ABSTRACT
Most Pseudomonas aeruginosa strains produce bacteriocins derived from contractile or noncontractile phage tails known as R- and F-type pyocins, respectively. These bacteriocins possess strain-specific bactericidal activity against P. aeruginosa and likely increase evolutionary fitness through intraspecies competition. R-type pyocins have been studied extensively and show promise as alternatives to antibiotics. Although they have similar therapeutic potential, experimental studies on F-type pyocins are limited. Here, we provide a bioinformatic and experimental investigation of F-type pyocins. We introduce a systematic naming scheme for genes found in R- and F-type pyocin operons and identify 15 genes invariably found in strains producing F-type pyocins. Five proteins encoded at the 3′ end of the F-type pyocin cluster are divergent in sequence and likely determine bactericidal specificity. We use sequence similarities among these proteins to define eleven distinct F-type pyocin groups, five of which had not been previously described. The five genes encoding the variable proteins associate in two modules that have clearly reassorted independently during the evolution of these operons. These proteins are considerably more diverse than the specificity-determining tail fibers of R-type pyocins, suggesting that F-type pyocins may have emerged earlier. Experimental studies on six F-type pyocin groups show that each displays a distinct spectrum of bactericidal activity. This activity is strongly influenced by the lipopolysaccharide O-antigen type, but other factors also play a role. F-type pyocins appear to kill as efficiently as R-type pyocins. These studies set the stage for the development of F-type pyocins as antibacterial therapeutics.
IMPORTANCE Pseudomonas aeruginosa is an opportunistic pathogen that causes antibiotic-resistant infections with high mortality rates, particularly in immunocompromised individuals and cystic fibrosis patients. Due to the increasing frequency of multidrug-resistant P. aeruginosa infections, there is great need for the development of alternative therapeutics. In this study, we investigate one such potential therapeutic: F-type pyocins, which are bacteriocins naturally produced by P. aeruginosa that resemble noncontractile phage tails. We show that they are potent killers of P. aeruginosa and identify their probable bactericidal specificity determinants, which opens up the possibility of engineering them to precisely target strains of pathogenic bacteria. The resemblance of F-type pyocins to well-characterized phage tails will greatly facilitate their development into effective antibacterials.
KEYWORDS: F-type pyocin, bacteriophage, Pseudomonas aeruginosa, R-type pyocin, bacteriocin, antibiotic alternative
INTRODUCTION
With increasing antibiotic resistance, there is a strong incentive to identify alternative antibacterial therapeutics. To this end, interest in using phages or parts of phages to treat bacterial infections has greatly increased in recent years (1), and phage treatments have proven effective in clearing bacterial infections in humans (2–4). This success notwithstanding, there are potential drawbacks to phage therapy, including the possibility that introduced phages may acquire and transmit virulence or antibiotic resistance genes (5, 6). To circumvent these problems, the therapeutic potential of phage tail-like bacteriocins, also referred to as tailocins, is also being explored. Tailocin-encoding operons, which are found in many diverse bacterial species, are likely derived from prophages. The utility of tailocins as antibacterials has been amply demonstrated (7, 8). Like phages, tailocins are highly specific for their target organism, but they possess additional advantages. A single tailocin type can be engineered to kill a variety of bacterial species (9, 10), and tailocins can be efficiently produced in easily cultured organisms, such as Escherichia coli (11) or Bacillus subtilis (12). Here, we provide a detailed investigation of a group of tailocins produced by Pseudomonas aeruginosa that are related to noncontractile phage tails.
The tailocins of P. aeruginosa, discovered many decades ago (13), fall into two types known as F-type pyocins and R-type pyocins. All P. aeruginosa strains possess a gene cluster located between the trpE and trpG genes encoding F-type pyocins, R-type pyocins, or both types of pyocins. R-type pyocins, which are related to contractile-tailed phages, such as E. coli phage P2 (14), have been extensively studied. These entities are produced by different strains of P. aeruginosa and have the ability to kill other strains of the same species. R-type pyocins bind specifically to target strains, and then puncture their inner membrane, leading to rapid cell death (15). Derivatives of R-type pyocins with engineered tail fibers are able to kill other species of bacteria, such as E. coli and Yersinia pestis, and these engineered variants have shown efficacy in preventing and/or ameliorating infection in animal models (8, 11, 16, 17). F-type pyocins, which are related to noncontractile tailed phages, such as E. coli phage lambda (14), have been studied much less than the R-type. Although encoded in more than half of P. aeruginosa strains (18), no experimental work has been published on F-type pyocins since 1981 (19). The activities of F-type pyocins produced by five different strains have been described in the literature (19–22), each of which killed distinct sets of P. aeruginosa strains. However, the sequences of the operons encoding only two of these are known. Based on genome sequencing data, four further groups have been defined (18), but neither the production nor the activity of these groups was assessed. The mechanism of action and killing specificity determinants of F-type pyocins have not been defined.
Given the potential importance of tailocins in treating bacterial infections and the relative dearth of information pertaining to F-type pyocins, we undertook a comprehensive investigation of F-type pyocins encoded in a large number of P. aeruginosa strains. The goals of this study were to bioinformatically characterize F-type pyocin operons and correlate sequence diversity with the killing spectra of defined F-type pyocin groups. Through this process, we have identified 11 distinct groups of F-type pyocins and their likely specificity determinants. We conclude that these F-type pyocins have the potential to be engineered as highly effective antibacterial therapeutics.
RESULTS
Selection of P. aeruginosa strains for this study.
To gain an understanding of the diversity of R-type and F-type pyocins produced by P. aeruginosa, we produced lysates of 88 diverse strains (diversity was evaluated by isolation location and multilocus sequence typing) from our collection (23) by treating cultures with mitomycin C, which induces pyocin production and cell lysis (24). Each of these lysates was examined by transmission electron microscopy (TEM), and those displaying abundant levels of R- and/or F-type pyocins were further analyzed (Fig. 1). Ultimately, a set of 30 strains was chosen that produced only F-type pyocins (n = 8), only R-type pyocins (n = 9), or both R- and F-type (n = 13) pyocins. This set contained clinical and environmental strains from seven different countries, collected over a few decades (see Table S1 in the supplemental material). Of the 30 strains, 28 were sequenced, assembled, and annotated in this study (annotated genomes of strains PAO1 and PA14 were obtained from the Pseudomonas Genome Database [25]).
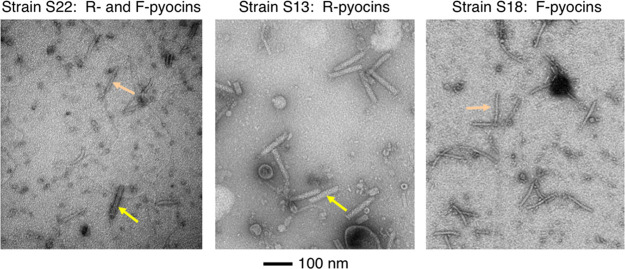
FIG 1.
TEM images of lysates of cells producing R- and F-type pyocins. A lysate of strain S22 (left panel), which produces both R- and F-type pyocins; a lysate of strain S13 (middle panel), which produces just R-type pyocins; and a lysate of strain S18 (right panel), which produces just F-type pyocins, are shown. R-type pyocin particles are indicated by yellow arrows, and F-type pyocin particles are indicated by orange arrows. Grids were negatively stained with uranyl acetate. The scale bar shown applies to all three micrographs.
Conserved features of R/F-type pyocin clusters.
The R- and F-type pyocin gene clusters are invariably found between the trpE and trpG genes in P. aeruginosa (14). To locate these gene clusters in each genome that was sequenced in this study, the regions between gene trpE and trpG were extracted and analyzed (Fig. 2a). Each of the 30 sequenced genomes was found to encode an F-type or R-type pyocin or both corresponding with the observed production of pyocin particles observed by electron microscopy. In total, 23 R-type and 21 F-type pyocin gene clusters were present in our analyses. The gene content of the pyocin clusters was constant across all the strains. We observed 8 genes, designated pyoRF1 to pyoRF8, which were found in all clusters, as well as 15 genes specific to R-type pyocins (pyoR1 to pyoR15) and 15 genes specific to F-type pyocins (pyoF1 to pyoF15) (Fig. 2a and Table 1). The pyoRF1 and pyoRF2 genes encode the PrtN activator and PrtR repressor, respectively. These proteins regulate expression of the cluster in response to DNA damage as previously described (24). The pyoRF3 and pyoRF4 gene products are uncharacterized, but their predicted functions suggest a role in regulating expression of the gene cluster. Analysis by HHpred indicates that the pyoRF3 gene encodes a putative zinc-binding transcription factor and pyoRF4 encodes a putative transcription antiterminator protein similar to gpQ of phage lambda (26). Homologs of PyoRF3 are found in more than 100 phage and prophage genomes, while homologs of PyoRF4 are found in a much smaller number of phages and prophages. The pyoRF5 to pyoRF8 genes encode a complete set of phage-like lysis genes, including a peptidoglycan hydrolase, holin, and Rz- and Rz1-like spannins (27). In operons encoding only F-type pyocins all eight pyoRF genes precede the genes encoding the F-type pyocin-specific genes. In clusters encoding just R-type pyocins or those encoding both R- and F-type pyocins, the R-type pyocin-specific genes are inserted within the lysis gene cluster between pyoRF5 and pyoRF6 (Fig. 2a).
