Significance
A unique anti-CD81 antibody inhibits human breast cancer metastases in xenograft models, and cell migration and invasion in vitro. Here we analyzed the contribution of structural attributes of CD81 to the metastatic process. We found that the antibody binds to a unique sequence of amino acids in the large extracellular loop of the CD81 molecule. The intracellular domains of CD81 are not essential for the antimetastatic effects induced by the antibody. Rather, the antibody mediates its effects by cross-linking CD81 molecules and recruiting partner proteins in the membrane including integrin β5, integrin αV, and the transferrin receptor 1.
Keywords: immunotherapy, breast cancer, invasion, migration, tetraspanins
Abstract
Metastases are reduced in CD81KO mice. In addition, a unique anti-CD81 antibody, 5A6, inhibits metastasis in vivo and invasion and migration in vitro. Here, we probed the structural components of CD81 required for the antimetastatic activity induced by 5A6. We found that the removal of either cholesterol or the intracellular domains of CD81 did not affect inhibition by the antibody. We show that the uniqueness of 5A6 is due not to increased affinity but rather to its recognition of a specific epitope on the large extracellular loop of CD81. Finally, we present a number of CD81 membrane-associated partners that may play a role in mediating the 5A6 antimetastatic attributes, including integrins and transferrin receptors.
Recently, we demonstrated that tetraspanin CD81 is a novel immunotherapeutic target in human breast cancer cells (1). Studies have shown that CD81 expression in human breast cancer is associated with reduced overall survival (2, 3). We targeted CD81 using an antibody that inhibited the growth of human breast cancer cell lines. Moreover, our antibody reduced metastasis in a patient-derived xenograft. In mice, CD81 affects the adaptive and innate immune systems of the host. We previously demonstrated that both T regulatory cells and myeloid-derived suppressor cells are impaired in CD81KO mice (4). Consequently, tumor growth and metastasis are inhibited in CD81KO mice (1, 5, 6).
CD81 belongs to an evolutionarily conserved tetraspanin family, members of which play a role in cell–cell interactions (7, 8). Tetraspanin family members associate with other cell surface proteins and modulate their functions (9–11). Tetraspanins have no transligands; therefore, antibodies are used to identify their associated partner proteins and to induce functional consequences upon engagement (12). Interestingly, antibodies against the same tetraspanin molecule differ in their ability to induce functional outcomes (1). Specifically, among the various anti-CD81 antibodies, only 5A6 blocks the migration and invasion of MDA-MB-231 human breast cancer cells in vitro and inhibits their metastasis in a xenograft model (1).
Here, we investigated how the structural domains of CD81 contribute to the functional effects of the 5A6 antibody. To this end, we generated CD81KO MDA-MB-231 cells and transfected these CD81KO cells with modified CD81 constructs to delineate the interaction of 5A6 with the defined domains of its target.
Here, we demonstrated that 5A6 inhibits MDA-MB-231 cells mutated at CD81 residues 18 and 219, which anchor cholesterol in the transmembrane cavity. Moreover, we demonstrated that the intracellular domains of CD81 were not required for the inhibitory effects of the antibody. We also showed that the antimetastatic effects of 5A6 are not due to its avidity of interaction with its target but rather to its recognition of a specific epitope structure. Finally, we identified and tested several CD81 membrane-associated partner protein candidates for their role in mediating the 5A6 antibody antimetastatic effects.
Results
Triggering CD81 by 5A6 Inhibits the Motility and Invasiveness of MDA-MB-231 Human Breast Cancer Cells.
Previously, we demonstrated that among the available anti-CD81 antibodies, only 5A6 inhibits the migration and invasion of MDA-MB-231 triple-negative breast cancer cells (1). Indeed, 5A6 significantly reduced cell migration from the top transwells to the lower chambers by 92 to 95% (Fig. 1A). Cell migration rate is an important indicator of metastatic aggressiveness. Therefore, we evaluated the effects of 5A6 on the migration of MDA-MB-231 cells on several substrates. The total migration distance and motility were both significantly reduced in the presence of 5A6 on collagen IV (Fig. 1B), laminin, and gelatin, but not on poly-L-lysine (SI Appendix, Fig. S1 A–C, respectively). However, it is noteworthy that while 5A6 profoundly inhibited migration, it had no impact on MDA-MB-231 cell adhesion and colony formation (SI Appendix, Fig. S1D).
Fig. 1.
The antihuman CD81 antibody 5A6 blocks breast cancer cell migration, motility, invasion, and ECM degradation, and its inhibitory effect of 5A6 is specific to CD81. (A–E and H–J) MDA-MB-231 cells were treated with 5A6 (10 µg/mL). (A, H, and I) 5A6 blocks MDA-MB-231 cell migration from the top transwell to the bottom chamber. Cells were imaged at 5× magnification after 48 h using a fluorescent microscope. Data is presented as percent of MDA-MB-231 cells per field after 48 h (n = 6), counted using Image-Pro Plus 6.0 software. (B) 5A6 reduces MDA-MB-231 cell motility on collagen IV. Time lapse 10-min interval tracing over 10 h is presented as Delta XY movement from frame 1, total movement distance (P = 0.0066), and movement distance per frame (P = 0.007). (C) 5A6 reduces degradation of DQ Fluorescein-conjugated Collagen IV by MDA-MB-231 cells. Cells were stained with DAPI and imaged at 10× magnification after 24 h using a fluorescent microscope. Degraded areas (integrated optical density of fluorescein) were analyzed using Image-Pro Plus 6.0 software. (D, H, and J) 5A6 blocks invasion to the extracellular matrix (ECM). Cells were imaged at 5× magnification after 48 h using a fluorescent microscope. Data is presented as the percentage of invaded areas after 48 h of ECM addition (n = 9) and analyzed using Image-Pro Plus 6.0 software. Blue line defines the core area, and the yellow line demarcates the invaded area. (E) 5A6 blocks the degradation of DQ Fluorescein-conjugated Collagen IV in invading MDA-MB-231 3D spheres. Cells were imaged at 5× magnification after 48 h using a fluorescent microscope. Data is presented after 48 h of ECM addition (n = 9), degraded areas (IOD of fluorescein) were analyzed using Image-Pro Plus 6.0 software. Data is presented as present change, and shown as Mean ± SEM, t-test, P < 0.05*, P < 0.01**, P < 0.0001***. (F) CD81 expression on the indicated cells was analyzed by staining with JS81-APC. (G) MDA-MB-231CD81KO cell migration from the top transwell to the bottom chamber, and invasion to the extracellular matrix (ECM) are not affected by the absence of CD81 expression. (I and J) Data is presented as present change, and shown as Mean ± SEM, one-way ANOVA, significance is marked for the comparison between the groups in presence of 5A6; P < 0.05*, P < 0.01**, P < 0.0001***.
