Abstract
Context
Sticky trap catches of agricultural pests can be employed for early hotspot detection, identification, and estimation of pest presence in greenhouses or in the field. However, manual procedures to produce and analyze catch results require substantial time and effort. As a result, much research has gone into creating efficient techniques for remotely monitoring possible infestations. A considerable number of these studies use Artificial Intelligence (AI) to analyze the acquired data and focus on performance metrics for various model architectures. Less emphasis, however, was devoted to the testing of the trained models to investigate how well they would perform under practical, in-field conditions.
Objective
In this study, we showcase an automatic and reliable computational method for monitoring insects in witloof chicory fields, while shifting the focus to the challenges of compiling and using a realistic insect image dataset that contains insects with common taxonomy levels.
Methods
To achieve this, we collected, imaged, and annotated 731 sticky plates - containing 74,616 bounding boxes - to train a YOLOv5 object detection model, concentrating on two pest insects (chicory leaf-miners and wooly aphids) and their two predatory counterparts (ichneumon wasps and grass flies). To better understand the object detection model's actual field performance, it was validated in a practical manner by splitting our image data on the sticky plate level.
Results and conclusions
According to experimental findings, the average mAP score for all dataset classes was 0.76. For both pest species and their corresponding predators, high mAP values of 0.73 and 0.86 were obtained. Additionally, the model accurately forecasted the presence of pests when presented with unseen sticky plate images from the test set.
Significance
The findings of this research clarify the feasibility of AI-powered pest monitoring in the field for real-world applications and provide opportunities for implementing pest monitoring in witloof chicory fields with minimal human intervention.
Keywords: Insect recognition, Convolutional neural networks, Pest management, Automatic monitoring
Graphical abstract
Highlights
-
•
Successful detection and classification of insects from sticky trap images using YOLOv5.
-
•
A data-centric approach to insect monitoring is key in developing robust insect detection models.
-
•
Image tiling of high-resolution sticky plate images is important for detecting small insects.
-
•
Classification of insects with shared taxonomy levels is representative for the real situation in the field.
1. Introduction
Witloof chicory (Cichorium intybus L. var. foliosum) is an etiolated vegetable with white-yellow leaves. It is a specialty crop characterized by its unique bitter taste and crunchiness. Due to its biennial life cycle, witloof chicory is produced in two stages. In the first stage of growth the plant develops a deep taproot and produces a rosette of leaves on a short stem. In the second stage, the vernalized taproot is “forced” to produce an etiolated apical bud in a dark, humid environment. Nowadays, witloof chicory is mostly grown hydroponically in a multilayer system in dark growth chambers (De Rijck et al., 1994; van Kruistum et al., 1997). The combination of hydroponic forcing systems with the introduction of hybrid cultivars and a well-balanced long-term root storage program enables chicon production all year round (De Proft et al., 2003). The production of witloof chicory is mainly localized in Western Europe, with the Netherlands, Belgium, France, and Germany as main producers. However, the chicory market is global, as chicons are exported to distant markets such as the USA, Canada, Japan, Qatar, and Israel, adding to a world trade value of 102 million US dollar in 2019 (OEC, 2021).
Witloof chicory has two major pest insects that cause serious damage: 1) The chicory leaf-miner (Phytomyza cichorii S) and 2) the wooly lettuce root aphid (Pemphigus bursarius). This fly is barely a few millimeters large, has a wingspan of 3.5 mm, dark legs with yellowish knees, yellow and black stripes, and is recognizable by a specific crossvein ("dm-cu") formation in its wings. Chicory leaf miners have three generations per year. One in July, one in August, and one in October. The fly lays eggs near the leaf stem in its first generation cycle. The larvae eat on the chicory plant's leaf, stem, and root. Pupation takes place inside the leaf or the root (Casteels and de Clercq, 1994; Coman and Rosca, 2011; Spencer, 1966). The infestation time determines the damage. Late summer larvae travel to the leaf collar and root collar, joining chicory roots collected from the farm. They graze on crops and leave apparent mining tunnels during crop cultivation. Deeper mines in the crop make it unfit for sale, requiring more head cleaning and reducing production. In recent years, the chicory leaf-miner has been attacking the roots' growth point quickly after sowing and over the summer, leaving the crop deformed or missing white chicory heart (etiolated bud) (Bunkens and Maenhout, 2019; de Lange, 2008).
The second major pest is the wooly lettuce root aphid, (Pemphigus bursarius L) also known as the “poplar-lettuce gall aphid”. This aphid is heteroecious and migrates in spring (winged fundatrigenia) from poplar trees (Populus nigra L.), the primary host, to ground and underground parts of Asteraceae plants - the secondary host. Wingless virginogenia—female aphids that give birth to surviving young by parthenogenesis—can multiply quickly (Hałaj and Osiadacz, 2013; Leclant, 1998). Phillips et al. (2000) describe how the lettuce root aphid can adapt for overwintering and thus survive underground without the need to migrate. P. bursarius apterae (wingless lettuce root aphids) are regularly observed on the roots causing a severe wilting of the leaves and seriously reducing the root production. This problem particularly occurs during warm and dry periods that favor aphid multiplication (Benigni et al., 2016).
