Significance
Dendritic cells (DCs) are a major force in medicine with the ability to induce antigen-specific immune responses. However, transfection of DCs is challenging, and current nanoparticle (NP)-based delivery systems have had limited success in facilitating tissue-mediated delivery to lymphoid organs following systemic administration. We report the design of biodegradable polymeric nanocarriers that enable targeted mRNA delivery to splenic DCs following systemic administration without the need for targeting ligands. We also demonstrate codelivery of adjuvants along with mRNA to trigger costimulatory signaling of DCs to activate antigen-specific CD8+ T cells. The NP design enables robust antitumor immune responses in multiple murine tumor models. This technology is an advancement that validates systemically delivered polymeric mRNA-based cancer vaccines with broad applications.
Keywords: mRNA, nanoparticle, delivery, cancer, vaccine
Abstract
Nanoparticle (NP)-based mRNA cancer vaccines hold great promise to realize personalized cancer treatments. To advance this technology requires delivery formulations for efficient intracellular delivery to antigen-presenting cells. We developed a class of bioreducible lipophilic poly(beta-amino ester) nanocarriers with quadpolymer architecture. The platform is agnostic to the mRNA sequence, with one-step self-assembly allowing for delivery of multiple antigen-encoding mRNAs as well as codelivery of nucleic acid–based adjuvants. We examined structure–function relationships for NP-mediated mRNA delivery to dendritic cells (DCs) and identified that a lipid subunit of the polymer structure was critical. Following intravenous administration, the engineered NP design facilitated targeted delivery to the spleen and preferential transfection of DCs without the need for surface functionalization with targeting ligands. Treatment with engineered NPs codelivering antigen-encoding mRNA and toll-like receptor agonist adjuvants led to robust antigen-specific CD8+ T cell responses, resulting in efficient antitumor therapy in in vivo models of murine melanoma and colon adenocarcinoma.
In recent years, the field of immunotherapy and the use of immune-checkpoint inhibitors has revolutionized treatment of many cancers and led to approvals by the FDA of novel therapeutics for various tumor types (1, 2). However, the diversity of tumor epitopes between individuals makes each patient’s tumor unique; thus, the development of personalized cancer therapy technologies would add an important tool to the immunotherapy toolbox (3). The availability of personalized diagnostics using sequencing and bioinformatics technologies has made it possible in a timely and cost-effective manner to identify tumor-specific mutations, referred to as neoantigens, in individual patients (4, 5). A promising strategy to target these neoantigens is delivery of neoantigen-encoded mRNA as a cancer vaccine. In this approach, neoantigen mRNA needs to be delivered to antigen-presenting cells (APCs), such as dendritic cells (DCs), to induce a tumor-specific T cell response. mRNA vaccine technologies offer many benefits over conventional vaccine approaches, including high potency, potential for low-cost manufacturing, capacity for rapid development, and improved safety (6).
The main challenge to realizing the potential of mRNA cancer vaccines is the requirement for delivery materials to facilitate efficient intracellular mRNA delivery to large numbers of DCs. The use of lipid nanoparticles (LNPs) for delivery of mRNA encoding the spike protein of the SARS-CoV-2 virus has provided safe and powerful vaccine technologies against COVID-19 (7–9), demonstrating the great potential and increasing public acceptance of mRNA vaccines. In these prophylactic vaccine platforms, mRNA-LNPs are delivered intramuscularly to induce humoral immunity. A limitation of this strategy for cancer vaccine applications is that skeletal muscle contains very few DCs (10, 11), and transfected DCs are restricted to the local site of injection and must migrate to a lymphoid organ for T cell activation. To address these limitations and broaden this technology for therapeutic cancer vaccines, technologies facilitating delivery to a DC- and T cell–rich lymphoid organ, such as the spleen, via systemic administration would be preferred to maximize cellular immunity. In order to effectively treat tumors, a potent and systemic cytotoxic T cell response is critical. Intravenous administration of mRNA cancer vaccines has been shown to result in a more robust antigen-specific CD8+ T cell response compared to local routes of injection (12, 13), and such administration routes are easily and regularly performed for cancer patients. However, there are concerns about systemic toxicity associated with intravenous administration (14). Thus, cell target–specific mRNA delivery is necessary both for efficacy and to minimize any risks of adverse side effects. The use of RNA therapeutics for systemic delivery had a major breakthrough in 2018 when the first RNA interference technology using an LNP formulation for siRNA delivery received its first FDA approval (15, 16). However, current delivery technologies for RNA therapeutics have had limited success to facilitate extrahepatic transfection. Thus, new nanoparticle (NP) formulations are needed to achieve specific transfection in organs beyond the liver following systemic administration.
Biodegradable polymeric NPs represent a promising class of delivery vehicles for clinical application since they offer scalable production and high safety (17). Poly(beta-amino ester)s (PBAE)s are biodegradable cationic polymers that spontaneously self-assemble with anionic nucleic acids into NPs in aqueous solutions. PBAEs have several advantageous characteristics, including positive charge for efficient binding of RNA therapeutics (18, 19), high buffering capacity in an acidic environment for endosomal escape (20, 21), and hydrolytic degradation into nontoxic by-products under aqueous conditions, that make them an attractive delivery material (22–26). PBAEs have shown promise for mRNA delivery; Patel et al. reported the design of hyperbranched PBAE NPs, which facilitated efficient transfection of lung epithelial cells following nebulized administration (27), and a recent report demonstrates the capacity of PBAE-mediated delivery of mRNA to nonliver and nonlung targets (20). However, for cancer vaccine applications, new PBAE architectures are needed for efficient systemic delivery and transfection targeted to DCs.
In addition to DC transfection with antigen-encoding mRNA, DC activation is required to boost the immune response for potent cancer treatment. One of the most promising classes of adjuvants for inducing T cell immunity is toll-like receptor (TLR) agonists, which stimulate DCs and induce expression of costimulatory molecules and secretion of cytokines that drive T cell responses (28, 29). Adjuvants of interest for cancer vaccine formulations include the TLR3 agonist polyinosinic-polycytidylic acid (poly(I:C)) and the TLR9 agonist CpG oligodeoxynucleotide (CpG ODN). In addition to being a TLR3 agonist, poly(I:C) can also activate cytosolic RIG-I (30), inducing a type 1 interferon response and leading to activation of cytotoxic T cells. The controlled biodistribution and cellular uptake provided by NP-mediated delivery are beneficial as a lower encapsulated adjuvant dose is required compared to methods that rely on unassisted uptake of the adjuvant, which reduces the risk of systemic side effects. mRNA cancer vaccine technologies may be used in combination with clinically approved and emerging immunotherapy treatments, such as immune-checkpoint blockade, for synergistic effect. It was reported in a clinical study treating patients with stage IV melanoma that the antitumor T cell response was broadened with combinational treatment of a T cell induced vaccine and the immune checkpoint inhibitor pembrolizumab (anti-PD-1) (31). Their synergistic effects might also be beneficial in preventing T cell exhaustion, which is a major hurdle to eliciting potent antitumor immunity (32).
Here, we present a structural design of bioreducible lipophilic PBAE NPs that address the major challenges for mRNA cancer vaccines by enabling efficient intracellular mRNA delivery to DCs following systemic administration. We used combinatorial library synthesis to generate polymeric nanocarriers and demonstrated that both the incorporation of lipid subunit and endcap modifications of the polymer structure tuned intracellular trafficking in DCs. Additionally, we incorporated disulfide bonds in the backbone structure for environmentally triggered biodegradation upon entry into the reducing environment of the cytosol (33, 34). We demonstrated that the engineered bioreducible lipophilic PBAE NPs facilitated preferential delivery to splenic DCs following systemic administration without the need for any surface functionalization, such as PEGylation or ligand modification–mediated uptake (35). The engineered NP formulation for DC transfection delivering antigen–mRNA along with either CpG ODN or poly(I:C) was further examined for therapeutic efficacy following systemic administration using multiple in vivo murine tumor models and was found to facilitate a robust antigen-specific CD8+ T cell response for antitumor treatment.
Results
Design, Synthesis, and Characterization of Polymeric NPs.
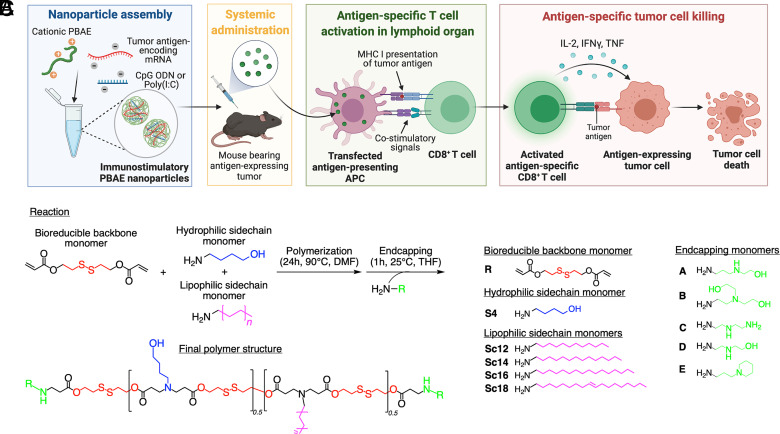
We engineered a polymeric NP platform composed of PBAEs to facilitate efficient and targeted mRNA delivery to DCs following systemic administration and explored its potential as a cancer vaccine (Fig. 1A). In a two-step synthesis using Michael addition, we combined diacrylate backbone, amine side-chain, and endcap monomers to form the final PBAE structure (Fig. 1B). In addition to the biodegradable ester bonds in the backbone structure, we incorporated disulfide bonds to create bioreducible (R) polymers to enable environmentally triggered rapid degradation in the reducing environment of the cytosol, which also increases the safety of the polymers (Fig. 1C) (18, 36). As side-chains, we combined hydrophilic (S4) and lipophilic monomers (ScX, where X is the number of carbons in the alkyl tail) to create amphipathic polymers.
