Abstract
Exosomal circRNA serves a novel genetic information molecule, facilitating communication between tumor cells and microenvironmental cells, such as immune cells, fibroblasts, and other components, thereby regulating critical aspects of cancer progression including immune escape, tumor angiogenesis, metabolism, drug resistance, proliferation and metastasis. Interestingly, microenvironment cells have new findings in influencing tumor progression and immune escape mediated by the release of exosomal circRNA. Given the intrinsic stability, abundance, and broad distribution of exosomal circRNAs, they represent excellent diagnostic and prognostic biomarkers for liquid biopsy. Moreover, artificially synthesized circRNAs may open up new possibilities for cancer therapy, potentially bolstered by nanoparticles or plant exosome delivery strategies. In this review, we summarize the functions and underlying mechanisms of tumor cell and non-tumor cell-derived exosomal circRNAs in cancer progression, with a special focus on their roles in tumor immunity and metabolism. Finally, we examine the potential application of exosomal circRNAs as diagnostic biomarkers and therapeutic targets, highlighting their promise for clinical use.
Keywords: Exosome, circRNA, Tumor immunity, Tumor metabolism, Biomarker, Cancer therapy
Introduction
Exosomes, the smallest component of extracellular vesicles (EVs), play a critical role in the tumor microenvironment (TME) by regulating its various components including tumor tissues, immune cells, stromal cells, endothelial cells and others. They carry biologically active molecules such as proteins, nucleic acids, lipids, and metabolites, which can have significant effects on the immune system, tumor metabolism, and drug resistance, thereby influencing the development of tumors [1–3]. TME-based components, particularly immune checkpoint inhibitors, have revolutionized cancer therapy [4, 5]. Exosomes play a crucial role in facilitating cell-to-cell communication between parent cells and recipient cells, thereby making a significant contribution to this field [6]. There is growing evidence that circRNAs, a type of non-coding RNA, play a crucial role in regulating cancer [7]. These circRNAs have unique advantages, such as their high stability and abundant content due to their circular structure, and the ability to be packaged into exosomes for intercellular transport. This leads to the formation of “exosomal circRNA,” which is emerging as an important biomarker and therapeutic target for cancer. However, the exact functions and mechanisms of exosomal circRNAs in tumor progression are not yet fully understood. Recent research shows that the expression of various exosomal circRNAs in the TME is often dysregulated. These exosomal circRNAs show great potential as diagnostic or prognostic biomarkers for cancer, due to their strong stability, long half-life, and resistance to degradation.
In this review, we focus on the roles and mechanisms of tumor and non-tumor cells-derived exosomal circRNAs in communicating with various cells in the TME, and their involvement in cancer-related processes such as immunity, angiogenesis, metabolism, drug resistance, proliferation and metastasis. Finally, we discuss recent advances in the use of exosomal circRNAs as cancer diagnostic biomarkers and new therapeutic strategies.
Exosome and exosomal circRNAs
Exosome biogenesis
Exosomes are composed of transmembrane lipid bilayers and various substances, including proteins, lipids, nucleic acids, and metabolites. These small vesicles are present in a range of body fluids, including blood, urine, thoracoabdominal fluid, cerebrospinal fluid, and saliva [8]. The biogenesis of exosomes occurs through the inward growth of the plasma membrane and the formation of multivesicular bodies (MVBs) [9]. Exosomes deliver proteins and nucleic acids to recipient cells through direct fusion with the cell membrane or various endocytic pathways, including phagocytosis, micropinocytosis, clathrin-dependent endocytosis, caveolin-mediated endocytosis, lipid raft-mediated endocytosis, and receptor-ligand interactions. Recent studies have found that many of the RNAs found in exosomes are different from those in their parent cells, which may be due to selective packaging of exosomes [10]. This highlights the complex and dynamic nature of exosome formation and contents.
Exosomal circRNA selectivity
CircRNAs are formed by reverse splicing of pre-mRNAs [11]. The biogenesis of circRNAs can be promoted by reverse repeat elements such as Alu and RNA-binding proteins such as QKI. These circRNAs have been found to accumulate in cells and have potential as biomarkers and therapeutic targets for various diseases, including cancer [12]. CircRNAs are abundant and stably expressed in exosomes [13], and some studies have shown that they can be packaged and function in exosomes. For example, circ_0088300 has been shown to be driven by KHDRBS3 and packaged into exosomes [14], and hnRNPA2B1 has been reported to package small RNA molecules, including circNEIL3 and circCCAR1, into exosomes [15–18]. However, the mechanism behind the selective packaging of specific circRNAs into exosomes is not yet clear and requires further investigation. Exosomal circRNAs have been shown to typically function as miRNA sponges or stabilizers [19, 20] and protein sponges or decoys [21]. They can also regulate parental gene transcription or serve as translation templates [22].
Tumor-derived exosomal circRNAs in tumor progression
Mounting evidence highlights that certain exosomal circRNAs are expressed abnormally in cancer cells and tissues and can have an impact on tumor progression through mechanisms such as immune evasion, stimulation of angiogenesis, metabolic reprogramming, drug resistance, proliferation and metastasis.
Tumor immunity
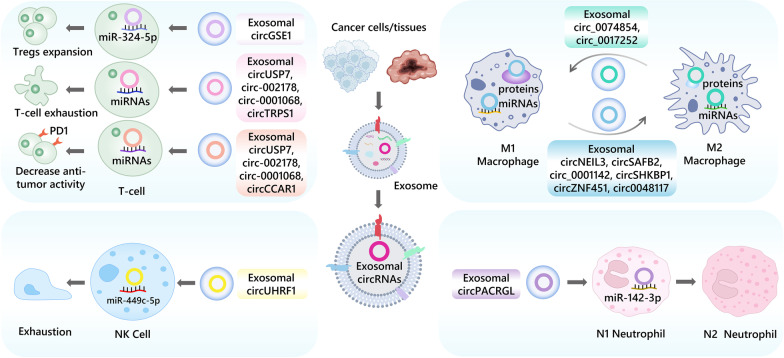
The process of tumor immunity is a multifaceted and dynamic phenomenon that involves the participation of diverse immune cells and active molecules within the body. This intricate process encompasses various stages, including antigen presentation by Dendritic (DC) cells, T lymphocytes (T cell) activation, and anti-tumor activity of innate immune cells such as macrophages. However, tumor cells can evade the host immune response by means of several strategies, including the inhibition of tumor antigens, suppression of regulatory immune cells, and the secretion of immunosuppressive molecules [23]. In tumor tissues, communication between tumor cells and immune cells occurs through exosomes, which foster environment conducive to tumor growth [24]. Currently, there has been significant headway and progress in the exploration of exosomal circRNAs in regulating tumor immunity (Fig. 1 and Table 1).
Fig. 1.
The role of tumor-derived exosomal circRNAs in immune cells, including T cell, macrophage, NK cell and neutrophil
Table 1.
Tumor-derived exosomal circRNAs in tumor progression
| Exosomal circRNAs | Parent cell | Target cell | Expression | Signaling pathway | Functions | References |
|---|---|---|---|---|---|---|
| Tumor immunity | ||||||
| circCCAR1 | Hepatocellular carcinoma cells | CD8 + T cell | ↑ | circCCAR1/PD1 deubiquitination | CD8 + T cell dysfunction, resistance to anti-PD1 immunotherapy | [18] |
| circUSP7 | Non-small cell lung cancer cells | CD8 + T cell | ↑ | circUSP7/miR-934/SHP2 | CD8 + T cell dysfunction, resistance to anti-PD1 immunotherapy | [26] |
| circRNA-002178 | Lung adenocarcinoma cells | CD8 + T cell | ↑ | circRNA-002178/miR-34/PDL1/PD1 | Immune escape, T-cell exhaustion | [29] |
| circTRPS1 | Bladder cancer cells | CD8 + T cell | ↑ | circTRPS1/miR141-3p/GLS1 | CD8 + T cell exhaustion, modulate intracellular ROS balance | [99] |
| circGSE1 | Hepatocellular carcinoma cells | Tregs | ↑ | circGSE1/miR-324-5p/TGFBR1/Smad3 | Immune escape, Tregs expansion | [28] |
| circ-0001068 | Ovarian cancer cells | Jurkat T cell | ↑ | circ-0001068/miR-28-5p/PD1 | Immune escape, induce PD1 expression | [49] |
| circSAFB2 | Renal cell carcinoma cells | Macrophages | ↑ | circSAFB2/miR-620/JAK1/STAT3 | Immune escape, promote M2 macrophage polarization | [31] |
| circNEIL3 | Glioma cells | Macrophages | ↑ | circNEIL3/HECTD4/IGF2BP3 | Promote M2 macrophage polarization | [17] |
| circ_0001142 | Breast cancer cells | Macrophages | ↑ | circ_0001142/miR-361-3p/PIK3CB | Promote M2 macrophage polarization | [32] |
| circSHKBP1 | Non-small cell lung cancer cells | Macrophages | ↑ | circSHKBP1/miR-1294/PKM2 | Promote M2 macrophage polarization | [98] |
| circZNF451 | Lung adenocarcinoma cells | Macrophages | ↑ | circZNF451/FXR1-ELF4-IRF4 | Induce the pro-inflammatory phenotype of macrophages, CD8 + T cell dysfunction, resistance to anti-PD1 immunotherapy | [33] |
| circ0048117 | Esophageal squamous cell carcinoma | Macrophages | ↑ | circ-0048117/miR-140/TLR4 | Promote M2 macrophage polarization | [34] |
| circRNA cSERPINE2 | Breast cancer cells | Macrophages | ↑ | cSERPINE2/MALT1/NF-κB/IL-6 | Promotes IL-6 secretion, proliferation and invasion of BC cells and TAM recruitment | [37] |
| hsa_circ_0074854 | Hepatocellular carcinoma cells | Macrophages | ↓ | hsa_circ_0074854/HUR | Suppress macrophage M2 polarization | [35] |
| hsa_circ_0017252 | Gastric cancer cells | Macrophages | ↓ | hsa_circ_0017252/miR-17-5p/DUSP2 | Suppress macrophage M2 polarization | [36] |
| circUHRF1 | Hepatocellular carcinoma cells | NK cells | ↑ | circUHRF1/miR-449c-5p/TIM-3 | Immune escape, NK cell dysfunction, resistance to anti-PD1 immunotherapy | [39] |
| circPACRGL | Colorectal cancer cells | Neutrophils | ↑ | circPACRGL/miR-142-3p/miR-506-3p-TGF-β1 | Promote N2 neutrophil polarization | [46] |
| Tumor angiogenesis | ||||||
| circSHKBP1 | Gastric cancer cells | HUVECs | ↑ | circSHKBP1/miR-582-3p/HUR/VEGF and inhibit HSP90 degradation | Promote GC cell angiogenesis, proliferation, migration and invasion | [56] |
| circ29 | Gastric cancer cells | HUVECs | ↑ | circ29/miR-29a/VEGF | Impair HUVEC proliferation, migration and tube formation | [57] |
| circ_0007334 | Colorectal cancer cells | HUVECs | ↑ | circ_0007334/miR-577/KLF12 | Promote cell viability, colony formation, migration, invasion and angiogenesis of CRC cells | [58] |
| circRNA-100338 | Hepatocellular carcinoma cells | HUVECs | ↑ | circRNA-100338/VE-cadherin | Promote cell proliferation, angiogenesis, permeability, and vasculogenic mimicry (VM) formation ability of HUVEC | [59] |
| circKIF18A | Glioblastoma multiforme cells | Human brain microvessel endothelial cells | ↑ | circKIF18A/FOXC2/PI3K/AKT | Promote angiogenesis in GBM | [60] |
| circHIPK3 | Breast cancer cells | Human endothelial cells | ↑ | circHIPK3/miR-124-3p/MTDH | Promote tube formation in ECs | [61] |
| circ-CCAC1 | Cholangiocarcinoma cells | Endothelial monolayer cells | ↑ | circ-CCAC1/miR-514a-5p/YY1 | Disrupt endothelial barrier integrity and induces angiogenesis | [62] |
| Tumor metabolism | ||||||
| circ_0008928 | CDDP-resistant cells | _ | ↑ | circ_0008928/miR-488/HK2 | Promote CDDP drug resistance, glycolysis metabolism, proliferation, migration, invasion in NSCLC | [67] |
| ciRS-122 | Oxaliplatin-resistant cells | Oxaliplatin- sensitive cells | ↑ | ciRS-122/miR-122/PKM2 | Promote glycolysis and transmits resistance to oxaliplatin | [71] |
| circNRIP1 | Gastric cancer cells | Gastric cancer cells | ↑ | circNRIP1/miR-149-5p/AKT1/mTOR | Promote GC metastasis by altering metabolism and autophagy | [74] |
| circFBLIM1 | Hepatocellular carcinoma cells | Hepatocellular carcinoma cells | ↑ | circFBLIM1/miR-338/LRP6 | Promote HCC progression and glycolysis | [76] |
| circ-MEMO1 | Non-small cell lung cancer cells | Non-small cell lung cancer cells | ↑ | circ-MEMO1/miR-101-3p/KRAS | Accelerate the proliferation, cell cycle progression, and glycolytic metabolism in NSCLC | [77] |
| circ-ZNF652 | Hepatocellular carcinoma cells | Hepatocellular carcinoma cells | ↑ | circ-ZNF652/miR-29a-3p/GUCD1 | Promote HCC progression, migration, invasion and glycolysis | [78] |
| circ_0072083 | TMZ-resistant cells | TMZ-sensitive cells | ↑ | circ_0072083/miR-1252-5p/NANOG/ALKBH5 | Promote TMZ resistance in glioma | [79] |
| circ93 | Lung adenocarcinoma cells | Lung adenocarcinoma cells | ↑ | cir93/FABP3/AA | Essential for desensitizing LUAD cells to ferroptosis | [97] |
| Tumor proliferation and metastasis | ||||||
| circIFT80 | Colorectal cancer cells | Colorectal cancer cells | ↑ | circIFT80/miR-1236-3p/HOXB7 | Promote CRC growth, proliferation, migration and invasion by activating EMT transcription factors | [100] |
| circSETDB1 | Hypoxic cells | Normoxic cells | ↑ | circSETDB1/miR-7/Sp1 | Promote aggressive growth and EMT in lung adenocarcinoma | [101] |
| circ-133 | Hypoxic cells | Normoxic cells | ↑ | circ-133/miR-133a/GEF-H1/RhoA | Promote CRC cell migration | [102] |
| circRNA_PVT1 | Cervical cancer cells | Cervical cancer cells | ↑ | circRNA_PVT1/miR-1286 | Induce migration and invasion of cervical cancer cells, promote pulmonary metastasis | [103] |
| circ-0004277 | Hepatocellular carcinoma cells | Surrounding normal cells | ↑ | circ-0004277/ZO-1 | Promote HCC EMT progression | [104] |
| circWHSC1 | Ovarian cancer cells | Peritoneal mesothelial cells | ↑ | circWHSC1/miR-145/MUC1 and circWHSC1/miR-1182/hTERT | Promote ovarian cancer occurrence and metastasis | [105] |
| circ-PDE8A | Pancreatic ductal adenocarcinoma cells | Pancreatic ductal adenocarcinoma cells | ↑ | circ-PDE8A/miR-338/MACC1/MET | Promote invasive growth of PDAC cells | [106] |
| hsa_circ_0000437 | Gastric cancer cells | Human lymphatic endothelial cells | ↑ | hsa_circ_0000437/HSPA2-ERK | Promote lymph node metastasis, invasion, migration and tube formation | [110] |
| circ_0026611 | Esophageal squamous carcinoma cells | Human lymphatic endothelial cells | ↑ | circ_0026611/NAA10/PROX1 | Promote lymphangiogenesis | [111] |
| circZNF652 | Glioblastoma cells | Glioblastoma cells | ↑ | circZNF652/miR-486-5p/SERPINE1 | Promote GBM proliferation, migration and EMT malignant phenotypes | [112] |
| circPTGR1 | Higher metastatic HCC cells | HCC cells with low or no metastatic potential | ↑ | circPTGR1/miR-449a/MET | Promote the increased migration and invasion capacity of HCC cells | [113] |
| circ-0072088 | Hepatocellular carcinoma cells | Hepatocellular carcinoma cells | ↑ | circ-0072088/miR-375/MMP-16 | Suppress HCC metastasis | [114] |
| circPUM1 | Ovarian cancer cells | Peritoneum | ↑ | circPUM1/miR-615-5p/NF-kB and circPUM1/miR-6753-5p/MMP2 | Promote ovarian cancer proliferation, migration and invasion | [115] |
| Drug resistance | ||||||
| circATG4B | Oxaliplatin-resistant CRC cells | CRC cells | ↑ | circATG4B-222aa/TMED10/ATG4B | Increase autophagy followed by induction of chemoresistance | [116] |
| circ_0000337 | CDDP-resistant cells | CDDP-sensitive cells | ↑ | circ_0000337/miR-377-3p/JAK2 | Promote CDDP-resistant metastasis, cell proliferation, and metastasis in esophageal cancer | [117] |
| circVMP1 | CDDP-resistant cells | CDDP-sensitive cells | ↑ | circVMP1/miR-524-5p-METTL3/SOX2 | Promote CDDP-resistant metastasis in NSCLC | [118] |
| circWDR62 | TMZ-resistant cells | TMZ-sensitive cells | ↑ | circWDR62/miR-370-3p/MGMT | Promote TMZ-resistant metastasis in gliomas, prognostic markers | [119] |
| circDNER | PTX-resistant cells | PTX-sensitive cells | ↑ | circDNER/miR-139-5p/ITGB8 | Associated with PTX resistance in lung cancer, prognostic markers | [120] |
| circDLGAP4 | Doxorubicin-resistant cells | Doxorubicin- sensitive cells | ↑ | circDLGAP4/miR-143/HK2 | Promote doxorubicin resistance transmission, glycolysis, proliferation and invasion in NB | [121] |
| circZNF91 | Hypoxic PC cells | Normoxic PC cells | ↑ | circZNF91/miR-23b-3p/SIRT1/HIF-1α | Enhance GEM resistance of PC cells | [122] |
T lymphocytes are crucial cells in the adaptive immune response, serving as primary cells in the anti-tumor immune response [25]. In certain instances, exosomal circRNAs have exhibited regulatory effects on CD8 + T cells leading to dysfunction and reduced immune response. For example, the hepatocellular carcinoma (HCC) exosomal circCCAR1 can be delivered to CD8 + T cells and stabilize PD1 expression to promote CD8 + T cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma [18]. In non-small cell lung cancer (NSCLC), exosomal circUSP7 inhibits the secretion of IFN-γ, TNF-α and tumor necrosis factors by CD8 + T cells dysfunction through miR-934/SHP2 axis, and reducing immune response [26]. Regulatory T cells (Tregs), a subset of T cells, can be recruited by tumor tissue and TME to promote and facilitate their proliferation, differentiation and secretion of immunosuppressive factors [27]. Exosomal circGSE1 induces Tregs amplification by targeting the miR-324-5p/TGFBR1/Smad3 axis, leading to promote hepatocellular carcinoma progression [28]. Interestingly, due to the ability of communication between tumor cells and immune cells, exosomal circRNA can modulate the role of T cells with direct or indirect pathway simultaneously. CircRNA-002178 could enhance PDL1 expression via sponging miR-34a in lung adenocarcinoma (LUAD) cells and also be delivered into T cells through tumor-secreted exosomes to promote PD1 expression via sequestering miR-28-5p, thereby inducing CD8 + T cell depletion and inhibiting their anti-tumor activity [29].
