Abstract
Cell polarization is critical for lineage segregation and morphogenesis during mammalian embryogenesis. However, the processes and mechanisms that establish cell polarity in the mammalian embryo are not well understood. Recent studies suggest that unique regulatory mechanisms are deployed by the mouse embryo to establish cell polarization. In this review, we discuss the current understanding of cell polarity establishment, focusing on the formation of the apical domain in the mouse embryo. We will also discuss outstanding questions and possible directions for future study.
Keywords: Mouse embryo, De novo cell polarity establishment, Apical domain, Par complex, Actomyosin, Protein clustering
Introduction
Similar to many other embryonic model systems, mammalian embryos establish cellular polarization that, as development proceeds, dynamically changes its form to execute critical cellular and developmental functions. In mammalian embryos, cell polarization is first established at the cleavage stage of development. At this time, embryos are free-floating in the oviduct prior to implantation in the uterus, and the cells of the embryo undergo equatorial partitions without a net increase in embryo volume. Such a polarity pattern is rudimentary, with the formation of just two complementary polarity domains localized to opposite sides of the cell; cell-contacts define the basal domain, and apical proteins centered to the cell-contact free surface form the apical domain [1] (Figure 1). Formation of such a polarity pattern in the mammalian embryo has long been recognized as important for the ensuing lineage diversification process [2,3], and still, the molecular mechanisms establishing cell polarization have not been well addressed until recently.
Figure 1. Apico-basal cell polarization established at the 8-cell stage in the mouse embryo.

The basal domain is defined by cell–cell contacts enriched by adherens junction proteins; the apical domain is enriched by apical polarity proteins, such as Ezrin, Par complex, and Jam-1. The apical domain is surrounded by an actomyosin ring on the outside. Microtubules emanate from the nucleus towards the apical domain.
As a model for studying cell polarization, the mouse embryo provides a reductive in vivo system, with advantages, such as ease of imaging and embryological manipulation, significantly facilitating the application of molecular and biophysical techniques. However, unlike other model systems, the mammalian embryo, at this early stage, is remarkably robust to genetic perturbation, with only a handful of gene knockouts showing polarity phenotypes. This is largely due to compensation between maternal and zygotic transcripts, as well as isoforms encoding proteins of similar function. As a result, most of our knowledge of cell polarity regulation in the mammalian embryo comes from comparative approaches, characterizing the dynamics, and roles of homologous polarity regulators identified in other model systems. Nevertheless, these studies reveal the unique regulatory mechanisms deployed by mammalian embryos to establish cell polarization. In this review, we summarize our current understanding of cell polarity regulation in the mammalian embryo, using the mouse embryo as a model, with a particular focus on the processes forming the apical domain.
Similarities and distinctions of the role of molecular regulatory modules in building the apical domain
In the mouse embryo, the formation of the apical domains begins at the 8-cell stage. At this time, adjacent blastomeres juxtapose closer, cell–cell contact length becomes more extensive, and cell-junction proteins sort exclusively to the cell-contacts—a process known as compaction [1,4]. On the cell-contact free surface, a specialized membranous structure grows, which is referred to as the apical domain. The apical domain is enclosed by a dense actin-myosin enriched ring, inside which a plethora of evolutionarily conserved epithelial apical polarity proteins localize, including villus associated ERM family members Ezrin/Raxin, universal polarity machinery Par6-aPKC complex, Crumbs complex, and some (but not all) tight junction proteins, such as Jam-1 [5] (Figure 1).
The formation of the apical domain has several cellular consequences. First, the apical domain tethers fate regulators, such as activators of Hippo signaling, to modulate and receive feedback from the transcription to dictate trophectoderm fate [6]. Second, the apical domain organizes the underlying microtubule network, leading to the apical polarization of endocytic vesicles [7] and mRNA transcripts, encoding crucial lineage specifiers, to consolidate lineage identities [8] (Figure 1). Third, the apical domain orients the spindle apicobasally during mitosis, producing polar (with an apical domain) and apolar (without an apical domain) daughter cells—a process known as asymmetric cell division [3]. This process is akin to that characterized in neuronal cells [9], although it is not clear whether the same set of polarity regulators are involved in both these systems. Finally, the establishment of the apical domain at the 8-cell stage initiates the epithelialization process in the outer trophectodermal layer that allows epithelial junctional proteins to be progressively assembled during the following stages, laying the foundation for the blastocyst formation [10,11].
