Abstract
Ralstonia solanacearum is one of the major plant pathogens causing bacterial wilt disease in a variety of plant species. In Vietnam, according to our knowledge, we first discovered R. pseudosolanacearum, which is one of four phylotypes of R. solanacearum, as a causal agent wilting in cucumber (Cucumis sativus). Due to the latent infection of R. pseudosolanacearum and its heterogenous species complex, controlling the disease becomes difficult.Therefore, the study of R. pseudosolanacearum has great significance to generate effective disease management and treatment. Here, we assembled the isolate R. pseudosolanacearum strain T2C-Rasto, which possessed 183 contigs with 67.03% GC content of 5,628,295 bp in. This assembly included 4,893 protein sequences, 52 tRNA genes, and 3 rRNA genes. In addition, the virulence genes involved in the colonization of the bacterium and wilting to the host were defined in twitching motility (pilT, pilJ, pilH and pilG), chemotaxis (cheA and cheW), type VI secretion system (ompA, hcp, paar, tssB, tssC, tssF, tssG, tssK, tssH, tssJ, tssL and tssM), type III secretion system (hrpB and hrpF).
Keywords: Agriculture, Bacterial wilt disease, Cucumis sativus, Draft genome sequencing, Ralstonia sp
Specifications Table
| Subject | Biology |
| Specific subject area | Microbiology, Genomics, Biotechnology |
| Type of data | Genomic sequence, predicted genes and annotation |
| How the data were acquired | Whole genome sequencing using Illumina MiniSeq Sequencing |
| Data format | Raw and analysed |
| Description of data collection | R. pseudosolanacearum strains T2C-Rasto, isolated from wilt Cucumis sativus, in An Giang province, Mekong Delta - southwest Vietnam. The total DNA was extracted, followed by library preparation sequenced on Illumina MiniSeq platform. Reads were assembled and annotated. |
| Data source location | Town/City: Chau Thanh, An Giang Country: Vietnam Latitude and longitude for collected samples: 10°27′00.0"N 105°19′25.3"E |
| Data accessibility | Raw read is available in the SRA database under the Bioproject PRJNA748156 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA748156) and assembled draft genome sequence (.fasta) has been deposited at GenBank under the accession number JAHWRH010000000 (https://www.ncbi.nlm.nih.gov/nuccore/JAHWRH000000000). The annotation result and supplemental data are available here: https://data.mendeley.com/datasets/7whx24cxwg/draft?a=284df775-1bb0-4e48-b51a-5b6148062612 |
Value of the Data
-
•
Bacteria wilt caused by R. pseudosolanacearum strains T2C-Rasto is one of the most damaging diseases for cucumber production, its genetic characteristics will provide useful data for disease management and further examination on treatment.
-
•
Data could be of interest from groups focus on the evaluation of bacterial antagonists of Ralstonia solanacearum.
-
•
Data can serve future analyses of the relationship between microorganism profiles cause cucumber wilt and risks of disease outbreak due to antibiotic resistance genes spread.
1. Objective
Cucumber bacterial wilt caused by R. solanacearum in Vietnam has not been reported before. Moreover, the high genetic diversity of the R. solanacearum raises issues for assessing the outcome of the treatment [1,2], particularly biological control strategies and resistant varieties. Therefore, understanding the genetic information of R. solanacearum is necessary, such as virulence and resistance genes, to be able to devise an effective treatment and prevention measure. We hereby provide the first draft genome of R. pseudosolanacearum T2C-Rasto.
2. Data Description
In Vietnam, Cucumis sativus is one of the largely consumed vegetables with the characteristic of being a short-term crop allowing farmers to grow multiple rounds of crops in a year, thus providing a stable income [3,4]. However, C. sativus wilt disease is caused by the bacterium, R. solanacearum, which has been threatening the huge losses of cucumber production. R. solanacearum is a heterogeneous species complex inducing postharvest disease known as foliage symptoms with rapid wilting of leaves and stems, particularly during the warmest season. Due to the latent infection of R. solanacearum during the preharvest stage, it becomes difficult to control the disease by chemical or physical treatment. The symptom onset appears on day 10 and then rapidly expands to entire the field within 5 days. Eventually, plants fail to recover, become yellow and brown necrotic and die following 3 days later (Fig. 1). Here, we present the draft genome sequences of R. solanacearum obtained from bacterial wilt diseased on Cucumis sativus in Vietnam.
