Abstract
Actin cytoskeleton predominantly regulates the formation and maintenance of synapses by controlling dendritic spine morphology and motility. To visualize actin dynamics, actin molecules can be labeled by genetically fusing fluorescent proteins to actin monomers, actin-binding proteins, or single-chain anti-actin antibodies. In the present study, we compared the dendritic effect of EGFP-actin, LifeAct-TagGFP2 (LifeAct-GFP), and Actin-Chromobody-TagGFP2 (AC-GFP) in mouse cultured hippocampal neurons using unbiased quantitative methods. The actin-binding probes LifeAct-GFP and AC-GFP showed similar affinity to F-actin, but in contrast to EGFP-actin, they did not reveal subtle changes in actin remodeling between mushroom-shaped spines and filopodia. All tested actin probes colocalized with phalloidin similarly; however, the enrichment of LifeAct-GFP in dendritic spines was remarkably lower compared with the other constructs. LifeAct-GFP expression was tolerated at a higher expression level compared with EGFP-actin and AC-GFP with only subtle differences identified in dendritic spine morphology and protrusion density. While EGFP-actin and LifeAct-GFP expression did not alter dendritic arborization, AC-GFP–expressing neurons displayed a reduced dendritic tree. Thus, although all tested actin probes may be suitable for actin imaging studies, certain limitations should be considered before performing experiments with a particular actin-labeling probe in primary neurons.
INTRODUCTION
Actin is a key cytoskeletal element in mammalian cells, involved in many cellular mechanisms (Campellone and Welch, 2010). Neurons, in particular, develop actin-rich growth cones during neurite outgrowth and special postsynaptic structures that are shaped by actin, called dendritic spines. These structures are small protrusions on the dendrites, receiving the majority of glutamatergic synaptic inputs in the brain (Yuste, 2010). Spines usually consist of a bulbous head region where the synaptic receptors and the postsynaptic density accumulates, and a thin neck region that separates the head from the dendritic shaft. Connected to synaptic activity, they can change their morphology, which is a phenomenon called structural plasticity (Arellano et al., 2007; Kasai et al., 2010; Amtul and Atta-Ur-Rahman, 2015; Bosch and Hayashi, 2015). Spines develop from dendritic filopodia, which are thin protrusions on the shaft without an established synaptic connection (Yuste and Bonhoeffer, 2004; Kayser et al., 2008; Ozcan, 2017). Structural remodeling of filopodia and spines underlies changes in synaptic strength on the cellular level. Activity-dependent alteration in synaptic strength is, in turn, the cellular basis of learning and memory, placing these protrusions in the focus of neuroscience research (Malenka and Bear, 2004; Nägerl et al., 2004; Lisman et al., 2012; Nishiyama and Yasuda, 2015).
In the spine heads, actin is present in branched filamentous polymers (F-actin) and globular monomers (G-actin; Hotulainen and Hoogenraad, 2010; Konietzny et al., 2017). Although straight filaments are also present in the spine head, they are more common in the neck part, or in immature filopodia (Korobova and Svitkina, 2010). Actin remodeling is tightly regulated in connection with synaptic plasticity and is therefore important in learning and memory as well as in neuronal network functions (Schubert and Dotti, 2007; Honkura et al., 2008; Rudy, 2015). This is in accordance with the accumulating evidence that abnormal actin regulation in dendritic spines plays an important role in different neurological diseases and thus research of actin regulation in dendritic spines is crucial in understanding these pathologies (Joensuu et al., 2018; Pelucchi et al., 2020).
Fluorescent imaging of actin in living cells is a vital tool to investigate cytoskeletal changes in dendritic spines (Koskinen et al., 2012). A plethora of fluorescent probes is already available, from cell penetrating small molecules that label filamentous actin in every cell, such as SiR-actin or phalloidin (Lukinavičius et al., 2014), to genetically encoded fusion proteins that are expressed in individual cells (Belin et al., 2014; Melak et al., 2017). In case of low density or nonoverlapping cells, general actin labeling may be useful, whereas for the intermingling network of neurites in neuronal tissue or cell cultures, it is often more desirable to label only a limited number of neurons.