Tumor cells’ invasiveness is dependent, in part, on their ability to degrade the extracellular matrix (ECM) (13, 14). To test the effect of 5A6 on ECM degradation, we performed a quenched collagen IV assay. In the absence of the antibody, MDA-MB-231 cells degraded collagen-IV, as evidenced by the high fluorescein release; however, 5A6 significantly blocked this collagen-IV degradation (Fig. 1C). As previously described (1), we confirmed that 5A6 blocked the invasion of MDA-MB-231 cells into the ECM (Fig. 1D). Next, we used ECM containing quenched collagen-IV in a 3D sphere invasion assay to evaluate ECM degradation. Again, 5A6 drastically reduced the degradation of collagen IV (Fig. 1E). 5A6 also impaired the degradation of fluorescein-conjugated gelatin, an additional component of ECM, as indicated by the dark areas (SI Appendix, Fig. S1E).
The Inhibitory Effect of 5A6 Is Specific to CD81.
To determine the specificity of 5A6, we first knocked out CD81 using CRISPR/Cas9 gRNAs (SI Appendix, Table S3). Next, we reconstituted CD81 expression in MDA-MB-231CD81KO cells (MDA-MB-231 CD81REC). As expected, MDA-MB-231 CD81REC cells regained binding of 5A6 to CD81 (Fig. 1F). Importantly, 5A6 ability to inhibit invasion and migration was restored (Fig. 1 H–J and SI Appendix, Fig. S1 G and H), confirming that CD81 is indeed the relevant target of this antibody, leading to an antimetastatic effect. Surprisingly, lack of CD81 had no impact on the ability of cells to migrate and invade the ECM (Fig. 1G). Additionally, 5A6 is known to trigger cell death in human lymphoma B cells (5), we wondered whether 5A6 is also cytotoxic to MDA-MB-231 cells. However, cell proliferation assays revealed that parental and MDA-MB-231CD81KO cells proliferated similarly in the absence and presence of 5A6 (SI Appendix, Fig. S1F). 5A6 ability to reduce ECM degradation is not justifiable by alterations in the expression or secretion of any of the tested metalloproteases (MMPs) (SI Appendix, Fig. S1I).
The Unique Effects of 5A6 Are Not Explained by Its High Avidity.
Among anti-CD81 antibodies, only 5A6 inhibits the metastatic behavior of MDA-MB-231 cells (1). Therefore, we investigated whether the inhibitory effects of 5A6 were due to its higher avidity. First, we compared the affinity of anti-CD81 antibodies for the soluble human GST-CD81-LEL fusion protein (15) using real-time kinetic characterization of the interaction via Octet Qk (ForteBio). The calculated association and dissociation of these antibodies to the soluble human GST-CD81-LEL fusion protein demonstrated that the affinity of 5A6 was equivalent to that of JS81, an antibody without an antimetastatic effect and that the affinities of other non-antimetastatic anti-CD81 antibodies, 1D6 and 1.3.3.22, were also similar in the picomole range (SI Appendix, Fig. S2A). Interestingly, Hasezaki et al. determined that the affinity of JS81 for full-length CD81 embedded in nanodiscs was higher than that of 5A6 (16). Therefore, we compared the avidity of the binding of antibodies to cell surface-expressed CD81 using a competition assay. MDA-MB-231 cells were incubated with a fixed concentration of 5A6-Alexa-488 in the presence of increasing concentrations of unlabeled competing anti-hCD81 monoclonal antibodies (mAbs). Interestingly, JS81 competed with 5A6 (SI Appendix, Fig. S2B). Thus, the functional consequences of 5A6 engagement with cell surface-expressed CD81 were not due to its higher avidity of interaction.
The CD81 Epitope of 5A6 Is Unique among Anti-CD81 Antibodies.
The anti-CD81 antibodies competed for binding to CD81, but only 5A6 inhibited the motility and invasion of aggressive breast cancer cells. To pinpoint the 5A6 epitope, we focused on four amino acids that differ between human and monkey CD81, as this antibody binds to human, but not monkey CD81. In contrast, other candidate anti-CD81 antibodies recognize both human and monkey CD81 (15). First, we produced and purified three GST-CD81-LEL soluble fusion proteins with missense mutations. FPLC-purified proteins were analyzed for real-time kinetic interactions with 5A6 using Octet Qk. The affinity of 5A6 for T163A and D196E was in the picomolar range, similar to its binding to native GST-CD81 LEL. In contrast, 5A6 did not bind to F186L or E188K, suggesting that F186 and/or E188 were included in the 5A6-binding epitope (Fig. 2A). Next, we reconstituted MDA-MB-231CD81KO with mutated CD81. The analysis of the interaction of JS81 and 5A6 with these newly generated cell lines revealed that JS81 bound to all cells expressing the mutated proteins, whereas 5A6 showed a significant reduction in binding to F186L and E188K CD81 cell lines (Fig. 2B and SI Appendix, Fig. S2C). Finally, to ensure that the overexpression of CD81 in the reconstituted cells did not affect the binding of antibodies, we knocked in (KI) human CD81 exons 6/7 or exons 6/7 containing mutations in the mouse breast cancer cell line 4T1. The binding of 5A6 to cells expressing F186L and E188K was significantly reduced, whereas JS81 bound to all cell lines expressing the mutated amino acid residues (Fig. 2C and SI Appendix, Fig. S2D). Interestingly, 5A6 bound to cells expressing the single mutations F186L and E188K, suggesting that the antibody epitope includes all three residues (186 to 188). The invasion blockage capability upon treatment with 5A6 was affected by the insufficient binding of 5A6 to the MDA-MB-231 cells expressing F186L and E188K mutated CD81 (SI Appendix, Fig. S2E).
Fig. 2.