Fig. 1.
Bioreducible lipophilic PBAE–mRNA nanoparticles (NPs) as cancer vaccine. (A) Schematic of the mRNA-based cancer vaccine technology using polymeric (PBAE) NPs. (B) Reaction scheme for bioreducible lipophilic PBAEs. The bioreducible diacrylate backbone monomer (R) is polymerized with a 1:1 mixture of a hydrophilic amine sidechain monomer (S4) and a lipophilic amine side chain monomer (Sc12-18) via Michael addition. The obtained diacrylate-terminated random copolymer is endcapped with an amine-containing monomer (A–E) to form the final polymer structure. (C) Monomers used in the combinatorial library synthesis to form bioreducible lipophilic PBAEs.
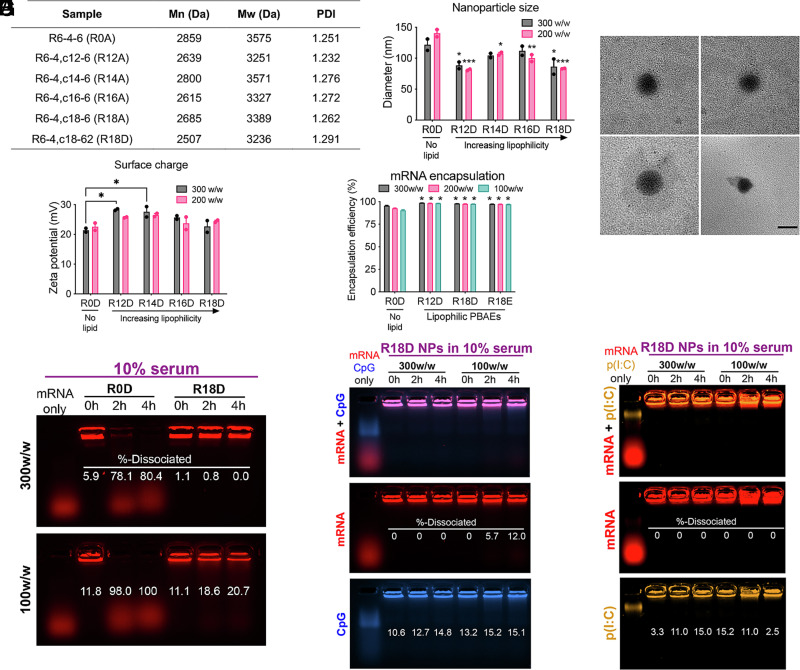
We assessed polymer molecular weights using gel permeation chromatography (GPC), and they remained approximately the same (2.5 to 2.8 kDa) after changing either the lipophilicity (R0A through R18A) or the endcap molecule (R18A compared to R18D) (Fig. 2A). We used dynamic light scattering (DLS) to analyze NP size as a function of lipophilicity (Fig. 2B). The incorporation of a lipid subunit decreased the hydrodynamic diameter of the NPs from 140 ± 6 nm for R0D NPs (no lipid; 200 w/w) down to 81 ± 1 nm for R12D NPs (Sc12 lipid; 200 w/w), likely due to increased hydrophobic interactions with the mRNA cargo forming more condensed NPs. However, increasing the length of the alkyl side-chain of the lipophilic subunit did not decrease the NP size further. We performed transmission electron microscopy (TEM) to visualize R18D mRNA-NPs, which demonstrated that the self-assembly resulted in spherical NPs of approximately 100 nm in diameter (Fig. 2C). All NPs of varying lipophilicity are cationic, with a surface charge in the range of +20 to +30 mV. The surface charge was statistically higher for lipophilic NP formulations using R12D and R14D nanocarriers compared to the nonlipophilic formulation using R0D (Fig. 2D) (*P < 0.05; n = 2). In addition to efficiently presenting tumor antigens, DCs need to become activated to induce a robust cytotoxic T cell response against the cancer cells (5). Our cationic nanocarriers can be used for codelivery of antigen-encoding mRNA and nucleic acid–based adjuvants. Accordingly, we examined the hydrodynamic diameter and surface charge for R18D NPs coencapsulating mRNA and the adjuvants CpG or poly(I:C). No statistical changes in NP size or surface charge were observed when the NPs were coencapsulating CpG or poly(I:C) compared to mRNA only (SI Appendix, Fig. S1). Moreover, using the RiboGreen assay, we observed improved mRNA encapsulation for lipophilic PBAE NPs compared to NPs with no lipid subunit (*P < 0.05; n = 3). The mRNA encapsulation efficiency for lipophilic PBAE NPs was above 97% even for formulations with lower polymer/mRNA ratios. As we sought to use the engineered NPs for systemic administration in order to induce the highest possible production of the encoded antigens, NP stability in the presence of serum is a key parameter. Previous electrostatically formed NP formulations composed of cationic polymeric nanocarriers have had limited success following systemic administration mainly due to insufficient stability in the presence of anionic serum proteins that readily dissociate the formulations (37, 38). To examine whether the presence of the lipophilic subunit would influence NP stability, we incubated NPs in media with 10% serum over 4 h (Fig. 2F). The PBAE NPs without a lipophilic subunit (R0D NPs) completely dissociated (100% release) when formed at 100 w/w, and 80% of the mRNA load was released over 4 h for the 300 w/w formulation. On the other hand, mRNA within lipophilic R18D NPs remained securely encapsulated in the presence of serum. Even when formulated at a lower polymer/mRNA ratio of 100 w/w, only 21% of mRNA was released over 4 h. Thus, in addition to electrostatic interactions, the lipophilic PBAE nanocarrier design facilitates hydrophobic interactions within the NP. This improved NP stability is a key aspect for systemic delivery technologies to ensure that the formulation does not dissociate prior to reaching the target site.
Fig. 2.
Characterization of bioreducible lipophilic poly(beta-amino ester) (PBAE) polymers and mRNA nanoparticles (NPs). (A) Molecular weights of PBAEs of varying lipophilicity assessed by GPC. (B) Hydrodynamic diameter of PBAE NPs formed at a 300 or 200 w/w ratio of polymer to mRNA assessed via DLS (n = 2). Significance indicates comparison to nonlipophilic PBAE nanoparticles (R0D) at the respective w/w ratio. (C) Representative TEM images of R18D mRNA NPs (Scale bar, 200 nm). (D) Surface charge of mRNA PBAE NPs in PBS (n = 2). (E) Encapsulation efficiency of mRNA assessed by the RiboGreen assay (n = 3). Significance indicates comparison to nonlipophilic PBAE nanoparticles (R0D) at the respective w/w ratio. (F) Encapsulation and dissociation of fluorescently labeled mRNA for nonlipophilic (R0D) and lipophilic (R18D) NPs formed at 300 and 100 w/w ratios after incubation in 10% serum over 4 h assessed by a gel electrophoresis assay. (G) mRNA and CpG ODN, (H) mRNA and poly(I:C) dissociation in 10% serum from R18D-based NPs formed at 300 and 100 w/w ratios over 4 h, respectively. Error bars represent SEM. *P < 0.05, **P < 0.01, and ***P < 0.001.
We also evaluated the ability of the lipophilic nanocarrier R18D to coencapsulate mRNA and CpG or poly(I:C) over time using a gel electrophoresis assay. We prepared R18D NPs carrying fluorescently labeled mRNA and either CpG or poly(I:C) at 300 w/w and 100 w/w and incubated them in media with 10% serum over 4 h. The majority of the mRNA and adjuvant remained encapsulated in the R18D NPs, indicating that they are stably bound in the particles. The highest observed dissociation was 12.0% and 15.2% for mRNA and CpG, respectively (Fig. 2G), and 15.2% for poly(I:C) (Fig. 2H).
In Vitro Evaluation of mRNA Delivery to DCs.
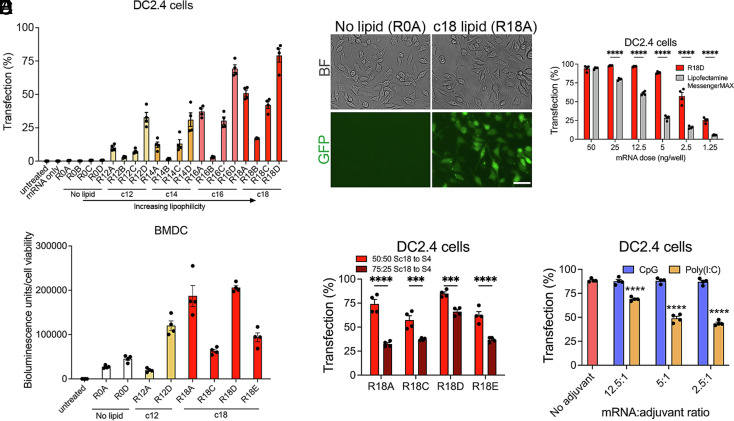
We evaluated the PBAE NP library for transfection efficiency and toxicity using the murine DC2.4 cell line and primary murine Bone marrow–derived dendritic cells (BMDCs). In these high-throughput in vitro screens, NPs delivered eGFP mRNA, and transfection was assessed quantitatively by flow cytometry (Fig. 3A) and qualitatively by fluorescence microscopy (Fig. 3B) 24 h posttreatment. We evaluated cell viability using the MTS assay 24 h posttreatment (SI Appendix, Fig. S2A). The percent of transfected cells generally increased as polymer lipophilicity increased, with nonlipophilic NPs (R0A-D) providing little to no transfection (Fig. 3 A and B). In each lipophilic PBAE series (R12, R14, R16, and R18), the PBAE with the D endcap monomer facilitated the highest level of transfection (Fig. 3A). The most lipophilic of those, R18D, achieved the highest level of transfection. Structures with the B endcap monomer showed very low transfection efficacy, indicating that the secondary amine in the A, C, and D endcaps is preferable to the tertiary amine and additional hydroxyl group in the B endcap. These studies highlight the utility of high-throughput in vitro valuation of polymer libraries with differential structure for mRNA delivery, as small, seemingly minor changes to single atoms in the side chain or end-group of the polymer lead to dramatic functional differences in transfection.