As far as innate immune, macrophages are often divided into two phenotypes: M1, which demonstrates anti-tumor properties, and M2, which is pro-tumor. Exosomal circRNAs are shown to regulate macrophage polarization and thus contribute to tumor progression and immune escape [30]. For instance, exosomal circSAFB2 facilitates M2 macrophage polarization in renal cell carcinoma (RCC), leading to increased immune escape and metastasis through miR-620/JAK1/STAT3 axis [31]. In glioma, after circNEIL3 packaged into exosomes by hnRNPA2B1 transmitted to infiltrated tumor-associated macrophages (TAMs), TAMs are endued immunosuppressive properties by stabilizing IGF2BP3 [17]. Exosomal circ_0001142 from breast cancer (BC) is released into macrophages, inducing M2 polarization and interfering with autophagy by targeting the miR-361-3p/PIK3CB pathway under the condition of endoplasmic reticulum (ER) stress [32]. Similarly, exosomal circZNF451 in LUAD promotes ubiquitination of FXR1 and activates the ELF4-IRF4 pathway to induce macrophage polarization and CD8 + T cell dysfunction [33], exosomal circ0048117 under hypoxic conditions can be delivered from esophageal squamous cell carcinoma (ESCC) cells to macrophages and cause M2-type polarization [34]. In contrast, low levels of hsa_circ_0074854 inhibit migration and invasion of HCC cells by interacting with human antigen R (HuR), while inhibiting M2 polarization [35]. Exosomal hsa_circ_0017252 secreted by gastric cancer (GC) cells also effectively inhibits M2 polarization of macrophages and GC invasion by sponging miR-17-5p [36]. It is noteworthy that exosomal circRNA cSERPINE2 in BC cells can cause high IL-6 secretion in TAM through the MALT1-NF-κB-IL-6 axis, while increased IL-6 could positively feedback to enhance ELF4A3 and CCL2 levels to promote cSERPINE2 synthesis and recruitment of TAMs [37].
Natural killer (NK) cells are an essential subpopulation of innate lymphocytes that serve a crucial function in cancer defense [38]. A study has demonstrated that exosomal circUHRF1 is highly expressed in caner tissues and plasma exosome of patients with HCC, while the number of NK cells in peripheral blood is decreased. It induces NK cells exhaustion and reduced secretion of IFN-γ and TNF-α through the degradation of miR-449c-5p and upregulation of TIM-3 expression, which then inhibits HCC progression [39]. Although limited research exists regarding the role of exosomal circRNA in NK cells, this study highlights the impact that exosomal circRNA has on the outcome of cancer, such as HCC, through its influence on NK cell function and exhaustion (Fig. 1).
Neutrophils serve a crucial role in tumor immunosuppression and can contribute to the tumor progression [40]. Tumor-associated neutrophils (TANs) have been identified as either an N1 (anti-tumor) or N2 (pro-tumor) phenotype [41]. Notably, exosomes have been observed to promote the polarization of neutrophils to N2 phenotype and facilitate the migration of neutrophils [42]. Exosome can also mediate the communication between tumor cells and neutrophils impact tumor progression [43–45]. For instance, exosomal circPACRGL derived from colorectal cancer (CRC) influences the progression of CRC through the miR-142-3p/miR-506-3p-TGF-β1 axis, while also regulating the differentiation of N1-N2 neutrophils. In contrast, the proportion of N2 neutrophils is significantly decreased in circPACRGL knockdown cells [46] (Fig. 1).
Immune checkpoints are critical mechanisms that allow tumors to escape immune attack, mediated through the expression of immune checkpoint proteins such as CTLA4, PD1/PDL1, and TIM-3 [47]. Tumor cells can achieve immune escape by increasing the expression of immune checkpoint proteins such as PD1 and PDL1 [48]. Exosomal circRNAs have been confirmed to play a role in the interaction between tumor cells and immune cells, affecting the expression of immune checkpoint proteins and regulating the anti-tumor activity of immune cells (Fig. 1). For example, circ-0001068 in ovarian cancer can be delivered to T cells via exosomes and targets miR-28-5p as well as induces the expression of PD1, causing immune escape of T cells [49]. Exosomal circCCAR1 stabilizes PD1 expression in CD8 + T cells, causing anti-PD1 resistance in HCC [18].
Collectively, based on the characters of abundance and intercellular communication, exosomal circRNAs involve in remodeling the TME directly or indirectly, and in turn regardless the immune cells or immune checkpoint molecules are both almost affected. In light of these findings, it is critical to explore new targets for immunotherapy based on exosomal circRNAs. However, much remains unknown about the relations between exosomal circRNAs and the immune system, particularly with regard to interactions with various immune cells. Further investigation is necessary to understand the impact of exosomal circRNAs on immune function in the context of cancer.
Tumor angiogenesis
Angiogenesis, the formation of new blood vessels, is a key feature of cancer and a crucial pathway in tumor development [50]. The activation and production of multiple angiogenic factors in TME and the rapid growth of tumor cells contribute to the development of vascular networks [51–54]. Vascular endothelial growth factor (VEGF) is a commonly produced angiogenic factor that plays an important role in physiological and pathological angiogenesis [55]. Xie et al. have demonstrated that circSHKBP1 is up-regulated in serum exosomes of gastric cancer patients, and it is correlated with vascular infiltration and TNM stage (Fig. 2 and Table 1). Mechanistically, circSHKBP1 promotes GC progression by upregulating HUR and VEGF through sponge miR-582-3p and decoy HSP90 by competing with STUB [56]. Moreover, exosomal circ29 [57], circ_0007334 [58] and circRNA-100338 [59] are released by tumor cells into human umbilical vein endothelial cell (HUVEC) and promote angiogenesis. Furthermore, glioblastoma-associated microglia-derived exosomal circKIF18A regulates nuclear translocation in human brain microvascular endothelial cells (hBMECs) and promotes angiogenesis in glioblastoma multiforme (GBM) [60]. In addition, many other exosomal circRNAs have been shown to promote angiogenesis [61, 62].
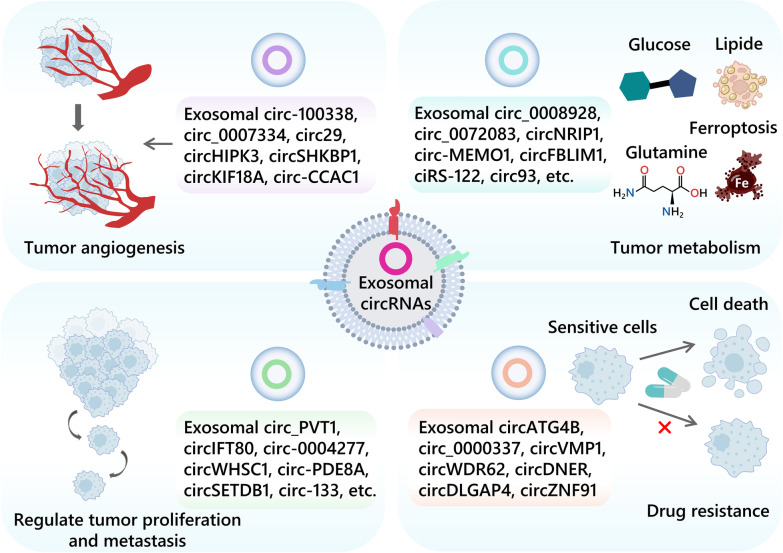
Fig. 2.
The roles of tumor-derived exosomal circRNAs in the tumor angiogenesis, metabolism, migration, metastasis and drug resistance
These findings indicate that exosomal circRNAs have the ability to boost the process of angiogenesis, thereby influencing the progression and inflation of tumor, and making them potential targets for anti-angiogenic therapies.
Tumor metabolism
Metabolism reprogramming and dysregulation accompany with tumor progression and provide the essential source to the tumor cell growth and division [63]. In a comparative study of HCC gene mutations, a cluster of metabolic disease-associated HCC was determined, which was intimately linked with metabolic risk factors such as diabetes and obesity [64]. In fact, there are many cancers associated with the metabolic disease and exosomal circRNA involves in the cancer metabolism.
In the hypoxic TME, malignant tumors usually rely on aerobic glycolysis to produce ATP, leading to resistance to chemotherapy and rapid growth, which is commonly referred to as the Warburg effect. Tumor tissue highly expresses HK2, which acts as the first rate-limiting enzyme of the glycolytic pathway to drive growth. An HK2 inhibitor with a glucose-like structure, 2-deoxy-D-glucose (2-DG), competes with glucose for hexokinase to inhibit glycolysis and synergistically enhances the anticancer effect with sorafenib [65, 66]. Circ_0008928 highly expressed in serum exosomes and cells of cisplatin-resistant NSCLC patients can sponge miR-488 and upregulate HK2 to promote glycolysis and tumor progression. The combination of si-circ_0008928 and 2-DG may lead to a better therapeutic outcome for cisplatin-resistant NSCLC [67]. Pyruvate kinase type M2 (PKM2), which is highly expressed in cancer cells, catalyzes the final reaction in glycolysis by transferring phosphoenolpyruvate (PEP) to ADP to produce ATP and pyruvate [68], and also strongly associated with chemoresistance in many types of cancer [69, 70]. Exosomal ciRS-122 from oxaliplatin-resistant CRC cells can be delivered to sensitive cells and target miR-122 to increase PKM2 expression levels, further promoting glycolysis and drug resistance (Fig. 2 and Table 1) [71]. Lactate dehydrogenase A (LDHA) is another important player in the Warburg effect. Phosphorylation of the LDHA residue (Tyr142) promotes the conversion of NADH to NAD + and the generation of pyruvate and lactate, which are associated with multiple cancer-promoting mechanisms. CircFOXK2 can encode FOXK2-142aa to promote LDHA phosphorylation while regulating the miR-484/Fis1 pathway inducing mitochondrial division, and ultimately activating aerobic glycolysis and lung metastasis in HCC. This shows the important value of circRNA-translated proteins in cancer metabolism [72]. The AKT/mTOR pathway is well-known that plays a significant role in regulating tumor metabolic homeostasis, including the Warburg effect, which ultimately facilitates the growth and spread of cancerous cells [73]. The upregulation of circNRIP1 in GC alters glucose metabolism and autophagy via the miR-149-5p/AKT/mTOR axis, promoting tumor metastasis via exosomal communication. Targeting circNRIP1 provides new therapeutic insights for gastric cancer [74]. Notably, circRNAs can also influence the Warburg effect by regulating the expression of transcription factors (TFs). C-myc, one of the crucial TFs that regulates the Warburg effect, is ubiquitinated and degraded by the circ-FBXW7-encoded protein FBXW7-185aa, which provides novel insights regarding the function of circRNAs in glycolysis [75]. In addition, many other exosomal circRNAs, such as exosomal circFBLIM1, exosomal circ-MEMO1, exosomal circ-ZNF652, have been found to influence tumor progression through promotion of glycolysis [76–78]. The Warburg effect also results in the release of exosomal circ_0072083 from drug-resistant cells in gliomas, contributing to increased Temozolomide (TMZ) resistance. It may be a new target for the treatment of TMZ resistance in gliomas [79].
Cholesterol is an essential lipid for cellular function. Abnormalities in cholesterol metabolism such as upregulation of cholesterol synthesis, increased cholesterol uptake and abnormal accumulation of large amounts of metabolites can occur in tumor tissue to promote cancer cell proliferation and metastasis [80, 81]. Squalene epoxidase (SQLE) is a monooxygenase that catalyzes the conversion of squalene to 2,3-epoxy squalene in the cholesterol synthesis pathway. And high expression of SQLE is associated with poor prognosis in breast and hepatocellular carcinoma. Qian et al. have confirmed that circ_0000182, which is highly expressed in gastric adenocarcinoma, can target miR-579-3p and promote SQLE expression, cholesterol synthesis and cell proliferation through a ceRNA mechanism, suggesting the potential application of targeting circRNA in tumor cholesterol metabolism [82]. A novel hypoxia-responsive circINSIG1-encoded protein, circINSIG1-121, which promotes ubiquitin-dependent degradation of INSIG1 to induce cholesterol biosynthesis for CRC proliferation and metastasis, providing a new way to consider the crosstalk between hypoxia and cholesterol metabolism, as well as a promising therapeutic target [83]. In addition, excess fatty acids (FAs) in the body can also stimulate ER stress, inducing hepatocellular damage, liver fibrosis and cirrhosis, and accelerating the development of hepatocellular carcinoma [84]. While combining conventional immune checkpoint inhibitors with mitochondrial fatty acid transporter inhibitors (e.g., etomoxir) can reverse the metabolic reprogramming of DC cells and restore the immune killing effect of T cells, which is an ideal medicine to target abnormal lipid metabolism [85]. Furthermore, it has been revealed that circ-DB is highly expressed in HCC patients with a high percentage of body fat and is associated with poor prognosis, suggesting that circ-DB may promote HCC progression through fatty acid synthesis (FAS), providing a new therapeutic target for targeting tumor lipid metabolism [86].
