Significance
This study examines how interleukin (IL)-11 regulates inflammatory cell trafficking to the CNS in RRMS (relapsing–remitting multiple sclerosis). Single-cell RNA sequencing of blood-derived cells found that IL-11 induces NLRP3 inflammasome–related genes in monocytes. NLRP3 inflammasome induces secretion of IL-1b, which mediates monocyte trafficking to the CNS. A similar pattern of DEGs (differentiallyexpressed genes) was found in monocytes from the RRMS CSF (cerebrospinalfluid). αIL-11 mAb treatment of mice with an animal model of MS decreased the expression of inflammasome-related genes and proteins in CNS-derived monocytes. Nucleotide-binding domain, leucine-rich–containing family, pyrin domain–containing-3 (NLRP3) inflammasome inhibitors and αIL-11 mAb are proposed as an effective therapy for RRMS.
Keywords: multiple sclerosis, IL-11, NLRP3 inflammasome, EAE, monocyte
Abstract
The objective of this study is to examine IL-11-induced mechanisms of inflammatory cell migration to the central nervous system (CNS). We report that IL-11 is produced at highest frequency by myeloid cells among the peripheral blood mononuclear cell (PBMC) subsets. Patients with relapsing–remitting multiple sclerosis (RRMS) have an increased frequency of IL-11+ monocytes, IL-11+ and IL-11R+ CD4+ lymphocytes, and IL-11R+ neutrophils in comparison to matched healthy controls. IL-11+ and granulocyte-macrophage colony-stimulating factor (GM-CSF)+ monocytes, CD4+ lymphocytes, and neutrophils accumulate in the cerebrospinal fluid (CSF). The effect of IL-11 in-vitro stimulation, examined using single-cell RNA sequencing, revealed the highest number of differentially expressed genes in classical monocytes, including up-regulated NFKB1, NLRP3, and IL1B. All CD4+ cell subsets had increased expression of S100A8/9 alarmin genes involved in NLRP3 inflammasome activation. In IL-11R+-sorted cells from the CSF, classical and intermediate monocytes significantly up-regulated the expression of multiple NLRP3 inflammasome–related genes, including complement, IL18, and migratory genes (VEGFA/B) in comparison to blood-derived cells. Therapeutic targeting of this pathway with αIL-11 mAb in mice with RR experimental autoimmune encephalomyelitis (EAE) decreased clinical scores, CNS inflammatory infiltrates, and demyelination. αIL-11 mAb treatment decreased the numbers of NFκBp65+, NLRP3+, and IL-1β+ monocytes in the CNS of mice with EAE. The results suggest that IL-11/IL-11R signaling in monocytes represents a therapeutic target in RRMS.
A significant increase in IL-11 and IL-17A serum levels during clinical exacerbations and accumulation of IL-11+CD4+ cells in active brain multiple sclerosis (MS) lesions (1) prompted studies of its role in the development of the autoimmune response in MS. Our previous study (2) reported that IL-11 is the most up-regulated cytokine in the serum and CSF (cerebrospinal fluid) of patients with a first clinical presentation of MS, along with up-regulated Th1 and Th17 cytokines. IL-11 induced Th17 cell differentiation, and the secretion of Th17-promoting cytokines IL-1β and IL-23 by monocytes. It induced worsening of EAE (experimental autoimmune encephalomyelitis), and passive transfer of IL-11-stimulated encephalitogenic CD4+ cells, which express IL-17, IFN (interferon)-γ and GM-CSF, resulted in severe EAE (1). Previous reports on GM-CSF-regulated IL-1β monocyte secretion proposed IL-1β as a key mediator of the inflammatory cell migration across the blood–brain barrier (BBB) (3, 4). However, the role of IL-11 and NLRP3 inflammasome activation in inducing IL-1β+ monocyte migration in MS has not been elucidated.
IL-11 has been implicated as a proinflammatory cytokine in several inflammatory (5, 6) and autoimmune diseases (7, 8). IL-11 is highly produced by stromal fibroblasts (synoviocytes), monocytes, macrophages, and endothelial cells (ECs) in patients with rheumatoid arthritis (RA). In-vitro experiments demonstrated an IL-11-induced EC and fibroblast migration (9). IL-11 is also highly up-regulated in the colonic mucosa in ulcerative colitis (UC) and Crohn’s disease (CD) (10) where it is suggested as a therapeutic target (10).
IL-11 binds to IL-11R, followed by IL-11/IL11R complex high-affinity binding to the ubiquitously expressed gp130 signal transducer (IL6ST), which is shared by multiple IL-6 cytokine family members (11). IL-11 signaling activates NFκB and JAK/STAT, which mediate proinflammatory cytokine secretion, and the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase (ERK) pathway (9, 11, 12) involved in cell migration. In-vitro and in-vivo studies have demonstrated that IL-11 can signal in an autocrine fashion, whereby IL-11 induces its own receptor expression and secretion (13, 14). This cytokine signals through membrane-bound IL-11R, and in trans via soluble or antigen-presenting cell (APC)-expressing IL-11/IL-11R complex, which binds to gp130 on activated cells (15), thus broadening cell targets for IL-11 signaling. Proteolytic shedding of IL-11R by ADAM9 and ADAM10/ADAMTS10 supplies it for the formation of IL-11/IL-11R complexes that enhance gp130 signaling (16).
Results
The Frequency of IL-11+ Monocytes, IL-11+ and IL-11R+CD4+ Cells, and IL-11R+ Neutrophils Is Increased in RRMS (Relapsing–Remitting Multiple Sclerosis).
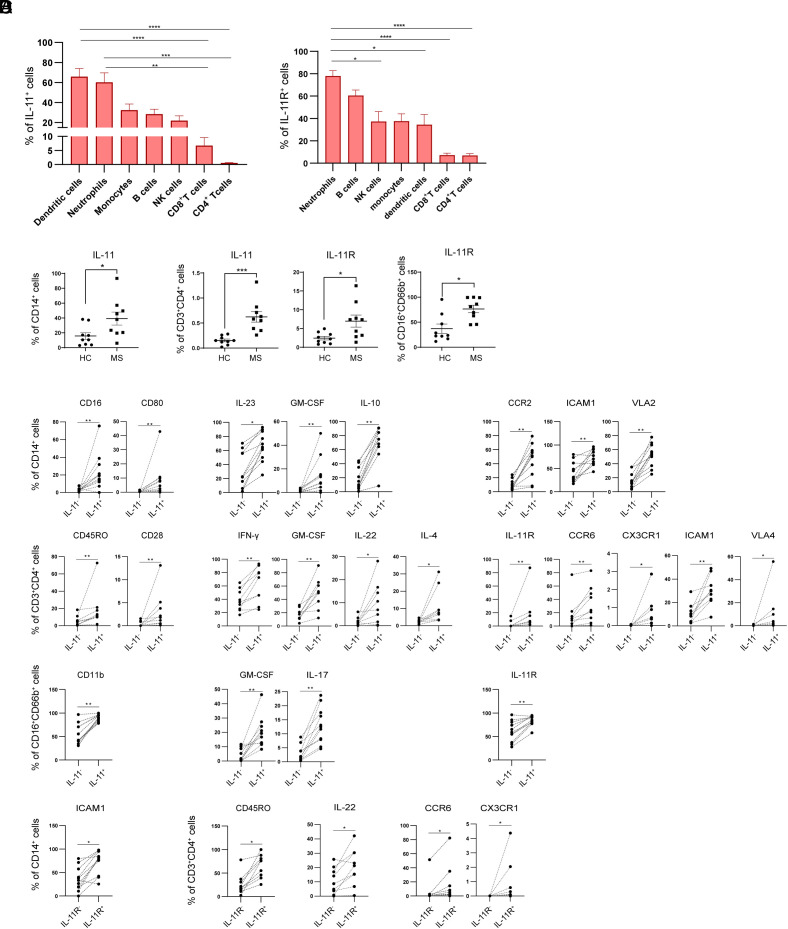
In order to determine cellular sources and targets of IL-11 in the autoimmune response, we examined the frequency of IL-11+ and IL-11R+ cell subsets in PBMCs from 14 untreated RRMS patients and 14 matched HCs (healthy controls) (SI Appendix, Table S1). The study examined myeloid cells (CD14+ monocytes, CD11c+ DCs, CD66+CD16+ neutrophils); CD56+ NK cells; and CD4+, CD8+, and CD19+ lymphocytes. The frequency of IL-11+ cells was highest in CD11c+CD14- DCs, CD66b+CD16+ neutrophils, and CD14+ monocytes in RRMS (Fig. 1A). The frequency of IL-11RA+ cells in PBMCs from RRMS patients was highest in neutrophils, please see representative staining in SI Appendix, Fig. S1.
