were divided into low- and high-expression groups according to median cutoffs. Table 1, Table 2 show the association between KYNU expression and clinical characteristics in both the TCGA and METABRIC cohorts. KYNU was associated with estrogen receptor (ER) status, progestogen receptor (PR) status, HER2 status, age, AJCC stage and tumor grade. In the TCGA database, we further explored the expression pattern of KYNU across several molecular and clinical characteristics (Fig. 1A). Accordingly, KYNU was enriched in triple-negative breast cancer (TNBC) patients compared to non-TNBC patients, and it was upregulated in tumor tissues, ER-negative and PR-negative groups, while downregulated in HER2-negavtive group. Importantly, in METABRIC database these results were also verified with the result that KYNU was enriched in TNBC group, ER-negative group, PR-negative group, and HER2-positive group (Fig. 1B). We observed KYNU was also enriched in basal-like and HER2-enriched subtype and higher tumor grade (Fig. 1C and D). Also, these findings were validated with independent microarray datasets from the GOBO database (n = 1881) [40], and a correlation analysis revealed a strong correlation between KYNU expression and immune response gene module, suggesting KYNU might play an important role in immune functions (Fig. 1).
Table 1.
Association between KYNU mRNA expression and clinicopathologic characteristics in TCGA cohort.
| Expression |
P-value | |||
|---|---|---|---|---|
| Total (n = 1090) | KYNU high (n = 545) | KYNU low (n = 545) | ||
| Age (years) | ||||
| ≥55 | 517 (47.4%) | 264 (48.4%) | 253 (46.4%) | 0.544 |
| <55 | 573 (52.6%) | 281 (51.6%) | 292 (53.6%) | |
| T stage | ||||
| T1 | 279 (25.6%) | 149 (27.3%) | 130 (23.9%) | 0.078 |
| T2 | 631 (57.9%) | 320 (58.7%) | 311 (57.1%) | |
| T3 | 137 (12.6%) | 55 (10.1%) | 82 (15.0%) | |
| T4 | 40 (3.7%) | 19 (3.5%) | 21 (3.9%) | |
| Unknown | 3 (0.3%) | 2 (0.4%) | 1 (0.2%) | |
| N stage | ||||
| N0 | 514 (47.2%) | 255 (46.8%) | 259 (47.5%) | 0.563 |
| N1 | 360 (33.0%) | 177 (32.5%) | 183 (33.6%) | |
| N2 | 120 (11.0%) | 61 (11.2%) | 59 (10.8%) | |
| N3 | 76 (7.0%) | 44 (8.1%) | 32 (5.9%) | |
| Unknown | 20 (1.8%) | 8 (1.5%) | 12 (2.2%) | |
| M stage | ||||
| M0 | 907 (83.2%) | 460 (84.4%) | 447 (82.0%) | 0.492 |
| M1 | 22 (2.0%) | 9 (1.7%) | 13 (2.4%) | |
| Unknown | 161 (14.8%) | 76 (13.9%) | 85 (15.6%) | |
| AJCC stage | ||||
| I | 181 (16.6%) | 95 (17.4%) | 86 (15.8%) | 0.849 |
| II | 621 (57.0%) | 309 (56.7%) | 312 (57.2%) | |
| III | 250 (22.9%) | 122 (22.4%) | 128 (23.5%) | |
| IV | 20 (1.8%) | 9 (1.7%) | 11 (2.0%) | |
| Unknown | 18 (1.7%) | 10 (1.8%) | 8 (1.5%) | |
| ER status | ||||
| Negative | 236 (21.7%) | 161 (29.5%) | 75 (13.8%) | <0.001 |
| Positive | 803 (73.7%) | 357 (65.5%) | 446 (81.8%) | |
| Unknown | 51 (4.7%) | 27 (5.0%) | 24 (4.4%) | |
| PR status | ||||
| Negative | 343 (31.5%) | 208 (38.2%) | 135 (24.8%) | <0.001 |
| Positive | 694 (63.7%) | 310 (56.9%) | 384 (70.5%) | |
| Unknown | 53 (4.9%) | 27 (5.0%) | 26 (4.8%) | |
| HER2 status | ||||
| Negative | 895 (82.1%) | 435 (79.8%) | 460 (84.4%) | <0.001 |
| Positive | 168 (15.4%) | 104 (19.1%) | 64 (11.7%) | |
| Unknown | 27 (2.5%) | 6 (1.1%) | 21 (3.9%) | |
Table 2.