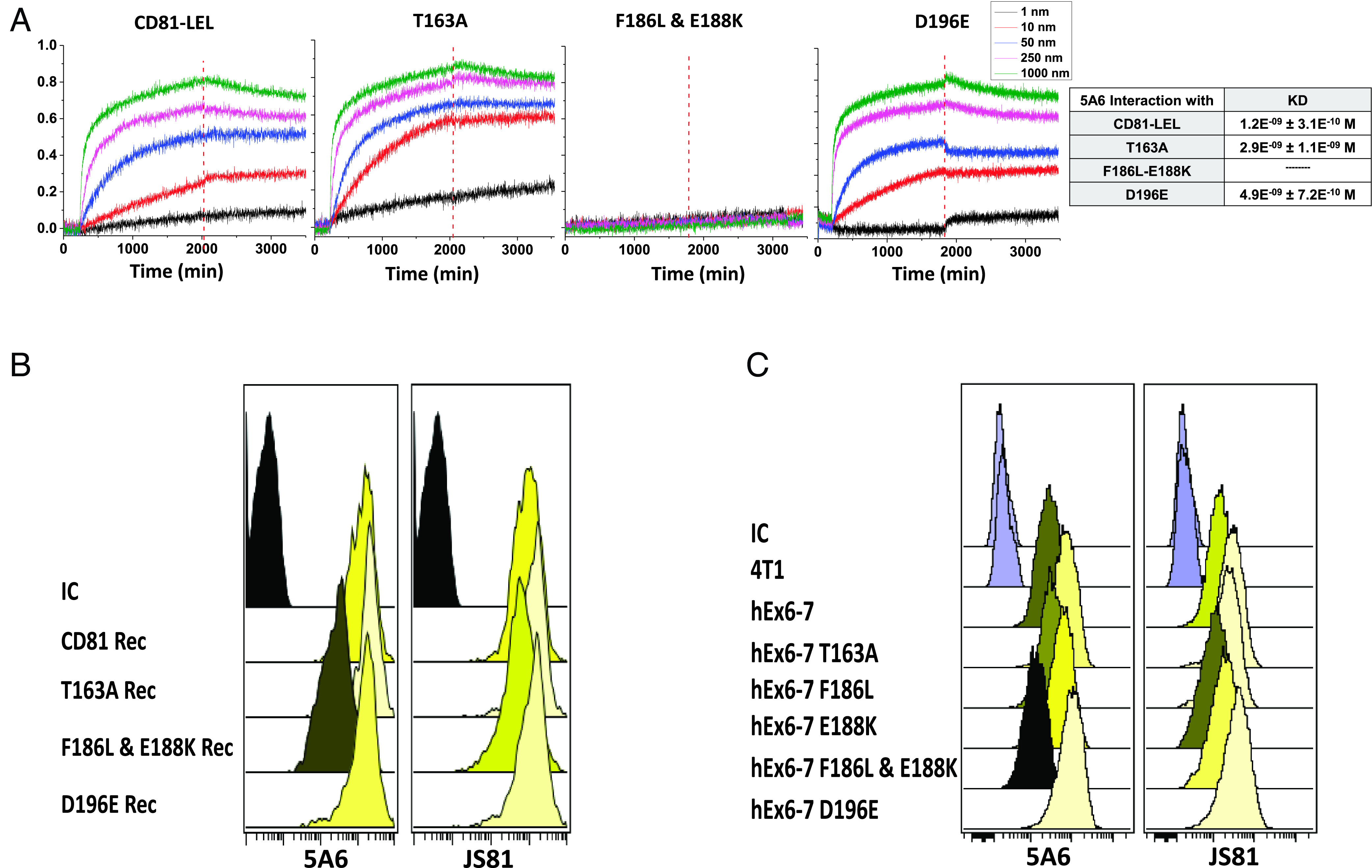
Identification of the 5A6 epitope. (A) Binding kinetics of 5A6 to the indicated GST-CD81-LEL soluble proteins. (B and C) Binding of 5A6 and JS81 to cells expressing the indicated CD81 proteins expressed on (B) MDA-MB-231 cells, and (C) on 4T1 cells into which the indicated mutated human CD81 exons 6/7 were knocked in. Binding of the antibodies was analyzed by flow cytometry.
The 5A6 Fab Does Not Inhibit the Invasion of MDA-MB-231 Cells.
It is of note, the 3D structure of the interaction between 5A6 Fab and CD81-LEL showed that E188 contacts the light chain of the antibody fragment. To test whether 5A6 Fab has a functional effect on cell surface-expressed CD81, we generated Fab and F(ab’)2 antibody fragments (SI Appendix, Fig. S3A). As expected, the purified proteins bound to MDA-MB-231 cells, as determined by flow cytometry (SI Appendix, Fig. S3B). 5A6 F(ab’)2 effectively inhibited the invasion and migration of MDA-MB-231 cells (SI Appendix, Fig. S3 C and D). In contrast, Fab did not inhibit invasion. Moreover, cross-linking the fragment with an antilight chain antibody did not induce an inhibitory effect, even in the absence of the cross-linking antibody (SI Appendix, Fig. S3 C and D). Interestingly, Fab had some effect on cell migration, which did not increase upon cross-linking.
5A6 Inhibits Motility and Invasiveness Even when CD81 Is Mutated in Cholesterol-Binding Residues.
The complete crystal structure of CD81 revealed the presence of cholesterol in the transmembrane cavity, forming close contact with amino acids N18 and E219 (17), and mutating these residues reduced embedded cholesterol (17). Additional molecular dynamics simulation studies have suggested that cholesterol in the transmembrane cavity affects the conformation of the LEL of the molecule, particularly the interaction of CD81 with CD19 (17, 18). Because cholesterol can affect the conformation of LEL, we generated individual and double N18A/E219A mutant constructs and expressed them in MDA-MB-231CD81KO cells. Cells with the double mutant construct expressed CD81 at a level comparable to that in the reconstituted MDA-MB-231 CD81REC cells (Fig. 3A). The analysis of the effect of 5A6 revealed that the antibody inhibited the migration and invasion of N18A/E219A-mutated MDA-MB-231 cells (Fig. 3 B and C and SI Appendix, Fig. S4 B and C). Notably, these mutations did not affect cell proliferation (SI Appendix, Fig. S4A).
Fig. 3.

MDA-MB-231 cells that are mutated in CD81 cholesterol-binding residues, or lack their intracellular domains are sensitive to 5A6. (A) Surface CD81 expression in MDA-MB-231 cells expressing the indicated mutations. (B, C, E, F, I, and J) The indicated MDA-MB-231 cells were incubated in the presence and absence of 10 µg/mL 5A6 for 48 h. (B, E, and I) Migration of the indicated cells (n = 6), and (C, F, and J) Invasion of the indicated cells (n = 9). Images were taken after 48 h, then quantified using Image-Pro Plus 6.0 software. Data is presented as present change, and shown as Mean ± SEM, one-way ANOVA, significance is marked for the comparison between the groups in presence of 5A6; P < 0.05*, P < 0.01**, P < 0.0001***. (G) Schema of CD81 and constructs lacking (D) the indicating intracellular domains. (H) MDA-MB-231 cells express the indicated CD81 proteins on their surface, cells were stained with JS81-APC and analyzed by flow cytometry.