Fig. 1.
The pictures of non-infected (A) and infected (B) Cucumis sativus.
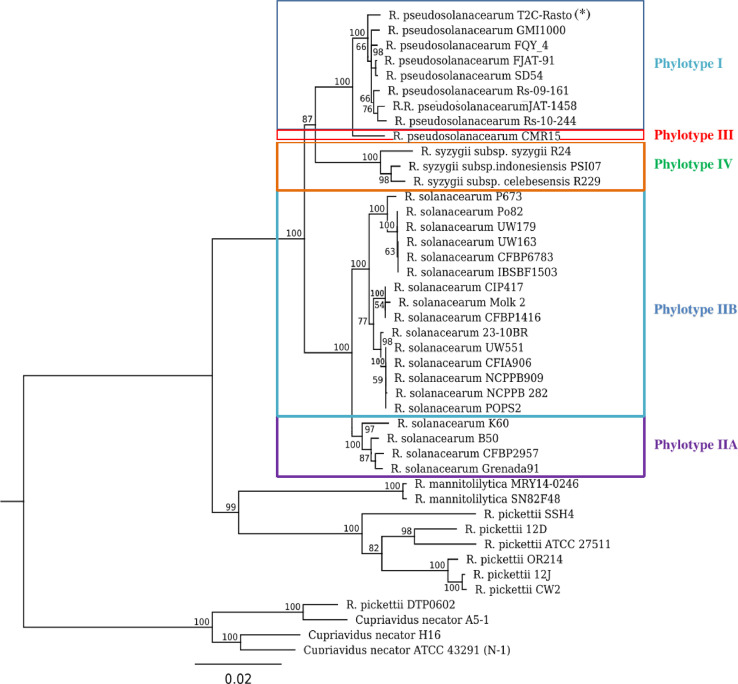
The statistics of the sequencing and assembly data are presented in Table 1. The cleaned reads obtained by removing adapters and low-quality sequences were used for further analysis. The sequence of six housekeeping genes was used to create the phylogenetic tree (Fig. 2) and identify the phylotype of T2C-Rasto isolated [5]. The phylogenomic analysis demonstrated isolate T2C-Rasto was R. pseudosolanacearum in phylotype I found predominantly in Asia [6,7]. The draft genome sequence of isolate T2C-Rasto was 5,628,295 bp with an average GC content of 67.03% over 183 contigs at least length of 200bp. The total number of genes after annotation was 5,077, of which 4,893 were coding sequences, 3 rRNA and 52 tRNA genes.
Table 1.
Genome features of R. pseudosolanacearum T2C-Rasto.
| Attribute | R. pseudosolanacearum T2C-Rasto value |
|---|---|
| Number of raw paired end (PE) reads | 8,145,376 |
| Number of cleaned PE reads | 7,904,919 |
| Total number of cleaned bases | 1,183,199,150 |
| Length of the consensus sequence | 5,628,295 |
| GC content | 67.03 |
| Genome coverage | 382 |
| No. of contigs (>= 200 bp) | 183 |
| N50 | 105,684 |
| Predicted coding genes | 4,893 |
| Number of tRNA genes | 52 |
| Number of 5S, 16S and 23S rRNA genes | 1, 1, 1 (5S, 16S, 23S) |
| GenBank accession | JAHWRH000000000 |
Fig. 2.
Neighbour-Joining phylogenetic trees based on concatenation of nucleotide acid sequences of gdhA, mutS, adk, leuS, rplB and gyrB[5] using the Tamura-Nei model, 1000 bootstrap replicates were computed. There were three strains of Cupriavidus sp used as an outgroup. Numbers above branches are bootstrap percentages.