Given their negative impact on the yield, an efficient control strategy against these pests would provide a significant economic added value. A screening performed by Flemish research stations Inagro and Praktijkpunt Landbouw Vlaams-Brabant on the 20 fields that were monitored for the observations and warnings showed that no plot is free of damage at the growing point. On average, 18% of the chicory roots showed mining corridors of the chicory leaf-miner. In 7% of the roots, the growing point was seriously affected, which means that chicory heads that grew on these roots were unfit for sale. Moreover, the appearance of pest insects in the crop can inhibit transport to other countries (Kamiji and Iwaizumi, 2013). Consequently, the loss caused by chicory leaf-miners can be more than 30% of the production value, involving loss of production, loss of quality, increased labor costs and loss of exports.
Manual monitoring systems have been used to evaluate and limit pest presence. Yellow water traps (height: 10 cm, width: 40 cm and length: 60 cm) are often placed on a chicory field. They are filled with water with a small quantity of antifreeze and a drop of detergent to conserve and lower the surface tension, respectively. Captured insects are usually collected on a weekly basis. This labor-intensive procedure involves several steps: 1) the traps are emptied through a filter, 2) the insects are collected and carefully transferred into a labelled cup, and finally, 3) transported back to the lab for the actual leaf miner count under a microscope. All traps need to be refilled and cleaned thoroughly to remove old catches. In the laboratory, the contents of the labelled cup with captured insects are first rinsed and placed in dishes which are placed under a microscope. Insects are then observed one by one to count chicory leaf-miners and wooly aphids. Observations on any given plot are considered valid within a 10 km radius, and with a careful choice of plots a large region can be studied annually. However, the installation and regular collection of all traps, followed by the insect counting and identification is very labor-intensive.
These time- and labor-demanding procedures ask for more efficient approaches to enable accurate and timely monitoring of large areas of crop production. Recently, several automatic monitoring systems have been proposed, benefiting from advancements in the field of Artificial Intelligence (AI) (Barbedo, 2020; Li et al., 2019, Li et al., 2021; Rustia et al., 2021a; Wang et al., 2020c). Wang et al., 2020a, Wang et al., 2020b, Wang et al., 2020c obtained a mean Average Precision (mAP) score of 63.54% using YOLOv3 (Redmon and Farhadi, 2018) while detecting and classifying pest species from the images in their “Pest24” dataset. The dataset consisted of 25,378 annotated images coming from 24 pest species that were all collected by an automatic imaging trap. Rustia et al. (2021) designed a multi-stage deep learning method including object detection, insect vs. non-insect separation and a multi-class insect classification for insect identification. Data were acquired by multiple wireless imaging devices installed in greenhouses – under varying lighting environments - and their classification model reached average F1-scores of up to 0.92. Hong et al. (2021) developed an AI-based pest counting method for monitoring the black pine bast scale (M. thungergianae) which reached a counting accuracy of 95%. No previous studies were found for detecting and monitoring chicory leaf-miners or wooly lettuce root aphids. While AI systems show great promise for automatic insect recognition and monitoring from images, they have not yet resulted in many robust and field-ready insect monitoring systems (SmartProtect, 2022). In several reported studies, the used datasets consist of close-up and very high quality images taken from professional photographers or mined from the web, which are not representative for the challenging environments found in the field (Cheng et al., 2017; Kasinathan et al., 2021; Li and Yang, 2020; Nanni et al., 2020; Pattnaik et al., 2020; Wang et al., 2017; Wang et al., 2020b). Imaging systems that can automatically take a high-quality snapshot of a single insect are too difficult or inefficient to build, especially for small or flying insects (Barbedo, 2020). A more practical approach is to capture a surface covered with multiple (trapped) insects using a single image acquired within a trap in the field (Ding and Taylor, 2016; Jiao et al., 2020; Rustia et al., 2021; Xia et al., 2015; Zhong et al., 2018). Smaller tiles of individual insect images are then extracted from the full image for further analysis. In many cases, the insect monitoring systems are focused on the detection of a single pest (Ding and Taylor, 2016; Hong et al., 2021; Nazri et al., 2018; Roosjen et al., 2020), ignoring the potential presence of predatory or pollinating species that could provide valuable information about a field’s ecosystem and thus, create more added value to growers. Besides limitations in the scope of the insect monitoring application, AI practitioners often fail to apply strict validation procedures to challenge their models and fall into known methodological pitfalls like “data leakage” (Kapoor and Narayanan, 2022). In our previous research (Kalfas et al., 2021; Kalfas et al., 2022), we showed that model performance can be highly overestimated when weak validation procedures – like random data splitting- are followed. Despite taking the above limitations into consideration, other studies still only focused on datasets where insect classes were quite broad – insect order or family level, quite dissimilar and thus, relatively easy to classify (Rustia et al., 2021b; Wang et al., 2020c).
Therefore, the goal of this study is to assist towards shifting the focus of insect monitoring systems from model-centric to data-centric approaches. Moreover, we aim to showcase a robust computational approach towards insect monitoring in witloof chicory fields. To this end, we trained an object detection model on a dataset of insect images collected from witloof chicory fields whilst specifically focusing on two pest insects (chicory leaf-miner and wooly aphid) and their two predatorial counterparts (ichneumon wasp and grass fly). The object detection model was validated in a practice-oriented way to help uncover its true performance in field conditions. The pest insects were also classified against broader insect classes from common levels of taxonomy, which is expected to be critical for any insect monitoring system to be deployed in the field.