Expression of fluorescently labeled actin monomers is a widely used method in neuronal actin imaging studies (Star et al., 2002; Hotulainen et al., 2009; Koskinen et al., 2012; Melak et al., 2017); however, introducing new actin monomers might imbalance the F-G-actin treadmilling, leading to alterations in actin-dependent processes (Koskinen and Hotulainen, 2014). Furthermore, fluorescent label can interfere with formin-mediated actin incorporation into the contractile rings during cytokinesis (Chen et al., 2012).
Actin-binding protein probes aim to circumvent this disadvantage by recognizing endogenous actin. Most of these molecules consist of sequences from actin-binding proteins like F-tractin (Johnson and Schell, 2009), utrophin (Patel et al., 2017), and LifeAct (Riedl et al., 2008). Detailed analysis has been previously published regarding the negative effects of utrophin on dendritic morphology, as well as the reliability of F-tractin to label endogenous actin cytoskeleton in neuronal cells (Johnson and Schell, 2009; Melak et al., 2017; Patel et al., 2017). The effects of LifeAct on dendritic morphology have been described rather controversially. LifeAct-GFP has been successfully used in experiments with living neurons (Belin et al., 2014; Panza et al., 2015; Wegner et al., 2017); however, it was reported to have dose-dependent negative effects on various cell types (Flores et al., 2019; Xu and Du, 2021), and certain actin structures are excluded from labeling (Munsie et al., 2009, Kumari et al., 2020). This could be explained by the competition of LifeAct with other actin-binding regulatory proteins (Belyy et al., 2020). An additional but much less investigated actin-labeling tool in neurons is actin-chromobody (AC), a small single-chain camelid antibody specific for actin (Rocchetti et al., 2014; Traenkle and Rothbauer, 2017). LifeAct and AC were shown to cause spine loss upon strong overexpression in vivo, but they have not been yet quantitatively compared and analyzed regarding their effects on dendritic branching, spine morphology, and motility under more physiological expression levels (Wegner et al., 2017).
In the present study, we compared the usability of LifeAct-GFP and AC-GFP with EGFP-actin in cultured neurons. We report that AC-GFP and LifeAct-GFP have similar relative affinity for actin in spines but only EGFP-actin can reveal small differences in actin dynamics between motile filopodial and more mature mushroom-shaped dendritic protrusions. Based on unbiased quantitative morphological evaluations, the tested actin probes show similar colocalization index with phalloidin, but LifeAct is less enriched in dendritic spines. While transfected neurons can tolerate varying expression of LifeAct without severe morphological consequences, overexpression of AC-GFP for 24 h results in decreased dendritic arborization and increased density of thin spines. These results extend previous in vitro and in vivo observations on LifeAct and AC, and provide a direct comparison of their suitability with EGFP-actin to analyze dendritic spine morphology.
RESULTS AND DISCUSSION
EGFP-actin expression reveals differences in actin remodeling between thin and mushroom spines
To quantitatively compare the three different actin probes, we prepared hippocampal neurons from CD1 wild-type murine embryos and let them develop for 12–13 days in vitro (DIV). By this age, cultured neurons extend their neurites, establish synaptic connections, and modify their dendritic spines according to cellular and network activity (Dotti et al., 1988; Yuste, 2010).
We transfected the cells with plasmids encoding EGFP-tagged actin (EGFP-actin), AC (AC-GFP), or LifeAct (LifeAct-GFP). Besides the actin-labeling constructs, mCherry was also coexpressed, providing the possibility to detect spine shape. Following transfection (24 h), time-lapse recordings were taken in every second (Figure 1A, and Supplemental Video 1). As both AC-GFP and LifeAct-GFP label actin in a noncovalent manner, we tested their relative affinity to actin, by measuring fluorescence recovery after photobleaching (FRAP method) in control cells and with a Jasplakinolide-stabilized actin cytoskeleton.