In addition, the genome contained genes encoding the components of type VI secretion system (T6SS) involved in both host manipulation and interbacterial competition to prevent self-intoxication of the host [8], which are baseplates components (tssA, tssF, tssG, tssK, tssJ, tssL and tssM), the sheath components (tssB, tssC), T6SS effectors (ompA, hcp and paar) and accessory proteins (tssH). The presence of hrpB and hrpF genes encoding the type III secretions systems (T3SS) were also detected, which enables a bacterium to deliver pathogenicity proteins into plant cells [9,10]. Furthermore, the genes identified for chemotaxis (cheA and cheW) and twitching motility (pilT, pilJ, pilH and pilG) were found in T2C-Rasto playing important role in locating and biofilm formation [11,12].
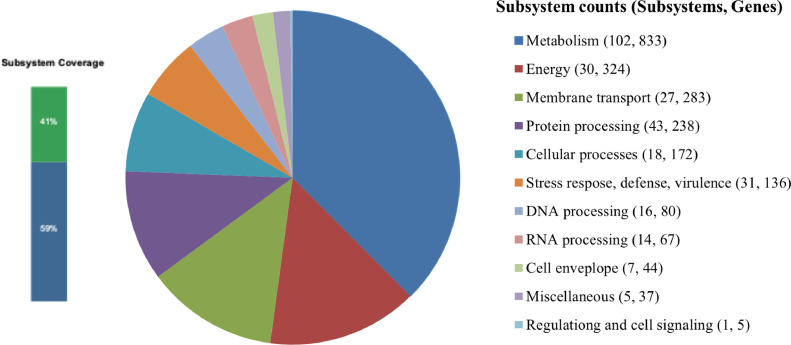
Annotation with the PATRIC server [13] revealed 294 subsystems. An overview of the distribution genes assigned to subsystem categories for the draft genome sequence of R. pseudosolanacearum strain T2C-Rasto generated by PATRIC is shown in Fig. 3.
Fig. 3.
Distribution predicted genes of R. pseudosolanacearum T2C-Rasto assigned to subsystem categories. The subsystem coverage bar chart depicts the percentage of features found in PATRIC subsystem (green color) and features not assigned to a subsystem (blue color).
3. Experimental Design, Materials and Methods
3.1. Sample Preparation and Genome Sequencing
Cucumber stem samples collected at the cucumber garden have just begun to suffer from wilt disease in An Giang province, Vietnam. The pure strain was isolated from the milky-white suspension found inside the wilt Cucumber stem and cultured on TZCA medium at 28°C for 48 hours. Through subsequent biochemical identification (Supplemental File 1), and 16S rRNA gene sequence analysis, we determined four strains belong to Enterobacter sp, Stenotrophomonas sp, Paenibacillus sp, Ralstonia sp (Supplemental File 2-5).The challenge infection with Ralstonia sp strain on cucumber plants under greenhouse conditions revealed that Ralstonia sp T2C-Rasto has the strongest ability to cause wilt disease on cucumbers with 100% death rate on day 10. Therefore, Ralstonia sp strain T2C-Rasto was selected for further genomic analysis. Genomic DNA was extracted using a QIAamp DNA Mini Kit (Qiagen, Germany) and concentration was determined using a Qubit 4.0 Fluorometer (Invitrogen, USA). Sequencing libraries were constructed using the NEBNext® Ultra™ II DNA Library Prep Kit with a sequencing length of 2 × 150 bp (paired-end read sequencing) and evaluated by capillary electrophoresis (Bioanalyzer, Agilent) (Supplemental File 1). Whole genome sequence was performed using Illumina MiniSeq platform with V3 chemistry (Illumina, San Diego, CA).
3.2. Genome Assembly and Annotation
The raw reads were quality controlled using FastQC (version 0.11.5 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Trimmomatic (version 0.36) [14] was used to trim and remove the adapter sequence using the following parameters ILLUMINACLIP:TruSeq3-PE-2.fa:2:30:10 LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36. To merge the trimmed reads, the BBMerge [15] from bbtools software suite was used and assembled using Unicycler (version 0.4.4) [16], with default settings. The genome statistics and annotation of the R. pseudosolanacearum T2C-Rasto were determined by using NCBI Prokaryotic Genomes Automatic Annotation Pipeline [17] and PATRIC [13]. The phylogenetic tree was generated based on the six house-keeping genes (gdhA, mutS, adk, leuS, rplB and gyrB) [5] and other reference genomes downloaded from NCBI and constructed in MEGA 6 using the Tamura-Nei model with 1000 bootstraps. The sequencing depth was computed by in-house script and report based on the depth of coverage of longest contig.
