Abstract
Vaccines are regarded as the most cost-effective countermeasure against infectious diseases. One challenge often affecting vaccine development is antigenic diversity or pathogen heterogeneity. Different strains produce immunologically heterogenous virulence factors, therefore an effective vaccine needs to induce broad-spectrum host immunity to provide cross protection. Recent advances in genomics and proteomics, particularly computational biology and structural biology, establishes structural vaccinology and highlights the feasibility of developing effective and precision vaccines. Here, we introduce the epitope- and structure-based vaccinology platform multiepitope-fusion-antigen (MEFA), and provide instructions to generate multivalent MEFA immunogens for vaccine development. Conceptually, MEFA combines epitope vaccinology and structural vaccinology to enable a protein immunogen to present heterogenous antigenic domains (epitopes) and to induce broadly protective immunity against different virulence factors, strains or diseases. Methodologically, the MEFA platform first identifies a safe, structurally stable and strongly immunogenic backbone protein and immunodominant (ideally neutralizing or protective) epitopes from heterogenous strains or virulence factors of interest. Then, assisted with protein modeling and molecule dynamic simulation, MEFA integrates heterogeneous epitopes into a backbone protein via epitope substitution for a multivalent MEFA protein and mimics epitope native antigenicity. Finally, the MEFA protein is examined for broad immunogenicity in animal immunization, and assessed for potential application for multivalent vaccine development in preclinical studies.
Keywords: MEFA (multiepitope-fusion-antigen), vaccinology platform, multivalent vaccine, epitopes, structural vaccinology
1. Introduction
Infectious diseases are a leading cause of death and a major threat to global health [1,2]. Vaccines are considered the most cost-effective countermeasure to reduce human mortality and morbidity, to eliminate pandemic or epidemic risk, and to lower antibiotic use. Success in vaccine and vaccination against infectious diseases includes the eradication of smallpox and the control of polio, measles, mumps and diphtheria. However, major challenges remain in developing vaccines for other diseases including human immunodeficiency virus (HIV), diarrhea caused by viral, bacterial or parasitic pathogens, tuberculosis (TB) and malaria which continuously claim millions of human lives annually [3,4]. While a lack of clear understanding of host pathogen interactions and difficulties in identifying immune biomarkers or immune correlates of protection are commonly major challenges in vaccine development, antigenic diversities and immunological heterogeneity among pathogen strains or virulence factors have also severely hampered development of effective vaccines against certain infectious diseases. Strategies including conserved antigen and cocktail vaccines have attempted to overcome antigenic diversity or immunological heterogeneity, but they often encounter bottlenecks at improving protective efficacy. Different from conventional approaches, we introduce a new vaccinology platform called multiepitope-fusion-antigen, MEFA [5,6], to construct multivalent immunogens and to develop broadly protective vaccines. Benefiting from recent advance in genomics and proteomics, particularly computational biology and structural biology, MEFA combines epitope vaccinology and structural vaccinology concepts to generate epitope- and structure-based multivalent protein immunogens for developing cross protective vaccines against heterogeneous strains, pathogens or diseases [7].
The MEFA platform enables integration of foreign epitopes, ideally protective or neutralizing epitopes (epitopes induce protective immune responses) of various pathogenic strains or virulence factors, into a backbone protein and mimicking of epitope native antigenicity, thus constructs a multivalent immunogen for broadly protective immunity. In principle, a protective epitope, if it retains native antigenicity after being integrated into a backbone protein, induces protective host immunity against the representing virulence factor or strain. Therefore, an MEFA immunogen carrying multiple protective epitopes from different strains serves as a multivalent antigen for a cross protective vaccine. By substituting backbone protein epitopes with protective epitopes (of heterogeneous strains or virulence factors) of interest, we produce a chimeric multivalent immunogen (Fig. 1), and by applying protein modeling and molecular dynamic simulation, we mimic epitope native antigenicity. Mechanistically, MEFA vaccinology platform combines novel in silico analyses with conventional in vivo empirical studies to accelerate development of broadly protective multivalent vaccines. First, an MEFA backbone immunogen and epitopes of interest (from heterogeneous strains or virulence factors) are identified, and an MEFA immunogen is constructed in silico, followed by synthesis and cloning of an MEFA gene into an expression vector. Then, the MEFA protein is expressed, extracted and used for animal immunization studies. Finally, the MEFA protein-induced broad-spectrum immune responses are measured, and application of an MEFA immunogen for multivalent vaccine development is evaluated in preclinical investigation prior to human subject safety, immunogenicity and efficacy studies.
Figure 1.
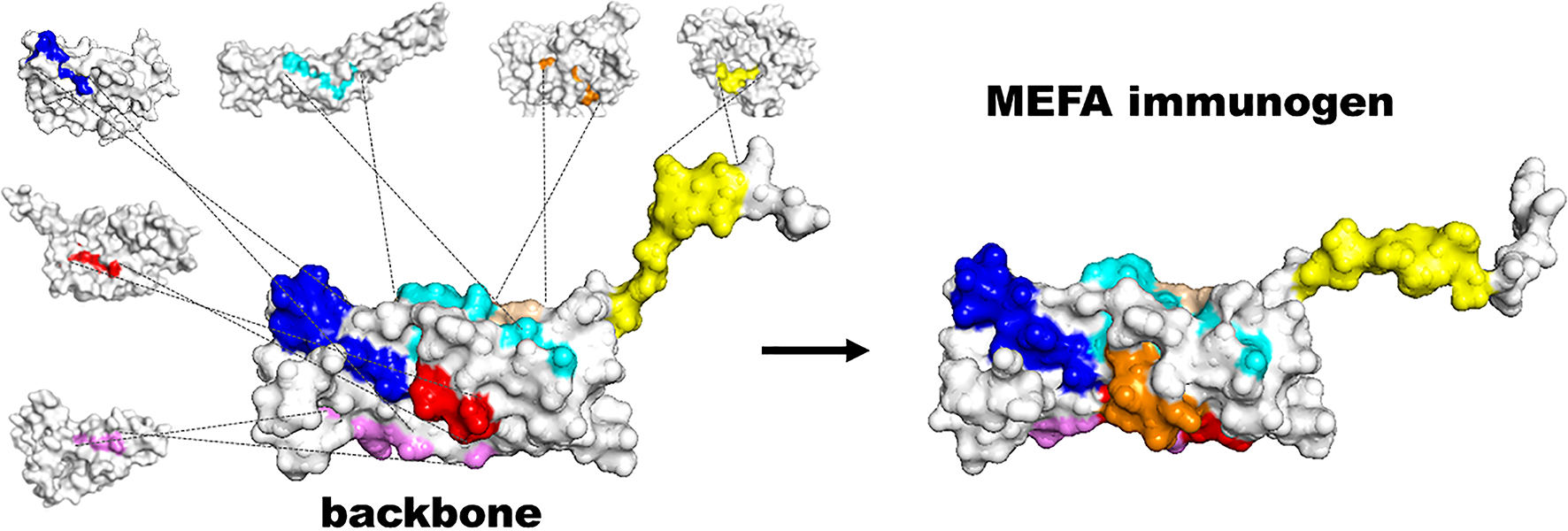
Illustration of constructing a multivalent MEFA immunogen by using the MEFA vaccinology platform. Surface-exposed epitopes of a backbone immunogen are substituted with protective or immunodominant epitopes from heterogeneous virulence factors or strains of interest to constrict an MEFA immunogen.