Furthermore, we depleted total cellular cholesterol in MDA-MB-231 cells by incubation with 2 mM MβCD for 48 h. MβCD depleted 30 to 40% of total cellular cholesterol (SI Appendix, Fig. S4E) and had no impact on cell proliferation (SI Appendix, Fig. S4D). Total cholesterol depletion reduced cell migration and invasion (Fig. 3 E and F and SI Appendix, Fig. S4 F and G); however, 5A6 completely inhibited the migration and invasion of cholesterol-depleted cells (Fig. 3 E and F and SI Appendix, Fig. S4 F and G).
The Intracellular Domains of CD81 Are not Required for the Inhibitory Effect of 5A6.
In B cells, 5A6 induces a downstream signaling cascade that depends on the intracellular domain of CD81 (19). To determine whether these domains are required for the inhibitory effect of 5A6, we generated MDA-MB-231 cells that lacked either the 5′, 3′, or both cytoplasmic 5′ and 3′ domains (Fig. 3G). These newly generated cells expressed and trafficked CD81 to the cell surface (Fig. 3H) and proliferated at a rate comparable to that of parental MDA-MB-231 cells (SI Appendix, Fig. S5A). Remarkably, 5A6 inhibited the migration and invasion of cells lacking their cytoplasmic domains as effectively as it inhibited parental MDA-MB-231 cells (Fig. 3 I and J and SI Appendix, Fig. S5 B–E). Because the inhibitory effects of 5A6 do not depend on the cytoplasmic domains of CD81, it is highly likely that the antibody either disrupts the interaction of CD81 with a membrane-anchored partner protein and/or affects the transmembrane domains.
Identification of Membrane CD81-Associated Proteins.
The antimetastatic effects of 5A6 may be mediated by CD81 membrane-associated partner proteins; therefore, we coimmunoprecipitated CD81-associated proteins using antibody 5A6. Cells were lysed using Brij-98 to maintain the association of CD81 with partner proteins (20, 21), and the peptides were analyzed by mass spectroscopy (MS). A comparative MS Heat-Map showed CD9 as the most abundant membrane protein that interacts with CD81, followed by transferrin receptor 1, different integrins, and other tetraspanins such as CD63 and CD151 (Dataset S1).
Tetraspanins CD9, CD63, and CD151 Are Not Required for the Inhibitory Effect of 5A6.
CD81 partners with its family members to form tetraspanin-enriched microdomains (TEMs) (9–11, 22). CD9, CD81, and CD151 have been shown to affect the motility and invasiveness of MDA-MB-231 cells (23). Tetraspanin CD151 was originally discovered by an antimetastatic antibody, as it increased cell migration and invasion (24), and CD63 showed antimetastatic properties in melanoma and colon carcinoma (25, 26). Collectively, these tetraspanins represent candidates that could facilitate the antimetastatic effect of 5A6. To test this hypothesis, we knocked out CD9, CD63, or CD151 in MDA-MB-231 cells using CRISPR/Cas9 (Fig. 4A). These newly generated cell lines proliferated similarly to parental cells (SI Appendix, Fig. S6A). Surprisingly, 5A6 inhibited their migration and invasion (Fig. 4 B and C and SI Appendix, Fig. S6 B and C), showing a lack of dependence on these other tetraspanins for the antimetastatic effect of the 5A6 antibody.
Fig. 4.
MDA-MB-231 cells lacking CD9, CD63, and CD151 are inhibited by 5A6. (A) Cell surface expression of the indicated tetraspanins. (B and C) The cells were incubated in the presence or absence of 10 µg/mL 5A6 for 48 h. (B) Data is presented as present change, and shown as Mean ± SEM, one-way ANOVA, significance is marked for the comparison between the groups in presence of 5A6; P < 0.05*, P < 0.01**, P < 0.0001*** for cell migration (n = 6) (Middle) and (C) cell invasion (n = 9) (Lower). Blue line defines the core area, and the yellow line demarcates the invaded area.
Transferrin Receptor 1 and Integrins Alpha-6, Alpha-v, and Beta5 May be Involved in Mediating the Inhibitory Impact of 5A6 on Metastasis.
The MS heatmap presented integrins and transferrin receptor 1 as promising candidates for mediating 5A6 antimetastatic effects on MDA-MB-231 cells (Dataset S1). The interaction of different integrins with tetraspanins and their involvement in tumor metastasis are gaining interest (27–29), and transferrin receptor 1 has also been noted to be an antimetastatic therapeutic target (30, 31). We knocked out the integrins expressed in MDA-MB-231 cells and transferrin receptor 1 using CRISPR/CAS9 (SI Appendix, Fig. S7A). CD81 expression level was not affected by the KOs. The inhibition of invasion by β1 KO cells was not testable because the cells were unable to create a sphere or invade. In addition, the invasion ability of cells lacking integrins α3 and α5 was impaired. Nevertheless, 5A6 completely prevented invasion of these KO cell lines (SI Appendix, Fig. S7 B–D).
The lack of integrins β5 and α6, but not other integrins, partially reduced the ability of 5A6 to block cell migration (Fig. 5 A and B and SI Appendix, Fig. S7D). The lack of integrins β3 and α5 decreased the migration of the cell line yet did not prevent the ability of 5A6 to block migration (Fig. 5 A and B and SI Appendix, Fig. S7D). The absence of α2 or α6 had a minor impact on invasion; However, 5A6 effects were significantly impaired in integrin β5 and αv KO cell lines (Fig. 5 A and B and SI Appendix, Fig. S7D).
Fig. 5.

MDA-MB-231 cells lacking integrin β5, integrin αv or transferrin receptor 1 show partial impact of 5A6 on invasion. Migration and invasion of the KO cell lines of (A) β Integrin (B) α Integrin and (C) transferrin receptor 1, in the presence or absence of 10 µg/mL 5A6. Cells were imaged at 5× magnification after 48 h using a fluorescent microscope, then quantified using Image-Pro Plus 6.0 software. In invasion images, the blue line defines the core area, and the yellow line demarcates the invaded area. Data is presented as present change, and shown as Mean ± SEM, one-way ANOVA, significance is marked for the comparison between the groups in presence of 5A6; P < 0.05*, P < 0.01**, P < 0.0001***. Cell migration (n = 6) and cell invasion (n = 9).
The absence of transferrin receptor-1 in MDA-MB-231 cells increased the baseline cell invasiveness. However, interestingly, 5A6 antimetastatic effects in transferrin receptor-1 KO cells were significantly obstructed, making this transferrin receptor the most dominant candidate for facilitating 5A6 antimetastatic effects in these triple-negative breast cancer cells (Fig. 5C and SI Appendix, Fig. S7D).
