ABSTRACT
Cells stabilize intracellular inorganic phosphate (Pi) to compromise between large biosynthetic needs and detrimental bioenergetic effects of Pi. Pi homeostasis in eukaryotes uses Syg1/Pho81/Xpr1 (SPX) domains, which are receptors for inositol pyrophosphates. We explored how polymerization and storage of Pi in acidocalcisome-like vacuoles supports Saccharomyces cerevisiae metabolism and how these cells recognize Pi scarcity. Whereas Pi starvation affects numerous metabolic pathways, beginning Pi scarcity affects few metabolites. These include inositol pyrophosphates and ATP, a low-affinity substrate for inositol pyrophosphate-synthesizing kinases. Declining ATP and inositol pyrophosphates may thus be indicators of impending Pi limitation. Actual Pi starvation triggers accumulation of the purine synthesis intermediate 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), which activates Pi-dependent transcription factors. Cells lacking inorganic polyphosphate show Pi starvation features already under Pi-replete conditions, suggesting that vacuolar polyphosphate supplies Pi for metabolism even when Pi is abundant. However, polyphosphate deficiency also generates unique metabolic changes that are not observed in starving wild-type cells. Polyphosphate in acidocalcisome-like vacuoles may hence be more than a global phosphate reserve and channel Pi to preferred cellular processes.
KEYWORDS: Saccharomyces cerevisiae, acidocalcisome, phosphate signalling, polyphosphate, SPX domains
INTRODUCTION
Inorganic phosphate (Pi) is an essential nutrient and is a component of lipids and nucleic acids, controls the activity of proteins through covalent modification, and serves as an energy transducer when integrated into nucleotides, driving many endergonic biochemical reactions. Therefore, perturbed Pi homeostasis strongly affects growth and development of various living organisms (1–4). Eukaryotic cells have developed multilayered systems to maintain cytosolic Pi concentration within a suitable range despite fluctuating environmental conditions. Shortage of intracellular Pi was proposed to be signaled through a dedicated signaling pathway for intracellular phosphate reception and signaling (INPHORS), where the level of cytosolic Pi is translated into changes of inositol pyrophosphates (PP-IPs), which then bind to Syg1/Pho81/Xpr1 (SPX) receptor domains (5, 6). These domains form part of or interact with numerous cellular proteins that control transcription or mediate uptake, secretion, storage, and recycling of phosphate (7, 8). It is expected that INPHORS coordinates these systems such that cytosolic Pi concentration is maintained in a viable range.
In Saccharomyces cerevisiae, INPHORS triggers the PHO pathway, the transcriptional phosphate starvation response that controls expression of genes for Pi scavenging, uptake, and recycling (9). The PHO pathway in yeast is controlled through the transcription factor Pho4, which accumulates in the nucleus after Pi starvation and interacts with another transcription factor, Pho2, to activate many Pi-dependent genes. The interaction between Pho2 and Pho4 is impaired by phosphorylation of Pho4 through Pho85/Pho80 kinase (10, 11). Early studies postulated that Pi starvation triggers an increase in 1-IP7, which should inactivate Pho85/Pho80 kinase by direct interaction with a central part of its regulatory subunit Pho81 (12, 13). However, several subsequent studies failed to confirm the starvation-induced increase in IP7 and instead showed a strong decline in the inositol phosphate pools after Pi starvation (5, 9, 14). These qualitatively different results on the inositol pyrophosphate pools and additional characterization of Pho81 led to a revised model of PHO pathway regulation in which 1,5-IP8 controls Pho81 through its SPX domain. The decline of 1,5-IP8, which is triggered by Pi starvation, allows Pho81 to bind Pho85/Pho80 more tightly and inhibit its kinase activity, favoring dephosphorylation and activation of Pho4. Pho4-dependent transcription depends on the interaction of Pho4 with Pho2, which is stimulated by Pho4 dephosphorylation (9–11). This interaction is also stimulated by an intermediate of purine synthesis, 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), but it is unknown whether AICAR levels respond to Pi availability (15–17).
Many organisms developed strategies to cope with a gradual decline (scarcity) or final absence (starvation) of phosphate, which then becomes growth limiting (1–4). Budding yeast induces its starvation response in at least two stages depending on the degree of Pi depletion (18–20). Phosphate scarcity activates Pi scavenging from the environment and maintains growth. Acid phosphatases, such as Pho5, are expressed and secreted to liberate Pi from external Pi-containing molecules (21). As a further measure to facilitate Pi scavenging from the environment, low-affinity transporters are degraded and replaced by high-affinity transporters, such as Pho84 (22–25). Under complete Pi starvation, when Pi can no longer be obtained from the environment, yeast adopts different strategies. Cells recycle Pi by decomposing various intracellular organic molecules, including lipids, nucleotides, and even subcellular organelles (26–30). In addition, cells stop the cell cycle, saving Pi that would otherwise be used for nucleic acid and phospholipid duplication (31, 32).
In addition to scavenging and recycling, yeast cells also maintain a large store of phosphate inside their vacuoles (33, 34). These vacuoles share key features of acidocalcisomes, a class of conserved lysosome-like organelles that occur in all eukaryotic kingdoms (35, 36). These features include high lumenal concentrations of divalent cations, inorganic polyphosphates (poly[P]s), and basic amino acids. Their high poly(P) content is of potential relevance to phosphate homeostasis (18, 37, 38). This polymer can accumulate to the equivalent of hundreds of millimolar phosphate units in an osmotically almost inert form and is considered an efficient storage form of Pi (34, 39). Vacuolar poly(P) is produced from cytosolic ATP by the vacuolar transport chaperone (VTC) complex, which at the same time translocates the nascent poly(P) chain into the lumen of the organelle (40–42). Poly(P) can be hydrolyzed by polyphosphatases in the lumen of the organelle (43–46). It is assumed that the liberated Pi is released back into the cytosol to support metabolism. Therefore, poly(P) was proposed to act as a buffer system for cytosolic Pi (18, 47, 48). Synthesis of poly(P) by VTC is stimulated by inositol pyrophosphates (5, 49, 50). The levels of all these compounds (in yeast: 1-IP7, 5-IP7, and 1,5-IP8) increase in abundance with increasing Pi availability (9, 14) and will hence favor accumulation and storage of Pi in the form of poly(P) only if Pi is abundant. How the turnover of poly(P) inside the organelle is regulated and coordinated with the release of Pi into the cytosol is unknown.
To explore mechanisms that contribute to an early recognition of forthcoming Pi limitation, we explored the metabolic consequences experienced by yeast cells that are at the brink of phosphate limitation, that is, where Pi becomes scarce but not yet limiting for growth. Furthermore, we analyzed the metabolic significance of acidocalcisome-like vacuoles and their poly(P) pool.
RESULTS
Optimization of Pi starvation conditions for metabolomic analysis.
Yeast cells distinguish scarcity of Pi (where they induce genes to optimize Pi scavenging and maintain normal growth) from Pi starvation that reduces growth and induces genes to facilitate Pi recycling from internal sources (6, 18–20, 24, 51, 52). We used nontargeted metabolomic analysis to explore whether and how cells react to beginning Pi scarcity in comparison to profound Pi starvation. To this end, we first identified conditions that bring our strain background to the brink of Pi limitation of growth. Batch cultures of cells were grown logarithmically overnight until the optical density at 600 nm (OD600) was around 1 (4.6 × 107 cells/mL). A small inoculum of these logarithmically growing cells was transferred into synthetic complete (SC) liquid medium containing 0 mM to 10 mM Pi (OD600 = 0.05; 2.3 × 106 cells/mL), and the OD600 was followed over 24 h. Compared to cells growing in high-Pi medium (10 mM Pi), growth was normal at 1 mM and 0.5 mM Pi, and we observed only a mild retardation of growth in 0.25 mM and 0.1 mM Pi. Incubation in 0 mM Pi arrested growth (Fig. 1A). We measured the activity of secreted acid phosphatase, which is activated by Pi starvation (21), as a readout of the PHO pathway. In Pi-free medium, the activity of secreted acid phosphatase gradually increased up to 8 h and then maintained a similar level up to 24 h, suggesting that the PHO pathway was maximally activated at 8 h (Fig. 1B). In 0.1 mM and 0.25 mM Pi, the activity of acid phosphatase was partially induced at 8 h but fully induced at 24 h, probably reflecting the gradual depletion of Pi from the medium over that period. In 0.5 mM Pi medium, acid phosphatase activity increased to half of the maximal value on 0 mM Pi. There was no significant growth delay under this condition, indicating that the cells managed to compensate the limited availability of Pi, probably through induction of the PHO pathway (Fig. 1A). To assess the Pi consumption of the cells under each condition, we monitored the amounts of Pi remaining in the medium using the malachite green assay (Fig. 1C). In 10 mM Pi, more than 90% of Pi remained even after 8 h of incubation. In 0.5 mM Pi medium, 70% of the Pi was consumed after 6 h, and none remained after 8 h. We monitored this induction by tagging the Pho4 transcription factor with green fluorescent protein (GFP), which is translocated into the nucleus when cells lack Pi. Pho4-GFP was predominantly in the nucleus after 8 h of incubation without Pi (Fig. 1D), and 0.5 mM Pi led to partial nuclear accumulation of Pho4-GFP, suggesting that the PHO pathway was moderately activated. We further measured the transcript level of the PHO pathway marker genes Pho5 and Pho84 using quantitative real-time reverse transcription-PCR (qRT-PCR). The gene expression of Pho5 and Pho84 was highly induced after 8 h of Pi starvation (Fig. 1E and F). Under Pi scarcity (0.5 mM Pi), however, only Pho84 showed a mild, yet statistically significant, induction, whereas induction of Pho5 remained below 1% of the maximal value and was not significant. In agreement with earlier studies (18, 53), this suggests that partial induction of the PHO pathway allows the cells to compensate for reduced availability of Pi to a level that supports normal growth.
FIG 1.
Response of S. cerevisiae under different Pi starving conditions. (A) Growth curves of yeast cells in synthetic complete medium supplemented with different concentrations of Pi from 10 mM to 0 mM. Cells were inoculated at an OD600 of 0.05 and cultured for 24 h. The means of triplicates are shown with standard deviation. (B) Acid phosphatase activities of yeast cells grown as in A. The means of triplicates are shown with standard deviation. (C) Concentrations of remaining Pi in the medium during cell growth. Pi concentration in the medium was monitored every 2 h for 8 h using the malachite green assay. The means of triplicates are shown with standard deviation; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant by Student’s t test; nd, not detected. (D) Fluorescence microscopy of live yeast cells producing Pho4 genomically tagged with GFP as the sole source of this protein. Cells were incubated for 8 h in 10 mM, 0.5 mM, and 0 mM Pi medium as in A before observation. (E and F) Relative gene expression levels of PHO5 (E) and PHO84 (F). Cells were grown in 10 mM, 0.5 mM, and 0 mM Pi medium for 8 h and harvested for RNA extraction and qRT-PCR. Fold change values were normalized with internal control TAF10. The means of three biological replicates are shown with standard deviation; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant by Student’s t test. (G) Polyphosphate levels in different Pi-containing medium. Cells were incubated for 8 h as in A and harvested for polyphosphate measurement. The means of triplicates are shown with standard deviation; ***, P < 0.001; **, P < 0.01; *, P < 0.05 by Student’s t test; a.u., arbitrary units.
Under Pi-limiting conditions, vacuolar poly(P) is degraded (18, 34, 45, 46, 54). It is assumed that resulting Pi is exported from the vacuoles to replenish the cytosolic Pi pool. In 0.25 mM, 0.125 mM, and 0 mM Pi, poly(P) was completely degraded (Fig. 1G). However, after 8 h of growth in 0.5 mM Pi, cells retained 40% of poly(P) compared to Pi-replete conditions. We hence chose the described scheme of 8 h of growth in 0.5 mM Pi as a condition for metabolomic analysis under Pi limitation because it partially activates the PHO pathway, has no significant effect on cell growth, and allows partial maintenance of the vacuolar poly(P) pool.
Complete Pi starvation induces broad metabolic changes.
For metabolome analyses, logarithmically growing cells were transferred into medium providing abundant Pi (10 mM), Pi scarcity (0.5 mM), or Pi starvation (0 mM Pi). After 8 h of incubation in these medium formulations, 0.5 OD600 units of cells were harvested using vacuum filtration through a polytetrafluoroethylene polymer (PTFE) membrane and immediately frozen in liquid nitrogen (55). To analyze the metabolic effects of Pi limitation, untargeted metabolomic analyses were conducted through hydrophilic interaction liquid chromatography (LC)-mass spectrometry (HILIC-MS). A total of 169 metabolites were identified. Individual metabolic features were normalized by the median of each sample, transformed to log10, and centralized to the mean by an autoscaling method for further statistical analyses. Partial least-squares discriminate analysis (PLS-DA) was performed to condense the metabolomic data into a simple plot, allowing easy comparison of overall metabolic features (Fig. 2A). This revealed a clear separation of the three growth conditions. Component 1, which comprises the largest difference of the total variance in metabolites (53.3%), placed the 0 mM Pi samples far from 10 mM and 0.5 mM Pi samples, suggesting that the metabolic changes caused by Pi starvation were greater than by Pi limitation (Fig. 2A). In the loading plot, which visualizes the contribution of individual metabolites to components 1 and 2, most phosphate-containing metabolites (red open circle) negatively contributed to component 1, indicating that they decreased under Pi starvation (Fig. 2B). Only two phosphate-containing metabolites, AICAR and 3′,5′-cyclic GMP (3′,5′-cGMP), positively contributed to component 1. For further analysis, we compared variable importance in projection (VIP) scores of each metabolite, which represent the contribution of variables to the PLS-DA model encompassing 10 mM, 0.5 mM, and 0 mM Pi data. ATP showed the highest VIP value among the top 20 most influential metabolites (Fig. 2C), underscoring an interrelation of Pi and ATP metabolism (56).
