ABSTRACT
Lodderomyces elongisporus is a recently emerging yeast pathogen predominantly reported in adult patients who had immunosuppression and/or intravenous access devices. Here, we report a fungemia outbreak caused by L. elongisporus in a neonatal intensive care unit (NICU) in Delhi, India, from September 2021 to February 2022. All 10 neonates had low birthweight, and nine of the patients survived after amphotericin B treatment. Whole-genome sequence analyses of the patient isolates as well as those from other sources in India grouped them into two clusters: one cluster consists of isolates exclusively from stored apples and the other cluster includes isolates from patients, clinical environments, and stored apples. All outbreak strains from patients were closely related to each other and showed highly similar heterozygosity patterns across all 11 major scaffolds. While overall very similar, strains from the inanimate environment of the same neonatal intensive care unit showed loss of heterozygosity at scaffold 2 (NW_001813676) compared to the patient strains. Interestingly, evidence for recombination was found in all samples. All clinical strains were susceptible to 10 tested antifungal drugs, and comparisons with strains with high fluconazole MICs derived from the surface of stored apples revealed significant genome divergence between the clinical and apple surface strains, including 119 nonsynonymous single nucleotide polymorphisms (SNPs) in 24 triazole resistance-related genes previously found in other Candida spp. Together, our results indicate significant diversity, recombination, and persistence in the hospital setting and a high rate of evolution in this emerging yeast pathogen.
KEYWORDS: L. elongisporus, neonate, loss of heterozygosity, phenotypic divergence, ploidy, ascospore, sodium hypochlorite
INTRODUCTION
Invasive fungal infections in neonatal intensive care units (NICUs) have increased significantly worldwide, representing an important infective complication in hospitalized patients. Among hospital-born infants, hospital-acquired infections account for an estimated 4% to 56% of all deaths in the neonatal period, depending on the study population and geographical area (1, 2). Although nosocomial bloodstream infections (BSIs) by bacteria are still more frequent, the frequency of fungal sepsis caused by species of Candida and other fungi is increasingly recognized as an important cause of morbidity and mortality in neonatal intensive care units (NICUs) (3–5). In the last 2 decades, several outbreaks of nosocomial BSIs due to rare and emerging yeasts have been reported worldwide, stressing the importance of early detection and timely interventions to prevent and control outbreaks (6–9). Lodderomyces elongisporus is an uncommon cause of fungemia, predominantly reported in adult patients who had immunosuppression and/or intravenous access devices (10–12). This yeast was considered a teleomorph of Candida parapsilosis until sequencing of the 18S rRNA gene differentiated it as a unique species (13). Here, we report an outbreak of fungemia in a NICU involving 10 neonates in a multispecialty hospital in Delhi, India. We used whole-genome sequencing to assess the genetic diversity and relationships among strains obtained from the neonates as well as from the inanimate environment of the hospitals and stored apples from northern India (14).
RESULTS
Outbreak investigations.
From September 2021 to February 2022, L. elongisporus fungemia occurred in 10 neonates in the 24-bed-capacity NICU for hospital-born neonates of a multispecialty hospital in Delhi, India. Cases of bloodstream infections (BSIs) with this yeast were defined as the isolation of L. elongisporus from blood culture of peripheral or central vein blood samples in patients with signs and symptoms of sepsis.
The first case of fungal sepsis occurred in a preterm infant with very low birthweight (<1,500 g) who developed thrombocytopenia and signs of sepsis. The neonate received vancomycin, and meropenem along with fluconazole (FLU) (loading dose of 12 mg/kg of body weight followed by 6 mg/kg of body weight). The patient’s blood culture was positive for the yeast on day 9 after birth. The yeast was identified as L. elongisporus, FLU administration was stopped, and liposomal amphotericin B (AMB) was started at a dose of 1 mg/kg of body weight intravenously for 14 days. Overall, 10 cases were identified in neonates hospitalized in the NICU during the period of 6 months. The first four cases (cases 1 to 4) occurred within 2 weeks in September 2021. After the second case of L. elongisporus was identified, infection prevention practices were evaluated with the health care workers to mitigate further transmission of the yeast. The quality and frequency of hand hygiene and environmental cleaning were emphasized to the health care staff. Further, two NICU rooms that accommodated L. elongisporus-infected neonates were vacated for enhanced cleaning of the rooms. Meanwhile, the neonates were kept in the other designated room in the NICU until the cleaning procedures were satisfactorily completed, which was followed by shifting the neonates to the previous rooms. Subsequently, a reduction in the number of cases was observed, resulting in one case each of fungemia due to L. elongisporus in the next 2 months. However, between 31 January and 21 February 2022, a sudden clustering of four cases occurred, and at this point environmental sampling of two sites, i.e., the railing of the neonate open care warmer and its temperature control panel, yielded L. elongisporus. Notably, both environmental sites colonized by L. elongisporus were cocolonized by C. parapsilosis. Hand swabs of health care workers were found to be negative for any yeast species. Further outbreak investigations were accompanied by strict infection prevention and control practices. Specifically, hand hygiene was practiced before and after each patient contact and before donning or doffing personal protection equipment, and sterilization and daily disinfection were performed for personal protection equipment. The outbreak was contained, and until now (February 2023), no new case of L. elongisporus fungemia has been detected in the NICU.
Clinical details.
Clinical data were extracted from medical records, and the details are described in Table 1. Age at onset of fungemia ranged from 7 to 35 days. All neonates presented a nonspecific sign as increased frequency of apnea episodes; thrombocytopenia (platelet counts of <100 × 109/L) was the early laboratory finding in six neonates. The mean gestational age was 32.4 weeks, and the mean birthweight was 1.1 kg. The risk factors included preterm birth and low/very low birthweight. All neonates were treated with AMB and improved and were discharged, except one who died.
TABLE 1.
Clinical details of 10 neonates with Lodderomyces elongisporus fungemia in a neonatal intensive care unit, Indiaa
| Patient | Isolate ID | DOB/DOA (day-mo-yr) | Date of blood culture (day-mo-yr) | Clinical details | Outcome | Antifungal therapy |
|---|---|---|---|---|---|---|
| 1 | 906-P-21 | 07-09-2021 | 16-09-2021 | PT, VLBW, thrombocytopenia | Survived | FLU and AMB |
| 2 | 950-P-21 | 02-09-2021 | 24-09-2021 | PT, IUGR, thrombocytopenia | Survived | FLU and AMB |
| 3 | 963-P-21 | 18-09-2021 | 28-09-2021 | PT, VLBW, severe asphyxia | Survived | FLU and AMB |
| 979-P-21-S62 | 30-09-2021 | |||||
| 979-P-21-S87 | 19-10-2021 | |||||
| 1023-P-21 | 28-10-2021 | |||||
| 4 | 972-P-21 | 17-09-2021 | 30-09-2021 | PT, ELBW, thrombocytopenia, hypoglycemia | Survived | FLU and AMB |
| 5 | 1003-P-21 | 24-09-2021 | 12-10-2021 | Term, hypoglycemia, thrombocytopenia | Survived | FLU and AMB |
| 6 | 1107-P-21 | 01-11-2021 | 16-11-2021 | LBW, PT, thrombocytopenia | Survived | FLU and AMB |
| 7 | 89-P-22 | 2-01-2022 | 31-01-2022 | VLBW, PT | Died | FLU and AMB |
| 8 | 144-P-22 | 01-02-2022 | 07-02-2022 | Term, LBW, sepsis, thrombocytopenia | Survived | FLU and AMB |
| 9 | 147-P-22 | 03-02-2022 | 09-02-2022 | VLBW, severe asphyxia at birth | Survived | FLU and AMB |
| 10 | 160-P-22 | 12-02-2022 | 21-02-2022 | VLBW, PT | Survived | FLU and AMB |
ID, identifier; DOA, date of admission; DOB, date of birth; VLBW, very low birthweight; IUGR, intrauterine growth restriction; PT, preterm; ELBW, extremely low birthweight; LBW, low birthweight; FLU, fluconazole; AMB; amphotericin B.