2. Materials and methods
2.1. Data collection
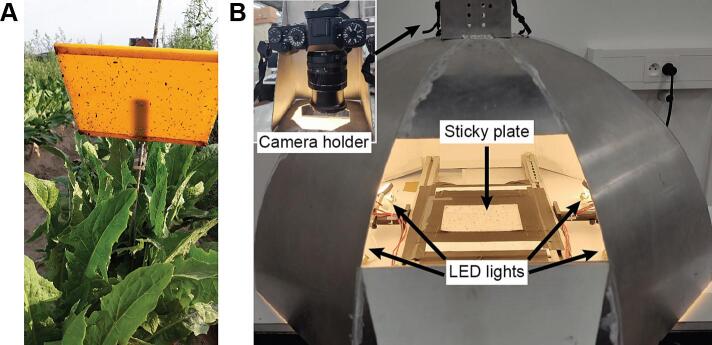
Insect populations were sampled with sticky plate traps from witloof chicory fields situated in various locations across Belgium (Herent, Braine-l'Alleud, Kampenhout, Merchtem, Racour). The data collection period for this study lasted from the beginning of May until the end of October in years 2019, 2020 and 2021. Three traps were installed in each field to capture the insect population variability efficiently. Each trap consisted of a yellow plastic holder (Goudval©, De Groene Vlieg, 2022) known to attract several flying pests due to phototaxis and a sticky plate (Fig. 1A). The holders are made of a firm yellow plastic that is fixed on a metal rod which is hammered into the ground. The sticky plates are transparent flexible plastic surfaces of 199 mm width and 148 mm height, covered with a thin layer of wet glue. They were placed at the same height on their respective holders, positioned in a straight line and distanced at approximately 20 m. The sticky traps were placed as low as possible to the ground on the ridge in between the chicory plants to trap flying insects. All sticky plates were replaced with new ones on a weekly basis. For this, the following steps were followed: First, we carefully removed the sticky plate from the holder and assigned a label to it (location, field position, date). Secondly, matt varnish was sprayed all over the surface of each plate from a 10-20 cm distance to better fix the insects and to decrease the stickiness of the plates, facilitating the collection and transportation. Thirdly, the plates were placed in holders with corresponding parcel codes, and lastly, stored and kept in a cool and dark room.
Fig. 1.
Illustrations of A) a sticky plate fixed on a holder in a witloof chicory field and B) the imaging setup.
Camera calibration was applied to remove lens distortion from the sticky plate images. After spatial distortion correction, the images were sent back to entomology experts for labelling. Entomology experts, using a microscope to inspect the insects on a given sticky plate, assigned “simplified” labels for insects belonging to our target classes, as shown in Fig. 2, which we will also use throughout this manuscript. The derivation of our simplified naming scheme from scientific names is illustrated in Fig. 3. The experts, using the online tool “Make Sense” (Skalski, 2019), created annotated bounding boxes on the digital plate images around each identified insect. Data acquired this way were stored and organized in a database that contained information for each sticky plate image (i.e., location, field position, date, and camera settings), the coordinates of insects’ bounding boxes and the insect classes. The total counts for each category are shown in Fig. 4.
Fig. 2.
Illustration of the 12 classification categories (4 images per category; each image is a 150x150 pixel area cut from a sticky plate image). Images of the two pest insect categories are marked with a red border.
Fig. 3.
Dendrogram illustrating the taxonomy levels for all the insect categories used.
Fig. 4.
Insect counts (in thousands) for each category in the dataset.
From a model-centric view misclassifications are often explained by the model complexity, its hyperparameters and the training procedure efficiency. Hence, many researchers employ a variety of models and training procedure optimizations until they find the “best” result. Moreover, as we have already shown in our previous research (Kalfas et al., 2021; Kalfas et al., 2022), several studies on insect recognition don’t apply any strict validation procedure to evaluate their models, which often leads to over-optimistic or unrealistic performance.
From a data-centric approach, wrong detections are assumed to be caused by either intra- or inter-class variability (Fig. 2 and Fig. 3), class imbalance (Fig. 4) or labelling errors.