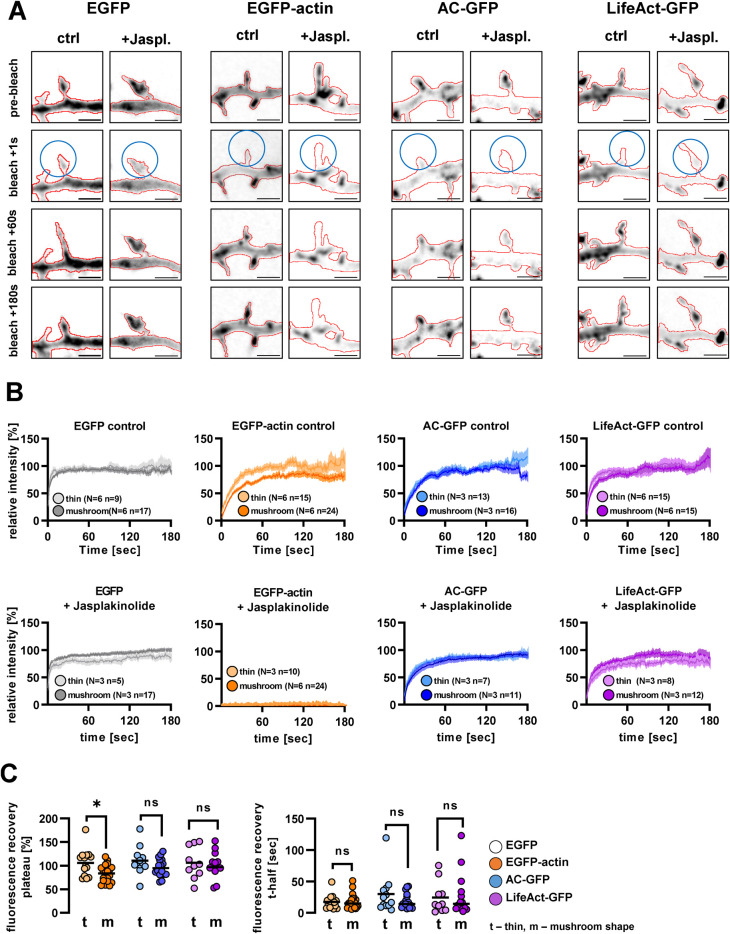
FIGURE 1:
GFP fluorescence recovery after bleaching in cells expressing different actin-labeling fusion proteins. (A) Representative live cell confocal image series of cells expressing EGFP, EGFP-actin, actin-chromobody-GFP or LifeAct-GFP. The GFP signal is shown in inverted grayscale; the outlines of the cell, based on mCherry, are highlighted by red lines. Blue circles show the bleached area. Jasplakinolide (5 µM) was applied for 7–10 min before imaging. Scale bar: 2 µm. (B) Fluorescence recovery curves after photobleaching in control (upper row) and 5 µM jasplakinolide-treated (lower row) cells. Fluorescence was measured in the mCherry positive area and normalized to the mean of the last 10 prebleach values in every spine. Thin and mushroom-shaped morphotypes were distinguished based on apparent spine morphology. (C) Fluorescence recovery plateau and t-half values of the analyzed control spines. Each point represents an individual spine. N, number of independent cultures; n, number of analyzed spines. Sample sizes and colors on panel C are identical to panel B. *, p < 0.05.
Movie S1.
GFP fluorescence recovery after bleaching in cells expressing different actin labelling fusion proteins. Representative live cell confocal time-lapse recordings of GFP (green) together with mCherry (magenta). 5 μM Jasplakinolide was applied for 7-10 min before imaging. Scalebar: 2 μm.
Both thin and mushroom-shaped protrusions were photobleached (see circled areas in Figure 1A) and analyzed separately (Figure 1, B and C). EGFP-expressing cells served as negative control as the fluorescent protein itself lacks actin-binding affinity. EGFP-actin, on the other hand, served as positive control, as it incorporates into the endogenous F-actin.
As expected, EGFP recovered almost immediately upon photobleaching, preventing recovery curve analysis. In the case of the actin probes, one-phase decay curves were fitted to individual fluorescence recovery data, then recovery half-time (t-half) and plateau values were extracted, representing the speed of recovery and the mobile actin fraction, respectively (Figure 1C). Because mushroom-shaped spines have a stable actin cytoskeletal core within the head region (Hlushchenko et al., 2016), we assumed that thin and mushroom-shaped protrusions differ in their ratio of mobile actin. Indeed, in EGFP-actin–expressing cells, plateau values were significantly smaller in mushroom than in thin spines, while recovery half-time values did not differ significantly. In contrast, neurons expressing AC-GFP or LifeAct-GFP had indistinguishable recovery curves between mushroom-shaped and thin protrusions (Figure 1, B and C).