As tight junction establishment commences, a set of molecular regulatory relationships, analogous to those found in epithelial tissue, are used to maintain cell polarization in the mouse embryo. Cdc42 is required for anchoring Par6-aPKC to the apical domain [3], adherens junction proteins residing in cell-contacts antagonize apical protein localization [12], and Par6-aPKC inhibits Lgl and Scrib to restrict their localization to the basal region [6,13]. In spite of extended mechanistic conservation of the pathways used to maintain cell polarity, the regulatory pathways establishing cell polarity appear distinct. This is illustrated by the relationships between polarity regulators, as well as the interplay between polarity regulators and signaling molecules. For example, the apical polarization of the ERM proteins at the 8-cell stage is established in parallel with the Par complex but is independent of the Par complex at later stages [14]. The Hippo signaling pathway can feed back onto the apical domain to reinforce cell polarization status; however, it is not involved in the establishment of cell polarization [15]. Thus, the mechanisms used for cell polarity maintenance are not mixed with those used for polarity establishment.
The precise regulatory mechanisms underlying the establishment of cell polarization to form the apical domain remain unclear but captivating. This is mainly because the formation of the apical domain is independent of many known polarity cues present in many other model systems. For instance, as shown in yeast and worm cells, microtubule mediated transport is used to deliver symmetry breaking cues for cortical domain segregation [16,17]. However, in the mouse embryo, disruption of the microtubule network using a high dose of Nocodazole and Colcemid has only a subtle effect on the formation of the apical domain [18]. The extracellular matrix (ECM) provides a crucial cue to direct cell polarity in epithelial cells; however, the 8-cell stage embryo does not have an ECM network. Key ECM isoforms of integrin, fibronectin, and collagen only become synthesized at a later stage of preimplantation development [19], making it unlikely that they play a critical role in apical domain formation. Cell–cell contact is often important for establishing cell polarity. As shown in Drosophila epithelial cells and the early embryo of Caenorhabditis elegans, cell-contacts can recruit adaptor proteins that function to exclude apical proteins from the cell-junctions and hence define the apical-basal boundaries [20,21]. Consequently, in these systems the depletion of key cell-contact associated proteins results in failure to establish apical-basal polarization. In the case of the mouse embryo, the role of cell-contact is instructive but dispensable; the presence of cell-contact negatively impacts the localization of the apical domain. However, the absence of cell-contact does not prevent the formation of the apical domain at the 8-cell stage. The apical domain is still established on time in embryos genetically depleted of E-cadherin; isolated 8-cell stage blastomeres, thus, deprived of any type of cell junction, are capable of developing the apical domain at a random location without obvious “positional memory” [3,22]. In these isolated cells, basal components such as E-cadherin do not show complementary distribution to the apical proteins. These observations suggest that (1) the apical domain at this stage is a specialized membranous unit, rather than a fully functional epithelial domain; (2) the regulatory mechanisms establishing the apical domain do not necessarily rely on the inhibitory signal from adherens junction. These results, taken together, suggest that the apical domain formation in the mouse embryo is a highly self-organized process and that the common polarity pathways used in other systems are nonessential. The question then arises, what is the dominant mechanism that the mouse embryo uses to construct the apical domain?
Symmetry breaking: implication of actin-dependent mechanisms?