Strain identification.
A total of 13 bloodstream strains from 10 neonates, including four strains from four serial blood culture specimens from a single case (obtained at 0, 2, 21, and 30 days), were identified as L. elongisporus by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) and confirmed by sequencing of internal transcribed spacer (ITS) regions of the ribosomal DNA (rDNA). All L. elongisporus strains on CHROMagar Candida medium showed green/blue color at 37°C after 48 h of incubation. Also, the Vitek-2 (bioMérieux, Marcy l’Etoile, France) commercial identification system misidentified L. elongisporus as C. parapsilosis with 91 to 96% probability.
Antifungal susceptibility testing (AFST).
We performed AFST of all L. elongisporus strains (13 bloodstream isolates and 2 inanimate-environment strains) from the present outbreak along with three inanimate-environment strains from two other hospitals in Delhi. All strains exhibited low MICs of azoles by the CLSI broth microdilution method (M27-A3) (15). Among azoles, FLU (MIC range, 0.25 to 1 mg/L), voriconazole (VRC; MIC range, <0.03 to 0.06 mg/L), and itraconazole (ITC; MIC range, 0.03 to 0.25 mg/L) showed low MICs. Also, low MICs were observed for AMB (0.125 to 0.5 mg/L), anidulafungin (AFG; <0.015 to 0.5 mg/L), micafungin (MFG; <0.015 to 0.125 mg/L), and 5-flucytosine (5-FC; 0.06 to 0.25 mg/L) (Table 2). We also compared AFST data of 13 L. elongisporus strains obtained previously from the surface of stored apples with those of the strains obtained in this study (14). Interestingly, the L. elongisporus strains recovered from the surface of stored apples showed 2- to 8-fold-higher fluconazole MICs (MIC range, 2 to 4 mg/L) than the clinical and inanimate-environment strains of L. elongisporus obtained in the present outbreak.
TABLE 2.
MIC distribution of Lodderomyces elongisporus strains isolated in the present outbreak including clinical and inanimate sources along with strains obtained from other sources against 10 antifungal drugs using CLSI-broth microdilution method
| L. elongisporus (no. of isolates) | MIC range (mg/L)a |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| FLU | ITC | VRC | ISA | POS | AMB | MFG | AFG | 5-FC | |
| Clinical, bloodstream isolates (n = 13) | 0.25–1 | 0.03–0.25 | <0.03–0.06 | <0.015 | <0.015–0.125 | 0.125–0.5 | <0.015–0.125 | <0.015–0.5 | 0.06–0.25 |
| Inanimate environment, NICU (neonate warmer; n = 2) | 0.5 | 0.03–0.06 | <0.03 | <0.015 | <0.015 | 0.25 | <0.015 | <0.015–0.5 | 0.06 |
| Inanimate environment of two hospitals, Delhi (floor; n = 3)b | 0.5 | 0.03–0.06 | <0.03 | <0.015 | <0.015 | 0.25 | <0.015 | <0.015–0.5 | 0.06 |
| Strains from surfaces of apples (n = 13)c | 2–4 | 0.25 | 0.03–0.12 | <0.015 | 0.06–0.5 | 0.01–0.5 | 0.01 | 0.01–0.5 | 0.06–0.12 |
Genome analysis.
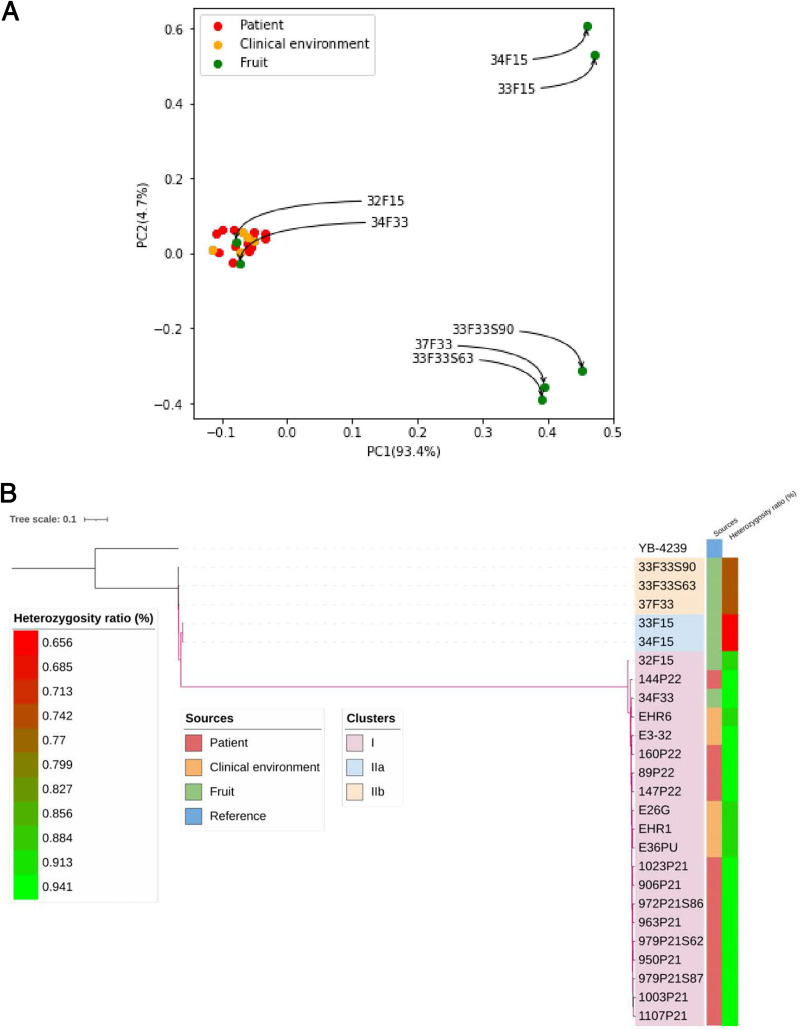
The genomes of 13 clinical strains and two inanimate-environment strains from the present outbreak were analyzed along with the seven L. elongisporus strains isolated from the surface of stored apples and three environmental strains obtained from the floor of two other hospitals (14, 16). The reference strain YB-4239 genome was included for single nucleotide polymorphism (SNP) calling and phylogenetic analysis (17). The isolates belonged to two clusters, cluster I and cluster II. All clinical and inanimate-hospital-environment strains of three hospitals were closely related (3,260- to 4,581-SNP difference) and comprised a tight genotype cluster I. Interestingly, two strains from individual apple surfaces (VPCI/34/F33/2020, and VPCI/32/F15/2020) clustered with clinical and inanimate-environment strains in cluster I (Fig. 1A). The remaining five strains obtained from surfaces of stored apples formed cluster II, comprising two subclusters (cluster IIa, n = 2; cluster IIb, n = 3; 4,984- to 5,387-SNP difference between subclusters IIa and IIb). Overall, the cluster II isolates from the surfaces of apples were genotypically divergent from cluster I strains, with 176,721 to 177,579 SNPs separating them (Fig. 1A and B). All L. elongisporus strains in cluster I (clinical, inanimate environment, and two fruit strains) had identical genotypes at the 24 drug-resistance-related genes (see Table S1 in the supplemental material). Interestingly, strains from surfaces of apples (cluster II) which were genetically distinct from the clinical and inanimate-environment strains (cluster I) had a total of 119 nonsynonymous substitutions in 24 triazole-resistance-related genes previously identified in Candida spp. (Table S1), including three mutations in ERG11 (Met15Leu, Ser183Thr, and Asn363Ile). Further, strains in cluster II from surfaces of apples which were genetically distinct from clinical and inanimate-environment strains showed partial or complete loss of heterozygosity (LOH) in scaffolds 1 (NW_001813675) and 6 (NW_001813680) (Fig. S1). Similarly, environmental strains obtained from the neonate warmer (n = 2; VPCI/E/HR6/2022 and VPCI/E/HR1/2022) and inanimate-environment strains (n = 2; VPCI/E/28G/2020 and VPCI/E/36PU/2020) from the floor of other hospitals also showed LOH in scaffold 2 (NW_001813676).
