Abstract
The spatiotemporal control of gene expression in complex multicellular organisms relies on noncoding regulatory sequences such as enhancers, which activate transcription of target genes often over large genomic distances. Despite the advances in the identification and characterization of enhancers, the principles and mechanisms by which enhancers select and control their target genes remain largely unknown. Here, we review recent interdisciplinary and quantitative approaches based on emerging techniques that aim to address open questions in the field, notably how regulatory information is encoded in the DNA sequence, how this information is transferred from enhancers to promoters, and how these processes are regulated in time.
Current Opinion in Genetics & Development 2023, 80:102052
This review comes from a themed issue on Genome Architecture and Expression
Edited by Giacomo Cavalli and Job Dekker
For complete overview of the section, please refer to the article collection, “Genome Architecture and Expression (2023)”
https://doi.org/10.1016/j.gde.2023.102052
0959–437X/© 2023 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
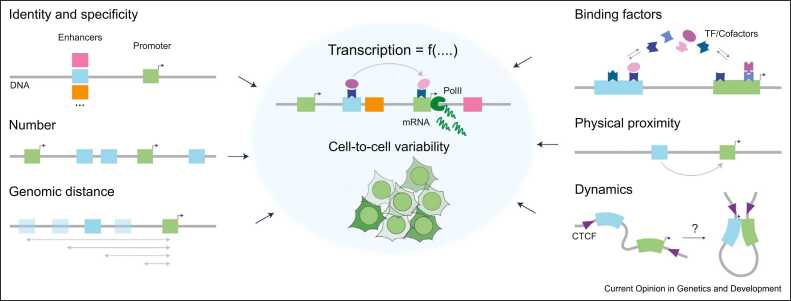
In metazoans, the spatiotemporal control of gene expression relies not only on promoters but also, and crucially so, on enhancers. These noncoding regulatory sequences are located outside promoter regions and activate target genes often over large genomic distances, which in mammals can extend over the megabase scale [1]. Despite the intervening distance, the regulatory role of enhancers is essential for accurate tissue- and developmental time-specific gene expression patterns. Genetic variation within these noncoding regions results in quantitative changes in gene expression and is a major driver of evolution, but also causal to developmental disorders and strongly associated with numerous human diseases [2]. However, despite our increasing ability to identify putative enhancer sequences on the basis of functional genomic annotations and genome-wide association studies, to date, it remains impossible to predict and causally explain distal genetic associations between enhancers and their target promoters from first principles. How enhancers control a promoter’s transcriptional level, and how this depends on their sequence, number, and genomic distance, remains to be understood, as well as how these dependencies relate to binding and interactions of transcription factors (TFs) and cofactors, as well as on modulations of enhancer–promoter (E–P) physical interactions (Figure 1).
Figure 1.
Enhancer-promoter (E-P) communication and transcriptional outputs depend on a number of variables, including enhancer and promoter sequence, number, and mutual genomic distance, in a way that depends on binding of TFs and interactions with cofactors, the physical proximity between regulatory sequences and their dynamics.
Several excellent reviews have covered recent progress in enhancer biology 1, 3, 4. Here, we focus specifically on emerging evidence from highly diverse approaches, which strongly suggest that a truly quantitative understanding of E–P communication requires understanding the spatiotemporal dynamics of the underlying molecular processes, which are highly stochastic and dynamic in individual living cells. We notably review technical and methodological developments bridging across biochemistry, molecular biology, and biophysics that have recently enabled exciting insights, and outline future challenges.
How are regulatory information and specificity encoded in enhancer and promoter sequences?
One key question is how an enhancer’s regulatory activity and its specificity for selected promoters are encoded in its DNA sequence. While it is well established that the activity of an enhancer depends on its ability to recruit combinations of TFs [5], it remains unclear how enhancer activity is modulated by TF binding site (TFBS) number, affinity, and arrangement (i.e. enhancer grammar) [6], and how it depends on covalent DNA or histone modifications as well as TF interactions with cofactors. The last few years have seen major advances in methods for the identification of putative regulatory sequences genome-wide [7] (some of which can be deployed at single-cell resolution [8]), the synthesis and/or mutagenesis of endogenous or ectopic sequences [9], and the massively parallel detection of their effects on transcriptional reporters [10]. Coupled with machine learning algorithms, these methods have given considerable insights into TFBS usage within enhancer regions and at least in some contexts have enabled the design of artificial enhancers with tissue-specific activity 11••, 12••.
While such approaches have unlocked the potential to reveal quantitative sequence determinants of intrinsic enhancer activity, it remains unclear if an enhancer can activate any promoter, or rather only a compatible subset of promoters. Answering this fundamental question requires measuring the transcriptional output of large numbers of E–P combinations in parallel. Initial experiments based on transiently expressed combinatorial libraries of enhancer-reporter pairs have suggested that housekeeping and developmentally regulated promoters are activated by distinct sets of enhancers in D. melanogaster cells [13]. However, in mammalian cells, a rather broad compatibility was observed, with quantitative rather than qualitative differences in E–P communication 14••, 15••, 16••. This is in line with the earlier finding that once randomly inserted into the mouse genome, ectopic promoter sequences are activated by the (unrelated) surrounding endogenous-regulatory landscapes 17, 18. To what extent these results can be extrapolated into general principles of E–P communication in a chromosomal context is unclear. Moving the field forward will critically depend on the development of large-scale reporter assays allowing to study E–P communication in an endogenous chromatin environment, possibly taking advantage of experimental setups where genomic sequence can be engineered bottom-up 19, 20, 21 and chromatin states and/or recruitment of TFs and cofactors can be artificially modulated 22, 23, 24, 25, 26, 27•, 28. Coupled to increasingly more refined ‘explainable’ artificial intelligence approaches [29], such experiments will allow disentangling regulatory layers and unravel quantitative contributions of enhancer/promoter sequence, TFBS usage, and chromatin-mediated effects to E–P communication.
Support for a critical role of chromatin effects in E–P communication is also supported by recent studies that measured TF and nucleosome occupancy with single-molecule resolution using DNA methyltransferase footprinting followed by high-throughput sequencing. These studies revealed that nucleosome eviction following TF binding to enhancer DNA often results in indirect binding cooperativity (e.g. not mediated by direct protein–protein interactions) 30, 31. The single-molecule resolution of such assays was also key to demonstrate that CpG methylation within enhancer regions directly prevents binding of at least some TFs [32]. Coupled to long-read DNA sequencing, single-molecule footprinting methods hold great promise for revealing correlations between TFBS occupancy at distal promoter and enhancer sites [33] and downstream transcription events [34], and even inferring their temporal order [35]. It will be exciting to see how such assays can be combined in the future with atomic models of molecular interactions provided by cryo-electron microscopy (cryo-EM) [36] as well as fluorescent microscopy-based measurements of the rates and stoichiometries of such interactions in single living cells 37, 38 to gain insight into the mechanistic bases of sequence determinants of E–P communication. Ultimately, studying E–P pairs in a genomic context will also be fundamental to reveal how their communication is influenced by the intervening genomic distance and their numbers [39]. This will lead to an understanding of how E–P interactions are modulated by additional regulatory elements and chromosome structure (see below).