2. Materials
2.1. Computation and programs
MEFA backbone protein selection, epitope identification, and protein modeling are carried out on standard desktop or laptop computers through open-access servers. Atomic molecular dynamics simulation requires a local server and remote access to license-based CHARMM server or open-source GROMACS-5.0.7 MD server.
Programs or servers that are applied for in silico analyses are:
IEDB (http://www.iedb.org/) or BepiPred (http://www.cbs.dtu.dk/services/BepiPred/).
ExPASy (https://www.expasy.org/).
PyMol (https://www.pymol.org/).
Phyre2 (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index).
GROMACS (http://www.gromacs.org/).
Or CHARMM (https://www.charmm.org/), and MMTSB toolset (http://blue11.bch.msu.edu/mmtsb/Main_Page).
2.2. A backbone protein and heterogeneous epitopes of interest
The MEFA platform requires a backbone protein and epitopes of heterogeneous strains or virulence factors of interest to construct an MEFA immunogen. A backbone protein should be safe and structurally stable, preferably a mutant key virulence factor which becomes avirulent after epitope substitution and induces protective immunity (housekeeping proteins may not be desirable since they potentially induce host immune tolerance or immunity against normal flora organisms). A backbone protein needs to be strongly immunogenic, possesses multiple well-separated continuous epitopes, and ideally exhibits systemic and mucosal adjuvanticity. Additionally, a backbone protein can be easily expressed in an expression vector and an Escherichia coli strain and extracted at a great purity and in high yield. In the case there is no suitable backbone proteins available from a target pathogen, mutant cholera toxin (CT) of Vibrio cholerae and heat-labile toxin (LT) of enterotoxigenic E. coli, which have been demonstrated with little or low enterotoxicity but strong immunogenicity and mucosal and systemic adjuvanticity [8–11], can serve as alternative backbone proteins.
Epitopes from heterogeneous strains or virulence factors of interest are presented on an identified backbone protein. Preferably, one epitope represents one strain or one virulence factor and induces protective immune responses against its representing strain or virulence factor. With several protective epitopes integrated into a backbone immunogen, an MEFA immunogen induces broad immunity against multiple heterogeneous strains or virulence factors. Neutralizing or functionally protective epitopes which have been characterized are available at databases including Immune Epitope Database - IEDB (www.iedb.org). However, in many cases, protective epitopes likely have not been identified. Alternatively, immunodominant epitopes which are identified in silico can be used initially. Eventually, as we recently described [12–15], immunodominant epitopes are screened in empirical studies and protective epitopes are selected for MEFA immunogen construction.
2.3. MEFA immunogen construction, expression and extraction
pET28a or other expression vectors, to clone an MEFA immunogen gene.
E. coli strain BL21-CodonPlus (DE3) or other E. coli strains, to host MEFA plasmids for MEFA protein expression.
LB agar plates and 2x YT medium broth.
Kanamycin, 30 μg per mL in final (for pET28a).
IPTG (isopropyl-β-D-thiogalactopyranoside).
B-PER, Bacterial Protein Extraction Reagent (in phosphate buffer; solution contains a proprietary, nonionic detergent in 20 mM Tris-HCl; pH 7.5).
Ni-NTA Agarose, Ni-NTA columns, wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, 0.05% Tween 20, pH 8.0), elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, 0.05% Tween 20, pH 8.0), to extract 6x His-tagged recombinant protein (see Note 1).
Spectrum Spectra/Por dialysis membrane tubing, at suitable pore sizes depending on MEFA protein molecular mass (5–10 kDa less than the MEFA protein molecule mass).
1x IB solubilization buffer: 50mM CAPS, pH 11.0, supplemented with 1% N-lauroylsarcosine and 1 mM DTT.
Protein refolding and dialysis buffer: 20 mM Tris-HCl, pH 8.5, supplemented with 0.1 mM DTT or without DTT.
2.4. Protein biochemical property and immunogenicity characterization
SDS-PAGE, for MEFA protein Coomassie blue stain or Western blot with specific antibodies.
Coomassie blue stain buffer (0.125% Coomassie blue R350, 50% methanol, 10% acetic acid), and destain buffer (20% methanol, 10% acetic acid, in ddH2O).
Protein immunogenicity studies: MEFA protein-induced T cell and/or B cell immune responses are examined in animal immunization studies. Antigen specific adaptive immunity and in vitro protective activities, which vary and are specific to the pathogen of interest, are measured to assess MEFA immunogen broad immunity (B cell epitopes are used for MEFA immunogen construction in this protocol).
Animal immunization: 8-week old mice, 25-gauge needle and 1-mL syringe, alum or incomplete Freund’s adjuvant; mouse fecal and serum samples collected for measuring antigen-specific antibody responses.
Antibody titration: 96-well microtiter plates, ELISA coating antigens specific to the virulence factors targeted by MEFA protein, HRP-conjugated goat-anti-mouse IgG and IgA secondary antibodies, 3,3’,5,5’-tetramethylbezidine (TMB) Microwell Peroxidase Substrate, a 96-well plate reader.
Antibody in vitro protection: assays to evaluate in vitro protection of MEFA-induced antibodies vary and are specific to pathogens or virulence factors of interest.
Preclinical challenge studies: if animal challenge models are available, immunizing animals with an MEFA protein to assess epitope-specific immunogenicity, then challenging the immunized animals with pathogen strains to measure cross protection and to evaluate potential application of an MEFA immunogen for multivalent vaccine development.
3. Methods
3.1. Backbone protein selection
Online programs that predict epitopes, locate epitope positions, image protein secondary structure and examine protein stability are applied to select a suitable backbone protein. B cell epitopes on a backbone protein, including epitope amino acid sequence, position and antigenic score, are predicted and analyzed by using immune epitope database and analysis resource server IEDB [16]. Backbone protein half-life and stability are evaluated with ExPASy ProtParam [17]. Protein homology/analogy recognition engine version 2.0 - Phyre2 [18] and PyMol are used to generate backbone protein 3-D structure images and to show epitope presentation on a backbone protein (see Note 2).
3.1.1. To predict backbone immunogen epitopes
Open IEDB www.iedb.org/.
Select B-cell Tools.
Click ‘Prediction of linear epitopes from protein sequence’ for B cell epitope prediction.
Enter protein Swiss-Prot ID or input a backbone protein amino acid sequence, select ‘Bepipred Linear Epitope Prediction 2.0’ for B-cell epitopes, and submit to proceed epitope prediction.
-
Identified epitopes are presented in a graph and tables (Fig. 2).
(see Note 3).
Figure 2.