Association between KYNU mRNA expression and clinicopathologic characteristics in METABRIC cohort.
| Expression |
P-value | |||
|---|---|---|---|---|
| Total (n = 1904) | KYNU high (n = 952) | KYNU low (n = 952) | ||
| Age (years) | ||||
| ≥55 | 952 (50.0%) | 535 (56.2%) | 417 (43.8%) | <0.001 |
| <55 | 952 (50.0%) | 417 (43.8%) | 535 (56.2%) | |
| Tumor size | ||||
| ≥2 cm | 592 (31.1%) | 298 (31.3%) | 294 (30.9%) | 0.857 |
| <2 cm | 1292 (67.9%) | 643 (67.5%) | 649 (68.2%) | |
| Unknown | 20 (1.1%) | 11 (1.2%) | 9 (0.9%) | |
| AJCC stage | ||||
| 0 | 4 (0.2%) | 3 (0.3%) | 1 (0.1%) | 0.004 |
| I | 475 (24.9%) | 217 (22.8%) | 258 (27.1%) | |
| II | 800 (42.0%) | 420 (44.1%) | 380 (39.9%) | |
| III | 115 (6.0%) | 71 (7.5%) | 44 (4.6%) | |
| IV | 9 (0.5%) | 2 (0.2%) | 7 (0.7%) | |
| Unknown | 501 (26.3%) | 239 (25.1%) | 262 (27.5%) | |
| Tumor Grade | ||||
| I | 165 (8.7%) | 55 (5.8%) | 110 (11.6%) | <0.001 |
| II | 740 (38.9%) | 279 (29.3%) | 461 (48.4%) | |
| III | 927 (48.7%) | 588 (61.8%) | 339 (35.6%) | |
| Unknown | 72 (3.8%) | 30 (3.2%) | 42 (4.4%) | |
| ER status | ||||
| Negative | 445 (23.4%) | 359 (37.7%) | 86 (9.0%) | <0.001 |
| Positive | 1459 (76.6%) | 593 (62.3%) | 866 (91.0%) | |
| PR status | ||||
| Negative | 895 (47.0%) | 558 (58.6%) | 337 (35.4%) | <0.001 |
| Positive | 1009 (53.0%) | 394 (41.4%) | 615 (64.6%) | |
| HER2 status | ||||
| Negative | 1668 (87.6%) | 771 (81.0%) | 897 (94.2%) | <0.001 |
| Positive | 236 (12.4%) | 181 (19.0%) | 55 (5.8%) | |
Fig. 1.
KYNU expression in different molecular subtypes and stage of transcriptional classification scheme in TCGA and METABRIC cohort. (*P < 0.05, **P < 0.01, ***P < 0.001, ***P < 0.0001).
3.2. KYNU was closely related to immune functions in breast cancer
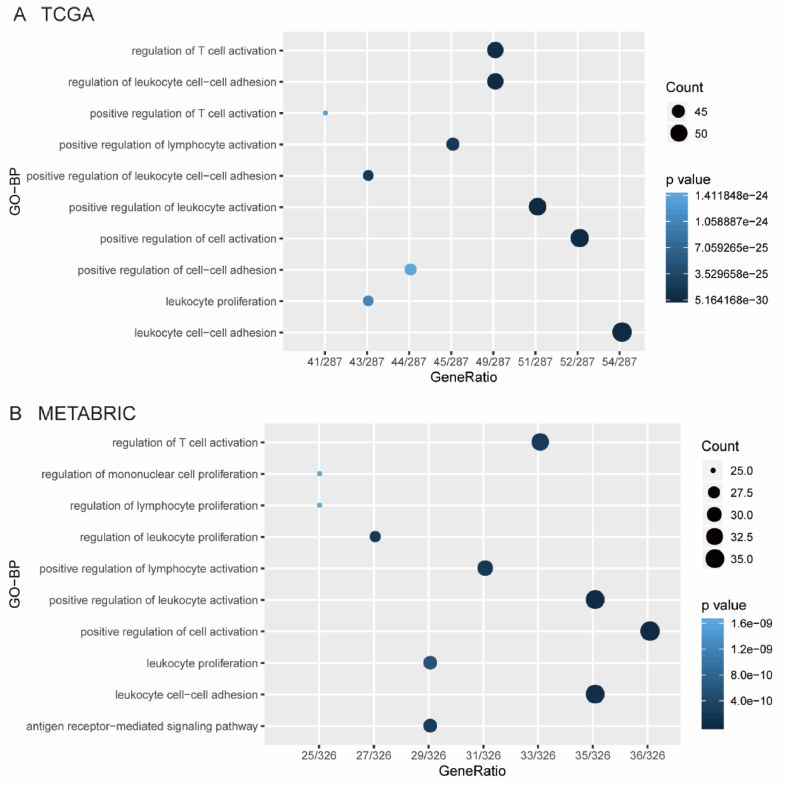
To further investigate the potential biological role of KYNU in BC, according to Spearman correlation analysis, we identified 304 genes and 378 genes that were strongly correlated with KYNU in BC (|R|>0.4 and P < 0.05) by TCGA and METABRIC datasets, respectively. Subsequently, the potential biological roles of KYNU were then determined using Gene Ontology (GO) functional enrichment analyses. According to GO analyses, genes correlated with KYNU mainly belonged to biological processes related to immunity, especially in T cell related immune biological processes (Fig. 2A and B), which agrees with the results from the 1881-sample microarray study mentioned above, Interestingly, when using increasing p-values to arrange gene functions, both the TCGA and the METABRIC databases showed that genes relevant to KYNU are mostly involved in immune-related functions (Fig. 2A and B).