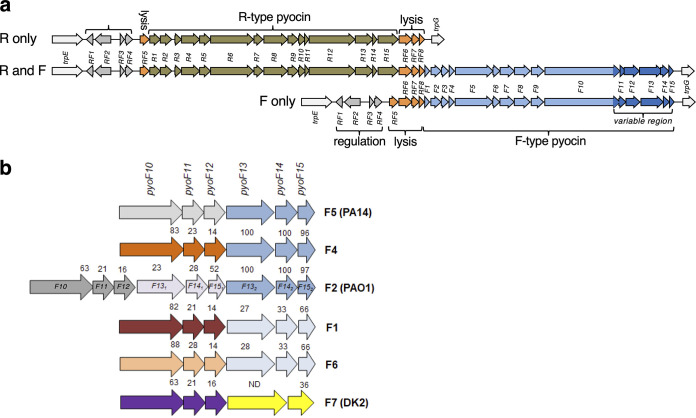
FIG 2.
R- and F-type pyocin operons. (a) Three types of R- or F-type pyocin operons are found in P. aeruginosa: operons encode just R-type pyocins (top), R- and F-type pyocins (middle), or just F-type pyocins (bottom). All three types share the same regulatory genes (pyoRF1 to pyoRF4) and lysis genes (pyoRF5 to pyoRF8). Genes unique to R-type pyocins (pyoR1 to pyoR15) and those unique to F-type pyocins (pyoF1 to pyoF15) are indicated. Genes in the F-type region that vary between groups and are involved in bactericidal specificity are shaded in darker blue. All three types of operons are located in the same position in the P. aeruginosa genome between the trpE and trpG genes. (b) A close-up of the variable region genes encoded at the 3′ end of the F-type pyocin clusters show the six different groups identified in our sequenced strains. The numbers above the genes indicate the percent pairwise sequence identity of the encoded protein with the homolog found in strain PA14 (group 5). Group F2 contains two copies of genes pyoF13 to pyoF15, but the two sets of encoded proteins are diverged in sequence. Proteins PyoF13, PyoF14, and PyoF15 are very closely related (>95% sequence identity) in groups F5, F4, and F2 (second group), as indicated by their shared coloring. The same proteins are highly similar in groups F1 and F6. For PyoF10 and PyoF13, sequence comparisons were carried out including only their variable C-terminal domains. P. aeruginosa strains where certain groups were previously identified are shown in parentheses.
TABLE 1.
Gene composition of the pyocin operon in P. aeruginosa strain PA14
| Gene | PA14 locus tag | Identity with HK022 homolog (%)a | Protein function |
|---|---|---|---|
| pyoRF1 | PA14_07950 | Activator (PrtN) | |
| pyoRF2 | PA14_07960 | Repressor (PrtR) | |
| pyoRF3 | PA14_07970 | Zinc finger transcription factorb | |
| pyoRF4 | PA14_07980 | Lambda gpQ-likeb | |
| pyoRF5 | PA14_07990 | Holinb | |
| pyoR1 | PA14_08000 | Tail terminatorb | |
| pyoR2 | PA14_08010 | Tail spike | |
| pyoR3 | PA14_08020 | Baseplate wedge 1c | |
| pyoR4 | PA14_08030 | Baseplate wedge 2c | |
| pyoR5 | PA14_08040 | Baseplate wedge 3c | |
| pyoR6 | PA14_08050 | Tail fiber | |
| pyoR7 | PA14_08060 | Tail fiber chaperone | |
| pyoR8 | PA14_08070 | Tail sheath | |
| pyoR9 | PA14_08090 | Tail tube | |
| pyoR10 | PA14_08100 | Tape measure chaperone | |
| pyoR11 | PA14_08110 | Tape measure chaperoned | |
| pyoR12 | PA14_08120 | Tape measure | |
| pyoR13 | PA14_08130 | Baseplate hub 1c | |
| pyoR14 | PA14_08140 | Baseplate LysM domain | |
| pyoR15 | PA14_08150 | Baseplate hub 2c | |
| pyoRF6 | PA14_08160 | Peptidoglycan hydrolase | |
| pyoRF7 | PA14_08180 | Spannin (Rz)b | |
| pyoRF8 | PA14_08190 | Spannin (Rz1)b | |
| pyoF1 | PA14_08200 | Unknown | |
| pyoF2 | PA14_08210 | 54 | Tail tube |
| pyoF3 | PA14_08220 | ND | Tape measure chaperone |
| pyoF4 | PA14_08230 | 36 | Tape measure chaperoned |
| pyoF5 | PA14_08240 | 35 | Tape measure |
| pyoF6 | PA14_08250 | 37 | Tail tip protein |
| pyoF7 | PA14_08260 | 46 | Tail tip protein |
| pyoF8 | PA14_08270 | 44 | Tail tip protein |
| pyoF9 | PA14_08280 | 49 | Tail tip protein |
| pyoF10 | PA14_08300 | 44 | Central fiber |
| pyoF11 e | NAf | 24 | Module 1 specificity determinant |
| pyoF12 e | NA | ND | Module 1 specificity determinant |
| pyoF13 | PA14_08310 | 20 | Module 2 specificity determinant |
| pyoF14 | PA14_08320 | Module 2 specificity determinant | |
| pyoF15 | PA14_08330 | Module 2 specificity determinant |
Values indicate the pairwise percent amino acid identity and are shown for tail morphogenesis proteins that are present in both E. coli phage HK022 and the F-type pyocin cluster. The genes encoding these proteins are arranged syntenically in the two clusters.
Functions were predicted using HHpred.
These myophage baseplate functions are defined by Buttner et al. (51).
The second tape measure chaperone encoding segment is appended to the first through a programmed translational frameshift (52). The annotation of this ORF is missed in many P. aeruginosa genomes.
These proteins, though clearly encoded in the PA14 genome, were not annotated.
NA, not applicable; ND, likely homologous proteins were present, but the pairwise sequence identity was below 20%.
Within the 23 R-type pyocin clusters in the genomes studied here, 13 of the 15 encoded proteins are highly conserved among the clusters, with at least 97% sequence identity between each gene product, as has been previously documented (17). The two proteins that vary significantly are PyoR6 and PyoR7, which encode the tail fiber and tail fiber chaperone, respectively. The tail fiber determines the specificity of R-type pyocins, and the chaperone is specific to its cognate fiber. We compared the fiber and chaperone proteins of each of our sequenced clusters to those of the characterized R-type pyocin types (17). Fiber sequences of the R2, R3, and R4 types are very similar to each other (>98% identical). Hence, we considered groups R2, R3, and R4 as one group and called it group R2, as was done in a previous study (28). For the R-type pyocin clusters sequenced here, 10 belonged to the R1 group, 10 belonged to the R2 group, and 2 belonged to the R5 group. Since R-type pyocins have been well characterized in previous studies (9, 10, 17), we focused the present investigation on the F-type pyocins.
Conserved proteins encoded in the F-type pyocin cluster.
The pyoF2 to pyoF10 genes encode confidently annotated functions required for formation of the F-type pyocin tube and tip (Table 1). The protein products of each of these genes are clearly homologous to phage tail proteins (29), and these proteins are very similar (~95% pairwise sequence identity) among the 21 F-type pyocin gene clusters that we have analyzed. Although the F-type pyocin genes are arranged in an order that is syntenic with the genome of the well-characterized E. coli phage lambda (14), only the tail tip and central fiber proteins (PyoF6 to PyoF10) of this phage share significant sequence identity with F-type pyocin proteins (31 to 38% sequence identity). The phage tail region with the greatest similarity to the F-type pyocin cluster across the tube and tail tip region is that from E. coli phage HK022. (Table 1). The HK022 proteins share 43% sequence identity, on average, to those of the F-type pyocin (Table 1). No prophage tail region was more closely related to the F-type pyocin cluster than phage HK022 across the whole cluster, though some P. aeruginosa prophages were more closely related to the 3′ end of the cluster, where genes encoding the tail tip proteins are located.
An unusual feature of F-type pyocin regions compared to phage tails is the lack of any protein with detectable similarity to a tail terminator. This protein is essential for phage tails because it is required to join the tail to the head (30). The tail terminator also prevents uncontrolled polymerization of the tails of some (30), but not all phages (31). Since F-type pyocins are not joined to a head, the tail terminator appears to be dispensable. The pyoF1 gene lies in the genomic position expected for a tail terminator gene. However, the 95-amino-acid protein encoded by this gene bears no detectable sequence similarity to tail terminators, has no homologs outside P. aeruginosa F-type pyocin clusters, and stop codons are observed in this open reading frame (ORF) in several strains. Thus, we conclude that this protein is likely not required for F-pyocin function, as was also concluded in a previous publication (12).
F-type pyocins can be grouped based on proteins encoded at the 3′ end of the cluster.
The host range specificity of phages is determined by proteins located at the tail tip, which are typically encoded by genes at the 3′ end of tail-encoding regions (29). The homologous proteins in the F-type pyocin are encoded by genes pyoF10 to pyoF15. Noncontractile phage tails resembling F-type pyocins possess a long (>700 residues) central fiber protein that projects directly below the tail tip. In phage lambda, this central fiber is encoded by the J gene, and a region within the last 250 residues of the gpJ protein mediates host cell specificity and surface binding (32, 33). The homologous protein in F-type pyocins is encoded by pyoF10. We observed that the first 1,160 residues of the PyoF10 proteins are highly conserved among F-type pyocins (>93% sequence identity), but the last 60 residues vary greatly, with pairwise identities in this region often ranging below 35% (see Fig. S1). This sequence variability is consistent with a role for the C terminus of PyoF10 in mediating host specificity.