As an important amino acid metabolism process in the human body, glutamine metabolism plays an important role in a variety of physiological processes such as cholesterol and vitamin synthesis, providing many essential nutrients to our body [63]. SLC1A5 variant is a mitochondrial glutamine transporter for cancer metabolic reprogramming [87]. It has been confirmed that circ_0000069, circ_0000808 and circ_0025033 all positively regulate SLC1A5 and glutamine metabolism through ceRNA mechanisms to promote cancer progression [88–90]. Additionally, Song et al. have found that circ_0067717 can promote CRC progression and glutamine metabolism by upregulating SLC7A5 through sponge miR-497-5p [91]. These are new evidence for the involvement of circRNAs in cancer amino acid metabolism and provide new molecular targets for anti-cancer therapies targeting glutamine metabolism.
Recently, it has been shown that the m6A-modified circRBM33-FMR1 complex promotes prostate cancer (PCa) progression by stabilizing PDHA1 mRNA to activate mitochondrial metabolism. More importantly, depletion of circRBM33 reduces ATP, acetyl CoA levels and the NADH/NAD + ratio, as well as increases the therapeutic sensitivity of androgen receptor signaling inhibitors (ARSIs), including enzalutamide and darolutamide. This suggests the important value of circRNA in regulating ARSI therapy, presenting a potential therapeutic target for PCa [92].
Exosomal circRNAs also regulate tumor metabolism by inducing ferroptosis. Ferroptosis is an iron dependent, non-apoptotic form of cell death, primarily due to an imbalance in the production and degradation of intracellular lipid reactive oxygen species, which is closely associated with cancer progression. Glutathione peroxidase 4 (GPX4) is a major scavenger of lipid peroxidation and is considered as a major regulator of ferroptosis [93]. Zhai et al. reported that inhibition of the circIDE/miR-19b-3p/RBMS1 axis in HCC upregulated GPX4 to reduce ferroptosis and promote tumor progression [94]. As an ferroptosis inducer, erastin acts on the cysteine glutamate reverse transporter (systemXc-) to induce cell death by promoting the accumulation of lipid reactive oxygen species (ROS) and reducing the antioxidant capacity of cells, leading to excessive total cellular lipid ROS [95]. CircRNA-ST6GALNAC6 increases the sensitivity of bladder cancer cells to erastin-induced ferroptosis by regulating the HSPB1/P38 axis, which is important for the development and application of ferroptosis intervention methods [96]. Interestingly, exosomal circ93 is critical in ferroptosis desensitization of LUAD through regulating arachidonic acid (AA), which consequently affects ferroptosis-related plasma membrane peroxidation. It suggests that blocking exosomal circ93 reverses ferroptosis desensitization and provides a new therapeutic target for LUAD [97].
Actually, tumor metabolism is accompanied with the tumor immunity, for instance, the NSCLC exosomal circSHKBP1 targets the miR-1294/PKM2 axis to promote glycolysis and macrophage M2 polarization for tumor progression [98]. In bladder cancer (BCa), the exosomal circTRPS1 regulates the intracellular ROS balance and CD8 + T cell exhaustion via the circTRPS1/miR141-3p/GLS1 axis-mediated glutamine metabolism [99].
In conclusion, exosomal circRNAs are strongly associated with multiple metabolic pathways and provide a favorable microenvironment for cancer cells growth and metastasis. Target exosomal circRNAs can help to reprogram and remodel the TME, making it an important consideration in tumor metabolism research. At the same time, many potential targets have been developed for cancer therapy.
Tumor proliferation and metastasis
Many exosomal circRNAs are involved in the proliferation and metastasis of multiple types of tumors (Fig. 2 and Table 1). Epithelial-mesenchymal transition (EMT) is a significant process of cellular plasticity that plays a role in tumor development, migration and metastasis. Increasing evidence has shown that exosomal circRNAs have an impact on tumor progression through their ability to regulate miRNAs and activate EMT. For example, the CRC serum exosomal circIFT80 promotes cell growth, proliferation, migration, and invasion by functioning as a ceRNA of miR-1236-3p and increasing the expression of HOXB7 [100]. Additionally, hypoxia-induced exosomal circSETDB1 expression enhances migration, invasion and proliferation of normoxic LUAD cells [101]. The hypoxic tumor-derived exosomal circ-133 promotes aggressive growth and EMT in CRC through the miR-133a/GEF-H1/RhoA axis [102]. Moreover, the exosomal circRNA_PVT1 in cervical cancer promotes metastasis by targeting miR-1286 and enhancing EMT [103]. Similarly, exosomal circ-0004277 in HCC stimulates the malignant phenotype and EMT of peripheral cells, promoting HCC invasion [104]. In addition, exosomal circWHSC1 of ovarian cancer cells can metastasize to peritoneal mesothelial cells and promote peritoneal spread and tumor metastasis [105]. Plasma exosomal circ-PDE8A plays an important role in tumor invasion in pancreatic ductal adenocarcinoma (PDAC) [106].
Lymphatic system plays a vital role in the transportation of fluids, biomolecules and cells between the peripheral tissues and the central circulation through its network of lymphatic vessels and lymph nodes. Lymph node metastasis (LNM) is an important step and is often associated with a poor prognosis [107]. Serum and plasma-derived exosomal hsa_circ_0026611 and hsa_circRNA_0056616 have been shown to be potential biomarkers of tumor lymph node metastasis and poor prognosis [108, 109]. In addition, the exosomal hsa_circ_0000437 has also been found to induce LNM via an HSPA2-ERK signaling pathway, independent of VEGF-C, presenting a novel target for GC therapy (Table 1) [110]. Furthermore, exosomal circ_0026611 has been demonstrated to promote lymph angiogenesis in ESCC by inhibiting PROX1 acetylation and ubiquitination, providing a new perspective on tumor lymph node metastasis [111].
Overall, exosomal circRNAs play a crucial role in regulating tumor proliferation and metastasis by sponging miRNAs and activating EMT [112–115]. And some exosomal circRNAs can promote lymph node metastasis to facilitate tumor progression. Understanding the underlying mechanism of exosomal circRNAs in tumor metastasis is crucial for the development of anti-metastasis therapy.
Drug resistance
Exosome-mediated intercellular communication has been found to be a novel mechanism of drug resistance in cancer. This phenomenon provides new avenues for exploring therapeutic targets for the treatment of cancer. There is evidence that certain exosomal circRNAs can contribute to drug resistance (Fig. 2 and Table 1). Interestingly, circATG4B-222aa [116], a protein encoded by circATG4B gene, has been shown to increase resistance to the drug oxaliplatin in colorectal cancer by blocking the interaction between TMED10 and ATG4B. Similarly, circ_0000337 has been identified as a potential target for drug resistance in esophageal cancer, as it promotes resistance to the drug cisplatin (CDDP) by interacting with miR-377-3p and JAK2 [117]. While, exosomal circVMP1 promotes CDDP resistance delivery in NSCLC [118].
In glioma, exosomal circWDR62 derived from temozolomide (TMZ)-resistant cells can be delivered to receptor-sensitive cells, conferring TMZ resistance and promoting tumor malignant phenotypes, including proliferation, migration, and invasion [119]. Moreover, exosomal circDNER has been found to increase paclitaxel resistance and tumorigenicity in lung cancer by sponging miR-139-5p, thus promoting ITGB8 expression [120]. Exosomes derived from drug-resistant neuroblastoma cells can deliver circDLGAP4 to drug-sensitive cells, resulting in increased doxorubicin resistance and glycolysis through the inhibition of miR-143 and the promotion of HK2 expression [121]. In addition, exosomal circRNAs regulated by hypoxia have also been shown to contribute to tumor progression and drug resistance. For instance, hypoxia-induced exosomal circZNF91 has been found to enhance glycolysis and resistance to the drug gemcitabine in pancreatic cancer (PC) cells by promoting the stability of HIF-1α protein through miR-23b-3p and SIRT1PC [122].
Overall, exosomal circRNAs can induce tumor cell resistance to chemotherapy or radiotherapy and transmit this resistance to sensitive cells. Therefore, these circRNAs could be crucial targets for overcoming drug resistance in cancer.
Non-tumor-derived exosomal circRNAs in tumor progression
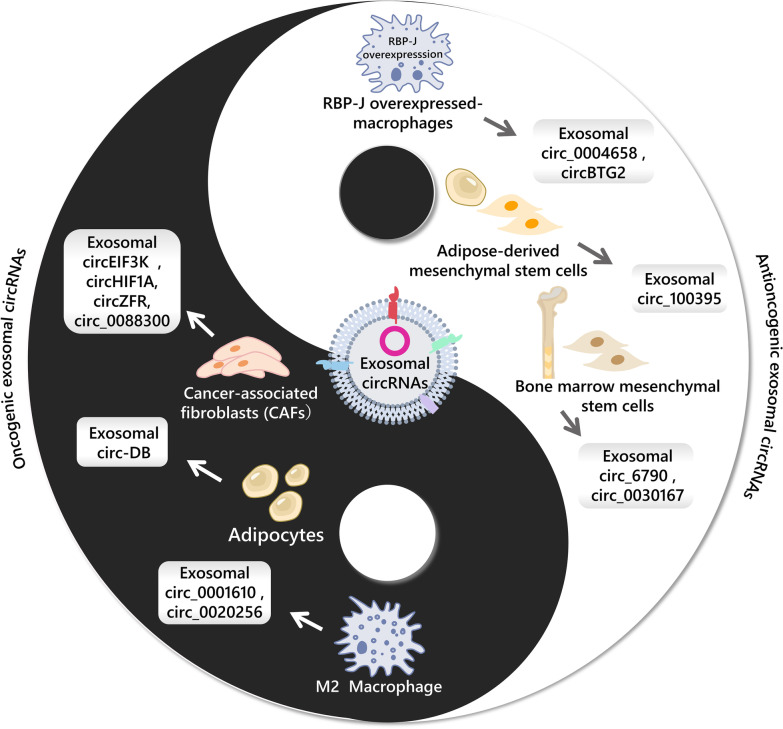
Cellular components in the tumor microenvironment communicate with tumor cells through exosomes and can impact tumor progression by releasing exosomal circRNAs (Fig. 3 and Table 2).
Fig. 3.
The roles of non-tumor-derived exosomal circRNAs in tumor progression
Table 2.
Non-tumor-derived exosomal circRNAs in tumor progression
| Exosomal circRNA | Parent cell | Target cell | Expression | Signaling pathway | Functions | References |
|---|---|---|---|---|---|---|
| circ_0020256 | M2 macrophages | Cholangiocarcinoma cells | ↑ | circ_0020256/miR-432-5p/E2F3 | Promote the proliferation, migration, and invasion of CCA cells | [123] |
| hsa_circ_0001610 | M2 macrophages | Endometrial cancer cells | ↑ | hsa_circ_0001610/miR-139-5p/cyclin B1 | Reduce radiosensitivity of EC cells | [124] |
| has_circ_0004658 | RBPJ-overexpressed macrophages | Hepatocellular carcinoma cells | ↓ | hsa_circ_0004658/miR-499b-5p/JAM3 | Inhibit HCC progression | [127] |
| circBTG2 | RBPJ-overexpressed macrophages | Glioma cells | ↑ | circBTG2/miR-25-3p/PTEN | Inhibit glioma progression | [128] |
| circEIF3K | CAFs | Colorectal cancer cells | ↑ | circEIF3K/miR-214/PD-L1 | Promote proliferation, invasion and tube formation of CRC cells | [136] |
| circHIF1A | CAFs | Breast cancer cells | ↑ | circHIF1A/miR-580-5p/CD44 | Promote breast cancer proliferation and stem cell stemness | [137] |
| circZFR | CAFs | Hepatocellular carcinoma cells | ↑ | circZFR/STAT3/NF-κB | Promote HCC growth and CDDP resistance | [138] |
| circ_0088300 | CAFs | Gastric cancer cells | ↑ | circ_0088300/miR-1305/JAK/STAT | Promote GC cell proliferation, migration and invasion | [14] |
| circ-DB | Adipocytes | Hepatocellular carcinoma cells | ↑ | circ-DB/miR-34a/USP7/Cyclin A2 | Promote HCC growth and reduce DNA damage | [86] |
| circ_6790 | Bone marrow mesenchymal stem cells | Pancreatic ductal adenocarcinoma cells | ↑ | circ_6790/CBX7/S100A11 | Hamper immune escape in PDAC cells | [141] |
| circ_0030167 | Bone marrow mesenchymal stem cells | Pancreatic cancer | ↑ | circ_0030167/miR-338-5p/Wif1/Wnt 8/β-catenin | Inhibit the stemness of PC cells and tumor progression | [142] |
| circ_100395 | Adipose-derived mesenchymal stem cells | Non-small cell lung cancer cells | ↓ | circ_100395/miR-141-3p/Hippo/YAP | Inhibit NSCLC malignant transformation | [143] |
Immune cell-derived exosomal circRNAs in tumor progression
Due to the two phenotypes of anti-tumor (M1) and pro-tumor (M2), exosomal circRNAs released by tumor-associated macrophages (TAM) also play different roles in cancer progression. For example, circ_0020256 in the M2 TAM-secreted exosomes regulates the proliferation, migration and invasion of cholangiocarcinoma cells by targeting the miR-432-5p/E2F3 axis [123]. Similarly, research has shown that hsa_circ_0001610 transferred from the M2 TAMs to endometrial cancer cells by exosomes, and impairs the radiosensitivity of the cancer cells [124]. The recombination signal-binding protein-Jκ (RBP-J) is a transcriptional regulator and a marker of Notch signaling activation [125]. The Notch-RBP-J signaling is associated with polarization of inflammatory macrophage [126]. Exosomal hsa_circ_0004658, produced by RBP-J overexpressing macrophages, has been shown to inhibit liver cancer progression through the miR-499b-5p/JAM3 pathway [127]. Moreover, exosomal circBTG2 from macrophages overexpressing RBP-J targets miR-25-3p/PTEN pathway to hinder glioma progression [128]. Furthermore, an interesting finding highlights that exosomal miR-628-5p from M1 polarized macrophages inhibits the modification of m6A circFUT8 and decelerates the progression of HCC. This reveals a novel mechanism by which immune cell-derived exosomal cargos regulate circRNA and downstream genes in tumor cells and also offers a new potential therapeutic target for HCC therapy [129].
Additionally, although reports regarding immune cell-derived exosomal circRNAs are infrequent, there are already many reports describing the important role of other cargos carried by exosomes such as non-coding RNAs and proteins in cancer progression. For example, miR-765, which is released from CD45RO-CD8 + T cell-derived exosomes, has been found to impede the progression of uterine corpus endometrial cancer (UCEC) via the PLP2/Notch signaling pathway [130]. And NK cell exosomal miR-186 inhibits neuroblastoma growth and impedes immune escape [131]. Furthermore, exosomal cargos derived from neutrophils, DC cells and mast cells, among others, have been demonstrated to influence tumor progression [42, 132, 133].