How is regulatory information conveyed from enhancers to promoters?
The exchange of regulatory information between an enhancer and a promoter is thought to require at least some degree of physical proximity in the three-dimensional (3D) space of the cell nucleus. E–P communication is indeed generally restricted within topologically associating domains (TAD) 40, 41, which are established as the loop-extrusion activity of the cohesin complex is arrested by CTCF molecules bound to DNA in a specific orientation [42]. Compartment-like interactions as well as cohesin/CTCF-independent tethering elements also contribute to establishing physical associations between active regulatory sequences 43, 44, 45. The notion that proximity is required for conveying regulatory information is also supported by the fact that E–P interaction probabilities measured with cross-linking-based chromosome conformation capture (3C) correlate quantitatively with the degree of promoter activity 46, 47, 48 and that cohesin-mediated CTCF loops can often modulate E–P communication 46, 49, 50, although the effect of TAD boundaries and CTCF sites might depend on the actual intrinsic activity of the enhancer 46, 51 or compatibility with the promoter.
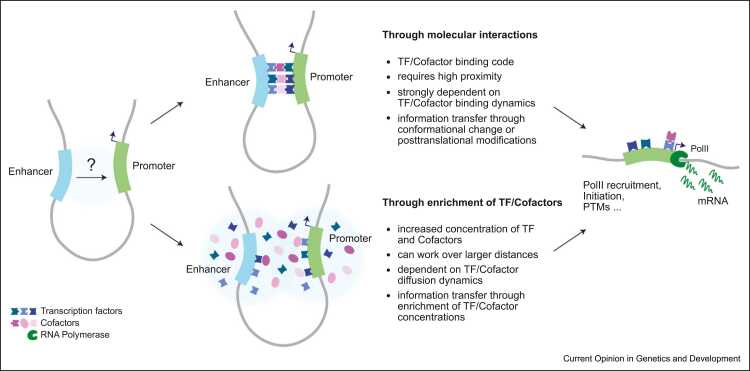
However, what level of ‘proximity’ is required and what molecular processes actually mediate regulatory exchange remains unclear. Spatial proximity is usually defined as the ability to form ligation products in 3C methods, but it is unclear at what spatial distance two genomic loci can be cross-linked and ligated, and whether this distance depends on local chromatin composition (e.g. active or repressive chromatin) [52]. Measurements of proximity-induced DNA methylation have shown that at least a fraction of cross-linking events detected by 3C methods occur in a range of a few nanometers [53]. Absolute quantification of physical distances between regulatory sequences may be enabled by optical methods, but is complicated by experimental uncertainty on distance measurements in fluorescence microscopy (approx. 50 nm in single-dye sequential FISH 54, 55, 56), which currently exceeds the scale of molecular interactions [57]. Indeed, although measurements of physical proximity based on 3C and optical methods often correlate well, discrepancies have been observed [58]. Recent developments in super-resolution microscopy start to bridge this gap achieving a localization precision in the range of 1 nm [59] and promise to yield more precise estimations of molecular proximity in the near future. However, since the mechanisms and factors that mediate E–P communication remain unclear, so do the physical scales that are the most relevant to study. If activation is mediated by direct TF-cofactor–RNA polymerase II (PolII) interactions, these are expected to take place in the few tens-of-nanometer range (as a reference, a single human Mediator/pre-initiation complex spans ∼25×10×10 nm based on recent cryo-EM structures [60]). If instead communication occurs through macromolecular complexes, phase-separated droplets nucleated by low-complexity protein domains 61, 62, or local diffusion of (post-translationally modified) transcriptional coactivators [63], it is possible that distances involved are much larger (in the range of hundreds of nm) (Figure 2). Both mechanisms, irrespective of whether they co-occur or are mutually exclusive, must result in transmission of information to the promoter that ultimately leads to PolII licensing into productive transcription [5].
Figure 2.
Two alternative (and not mutually exclusive) potential mechanisms of information transfer from enhancers to promoters. Information might be passed on to the promoter through direct interactions between TFs and cofactors, or through local enrichment of TFs and cofactors in the vicinity of the promoter. Both mechanisms must result in the transfer of information being used by the promoter to enhance PolII recruitment, post-transcriptional modifications, or events leading to transcriptional initiation or elongation (PTM=post-translational modifications).
Identifying cofactors involved in E–P communication (and in transcriptional regulation in general) has proven difficult because contrary to TFs, coactivators and corepressors do not bind DNA in a sequence-specific manner and no specific ‘interaction code’ with TFs has been identified so far [1]. The last few years have nonetheless seen exciting progress in the identification and characterization of transcriptional cofactors. Genetic screens and new methods for the multiplexed recruitment of cofactors 64, 65•• have revealed large numbers of protein domains with either activating or repressive effects on transcription. The large-scale, unbiased design of these assays has, for example, enabled the discovery of previously unknown amino acid compositional biases in protein fragments that show coactivator activity and a distinctive role of post-translational SUMOylation of transcriptional repressors [65]. Another step ahead in the study of cofactors is the recent development of inducible degradation methods and availability of small-molecule inhibitors, which complement gene knockout approaches by making it possible to study the acute effects of temporary depletion of essential cofactors [66]. Degradation of Mediator subunits was notably key to characterizing their cell-type-specific roles in transcriptional regulation [67] and a role in facilitating E–P interactions 68, 69. Interestingly, depletion and chemical inhibition have shown that chromatin remodeler cofactors, such as SWI/SNF (SWItch/Sucrose Non-Fermentable), not only regulate promoter expression by modulating TFBS accessibility in a TF-specific manner 70, 71, but can also regulate chromosome interactions by modulating accessibility of CTCF binding sites [70].
A better characterization of cofactors will likely enable a more functional, rather than purely physical understanding of E–P contacts. This would allow one to tag cofactors specifically involved in long-range communication with enzymes that promote proximity-mediated chemical modifications (e.g. through proximity-mediated biotinylation followed by mass spectrometry [72] or fluorescent complementation assays [73]). This could also account for the temporal aspect of such interactions. One interesting step in this direction was recently provided by proximity-assisted photoactivation [74], in which energy transfer between two rhodamine dyes allows the fluorescent detection of tagged molecules separated by a distance of few nanometers at single-molecule resolution. Finally, the exciting perspective to visualize physical proximity between genomic locations together with the factors that mediate functional communication might come from spatial sequencing techniques [75], especially combined with methods to fluorescently detect chromatin-binding proteins in situ [76]. Recent advances in correlative light and electron microscopy [77] also open up the possibility that functional interactions might become observable at nearly atomic resolution in a not-too-distant future.