FIG 1.
(A) Principal components unveil population structure of Lodderomyces elongisporus samples from three sources, i.e., clinical (patient), inanimate environment (clinical environment), and fruits (apple surface). (B) Genetic relatedness among 25 isolates and the reference strain inferred based on homozygous SNPs. Branches with bootstrap support over 0.95 are highlighted in purple. The inner color strips indicate the sources of the isolates. The outer color gradients display the heterozygosity ratio of each sample calculated by the number of heterozygous sites relative to reference genome size.
Loss of heterozygosity pattern of 25 Lodderomyces elongisporus strains including 13 clinical strains and two inanimate-environment strains from the present outbreak, seven strains isolated from the surface of stored apples, and three environmental strains obtained from the floor of two other hospitals over the 11 scaffolds. Samples are ordered based on isolation sources. Samples from patients, clinical environment, and fruit are marked by red, orange, and green labels. Purple and yellow blocks indicate heterozygous and homozygous regions, respectively. Download FIG S1, DOCX file, 0.5 MB (492KB, docx) .
Copyright © 2023 Yadav et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SNP difference at 24 antifungal resistance-related genes between 13 clinical isolates of Lodderomyces elongisporus along with five isolates from inanimate environment of three different hospitals and seven isolates from surfaces of apples. Download Table S1, XLSX file, 0.2 MB (238.3KB, xlsx) .
Copyright © 2023 Yadav et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Morphological characterization of Lodderomyces elongisporus.
On acetate ascospore agar, all bloodstream isolates of L. elongisporus developed abundant ascospores within 2 weeks of incubation at 37°C (Fig. S2). Also, a few strains from surfaces of apples (n = 3, VPCI/33F33/2020, VPCI/34F33/2020, and VPCI/37F33/2020) showed abundant ascospore formation after 3 weeks of incubation. Interestingly, one of the fruit strains (VPCI/34F33/2020) producing ascospores was genetically related to clinical strains in cluster I. In contrast, only one strain from the inanimate environment of the NICU developed a few (1 to 2/slide) ascospores after 3 weeks of incubation.
Ascospore formation in a clinical isolate (VPCI/160/P/2022) on ascospore agar containing 0.1% sodium acetate stained with lactophenol cotton blue at ×40 magnification showing one or two ascospores (white arrow) enclosed in an ascus. Download FIG S2, DOCX file, 5.1 MB (5.1MB, docx) .
Copyright © 2023 Yadav et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Determination of ploidy in Lodderomyces elongisporus.
We examined the genomic DNA content of 13 L. elongisporus isolates including two randomly selected clinical (VPCI/147/P/2022 and VPCI/160/P/2022) strains, all five strains from the inanimate environment of hospitals, and six strains recovered from the surface of apples using fluorescence-activated cell sorting (FACS) assays. Overall, most L. elongisporus strains had 2n DNA content (diploid), similar to the Candida albicans reference strain ATCC 90028, except two strains obtained from the open warmer of the NICU. These two strains recovered from the inanimate environment of the present outbreak showed distinct genomic DNA (aneuploid), between haploid and diploid (Fig. S3).
Histograms representing DNA content or cell cycle profile obtained by FACS. For comparison, diploid C. albicans ATCC 90038 was used as a reference strain. The x axis represents nuclear fluorescence, and the y axis represents cell number. Download FIG S3, DOCX file, 0.2 MB (249.1KB, docx) .
Copyright © 2023 Yadav et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Susceptibility to disinfectants.
We investigated the susceptibility pattern of four classes of disinfectants against all strains including low-level disinfectant (absolute ethanol) and high-level disinfectants, i.e., glutaraldehyde, fifth-generation quaternary ammonium compound, and sodium hypochlorite. We compared the activities of disinfectant on clinical strains, strains from the inanimate hospital environment, and strains obtained from the apple surfaces. Overall, all 25 L. elongisporus isolates showed low MICs against all disinfectants tested except sodium hypochlorite. Specifically, high MIC values (MIC value of ≥1%) were observed for sodium hypochlorite against the majority of clinical strains (85%), all inanimate-environment strains from hospitals, and a single fruit strain which was genetically related to clinical strains. Notably, for sodium hypochlorite, all inanimate-environment strains and 50% of clinical strains showed 2- to 4-fold-higher MICs (2% to 4%) than the recommended concentration (1%, equivalent 1,000 ppm) of sodium hypochlorite used in clinical practices for disinfection (18). In contrast, five fruit strains distinct from the outbreak cluster (cluster II) showed low sodium hypochlorite MICs (0.06% to 0.5%).
Time-kill kinetics.
As L. elongisporus strains exhibited high MIC values for sodium hypochlorite, we investigated the activity of sodium hypochlorite on yeast cells, using a time-kill curve assay. The time-kill kinetics profile of clinical and inanimate strains against 1% sodium hypochlorite showed that both types of strains had lower susceptibility to sodium hypochlorite than did strains from apple surfaces (Fig. S4). Further, after 8 h, inanimate-environment and clinical strains from cluster I that exhibited high MICs for sodium hypochlorite (MIC value of >1%) showed prominent growth in the presence of 1% sodium hypochlorite, with growths not significantly different (<2-fold difference; P > 0.05) from that of control (absence of sodium hypochlorite). In contrast, after 8 h, fruit strains treated with sodium hypochlorite showed prominent growth inhibition (>2-fold difference; P < 0.05) compared with untreated cells.
The graph illustrates the time-kill curve (optical density versus time plot) of three Lodderomyces elongisporus strains from clinical (VPCI/160/P/2022), neonate warmer (inanimate environment, VPCI/E/HR6/2022), and apple surface (VPCI/32F15/2020) sources in the presence of 1% (vol/vol) sodium hypochlorite. The untreated inoculum was used as a control. Download FIG S4, DOCX file, 0.07 MB (74.2KB, docx) .
Copyright © 2023 Yadav et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Further, the viability of all the three strains in the presence of 1% sodium hypochlorite was examined by spreading the inoculum on Sabouraud dextrose agar (SDA) plates after 2-h intervals up to 24 h of the incubation period at 37°C. Among the three isolates, the inanimate-environment strain grew luxuriantly in the presence of 1% sodium hypochlorite within 2 h. However, the strain from the surface of fruit was killed in the presence of 1% sodium hypochlorite after 2 h (Fig. S5).
Representation of viable cells on culture plates by in-use testing in the presence of 1% sodium hypochlorite. Time of incubation is depicted horizontally. Panels A (VPCI/160/P/2022; clinical), B (VPCI/32F15/2020; from apple surface), and C (inanimate environment VPCI/E/HR6/2022; neonate warmer) represent three Lodderomyces elongisporus strains in the absence of sodium hypochlorite. Panels D, E, and F represent the three L. elongisporus strains in the presence of 1% sodium hypochlorite in similar order. Download FIG S5, DOCX file, 0.4 MB (421.3KB, docx) .
Copyright © 2023 Yadav et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SEM of Lodderomyces elongisporus after 1% sodium hypochlorite treatment.