2.2. Imaging setup
Sticky plates with trapped insects on top of them were photographed using a custom-built imaging setup. As illustrated in Fig. 1, the setup is comprised of a metal dome frame, a sticky plate base, LED lamps and a camera holder. Four Sencys® LED lamps (5W, 345lm, 2700K, GU5.3) are positioned around the sticky plate base, pointing towards the dome’s inner walls which are painted white. This provides a homogeneous illumination of the sticky plate. A 24Mpixel digital camera (X-T1, Fujifilm®) camera is installed on the top opening of the dome 47 cm above the sticky plate base, while connected via a USB-C cable to a laptop computer. Images are acquired with a resolution of 6,240 by 4,160 pixels (24.5 pixels/mm) and saved locally to the laptop’s hard drive. The camera settings used to image all sticky plates are listed below:
-
•
Aperture: F/10
-
•
ISO: 160
-
•
Shutter speed: 1/15 s
-
•
Focal Length: 55 mm
-
•
White balance: “Incandescent”
2.3. Data splitting and image tiling
In order to perform data splitting in this study, all individual insect images were grouped in terms of the sticky plate image they belonged to, rather than the overall quantity of insects present in the entire dataset. This approach allowed us to validate the detection model by simulating its use in practice where the model would be given a sticky plate image to detect insects on. 80% of all sticky plate images were used for training and validating our model (60% training, 20% validation), while the remaining 20% were used as a held-out test set to evaluate its performance. The distribution and presence of insects belonging to a particular category varied across the sticky plates. However, the high-resolution sticky plate images require high computational power, and computer and GPU memory to fit into the training pipeline. Since this is not easily available, in literature it is common practice to downscale input images until they fit in memory. Unfortunately, this method throws away information that could be crucial for the classifier. To tackle this issue, we sliced our images in smaller tiles of a fixed size (512x512 pixels) which were then fed to the model for training and validation. This way, all insect images maintained their original pixel dimensions which is especially important for detecting small insects. To achieve this, we used the implementation of the Python library “impy” (Lucero, 2018) where the tiles are designed to fulfill two requirements: 1) have a user-defined size of 512x512 pixels and 2) contain the maximum number of complete bounding boxes. This way, image regions that did not contain any bounding box were not included in the data. The resulting tiles made up the final Train, Validation and Test sets that were used with the detection model (see Fig. 5 splitting).
Fig. 5.
A) Illustration of the procedure for separating data into training, validation and test sets, and tiling sticky plate images by finding regions of interest of a fixed size (512x512 pixels) that contain the maximum number of complete bounding boxes. B) Illustration of applying the YOLOv5 model on a sticky plate image using “Slicing Aided Hyper Inference” (SAHI).
2.4. Model configuration
Detection and classification of the insects on the sticky plates was performed by the pretrained model YOLOv5x (Jocher et al., 2020) which was fine-tuned on our custom dataset. This model was created by an open-source project known as “Ultralytics” (Jocher et al., 2020). Their framework provides four pre-trained neural network configurations that range in terms of complexity (“nano”, “small”, “medium”, “large”, “extra-large”). For this study, we opted for the “extra-large” option, or YOLOv5x, which demonstrated the largest promise in terms of detection and classification accuracy in the popular object detection benchmarking dataset, known as COCO (Lin et al., 2015), on which it was originally trained. In this version of the model, a Cross Stage Partial Network (CSPNet) (Wang et al., 2020a) with a Spatial Pyramid Pooling (SPP) layer is used as the model back-bone and a path aggregation network (PANet) as neck to boost information flow. The head in YOLOv5 achieves multi scale prediction by generating different output feature maps.
2.5. Training, inference and evaluation
The insect detection model was trained and tested using Ubuntu 20.04.4 LTS on an HP® Zbook 17 G6 with an Intel®Xeon® E-2286M (16 cores at 2.4GHz) CPU, 32 GB Memory, and an NVIDIA® Quadro RTX 5000 (16GB Memory; CUDA Version: 11.4) GPU.
The training procedure of the detection model ran for 150 epochs. The model was fed with batches of 16 tiles at a time which was motivated by the available GPU memory and training efficiency. Stochastic Gradient Descent (SGD) was used as the model’s optimizer with a momentum of 0.9. The default data augmentation techniques of YOLOv5 were applied during training to improve the model robustness. Predicted bounding boxes with intersection-over-union (IoU) ≥ 0.6 were considered positive while bounding boxes with IoU<0.6 were considered negative (Zhang et al., 2020).
To produce model predictions for a whole sticky plate we used “Slicing Aided Hyper Inference” (SAHI; Akyon et al., 2022). This technique can be applied on top of any object detector to improve the detection of small objects. To achieve this, SAHI slices the original image into smaller equally sized tiles. Some overlap among tiles is also used to handle objects located on the tile edges. In our case, each tile was 512x512 pixels and an overlap ratio of 20% was applied.
Evaluation of the model performance was performed based on standard metrics used in common object detection datasets like MS COCO (Lin et al., 2015). Precision measures the fraction of the target insects in the detected objects, while recall measures the fraction of detected target insects from all the target insects within one image. A Precision-Recall (PR) curve was used to evaluate the overall model performance for insect detection. Average Precision (AP) is the area under the PR curve for a given IoU threshold, and mAP is the mean AP for all insect classes in our dataset. The metrics are defined as follows:
| (1) |
| (2) |
| (3) |
where TP, TN, FP, and FN stand for true positive, true negative, false positive and false negative, respectively.
3. Results and discussion
3.1. Insect image database
The number of images collected for each class depended primarily on the respective insect’s presence on the sticky plates sampled from the fields (see Fig. 4). Insect labelling is performed by entomology experts and is a very labor-intensive procedure. In total, they created 74,616 bounding boxes from 731 sticky plates over 3 summer seasons. This procedure took multiple hours to complete since there are often hundreds of insects on a single plate. It is estimated that on average human annotators were required to spend 2-3 hours per sticky plate. In total, approximately 2,000 hours were needed for all 731 sticky plates. To optimize the workers’ workload, we marked some of our classes as “critical” and labelled all individual insects from those classes with a high priority compared to insects belonging to other classes. Classes marked as ‘critical’ were: “aphid”, “aphid-wooly”, “fly”, “fly-chicory”, “fly-grass” and “wasp”. Insects belonging to the remaining classes were labelled with a lower priority, leaving some individuals unlabeled if the expert’s workload was too high.