In addition to the last 60 residues of PyoF10, the other five proteins encoded at the 3′ end of the F-type pyocin cluster, PyoF11 to PyoF15, were found to vary considerably in sequence between different F-type pyocin clusters. Based on pairwise comparison of homologous proteins encoded in this region of the clusters (see Fig. S1), the F-type pyocin regions found in different genomes were divided into six groups—F1, F2, F4, F5, F6, and F7 (Fig. 2b). Regions were placed into the same group if each of their corresponding homologous proteins shared at least 90% sequence identity with all others in the group. The nomenclature used here extends from previous work where groups F1 to F3 were established based on differences in host killing specificity (19). We do not know whether any of the groups identified here match group F3 because no examples from this group have been sequenced. The groups that we called F4 and F6 have not been previously recognized, while group F5 and F7 were previously described in P. aeruginosa strains PA14 and DK2, respectively (18). The two most frequently occurring groups are F2 (11 members) and F7 (4 members). The F1, F5, and F6 groups were encoded only in pyocin clusters that also encoded R-type pyocins, while F4, F7, and F2 group clusters were found in the absence of R-type clusters except in two instances (both F2 group). Further bioinformatic comparisons described below compare representative protein sequences from each of the six F-type pyocin groups that we identified here.
PyoF11 and PyoF12 are newly recognized conserved proteins.
PyoF11 and PyoF12 are proteins of unknown function that are encoded in every F-type pyocin region. These are the most diverse proteins in the F-type pyocin clusters, often displaying pairwise sequence identities of <25% (see Fig. S1). Despite their diversity, the homologs of these proteins from the six groups could be convincingly aligned (see Fig. S2a and b). We used HMMer (34) to create Hidden Markov Model (HMM) profiles from the PyoF11 and PyoF12 alignments. Searching with these HMMs, we identified more than 50 occurrences each of pyoF11 and pyoF12 gene homologs in diverse phages and prophages. These genes often occur together and invariably lie immediately 3′ to the central fiber gene (homolog of pyoF10). In some phage genomes, the pyoF11 and pyoF12 genes are very likely the last genes in the tail operon as they are followed immediately by lysis genes (e.g., Burkholderia phage Bcep176 and Xanthomonas phage CP1). These observations suggest that PyoF11 and PyoF12 function in conjunction with the central fiber protein, possibly binding to it or acting as chaperones to aid in folding of the fiber. PyoF11 and PyoF12 had not been previously recognized as conserved proteins in the F-type pyocin cluster because these ORFs are not annotated as proteins in most P. aeruginosa genomes. This is likely a result of the lack of annotation of these genes in the PA14 genome, which is commonly used as the reference genome for genome assembly and annotation. The functions of PyoF11 and PyoF12 homologs in phages have never been investigated.
PyoF13, PyoF14, and PyoF15 are likely involved in host specificity.
In addition to the central fiber, most noncontractile tailed phages possess genes downstream of the central fiber gene that also encode cell surface receptor binding proteins These are known as “side fibers” in E. coli phage lambda (35). The PyoF13 proteins, which share a genomic position with the lambda side fibers, are likely involved in determining host range specificity, functioning as receptor binding proteins. A striking feature of the Pyo13 homologs is that their N termini (the first 140 residues) are very similar among the F-type pyocin groups with pairwise sequence identities ranging from 55 to 90%, while the pairwise identities in their C-terminal regions generally range between 20 and 35% (see Fig. S1 and S3). We surmise that the more conserved N terminus of PyoF13 mediates binding of this putative receptor binding protein to the F-pyocin tail tip, while the variable C terminus mediates cell surface binding. The fibers from different groups of R-type pyocins, which have been shown to determine bactericidal specificity (17), display the same type of conservation pattern with N-terminal regions (first 450 residues) displaying >95% pairwise sequence identity and C-terminal regions (last 250 residues) displaying pairwise sequence identities between 50 and 70% (see Fig. S4). In contrast to the F-type pyocins, there are only three distinct groups of R-type pyocins, as defined by fiber sequences, and there is much less variability.
Homologs of PyoF13, which share sequence similarity with its N-terminal region, are found in diverse phages and prophages and are located in similar genomic positions as pyoF13, downstream from the central fiber gene. Genes encoding homologs of PyoF14 (~100 residues) and PyoF15 (~75 residues) are also found in many phages and prophages, and they are invariably located downstream of pyoF13 homologs or genes encoding other putative phage receptor-binding proteins. The sequences of PyoF14 and PyoF15 are variable, mirroring the sequence variation seen in the C-terminal regions of PyoF13 (see Fig. S1). We expect that PyoF14 and PyoF15 are involved in host range specificity through interactions with PyoF13 or possibly by acting as chaperones for the assembly of PyoF13, as is required for phage-encoded receptor-binding proteins (36).
Two F-type pyocin groups deviate from the others in the pyoF13 to pyoF15 region. The F2 group has a complete duplication of this region so that it possesses two copies of each gene. The proteins encoded by the first copy, PyoF131 to PyoF151, are distinct in sequence compared to the homologs in other groups (see Fig. S1; see also Fig. S3). Conversely, PyoF132 to PyoF152 are very similar (>90% identical) to homologs found in groups F4 and F5. In contrast to all other groups, group F7 lacks a pyoF14 gene. Consistent with this absence, its PyoF13 homolog displays a C-terminal region that has no detectable sequence similarity to the other groups.
Characterization of F-type pyocin bactericidal specificity.
To determine whether our bioinformatic groupings of the F-type pyocin clusters correlate with bactericidal specificity, we examined the killing profiles of lysates produced from the 30 strains following induction by mitomycin C. Serial dilutions of lysates of each of the 30 strains were spotted onto lawns of the same 30 strains to produce an all-against-all matrix. Bactericidal activities were detected as zones of clearing on the bacterial lawn. Analysis of these data were complicated because P. aeruginosa produces other bactericidal entities in addition to R-type and F-type pyocins, including S-type pyocins (13) and bacteriophages. Since the presence of any of these can produce zones of clearing, further analyses were necessary to delineate the type of activity present. Testing serial dilutions of lysates allowed us to distinguish clearings produced by phages from those produced by pyocins (Fig. 3a). Due to their replicative nature, clearings resulting from phage lysates resolved into individual plaques upon dilution, while the clearings resulting from pyocins gradually disappeared without the appearance of individual plaques (Fig. 3a). Lysates were also spotted onto bacterial lawns containing proteinase K, which had been previously shown to eliminate clearings caused by protease-sensitive S-type pyocins without affecting the activity of R- or F-type pyocins (Fig. 3a) (13). The effect of proteinase K in inactivating S-type pyocins is clearly shown in the assay performed with a lysate made from a double mutant PA14 strain bearing mutations inactivating both R-type and F-type pyocins (Fig. 3a, bottom row). It can also be seen that PA14 single mutants expressing only either R- or F-type pyocins display bactericidal activity that is insensitive to proteinase K treatment (Fig. 3a).
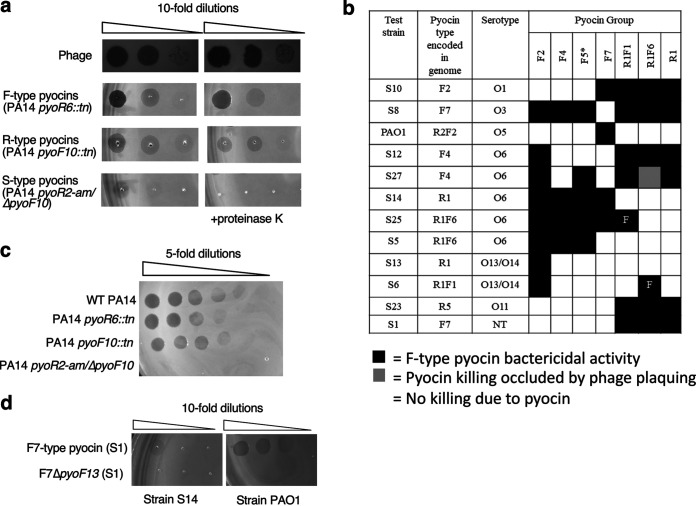
FIG 3.
Bactericidal activity of F-type pyocins. (a) Lysates containing phages, only R-type pyocins, only F-type pyocins, or only S-type pyocins were spotted onto a susceptible strain. Where indicated, proteinase K was also added to the plates. (b) The bactericidal activity of lysates containing F-type or R-type pyocins on a selected group of bacterial lawns is shown. These lawns were selected to emphasize the differences in specificity among the different groups. Black boxes denote strains killed by a given lysate while white boxes denote no killing. The gray box indicates a case where the killing by pyocins was occluded by phage plaquing. NT, nontypeable LPS serotype. Bactericidal activity of the F5 group was determined using a mutant strain of PA14 bearing a transposon insertion in the pyoR6 gene, so that the lysate contained only F-type pyocin particles (indicated by an asterisk). The F1- and F6-type pyocins were produced in strains that also produced R1-type pyocins. By comparing with a strain producing only R1-type pyocins, two strains killed only by these F-type pyocins could be identified (marked with “F”). (c) Lysates were produced from wild-type PA14 and strains bearing transposon insertions in either the pyoR6 or pyoF10 genes were spotted onto a lawn of strain S19. A lysate of a PA14 double mutant bearing an amber mutation in the pyoR2 gene, and an in-frame deletion mutation in the pyoF10 gene was also tested. (d) A lysate of F7-type pyocins produced from strain S1 was spotted on lawns of strains S14 or PAO1. The experiments depicted in panels c and d were performed with plates containing proteinase K so that S-type pyocin activity was eliminated.