FIG 2.
Partial least-squares discriminant analysis (PLS-DA) of S. cerevisiae metabolites under different Pi conditions. (A) Score plot of PLS-DA. Red, green, and blue dots indicate the replicates of yeast metabolomic data incubated in 10 mM, 0.5 mM, and 0 mM Pi medium, respectively. The shaded regions represent the 95% confidence intervals. (B) Loading plot of PLS-DA. Red dots indicate Pi-containing metabolites. (C) Variable of importance in projection (VIP) scores of the top 20 metabolites generated from PLS-DA. The color code indicates the relative abundance of each metabolite under different Pi conditions.
Pearson coefficients were calculated as a measure for the correlation between metabolites. Two metabolic groups (group 1 and group 2) were negatively correlated with each other (Fig. 3A). The relative abundance of metabolites in 0 mM Pi medium increased for group 1 and decreased for group 2 (Fig. 3B and C). Purine and pyrimidine pathway metabolites, nucleosides, and nucleobases increased (Table 1), mirrored by a decrease of nucleotides. Metabolites of the citrate cycle (TCA) increased, such as citric acid, isocitric acid, and oxoglutaric acid, whereas the levels of glycolytic intermediates declined, suggesting an altered strategy for energy production under Pi starvation. Nicotinate and nicotinamide metabolites also decreased. By contrast, metabolites of the tryptophan degradation pathway, which are involved in NAD de novo synthesis, accumulated.
FIG 3.
Correlation analysis of S. cerevisiae metabolites under different Pi starving conditions. (A) Clustered correlation heatmap between metabolites under different Pi conditions. The correlation matrix was generated by Pearson correlation coefficients, which are represented by a color code. Red and blue indicate positive and negative correlations, respectively. Two representative metabolic groups showing strong correlations are marked as group 1 and group 2. (B and C) Relative abundance of metabolites included in group 1 (B) and group 2 (C) under different Pi conditions. The profiles of individual metabolites are shown in gray.
TABLE 1.
List of metabolites in group 1 and group 2 from the correlation heatmap
| Group | Metabolic pathway | Metabolites |
|---|---|---|
| Group 1 | Purine | Deoxyadenosine, adenosine, AICAR, guanosine, inosine, xanthine, xanthosine |
| Pyrimidine | Cytosine, uridine | |
| TCA | Citric acid, isocitric acid, oxoglutaric acid | |
| Cysteine and methionine | S-Adenosylmethionine, 5′-methylthioadenosine | |
| Tryptophan | Kynurenine, kynurenic acid, xanthurenic acid, quinolinic acid | |
| Glycerolipid | Choline, glyceric acid | |
| Others | 5′-Deoxyadenosine, acetylcarnitine, cadaverine, glutamine, methylglutaric acid, N-acetylphenylalanine, N-methylglutamic acid, N-6-trimethyllysine, p-hydroxyphenylacetic acid, pantothenic acid, pyridoxal | |
| Group 2 | Purine | AMP, ADP, ADP ribose, ATP, dATP, GMP, GDP, guanine, IMP, hypoxanthine |
| Pyrimidine | CMP, CDP, dCDP, UMP, UDP, orotic acid | |
| Glycolysis | Acetyl-CoA, fructose-6-phosphate, glyceraldehyde-3-phosphate, glucose-6-phosphate, 2/3-phosphoglyceric acid, phosphoenolpyruvic acid | |
| Cysteine and methionine | 1-Aminocyclopropanecarboxylic acid, ophthalmic acid, S-adenosylhomocysteine, cystathionine, 3-sulfinoalanine, pyroglutamic acid | |
| Nicotinate and nicotinamide | NAD+, NADP+, nicotinic acid mononucleotide | |
| Glycerolipid | sn-Glycerol-3-phosphate, ethanolamine, glycerol | |
| Arginine | N-Acetylputrescine, proline, N-acetylglutamic acid, citrulline, dimethylarginine | |
| Others | Guaiacol, aminoisobutyric acid, 2-hydroxyglutaric acid, aspartic acid, glutamic acid, glycine, histidine, lysine, serine, threonine, mevalonic acid, N2-acetyllysine, TMP, UDP-hexose, UDP-N-acetylhexosamine |
Pathway analysis of Pi starvation.
To identify metabolites that accumulated differentially and in a statistically significant manner, we compared their relative abundance by volcano plots. Profound metabolic effects occurred under complete Pi starvation; 31 metabolites increased and 49 decreased in a statistically meaningful way, representing 47% of the detected metabolites (Fig. 4B and D; Data Set S1 in the supplemental material). Pathway analysis of these 80 metabolites identified the most affected pathways as purine metabolism, pyrimidine metabolism, nicotinate and nicotinamide metabolism, glycolysis/gluconeogenesis, citrate cycle, and cysteine and methionine metabolism, with a −log (P value) of >1.5 (Fig. 5B; Table 2).
FIG 4.
Metabolites differentially accumulated under Pi limitation or Pi starvation. Volcano plot analysis of changes after Pi limitation and Pi starvation. (A) Changes after Pi limitation (0.5 mM Pi). Red dots indicate differentially accumulated metabolites (fold change, |FC| > 1.5; P < 0.1). (B) Changes after Pi starvation (0 mM Pi). Red dots indicate differentially accumulated metabolites (fold change, |FC| > 2; P < 0.1). These metabolites are listed in Data Set S1. (C) Heatmap of Pi limitation. The list was selected by t test (P < 0.1), showing metabolites changing at least 1.5-fold under Pi limitation (0.5 mM Pi). (D) Heatmap of Pi starvation. Same as in C but shows metabolites changing at least 2-fold under Pi starvation (0 mM Pi). The relative abundance of metabolites is represented as log2 (fold change) through a color code.
FIG 5.
Pathway analysis of differentially accumulated metabolites under different Pi starving conditions. (A and B) Pathway analysis of differentially accumulated metabolites after Pi limitation (0.5 mM Pi) (A) and Pi starvation (0 mM Pi) (B). The size and color of the circle indicate impact value and P value, respectively. The annotated metabolic pathways have higher statistical significance (−log [P] > 1.5).
TABLE 2.
Six out of the top 15 metabolic sets are significantly affected under Pi starvation
| Pathwaya | Match statusb | P value | −log (P)c | Impactd |
|---|---|---|---|---|
| Purine metabolism | 17/62 | 0.0000056885 | 5.245 | 0.43543 |
| Pyrimidine metabolism | 9/34 | 0.0016028 | 2.7951 | 0.47488 |
| Glycolysis or gluconeogenesis | 6/24 | 0.013697 | 1.8634 | 0.41811 |
| Nicotinate and nicotinamide metabolism | 4/12 | 0.015489 | 1.81 | 0.50185 |
| Cysteine and methionine metabolism | 8/41 | 0.02079 | 1.6822 | 0.21078 |
| Citrate cycle (TCA cycle) | 5/20 | 0.024305 | 1.6143 | 0.2306 |
| Methane metabolism | 5/23 | 0.042866 | 1.3679 | 0.32774 |
| Lysine biosynthesis | 4/16 | 0.043511 | 1.3614 | 0.0 |
| Glycine, serine, and threonine metabolism | 6/32 | 0.052435 | 1.2804 | 0.30324 |
| Arginine biosynthesis | 4/18 | 0.064018 | 1.1937 | 0.14819 |
| Pentose phosphate pathway | 4/18 | 0.064018 | 1.1937 | 0.52896 |
| Glyoxylate and dicarboxylate metabolism | 5/26 | 0.068182 | 1.1663 | 0.26506 |
| Cyanoamino acid metabolism | 2/8 | 0.14937 | 0.82572 | 0.0 |
| Synthesis and degradation of ketone bodies | 1/3 | 0.23995 | 0.61987 | 0.0 |
| Beta-alanine metabolism | 2/11 | 0.24839 | 0.60487 | 0.0 |
The data correspond to Fig. 4B; bold font indicates significant pathways.
(Number of significant metabolites)/(total number of metabolites) in given pathway.
The top 15 metabolic pathways were ranked based on P value. The metabolic pathways with a −log (P) value of >1.5 were considered significantly altered in this analysis.
The impact value was calculated by pathway topology analysis.
Metabolites significantly affected by different Pi conditions. Download Data Set S1, XLSX file, 0.01 MB (15.4KB, xlsx) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Results of two-way ANOVA of wild-type and Δvtc4 cells under 2 h of Pi-replete (10 mM Pi) and Pi-starving conditions (0 mM Pi). Download Data Set S2, XLSX file, 0.01 MB (15.5KB, xlsx) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(i) Glycolysis and the TCA cycle. Pi starvation reduced the abundance of glycolysis intermediates 2- to 10-fold, including glucose-6-phosphate, fructose-6-phosphate, glyceraldehyde-3-phosphate, 2/3-phosphoglyceric acid, phosphoenolpyruvic acid, and acetyl-coenzyme A (acetyl-CoA) (Fig. S1A). In addition, ribose-5-phosphate, which is derived from glucose-6-phosphate via the oxidative pentose phosphate pathway (PPP), was reduced. The upstream TCA cycle metabolites, including citric acid, isocitric acid, and oxoglutaric acid, accumulated 2- to 10-fold, whereas later TCA cycle metabolites, including succinic acid and malic acid, did not change (Fig. S1B). Thus, Pi starvation has a strong impact on glycolysis and the TCA cycle.
Metabolic changes in energy metabolism pathways under different Pi conditions. (A and B) Metabolomic data of yeast under different Pi conditions are presented on energy metabolic pathways, including glycolysis (A) and the TCA cycle (B). Pi-containing substrates are marked as red. Gray indicates enzymes involved in the reaction. Different letters on the graph indicate a significant difference by Tukey-Kramer test (P < 0.05). Download FIG S1, TIF file, 0.7 MB (762.5KB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(ii) Nicotinate and nicotinamide metabolism. The Pi-containing metabolites nicotinic acid mononucleotide, NAD+, and NADP+ decreased 2- to 5-fold under Pi starvation (Fig. S2). By contrast, metabolites of NAD+ de novo synthesis, also known as the kynurenine pathway, accumulated more than 6-fold. Based on these results, it can be speculated that the impaired NAD+ synthesis from the kynurenine pathway will affect intracellular redox homeostasis.
Metabolic changes in nicotinate and nicotinamide pathway under different Pi conditions. Metabolomic data of yeast under different Pi conditions are presented on a metabolic pathway. Pi-containing substrates are marked as red. Gray indicates enzymes involved in the reaction. Different letters on the graph indicate a significant difference by Tukey-Kramer test (P < 0.05). Download FIG S2, TIF file, 0.8 MB (779.7KB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(iii) Cysteine and methionine metabolism. S-Adenosylmethionine (SAM) is the major methyl donor for modifications of various biomolecules, including proteins, DNA, RNA, and metabolites, producing S-adenosylhomocysteine (SAH) as a byproduct of the reaction. The relative abundance of SAM and of methionine salvage pathway metabolites increased while that of SAH decreased under Pi starvation (Fig. S3). Cystathionine, which can be produced from SAH via homocysteine, diminished under Pi starvation, whereas metabolites derived from cystathionine, such as glutathione and taurine, were not significantly affected. Pi starvation also affected another SAM-dependent branch, the synthesis of polyamines, because a byproduct of polyamine synthesis, 5′-methylthioadenosine (MTA), accumulated strongly. These results suggest that yeast cells change their strategy of SAM utilization under Pi deprivation. The reduction of nucleic acid synthesis, which accompanies growth arrest, may reduce consumption of SAM for nucleotide synthesis and promote accumulation of this compound.
Metabolic changes in methionine and cysteine metabolism pathways under different Pi conditions. Metabolomic data of yeast under different Pi conditions are presented on a metabolic pathway. Pi-containing substrates are marked as red. Gray indicates enzymes involved in the reaction. Different letters on the graph indicate a significant difference by Tukey-Kramer test (P < 0.05). Download FIG S3, TIF file, 0.8 MB (854.6KB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(iv) Purine and pyrimidine metabolism. Nucleosides and nucleobases, such as adenosine, inosine, xanthosine, guanosine, uridine, cytosine, and xanthine, significantly accumulated under Pi starvation (Fig. S4A and B), whereas Pi-containing nucleotides, nucleoside diphosphates, and nucleoside triphosphates decreased. By contrast, the amount of AICAR increased. This metabolic change may contribute to triggering the PHO pathway under Pi starvation in two ways. First, AICAR inhibits the production of IP8 (57), which itself is a potent suppressor of the PHO pathway (9). Second, AICAR stabilizes the interaction of the association of the transcription factors Pho4 and Pho2, which is necessary for full induction of the PHO pathway (17).
Metabolic changes in purine and pyrimidine metabolism pathways under different Pi conditions. (A and B) Metabolomic data of yeast under different Pi conditions are presented on a purine metabolic pathway (A) and a pyrimidine metabolic pathway (B). Pi-containing substrates are marked as red. Gray indicates enzymes involved in the reaction. Different letters on the graph indicate a significant difference by Tukey-Kramer test (P < 0.05). Download FIG S4, TIF file, 0.8 MB (826.8KB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Moderate Pi limitation causes few potentially diagnostic metabolic changes.