Fig. 2.
KYNU was closely related to immune functions in breast cancer. Gene ontology analysis showed that KYNU was mainly involved in immune response and inflammatory response in TCGA and METABRIC databases (A and B).
3.3. KYNU related immune responses
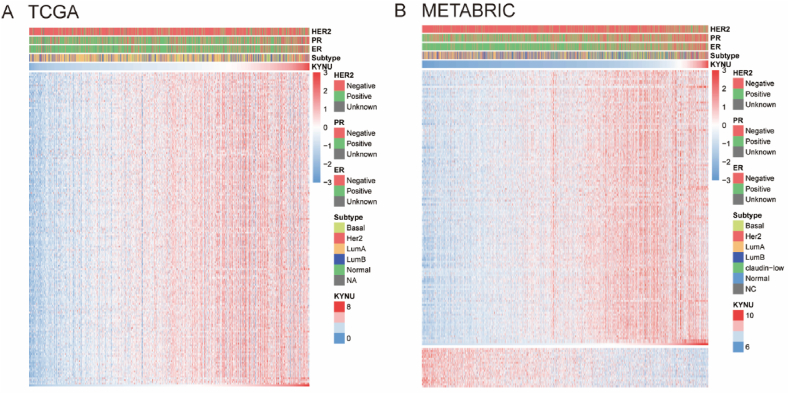
The Immunology Database and Analysis Portal (ImmPort) database was explored to further elucidate the potential role of KYNU in the immune response in BC. A total of 4723 immunologically related genes were collected from ImmPort database. A profile of KYNU and immune response was derived by finding the genes most strongly correlated with KYNU (Spearman |R| > 0.4, P < 0.05). We discovered that KYNU positively correlated with 182 and 141 immunologically related genes in TCGA and METABRIC, respectively, While only 0 and 21 genes related to immunology were negatively correlated with KYNU, respectively (Fig. 3A and B). In BC, KYNU is positively correlated with most relevant immune responses, whereas it is negatively correlated with only a few immune responses.
Fig. 3.
KYNU related immune responses. Most immune-related genes were positively correlated with KYNU expression in TCGA and METABRIC databases, while a small number of genes were negatively associated (A and B).
3.4. The relationship between KYNU and immune response
3.4.1. The relationship between checkpoint members and KYNU
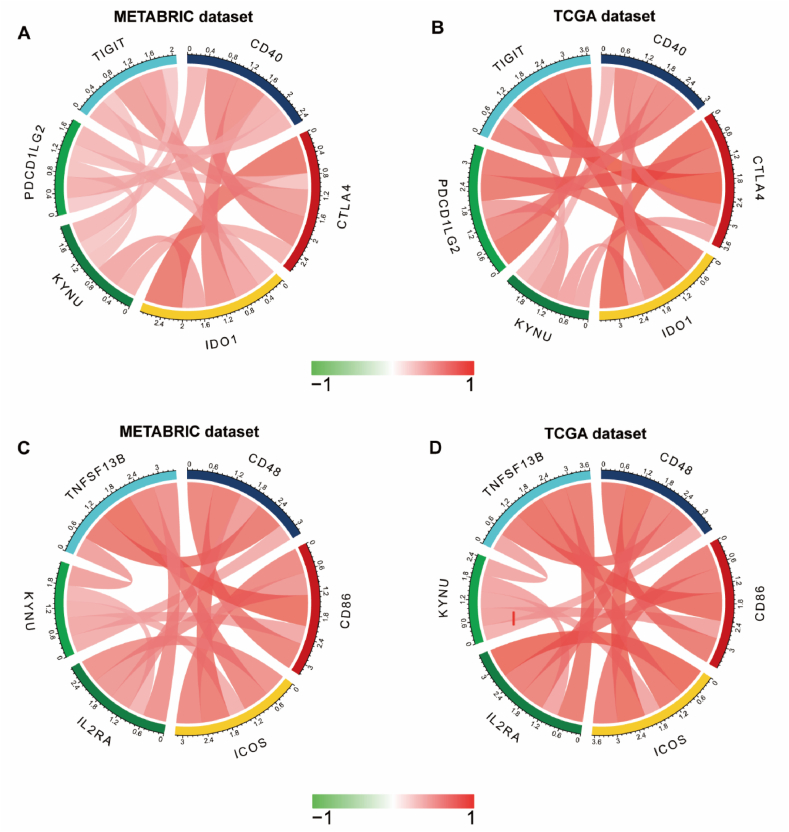
The association between KYNU and other checkpoint members was assessed in order to further explore the potential synergistic role of KYNU induced immune response in breast cancer. In both TCGA and METABRIC, we found strong correlations between KYNU and other checkpoint members (Fig. 4A–D). KYNU was positively correlated with PDCD1LG2 (PDL2) (TCGA: r = 0.53; METABRIC: r = 0.334), CTLA4 (TCGA: r = 0.53; METABRIC: r = 0.334), IDO1 (TCGA: r = 0.5; METABRIC: r = 0.486), TIGIT (TCGA: r = 0.457; METABRIC: r = 0.302), CD40 (TCGA: r = 0.397; METABRIC: r = 0.409), and other checkpoint members including CD48 (TCGA: r = 0.434; METABRIC: r = 0.457), CD86 (TCGA: r = 0.523; METABRIC: r = 0.481), ICOS (TCGA: r = 0.49; METABRIC: r = 0.444), IL2RA (TCGA: r = 0.502; METABRIC: r = 0.417), TNFSF13B (TCGA: r = 0.493; METABRIC: r = 0.531).