Several recent studies reveal that the establishment and maintenance of the apical domain are closely coupled to, and controlled by, the dynamic actin cytoskeleton rearrangements occurring circumferentially on the cell cortex [11,23]. As soon as the embryo enters the 8-cell stage, the motor protein myosin II becomes activated by phosphorylation of its light chain and subsequently associates with the actin filament [24]. A direct result of this, actomyosin contractility is initiated, a process most well-known for driving muscle contraction. The localization of actomyosin is not favored at cell-contacts, through a mechanism still to be discovered, leading to its apical polarization. This, in turn, causes a net increase in apical surface tension, overriding the tension found at cell junctions, and thus, driving embryo compaction [25]. As the embryo develops, the actomyosin cortex undergoes further remodeling to form an actin cap in the middle of the cell-contact free surface, at which time various apical polarity proteins are also enriched apically [23]. While this accumulation continues, the Par complex negatively regulates the actomyosin meshwork, excluding it from the center to the periphery of the growing apical patch. This results in a configuration whereby an actomyosin ring marks the maturation of the apical domain [23] (Figure 2). Consequently, embryos genetically depleted of both aPKC isoforms fail to clear the central actin, and thus, no matured apical domain can be formed [26].
Figure 2. Movements of actin and apical polarity protein during the apical domain formation.
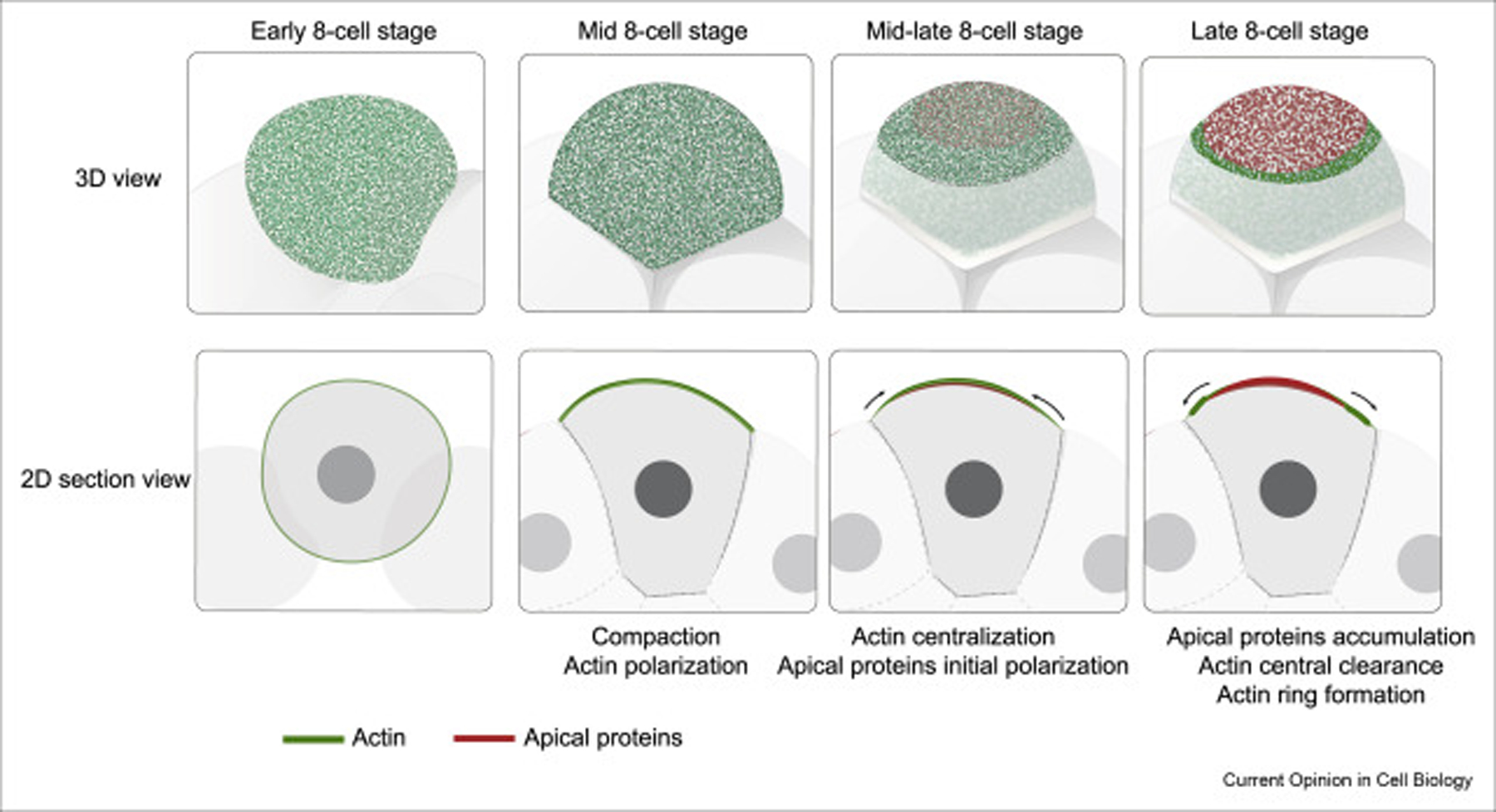
In the first few hours of apical domain formation, the actin becomes polarized to the cell-contact free surface, concurrent with cell compaction. As the actin becomes further centralized to the cell-contact free surface, the apical proteins also polarize to the center of the cell-contact free surface. The accumulation of apical proteins excludes actin to the periphery, to form the actomyosin ring.