Non-immune cell-derived exosomal circRNAs in tumor progression
One key component of the tumor microenvironment is cancer-associated fibroblasts (CAFs), which are widely distributed stromal cells that play a significant role in regulating tumor growth, metastasis, angiogenesis [134], and drug resistance [135]. For example, hypoxia-induced CAFs release exosomal circEIF3K, promoting CRC proliferation, invasion and tube formation through targeting miR-214/PDL1, potentially offering new therapy targets for CRC [136]. Additionally, hypoxic CAF-derived exosomal circHIF1A has been shown to play a role in the stem cell properties of breast cancer, suggesting that circHIF1A may serve as a therapeutic target [137]. Furthermore, research has demonstrated that CAF-derived exosomal circZFR inhibits STAT3/NF-κB pathway in liver cancer cells to promote tumor growth and drug resistance [138]. And CAF-derived circ_0088300, packaged into exosomes driven by KHDRBS3, is delivered to gastric cancer cells to target miR-1305/JAK/STAT and accelerate tumorigenesis [14]. These examples highlight how CAFs can facilitate the development of various tumors by creating a supportive microenvironment and interacting with cancer cells. However, further research is needed to fully understand the interaction between circRNAs and CAFs in influencing tumor progression.
Adipocytes, an important contributor to metabolism, supply various nutrients such as lipids and enzymes, to the tumor microenvironment and support tumor cells [139]. Adipocyte-secreted exosomal circ-DB has been shown to promote liver cancer growth by targeting deubiquitination-related USP7 and reducing DNA damage [86].
Mesenchymal stem cells (MSCs), an important component of the microenvironment, have both tumor-promoting and tumor-suppressive effects [140]. For example, exosomal circ_6790 produced by bone marrow mesenchymal stem cells (BM-MSCs) can be transferred to pancreatic ductal adenocarcinoma, altering DNA methylation of CBX7 and S100A11, and hindering immune escape [141]. Additionally, Yao et al. have confirmed that BM-MSC exosomal circ_0030167 targets miR-338-5p to inhibit the progression and stemness of PC cells, providing a potential target for PC therapy [142]. Moreover, exosomal circ_100395 from adipose-derived mesenchymal stem cells (AMSCs) regulates the Hippo/YAP signaling pathway, increasing LATS2 expression, and suppressing malignant transformation in NSCLC [143].
The delivery of exosomal circRNAs between tumor cells and microenvironmental cells has sparked considerable interest in field of tumor therapy and mechanisms. While current research has largely addressed the influence of tumor cell exosomal circRNAs on microenvironmental cells, we posit that exosomal circRNA derived from non-tumor cell sources can represent promising targets for novel therapeutic interventions in cancer progression.
Clinical significance
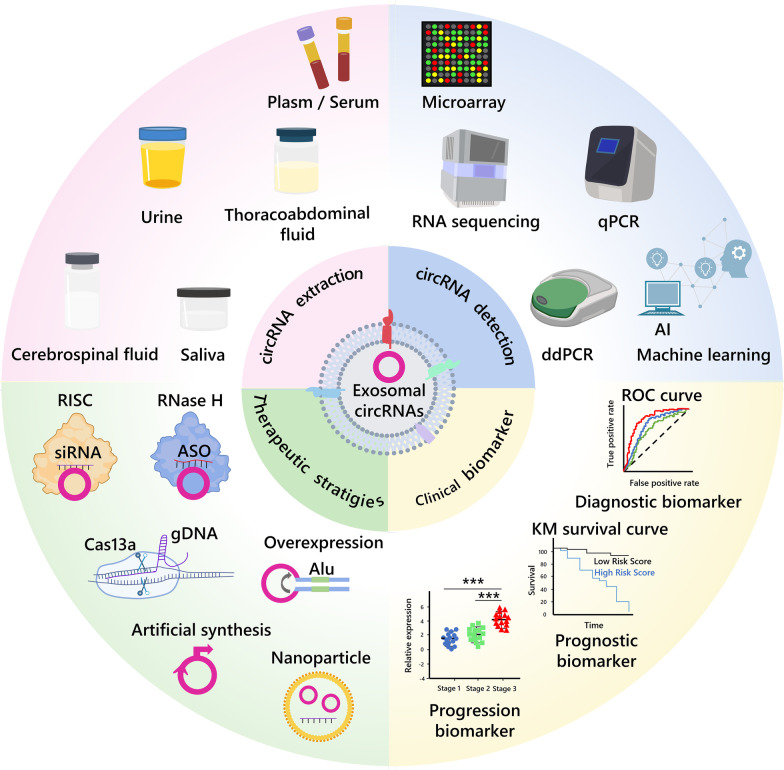
Exosomal circRNAs offer several advantages, such as high stability, abundance, and long half-life, and they are present in human body fluids such as plasma, serum, and urine [56, 144, 145] (Fig. 4). In addition to their aberrant expression in tumor tissues, exosomal circRNAs have been widely shown to be associated with the progression and clinical features of various cancers. They can serve as early diagnostic and prognostic markers for cancer [146, 147]. Currently, RNA microarrays or sequencing followed by real-time quantitative reverse transcription PCR (qRT-PCR) are commonly used to detect differentially expressed circRNAs, but this can be challenging for exosomal circRNAs with low abundance. To address this issue, droplet digital PCR (ddPCR), which offers high accuracy, sensitivity, reproducibility, and absolute quantification, is increasingly being used to detect exosomal circRNAs in liquid biopsy [148, 149]. In addition, machine learning is being used to improve the accuracy of clinical decisions, particularly when combined with ddPCR. For instance, He et al. have applied a machine learning approach to accurately detect five differential circRNAs in urine extracellular vesicles using ddPCR, which improved the diagnostic efficacy for high-grade prostate cancer [145]. Furthermore, new methods for detecting exosomal circRNAs based on biosensors, electrochemistry, and rolling loop amplification are attracting increasing attention [150–152].
Fig. 4.
Clinical applications of circRNAs and exosomal circRNAs in tumors, including circRNA extraction, detection, as clinical markers and therapeutic strategies
Exosomal circRNA functions as clinical biomarker
Increasing evidence suggests that exosomal circRNAs hold great promise as diagnostic and prognostic biomarkers for various types of tumors (Fig. 4 and Table 3) [153, 154]. For instance, a combination of exosomal hsa_circ_0001439, hsa_circ_0001492, and hsa_circ_0000896 demonstrated better diagnostic sensitivity and specificity for LUAD, with an AUC value of 0.805. Notably, the diagnostic sensitivity (73.2%, 66.1%,62.2%) and specificity (70.8%, 84%, 87.5%) of the three serum exosomal circRNAs were higher than those of serum circRNAs [155]. Hsa_circ_0055202, hsa_circ_0074920 and hsa_circ_0043722 have been confirmed to highly expressed in plasma exosomes of GBM patients by circRNA microarray technology. The area under the curve (AUC) of the triple combination is high at 0.988 and 0.925 in the training and validation cohorts, respectively [156]. Also, a recent study has shown that high expression of circMYC in serum exosomes of patients with nasopharyngeal carcinoma (NPC) is associated with tumor size, lymph node metastasis, TNM stage, survival rate, and disease recurrence, with an AUC of 0.945, granting a new biomarker for the diagnosis and prognosis of NPC [157]. Interestingly, machine learning algorithms have been used to construct a nine-circRNA combination classifier with an SVM model (BCExoC) to improve the diagnostic value for breast cancer, with AUCs of 0.83 and 0.80 in the training and validation cohorts, respectively [158]. These findings suggest that combining biomarkers with machine learning algorithms may be a crucial technique in future advancements to improve diagnostic efficacy.
Table 3.
Exosomal circRNAs as cancer biomarkers
| Exosomal circRNA | Type of disease | Source of exosome | Expression | Clinical relevance | Case number | AUC | Sensitivity and specificity | References |
|---|---|---|---|---|---|---|---|---|
| circPDLIM5, etc. | Prostate cancer | Urine | ↑ | _ | 263 in training cohort; 497 in validation cohort 1; 505 in validation cohort 2 | 0.820 in training cohort; 0.807 in validation cohort 1; 0.810 in validation cohort 2 | _ | [145] |
| circ_0047921, circ_0056285, circ_0007761 | Non-small cell lung cancer | Serum | ↓ ↓ ↑ | circ_0056285: clinical stages and lymph node metastasis | 120 in training set; 62 in validation set1; 63 in validation set2 | Combined:0.926 in training set;0.919 in validation set | _ | [153] |
| circ_0070396 | Hepatocellular cancer | Plasma | ↑ | AFP, AST, ALT, ALB, TBIL, DBIL | 111 | 0.8574 | Sensitivity:62.16% Specificity:98.15% | [154] |
| hsa_circ_0001439, hsa_circ_0001492, hsa_circ_0000896 | Lung adenocarcinoma | Serum | ↑ ↑ ↑ | TNM stage, tumor size, NSE, CEA | 134 | Combined:0.805 | Sensitivity:73.2%; 66.1%;62.2% Specificity:70.8%;84%;87.5% | [155] |
| hsa_circ_0055202, hsa_circ_0074920, hsa_circ_0043722 | Glioblastoma multiforme | Plasma | ↑ ↑ ↑ | _ | 120 | Combined:0.988 in training set; 0.925 in validation set | _ | [156] |
| circMYC | Radio-resistant nasopharyngeal carcinoma | Serum | ↑ | Tumor size, lymph node metastasis, TNM stage, survival rate, and disease recurrence | 210 | 0.945 | _ | [157] |
| BCExoC (hsa_circ_0002190, etc.) | Breast cancer | Plasma | ↑ | _ | 144 in training cohort; 101 in testing cohort; | SVM:0.83 in training cohort; 0.80 in testing cohort | _ | [158] |
However, to validate the clinical utility of exosomal circRNA biomarkers, more rigorous data and a large number of clinical samples are needed (Table 3). Furthermore, since biomarkers with high specificity and sensitivity are lacking, a combination of exosomal circRNAs with traditional biomarkers such as CEA and CA19-9 may yield better diagnostic outcomes. After large-scale sample collections, clinical cohort studies, and multi-center collaborative research, exosomal circRNAs may be considered promising diagnostic markers.
Given the potential value of exosomal circRNAs in tumor diagnosis and prognosis, there have been a variety of clinical trials targeting exosomal circRNAs as tumor biomarkers. Details of current data from both domestic and international clinical trials of circRNAs and exosomal circRNAs as diagnostic or prognostic biomarkers for cancer, including a variety of cancers such as cholangiocarcinoma, pancreatic cancer and prostate cancer, are summarized in Table 4. (data from https://www.chictr.org.cn/ and https://clinicaltrials.gov/) In addition, as emerging clinical biomarkers, most of the current clinical trials are in Phase 0 and some projects have not yet initiated recruitment of recipients. However, we believe that with the support of more and more research data, exosomal circRNAs will become very promising and reliable tumor biomarkers and will eventually go to the clinic.
Table 4.
Clinical trials with circRNAs and exosomal circRNAs as cancer diagnosis or prognosis biomarkers
| Registration number/NCT number | Study type | Study phase | Recruiting status | Tumor type | Sample name | Sample size |
|---|---|---|---|---|---|---|
| ChiCTR2300069863 | Observational study | 0 | Recruiting | Cholangiocarcinoma | Bile, serum | 320 |
| ChiCTR1900027419 | Basic science | 0 | Not yet recruiting | Lung cancer | Blood | 58 |
| ChiCTR1900024188 | Diagnostic test | 0 | Recruiting | Prostate cancer | Urine | 300 |
| ChiCTR1800019529 | Diagnostic test | Diagnostic new technique clinical study | Recruiting | Prostate cancer | Plasma | 200 |
| ChiCTR1800018038 | Diagnostic test | Diagnostic new technique clinical study | Not yet recruiting | Pancreatic cancer | Blood | 20 |
| NCT05771337 | Observational study | Not available | Not yet recruiting | Breast cancer | Blood | 80 |
| NCT04584996 | Observational study | Not available | Recruiting | Pancreatic cancer, biliary tract cancer | Blood, bile | 186 |
| NCT04464122 | Observational study | Not available | Recruiting | Neuroendocrine neoplasm | Not available | 60 |
| NCT03334708 | Observational study | Not available | Recruiting | Pancreatic cancer | Blood | 700 |
Therapeutic strategies based on exosomal circRNA
Exosomal circRNAs have been implicated in cancer, and targeting them for modulation has led to the development of novel therapeutic strategies for cancer. Here, we describe the therapeutic strategies and limitations of direct targeting of circRNAs as well as present the current status and prospects for nanoparticles and exosome-based delivery of circRNAs (Fig. 4).
Strategies for targeting circRNAs
Technologies such as small interfering RNA (siRNA), antisense oligonucleotide (ASO), and CRISPR cas13a have been designed to target oncogenic circRNAs. For example, in vivo treatment with circNRIP1 siRNA in Patient-derived tumor xenograft (PDX) mice significantly reduced tumor weight and volume [74]. Additionally, ASOs targeting circRHOBTB3 inhibited CRC growth and metastasis in vitro and in vivo [159]. CRISPR/Cas13a-mediated knockdown of hsa_circ_0000190(C190) inhibited NSCLC cell proliferation and migration in vitro and reduced tumor growth in vivo [160]. However, techniques based on silencing circRNAs, such as siRNA and ASO, usually inevitably cause off-target effects, resulting in inconsistent efficiency in silencing circRNAs. In addition, siRNAs with a high G/C base content may be less effective. As a Cas13 effector, the PspCas13b effector shows a higher specificity for Drosophila circRNA, but still produces off-target effects [161].
Conversely, overexpression vectors have been used to promote the production of tumor-suppressor circRNAs such as circFNDC3B, which inhibits angiogenesis and CRC progression [12, 162]. Notably, due to the specificity of the loop structure, there may be incorrect splicing or insertion and deletion of bases in the construction of circRNA overexpression vectors to cause failure or inefficient loop formation. The incorporation of a circularized expression framework into the circRNA vector is important for overexpression of circRNA. In addition, circRNA primers used for qPCR experiments must specifically and efficiently to amplify circRNAs rather than other transcripts in order to properly validate circRNA silencing and overexpression efficiency.
Intriguingly, a growing number of studies have confirmed that circRNAs can encode functional proteins and participate in cancer progression [163]. In order to increase the value of these circRNAs, researchers are working to synthesize circRNAs artificially to avoid the limitations of linear mRNAs. For example, Alex Wesselhoeft's team used an engineering approach to prepare a circRNA with the potential to efficiently encode proteins for efficient and stable expression in eukaryotic cells [164]. Excitingly, through optimizing circRNA translation conditions, Chen et al. have improved the yield of circRNA-translated proteins to hundreds of folds, enabling more sustainable expression of translated protein products for biological functions in vitro and in vivo, which holds tremendous promise for application [165]. A recent study reported that down-regulated expression of circ-INSR may be an important target in the process of cardiotoxicity and cardiac remodeling. The use of adeno-associated virus (AAV) and artificially prepared circ-INSR mimics prevents and reverses adriamycin-mediated cardiomyocyte death and improves cardiac function. It is significant for engineering circRNAs from basic to clinical [166]. Controversially, engineered circRNAs produced exogenously have been demonstrated to induce activation of the immune system in vivo [167, 168]. It is also a limitation of engineered circRNA. Through comparing the efficiency and immunogenicity of circRNAs prepared by various in vitro synthesis methods, Chen et al. have found that circRNAs synthesized by T4 RNA ligase have lower immunogenicity, which is an important foundation for the engineering of circRNAs in gene therapy [169]. Furthermore, the generation, purification, primary sequence, secondary structure and RNA modification of engineered circRNAs are all important factors affecting their immunogenicity [170]. A strategy of encapsulating synthetic circRNAs with RNA-binding proteins and splicing factors may be an effective means of suppressing innate immune activation [171].