Discussion
CD81 is widely expressed in normal human and mouse tissues (32, 33). In humans, overexpression of CD81 in melanoma, prostate, and breast cancers is associated with increased metastases (2, 3, 34). Moreover, downregulation of CD81 in human cancers correlates with reduced tumor growth, migration, and invasion (3, 35, 36). This is even more evident in CD81KO mice, in which tumor growth and metastasis are highly reduced (4). The involvement of CD81 in tumor promotion led our laboratory to explore the ability of antihuman CD81 antibodies to inhibit cancer processes. Our studies have shown that a particular antibody, 5A6, which was identified by its antiproliferative ability against B cell lymphoma (37), prolonged the survival of xenografts inoculated with lymphoma (5). 5A6 is also effective against human breast cancer cells, halted metastases of MDA-MB-231 cells, and patient-derived Triple-Negative Breast Cancer (TNBC) in immunodeficient mice (1). Remarkably, among a panel of antihuman CD81 mAbs, only 5A6 is effective against both lymphoid and breast cancer (1, 5). This unique activity of 5A6 is not explained by its higher affinity because all antibodies have comparable affinities (16). To prove that the anti-invasive effect of 5A6 was indeed specific for CD81, we used CRISPR/Ca9 to generate a CD81 knockout (KO) cell line that did not respond to the antibody, as expected. We restored the CD81 gene in this KO cell line and restored its anti-invasion sensitivity to 5A6. According to these findings, we propose that the control of metastasis is not dependent on the expression of CD81 itself, but rather on the intricate interaction between CD81 and the antibody 5A6. We hypothesize that the interplay between CD81 and 5A6 serves as a key trigger for the modulation of metastatic processes, perhaps by initiating the activation or deactivation of specific proteins which subsequently leads to the observed phenomena.
The interaction of 5A6 with tetraspanin CD81 inhibited cell migration and invasion and reduced ECM degradation. Specifically, 5A6 reduced cell motility on laminin, collagen IV, and gelatin in the parental MDA-MB-231 cells.
A clue to the special property of the 5A6 antibody is that it fails to recognize monkey CD81 (15), a feature not shared by other anti-CD81 antibodies, even though these antibodies compete for binding with 5A6 on human CD81. Human and monkey CD81 differ in just four amino acids in the LEL, at positions 163, 186, 188, and 196. Here, we show that among the CD81 antibodies, only 5A6 requires F186 and E188 amino acids to bind to the D helix of CD81-LEL. These results validated and further refined epitope mapping by Hasezaki et al. (16). Several NMR studies and molecular dynamic simulations have shown that Helix D of CD81 is the most flexible domain in LEL, and the binding of 5A6 to CD81 results in significant conformational changes in Helices C and D (17, 38–40). Finally, we showed that cross-linking of the CD81 target on MDA-MB-231 cells is necessary for the antimetastatic effect of 5A6, since F(ab’2), but not Fab, can mediate these effects.
Cholesterol is embedded in the intramembrane pocket of CD81 through its interaction with the amino acids N18 and E219. Cholesterol has been shown to regulate the conformation of CD81-LEL, being closed in its presence and open in its absence (17). CD81 is a key receptor for hepatitis C virus (HCV), and mutation of these cholesterol-interacting residues in Huh-7 hepatocytes reduces HCV infection (18). Therefore, we investigated whether cholesterol influences the antimetastatic response of MDA-MB-231 cells to 5A6. Interestingly, the absence of cholesterol had no effect on the ability of 5A6 to block the metastatic processes.
5A6 induces cytotoxicity in lymphoma B cells and Syk phosphorylation, which is dependent on the intracellular domain of CD81 (41). In MDA-MB-231 cells, 5A6 did not affect viability, yet we wondered whether the effects of the antibody required the intracellular domains of CD81. Interestingly, our results demonstrated that 5A6 effectively inhibited cell migration and invasion in the absence of intracellular domains of CD81.
Since the inhibitory effects of 5A6 do not depend on the cytoplasmic domains of CD81, its interaction with CD81 may hinge on one or more CD81 membrane-anchored partner proteins. Previously, we showed JAM-A as a CD81 partner protein partially involved in mediating the inhibitory effects of 5A6 (1), which altogether, motivated us to determine whether the antibody affects the interaction of CD81 with other functional transmembrane proteins. MS data revealed several candidates that might mediate the effects of the antibody, including other tetraspanins, integrins, and transferrin receptor 1. Tetraspanins are organized in TEMs and partake in a wide range of vital cellular processes (7, 32, 42–48). The other tetraspanins identified as CD81 membrane partner proteins include CD9, CD63, and CD151. These tetraspanins have been implicated in a wide variety of metastatic properties (49, 50). Therefore, we investigated the role of CD81 partners in the effects of the 5A6 antibody. However, MDA-MB-231 cells, in which these tetraspanins were knocked out, were still sensitive to 5A6.
Integrin subunits and complexes are known for their contribution to the metastatic processes, including in breast cancer models (27, 28, 51). Integrins are also well documented in interacting with different tetraspanins to mediate cell growth, proliferation, motility, migration, and invasion (23, 26, 29, 52). We identified integrins beta5 and alpha-v as partially impacting the invasion inhibitory effects of 5A6.
Finally, we tested the involvement of transferrin receptor 1 in facilitating 5A6 antimetastatic ability. Transferrin receptor 1 controls iron uptake, is associated with poor prognosis of cancer, and increases in malignant cells, making it a good candidate for cancer therapy (30, 31). Interestingly, transferrin receptor 1 was significantly important for 5A6 antimetastatic effects, as KO cells were uncontrollably invasive in the presence of 5A6.
In summary, we presented evidence that 5A6 exclusively and specifically reduces metastatic function by interacting with CD81. The effects of 5A6 were not mediated by the intracellular domains of CD81, presence of cholesterol, or interactions with other tetraspanins tested. Additionally, the antimetastatic capability of 5A6 is not based on its superior avidity or affinity for CD81. Rather, these effects require a specific epitope imparted by amino acids 186-188, and which is best facilitated by the cross-linking of the CD81 target. Finally, the 5A6 effects on CD81 are indirect and may be mediated by several membrane proteins, such as integrin β5, integrin αv, and transferrin receptor 1.
Material and Methods
Antibodies.
Specific antibodies used in this study are described in SI Appendix, Table S1.
Reagents.
Sources of reagent used in this study are detailed in SI Appendix, Table S2.
Culture of Cell Lines.