Due to the observational nature of the followed approach, the dataset is not well-balanced, containing thousands of images for some classes and only a couple of hundreds for others. The insect class with the fewest data was “fly-grass” with only 391 individual insect images in total, while the most populated class was “fly” with 18,389 images (see Fig. 4). The “fly” class is broader and contains insects from several fly genera, which explains the high sticky plate presence. The higher numbers of “fly-chicory” insects compared to “fly-grass” occurred naturally. Similarly, as expected, the broader “aphid” class contains more images (6,465) than “aphid-wooly” (711). Having fewer images for a given class is expected to affect model performance, which we discuss later. However, this also depends on the intra-class variability. In Fig. 2, we show that the intra-class variability can sometimes be as high as the inter-class variability. For example, the “wasp” class can contain individuals that vary significantly in size. This can lead to numerous classification challenges since small wasps could be confused with thrips and larger ones with flies. As discussed earlier, such challenges were avoided in most reported studies by including highly dissimilar insect classes with low intra-class variability, resulting in high theoretical classification rates which are difficult to hold in practice. While our approach may be less favorable from a machine learning point of view, it is more representative for the real situation that the classifier will be confronted with in practice.
3.2. Model performance
To validate our model in a practice-oriented way, we split our data on the sticky plate level. Therefore, individual insects that belonged to the same sticky plate image were kept together in the same dataset (training, validation, or testing). This allowed us to validate our model’s performance by emulating the way in which the model will be used in practice. Namely, it predicted a class for any given object it detected on unseen sticky plate images – data it never saw during training.
In Fig. 6, the precision recall (PR) curves and mAP scores of the model for test-set data from the two target pests (“aphid-wooly”, “fly-chicory) and their respective natural predators (“fly-grass”, “wasp”) are illustrated. The confusion matrix based on the test-set is illustrated in Fig. 7, where model predictions are compared to the reference labels that were created by the entomology experts. The confusion matrix was produced using the default YOLOv5 value for model confidence (0.25) and the same IoU that was used for training (0.6). The “background FP” column indicates false positives where the background was mistaken for a dataset class, while the “background FN” row denotes false negatives, namely, a dataset class mistaken for the background. Background images are either “objectless” images or with objects not belonging to the target class and they are added to a dataset to reduce FP numbers, as recommended by the authors of YOLOv5 (ultralytics/yolov5 - Tips For Best Training Results, 2022). Our ‘not-insect’ class serves this purpose since it contains either background images or pieces of insects or dirt. As expected, the highest cross-confusion for “background FP” was with the “not-insect” class (26%) followed by the “fly” class (21%). The “fly” class was the most populated in our dataset. It was characterized by a high intra-class variability and shared many visual similarities with other insects, like “fly-chicory” and “fly-grass”, creating cross-confusion with almost every other insect class. However, this could also indicate the existence of label errors for “fly” bounding boxes or flies without any label assignment which would be considered part of the background by the model. When training object detection models, it is recommended to label all objects in an image, if feasible.
Fig. 6.
Precision-recall curves for YOLOv5 for the two pests (‘aphid-wooly’, ‘fly-chicory’), their predatory counterparts (‘fly-grass’, ‘wasp’), and all classes.
Fig. 7.
Confusion matrix for YOLOv5 (“IoU”: 0.6, “confidence threshold”: 0.25).
