Abstract
This study presents the complete genome sequence of Streptomyces californicus TBG-201 isolated from the soil samples of Vandanam sacred groves in Alleppey District, Kerala, India. The organism has potent chitinolytic activity. The genome of S. californicus TBG-201 was sequenced using the Illumina HiSeq-2500 platform with 2 × 150bp pair-end protocol and assembled using Velvet version 1.2.10.0. The assembled genome has a 7.99 Mb total length, a G+C content of 72.60%, and 6683 protein-coding genes, 116 pseudogenes, 31 rRNAs, and 66 tRNAs. AntiSMASH analysis revealed abundant biosynthetic gene clusters, while the dbCAN meta server was used to detect carbohydrate-active enzyme coding genes. The NCBI Prokaryotic Genome Annotation Pipeline was used for genome annotation. The presence of numerous genes coding for chitin degradation indicates the chitinolytic ability of this strain. The genome data have been deposited in NCBI with the accession number JAJDST000000000.
Keywords: Streptomyces californicus, Draft-genome, Chitinase, Secondary metabolites, CAZyme, AntiSMASH
Specifications Table
WGS of Streptomyces californicus TBG-201: Specifications table
| Subject | Biological science |
| Specific subject area | Microbiology, Bacterial Genomics, Microbial biotechnology. |
| Type of data | Whole genome sequence data, predicted genes, and functional analysis of respective proteins, figure, table. |
| How the data were acquired | De novo sequencing was performed using the Illumina HiSeq-2500 sequencing platform. Genome assembled using ABySS v. 2.0.1, MaSuRCA v. 2.3.2, and Velvet v. 1.2.10. Genome annotation was done by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP). |
| Data format | Raw and analyzed. |
| Description of data collection | The modified CTAB method was used to extract the genomic DNA of S. californicus TBG-201, and a genomic library was prepared using the Illumina TruSeq Nano DNA Library Prep Kit. The genome was sequenced on the Illumina HiSeq-2500 platform, utilizing a 2 × 150bp pair-end protocol. Genome assembly was carried out using ABySS version 2.0.1, MaSuRCA version 2.3.2, and Velvet version 1.2.10. The NCBI Prokaryotic Genome Annotation Pipeline (PGAP) was employed to perform genome annotation, utilizing the best-placed reference protein set. Prediction of biosynthetic gene clusters was conducted using AntiSMASH, while identification of carbohydrate-active enzymes was performed using CAZy analysis via the dbCAN meta server. |
| Data source location |
|
| Data accessibility | Repository name: NCBI GenBank Bioproject: PRJNA772892 NCBI BioSample: SAMN22418706 NCBI GenBank Accession Number: JAJDST000000000 Assembly: ASM2064085v1 Direct URL to data: https://www.ncbi.nlm.nih.gov/nuccore/JAJDST000000000 All additional data and supplementary files may be accessed at Mendeley data: https://data.mendeley.com/datasets/fgtz42yfh7, DOI:10.17632/fgtz42yfh7.1 |
Value of the Data
-
•
The isolate S. californicus TBG-201 is a potent chitinase producer, which makes it a significant candidate for biotechnological applications. The genome contains genes coding for chitin degradation. The presence of the GH19 chitinase gene shows that it can produce family-19 chitinases, which are very similar to plant chitinase-C. Chitinase-19 has received much attention recently because of its potential use in the biocontrol of phytopathogens like insects and fungi.
-
•
Thirty-five biosynthetic gene clusters were identified from the genome using AntiSMASH, which suggests the potential of the organism to produce a wide range of secondary metabolites. Various carbohydrate-active enzymes were identified in the genome by CAZy analysis which provides an understanding of the organism's carbohydrate metabolism and potential biotechnological applications. The genome data can be used for elucidating specific genomic and functional analysis.
-
•
Whole genome sequence data of S. californicus TBG-201 can benefit researchers and scientists for functional genomics and enzyme research. The data also provide insights for the researchers on the potential applications of S. californicus TBG-201.
-
•
The genome sequence data of S. californicus TBG-201 can be primarily used for research on various biotechnological applications. The presence of several gene clusters, genes for chitin degradation, and other carbohydrate-active enzymes in the genome indicates the organism's ability to produce numerous secondary metabolites and degrade chitin and other complex carbohydrates which may be experimentally studied.