Fig. 1.
IL-11 and IL-11RA have the highest expression in myeloid cells. (A) The percentages of IL-11+ and IL-11R+ cells in 7 gated PBMC cell subsets from 14 RRMS patients. (B) The percentages of IL-11+ and IL-11R+ cells from 9 RRMS patients in comparison to matched HCs. (C) Phenotype of IL-11+ CD14+ monocytes, CD4+ cells, and neutrophils. (D) IL-11R+ CD14+ monocytes and CD4+ lymphocytes in 9 RRMS patients.
We next compared the frequency of IL-11+ and IL-11R+ cells in each cell subset between MS patients and HCs. RRMS patients had a significantly increased frequency of IL-11+CD14+ monocytes; IL-11+ and IL-11RA+ CD4+ cells; and IL-11R+ CD66b+CD16+ neutrophils (Fig. 1B) in comparison to HCs.
In order to characterize the phenotype of IL-11+ and IL-11R+ cells, we determined multiple marker expression in MS patients–derived IL-11+ and IL-11R+ monocytes, CD4+ lymphocytes, and neutrophils in comparison to the IL-11− and IL-11R− cells (SI Appendix, Table S2). IL-11+ monocytes had increased expression of CD16 and CD80; IL-23, GM-CSF, and IL-10; and surface CCR2, ICAM-1, and VLA-2. IL-11+ CD4+ cells had an increased expression of CD45RO, CD28; IFN-γ, GM-CSF, IL-22, and IL-4; and surface IL-11R, CCR6, CX3CR1, ICAM-1, and VLA-4, suggestive of their memory proinflammatory Th1/Th17 (TH1*) phenotype. In comparison to the other CD4+ cell lineages, IL-11+CD4+ cells have an increased coexpression of surface IL-11R, CXCR3, and CCR6 (SI Appendix, Fig. S2). IL-11+ neutrophils had an increased CD11b; GM-CSF and IL-17A; and increased IL-11R expression in comparison to the IL-11− neutrophils from MS patients (Fig. 1C). IL-11R+ RRMS monocytes had an increased ICAM1, while IL-11R+CD4+ cells had increased CD45RO, IL-22, CCR6, and CX3CR1 in comparison to the IL-11R− cells (Fig. 1D).
Consistent with previous reports on IL-11 autocrine signaling (13, 17), we found a high coexpression of IL-11R in IL-11+ monocytes (51%), CD4+ cells (22%), and neutrophils (87%), SI Appendix, Fig. S3.
IL-11- and IL-11R-Expressing Cells Accumulate in the CSF of RRMS Patients.
In order to assess the migratory capacity of the above phenotyped monocytes, CD4+ lymphocytes, and neutrophils, we compared their frequencies between CSF and matched PBMC samples from 9 RRMS patients (SI Appendix, Table S1). Representative staining is presented in SI Appendix, Fig. S4.
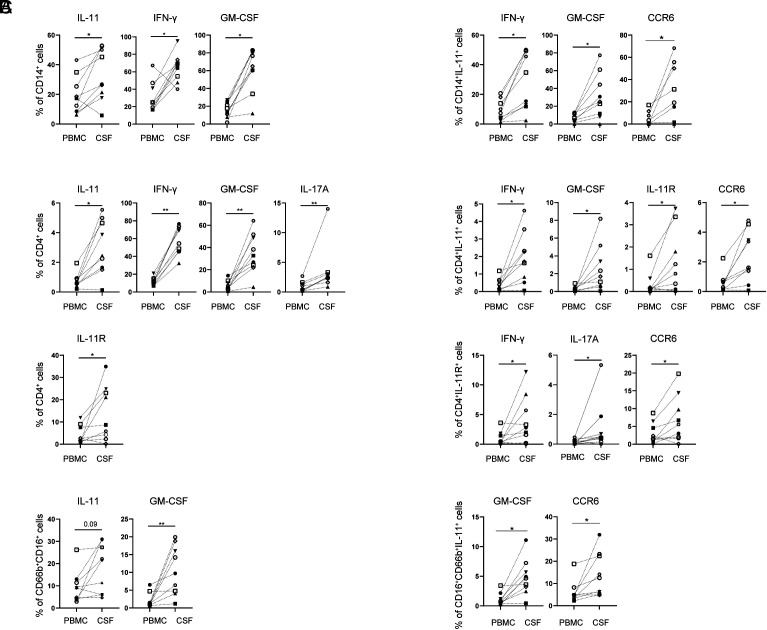
The frequency of IL-11+ (1.7-fold), IFN-γ+ (2.3-fold), and GM-CSF+ (3.6-fold) monocytes was increased in CSF (Fig. 2 A, Left). CSF-accumulated IL-11+ monocytes coexpressed IFN-γ+, GM-CSF+, and CCR6+ (Fig. 2 A, Right).
Fig. 2.
IL-11+ and GM-CSF+ monocytes, CD4+ cells, and neutrophils accumulate in the CSF of RRMS patients. (A) The frequency of cytokine-expressing CD14+ cells and IL-11+CD14+ monocytes coexpressing inflammatory markers; (B) CD4+ cells; and (C) neutrophils was determined in paired PBMC and CSF samples from 9 RRMS patients.
The frequency of IL-11+ and IL-11R+CD4+ cells (fourfold and 3.2-fold), IFN-γ+ (4.8-fold), GM-CSF+ (6.1-fold), and IL-17A+CD4+ cells (fourfold) was significantly increased in the CSF in comparison to matched PBMCs (Fig. 2 B, Left). CSF-enriched IL-11+CD4+ cells coexpressed IFN-γ, GM-CSF, IL-11R, and CCR6, while CD4+IL-11R+ cells had increased coexpression of IFN-γ, IL-17A, and CCR6 (Fig. 2 B, Right).