Fig. 4.
KYNU expression is correlated with immune checkpoint members in TCGA and METABRIC databases (A–D).
3.4.2. The relationship between KYNU and T cell immunity
GSVA analysis was performed to clarify the relationship between KYNU expression and Tcell immune response in BC. A strong positive correlation was found between KYNU and gene ontology related to immune responses of T cells (Fig. 5A). KYNU was positively correlated with regulation of T cell activation, alpha-beta T cell activation, alpha-beta T cell differentiation, T-helper 1 type immune response, T cell mediated cytotoxicity, and also T cell receptor signaling pathway. In addition, TCGA and METABRIC databases can be used to mutually verify these results (Fig. 5A). These findings implicated KYNU might play an inhibitory role in T cell immunity in breast tumor.
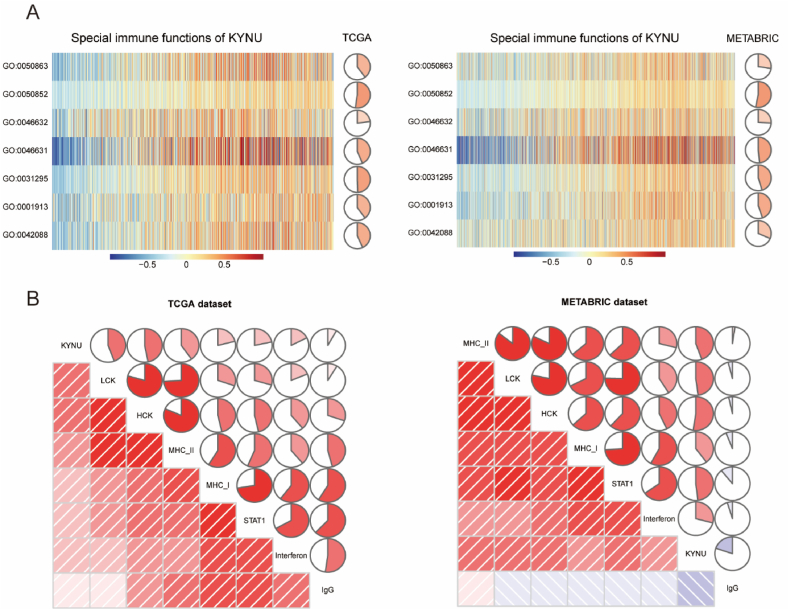
Fig. 5.
KYNU related cell immunity and inflammatory activities in breast cancer. The relationship between KYNU and cell immunity in TCGA and METABRIC datasets (A). The relationship between KYNU and inflammatory activities in TCGA and METABRIC datasets (B). GO:0001913: T cell mediated cytotoxicity; GO:0031295: T cell co-stimulation; GO:0042088: T-helper 1 type immune response; GO:0046631: alpha-beta T cell activation; GO:0046632: alpha-beta T cell differentiation; GO:0050852: T cell receptor signaling pathway; GO:0050863 regulation of T cell activation.
3.4.3. The relationship between KYNU and inflammatory activities
To further revealed inflammatory activities related with KYNU, we defined a total of 104 genes derived from seven clusters as metagenes through Gene Sets Variation Analysis (GSVA) [34]. Table S1 contains detailed gene lists for various types of inflammation and immune response. As shown in Fig. 5B, LCK, HCK, MHC-I, MHC-II, STAT1, and interferon showed positive correlations with KYNU, but not IgG. There was the strongest correlation between KYNU and HCK metagenes among these seven clusters. Furthermore, these findings can be validated in both TCGA and METABRIC cohorts independently. It was further confirmed that KYNU played an essential role in immune and inflammatory functions in BC as a result of these findings.