1. Objective
S. californicus TBG-201 was isolated in our laboratory from the soil samples of Vandanam sacred groves of Alleppey District in Kerala and was found to be a potent chitinase producer. The organism's whole genome was sequenced to understand better the genetic basis of the isolate's chitinolytic activity. The genome assembly was annotated using NCBI PGAP to identify the protein-coding genes, rRNAs, tRNAs, and pseudogenes. The biosynthetic gene clusters were identified using antiSMASH, which suggested the potential of the organism to produce a broad spectrum of secondary metabolites. The genes for carbohydrate-active enzymes were identified using CAZy analysis. Overall, the generation of this dataset was motivated by the need to understand the genetic basis of the chitinolytic activity of S. californicus TBG-201, which has potential biotechnological applications.
2. Data Description
Whole genome sequence data of the chitinolytic actinomycete, S. californicus TBG-201, is reported here. The pre-processing of data after quality control gave 3,976,878 reads with 555.71MB of base pairs for R1 and 503.25MB of base pairs for R2. The de novo assembly resulted in 50 scaffolds, 129 contigs, and an N50 value of 154,990. Velvet assembly was done using a k-mer value of 79, resulting in a genome with 7,994,281 base pairs with a genome coverage of 99.5x. The BUSCO score was C: 95.3% (S: 93.9%, D: 1.4%, F: 0.7%, M: 4.0%, N: 148). The sequence was deposited in GenBank under the accession number JAJDST000000000. The functional annotations and gene predictions using the NCBI prokaryotic genome annotation pipeline are available at GenBank. The general features of the genome assembly are given in Table 1. The genes coding for proteins associated with chitin degradation in the S. californicus TBG-201 genome, as obtained from NCBI PGAP annotation, are shown in Table 2.
Table 1.
The general characteristics of the S. californicus TBG-201 genome.
| Features | S. californicus TBG-201 |
|---|---|
| Total sequence length (bp) | 7,994,281bp |
| Total un-gapped length (bp) | 7,988,965bp |
| Number of scaffolds | 50 |
| Gaps between scaffolds | 0 |
| Scaffolds N50 | 1,079,985 |
| Scaffolds L50 | 3 |
| Number of contigs | 129 |
| Contig N50 | 154,990 |
| Contig L50 | 15 |
| G + C content (%) | 72.60% |
| Genes (Total) | 6,899 |
| CDSs (Total) | 6,799 |
| Genes (coding) | 6,683 |
| CDSs (with protein) | 6,683 |
| Genes (RNA) | 100 |
| rRNAs | 7, 10, 14 (5S, 16S, 23S) |
| Complete rRNAs | 5, 5 (5S, 23S) |
| Partial rRNAs | 2, 10, 9 (5S, 16S, 23S) |
| tRNAs genes | 66 |
| ncRNAs | 3 |
| Pseudo genes (total) | 116 |
| CRISPR Arrays | 2 |
| Number of component sequences (WGS) | 50 |
Table 2.
Genes for chitin degradation identified from S. californicus TBG-201 genome.