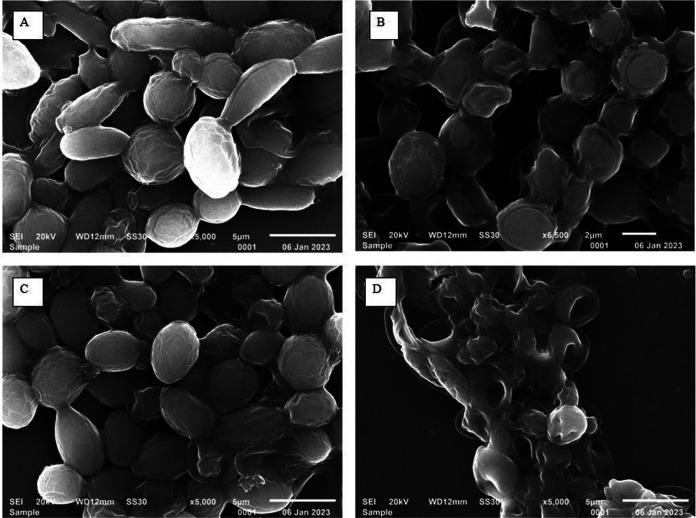
During adaptation to different microenvironments, morphological features of yeast cells may change. Therefore, we investigated the morphological changes among the clinical and inanimate-environment strains from hospitals in the presence of disinfectant using scanning electron microscopy (SEM). Scanning images of cells were captured before and after treatment with 1% sodium hypochlorite. Before treatment, the clinical strain showed ovoid budding yeast cells along with tube-like blastoconidia whereas cells of the inanimate-environment strain (NICU warmer) showed only spherical, ovoid yeast cells. After treatment of cell inoculum with 1% sodium hypochlorite, the damaging effect was prominent in the inanimate-environment strain compared to the clinical strain. Deep invaginations over cell surfaces were observed in the environmental strain whereas the clinical strain did not exhibit such changes (Fig. 2).
FIG 2.
Scanning electron micrograph of Lodderomyces elongisporus. (A) VPCI/160/P/2022 clinical strain without 1% sodium hypochlorite treatment. (B) VPCI/160/P/2022 clinical strain with 1% sodium hypochlorite. (C) SEM images of VPCI/E/HR6/2022 inanimate clinical environmental strain without 1% sodium hypochlorite treatment. (D) VPCI/E/HR6/2022 inanimate clinical environmental strain with 1% sodium hypochlorite.
Mode of reproduction.
L. elongisporus is diploid and has been described as sexual and homothallic (self-mating) (17). However, previous studies failed to identify mating-type-like (MTL) genes that were homologous to those in Candida spp. in L. elongisporus. Here, we used both the nucleotide and amino acid sequences of MTL genes in Candida albicans and C. parapsilosis to search for potential matches in our strains. Using TBLASTN searches, a strong significant match (5e−150) to MTLa1p of C. parapsilosis was found on scaffold 7 in all our strains as well as in the reference strain but no match was found for MTLa2p in the genomes of any of our sequenced strains or the reference strain. Adjacent to the MTLa1 locus on scaffold 7 of L. elongisporus, homologs to several other genes of the C. albicans MTLa locus were found, including RCY1, PIKa, OBPa, and PAPa. Interestingly, a very weak match (3e−04) was found for MTLα2p of C. albicans on scaffold 2 in all L. elongisporus strains. However, no match was found for MTLα1p in the genomes of any of our sequenced strains or the reference strain. Together, the results suggest that the L. elongisporus strains analyzed here each contained partial sequences of both the MTLa and MTLα loci found in several other related Candida species. Combined with the observation of ascospore formation in several of the strains, the genomic results suggest that L. elongisporus is capable of sexual reproduction via homothallism.
In homothallism, a single individual cell can complete sexual reproduction by itself. In haploid organisms, homothallic reproduction results in progeny with genotypes identical to those of parental strains, similar to those in clonal/asexual reproduction. However, in diploid organisms, homothallism can produce recombinant genotypes. To investigate whether there is evidence for recombination, we conducted a four-gamete test for our samples. In this test, assuming no parallel or reverse mutations, the presence of all four genotypes at two loci with two alleles each in the population would be consistent with recombination. Because L. elongisporus is a diploid, we used only homozygous SNPs for this test. In addition, only SNPs with the minor-frequency allele present in at least two strains were included for analysis. Our analyses revealed that in the total sample of 25 isolates, 0.96% (out of 16,653,581,253) of the analyzed SNP pairs were consistent with recombination. However, for the clinical environmental population and the patient population of L. elongisporus, 59.72% (out of 3,901,821) and 64.76% (out of 51,770,400) of the SNP pairs were consistent with recombination. In contrast, only 0.19% (out of 15,321,513,826) of the SNP pairs in the fruit sample were consistent with recombination. Together, the results suggest that L. elongisporus strains were undergoing infrequent sexual recombination in the fruit population but frequent recombination in the patient population and in the hospital environment.
DISCUSSION
We report the outbreak of L. elongisporus in a NICU and confirm the close genetic relationships among strains obtained from the neonates, those from their inanimate environment, and those from the surfaces of apples using whole-genome sequencing. The present outbreak of L. elongisporus infection affected primarily preterm neonates with low birthweight. Though overall infrequent, L. elongisporus infections have been mainly reported as sporadic cases occurring primarily in adult patients with intravenous drug use and in patients with cardiac-related conditions (10, 12, 19, 20). Invasive fungal infections caused by L. elongisporus have been reported from diverse geographical regions including countries in the Middle East and East Asia, as well as from India, Spain, Australia, North America, and Canada (21–23). Recently, L. elongisporus bloodstream infections were reported in three infants and five adults in a single center in North India. Among the pediatric cases, underlying diseases included congenital heart disease/atrophic kidney and tracheoesophageal fistula (22). Also, a single case in a pediatric patient with a ventricular septal defect was reported from Delhi, India (23).
L. elongisporus was first recovered from citrus concentrate in 1952 and named Saccharomyces elongisporus. This yeast was later renamed L. elongisporus by van der Walt and colleagues in 1966 (24). The epidemiologic clustering of L. elongisporus in 10 neonates in the span of 6 months suggested transmission of this yeast among the hospitalized neonates. The fact that the railing and the temperature panel of the open care warmer of the L. elongisporus-positive neonate also harbored this yeast suggested a potential link between the environmental strains and the neonates. Indeed, whole-genome sequence analyses revealed the overall high genome sequence similarity between strains from the inanimate environment of the hospital and those infecting the neonates. Further, two strains obtained from surfaces of apples also showed genetic relatedness with the patient and inanimate-clinical-environment sources in hospitals, suggesting L. elongisporus strains could be exchanged among patients and hospital environments and outside the clinical environments. Previously, strains of Candida auris isolated from surfaces of apples in Delhi, India, were found to be genetically linked to clinical strains, suggesting fruits as a possible source of transmission of the yeast and vice versa (14).
Surprisingly, we found evidence for recombination in all three ecological samples, especially those from the inanimate clinical environment and neonates. Indeed, the high genetic similarity among strains within both cluster I and cluster II initially suggested evidence of clonal expansion of one or a few strains in India. The low rates of SNP pairs showing evidence of recombination within both the total sample and the fruit sample were due to genetic structuring within these two samples where two divergent clusters were included in these analyses, contributing to a high number of SNPs being analyzed. The inclusion of divergent populations in the same four-gamete test would vastly increase the number of SNP pairs in the analyses (i.e., increased denominator) and, if there were no outcrossing and recombination between the divergent populations, would reduce the percentages of SNP pairs with evidence of recombination. Our observed results further support the genetic distinctiveness of clusters I and II.
The genome-wide heterozygosity patterns showed a unique loss of heterozygosity (LOH) for a large part of scaffold 2 (NW_001813676) in the strain obtained from the open warmer in the NICU. Such an observed LOH could be derived from sexual, asexual, or parasexual reproduction (25). In addition, neonatal environment strains exhibited aneuploidy. Notably, inanimate-environment strains exhibited resistance to a commonly used surface disinfectant, i.e., sodium hypochlorite (MIC, >1% or >1,000 ppm of active chlorine), used for disinfecting soiled surfaces. In the present study, the broth microdilution method employed for disinfectant testing used RPMI growth medium, which may consume some amounts of available chlorine. However, consistent results were obtained with repeated testing which also correlated with the time-kill assay. In health care settings various guidelines recommend 1,000 ppm of active chlorine for routine disinfection of patient care areas and equipment and higher concentration of 10,000 ppm for terminal cleaning against Candida species, including Candida auris (26–28). Also, a similar concentration of sodium hypochlorite is suggested for surface disinfection against multidrug-resistant bacteria like Acinetobacter baumannii and Klebsiella pneumoniae (29, 30). Exposure to antifungal drugs is known to create aneuploidy or genome instability in a diversity of fungal pathogens, causing drug resistance/tolerance (31). Indeed, disinfectants and antifungal drug exposure have likely been a driving force for selection or faster evolution of the inanimate environmental and clinical strains in this study.