By analyzing the activities of the 30 lysates on 30 strains in this manner described above, we detected more than 450 bactericidal combinations and found that greater than 90% were due to R- or F-type pyocins (see Fig. S5). All groups of R-type and F-type pyocins identified displayed bactericidal activities against multiple strains. Notably, the killing spectra of lysates were invariably the same if they contained pyocins of the same R- or F-type group (see Fig. S5). For example, lysates of four different strains encoding F7 pyocins all displayed bactericidal activity against the same 11 strains (note that in a single case the F-type pyocin activity was occluded by the presence of phage activity, as denoted by an orange color [see Fig. S5]). These results demonstrate that our classification of pyocins based on sequence analysis is predictive of biological activity. In Fig. 3b, a small subset of the bactericidal data is shown to emphasize the differences in the killing spectra of the F-type pyocin groups. No two groups kill exactly the same set of bacterial strains; however, considerable overlap exists between some groups like F4 and F5. We also noted that no strain was susceptible to an R- or F-type pyocin that was encoded in its own genome, which is consistent with previous observations that strains are resistant to their own pyocins (37). Since the F1 and F6 groups were encoded only in strains that also encoded R1 pyocins, the killing caused only by the F-type pyocins could not be discerned. However, comparison with results obtained using a strain encoding only an R1 pyocin suggested that strain S25 is susceptible to F1 pyocin since it was killed by a lysate containing F1 and R1 pyocins, but not by a lysate containing only R1 pyocin (Fig. 3b). By the same logic, strain S30 was found to be killed by F6 pyocin. The F5 group was found only in strains that also encode an R-type pyocin. To assess the activity of this group, we took advantage of a transposon insertion mutant of an essential R-type pyocin gene in PA14 (38) to detect the activity of the F5 pyocin alone (Fig. 3b).
F-type and R-type pyocin containing lysates display similar levels of bactericidal activity.
It was previously reported that one R-type pyocin particle is sufficient to kill a single cell, while up to 280 F-type pyocin particles are required to kill the same cell (21). This implies that an F-type pyocin containing lysate would have considerably less killing activity than an R-pyocin containing lysate. However, we observed many cases where lysates of F-type pyocins displayed levels of killing activity as high R-type pyocin lysates. Although R- and F-type pyocin lysates may contain different numbers of particles, we do not expect these numbers to deviate greatly since all pyocin operons utilize the same transcriptional regulatory region. To address this issue in a direct manner, we tested the bactericidal activity of lysates of two PA14 mutants, one of which carried a transposon insertion in an essential R-type pyocin gene (pyoR6) and one of which carried a similar insertion in an essential F-type pyocin gene (pyoF10). It can be seen that the bactericidal activity of these two lysates was the same (Fig. 3c). If we assume that the expression levels of R- and F-type pyocins in PA14 are similar, which is reasonable since their transcription is driven from the same promoter, then these results suggest that these two types of pyocins have similar levels of killing activity. We also observed that the same F-type pyocin lysate may display different levels of activity on different strains. For example, lysates of F7 group pyocins displayed >10-fold greater bactericidal activity on strain PAO1 as on strain S14 (Fig. 3d). The previously observed low activity of F-type pyocins may have resulted from the use of a non-optimal indicator strain. Overall, our data indicate that F-type pyocins have the potential to kill bacterial cells as efficiently as R-type pyocins. However, further quantitative testing of different R- and F-type pyocin groups on various strains will be required to definitively determine whether one type has greater bactericidal activity than the other.
The genes downstream of pyoF10 are required for bactericidal activity.
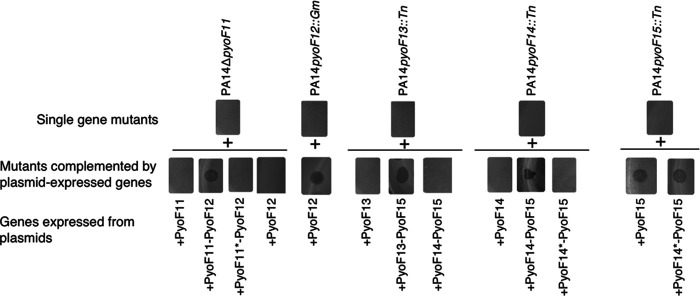
Although homologs of the proteins encoded at the 3′ end of the F-type pyocin cluster (PyoF11 to PyoF15) are encoded in phages and prophages, the roles of these proteins have never been investigated. To determine whether these proteins are essential for bactericidal activity, we tested the activity of F-type pyocin mutants in strain PA14 (group F5). We tested transposon insertion mutations in pyoF14, and pyoF15 from the PA14 nonredundant transposon mutant library (38). We constructed in-frame deletion mutations in pyoF11 and pyoF12 and a nonsense mutation in pyoF13. Mutations in each of these genes completely abrogated bactericidal activity, indicating that their protein products play essential roles in the production of functional F-type pyocin particles (Fig. 4). To ensure that the loss of activity resulting from these mutations was the result of abrogation of only the gene in which the mutation was located, each gene was cloned into a plasmid expression vector, and we determined whether mutations could be complemented by the plasmid-expressed genes. The pyoF12 mutant could be complemented by a plasmid expressing only pyoF12 (Fig. 4). However, complementation of the pyoF11 mutant required plasmid-based expression of both pyoF11 and pyoF12. A plasmid expressing only pyoF12 did not complement the pyoF11 mutant. We conclude that both pyoF11 and pyoF12 are essential for bactericidal activity and that the pyoF11 in-frame deletion mutation also causes loss of pyoF12 activity, possibly through a polarity effect. Through a similar series of plasmid-based complementation experiments, we determined that pyoF13, pyoF14, and pyoF15 are also essential for bactericidal activity and that polarity effects are also manifested in this group of genes (Fig. 4). For example, while the pyoF15 mutation could be complemented by expression of pyoF15 alone, complementation of the pyoF14 mutation required the expression of both pyoF14 and pyoF15.
FIG 4.
Plasmid-based complementation of pyoF11 to pyoF15 mutants. In the top row of experiments, lysates from PA14 strains with either deletions or insertions in the indicated genes were spotted onto lawns of P. aeruginosa strain S19. In the lower row of experiments, mutant strains were transformed with plasmids expressing the indicated proteins. Successful complementation by the plasmids is indicated by the appearance of a zone of clearing. Asterisks denote that the gene encoding the indicated protein contains a nonsense mutation.
Serotype correlates with F-type pyocin killing spectra.
The outer membrane lipopolysaccharide (LPS) of P. aeruginosa is composed of three domains: lipid A, core oligosaccharide and a long-chain polysaccharide O-antigen (39). Most P. aeruginosa strains produce two distinct forms of O-antigen; a homopolymer of d-rhamnose known as the common polysaccharide antigen, and a heteropolymer repeat of three to five distinct sugars known as the O-specific antigen (OSA), which forms the basis of P. aeruginosa serotyping. Previous studies showed that the OSA acts a receptor for some R-type pyocins, while it blocks killing by other R-type pyocins (28). To investigate the effect of the OSA on the activity of F-type pyocins, we experimentally determined the serotypes of the 30 strains used in this study by a slide agglutination assay. We observed a correlation between the serotype of a strain and its F-type pyocin susceptibility profile (Fig. 5a). For example, all three strains of O2 serotype were resistant to all F-type pyocins, while the four O5 strains were killed only by F7 pyocins. Among the eight O6 serotype strains, the F2 pyocin killed all, but the F4, F5, and F7 pyocins were unable to kill some of these strains (Fig. 5a). The resistance of O6 strains S12 and S27 to the activity of the F4 pyocin is expected as these strains encode an F4 pyocin. However, it is not clear why the F7 pyocin fails to kill O6 strains S12, S27, and S5 or why strain S12, alone among O6 strains, is resistant to F5 pyocin. Similarly, the F1 pyocin kills strain S25 but no other O6 strains, and the F6 pyocin kills strain S6 but no other O13/O14 strains. These data show that factors independent of OSA and pyocin type encoded within a strain contribute to F-type pyocin susceptibility.
FIG 5.