Fig. 3.
Transfection of dendritic cells (DCs) in vitro by bioreducible liphophilic PBAE mRNA nanoparticles (NPs). (A) Polymer library was evaluated for transfection of the murine dendritic cell line DC2.4 using mRNA encoding GFP. Cells were treated with NPs formed at 200 w/w and a dose of 50 ng mRNA/well, and transfection efficiency was assessed via flow cytometry after 24 h. (B) Representative bright-field (BF) and fluorescent microscopy images of DC2.4 cells transfected with nonlipophilic R0A or lipophilic R18A GFP mRNA NPs (Scale bar, 50 nm). (C) Transfection of DC2.4 cells by top-performing R18D NPs was assessed at various mRNA doses and compared to leading commercial mRNA transfection reagent Lipofectamine MessengerMAX. (D) A subset of the polymer library was evaluated on murine BMDCs using luciferase-encoding mRNA. Cells were treated with NPs at a dose of 25 ng mRNA/well, and bioluminescence activity was assessed after 24 h to determine transfection levels normalized to cell viability. (E) Polymers with the Sc18 monomer were synthesized with a 50:50 or 75:25 ratio of lipophilic side chain monomer Sc18 to hydrophilic side chain monomer S4. DC2.4 cells were treated with GFP mRNA NPs with varied lipophilicity at a dose of 25 ng mRNA/well, and transfection was assessed after 24 h. (F) The transfection efficiency in DC2.4s was examined following treatment with R18D NPs coencapsulating GFP mRNA and CpG or poly(I:C) adjuvants with varied mRNA to adjuvant ratios where the mRNA dose was kept constant (25 ng/well) after 24 h. Significance indicates comparison to no adjuvant control. Error bars represent SEM (n = 4). ***P < 0.001 and ****P < 0.0001.
R18D NPs were further examined in DC2.4 cells at a wide range of mRNA doses and compared to Lipofectamine MessengerMAX, a leading commercial mRNA transfection agent (Fig. 3C). R18D NPs performed significantly better than Lipofectamine MessengerMAX at all but the highest mRNA dose. Even at a low dose of 5 ng mRNA per well in a 96-well plate (50 ng mRNA/mL), 20 times lower than what is traditionally recommended for commercial transfection reagents, R18D NPs transfected 88.6% of cells, while Lipofectamine MessengerMAX transfected 28.7% of cells on average.
To better mimic in vivo DCs, we evaluated a subset of the PBAE NP library on primary murine BMDCs. In this screen, tested NP formulations delivered fLuc mRNA, and bioluminescence was assessed after 24 h to evaluate relative transfection (Fig. 3D). Similar to DC2.4 cells, BMDCs showed the highest level of transfection with the most lipophilic NPs, and R18D was a top-performing candidate. Importantly, the NPs caused no toxicity in the BMDCs (SI Appendix, Fig. S2B). We also compared R18D NPs to two leading commercial mRNA transfection reagents (SI Appendix, Fig. S3). R18D NPs facilitated significantly higher GFP-mRNA transfection, transfecting 17 to 20% of BMDCs, compared to Lipofectamine MessengerMAX and jetMESSENGER, which transfected 3.6% and 5.5% of BMDCs, respectively.
To further examine the effect of PBAE hydrophobicity on transfection efficiency, we synthesized PBAEs in the R18 series with a 75:25 molar ratio of lipophilic Sc18 monomer to hydrophilic S4 monomer and compared their in vitro efficacy to PBAEs synthesized with a 50:50 molar ratio of the two monomers (Fig. 3E). All PBAE NPs with a 50:50 ratio of Sc18 to S4 performed significantly better than their 75:25 counterparts with no differences in cell viability (SI Appendix, Fig. S2C). Codelivery of CpG did not influence mRNA transfection, whereas poly(I:C) did significantly reduce transfection in a dose-dependent fashion (Fig. 3F).
Effect of Polymer Structure on NP-Mediated Intracellular Trafficking.
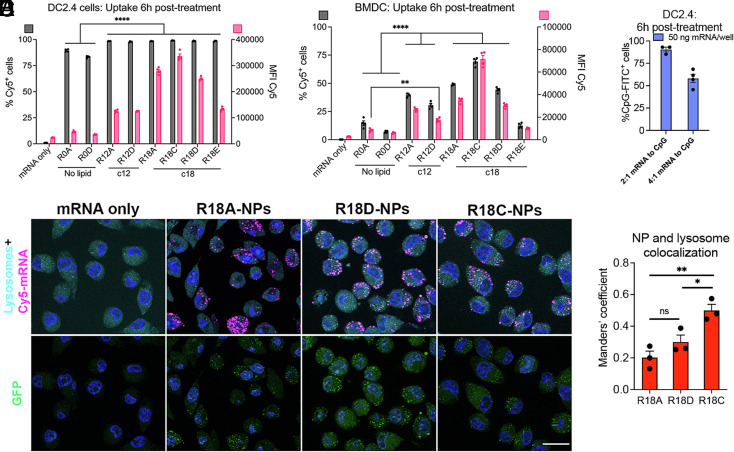
We used fluorescently labeled (Cy5) mRNA-NPs to facilitate mechanistic understanding of how the structural design of the nanocarrier influences intracellular trafficking. We first analyzed cellular uptake in DC2.4 cells, and all tested NPs were taken up by over 83% of cells, with lipophilic NPs demonstrating cellular uptake between 97% and 99% after 6 h (Fig. 4A). We also analyzed the mean fluorescence intensity of Cy5, which correlates with the average number of NPs taken up per cell and found that NPs with the longer alkyl side-chain (Sc18) were taken up more efficiently than NPs with a shorter alkyl side-chain (Sc12) (Fig. 4A). Both percent cellular uptake and fluorescence intensity of uptake were statistically higher for all lipophilic NPs compared to the NP formulations without a lipophilic subunit (****P < 0.0001; n = 4). We further examined NP uptake in BMDCs and found that increased lipophilicity of the nanocarrier improved cellular uptake (Fig. 4B). The NP uptake was statistically higher for the lipophilic NPs compared to NPs without a lipophilic subunit using the same endcap (R12A and R18A compared to R0A; R12D and R18D compared to R0D; ****P < 0.0001; n = 4). This finding demonstrates that the chemical structure of the lipophilic subunit is a key property for cellular uptake. The trend of improved cellular uptake with increased lipophilicity supports the higher in vitro transfection efficiency observed for lipophilic mRNA-NPs (Fig. 3 A and D). Additionally, endcap modifications also influenced cellular uptake, as shown for NP formulations using Sc18 as the lipophilic subunit, where the R18 nanocarrier with endcapping monomer C (R18C) facilitated the highest uptake (Fig. 4 A and B). The same trends between nanocarrier structure and NP uptake at 6 h posttreatment were also observed at 24 h posttreatment (SI Appendix, Fig. S4 A and B). Moreover, we prepared R18D NPs coencapsulating mRNA and FITC-labeled CpG. The uptake of CpG was 90% ± 3% and 58% ± 5% for NPs formulated at w/w ratios of mRNA to CpG of 2:1 and 4:1, respectively (Fig. 4C). CpG was still present in cells 24 h posttreatment (SI Appendix, Fig. S4C).
Fig. 4.
Cellular uptake and endosomal escape following mRNA nanoparticle (NP) design. (A) DC2.4 cells and (B) murine primary BMDCs were treated with NPs carrying Cy5-mRNA at a dose of 50 ng mRNA/well, and NP uptake was assessed at 6 h posttreatment by flow cytometry (n = 4). (C) DC2.4 cells were treated with R18D NPs coencapsulating mRNA at 50 ng mRNA/well and FITC-CpG, and uptake of CpG was assessed 6 h posttreatment by flow cytometry (n = 4). (D) Representative images of DC2.4 cells labeled with lysosome/endosome dye 6 h posttreatment with NPs carrying Cy5-labeled GFP-encoding mRNA to visualize cellular uptake, NP colocalization with endosomes/lysosomes, and GFP transfection (Scale bars, 20 μm). (E) Manders’ coefficient was determined using ImageJ to quantify the degree of colocalization between NPs and endosomes/lysosomes (n = 3). Error bars represent SEM. *P < 0.05, **P < 0.01, and ****P < 0.0001.