IL-11+ and GM-CSF+ neutrophils accumulate in the CSF (1.9 and 5.2-fold) (Fig. 2 C, Left), and IL-11+ neutrophils in the CSF coexpressed GM-CSF and CCR6 (Fig. 2 C, Right).
IL-11 Induces Proinflammatory Phenotype in Monocytes.
To characterize the in-vitro effect of IL-11, we stimulated separated CD14+ monocytes and found that IL-11 increased IL-23 and IL-1β secretion (SI Appendix, Fig S5 A and B), similar to a previous report (2). IL-11-stimulated monocytes in coculture with autologous CD4+ cells induced their GM-CSF production, resulting in the encephalitogenic CD4+ cell phenotype (SI Appendix, Fig S5C), (18).
To examine whether IL-11 induces inflammatory cell migration, we used a transwell system with human cerebral microvascular EC (hCMEC/d3) barrier. Fresh PBMCs from 3 HCs were pretreated with hrIL-11 in the absence or presence of anti-human IL-11 mAb for 24 hours (h), washed, and added to an upper chamber for a migration assay over 24 h. IL-11 stimulation increased the number of migrated cells, which was significantly decreased by αIL-11 mAb-pretreatment. The migrated CD14+ monocytes had an increased surface expression of IL-11R and HLA-DR in IL-11-stimulated cells, which was decreased by αIL-11 mAb (SI Appendix, Fig. S6).
IL-11 Induces NLRP3 Inflammasome Gene Expression in Monocytes.
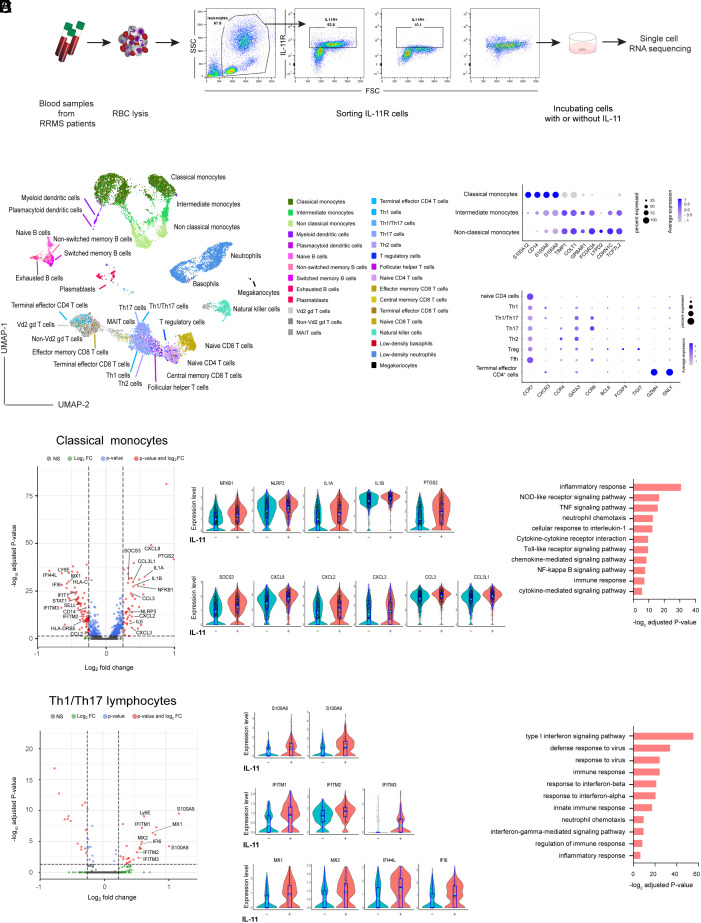
In order to examine the effects of IL-11/IL-11R signaling in RRMS in a comprehensive and unbiased fashion, we performed scRNAseq (single-cellRNA sequencing) of IL-11R+-sorted cells from 3 untreated RRMS patients that were stimulated with recombinant human (rh) IL-11 (100 ng/mL) over 1 h to identify early transcriptome changes in comparison to nonstimulated cells (Fig. 3A).
Fig. 3.
IL-11 induces the expression of NLPR3 inflammasome genes in monocytes. (A) Schematic representation of the experiment. Blood samples were sorted for IL-11R+ cells, incubated with/without IL-11 before scRNA sequencing (n = 3 RRMS patients). (B) The Uniform Manifold Approximation and Projection (UMAP) clustering in 29 immune cell types. (C) A cluster-defining gene expression in monocytes and CD4+ T cell subsets. (D) DEGs following IL-11 stimulation in classical monocytes, volcano plot. The expression level of some biologically relevant significantly up-regulated genes in classical monocytes, violin plots. Gene ontology and pathway analysis for IL-11-up-regulated genes in monocytes. (E) DEGs following IL-11 stimulation in Th1/Th17 CD4+ cells, volcano plot; up-regulated genes in Th1/Th17 cells in violin plots. Gene ontology and pathway analysis results for up-regulated genes in all CD4+ cell types.
In IL-11-stimulated/unstimulated blood samples, after filtering of low-quality cells and normalization, we had a total 14,846 cells (6,478 from IL-11-stimulated and 8,368 from nonstimulated samples). After performing unsupervised clustering, cells were annotated to 29 immune cell clusters (19). The UMAP projections of cells are presented in Fig. 3B. The frequency and number of cells for each cell type are listed in Dataset S1. The complete list of genes, defining 29 clusters, is presented in Dataset S2. A change in UMAP distribution between IL-11-stimulated and unstimulated cells is shown in SI Appendix, Fig. S7A.
The study of differentially expressed genes (DEGs) between IL-11-stimulated and nonstimulated cells found that classical monocytes have the highest number of DEGs (51 up-regulated and 79 down-regulated genes) among the 29 cell clusters. Further analysis focused on monocytes, which were divided into classical (inflammatory), intermediate, and nonclassical (patrolling) monocytes (19). The key genes identifying classical, intermediate, and nonclassical monocytes were consistent with published studies (20) (Fig. 3 C, Upper plot).
Following IL-11 stimulation, classical monocytes showed the highest number of DEGs, (Dataset S3), as presented in a volcano plot. The levels of expression of significantly changed (adjusted p value <0.05) relevant up-regulated genes are shown in violin plots (Fig. 3D). IL-11 stimulation increased the expression of NFKB1, NLRP3, IL1A, and IL1B genes, which are all associated with NLRP3 inflammasome activation (21). Increased expression of COX-2 (PTGS2), which is induced by IL-1β (22), IL6, and chemokine genes (CXCL8 (IL8), CXCL2, CXCL3, CCL3, and CCL3L1) involved in myeloid cell migration, suggests that IL-11 induces IL1B expression via NLRP3 inflammasome activation, which may mediate monocyte transendothelial migration (4, 23).