Therapeutic strategies using nanoparticle-delivered circRNAs
While most studies have used adenoviral and lentiviral vectors as viral delivery systems to target circRNAs, there are high costs and potential biosafety issues, as well as limited ability to carry genes. Nanoparticles are materials in the size range of 10 to 1000 nm, which have the advantages of low cost, ease for preparation, good biocompatibility, biodegradability and high safety compared to viral vectors, and play an important role in bioimaging, molecular detection, disease diagnosis and drug delivery [172, 173]. A variety of organic materials (such as phospholipid, polymers, etc.) and inorganic materials (such as gold and metal oxides.) can be used to produce nanoparticles to protect RNA from degradation [174]. Emerging evidence suggests that nanoparticle-based delivery systems for circRNAs may also be a promising therapeutic strategy. A lipid nanoparticle (LNP) system developed for in vitro and in vivo delivery of circRNA triggered adaptive immune activation and showed excellent antitumor efficacy in multiple mouse tumor models, offering new prospects for the development of cancer RNA vaccines [175]. Qu et al. have generated high concentrations and abundance of circRNA-RBD with antigenic potential encoding SARS-CoV-2-RBD in vitro using type I intron nuclease and T4 RNA ligase and have demonstrated that SARS-CoV-2 circRNA-RBD vaccine induces sustained humoral immune responses in mice and rhesus monkeys with LNP delivery system. It shows higher and more sustained antigen expression than the mRNA vaccine. In addition, the circRNA vaccine does not cause clinical signs of disease in rhesus monkeys and exhibits a good biosafety [176]. In addition, Du et al. have demonstrated that delivery of circ-Foxo3 plasmids coupled with gold nanoparticles in vivo can target Foxo3 and upregulate PUMA to induce apoptosis and enhance survival in mice. Nevertheless, more clinical trials are needed to evaluate its biosafety [177]. It is also possible to use nanoparticles to encapsulate siRNA to improve delivery efficiency. In the orthotopic tumor model and lung metastasis model mice, the poly lactic-co-glycolic acid (PLGA)-based si-cSERPINE2 nanoparticles effectively attenuated breast cancer progression without apparent systemic toxicity, hepatotoxicity and renotoxicity [37]. Meng et al. used nanoparticles made from PLGA and polyethylene glycol (PEG) chains to deliver si-circROBO1 in vitro and in vivo with good anticancer effects and no significant organ toxicity [178].
Nevertheless, we still need to develop more novel drug delivery systems with precision drug delivery capabilities to facilitate circRNAs clinical translation. As an advanced delivery system, the stimulus-responsive delivery system protects the precise delivery of RNA with dynamic activity compared to conventional non-stimulus-responsive nanocarriers. Some popular stimulus-responsive nanomaterials, including pH-responsive nanocarriers, enzyme-responsive nanocarriers, hypoxia-responsive nanomaterials, etc., are more suitable for tumor sites [179]. You et al. have reported a circ_0058051-siRNA delivery system based on superparamagnetic iron oxide nanoparticles (SPIONs) to protect it from degradation in serum and ultimately inhibit HCC progression in vivo with a good biosafety [180]. Stimulus-responsive nanoparticle-based systems are now widely used to deliver siRNA, miRNA, etc. [181, 182]. We believe that its application to the delivery of circRNAs will become a future research trend.
Therapeutic strategies using exosome-delivered circRNAs
Exosomes are 30–150 nm extracellular vesicles with an average size of 100 nm that have good stability, small size, and low toxicity, making them ideal drug delivery carriers [6]. Compared to synthetic carriers such as liposomes and nanoparticles, exosomes have attracted much attention in recent years because of their strong biocompatibility, low immunogenicity and high chemical stability [183, 184].
Engineered exosome-based therapies have been developed for various diseases, including cancer [185, 186]. Therapeutic strategies based on exosomal circRNAs are particularly promising. Wang et al. have confirmed that exosomal ciRS-122 promotes glycolysis and induces chemical resistance through the miR-122/PKM2 axis in CRC. Both in vivo and in vitro experiments have demonstrated that si-ciRS-122 delivered by exosomes reversed oxaliplatin resistance and glycolysis and inhibited tumor proliferation in CRC [71]. RGD-exo-circDIDO1 inhibited the development of GC and was found to have no potential toxic effects [187]. Engineered RVG-circSCMH1-EVs selectively delivered circSCMH1 to the brain, enhancing neuronal plasticity in mice and monkeys after stroke [188]. However, packaging and delivering large circRNAs into exosomes is challenging, and there is a need to develop more effective approaches.
Currently, a variety of exosome-delivered vaccines have entered clinical trials. For example, a trial of vaccination with dendritic cell-derived exosomes loaded with tumor antigens has recruited 41 participants and is currently in phase II clinical trials (NCT01159288). And another open dose-escalation clinical study of chimeric exosomal tumor vaccines for recurrent or metastatic bladder cancer is still recruiting and is currently in an early phase I clinical trial (NCT05559177). Some relevant clinical trials have also been registered in China (ChiCTR2100050939, ChiCTR2000034241). Although there are no clinical trials available, we believe that circRNA-based exosome delivery technology will shine in the field of cancer therapy in the future.
The last but not least, the delivery of drugs based on plant or food exosomes is a promising trend. They have the advantages of wider availability, lower cost and ease to access compared to cellular exosomes which are already used in a variety of diseases including cancer [189–191]. Currently, grape extract (NCT01668849), ginger and aloe exosomes (NCT03493984) have been approved for clinical trials. This is certainly the future direction of engineered exosomes.
Taken together, exosomal circRNAs-based therapeutic approaches hold great promise, and more research is needed to explore their potential as a therapeutic strategy for cancer. In addition, exosomal circRNA delivery technologies based on nanoparticles and plant exosomes are promising stars for tomorrow.
Conclusions and perspectives
Exosomal circRNAs have become a focal point of research due to their involvement in cancer progression, including immune escape, angiogenesis, metabolism, drug resistance, proliferation and metastasis. Both tumor and non-tumor derived extracellular circRNAs play a double-edged sword or “pro-and anti-cancer” role in cancer progression through mechanisms such as miRNA sponges, interactions with target proteins, protein scaffolds, translation proteins and small molecule peptides, indicating that these molecules have been providing insights into cancer pathogenesis and treatment. Moreover, increasing evidence suggests that extracellular circRNAs have the potential to become biomarkers for cancer diagnosis and prognosis, especially when combined with traditional tumor markers, which significantly improves the sensitivity and specificity of diagnosis and efficacy analysis. At the same time, the application of machine learning and artificial intelligence can establish a multi-molecule diagnosis panel system and platform based on extracellular circRNAs, providing ideal evidence for precise diagnosis and treatment of cancers. Exosomal circRNAs present amazing bidirectional regulating ability to the remodeling immunity microenvironment no matter in tumor cells or immune cells. And exosomal circRNAs involve in the reprogramming tumor metabolism such as glycolysis, lipid metabolism, amino acid metabolism, oxidative respiration through diverse enzymes, TFs, and signaling pathways. Especially, exosomal circRNAs can alter the condition of immune cell, immune checkpoint molecules and metabolism simultaneously. These pave a way for precise dynamic surveillance and target treatment.
However, the specific mechanisms by which components in the tumor microenvironment (TME) affect tumor cell fate through exosomal circRNAs remain unclear, and further investigation is needed to understand the unique functions of TME components. Additionally, several limitations remain about the use of exosomal circRNAs in body fluids as biomarkers for cancer diagnosis and prognosis, including a limited sample size, insufficient specimens representing various clinical stages and.differential diagnoses of cancer, as well as a lack of standardized and uniformly applied quality control evaluation systems. The engineering strategies for exosomal circRNAs also face limitations such as the size of circRNAs for effective packaging and delivery, as well as safety, pharmacokinetics, and pharmacodynamics considerations. In order to effectively target oncogenic circRNAs, it is imperative to develop knockdown or knockout strategies tailored to a specific circRNA. However, it is inevitable that off-target effects will occur. Furthermore, constructing overexpression vectors for cancer suppressor circRNAs may a challenge due to the specificity of the loop structure of circRNAs which may result in failed loop formation or inefficiency. Artificial preparation of circRNAs presents a promising approach to developing therapeutics; however, the issues of immunogenicity and standardization must be addressed. Another important direction is the engineering of circRNAs using exosomes from immune cells, which have potent anti-tumor activity. Incorporating stimulus-responsive nanocarriers with exosomes may enhance circRNA delivery, especially when adapted to the tumor microenvironment. In addition, circRNA delivery based on plant or food exosomes is a promising strategy due to its easy availability and low cost.
Technological advancements in the extraction and purification of exosomes are necessary to differentiate them from microvesicles, and to ensure their purity and specificity. Innovation in this field, such as the utilization of machine learning, droplet digital PCR, and artificial intelligence, may enhance our ability to identify the involvement of exosomal circRNAs in tumor diagnosis, ultimately leading to better clinical decision-making. Moreover, exosomal circRNAs have significant therapeutic potential, and manipulating exosomes presents new possibilities in the realm of cancer therapy. In conclusion, increasing evidence supports the promising clinical applications of exosomal circRNAs as both novel potential biomarkers and therapeutic targets.
Acknowledgements
Not applicable.
Abbreviations
- CircRNA
Circular RNA
- EVs
Extracellular vesicles
- TME
Tumor microenvironment
- MVBs
Multivesicular bodies
- DC cell
Dendritic cell
- T cell
T lymphocyte
- HCC
Hepatocellular carcinoma
- PD1
Programmed cell death protein 1
- NSCLC
Non-small cell lung cancer
- IFN-γ
Interferon-γ
- TNF-α
Tumor necrosis factor α
- MiRNA
MicroRNA
- Tregs
Regulatory T cells
- PDL1
Programmed cell death ligand 1
- LUAD
Lung adenocarcinoma
- RCC
Renal cell carcinoma
- TAMs
Tumor-associated macrophages
- BC
Breast cancer
- ER
Endoplasmic reticulum
- ESCC
Esophageal squamous cell carcinoma
- HuR
Human antigen R
- GC
Gastric cancer
- IL-6
Interleukin-6
- NK cells
Nature killer cells
- TAN
Tumor-associated neutrophil
- CRC
Colorectal cancer
- CTLA4
Cytotoxic T lymphocyte associate protein-4
- TIM-3
T-cell immunoglobulin and mucin domain 3
- VEGF
Vascular endothelial growth factor
- HUVEC
Human umbilical vein endothelial cell
- hBMECs
Human brain microvascular endothelial cells
- GBM
Glioblastoma multiforme
- HK2
Hexokinase 2
- 2-DG
2-Deoxy-D-glucose
- PKM2
Pyruvate kinase type M2
- PEP
Phosphoenolpyruvate
- LDHA
Lactate dehydrogenase A
- TF
Transcription factor
- TMZ
Temozolomide
- SQLE
Squalene epoxidase
- FAs
Fatty acids
- FAS
Fatty acid synthesis
- PCa
Prostate cancer
- ARSI
Androgen receptor signaling inhibitor
- GPX4
Glutathione peroxidase 4
- ROS
Reactive oxygen species
- AA
Arachidonic acid
- BCa
Bladder cancer
- EMT
Epithelial-mesenchymal transition
- PDAC
Pancreatic ductal adenocarcinoma
- LNM
Lymph node metastasis
- CDDP
Cisplatin
- PC
Pancreatic cancer
- HIF
Hypoxia-inducible factor
- RBP-J
Recombination signal-binding protein-Jκ
- UCEC
Uterine corpus endometrial cancer
- CAFs
Cancer-associated fibroblasts
- MSCs
Mesenchymal stem cells
- BM-MSCs
Bone marrow mesenchymal stem cells
- AMSCs
Adipose-derived mesenchymal stem cells
- qRT-PCR
Quantitative reverse transcription polymerase chain reaction
- ddPCR
Droplet digital polymerase chain reaction
- AUC
Area under the curve
- NPC
Nasopharyngeal carcinoma
- CEA
Carcinoembryonic antigen
- CA19-9
Carbohydrate antigen19-9
- siRNA
Small interfering RNA
- ASO
Antisense oligonucleotide
- CRISPR
Clustered regularly interspaced short palindromic repeats
- Cas13a
CRISPR-associated protein 13a
- PDX
Patient-derived tumor xenograft
- AAV
Adeno-associated virus
- LNP
Lipid nanoparticle
- PLGA
Poly lactic-co-glycolic acid
- PEG
Polyethylene glycol
- SPIONs
Superparamagnetic iron oxide nanoparticles
- RVG
Rabies virus glycoprotein
Author contributions
FZ and JJ searched the literature and wrote the first draft of the manuscript. HQ provided direction and guidance. WX and YY critically revised the work. All authors contributed to the article and approved the submitted version. All authors read and approved the final manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (NO.81971757), the Jiangsu Province's Major Project in Research and Development (BE2020680), Zhenjiang Key Laboratory of sEVs Foundation and Transformation Application High-tech Research, China (SS2018003), the Clinical Major Disease Project of Suzhou (LCZX202019), the Technology Development Project of Suzhou (SKY2021018), the Technology Project of Zhangjiagang (ZKHBZ2216), the Innovation Foundation for Medicine and Education Synergy of Jiangsu University (JDY2022020).
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fan Zhang and Jiajia Jiang contributed equally to this work
Contributor Information
Yongmin Yan, Email: yym@wjrmyy.cn.
Wenrong Xu, Email: icls@ujs.edu.cn.