Diploid yeasts such as C. albicans and Candida tropicalis often are heterozygous for many genetic variants across the genome (32). Nevertheless, studies have pointed out that regions of the genome can frequently become homozygous through sexual, parasexual, and asexual processes (24). Loss of heterozygosity events can be induced by stress; notably, several stressors including exposure to heat, an oxidizing agent such as hydrogen peroxide, and fluconazole in C. albicans and Cryptococcus neoformans serotype allodiploid (AD) hybrids increased LOH even at doses that did not inhibit growth (33, 34). Typically, in these cases, the LOH events were followed by the duplication of the remaining chromosome/chromosomal segments to restore diploidy. It is tempting to speculate that the environmental strains of L. elongisporus isolated in the present study exhibited LOH probably due to the presence of several stressors in the hospital environment, such as enhanced use of disinfectants. Regardless, the seemingly rapid genome change in the hospital environment suggests that L. elongisporus is capable of fast evolution and adaptation in stressed environments. Indeed, L. elongisporus strains have been recovered from various environmental sources including fresh fruit, fruit concentrates, soft drinks, and insects (10, 35, 36).
We found a range of unique SNPs within each of the strains, including those from patients. Some of these SNPs in strains from patients were likely de novo mutations accumulated during their asexual reproduction. Interestingly, our genome comparisons revealed a large number of mutations in genes involved in DNA repair, including both missense mutations and four base substitutions that caused predicted “stop_gained”/“stop_lost”/“start_lost”/“stop_gained” functions in four genes annotated to be involved in DNA repair in all our clinical strains compared to the reference strain NRRL YB-4239. Specifically, those included (i) a translation “stop_gained” A→T mutation at nucleotide position 516372 of scaffold NW_001813679 of gene LELG_04803 annotated as involved in “DNA repair” where all clinical strains were homozygous for the mutant nucleotide T at this position, (ii) a translation “stop_lost” A→T mutation at nucleotide position 391028 of scaffold NW_001813680 of gene LELG_04316 annotated as involved in “DNA repair” where all clinical strains were heterozygous A/T at this position, (iii) a translation “start_lost” T→C mutation at nucleotide position 901485 of scaffold NW_001813680 of gene LELG_04499 annotated as involved in “nucleotide excision repair” where all clinical strains were homozygous for mutant allele C/C at this position, and (iv) a translation “stop_gained” C→T mutation at nucleotide position of 185369 of scaffold NW_001813681 of gene LELG_03735 annotated as involved in “nucleotide excision repair” where all clinical strains were heterozygous C/T at this position (see Table S1 in the supplemental material). Together, these mutations could have contributed to the high number of unique SNPs observed for each strain.
In the present outbreak, infection was likely transmitted among neonates by contact with health care workers and fomites. Suboptimal adherence to infection prevention and control practices is usually a major contributing factor to outbreaks, which probably was the case in the present outbreak. Of neonates infected with L. elongisporus, the majority had very low birthweights and were born preterm. Prematurity and low birthweight are well-recognized risk factors for candidemia. All the neonates were empirically treated with fluconazole, and amphotericin B was instituted after the confirmation of L. elongisporus. In fact, all the bloodstream strains exhibited susceptibility to azoles, amphotericin B, and echinocandins. Interestingly, strains obtained from the natural environment, i.e., from the surfaces of apples (n = 7), showed 2- to 8-fold-increased MICs of azoles compared to clinical strains. Further, the strains obtained from surfaces of apples showed alterations in the azole target genes such as TAC1B, CDR1, and MDR1. These findings suggest that continued exposure to azoles in the environment may select azole-resistant strains of L. elongisporus. Finally, the multifaceted investigation of the L. elongisporus outbreak in the present study suggests that this yeast is among a growing list of fungi capable of causing severe infections among a diversity of human hosts (37). The genetic mechanisms underlying their adaptations to humans and hospital and natural environments warrant in-depth investigations and measures to contain their spread and persistence in clinics.
MATERIALS AND METHODS
Microbiological investigation.
Blood culture specimens obtained from all neonates flagged positive in the BacT/Alert 3D system (bioMérieux, Inc., Durham, NC). Subculture of the positive blood culture bottles was undertaken on Sabouraud dextrose agar (SDA), and the colony characteristics of L. elongisporus on SDA and CHROMagar Candida medium (Becton, Dickinson, Baltimore, MD, USA) were recorded at 37°C. Further, biochemical profiling of isolates using the Vitek-2 (bioMérieux, Marcy l’Etoile, France) commercial identification system was evaluated (38). The identification of isolates to species level was done by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS; Bruker Biotyper OC version 3.1; Daltonics, Bremen, Germany) (≥2 score) and confirmed by sequencing of internal transcribed spacer (ITS) regions of the ribosomal DNA (rDNA) (38).
Environmental sampling.
A total of 80 swabs (Himedia, Mumbai, India) were collected from the surrounding inanimate environments where L. elongisporus-positive neonates resided within the hospital. Environment samples were collected at the end of February 2022, after the sudden surge of four fungemia cases in the neonatal intensive care unit (NICU). We collected swab samples from all neonatal open care warmers including their surfaces, railings, monitoring panels attached to warmers, such as temperature display panels, pulse oximeter probes, bedsheets, and respiratory care equipment. Also, trays containing disposable supplies, adjacent floor trolleys, water sink, taps, the hand dryer, doorknobs, and weighing machines were thoroughly swabbed (16). Simultaneously, swabs from fingertips and finger webs of health care workers at the neonate ICU were also collected to check for colonization (39).
AFST.
A total of 31 L. elongisporus isolates recovered from clinical specimens (n = 13 BSIs) and inanimate-environment isolates (n = 2) from the NICU, from the floor of two other hospitals (n = 3), and from the surfaces of stored apples (n = 13) were subjected to antifungal susceptibility testing (AFST) by the CLSI broth microdilution method following method M27-A3 (15). The isolates were tested against 10 antifungal drugs of three classes, i.e., azoles, echinocandins, and amphotericin B, as detailed in Table 2. The antifungals tested were fluconazole (FLU; Sigma, St. Louis, MO, USA), itraconazole (ITC; Lee Pharma, Hyderabad, India), voriconazole (VRC; Pfizer, Groton, CT, USA), posaconazole (POS; Merck, Whitehouse Station, NJ, USA), isavuconazole (ISA; Basilea Pharmaceutical, Basel, Switzerland), 5-flucytosine (5-FC; Sigma), caspofungin (CFG; Sigma), micafungin (MFG; Sigma), anidulafungin (AFG; Sigma), and amphotericin B (AMB; Sigma). The drugs were tested for 10 (2-fold) dilutions, and the drug concentration ranges were as follows: FLU, 0.25 to 128 mg/L; ITC, VRC, and AMB, 0.03 to 16 mg/L; POS, ISA, AFG, MFG, and CFG, 0.015 to 8 mg/L; 5-FC, 0.125 to 64 mg/L. Candida krusei strain ATCC 6258 and Candida parapsilosis strain ATCC 22019 were used as quality control strains.
Whole-genome sequencing and phylogenetic analysis.