Parental and mutated MDA-MB-231 cells (American Type Culture Collection (ATCC), RRID:CVCL_0062) as well as parental and KI mouse breast cancer 4T1 (ATCC, RRID:CVCL_0125) cells were cultured in RPMI 1640 medium (Gibco, Cat# 11875). Phoenix-A cells (ATCC, RRID:CVCL_H716) were cultured in Dulbecco's Modified Eagle Medium (DMEM) medium (DMEM; Gibco 11965). The medium was supplemented with 10% bovine serum albumin (BSA) (HyClone), 1% sodium pyruvate, 1% L-glutamine (Cellgro), and 100 U/mL penicillin (Gibco). Cells were maintained in a humidified incubator with 5% CO2 at 37 °C.
Generation of KO Cell Lines.
CRISPR/Cas9 gRNAs (SI Appendix, Table S3) were used to generate MDA-MB-231 KO cells. The cells were transfected using Lipofectamine 2000 and a pSpCas9(BB)-2A-GFP (pX458) plasmid (Addgene, RRID:Addgene_101731) containing gRNAs. The KO cells were sorted using Aria flow cytometry (BD Biosciences, RRID:SCR_013311) and their lack of expression was verified using flow cytometry.
Mutant Human CD81 Constructs.
Lacking the intracellular domains.
The human CD81 cDNA was amplified using specific primers (SI Appendix, Table S3). The N’ domain excludes 10 amino-terminal residues, and the C’ domain ends at amino acid 228. The inserts were cleaved with EcoRI and XhoI and inserted into the multicloning site of pBabeMN IRES-GFP.
Cholesterol-binding mutants.
Residues involved in cholesterol binding (17) were mutated using QuikChange (site-directed mutagenesis kit; Stratagene) as recommended by the manufacturer, using specific primers (SI Appendix, Table S3).
Mutants of residues that differ between human and african green monkey.
Mutations in human CD81 cDNA inserted into pBabeMN IRES-GFP were generated using QuikChange™ (site-directed mutagenesis kit; Stratagene), according to the manufacturer’s instructions (see primers in SI Appendix, Table S3). These mutations were also generated in human CD81 exons 6 to 7 and inserted into the pENTR-donor plasmid (Addgene, RRID:Addgene_60605).
Generation of MDA-MB-231 CD81 Mutant Cell Lines.
The pBabeMN IRES-GFP constructs were transfected into the Phoenix (fNX)-Eco packaging cell line using the TransIT transfection reagent (Thermo Fisher). The retroviral supernatant (800 µL) plus polybrene (8 μg/mL) were used to infect 1 × 106 MDA-MB-231 CD81KO cells plated in 1 mL six-well plates. The infected cells were sorted after 48 h using an Aria flow cytometer (BD Biosciences), followed by two subsequent rounds of sorting, and then validated by flow cytometry.
Generation of Mouse 4T1 Cells KI with Human CD81 Exons 6 to 7.
4T1 cells were cotransfected with the pSpCas9(BB)-2A-GFP (pX458) (Addgene) plasmid containing gRNAs targeting upstream of exon 6 and downstream of exon 7, and with the pENTR-Donor plasmid containing human CD81 cDNA encoding exons 6 to 7 (hExon 6 to 7), flanked on both sides by 750bp of the mouse homology arms. 4T1 GFP-expressing cells were sorted after 36 h, and the 5A6 epitope KI cells were selected to bind the antihuman CD81 antibody (JS81-APC) without binding to antimouse CD81(Eat2-PE).
Flow Cytometry.
MDA-MB-231 cells and mutants were detached using 2 mM ethylenediaminetetraacetic acid (EDTA) and suspended single cells were centrifuged, resuspended in 1% BSA in phosphate-buffered saline (PBS), and stained at room temperature with fluorochrome-conjugated antibodies (SI Appendix, Table S1) for 15 min. Cells were washed twice in PBS, fixed in 2% paraformaldehyde, analyzed using a FACSCalibur flow cytometer (BD Biosciences), and analyzed using Cytobank (RRID:SCR_014043) and FlowJo software (RRID:SCR_008520).
Cell Viability.
MDA-MB-231 cells were plated in triplicates in 96 well plates starting at 10,000 cells/well in complete growth medium. The cells were then incubated at 37 °C with 5% CO2. Presto Blue (1:10) was added to the medium, followed by incubation in the dark for 2 h prior to measuring fluorescence (excitation 560 nm, emission 590 nm) using a SpectraMax Paradigm Plate Reader.
Transwell Migration and 3D Invasion Assays.
Experiments were performed as previously described (1). Cells migrating to the bottom chamber after 48 h were imaged, and cell counts per field were quantified using Image-Pro Plus 6.0.(RRID:SCR_007369)Invasion areas were imaged after 48 to 96 h, invasion areas were quantified at 48 h using Image-Pro Plus 6.0.
Time Lapse Cell Motility—Migration Velocity.
1 × 104 MDA-MB-231 Cells were plated on coverslips coated with collagen IV, laminin, gelatin, or L-lysine. Cells were allowed 18 h to attach and spread and then imaged using an ImageXpress Micro Confocal-Automated Microscope in the presence or absence of 10 μg/mL 5A6. Images were collected every 6 min for 12 h. MetaXpress software (RRID:SCR_016654) was used with the ‘Multi-Dimensional Motion Analysis’ addon App to present the motility direction and calculate the cell motility direction, mean velocity, and net travel distance.
ECM Degradation.
Coverslips were coated with a combination of 30 μL of Matrigel and 25 μg/mL of DQ-collagen IV or with one part of fluorescein-conjugated gelatin into eight parts of unlabeled gelatin/sucrose and left until solidification. 5 × 104 cells were plated on coated coverslips with culture medium and incubated in the presence or absence of 5A6 for 24 h at 37 °C. Fluorescein emitted by collagen IV was imaged using a fluorescence microscope. The degraded gelatin, presented as black areas, was imaged using confocal microscopy (LSM 880). The degraded areas were calculated using Image-Pro Plus 6.0.
MMPs Levels.
The MMP levels were evaluated in lysate (expression) and medium (secretion) of MDA-MB-231 cells in the presence or absence of 5A6. The MMPs were tested using the Quantibody® Human MMP Array kit. The kit contains an array of 10 human Matrix Metalloproteinase related proteins in 16 array sets. The experiments were held per the manufacture instructions.
Fast Protein Liquid Chromatography (FPLC) of GST-CD81 Large Extracellular Loop (LEL) Fusion Proteins.