In jasplakinolide-treated neurons, EGFP-actin signal did not recover while AC-GFP and LifeAct-GFP signals reappeared within the bleached protrusions (Figure 1, A and B, and Supplemental Video 1). Our results are in accordance with FRAP data obtained with F-tractin (Johnson and Schell, 2009), showing that fluorescence recovery of both actin-binding protein-based and nanobody-based probes is independent from actin remodeling and depends only on their affinity to F-actin, which is similar for AC-GFP and LifeAct-GFP. In addition, our results suggest that protrusion-specific differences in actin reorganization cannot be revealed when using actin-binding probes as opposed to directly incorporating actin monomers.
Genetically encoded actin-labeling probes do not alter filopodial motility
Dendritic filopodia are small filamentous structures whose protrusion and retraction are driven predominantly by their actin cytoskeleton (Kayser et al., 2008; Hotulainen and Hoogenraad, 2010; Ozcan, 2017). Therefore, we investigated how the different actin probes affect the dynamics of filopodial motility, by comparing the average displacement of intensity-weighted center of mass of selected segmented filopodia (Figure 2 and Supplemental Video 2).
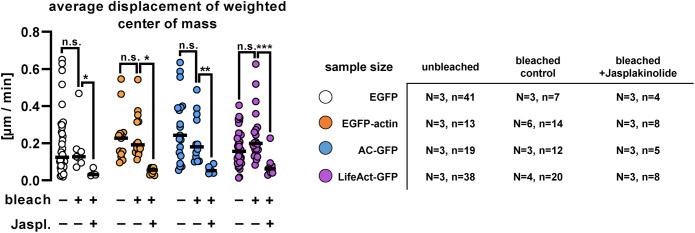
FIGURE 2:
Dendritic filopodial motility in cells expressing EGFP, EGFP-actin, actin-chromobody-GFP, or LifeAct-GFP. Average intensity-weighted center of mass displacement over 60-s time periods in nonbleached and bleached control vs. 5-µM jasplakinolide-treated cells. Each point represents an individual spine. Black lines show medians. Numbers of elements: N, independent cultures; n, analyzed filopodia. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Movie S2.
Filopodial motility in cells expressing different genetically encoded actin probes. Representative mCherry fluorescence confocal time-lapse recordings of filopodia. 5 μM Jasplakinolide was applied for 7-10 min before imaging. Red circles show current center of mass in every frame, yellow lines show the movement track. Scalebar: 1 μm.
We found no significant difference in center of mass displacement between actin probe expressing cells and EGFP-expressing control filopodia (Figure 2), even when the filopodia were bleached (see previous FRAP experiments and Figure 2). As expected, jasplakinolide treatment completely blocked motility in every transfected protrusion (Figure 2). Our data suggest that the tested actin-labeling proteins do not interfere with fast morphological changes in these temporal protrusions.
As expression levels of different actin probes might have dose-dependent effects (see Courtemanche et al., 2013 and Wegner et al., 2017), motility data were compared with somatic fluorescence levels of the investigated constructs. Care was taken to analyze only intact cells with nonfragmented axons and dendrites that contained dendritic spines. EGFP and LifeAct-GFP expression varied on a wide range of fluorescence intensity, whereas intact EGFP-actin and AC-GFP–expressing cells showed lower fluorescence intensities, indicating that high expression of these constructs in the soma might have detrimental effects on cell morphology and survival. We found a negative correlation between somatic fluorescence intensity and filopodial motility in LifeAct-GFP–expressing cells, while motility did not show relative expression dependence in any other experimental groups (Supplemental Figure 1A).