3.5. KYNU predicts a worse outcome in breast cancer
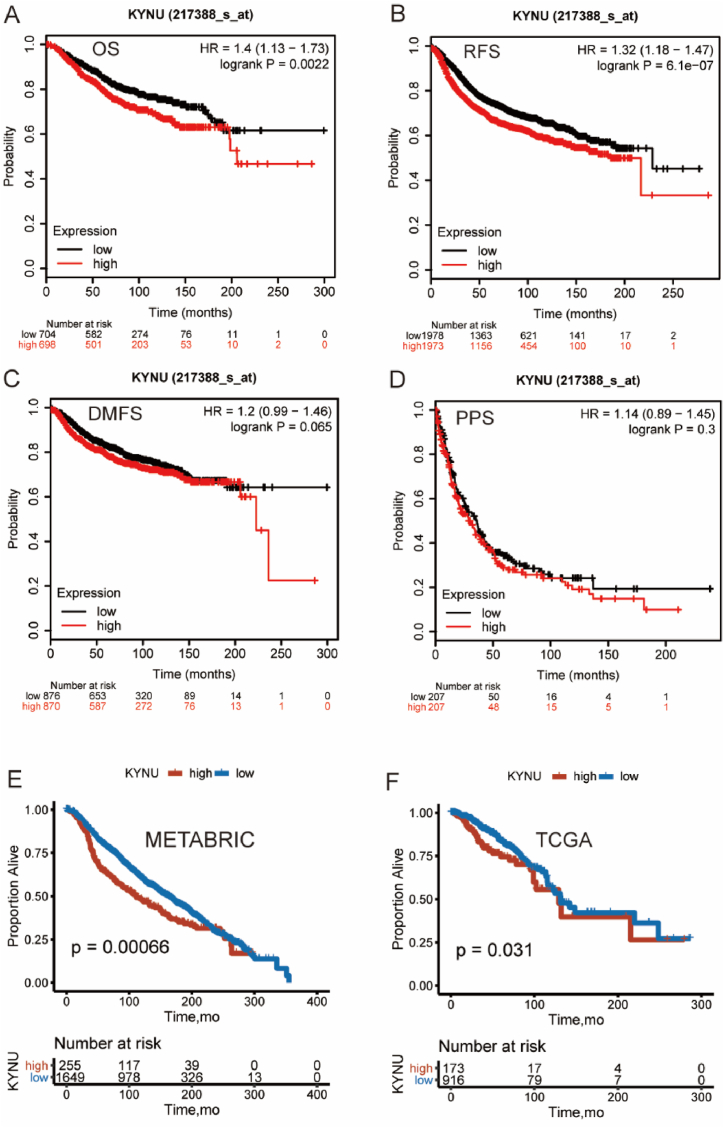
The prognostic value of KYNU was further examined because of its strong relationship with T cell immune response in breast cancer. Firstly, a total of 6243 breast cancer patients were analyzed using the KM-plotter database to determine if KYNU expression had a prognostic value. High KYNU expression was associated with poorer overall survival (OS), relapse-free survival (RFS), and distant metastasis-free survival (DMFS) (Fig. 6A–D). Secondly, both the TCGA and METABRIC datasets showed similar Kaplan-Meier curve patterns (Fig. 6E and F). In summary, these results suggested that KYNU was a negative prognostic indicator of BC due to suppressive effect on T-cell-related immune response.
Fig. 6.
Validation of Kaplan-Meier survival curves comparing the high and low expression of KYNU in KM-plotter database (n = 6243) (A–D), TCGA and METABRIC dataset (E–F). Hazard ratio: HR; 95% Confidence interval: 95% CI.
4. Discussion
As far as we know, our study, which included a total of 2994 samples, is the largest and most comprehensive research that has systematically investigated KYNU expression pattern, prognostic value, and the relationship between KYNU and immune infiltrates in breast cancer (BC). BCs with higher grades and degrees of malignancy had higher KYNU expression. Furthermore, we found that patients with higher KYNU expression had a shorter OS, RFS, and DMFS. However, our findings were contrary to the results of a previous study, which was the only study we could find focusing on the relationship between KYNU and BC [24]. This study examined KYNU expression by IHC in 137 primary breast cancer tissues and found that KYNU expression had a negative correlation with ER expression, PR expression and tumor grade; this suggested KYNU was a tumor suppressor in BC. As we know, IHC examinations are subjective visual interpretation; therefore, results can be affected by the experience of different pathologists, the quality of slides, and staining. Our data came from large datasets of RNA sequencing with many more samples, so our findings may be more effective and persuasive. Research about the relationship between aryl hydrocarbon receptor (AhR) depletion and expression of tumor-related genes in breast cancer reflected that KYNU expression was positively correlated with tumorigenic properties and tumor malignancy, which was consistent with our suggestions [23]. In addition, another study focusing on TDO2-AhR signaling provided evidence that KYNU expression was higher in suspended TNBC cell lines and ER-negative breast cancer cell lines [41].