In order to gain insight into the function of the genes up-regulated in monocytes following IL-11 stimulation, we performed gene ontology and pathway analysis (Fig. 3D and Dataset S4). IL-11 stimulation up-regulated “Tall-like receptor (TLR) signaling pathway” (IL6, CXCL8, CCL3L1, IL1B, CCL3, and NFKB1), “NF-kappa B signaling pathway” (NFKB1, NLRP3, IL1B, CXCL8, and PTGS2), “NLRP3 signaling pathway” (IL6, CXCL8, IL1B, NLRP3, CXCL2, NFKB1), “cellular response to IL-1” (IL6, CXCL8, CCL3L1, CCL3, NFKB1), and “neutrophil chemotaxis” (CXCL8, IL1B, CXCL3, and CCL3L1).
By annotating IL-11R+ cells to 29 clusters (19), CD4+ cells were divided into 8 cell subsets including naïve CD4+ T cells (CCR7), Th1 (CXCR3), Th1/Th17 (CXCR3, CCR6), Th17 (CCR6), Th2 (CCR4, GATA3), T regulatory cells (FOXP3, TIGIT), Tfh (BCL6), and terminal effector CD4+ T cells (GZMH, GNLY), (Fig. 3 C, Lower and plot Dataset S2).
Following IL-11 stimulation, Th1/Th17 cells showed the highest number of DEGs (26 up-regulated and 14 down-regulated genes) among the CD4+ cell subsets (Dataset S3). The significantly up-regulated genes are shown in volcano plots and their level of expression in violin plots, Fig. 3E. Most strikingly, we noted an increased expression of S100A8 and S100A9 alarmin or danger-associated molecular pattern genes in almost all CD4+ cell subsets (naïve, Th1/Th17, Th17, Th2 and Thf, and T cells, SI Appendix, Fig. S7B) following IL-11 stimulation (Dataset S3). The second group of genes up-regulated in most of the CD4+ cell clusters are type I IFN–induced genes STAT1 (in Th17 cells), MX1, and MX2 in Th1/Th17 cells and type II IFN–related genes (IFITM3, IF144, and IF144L) (Fig. 3E and Dataset S3).
Pathway enrichment analysis revealed that these genes are associated with “innate immune response (MX2, MX1, HLA-B, S100A9, S100A8),” “neutrophil chemotaxis (CXCL8, IL1B, CCL4L2, S100A9, S100A8),” “type I IFN signaling,” “IFN-gamma-mediated signaling pathway,” and neutrophil chemotaxis GOTERMs, involved in innate immune response activation and cell migration, Fig. 3E and Datasets S4 and S5.
Our results demonstrate that IL-11 induces a CD4+ cell expression of S100A8 and S100A9, which may induce NLRP3 inflammasome activation (24) and subsequent IL1B expression and migratory phenotype in monocytes.
CSF-Derived IL-11R+ Monocytes Up-Regulate Genes Involved in Transendothelial Migration.
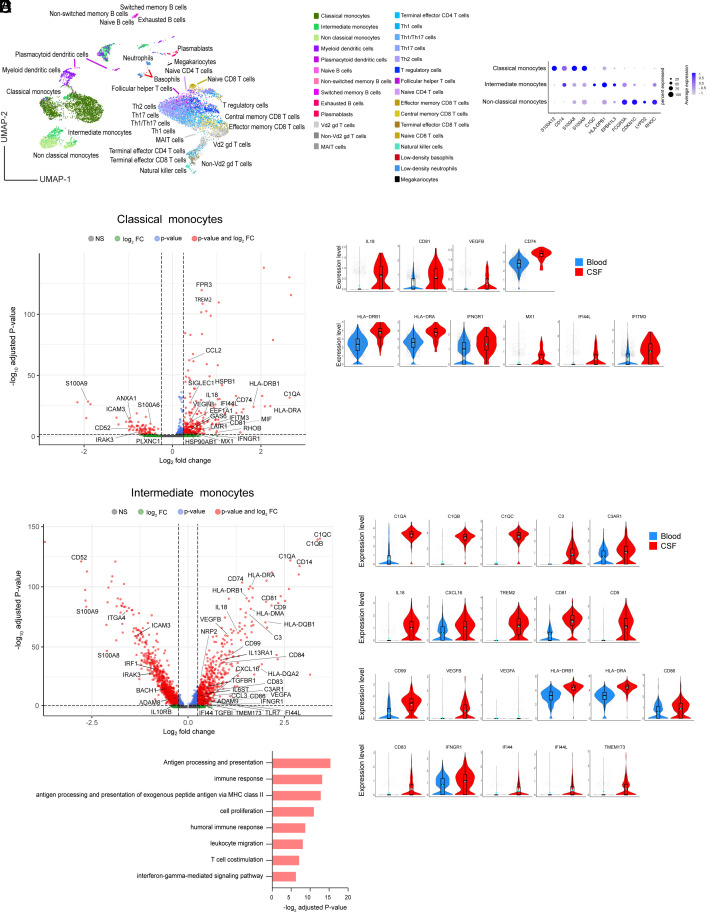
To characterize the transcriptome changes in IL-11R+ inflammatory cells that migrated to the CSF, we sorted IL-11R+ cells from CSF and paired blood samples from two untreated RRMS patients and submitted for scRNAseq without stimulation. We analyzed 12,381 cells (6.609 from CSF and 5,772 from blood) that were clustered in 29 immune cell subsets (19), UMAP plot in Fig. 4A and Dataset S6. We identified a difference in the distribution of IL-11R+-sorted cell subsets between CSF and blood SI Appendix, Fig. S8A. As expected, the frequency of memory T cell subsets, intermediate monocytes, and myeloid DCs (mDCs) was higher in CSF (25) (SI Appendix, Fig. S8B and Dataset S7).
Fig. 4.
CSF-derived IL-11R+ monocytes up-regulate genes involved in cell migration. (A) The UMAP cell clustering in 29 immune cell types. (B) A cluster-defining gene expression in monocyte subsets. (C) DEGs between CSF and blood samples in classical monocytes, volcano plot; up-regulated genes in CSF in classical monocytes, violin plots. (D) DEGs in intermediate monocytes, volcano plot; the expression level of selected up-regulated genes in CSF intermediate monocytes, violin plots. (E) Gene ontology and pathway analysis for up-regulated genes in all monocyte subsets.
In order to characterize the transcriptome of cells migrating to the CSF, we identified DEGs between the IL-11R+ CSF and blood cells in 29 immune cell subsets. Surprisingly, the highest number of DEGs was found in monocytes. The expression of genes defining monocyte subsets is presented in Fig. 4B. In CSF classical and intermediate monocytes, we found 249 and 486 up-regulated and 78 and 889 down-regulated genes, respectively, in comparison to blood (Dataset S8).
DEGs up-regulated in CSF classical monocytes include complement gene C1QA, inflammasome-induced IL18 (26), and genes involved in cell migration (CD81, VEGFA, and VEGFB). Similar to IL-11-stimulated CD4+ cells, CSF IL-11R+ classical monocytes up-regulate IFN type I (MX1, IFI44L)– and type II (IFNGR1, IFITM3)–related genes, (Fig. 4C), which regulate inflammasome activation (27). It is of note that the highest up-regulated genes in CSF cells belong to the HLA family, including DRA, DRB1, and CD74, involved in MHC class II antigen processing. Since those genes were not induced by the IL-11 in-vitro stimulation, we propose that they are up-regulated as classical monocytes differentiate into antigen-presenting mDCs and macrophages when they enter the CNS inflammatory tissue, and that they may not be related to IL-11 signaling. Classical monocytes are precursors for mDCs, whose DEGs between CSF and blood compartment reflect a similar pattern (111 up-regulated and 286 down-regulated genes, Dataset S8).