Actin-labeling probes exert distinct effects on dendritic spine morphology and density
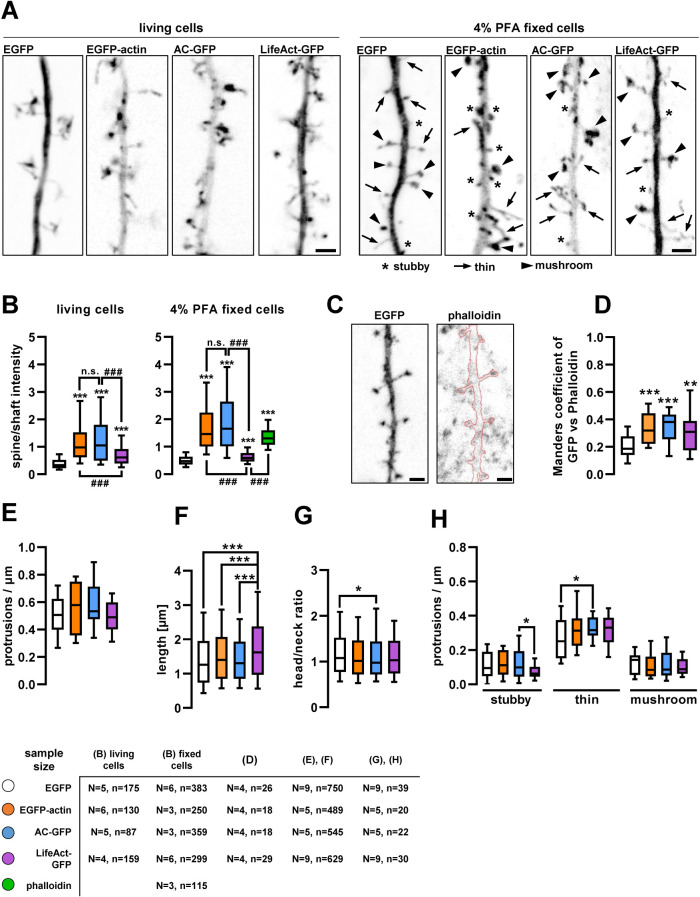
It has been shown that F-actin is enriched in dendritic spines (Korobova and Svitkina, 2010). To compare how reliably actin-labeling constructs highlight F-actin within dendritic spines, we systematically quantified the relative spine-to-shaft GFP fluorescence distribution and colocalization with phalloidin, an established probe for endogenous F-actin (Figure 3, A–D). Compared to cytoplasmic EGFP fluorescence distribution, which is determined by the difference of thickness between the spines and shaft, all actin-labeling probes in fixed cells showed increased spine-to-shaft fluorescence ratio (Figure 3B) and Manders colocalization coefficient with phalloidin (Figure 3D). Relative enrichment was also evident under living conditions, albeit to a lesser extent. In particular, LifeAct-GFP–expressing cells had a significantly lower level of relative enrichment in spines compared with EGFP-actin and AC-GFP (Figure 3, A and B; see the elevated LifeAct-GFP fluorescence intensity within the dendritic shaft). This is likely due to the known high background fluorescence level of LifeAct, originating from its affinity to G-actin (Melak et al., 2017).
FIGURE 3:
Dendritic spine localization of actin-labeling fusion proteins and their effect on dendritic spine morphology. (A) Representative max-projected inverted GFP fluorescence confocal images of dendritic segments. Scale bars: 2 µm. (B) Relative fluorescence intensity in dendritic spines compared with an adjacent shaft segment in live and fixed cell images. (C) Inverted EGFP and phalloidin-Alexa Fluor 633 signal projected from five adjacent focal planes, used for colocalization test. Red highlight indicates the dendritic outline on the phalloidin image. Scale bars: 2 µm. (D) Manders’ colocalization coefficients of the different actin-labeling proteins and EGFP with phalloidin. (E) Protrusion density of dendritic segments expressing EGFP or the actin-labeling fusion proteins. (F) Length and (G) head-to-neck ratio of individual dendritic spines of transfected cells. (H) Protrusion density of different morphotypic spines on transfected dendritic segments. Boxes show median and interquartile range; whiskers show 10–90%. Numbers of elements in B, F, and G: N, independent cultures; n, analyzed spines. In D and H: N, independent cultures; n, analyzed dendrites. *, p < 0.05; **, p < 0.01; ***, p < 0.001. ###, p < 0.001. In B, *** is compared with EGFP control, and ### is compared with the connected groups.
For morphometric analysis, fixed spines on high-resolution confocal images along secondary dendritic branches were categorized into three main morphotypes, in accordance with the literature: stubby, thin, and mushroom protrusions (see Supplemental Figure 2 and Figure 3A; Peters and Kaiserman-abflamof 1970; Bourne and Harris 2008; Rochefort and Konnerth 2012). LifeAct-GFP–expressing spines were significantly longer compared with all other conditions (Figure 3F), leading to a decreased ratio of short stubby spine morphotypes (Figure 3H). At the same time, head/neck ratio was slightly but significantly decreased in AC-GFP–expressing cells (Figure 3G) resulting in more thin protrusions while overall protrusion density was not affected (Figure 3E). Comparing somatic fluorescence intensity to morphometric data revealed that only EGFP-actin intensity was positively correlated with total protrusion density, while none of the other morphometric parameters depended on relative expression levels (Supplemental Figure 1, B–D).