How do stochastic molecular processes at enhancer–promoter interfaces mediate transcriptional regulation?
Irrespective of the exact nature of the biochemical reactions and molecular players mediating the transfer of regulatory information, many studies have shown that the physical distances of E–P pairs vary substantially from one cell to another within the same cell population or tissue 78, 79, 80. This suggests that if E–P communication requires physical proximity, it can generally only occur in a subset of cells at any given time. But it is unclear how often these windows of opportunity are created and how long they last in time within individual cells. This ultimately depends on how fast chromosome structure changes, especially within TADs where most E–P interactions occur in mammalian cells.
Since TADs are established by the loop-extrusion activity of cohesin, the dynamics of chromosome structure inside a TAD is expected to be linked to the kinetics of loop extrusion and of CTCF binding. A series of recent exciting studies based on quantitative microscopy of reconstituted protein complexes demonstrated that mammalian cohesin indeed extrudes loops of DNA and can be blocked by CTCF, and even measured rates of extrusion in vitro (∼1 kb/s) 81, 82, 83. Single-molecule tracking and analysis of protein mobility in vivo 84, 85 additionally suggested that cohesin resides on DNA for 5–20 min on average. Together, these studies suggested that the conformation of the chromatin fiber within a TAD might vary considerably over the timescale of minutes in living cells under the action of cohesin-mediated extrusion.
Analysis of the motion of genomic locations 86, 87, 88, 89 or chromatin-bound proteins [90] initially suggested that chromatin motion is subdiffusive and that promoters or enhancers explore the surrounding nuclear space in a way that might depend on their activity 86, 87. Only very recently, live-cell imaging studies provided insights into how CTCF and cohesin control the looping dynamics of chromosomal sequences located within or close to TAD boundaries. Analysis of the dynamics of pairs of genomic loci in mouse embryonic stem cells revealed that cohesin-mediated loops between pairs of convergent CTCF sites last ∼5–30 min on average 91••, 92••, which is a relatively small fraction of the cell cycle duration (∼12 hours in this cell type). One study estimated that in the absence of CTCF sites, two sequences within a TAD only spend around 5 min on average within ∼150 nm from each other (i.e. the upper range of estimated E–P communication distances, see above) [92]. Together with other recent studies of chromatin motion 93•, 94•, these new results suggest that windows of opportunities for regulatory exchange might be transient events occurring as enhancers and promoters explore the nuclear space in the context of a highly dynamic chromatin fiber.
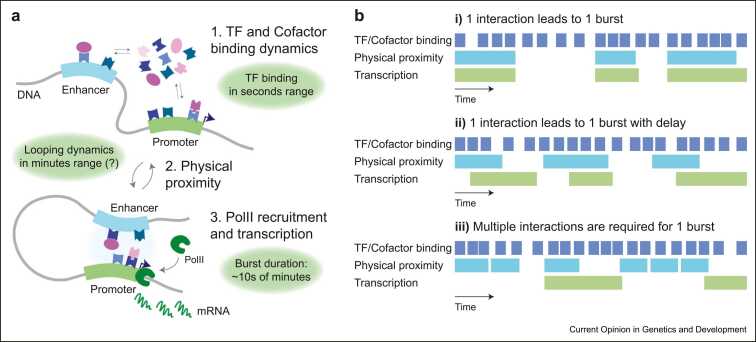
Interestingly, these experimental estimates for the timescales of CTCF looping and physical proximity represent an intermediate level between those of nucleosome remodeling and TF binding/unbinding, which typically occur on the second timescale, and those of promoter bursting, which in mammalian cells takes place over tens of minutes (Figure 3a) [95]. How these substantially disconnected timescales (at least in mammals; this might not be the case in budding yeast [96]) are integrated to elicit the correct transcription levels and cell-to-cell and temporal variability remains to be understood. Given the fast binding/unbinding kinetics of many TFs and the potentially longer timescales of E–P proximity, it is possible that time-averaged TFBS occupancies within enhancer and promoter regions are sufficient to enable the exchange of regulatory information throughout the duration of a contact. But even if this is the case, does a burst of transcription initiate every time that an enhancer is close enough to its target promoter that their transiently bound TFs can interact (Figure 3b, i)? Or does this happen with a delay, possibly due to the kinetics of intermediate regulatory steps (e.g. assembly or interactions with cofactors) (Figure 3b, ii)? Or rather does a burst of transcription only initiate after several contacts, for example, because of cooperativity in one or more sequential events occurring at the promoter (Figure 3b, iii)? Is every E–P pair actually expected to behave the same, or do alternative modes of signal integration [97] result in different behaviors at different E–P pairs? And how is this related to bursting kinetics at the promoter? Although the 1-contact-1-burst scenario has been observed in live Drosophila embryos, it is likely that it was favored by ectopically induced, unusually stable E–P interactions [98]. Live-cell imaging experiments in mammalian cells suggested a total absence of correlation between E–P and transcription bursts [99], but optical resolution limits discussed above urge caution on the interpretation of these results. Theoretical arguments that are compatible with scenarios ii) and iii) have been recently evoked to explain striking nonlinearities in how transcription levels depend on E–P contact probabilities 46, 100, although none has been formally proven. The rapid advance of super-resolution microscopy techniques that are compatible with live-cell approaches 101••, 102 will likely provide measurements of E–P proximity with increased accuracy together with single-molecule-resolved binding/unbinding of TFs and cofactors [96], thus allowing for a more precise assessment of the temporal order of events at E–P interfaces over timescales ranging from a fraction of a second to minutes and hours.
Figure 3.
Temporal dynamics of E–P communication. (a) Transcription initiation and the resulting transcriptional bursts are preceded by multiple processes, notably fast TF and cofactor binding (typically in the second to tens-of-seconds range) and looping of the chromatin fiber that establishes physical proximity, recently suggested to occur on the order of minutes. (b) While TF-binding dynamics are fast and occur frequently, possibly enabling continuous occupancy of TFBS at the enhancer and the promoter, E–P proximity is presumably longer and occurs less frequently, leading to potential models of E–P communication in time: i) one interaction might lead to one event of transcription initiation, ii) possibly with a delay, or alternatively more complicated scenarios iii) where one initiation event can only be achieved by multiple consecutive interaction events.
We would finally like to note that mathematical and physical models have been instrumental to push forward our understanding of how stochastic and dynamic regulatory processes in single cells convert regulatory inputs into transcriptional outputs. To cite just a few, models have provided mechanistic bases for the processes that shape chromosome organization and dynamics [42] as well as alternative hypotheses on how transcription bursts are generated [97] and connected to E–P interactions 46, 100. Further, they have allowed to quantify how regulatory information is processed by E–P modules into cell states [103]. We expect that the role of models will become even more prominent in the future as the data in the field become more and more quantitative.