The fusion proteins were cloned, expressed, and purified as previously described (15). Next, we purified the dimer GST-LEL fused proteins using FPLC. Briefly, we fractionated the samples (150 μL) based on size using the Amersham Biosciences AKTA FPLC System and Superdex 200 Increase 10/30 0GL columns (Amersham Biosciences, Piscataway, NJ, USA). Fractions were collected, and dimers were identified and verified by comparison with a calibrator. Next, the dimer fractions were collected and concentrated using Amicon Ultra Filters (0.5 mL 3 K (Sigma-Aldrich). Purified GST-LEL samples were validated by non-reduced western blotting.
Octet Qk (ForteBio) Real-Time Kinetic Characterization.
The interaction between 1 nM and 1,000 nM GST-LELs (analyte) and the antihuman CD81 antibodies (ligands) at 10 μg/mL, was performed using the Octet Qk (ForteBio) with Anti-Mouse IgG Capture biosensors at 25 °C and the ForteBio Sample dilution buffer, which was also used for washing and dissociation. The ligands were immobilized on the biosensor tip surfaces for 600 s. The biosensors saturated with the ligands were incubated with the analytes for 1,800 s, followed by a 1,800-s dissociation period by moving the sensors to the kinetic buffer (SI Appendix, Table S4 details the program detailed steps).
Antibody Competition Assay to Cell Surface CD81.
5A6-Alexa 488 (0.2 μg/mL) was mixed with increased concentrations (0 to 2.5 μg/mL) of the unconjugated antihuman CD81 antibodies and added for 20 min to MDA-MB-231 cells at RT, followed by three washes with PBS. The binding of Alexa-488-conjugated 5A6 was analyzed using a FACSCalibur flow cytometer (BD Biosciences). The relative MFI was calculated in the absence or presence of competing antibodies.
Production of 5A6 Fab and F(ab’)2.
Fragmentation of 5A6 (mouse IgG1) was performed using the PierceTM Mouse IgG1 Fab and F(ab’)2 Preparation Kit, according to the manufacturer’s protocol. Following digestion, the fragments were purified using the NAb Protein A Plus Spin Column provided with the kit.
Cholesterol Depletion.
3 × 106 MDA-MB-231 cells in 6 well plates were treated with 0.5 ug/mL Methyl-β-Cyclodextrins (MβCD) for 48 h, cholesterol level was then determined using the Cholesterol Assay Kit (Cell-Based), Abcam, as recommended by the manufacturer. The cells were washed three times with Cholesterol Detection Wash Buffer, followed by the addition of 100 μL of Filipin III (1:100) diluted in the Cholesterol Detection Assay Buffer. Cells were incubated in the dark for 30 to 60 min and washed twice with wash buffer, and then analyzed using a SpectraMax Paradigm Plate Reader using excitation at 380 nm and emission at 470 nm.
Statistical Analysis.
All experiments were conducted in triplicates and independently repeated at least three times for validation. The results are presented as the mean with the SEM of the independent experiments. Data were analyzed using Prism 6.0 (GraphPad Software, RRID:SCR_002798) by unpaired t-test, for instance in total distance motility and ECM degradation, or by one-way ANOVA (ANOVA) when more than two groups were compared, as for the migration assays, invasion assays, and time lapse motility. Statistical significance was set at P < 0.05.
Data Access Statement.
The data used in this study are mostly available in the article tables and figures. More access is required to contact the corresponding author, in accordance with the data-sharing policies of the Cancer Research Journal.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We thank Felipe Vences-Catalán for his help setting up the initial experimental system, and Debra K. Czerwinski for her help analyzing flow data.
Author contributions
N.A.-S., R.L., and S.L. designed research; N.A.-S. and C.-C.K. performed research; N.A.-S., C.-C.K., and W.J. contributed new reagents/analytic tools; N.A.-S. analyzed data; and N.A.-S., R.L., and S.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: J.M.G., The University of New Mexico; and A.v.S., Radboud Institute for Molecular Life Sciences.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
Supporting Information
References
- 1.Vences-Catalan F., et al. , Targeting the tetraspanin CD81 reduces cancer invasion and metastasis. Proc. Natl. Acad. Sci. U.S.A. 118, e2018961118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang N., Zuo L., Zheng H., Li G., Hu X., Increased expression of CD81 in breast cancer tissue is associated with reduced patient prognosis and increased cell migration and proliferation in MDA-MB-231 and MDA-MB-435S human breast cancer cell lines in vitro. Med. Sci. Monit. 24, 5739–5747 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y., Qian H., Xu A., Yang G., Increased expression of CD81 is associated with poor prognosis of prostate cancer and increases the progression of prostate cancer cells in vitro. Exp. Ther. Med. 19, 755–761 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vences-Catalan F., et al. , Tetraspanin CD81 promotes tumor growth and metastasis by modulating the functions of T regulatory and myeloid-derived suppressor cells. Cancer Res. 75, 4517–4526 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Vences-Catalan F., et al. , CD81 is a novel immunotherapeutic target for B cell lymphoma. J. Exp. Med. 216, 1497–1508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vences-Catalan F., et al. , Tetraspanin CD81, a modulator of immune suppression in cancer and metastasis. Oncoimmunology 5, e1120399 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy S., Shoham T., The tetraspanin web modulates immune-signalling complexes. Nat. Rev. Immunol. 5, 136–148 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Termini C. M., Gillette J. M., Tetraspanins function as regulators of cellular signaling. Front. Cell Dev. Biol. 5, 34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy S., Shoham T., Protein-protein interactions in the tetraspanin web. Physiology (Bethesda) 20, 218–224 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Charrin S., et al. , Multiple levels of interactions within the tetraspanin web. Biochem. Biophys. Res. Commun. 304, 107–112 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Le Naour F., Andre M., Boucheix C., Rubinstein E., Membrane microdomains and proteomics: Lessons from tetraspanin microdomains and comparison with lipid rafts. Proteomics 6, 6447–6454 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Boucheix C., Rubinstein E., Tetraspanins. Cell Mol. Life Sci. 58, 1189–1205 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poltavets V., Kochetkova M., Pitson S. M., Samuel M. S., The role of the extracellular matrix and its molecular and cellular regulators in cancer cell plasticity. Front. Oncol. 8, 431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garde A., Sherwood D. R., Fueling cell invasion through extracellular matrix. Trends Cell Biol. 31, 445–456 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Higginbottom A., et al. , Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74, 3642–3649 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasezaki T., Yoshima T., Mattsson M., Sarnefalt A., Takubo K., A monoclonal antibody recognizing a new epitope on CD81 inhibits T-cell migration without inducing cytokine production. J. Biochem. 