Intermediate monocytes (CD14++CD16+) patrol the luminal side of the blood vessel and are enriched in the CSF in MS (28). This monocyte subset also expresses increased levels of complement genes (C1QA, C1QB C1QC, C3, and C3AR1), which play a role in the inflammasome activation and disruption of BBB (29); inflammasome-induced IL18; and genes regulating cell migration (CD81, CD9, CD99, VEGFA, and VEGFB), across the BBB (30). Notably, intermediate monocytes in CSF had up-regulated chemokine CXCL16 gene, implicated in the NLRP3-mediated APC migration to the CNS (31). Similar to classical monocytes, intermediate monocytes up-regulate the expression of multiple HLA genes, as well as CD86 costimulatory and CD83 (mature DC marker) genes, and type I IFN (IFI44, IFI44L, and TMEM173 (STING)– and type II IFN (IFNGR1)–related genes (Fig. 4D). Finally, CSF intermediate monocytes up-regulated gene for ADAM9 protease that cleaves IL-11R from the cell surface (16), which can contribute to IL11/IL-11R complex transsignaling in a large number of cells expressing gp130.
Gene ontology search for the significantly up-regulated genes showed that CSF IL-11R+ monocytes up-regulated genes are associated with biological processes such as “immune response (C1QC, C3, CCL2, CCL3, IL18),” “leukocyte migration (CD74, CD84, ITGAM, MIF, C3AR1),” and “IFN gamma–mediated signaling pathway” (Fig. 4E and Dataset S9).
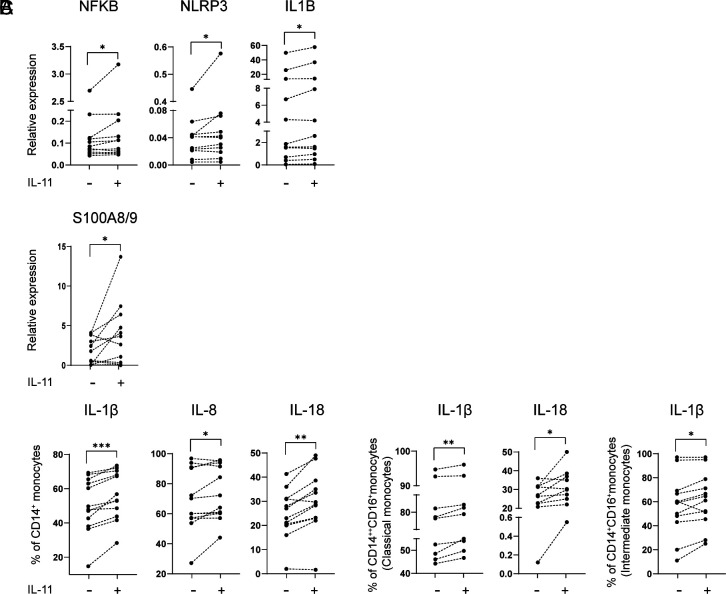
Validation study of DEGs identified in monocytes and CD4+ cells following IL-11 in-vitro stimulation and in IL-11R+ CSF vs. blood-derived cells was performed in an independent cohort of 11 RRMS patients. Gene expression measurement in sorted RRMS monocytes, stimulated with IL-11, confirmed a significantly increased NFKB, NLRP3, IL1B, and IL18 gene expression (Fig. 5A). Flow cytometry study of the PBMCs from the same patients confirmed an increased expression of S100A8/9 in gated CD4+ T cells, Fig. 5B. CD14+-gated monocytes had an increased expression of IL-1b, IL-8 (CXCL8), and IL-18; classical monocytes had an increased IL-1β and IL-18; while intermediate monocytes had an increased IL-1β following IL-11 in vitro stimulation (48 h), Fig. 5C (SI Appendix, Table S2).
Fig. 5.
IL-11 induces NLPR3 inflammasome–related proteins in RRMS. (A) Magnetic bead–separated monocytes from RRMS patients (n = 11) were treated with rhIL-11, and the expression of indicated genes was measured by qRT-PCR. (B and C) PBMCs from the same patients were treated with rhIL-11, and the expression of S100A8/9 was measured in gated CD3+CD4+ T cells and intracellular cytokines in monocyte subsets.
Anti-(α)IL-11 mAb Suppresses RREAE via Inhibition of NLRP3 Inflammasome Activation in Monocytes.
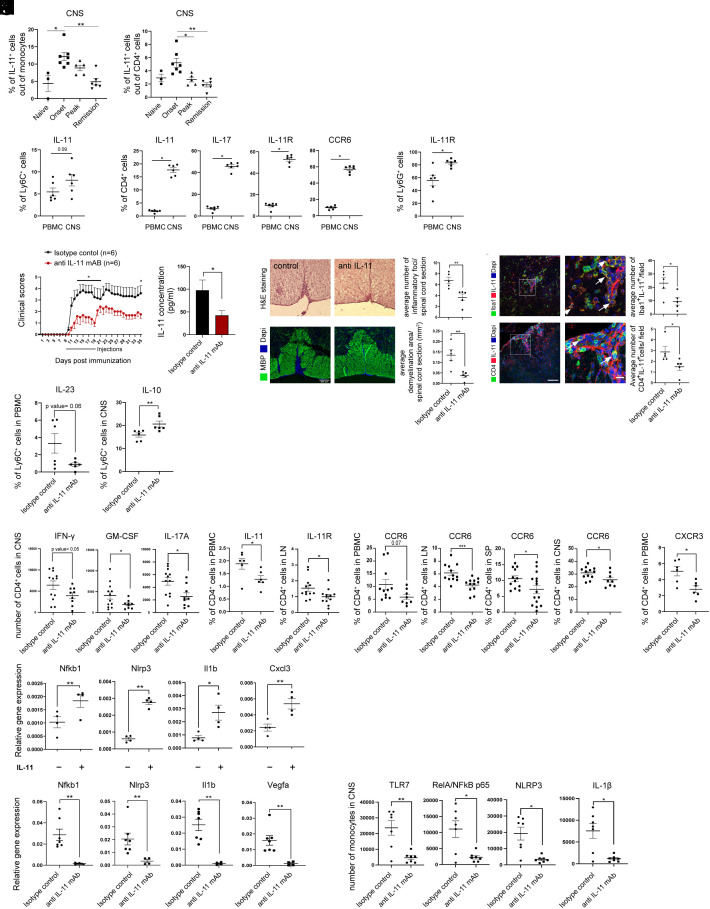
In order to longitudinally study the role of IL-11-producing cells in RREAE, and to characterize them as a potential treatment target, we monitored their frequencies in the CNS of mice with proteolipid protein (PLP)139-151–induced RREAE. A longitudinal flow cytometry study showed an increased frequency of IL-11+Ly6C+ monocytes and IL-11+CD4+ cells in the CNS at the onset of disease [10 days post immunization (d.p.i)] (Fig. 6A). Similar to the CSF studies in RRMS, we found increased frequencies of IL-11+CD11b+Ly6C+Ly6G- monocytes; IL-11+, IL-17+, IL-11R+, and CCR6+ CD4+ cells; and IL-11R+Ly6G+ neutrophils in the CNS of mice with RREAE in comparison to matched PBMCs (Fig. 6B). The CNS accumulation of CCR6+CD4+ cells is consistent with the in-vitro effect of IL-11, which dose-dependently increased CCR6 expression n CD4+ cells (1).