References
- 1.He G, Peng X, Wei S, Yang S, Li X, Huang M, et al. Exosomes in the hypoxic TME: from release, uptake and biofunctions to clinical applications. Mol Cancer. 2022;21:19. doi: 10.1186/s12943-021-01440-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Downs-Canner SM, Meier J, Vincent BG, Serody JS. B cell function in the tumor microenvironment. Annu Rev Immunol. 2022;40:169–193. doi: 10.1146/annurev-immunol-101220-015603. [DOI] [PubMed] [Google Scholar]
- 3.Lian X, Yang K, Li R, Li M, Zuo J, Zheng B, et al. Immunometabolic rewiring in tumorigenesis and anti-tumor immunotherapy. Mol Cancer. 2022;21:27. doi: 10.1186/s12943-021-01486-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petroni G, Buqué A, Coussens LM, Galluzzi L. Targeting oncogene and non-oncogene addiction to inflame the tumour microenvironment. Nat Rev Drug Discov. 2022;21:440–462. doi: 10.1038/s41573-022-00415-5. [DOI] [PubMed] [Google Scholar]
- 5.Wu M, Huang Q, Xie Y, Wu X, Ma H, Zhang Y, et al. Improvement of the anticancer efficacy of PD-1/PD-L1 blockade via combination therapy and PD-L1 regulation. J Hematol Oncol. 2022;15:24. doi: 10.1186/s13045-022-01242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paskeh MDA, Entezari M, Mirzaei S, Zabolian A, Saleki H, Naghdi MJ, et al. Emerging role of exosomes in cancer progression and tumor microenvironment remodeling. J Hematol Oncol. 2022;15:83. doi: 10.1186/s13045-022-01305-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188–206. doi: 10.1038/s41571-021-00585-y. [DOI] [PubMed] [Google Scholar]
- 8.Preethi KA, Selvakumar SC, Ross K, Jayaraman S, Tusubira D, Sekar D. Liquid biopsy: exosomal microRNAs as novel diagnostic and prognostic biomarkers in cancer. Mol Cancer. 2022;21:54. doi: 10.1186/s12943-022-01525-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu W, Liu C, Bi ZY, Zhou Q, Zhang H, Li LL, et al. Comprehensive landscape of extracellular vesicle-derived RNAs in cancer initiation, progression, metastasis and cancer immunology. Mol Cancer. 2020;19:102. doi: 10.1186/s12943-020-01199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Liu S, Zhang L, Issaian A, Hill RC, Espinosa S, et al. A unified mechanism for intron and exon definition and back-splicing. Nature. 2019;573:375–380. doi: 10.1038/s41586-019-1523-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 13.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi H, Huang S, Qin M, Xue X, Guo X, Jiang L, et al. Exosomal circ_0088300 derived from cancer-associated fibroblasts acts as a miR-1305 sponge and promotes gastric carcinoma cell tumorigenesis. Front Cell Dev Biol. 2021;9:676319. doi: 10.3389/fcell.2021.676319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Pan Z, Zhao R, Li B, Qi Y, Qiu W, Guo Q, et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol Cancer. 2022;21:16. doi: 10.1186/s12943-021-01485-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Z, Chen G, Zhao Y, Gao H, Li L, Yin Y, et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol Cancer. 2023;22:55. doi: 10.1186/s12943-023-01759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Cao Q, Shi Y, Wu X, Mi Y, Liu K, et al. Identification of low-dose radiation-induced exosomal circ-METRN and miR-4709-3p/GRB14/PDGFRα pathway as a key regulatory mechanism in Glioblastoma progression and radioresistance: functional validation and clinical theranostic significance. Int J Biol Sci. 2021;17:1061–1078. doi: 10.7150/ijbs.57168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S, Huang V, Xu X, Livingstone J, Soares F, Jeon J, et al. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176:831–843.e822. doi: 10.1016/j.cell.2019.01.025. [DOI] [PubMed] [Google Scholar]
- 21.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang X, Deng Z, Ding P, Qiang W, Lu Y, Gao S, et al. A novel protein encoded by circHNRNPU promotes multiple myeloma progression by regulating the bone marrow microenvironment and alternative splicing. J Exp Clin Cancer Res. 2022;41:85. doi: 10.1186/s13046-022-02276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuo B, Chen Z, Dang Q, Chen C, Zhang H, Hu S, et al. Roles of exosomal circRNAs in tumour immunity and cancer progression. Cell Death Dis. 2022;13:539. doi: 10.1038/s41419-022-04949-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuan Y, Li H, Pu W, Chen L, Guo D, Jiang H, et al. Cancer metabolism and tumor microenvironment: fostering each other? Sci China Life Sci. 2022;65:236–279. doi: 10.1007/s11427-021-1999-2. [DOI] [PubMed] [Google Scholar]
- 25.Watowich MB, Gilbert MR, Larion M. T cell exhaustion in malignant gliomas. Trends Cancer. 2023;9:270–292. doi: 10.1016/j.trecan.2022.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen SW, Zhu SQ, Pei X, Qiu BQ, Xiong D, Long X, et al. Cancer cell-derived exosomal circUSP7 induces CD8+ T cell dysfunction and anti-PD1 resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol Cancer. 2021;20:144. doi: 10.1186/s12943-021-01448-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang YA, Li XL, Mo YZ, Fan CM, Tang L, Xiong F, et al. Effects of tumor metabolic microenvironment on regulatory T cells. Mol Cancer. 2018;17:168. doi: 10.1186/s12943-018-0913-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang M, Huang X, Huang N. Exosomal circGSE1 promotes immune escape of hepatocellular carcinoma by inducing the expansion of regulatory T cells. Cancer Sci. 2022;113:1968–1983. doi: 10.1111/cas.15365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11:32. doi: 10.1038/s41419-020-2230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Colin S, Chinetti-Gbaguidi G, Staels B. Macrophage phenotypes in atherosclerosis. Immunol Rev. 2014;262:153–166. doi: 10.1111/imr.12218. [DOI] [PubMed] [Google Scholar]
- 31.Huang X, Wang J, Guan J, Zheng Z, Hao J, Sheng Z, et al. Exosomal Circsafb2 reshaping tumor environment to promote renal cell carcinoma progression by mediating M2 macrophage polarization. Front Oncol. 2022;12:808888. doi: 10.3389/fonc.2022.808888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu C, Shi W, Hu W, Zhao Y, Zhao X, Dong F, et al. Endoplasmic reticulum stress promotes breast cancer cells to release exosomes circ_0001142 and induces M2 polarization of macrophages to regulate tumor progression. Pharmacol Res. 2022;177:106098. doi: 10.1016/j.phrs.2022.106098. [DOI] [PubMed] [Google Scholar]
- 33.Gao J, Ao YQ, Zhang LX, Deng J, Wang S, Wang HK, et al. Exosomal circZNF451 restrains anti-PD1 treatment in lung adenocarcinoma via polarizing macrophages by complexing with TRIM56 and FXR1. J Exp Clin Cancer Res. 2022;41:295. doi: 10.1186/s13046-022-02505-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Q, Wang X, Zhu J, Fei X, Chen H, Li C. Hypoxic tumor-derived exosomal Circ0048117 facilitates M2 macrophage polarization acting as miR-140 sponge in esophageal squamous cell carcinoma. Onco Targets Ther. 2020;13:11883–11897. doi: 10.2147/OTT.S284192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, et al. Downregulation of hsa_circ_0074854 suppresses the migration and invasion in hepatocellular carcinoma via interacting with HuR and via suppressing exosomes-mediated macrophage M2 polarization. Int J Nanomed. 2021;16:2803–2818. doi: 10.2147/IJN.S284560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song J, Xu X, He S, Wang N, Bai Y, Li B, et al. Exosomal hsa_circ_0017252 attenuates the development of gastric cancer via inhibiting macrophage M2 polarization. Hum Cell. 2022;35:1499–1511. doi: 10.1007/s13577-022-00739-9. [DOI] [PubMed] [Google Scholar]
- 37.Zhou B, Mo Z, Lai G, Chen X, Li R, Wu R, et al. Targeting tumor exosomal circular RNA cSERPINE2 suppresses breast cancer progression by modulating MALT1-NF- J Exp Clin Cancer Res. 2023;42:48. doi: 10.1186/s13046-023-02620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laskowski TJ, Biederstädt A, Rezvani K. Natural killer cells in antitumour adoptive cell immunotherapy. Nat Rev Cancer. 2022;22:557–575. doi: 10.1038/s41568-022-00491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19:110. doi: 10.1186/s12943-020-01222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geh D, Leslie J, Rumney R, Reeves HL, Bird TG, Mann DA. Neutrophils as potential therapeutic targets in hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2022;19:257–273. doi: 10.1038/s41575-021-00568-5. [DOI] [PubMed] [Google Scholar]
- 41.Tie Y, Tang F, Wei YQ, Wei XW. Immunosuppressive cells in cancer: mechanisms and potential therapeutic targets. J Hematol Oncol. 2022;15:61. doi: 10.1186/s13045-022-01282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyagi A, Wu SY, Sharma S, Wu K, Zhao D, Deshpande R, et al. Exosomal miR-4466 from nicotine-activated neutrophils promotes tumor cell stemness and metabolism in lung cancer metastasis. Oncogene. 2022;41:3079–3092. doi: 10.1038/s41388-022-02322-w. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Leal AC, Mizurini DM, Gomes T, Rochael NC, Saraiva EM, Dias MS, et al. Tumor-derived exosomes induce the formation of neutrophil extracellular traps: implications for the establishment of cancer-associated thrombosis. Sci Rep. 2017;7:6438. doi: 10.1038/s41598-017-06893-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singhto N, Thongboonkerd V. Exosomes derived from calcium oxalate-exposed macrophages enhance IL-8 production from renal cells, neutrophil migration and crystal invasion through extracellular matrix. J Proteom. 2018;185:64–76. doi: 10.1016/j.jprot.2018.06.015. [DOI] [PubMed] [Google Scholar]
- 45.Zhang X, Shi H, Yuan X, Jiang P, Qian H, Xu W. Tumor-derived exosomes induce N2 polarization of neutrophils to promote gastric cancer cell migration. Mol Cancer. 2018;17:146. doi: 10.1186/s12943-018-0898-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 2020;19:117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang X, Liu G, Li Y, Pan Y. Immune checkpoint: the novel target for antitumor therapy. Genes Dis. 2021;8:25–37. doi: 10.1016/j.gendis.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Li CW, Li X, Ding Q, Guo L, Liu S, et al. MET inhibitors promote liver tumor evasion of the immune response by stabilizing PDL1. Gastroenterology. 2019;156:1849–1861.e1813. doi: 10.1053/j.gastro.2019.01.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Yao Y, Jin M. Circ-0001068 is a novel biomarker for ovarian cancer and inducer of PD1 expression in T cells. Aging (Albany NY) 2020;12:19095–19106. doi: 10.18632/aging.103706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang M, Lin Y, Wang C, Deng L, Chen M, Assaraf YG, et al. New insights into antiangiogenic therapy resistance in cancer: Mechanisms and therapeutic aspects. Drug Resist Updates. 2022;64:100849. doi: 10.1016/j.drup.2022.100849. [DOI] [PubMed] [Google Scholar]
- 51.Das A, Ash D, Fouda AY, Sudhahar V, Kim YM, Hou Y, et al. Cysteine oxidation of copper transporter CTR1 drives VEGFR2 signalling and angiogenesis. Nat Cell Biol. 2022;24:35–50. doi: 10.1038/s41556-021-00822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JW, Hur J, Kwon YW, Chae CW, Choi JI, Hwang I, et al. KAI1(CD82) is a key molecule to control angiogenesis and switch angiogenic milieu to quiescent state. J Hematol Oncol. 2021;14:148. doi: 10.1186/s13045-021-01147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bae E, Huang P, Müller-Greven G, Hambardzumyan D, Sloan AE, Nowacki AS, et al. Integrin α3β1 promotes vessel formation of glioblastoma-associated endothelial cells through calcium-mediated macropinocytosis and lysosomal exocytosis. Nat Commun. 2022;13:4268. doi: 10.1038/s41467-022-31981-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jaykumar AB, Plumber S, Barry DM, Binns D, Wichaidit C, Grzemska M, et al. WNK1 collaborates with TGF-β in endothelial cell junction turnover and angiogenesis. Proc Natl Acad Sci U S A. 2022;119:e2203743119. doi: 10.1073/pnas.2203743119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Apte RS, Chen DS, Ferrara N. VEGF in signaling and disease: beyond discovery and development. Cell. 2019;176:1248–1264. doi: 10.1016/j.cell.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020;19:112. doi: 10.1186/s12943-020-01208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S, Li J, Zhang H, Zhang Y, Wang X, Yang H, et al. Gastric cancer derived exosomes mediate the delivery of circRNA to promote angiogenesis by targeting miR-29a/VEGF axis in endothelial cells. Biochem Biophys Res Commun. 2021;560:37–44. doi: 10.1016/j.bbrc.2021.04.099. [DOI] [PubMed] [Google Scholar]
- 58.Bai L, Gao Z, Jiang A, Ren S, Wang B. Circular noncoding RNA circ_0007334 sequestrates miR-577 to derepress KLF12 and accelerate colorectal cancer progression. Anticancer Drugs. 2022;33:e409–e422. doi: 10.1097/CAD.0000000000001221. [DOI] [PubMed] [Google Scholar]
- 59.Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res. 2020;39:20. doi: 10.1186/s13046-020-1529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiang Y, Zhao J, Xu J, Zhang H, Zhou J, Li H, et al. Glioblastoma-associated microglia-derived exosomal circKIF18A promotes angiogenesis by targeting FOXC2. Oncogene. 2022;41:3461–3473. doi: 10.1038/s41388-022-02360-4. [DOI] [PubMed] [Google Scholar]
- 61.Shi P, Liu Y, Yang H, Hu B. Breast cancer derived exosomes promoted angiogenesis of endothelial cells in microenvironment via circHIPK3/miR-124-3p/MTDH axis. Cell Signal. 2022;95:110338. doi: 10.1016/j.cellsig.2022.110338. [DOI] [PubMed] [Google Scholar]
- 62.Xu Y, Leng K, Yao Y, Kang P, Liao G, Han Y, et al. A circular RNA, cholangiocarcinoma-associated circular RNA 1, contributes to cholangiocarcinoma progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology. 2021;73:1419–1435. doi: 10.1002/hep.31493. [DOI] [PubMed] [Google Scholar]
- 63.Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol. 2019;12:90. doi: 10.1186/s13045-019-0776-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shimada S, Mogushi K, Akiyama Y, Furuyama T, Watanabe S, Ogura T, et al. Comprehensive molecular and immunological characterization of hepatocellular carcinoma. EBioMedicine. 2019;40:457–470. doi: 10.1016/j.ebiom.2018.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L, Yang Q, Peng S, Liu X. The combination of the glycolysis inhibitor 2-DG and sorafenib can be effective against sorafenib-tolerant persister cancer cells. Oncol Targets Ther. 2019;12:5359–5373. doi: 10.2147/OTT.S212465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tomizawa M, Shinozaki F, Motoyoshi Y, Sugiyama T, Yamamoto S, Ishige N. 2-Deoxyglucose and sorafenib synergistically suppress the proliferation and motility of hepatocellular carcinoma cells. Oncol Lett. 2017;13:800–804. doi: 10.3892/ol.2016.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi Q, Ji T, Ma Z, Tan Q, Liang J. Serum exosomes-based biomarker circ_0008928 regulates cisplatin sensitivity, tumor progression, and glycolysis metabolism by miR-488/HK2 axis in cisplatin-resistant nonsmall cell lung carcinoma. Cancer Biother Radiopharm. 2021. [DOI] [PubMed]
- 68.Hsu MC, Hung WC. Pyruvate kinase M2 fuels multiple aspects of cancer cells: from cellular metabolism, transcriptional regulation to extracellular signaling. Mol Cancer. 2018;17:35. doi: 10.1186/s12943-018-0791-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Icard P, Simula L, Fournel L, Leroy K, Lupo A, Damotte D, et al. The strategic roles of four enzymes in the interconnection between metabolism and oncogene activation in non-small cell lung cancer: therapeutic implications. Drug Resist Updates. 2022;63:100852. doi: 10.1016/j.drup.2022.100852. [DOI] [PubMed] [Google Scholar]