We sequenced whole genomes from each selected isolate (n = 13 clinical isolates, n = 2 environmental isolates) obtained from the present outbreak. In addition, we selected 10 L. elongisporus isolates, seven from the surface of stored apples and three from the floor of two other hospitals in Delhi, using a NextSeq sequencer. Whole-genome sequence libraries were prepared using the NEBNext Ultra II DNA FS kit (New England Biolabs, Ipswich, MA, USA). Whole-genome sequencing data of 25 Indian samples were first filtered for low-quality reads and adapters using Trimmomatic v0.39 (40). Then, clean reads were aligned with the genome of L. elongisporus strain YB-4239 (GCF_000149685.1_ASM14968v1_genomic.fna) using BWA-MEM v0.7.17 (41). Postalignment processing was performed using SAMtools v1.13 and Picard tools v2.26.3 (http://broadinstitute.github.io/picard/). Variant identification was conducted using GATK v4.2 and GATK tools HaplotypeCaller, CombineGVCFs, GenotypeGVCFs, SelectVariants, and Variant Filtration (42). Single nucleotide polymorphisms (SNPs) were selected by removing SNP loci that failed to meet the following hard thresholds “QualByDepth (QD) < 2.0 ‖ FisherStrand (FS) > 60.0 ‖ RMSMappingQuality (MQ) < 40.0 ‖ StrandOddsRatio (SOR) > 4.0”. The specific filters are as follows: QD: the variant confidence normalized by unfiltered depth of variant samples; FS: strand bias estimated using Fisher’s exact test; MQ: root mean square of the mapping quality of reads across all samples; SOR: strand bias estimated by the symmetric odds ratio test. Phylogenetic analysis was conducted on homozygous loci containing SNPs in at least one sample (43). Maximum likelihood phylogeny was inferred with the GTR+CAT model based on 244,443 homozygous loci using FastTree v 2.1.11 (44). We deposited FASTQ files for all isolates at GenBank under BioProject no. PRJNA889843.
Four-gamete test for evidence of recombination.
To infer whether recombination has occurred in our samples, four-gamete tests were conducted, following procedures described in reference 40. Specifically, strains were first grouped based on isolation sources, i.e., clinical environment (5 isolates), patients (13 isolates), fruit (7 isolates), and the total population (all 25 isolates). Then, pairs of homozygous SNP loci with the minor allele present in at least 2 strains of each group were compared. Here, for any two SNP sites containing both the reference allele (0) and an alternative allele (1), if all four possible combinations, i.e., 00, 01, 10, and 11, were identified, we considered the SNP pair consistent with recombination.
Morphological characterization of Lodderomyces elongisporus.
To check any morphological difference between L. elongisporus strains on the basis of their source of isolation, all were subcultured on ascospore agar containing 0.1% sodium acetate and incubated at 37°C for ≥21 days. Cells were monitored under an optical microscope (Nikon H600L; Japan) at ×40 magnification.
Determination of ploidy of Lodderomyces elongisporus strains.
A total of 13 L. elongisporus isolates including two clinical strains (VPCI/147/P/2022 and VPCI/160/P/2022), all five strains from the inanimate environment, and six strains recovered from the surface of fruits were subjected to fluorescence-activated cell sorting (FACS) (FACSAria III; BD Biosciences, USA). Candida albicans (ATCC 90028) was used as a reference for the diploid strain. Sample preparation and analysis were done as described previously (14). FlowJo 7.8 software was used to interpret the FACS results.
Activity of disinfectants against Lodderomyces elongisporus.
A disinfectant activity assay was done to compare the activities of disinfectants on clinical strains, strains from the inanimate environment, and strains from the apple surfaces. In this study, four disinfectants were used, which were the intermediate-level disinfectant absolute ethanol (Changshu Hongsheng Fine Chemical, China) and high-level disinfectants glutaraldehyde (2.45% [wt/vol]; Sceptre Medical India, Delhi, India), fifth-generation quaternary ammonium compound (13.6% [wt/vol] Mi-Fog [Faith Microsolution, Delhi, India]), and sodium hypochlorite containing 10% available chlorine (Ebele Cosmeceuticals, India). The broth microdilution method was used for determining MICs of the disinfectants against 25 strains of L. elongisporus. All the disinfectants were tested for 10 (2-fold) dilutions, and the drug concentration ranges were 0.06 to 32% for sodium hypochlorite, corresponding to 6 to 3,200 ppm of available chlorine, and 0.19 to 100% for ethanol, quaternary ammonium compound, and glutaraldehyde. MIC endpoints for disinfectants were defined as the lowest concentration that inhibited 100% of the growth as read visually at 24 h of incubation at 37°C.
Time-kill assay of Lodderomyces elongisporus against disinfectant (sodium hypochlorite).
Three isolates including one from each source, i.e., clinical (VPCI/160/P/2022), inanimate environment (VPCI/E/HR6/2022), and apple surface (VPCI/32F15/2020), were grown in SD broth at 37°C for 16 h. Briefly, an inoculum of 1 × 108 cells (optical density at 625 nm [OD625] = 0.1) from an overnight culture was inoculated again into SD broth for 3 h. After 3 h of incubation, suspensions were supplemented with 1% sodium hypochlorite (vol/vol; recommended concentration of sodium hypochlorite). An untreated inoculum of yeast cells was used as a control. Further, a 200-μL inoculum of both treated and untreated cells was dispensed into a 96-well microtiter plate. The plate was incubated inside a microplate reader (Infinite 200 Pro; Tecan, Switzerland) with constant agitation at 200 rpm at 37°C. The growth turbidity was measured at a regular interval of 2 h for 24 h at an optical density at 530 nm. The growth curve was plotted against OD530 versus time by using GraphPad Prism 4. In addition, in-use testing was performed for the same three isolates (45). An inoculum with an OD625 of 0.1 of each isolate was transferred to a 15-mL solution of 1% sodium hypochlorite and incubated at 37°C. The kill rate of L. elongisporus strains in the presence of sodium hypochlorite solution was determined by examining the viability of fungal cells after 0-, 2-, 4-, 6-, 8-, and 24-h periods of contact with the 1% sodium hypochlorite solution. To examine the viable cells in suspension, 100 μL of inoculum after every 2 h was taken and subcultured on SDA plates and images of culture plates were captured by a gel imager (Bio-Rad, CA, USA) after 24 h of incubation.
Scanning electron microscopy of Lodderomyces elongisporus.
To evaluate the effect of sodium hypochlorite on the surface of L. elongisporus cells, scanning electron microscopy (SEM) was done. One clinical strain (VPCI/160/P/2022) and one strain from the inanimate environment (VPCI/E/HR6/2022) were selected for this purpose. Microbial samples for SEM analysis were prepared as described previously (46). A single colony of both selected strains was subcultured on SDA followed by inoculation onto the yeast extract-peptone-dextrose (YPD) broth incubated at 37°C for 17 h in a shaking incubator at 200 rpm. An inoculum with 1 × 108 cells (OD600 = 0.1) from overnight culture was transferred in YPD broth. A prepared volume of cell suspension was further divided into two equal volumes. One was supplemented with 1% (vol/vol) sodium hypochlorite, and further, both the treated and the untreated suspension were incubated for 24 h at 37°C. Subsequently, both treated and untreated cells were fixed with 2.5% glutaraldehyde (Sigma-Aldrich) in phosphate-buffered saline (PBS; pH 6; Sigma-Aldrich) for 16 h at 4°C. After primary fixation, cells were washed twice with PBS and then fixed with 1% osmium tetroxide for 3 h at 4°C. The fixed cells were dehydrated by using increasing concentrations of ethanol from 30% to 100% followed by drying. The samples (treated and untreated) were further sputter coated with gold (JEC 300 sputter coater) and analyzed under a scanning electron microscope (JEOL JSM 6610LV; Japan) with a ×5,000 magnification.
Data availability.
The genome sequences of all 25 strains analyzed in the present study are accessible through BioProject number PRJNA889843.
ACKNOWLEDGMENTS
Anuradha Chowdhary is a fellow of the Canadian Institute for Advanced Research (CIFAR). Anamika Yadav acknowledges the University Grants Commission (file no. 16-9/2019), 596/Govt of India (GoI), New Delhi, and Kusum Jain acknowledges the Council of Scientific & Industrial Research, GoI [F. No. 08/728(0001)/(2019-EMR-I)] for grant of fellowships. Jianping Xu acknowledges a research grant from McMaster University Global Science Initiative (2020-03).
We thank Dhiraj Kumar and Shilpa Sharma, Cellular Immunology Group, International Centre for Genetic Engineering and Biotechnology (ICGEB), New Delhi, for providing assistance in FACS experiments. We thank the University Science Instrumentation Center (USIC), University of Delhi (North Campus), for the SEM facility.