167, 399–409 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Zimmerman B., et al. , Crystal structure of a full-length human tetraspanin reveals a cholesterol-binding pocket. Cell 167, 1041–1051.e1011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palor M., Cholesterol sensing by CD81 is important for hepatitis C virus entry. J. Biol. Chem. 295, 16931–16948 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coffey G. P., et al. , Engagement of CD81 induces ezrin tyrosine phosphorylation and its cellular redistribution with filamentous actin. J. Cell Sci. 122, 3137–3144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tham T. N., et al. , Tetraspanin CD81 is required for Listeria monocytogenes invasion. Infect. Immun. 78, 204–209 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vences-Catalan F., Rajapaksa R., Levy S., Santos-Argumedo L., The CD19/CD81 complex physically interacts with CD38 but is not required to induce proliferation in mouse B lymphocytes. Immunology 137, 48–55 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shoham T., Rajapaksa R., Kuo C. C., Haimovich J., Levy S., Building of the tetraspanin web: Distinct structural domains of CD81 function in different cellular compartments. Mol. Cell Biol. 26, 1373–1385 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gustafson-Wagner E., Stipp C. S., The CD9/CD81 tetraspanin complex and tetraspanin CD151 regulate alpha3beta1 integrin-dependent tumor cell behaviors by overlapping but distinct mechanisms. PLoS One 8, e61834 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng D., et al. , Key role of CD151-integrin complex in lung cancer metastasis and mechanisms involved. Curr. Med. Sci. 40, 1148–1155 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Kondoh M., Ueda M., Ichihashi M., Mishima Y., Decreased expression of human melanoma-associated antigen ME491 along the progression of melanoma pre-canceroses to invasive and metastatic melanomas. Melanoma Res. 3, 241–245 (1993). [PubMed] [Google Scholar]
- 26.Sordat I., et al. , Complementary DNA arrays identify CD63 tetraspanin and alpha3 integrin chain as differentially expressed in low and high metastatic human colon carcinoma cells. Lab. Invest. 82, 1715–1724 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Hamidi H., Ivaska J., Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 18, 533–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taherian A., Li X., Liu Y., Haas T. A., Differences in integrin expression and signaling within human breast cancer cells. BMC Cancer 11, 293 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erfani S., Hua H., Pan Y., Zhou B. P., Yang X. H., The context-dependent impact of integrin-associated CD151 and other tetraspanins on cancer development and progression: A class of versatile mediators of cellular function and signaling, Tumorigenesis and Metastasis. Cancers (Basel) 13, 2005 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Candelaria P. V., Leoh L. S., Penichet M. L., Daniels-Wells T. R., Antibodies targeting the transferrin receptor 1 (TfR1) as direct anti-cancer agents. Front. Immunol. 12, 607692 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao C., et al. , Transferrin receptor regulates malignancies and the stemness of hepatocellular carcinoma-derived cancer stem-like cells by affecting iron accumulation. PLoS One 15, e0243812 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charrin S., Jouannet S., Boucheix C., Rubinstein E., Tetraspanins at a glance. J. Cell Sci. 127, 3641–3648 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Shoham T., et al. , The tetraspanin CD81 regulates the expression of CD19 during B cell development in a postendoplasmic reticulum compartment. J. Immunol. 171, 4062–4072 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Hong I. K., et al. , The tetraspanin CD81 protein increases melanoma cell motility by up-regulating metalloproteinase MT1-MMP expression through the pro-oncogenic Akt-dependent Sp1 activation signaling pathways. J. Biol. Chem. 289, 15691–15704 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizoshiri N., et al. , The tetraspanin CD81 mediates the growth and metastases of human osteosarcoma. Cell Oncol. (Dordr) 42, 861–871 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Park H. S., et al. , Suppression of CD81 promotes bladder cancer cell invasion through increased matrix metalloproteinase expression via extracellular signal-regulated kinase phosphorylation. Investig. Clin. Urol. 60, 396–404 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oren R., Takahashi S., Doss C., Levy R., Levy S., TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Mol. Cell Biol. 10, 4007–4015 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Homsi Y., Lang T., The specificity of homomeric clustering of CD81 is mediated by its delta-loop. FEBS Open Bio. 7, 274–283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Susa K. J., Seegar T. C., Blacklow S. C., Kruse A. C., A dynamic interaction between CD19 and the tetraspanin CD81 controls B cell co-receptor trafficking. Elife 9, e52337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt T. H., Homsi Y., Lang T., Oligomerization of the Tetraspanin CD81 via the flexibility of its delta-Loop. Biophys. J. 110, 2463–2474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coffey G. P., et al. , Engagement of CD81 induces ezrin tyrosine phosphorylation and its cellular redistribution with filamentous actin. J. Cell Sci. 122, 3137–3144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuidscherwoude M., et al. , The tetraspanin web revisited by super-resolution microscopy. Sci. Rep. 5, 12201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Deventer S. J., Dunlock V. E., van Spriel A. B., Molecular interactions shaping the tetraspanin web. Biochem. Soc. Trans. 45, 741–750 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Yanez-Mo M., Barreiro O., Gordon-Alonso M., Sala-Valdes M., Sanchez-Madrid F., Tetraspanin-enriched microdomains: A functional unit in cell plasma membranes. Trends Cell Biol. 19, 434–446 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Aleckovic M., McAllister S. S., Polyak K., Metastasis as a systemic disease: Molecular insights and clinical implications. Biochim. Biophys. Acta Rev. Cancer 1872, 89–102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Deventer S., Arp A. B., van Spriel A. B., Dynamic plasma membrane organization: A complex symphony. Trends Cell Biol. 31, 119–129 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Monk P. N., Partridge L. J., Tetraspanins: Gateways for infection. Infect. Disord. Drug. Targets 12, 4–17 (2012). [DOI] [PubMed] [Google Scholar]
- 48.Balise V. D., Saito-Reis C. A., Gillette J. M., Tetraspanin scaffold proteins function as key regulators of hematopoietic stem cells. Front. Cell Dev. Biol. 8, 598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hemler M. E., Tetraspanin proteins promote multiple cancer stages. Nat. Rev. Cancer 14, 49–60 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Hemler M. E., Targeting of tetraspanin proteins–potential benefits and strategies. Nat. Rev. Drug. Discov. 7, 747–758 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamidi H., Ivaska J., Author correction: Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 19, 179 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Yu J., et al. , The CD9, CD81, and CD151 EC2 domains bind to the classical RGD-binding site of integrin alphavbeta3. Biochem. J. 474, 589–596 (2017). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
All study data are included in the article and/or supporting information.