| Enzyme | GenBank Accession | Product Name |
|---|---|---|
| Chitinases | MCC0576132.1 | GH18, Chitinase D- Exochitinase |
| MCC0576640.1 | GH18 type II chitinase C- Endochitinase and CBM_2 | |
| MCC0577086.1 | GH18 type II chitinase C- Endochitinase and CBM_2 | |
| MCC0578779.1 | GH18 type II chitinases ChiA, ChiC and ChiC_BD | |
| MCC0576765.1 | GH18 chitinase D- Exochitinase and CBM_4_9 | |
| MCC0577358.1 | GH18 type II chitinases- Endochitinase | |
| MCC0577439.1 | GH18 Chitinase D Exochitinase and CBM_4_9 | |
| MCC0580136.1 | GH19, chitinase class I and ChiC_BD | |
| MCC0580137.1 | GH19, chitinase class I and ChiC_BD | |
| Deacetylases | MCC0574417.1 | CE4- polysaccharide deacetylase |
| MCC0574556.1 | CE4- polysaccharide deacetylase | |
| MCC0575334.1 | CE4- polysaccharide deacetylase | |
| MCC0577008.1 | CE4- NodB_like_6s_7s domain-containing- polysaccharide deacetylase | |
| MCC0577677.1 | CE4- polysaccharide deacetylase | |
| MCC0577144.1 | CE4- polysaccharide deacetylase | |
| MCC0578155.1 | N-acetylglucosamine-6-phosphate deacetylase | |
| N-acetyl glucosaminidase (NAGase) | MCC0579983.1 | GH20 beta-N-acetyl glucosaminidase domain-containing protein |
| MCC0574893.1 | GH20- Chitobiases, beta-N-acetyl hexosaminidase | |
| MCC0577590.1 | GH20 glycosyl hydrolase | |
| β Galactosidase | MCC0576278.1 | GH 2- beta-galactosidase |
| MCC0579319.1 | GH3- beta-galactosidase | |
| β Glucosidase | MCC0574707.1 | GH 3- Periplasmic beta-glucosidase |
| MCC0576914.1 | GH3, Periplasmic beta-glucosidase, CBM_11 | |
| MCC0577205.1 | GH3, Periplasmic beta-glucosidase | |
| MCC0578016.1 | GH3 Periplasmic beta-glucosidase and CBM6 | |
| MCC0574753.1 | beta-glucosidase | |
| MCC0575370.1 | beta-glucosidase | |
| MCC0580340.1 | GH1 beta-glucosidase | |
| Chitosanase | MCC0577157.1 | GH5 glycosyl hydrolase |
| MCC0575462.1 | GH5 protein- endoglucanase/ cellulase | |
| Glucokinase | MCC0575527.1 | ROK family glucokinase |
| MCC0579445.1 | ROK family glucokinase | |
| Glucosamine 6-phosphate deaminase | MCC0577204.1 | glucosamine-6-phosphate deaminase |
| Lytic chitin monooxygenase | MCC0574750.1 | lytic polysaccharide monooxygenase |
| MCC0574783.1 | lytic polysaccharide monooxygenase | |
| MCC0576047.1 | lytic polysaccharide monooxygenase | |
| MCC0577833.1 | lytic polysaccharide monooxygenase | |
| MCC0579060.1 | lytic polysaccharide monooxygenase | |
| Chitinase sensor kinase | MCC0575518.1 | two-component sensor histidine kinase |
| MCC0577752.1 | two-component sensor histidine kinase | |
| MCC0578211.1 | two-component sensor histidine kinase | |
| MCC0580129.1 | two-component sensor histidine kinase | |
| Two-component system response regulator protein | MCC0574951.1 | two-component system response regulator MtrA |
| MCC0577530.1 | two-component system response regulator AfsQ1 |
The annotation of the constitutive modules of CAZymes from the gene sequence is primarily used to assess and identify an organism's capacity to produce complex carbohydrate-degrading enzymes. The meta server dbCAN combines three cutting-edge tools for CAZome annotation: (i) HMMER search against the dbCAN HMM (hidden Markov model) database; (ii) DIAMOND search against the CAZy pre-annotated CAZyme sequence database; and (iii) Hotpep search against the conserved CAZyme short peptide database. The three methods' outputs were combined to get the best possible results from automated CAZyme annotation. Only the ones detected by at least two methods were selected and given in Table 3.
Table 3.
CAZy count of S. californicus TBG-201.