All other authors declare no potential conflicts of interest. We alone are responsible for the content and writing of the paper.
Anamika Yadav and Peeyush Jain contributed to sample collection and processing, and Anamika Yadav and Kusum Jain conducted the experiments. Yue Wang performed whole-genome sequence analysis; Aditi Singh helped in sample collection. Anuradha Chowdhary designed the study, analyzed the data, and drafted the manuscript; Jianping Xu contributed to data interpretation and manuscript writing and reviewing; Ashutosh Singh and Peeyush Jain analyzed the data and reviewed the manuscript. All authors have read and agreed to the final version of the manuscript.
Contributor Information
Jianping Xu, Email: jpxu@mcmaster.ca.
Anuradha Chowdhary, Email: chowdhary.anuradha@gmail.com.
Alexandre Alanio, Institut Pasteur.
REFERENCES
- 1.World Health Organization. 2020. WHO global report on the epidemiology and burden of sepsis: current evidence, identifying gaps and future directions. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Roble AK, Ayehubizu LM, Olad HM. 2022. Neonatal sepsis and associated factors among neonates admitted to neonatal intensive care unit in general hospitals, Eastern Ethiopia 2020. Clin Med Insights Pediatr 16:11795565221098346. doi: 10.1177/11795565221098346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delaloye J, Calandra T. 2014. Invasive candidiasis as a cause of sepsis in the critically ill patient. Virulence 5:161–169. doi: 10.4161/viru.26187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleischmann C, Reichert F, Cassini A, Horner R, Harder T, Markwart R, Tröndle M, Savova Y, Kissoon N, Schlattmann P, Reinhart K, Allegranzi B, Eckmanns T. 2021. Global incidence and mortality of neonatal sepsis: a systematic review and meta-analysis. Arch Dis Child 106:745–752. doi: 10.1136/archdischild-2020-320217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uttam KG, Gupta P, Poddar S. 2022. Fungal sepsis in a tertiary neonatal intensive care unit: a cross-sectional study. Paediatric Infect Dis 4:33–37. doi: 10.5005/jp-journals-10081-1320. [DOI] [Google Scholar]
- 6.Vaux S, Criscuolo A, Desnos-Ollivier M, Diancourt L, Tarnaud C, Vandenbogaert M, Brisse S, Coignard B, Dromer F, Geotrichum Investigation Group . 2014. Multicenter outbreak of infections by Saprochaete clavata, an unrecognized opportunistic fungal pathogen. mBio 5:e02309-14. doi: 10.1128/mBio.02309-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhary A, Stielow JB, Upadhyaya G, Singh PK, Singh A, Meis JF. 2020. Candida blankii: an emerging yeast in an outbreak of fungaemia in neonates in Delhi, India. Clin Microbiol Infect 26:648.e5–648.e8. doi: 10.1016/j.cmi.2020.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhary A, Sharada K, Singh PK, Bhagwani DK, Kumar N, de Groot T, Meis JF. 2020. Outbreak of Dirkmeia churashimaensis fungemia in a neonatal intensive care unit, India. Emerg Infect Dis 26:764–768. doi: 10.3201/eid2604.190847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow NA, Chinn R, Pong A, Schultz K, Kim J, Gade L, Jackson BR, Beer KD, Litvintseva AP. 2020. Use of whole-genome sequencing to detect an outbreak of Malassezia pachydermatis infection and colonization in a neonatal intensive care unit-California, 2015-2016. Infect Control Hosp Epidemiol 41:851–853. doi: 10.1017/ice.2020.73. [DOI] [PubMed] [Google Scholar]
- 10.Daveson KL, Woods ML. 2012. Lodderomyces elongisporus endocarditis in an intravenous drug user: a new entity in fungal endocarditis. J Med Microbiol 61:1338–1340. doi: 10.1099/jmm.0.047548-0. [DOI] [PubMed] [Google Scholar]
- 11.Taj-Aldeen SJ, AbdulWahab A, Kolecka A, Deshmukh A, Meis JF, Boekhout T. 2014. Uncommon opportunistic yeast bloodstream infections from Qatar. Med Mycol 52:552–556. doi: 10.1093/mmycol/myu016. [DOI] [PubMed] [Google Scholar]
- 12.Thompson CM, Warner N, Hurt CB, Alby K, Miller MB. 2021. Closing the brief case: a case of prosthetic valve endocarditis due to Lodderomyces elongisporus. J Clin Microbiol 59:e01227-20. doi: 10.1128/JCM.01227-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James SA, Collins MD, Roberts IN. 1994. The genetic relationship of Lodderomyces elongisporus to other ascomycete yeast species as revealed by small-subunit rRNA gene sequences. Lett Appl Microbiol 19:308–311. doi: 10.1111/j.1472-765x.1994.tb00462.x. [DOI] [PubMed] [Google Scholar]
- 14.Yadav A, Jain K, Wang Y, Pawar K, Kaur H, Sharma KK, Tripathy V, Singh A, Xu J, Chowdhary A. 2022. Candida auris on apples: diversity and clinical significance. mBio 13:e0051822. doi: 10.1128/mbio.00518-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CLSI. 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts, 3rd ed. M27-A3. CLSI, Wayne, PA. [Google Scholar]
- 16.Yadav A, Singh A, Wang Y, Haren MHV, Singh A, de Groot T, Meis JF, Xu J, Chowdhary A. 2021. Colonisation and transmission dynamics of Candida auris among chronic respiratory diseases patients hospitalised in a chest hospital, Delhi, India: a comparative analysis of whole genome sequencing and microsatellite typing. J Fungi (Basel) 7:81. doi: 10.3390/jof7020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CDC. 2008. Guideline for disinfection and sterilization in healthcare facilities. CDC, Atlanta, GA. [Google Scholar]
- 19.Ahmad S, Khan ZU, Johny M, Ashour NM, Al-Tourah WH, Joseph L, Chandy R. 2013. Isolation of Lodderomyces elongisporus from the catheter tip of a fungemia patient in the Middle East. Case Rep Med 2013:560406. doi: 10.1155/2013/560406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Obaid K, Ahmad S, Joseph L, Khan Z. 2018. Lodderomyces elongisporus: a bloodstream pathogen of greater clinical significance. New Microbes New Infect 26:20–24. doi: 10.1016/j.nmni.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dear T, Joe Yu Y, Pandey S, Fuller J, Devlin MK. 2021. The first described case of Lodderomyces elongisporus meningitis. J Assoc Med Microbiol Infect Dis Can 6:221–228. doi: 10.3138/jammi-2021-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaur H, Gupta P, Shankarnarayan S, Pandey A, Ghosh A, Chakrabarti A, Rudramurthy S. 2022. P142 Lodderomyces elongisporus: an emerging cause of fungemia. Med Mycol 60(Suppl 1):myac072P142. doi: 10.1093/mmy/myac072.P142. [DOI] [Google Scholar]
- 23.Gourav S, Singh G, Xess I, Xess AB, Yadav RK, Pandey M, Sachdev J, Ramakrishnan S, Iram A, Rana B. 2022. P200 Lodderomyces elongisporus fungemia in a late post-operative patient with ventricular septal defect: case report. Med Mycol 60(Suppl 1):myac072P200. doi: 10.1093/mmy/myac072.P200. [DOI] [Google Scholar]
- 24.Kurtzman CP. 2011. Lodderomyces van der Walt (1971), p 561–563. In Kurtzman CP, Fell JW, Boekhout T (ed), The yeasts: a taxonomic study, 5th ed. Elsevier, Amsterdam, Netherlands. [Google Scholar]
- 25.Xu J. 2021. Is natural population of Candida tropicalis sexual, parasexual, and/or asexual? Front Cell Infect Microbiol 11:751676. doi: 10.3389/fcimb.2021.751676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Public Health England. 2017. Guidance for the laboratory investigation, management and infection prevention and control for cases of Candida auris, v2.0. Public Health England, London, United Kingdom. [Google Scholar]
- 27.Ong CW, Chen SC, Clark JE, Halliday CL, Kidd SE, Marriott DJ, Marshall CL, Morris AJ, Morrissey CO, Roy R, Slavin MA, Stewardson AJ, Worth LJ, Heath CH, Australian and New Zealand Mycoses Interest Group (ANZMIG), Healthcare Infection Control Special Interest Group (HICSIG), Australasian Society for Infectious Diseases (ASID) . 2019. Diagnosis, management and prevention of Candida auris in hospitals: position statement of the Australasian Society for Infectious Diseases. Intern Med J 49:1229–1243. doi: 10.1111/imj.14612. [DOI] [PubMed] [Google Scholar]