Our results are in line with previous work using AAV-mediated and long-term, hSyn promoter-driven expression of LifeAct and AC in vivo as well as in primary cell cultures (Wegner et al., 2017). Regardless that we used 24-h-long survival time of liposome-based transfection and a general CMV promoter, our more detailed quantitative analysis indicates that moderate level expression of EGFP-actin, LifeAct-GFP, or AC-GFP does not result in drastic changes of dendritic morphology.
AC expression impairs dendritic arborization within 24 h
Besides dendritic spine morphology, we also investigated the intracellular distribution of the tested actin probes within transfected dendrites. All constructs were present in both the somatodendritic and axonal compartments (Figure 4A). We did not observe striking differences in axonal or growth cone morphology between the different groups.
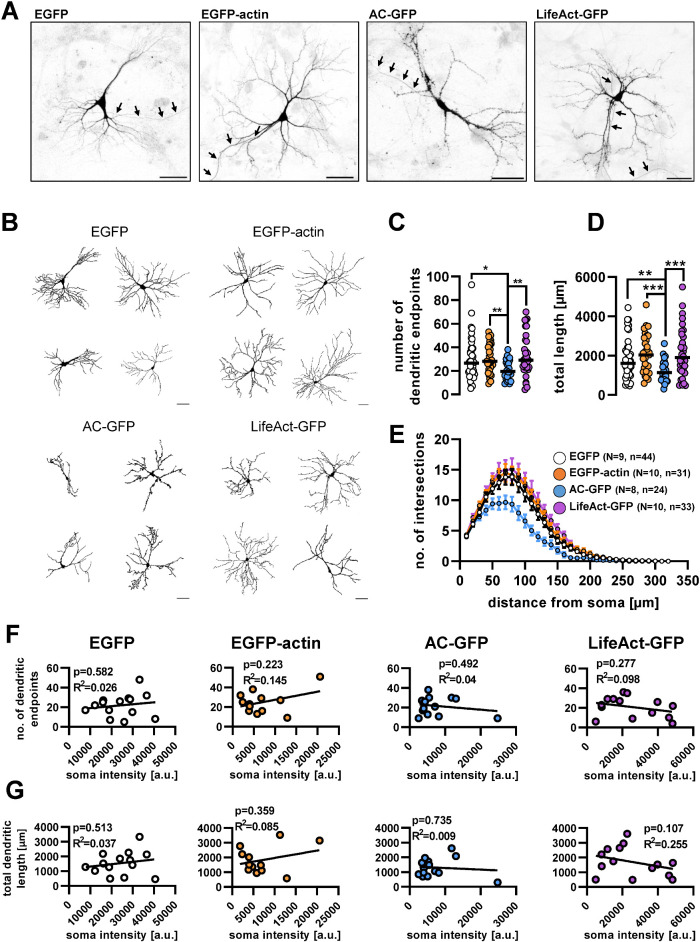
FIGURE 4:
Cellular distribution of actin-labeling fusion proteins and their effect on dendritic morphology. (A) Confocal images of cultured hippocampal neurons expressing actin-labeling fusion proteins. The images are shown as inverted GFP fluorescence. Arrows point at the axons. Scale bar: 50 µm. (B) Representative dendritic outlines used for morphometric analysis. Scale bar: 50 µm. (C) Number of dendritic endpoints and (D) total dendritic length of neurons expressing the different fusion proteins. Each data point presents an individual cell; black lines show median. (E) Number of dendritic intersections with increasing radius circles starting from the soma. Data presented as mean ± SEM. (F) Number of dendritic endpoints and (G) total dendritic length vs. GFP intensity in the soma. In F and G each circle represents an individual cell. Numbers of elements: N, independent cultures; n, individual cells. **, p < 0.01; ***, p < 0.001.