Although the molecular interactions responsible for apical domain formation—the clearance of actin and the formation of actin ring—are well-characterized, the mechanisms driving the initial recruitment of the apical proteins are less clear. In spite of the importance of polarized actomyosin in compaction, the contractile property of actomyosin seems to be unnecessary for apical domain formation; blocking actomyosin contractility has little effect on the apical domain morphology, albeit abolishing cell compaction [23]. This result, together with the fact that the movements of the apical proteins and actomyosin complex are not completely synchronized, imply that the initial polarization of the apical proteins is independent of actomyosin contractility. This is different from the working model in the C. elegans zygote, in which actomyosin contraction generates cortical flows, acting as an advective platform to bring the PAR3-PAR6-aPKC complex anteriorly from the posterior end [27,28]. Nevertheless, a low level of contractile actomyosin activity is important for the formation of the outer actomyosin boundary, as the global elevation of actomyosin activity by pharmacological treatment or by cytokine signaling disrupted the actomyosin boundary and resulted in the expansion of the apical domain [11,23]. In contrast, while the apical protein polarization does not appear to be dependent on actomyosin contractility, the organization of apical proteins is sensitive to the topology of the actin network. Depolymerization of actin or myosin filaments, or perturbing the activity of the actin remodelers Arp2/3 and MLCK, consistently abolished the apical domain formation process [23,29]. These results suggest that actin network remodeling during the formation of the actin cap may provide a permissive environment for the apical proteins to localize to the center. This notion could be supported by other membrane protein systems.
The coordinated movements between the actin cortex and the apical proteins mirror the processes that lead to the formation of a membrane receptor patch following T-cell receptor activation or the formation of a continuous adherens junction belt at the interface of neighboring cells [30,31] (Figure 3). In these scenarios, membrane receptor activation leads to the clustering of the receptors, a process referred to as “phase separation” [32]. These micro-clusters are then delivered to the center by the actin cytoskeleton, accompanied by centralization of the actin network and actin rearrangement to the periphery of the membrane receptor cluster [30]. Theoretical calculations suggest that, at micro-scale, clustering of the membrane proteins could help to minimize local free energy [33]; and at the macro-scale, the centralization of ligand-binding proteins can reduce the free energy of the system [34]. Studies of membrane protein clustering suggest that the membrane clustering is closely coupled to actin dynamics. Specifically, the formation of micro-clusters would require physical attachment to short, dynamic actin filaments underneath [35], whereas the conjugation of micro-clusters would require actin movements over a large-distance. This would involve the activity of motor proteins and the coordinated action of different actin-associated proteins [33]. The actin dynamics behind these processes may be analogous to apical domain formation in the mouse embryo. Although apical proteins do not bind to a ligand, they are often subjected to posttranslational modifications that change their binding affinity to membrane lipids during their polarization process [36]. Such chemical reactions may allow the formation of apical clusters. Although myosin contractility is not required, the long-range movement of actin could be driven by actin polymerization–depolymerization cycles (actin treadmilling [37]) or by capping and nucleation factors [38]. This could be one of the many explanations for the requirement of nucleation factors, such as Arp2/3 and actin filaments during apical domain formation. Close examination of apical protein conformation, dynamics, and their relationship with actin regulators would be necessary to effectively test the validity of this model.