| CAZy Function class | CAZy Family (No.) |
|---|---|
| Auxiliary activity | AA10 (5), AA3 (1), AA5 (1) |
| Carbohydrate-binding module | CBM11 (1), CBM12 (3), CBM13 (5), CBM16 (2), CBM2 (2), CBM20 (1), CBM25 (1), CBM32 (9), CBM35 (2), CBM41 (1), CBM42 (1), CBM48 (5), CBM0 (1), CBM6 (1), CBM5 (4), CBM50 (8), CBM51 (1) |
| Carbohydrate esterase | CE14 (5), CE4 (5), CE9 (1) |
| Glycoside hydrolases | GH0 (2), GH1 (3), GH101 (1), GH109 (1), GH114 (1), GH135 (1), GH136 (1), GH13 (13), GH15 (2), GH154 (1), GH16 (2), GH171 (1), GH18 (7), GH19 (2), GH2 (1), GH20 (2), GH23 (7), GH25 (2), GH29 (1), GH3 (3), GH31 (1), GH33 (1), GH35 (1), GH4 (2), GH43 (1), GH5 (2), GH6 (2), GH64 (2), GH65 (1), GH77 (1), GH81 (1), GH84 (1), GH87 (2), GH92 (1) |
| Glycosyl transferases | GT1 (5), GT2 (28), GT20 (1), GT28 (2), GT35 (1), GT39 (1), GT4 (13), GT51 (4), GT81 (1), GT83 (3), GT87 (1) |
| Polysaccharide lyases | PL31 (1), PL8 (1) |
Thirty-five biosynthetic gene clusters, including those for antibiotics, melanin, antifungal compounds, siderophore, geosmin, carotenoid, osmolyte, and terpenes, were identified using the AntiSMASH tool (Table 4). Many of them codes for secondary metabolites that have less than 20% similarity to known compounds. That indicates the novelty of metabolites offering the possibility of discovering new bioactive compounds.
Table 4.
Secondary metabolite clusters of S. californicus TBG-201 as determined by antiSMASH.
| Region | Type | The most similar known cluster | Similarity % |
|---|---|---|---|
| Region 3.1 | T1PKS, NRPS | Kanamycin | 2% |
| Region 3.2 | Phosphonate | Rhizocticin A | 9% |
| Region 4.1 | Siderophore | Ficellomycin | 3% |
| Region 5.1 | NRPS, T3PKS | Tetronasin | 11% |
| Region 5.2 | Melanin | Melanin | 100% |
| Region 5.3 | NRPS | Ibomycin | 7% |
| Region 5.4 | NRPS, T1PKS | SGR PTMs | 100% |
| Region 6.1 | Lanthipeptide-class-ii and iii | - | - |
| Region 6.2 | Siderophore | Desferrioxamin B | 100% |
| Region 6.3 | Thiopeptide, LAP | - | - |
| Region 7.1 | NRPS | Kanamycin | 1% |
| Region 7.2 | RiPP-like | - | - |