Few significant changes (−log [P] > 1) occurred under Pi-limiting conditions; only 17 metabolites increased and 10 decreased more than 1.5-fold (Fig. 4A and C and 5A; Data Set S1). ATP decreased in a statistically significant manner, which suggests that the ATP level is more sensitive to Pi availability than most other metabolites, making ATP a bona fide early indicator of Pi scarcity. In line with this, one class of key enzymes for signaling the intracellular Pi state, IP6 kinases, have an unusually high Km for ATP, which is close to the normal cellular concentration of this compound (58). To test this possibility, we measured the products of these enzymes, inositol pyrophosphates, under different Pi conditions using capillary electrophoresis-coupled electrospray ionization mass spectrometry (CE-ESI-MS). Under Pi starvation, 1,5-IP8 was not detected at all, 5-IP7 and 1-IP7 decreased by 80%, and IP6 decreased by 60% (Fig. 6A to D). In contrast to the PP-IPs, the decrease of IP6 occurred only with a lag phase of 2 h (Fig. S8A to D). Even under mild Pi scarcity, all four compounds significantly declined in comparison with Pi-replete conditions, by 60% for 1,5-IP8, 30% for 5-IP7, and 50% for 1-IP7 and IP6. This is consistent with the hypothesis that even moderate decreases in ATP levels under Pi limitation can be translated into decreased PP-IP levels, which then activate SPX domain-based signaling to stabilize cytosolic Pi.
FIG 6.
Inositol pyrophosphate profiles under different Pi conditions. (A to D) Inositol pyrophosphate levels of yeast cells grown in 10 mM, 0.5 mM, and 0 mM Pi medium for 8 h. The data were normalized by the number of cells. The amount of each inositol pyrophosphate in 10 mM Pi medium was set to 1. The means of triplicates are shown with standard deviation; ***, P < 0.001; **, P < 0.01; *, P < 0.05 by Student’s t test; nd, not detected; a.u., arbitrary unit.
Inositol pyrophosphate profiles. Time course analysis under Pi starvation and Δvtc4 under Pi-replete conditions. (A to D) Time course of PP-IP levels of wild-type yeast cells transferred to 0 mM Pi medium for 8 h. Logarithmically growing yeast cells in 10 mM Pi medium were transferred into 0 mM Pi medium. In contrast to the direct extraction of the cultures with perchloric acid in Fig. 6, cells were recovered by filtration after the indicated periods of Pi starvation. The filters were extracted with perchloric acid, and the inositol pyrophosphates were quantified by CE-MS. Note that the filtration method used here is less efficient for the small quantities of remaining PP-IPs after starvation. The data were normalized by the number of cells per sample. The concentration of each inositol pyrophosphate in 10 mM Pi medium was set to 1. (E to H) Wild-type and Δvtc4 yeast cells were logarithmically grown in 10 mM Pi medium overnight, harvested by centrifugation, and subjected to perchloric acid extraction and PP-IP analysis as in A. The data were normalized by the number of cells. The amount of each inositol pyrophosphate in wild-type cells was set to 1. Means with standard deviations are shown; n = 3; a.u., arbitrary units; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant by Student’s t test. Download FIG S8, TIF file, 0.5 MB (540KB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Poly(P) in acidocalcisome-like vacuoles contributes to Pi homeostasis even under Pi-replete conditions.
Yeast cells contain acidocalcisome-like vacuoles, which can convert the γ-phosphate from ATP into inorganic polyphosphate. Thereby, they can store phosphate units at concentrations in the hundreds of millimolar in an osmotically inactive form (40, 41). At the same time, vacuoles contain polyphosphatases and Pi exporters, which can hydrolyze poly(P) and export the liberated Pi to the cytosol (46). This system is powerful enough to influence Pi homeostasis of the cells and the Pi starvation response when it is dysregulated (18, 37, 38, 54). To investigate the metabolic role of vacuolar polyphosphates, we analyzed the metabolic profiles of the Δvtc4 mutant, which lacks the poly(P)-synthesizing complex VTC, under Pi-replete conditions and Pi starvation. To maximize the chance of observing poly(P)-dependent differences, we restricted the starvation period to 2 h because wild-type cells mobilize their poly(P) pool over the first 2 to 3 h of Pi starvation (18–20). A PLS-DA plot showed that the metabolic features of Δvtc4 cells were clearly distinct from those of the wild type under Pi-rich conditions and even more under Pi starvation (Fig. 7A). The loading plot of PLS-DA revealed that almost all phosphate-containing metabolites (red open circles) were abundant under Pi-rich conditions, except AICAR, consistent with the results above (Fig. 2B and 7B).
FIG 7.
Multivariate statistical analysis of metabolite profiling data from wild-type and Δvtc4 cells under 10 mM and 0 mM Pi conditions. (A) Score plot of partial least-squares discriminant analysis (PLS-DA). Red, green, blue, and light blue indicate the replicates of metabolomic data from wild-type 10 mM Pi, wild-type 0 mM Pi, Δvtc4 10 mM Pi, and Δvtc4 0 mM Pi, respectively. The shaded regions represent the 95% confidence intervals. (B) Loading plot of PLS-DA. Red dots indicate Pi-containing metabolites.
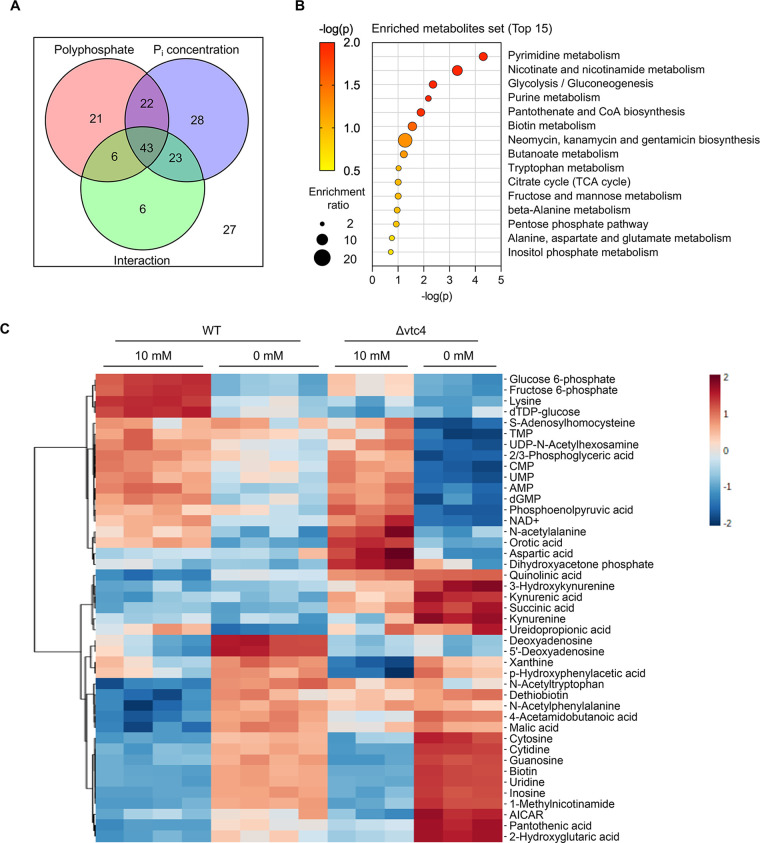
To analyze how polyphosphate synthesis and Pi concentration in the medium affect metabolic features, a two-way analysis of variance (ANOVA) was performed. The relative abundances of 92, 116, and 78 metabolic features were affected by poly(P), Pi concentration, and their interaction, respectively (Fig. 8A; Data Set S2). A total of 65 metabolites were affected by both poly(P) and Pi, of which 66% (43 features) were additionally affected by their interaction. Metabolite set enrichment analysis was performed using these 43 metabolites. Most metabolic pathways affected by Pi starvation shown in the previous pathway analysis, such as pyrimidine metabolism, nicotinate and nicotinamide metabolism, glycolysis, and purine metabolites, were again ranked statistically high, showing consistency between the analyses (Fig. 8B; Table 3). The changes of these 43 metabolites were visualized by heatmap analysis (Fig. 8C). Δvtc4 cells showed much more pronounced changes than wild-type cells in several respects, including the decrease of Pi-containing purines and pyrimidines (CMP, UMP, AMP, and dGMP) (Fig. 8C; Fig. S5A and B), the increase of nucleosides and nucleobases (cytosine, cytidine, guanosine, uridine, and inosine) (Fig. 8C; Fig. S7 and S8), and the reduction of NAD+ and NADP+ (Fig. S6).
FIG 8.
Interrelated effect of polyphosphate and Pi starvation on metabolic pathways. (A) Summary of two-way ANOVA analysis (adjusted P value of <0.05). Red, blue, and green represent the metabolites affected by polyphosphate (wild type and Δvtc4), Pi concentration (10 mM and 0 mM Pi), and interaction between both (polyphosphate and Pi concentration), respectively. (B) Metabolite set enrichment analysis of 43 metabolites simultaneously affected by polyphosphate, Pi concentration, and their interaction. The top 15 metabolite sets were selected based on P value. (C) A heatmap was generated based on the list of 43 metabolites affected by Pi concentration, poly(P), and their interaction from a two-way ANOVA. Features were clustered by Euclidean distance using Ward’s clustering method. The color code indicates the normalized intensity of metabolic features.
TABLE 3.
Six out of the top 15 pathways emerging from metabolite set enrichment analysis are significant
| Metabolite seta | Total | Hits | Expected | P valueb |
|---|---|---|---|---|
| Pyrimidine metabolism | 39 | 7 | 1.05 | 0.0000494 |
| Nicotinate and nicotinamide metabolism | 15 | 4 | 0.404 | 0.0005 |
| Glycolysis/gluconeogenesis | 26 | 4 | 0.7 | 0.00441 |
| Purine metabolism | 65 | 6 | 1.75 | 0.00666 |
| Pantothenate and CoA biosynthesis | 19 | 3 | 0.512 | 0.013 |
| Biotin metabolism | 10 | 2 | 0.269 | 0.0278 |
| Neomycin, kanamycin, and gentamicin biosynthesis | 2 | 1 | 0.0539 | 0.0532 |
| Butanoate metabolism | 15 | 2 | 0.404 | 0.0596 |
| Tryptophan metabolism | 41 | 3 | 1.1 | 0.0954 |
| Citrate cycle (TCA cycle) | 20 | 2 | 0.539 | 0.0991 |
| Fructose and mannose metabolism | 20 | 2 | 0.539 | 0.0991 |
| β-Alanine metabolism | 21 | 2 | 0.566 | 0.108 |
| Pentose phosphate pathway | 22 | 2 | 0.593 | 0.117 |
| Alanine, aspartate, and glutamate metabolism | 28 | 2 | 0.754 | 0.173 |
| Inositol phosphate metabolism | 30 | 2 | 0.808 | 0.192 |
The data correspond to Fig. 8B; bold font indicates significant pathways.
The top 15 metabolite sets were ranked based on P value. Metabolic sets with a P value of <0.05 were considered significantly altered.
Metabolic changes in purine and pyrimidine metabolism pathways under Pi starvation in wild-type and Δvtc4 cells. (A and B) Metabolomic data of yeast under Pi starvation in wild-type and Δvtc4 cells are presented on a purine metabolic pathway (A) and a pyrimidine metabolic pathway (B). Pi-containing substrates are marked as red. Gray indicates enzymes involved in the reaction. Different letters on the graph indicate a significant difference by Tukey-Kramer test (P < 0.05). Download FIG S5, TIF file, 0.9 MB (969.7KB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic changes in nicotinate and nicotinamide pathways under Pi starvation in wild-type and Δvtc4 cells. Metabolomic data of yeast under Pi starvation in wild-type and Δvtc4 cells are presented in a metabolic pathway. Pi-containing substrates are marked as red. Gray indicates enzymes involved in the reaction. Different letters on the graph indicate a significant difference by Tukey-Kramer test (P < 0.05). Download FIG S6, TIF file, 0.8 MB (870.8KB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic changes in glycolysis under Pi starvation in wild-type and Δvtc4 cells. Metabolomic data of yeast under Pi starvation in wild-type and Δvtc4 cells are presented in a metabolic pathway. Pi-containing substrates are marked as red. Gray indicates enzymes involved in the reaction. Different letters on the graph indicate a significant difference by Tukey-Kramer test (P < 0.05). Download FIG S7, TIF file, 1.2 MB (1.2MB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Δvtc4 cells grown on Pi-replete medium showed numerous metabolic features of Pi-starved wild-type cells. They did not, however, simply phenocopy a Pi starvation response at a reduced scale. Intermediates of tryptophan degradation, such as kynurenine, kynurenic acid, 3-hydroxykynurenine, and the NAD+ precursor quinolinic acid, which did not change significantly in Pi-starved wild-type cells, were increased in Δvtc4 cells already under Pi-replete conditions and increased further (up to 5-fold) after Pi starvation (Fig. 8C; Fig. S6). In Δvtc4 cells, the later glycolytic intermediates dihydroxyacetone phosphate and phosphoenolpyruvate were more abundant than in wild-type cells in Pi-replete medium, and 2/3-phosphoglyceric acid underwent a much more pronounced reduction than in wild-type cells after Pi starvation (Fig. 8C; Fig. S7). These results indicate that poly(P) synthesis by VTC has a significant effect on the metabolic profile of cells. Poly(P) synthesis dampens the metabolic consequences of Pi starvation, which is consistent with its proposed role as a Pi reserve, but it also has, so far, unrecognized metabolic functions under Pi-replete conditions, as indicated by metabolic features of Δvtc4 cells that cannot be recapitulated by Pi scarcity or Pi starvation of wild-type cells.