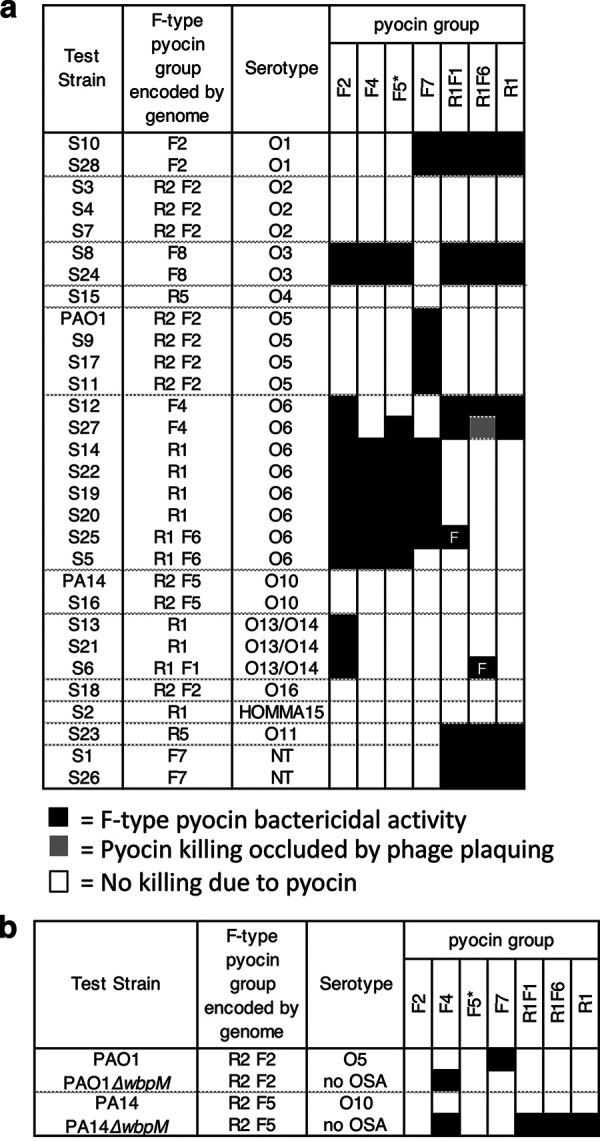
(a) Effect of LPS serotype on bactericidal activity of F-type pyocins. The bactericidal activity of F-type pyocins on a group of bacterial lawns arranged by their serotypes is shown. Black boxes denote strains killed by a given F-type pyocin, while white boxes denote no killing. The gray box indicates a case where the killing by pyocins was occluded by phage plaquing. Bactericidal activity of the F5 group was determined using a mutant strain of PA14 bearing a transposon insertion in the pyoR3 gene, so that the lysate contained only F-type pyocin particles (indicated by an asterisk). The F1- and F6-type pyocins were produced in strains that also produced R1-pyocins. By comparing with a strain producing only R1-type pyocins, two strains killed only by these F-type pyocins could be identified (marked with “F”). NT, nontypeable serotype. (b) The indicated F-type pyocin containing lysates were tested against mutants that lack OSA (ΔwbpM).
To directly assess the role of OSA in F-type pyocin activity, we tested ΔwbpM mutant strains, which lack OSA in strains PAO1 and PA14 (Fig. 5b). The F7-type pyocin is active against PAO1 but was unable to kill the PAO1ΔwbpM mutant, suggesting that this pyocin uses the OSA as a receptor. In contrast, the ΔwbpM mutants of PAO1 and PA14 became susceptible to the F4 group, though the wild-type strains were not. In this case, the OSA appears to block the pyocin from contacting its receptor. The F2 and F5 groups, which are unable to kill PAO1 or PA14, were also not able to kill the mutants lacking OSA. Strains producing the F1 and F6 group pyocins were unable to kill PAO1 with or without OSA, but PA14ΔwbpM did become susceptible to killing. However, this effect may have been due to the R1 pyocins produced by these strains. From these experiments with strains lacking OSA, it is clear that the presence of OSA affects the bactericidal activity of the F4 and F7 groups, while the data are inconclusive for the other groups.
Discovery of new groups of F-type pyocins.
To determine whether this collection of F-type pyocin described above encompassed the full diversity of F-type pyocins found across the P. aeruginosa species, we performed BLAST searches against all P. aeruginosa genomes in the NCBI database using a PyoF13 sequence as the query with the goal of identifying homologs with distinct sequences (i.e., share less than 90% sequence identity with those in our established F-type pyocin groups). PyoF13 was chosen for these searches because it is highly conserved among the F-type pyocin operons in its N-terminal region, yet its C-terminal region varies depending on the pyocin group. We discovered three PyoF13 homolog families encoded in F-type pyocin operons that shared less than 70% sequence identity to any other PyoF13 group. F-type pyocins encoding these newly identified PyoF13 varieties were defined as groups F8, F9, and F10. The F9 group is identical to a previously identified group designated the PA7 group (18). Another identified group, called F11, possessed PyoF13, PyoF14, and PyoF15 homologs that are greater than 95% identical to group F22, F4 and F5, but the PyoF10, PyoF11, and PyoF12 were unique. Finally, group F12 combined PyoF10 to PyoF12 homologs that were 99% identical to group F11 with PyoF13 to PyoF15 homologs that were greater than 95% identical to group F10 (Fig. 6, S1). Group F12 was previously identified in P. aeruginosa strain M18 (18). The sequences of proteins PyoF10 to PyoF15 for all 11 F-type pyocin groups can be found in the supplemental material.
FIG 6.
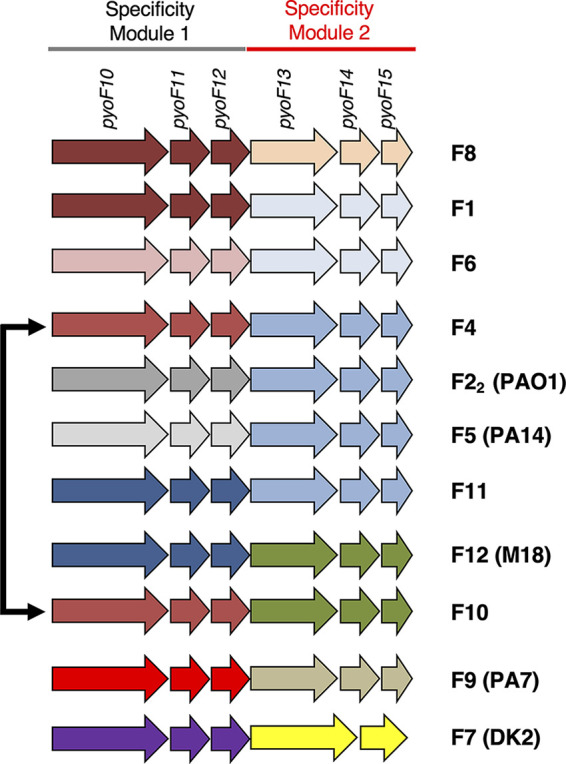
All F-type pyocin groups. A close-up of genes encoded at the 3′ end of F-type pyocin clusters shows the 11 different groups identified in our sequenced strains and in the database. Groups of genes are colored the same if the proteins they encode display >90% sequence identity. The extent of specificity modules 1 and 2 is shown at the top, as are the names of the genes in these regions. P. aeruginosa strains where certain groups were previously identified are shown in parentheses. The black arrow indicates that F4 and F10 share identical module 1 proteins.
Pairwise sequence comparisons among all the F-type pyocin groups that we have identified strongly supports the existence of two distinct specificity modules in F-type pyocins (see Fig. S1). Whenever the C-terminal regions of PyoF10 proteins in two groups are highly similar (>90% identity), then the PyoF11 and PyoF12 proteins are also highly similar. Similarly, when two groups have PyoF13 proteins with highly similar C-terminal regions, then the PyoF14 and PyoF15 proteins are also highly similar. Therefore, we have designated regions encoding the C terminus of PyoF10, PyoF11, and PyoF12 as specificity module 1 and regions encoding PyoF13, PyoF14, and PyoF15 as specificity module 2. Among our full set of pyocin groups, we observed three instances where the same specificity module 1 region assorted with different specificity module 2 regions (Fig. 6). We also observed three cases where identical specificity module 2 regions assorted with different specificity module 1 regions. These data indicate that recombination events have occurred between different F-type pyocin operons.
DISCUSSION
This study provides a comprehensive analysis of F-type pyocin operons present in P. aeruginosa strains. We have defined the conserved genes in these operons and introduced a systematic naming system for them. The initial 21 F-type clusters examined in strains from our collection were categorized into six different groups, two of which were previously known (F1 and F2) and four of which were named in this study (groups F4 to F7). The killing specificity of each of these groups was shown to be distinct. An additional five F-type pyocin groups were discovered bioinformatically, but the killing specificity of these groups remains to be tested. Importantly, we identified two highly diverse F-type pyocin genes, pyoF11 and pyoF12, which are not annotated as genes in many P. aeruginosa strains and yet are essential for bactericidal activity. The sequence diversity in these two genes contributes to defining the F-type pyocin groups. Another important finding is that there are no genes in the F-type pyocin operons that are not also found in the genomes of phages or prophages. Thus, the ability of these pyocins to efficiently kill bacteria while isolated phage tails do not must be due to sequence modifications within their phage-derived proteins and not to the presence of unique toxin-encoding genes (unless such genes are encoded elsewhere in the P. aeruginosa genome). Fully active R-type pyocins have been produced heterologously in E. coli from a plasmid vector, including only genes from the R-type pyocin operon, indicating that their toxicity does not rely on genes outside this region (11).
Analysis of the F-type pyocin operons clearly indicates the genes that are involved in killing specificity. The proteins encoded by the pyoF2 to pyoF9 genes are highly similar in all the F-type pyocin groups. Divergence among the groups begins with the last 60 residues of PyoF10 and extends through PyoF15. We defined the F-type pyocin groups according to sequence identity among these proteins (Fig. 6). Since F-type pyocins within the same group invariably displayed the same killing spectra (see Fig. S5), we conclude that some or all of the pyoF10 to pyoF15 region determines killing specificity. By examining the patterns of recombination among the specificity genes, we defined two specificity modules: module 1 (pyoF10 to pyoF12) and module 2 (pyoF13 to pyoF15). The occurrence of highly similar module 1 regions with distinct module 2 regions and vice versa in different F-type pyocin groups indicates that the two modules act independently of one another (Fig. 6). This conclusion is supported by the appearance of pyoF11 and pyoF12 homologs without adjacent pyoF13 to pyoF15 homologs and vice versa in phages and prophages. Since these families of proteins have not been characterized, defining their roles in host specificity will be an important goal for further study.