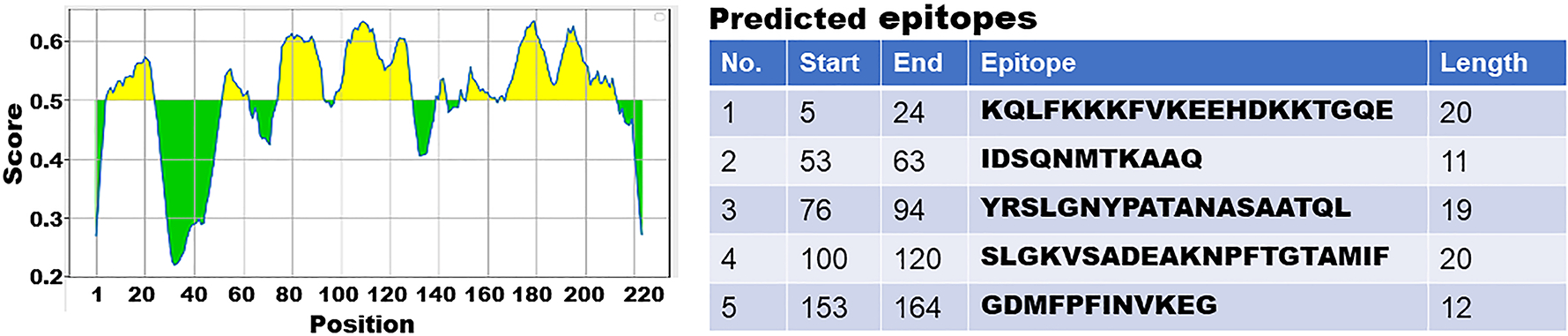
Illustration of in silico identifying B cell immunodominant epitopes with IEDB program. Left: a diagram to show in silico predicted epitopes, with antigenic scores and locations, from a backbone protein. Right: a table to indicate epitope position, sequence and length.
3.1.2. To examine backbone protein stability
Open ProtParam tool link at ExPASy (https://web.expasy.org/ProtParam/).
Enter backbone protein Swiss-Prot ID or accession number (if protein is deposited in ExPASy database); alternatively, input protein amino acid sequence as directed.
Press ‘computer parameters’ command to proceed computation.
-
Retrieve data of protein molecular weight, extinction coefficients, estimated protein half-life, and protein instability index.
(see Note 4).
3.1.3. To generate backbone protein model and to illustrate protein 3-D images and epitopes
Open Phyre2 site http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index.
Type in your e-mail address (to receive analysis updates) in a designate box shown on the window.
Input protein amino acid sequence and name your optional job description.
Select ‘Intensive’ as the modeling and analysis mode.
Press ‘Phyre Search’.
Receive e-mail notices of computation updates, results in a ‘xxx.pdb file format’ (xxx is the name of your optional job description; the confidence in the intensive model should be greater than 90%), and a link to view your protein model and 3-D structure image.
Download your ‘xxx.pdb’ file from Phyre2 site, view protein structure, and further analyze ‘xxx.pdb’ data for 3-D structure with PyMol/ EdiPyMol.
Download PyMol (https://www.pymol.org/).
Open your Phyre2 file ‘xxx.pdb’ through PyMol program by using ‘File’ → ‘Open’ → ‘Downloads’ by selecting your ‘xxx.pdb file.
Select the ‘Display’ tab on the top to view protein amino acid sequence.
Select the ‘S’ tab to show ‘surface’ at the right for display type of the secondary structure in a 3-D model, and the ‘C’ for a uniform color for the backbone protein model (Fig. 3A).
-
Mark epitope amino acid sequences in different colors to image the location and surface exposure of each epitope (Fig. 3B).
(see Note 5).
Figure 3.
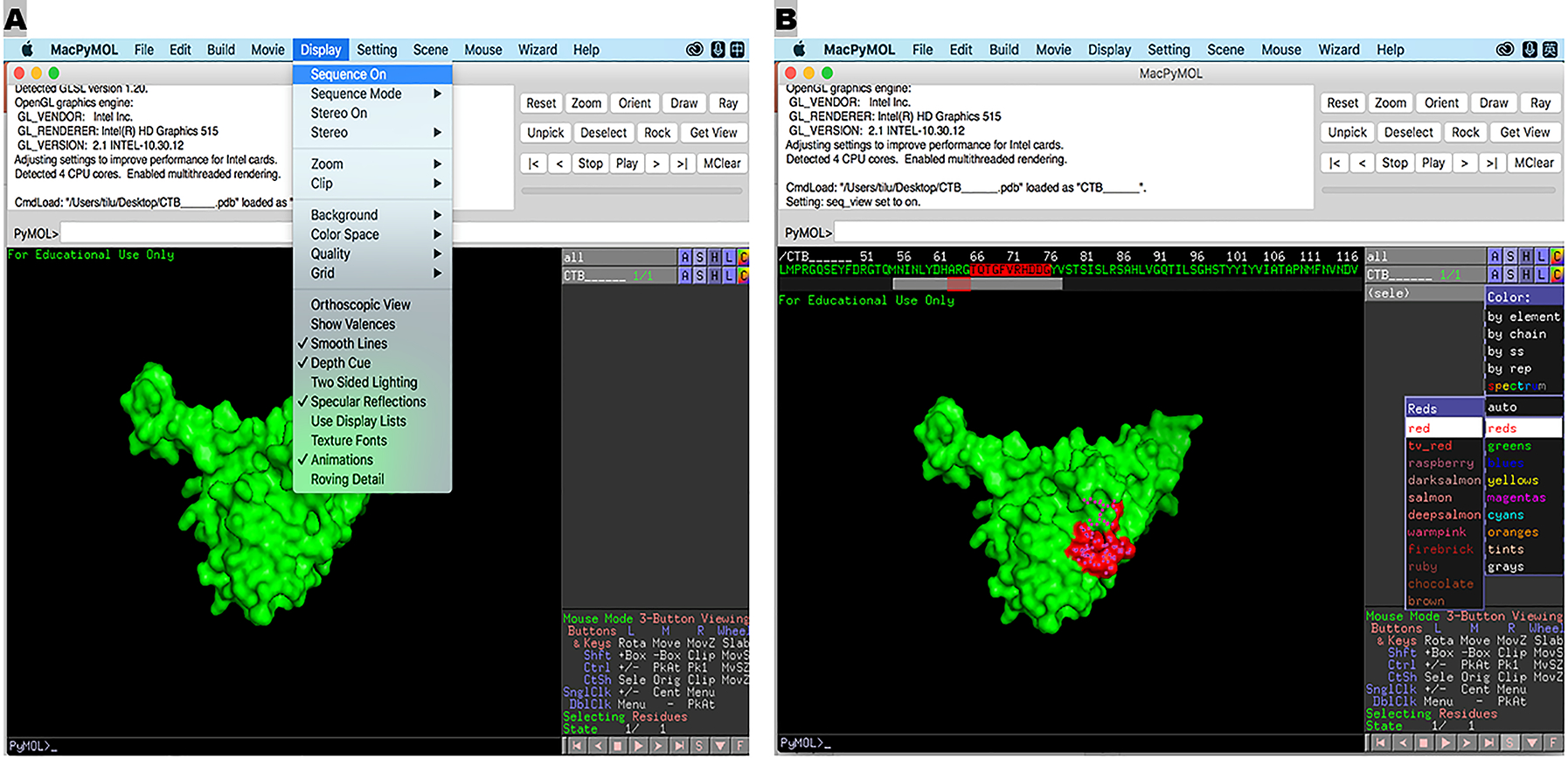
Illustration of a backbone protein 3-D model (A) and position of a backbone epitope (B), by PyMol program.