DISCUSSION
Our results extend earlier studies of Pi starvation (56, 59, 60). In agreement with these studies, we observed reductions in nucleotides and late glycolytic intermediates and increased nucleoside and nucleobase levels. These changes can be explained by simple mass action (60, 61) because they reduce phosphate-containing metabolites under conditions of intracellular Pi shortage. By contrast, early TCA cycle metabolites, such as citric acid, isocitric acid, and oxoglutaric acid, strongly increased during Pi starvation. Furthermore, oxygen consumption of yeast increases after Pi starvation (62). This suggests that mitochondrial respiration may become activated as an alternative mechanism for energy production because it fixes less Pi in metabolic intermediates than ATP production based on glycolysis. In line with this, we made the side observation that Pi starvation also caused mitochondrial fragmentation (data not shown). Mitochondria fragment in medium favoring respiration, such as nonfermentable carbon sources or glucose-limited medium (63, 64).
Following Pi starvation, two Pi-containing metabolites increased, AICAR and cGMP (Fig. 2B; Fig. S4A in the supplemental material). Little is known about the roles of cGMP in yeast so far (65); however, it provides a potential link to protein kinase A signaling, which is involved in Pi homeostasis and signaling through Pi transporters in yeast (66–72). cGMP can also inhibit DNA polymerase (73), which might reduce Pi consumption by cells and avoid Pi depletion during S phase (47). AICAR is an intermediate of de novo purine biosynthesis and activates a master regulator of energy homeostasis, AMP-activated protein kinase (AMPK), in mammalian cells (74), but the yeast AMPK Snf1 does not depend on it. Snf1 is activated by ADP (17, 75). After a block in nucleoside synthesis, accumulating AICAR stimulates the PHO transcription pathway by stabilizing the interaction of the responsible transcription factors Pho4 and Pho2 (17). Furthermore, AICAR reduces the activity of mammalian PPIP5 kinase (57), suggesting that it might contribute to the decline of IP7 and IP8 after Pi starvation that also occurs in these cells (76). However, it has remained unknown how AICAR behaves under Pi starvation. Our analysis now shows that AICAR accumulates during Pi starvation. AICAR accumulation should be favored by the decrease of ADP and ATP levels following Pi starvation because these nucleotides exert feedback inhibition on the first step in purine synthesis and thereby on AICAR synthesis (77). Thus, AICAR may promote expression of PHO genes following Pi starvation. However, under Pi scarcity, when ATP decreases less severely than under Pi starvation, AICAR did not increase. This corresponds to the only partial activation of the PHO pathway under Pi scarcity. We hence propose that an increase in AICAR may contribute to switching the PHO pathway from partial to full activation when cells transit from Pi scarcity to starvation.
Glucose-6-phosphate is used by the pentose phosphate pathway (PPP) to convert NADP+ to NADPH, which is essential for cellular redox homeostasis (78, 79). Although NADPH was not detected in our metabolomic analysis, NADP+ decreased, and we hence assume that NADPH should decrease as well. NAD+, nicotinic acid mononucleotide, and nicotinic acid, which are precursors of NADP+, were all reduced by Pi starvation, but intermediates of the kynurenine pathway, also known as the de novo NAD+ synthetic pathway, were significantly accumulated (Fig. S2). These changes may result from the accumulation of AICAR, which stimulates the expression of enzymes involved in the kynurenine pathway (80). The last metabolite of the kynurenine pathway, quinolinic acid, is converted to nicotinic acid mononucleotide by consuming phosphoribosyl pyrophosphate (PRPP). Because PRPP is produced from ribose-5-phosphate, this reaction may be impaired through the decrease in ribose-5-phosphate under Pi starvation, favoring the observed accumulation of quinolinic acid. In addition, the PHO pathway directly promotes the catabolism of NAD+ by inducing the vacuolar phosphatase Pho8, which removes Pi from nicotinic acid mononucleotide and nicotinamide mononucleotide (81). The decrease in the NAD+ and NADP+ pools is expected to affect intracellular redox homeostasis. This may increase the dependence of cells on the oxidative stress response for surviving Pi starvation, which had been previously observed (62). In line with this, cells with perturbed Pi and inositol pyrophosphate homeostasis induce the environmental stress response (82, 83).
We observed increased SAM and decreased SAH under Pi starvation. SAM is synthesized from methionine and ATP, releasing Pi and PPi. SAM provides methyl groups for methyl transfer reactions, generating SAH as a byproduct (84). Histone methylation affects global gene expression patterns by changing the structure of chromatin through interactions with various chromatin remodeling factors and transcription regulators (85, 86). The expression of PHO genes is also under the control of histone methylation. Expression of Pho5 and Pho84 is induced in the Δset1 mutant, which affects methylation of Lys4 of histone H3 (87, 88). In addition, the methyltransferase Hmt1 promotes expression of several Pi-responsive genes (89). Thus, the changes of SAM following Pi starvation might alter the intracellular methylation status and thereby provide a further route of input for Pi-dependent gene expression.
An interesting question is how cells distinguish Pi scarcity from Pi starvation. This is challenging because Pi scarcity can be corrected by cells through partial induction of the PHO pathway. The resulting improved capacity for Pi scavenging (e.g., through expression of high-affinity Pi transporters) apparently allows them to maintain sufficient metabolic performance to support normal growth. Furthermore, positive feedback loops involving Spl2, a small regulator of the Pi transporter Pho90, may stably commit cells to activation of the starvation program, even if this reestablishes sufficient intracellular Pi supply (20, 51, 52). Nevertheless, the Pi starvation program is not launched in a simple all-or-none fashion. Certain genes are activated at different levels of Pi shortage, as exemplified by the gene for the high-affinity transporter Pho84, which is activated earlier than the secreted acid phosphatase Pho5 (18–20). Our analyses of the low-Pi state, which were performed in cells that induced Pho84 but not Pho5, provide hints on metabolic changes that might be used by cells to distinguish Pi scarcity from Pi starvation.
Under Pi scarcity, few metabolites changed in a statistically meaningful way, but the levels of ATP, ADP, and PP-IPs decreased by 30 to 50% compared to Pi-replete conditions. Known properties of the enzymes involved in the production of ATP and in the synthesis of PP-IPs support the following working hypothesis on the reasons for these declines (Fig. 9). Glycolysis and oxidative phosphorylation both contain enzymes that use Pi as a substrate, glyceraldehyde-3-phosphate dehydrogenase and F-ATPase, respectively. Both enzymes have Km values for Pi of around 1.5 to 2 mM (90, 91), rendering them susceptible to declines of Pi below the 1 to 2 mM threshold that cells normally maintain on Pi-replete medium. This can reduce ATP production by both pathways following Pi scarcity. PP-IP production is sensitive to both ATP and Pi. The IP6 kinases synthesizing inositol pyrophosphates have Km values for ATP of 1 to 2 mM, which is in the range of the cellular ATP concentration under Pi-replete conditions (58). While PPIP5 kinase activity, with its Km for ATP of 0.1 mM, is shielded from changes in ATP and weakly stimulated by Pi (92, 93), the opposing phosphatase activity of this bifunctional enzyme is increasingly inhibited in the Pi range from 0.1 to 2 mM, covering the normal cellular concentration range (93). The combination of these enzymatic properties may allow an even moderate decrease in Pi to significantly reduce PP-IP levels. As we recently observed that the PHO pathway is repressed by PP-IPs through the SPX domain of Pho81 (9), we propose that this decline of PP-IPs following Pi scarcity reduces Pho85/Pho80 activity and promotes partial Pho4 nuclear translocation and partial activation of the PHO pathway, allowing cells to maintain normal growth.
FIG 9.

Working hypothesis on the translation of cytosolic Pi concentration into changes of PP-IPs. The scheme illustrates the inhibitory (red) and stimulatory (green) influences of Pi on key enzymes of ATP and PP-IP production, which are postulated to result from the high Km and half-maximal inhibitory concentration (IC50) values of GAP-DH, F-ATPase, IP6 kinase, and PPIP5 kinase. Details are discussed in the main text.
Yeast stores Pi in the form of polymers in acidocalcisome-like vacuoles. Under Pi-limiting conditions, polyphosphatases degrade polyphosphate and liberate Pi, which could potentially be brought back to the cytosol through the vacuolar Pi transporter Pho91 (34, 45, 46). In this way, the vacuolar poly(P) pool might buffer the cytosol against sudden drops in Pi and delay the onset of the Pi starvation response (6, 18). In line with this, our metabolomic analysis revealed exaggerated metabolic changes when the poly(P)-deficient Δvtc4 mutant was starved for Pi. This supports a significant role of acidocalcisome-like vacuoles in Pi homeostasis, which may provide the proposed buffer for cytosolic Pi. Such a buffer is of obvious relevance for an ordered transition into Pi starvation and cell cycle arrest (48, 94). Surprisingly, metabolic features of Pi starvation were observed in the Δvtc4 mutant already under high Pi conditions. This phenotype may again reflect the buffering function of poly(P). This function may become important even on high-Pi medium because the duplication of all nucleic acids and phospholipids generates a very high need for Pi in S phase. It has been proposed that this need can transiently exceed the maximal import capacity of cells, necessitating the vacuolar poly(P) store to cover the deficit (47, 48, 54, 95). In line with this, we observed that the amounts of 1,5-IP8, 5-IP7, and 1-IP7 were reduced in the Δvtc4 mutant in high-Pi medium (Fig. S8E to H). An explanation of the starvation features of Δvtc4 cells from this perspective is, however, only partially satisfactory for our data set because Δvtc4 cells on high-Pi medium shows numerous, but not all, features associated with Pi starvation. The trend for some metabolites was even inversed, such as for adenosine, guanosine, orotic acid, acetyl-CoA, and phosphoenolpyruvate. This suggests that poly(P) may have additional metabolic functions that go beyond those of a Pi buffer for the cytosol. There is potential for this because poly(P) has a significant role for the storage of cations, such as for Zn2+, Ca2+, Mn2+, and Mg2+ (96–99), and also for cation uptake, as shown for Mg2+ (100). Furthermore, poly(P) may affect cellular signaling, influencing the stress response (101–104). Here, it may even have direct impact, such as through polyphosphorylation of lysine residues, which modifies yeast topoisomerase 1 (Tpo1) and nuclear signal recognition factor 1 (Nsr1) (105, 106).
In sum, our observations favor a model where lack of Pi induces differential metabolic changes, which together promote the Pi starvation response. Beginning Pi scarcity could be diagnosed through moderate declines in ATP and inositol pyrophosphates, leading the cells to partially activate Pi scavenging systems and maintain normal growth. Profound Pi starvation entails numerous additional metabolic changes, such as through AICAR, SAM, and strong reductions in ATP and inositol pyrophosphates. These changes may fully stimulate the transcriptional Pi starvation and stress responses in a combinatorial manner. We consider the latter point as an attractive potential solution of the specificity problem that is inherent in the task of measuring a very abundant metabolite. Nuclear magnetic resonance (NMR) studies of yeast found total Pi concentrations in the cell to be around 20 mM, of which approximately one-fourth is cytosolic (38, 107). This allows for the estimation of cytosolic Pi under Pi-replete conditions at 5 mM, declining to 1 mM after Pi starvation. Given that Pi is present in the cytosol in millimolar concentrations, it is difficult to envision that it be “measured” by specific binding to a very low-affinity receptor, which might be susceptible to competition by numerous other compounds. Coincidence detection through a network of Pi-dependent metabolic reactions could, however, generate such specificity, even for a highly abundant ligand such as Pi. Therefore, we favor this model of Pi detection.
MATERIALS AND METHODS
Yeast strains.
The S. cerevisiae strains used in this study are listed in Table 4. Endogenous GFP tagging was performed as described previously (108). The yEGFP-CaURA3 was PCR amplified from the plasmid pKT209 by introducing 40-bp homology before and after the stop codon of the Pho4 gene. Gene deletion was conducted based on the CRISPR-Cas9 system as described previously (109). The single guide RNA (sgRNA) was cloned into the sgRNA expression vector and cotransformed into yeast cells with hybridized double-stranded oligonucleotides, which contain 40-bp homology of each side before the start codon and after the stop codon of the Vtc4 gene as the templates for homologous recombination. After transformation, positive colonies were selected by colony PCR and sequencing. PCR primers used for genetic manipulation are listed in Table 5.
TABLE 4.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Euroscarf |
| BY4741 Pho4-yEGFP | MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; Pho4-yEGFP::CaURA | This study |
| BY4741 vtc4Δ | MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0; vtc4Δ | This study |
TABLE 5.
Primers used in this study
| Namea | Sequence | Purpose |
|---|---|---|
| Pho4-yEGFP_f | CTGCCGGTACATCCGTCACCTACAGCAGAACGTGAGCACGggtgacggtgctggttta | Pho4-yEGFP tagging |
| Pho4-yEGFP_r | AGTCCGATATGCCCGGAACGTGCTTCCCATTGGTGCACGGtcgatgaattcgagctcg | |
| Pho4-yEGFP_colony_f | CCGCTCGCACGGAAATATTT | |
| Pho4-yEGFP_colony_r | ACTAAGGTATCACCTTCAAAC | |
| Vtc4_guide_f | CGAAGATAACGACTTCGATGGTTTTAGAG | Vtc4 deletion |
| Vtc4_guide_r | CTAGCTCTAAAACCATCGAAGTCGTTATCTTCGACGT | |
| Vtc4_HR_f | AAATCGGCCAATAAAAGAGCATAACAAGGCAGGAACAGCTATACACAGCGTGTTTTTTTTACTGTATAATTAAGTAATAA | |
| Vtc4_HR_r | TTATTACTTAATTATACAGTAAAAAAAACACGCTGTGTATAGCTGTTCCTGCCTTGTTATGCTCTTTTATTGGCCGATTT | |
| Vtc4_colony_f | GACGGAGAGCTACTGACTTGT | |
| Vtc4_colony_r | TGTGATGGTGACGATGGCATG | |
| Taf10_qRT_f | ATATTCCAGGATCAGGTCTTCCGTAGC | qRT-PCR |
| Taf10_qRT_r | GTAGTCTTCTCATTCTGTTGATGTTGTTGTTG | |
| Pho5_qRT_f | GGGCAACACTTTCCACAGAT | |
| Pho5_qRT_r | CAATTGGAACAACAGCATCG | |
| Pho84_qRT_f | TTCTGCTGCATCTGGTAAGG | |
| Pho84_qRT_r | TCCATGACGTGAGGTAACCA |
f, forward; r, reverse; HR: homologous recombination.