The strong sequence similarity between most of the F-type pyocin genes implies that all the groups are descended from a common ancestor that likely arose from a defective prophage. However, it is also clear that some type of horizontal gene transfer mechanism has been responsible for the evolution of the specificity regions, which are comprised of different combinations of specificity modules 1 and 2. These reassortments could be caused by phages carrying genes that are similar to these F-type pyocin genes occasionally recombining with the homologous F-type pyocin genes. With respect to the evolution of the F- and R-type pyocins as a whole, it is relevant that the F-type display considerably more divergence among their specificity-determining genes compared to the R-type (see Fig. S1 and S4). This suggests that the F-type pyocins may have arisen first and thus have had more time to diverge. Also supporting the possibility that the F-type pyocin operon appeared first is that the R-type pyocin genes are inserted in the middle of the lysis genes, which comprise an intact lysis cassette in strains possessing only an F-type operon.
Our examination of the activity of the different F-type pyocin groups on 12 different serotypes of P. aeruginosa revealed a clear correlation between bactericidal activity and O-antigen serotypes (Fig. 5a). Further supporting a role for the OSA in the activity of at least some F-type pyocins is the fact that activity of the F7 group required the presence of OSA, while group F4 activity was blocked by OSA (Fig. 5b). While the OSA serotype clearly influences F-type pyocin host recognition, this is not the only determining factor. For example, the F7 pyocin is able to kill some but not all strains with the O6 serotype. In addition, F-type pyocins of a given type were consistently unable to kill strains encoding the same type of F-type pyocin, regardless of serotype. The mechanism of this self-immunity is not known. Many bacteriocins, such as S-type pyocins and colicins, are encoded with specific immunity proteins (13). However, there is no obvious immunity protein candidate encoded within F-type pyocin clusters as each gene is homologous to phage tail proteins. It is possible that no specific immunity proteins exist for R-type or F-type pyocins. Rather, strains may have evolved to resist their resident R- and F-type pyocins by altering their cell surface in subtle ways undetectable by the antibodies used in serotyping.
Overall, our study shows that F-type pyocins are produced by a large number of P. aeruginosa strains, they all possess antimicrobial properties, and they are promising candidates to study for the development of new therapeutics. Our identification of the specificity determinants of F-type pyocins points the way toward precisely engineering their killing as has been done with the contractile R-type pyocins and noncontractile tailocins of Listeria (9, 10, 12).
MATERIALS AND METHODS
Whole-genome sequencing.
Genomic DNA was isolated using a genomic DNA extraction kit (Bio Basic Inc.). Next-generation whole-genome sequencing was performed by the Donnelly Sequencing Center, University of Toronto, using Illumina HiSeq2500. De novo assembly of reads into contigs was performed using Velvet version 2.2.5 using default settings with optimized k-mer lengths of 29 to 31 bp (40). Genes trpE and trpG were located, and the region between these genes was analyzed by using Geneious (41). BLAST searches with conserved F- and R-type pyocin proteins showed that these entities were not encoded in other genomic regions. The sequences of the R- and F-type pyocin regions discussed here have been uploaded to GenBank under accession numbers OQ870446 to OQ870473.
Bioinformatic analysis.
Most of the bioinformatic analyses, including BLAST (42) searches and genome synteny analyses, were carried out on a custom database comprised of 755 tailed phage genomes and 2,119 bacterial genomes downloaded from the National Center for Biotechnology Information (NCBI) RefSeq database in April 2013. This database contains diverse phage and bacterial species but was small enough to allow manual analysis of the protein sequences and the genomic context of genes encoding proteins related to pyocin proteins. This study was aided by a synteny viewing and phage gene annotation toolkit developed in our laboratory, which will be described in detail elsewhere. Sequence alignment analysis was performed in Jalview (43). To identify protein sequences similar to less frequently occurring proteins found in the pyocin cluster (e.g., PyoR1, PyoF11, and PyoF12), alignments were constructed of the pyocin proteins. HMMER3 (34) was then used to create Hidden Markov Models (HMMs). These HMMs were used to detect proteins similar to a given pyocin protein. The genome context of genes encoding these similar proteins within phage genomes was assessed to support a conclusion that the pyocin protein possesses the same function as the phage protein. BLAST searches to identify new groups of F-type pyocins were carried out against all P. aeruginosa genomes available at NCBI in April 2018.
HHpred searches were carried out using the online server (https://toolkit.tuebingen.mpg.de/hhpred) (44). HMM-based searches were carried out using HMMer (34) and analyzed by searching the Pfam (45) and TIGRfam (www.jcvi.org/cgi-bin/tigrfams/index.cgi) databases.
Transmission electron microscopy.
A continuous carbon film-coated TEM grid was made hydrophilic by glow discharge. Then, 5 μL of sample was applied to the surface of the grid and left for absorption for 2 min. Excess sample was blotted away using the corner of a filter paper. The grid was washed three times with water and stained with 2% (wt/vol) uranyl acetate. Grids were examined with a Hitachi H-7000 microscope.
Assays of pyocin and phage bactericidal activity.
To generate lysates containing pyocins and/or phages, 5-mL culture samples started from overnight cultures were grown in LB at 30°C until the cells reached an optical density at 600 nm of 0.4. Mitomycin C was then added to a final concentration of 2 μg/mL, and shaking at 30°C was resumed for 3 h or until cell lysis occurred. Chloroform was added to all induced cultures (1 to 2 drops/mL) to ensure maximum bacterial lysis. In experiments testing complementation from plasmids, 0.2% arabinose was added to cells after 1 h of growth at 30°C to induce the expression of proteins from the plasmid prior to addition to mitomycin C to induce F-type pyocin induction from the genome. After lysis, cultures were incubated at room temperature with DNase (10 μg/mL) for 30 min prior to centrifugation at 10,000 rpm for 10 min. For activity assays, 2-μL volumes of dilutions of these lysates were spotted onto lawns of P. aeruginosa strains. Lawns of strains to be tested were made by adding 150 μL of overnight culture to 3 mL of molten 0.7% top agar, which was immediately poured onto an LB agar plate and allowed to harden. To distinguish S-type pyocin activity, duplicate lawns were poured containing proteinase K (100 μg/mL). At least three biological replicates were performed for each strain and lysate combination. A lysate was scored as positive if it displayed a strong zone of cell growth inhibition in every assay. There was a range of activity in positive lysates with some displaying moderate zones of growth inhibition even when diluted 102-fold and others displaying strong activity only when undiluted. Lysates scored as negative displayed a barely discernible or no zone of growth inhibition in all replicate assays.
Serotyping of P. aeruginosa strains.
Strains were serotyped using the slide agglutination method using commercial antisera (MAST Diagnostics) against all 20 P. aeruginosa serotypes recognized by the International Antigenic Typing Scheme (39).
Construction of mutations in pyoF11 and pyoF12.
The sequences of all primers used for mutagenesis and plasmid construction can be found in Table S2.
To construct a mutation in the PA14 pyoF11 gene, the gene with upstream (500 bp) and downstream (396 bp) flanking sequences was PCR amplified from a single PA14 colony using primers PA14pyo11rec-F and PA14pyoF11rec-R. Flanking sequences were included to promote homologous recombination into the pyoF11 locus. The PCR product generated was cloned into the NcoI and HindIII sites of pHERD30T (46) and ligated with T4 DNA ligase to create plasmid 30T-PA14pyoF11-rec. To make a construct with an in-frame deletion in pyoF11, primers PA14pyoF11-delF and PA14pyoF11-delR were used to PCR amplify all of 30T-PA14pyoF11-rec except for a 201-bp region in the middle of the pyoF11 gene. Primer PA14pyoF11-delR amplified the first 72 pyoF11 nucleotides and its upstream flanking sequence in one direction, while primer PA14pyoF11-delF amplified the last 84 nucleotides of pyoF11 and the downstream sequence in opposite direction, to produce a linearized PCR product which was then religated to create 30T-PA14pyoF11-del. PA14 transformed with plasmid 30T-PA14pyoF11-del was grown serially for 2 to 3 days to allow for recombination between the plasmid and genomic locus. Subsequently, to select for cells bearing the pyoF11 deletion, cells were transformed with a derivative of pHERD20T (46) expressing a CRISPR RNA molecule (crRNA, primers PA14CRpyoF11-del-F and PA14CRpyoF11-del-R) targeting the deleted region of pyoF11. In this way, the native CRISPR-Cas system of PA14 mediates genome cleavage and cell death to strains bearing the wild-type pyoF11 gene. A similar approach to constructing genomic mutations in P. aeruginosa has been previously described (47). To cure mutants of plasmids, strains were streaked onto LB plates without any antibiotics. This was repeated 2 times, then colonies displaying no antibiotic resistance were identified. Deletion mutations were confirmed by PCR.