3.2. Identification of epitopes from target heterogeneous strains or virulence factors
With a desirable backbone immunogen selected, epitopes from the heterogeneous strains or virulence factors of interest must be identified. Protective epitopes are ideal for MEFA immunogen construction. Having epitope native antigenicity retained, an MEFA immunogen carrying multiple protective epitopes of various strains or virulence factors induces broadly protective immune responses. As pointed out in 3.1, protective epitopes from well characterized pathogens are deposited at IEDB (www.iedb.org) and can be used directly for MEFA immunogen construction. For strains or virulence factors without protective epitopes being identified, immunodominant epitopes in silico identified can be used first; eventually protective epitopes selected by screening immunodominant ones in empirical studies are included in an MEFA immunogen.
3.2.1. To predict epitopes of heterogeneous strains or virulence factors of interest
Immunodominant linear epitopes from each virulence factor or strain are predicted with IEDB, the same as backbone protein epitope predication described in 3.1.1, with the only difference is the input of different amino acid sequences.
3.2.2. To identify protective epitopes
Immunodominant epitopes identified in silico may not necessarily induce protective immune responses. Practically, empirical studies are followed to screen immunodominant epitopes and to identify the protective epitopes. A panel of immunodominant epitopes from a virulence factor or strain is selected (based on antigenic scores), and each epitope is genetically fused to a carrier protein for epitope fusions (to mimic epitope native antigenicity). With each epitope fusion gene cloned in an expression vector, individual epitope fusion proteins are extracted and used for mouse immunization. As we described [12–14], epitope fusion-induced immune responses are measured in ELISAs with specific coating antigens to confirm immune dominance, and then examined for protection against the virulence factor or disease with in vitro assays. Epitopes in the fusion proteins that induce protective immune responses are determined as protective.
3.3. MEFA immunogen construction
Having a backbone protein and protective (or immunodominant) epitopes from heterogeneous strains or virulence factors identified, we are ready in silico to construct and to optimize an MEFA immunogen. Substitution of backbone epitopes with selecting protective epitopes (or immunodominant epitopes if protective ones are not identified) is carried out either sequentially, substituting one epitope at a time, or with all epitopes replaced at once and modified afterwards.
To substitute epitopes sequentially, we select an epitope of interest that possesses a similar length and antigenic score as a backbone epitope, proceed the substitution, construct a chimeric protein, and examine the resultant protein comparatively with the backbone protein for protein stability in ExPASy as in 3.1.2 and for epitope presentation with Phyre2 and PyMol as in 3.1.3. If the top ranked epitope from a virulence factor or strain does not match well with a backbone epitope, we select the second or the third ranked epitope whichever shows better match with the backbone epitope. With assessment of no major alteration at protein stability and secondary structure, we proceed substitution of the next epitope and continue the process till all epitopes are substituted. Alternatively, based on epitope length and antigenic score, we pairwise epitopes of interest with backbone epitopes and complete epitope substitution at once. The resultant MEFA protein is examined with ExPASy, Phyre2 and PyMol for MEFA protein stability, protein secondary structure, and epitope presentation. If protein stability and structure are altered significantly, we adjust epitope pairing and optimize epitope substitution.
MEFA protein is further characterized with molecular dynamic simulation program from GROMACS or CHARMM as we described previously [6]. With comparative analyses of protein dynamic properties, including protein structure stability, conformational flexibility and solvent-accessible surface, of an MEFA protein with a backbone protein, MEFA protein overall structure and stability similarity is examined. With analyses of dynamic simulation focusing on an epitope on the MEFA versus its native format on a virulence factor protein, mimicking of epitope native antigenicity is evaluated.
3.3.1. GROMACS to characterize an MEFA protein
Download and install SSH client from link https://www.netsarang.com/en/all-downloads/ (Xshell for Windows OS); log in from your desktop as an SSH client to connect remote server (Linux OS) and execute commands (note: more information at https://netsarang.atlassian.net/wiki/spaces/ENSUP/pages/31555780/Getting+Started).
Download and install GROMACS (http://www.gromacs.org/) and FFTW (version 3.3.8) (http://www.fftw.org/download.html).
Input ‘MEFA protein (or backbone or each virulence factor).pdb’ file into Xshell generated from Phyre2 under your server account.
Generate an MEFA protein topology form the input pdb.file, with command ‘gmx pdb2gmx -f MEFA protein.pdb -o mefa_processed.gro -water spce’, with the selection of ‘GROMOS96 53a6 force field’.
Select periodic boundary conditions (PBC) at ‘cubic’ shape and ‘1.0 nm’ parameters with ‘gmx editconf -f mefa_processed.gro -o mefa_newbox.gro -c -d 1.0 -bt cubic’.
Enter solvent water into the simulation box with ‘gmx solvate -cp mefa_newbox.gro -cs spc216.gro -o mefa_solv.gro -p topol.top’.
Conduct system neutralization with command ‘gmx grompp -f ions.mdp -c mefa_solv.gro -p topol.top -o ions.tpr’, add counter ions through ‘genion’ with ‘gmx genion -s ions.tpr -o mefa_solv_ions.gro -p topol.top -pname NA -nname CL -np 9’.
Minimize energy with commands ‘gmx grompp -f minim.mdp -c mefa_solv_ions.gro -p topol.top -o em.tpr’ and ‘gmx mdrun -v -deffnm em’.
Equilibrate NVT with ‘gmx grompp -f nvt.mdp -c em.gro -p topol.top -o nvt.tpr’ and ‘gmx mdrun -deffnm nvt’.
Equilibrate NPT with command ‘gmx grompp -f npt.mdp -c nvt.gro -t nvt.cpt -p topol.top -o npt.tpr gmx mdrun -deffnm npt’.
Carry out molecule dynamics (MD) simulations, with ‘gmx grompp -f md.mdp -c npt.gro -t npt.cpt -p topol.top -o md_0_1.tpr’ and ‘gmx mdrun -deffnm md_0_1’.
Calculate Cα root-mean-square deviation (RMSD) and root-mean-square fluctuations (RMSF) (Fig. 4), with commands ‘gmx trjconv -s md_0_1.tpr -f md_0_1.xtc -o md_0_1_noPBC.xtc -pbc mol -ur compact’, ‘gmx rms -s md_0_1.tpr -f md_0_1.trr -o rmsd.xvg’ and ‘gmx rmsf -s md_0_1.tpr -f md_0_1.xtc -o rmsf.xvg’ respectively.
Calculate solvent-accessible surface areas (SASA) (Fig. 5) with ‘gmx sasa -s md_0_1.tpr -f md_0_1.trr -o area.xvg -or -oa’.
-
Compare results of the MEFA versus backbone, and each epitope on the MEFA versus the native epitope on each virulence factor, to evaluate MEFA protein construction in silico (if overall protein structure or epitope native antigenicity is significantly altered, MEFA is optimized by modifying epitope position or selecting a different epitope from the representing virulence factor).
(see Note 6).
Figure 4.