Synthetic complete (SC) medium was prepared using yeast nitrogen base without phosphate (Formedium, UK). The desired phosphate concentration was adjusted by adding KH2PO4. The potassium concentration was controlled by adding KCl instead of KH2PO4.
Media and cell growth.
All media were prepared with ultrapure, UV-treated water from an ultrapurification system (SG, Germany). SC medium was prepared using yeast nitrogen base without phosphate (Formedium, UK). The phosphate concentration was adjusted by adding KH2PO4, and the potassium concentration was kept constant by substituting KCl for KH2PO4.
For assays of growth, acid phosphatase, and malachite green, yeast cells were logarithmically grown overnight in 50 mL of SC medium containing 10 mM Pi up to an OD600 of 1 (4.6 × 107 cells/mL). Cells were sedimented in a table-top centrifuge (3,000 × g) and washed with SC medium containing different concentrations of Pi. After two washing steps, the OD600 was measured, and cells were inoculated in 100-mL Erlenmeyer flasks containing 20 mL of SC medium with the desired Pi concentration (OD600 = 0.05; 2.3 × 106 cells/mL). Cells were incubated at 30°C with shaking at 210 rpm in a shaking incubator (Climo-shaker ISFL-X, Kühner, Switzerland). To assess growth, the OD600 was monitored at different time points (0, 2, 4, 8, and 24 h).
For microscopic analysis, RNA extraction for qRT-PCR, poly(P) measurement, inositol pyrophosphate extraction, and yeast metabolite extraction, yeast cells were prepared in the same manner as described above except that they were transferred into 0 mM Pi medium after the washing steps (OD600 = 0.2; 9.2 × 106 cells/mL).
Acid phosphatase assay.
An acid phosphatase assay was performed as previously described (110). Cells were grown in the same manner for growth assays as described above. At each time point, 0.2 OD600 units of cells (9.2 × 106 cells) were harvested by centrifugation in a bench-top centrifuge (3,000 × g) and resuspended in 250 μL of 0.1 M sodium acetate (pH 4.2) and 250 μL of freshly prepared 9 mg/mL p-nitrophenyl phosphate. The mixture was incubated at 37°C for 9 min, and 800 μL of 1.4 M Na2CO3 was added to stop the reaction. After centrifugation, the OD420 was measured from the supernatant as acid phosphatase activity.
Malachite green assay.
Logarithmically grown cells were inoculated into SC medium containing different concentrations of Pi as described above. At different time points (0, 2, 4, 6, and 8 h), 1 mL of cell culture was transferred into a microcentrifuge tube and sedimented by centrifugation at 13,000 × g for 1 min. The supernatant was transferred into a new tube and diluted with Pi-free SC medium to be within a linear range of detection (200-fold dilution for 10 mM samples, 10-fold dilution for 0.5 mM samples, and no dilution for 0 mM samples). Fifty microliters of diluted samples was mixed with 32 μL of 0.1 mM malachite green solution containing 0.35% (mass/vol) of polyvinyl alcohol (molecular mass 85,000 to 124,000 Da) and 43 μL of 4.48 mM ammonium molybdate solution containing 12.5% (vol/vol) H2SO4. After incubation for 15 min at room temperature, the absorbance was measured at 620 nm on a SpectraMax M3 plate reader (Molecular Devices, USA) in a 96-well clear plate with a flat bottom.
Fluorescence microscopy.
Cells in the logarithmic phase were inoculated in SC medium and grown as described for the growth assay above. Fluorescence images were obtained with a Nikon Eclipse Ti2/Yokogawa CSU-X1 spinning-disk microscope with two Prime BSI scientific complementary metal oxide semiconductor (sCMOS) cameras (Teledyne Photometrics, USA), a LightHub Ultra laser light (Omicron Laserage, Germany), and an Apo total internal-reflection fluorescence (TIRF) ×100/1.49 oil lens (Nikon, Japan). Experiments were repeated at least three times. Representative images are shown in the figures.
RNA extraction and qRT-PCR.
Total RNA was extracted from 10 OD600 units of yeast cells (4.6 × 108 cells) with RNeasy kits (Qiagen, Germany) according to the manufacturer’s instructions. One microgram of total RNA was used for cDNA synthesis using RevertAid reverse transcriptase (Thermo Fisher Scientific, USA). Gene expression levels were quantitatively monitored using real-time PCR (LightCycler 480, Roche, Switzerland) with SYBR Green I master mix (Roche, Switzerland). Gene expression was normalized by using TATA-binding protein-associated factor Taf10 transcript as an internal control. Primers used for qRT-PCR are listed in Table 5. The mean and standard deviation values of gene expression were calculated from three biological replicates with three technical replicates.
Poly(P) measurement.
Poly(P) levels were evaluated from cells using the direct 4′-6-diamidino-2-phenylindole (DAPI) assay (62). Cells were logarithmically grown in Pi-rich SC medium and transferred to SC medium containing different concentrations of Pi, as described above. After 8 h of incubation, 0.5 OD600 units of cells (2.3 × 107 cells) were harvested by centrifugation in a bench-top centrifuge (3,000 × g) and washed with 50 mM HEPES-KOH (pH 7.5). The cell pellet was resuspended in 400 μL of DAPI buffer containing 20 mM HEPES-KOH (pH 6.8), 150 mM KCl, and 10 μM DAPI. After two rounds of freeze-thaw in liquid nitrogen, samples were centrifuged for 2 min at 13,000 rpm. The supernatant was diluted with DAPI buffer (1:20), and 200 μL of diluted sample was transferred to black 96-well polypropylene plates. DAPI fluorescence was measured with a SpectraMax Gemini EM microplate reader (Molecular Devices, USA), with excitation and emission set to 420 nm and 550 nm, respectively (111, 112).
Extraction of inositol pyrophosphates for quantification.
To extract PP-IPs, 1 mL of cell culture grown in the SC medium containing different Pi levels (10 mM, 0.5 mM, and 0 mM) for 8 h was transferred into a microcentrifuge tube and mixed with 100 μL of 11 M perchloric acid. The mixture was snap-frozen in liquid nitrogen, thawed again, and centrifuged (3 min, 13,000 rpm, 4°C). The supernatant was transferred into a new tube. Titanium dioxide (TiO2) beads (GL Sciences, Japan) were washed twice with water and 1 M perchloric acid (1.5 mg of beads/sample) and were added to the perchloric acid extract from the cells. The sample with TiO2 beads was gently rotated for 15 min at 4°C and centrifuged (1 min, 13,000 rpm, 4°C). The beads were washed twice using 500 μL of 1 M perchloric acid and were resuspended in 300 μL of 3% (vol/vol) NH4OH by gentle rotation at room temperature. After centrifugation (1 min, 13,000 rpm), the eluents were transferred into new tubes and completely dried in a SpeedVac (Labogene, Denmark) at 42°C.
For PP-IP extractions (Fig. S8A and B), 4 OD600 units of cells (1.38 × 108 cells) were harvested using rapid vacuum filtration through a PTFE membrane filter (1.2 μm; Piper Filter GmbH, Germany) (55). Yeast cells on the PTFE membrane were resuspended in 400 μL of 1 M perchloric acid, and the samples were lysed by vortexing with glass beads (0.25 to 0.5 mm) for 10 min at 4°C. After centrifugation (3 min, 13,000 rpm, 4°C), the cleared supernatant was transferred into a new tube, and 4 mg of prewashed TiO2 beads was added. The rest of the analysis was performed as described above.
Analysis of inositol pyrophosphates.
The amounts of PP-IPs were quantified by capillary electrophoresis coupled to electrospray ionization mass spectrometry (CE-ESI-MS) as described previously (113–115). CE-ESI-MS analysis was performed on an Agilent 7100 CE coupled to a triple quadrupole mass spectrometer (QqQ MS) Agilent 6495c, equPP-IPed with an Agilent Jet Stream (AJS) ESI source. Stable CE-ESI-MS coupling was enabled by a commercial sheath liquid coaxial interface, with an isocratic LC pump constantly delivering the sheath liquid.
All experiments were performed with a bare fused silica capillary with a length of 100 cm and a 50-μm internal diameter (i.d.); 35 mM ammonium acetate titrated by ammonia solution to pH 9.7 was used as background electrolyte (BGE). The sheath liquid was composed of a water:isopropanol (1:1) mixture, with a flow rate of 10 μL/min. Fifteen microliters of IP extracts was spiked with 0.75 μL of isotopic internal standards mixture (2 μM [13C6]1,5-IP8, 10 μM [13C6]5-IP7, 10 μM [13C6]1-IP7, and 40 μM [13C6]IP6). Samples were introduced by applying 10,000 Pa of pressure for 10 s (20 nL). A separation voltage of 30 kV was applied over the capillary, generating a constant CE current at around 19 μA.
The MS source parameter settings were as follows: nebulizer pressure was 8 lb/in2, sheath gas temperature was 175°C with a flow rate at 8 L/min, gas temperature was 150°C with a flow rate of 11 L/min, and the capillary voltage was −2,000 V with a nozzle voltage of 2,000 V. Negative high pressure radio frequency and low pressure radio frequency (ion funnel parameters) was 70 V and 40 V, respectively. Parameters for multiple reaction monitoring (MRM) transitions are indicated in Table 6.
TABLE 6.
Parameters for MRM transitions
| Compound | Precursor ion | Product ion | Dwell | CE (V) | Cell acceleration (V) | Polarity |
|---|---|---|---|---|---|---|
| [13C6]IP8 | 411.9 | 362.9 | 80 | 10 | 1 | Negative |
| IP8 | 408.9 | 359.9 | 80 | 10 | 1 | Negative |
| [13C6]IP7 | 371.9 | 322.9 | 80 | 10 | 1 | Negative |
| IP7 | 368.9 | 319.9 | 80 | 10 | 1 | Negative |
| [13C6]IP6 | 331.9 | 486.9 | 80 | 17 | 1 | Negative |
| IP6 | 328.9 | 480.9 | 80 | 17 | 1 | Negative |
Preparation of yeast metabolite extracts.
Log-phase yeast cells grown in synthetic complete (SC) medium (0.5 OD600 units) were collected by the vacuum filtration method using a PTFE membrane filter (1.2 μm; Piper Filter GmbH, Germany) (55). Yeast cells on PTFE membranes were resuspended with a methanol:water (4:1, vol/vol) mixture. The samples were homogenized with a Cryolys Precellys 24 sample homogenizer (Bertin Technologies, USA) with ceramic beads. Homogenized extracts were centrifuged for 15 min at 4,000 × g at 4°C. The precipitated protein pellets were used to measure total protein concentration, and the supernatant was collected and evaporated in a vacuum concentrator (LabConco, USA). Dried sample extracts were resuspended in a methanol:water (4:1, vol/vol) mixture based on the total protein content.
LC-MS.
Untargeted metabolite profiling was performed by hydrophilic interaction liquid chromatography coupled to tandem mass spectrometry (HILIC-MS/MS) in both positive and negative ionization modes using a 6495 triple quadrupole system (QqQ) interfaced with a 1290 ultrahigh-performance LC (UHPLC) system (Agilent Technologies, USA) (116). In positive mode, the chromatographic separation was performed in an Acquity BEH Amide, 1.7-μm, 100 mm × 2.1 mm i.d. column (Waters, USA). The mobile phase was composed of A (20 mM ammonium formate and 0.1% formic acid in water) and B (0.1% formic acid in acetonitrile). The linear gradient elution from 95% B (0 to 1.5 min) down to 45% B was applied (1.5 to 17 min), and these conditions were held for 2 min. Initial chromatographic conditions were then maintained as a postrun during 5 min for column reequilibration. The flow rate was 400 μL/min, column temperature was 25°C, and the sample injection volume was 2 μL. ESI source conditions were set as follows: dry gas temperature of 290°C, nebulizer of 35 lb/in2 and flow of 14 L/min, sheath gas temperature of 350°C and flow of 12 L/min, nozzle voltage of 0 V, and capillary voltage of 2,000 V. Dynamic multiple reaction monitoring (DMRM) was used as acquisition mode with a total cycle time of 600 ms. Optimized collision energies for each metabolite were applied. In negative mode, a SeQuant ZIC-pHILIC (100-mm, 2.1-mm i.d., and 5-μm particle size; Merck, Germany) column was used. The mobile phase was composed of A (20 mM ammonium acetate and 20 mM ammonium hydroxide in water, pH 9.7) and B (100% acetonitrile). The linear gradient elution was from 90% (0 to 1.5 min) to 50% B (8 to 11 min) down to 45% B (12 to 15 min). Finally, the initial chromatographic conditions were established as a postrun during 9 min for column reequilibration. The flow rate was 300 μL/min, the column temperature was 30°C, and the sample injection volume was 2 μL. ESI source conditions were set as follows: dry gas temperature of 290°C and flow of 14 L/min, sheath gas temperature of 350°C, nebulizer of 45 lb/in2 and flow of 12 L/min, nozzle voltage of 0 V, and capillary voltage of −2,000 V. DMRM was used as an acquisition mode, with a total cycle time of 600 ms. Optimized collision energies for each metabolite were applied.