| Region 7.3 | Other, NRPS | Mitomycin | 16% |
| Region 7.4 | NRPS-like, ladderane | Atratumycin | 39% |
| Region 7.5 | Terpene | Hopene | 69% |
| Region 7.6 | NRPS-like, NRPS | Viomycin | 100% |
| Region 8.1 | Butyrolactone | Coelimycin P1 | 12% |
| Region 8.2 | Terpene | Geosmin | 100% |
| Region 8.3 | NRPS | Streptobactin | 94% |
| Region 8.4 | NRPS | Coelichelin | 81% |
| Region 8.5 | T3PKS | Herboxidiene | 6% |
| Region 8.6 | NRPS-like | - | - |
| Region 9.1 | Terpene | - | - |
| Region 9.2 | Lanthipeptide-class-iii | AmfS | 100% |
| Region 9.3 | T1PKS | - | - |
| Region 9.4 | Melanin | Melanin | 100% |
| Region 9.5 | Lanthipeptide-class-i | - | - |
| Region 10.1 | Terpene | Isorenieratene | 100% |
| Region 11.1 | Ectoine | Ectoine | 100% |
| Region 11.2 | T2PKS | Griseorhodin A | 100% |
| Region 12.1 | Butyrolactone, Ectoine | Showdomycin | 47% |
| Region 12.2 | Lasso peptide | Keywimysin | 100% |
| Region 12.3 | Lanthipeptide-class-i | - | - |
| Region 12.4 | NRPS-like | WS9326 | 7% |
| Region 12.5 | RRE-containing | - | - |
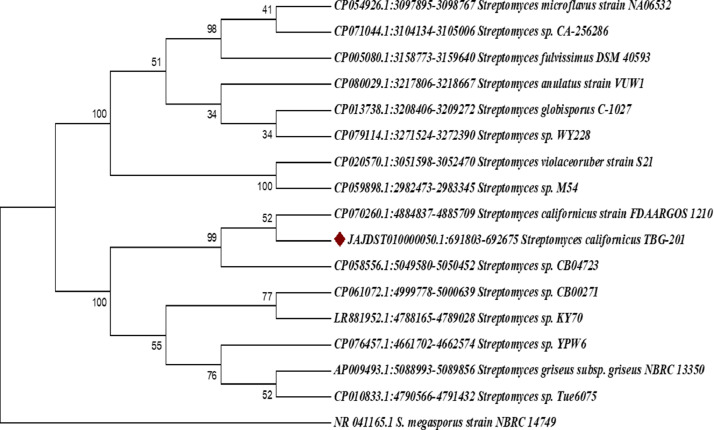
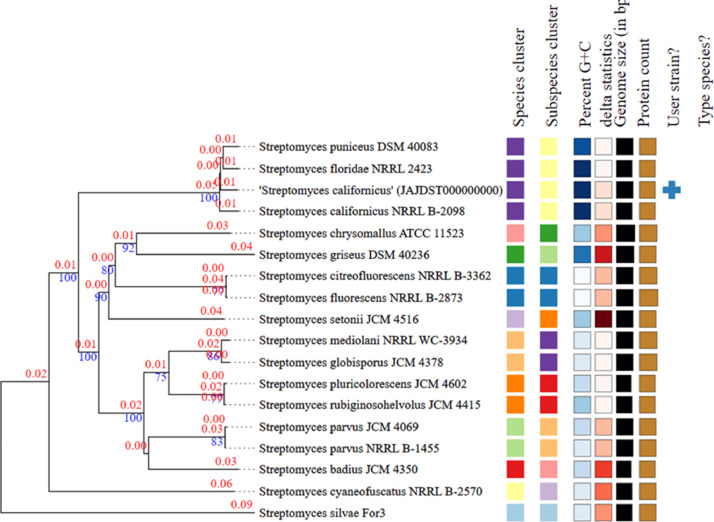
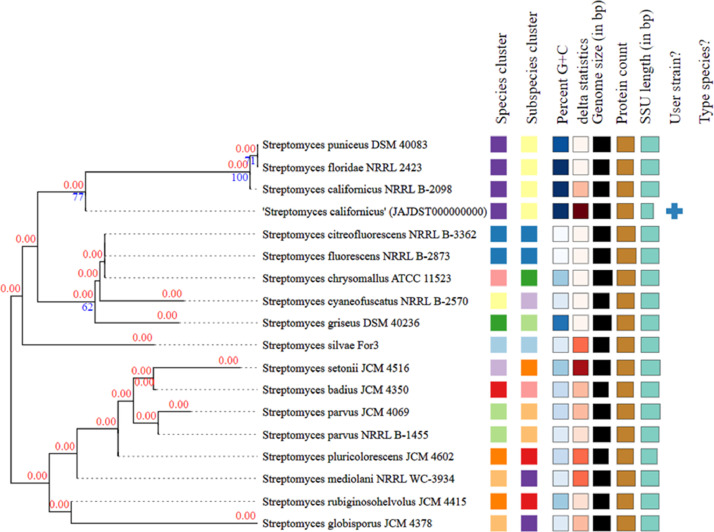
The neighbor-joining tree based on 16S rDNA gene sequences shows that the strain TBG-201 is highly similar to S. californicus strain FDAARGOS 1210 (Fig. 1). To confirm the taxonomic identity of strain TBG-201, digital DNA-DNA hybridization (dDDH) was done. The dDDH values d4 for S. puniceus strain DSM 40083 and S. floridae NRRL 2423 are 88.4% for both. S. puniceus [1] and S. floridae [2] are synonyms for S. californicus [3]. The strain TBG-201 (JAJDST000000000) belongs to the known species S. californicus (Fig. 2, Fig. 3). The average nucleotide identity (ANI) value of S. californicus TBG-201 was found to be 98.65% with S. californicus strain FDAARGOS_1210 and 97.69% with Streptomyces sp. CB04723, the closest phylogenetic neighbors. These values are higher than the generally accepted species threshold level of 96%, indicating that the strain TBG-201 (JAJDST000000000) belongs to the known species S. californicus.