- 70.Wang L, Fu B, Hou DY, Lv YL, Yang G, Li C, et al. PKM2 allosteric converter: a self-assembly peptide for suppressing renal cell carcinoma and sensitizing chemotherapy. Biomaterials. 2023;296:122060. doi: 10.1016/j.biomaterials.2023.122060. [DOI] [PubMed] [Google Scholar]
- 71.Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539–555. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng J, Yan X, Lu T, Song W, Li Y, Liang J, et al. CircFOXK2 promotes hepatocellular carcinoma progression and leads to a poor clinical prognosis via regulating the Warburg effect. J Exp Clin Cancer Res. 2023;42:63. doi: 10.1186/s13046-023-02624-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pi M, Kuang H, Yue C, Yang Q, Wu A, Li Y, et al. Targeting metabolism to overcome cancer drug resistance: a promising therapeutic strategy for diffuse large B cell lymphoma. Drug Resist Updates. 2022;61:100822. doi: 10.1016/j.drup.2022.100822. [DOI] [PubMed] [Google Scholar]
- 74.Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lai Z, Wei T, Li Q, Wang X, Zhang Y, Zhang S. Exosomal circFBLIM1 promotes hepatocellular carcinoma progression and glycolysis by regulating the miR-338/LRP6 axis. Cancer Biother Radiopharm. 2020. [DOI] [PubMed]
- 77.Ding C, Xi G, Wang G, Cui D, Zhang B, Wang H, et al. Exosomal circ-MEMO1 promotes the progression and aerobic glycolysis of non-small cell lung cancer through targeting MiR-101-3p/KRAS axis. Front Genet. 2020;11:962. doi: 10.3389/fgene.2020.00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y, Zang H, Zhang X, Huang G. Exosomal circ-ZNF652 promotes cell proliferation, migration, invasion and glycolysis in hepatocellular carcinoma via miR-29a-3p/GUCD1 axis. Cancer Manag Res. 2020;12:7739–7751. doi: 10.2147/CMAR.S259424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ding C, Yi X, Chen X, Wu Z, You H, Chen X, et al. Warburg effect-promoted exosomal circ_0072083 releasing up-regulates NANGO expression through multiple pathways and enhances temozolomide resistance in glioma. J Exp Clin Cancer Res. 2021;40:164. doi: 10.1186/s13046-021-01942-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brendolan A, Russo V. Targeting cholesterol homeostasis in hematopoietic malignancies. Blood. 2022;139:165–176. doi: 10.1182/blood.2021012788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang B, Song BL, Xu C. Cholesterol metabolism in cancer: mechanisms and therapeutic opportunities. Nat Metab. 2020;2:132–141. doi: 10.1038/s42255-020-0174-0. [DOI] [PubMed] [Google Scholar]
- 82.Qian CJ, Zhou YX, Wu LK, Wang YC, Teng XS, Yao J. Circ_0000182 promotes cholesterol synthesis and proliferation of stomach adenocarcinoma cells by targeting miR-579-3p/SQLE axis. Discov Oncol. 2023;14:22. doi: 10.1007/s12672-023-00630-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xiong L, Liu HS, Zhou C, Yang X, Huang L, Jie HQ, et al. A novel protein encoded by circINSIG1 reprograms cholesterol metabolism by promoting the ubiquitin-dependent degradation of INSIG1 in colorectal cancer. Mol Cancer. 2023;22:72. doi: 10.1186/s12943-023-01773-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H, Guan T, Qin S, Xu Q, Yin L, Hu Q. Natural products in pursuing novel therapies of nonalcoholic fatty liver disease and steatohepatitis. Drug Discov Today. 2023;28:103471. doi: 10.1016/j.drudis.2022.103471. [DOI] [PubMed] [Google Scholar]
- 85.Zhao J, Lee K, Toh HC, Lam KP, Neo SY. Unravelling the role of obesity and lipids during tumor progression. Front Pharmacol. 2023;14:1163160. doi: 10.3389/fphar.2023.1163160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38:2844–2859. doi: 10.1038/s41388-018-0619-z. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87.Yoo HC, Park SJ, Nam M, Kang J, Kim K, Yeo JH, et al. A variant of SLC1A5 is a mitochondrial glutamine transporter for metabolic reprogramming in cancer cells. Cell Metab. 2020;31:267–283.e212. doi: 10.1016/j.cmet.2019.11.020. [DOI] [PubMed] [Google Scholar]
- 88.Yang L, Wang L, Wu J, Wang Y. Circ_0000069 contributes to the growth, metastasis and glutamine metabolism in renal cell carcinoma (RCC) via regulating miR-125a-5p-dependent SLC1A5 expression. Transpl Immunol. 2023;77:101764. doi: 10.1016/j.trim.2022.101764. [DOI] [PubMed] [Google Scholar]
- 89.Cai Y, Dong Z, Wang J. Circ_0000808 promotes the development of non-small cell lung cancer by regulating glutamine metabolism via the miR-1827/SLC1A5 axis. World J Surg Oncol. 2022;20:329. doi: 10.1186/s12957-022-02777-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma H, Qu S, Zhai Y, Yang X. circ_0025033 promotes ovarian cancer development via regulating the hsa_miR-370-3p/SLC1A5 axis. Cell Mol Biol Lett. 2022;27:94. doi: 10.1186/s11658-022-00364-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Song M, Liu J. Circ_0067717 promotes colorectal cancer cell growth, invasion and glutamine metabolism by serving as a miR-497-5p sponge to upregulate SLC7A5. Histol Histopathol. 2023;38:53–64. doi: 10.14670/HH-18-494. [DOI] [PubMed] [Google Scholar]
- 92.Zhong C, Long Z, Yang T, Wang S, Zhong W, Hu F, et al. M6A-modified circRBM33 promotes prostate cancer progression via PDHA1-mediated mitochondrial respiration regulation and presents a potential target for ARSI therapy. Int J Biol Sci. 2023;19:1543–1563. doi: 10.7150/ijbs.77133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Conche C, Finkelmeier F, Pešić M, Nicolas AM, Böttger TW, Kennel KB, et al. Combining ferroptosis induction with MDSC blockade renders primary tumours and metastases in liver sensitive to immune checkpoint blockade. Gut. 2023. [DOI] [PMC free article] [PubMed]
- 94.Zhai H, Zhong S, Wu R, Mo Z, Zheng S, Xue J, et al. Suppressing circIDE/miR-19b-3p/RBMS1 axis exhibits promoting-tumour activity through upregulating GPX4 to diminish ferroptosis in hepatocellular carcinoma. Epigenetics. 2023;18:2192438. doi: 10.1080/15592294.2023.2192438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Y, Zeng X, Lu D, Yin M, Shan M, Gao Y. Erastin induces ferroptosis via ferroportin-mediated iron accumulation in endometriosis. Hum Reprod. 2021;36:951–964. doi: 10.1093/humrep/deaa363. [DOI] [PubMed] [Google Scholar]
- 96.Wang L, Wu S, He H, Ai K, Xu R, Zhang L, et al. CircRNA-ST6GALNAC6 increases the sensitivity of bladder cancer cells to erastin-induced ferroptosis by regulating the HSPB1/P38 axis. Lab Invest. 2022;102:1323–1334. doi: 10.1038/s41374-022-00826-3. [DOI] [PubMed] [Google Scholar]
- 97.Zhang X, Xu Y, Ma L, Yu K, Niu Y, Xu X, et al. Essential roles of exosome and circRNA_101093 on ferroptosis desensitization in lung adenocarcinoma. Cancer Commun (Lond) 2022;42:287–313. doi: 10.1002/cac2.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen W, Tang D, Lin J, Huang X, Lin S, Shen G, et al. Exosomal circSHKBP1 participates in non-small cell lung cancer progression through PKM2-mediated glycolysis. Mol Ther Oncolytics. 2022;24:470–485. doi: 10.1016/j.omto.2022.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang C, Wu S, Mou Z, Zhou Q, Dai X, Ou Y, et al. Exosome-derived circTRPS1 promotes malignant phenotype and CD8+ T cell exhaustion in bladder cancer microenvironments. Mol Ther. 2022;30:1054–1070. doi: 10.1016/j.ymthe.2022.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Feng W, Gong H, Wang Y, Zhu G, Xue T, Wang Y, et al. circIFT80 functions as a ceRNA of miR-1236-3p to promote colorectal cancer progression. Mol Ther Nucleic Acids. 2019;18:375–387. doi: 10.1016/j.omtn.2019.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 101.Xu L, Liao WL, Lu QJ, Zhang P, Zhu J, Jiang GN. Hypoxic tumor-derived exosomal circular RNA SETDB1 promotes invasive growth and EMT via the miR-7/Sp1 axis in lung adenocarcinoma. Mol Ther Nucleic Acids. 2021;23:1078–1092. doi: 10.1016/j.omtn.2021.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 102.Yang H, Zhang H, Yang Y, Wang X, Deng T, Liu R, et al. Hypoxia induced exosomal circRNA promotes metastasis of colorectal cancer via targeting GEF-H1/RhoA axis. Theranostics. 2020;10:8211–8226. doi: 10.7150/thno.44419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang H, Wei M, Kang Y, Xing J, Zhao Y. Circular RNA circ_PVT1 induces epithelial-mesenchymal transition to promote metastasis of cervical cancer. Aging (Albany NY) 2020;12:20139–20151. doi: 10.18632/aging.103679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu C, Su Y, Liu L, Wang S, Liu Y, Wu J. Circular RNA hsa_circ_0004277 stimulates malignant phenotype of hepatocellular carcinoma and epithelial-mesenchymal transition of peripheral cells. Front Cell Dev Biol. 2020;8:585565. doi: 10.3389/fcell.2020.585565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zong ZH, Du YP, Guan X, Chen S, Zhao Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer Res. 2019;38:437. doi: 10.1186/s13046-019-1437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237–250. doi: 10.1016/j.canlet.2018.04.035. [DOI] [PubMed] [Google Scholar]
- 107.Dieterich LC, Tacconi C, Ducoli L, Detmar M. Lymphatic vessels in cancer. Physiol Rev. 2022;102:1837–1879. doi: 10.1152/physrev.00039.2021. [DOI] [PubMed] [Google Scholar]
- 108.Liu S, Lin Z, Rao W, Zheng J, Xie Q, Lin Y, et al. Upregulated expression of serum exosomal hsa_circ_0026611 is associated with lymph node metastasis and poor prognosis of esophageal squamous cell carcinoma. J Cancer. 2021;12:918–926. doi: 10.7150/jca.50548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.He F, Zhong X, Lin Z, Lin J, Qiu M, Li X, et al. Plasma exo-hsa_circRNA_0056616: a potential biomarker for lymph node metastasis in lung adenocarcinoma. J Cancer. 2020;11:4037–4046. doi: 10.7150/jca.30360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shen X, Kong S, Ma S, Shen L, Zheng M, Qin S, et al. Hsa_circ_0000437 promotes pathogenesis of gastric cancer and lymph node metastasis. Oncogene. 2022;41:4724–4735. doi: 10.1038/s41388-022-02449-w. [DOI] [PubMed] [Google Scholar]
- 111.Yao W, Jia X, Zhu L, Xu L, Zhang Q, Xia T, et al. Exosomal circ_0026611 contributes to lymphangiogenesis by reducing PROX1 acetylation and ubiquitination in human lymphatic endothelial cells (HLECs) Cell Mol Biol Lett. 2023;28:13. doi: 10.1186/s11658-022-00410-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu L, Xiao S, Wang Y, Zhu Z, Cao Y, Yang S, et al. Identification of a novel circular RNA circZNF652/miR-486-5p/SERPINE1 signaling cascade that regulates cancer aggressiveness in glioblastoma (GBM) Bioengineered. 2022;13:1411–1423. doi: 10.1080/21655979.2021.2018096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo J, et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. 2019;40:432–445. doi: 10.1016/j.ebiom.2018.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lin Y, Zheng ZH, Wang JX, Zhao Z, Peng TY. Tumor cell-derived exosomal Circ-0072088 suppresses migration and invasion of hepatic carcinoma cells through regulating MMP-16. Front Cell Dev Biol. 2021;9:726323. doi: 10.3389/fcell.2021.726323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Guan X, Zong ZH, Liu Y, Chen S, Wang LL, Zhao Y. circPUM1 promotes tumorigenesis and progression of ovarian cancer by sponging miR-615-5p and miR-6753-5p. Mol Ther Nucleic Acids. 2019;18:882–892. doi: 10.1016/j.omtn.2019.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pan Z, Zheng J, Zhang J, Lin J, Lai J, Lyu Z, et al. A novel protein encoded by exosomal CircATG4B induces oxaliplatin resistance in colorectal cancer by promoting autophagy. Adv Sci (Weinh) 2022;9:e2204513. doi: 10.1002/advs.202204513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zang R, Qiu X, Song Y, Wang Y. Exosomes mediated transfer of Circ_0000337 contributes to cisplatin (CDDP) resistance of esophageal cancer by regulating JAK2 via miR-377-3p. Front Cell Dev Biol. 2021;9:673237. doi: 10.3389/fcell.2021.673237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xie H, Yao J, Wang Y, Ni B. Exosome-transmitted circVMP1 facilitates the progression and cisplatin resistance of non-small cell lung cancer by targeting miR-524-5p-METTL3/SOX2 axis. Drug Deliv. 2022;29:1257–1271. doi: 10.1080/10717544.2022.2057617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Geng X, Zhang Y, Lin X, Zeng Z, Hu J, Hao L, et al. Exosomal circWDR62 promotes temozolomide resistance and malignant progression through regulation of the miR-370-3p/MGMT axis in glioma. Cell Death Dis. 2022;13:596. doi: 10.1038/s41419-022-05056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li J, Zhu T, Weng Y, Cheng F, Sun Q, Yang K, et al. Exosomal circDNER enhances paclitaxel resistance and tumorigenicity of lung cancer via targeting miR-139-5p/ITGB8. Thorac Cancer. 2022;13:1381–1390. doi: 10.1111/1759-7714.14402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tan WQ, Yuan L, Wu XY, He CG, Zhu SC, Ye M. Exosome-delivered circular RNA DLGAP4 induces chemoresistance via miR-143-HK2 axis in neuroblastoma. Cancer Biomark. 2022;34:375–384. doi: 10.3233/CBM-210272. [DOI] [PubMed] [Google Scholar]
- 122.Zeng Z, Zhao Y, Chen Q, Zhu S, Niu Y, Ye Z, et al. Hypoxic exosomal HIF-1α-stabilizing circZNF91 promotes chemoresistance of normoxic pancreatic cancer cells via enhancing glycolysis. Oncogene. 2021;40:5505–5517. doi: 10.1038/s41388-021-01960-w. [DOI] [PubMed] [Google Scholar]
- 123.Chen S, Chen Z, Li Z, Li S, Wen Z, Cao L, et al. Tumor-associated macrophages promote cholangiocarcinoma progression via exosomal Circ_0020256. Cell Death Dis. 2022;13:94. doi: 10.1038/s41419-022-04534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gu X, Shi Y, Dong M, Jiang L, Yang J, Liu Z. Exosomal transfer of tumor-associated macrophage-derived hsa_circ_0001610 reduces radiosensitivity in endometrial cancer. Cell Death Dis. 2021;12:818. doi: 10.1038/s41419-021-04087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tanigaki K, Kuroda K, Han H, Honjo T. Regulation of B cell development by Notch/RBP-J signaling. Semin Immunol. 2003;15:113–119. doi: 10.1016/S1044-5323(03)00008-3. [DOI] [PubMed] [Google Scholar]
- 126.Xu H, Zhu J, Smith S, Foldi J, Zhao B, Chung AY, et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol. 2012;13:642–650. doi: 10.1038/ni.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang L, Zhang J, Li P, Li T, Zhou Z, Wu H. Exosomal hsa_circ_0004658 derived from RBPJ overexpressed-macrophages inhibits hepatocellular carcinoma progression via miR-499b-5p/JAM3. Cell Death Dis. 2022;13:32. doi: 10.1038/s41419-021-04345-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shi L, Cao Y, Yuan W, Guo J, Sun G. Exosomal circRNA BTG2 derived from RBP-J overexpressed-macrophages inhibits glioma progression via miR-25-3p/PTEN. Cell Death Dis. 2022;13:506. doi: 10.1038/s41419-022-04908-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang L, Yi X, Xiao X, Zheng Q, Ma L, Li B. Exosomal miR-628-5p from M1 polarized macrophages hinders m6A modification of circFUT8 to suppress hepatocellular carcinoma progression. Cell Mol Biol Lett. 2022;27:106. doi: 10.1186/s11658-022-00406-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhou WJ, Zhang J, Xie F, Wu JN, Ye JF, Wang J, et al. CD45RO-CD8+ T cell-derived exosomes restrict estrogen-driven endometrial cancer development via the ERβ/miR-765/PLP2/Notch axis. Theranostics. 2021;11:5330–5345. doi: 10.7150/thno.58337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Neviani P, Wise PM, Murtadha M, Liu CW, Wu CH, Jong AY, et al. Natural killer-derived exosomal miR-186 inhibits neuroblastoma growth and immune escape mechanisms. Cancer Res. 2019;79:1151–1164. doi: 10.1158/0008-5472.CAN-18-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen S, Lv M, Fang S, Ye W, Gao Y, Xu Y. Poly(I:C) enhanced anti-cervical cancer immunities induced by dendritic cells-derived exosomes. Int J Biol Macromol. 2018;113:1182–1187. doi: 10.1016/j.ijbiomac.2018.02.034. [DOI] [PubMed] [Google Scholar]