Ethics Statements
This work did not involve human subjects, animal experiments and data collected from social media platforms.
CRediT authorship contribution statement
Thanh Binh Le: Conceptualization, Investigation, Writing – original draft. Minh Ngoc Truong: Investigation. Ba Tho Nguyen: Investigation, Resources. Dinh Quang Vo: Data curation, Visualization. Trang Thi Phuong Phan: Conceptualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2023.109252.
Appendix. Supplementary materials
Data Availability
T2C-Ralsto Genome (Original data) (Mendeley Data).
References
- 1.Truong H.T.H., Duong T.T., Nguyen T.T.H., Nguyen T.T.T., Nguyen L.H.K., Bui T.T.Q. Aggressiveness and genetic diversity of Ralstonia solanacearum strains from tomato in Vietnam. Indian Phytopathol. 2018;71(4):599–610. doi: 10.1007/s42360-018-0084-1. [DOI] [Google Scholar]
- 2.Demirjian C., et al. Study of natural diversity in response to a key pathogenicity regulator of Ralstonia solanacearum reveals new susceptibility genes in Arabidopsis thaliana. Mol. Plant Pathol. 2022;23(3):321–338. doi: 10.1111/mpp.13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nien N.C. 2016. Survey, Collection and Characterization of Indigenous and Non-Indigenous Cucurbits in Vietnam. PhD Diss. [Google Scholar]
- 4.Huong P.T.T., Everaarts A.P., Neeteson J.J., Struik P.C. Vegetable production in the Red River Delta of Vietnam. II. Profitability, labour requirement and pesticide use. NJAS - Wageningen J. Life Sci. 2013;67:37–46. doi: 10.1016/j.njas.2013.09.003. [DOI] [Google Scholar]
- 5.Zhang Y., Qiu S. Phylogenomic analysis of the genus Ralstonia based on 686 single-copy genes. Int. J. Gen. Mol. Microbiol. 2016;109(1):71–82. doi: 10.1007/s10482-015-0610-4. Antonie van Leeuwenhoek. [DOI] [PubMed] [Google Scholar]
- 6.Genin S., Denny T.P. Pathogenomics of the ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 2012;50(April):67–89. doi: 10.1146/annurev-phyto-081211-173000. [DOI] [PubMed] [Google Scholar]
- 7.Prior P., Fegan M. Recent developments in the phylogeny and classification of ralstonia solanacearum. Acta Hortic. 2005;695:127–136. doi: 10.17660/ActaHortic.2005.695.14. [DOI] [Google Scholar]
- 8.Bernal P., Llamas M.A., Filloux A. Type VI secretion systems in plant-associated bacteria. Environ. Microbiol. 2018;20(1):1–15. doi: 10.1111/1462-2920.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asolkar T., Ramesh R. Identification of virulence factors and type III effectors of phylotype I, Indian Ralstonia solanacearum strains Rs-09-161 and Rs-10-244. J. Genet. 2018;97(1):55–66. doi: 10.1007/s12041-018-0894-z. [DOI] [PubMed] [Google Scholar]
- 10.Meyer D., et al. PopF1 and PopF2, two proteins secreted by the Type III protein secretion system of Ralstonia solanacearum, are translocators belonging to the HrpF/NopX family. J. Bacteriol. 2006;188(13):4903–4917. doi: 10.1128/JB.00180-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corral J., Sebastià P., Coll N.S., Barbé J., Aranda J., Valls M. Twitching and swimming motility play a role in ralstonia solanacearum pathogenicity. mSphere. 2020;5(2):1–16. doi: 10.1128/msphere.00740-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao J., Allen C. Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J. Bacteriol. 2006;188(10):3697–3708. doi: 10.1128/JB.188.10.3697-3708.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wattam A.R., et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42(D1):581–591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bushnell B., Rood J., Singer E. BBMerge – Accurate paired shotgun read merging via overlap. PLoS One. 2017;12(10):1–15. doi: 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13(6):1–22. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatusova T., et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
T2C-Ralsto Genome (Original data) (Mendeley Data).