We used apotome microscopy to visualize cellular uptake and trafficking of Cy5-mRNA NPs and GFP-mRNA transfection 6 h posttreatment (Fig. 4D). We evaluated endosomal escape by quantifying the colocalization of endosomes/lysosomes stained with a LysoTracker dye and Cy5-NPs, with lower colocalization corresponding to more effective endosomal escape. The calculated Manders’ coefficient of colocalization demonstrated that R18A and R18D NPs facilitated significantly higher endosomal escape than R18C NPs (*P < 0.05 and **P < 0.01; n = 3) (Fig. 4E). Thus, the incorporation of a hydroxyl group (endcapping monomers A and D) instead of an additional amine group (endcapping monomer C) as endcap modification promoted endosomal escape. The lower level of endosomal escape for R18C NPs explains why their higher cellular uptake did not result in higher mRNA transfection. Interestingly, R18D exhibited significantly higher transfection than R18A (****P < 0.0001; n = 4) (Fig. 3A) despite both structures demonstrating similarly high levels of cellular uptake and endosomal escape in DC2.4 cells. This discrepancy is likely due to the difference in cell viability, as R18A NPs were significantly more toxic than R18D NPs in DC2.4 cells (*P < 0.05; n = 4) (SI Appendix, Fig. S2A). Taken together, these results demonstrate that both the hydrophobicity of the polymer side-chains and the chemistry of the endcaps influence NP-mediated intracellular trafficking in DCs.
Targeted Transfection of Splenic DCs In Vivo.
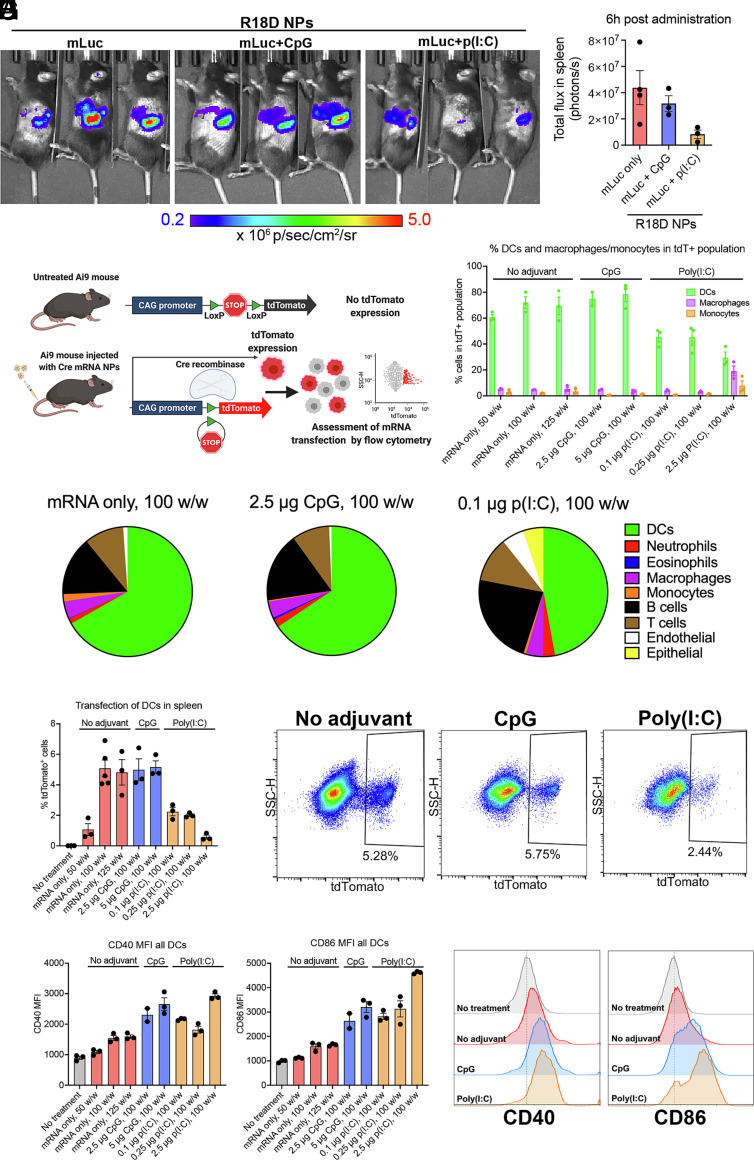
We assessed the in vivo transfection following systemic administration of fLuc mRNA NPs (10 μg mRNA/mouse) by whole animal bioluminescence imaging (SI Appendix, Fig. S5). We included NPs with the lipophilic side chains Sc16 and Sc18 and the endcaps A, C, and D to explore the influence of side chain hydrophobicity and endcap chemistry. These formulations all showed high in vitro transfection in both immortalized and primary murine DCs. These structural designs of the bioreducible lipophilic PBAE NPs facilitated, almost exclusively, transfection localized to the spleen (SI Appendix, Fig. S5A). The NPs with the more hydrophobic side chains (Sc18 compared to Sc16) promoted NP-mediated transfection in the spleen, whereas the endcaps with the same base polymer chemistry showed a similar level of transfection of splenocytes (SI Appendix, Fig. S5B). Based on in vitro experiments for transfection and biocompatibility as well as the in vivo spleen-targeted transfection, we selected the top-performing NP formulation R18D for further in vivo studies. We first assessed its time window for transfection in vivo following systemic administration of fLuc mRNA NPs by further whole animal bioluminescence imaging (SI Appendix, Fig. S6). As previously demonstrated, the structural design of the R18D NPs facilitated targeted transfection in the spleen (SI Appendix, Fig. S6B), and peak expression was between 2 and 6 h, with continuing expression up to 48 h postadministration (SI Appendix, Fig. S6C). We similarly evaluated in vivo transfection for R18D mRNA-NPs codelivering no adjuvant, 2.5 μg CpG ODN, or 0.1 μg poly(I:C)/mouse along with 10 μg mRNA/mouse at a polymer-to-nucleic acid ratio of 100 w/w 6 h postadministration (Fig. 5A). Congruent with in vitro results, codelivery of CpG did not significantly reduce the total bioluminescence in the spleen, while poly(I:C) reduced the bioluminescence signal by approximately fivefold compared to the no adjuvant group (Fig. 5B). However, all groups, with and without adjuvants, showed localized transfection in the spleen.
Fig. 5.
In vivo transfection in spleen following systemic administration of R18D mRNA nanoparticles (NPs). (A) R18D NPs carrying luciferase mRNA (mLuc) (10 μg/mouse) and CpG (2.5 μg/mouse) or poly(I:C) (0.1 μg/mouse) were assembled at a polymer-to-nucleic acid ratio of 100 w/w and administered intravenously to C57BL/6J mice. Whole-animal bioluminescence imaging was performed 6 h after administration. (B) Image analysis was used to assess total flux in the spleen. (C) Schematic of the Ai9 mouse model used to assess transfected cell types in vivo following systemic administration of mRNA NPs carrying Cre mRNA. Cells that are transfected undergo Cre recombinase–mediated recombination, resulting in tdTomato expression that is detected by flow cytometry. (D–H) R18D Cre mRNA NPs were administered intravenously to Ai9 mice at 10 μg mRNA/mouse, and tdTomato expression in key cell populations in the spleen was assessed after 24 h. (D) Percent of all tdTomato+ (tdT+) cells in the spleen that are DCs, macrophages, or monocytes. (E) Pie charts indicating average share of transfected cells in the spleen belonging to each cell population shown for NP treatments carrying no adjuvant, 2.5 μg CpG/mouse, or 0.1 μg poly(I:C)/mouse. (F) Percent of DCs in the spleen that are transfected. (G) Representative flow cytometry plots showing transfected tdTomato+ DCs treated with mRNA-NP formulations coencapsulating no adjuvant, 2.5 μg CpG, or 0.1 μg poly(I:C). (H) Geometric mean fluorescent intensity (MFI) of CD40 and CD86 expression in all splenic DCs. (I) Representative histograms of CD40 and CD86 expression in no-treatment control, and following NP treatment coencapsulating no adjuvant, 2.5 μg CpG/mouse, or 0.1 μg poly(I:C)/mouse. Error bars represent SEM.
In addition to organ-level targeting, we characterized the specific cell populations in the spleen that are targeted for transfection using the Ai9 mouse model. Upon successful delivery of Cre mRNA, cells undergo Cre recombinase–mediated recombination, resulting in tdTomato expression (39) (Fig. 5C). We assessed transfection in splenic cell populations 24 h posttreatment with systemically administered R18D NPs encapsulating Cre mRNA alone at varying w/w ratios or codelivering Cre mRNA with CpG or poly(I:C) at varying adjuvant doses. R18D NPs formed at 50 w/w were less effective than their 100 and 150 w/w counterparts, in terms of both overall splenocyte transfection (SI Appendix, Fig. S7A) and transfection of DCs (Fig. 5F). R18D NPs facilitated transfection of approximately 5% of DCs in the spleen (Fig. 5 F and G and SI Appendix, Fig. S7D), and DCs represented the largest share of transfected cells in the spleen, accounting for approximately 70% of all transfected cells, demonstrating cell specificity of transfection (Fig. 5 D and E and SI Appendix, Fig. S7E). For R18D NPs codelivering poly(I:C), DCs represented a smaller share of transfected cells, although still the largest of any cell population, at approximately 45%, with about 2% of splenic DCs transfected for the two lower poly(I:C) doses (Fig.5 D–G and SI Appendix, Fig. S7 D and E). Macrophages and monocytes represented a small percentage of the transfected splenocytes, approximately 4 to 5% and 1 to 2%, respectively (Fig. 5 D and E and SI Appendix, Fig. S7E). We assessed the expression of DC activation markers, CD40 and CD86, in the transgenic Ai9 mouse model. The mRNA NPs on their own led to minimal upregulation of CD40 and CD86, but codelivery of CpG or poly(I:C) adjuvants led to a substantial increase in expression of those activation markers (Fig. 5 H and I and SI Appendix, Fig. S7 B and C). To balance transfection and immunostimulation, the lowest CpG and poly(I:C) doses of 2.5 μg (0.4 nmol) and 0.1 μg per mouse, respectively, were selected for in vivo therapeutic studies. These doses are significantly lower than conventional doses for vaccine adjuvants, which are typically reported to be administered at doses of 10 to 50 μg/mouse for both CpG (40–47) and poly(I:C) (48–53).