Conclusions and outlook
Understanding how genomic sequence determines gene expression programs and their dynamics during development and homeostasis relies on our ability to understand E–P communication from a quantitative point of view, that is, to a level that allows us to predict how sequence mutations translate into changes of transcription levels and their cell-to-cell variability. Over the last few years, we have witnessed spectacular progress in this direction, thanks to the fast-paced rate of technological development and the advent of quantitative methods for the measurements of biological processes. It will be exciting to see how these methods can be coupled to another emerging family of approaches relying on the ‘bottom-up’ generation of regulatory landscapes with controllable levels of complexity 20, 21, 46, 47. By reducing the number of biological degrees of freedom to an amenable amount, such genome engineering approaches have the potential to disentangle regulatory layers and push the field forward toward the identification of mechanistic regulatory principles of E–P communication. We very much look forward to the next phase of experimental and theoretical studies, which holds great promise to open exciting insights into the molecular mechanisms of long-range transcriptional regulation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Ilya Flyamer for critical reading of the paper. P.M. was supported by a Marie Skłodowska-Curie Innovative Training Network (Grant no. 813327 ‘ChromDesign’). Research in the Giorgetti laboratory is funded by the Novartis Foundation, the European Research Council (Grant no. 759366, ‘BioMeTre’), Marie Skłodowska-Curie Innovative Training Networks (Grant nos. 813327 ‘ChromDesign’ and 813282 ‘PEP-NET’) under the European Union’s Horizon 2020 research and innovation program, and the Swiss National Science Foundation (Grant no. 310030_192642).
Data Availability
No data were used for the research described in the article.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
-
•
of special interest
-
••
of outstanding interest
- 1.Kim S., Wysocka J. Deciphering the multi-scale, quantitative cis-regulatory code. Mol Cell. 2023;83:373–392. doi: 10.1016/j.molcel.2022.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Claringbould A., Zaugg J.B. Enhancers in disease: molecular basis and emerging treatment strategies. Trends Mol Med. 2021;27:1060–1073. doi: 10.1016/j.molmed.2021.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Karpinska M.A., Oudelaar A.M. The role of loop extrusion in enhancer-mediated gene activation. Curr Opin Genet Dev. 2023;79 doi: 10.1016/j.gde.2023.102022. [DOI] [PubMed] [Google Scholar]
- 4.Pachano T., Haro E., Rada-Iglesias A. Enhancer-gene specificity in development and disease. Development. 2022;149 doi: 10.1242/dev.186536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Field A., Adelman K. Evaluating enhancer function and transcription. Annu Rev Biochem. 2020;89:213–234. doi: 10.1146/annurev-biochem-011420-095916. [DOI] [PubMed] [Google Scholar]
- 6.Jindal G.A., Farley E.K. Enhancer grammar in development, evolution, and disease: dependencies and interplay. Dev Cell. 2021;56:575–587. doi: 10.1016/j.devcel.2021.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasperini M., Hill A.J., McFaline-Figueroa J.L., Martin B., Kim S., Zhang M.D., Jackson D., Leith A., Schreiber J., Noble W.S., et al. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell. 2019;176:377–390. doi: 10.1016/j.cell.2018.11.029. .e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark S.J., Argelaguet R., Kapourani C.-A., Stubbs T.M., Lee H.J., Alda-Catalinas C., Krueger F., Sanguinetti G., Kelsey G., Marioni J.C., et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03149-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Findlay G.M., Boyle E.A., Hause R.J., Klein J.C., Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnold C.D., Gerlach D., Stelzer C., Boryń Ł.M., Rath M., Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- 11••.de Almeida B.P., Reiter F., Pagani M., Stark A. DeepSTARR predicts enhancer activity from DNA sequence and enables the de novo design of synthetic enhancers. Nat Genet. 2022;54:613–624. doi: 10.1038/s41588-022-01048-5. [DOI] [PubMed] [Google Scholar]; Together with Ref. [12], this study provides a striking example of deep-learning guided strategy for enhancer grammar inference. Learning was done on STARR-seq libraries and generated models were then used to inform the design of synthetic enhancer sequences that were experimentally tested for transcriptional activity in Drosophila cells.
- 12••.Taskiran I.I., Spanier K.I., Christiaens V., Mauduit D., Aerts S. Cell type directed design of synthetic enhancers. bioRxiv. 2022 doi: 10.1101/2022.07.26.501466. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Ref. [11], this study provides a striking example of deep-learning guided strategy for enhancer grammar inference. This study employed deep learning approaches to learn transcription levels from endogenous DNA sequence in Drosophila. Models were then used to inform the design of short (∼50 bp) synthetic enhancers whose functionality can be modulated in strength and cell-type specificity.
- 13.Haberle V., Arnold C.D., Pagani M., Rath M., Schernhuber K., Stark A. Transcriptional cofactors display specificity for distinct types of core promoters. Nature. 2019;570:122–126. doi: 10.1038/s41586-019-1210-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14••.Sahu B., Hartonen T., Pihlajamaa P., Wei B., Dave K., Zhu F., Kaasinen E., Lidschreiber K., Lidschreiber M., Daub C.O., et al. Sequence determinants of human gene regulatory elements. Nat Genet. 2022;54:283–294. doi: 10.1038/s41588-021-01009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reports massive parallel reporter assays with fully synthetic enhancer and promoter sequences. They found that spacing between TFBS, TFBS motif, and orientation, as well as their combination, is a weak determinant of the activity of single TFs, but that many TFs interact in a additive manner. This study also argues against a model of E–P communication that is mediated by specific TF–TF interactions.
- 15••.Martinez-Ara M., Comoglio F., van Arensbergen J., van Steensel B. Systematic analysis of intrinsic enhancer-promoter compatibility in the mouse genome. Mol Cell. 2022;82:2519–2531. doi: 10.1016/j.molcel.2022.04.009. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Here, E–P compatibility rules in mammalian cells were investigated with a high-throughput combinatorial reporter assay that samples thousands ofE–P combinations from a 3-Mb region in the mouse genome, revealing quantitative differences in compatibility levels. Interestingly, comparing the compatibility of individual E–P pairs to their contact probability in the mouse genome revealed no correlation, suggesting that genome topology and E–P compatibility are independent mechanisms of transcriptional regulation.
- 16••.Bergman D.T., Jones T.R., Liu V., Ray J., Jagoda E., Siraj L., Kang H.Y., Nasser J., Kane M., Rios A., et al. Compatibility rules of human enhancer and promoter sequences. Nature. 2022;607:176–184. doi: 10.1038/s41586-022-04877-w. [DOI] [PMC free article] [PubMed] [Google Scholar]; Combinations of 1'000 × 1'000 human enhancer and promoter fragments were assayed in massively parallel reporter assays. Most of the transcriptional activity of E–P pairs could be explained by the product of individual enhancer and promoter intrinsic activities, and broad levels of E–P compatibility were detected, with only mild sequence specificity.