Fig. 6.
αIL-11 mAb suppresses RREAE via inhibition of NLRP3 inflammasome activation. (A) A time course flow cytometry study during the RREAE (6 mice per group). (B) The frequencies of IL-11+ and IL-11R+ cells in PBMC and CNS-infiltrating cells from the same mice, 21 d.p.i. (C) Twelve SJL mice were immunized with PLP139-151 to induce RREAE and were treated with αIL-11 mAb or IgG2A isotype control i.p starting at the onset of disease for 10 consecutive days. The presented data are representative of three independent experiments with at least 6 mice per group. (D) IL-11 was measured in plasma on day 21 p.i. (n = 14 per group). (E) On 18 d.p.i., H&E and myelin basic protein (MBP) staining quantified inflammatory foci and demyelinated areas. (F) Number of IL-11+Iba-1+ cells and IL-11+CD4+ cells (arrows) averaged from at least 8 microscope fields within the lesion (n = 5 mice per group, scale bar, 100 µm; 10 µm for higher magnification). (G–K) At day 21 p.i., RREAE mice treated with αIL-11mAb or isotype control were killed, and cell suspension from PBMCs, LN, spleen, and CNS infiltrates was used for flow cytometry (n = 6 or 12 per group). (G) Intracellular cytokine staining in PBMCs and CNS (n = 6). (H) CD4+ cells from CNS infiltrates. (I) The frequency of IL-11+CD4+ (PBMC) and IL-11R+CD4+ (LN) cells (n = 6 for IL-11 and n = 12 for IL-11R). (J) The percentages of CCR6+CD4+ cells in PBMCs, LNs, spleen, and CNS, and (K) CXCR3+CD4+ cells in PBMCs. (L) IL-11R+ monocytes were isolated from the spleen of RREAE mice and treated with rmIL-11. Inflammasome-related gene expression was determined by RT-PCR (n = 4). (M) IL-11R+ monocytes from the spleen of isotype-control or αIL-11 mAb–treated mice with RREAE, the expression of NLRP3-related genes (n = 7), and the number of monocytes expressing inflammasome-related proteins in the CNS infiltrates (n = 7).
The therapeutic effect of αIL-11 mAb was tested in mice with RREAE in comparison to an IgG2 isotype control administered daily starting at the onset of clinical disease (day 10 p.i.) for 10 consecutive days. The inhibitory effect of this antibody on the expression of IL-11-induced pSTAT3, pERK1/2, and pp65 NFB and on the IL-11-induced mouse monocyte migration is presented in SI Appendix, Fig. S9. This treatment decreased clinical scores throughout the 35-day experiment, with a significant decrease on days 11 to 19 p.i., Fig. 6C. As expected, serum IL-11 levels were decreased in treated mice (Fig. 6D).
The CNS infiltrates and the demyelinated area were decreased in αIL-11 mAb–treated mice (Fig. 6E). The numbers of IL-11+Iba-1+-infiltrating monocyte–derived macrophages and IL-11+ CD4+ T cells were decreased in the spinal cords of treated mice (Fig. 6F).
In order to examine the therapeutic mechanisms of αIL-11 mAb, we killed the mice at day 21 p.i. and harvested PBMC, lymph node (LN), spleen, and CNS mononuclear cells for flow cytometry study (SI Appendix, Table S2). Consistent with human in-vitro studies (SI Appendix, Fig. S5), αIL-11 mAb decreased the frequency of IL-23+ Ly6C+CD11b+Ly6G− monocytes in PBMCs (Fig. 6G). In CD4+ cells, αIL-11 mAb treatment decreased the numbers of IFN-γ+, GM-CSF+, and IL-17A+ cells in the CNS (Fig. 6H). In peripheral immune organs, the treatment decreased the frequency of IL-11+CD4+ cells in PBMCs and IL-11R+CD4+ cells in LNs (Fig. 6I). It also decreased CCR6+CD4+ cells in PBMCs, LN, spleen, and CNS, and CXCR3+CD4+ cells in PBMCs (Fig. 6 J and K), suggestive of therapeutic effect on the peripheral immune cells, which decreased their migration to the CNS.
In order to validate human scRNAseq data from the in-vitro IL-11-stimulated cells, we tested the expression of several biologically relevant genes in IL-11R+ monocytes from the spleen of RREAE mice. The IL-11 stimulation significantly increased NFKB1, NLRP3, IL1B, and CXCL3 gene expression (Fig. 6L).
Further validation of scRNAseq human results was performed in RREAE mice following αIL-11 mAb treatment. αIL-11 mAb decreased the gene expression of NFKB, NLRP3, IL1B, and VEGFA in sorted IL-11R+ Ly6C+ monocytes, and the numbers of TLR7+, NFκB p65+, NLRP3+, and IL-1b+ monocytes in the CNS infiltrates of treated mice, Fig. 6M (SI Appendix, Table S2).
Discussion
In this study, we investigated the role of IL-11 signaling in the immunopathogenesis of RRMS and EAE. We found that IL-11 is expressed at highest frequency in DCs, neutrophils, and monocytes and IL-11R in neutrophils, highlighting the role of IL-11 signaling in myeloid cells, which facilitate BBB disruption and the inflammatory cell migration to the CNS (32). RRMS patients have an increased expression of IL-11+CD14+ monocytes, IL-11+ and IL-11R+ CD4+ cells, and IL-11R+ neutrophils in comparison to matched HCs.
The CSF studies revealed an accumulation of IL-11+ and GM-CSF+ monocytes, CD4+ cells, and neutrophils. Consistent with those results, our prior study reported that the IL-11-induced encephalitogenic CD4+ cells were IFN-γ+ and GMCSF+ (1), indicating a pathogenic role of IL-11 in the CNS accumulation of those effector cells (33).
scRNAseq studies of IL-11-stimulated IL-11R+ cells from blood of RRMS patients revealed the highest number of DEGs in classical monocytes including NFKB1, NLRP3, and IL1B, which are involved in NLRP3 inflammasome activation (21). The role of activated NLRP3 inflammasome in MS is supported by increased NLRP3 inflammasome–induced caspase-1 and IL-18 gene expression in PBMCs (34, 35) and caspase-1 and IL-1β proteins in CSF of RRMS patients (26). NLRP3 KO mice are resistant to EAE due to decreased APC migration and their CXCL16 chemokine expression, with subsequent decrease in Th1 and Th17 cell CNS migration (31).