As expected, we found that in breast cancer KYNU expression was closely associated with immune-related biological processes, especially T cell-related ones. Besides, KYNU was mainly positively correlated with genes relevant to immune responses in BC patients. Moreover, KYNU was positively correlated with several immune checkpoint members, including CTLA4, PDCD1LG2, IDO1, CD40, TIGIT, ICOS, CD86, IL2RA, CD48, and TNFSF13B. Numerous studies are focusing on the immunotherapy efficacy of targeting immune checkpoints like CTLA-4, PD-1, and PD-L1 in breast cancer patients. CTLA4 is expressed by activated T cells and regulatory T cells (Tregs). In 1995, CTLA4 was first reported as a negative regulator of T cell activation [42,43]. CTLA4 could inhibit T cell activation by the combination ofCD80 (B7-1) and CD86 (B7-2) on antigen presenting cells (APCs) with greater affinity than CD28, which breaks the co-stimulation necessary for T cell activation [44]. Besides, when bound to CTLA4 on Tregs, CD86 could be removed from the surface of APCs and transferred to the Tregs to inhibit T cell activation [45]. Blocking both processes with CTLA4 blockers tends to be an effective way to treat cancer. Currently, Anti-CTLA4 antibodies such as ipilimumab and tremelimumab are currently being used in clinical trials in TNBC patients [46]. PD-L2 is an immune checkpoint receptor ligand encoded by PDCD1LG2 gene [47]. PD-L2 is expressed on the surface of professional APCs, including DCs and macrophages, as well as CD4+ and CD8+ T cells [[48], [49], [50]]. As well as PD-L1, PD-L2 could inhibit T cell receptor-mediated immune cell activation by engagement with PD1 [48], and play a crucial role in immune tolerance and autoimmunity [51]. Anti-PD1 and anti-PD-L1 antibodies are being clinically tested in multiple cancers, including BC. Although PD-L2 blockers are still under study, they are definitely a certain promising immunotherapeutic target. These findings show that KYNU has the potential to be an important immunotherapy target.
Importantly, KYNU was significantly correlated withIDO1, the upstream enzyme of KYNU in the kynurenine pathway. A large quantity of evidence supports the hypothesis that IDO1 contributes to immunosuppression. High IDO1 expression that catabolizes tryptophan (Trp) to kynurenine could deplete tryptophan. Consequently, the mTORC1 and PKC signaling pathways inside effector T cell would be inhibited, resulting in effector T cells suppression [52]. Furthermore, kynurenine, the metabolites of Trp through the kynurenine pathway (KP), has the ability to stimulate Foxp3+ Tregs to suppress T cell immune activity [53]. In early-stage cancer clinical trials, antibodies targeting IDO1 alone produced promising results, but a phase III trial did not produce positive results [54]. Hence, targeting IDO1 alone is not sufficient to combat tumor immune invasion. More importantly, in a previous study, it was found that the KYNU expression pattern in BC was similar to IDO1 and TDO2 [55]. As derived from our data exploration, KYNU had a close correlation with T cell-related immune functions and MHC-related modules, as well as several checkpoints related to T cell activities. KYNU might modulate the tumor immune microenvironment through regulation of Trp catabolism, based on these studies. Furthermore, KYNU might act synergistically with CTLA4, PDL2, IDO1, and other immune checkpoints, which supports the concept of combining cancer immunotherapy by targeting KYNU along with other checkpoints. However, the precise molecular mechanism by which KYNU regulates immune activity in tumors remains unknown. Studies looking deeper into these mechanisms are warranted and necessary.
Funding
This work was supported by basic scientific research project of Beijing University of Traditional Chinese Medicine in 2019 (Grant No. 2019-JYB-JS-108). Declaration of interest No potential conflicts of interest were disclosed.
Author contributions
Wei Li conceived and designed the experiments Yiliang Li performed the experiments Yiliang Li analyzed and interpreted the data Lina Zhao, Mengyu Wang and Chen Liang contributed reagents, materials, analysis tools or data Yiliang Li and Wei Li wrote the paper.
Availability of data and materials
The datasets generated and/or analyzed during the current study are publicly available in the TCGA data portal (https://portal.gdc.cancer.gov/) and cbioportal database (https://www.cbioportal.org/).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We wish to thank TCGA project organizers as well as all study participants.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17216.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bray F., Ferlay J., Soerjomataram I., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018 Nov;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA A Cancer J. Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. 2019. [DOI] [PubMed] [Google Scholar]
- 3.Waks A.G., Winer E.P. Breast cancer treatment: a review. JAMA. 2019 Jan 22;321(3):288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 4.Driessens G., Crosignani S., Detheux M., et al. Preclinical assessment of a novel small molecule inhibitor of indoleamine 2,3-dioxygenase 1 (IDO1) J. ImmunoTher. Canc. 2014;2(Suppl 3) [Google Scholar]