- 17.Ruf S., Symmons O., Uslu V.V., Dolle D., Hot C., Ettwiller L., Spitz F. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat Genet. 2011;43:379–386. doi: 10.1038/ng.790. [DOI] [PubMed] [Google Scholar]
- 18.Anderson E., Devenney P.S., Hill R.E., Lettice L.A. Mapping the Shh long-range regulatory domain. Development. 2014;141:3934–3943. doi: 10.1242/dev.108480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galupa R., Alvarez-Canales G., Borst N.O., Fuqua T., Gandara L., Misunou N., Richter K., Alves M.R.P., Karumbi E., Perkins M.L., et al. Enhancer architecture and chromatin accessibility constrain phenotypic space during Drosophila development. Dev Cell. 2023;58:51–62. doi: 10.1016/j.devcel.2022.12.003. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brosh R., Coelho C., Ribeiro-dos-Santos A.M., Ellis G., Hogan M.S., Ashe H.J., Somogyi N., Ordoñez R., Luther R.D., Huang E., et al. Synthetic regulatory genomics uncovers enhancer context dependence at the Sox2 locus. Mol Cell. 2023;7:1140–1152. doi: 10.1016/j.molcel.2023.02.027. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blayney J.W., Kassouf M., Francis H., Camellato B.R., Stolper R., Higgs D.R., Mitchell L., Boeke J. Super-enhancers require a combination of classical enhancers and novel facilitator elements to drive high levels of gene expression. bioRxiv. 2022 doi: 10.1101/2022.06.20.496856. [DOI] [Google Scholar]
- 22.Policarpi C., Munafò M., Tsagkris S., Carlini V., Hackett J.A. Systematic epigenome editing captures the context-dependent instructive function of chromatin modifications. bioRxiv. 2022 doi: 10.1101/2022.09.04.506519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alerasool N., Leng H., Lin Z.-Y., Gingras A.-C., Taipale M. Identification and functional characterization of transcriptional activators in human cells. Mol Cell. 2022;82:677–695. doi: 10.1016/j.molcel.2021.12.008. e7. [DOI] [PubMed] [Google Scholar]
- 24.Neumayr C., Haberle V., Serebreni L., Karner K., Hendy O., Boija A., Henninger J.E., Li C.H., Stejskal K., Lin G., et al. Differential cofactor dependencies define distinct types of human enhancers. Nature. 2022;606:406–413. doi: 10.1038/s41586-022-04779-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukund A.X., Tycko J., Allen S.J., Robinson S.A., Andrews C., Ludwig C.H., Spees K., Bassik M.C., Bintu L. High-throughput functional characterization of combinations of transcriptional activators and repressors. bioRxiv. 2022 doi: 10.1101/2022.12.20.521091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naqvi S., Kim S., Hoskens H., Matthews H.S., Spritz R.A., Klein O.D., Hallgrímsson B., Swigut T., Claes P., Pritchard J.K., et al. Precise modulation of transcription factor levels reveals drivers of dosage sensitivity. Nat Genet. 2023;55:841–851. doi: 10.1038/s41588-023-01366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Noviello G., Gjaltema R.A.F., Schulz E.G. CasTuner: a degron and CRISPR/Cas-based toolkit for analog tuning of endogenous gene expression. bioRxiv. 2022 doi: 10.1101/2022.10.05.511019. [DOI] [PMC free article] [PubMed] [Google Scholar]; The method developed here allows the fine-tuning of endogenous gene expression by targeting dCas9 fused to FKBP degron domain to endogenous promoters. By titrating the small molecule dTAG-13 to induce degradation of the FKBP-fusion protein, expression levels at targeted promoters can be tuned with a high homogeneity across a cell population.
- 28.Zhu F., Farnung L., Kaasinen E., Sahu B., Yin Y., Wei B., Dodonova S.O., Nitta K.R., Morgunova E., Taipale M., et al. The interaction landscape between transcription factors and the nucleosome. Nature. 2018;562:76–81. doi: 10.1038/s41586-018-0549-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novakovsky G., Dexter N., Libbrecht M.W., Wasserman W.W., Mostafavi S. Obtaining genetics insights from deep learning via explainable artificial intelligence. Nat Rev Genet. 2023;24:125–137. doi: 10.1038/s41576-022-00532-2. [DOI] [PubMed] [Google Scholar]
- 30.Sönmezer C., Kleinendorst R., Imanci D., Barzaghi G., Villacorta L., Schübeler D., Benes V., Molina N., Krebs A.R. Molecular co-occupancy identifies transcription factor binding cooperativity in vivo. Mol Cell. 2021;81:255–267. doi: 10.1016/j.molcel.2020.11.015. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rao S., Ahmad K., Ramachandran S. Cooperative binding between distant transcription factors is a hallmark of active enhancers. Mol Cell. 2021;81:1651–1665. doi: 10.1016/j.molcel.2021.02.014. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Kreibich E., Kleinendorst R., Barzaghi G., Kaspar S., Krebs A.R. Single-molecule footprinting identifies context-dependent regulation of enhancers by DNA methylation. Mol Cell. 2023;83:787–802. doi: 10.1016/j.molcel.2023.01.017. e9. [DOI] [PubMed] [Google Scholar]; Dual-enzyme single-molecule footprinting performed on endogenous mouse regulatory sequences demonstrated that although the majority of enhancers is not sensitive to DNA methylation, CpG methylation can directly inhibit the binding of several TFs whose TFBS define a class of methylation-sensitive regulatory elements.