- 28.Malyshev D, Jones IA, McKracken M, Öberg R, Harper GM, Joshi LT, Andersson M. 2023. Hypervirulent R20291 Clostridioides difficile spores show disinfection resilience to sodium hypochlorite despite structural changes. BMC Microbiol 23:59. doi: 10.1186/s12866-023-02787-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu WL, Liang HW, Lee MF, Lin HL, Lin YH, Chen CC, Chang PC, Lai CC, Chuang YC, Tang HJ. 2014. The impact of inadequate terminal disinfection on an outbreak of imipenem-resistant Acinetobacter baumannii in an intensive care unit. PLoS One 9:e107975. doi: 10.1371/journal.pone.0107975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang C, Tao S, Yuan J, Li X. 2022. Effect of sodium hypochlorite on biofilm of Klebsiella pneumoniae with different drug resistance. Am J Infect Control 50:922–928. doi: 10.1016/j.ajic.2021.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Fisher MC, Alastruey-Izquierdo A, Berman J, Bicanic T, Bignell EM, Bowyer P, Bromley M, Brüggemann R, Garber G, Cornely OA, Gurr SJ, Harrison TS, Kuijper E, Rhodes J, Sheppard DC, Warris A, White PL, Xu J, Zwaan B, Verweij PE. 2022. Tackling the emerging threat of antifungal resistance to human health. Nat Rev Microbiol 20:557–571. doi: 10.1038/s41579-022-00720-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alabrudzinska M, Skoneczny M, Skoneczna A. 2011. Diploid-specific genome stability genes of S. cerevisiae: genomic screen reveals haploidization as an escape from persisting DNA rearrangement stress. PLoS One 6:e21124. doi: 10.1371/journal.pone.0021124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forche A, Abbey D, Pisithkul T, Weinzierl MA, Ringstrom T, Bruck D, Petersen K, Berman J. 2011. Stress alters rates and types of loss of heterozygosity in Candida albicans. mBio 2:e00129-11. doi: 10.1128/mBio.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong K, You M, Xu J. 2019. Genetic changes in experimental populations of a hybrid in the Cryptococcus neoformans species complex. Pathogens 9:3. doi: 10.3390/pathogens9010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji ZH, Jia JH, Bai FY. 2009. Four novel Candida species in the Candida albicans/Lodderomyces elongisporus clade isolated from the gut of flower beetles. Antonie Van Leeuwenhoek 95:23–32. doi: 10.1007/s10482-008-9282-7. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen NH, Suh SO, Blackwell M. 2007. Five novel Candida species in insect-associated yeast clades isolated from Neuroptera and other insects. Mycologia 99:842–858. doi: 10.3852/mycologia.99.6.842. [DOI] [PubMed] [Google Scholar]
- 37.Xu J. 2022. Assessing global fungal threats to humans. mLife 1:223–240. doi: 10.1002/mlf2.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kathuria S, Singh PK, Sharma C, Prakash A, Masih A, Kumar A, Meis JF, Chowdhary A. 2015. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization–time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol 53:1823–1830. doi: 10.1128/JCM.00367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Proctor DM, Dangana T, Sexton DJ, Fukuda C, Yelin RD, Stanley M, Bell PB, Baskaran S, Deming C, Chen Q, Conlan S, Park M, NISC Comparative Sequencing Program, Welsh RM, Vallabhaneni S, Chiller T, Forsberg K, Black SR, Pacilli M, Kong HH, Lin MY, Schoeny ME, Litvintseva AP, Segre JA, Hayden MK. 2021. Integrated genomic, epidemiologic investigation of Candida auris skin colonization in a skilled nursing facility. Nat Med 27:1401–1409. doi: 10.1038/s41591-021-01383-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997. [q-bio.GN].
- 42.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mixão V, Saus E, Boekhout T, Gabaldón T. 2021. Extreme diversification driven by parallel events of massive loss of heterozygosity in the hybrid lineage of Candida albicans. Genetics 217:iyaa004. doi: 10.1093/genetics/iyaa004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta AK, Ahmad I, Summerbell RC. 2002. Fungicidal activities of commonly used disinfectants and antifungal pharmaceutical spray preparations against clinical strains of Aspergillus and Candida species. Med Mycol 40:201–208. doi: 10.1080/mmy.40.2.201.208. [DOI] [PubMed] [Google Scholar]
- 46.Staniszewska M, Bondaryk M, Swoboda-Kopec E, Siennicka K, Sygitowicz G, Kurzatkowski W. 2013. Candida albicans morphologies revealed by scanning electron microscopy analysis. Braz J Microbiol 44:813–821. doi: 10.1590/S1517-83822013005000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Loss of heterozygosity pattern of 25 Lodderomyces elongisporus strains including 13 clinical strains and two inanimate-environment strains from the present outbreak, seven strains isolated from the surface of stored apples, and three environmental strains obtained from the floor of two other hospitals over the 11 scaffolds. Samples are ordered based on isolation sources. Samples from patients, clinical environment, and fruit are marked by red, orange, and green labels. Purple and yellow blocks indicate heterozygous and homozygous regions, respectively. Download FIG S1, DOCX file, 0.5 MB (492KB, docx) .
Copyright © 2023 Yadav et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SNP difference at 24 antifungal resistance-related genes between 13 clinical isolates of Lodderomyces elongisporus along with five isolates from inanimate environment of three different hospitals and seven isolates from surfaces of apples. Download Table S1, XLSX file, 0.2 MB (238.3KB, xlsx) .
Copyright © 2023 Yadav et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Ascospore formation in a clinical isolate (VPCI/160/P/2022) on ascospore agar containing 0.1% sodium acetate stained with lactophenol cotton blue at ×40 magnification showing one or two ascospores (white arrow) enclosed in an ascus. Download FIG S2, DOCX file, 5.1 MB (5.1MB, docx) .
Copyright © 2023 Yadav et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Histograms representing DNA content or cell cycle profile obtained by FACS. For comparison, diploid C. albicans ATCC 90038 was used as a reference strain. The x axis represents nuclear fluorescence, and the y axis represents cell number. Download FIG S3, DOCX file, 0.2 MB (249.1KB, docx) .
Copyright © 2023 Yadav et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The graph illustrates the time-kill curve (optical density versus time plot) of three Lodderomyces elongisporus strains from clinical (VPCI/160/P/2022), neonate warmer (inanimate environment, VPCI/E/HR6/2022), and apple surface (VPCI/32F15/2020) sources in the presence of 1% (vol/vol) sodium hypochlorite. The untreated inoculum was used as a control. Download FIG S4, DOCX file, 0.07 MB (74.2KB, docx) .
Copyright © 2023 Yadav et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Representation of viable cells on culture plates by in-use testing in the presence of 1% sodium hypochlorite. Time of incubation is depicted horizontally. Panels A (VPCI/160/P/2022; clinical), B (VPCI/32F15/2020; from apple surface), and C (inanimate environment VPCI/E/HR6/2022; neonate warmer) represent three Lodderomyces elongisporus strains in the absence of sodium hypochlorite. Panels D, E, and F represent the three L. elongisporus strains in the presence of 1% sodium hypochlorite in similar order. Download FIG S5, DOCX file, 0.4 MB (421.3KB, docx) .
Copyright © 2023 Yadav et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The genome sequences of all 25 strains analyzed in the present study are accessible through BioProject number PRJNA889843.