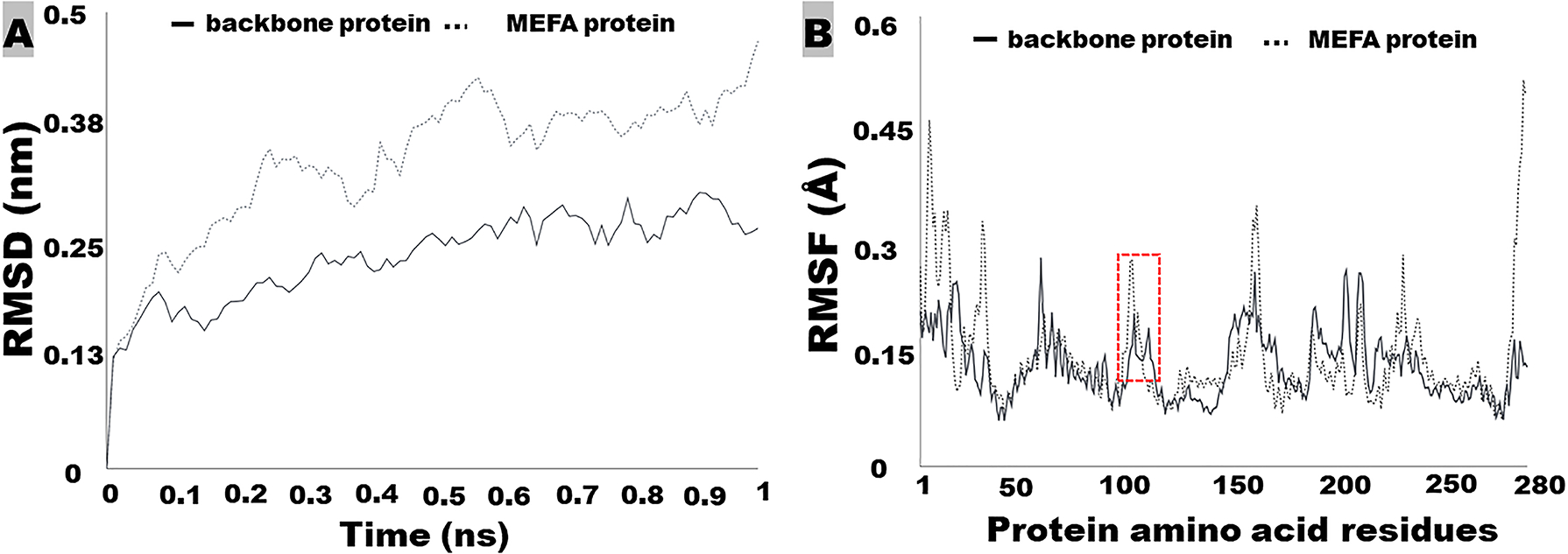
GROMOS molecule dynamic simulation to show rood-mean-square deviation - RMSD (A) and root-mean-square fluctuation – RMSF (B) of a backbone protein and an MEFA immunogen protein. RMSD and RMSF are used to assess structural and antigenic similarity between a backbone protein and an MEFA protein, or between an epitope on the MEFA and the epitope on the native virulence factor protein. Boxed is a backbone protein epitope and a substituting epitope on an MEFA protein.
Figure 5.
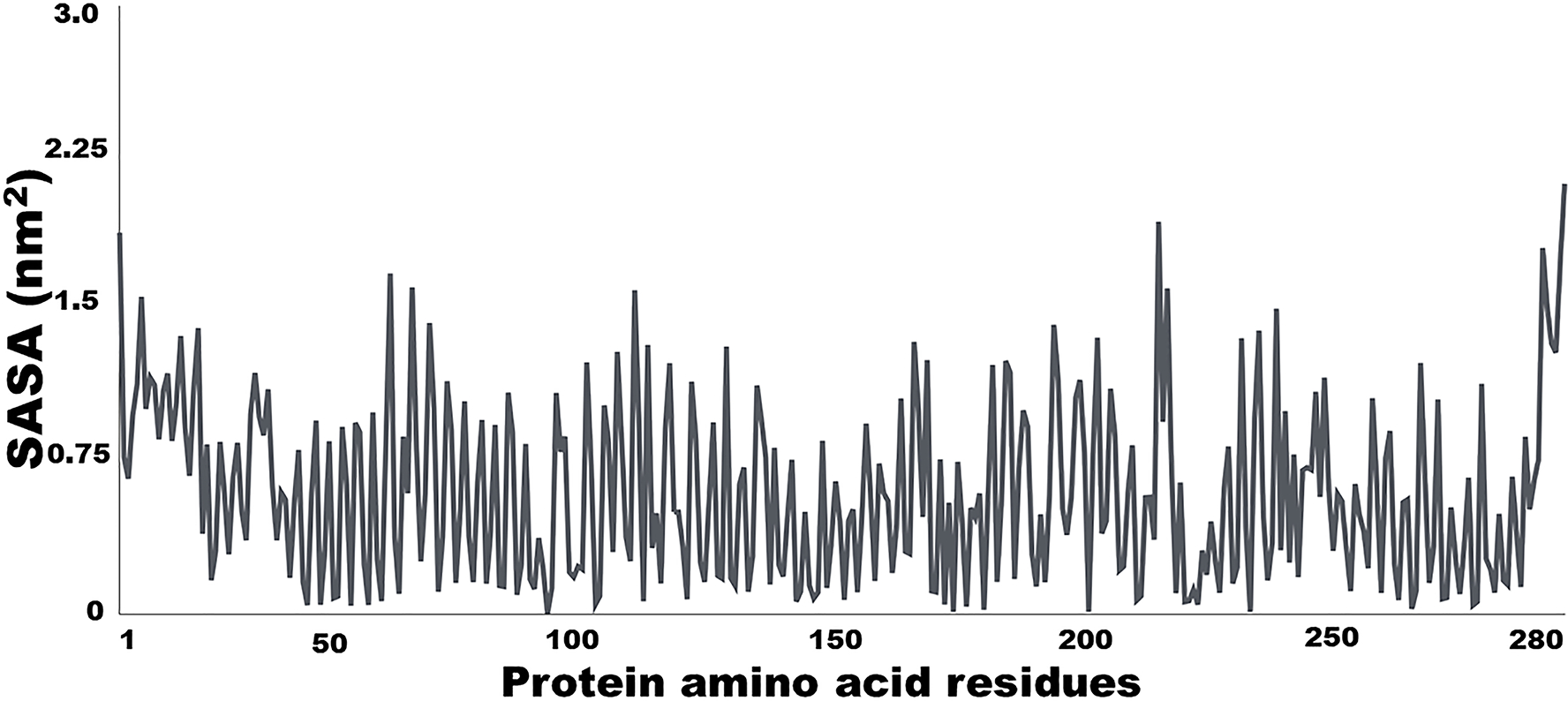
GROMOS molecule dynamic simulation to show solvent average surface area (SASA) of a backbone protein (or an MEFA immunogen protein, or the epitope of interest on an MEFA protein or native virulence factor protein). SASA is used to assess structural and antigenic similarity between a backbone protein and an MEFA protein, or between an epitope on the MEFA and the epitope on the native virulence factor protein.