Figure 3. Similarities of apical domain to the membrane receptor relocation scenarios.

(a) During adhesive receptor activation, the receptors first form micro-clusters, which are then delivered to the center of the cell-surface. T-cell receptor scheme adapted from Yu et al., 2013. Adherens Junction formation scheme adapted from Engl et al., 2014. The accumulation of the receptor in the center of the cell membrane excludes actin from the outside. (b) Theoretical calculations suggest that the centralization of membrane receptors helps to minimize free energy per bond as a result of reducing the bond density. Scheme adapted from Schmidt et al., 2015 [34].
Challenges and perspectives
The formation of the apical domain is becoming one of the fascinating models in which to study mechanisms of de novo establishment of cell polarization. Although certain molecular characterizations have been carried out, we still lack an understanding of the biochemical and physical principles underlying the dynamics of symmetry breaking and boundary formation. Recently developed in vitro reconstruction systems reveal unique features of the cytoskeletal network (both actin and microtubules), and of many adhesive membrane proteins. These systems will be instrumental for understanding the pattern of membrane protein and actin network formation in developmental processes, such as the formation of the apical domain. Imaging techniques, such as Fluorescence Correlation Spectroscopy (FCS), Fluorescence Resonance Energy Transfer (FRET), and Optogenetics, have been successfully adapted to the mouse embryo, allowing more accurate functional studies and quantitative analysis [23,39]. Other imaging techniques, such as TIRF and light-sheet microscopy, also have the potential to be applied to studies of cell polarity in the mouse embryo. These new cutting-edge approaches show great promise for uncovering the principles of mammalian embryo cortical polarization.
Acknowledgement
We thank Christos Kyprianou and Hannah Mudge for their feedback on the manuscript. M.Z.G receives funding from Wellcome Trust, ERC, and Leverhulme Trust. M.Z is funded by Leverhulme Trust.
Footnotes
Conflict of interest statement
Nothing declared.
Papers of particular interest, published within the period of review, have been highlighted as:
of special interest
of outstanding interest
References
- 1.Fleming TP, Johnson MH: From egg to epithelium. Annu Rev Cell Biol 1988, 4:459–485. [DOI] [PubMed] [Google Scholar]
- 2.Johnson MH, Ziomek CA: The foundation of two distinct cell lineages within the mouse morula. Cell 1981, 24:71–80. [DOI] [PubMed] [Google Scholar]
- 3. Korotkevich E, Niwayama R, Courtois A, Friese S, Berger N, Buchholz F, Hiiragi T: The apical domain is required and sufficient for the first lineage segregation in the mouse embryo. Dev Cell 2017, 40:235–247. e237. • This work provides key evidence to show that the presence of apical domain is sufficient to specify trophectdodermal cell fate. It also shows a requirement of Cdc42 in maintaining apical domain organization.