The efficient intracellular delivery to DCs observed in vivo for the R18D NPs would have ramifications on other routes of administration; thus, we compared the in vivo transfection following systemic administration to that following intramuscular (I.M.) and subcutaneous (S.Q.) administration (SI Appendix, Fig. S8). However, the engineered NP formulation facilitated much higher transfection following systemic administration than the other routes examined. Thus, modifications to the design principles reported here would be needed to expand the technology to other routes of administration.
In Vivo Therapeutic Tumor Vaccination.
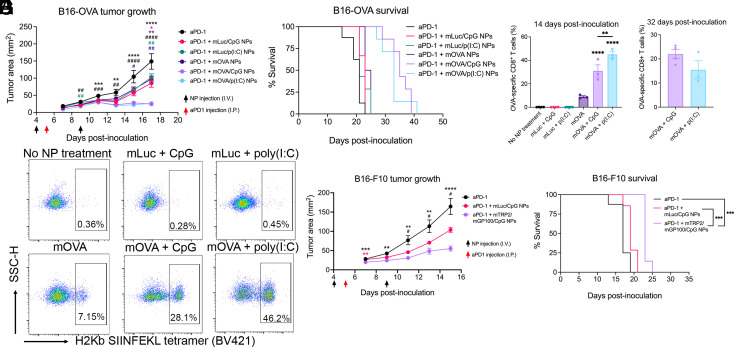
We first evaluated the vaccine platform in the B16-F10-OVA mouse melanoma model, which expresses ovalbumin (OVA) as a tumor antigen, in combination with immune checkpoint blockade (Fig. 6A). R18D NPs coencapsulating OVA mRNA (mOVA) or fLuc mRNA (mLuc, as an irrelevant mRNA control) and CpG, poly(I:C), or no adjuvant were administered. The mOVA/CpG and mOVA/poly(I:C) NP treatments resulted in a statistically significant decrease in tumor burdens, completely halting tumor growth for over a week after the final vaccination compared to the aPD-1-only control (Fig. 6A and SI Appendix, Fig. S9). The mOVA/CpG and mOVA/poly(I:C) NP treatments also prolonged survival from a median survival of 21 d for the aPD-1 group to 35 and 33 d, respectively (P = 0.0002) (Fig. 6B). Additionally, we assessed the presence of OVA-specific CD8+ T cells in the blood 14 d postinoculation using a tetramer stain for the H-2Kb restricted OVA SIINFEKL epitope (Fig. 6 C and E). Controls without OVA mRNA generated almost no antigen-specific CD8+ T cells, while in mice treated with OVA mRNA NPs with no adjuvant, 8.9% of CD8+ T cells in circulation were found to be OVA-specific. The percent of OVA-specific CD8+ T cells in circulation substantially increased with the inclusion of CpG or poly(I:C) to 31.0% and 45.1%, respectively, confirming that an immunostimulatory component is required for the NPs to elicit robust antigen-specific T cell proliferation. We assessed OVA-specific T cells again 32 d postinoculation in surviving mice treated with mOVA/CpG and mOVA/poly(I:C) NPs to analyze the long-term CD8+ T cell response and found that there were still 22.0% and 15.5% OVA-specific CD8+ T cells, respectively.
Fig. 6.
In vivo therapeutic efficacy of PBAE mRNA nanoparticle (NP) vaccination in B16-OVA and B16-F10 mouse melanoma models. (A–E) 3 × 105 B16-OVA cells were inoculated subcutaneously on day 0, and R18D NPs encapsulating luciferase-encoding mRNA or OVA-encoding mRNA were administered intravenously (I.V.) on days 4 and 9 at 10 μg mRNA/mouse and 2.5 μg CpG/mouse or 0.1 μg poly(I:C)/mouse (n = 7 to 8 mice/group). Then, 200 μg of aPD-1 was injected intraperitoneally (I.P.) on day 5. (A) Tumor growth measurements showing in vivo therapeutic effects between treatments. *P < 0.05, **P < 0.01, and ****P < 0.0001 for comparison between aPD-1 + mOVA/CpG NP treatment and respective controls (indicated by color). #P < 0.05, ##P < 0.01, and ####P < 0.0001 for comparison between aPD-1 + mOVA/p(I:C) NP treatment and respective controls (indicated by color). (B) Mice were euthanized once tumors reached 200 mm2, and survival curves are shown. (C) Mice were bled on day 14 postinoculation, and the percent of OVA-specific CD8+ T cells out of total CD8+ T cells was assessed using H2Kb-SIINFEKL tetramer staining. Significance indicates comparison of mOVA/CpG and mOVA/p(I:C) NP treatment compared to all respective controls. (D) On day 32 postinoculation, surviving mice in mOVA/CpG and mOVA/p(I:C) NP treatment groups were bled, and the presence of OVA-specific CD8+ T cells was assessed via tetramer staining. (E) Representative flow cytometry plots showing H2Kb-SIINFEKL tetramer staining in CD3+ CD8+ cells on day 14. (F and G) 3 × 105 B16-F10 cells were inoculated subcutaneously on day 0, and R18D NPs encapsulating luciferase mRNA or a 1:1 mixture of TRP2 and GP100-encoding mRNA (10 μg total mRNA/mouse) and CpG were administered following the previously described treatment scheme (n = 7 to 8 mice/group). (F) Tumor growth measurements showing in vivo therapeutic effects between treatments. **P < 0.01, ***P < 0.001, and ****P < 0.0001 for comparison between aPD-1 + mTRP2/mGP100/CpG NP treatment and aPD-1 control (black) or aPD-1 + mLuc/CpG NP group (pink). #P < 0.05 for comparison between the aPD-1 + mLuc/CpG NP group and aPD-1 group. (G) Mice were euthanized once tumors reached 200 mm2, and survival curves are shown. Error bars represent SEM.
We next assessed the efficacy of the vaccination platform in B16-F10 murine melanoma cells that do not express the immunogenic OVA antigen. We opted to deliver mRNA encoding two well-established melanoma-associated antigens, tyrosinase-related protein 2 (TRP2) and glycoprotein 100 (GP100) (54). As TRP2 and GP100 are self-antigens, they are typically difficult to vaccinate against (55), so we sought to explore whether the NP vaccine platform could be potent enough to exert a therapeutic effect in this model. Following the results from the B16-F10-OVA study in which mOVA/CpG-treated mice had a slightly longer median survival time and a more robust long-term CD8+ T cell response compared to the mOVA/poly(I:C) group, we elected to move forward with the CpG adjuvant for subsequent in vivo tumor studies. Treatment with R18D NPs coencapsulating a 1:1 weight ratio of TRP2 and GP100 mRNA, in combination with CpG, led to a significant reduction in tumor burden compared to the aPD-1 only control (Fig. 6F and SI Appendix, Fig. S10). Median survival was significantly extended (P = 0.0001) from 17 d in the aPD-1 control group to 23 d in the full treatment group (Fig. 6G).
We thereafter examined the efficacy of the engineered PBAE NP vaccine in the murine colon carcinoma MC38-OVA model (Fig. 7A). Treatment with mOVA/CpG-NPs significantly reduced tumor burden compared to both controls on days 11 to 17, and 50% of mice completely cleared their tumors and were long-term survivors compared to only 14.3% and 28.6% in the mLuc/CpG and aPD-1-only groups, respectively (Fig. 7B). On day 65, all long-term survivors, in addition to seven age-matched mice, were rechallenged with MC38-OVA cells on the opposite flank. All long-term survivors completely rejected the rechallenge, indicating the generation of antigen-specific memory. We also assessed OVA-specific CD8+ T cells in the blood 21 d postinoculation (Fig. 7 C–E). There was a remarkable antigen-specific CD8+ T cell response, with 64.1% of CD8+ T cells in the blood being OVA-specific in mOVA/CpG NP-treated mice, compared to approximately 1% in the controls (Fig. 7 C and E). Additionally, mOVA/CpG NP-treated mice had a higher percentage of CD8+ T cells out of all CD3+ T cells in the blood compared to both controls (Fig. 7D).
Fig. 7.
In vivo therapeutic efficacy of PBAE mRNA nanoparticle (NP) vaccination in the MC38-OVA mouse colon carcinoma model. (A) 1 × 106 MC38-OVA cells were inoculated subcutaneously in the right flank of C57BL/6J mice on day 0, and R18D NPs encapsulating luciferase-encoding mRNA or OVA-encoding mRNA (10 μg mRNA/mouse) and CpG (2.5 μg/mouse) were administered intravenously on days 9 and 14 (n = 7 to 8 mice/group). Then, 200 μg of aPD-1 was injected intraperitoneally on day 10. Tumor growth measurements showing the in vivo therapeutic effects between the treatment groups. Significance indicates comparison of the aPD-1 + mOVA/CpG NP treatment group to the aPD-1 group (black) or aPD-1 + mLuc/CpG NP group (pink). (B) Mice were euthanized once tumors reached 200 mm2, and survival curves are shown. (C) Four mice were randomly selected from each group to be bled on day 21 postinoculation, and the percent of OVA-specific CD8+ T cells out of total CD8+ T cells in the blood was assessed using H2Kb SIINFEKL tetramer staining. (D) The percent of CD8+ T cells out of total CD3+ T cells in blood is shown. (E) Representative flow cytometry plots showing BV421 H2Kb SIINFEKL tetramer staining in CD3+ CD8+ cells in all groups. Error bars represent SEM.