The model was able to correctly identify the target pests with a higher mAP score (“aphid-wooly”: 0.732, “fly-chicory”: 0.863) than their predators (“fly-grass”: 0.609, “wasp”: 0.675; Fig. 6). The “fly-chicory” class was one of the most populated in our dataset. Our trained YOLOv5 model was able to correctly classify “fly-chicory” insect images with 88% accuracy, while only 7% of its reference detections were confused with detections belonging to the parent class “fly” (Fig. 3). Only a limited amount of data was available for the “aphid-wooly” and “fly-grass” classes since their presence was quite limited when we compiled our dataset. Still, the model correctly identified 72% of reference “aphid-wooly” bounding boxes and misclassified 14% as “aphid” which is its parent class. The model performed relatively poorly (50% accuracy) when classifying “fly-grass” images and misclassified 26% of all reference “fly-grass” images into the parent class “fly”. This may partly be attributed to the low number of “fly-grass” images which were the least among all classes. By establishing a different labelling protocol that focuses more on the under-represented classes, model performance is expected to rise. For example, the use of sticky plates in a controlled environment with only one given target insect present. Custom sticky plates that contain known insect types are also easier to annotate since they do not require expert knowledge in determining each individual insect’s type. The model accuracy for the “wasp” class detections was 66% (Fig. 7). One reason for this could be the high intra-class variability of “wasp”. Insects belonging to that class varied greatly in size and shape, leading to model misclassifications with multiple other insect classes such as ‘thrip’, ‘not-insect’, ‘mosquito’, “fly”, “cicada” and “beetle”. A suggested approach to tackle this issue would be to subdivide the class based on different genera or species. This would allow to investigate misclassifications with other insects further or detect annotation errors. A more general alternative solution would be to use a hierarchical classification approach that incorporates taxonomy levels as labels for different models. This method involves training a model to classify at higher levels of the taxonomic hierarchy (e.g., Insects vs Araneae; see Fig. 3) and using the output of this classification to guide the input for models at lower levels of the taxonomic hierarchy. Essentially, this approach would involve using a series of nested models, each of which is trained to classify at a specific level of the taxonomic dendrogram (Fig. 3). While this approach could prove effective, it would be impractical to implement in practice due to the need for training and maintaining multiple models at each branch. 3.3 Towards deploying the model in the field
In Fig. 8, the reference number of insects is plotted against the model predicted number per sticky-plate in the test-set. The model was able to estimate the correct number of insects quite accurately, showing a Pearson’s R correlation coefficient of 0.96 and 0.88 for “aphid-wooly” and “fly-chicory” numbers, respectively. In a second model evaluation towards field deployment, we compared the reference number of the two pests against the pest presence predicted by the model on a weekly basis for a chosen location and time period included in our test data (Fig. 9). Again, the model was able to closely follow the evolution of the pests’ presence throughout the chosen season, indicating that it could serve as a reliable insect monitoring tool for estimating pest presence in witloof chicory fields. Since all predictions could be generated immediately upon imaging a sticky plate, the existing delays related to manual counting, identifying, and reporting pest presence would be eliminated.
Fig. 8.
Number of insect individuals per sticky plate for the two pests according to human annotators (reference) vs number predicted by YOLOv5. Pearson’s correlation coefficient is denoted with “r” in each figure. A regression line and translucent confidence intervals bands are shown in dark and light blue colors, respectively. The 1:1 line is shown in red color.
Fig. 9.
Pest presence per week (counts) according to human annotators (reference) vs. as predicted by YOLOv5. No available data existed for weeks 25 and 35 in the test set. Counts were averaged by the number of available sticky plates per week. All weeks had one sticky plate available in the test set except for week 24 (2), week 32 (2) and week 36 (3).
4. Conclusions
Crop production is facing new challenges by a changing climate, unstable economies, and a fast-paced global trade. Insect populations are sensitive to these changes with their numbers fluctuating and new species being rapidly introduced to remote regions due to human activity. To cope with these challenges, farmers demand for tools to monitor insect presence and especially pest activity. To this end, we investigated the potential of a widely used object detection model - YOLOv5 - for insect classification in images from sticky plate insect traps. Challenges in compiling an insect dataset and evaluating the detection model in a practice-oriented and data-centric way were discussed, such as the presence of non-target insects belonging to the same order, family, or genus as the target insects.
Experimental results indicated an mAP score of 0.76 across all dataset classes. High mAP scores of 0.73 and 0.86 were obtained for both pest species (wooly aphid and chicory leaf-miner) and their respective predators: grass-fly (mAP=0.61) and wasp (mAP=0.67). Model evaluation focusing on the model’s future field deployment showed that it was able to accurately estimate pest presence when given novel individual sticky plate images and when aggregating test images on a weekly basis.
Consequently, an imaging system that resembles the one employed in this study by leveraging standardized illumination conditions during image acquisition could incorporate the proposed model for automatically and reliably identifying insects in witloof chicory fields with minimal human intervention. Using transfer learning techniques and further data collection, the same model can be adapted to function with new data from a different camera system, or it can serve as a "warm-start" for a new model. This opens opportunities for automated insect monitoring in witloof chicory fields.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank Frank Mathijs of the MeBioS technical staff for his invaluable help to materialize many of the ideas for this project. We also thank Sarah Fonteyn, Leen Coremans, Rana Yurduseven, Laura Vercammen and all other collaborators from Praktijkpunt Landbouw Vlaams-Brabant who helped tremendously with their efforts in the insect annotation work. This research was conducted with the financial support of VLAIO (Flanders Innovation & Entrepreneurship) (project HBC.2016.0795) and the Horizon 2020 Research and Innovation Programme, Grant Agreement no. 862563.
Data availability
Data will be made available on request.