Fig. 1.
Evolutionary relationships of taxa of S. californicus TBG-201 based on 16S rDNA. Using the Neighbor-Joining method, the evolutionary history was deduced [4]. The bootstrap consensus tree inferred from 1000 replicates represents the evolutionary history. Next to each branch is the percentage of replicate trees in which the related taxa grouped together in the bootstrap test (1000 replicates) [5]. The evolutionary distances were calculated using the Jukes-Cantor method [6] and are in the units of the number of base substitutions per site. The analysis involved 17 nucleotide sequences. Codon positions included were 1st+2nd+3rd and noncoding. For each sequence pair, the ambiguous positions were eliminated. There were a total of 1474 positions in the final dataset.
Fig. 2.
TYGS GBDP phylogeny of S. californicus TBG-201 based on genome data. The phylogenetic tree for strain TBG-201 was inferred using FastME 2.1.6.1 [7]. GBDP pseudo-bootstrap support values above 60% from 100 replications with branch support of 78.7% are shown above the branches. The tree has a δ statistics value of 0.096 - 0.221.
Fig. 3.
TYGS GBDP Phylogeny of S. californicus TBG-201 based on 16s data. The phylogenetic tree was constructed with FastME 2.1.6.1 [7] using GBDP distances assessed from 16S rDNA gene sequences. The digits above branches represent the GBDP pseudo-bootstrap support value > 60% from 100 replications, with average branch support of 46,1%. δ statistics value of the tree was found to be 0.264 - 0.446.
3. Experimental Design, Materials and Methods
3.1. Culture maintenance
S. californicus TBG-201 was grown and maintained on ISP2 agar media (Yeast extract Malt extract agar) at 28 ± 2°C. Stock cultures were maintained at -80°C in a 50% glycerol stock.
3.2. Genomic DNA extraction
S. californicus TBG-201, grown in YEME Medium with 34% sucrose and 0.5% glycine, was used to isolate high molecular weight genomic DNA for whole genome sequencing. The organism was incubated at 28 ± 2°C at 180 rpm for five days, and the genomic DNA was extracted using the CTAB method [8].
3.3. Genome sequencing, data pre-processing, and De Novo assembly
Library preparation was done using the Illumina TruSeq Nano DNA Library Prep Kit (Nextera mate-pair library prep kit). The Illumina HiSeq 2500 sequencing platform with a 2 × 150bp pair-end protocol was used for doing the De novo sequencing of the genome. A fastq quality check was carried out for average base content per read, base quality score distribution, and G+C distribution in the reads. The fastq files were pre-processed using AdapterRemovalV2 v2.3.1 (https://github.com/mikkelschubert/adapterremoval) and filtering out the reads with an average quality score of less than 30 from the paired-end reads using Cutadapt v1.8 [9]. FastUniq v1.1 (https://sourceforge.net/projects/fastuniq/files/ ) was used to remove the duplicate reads [10]. De novo Assembly was done using AbySS v2.0.1 (https://github.com/bcgsc/abyss), MaSuRCA v2.3.2 (http://www.genome.umd.edu/masurca.html), SPades, and Velvet v1.2.10 (http://www.mybiosoftware.com/velvet-1-1-07-sequence-assembler-short-reads.html) [11]. BUSCO v2 (http://busco.ezlab.org/) was used to check if assembled contigs have conserved genes [12].
3.4. Sequence submission to NCBI, annotation, and analysis
The genome sequence was submitted to the NCBI through its genome submission portal (https://submit.ncbi.nlm.nih.gov/subs/genome/). The genome annotation was done by NCBI Prokaryotic Genome Annotation Pipeline (PGAP) using the best-placed reference protein set, the GeneMarkS-2+ annotation method [13]. The annotated genes were searched manually to identify the genes involved in chitin degradation. Carbohydrate-active enzymes (CAZyme) annotation was performed using the dbCAN meta server (https://bcb.unl.edu/dbCAN2/blast.php) [14]. The presence of biosynthetic gene clusters (BGCs) in the genome was predicted using the AntiSMASH 6.0.1 server (https://antismash.secondarymetabolites.org/#!/start) [15].