- 33.Stergachis A.B., Debo B.M., Haugen E., Churchman L.S., Stamatoyannopoulos J.A. Single-molecule regulatory architectures captured by chromatin fiber sequencing. Science. 2020;368:1449–1454. doi: 10.1126/science.aaz1646. [DOI] [PubMed] [Google Scholar]
- 34.Krebs A.R., Imanci D., Hoerner L., Gaidatzis D., Burger L., Schübeler D. Genome-wide single-molecule footprinting reveals high RNA Polymerase II turnover at paused promoters. Mol Cell. 2017;67:411–422. doi: 10.1016/j.molcel.2017.06.027. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Battaglia S., Dong K., Wu J., Chen Z., Najm F.J., Zhang Y., Moore M.M., Hecht V., Shoresh N., Bernstein B.E. Long-range phasing of dynamic, tissue-specific and allele-specific regulatory elements. Nat Genet. 2022;54:1504–1513. doi: 10.1038/s41588-022-01188-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses single-molecule methylation footprinting coupled with Cas9-mediated sequence enrichment followed by Nanopore sequencing to profile open chromatin regions across immune loci of human cells. Long read length (∼100 kb) generated allows interrogation of correlation between activities of distal regulatory elements, such as enhancers and promoters
- 36.Michael A.K., Thomä N.H. Reading the chromatinized genome. Cell. 2021;184:3599–3611. doi: 10.1016/j.cell.2021.05.029. [DOI] [PubMed] [Google Scholar]
- 37.Li J., Dong A., Saydaminova K., Chang H., Wang G., Ochiai H., Yamamoto T., Pertsinidis A. Single-molecule nanoscopy elucidates RNA Polymerase II transcription at single genes in live cells. Cell. 2019;178:491–506. doi: 10.1016/j.cell.2019.05.029. e428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balzarotti F., Eilers Y., Gwosch K.C., Gynnå A.H., Westphal V., Stefani F.D., Elf J., Hell S.W. Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science. 2017;355:606–612. doi: 10.1126/science.aak9913. [DOI] [PubMed] [Google Scholar]
- 39.Osterwalder M., Barozzi I., Tissières V., Fukuda-Yuzawa Y., Mannion B.J., Afzal S.Y., Lee E.A., Zhu Y., Plajzer-Frick I., Pickle C.S., et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 2018;554:239–243. doi: 10.1038/nature25461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., van Berkum N.L., Meisig J., Sedat J., et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fudenberg G., Imakaev M., Lu C., Goloborodko A., Abdennur N., Mirny L.A. Formation of chromosomal domains by loop extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsieh T.-H.S., Cattoglio C., Slobodyanyuk E., Hansen A.S., Darzacq X., Tjian R. Enhancer–promoter interactions and transcription are largely maintained upon acute loss of CTCF, cohesin, WAPL or YY1. Nat Genet. 2022;54:1919–1932. doi: 10.1038/s41588-022-01223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Batut P.J., Bing X.Y., Sisco Z., Raimundo J., Levo M., Levine M.S. Genome organization controls transcriptional dynamics during development. Science. 2022;375:566–570. doi: 10.1126/science.abi7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L.-F., Long H.K., Park M., Swigut T., Boettiger A.N., Wysocka J. Structural elements promote architectural stripe formation and facilitate ultra-long-range gene regulation at a human disease locus. Mol Cell. 2023;83:1446–1461.e6. doi: 10.1016/j.molcel.2023.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Zuin J., Roth G., Zhan Y., Cramard J., Redolfi J., Piskadlo E., Mach P., Kryzhanovska M., Tihanyi G., Kohler H., et al. Nonlinear control of transcription through enhancer–promoter interactions. Nature. 2022;604:571–577. doi: 10.1038/s41586-022-04570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rinzema N.J., Sofiadis K., Tjalsma S.J.D., Verstegen M.J.A.M., Oz Y., Valdes-Quezada C., Felder A.-K., Filipovska T., van der Elst S., de Andrade dos Ramos Z., et al. Building regulatory landscapes reveals that an enhancer can recruit cohesin to create contact domains, engage CTCF sites and activate distant genes. Nat Struct Mol Biol. 2022;29:563–574. doi: 10.1038/s41594-022-00787-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kane L., Williamson I., Flyamer I.M., Kumar Y., Hill R.E., Lettice L.A., Bickmore W.A. Cohesin is required for long-range enhancer action at the Shh locus. Nat Struct Mol Biol. 2022;29:891–897. doi: 10.1038/s41594-022-00821-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakraborty S., Kopitchinski N., Zuo Z., Eraso A., Awasthi P., Chari R., Mitra A., Tobias I.C., Moorthy S.D., Dale R.K., et al. Enhancer–promoter interactions can bypass CTCF-mediated boundaries and contribute to phenotypic robustness. Nat Genet. 2023;55:280–290. doi: 10.1038/s41588-022-01295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang H., Zhu Q., Jussila A., Han Y., Bintu B., Kern C., Conte M., Zhang Y., Bianco S., Chiariello A.M., et al. CTCF mediates dosage- and sequence-context-dependent transcriptional insulation by forming local chromatin domains. Nat Genet. 2021;53:1064–1074. doi: 10.1038/s41588-021-00863-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fulco C.P., Nasser J., Jones T.R., Munson G., Bergman D.T., Subramanian V., Grossman S.R., Anyoha R., Doughty B.R., Patwardhan T.A., et al. Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nat Genet. 2019;51:1664–1669. doi: 10.1038/s41588-019-0538-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gavrilov A., Razin S.V., Cavalli G. In vivo formaldehyde cross-linking: it is time for black box analysis. Brief Funct Genom. 2015;14:163–165. doi: 10.1093/bfgp/elu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Redolfi J., Zhan Y., Valdes-Quezada C., Kryzhanovska M., Guerreiro I., Iesmantavicius V., Pollex T., Grand R.S., Mulugeta E., Kind J., et al. DamC reveals principles of chromatin folding in vivo without crosslinking and ligation. Nat Struct Mol Biol. 2019;26:471–480. doi: 10.1038/s41594-019-0231-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bintu B., Mateo L.J., Su J.-H., Sinnott-Armstrong N.A., Parker M., Kinrot S., Yamaya K., Boettiger A.N., Zhuang X. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science. 2018;362 doi: 10.1126/science.aau1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mateo L.J., Murphy S.E., Hafner A., Cinquini I.S., Walker C.A., Boettiger A.N. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature. 2019;568:49–54. doi: 10.1038/s41586-019-1035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gizzi A.M.C., Cattoni D.I., Fiche J.-B., Espinola S.M., Gurgo J., Messina O., Houbron C., Ogiyama Y., Papadopoulos G.L., Cavalli G., et al. Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms. Mol Cell. 2019;74:212–222. doi: 10.1016/j.molcel.2019.01.011. e5. [DOI] [PubMed] [Google Scholar]
- 57.Brandão H.B., Gabriele M., Hansen A.S. Tracking and interpreting long-range chromatin interactions with super-resolution live-cell imaging. Curr Opin Cell Biol. 2021;70:18–26. doi: 10.1016/j.ceb.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Acuña L.I.G., Flyamer I., Boyle S., Friman E.T., Bickmore W.A. Transcription decouples estrogen-dependent changes in enhancer-promoter contact frequencies and physical proximity. bioRxiv. 2023 doi: 10.1101/2023.03.29.534720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ostersehlt L.M., Jans D.C., Wittek A., Keller-Findeisen J., Inamdar K., Sahl S.J., Hell S.W., Jakobs S. DNA-PAINT MINFLUX nanoscopy. Nat Methods. 2022;19:1072–1075. doi: 10.1038/s41592-022-01577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rengachari S., Schilbach S., Aibara S., Dienemann C., Cramer P. Structure of the human Mediator–RNA polymerase II pre-initiation complex. Nature. 2021;594:129–133. doi: 10.1038/s41586-021-03555-7. [DOI] [PubMed] [Google Scholar]
- 61.Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A., Phase A. Separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chong S., Dugast-Darzacq C., Liu Z., Dong P., Dailey G.M., Cattoglio C., Heckert A., Banala S., Lavis L., Darzacq X., et al. Imaging dynamic and selective low-complexity domain interactions that control gene transcription. Science. 2018;361:eaar2555. doi: 10.1126/science.aar2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karr J.P., Ferrie J.J., Tjian R., Darzacq X. The transcription factor activity gradient (TAG) model: contemplating a contact-independent mechanism for enhancer–promoter communication. Genes Dev. 2022;36:7–16. doi: 10.1101/gad.349160.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tycko J., DelRosso N., Hess G.T., Aradhana, Banerjee A., Mukund A., Van M.V., Ego B.K., Yao D., Spees K., et al. High-throughput discovery and characterization of human transcriptional effectors. Cell. 2020;183:2020–2035. doi: 10.1016/j.cell.2020.11.024. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65••.DelRosso N, Tycko J, Suzuki P, Andrews C, Aradhana, Mukund A, Liongson I, Ludwig C, Spees K, Fordyce P, et al. Large-scale mapping and mutagenesis of human transcriptional effector domains. Nature. 2023;616:365–372. doi: 10.1038/s41586-023-05906-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; Highly parallel assays where protein fragments are recruited to a reporter promoter and their sequences are identified by high-throughput sequencing revealed amino acid sequence that can activate or repress transcription. Sequence analysis revealed composition biases of activators and repressors and a widespread role of SUMOylation in transcriptional repressors.