3.3.2. CHARMM to characterize MEFA protein
Alternatively, an MEFA protein is examined with license based CHARMM, by constructing protein comparative modeling using Rosetta [19–21], carrying out molecular dynamics (MD) simulation, and analyzing protein secondary structure.
Download and install MMTSB toolset (http://blue11.bch.msu.edu/mmtsb/Main_Page).
Using ‘convpdb.pl’ from MMTSB toolset to clean up the PDB file with ‘convpdb.pl -segnames model_pdb.pdb > ms.pdb’. Here ms.pdb is the final clean PDB file for CHARMM.
Download and install CHARMM (https://www.charmm.org/).
Prepare protein structure in CHARMM format ‘charmm pdbid=ms < mkprotein.inp’.
Prepare water box for protein solvation ‘charmm WaterDepth=8 target=protein cubic=1 < mkwaterbox.inp’.
Set protein solvation and neutralizing charge ‘charmm target=protein < solvation.inp’.
Apply 5.0 ns equilibrium in NVT, harmonic positional restrains on heavy atoms but gradually reduced ‘charmm < nvt_equil.5ns.inp > npt_equil.5ns.log’
Conduct the first 50 ns production simulation ‘charmm-gpu < nvt_prod.0.inp > nvt_prod.0.log’ (see Note 7).
Get protein dcd out of water box ‘charmm < get-dcd-nvt_prod.0.inp’ and ‘charmm cycle=1 < get-dcd-nvt_prod.restart.inp’ for additional cycle.
Calculate protein backbone and Cα RMSD ‘charmm dcdin=0 < anal-rms-asa.inp’, and ‘charmm dcdin=1 < anal-rms-asa.inp’ for additional dcd file.
Calculate accessible surface areas (ASA) ‘charmm dcdin=0 < anal-cs*.inp’, and ‘charmm dcdin=1 < anal-cs*.inp’ for additional dcd file. Here the anal-cs*.inp files are pre-defined. It’s based on different protein segments (i.e., a range of residue id for epitope region).
-
Normalize ASA into the relative ASA. The calculation using the ASA of each epitope region divided by total ASA.
(see Note 8).
3.4. MEFA protein expression, extraction, and immunogenicity characterization
After protein structure stability and epitope antigenicity being assessed and verified in silico, we synthesize an MEFA immunogen gene and clone the gene for protein expression and extraction. The MEFA gene can be inserted into pET28a expression vector directly at synthesis, or with two restriction enzyme sites flanked at the MEFA gene for cloning into pET28a vector, for expression of 6xHis-tagged or tag-less recombinant MEFA protein in E. coli strain BL21 (DE3), by following standard molecular biology protocols [22]. MEFA protein expression and extraction are verified in SDS PAGE with Coomassie blue stain and Western blot with specific antibodies; MEFA protein broad immunogenicity is confirmed in mouse immunization.
3.4.1. MEFA protein expression and extraction
Recombinant 6xHis-tag MEFA protein is purified with Ni-NTA agarose and column (www.qiagen.com), whereas tag-less MEFA protein expressed in inclusion body is extracted with B-PER™ Bacterial Protein Extraction Reagent (https://www.thermofisher.com).
Pick a colony from overnight growth at LB/kan agar plate, culture in 5–10 ml 2× YT medium supplemented with kanamycin (30 μg/mL) at 37°C overnight on a shaker (200 rpm).
Transfer 2–3 mL bacteria growth to 200 mL 2× YT medium supplemented with kanamycin (30 μg/mL) in a 500 mL flask, culture at 37°C on a shaker (200 rpm) till the OD600 reaches 0.5 – 0.7.
Add IPTG (0 – 1 mM; optimal concentration varies for different proteins), culture for 4 more hours, and transfer into 250 mL high-speed centrifuge bottle.
Harvest bacteria by centrifugation for 15 min at 17,000 × g (12,000 rpm with rotor F21-8×50Y), discard supernatant.
Freeze the pellet in a −80°C freezer overnight.
Thaw the frozen pellet and add 10 mL B-PER reagent (in phosphate buffer to lyse bacteria. Vortex or pipet up and down until suspension is homogenous, incubate for 30 min at room temperature (RT) on the shaker (100 rpm).
Sonicate bacteria lysate for 5 min (15 sec on, 15 sec off, at 30% amplitude; on ice).
Centrifuge bacteria lysate at 17,000 × g at 4°C for 15 min, resuspend pellet in 10–20 mL 1x phosphate buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4).
To extract 6xHis tag MEFA protein: gently mix suspension with 5 – 10 mL Ni-NTA agarose resin by rotating at 4°C overnight.
Apply suspension to a 100 mL column, flow through, wash 4x with 100 mL wash buffer, elute 4x with 100 mL elution buffer to collect 6 x His tag MEFA protein, run samples (20 μl) on SDS-PAGE. Use Amico or equivalent spin column (at a suitable pore size), or spectrum tubing with polyethylene glycol (PEG) at 4°C, to concentrate MEFA protein, store MEFA protein at −80°C (ready for immunization).
To extract tag-less MEFA inclusion body protein, homogenize suspension from Step 8 by passing up and down with 20 mL syringe (use 18G needle).
Add 100 mL PBS, vortex, centrifuge at 17,000 × g at 4°C for 15 min, collect pellet.
Repeat step 12 twice to wash protein pellet.
Dissolve the purified inclusion body protein pellet in 5–10 mL PBS.
Transfer resuspended protein to 1.5 mL micro centrifuge tube (1 mL per tube), centrifuge with a bench top microcentrifuge at 16,200 × g at RT for 10 min, discard supernatant.
(To refold purified inclusion body protein) Suspend and solubilize protein pellet with 1 mL1x IB solubiliation buffer (freshly made) by vigorously pipetting and vortex; incubate suspension on a shaker (100 rpm) at RT for 1 -2 h.
Centrifuge at 16,200 × g at RT for 10 min and collect supernatant.
Check protein with SDS-PAGE, proceed one-step protein dialysis and refolding next.
To dialyze and refold protein, transfer all supernatant into dialysis tubing membrane with a pore size 5–10 kDa less than the MEFA protein molecular mass.
Prepare dialysis buffer for a minimum of two buffer exchanges, for a volume of greater than 50 times of the protein suspension sample volume (for an example, 500 mL dialysis and refolding buffer for 10 mL protein suspension).
Dialyze for 4 h at 4°C and continue with two additional buffer exchanges using dialysis buffer without DTT; If visible precipitation occurs following dialysis, centrifuge the dialysis protein solution at 16,200 × g for 10 min at 4°C.
Collect clear supernatant containing refolded MEFA protein, aliquot, measure protein concentration, and store MEFA protein at −80°C.
Characterize refolded protein in ELISAs, SDS-PAGE Coomassie blue stain (incubation for 1 h in stain buffer, and then in destain buffer overnight; room temperature (50 rpm shaker), and Western blot with MEFA protein specific antibodies.
3.4.2. MEFA protein immunization and MEFA-specific antibody titration
Eight-week old female BALB/c mice, 8–10 per group, are commonly used for immunization with MEFA protein to examine broad immunogenicity (see Note 9).