- 4.Pratt HP, Ziomek CA, Reeve WJ, Johnson MH: Compaction of the mouse embryo: an analysis of its components. J Embryol Exp Morphol 1982, 70:113–132. [PubMed] [Google Scholar]
- 5.Leung CY, Zhu M, Zernicka-Goetz M: Polarity in cell-fate acquisition in the early mouse embryo. Curr Top Dev Biol 2016, 120:203–234. [DOI] [PubMed] [Google Scholar]
- 6.Hirate Y, Hirahara S, Inoue K, Suzuki A, Alarcon VB, Akimoto K, Hirai T, Hara T, Adachi M, Chida K, et al. : Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr Biol 2013, 23:1181–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleming TP, Cannon PM, Pickering SJ: The cytoskeleton, endocytosis and cell polarity in the mouse preimplantation embryo. Dev Biol 1986, 113:406–419. [DOI] [PubMed] [Google Scholar]
- 8.Skamagki M, Wicher KB, Jedrusik A, Ganguly S, Zernicka-Goetz M: Asymmetric localization of Cdx2 mRNA during the first cell-fate decision in early mouse development. Cell Rep 2013, 3:442–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venkei ZG, Yamashita YM: Emerging mechanisms of asymmetric stem cell division. J Cell Biol 2018, 217:3785–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vinot S, Le T, Ohno S, Pawson T, Maro B, Louvet-Vallee S: Asymmetric distribution of PAR proteins in the mouse embryo begins at the 8-cell stage during compaction. Dev Biol 2005, 282:307–319. [DOI] [PubMed] [Google Scholar]
- 11. Zenker J, White MD, Gasnier M, Alvarez YD, Lim HYG, Bissiere S, Biro M, Plachta N: Expanding actin rings zipper the mouse embryo for blastocyst formation. Cell 2018, 173: 776–791. e717. • This paper illustrates that the zippering of adjacent apical domain boundaries facilitates tight junction formation in the morula stage mouse embryo.
- 12.Stephenson RO, Yamanaka Y, Rossant J: Disorganized epithelial polarity and excess trophectoderm cell fate in preimplantation embryos lacking E-cadherin. Development 2010, 137:3383–3391. [DOI] [PubMed] [Google Scholar]
- 13.Kono K, Tamashiro DA, Alarcon VB: Inhibition of RHO-ROCK signaling enhances ICM and suppresses TE characteristics through activation of Hippo signaling in the mouse blastocyst. Dev Biol 2014, 394:142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirate Y, Hirahara S, Inoue K, Kiyonari H, Niwa H, Sasaki H: ParaPKC-dependent and -independent mechanisms cooperatively control cell polarity, Hippo signaling, and cell positioning in 16-cell stage mouse embryos. Dev Growth Differ 2015, 57:544–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frum T, Murphy TM, Ralston A: HIPPO signaling resolves embryonic cell fate conflicts during establishment of pluripotency in vivo. Elife 2018, 7. • This paper demonstrates a feedback loop between Hippo signalling and cell polarity, after the apical domain has been formed.
- 16.Motegi F, Zonies S, Hao Y, Cuenca AA, Griffin E, Seydoux G: Microtubules induce self-organization of polarized PAR domains in Caenorhabditis elegans zygotes. Nat Cell Biol 2011, 13:1361–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang F, Martin SG: Shaping fission yeast with microtubules. Cold Spring Harb Perspect Biol 2009, 1:a001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maro B, Pickering SJ: Microtubules influence compaction in preimplantation mouse embryos. J Embryol Exp Morphol 1984, 84:217–232. [PubMed] [Google Scholar]
- 19.Johnson MH: From mouse egg to mouse embryo: polarities, axes, and tissues. Annu Rev Cell Dev Biol 2009, 25: 483–512. [DOI] [PubMed] [Google Scholar]
- 20.Klompstra D, Anderson DC, Yeh JY, Zilberman Y, Nance J: An instructive role for C. elegans E-cadherin in translating cell contact cues into cortical polarity. Nat Cell Biol 2015, 17: 726–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.St Johnston D, Ahringer J: Cell polarity in eggs and epithelia: parallels and diversity. Cell 2010, 141:757–774. [DOI] [PubMed] [Google Scholar]
- 22.Anani S, Bhat S, Honma-Yamanaka N, Krawchuk D, Yamanaka Y: Initiation of Hippo signaling is linked to polarity rather than to cell position in the pre-implantation mouse embryo. Development 2014, 141:2813–2824. [DOI] [PubMed] [Google Scholar]
- 23. Zhu M, Leung CY, Shahbazi MN, Zernicka-Goetz M: Actomyosin polarisation through PLC-PKC triggers symmetry breaking of the mouse embryo. Nat Commun 2017, 8:921. •• Whether and how apical domain establishment is regulated by cytoskeleton dynamics remains unclear. This work provides detailed examination of the requirement of actin network to apical domain formation and reveals the upstream regulators of actin dynamics during apical domain formation.