Data preprocessing.
Raw LC-MS/MS data were processed using the Agilent Quantitative analysis software (version B.07.00 MassHunter, Agilent Technologies, USA). Relative quantification of metabolites was based on extracted ion chromatogram (EIC) areas for the monitored MRM transitions. The obtained results were exported to R software (http://cran.r-project.org/), and signal intensity drift correction was done within the LOWESS/Spline normalization program followed by noise filtering (Coefficient of Variance [Quality Control features] of >30%).
Statistical analysis of metabolite profiling.
Statistical analyses of metabolomic data were performed by MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/) (117). Before analysis, signal intensity data were median normalized, log transformed, and mean centered using the autoscaling method. PLS-DA of the first metabolic profiling with different Pi conditions was conducted by considering the Pi concentration order. The heatmap of the correlation matrix between metabolites of different Pi conditions was calculated by the Pearson r correlation coefficient. Volcano plot analysis was performed by a two-sample t test. The metabolites showing a P value of <0.1 with an absolute value fold change (|FC|) of >1.5 (10 mM Pi versus 0.5 mM Pi) or |FC| of >2 (10 mM Pi versus 0 mM Pi) were considered statistically meaningful metabolites. Results of the volcano plot analysis were exported and visualized with GraphPad Prism 9 (GraphPad Software, USA). For the second metabolic profiling analysis, using wild-type and Δvtc4 cells, the prominent outliers from the PLS-DA were removed before further analyses. A two-way ANOVA followed by false discovery rate correction (P < 0.05) was performed to investigate metabolite variabilities between two different factors, genotype (wild type and Δvtc4), and Pi conditions (10 mM Pi and 0 mM Pi) and their interaction. A hierarchical clustering heatmap was generated using the Euclidean distance measure with Ward’s clustering method. The 50 most significantly changed metabolites according to the ANOVA were selected for visualization.
Pathway analysis and metabolite set enrichment analysis.
Pathway analysis was performed using MetaboAnalyst 5.0 based on the metabolites that statistically significantly increased or decreased under 0.5 mM Pi (|FC| > 1.5; P < 0.1) or 0 mM Pi (|FC| > 2; P < 0.1) conditions compared to under 10 mM Pi conditions. Hypergeometric test and relative betweenness centrality were used for the enrichment method and topology analysis, respectively. Metabolite set enrichment analysis was performed by MetaboAnalyst 5.0 based on 84 metabolite sets of KEGG human metabolic pathways. Results of pathway analysis and metabolite set enrichment analysis were exported and visualized with GraphPad Prism 9.
ACKNOWLEDGMENTS
We thank the metabolomics facility of University of Lausanne for support with the metabolite analyses. This project was funded through grants from the SNSF (CRSII5_170925, 173915, and 177127 to AM), the ERC (788442 to A.M.) and HFSP (LT000588/2019 to G.-D.K.).
Contributor Information
Andreas Mayer, Email: andreas.mayer@unil.ch.
James W. Kronstad, University of British Columbia
REFERENCES
- 1.Paz-Ares J, Puga MI, Rojas-Triana M, Martinez-Hevia I, Diaz S, Poza-Carrión C, Miñambres M, Leyva A. 2022. Plant adaptation to low phosphorus availability: core signaling, crosstalks and applied implications. Mol Plant 15:104–124. doi: 10.1016/j.molp.2021.12.005. [DOI] [PubMed] [Google Scholar]
- 2.Bhalla K, Qu X, Kretschmer M, Kronstad JW. 2021. The phosphate language of fungi. Trends Microbiol 30:338–349. doi: 10.1016/j.tim.2021.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fabiańska I, Bucher M, Häusler RE. 2019. Intracellular phosphate homeostasis—a short way from metabolism to signaling. Plant Sci 286:57–67. doi: 10.1016/j.plantsci.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 4.Michigami T, Kawai M, Yamazaki M, Ozono K. 2018. Phosphate as a signaling molecule and its sensing mechanism. Physiol Rev 98:2317–2348. doi: 10.1152/physrev.00022.2017. [DOI] [PubMed] [Google Scholar]
- 5.Wild R, Gerasimaite R, Jung J-Y, Truffault V, Pavlovic I, Schmidt A, Saiardi A, Jessen HJ, Poirier Y, Hothorn M, Mayer A. 2016. Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352:986–990. doi: 10.1126/science.aad9858. [DOI] [PubMed] [Google Scholar]
- 6.Austin S, Mayer A. 2020. Phosphate homeostasis—a vital metabolic equilibrium maintained through the INPHORS signaling pathway. Front Microbiol 11:1367. doi: 10.3389/fmicb.2020.01367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Secco D, Wang C, Arpat BA, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J. 2012. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol 193:842–851. doi: 10.1111/j.1469-8137.2011.04002.x. [DOI] [PubMed] [Google Scholar]
- 8.Secco D, Wang C, Shou H, Whelan J. 2012. Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett 586:289–295. doi: 10.1016/j.febslet.2012.01.036. [DOI] [PubMed] [Google Scholar]
- 9.Chabert V, Kim G, Qiu D, Michaillat-Mayer L, Jessen HJ, Mayer A. 2023. Inositol pyrophosphate dynamics in yeast reveals control of the PHO starvation program through 1,5-IP8 and the SPX domain of the CDK inhibitor Pho81. bioRxiv. doi: 10.1101/2023.02.14.528555. [DOI] [PMC free article] [PubMed]
- 10.Komeili A, O'Shea EK. 1999. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 11.O'Neill EM, Kaffman A, Jolly ER, O'Shea EK. 1996. Regulation of PHO4 nuclear localization by the PHO80-PHO85 cyclin-CDK complex. Science 271:209–212. doi: 10.1126/science.271.5246.209. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y-S, Huang K, Quiocho FA, O'Shea EK. 2008. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat Chem Biol 4:25–32. doi: 10.1038/nchembio.2007.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee Y-S, Mulugu S, York JD, O'Shea EK. 2007. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lonetti A, Szijgyarto Z, Bosh D, Loss O, Azevedo C, Saiardi A. 2011. Identification of an evolutionary conserved family of inorganic polyphosphate endopolyphosphatases. J Biol Chem 286:31966–31974. doi: 10.1074/jbc.m111.266320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gauthier S, Coulpier F, Jourdren L, Merle M, Beck S, Konrad M, Daignan-Fornier B, Pinson B. 2008. Co-regulation of yeast purine and phosphate pathways in response to adenylic nucleotide variations. Mol Microbiol 68:1583–1594. doi: 10.1111/j.1365-2958.2008.06261.x. [DOI] [PubMed] [Google Scholar]
- 16.Hürlimann HC, Laloo B, Simon-Kayser B, Saint-Marc C, Coulpier F, Lemoine S, Daignan-Fornier B, Pinson B. 2011. Physiological and toxic effects of purine intermediate 5-amino-4-imidazolecarboxamide ribonucleotide (AICAR) in yeast. J Biol Chem 286:30994–31002. doi: 10.1074/jbc.M111.262659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinson B, Vaur S, Sagot I, Coulpier F, Lemoine S, Daignan-Fornier B. 2009. Metabolic intermediates selectively stimulate transcription factor interaction and modulate phosphate and purine pathways. Genes Dev 23:1399–1407. doi: 10.1101/gad.521809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas MR, O'Shea EK. 2005. An intracellular phosphate buffer filters transient fluctuations in extracellular phosphate levels. Proc Natl Acad Sci USA 102:9565–9570. doi: 10.1073/pnas.0501122102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gurvich Y, Leshkowitz D, Barkai N. 2017. Dual role of starvation signaling in promoting growth and recovery. PLoS Biol 15:e2002039. doi: 10.1371/journal.pbio.2002039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vardi N, Levy S, Gurvich Y, Polacheck T, Carmi M, Jaitin D, Amit I, Barkai N. 2014. Sequential feedback induction stabilizes the phosphate starvation response in budding yeast. Cell Rep 9:1122–1134. doi: 10.1016/j.celrep.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Oshima Y. 1997. The phosphatase system in Saccharomyces cerevisiae. Genes Genet Syst 72:323–334. doi: 10.1266/ggs.72.323. [DOI] [PubMed] [Google Scholar]
- 22.Samyn DR, Persson BL. 2016. Inorganic phosphate and sulfate transport in S. cerevisiae. Adv Exp Med Biol 892:253–269. doi: 10.1007/978-3-319-25304-6_10. [DOI] [PubMed] [Google Scholar]
- 23.Ghillebert R, Swinnen E, Snijder PD, Smets B, Winderickx J. 2011. Differential roles for the low-affinity phosphate transporters Pho87 and Pho90 in Saccharomyces cerevisiae. Biochem J 434:243–251. doi: 10.1042/BJ20101118. [DOI] [PubMed] [Google Scholar]
- 24.Levy S, Kafri M, Carmi M, Barkai N. 2011. The competitive advantage of a dual-transporter system. Science 334:1408–1412. doi: 10.1126/science.1207154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wykoff DD, O'Shea EK. 2001. Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159:1491–1499. doi: 10.1093/genetics/159.4.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy VS, Singh AK, Rajasekharan R. 2008. The Saccharomyces cerevisiae PHM8 gene encodes a soluble magnesium-dependent lysophosphatidic acid phosphatase. J Biol Chem 283:8846–8854. doi: 10.1074/jbc.M706752200. [DOI] [PubMed] [Google Scholar]
- 27.Yadav KK, Singh N, Rajasekharan R. 2015. PHO4 transcription factor regulates triacylglycerol metabolism under low-phosphate conditions in Saccharomyces cerevisiae. Mol Microbiol 98:456–472. doi: 10.1111/mmi.13133. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y-F, Létisse F, Absalan F, Lu W, Kuznetsova E, Brown G, Caudy AA, Yakunin AF, Broach JR, Rabinowitz JD. 2013. Nucleotide degradation and ribose salvage in yeast. Mol Syst Biol 9:665. doi: 10.1038/msb.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gresham D, Boer VM, Caudy A, Ziv N, Brandt NJ, Storey JD, Botstein D. 2011. System-level analysis of genes and functions affecting survival during nutrient starvation in Saccharomyces cerevisiae. Genetics 187:299–317. doi: 10.1534/genetics.110.120766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebrahimi M, Habernig L, Broeskamp F, Aufschnaiter A, Diessl J, Atienza I, Matz S, Ruiz FA, Büttner S. 2021. Phosphate restriction promotes longevity via activation of autophagy and the multivesicular body pathway. Cells 10:3161. doi: 10.3390/cells10113161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smets B, Ghillebert R, Snijder PD, Binda M, Swinnen E, Virgilio CD, Winderickx J. 2010. Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae. Curr Genet 56:1–32. doi: 10.1007/s00294-009-0287-1. [DOI] [PubMed] [Google Scholar]
- 32.Swinnen E, Rosseels J, Winderickx J. 2005. The minimum domain of Pho81 is not sufficient to control the Pho85-Rim15 effector branch involved in phosphate starvation-induced stress responses. Curr Genet 48:18–33. doi: 10.1007/s00294-005-0583-3. [DOI] [PubMed] [Google Scholar]
- 33.Urech K, Dürr M, Boller T, Wiemken A, Schwencke J. 1978. Localization of polyphosphate in vacuoles of Saccharomyces cerevisiae. Arch Microbiol 116:275–278. doi: 10.1007/BF00417851. [DOI] [PubMed] [Google Scholar]
- 34.Hürlimann HC, Stadler-Waibel M, Werner TP, Freimoser FM. 2007. Pho91 Is a vacuolar phosphate transporter that regulates phosphate and polyphosphate metabolism in Saccharomyces cerevisiae. Mol Biol Cell 18:4438–4445. doi: 10.1091/mbc.e07-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lander N, Cordeiro C, Huang G, Docampo R. 2016. Polyphosphate and acidocalcisomes. Biochem Soc Trans 44:1–6. doi: 10.1042/BST20150193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Docampo R, Moreno SNJ. 2011. Acidocalcisomes. Cell Calcium 50:113–119. doi: 10.1016/j.ceca.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desfougères Y, Gerasimaitė RU, Jessen HJ, Mayer A. 2016. Vtc5, a novel subunit of the vacuolar transporter chaperone complex, regulates polyphosphate synthesis and phosphate homeostasis in yeast. J Biol Chem 291:22262–22275. doi: 10.1074/jbc.M116.746784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Auesukaree C, Homma T, Tochio H, Shirakawa M, Kaneko Y, Harashima S. 2004. Intracellular phosphate serves as a signal for the regulation of the PHO pathway in Saccharomyces cerevisiae. J Biol Chem 279:17289–17294. doi: 10.1074/jbc.M312202200. [DOI] [PubMed] [Google Scholar]
- 39.Wiemken A, Matile P, Moor H. 1970. Vacuolar dynamics in synchronously budding yeast. Arch Mikrobiol 70:89–103. doi: 10.1007/BF00412200. [DOI] [PubMed] [Google Scholar]
- 40.Hothorn M, Neumann H, Lenherr ED, Wehner M, Rybin V, Hassa PO, Uttenweiler A, Reinhardt M, Schmidt A, Seiler J, Ladurner AG, Herrmann C, Scheffzek K, Mayer A. 2009. Catalytic core of a membrane-associated eukaryotic polyphosphate polymerase. Science 324:513–516. doi: 10.1126/science.1168120. [DOI] [PubMed] [Google Scholar]