IL-11 induced the expression of S100A8 and S100A9 genes in almost all CD4+ cell subsets. These alarmins bind TLR and RAGE receptors and induce NLRP3 inflammasome activation via NFκB signaling (24). Thus, secreted S100A8/9 act as an upstream signal for many inflammatory pathways and are proposed as serum biomarker of several autoimmune and inflammatory diseases (RA, lupus, and IBD) (36). The S100A8/S100A9 serum levels were increased in RRMS in comparison to HCs, and serum and CSF levels were increased in clinical relapses (37, 38). We therefore propose that IL-11-induced upregulation of S100A8 and S100A9 in CD4+ T cells activates NLRP3 inflammasome in monocytes.
scRNAseq studies of CSF vs. peripheral blood IL-11R+-sorted cells also revealed the highest number of DEGs in classical and intermediate monocytes with upregulation of multiple complement genes, inflammasome-induced IL18, multiple chemokines (CXCL16), and genes mediating monocyte migration (CD81, CD84, CD9, CD99, VEGFA, and VEGFB). Given the role of NLRP3 inflammasome in the induction of IL-1β, IL-8, IL-6, TNFα, IL-18, inflammatory mediator (COX-2), and angiogenic factors (VEGFA, VEGFB) (30, 39, 40), we propose that a DEG pattern induced by in-vitro IL-11 stimulation of peripheral blood cells is consistent with DEGs in CSF vs. blood IL-11R+ cells involved in inflammatory cell migration.
Similar to IL-11-stimulated peripheral CD4+ cells, CSF IL-11R+ classical and intermediate monocytes up-regulate IFN type I (MX1, IFI44L)– and type II (IFNGR1, IFITM3)–related genes, which regulate the NLRP3 inflammasome activation (27), as reported in studies of EAE (41) and RRMS (42).
While neutrophils have high IL-11R expression, they do not migrate to the CNS due to their short life span (43). We propose that monocytes orchestrate neutrophil chemotaxis via IL-8 and neutrophil adhesion to the EC barrier and increase BBB permeability for the inflammatory cell migration.
αIL-11 mAb treatment of RREAE suppressed clinical disease and decreased CNS inflammatory infiltrates and demyelination, similar to the report of decreased EAE in IL-11RKO mice (44). Moreover, αIL-11 mAb treatment decreased the frequency of CNS-infiltrating TLR7+, NFκB+, NLRP3+, and IL-1β+ Ly6C+ monocytes. The results suggest that the therapeutic effect of αIL-11 mAb is mediated via suppression of NLRP3 inflammasome activation in peripheral monocytes and subsequent inflammatory cell migration to the CNS.
Several older studies have reported contradictory results regarding the antiinflammatory effects of IL-11, which were explained by the use of human IL-11 in the mouse system, with blocking effect on the mouse IL-11R signaling (45). In contrast, most recent studies consistently demonstrate proinflammatory role of IL-11 in the inflammatory-fibrotic pulmonary, cardiac, and renal diseases (17, 45, 46).
Several FDA-approved disease-modifying therapies suppress IL-11 serum levels (alemtuzumab) (47) and its PBMC gene expression (IFNb-1a) (48). However, selective targeting of this pathway with αIL-11 mAb may be better tolerated, as multiple studies of αIL-11 mAb in animal models of cardiac and pulmonary fibrosis, UC, and RA had good tolerability (45, 46). Phase I clinical trial of αIL-11 mAb (NCT05740475) and αIL-11R mAb (NCT05331300) in pulmonary fibrosis is currently in progress. Targeting inflammasome with currently available NLRP3 inhibitors (MCC950), caspase-1 inhibitors, or inflammasome assembly inhibitors (glyburide) may provide a treatment approach, as inflammasomes bridge innate and adaptive immune response (49).
Our study indicates that IL-11 may represent an unique cytokine in the NLRP3 inflammasome activation, whose therapeutic targeting may be effective at the onset of the disease or as a relapse treatment and may complement existing immunomodulatory therapies for RRMS.
Methods
Study Subjects.
The human study was approved by the Thomas Jefferson University and University of Hasselt Institutional Review Board. Fifty-four RRMS patients (50) and 17 HCs were enrolled in the study after signing IRB consent at Thomas Jefferson University and University of Hasselt.
Flow Cytometry.
The list of antibodies is in SI Appendix, Table S2.
scRNAseq.
Sorted IL-11R+ cells from blood of 3 RRMS patients were unstimulated or stimulated with IL-11. IL-11R+ cells were sorted from the CSF and blood samples from 2 RRMS patients.
Quantitative Real-Time-PCR (qRT-PCR).
Primers are shown in SI Appendix, Table S2.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Acknowledgments
We thank the study subjects and Dr. Li, Tsygankova, Yang, and Gonzalez for work on experiments and data analysis; Ms. McGuire, Marcello, Boncompagni, Sweeney, and Paglione for clinical data; and Ms. Regan for editorial assistance. The study was supported by NIH 1R01AI131238-01A1 and PA Cure SAP4100083100 grant to S.M.-P. and R01-AI029564 to J.P.T.
Author contributions
M. Seyesadr and S.M.-P. designed research; M. Seyesadr, Y.W., M.E., S.S.G., S.J., G.D., E.K., D.H., X.Z., and T.H.K. performed research; U.L., M. Su, J.P.T., B.B., A.R., and D.M. contributed new reagents/analytic tools; M. Seyesadr, Y.W., I.C., J.A.W., and J.G. analyzed data; and M. Seyesadr and S.M.-P. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information. Raw and processed scRNAseq data used in the study are available at the National Center for Biotechnology Information Gene Expression Omnibus database under the repository accession number GSE233917 (51).