Finally, we evaluated the alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels in serum to assess the safety of the engineered R18D NPs delivering mRNA alone or in combination with the adjuvants CpG or poly(I:C). The activity levels of ALT and AST used as biomarkers for liver health were within the tolerated level after both 24 and 72 h (SI Appendix, Fig. S11 A and B). After 24 h, a slight increase in ALT activity was observed when codelivering mRNA with CpG, and an increase in AST activity was observed when codelivering mRNA with either CpG or poly(I:C). However, by 72 h, their activities were down to the same level as the untreated controls. Serum levels of cytokines associated with cytokine release syndrome were also evaluated. As expected, for the NPs codelivering adjuvants, certain cytokines had somewhat increased levels at 24 h, but they had already decreased to the same level as the no-treatment controls after 72 h (SI Appendix, Fig. S11C). Taken together, these data demonstrate that the engineered structural design of bioreducible lipophilic PBAE NPs used for codelivery of mRNA along with CpG or poly(I:C) can be used for safe and robust antitumor vaccination.
Discussion
We synthesized a library of bioreducible lipophilic PBAE structures to facilitate fundamental understanding of structure–function relationships for efficient intracellular delivery to DCs. We determined that inclusion of a lipophilic subunit in the PBAE backbone is critical for DC transfection, as PBAE NPs with only the hydrophilic subunit S4 resulted in very minimal mRNA transfection in vitro. Moreover, the length of the lipophilic subunit strongly affects cellular uptake and transfection. Polymers with increased lipophilicity showed greater cellular uptake and improved mRNA transfection. One potential mechanism contributing to the enhanced cellular uptake of the lipophilic NPs by DCs may be that many innate immune receptors have evolved to recognize hydrophobic portions of molecules (56, 57). Thus, particles with more hydrophobic surfaces may have increased cell uptake by DCs due to interaction with these receptors (56). In addition to the lipophilic subunit, we found that the chemistry of the endcap molecule influences uptake and subsequent endosomal escape. Within the R18 polymer series (most hydrophobic), R18C NPs facilitated the highest uptake. However, R18A and R18D NPs provided higher endosomal escape compared to R18C, suggesting that the hydroxyl group in endcap monomers A and D is preferable to the additional amine group in endcap monomer C for promoting endosomal escape. Balancing uptake, endosomal escape, and cellular biocompatibility, R18D NPs facilitated the highest in vitro DC transfection among all structures in the library. Additionally, R18D NPs transfected DC2.4 cells and murine BMDCs at significantly higher levels in vitro compared to leading commercial mRNA transfection reagents, even at very low mRNA doses.
When the engineered bioreducible lipophilic NPs were used for systemic administration in vivo, they were found to transfect the spleen with a high degree of specificity, largely avoiding the liver, which is usually a major site of NP accumulation and transfection for many lipid- and polymer-based NPs (58, 59). Increased hydrophobicity of the lipophilic side chain of the polymeric nanocarrier promoted spleen-targeted transfection. Furthermore, within the spleen, the R18D NPs facilitated preferential transfection of DCs over other cell types, including macrophages. This is in contrast to most nonviral gene delivery platforms, in which macrophages and monocytes rapidly phagocytose NPs upon systemic administration and represent a large proportion of transfected cells (60), and even LNPs that can transfect nonmacrophage populations largely target hepatocytes (59). Previous work has generally required surface functionalization with DC-targeting ligands to achieve such specificity (61, 62). For a cancer vaccine application, delivery to DCs over other APCs, namely monocytes and macrophages, is preferred as DCs are much more potent in initiating a T cell response (63).
We also demonstrated the ability of the engineered polymeric NPs to coencapsulate and deliver the nucleic acid–based TLR agonists CpG and poly(I:C) along with mRNA. By codelivering mRNA and adjuvant within the same PBAE NPs, we ensure that any DC that is transfected with antigen-encoding mRNA also receives a danger signal, resulting in its activation. Codelivery of poly(I:C) reduced mRNA transfection both in vitro and in vivo, likely due to the immunostimulatory effects of poly(I:C). Previous work has shown that increased immunogenicity of mRNA formulations has a negative impact on mRNA translation. Innate immune sensing of unmodified mRNA leads to reduced translation efficiency, and nucleoside modification has been widely employed to reduce the inherent immunogenicity of mRNA and, thereby, improve transfection (64, 65). Contamination with double-stranded RNA (dsRNA) has also been implicated in poor mRNA translation due to activation of TLR3 and RIG-I (66). Poly(I:C) is a synthetic analog of dsRNA and activates the same receptors, so a similar mechanism is likely responsible for the decreased mRNA expression observed with poly(I:C) codelivery. Additionally, codelivery of certain adjuvants, such as STING agonist R848 (67) and TLR4 agonist LPS (68), has been shown to reduce mRNA expression both in vitro and in vivo. Unlike TLR3 agonist poly(I:C), codelivery of TLR9 agonist CpG did not result in significantly decreased mRNA transfection in vitro or in vivo. This discrepancy may be due, in part, to the variable expression of TLR3 and TLR9. TLR3 is expressed more broadly than TLR9, so poly(I:C) may stimulate the innate immune system more strongly and across more cell types, resulting in dampened mRNA translation (69). It should be noted that these results were observed for codelivery of CpG and poly(I:C) with GFP, luciferase, and Cre-encoding mRNA, indicating that this phenomenon is not sequence specific. In future cancer vaccine design, adjuvanticity and translation efficiency will have to be carefully balanced to garner optimal antigen expression while inducing a robust CD8+ T cell response. In vivo, R18D mRNA-NPs codelivering CpG or poly(I:C) were able to activate splenic DCs at doses of 2.5 μg CpG or 0.1 μg poly(I:C) per mouse, substantially lower than previously reported doses, which are typically in the range of 10 to 50 μg/mouse for both CpG (40–47) and poly(I:C) (48–53). The engineered NPs facilitated highly targeted delivery to splenic DCs with efficient cellular internalization. As a result, the reported NP design enables a therapeutic effect with much lower adjuvant doses and avoids delivery to off-target cells, potentially reducing the risk of systemic side effects.
Finally, we demonstrated therapeutic efficacy of the engineered PBAE NP vaccine platform in multiple in vivo tumor models in combination with immune checkpoint blockade. R18D NPs codelivering antigen mRNA and CpG or poly(I:C) showed efficacy in eliciting an antitumor response in B16-F10-OVA and B16-F10 murine melanoma models and MC38-OVA murine colon carcinoma. In the B16-F10-OVA and MC38-OVA tumor models, we observed a dramatic antigen-specific CD8+ T cell response following treatment with NPs codelivering antigen mRNA and adjuvant, resulting in reduced tumor burdens and prolonged median survival. We also assessed the efficacy of the R18D NP platform in treating B16-F10 by targeting established melanoma self-antigens, TRP2 and GP100 (54). Tumor-associated self-antigens are typically difficult to vaccinate against due to immune tolerance (55), but our NP platform was able to mediate a significant reduction in tumor size and prolonged survival. Additionally, we demonstrated that this platform induces long-term systemic immunity in the MC38-OVA model. Moreover, treatment with the R18D NPs does not cause a risk of systemic toxicity. This is likely due to the targeted delivery to splenic DCs facilitated by the structural design of the engineered NPs and biodegradability of the polymeric nanocarrier, which undergoes degradation into safe by-products under physiological conditions (22).
Taken together, the PBAE NP-mRNA vaccine showed efficacy in treating three different in vivo tumor models by incorporating low adjuvant doses and targeting clinically relevant antigens, demonstrating the translational promise of this platform and potential for application to neoantigen vaccines. Compared to virus-based strategies or ex vivo engineered cell-based immuno-oncology strategies, the biodegradable polymeric NP vaccine platform discussed here overcomes many safety, manufacturing, and scalability challenges. Compared to traditional and emerging LNP technology, safe, efficient, and specific combination mRNA delivery to splenic DCs in vivo was achieved without the need for PEGylation or targeting ligands. This biotechnology platform is a simple, modular, and scalable method for in vivo production of cancer antigen–specific CD8+ T cells that sidesteps the many challenges of alternative viral and/or ex vivo cellular engineering strategies. These results show tremendous promise for the use of bioreducible lipophilic PBAE NPs as a modular genetic vaccine.
Materials and Methods
Expanded Materials and Methods are available in SI Appendix.
Monomer and Polymer Synthesis.
The bioreducible monomer 2,2′-disulfanediylbis(ethane-2,1-diyl) diacrylate (R) was synthesized as previously described (23, 70). Bioreducible lipophilic PBAEs were synthesized via the Michael addition reaction. The obtained polymers were purified by precipitation in a 1:1 mixture of diethyl ether and hexane and two washes.
NP Preparation.
NPs were formed by mixing polymer and nucleic acid cargo in 25 mM sodium acetate buffer (NaAc, pH 5) and allowed to self-assemble into NPs for 6 min at room temperature.
Polymer and NP Characterization.
Polymer molecular weight was measured using GPC relative to linear polystyrene standards (Waters). The hydrodynamic diameter of the NPs in 1× PBS was measured by DLS using a Zetasizer Pro (Malvern Panalytical). Zeta potential was measured via electrophoretic mobility using the same instrument to characterize the surface charge of the NPs. NP size and morphology were visualized by TEM using a Talos L120C microscope (Thermo Scientific).
Cell Culture and Cell Line Preparation.