Mix and emulsify 50 μg MEFA protein (in 50 μL PBS) with an equal amount of incomplete Freund’s adjuvant (IFA); 50 μL PBS mixed with IFA as a negative control.
Inject each mouse with MEFA protein (or PBS for the control group) intraperitoneally (IP) [ or intramuscularly (IM), subcutaneously (SC), or intradermally (ID)].
Two booster injections (of the same dose and route of the primary) are followed at the interval of two weeks.
Collect mouse serum and fecal samples two weeks after the final booster (see Note 10)
Store serum and fecal supernatant at −80°C until the use for antibody titration and antibody neutralization studies.
To titrate antigen-specific antibody responses, coat 96-well microtiter 2HB ELISA plates with epitope specific antigen (50 to 100 ng of each virulence factor protein in 100 μL 0.05 M carbonate-bicarbonate buffer, pH 9.6; per well) overnight at 4°C.
Discard coating reagent, wash wells three times with PBST (PBS with 0.05% Tween 20), 150 μL PBST per well.
Block uncoated sites with 5% nonfat milk (in PBST), 150 mL per well, for 1 h at 37°C.
Discard blocking buffer, wash 3x with PBST.
Add two-fold serial dilution of mouse serum samples (1:200 initial dilution) or fecal suspension samples (1:50 initial dilution), 100 μl per well, in triplicate, incubate 1 h at 37°C.
Wash wells with PBST three times and one time with PBS, add HRP-conjugated goat-anti-mouse IgG (1:5.000 dilution) or IgA (1:3000 dilution), 100 μL per well, incubate for 1 h at 37°C.
Wash wells with PBST three times, add 3,3’,5,5’-tetramethylbenzidine (TMB; 0.4 g/L in an organic base) Microwell Peroxidase Substrate System, 100 μl per well, incubate at room temperature for 25 min.
Read optical density at OD650 in a plate reader.
-
Calculate OD readings into log10, by multiplying the highest dilution that produces OD readings greater than 0.3 (after subtraction of the background readings) by the adjusted OD and converting to a log10 scale.
(see Note 11).
3.4.3. MEFA protein-induced antibody in vitro and in vivo protective activities
MEFA-induced antibody protective activities are examined with in vitro assays, or in vivo if animal challenge models are available. These assays vary and are pathogen or disease specific.
4. Notes
The protocols for in silico epitope identification, protein stability assessment, protein 3-D image creation and molecule dynamic simulation provided in this chapter are basic and simplified. Detailed instructions to run programs and to analyze data are available from individual program home page. While most programs are user friendly, molecule dynamic simulation from CHARMM and GROMCS is comprehensive and requires specialized computer-related skills. Other online programs can be used alternatively or synergistically to identify consensus epitopes and to enhance protein structure and stability characterization. Additionally, program in MacOS version and WinOS version can be slightly different. Native antigenicity of epitopes on MEFA is assessed comparatively with their original format on individual representing virulence factor proteins, based on epitope dynamic property, structure conformational flexibility of residues, accessible surface areas, epitope antigenic scores as well as visual assessment of epitope conformation based on 3-D structure. Eventually, in silico constructed MEFA immunogens are validated in empirical studies for broad immunogenicity and potential application of multivalent vaccine development.
Mice are used here to initially confirm MEFA protein broad immunogenicity. Other animal species, if they are relevant to the disease of interest, will be preferred for immunogenicity studies. Since B cell epitopes are discussed, only MEFA-induced antibody responses, but not T cell epitopes and T cell responses, are included in this chapter. Additionally, in vitro antibody neutralization assays are specific to each virulence factor or disease, whereas suitable animal challenge model may not exist for many pathogens and diseases, thus no protocols for antibody functional assays and animal challenge studies are provided. A suitable animal challenge model uses the animal species that are naturally susceptible to the pathogen and mimics disease progression, and after infection develop similar disease outcomes and similar levels of immune responses as humans [23–26]. Therefore, vaccine efficacy can be evaluated preclinically by immunizing the animals and then infecting the immunized animals with a pathogen. Eventually, vaccine candidacy is examined in human subject studies. Control human infection model (CHIM) [27], which directly tests immunogenicity, safety and efficacy of vaccine candidates in volunteers, however, enables to skip the use of animal challenge models and to accelerate vaccine development.
Other wash and elution buffers can be found at QIAGEN home page.
Protein secondary structure can be generated with CHARMM and ExPASy as well.
IEDB predicts T-cell epitopes as well; instructional details for epitope prediction are at IEDB webpage www.iedb.org/.
A protein is classified stable if an instability index is less than 40. Instructions for protein stability assessment are available at ExPASy home page.
Phyre2 and PyMol program updates and online assistance are at Phyre2 and PyMol webpages.
A constructed MEFA protein is expected to show protein structure and stability similar as the backbone protein; epitopes of interest on the MEFA protein display similar structural conformation and solvent-accessible surface areas as they are on virulence factor proteins.
Depending on convergence, one may increase the simulation length, for another 50 ns production simulation with ‘charmm-gpu cycle=1 < nvt_prod.restart.inp > nvt_prod.1.log’. The index number can be changed to extend another 50 ns until the simulation is converged.
Backbone or MEFA model was first solvated in a cubic box of TIP3P water, with the box size was set at 69 Å. Sodium ions were added to neutralize the total charge of the system. Energy minimization was first performed to remove improper molecular arrangement. Followed by 5.0 nanosecond (ns) equilibrium, harmonic positional restrains were applied on heavy atoms but gradually reduced to equilibrate the protein structures and water orientation. Finally, the production simulation was performed for a total simulation length of 350 ns. In the production run, NPT simulation was performed in Langevin dynamics with the constant temperature of 298 K and the constant pressure at 1.0 atm. Timestep 2.0 fs was used with the SHAKE algorithm applied to all hydrogen-containing bonds. Particle mesh Ewald was utilized for electrostatics cutoff of 13 Å. The cutoff of van der Waals interactions is 13 Å, with a switching function between 12 and 13 Å was used. The time evolution of the backbone or MEFA RMSD was first calculated to benchmark the convergence of simulation. The solvent accessible surface area (ASA) was calculated using 1.4 Å as the size of water probe and was further normalized into the relative ASA by calculating ASA for each epitope region. The relative ASA is a normalized calculation using each epitope region divided by total ASA.
To avoid gender bias, both sexes, in equal distribution, are used.
Fecal pellets are suspended in fecal reconstitution buffer: 10 mM Tris, 100 mM NaCl, 0.05% Tween-20, 5 mM sodium azide, pH 7.4, supplemented with protease inhibitor phenylmethylsulfonyl fluoride (0.5 mM final concentration); 1 g fecal pellet in 5 ml buffer; spin at 13,000 g at room temperature for 15 min to collect supernatant.
Other adjuvants including double mutant heat-labile toxin of enterotoxigenic E. coli (dmLT) can also be used for mouse immunization under parenteral or mucosal routes. The amount of each coating antigen and secondary dilutions are optimized in standard checkboard test.
Acknowledgment:
This work is supported by NIH R01AI121067-01A1 and University of Illinois at Urbana-Champaign. We also thank Shuangqi Wang, Ti Lu and Ipshita Upadhyay for technical assistance.