Supporting Information
References
- 1.Zhang X., et al. , IL-11 induces encephalitogenic TH17 cells in multiple sclerosis and experimental autoimmune encephalomyelitis. J. Immunol. 203, 1142–1150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X., et al. , IL-11 induces Th17 cell responses in patients with early relapsing-remitting multiple sclerosis. J. Immunol. 194, 5139–5149 (2015), 10.4049/jimmunol.1401680. [DOI] [PubMed] [Google Scholar]
- 3.Croxford A. L., et al. , The cytokine GM-CSF drives the inflammatory signature of CCR2+ monocytes and licenses autoimmunity. Immunity 43, 502–514 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Levesque S. A., et al. , Myeloid cell transmigration across the CNS vasculature triggers IL-1beta-driven neuroinflammation during autoimmune encephalomyelitis in mice. J. Exp. Med. 213, 929–949 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang W., et al. , Targeted expression of IL-11 in the murine airway causes lymphocytic inflammation, bronchial remodeling, and airways obstruction. J. Clin. Invest. 98, 2845–2853 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson R. B., Wood N., Serio F. G., Interleukin-11 and IL-17 and the pathogenesis of periodontal disease. J. Periodontol. 75, 37–43 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Mima T., et al. , Interleukin 11 and paired immunoglobulin-like type 2 receptor alpha expression correlates with the number of joints with active arthritis in systemic juvenile idiopathic arthritis. Ann. Rheum. Dis. 68, 286–287 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Ameglio F., et al. , Interleukin-11 production is increased in organ cultures of lesional skin of patients with active plaque-type psoriasis as compared with nonlesional and normal skin. Similarity to interleukin-1 beta, interleukin-6 and interleukin-8. Arch. Dermatol. Res. 289, 399–403 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Elshabrawy H. A., et al. , IL-11 facilitates a novel connection between RA joint fibroblasts and endothelial cells. Angiogenesis 21, 215–228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim W. W., et al. , Transgenic interleukin 11 expression causes cross-tissue fibro-inflammation and an inflammatory bowel phenotype in mice. PLoS One 15, e0227505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Metcalfe R. D., Putoczki T. L., Griffin M. D. W., Structural understanding of interleukin 6 family cytokine signaling and targeted therapies: Focus on interleukin 11. Front. Immunol. 11, 1424 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenkins B. J., et al. , Pathologic consequences of STAT3 hyperactivation by IL-6 and IL-11 during hematopoiesis and lymphopoiesis. Blood 109, 2380–2388 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Onnis B., Fer N., Rapisarda A., Perez V. S., Melillo G., Autocrine production of IL-11 mediates tumorigenicity in hypoxic cancer cells. J. Clin. Invest. 123, 1615–1629 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng B., et al. , Similarities and differences between IL11 and IL11RA1 knockout mice for lung fibro-inflammation, fertility and craniosynostosis. Sci. Rep. 11, 14088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murakami M., Kamimura D., Hirano T., Pleiotropy and specificity: Insights from the interleukin 6 family of cytokines. Immunity 50, 812–831 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Lokau J., et al. , Proteolytic cleavage governs interleukin-11 trans-signaling. Cell Rep. 14, 1761–1773 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Schafer S., et al. , IL-11 is a crucial determinant of cardiovascular fibrosis. Nature 552, 110–115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El-Behi M., et al. , The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat. Immunol. 12, 568–575 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monaco G., et al. , RNA-Seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 26, 1627–1640.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong K. L., et al. , Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood 118, e16–e31 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Guo H., Callaway J. B., Ting J. P., Inflammasomes: Mechanism of action, role in disease, and therapeutics. Nat. Med. 21, 677–687 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W., et al. , Cyclooxygenase-2 is up-regulated by interleukin-1 beta in human colorectal cancer cells via multiple signaling pathways. Cancer Res. 63, 3632–3636 (2003). [PubMed] [Google Scholar]
- 23.Boyette L. B., et al. , Phenotype, function, and differentiation potential of human monocyte subsets. PLoS One 12, e0176460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simard J. C., et al. , S100A8 and S100A9 induce cytokine expression and regulate the NLRP3 inflammasome via ROS-dependent activation of NF-kappaB(1.). PLoS One 8, e72138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schafflick D., et al. , Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis. Nat. Commun. 11, 247 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gharagozloo M., et al. , NLR-dependent regulation of inflammation in multiple sclerosis. Front. Immunol. 8, 2012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopitar-Jerala N., The role of interferons in inflammation and inflammasome activation. Front. Immunol. 8, 873 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waschbisch A., et al. , Pivotal role for CD16+ monocytes in immune surveillance of the central nervous system. J. Immunol. 196, 1558–1567 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Hajishengallis G., Reis E. S., Mastellos D. C., Ricklin D., Lambris J. D., Novel mechanisms and functions of complement. Nat. Immunol. 18, 1288–1298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Girolamo F., Coppola C., Ribatti D., Trojano M., Angiogenesis in multiple sclerosis and experimental autoimmune encephalomyelitis. Acta Neuropathol. Commun. 2, 84 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoue M., Williams K. L., Gunn M. D., Shinohara M. L., NLRP3 inflammasome induces chemotactic immune cell migration to the CNS in experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U.S.A. 109, 10480–10485 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rumble J. M., et al. , Neutrophil-related factors as biomarkers in EAE and MS. J. Exp. Med. 212, 23–35 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou L., et al. , SerpinB1 controls encephalitogenic T helper cells in neuroinflammation. Proc. Natl. Acad. Sci. U.S.A. 116, 20635–20643 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang W. X., Huang P., Hillert J., Increased expression of caspase-1 and interleukin-18 in peripheral blood mononuclear cells in patients with multiple sclerosis. Mult. Scler. 10, 482–487 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Piancone F., La Rosa F., Marventano I., Saresella M., Clerici M., The role of the inflammasome in neurodegenerative diseases. Molecules 26, 953 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S., et al. , S100A8/A9 in inflammation. Front. Immunol. 9, 1298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogumil T., Rieckmann P., Kubuschok B., Felgenhauer K., Brück W., Serum levels of macrophage-derived protein MRP-8/14 are elevated in active multiple sclerosis. Neurosci. Lett. 247, 195–197 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Berg-Hansen P., Vandvik B., Fagerhol M., Holmøy T., Calprotectin levels in the cerebrospinal fluid reflect disease activity in multiple sclerosis. J. Neuroimmunol. 216, 98–102 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Marneros A. G., NLRP3 inflammasome blockade inhibits VEGF-A-induced age-related macular degeneration. Cell Rep. 4, 945–958 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Argaw A. T., Gurfein B. T., Zhang Y., Zameer A., John G. R., VEGF-mediated disruption of endothelial CLN-5 promotes blood-brain barrier breakdown. Proc. Natl. Acad. Sci. U.S.A. 106, 1977–1982 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inoue M., et al. , Interferon-beta therapy against EAE is effective only when development of the disease depends on the NLRP3 inflammasome. Sci. Signal 5, ra38 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malhotra S., et al. , NLRP3 inflammasome as prognostic factor and therapeutic target in primary progressive multiple sclerosis patients. Brain 143, 1414–1430 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Naegele M., et al. , Neutrophils in multiple sclerosis are characterized by a primed phenotype. J. Neuroimmunol. 242, 60–71 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Figueiredo C. A., et al. , Optimal attenuation of experimental autoimmune encephalomyelitis by intravenous immunoglobulin requires an intact interleukin-11 receptor. PLoS One 9, e101947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cook S. A., Schafer S., Hiding in plain sight: Interleukin-11 emerges as a master regulator of fibrosis, tissue integrity, and stromal inflammation. Annu. Rev. Med. 71, 263–276 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Ng B., et al. , Interleukin-11 is a therapeutic target in idiopathic pulmonary fibrosis. Sci. Transl. Med. 11, eaaw1237 (2019). [DOI] [PubMed] [Google Scholar]
- 47.Zhang X., et al. , Differential reconstitution of T cell subsets following immunodepleting treatment with alemtuzumab (anti-CD52 monoclonal antibody) in patients with relapsing-remitting multiple sclerosis. J. Immunol. 191, 5867–5874 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Tao Y., et al. , The role of endogenous IFN-beta in the regulation of Th17 responses in patients with relapsing-remitting multiple sclerosis. J. Immunol. 192, 5610–5617 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Swanson K. V., Deng M., Ting J. P., The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 19, 477–489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson A. J., et al. , Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 17, 162–173 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Seyedsadr M., Wang Y., Wrobel J. A., Garifallou J., IL-11 induces NLRP3 inflammasome activation. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE233917. Deposited 1 June 2023. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Data Availability Statement
All study data are included in the article and/or supporting information. Raw and processed scRNAseq data used in the study are available at the National Center for Biotechnology Information Gene Expression Omnibus database under the repository accession number GSE233917 (51).