- 5.Zhai L., Spranger S., Binder D.C., et al. Molecular pathways: targeting IDO1 and other tryptophan dioxygenases for cancer immunotherapy. Clin. Cancer Res. 2015 Dec 15;21(24):5427–5433. doi: 10.1158/1078-0432.CCR-15-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheong J.E., Sun L. Targeting the IDO1/TDO2-KYN-AhR pathway for cancer immunotherapy - challenges and opportunities. Trends Pharmacol. Sci. 2018 Mar;39(3):307–325. doi: 10.1016/j.tips.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Badawy A.A.B. Kynurenine pathway of tryptophan metabolism: regulatory and functional aspects. Int. J. Tryptophan Res. : IJTR. 2017;10 doi: 10.1177/1178646917691938. 1178646917691938-1178646917691938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koushik S.V., Sundararaju B., McGraw R.A., et al. Cloning, sequence, and expression of kynureninase from Pseudomonas fluorescens. Arch. Biochem. Biophys. 1997 Aug 15;344(2):301–308. doi: 10.1006/abbi.1997.0220. [DOI] [PubMed] [Google Scholar]
- 9.Triplett T.A., Garrison K.C., Marshall N., et al. Reversal of indoleamine 2,3-dioxygenase-mediated cancer immune suppression by systemic kynurenine depletion with a therapeutic enzyme. Nat. Biotechnol. 2018;36(8):758–764. doi: 10.1038/nbt.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puccetti P., Fallarino F., Italiano A., et al. Accumulation of an endogenous tryptophan-derived metabolite in colorectal and breast cancers. PLoS One. 2015;10(4) doi: 10.1371/journal.pone.0122046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spranger S., Spaapen R.M., Zha Y., et al. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci. Transl. Med. 2013 Aug 28;5(200) doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mondanelli G., Bianchi R., Pallotta M.T., et al. A relay pathway between arginine and tryptophan metabolism confers immunosuppressive properties on dendritic cells. Immunity. 2017 Feb 21;46(2):233–244. doi: 10.1016/j.immuni.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harden J.L., Lewis S.M., Lish S.R., et al. The tryptophan metabolism enzyme L-kynureninase is a novel inflammatory factor in psoriasis and other inflammatory diseases. J. Allergy Clin. Immunol. 2016;137(6):1830–1840. doi: 10.1016/j.jaci.2015.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y.-J., Chang W.-A., Wu L.-Y., et al. Systematic analysis of differential expression profile in rheumatoid arthritis chondrocytes using next-generation sequencing and bioinformatics approaches. Int. J. Med. Sci. 2018;15(11):1129–1142. doi: 10.7150/ijms.27056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberson E.D.O., Liu Y., Ryan C., et al. A subset of methylated CpG sites differentiate psoriatic from normal skin. J. Invest. Dermatol. 2012;132(3 Pt 1):583–592. doi: 10.1038/jid.2011.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwarcz R., Stone T.W. The kynurenine pathway and the brain: challenges, controversies and promises. Neuropharmacology. 2017 Jan;112(Pt B):237–247. doi: 10.1016/j.neuropharm.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson A.-S., Owe-Larsson B., Asp L., et al. Activation of kynurenine pathway in ex vivo fibroblasts from patients with bipolar disorder or schizophrenia: cytokine challenge increases production of 3-hydroxykynurenine. J. Psychiatr. Res. 2013;47(11):1815–1823. doi: 10.1016/j.jpsychires.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Ashenagar M.S., Tabuchi M., Kinoshita K., et al. Gene expression in the adrenal glands of three spontaneously hypertensive rat substrain. Mol. Med. Rep. 2010 Mar-Apr;3(2):213–222. doi: 10.3892/mmr_00000242. [DOI] [PubMed] [Google Scholar]
- 19.Lauvrak S.U., Munthe E., Kresse S.H., et al. Functional characterisation of osteosarcoma cell lines and identification of mRNAs and miRNAs associated with aggressive cancer phenotypes. Br. J. Cancer. 2013;109(8):2228–2236. doi: 10.1038/bjc.2013.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H., Lu D., Liu X., et al. Survival-related risk score of lung adenocarcinoma identified by weight gene co-expression network analysis. Oncol. Lett. 2019;18(5):4441–4448. doi: 10.3892/ol.2019.10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkateswaran N., Lafita-Navarro M.C., Hao Y.H., et al. MYC promotes tryptophan uptake and metabolism by the kynurenine pathway in colon cancer. Genes Dev. 2019 Sep 1;33(17–18):1236–1251. doi: 10.1101/gad.327056.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ci C., Wu C., Lyu D., et al. Downregulation of kynureninase restrains cutaneous squamous cell carcinoma proliferation and represses the PI3K/AKT pathway. Clin. Exp. Dermatol. 2019 doi: 10.1111/ced.14072. [DOI] [PubMed] [Google Scholar]
- 23.Goode G., Pratap S., Eltom S.E. Depletion of the aryl hydrocarbon receptor in MDA-MB-231 human breast cancer cells altered the expression of genes in key regulatory pathways of cancer. PLoS One. 2014;9(6) doi: 10.1371/journal.pone.0100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y., Feng X., Lai J., et al. A novel role of kynureninase in the growth control of breast cancer cells and its relationships with breast cancer. J. Cell Mol. Med. 2019;23(10):6700–6707. doi: 10.1111/jcmm.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Al-Mansoob M., Gupta I., Stefan Rusyniak R., et al. KYNU, a novel potential target that underpins CD44-promoted breast tumour cell invasion. J. Cell Mol. Med. 2021 Mar;25(5):2309–2314. doi: 10.1111/jcmm.16296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li R., Qu H., Wang S., et al. GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics. 2018;34(14):2515–2517. doi: 10.1093/bioinformatics/bty124. [DOI] [PubMed] [Google Scholar]