3.5. Phylogenetic and comparative genomic analysis
The gene sequence encoding the 16S rDNA of S. californicus TBG-201 was retrieved from GenBank. The NCBI BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to retrieve closely related sequences from GenBank, and similar sequences were then aligned using the ClustalW. MEGA6 was used to construct the evolutionary tree [16]. Type Strain Genome Server (TYGS) (http://tygs.dsmz.de) was used for whole genome-based taxonomy analysis [17]. The average Nucleotide Identity (ANI) value was calculated using CJ Bioscience's online Average Nucleotide Identity calculator that uses the OrthoANIu algorithm (https://www.ezbiocloud.net/tools/ani) [18].
Ethics Statements
Not applicable.
CRediT authorship contribution statement
Kumaradasan Sreelatha Deepthi: Methodology, Formal analysis, Investigation, Writing – original draft. Sajna Salim: Resources, Validation. Anandhavally Satheesan Anugraha: Writing – review & editing. Shiburaj Sugathan: Conceptualization, Funding acquisition, Project administration, Supervision.
Declaration of Competing Interest
The authors of this paper state that they do not have any financial or personal interest that could have influenced their work or created a conflict of interest.
Acknowledgements
Authors acknowledge DBT for the Grant received under Project (B5): Characterisation, recombinant expression, process scale-up and validation of selected hydrolases from native actino-bacteria for commercial exploitation (BT/PR12720/COE/34/21/2015). The authors acknowledge AgriGenome Labs Private Limited, Kakkanad, Kerala, India (www.aggenome.com), as the service provider for genome sequencing.
Data Availability
References
- 1.Patelski R.A. National Museum of Natural History. Smithsonian Institution; 2023. Streptomyces puniceus. Integrated Taxonomic Information System (ITIS), Checklist dataset, 1951. [DOI] [Google Scholar]
- 2.Bartz Q.R., Ehrlich J., Mold J.D., Penner M.A., Smith R.M. Viomycin, a new tuberculostatic antibiotic. Am. Rev. Tuberculosis. 1951;63:4–6. doi: 10.1164/art.1951.63.1.4. [DOI] [PubMed] [Google Scholar]
- 3.Waksman S.A., Curtis R.E. The actinomyces of the soil. Soil Sci. 1916;1:99–134. doi: 10.1097/00010694-191602000-00001. [DOI] [Google Scholar]
- 4.Saitou N., Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evolut. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 5.J. Felsenstein, Confidence limits on phylogenies: an approach using the bootstrap, evolution. 39 (1985) 783-791. 10.2307/2408678. [DOI] [PubMed]
- 6.T.H. Jukes, C.R. Cantor, Evolution of protein molecules, mammalian protein metabolism. 3 (1969) 121-132. 10.1016/B978-1-4832-3211-9.50009-7. [DOI]
- 7.Lefort V., Desper R., Gascuel O. FastME 2.0: a comprehensive, accurate, fast distance-based phylogeny inference program. Mol. Biol. Evolut. 2015;32:2798–2800. doi: 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kieser T., Bibb M.J., Butter M.J., Chater K.F., Hopwood D.A. The John Innes Foundation; Norwich, United Kingdom: 2000. Practical Streptomyces Genetics: A Laboratory Manual. [Google Scholar]
- 9.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 10.Xu H., Luo X., Qian J., Pang X., Song J., Qian G., Chen S. FastUniq: a fast de novo duplicate removal tool for paired short reads. PloS one. 2012;7:52249. doi: 10.1371/journal.pone.0052249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simao F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 13.Li W., O'Neill K.R., Haft D.H., DiCuccio M., Chetvernin V., Badretdin A., Thibaud-Nissen F. RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2021;49:1020–1028. doi: 10.1093/nar/gkaa1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H., Yohe T., Huang L., Entwistle S., Wu P., Yang Z., Yin Y. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:95–101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blin K., Shaw S., Kloosterman A.M., Charlop-Powers Z., Van Wezel G.P., Medema M.H., Weber T. AntiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021;49:29–35. doi: 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evolut. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meier-Kolthoff J.P., Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019;10:1–10. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon S.H., Ha S.M., Lim J., Kwon S., Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.