5. References:
- 1.Collaborators GD, Collaborators GH (2017) Global, regional, and national disability-adjusted life-years (DALYs) for 333 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 390 (10100):1260–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global health estimates 2016: Deaths by cause, age, sex, by country and by region, 2000 – 2016 (2018) https://www.who.int/healthinfo/global_burden_disease/estimates/en/index1.html.
- 3.Nabel GJ (2013) Designing tomorrow’s vaccines. New Engl J Med 368 (6):551–560. doi: 10.1056/NEJMra1204186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mascola JR, Ahmed R (2015) Vaccines: Vaccines for cancer and infectious diseases. Curr Opin Immunol 35:V–Vii. doi: 10.1016/j.coi.2015.07.009 [DOI] [PubMed] [Google Scholar]
- 5.Ruan X DA Sack, W. Zhang (2015) Genetic fusions of a CFA/I/II/IV MEFA (multiepitope fusion antigen) and a toxoid fusion of heat-stable toxin (STa) and heat-labile toxin (LT) of enterotoxigenic Escherichia coli (ETEC) retain broad anti-CFA and antitoxin antigenicity. PLoS One 10 (3):e0121623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan Q, Lee KH, Nandre RM, Garcia C, Chen J, Zhang W (2017) MEFA (multiepitope fusion antigen)-Novel Technology for Structural Vaccinology, Proof from Computational and Empirical Immunogenicity Characterization of an Enterotoxigenic Escherichia coli (ETEC) Adhesin MEFA. J Vaccines Vaccin 8 (4):367. doi: 10.4172/2157-7560.1000367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seo H, Duan Q, Zhang W (2020) Vaccines against gastroenteritis, current progress and challenges. Gut Microbes 11 (6):1486–1517. doi: 10.1080/19490976.2020.1770666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lycke N, Holmgren J (1986) Strong adjuvant properties of cholera toxin on gut mucosal immune responses to orally presented antigens. Immunology 59 (2):301–308 [PMC free article] [PubMed] [Google Scholar]
- 9.Clements JD, Hartzog NM, Lyon FL (1988) Adjuvant activity of Escherichia coli heat-labile enterotoxin and effect on the induction of oral tolerance in mice to unrelated protein antigens. Vaccine 6 (3):269–277 [DOI] [PubMed] [Google Scholar]
- 10.Lycke N (2005) Targeted vaccine adjuvants based on modified cholera toxin. Current Mol Med 5 (6):591–597 [DOI] [PubMed] [Google Scholar]
- 11.Freytag LC, Clements JD (2005) Mucosal adjuvants. Vaccine 23 (15):1804–1813. doi: 10.1016/j.vaccine.2004.11.010 [DOI] [PubMed] [Google Scholar]
- 12.Huang JC, Duan QD, Zhang WP (2018) Significance of enterotoxigenic Escherichia coli (ETEC) heat-labile toxin (LT) enzymatic subunit epitopes in LT enterotoxicity and immunogenicity. Appl Environ Microbiol 84 (15):e00849–00818. doi:UNSP e00849-18 10.1128/AEM.00849-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu T, Moxley RA, Zhang W (2019) Mapping the neutralizing epitopes of enterotoxigenic Escherichia coli (ETEC) K88 (F4) fimbrial adhesin and major subunit FaeG. Appl Environ Microbiol 85 (11):e00329–00319. doi: 10.1128/AEM.00329-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu T, Seo H, Moxley RA, Zhang W (2019) Mapping the neutralizing epitopes of F18 fimbrial adhesin subunit FedF of enterotoxigenic Escherichia coli (ETEC). Vet Microbiol 230:171–177. doi: 10.1016/j.vetmic.2019.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu T, Moxley RA, Zhang W (2020) Application of a novel epitope and structure vaccinology-assisted fimbria-toxin multiepitope fusion antigen of enterotoxigenic Escherichia coli for multivalent vaccine development against porcine post-weaning diarrhea. Appl Environ Microbiol. doi: 10.1128/AEM.00274-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, Peters B (2019) The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res 47 (D1):D339–D343. doi: 10.1093/nar/gky1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H (2012) ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res 40 (W1):W597–W603. doi: 10.1093/nar/gks400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protocol 10 (6):845–858. doi: 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chivian D, Baker D (2006) Homology modeling using parametric alignment ensemble generation with consensus and energy-based model selection. Nucleic Acids Res 34 (17). doi:ARTN e112 10.1093/nar/gkl480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, Pei JM, Kim D, Kellogg E, DiMaio F, Lange O, Kinch L, Sheffler W, Kim BH, Das R, Grishin NV, Baker D (2009) Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins-Structure Function and Bioinformatics 77:89–99. doi: 10.1002/prot.22540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song JS, Liu YQ, Liu CZ, Xie JP, Ma LX, Wang LP, Zheng YY, Ma ZB, Yang H, Chen X, Shi GX, Li SL, Zhao JP, Han JX, Wang YX, Liu JP, Zhu J (2013) [Cumulative analgesic effects of EA stimulation of sanyinjiao (SP 6) in primary dysmenorrhea patients: a multicenter randomized controlled clinical trial]. Zhen ci yan jiu = Acupuncture research / [Zhongguo yi xue ke xue yuan Yi xue qing bao yan jiu suo bian ji] 38 (5):393–398 [PubMed] [Google Scholar]
- 22.Ausubel FM, Brent R, Kingston RK, Moore DD, Seidman JG, Smith JA, and Struhl K (1999) Short Protocols in Molecular Biology. 4th edition edn. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 23.Spira WM, Sack RB, Froehlich JL (1981) Simple adult rabbit model for Vibrio cholerae and enterotoxigenic Escherichia coli diarrhea. Infect Immun 32 (2):739–747. doi: 10.1128/IAI.32.2.739-747.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerdts V, Wilson HL, Meurens F, van Drunen Littel-van den Hurk S, Wilson D, Walker S, Wheler C, Townsend H, Potter AA (2015) Large animal models for vaccine development and testing. ILAR J 56 (1):53–62. doi: 10.1093/ilar/ilv009 [DOI] [PubMed] [Google Scholar]
- 25.Swearengen JR (2018) Choosing the right animal model for infectious disease research. Animal Model Exp Med 1:100–108. doi: 10.1002/ame2.12020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramin Sedaghat Herati EJW (2018) What is the predictive value of animal models for vaccine efficacy in humans. Cold Spring Harb Perspect Biol 10:a031583. doi: 10.1101/cshperspect.a031583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon SB, Rylance J, Luck A, Jambo K, Ferreira DM, Manda-Taylor L, Bejon P, Ngwira B, Littler K, Seager Z, Gibani M, Gmeiner M, Roestenberg M, Mlombe Y, Wellcome Trust Cwp (2017) A framework for Controlled Human Infection Model (CHIM) studies in Malawi: Report of a Wellcome Trust workshop on CHIM in Low Income Countries held in Blantyre, Malawi. Wellcome Open Res 2:70. doi: 10.12688/wellcomeopenres.12256.1 [DOI] [PMC free article] [PubMed] [Google Scholar]