- 24.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR: Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol 2009, 10:778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maitre JL, Niwayama R, Turlier H, Nedelec F, Hiiragi T: Pulsatile cell-autonomous contractility drives compaction in the mouse embryo. Nat Cell Biol 2015, 17:849–855. [DOI] [PubMed] [Google Scholar]
- 26.Maitre JL, Turlier H, Illukkumbura R, Eismann B, Niwayama R, Nedelec F, Hiiragi T: Asymmetric division of contractile domains couples cell positioning and fate specification. Nature 2016, 536:344–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Munro E, Nance J, Priess JR: Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell 2004, 7:413–424. [DOI] [PubMed] [Google Scholar]
- 28.Goehring NW, Trong PK, Bois JS, Chowdhury D, Nicola EM, Hyman AA, Grill SW: Polarization of PAR proteins by advective triggering of a pattern-forming system. Science 2011, 334: 1137–1141. [DOI] [PubMed] [Google Scholar]
- 29.Sun SC, Wang QL, Gao WW, Xu YN, Liu HL, Cui XS, Kim NH: Actin nucleator Arp2/3 complex is essential for mouse preimplantation embryo development. Reprod Fertil Dev 2013, 25: 617–623. [DOI] [PubMed] [Google Scholar]
- 30.Yu Y, Smoligovets AA, Groves JT: Modulation of T cell signaling by the actin cytoskeleton. J Cell Sci 2013, 126:1049–1058. [DOI] [PubMed] [Google Scholar]
- 31.Engl W, Arasi B, Yap LL, Thiery JP, Viasnoff V: Actin dynamics modulate mechanosensitive immobilization of E-cadherin at adherens junctions. Nat Cell Biol 2014, 16:587–594. [DOI] [PubMed] [Google Scholar]
- 32.Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD: Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 2016, 352:595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johannes L, Pezeshkian W, Ipsen JH, Shillcock JC: Clustering on membranes: fluctuations and more. Trends Cell Biol 2018, 28:405–415. • This article comprehensively summarizes the mechanisms regulating surface protein clustering over different cellular scales.
- 34.Schmidt D, Bihr T, Fenz S, Merkel R, Seifert U, Sengupta K, Smith AS: Crowding of receptors induces ring-like adhesions in model membranes. Biochim Biophys Acta 2015, 1853:2984–2991. [DOI] [PubMed] [Google Scholar]
- 35.Gowrishankar K, Ghosh S, Saha S, Rumamol C, Mayor S, Rao M: Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell 2012, 149: 1353–1367. [DOI] [PubMed] [Google Scholar]
- 36.Liu H, Wu Z, Shi X, Li W, Liu C, Wang D, Ye X, Liu L, Na J, Cheng H, et al. : Atypical PKC, regulated by Rho GTPases and Mek/Erk, phosphorylates Ezrin during eight-cell embryocompaction. Dev Biol 2013, 375:13–22. [DOI] [PubMed] [Google Scholar]
- 37.Allard J, Mogilner A: Traveling waves in actin dynamics and cell motility. Curr Opin Cell Biol 2013, 25:107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Durre K, Keber FC, Bleicher P, Brauns F, Cyron CJ, Faix J, Bausch AR: Capping protein-controlled actin polymerization shapes lipid membranes. Nat Commun 2018, 9:1630. • This paper uses an in vitro reconstruction system to show that the actin turnover- controlled by the Arp2/3-mediated actin polymerization, and capping proteins-regulated actin depolymerization- is sufficient to break the symmetry of actin cortex to induce the clustering of actin polymers.
- 39.White MD, Angiolini JF, Alvarez YD, Kaur G, Zhao ZW, Mocskos E, Bruno L, Bissiere S, Levi V, Plachta N: Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell 2016, 165:75–87. [DOI] [PubMed] [Google Scholar]