- 41.Gerasimaitė R, Sharma S, Desfougères Y, Schmidt A, Mayer A. 2014. Coupled synthesis and translocation restrains polyphosphate to acidocalcisome-like vacuoles and prevents its toxicity. J Cell Sci 127:5093–5104. doi: 10.1242/jcs.159772. [DOI] [PubMed] [Google Scholar]
- 42.Ogawa N, DeRisi J, Brown PO. 2000. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol Biol Cell 11:4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sethuraman A, Rao NN, Kornberg A. 2001. The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 98:8542–8547. doi: 10.1073/pnas.151269398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumble KD, Kornberg A. 1996. Endopolyphosphatases for long chain inorganic polyphosphate in yeast and mammals. J Biol Chem 271:27146–27151. doi: 10.1074/jbc.271.43.27146. [DOI] [PubMed] [Google Scholar]
- 45.Gerasimaitė R, Mayer A. 2017. Ppn2, a novel Zn2+-dependent polyphosphatase in the acidocalcisome-like yeast vacuole. J Cell Sci 130:1625–1636. doi: 10.1242/jcs.201061. [DOI] [PubMed] [Google Scholar]
- 46.Gerasimaitė R, Mayer A. 2016. Enzymes of yeast polyphosphate metabolism: structure, enzymology and biological roles. Biochem Soc Trans 44:234–239. doi: 10.1042/BST20150213. [DOI] [PubMed] [Google Scholar]
- 47.Bru S, Martínez-Laínez JM, Hernández-Ortega S, Quandt E, Torres-Torronteras J, Martí R, Canadell D, Ariño J, Sharma S, Jiménez J, Clotet J. 2016. Polyphosphate is involved in cell cycle progression and genomic stability in Saccharomyces cerevisiae. Mol Microbiol 101:367–380. doi: 10.1111/mmi.13396. [DOI] [PubMed] [Google Scholar]
- 48.Bru S, Samper-Martín B, Quandt E, Hernández-Ortega S, Martínez-Laínez JM, Garí E, Rafel M, Torres-Torronteras J, Martí R, Ribeiro MPC, Jiménez J, Clotet J. 2017. Polyphosphate is a key factor for cell survival after DNA damage in eukaryotic cells. DNA Repair (Amst) 57:171–178. doi: 10.1016/j.dnarep.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 49.Gerasimaite R, Pavlovic I, Capolicchio S, Hofer A, Schmidt A, Jessen HJ, Mayer A. 2017. Inositol pyrophosphate specificity of the SPX-dependent polyphosphate polymerase VTC. ACS Chem Biol 12:648–653. doi: 10.1021/acschembio.7b00026. [DOI] [PubMed] [Google Scholar]
- 50.Auesukaree C, Tochio H, Shirakawa M, Kaneko Y, Harashima S. 2005. Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. J Biol Chem 280:25127–25133. doi: 10.1074/jbc.M414579200. [DOI] [PubMed] [Google Scholar]
- 51.Wykoff DD, Rizvi AH, Raser JM, Margolin B, O'Shea EK. 2007. Positive feedback regulates switching of phosphate transporters in S. cerevisiae. Mol Cell 27:1005–1013. doi: 10.1016/j.molcel.2007.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vardi N, Levy S, Assaf M, Carmi M, Barkai N. 2013. Budding yeast escape commitment to the phosphate starvation program using gene expression noise. Curr Biol 23:2051–2057. doi: 10.1016/j.cub.2013.08.043. [DOI] [PubMed] [Google Scholar]
- 53.Springer M, Wykoff DD, Miller N, O'Shea EK. 2003. Partially phosphorylated Pho4 activates transcription of a subset of phosphate-responsive genes. PLoS Biol 1:E28. doi: 10.1371/journal.pbio.0000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neef DW, Kladde MP. 2003. Polyphosphate loss promotes SNF/SWI- and Gcn5-dependent mitotic induction of PHO5. Mol Cell Biol 23:3788–3797. doi: 10.1128/MCB.23.11.3788-3797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bordag N, Janakiraman V, Nachtigall J, Maldonado SG, Bethan B, Laine J-P, Fux E. 2016. Fast filtration of bacterial or mammalian suspension cell cultures for optimal metabolomics results. PLoS One 11:e0159389. doi: 10.1371/journal.pone.0159389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boer VM, de Winde JH, Pronk JT, Piper MDW. 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J Biol Chem 278:3265–3274. doi: 10.1074/jbc.M209759200. [DOI] [PubMed] [Google Scholar]
- 57.Choi K, Mollapour E, Choi JH, Shears SB. 2008. Cellular energetic status supervises the synthesis of bis-diphosphoinositol tetrakisphosphate independently of AMP-activated protein kinase. Mol Pharmacol 74:527–536. doi: 10.1124/mol.107.044628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voglmaier SM, Bembenek ME, Kaplin AI, Dormán G, Olszewski JD, Prestwich GD, Snyder SH. 1996. Purified inositol hexakisphosphate kinase is an ATP synthase: diphosphoinositol pentakisphosphate as a high-energy phosphate donor. Proc Natl Acad Sci USA 93:4305–4310. doi: 10.1073/pnas.93.9.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boer VM, Crutchfield CA, Bradley PH, Botstein D, Rabinowitz JD. 2010. Growth-limiting intracellular metabolites in yeast growing under diverse nutrient limitations. Mol Biol Cell 21:198–211. doi: 10.1091/mbc.e09-07-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klosinska MM, Crutchfield CA, Bradley PH, Rabinowitz JD, Broach JR. 2011. Yeast cells can access distinct quiescent states. Genes Dev 25:336–349. doi: 10.1101/gad.2011311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gupta R, Laxman S. 2021. Cycles, sources, and sinks: conceptualizing how phosphate balance modulates carbon flux using yeast metabolic networks. eLife 10:225. doi: 10.7554/eLife.63341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petti AA, Crutchfield CA, Rabinowitz JD, Botstein D. 2011. Survival of starving yeast is correlated with oxidative stress response and nonrespiratory mitochondrial function. Proc Natl Acad Sci USA 108:E1089–E1098. doi: 10.1073/pnas.1101494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Visser W, van Spronsen EA, Nanninga N, Pronk JT, Kuenen JG, van Dijken JP. 1995. Effects of growth conditions on mitochondrial morphology in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek 67:243–253. doi: 10.1007/BF00873688. [DOI] [PubMed] [Google Scholar]
- 64.Zheng F, Jia B, Dong F, Liu L, Rasul F, He J, Fu C. 2019. Glucose starvation induces mitochondrial fragmentation depending on the dynamin GTPase Dnm1/Drp1 in fission yeast. J Biol Chem 294:17725–17734. doi: 10.1074/jbc.RA119.010185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cardarelli S, Giorgi M, Poiana G, Biagioni S, Saliola M. 2019. Metabolic role of cGMP in S. cerevisiae: the murine phosphodiesterase-5 activity affects yeast cell proliferation by altering the cAMP/cGMP equilibrium. FEMS Yeast Res 19:foz016. doi: 10.1093/femsyr/foz016. [DOI] [PubMed] [Google Scholar]
- 66.Zeebroeck GV, Demuyser L, Zhang Z, Cottignie I, Thevelein JM. 2021. Nutrient sensing and cAMP signaling in yeast: G-protein coupled receptor versus transceptor activation of PKA. Microb Cell 8:17–27. doi: 10.15698/mic2021.01.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conrad M, Schothorst J, Kankipati HN, Zeebroeck GV, Rubio-Texeira M, Thevelein JM. 2014. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 38:254–299. doi: 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Samyn DR, Ruiz-Pávon L, Andersson MR, Popova Y, Thevelein JM, Persson BL. 2012. Mutational analysis of putative phosphate- and proton-binding sites in the Saccharomyces cerevisiae Pho84 phosphate:H+ transceptor and its effect on signalling to the PKA and PHO pathways. Biochem J 445:413–422. doi: 10.1042/BJ20112086. [DOI] [PubMed] [Google Scholar]
- 69.Popova Y, Thayumanavan P, Lonati E, Agrochão M, Thevelein JM. 2010. Transport and signaling through the phosphate-binding site of the yeast Pho84 phosphate transceptor. Proc Natl Acad Sci USA 107:2890–2895. doi: 10.1073/pnas.0906546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lundh F, Mouillon J-M, Samyn D, Stadler K, Popova Y, Lagerstedt JO, Thevelein JM, Persson BL. 2009. Molecular mechanisms controlling phosphate-induced downregulation of the yeast Pho84 phosphate transporter. Biochemistry 48:4497–4505. doi: 10.1021/bi9001198. [DOI] [PubMed] [Google Scholar]
- 71.Giots F, Donaton M, Thevelein JM. 2003. Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol 47:1163–1181. doi: 10.1046/j.1365-2958.2003.03365.x. [DOI] [PubMed] [Google Scholar]
- 72.Eckstein H, Flügge B. 1999. Guanosine 3′:5′-cyclic monophosphate-dependent particulate protein kinase activity from yeast (Saccharomyces cerevisiae). Z Naturforsch C J Biosci 54:84–93. doi: 10.1515/znc-1999-1-215. [DOI] [PubMed] [Google Scholar]
- 73.Eckstein H. 1981. Inhibition by cyclic guanosine 3′:5′-monophosphate of the soluble DNA polymerase activity, and of partially purified DNA polymerase A (DNA polymerase I) from the yeast Saccharomyces cerevisiae. Z Naturforsch C J Biosci 36:813–819. doi: 10.1515/znc-1981-9-1020. [DOI] [PubMed] [Google Scholar]
- 74.Corton JM, Gillespie JG, Hawley SA, Hardie DG. 1995. 5-Aminoimidazole-4-carboxamide ribonucleoside. A specific method for activating AMP-activated protein kinase in intact cells? Eur J Biochem 229:558–565. doi: 10.1111/j.1432-1033.1995.tb20498.x. [DOI] [PubMed] [Google Scholar]
- 75.Mayer FV, Heath R, Underwood E, Sanders MJ, Carmena D, McCartney RR, Leiper FC, Xiao B, Jing C, Walker PA, Haire LF, Ogrodowicz R, Martin SR, Schmidt MC, Gamblin SJ, Carling D. 2011. ADP regulates SNF1, the Saccharomyces cerevisiae homolog of AMP-activated protein kinase. Cell Metab 14:707–714. doi: 10.1016/j.cmet.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Desfougères Y, Wilson MSC, Laha D, Miller GJ, Saiardi A. 2019. ITPK1 mediates the lipid-independent synthesis of inositol phosphates controlled by metabolism. Proc Natl Acad Sci USA 93:201911431. doi: 10.1073/pnas.1911431116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rébora K, Desmoucelles C, Borne F, Pinson B, Daignan-Fornier B. 2001. Yeast AMP pathway genes respond to adenine through regulated synthesis of a metabolic intermediate. Mol Cell Biol 21:7901–7912. doi: 10.1128/MCB.21.23.7901-7912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stincone A, Prigione A, Cramer T, Wamelink MMC, Campbell K, Cheung E, Olin-Sandoval V, Grüning N-M, Krüger A, Alam MT, Keller MA, Breitenbach M, Brindle KM, Rabinowitz JD, Ralser M. 2015. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol Rev 90:927–963. doi: 10.1111/brv.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krüger A, Grüning N-M, Wamelink MMC, Kerick M, Kirpy A, Parkhomchuk D, Bluemlein K, Schweiger M-R, Soldatov A, Lehrach H, Jakobs C, Ralser M. 2011. The pentose phosphate pathway is a metabolic redox sensor and regulates transcription during the antioxidant response. Antioxid Redox Sign 15:311–324. doi: 10.1089/ars.2010.3797. [DOI] [PubMed] [Google Scholar]
- 80.Pinson B, Ceschin J, Saint-Marc C, Daignan-Fornier B. 2019. Dual control of NAD+ synthesis by purine metabolites in yeast. eLife 8:e43808. doi: 10.7554/eLife.43808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu S-P, Lin S-J. 2011. Phosphate-responsive signaling pathway is a novel component of NAD+ metabolism in Saccharomyces cerevisiae. J Biol Chem 286:14271–14281. doi: 10.1074/jbc.M110.217885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morrissette VA, Rolfes RJ. 2020. The intersection between stress responses and inositol pyrophosphates in Saccharomyces cerevisiae. Curr Genet 10:R57–R10. doi: 10.1007/s00294-020-01078-8. [DOI] [PubMed] [Google Scholar]
- 83.Steidle EA, Morrissette VA, Fujimaki K, Chong L, Resnick AC, Capaldi AP, Rolfes RJ. 2019. The InsP7 phosphatase Siw14 regulates inositol pyrophosphate levels to control localization of the general stress response transcription factor Msn2. J Biol Chem 295:2043–2056. doi: 10.1074/jbc.RA119.012148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, McCann PP. 1996. S-Adenosylmethionine and methylation. FASEB J 10:471–480. doi: 10.1096/fasebj.10.4.8647346. [DOI] [PubMed] [Google Scholar]
- 85.Hyun K, Jeon J, Park K, Kim J. 2017. Writing, erasing and reading histone lysine methylations. Exp Mol Med 49:e324. doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kouzarides T. 2002. Histone methylation in transcriptional control. Curr Opin Genet Dev 12:198–209. doi: 10.1016/S0959-437X(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 87.Wang S-S, Zhou BO, Zhou J-Q. 2011. Histone H3 lysine 4 hypermethylation prevents aberrant nucleosome remodeling at the PHO5 promoter. Mol Cell Biol 31:3171–3181. doi: 10.1128/MCB.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Carvin CD, Kladde MP. 2004. Effectors of lysine 4 methylation of histone H3 in Saccharomyces cerevisiae are negative regulators of PHO5 and GAL1-10. J Biol Chem 279:33057–33062. doi: 10.1074/jbc.M405033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chia SZ, Lai Y-W, Yagoub D, Lev S, Hamey JJ, Pang CNI, Desmarini D, Chen Z, Djordjevic JT, Erce MA, Hart-Smith G, Wilkins MR. 2018. Knockout of the Hmt1p arginine methyltransferase in Saccharomyces cerevisiae leads to the dysregulation of phosphate-associated genes and processes. Mol Cell Proteomics 17:2462–2479. doi: 10.1074/mcp.RA117.000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Matsuno-Yagi A, Hatefi Y. 1988. Role of energy in oxidative phosphorylation. J Bioenerg Biomembr 20:481–502. doi: 10.1007/BF00762205. [DOI] [PubMed] [Google Scholar]