- 133.Xiao H, Lässer C, Shelke GV, Wang J, Rådinger M, Lunavat TR, et al. Mast cell exosomes promote lung adenocarcinoma cell proliferation-role of KIT-stem cell factor signaling. Cell Commun Signal. 2014;12:64. doi: 10.1186/s12964-014-0064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bu L, Yonemura A, Yasuda-Yoshihara N, Uchihara T, Ismagulov G, Takasugi S, et al. Tumor microenvironmental 15-PGDH depletion promotes fibrotic tumor formation and angiogenesis in pancreatic cancer. Cancer Sci. 2022;113:3579–3592. doi: 10.1111/cas.15495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shi L, Zhu W, Huang Y, Zhuo L, Wang S, Chen S, et al. Cancer-associated fibroblast-derived exosomal microRNA-20a suppresses the PTEN/PI3K-AKT pathway to promote the progression and chemoresistance of non-small cell lung cancer. Clin Transl Med. 2022;12:e989. doi: 10.1002/ctm2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang K, Zhang J, Bao C. Exosomal circEIF3K from cancer-associated fibroblast promotes colorectal cancer (CRC) progression via miR-214/PD-L1 axis. BMC Cancer. 2021;21:933. doi: 10.1186/s12885-021-08669-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhan Y, Du J, Min Z, Ma L, Zhang W, Zhu W, et al. Carcinoma-associated fibroblasts derived exosomes modulate breast cancer cell stemness through exonic circHIF1A by miR-580-5p in hypoxic stress. Cell Death Discov. 2021;7:141. doi: 10.1038/s41420-021-00506-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhou Y, Tang W, Zhuo H, Zhu D, Rong D, Sun J, et al. Cancer-associated fibroblast exosomes promote chemoresistance to cisplatin in hepatocellular carcinoma through circZFR targeting signal transducers and activators of transcription (STAT3)/nuclear factor-kappa B (NF-κB) pathway. Bioengineered. 2022;13:4786–4797. doi: 10.1080/21655979.2022.2032972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Maguire OA, Ackerman SE, Szwed SK, Maganti AV, Marchildon F, Huang X, et al. Creatine-mediated crosstalk between adipocytes and cancer cells regulates obesity-driven breast cancer. Cell Metab. 2021;33:499–512.e496. doi: 10.1016/j.cmet.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hochheuser C, Windt LJ, Kunze NY, de Vos DL, Tytgat GAM, Voermans C, et al. Mesenchymal stromal cells in neuroblastoma: exploring crosstalk and therapeutic implications. Stem Cells Dev. 2021;30:59–78. doi: 10.1089/scd.2020.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gao G, Wang L, Li C. Circ_0006790 carried by bone marrow mesenchymal stem cell-derived exosomes regulates S100A11 DNA methylation through binding to CBX7 in pancreatic ductal adenocarcinoma. Am J Cancer Res. 2022;12:1934–1959. [PMC free article] [PubMed] [Google Scholar]
- 142.Yao X, Mao Y, Wu D, Zhu Y, Lu J, Huang Y, et al. Exosomal circ_0030167 derived from BM-MSCs inhibits the invasion, migration, proliferation and stemness of pancreatic cancer cells by sponging miR-338-5p and targeting the Wif1/Wnt8/β-catenin axis. Cancer Lett. 2021;512:38–50. doi: 10.1016/j.canlet.2021.04.030. [DOI] [PubMed] [Google Scholar]
- 143.Zhang C, Cao J, Lv W, Mou H. CircRNA_100395 carried by exosomes from adipose-derived mesenchymal stem cells inhibits the malignant transformation of non-small cell lung carcinoma through the miR-141-3p-LATS2 Axis. Front Cell Dev Biol. 2021;9:663147. doi: 10.3389/fcell.2021.663147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu H, et al. Circular RNAs in body fluids as cancer biomarkers: the new frontier of liquid biopsies. Mol Cancer. 2021;20:13. doi: 10.1186/s12943-020-01298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.He YD, Tao W, He T, Wang BY, Tang XM, Zhang LM, et al. A urine extracellular vesicle circRNA classifier for detection of high-grade prostate cancer in patients with prostate-specific antigen 2–10 ng/mL at initial biopsy. Mol Cancer. 2021;20:96. doi: 10.1186/s12943-021-01388-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Roy S, Kanda M, Nomura S, Zhu Z, Toiyama Y, Taketomi A, et al. Diagnostic efficacy of circular RNAs as noninvasive, liquid biopsy biomarkers for early detection of gastric cancer. Mol Cancer. 2022;21:42. doi: 10.1186/s12943-022-01527-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Huang Y, Qin S, Gu X, Zheng M, Zhang Q, Liu Y, et al. Comprehensive assessment of serum hsa_circ_0070354 as a novel diagnostic and predictive biomarker in non-small cell lung cancer. Front Genet. 2021;12:796776. doi: 10.3389/fgene.2021.796776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hu X, Wu D, He X, Zhao H, He Z, Lin J, et al. circGSK3β promotes metastasis in esophageal squamous cell carcinoma by augmenting β-catenin signaling. Mol Cancer. 2019;18:160. doi: 10.1186/s12943-019-1095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhou C, Liu HS, Wang FW, Hu T, Liang ZX, Lan N, et al. circCAMSAP1 promotes tumor growth in colorectal cancer via the miR-328-5p/E2F1 axis. Mol Ther. 2020;28:914–928. doi: 10.1016/j.ymthe.2019.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zhang B, Chen M, Cao J, Liang Y, Tu T, Hu J, et al. An integrated electrochemical POCT platform for ultrasensitive circRNA detection towards hepatocellular carcinoma diagnosis. Biosens Bioelectron. 2021;192:113500. doi: 10.1016/j.bios.2021.113500. [DOI] [PubMed] [Google Scholar]
- 151.Dong J, Zeng Z, Sun R, Zhang X, Cheng Z, Chen C, et al. Specific and sensitive detection of CircRNA based on netlike hybridization chain reaction. Biosens Bioelectron. 2021;192:113508. doi: 10.1016/j.bios.2021.113508. [DOI] [PubMed] [Google Scholar]
- 152.Cheng L, Yang F, Zhao Y, Liu Z, Yao X, Zhang J. Tetrahedron supported CRISPR/Cas13a cleavage for electrochemical detection of circular RNA in bladder cancer. Biosens Bioelectron. 2023;222:114982. doi: 10.1016/j.bios.2022.114982. [DOI] [PubMed] [Google Scholar]
- 153.Xian J, Su W, Liu L, Rao B, Lin M, Feng Y, et al. Identification of three circular RNA cargoes in serum exosomes as diagnostic biomarkers of non-small-cell lung cancer in the chinese population. J Mol Diagn. 2020;22:1096–1108. doi: 10.1016/j.jmoldx.2020.05.011. [DOI] [PubMed] [Google Scholar]
- 154.Lyu L, Yang W, Yao J, Wang H, Zhu J, Jin A, et al. The diagnostic value of plasma exosomal hsa_circ_0070396 for hepatocellular carcinoma. Biomark Med. 2021;15:359–371. doi: 10.2217/bmm-2020-0476. [DOI] [PubMed] [Google Scholar]
- 155.Kang Y, You J, Gan Y, Chen Q, Huang C, Chen F, et al. Serum and serum exosomal CircRNAs hsa_circ_0001492, hsa_circ_0001439, and hsa_circ_0000896 as diagnostic biomarkers for lung adenocarcinoma. Front Oncol. 2022;12:912246. doi: 10.3389/fonc.2022.912246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Xia D, Gu X. Plasmatic exosome-derived circRNAs panel act as fingerprint for glioblastoma. Aging (Albany NY) 2021;13:19575–19586. doi: 10.18632/aging.203368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Luo Y, Ma J, Liu F, Guo J, Gui R. Diagnostic value of exosomal circMYC in radioresistant nasopharyngeal carcinoma. Head Neck. 2020;42:3702–3711. doi: 10.1002/hed.26441. [DOI] [PubMed] [Google Scholar]
- 158.Lin L, Cai GX, Zhai XM, Yang XX, Li M, Li K, et al. Plasma-derived extracellular vesicles circular RNAs serve as biomarkers for breast cancer diagnosis. Front Oncol. 2021;11:752651. doi: 10.3389/fonc.2021.752651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chen C, Yu H, Han F, Lai X, Ye K, Lei S, et al. Tumor-suppressive circRHOBTB3 is excreted out of cells via exosome to sustain colorectal cancer cell fitness. Mol Cancer. 2022;21:46. doi: 10.1186/s12943-022-01511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ishola AA, Chien CS, Yang YP, Chien Y, Yarmishyn AA, Tsai PH, et al. Oncogenic circRNA C190 promotes non-small cell lung cancer via modulation of the EGFR/ERK pathway. Cancer Res. 2022;82:75–89. doi: 10.1158/0008-5472.CAN-21-1473. [DOI] [PubMed] [Google Scholar]
- 161.Ai Y, Liang D, Wilusz JE. CRISPR/Cas13 effectors have differing extents of off-target effects that limit their utility in eukaryotic cells. Nucleic Acids Res. 2022;50:e65. doi: 10.1093/nar/gkac159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zeng W, Liu Y, Li WT, Li Y, Zhu JF. CircFNDC3B sequestrates miR-937-5p to derepress TIMP3 and inhibit colorectal cancer progression. Mol Oncol. 2020;14:2960–2984. doi: 10.1002/1878-0261.12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fan X, Yang Y, Chen C, Wang Z. Pervasive translation of circular RNAs driven by short IRES-like elements. Nat Commun. 2022;13:3751. doi: 10.1038/s41467-022-31327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chen R, Wang SK, Belk JA, Amaya L, Li Z, Cardenas A, et al. Engineering circular RNA for enhanced protein production. Nat Biotechnol. 2023;41:262–272. doi: 10.1038/s41587-022-01393-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lu D, Chatterjee S, Xiao K, Riedel I, Huang CK, Costa A, et al. A circular RNA derived from the insulin receptor locus protects against doxorubicin-induced cardiotoxicity. Eur Heart J. 2022;43:4496–4511. doi: 10.1093/eurheartj/ehac337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, et al. Sensing self and foreign circular RNAs by intron identity. Mol Cell. 2017;67:228–238.e225. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, et al. N6-methyladenosine modification controls circular RNA immunity. Mol Cell. 2019;76:96–109.e109. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Liu CX, Guo SK, Nan F, Xu YF, Yang L, Chen LL. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol Cell. 2022;82:420–434.e426. doi: 10.1016/j.molcel.2021.11.019. [DOI] [PubMed] [Google Scholar]
- 170.Tai J, Chen YG. Differences in the immunogenicity of engineered circular RNAs. J Mol Cell Biol. 2023. [DOI] [PMC free article] [PubMed]
- 171.Holdt LM, Kohlmaier A, Teupser D. Circular RNAs as therapeutic agents and targets. Front Physiol. 2018;9:1262. doi: 10.3389/fphys.2018.01262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li C, Wang Y, Li PF, Fu Q. Construction of rolling circle amplification products-based pure nucleic acid nanostructures for biomedical applications. Acta Biomater. 2023;160:1–13. doi: 10.1016/j.actbio.2023.02.005. [DOI] [PubMed] [Google Scholar]
- 173.Liu GW, Guzman EB, Menon N, Langer RS. Lipid nanoparticles for nucleic acid delivery to endothelial cells. Pharm Res. 2023;40:3–25. doi: 10.1007/s11095-023-03471-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.He AT, Liu J, Li F, Yang BB. Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal Transduct Target Ther. 2021;6:185. doi: 10.1038/s41392-021-00569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Li H, Peng K, Yang K, Ma W, Qi S, Yu X, et al. Circular RNA cancer vaccines drive immunity in hard-to-treat malignancies. Theranostics. 2022;12:6422–6436. doi: 10.7150/thno.77350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Qu L, Yi Z, Shen Y, Lin L, Chen F, Xu Y, et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell. 2022;185:1728–1744.e1716. doi: 10.1016/j.cell.2022.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Meng H, Li R, Xie Y, Mo Z, Zhai H, Zhang G, et al. Nanoparticles mediated circROBO1 silencing to inhibit hepatocellular carcinoma progression by modulating miR-130a-5p/CCNT2 Axis. Int J Nanomed. 2023;18:1677–1693. doi: 10.2147/IJN.S399318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yu C, Li L, Hu P, Yang Y, Wei W, Deng X, et al. Recent advances in stimulus-responsive nanocarriers for gene therapy. Adv Sci (Weinh) 2021;8:2100540. doi: 10.1002/advs.202100540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.You S, Luo Z, Cheng N, Wu M, Lai Y, Wang F, et al. Magnetically responsive nanoplatform targeting circRNA circ_0058051 inhibits hepatocellular carcinoma progression. Drug Deliv Transl Res. 2023;13:782–794. doi: 10.1007/s13346-022-01237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lu HH, Huang CH, Shiue TY, Wang FS, Chang KK, Chen Y, et al. Highly efficient gene release in spatiotemporal precision approached by light and pH dual responsive copolymers. Chem Sci. 2019;10:284–292. doi: 10.1039/C8SC01494A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wu J, Huang J, Kuang S, Chen J, Li X, Chen B, et al. Synergistic MicroRNA therapy in liver fibrotic rat using MRI-visible nanocarrier targeting hepatic stellate cells. Adv Sci (Weinh) 2019;6:1801809. doi: 10.1002/advs.201801809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Xu Z, Zeng S, Gong Z, Yan Y. Exosome-based immunotherapy: a promising approach for cancer treatment. Mol Cancer. 2020;19:160. doi: 10.1186/s12943-020-01278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shao J, Zaro J, Shen Y. Advances in exosome-based drug delivery and tumor targeting: from tissue distribution to intracellular fate. Int J Nanomed. 2020;15:9355–9371. doi: 10.2147/IJN.S281890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chen J, Cao F, Cao Y, Wei S, Zhu X, Xing W. Targeted therapy of lung adenocarcinoma by the nanoplatform based on milk exosomes loaded with paclitaxel. J Biomed Nanotechnol. 2022;18:1075–1083. doi: 10.1166/jbn.2022.3278. [DOI] [PubMed] [Google Scholar]
- 186.Wu P, Tang Y, Jin C, Wang M, Li L, Liu Z, et al. Neutrophil membrane engineered HucMSC sEVs alleviate cisplatin-induced AKI by enhancing cellular uptake and targeting. J Nanobiotechnol. 2022;20:353. doi: 10.1186/s12951-022-01574-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Guo Z, Zhang Y, Xu W, Zhang X, Jiang J. Engineered exosome-mediated delivery of circDIDO1 inhibits gastric cancer progression via regulation of MiR-1307-3p/SOCS2 Axis. J Transl Med. 2022;20:326. doi: 10.1186/s12967-022-03527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yang L, Han B, Zhang Z, Wang S, Bai Y, Zhang Y, et al. Extracellular vesicle-mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation. 2020;142:556–574. doi: 10.1161/CIRCULATIONAHA.120.045765. [DOI] [PubMed] [Google Scholar]
- 189.Chen Q, Zu M, Gong H, Ma Y, Sun J, Ran S, et al. Tea leaf-derived exosome-like nanotherapeutics retard breast tumor growth by pro-apoptosis and microbiota modulation. J Nanobiotechnol. 2023;21:6. doi: 10.1186/s12951-022-01755-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Seo K, Yoo JH, Kim J, Min SJ, Heo DN, Kwon IK, et al. Ginseng-derived exosome-like nanovesicles extracted by sucrose gradient ultracentrifugation to inhibit osteoclast differentiation. Nanoscale. 2023;15:5798–5808. doi: 10.1039/D2NR07018A. [DOI] [PubMed] [Google Scholar]
- 191.Pomatto MAC, Gai C, Negro F, Massari L, Deregibus MC, Grange C, et al. Plant-derived extracellular vesicles as a delivery platform for RNA-based vaccine: feasibility study of an oral and intranasal SARS-CoV-2 vaccine. Pharmaceutics. 2023;15:974. doi: 10.3390/pharmaceutics15030974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.