- 27.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritchie M.E., Phipson B., Wu D., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015 Apr 20;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J., Lichtenberg T., Hoadley K.A., et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416. doi: 10.1016/j.cell.2018.02.052. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Curtis C., Shah S.P., Chin S.-F., et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu G., Wang L.-G., Han Y., et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S B., S A., L G., et al. ImmPort: disseminating data to the public for the future of immunology. Immunol. Res. 2014;58:234–239. doi: 10.1007/s12026-014-8516-1. [DOI] [PubMed] [Google Scholar]
- 33.S H., R C., GSVA J.G. Gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.A R., H U., L P., et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11(2):R15. doi: 10.1186/bcr2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ru B., Wong C.N., Tong Y., et al. TISIDB: an integrated repository portal for tumor–immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 36.Wickham H. ggplot2. Wiley Interdisc. Rev.: Comput. Stat. 2011;3(2):180–185. [Google Scholar]
- 37.Robin X., Turck N., Hainard A., et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu Z., Gu L., Eils R., et al. Circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30(19):2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 39.Friendly M. Corrgrams: exploratory displays for correlation matrices. Am. Statistician. 2002;56(4):316–324. [Google Scholar]
- 40.Ringnér M., Fredlund E., Häkkinen J., et al. GOBO: gene expression-based outcome for breast cancer online. PLoS One. 2011;6(3) doi: 10.1371/journal.pone.0017911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Amato N.C., Rogers T.J., Gordon M.A., et al. A TDO2-AhR signaling axis facilitates anoikis resistance and metastasis in triple-negative breast cancer. Cancer Res. 2015 Nov 1;75(21):4651–4664. doi: 10.1158/0008-5472.CAN-15-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tivol E.A., Borriello F., Schweitzer A.N., et al. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity. 1995 Nov;3(5):541–547. doi: 10.1016/1074-7613(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 43.Waterhouse P., Penninger J.M., Timms E., et al. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science. 1995 Nov 10;270(5238):985–988. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 44.Krummel M.F., Allison J.P. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 1995 Aug 1;182(2):459–465. doi: 10.1084/jem.182.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohue Y., Nishikawa H. Regulatory T (Treg) cells in cancer: can Treg cells be a new therapeutic target? Cancer Sci. 2019 Jul;110(7):2080–2089. doi: 10.1111/cas.14069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X., Qi Y., Kong X., et al. Immunological therapy: a novel thriving area for triple-negative breast cancer treatment. Cancer Lett. 2019 Feb 1;442:409–428. doi: 10.1016/j.canlet.2018.10.042. [DOI] [PubMed] [Google Scholar]
- 47.Latchman Y., Wood C.R., Chernova T., et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2001 Mar;2(3):261–268. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 48.Sharpe A.H., Wherry E.J., Ahmed R., et al. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat. Immunol. 2007 Mar;8(3):239–245. doi: 10.1038/ni1443. [DOI] [PubMed] [Google Scholar]
- 49.Lesterhuis W.J., Steer H., Lake R.A. PD-L2 is predominantly expressed by Th2 cells. Mol. Immunol. 2011 Oct;49(1–2):1–3. doi: 10.1016/j.molimm.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 50.Messal N., Serriari N.E., Pastor S., et al. PD-L2 is expressed on activated human T cells and regulates their function. Mol. Immunol. 2011 Sep;48(15–16):2214–2219. doi: 10.1016/j.molimm.2011.06.436. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y., Chung Y., Bishop C., et al. Regulation of T cell activation and tolerance by PDL2. Proc. Natl. Acad. Sci. U. S. A. 2006 Aug 1;103(31):11695–11700. doi: 10.1073/pnas.0601347103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li F., Zhang R., Li S., et al. IDO1: an important immunotherapy target in cancer treatment. Int. Immunopharm. 2017 Jun;47:70–77. doi: 10.1016/j.intimp.2017.03.024. [DOI] [PubMed] [Google Scholar]
- 53.Fox E., Oliver T., Rowe M., et al. Indoximod: an immunometabolic adjuvant that empowers T cell activity in cancer. Front. Oncol. 2018;8:370. doi: 10.3389/fonc.2018.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garber K. American Association for the Advancement of Science; 2018. A New Cancer Immunotherapy Suffers a Setback. [DOI] [PubMed] [Google Scholar]
- 55.Qiang Liu J.Z., Kong Xiangyi, Wang Xiangyu, Wang Zhongzhao, Fang Yi, Wang Jing. Comprehensive analysis of the expression and prognosis for TDO2 in breast cancer. Mol. Ther.: Oncol. 2020 June 26;17:153–168. doi: 10.1016/j.omto.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are publicly available in the TCGA data portal (https://portal.gdc.cancer.gov/) and cbioportal database (https://www.cbioportal.org/).