- 91.Byers LD. 1982. Glyceraldehyde-3-phosphate dehydrogenase from yeast. Methods Enzymol 89:326–335. doi: 10.1016/s0076-6879(82)89059-9. [DOI] [PubMed] [Google Scholar]
- 92.Weaver JD, Wang H, Shears SB. 2013. The kinetic properties of a human PPIP5K reveal that its kinase activities are protected against the consequences of a deteriorating cellular bioenergetic environment. Biosci Rep 33:e00022. doi: 10.1042/BSR20120115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gu C, Nguyen H-N, Hofer A, Jessen HJ, Dai X, Wang H, Shears SB. 2017. The significance of the bifunctional kinase/phosphatase activities of PPIP5Ks for coupling inositol pyrophosphate cell-signaling to cellular phosphate homeostasis. J Biol Chem 292:4544–4555. doi: 10.1074/jbc.M116.765743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Menoyo S, Ricco N, Bru S, Hernández-Ortega S, Escoté X, Aldea M, Clotet J. 2013. Phosphate-activated cyclin-dependent kinase stabilizes G1 cyclin to trigger cell cycle entry. Mol Cell Biol 33:1273–1284. doi: 10.1128/MCB.01556-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pondugula S, Neef DW, Voth WP, Darst RP, Dhasarathy A, Reynolds MM, Takahata S, Stillman DJ, Kladde MP. 2009. Coupling phosphate homeostasis to cell cycle-specific transcription: mitotic activation of Saccharomyces cerevisiae PHO5 by Mcm1 and Forkhead proteins. Mol Cell Biol 29:4891–4905. doi: 10.1128/MCB.00222-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dunn T, Gable K, Beeler T. 1994. Regulation of cellular Ca2+ by yeast vacuoles. J Biol Chem 269:7273–7278. doi: 10.1016/S0021-9258(17)37279-4. [DOI] [PubMed] [Google Scholar]
- 97.Beeler T, Bruce K, Dunn T. 1997. Regulation of cellular Mg2+ by Saccharomyces cerevisiae. Biochim Biophys Acta 1323:310–318. doi: 10.1016/S0005-2736(96)00199-X. [DOI] [PubMed] [Google Scholar]
- 98.Cyert MS, Philpott CC. 2013. Regulation of cation balance in Saccharomyces cerevisiae. Genetics 193:677–713. doi: 10.1534/genetics.112.147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kulakovskaya T. 2018. Inorganic polyphosphates and heavy metal resistance in microorganisms. World J Microbiol Biotechnol 34:139. doi: 10.1007/s11274-018-2523-7. [DOI] [PubMed] [Google Scholar]
- 100.Klompmaker SH, Kohl K, Fasel N, Mayer A. 2017. Magnesium uptake by connecting fluid-phase endocytosis to an intracellular inorganic cation filter. Nat Commun 8:1879. doi: 10.1038/s41467-017-01930-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ryazanova LP, Ledova LA, Andreeva NA, Zvonarev AN, Eldarov MA, Kulakovskaya TV. 2020. Inorganic polyphosphate and physiological properties of Saccharomyces cerevisiae yeast overexpressing Ppn2. Biochemistry (Mosc) 85:516–522. doi: 10.1134/S0006297920040124. [DOI] [PubMed] [Google Scholar]
- 102.Trilisenko L, Zvonarev A, Valiakhmetov A, Penin AA, Eliseeva IA, Ostroumov V, Kulakovskiy IV, Kulakovskaya T. 2019. The reduced level of inorganic polyphosphate mobilizes antioxidant and manganese-resistance systems in Saccharomyces cerevisiae. Cells 8:461. doi: 10.3390/cells8050461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McCarthy L, Abramchuk I, Wafy G, Denoncourt A, Lavallée-Adam M, Downey M. 2022. Ddp1 cooperates with Ppx1 to counter a stress response initiated by nonvacuolar polyphosphate. mBio 13:e0039022. doi: 10.1128/mbio.00390-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McCarthy L, Bentley-DeSousa A, Denoncourt A, Tseng Y-C, Gabriel M, Downey M. 2019. Proteins required for vacuolar function are targets of lysine polyphosphorylation in yeast. FEBS Lett 594:21–30. doi: 10.1002/1873-3468.13588. [DOI] [PubMed] [Google Scholar]
- 105.Azevedo C, Livermore T, Saiardi A. 2015. Protein polyphosphorylation of lysine residues by inorganic polyphosphate. Mol Cell 58:71–82. doi: 10.1016/j.molcel.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 106.Azevedo C, Singh J, Steck N, Hofer A, Ruiz FA, Singh T, Jessen HJ, Saiardi A. 2018. Screening a protein array with synthetic biotinylated inorganic polyphosphate to define the human polyP-ome. ACS Chem Biol 13:1958–1963. doi: 10.1021/acschembio.8b00357. [DOI] [PubMed] [Google Scholar]
- 107.Zhang J, Sassen T, ten Pierick A, Ras C, Heijnen JJ, Wahl SA. 2015. A fast sensor for in vivo quantification of cytosolic phosphate in Saccharomyces cerevisiae. Biotechnol Bioeng 112:1033–1046. doi: 10.1002/bit.25516. [DOI] [PubMed] [Google Scholar]
- 108.Sheff MA, Thorn KS. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 109.Laughery MF, Hunter T, Brown A, Hoopes J, Ostbye T, Shumaker T, Wyrick JJ. 2015. New vectors for simple and streamlined CRISPR-Cas9 genome editing in Saccharomyces cerevisiae: vectors for simple CRISPR-Cas9 genome editing in yeast. Yeast 32:711–720. doi: 10.1002/yea.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ehrensberger AH, Kornberg RD. 2011. Isolation of an activator-dependent, promoter-specific chromatin remodeling factor. Proc National Acad Sci USA 108:10115–10120. doi: 10.1073/pnas.1101449108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kulakova AN, Hobbs D, Smithen M, Pavlov E, Gilbert JA, Quinn JP, McGrath JW. 2011. Direct quantification of inorganic polyphosphate in microbial cells using 4′-6-diamidino-2-phenylindole (DAPI). Environ Sci Technol 45:7799–7803. doi: 10.1021/es201123r. [DOI] [PubMed] [Google Scholar]
- 112.Omelon S, Georgiou J, Habraken W. 2016. A cautionary (spectral) tail: red-shifted fluorescence by DAPI-DAPI interactions. Biochem Soc Trans 44:46–49. doi: 10.1042/BST20150231. [DOI] [PubMed] [Google Scholar]
- 113.Qiu D, Wilson MS, Eisenbeis VB, Harmel RK, Riemer E, Haas TM, Wittwer C, Jork N, Gu C, Shears SB, Schaaf G, Kammerer B, Fiedler D, Saiardi A, Jessen HJ. 2020. Analysis of inositol phosphate metabolism by capillary electrophoresis electrospray ionization mass spectrometry. Nat Commun 11:6035. doi: 10.1038/s41467-020-19928-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Qiu D, Eisenbeis VB, Saiardi A, Jessen HJ. 2021. Absolute quantitation of inositol pyrophosphates by capillary electrophoresis electrospray ionization mass spectrometry. J Vis Exp. doi: 10.3791/62847. [DOI] [PubMed] [Google Scholar]
- 115.Riemer E, Qiu D, Laha D, Harmel RK, Gaugler P, Gaugler V, Frei M, Hajirezaei M-R, Laha NP, Krusenbaum L, Schneider R, Saiardi A, Fiedler D, Jessen HJ, Schaaf G, Giehl RFH. 2021. ITPK1 is an InsP6/ADP phosphotransferase that controls phosphate signaling in Arabidopsis. Mol Plant 14:1864–1880. doi: 10.1016/j.molp.2021.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gallart-Ayala H, Konz I, Mehl F, Teav T, Oikonomidi A, Peyratout G, van der Velpen V, Popp J, Ivanisevic J. 2018. A global HILIC-MS approach to measure polar human cerebrospinal fluid metabolome: exploring gender-associated variation in a cohort of elderly cognitively healthy subjects. Anal Chim Acta 1037:327–337. doi: 10.1016/j.aca.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 117.Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, Gauthier C, Jacques P-É, Li S, Xia J. 2021. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 49:gkab382. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Metabolites significantly affected by different Pi conditions. Download Data Set S1, XLSX file, 0.01 MB (15.4KB, xlsx) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Results of two-way ANOVA of wild-type and Δvtc4 cells under 2 h of Pi-replete (10 mM Pi) and Pi-starving conditions (0 mM Pi). Download Data Set S2, XLSX file, 0.01 MB (15.5KB, xlsx) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic changes in energy metabolism pathways under different Pi conditions. (A and B) Metabolomic data of yeast under different Pi conditions are presented on energy metabolic pathways, including glycolysis (A) and the TCA cycle (B). Pi-containing substrates are marked as red. Gray indicates enzymes involved in the reaction. Different letters on the graph indicate a significant difference by Tukey-Kramer test (P < 0.05). Download FIG S1, TIF file, 0.7 MB (762.5KB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic changes in nicotinate and nicotinamide pathway under different Pi conditions. Metabolomic data of yeast under different Pi conditions are presented on a metabolic pathway. Pi-containing substrates are marked as red. Gray indicates enzymes involved in the reaction. Different letters on the graph indicate a significant difference by Tukey-Kramer test (P < 0.05). Download FIG S2, TIF file, 0.8 MB (779.7KB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic changes in methionine and cysteine metabolism pathways under different Pi conditions. Metabolomic data of yeast under different Pi conditions are presented on a metabolic pathway. Pi-containing substrates are marked as red. Gray indicates enzymes involved in the reaction. Different letters on the graph indicate a significant difference by Tukey-Kramer test (P < 0.05). Download FIG S3, TIF file, 0.8 MB (854.6KB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic changes in purine and pyrimidine metabolism pathways under different Pi conditions. (A and B) Metabolomic data of yeast under different Pi conditions are presented on a purine metabolic pathway (A) and a pyrimidine metabolic pathway (B). Pi-containing substrates are marked as red. Gray indicates enzymes involved in the reaction. Different letters on the graph indicate a significant difference by Tukey-Kramer test (P < 0.05). Download FIG S4, TIF file, 0.8 MB (826.8KB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Inositol pyrophosphate profiles. Time course analysis under Pi starvation and Δvtc4 under Pi-replete conditions. (A to D) Time course of PP-IP levels of wild-type yeast cells transferred to 0 mM Pi medium for 8 h. Logarithmically growing yeast cells in 10 mM Pi medium were transferred into 0 mM Pi medium. In contrast to the direct extraction of the cultures with perchloric acid in Fig. 6, cells were recovered by filtration after the indicated periods of Pi starvation. The filters were extracted with perchloric acid, and the inositol pyrophosphates were quantified by CE-MS. Note that the filtration method used here is less efficient for the small quantities of remaining PP-IPs after starvation. The data were normalized by the number of cells per sample. The concentration of each inositol pyrophosphate in 10 mM Pi medium was set to 1. (E to H) Wild-type and Δvtc4 yeast cells were logarithmically grown in 10 mM Pi medium overnight, harvested by centrifugation, and subjected to perchloric acid extraction and PP-IP analysis as in A. The data were normalized by the number of cells. The amount of each inositol pyrophosphate in wild-type cells was set to 1. Means with standard deviations are shown; n = 3; a.u., arbitrary units; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant by Student’s t test. Download FIG S8, TIF file, 0.5 MB (540KB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic changes in purine and pyrimidine metabolism pathways under Pi starvation in wild-type and Δvtc4 cells. (A and B) Metabolomic data of yeast under Pi starvation in wild-type and Δvtc4 cells are presented on a purine metabolic pathway (A) and a pyrimidine metabolic pathway (B). Pi-containing substrates are marked as red. Gray indicates enzymes involved in the reaction. Different letters on the graph indicate a significant difference by Tukey-Kramer test (P < 0.05). Download FIG S5, TIF file, 0.9 MB (969.7KB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic changes in nicotinate and nicotinamide pathways under Pi starvation in wild-type and Δvtc4 cells. Metabolomic data of yeast under Pi starvation in wild-type and Δvtc4 cells are presented in a metabolic pathway. Pi-containing substrates are marked as red. Gray indicates enzymes involved in the reaction. Different letters on the graph indicate a significant difference by Tukey-Kramer test (P < 0.05). Download FIG S6, TIF file, 0.8 MB (870.8KB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolic changes in glycolysis under Pi starvation in wild-type and Δvtc4 cells. Metabolomic data of yeast under Pi starvation in wild-type and Δvtc4 cells are presented in a metabolic pathway. Pi-containing substrates are marked as red. Gray indicates enzymes involved in the reaction. Different letters on the graph indicate a significant difference by Tukey-Kramer test (P < 0.05). Download FIG S7, TIF file, 1.2 MB (1.2MB, tif) .
Copyright © 2023 Kim et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.