- 66.de Wit E., Nora E.P. New insights into genome folding by loop extrusion from inducible degron technologies. Nat Rev Genet. 2022;24:73–85. doi: 10.1038/s41576-022-00530-4. [DOI] [PubMed] [Google Scholar]
- 67.Jaeger M.G., Schwalb B., Mackowiak S.D., Velychko T., Hanzl A., Imrichova H., Brand M., Agerer B., Chorn S., Nabet B., et al. Selective Mediator dependence of cell-type-specifying transcription. Nat Genet. 2020;52:719–727. doi: 10.1038/s41588-020-0635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haarhuis J.H.I., van der Weide R.H., Blomen V.A., Flach K.D., Teunissen H., Willems L., Brummelkamp T.R., Rowland B.D., de Wit E. A Mediator-cohesin axis controls heterochromatin domain formation. Nat Commun. 2022;13 doi: 10.1038/s41467-022-28377-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ramasamy S., Aljahani A., Karpinska M., Cao N., Cruz N., Oudelaar A.M. The Mediator complex regulates enhancer-promoter interactions. bioRxiv. 2022 doi: 10.1101/2022.06.15.496245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barisic D., Stadler M.B., Iurlaro M., Schübeler D. Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors. Nature. 2019;569:136–140. doi: 10.1038/s41586-019-1115-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin B.J.E., Ablondi E.F., Goglia C., Adelman K. Global identification of direct SWI/SNF targets reveals compensation by EP400. bioRxiv. 2023 doi: 10.1101/2023.03.07.531379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Göös H., Kinnunen M., Salokas K., Tan Z., Liu X., Yadav L., Zhang Q., Wei G.-H., Varjosalo M. Human transcription factor protein interaction networks. Nat Commun. 2022;13 doi: 10.1038/s41467-022-28341-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tebo A.G., Gautier A. A split fluorescent reporter with rapid and reversible complementation. Nat Commun. 2019;10 doi: 10.1038/s41467-019-10855-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74•.Graham T.G., Ferrie J.J., Dailey G.M., Tjian R., Darzacq X. Detecting molecular interactions in live-cell single-molecule imaging with proximity-assisted photoactivation (PAPA) eLife. 2022;11 doi: 10.7554/eLife.76870. [DOI] [PMC free article] [PubMed] [Google Scholar]; Extending the detection range of proximity-mediated biotinylation, protein complementation assays and Förster resonance energy transfer, this new technique allows detecting single protein–protein interactions in living cells by fluorescence imaging of a receiver fluorophore that gets activated when in physical proximity with a donor fluorophore.
- 75.Nguyen H.Q., Chattoraj S., Castillo D., Nguyen S.C., Nir G., Lioutas A., Hershberg E.A., Martins N.M.C., Reginato P.L., Hannan M., et al. 3D mapping and accelerated super-resolution imaging of the human genome using in situ sequencing. Nat Methods. 2020;17:822–832. doi: 10.1038/s41592-020-0890-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lu T., Ang C.E., Zhuang X. Spatially resolved epigenomic profiling of single cells in complex tissues. Cell. 2022;185:4448–4464. doi: 10.1016/j.cell.2022.09.035. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Müller T.G., Zila V., Peters K., Schifferdecker S., Stanic M., Lucic B., Laketa V., Lusic M., Müller B., Kräusslich H.-G. HIV-1 uncoating by release of viral cDNA from capsid-like structures in the nucleus of infected cells. eLife. 2021;10 doi: 10.7554/eLife.64776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nagano T., Lubling Y., Stevens T.J., Schoenfelder S., Yaffe E., Dean W., Laue E.D., Tanay A., Fraser P. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502:59–64. doi: 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Giorgetti L., Galupa R., Nora E.P., Piolot T., Lam F., Dekker J., Tiana G., Heard E. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell. 2014;157:950–963. doi: 10.1016/j.cell.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Finn E.H., Misteli T. Molecular basis and biological function of variability in spatial genome organization. Science. 2019;365 doi: 10.1126/science.aaw9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davidson I.F., Bauer B., Goetz D., Tang W., Wutz G., Peters J.-M. DNA loop extrusion by human cohesin. Science. 2019;366:1338–1345. doi: 10.1126/science.aaz3418. [DOI] [PubMed] [Google Scholar]
- 82.Kim Y., Shi Z., Zhang H., Finkelstein I.J., Yu H. Human cohesin compacts DNA by loop extrusion. Science. 2019;366:1345–1349. doi: 10.1126/science.aaz4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Golfier S., Quail T., Kimura H., Brugués J. Cohesin and condensin extrude loops in a cell-cycle dependent manner. eLife. 2020;9 doi: 10.7554/eLife.53885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wutz G., Ladurner R., St, Hilaire B.G., Stocsits R.R., Nagasaka K., Pignard B., Sanborn A., Tang W., Várnai C., Ivanov M.P., et al. ESCO1 and CTCF enable formation of long chromatin loops by protecting cohesinSTAG1 from WAPL. eLife. 2020;9 doi: 10.7554/eLife.52091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cattoglio C., Pustova I., Walther N., Ho J.J., Hantsche-Grininger M., Inouye C.J., Hossain M.J., Dailey G.M., Ellenberg J., Darzacq X., et al. Determining cellular CTCF and cohesin abundances to constrain 3D genome models. eLife. 2019;8 doi: 10.7554/eLife.40164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gu B., Swigut T., Spencley A., Bauer M.R., Chung M., Meyer T., Wysocka J. Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science. 2018;359:1050–1055. doi: 10.1126/science.aao3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Germier T., Kocanova S., Walther N., Bancaud A., Shaban H.A., Sellou H., Politi A.Z., Ellenberg J., Gallardo F., Bystricky K. Real-time imaging of a single gene reveals transcription-initiated local confinement. Biophys J. 2017;113:1383–1394. doi: 10.1016/j.bpj.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Masui O., Bonnet I., Le Baccon P., Brito I., Pollex T., Murphy N., Hupé P., Barillot E., Belmont A.S., Heard E. Live-cell chromosome dynamics and outcome of X chromosome pairing events during ES cell differentiation. Cell. 2011;145:447–458. doi: 10.1016/j.cell.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Khanna N., Zhang Y., Lucas J.S., Dudko O.K., Murre C. Chromosome dynamics near the sol-gel phase transition dictate the timing of remote genomic interactions. Nat Commun. 2019;10 doi: 10.1038/s41467-019-10628-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nozaki T., Imai R., Tanbo M., Nagashima R., Tamura S., Tani T., Joti Y., Tomita M., Hibino K., Kanemaki M.T., et al. Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol Cell. 2017;67:282–293. doi: 10.1016/j.molcel.2017.06.018. e7. [DOI] [PubMed] [Google Scholar]