References
- Akyon F.C., Altinuc S.O., Temizel A. 2022. Slicing Aided Hyper Inference and Fine-tuning for Small Object Detection. [Google Scholar]
- Barbedo J.G.A. Detecting and classifying pests in crops using proximal images and machine learning: a review. Ai. 2020;1(2):312–328. doi: 10.3390/ai1020021. Available at: [DOI] [Google Scholar]
- Benigni M., et al. Control of root aphid (Pemphigus bursarius L.) in witloof chicory culture (Cichorium intybus L. var. foliosum) Crop Protect. [Preprint] 2016 doi: 10.1016/j.cropro.2016.07.027. Available at: [DOI] [Google Scholar]
- Bunkens K., Maenhout P. Alternatieven voor bestrijding mineervlieg hoognodig. 2019;21(13 December 2019):24–25. [Google Scholar]
- Casteels H., de Clercq R. Phenological observations on the witloof chicory fly Napomyza cichorii Spencer in Belgium during the decade 1984-1993. Parasitica. 1994;1 [Google Scholar]
- Cheng X., et al. Pest identification via deep residual learning in complex background. Comput. Electron. Agric. 2017;141:351–356. doi: 10.1016/j.compag.2017.08.005. Available at: [DOI] [Google Scholar]
- Coman M., Rosca I. Biology and life-cycle of leafminer Napomyza (Phytomyza) gymnostoma Loew., A new pest of Allium plants in Romania. South Western J. Hortic. Biol. Environ. 2011;2:57–64. [Google Scholar]
- de Lange J. 2008. Bestrijding Witlofmineervlieg in Witlof 2006-2007; p. 0228. [Google Scholar]
- De Proft M., Van Stallen N., Veerle N. Breeding and cultivar identification of Cichorium intybus L. var. foliosum Hegi Introduction : history of chicory breeding. Mol. Biol. 2003;2003(January 2003):83–90. [Google Scholar]
- De Rijck G., Schrevens E., De Proft M. Cultivation of Chicory Plants in Hydroponics. Acta Horticult. [Preprint] 1994 doi: 10.17660/actahortic.1994.361.21. Available at: [DOI] [Google Scholar]
- Ding W., Taylor G. Automatic moth detection from trap images for pest management. Comput. Electron. Agricult. [Preprint] 2016 doi: 10.1016/j.compag.2016.02.003. Available at: [DOI] [Google Scholar]
- Hałaj R., Osiadacz B. European gall-forming Pemphigus (Aphidoidea: Eriosomatidae) Zool. Anzeiger [Preprint] 2013 doi: 10.1016/j.jcz.2013.04.002. Available at: [DOI] [Google Scholar]
- Hong S.J., et al. Automatic pest counting from pheromone trap images using deep learning object detectors for matsucoccus thunbergianae monitoring. Insects. 2021;12(4) doi: 10.3390/insects12040342. Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao L., et al. AF-RCNN: An anchor-free convolutional neural network for multi-categories agricultural pest detection. Comput. Electron. Agric. 2020;174(April) doi: 10.1016/j.compag.2020.105522. Available at: [DOI] [Google Scholar]
- Jocher G., Stoken A., Borovec J., Chaurasia A., Changyu L., Laughing V., Hogan A., Hajek J., Diaconu L., Kwon Y., et al. 2020. ultralytics/yolov5: v6.1 - TensorRT, TensorFlow Edge TPU and OpenVINO Export and Inference, github. Available at: [DOI] [Google Scholar]
- Kalfas I., De Ketelaere B., Saeys W. Towards in-field insect monitoring based on wingbeat signals: The importance of practice oriented validation strategies. Comput. Electron. Agric. 2021;180(September) doi: 10.1016/j.compag.2020.105849. Available at: [DOI] [Google Scholar]
- Kalfas I., et al. Optical Identification of Fruitfly Species Based on Their Wingbeats Using Convolutional Neural Networks. Front. Plant Sci. 2022;13(June) doi: 10.3389/fpls.2022.812506. Available at: 10.3389/fpls.2022.812506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiji T., Iwaizumi R. 植物防疫所調査研究報告 = Research Bulletin of the Plant Protection Service Japan. 2013. Four Species of Agromyzidae (Diptera) Intercepted by Japanese Import Plant Quarantine. [Google Scholar]
- Kapoor S., Narayanan A. Leakage and the Reproducibility Crisis in ML-Based Science. 2022. http://arxiv.org/abs/2207.07048 Available at: [DOI] [PMC free article] [PubMed]
- Kasinathan T., Singaraju D., Uyyala S.R. Insect classification and detection in field crops using modern machine learning techniques. Inform. Process. Agricul. 2021;8(3):446–457. doi: 10.1016/j.inpa.2020.09.006. Available at: [DOI] [Google Scholar]
- Leclant F. Vol. 507. 1998. Les Pemphigiens du Peuplier et la Gallogenese. A la Decouverte des Pucerons Provoquant des Galles Sur les Peupliers; pp. 15–19. [Google Scholar]
- Li Y., Yang J. Few-shot cotton pest recognition and terminal realization. Comput. Electron. Agric. 2020;169(November 2019) doi: 10.1016/j.compag.2020.105240. Available at: [DOI] [Google Scholar]
- Li R., et al. A coarse-to-fine network for aphid recognition and detection in the field. Biosyst. Eng. 2019;187:39–52. doi: 10.1016/j.biosystemseng.2019.08.013. Available at: [DOI] [Google Scholar]
- Li W., et al. Classification and detection of insects from field images using deep learning for smart pest management: A systematic review. Ecol. Inform. 2021;66(October) doi: 10.1016/j.ecoinf.2021.101460. Available at: [DOI] [Google Scholar]
- Lin T.-Y., et al. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition [Preprint] 2015. Microsoft COCO: Common Objects in Context. [Google Scholar]
- Lucero R.A.L. Impy (Images in python), Github. 2018. https://github.com/lozuwa/impy Available at: Accessed: 1 February 2022.