Murine DC2.4 cells were cultured in RPMI 1640 media (Gibco) supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin, 10 mM HEPES, 1X nonessential amino acids, and 50 μM beta-mercaptoethanol. BMDCs were generated from bone marrow isolated from C57BL/6J mice (Jackson Laboratory; Bar Harbor, ME). MC38-OVA cells were cultured in RPMI 1640 supplemented with 10% FBS and 1% penicillin/streptomycin.
In Vitro mRNA Transfection.
Unless stated otherwise, all mRNA used was purchased from TriLink Biotechnologies with 5-methoxyuridine modification and CleanCap technology. NPs were formulated as described above with eGFP mRNA [and CpG or Poly(I:C) if indicated] and then added to the cells. For cellular uptake studies, 20% of the total mRNA was replaced with Cy5-labeled eGFP mRNA (TriLink Biotechnologies). After 6 h (for uptake experiments) or 24 h (for transfection experiments), transfection/uptake was evaluated via flow cytometry. The MTS CellTiter 96 Aqueous One (Promega) cell proliferation assay was performed 24 h posttransfection according to the manufacturer's instructions as a measure of cell viability.
Encapsulation Efficiency and NP Stability.
mRNA encapsulation efficiency was assessed using the RiboGreen RNA assay (Invitrogen) following the manufacturer’s protocols. The gel electrophoresis assay was performed to examine NP stability when incubated in 10% serum for 4 h.
Characterization of Intracellular Trafficking.
Endosomal escape of NPs was studied in DC2.4 cells using immunofluorescence staining. Cells were plated onto coverslips in 12-well plates and grown overnight. NPs were prepared with 20% Cy5-mRNA and 80% unlabeled mRNA and incubated with cells for 6 h. Cells were washed with Phosphate-Buffered Saline (PBS) and then stained for 30 min with Hoechst 33,342 (Thermo Fisher Scientific) nuclear stain at 1:5000 dilution and Cell Navigator Lysosome Staining dye (AAT Bioquest) at 1:2500 dilution in complete media. Colocalization of Cy5-NPs and LysoTracker stain was quantified by calculating the Manders’ coefficient in acquired images using ImageJ.
NP Formulation for In Vivo Transfection.
All animal work was performed in strict adherence to the policies and guidelines of the Johns Hopkins University Animal Care and Use Committee. NPs for in vivo mRNA delivery were formulated at 100 w/w unless stated otherwise. All in vivo transfections utilized the R18D polymer (BR6-S4, Sc18 [50:50]-D).
In Vivo Bioluminescence Transfection Studies.
NPs encapsulating fLuc mRNA were formulated as described above and administered to 6- to 7-wk-old male BALB/c or C57BL/6J mice via retro-orbital injection. Whole-body bioluminescence of BALB/c or shaved C57BL/6J mice was assessed at 6 h postinjection (or at prespecified timepoints up to 96 h for time course study). For timecourse studies, animals were humanely killed immediately after whole-body imaging via cervical dislocation, and selected organs were extracted, submerged in D-luciferin solution (250 μg/mL), and imaged with IVIS.
Cre mRNA Delivery to Ai9 Mice.
Ai9 mice were purchased from Jackson Laboratory (JAX stock #007909) and bred in the Johns Hopkins animal facility (39). NPs encapsulating Cre mRNA and adjuvants (if indicated) were administered to Ai9 mice via tail vein injections, and tdTomato expression following Cre-Lox recombination was allowed to accumulate for 24 h, at which point animals were euthanized via cervical dislocation. Surface staining of cells with fluorescent antibodies was then performed using the antibodies and dilutions listed in SI Appendix, Table S1.
Tumor Vaccination and Safety Studies.
For both B16-F10-OVA and B16-F10 tumor studies, 3 × 105 cells in 50 μL RPMI 1640 media were inoculated subcutaneously in the right flank of C57BL/6J mice on day 0. For MC38-OVA studies, 1 × 106 cells in 50 μL RPMI 1640 media were inoculated subcutaneously in the right flank of C57BL/6J mice on day 0. R18D NPs encapsulating fLuc mRNA (as an irrelevant mRNA control) or OVA mRNA were administered intravenously by retro-orbital injections on days 4 and 9 for the B16-F10-OVA study or days 9 and 14 for the MC38-OVA study at 10 μg mRNA/mouse and 2.5 μg CpG or 0.1 μg poly(I:C) for adjuvant groups (n = 7 to 8 mice/group). For the B16-F10 study, instead of OVA mRNA, a 1:1 mixture (each at 5 μg/mouse) of custom-synthesized TRP2 mRNA (NCBI gene accession number: NM_021882) and GP100 mRNA (NCBI gene accession number: NM_010024) (TriLink Biotechnologies) was used, and NPs were administered intravenously by retro-orbital injection on days 4 and 9. Then, 200 μg of aPD-1 was injected intraperitoneally on day 5 for B16 studies or day 10 for the MC38-OVA study.
Statistical Analysis.
All experiments were performed with n = 4 replicates unless otherwise stated. Bar graphs indicate mean ± SEM n.s. (not significant) indicates P > 0.05, * indicates P < 0.05, ** indicates P < 0.01, *** indicates P < 0.001, and **** indicates P < 0.0001. For studies with two variables, two-way ANOVA was used with recommended post hoc tests. For studies with only one variable, one-way ANOVA was used with recommended post hoc tests. For tumor studies, one-way ANOVA with post hoc Tukey’s test was used to compare tumor sizes on each day, and a log rank (Mantel–Cox) test was used to assess significance between survival curves. All statistics were performed using statistical analysis software modules in GraphPad Prism 9 (GraphPad Software, Inc.), and P < 0.05 was considered statistically significant.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
This work was supported in part by grants from the NIH (R01CA228133, P41EB028239, and R37CA246699), the Goldhirsh-Yellin Foundation, and funding from the Johns Hopkins University (JHU) Bloomberg-Kimmel Institute for Cancer Immunotherapy. E.B.-A. thanks National Cancer Institute of the NIH (F31CA250367) for fellowship support. J.K. was supported by the Swedish Research Council International Postdoc grant (2016-06675). BioRender.com is acknowledged for figure preparation.
Author contributions
E.B.-A., J.K., D.M.P., and J.J.G. designed research; E.B.-A., J.K., S.H., H.Y., and S.Y.T. performed research; E.B.-A., J.K., S.Y.T., D.M.P., and J.J.G. analyzed data; and E.B.-A., J.K., S.Y.T., D.M.P., and J.J.G. wrote the paper.
Competing interests
D.M.P. is a consultant for Compugen, Shattuck Labs, Tempest, Immunai, Bristol-Myers Squibb, Amgen, Janssen, Astellas, Rockspring Capital, Immunomic, and Dracen. J.J.G. is a cofounder, manager, and CTO of Dome Therapeutics; cofounder, board member, and CSO of Cove Therapeutics; cofounder and manager of OncoSwitch Therapeutics; cofounder of WyveRNA Therapeutics; scientific advisory board member of Mana Bio; and board member of VasoRx. S.Y.T. is a cofounder and manager of OncoSwitch Therapeutics. D.M.P. owns founder’s equity in manaT Holdings, LLC, Trex, Jounce, Anara, Tizona, Tieza, and RAPT. J.J.G. owns equity in Dome, Cove, OncoSwitch, WyveRNA, Mana Bio, and VasoRx. S.Y.T. owns equity in OncoSwitch. E.B.-A., J.K., S.Y.T., and J.J.G. are a coinventors on patents filed by Johns Hopkins University related to technologies discussed in the manuscript. D.M.P. receives research funding from Compugen, Bristol-Myers Squibb, and Anara and royalties on patents licensed by Compugen, BMS, and Immunomic.