- 91••.Gabriele M., Brandão H.B., Grosse-Holz S., Jha A., Dailey G.M., Cattoglio C., Hsieh T.-H.S., Mirny L., Zechner C., Hansen A.S. Dynamics of CTCF- and cohesin-mediated chromatin looping revealed by live-cell imaging. Science. 2022;376:496–501. doi: 10.1126/science.abn6583. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Ref. [92], this study presents the first measurements of cohesin-mediated CTCF looping in single living cells. CTCF-anchored loops between sites separated by ∼500 kb at an endogenous locus were found to last ∼10–30 min, for a total of ∼3–6% of time spent in the looped state. These results suggest that CTCF loops are rare and transient compared to the duration of the cell cycle and that the conformation of chromatin inside a TAD is highly dynamic.
- 92••.Mach P., Kos P.I., Zhan Y., Cramard J., Gaudin S., Tünnermann J., Marchi E., Eglinger J., Zuin J., Kryzhanovska M., et al. Cohesin and CTCF control the dynamics of chromosome folding. Nat Genet. 2022;54:1907–1918. doi: 10.1038/s41588-022-01232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; Together with Ref. [91], this study presents the first measurements of cohesin-mediated CTCF looping in single living cells. CTCF-anchored loops spanning 150 kb within an ectopic location were found to last ∼5–15 min, for a total of ∼20–30% of time spent in the looped state. These results are in excellent agreement with Ref. [85] considering the different genomic distance and CTCF sites and also suggest that the conformation of chromatin inside a TAD is highly dynamic.
- 93•.Keizer V.I.P., Grosse-Holz S., Woringer M., Zambon L., Aizel K., Bongaerts M., Delille F., Kolar-Znika L., Scolari V.F., Hoffmann S., et al. Live-cell micromanipulation of a genomic locus reveals interphase chromatin mechanics. Science. 2022;377:489–495. doi: 10.1126/science.abi9810. [DOI] [PubMed] [Google Scholar]; Using controlled magnetic forces to manipulate the motion of a genetic locus in living mouse stem cells, the authors showed that chromosome mobility in living cells is less influenced by nuclear constraints than previously thought. This enabling technology can potentially be used in the future to study the mechanics of other processes in combination with the motion of a single locus in living cells.
- 94•.Brückner D.B., Chen H., Barinov L., Zoller B., Gregor T. Stochastic motion and transcriptional dynamics of pairs of distal DNA loci on a compacted chromosome. bioRxiv. 2023 doi: 10.1101/2023.01.18.524527. [DOI] [PMC free article] [PubMed] [Google Scholar]; Live-cell imaging of ectopic E–P pairs in Drosophila embryos provides the first measurements of chromosome looping as a function of the intervening genomic distance. Genomic loci were found to undergo fast subdiffusive dynamics, while being constrained to a compact 3D organization of the chromatin fiber. Together with Refs. [85,86], the results point to a striking similarity of chromatin dynamics across organisms.
- 95.Coulon A., Chow C.C., Singer R.H., Larson D.R. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat Rev Genet. 2013;14:572–584. doi: 10.1038/nrg3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Donovan B.T., Huynh A., Ball D.A., Patel H.P., Poirier M.G., Larson D.R., Ferguson M.L., Lenstra T.L. Live-cell imaging reveals the interplay between transcription factors, nucleosomes, and bursting. EMBO J. 2019;38:e100809. doi: 10.15252/embj.2018100809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97•.Lammers N.C., Kim Y.J., Zhao J., Garcia H.G. A matter of time: using dynamics and theory to uncover mechanisms of transcriptional bursting. Curr Opin Cell Biol. 2020;67:147–157. doi: 10.1016/j.ceb.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Here, the authors propose different mathematical models of how the integrations of time scales of molecular processes involved in transcriptional regulation (TF/Cofactor binding, E–P contacts and transcriptional bursting) might result in transcriptional bursting.
- 98.Chen H., Levo M., Barinov L., Fujioka M., Jaynes J.B., Gregor T. Dynamic interplay between enhancer–promoter topology and gene activity. Nat Genet. 2018;50:1296–1303. doi: 10.1038/s41588-018-0175-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alexander J.M., Guan J., Li B., Maliskova L., Song M., Shen Y., Huang B., Lomvardas S., Weiner O.D. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. eLife. 2019;8 doi: 10.7554/eLife.41769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xiao J.Y., Hafner A., Boettiger A.N. How subtle changes in 3D structure can create large changes in transcription. eLife. 2021;10 doi: 10.7554/eLife.64320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101••.Deguchi T., Iwanski M.K., Schentarra E.-M., Heidebrecht C., Schmidt L., Heck J., Weihs T., Schnorrenberg S., Hoess P., Liu S., et al. Direct observation of motor protein stepping in living cells using MINFLUX. Science. 2023;379:1010–1015. doi: 10.1126/science.ade2676. [DOI] [PMC free article] [PubMed] [Google Scholar]; MINFLUX nanoscopy was used to track the dynamics of kinesin-1 stepping on microtubules in living cells at the single-molecule level. This is one of the first examples of how single proteins can be followed with nanometer spatial and millisecond temporal resolution in live-cell imaging.
- 102.Cao B., Coelho S., Li J., Wang G., Pertsinidis A. Volumetric interferometric lattice light-sheet imaging. Nat Biotechnol. 2021;39:1385–1393. doi: 10.1038/s41587-021-01042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tkačik G., Gregor T. The many bits of positional information. Development. 2021;148 doi: 10.1242/dev.176065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data were used for the research described in the article.