To construct the pyoF12 mutant, primer pairs SS633 and SS635, and SS636 and SS634 were used to amplify 600 bp upstream and downstream of gene PA14pyoF12, respectively. Primer pair SS545 and SS546 was used to amplify the gentamicin resistance cassette from plasmid pUCP20gm (48). Vector p15TV-L (Addgene catalogue number 26093) was digested with BseRI. The three amplified fragments and the linearized vector were assembled into a construct using Gibson assembly (49). The constructed plasmid was used as a template to amplify a DNA fragment containing the gentamicin-resistant cassette flanked by upstream and downstream regions of pyoF12 (600up-pyoF12::Gm-600down) using the primers SS639 and SS640. This fragment was used to replace gene pyoF12 with the gentamicin resistance cassette using the lambda Red recombinase system (50).
To construct the pyoF13 mutant, the PA14 pyoF13 gene with upstream (400 bp) and downstream (350 bp) flanking sequences was PCR amplified from a single colony of PA14 strain using the primers PA14pyoF13-rec-F and PA14pyoF13-rec-R. Flanking sequences were included to promote homologous recombination into the pyoF13 locus. The PCR product generated was cloned into the EcoRI and HindIII sites of pHERD20T and to create plasmid 20T-PA14pyoF13-rec. Using site-directed mutagenesis, the valine encoding codon (ninth amino acid) of the pyoF13 gene was replaced with a TAG stop codon (using primers PA14pyoF13sdm-F and PA14pyoF13sdm-R), which was recombined into PA14 chromosome. To select for cells that recombined correctly, crRNA targeting wild-type pyoF13 expressed from pHERD30T (constructed using the primers PA14CRpyoF13-sdm-F and PA14CRpyoF13-sdm-R) was used to select for mutants, as described above.
Construction of plasmids expressing pyoF11 to pyoF15 genes.
Plasmids to express F-type pyocin proteins encoded at the 3′ end of the operon were constructed as described below.
To construct a plasmid expressing PyoF11, primer pairs PA14pyoF11-For and PA14pyoF11-Rev were used to amplify the PA14 pyoF11 gene. The resulting PCR product was digested at NcoI and HindIII restriction sites and ligated into the pHERD20T plasmid. All constructs here and below were verified by sequencing.
To construct a plasmid expressing PyoF11 and PyoF12, primers 20T-gib-For and 20T-gib-Rev were used to produce a linearized pHERD-20T through PCR to achieve an in-frame deletion of the multiple cloning sites in the plasmid. Primers PA14pyoF11gib-For and PA14pyoF12gib-Rev amplified pyoF11 to the end of pyoF12 gene. These primers were designed to have at least 25-bp overlapping sequence with the linearized pHERD20T plasmid DNA. Both plasmid and genomic PCR products were joined by Gibson assembly.
To construct a plasmid expressing PyoF13, primer pairs PA14pyoF13-For and PA14pyoF13-Rev, were used to amplify the PA14 pyoF13 gene. The resulting DNA fragment was digested with EcoRI and HindIII and ligated into the same restriction sites of the pHERD20T plasmid.
Similarly, to construct a plasmid expressing PyoF13 to PyoF15, PA14 pyoF13 to pyoF15 genes were amplified using primers PA14pyoF13-For and PA14pyoF15-Rev. The resulting fragment was digested with EcoRI and HindIII and ligated into the same restriction sites of the pHERD20T plasmid. To construct a plasmid expressing Pyo14, the primers PA14-pyoF14-For and PA14-pyoF14-Rev were used to amplify the PA14 pyoF14 gene and digested with NcoI and HindIII restriction enzymes. Digested products were then ligated into the same restriction sites (NcoI, HindIII) of the pHERD20T plasmid. To construct a plasmid expressing PyoF14 and PyoF15, the primers PA14pyoF14-For and PA14pyoF15-Rev were used to amplify the PA14 pyoF14 and pyoF15 genes. The resulting PCR product was digested and ligated into pHERD20T at the NcoI and HindIII restriction sites. Finally, to construct a plasmid expressing PyoF15, the primer pair PA14pyoF15-For and PA14pyoF15-Rev was used to amplify the PA14 pyoF15 gene. The resulting fragment was digested with NcoI and HindIII and ligated into the same restriction sites of the pHERD20T plasmid.
Construction of a pyoF10 mutant in an R-type pyocin mutant background (PA14pyoR2-amΔpyoF10).
The PA14 pyoR1 and pyoR2 genes were PCR amplified from the start of pyoR1 to the end of pyoR2 using the primers PA14pyoR1rec-F and PA14pyoR2rec-R. The PCR product generated was cloned into the NcoI and HindIII sites of pHERD20T to create plasmid 20T-PA14pyoR2-rec. Using site-directed mutagenesis, the methionine encoding codon (11th amino acid) of the pyoR2 gene was replaced with a TAG stop codon using the primers PA14pyoR2sdm-F and PA14pyoR2sdm-R, which was recombined into PA14 chromosome. To select for cells that recombined correctly, crRNA targeting wild-type pyoR2 expressed from pHERD30T.
To create a pyoF10 mutant, a part of the PA14 pyoF10 gene from amino acids 111 to 710 (1,800 bp) was PCR amplified from a single colony of PA14 strain using the primers PA14pyoF10-rec-F and PA14pyoF10-rec-R. The PCR product generated was cloned into the NcoI and HindIII sites of pHERD20T to create plasmid 20T-PA14pyoF10-rec. To make a construct with an in-frame deletion in pyoF10, the primers PA14pyoF10-delF and PA14pyoF10-delR were used to PCR amplify all of 20T-PA14pyoF10-rec except for the 801-bp region in the middle of the pyoF10 gene. Primer PA14pyoF10-delR amplified the first upstream 495 bp of pyoF10 in one direction, while primer PA14pyoF10-delF amplified the last 468 bp of pyoF10 in the opposite direction to produce a linearized PCR product which was then religated to create 20T-PA14pyoF10-del. This was recombined in the chromosome of PA14pyoR2-am. crRNA targeting wild-type pyoF10 expressed from pHERD30T was used to select for mutant strains that recombined correctly (constructed from the primers PA14CRpyoF10-del-F and PA14CRpyoF10-del-R).
Data availability.
The sequences of the R- and F-type pyocin regions discussed here have been uploaded to GenBank under accession numbers OQ870446 to OQ870473.
ACKNOWLEDGMENTS
This study was supported by operating grants from the Canadian Institutes of Health Research (CIHR) to A.R.D. (XNE-86943 and FDN-15427), K.L.M. (MOP-136845), and A.W.E. (PHT-148819). A.R.D. is also supported by a Tier 1 Canada Research Chair (950-232058). S.S. was supported by a CIHR Doctoral Scholarship.
Footnotes
Supplemental material is available online only.
Contributor Information
Alan R. Davidson, Email: alan.davidson@utoronto.ca.
George O’Toole, Geisel School of Medicine at Dartmouth.
REFERENCES
- 1.Vandenheuvel D, Lavigne R, Brussow H. 2015. Bacteriophage therapy: advances in formulation strategies and human clinical trials. Annu Rev Virol 2:599–618. doi: 10.1146/annurev-virology-100114-054915. [DOI] [PubMed] [Google Scholar]
- 2.Chan BK, Turner PE, Kim S, Mojibian HR, Elefteriades JA, Narayan D. 2018. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol Med Public Health 2018:60–66. doi: 10.1093/emph/eoy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dedrick RM, Guerrero-Bustamante CA, Garlena RA, Russell DA, Ford K, Harris K, Gilmour KC, Soothill J, Jacobs-Sera D, Schooley RT, Hatfull GF, Spencer H. 2019. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med 25:730–733. doi: 10.1038/s41591-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, Barr JJ, Reed SL, Rohwer F, Benler S, Segall AM, Taplitz R, Smith DM, Kerr K, Kumaraswamy M, Nizet V, Lin L, McCauley MD, Strathdee SA, Benson CA, Pope RK, Leroux BM, Picel AC, Mateczun AJ, Cilwa KE, Regeimbal JM, Estrella LA, Wolfe DM, Henry MS, Quinones J, Salka S, Bishop-Lilly KA, Young R, Hamilton T. 2017. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother 61:e00954-17. doi: 10.1128/AAC.00954-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loc-Carrillo C, Abedon ST. 2011. Pros and cons of phage therapy. Bacteriophage 1:111–114. doi: 10.4161/bact.1.2.14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Principi N, Silvestri E, Esposito S. 2019. Advantages and limitations of bacteriophages for the treatment of bacterial infections. Front Pharmacol 10:513. doi: 10.3389/fphar.2019.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scholl D. 2017. Phage tail-like bacteriocins. Annu Rev Virol 4:453–467. doi: 10.1146/annurev-virology-101416-041632. [DOI] [PubMed] [Google Scholar]