Footnotes
This article is a PNAS Direct Submission. D.J.M. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Blass E., Ott P. A., Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat. Rev. Clin. Oncol. 18, 215–229 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whiteside T. L., Demaria S., Rodriguez-Ruiz M. E., Zarour H. M., Melero I., Emerging opportunities and challenges in cancer immunotherapy. Clin. Cancer Res. 22, 1845–1855 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vormehr M., Türeci Ö., Sahin U., Harnessing tumor mutations for truly individualized cancer vaccines. Annu. Rev. Med. 70, 395–407 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Wang R. F., Wang H. Y., Immune targets and neoantigens for cancer immunotherapy and precision medicine. Cell Res. 27, 11–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Z., Ott P. A., Wu C. J., Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat. Rev. Immunol. 18, 168–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardi N., Hogan M. J., Porter F. W., Weissman D., mRNA vaccines-a new era in vaccinology. Nat. Rev. Drug Discov. 17, 261–279 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polack F. P., et al. , Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baden L. R., et al. , Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 384, 403–416 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin M. D., et al. , COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 15, 646–655 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Heine A., Juranek S., Brossart P., Clinical and immunological effects of mRNA vaccines in malignant diseases. Mol. Cancer 20, 52 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang F., Loré K., Local innate immune responses in the vaccine adjuvant-injected muscle. Clin. Transl. Immunology 5, e74 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sayour E. J., et al. , Systemic activation of antigen-presenting cells via RNA-loaded nanoparticles. OncoImmunology 6, e1256527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broos K., et al. , Particle-mediated intravenous delivery of antigen mRNA results in strong antigen-specific T-cell responses despite the induction of Type I interferon. Mol. Ther. Nucleic Acids 5, e326 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J., Eygeris Y., Gupta M., Sahay G., Self-assembled mRNA vaccines. Adv. Drug Deliv. Rev. 170, 83–112 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams D., et al. , Patisiran, an RNAi therapeutic, for hereditary transthyretin amyloidosis. N. Engl. J. Med. 379, 11–21 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Akinc A., et al. , The onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnology 14, 1084–1087 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Karlsson J., Vaughan H. J., Green J. J., Biodegradable polymeric nanoparticles for therapeutic cancer treatments. Annu. Rev. Chem. Biomol. Eng. 9, 105–127 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Bertoni H., et al. , Bioreducible polymeric nanoparticles containing multiplexed cancer stem cell regulating miRNAs inhibit glioblastoma growth and prolong survival. Nano Lett. 18, 4086–4094 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karlsson J., et al. , Engineered nanoparticles for systemic siRNA delivery to malignant brain tumours. Nanoscale 11, 20045–20057 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rui Y., et al. , High-throughput and high-content bioassay enables tuning of polyester nanoparticles for cellular uptake, endosomal escape, and systemic in vivo delivery of mRNA. Sci. Adv. 8, eabk2855 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunshine J. C., Peng D. Y., Green J. J., Uptake and transfection with polymeric nanoparticles are dependent on polymer end-group structure, but largely independent of nanoparticle physical and chemical properties. Mol. Pharmaceut. 9, 3375–3383 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlsson J., Rhodes K. R., Green J. J., Tzeng S. Y., Poly(beta-amino ester)s as gene delivery vehicles: Challenges and opportunities. Expert Opin. Drug Deliv. 17, 1395–1410 (2020), 10.1080/17425247.2020.1796628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozielski K. L., Tzeng S. Y., Hurtado De Mendoza B. A., Green J. J., Bioreducible cationic polymer-based nanoparticles for efficient and environmentally triggered cytoplasmic siRNA delivery to primary human brain cancer cells. ACS Nano 8, 3232–3241 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zamboni C. G., et al. , Polymeric nanoparticles as cancer-specific DNA delivery vectors to human hepatocellular carcinoma. J. Control. Release 263, 18–28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sunshine J., et al. , Small-molecule end-groups of linear polymer determine cell-type gene-delivery efficacy. Adv. Mater. 21, 4947–4951 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tzeng S. Y., et al. , Non-viral gene delivery nanoparticles based on Poly(β-amino esters) for treatment of glioblastoma. Biomaterials 32, 5402–5410 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel A. K., et al. , Inhaled nanoformulated mRNA polyplexes for protein production in lung epithelium. Adv. Mater. 31, e1805116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwasaki A., Medzhitov R., Toll-like receptor control of the adaptive immune responses. Nat. Immunol. 5, 987–995 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Maisonneuve C., Bertholet S., Philpott D. J., De Gregorio E., Unleashing the potential of NOD- and toll-like agonists as vaccine adjuvants. Proc. Natl. Acad. Sci. U.S.A. 111, 12294–12299 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gale E. C., et al. , A nanoparticle platform for improved potency, stability, and adjuvanticity of Poly(I:C). Adv. Therapeut. 3, 1900174–1900174 (2020). [Google Scholar]
- 31.Ott P. A., et al. , An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLane L. M., Abdel-Hakeem M. S., Wherry E. J., CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 37, 457–495 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Meng F., Hennink W. E., Zhong Z., Reduction-sensitive polymers and bioconjugates for biomedical applications. Biomaterials 30, 2180–2198 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Manickam D. S., et al. , Effect of innate glutathione levels on activity of redox-responsive gene delivery vectors. J. Control. Release 141, 77–84 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suk J. S., Xu Q., Kim N., Hanes J., Ensign L. M., PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 99, 28–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell M. J., et al. , Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discovery 20, 101–124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanco E., Shen H., Ferrari M., Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 33, 941–951 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y., Ye M., Xie R., Gong S., Enhancing the in vitro and in vivo stabilities of polymeric nucleic acid delivery nanosystems. Bioconjugate Chem. 30, 325–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madisen L., et al. , A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Callmann C. E., et al. , Tumor cell lysate-loaded immunostimulatory spherical nucleic acids as therapeutics for triple-negative breast cancer. Proc. Natl. Acad. Sci. U.S.A. 117, 17543–17550 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuai R., Ochyl L. J., Bahjat K. S., Schwendeman A., Moon J. J., Designer vaccine nanodiscs for personalized cancer immunotherapy. Nat. Mater. 16, 489–496 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu B., et al. , Equipping cancer cell membrane vesicles with functional DNA as a targeted vaccine for cancer immunotherapy. Nano Lett. 21, 9410–9418 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Nanishi E., et al. , An aluminum hydroxide:CpG adjuvant enhances protection elicited by a SARS-CoV-2 receptor binding domain vaccine in aged mice. Sci. Transl. Med. 14, eabj5305 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ni Q., et al. , A bi-adjuvant nanovaccine that potentiates immunogenicity of neoantigen for combination immunotherapy of colorectal cancer. Sci. Adv. 6, eaaw6071 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shih T.-Y., Najibi A. J., Bartlett A. L., Li A. W., Mooney D. J., Ultrasound-triggered release reveals optimal timing of CpG-ODN delivery from a cryogel cancer vaccine. Biomaterials 279, 121240 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Wang H., et al. , Biomaterial-based scaffold for in situ chemo-immunotherapy to treat poorly immunogenic tumors. Nat. Commun. 11, 5696 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., et al. , Supramolecular assembled programmable nanomedicine as in situ cancer vaccine for cancer immunotherapy. Adv. Mater. 33, 2007293 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Da Silva C. G., et al. , Co-delivery of immunomodulators in biodegradable nanoparticles improves therapeutic efficacy of cancer vaccines. Biomaterials 220, 119417 (2019). [DOI] [PubMed] [Google Scholar]
- 49.He X., et al. , HPV-associated tumor eradication by vaccination with synthetic short peptides and particle-forming liposomes. Small 17, 2007165 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim S.-Y., et al. , Synthetic vaccine nanoparticles target to lymph node triggering enhanced innate and adaptive antitumor immunity. Biomaterials 130, 56–66 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Nakamura T., Haloho S. E. E., Harashima H., Intravenous liposomal vaccine enhances CTL generation, but not until antigen presentation. J. Control. Release 343, 1–12 (2022). [DOI] [PubMed] [Google Scholar]
- 52.Qin H., et al. , Development of a cancer vaccine using in vivo click-chemistry-mediated active lymph node accumulation for improved immunotherapy. Adv. Mater. 33, 2006007 (2021). [DOI] [PubMed] [Google Scholar]
- 53.Roy D. G., et al. , Adjuvant oncolytic virotherapy for personalized anti-cancer vaccination. Nat. Commun. 12, 2626 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kirkin A. F., Dzhandzhugazyan K., Zeuthen J., Melanoma-associated antigens recognized by cytotoxic T lymphocytes. APMIS 106, 665–679 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Pedersen S. R., Sørensen M. R., Buus S., Christensen J. P., Thomsen A. R., Comparison of vaccine-induced effector CD8 T cell responses directed against self- and non–self-tumor antigens: Implications for cancer immunotherapy. J. Immunol. 191, 3955–3967 (2013). [DOI] [PubMed] [Google Scholar]
- 56.Bachmann M. F., Jennings G. T., Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 10, 787–796 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Seong S.-Y., Matzinger P., Hydrophobicity: An ancient damage-associated molecular pattern that initiates innate immune responses. Nat. Rev. Immunol. 4, 469–478 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y.-N., Poon W., Tavares A. J., McGilvray I. D., Chan W. C. W., Nanoparticle–liver interactions: Cellular uptake and hepatobiliary elimination. J. Control. Release 240, 332–348 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Samaridou E., Heyes J., Lutwyche P., Lipid nanoparticles for nucleic acid delivery: Current perspectives. Adv. Drug Deliv. Rev. 154–155, 37–63 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Wilhelm S., et al. , Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 1, 16014 (2016). [Google Scholar]
- 61.Tacken P. J., de Vries I. J. M., Torensma R., Figdor C. G., Dendritic-cell immunotherapy: From ex vivo loading to in vivo targeting. Nat. Rev. Immunol. 7, 790–802 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Phua K. K. L., Towards targeted delivery systems: Ligand conjugation strategies for mRNA nanoparticle tumor vaccines. J. Immunol. Res. 2015, 680620 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pozzi L.-A.M., Maciaszek J. W., Rock K. L., Both dendritic cells and macrophages can stimulate naive CD8 T cells in vivo to proliferate, develop effector function, and differentiate into memory cells. J. Immunol. 175, 2071–2081 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Anderson B. R., et al. , Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 38, 5884–5892 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karikó K., Buckstein M., Ni H., Weissman D., Suppression of RNA recognition by toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005). [DOI] [PubMed] [Google Scholar]
- 66.Barbier A. J., Jiang A. Y., Zhang P., Wooster R., Anderson D. G., The clinical progress of mRNA vaccines and immunotherapies. Nat. Biotechnol. 40, 840–854 (2022). [DOI] [PubMed] [Google Scholar]
- 67.Islam M. A., et al. , Adjuvant-pulsed mRNA vaccine nanoparticle for immunoprophylactic and therapeutic tumor suppression in mice. Biomaterials 266, 120431 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lokugamage M. P., et al. , Mild innate immune activation overrides efficient nanoparticle-mediated RNA delivery. Adv. Mater. 32, 1904905 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hafner A. M., Corthésy B., Merkle H. P., Particulate formulations for the delivery of poly(I:C) as vaccine adjuvant. Adv. Drug Deliv. Rev. 65, 1386–1399 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Karlsson J., et al. , Photocrosslinked bioreducible polymeric nanoparticles for enhanced systemic siRNA delivery as cancer therapy. Adv. Funct. Mater. 31, 2009768 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.