- Nanni L., Maguolo G., Pancino F. Insect pest image detection and recognition based on bio-inspired methods. Ecol. Inform. 2020;57(January) doi: 10.1016/j.ecoinf.2020.101089. Available at: [DOI] [Google Scholar]
- Nazri A., Mazlan N., Muharam F. Research article Penyek: Automated brown planthopper detection from imperfect sticky pad images using deep convolutional neural network. PLoS One. 2018;13(12):1–13. doi: 10.1371/journal.pone.0208501. Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- OEC Witloof chicory, fresh or chilled, Observatory of Economic Complexity. 2021. https://oec.world/en/profile/hs92/witloof-chicory-fresh-or-chilled Available at: Accessed: 13 June 2021.
- Pattnaik G., Shrivastava V.K., Parvathi K. Transfer Learning-Based Framework for Classification of Pest in Tomato Plants. Appl. Artif. Intell. 2020;00(00):981–993. doi: 10.1080/08839514.2020.1792034. Available at: [DOI] [Google Scholar]
- Phillips S.W., Bale J.S., Tatchell G.M. Overwintering adaptations in the lettuce root aphid Pemphigus bursarius (L.) J. Insect Physiol. 2000 doi: 10.1016/S0022-1910(99)00188-2. [Preprint]. Available at: [DOI] [PubMed] [Google Scholar]
- Redmon J., Farhadi A. 2018. YOLOv3: An Incremental Improvement. [Google Scholar]
- Roosjen P.P.J., et al. Deep learning for automated detection of Drosophila suzukii: potential for UAV-based monitoring. Pest Manag. Sci. 2020;76(9):2994–3002. doi: 10.1002/ps.5845. Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustia D.J.A., Lu C.Y., et al. Online semi-supervised learning applied to an automated insect pest monitoring system. Biosyst. Eng. 2021;208(1):28–44. doi: 10.1016/j.biosystemseng.2021.05.006. Available at: [DOI] [Google Scholar]
- Rustia D.J.A., Chao J.J., et al. Automatic greenhouse insect pest detection and recognition based on a cascaded deep learning classification method. J. Appl. Entomol. 2021;145(3):206–222. doi: 10.1111/jen.12834. Available at: [DOI] [Google Scholar]
- Rustia D.J.A., Chao J.J., et al. Automatic greenhouse insect pest detection and recognition based on a cascaded deep learning classification method. J. Appl. Entomol. 2021;145(3):206–222. doi: 10.1111/JEN.12834. Available at: [DOI] [Google Scholar]
- Skalski P. Make Sense, Github. 2019. https://github.com/SkalskiP/make-sense/ Available at:
- SmartProtect E.P. SmartProtect Platform. 2022. https://platform.smartprotect-h2020.eu/en/search?applications%5B0%5D=4 Available at: Accessed: 17 August 2022.
- Spencer K.A. Proceedings of the Royal Entomological Society of London. Series B, Taxonomy [Preprint] 1966. A clarification of the genus Napomyza Westwood (Diptera: Agromyzidae) Available at: [DOI] [Google Scholar]
- ultralytics/yolov5 - Tips For Best Training Results 2022. https://docs.ultralytics.com/tutorials/training-tips-best-results/ Available at: Accessed: 6 June 2022.
- van Kruistum G., et al. Productie van witlof en roodlof. Teelthandleid. nr. 1997;79(79):226. [Google Scholar]
- Vlieg De Groene. 2022. https://www.degroenevlieg.nl/producten/witlofmineervlieg/ Available at: Accessed: 1 June 2022.
- Wang R.J., et al. A crop pests image classification algorithm based on deep convolutional neural network. Telkomnika (Telecommun. Comput. Electron. Contr.) 2017;15(3):1239–1246. 10.12928/TELKOMNIKA.v15i3.5382 Available at: [Google Scholar]
- Wang C.Y., et al. IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops, 2020-June. 2020. CSPNet: A new backbone that can enhance learning capability of CNN; pp. 1571–1580. Available at: [DOI] [Google Scholar]
- Wang J., et al. Common pests image recognition based on deep convolutional neural network. Comput. Electron. Agric. 2020;179(June) doi: 10.1016/j.compag.2020.105834. Available at: [DOI] [Google Scholar]
- Wang Q.J., et al. Pest24: A large-scale very small object data set of agricultural pests for multi-target detection. Comput. Electron. Agric. 2020;175(November 2019) doi: 10.1016/j.compag.2020.105585. Available at: [DOI] [Google Scholar]
- Xia C., et al. Automatic identification and counting of small size pests in greenhouse conditions with low computational cost. Ecol. Inform. 2015;29(P2):139–146. doi: 10.1016/j.ecoinf.2014.09.006. Available at: [DOI] [Google Scholar]
- Zhang J., et al. Multi-class object detection using faster R-CNN and estimation of shaking locations for automated shake-and-catch apple harvesting. Comput. Electron. Agricult. [Preprint] 2020 doi: 10.1016/j.compag.2020.105384. Available at. [DOI] [Google Scholar]
- Zhong Y., et al. A vision-based counting and recognition system for flying insects in intelligent agriculture. Sensors (Switzerland) 2018;18(5) doi: 10.3390/s18051489. Available at: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.