- 8.Scholl D, Martin DW, Jr.. 2008. Antibacterial efficacy of R-type pyocins towards Pseudomonas aeruginosa in a murine peritonitis model. Antimicrob Agents Chemother 52:1647–1652. doi: 10.1128/AAC.01479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gebhart D, Lok S, Clare S, Tomas M, Stares M, Scholl D, Donskey CJ, Lawley TD, Govoni GR. 2015. A modified R-type bacteriocin specifically targeting Clostridium difficile prevents colonization of mice without affecting gut microbiota diversity. mBio 6:e02368-14. doi: 10.1128/mBio.02368-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scholl D, Cooley M, Williams SR, Gebhart D, Martin D, Bates A, Mandrell R. 2009. An engineered R-type pyocin is a highly specific and sensitive bactericidal agent for the food-borne pathogen Escherichia coli O157:H7. Antimicrob Agents Chemother 53:3074–3080. doi: 10.1128/AAC.01660-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ritchie JM, Greenwich JL, Davis BM, Bronson RT, Gebhart D, Williams SR, Martin D, Scholl D, Waldor MK. 2011. An Escherichia coli O157-specific engineered pyocin prevents and ameliorates infection by E. coli O157:H7 in an animal model of diarrheal disease. Antimicrob Agents Chemother 55:5469–5474. doi: 10.1128/AAC.05031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee G, Chakraborty U, Gebhart D, Govoni GR, Zhou ZH, Scholl D. 2016. F-Type Bacteriocins of Listeria monocytogenes: a new class of phage tail-like structures reveals broad parallel coevolution between tailed bacteriophages and high-molecular-weight bacteriocins. J Bacteriol 198:2784–2793. doi: 10.1128/JB.00489-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michel-Briand Y, Baysse C. 2002. The pyocins of Pseudomonas aeruginosa. Biochimie 84:499–510. doi: 10.1016/s0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama K, Takashima K, Ishihara H, Shinomiya T, Kageyama M, Kanaya S, Ohnishi M, Murata T, Mori H, Hayashi T. 2000. The R-type pyocin of Pseudomonas aeruginosa is related to P2 phage, and the F-type is related to lambda phage. Mol Microbiol 38:213–231. doi: 10.1046/j.1365-2958.2000.02135.x. [DOI] [PubMed] [Google Scholar]
- 15.Uratani Y, Hoshino T. 1984. Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J Bacteriol 157:632–636. doi: 10.1128/jb.157.2.632-636.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholl D, Gebhart D, Williams SR, Bates A, Mandrell R. 2012. Genome sequence of Escherichia coli O104:H4 leads to rapid development of a targeted antimicrobial agent against this emerging pathogen. PLoS One 7:e33637. doi: 10.1371/journal.pone.0033637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams SR, Gebhart D, Martin DW, Scholl D. 2008. Retargeting R-type pyocins to generate novel bactericidal protein complexes. Appl Environ Microbiol 74:3868–3876. doi: 10.1128/AEM.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghequire MG, De Mot R. 2014. Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol Rev 38:523–568. doi: 10.1111/1574-6976.12079. [DOI] [PubMed] [Google Scholar]
- 19.Kuroda K, Kageyama M. 1981. Comparative study of F-type pyocins of Pseudomonas aeruginosa. J Biochem 89:1721–1736. doi: 10.1093/oxfordjournals.jbchem.a133372. [DOI] [PubMed] [Google Scholar]
- 20.Govan JR. 1974. Studies on the pyocins of Pseudomonas aeruginosa: production of contractile and flexuous pyocins in Pseudomonas aeruginosa. J Gen Microbiol 80:17–30. doi: 10.1099/00221287-80-1-17. [DOI] [PubMed] [Google Scholar]
- 21.Kuroda K, Kageyama M. 1979. Biochemical properties of a new flexuous bacteriocin, pyocin F1, produced by Pseudomonas aeruginosa. J Biochem 85:7–19. doi: 10.1093/oxfordjournals.jbchem.a132332. [DOI] [PubMed] [Google Scholar]
- 22.Takeya K, Minamishima Y, Amako K, Ohnishi Y. 1967. A small rod-shaped pyocin. Virology 31:166–168. doi: 10.1016/0042-6822(67)90021-9. [DOI] [PubMed] [Google Scholar]
- 23.Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, Maxwell KL. 2016. Prophages mediate defense against phage infection through diverse mechanisms. ISME J 10:2854–2866. doi: 10.1038/ismej.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsui H, Sano Y, Ishihara H, Shinomiya T. 1993. Regulation of pyocin genes in Pseudomonas aeruginosa by positive (prtN) and negative (prtR) regulatory genes. J Bacteriol 175:1257–1263. doi: 10.1128/jb.175.5.1257-1263.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winsor GL, Lam DK, Fleming L, Lo R, Whiteside MD, Yu NY, Hancock RE, Brinkman FS. 2011. Pseudomonas Genome Database: improved comparative analysis and population genomics capability for Pseudomonas genomes. Nucleic Acids Res 39:D596–D600. doi: 10.1093/nar/gkq869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deighan P, Hochschild A. 2007. The bacteriophage λQ anti-terminator protein regulates late gene expression as a stable component of the transcription elongation complex. Mol Microbiol 63:911–920. doi: 10.1111/j.1365-2958.2006.05563.x. [DOI] [PubMed] [Google Scholar]
- 27.Young R. 2014. Phage lysis: three steps, three choices, one outcome. J Microbiol 52:243–258. doi: 10.1007/s12275-014-4087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kohler T, Donner V, van Delden C. 2010. Lipopolysaccharide as shield and receptor for R-pyocin-mediated killing in Pseudomonas aeruginosa. J Bacteriol 192:1921–1928. doi: 10.1128/JB.01459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davidson AR, Cardarelli L, Pell LG, Radford DR, Maxwell KL. 2012. Long noncontractile tail machines of bacteriophages. Adv Exp Med Biol 726:115–142. doi: 10.1007/978-1-4614-0980-9_6. [DOI] [PubMed] [Google Scholar]
- 30.Pell LG, Liu A, Edmonds L, Donaldson LW, Howell PL, Davidson AR. 2009. The X-ray crystal structure of the phage lambda tail terminator protein reveals the biologically relevant hexameric ring structure and demonstrates a conserved mechanism of tail termination among diverse long-tailed phages. J Mol Biol 389:938–951. doi: 10.1016/j.jmb.2009.04.072. [DOI] [PubMed] [Google Scholar]
- 31.Auzat I, Petitpas I, Lurz R, Weise F, Tavares P. 2014. A touch of glue to complete bacteriophage assembly: the tail-to-head joining protein (THJP) family. Mol Microbiol 91:1164–1178. doi: 10.1111/mmi.12526. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Hofnung M, Charbit A. 2000. The C-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J Bacteriol 182:508–512. doi: 10.1128/JB.182.2.508-512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werts C, Michel V, Hofnung M, Charbit A. 1994. Adsorption of bacteriophage lambda on the LamB protein of Escherichia coli K-12: point mutations in gene J of lambda responsible for extended host range. J Bacteriol 176:941–947. doi: 10.1128/jb.176.4.941-947.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, Eddy SR. 2015. HMMER web server: 2015 update. Nucleic Acids Res 43:W30–W38. doi: 10.1093/nar/gkv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hendrix RW, Duda RL. 1992. Bacteriophage lambda PaPa: not the mother of all lambda phages. Science 258:1145–1148. doi: 10.1126/science.1439823. [DOI] [PubMed] [Google Scholar]
- 36.North OI, Sakai K, Yamashita E, Nakagawa A, Iwazaki T, Buttner CR, Takeda S, Davidson AR. 2019. Phage tail fibre assembly proteins employ a modular structure to drive the correct folding of diverse fibres. Nat Microbiol 4:1645–1653. doi: 10.1038/s41564-019-0477-7. [DOI] [PubMed] [Google Scholar]
- 37.Gillies RR, Govan JR. 1966. Typing of Pseudomonas pyocyanea by pyocine production. J Pathol Bacteriol 91:339–345. doi: 10.1002/path.1700910207. [DOI] [PubMed] [Google Scholar]
- 38.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lam JS, Taylor VL, Islam ST, Hao Y, Kocincova D. 2011. Genetic and functional diversity of Pseudomonas aeruginosa lipopolysaccharide. Front Microbiol 2:118. doi: 10.3389/fmicb.2011.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Koonin EV. 1998. Iterated profile searches with PSI-BLAST: a tool for discovery in protein databases. Trends Biochem Sci 23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 43.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. 2009. Jalview version 2: a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V. 2018. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 45.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. 2010. The Pfam protein families database. Nucleic Acids Res 38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu D, Damron FH, Mima T, Schweizer HP, Yu HD. 2008. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl Environ Microbiol 74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pawluk A, Shah M, Mejdani M, Calmettes C, Moraes TF, Davidson AR, Maxwell KL. 2017. Disabling a type I-E CRISPR-Cas nuclease with a bacteriophage-encoded anti-CRISPR protein. mBio 8:e01751-17. doi: 10.1128/mBio.01751-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiang P, Burrows LL. 2003. Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa. J Bacteriol 185:2374–2378. doi: 10.1128/JB.185.7.2374-2378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 50.Lesic B, Rahme LG. 2008. Use of the lambda Red recombinase system to rapidly generate mutants in Pseudomonas aeruginosa. BMC Mol Biol 9:20. doi: 10.1186/1471-2199-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buttner CR, Wu Y, Maxwell KL, Davidson AR. 2016. Baseplate assembly of phage Mu: defining the conserved core components of contractile-tailed phages and related bacterial systems. Proc Natl Acad Sci USA 113:10174–10179. doi: 10.1073/pnas.1607966113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu J, Hendrix RW, Duda RL. 2014. Chaperone-protein interactions that mediate assembly of the bacteriophage lambda tail to the correct length. J Mol Biol 426:1004–1018. doi: 10.1016/j.jmb.2013.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Download jb.00029-23-s0001.doc, DOC file, 0.07 MB (67.5KB, doc)
Tables S1 and S2 and Fig. S1 to S5. Download jb.00029-23-s0002.pdf, PDF file, 2.74 MB (2.7MB, pdf)
Data Availability Statement
The sequences of the R- and F-type pyocin regions discussed here have been uploaded to GenBank under accession numbers OQ870446 to OQ870473.