As Patel and colleagues reported that certain actin-binding protein-based probes in developing neurons influence dendritic morphology (Patel et al., 2017), we analyzed whether dendritic arborization is altered within 24 h after the transfection of the tested probes. Confocal images were quantitatively evaluated using the Ilastik machine learning program to segment dendritic trees and dendritic endpoints (Berg et al., 2019). Examples of the analyzed dendritic outlines from each tested group are shown in Figure 4B. According to our data, neither EGFP-actin nor LifeAct-GFP expression altered dendritic endpoint number, total dendrite length, or the extent of dendritic arborization when compared with the EGFP control (Figure 4, C–E), consistent with the existing literature (Belin et al., 2014; Patel et al., 2017). We did not find a significant correlation between the fluorescence intensity of the soma and dendritic morphological parameters in any cases (Figure 4, F and G).
On the other hand, AC-GFP–expressing cells had significantly fewer dendritic endpoints (Figure 4C) and shorter total dendritic length (Figure 4D) than control, EGFP-expressing neurons. In addition, AC-GFP–expressing neurons had significantly shorter dendrites and fewer branches compared with the other constructs (Figure 4E). Although this fusion protein, when expressed in developing zebrafish embryos at various stages of development, did not grossly interfere with cell function (Panza et al., 2015; Melak et al., 2017), in the absence of a detailed analysis of neuronal morphology, altered dendritic arborization in fish embryos cannot be excluded.
Overall, our results support the view that all tested genetically encoded actin probes are generally appropriate to visualize dendritic actin cytoskeleton structures in cultured neurons but with certain limitations. EGFP-actin, similarly to phalloidin, is highly enriched in spines but its expression can change spine density in a dose-dependent manner. While it reveals small differences in actin dynamics between filopodia and mushroom spines, the interpretation is limited by its previously reported interference with formin-mediated actin remodeling (Chen et al., 2012). AC-GFP also colocalizes with phalloidin and shows enrichment in spines; however, it has adverse effects on dendritic morphology. LifeAct-GFP expression can reach a higher fluorescence level than the previous two constructs without affecting dendritic morphology. On the other hand, it shows a higher fluorescence background compared with the other actin probes studied and can affect dendritic filopodial motility.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Animal handling
Wild-type CD1 mice were housed in the animal facility at 22 ± 1°C with 12-h light/dark cycles and ad libitum access to food and water. All experiments complied with local guidelines and regulations for the use of experimental animals, in agreement with European Union and Hungarian legislation.
Cell cultures
Embryonic hippocampal cultures were prepared from CD1 mice on embryonic day 17-18, according to Morales et al. (2021). Cells were seeded onto poly-l-lysine (Sigma-Aldrich; #P5899) and laminin- (RandD Systems; #3446-005-01; 4–8 µg/cm2) coated glass-bottom Petri dishes (Greiner Bio-One; #627975) or onto PLL-laminin–coated glass coverslips (Marienfeld-Superior; #0111520) in 24-well plates (Greiner Bio-One; #66210). Glass coverslips had been previously plasma cleaned for 5 min each side using O2 plasma. Cell density was 1.4 × 105 cells per coverslip or compartment. The cultures were maintained for 12–13 d in vitro in 5% CO2 at 37°C before experiments.
The cells were initially seeded in NeuroBasal PLUS (ThermoFisher Scientific; #A35829-01) culture medium supplemented by 2% B27 PLUS (ThermoFisher Scientific; #A3582801), 5% fetal bovine serum (PAN Biotech; #P30-3309), 0.5 mM GlutaMAX (ThermoFisher Scientific; #35050-038), 40 μg/ml gentamicin (Sigma; #G1397), and 2.5 μg/ml amphotericin B (Life Technologies; #15290-026). On the fifth DIV, half of the medium was changed to BrainPhys (StemCell Technologies; #05790) supplemented with 2% SM1 (StemCell Technologie;s #05711), 40 μg/ml gentamicin, and 2.5 μg/ml amphotericin B medium. On DIV 9, one-third of the medium was again changed to BrainPhys supplemented with SM1, 40 μg/ml gentamicin and 2.5 μg/ml amphotericin B medium. Depending on glial confluency, on DIV4-6, 10 µM CAR (cytosine-arabinofuranozide; Sigma-Aldrich; #C6645) was added to the cultures.
Transfection and chemical treatment
Transfection was carried out on DIV 12-13 using lipofectamine STEM reagent (ThermoFisher Scientific; STEM00001) according to the manufacturer’s guide. The following plasmid constructs were used: pmCherry-N1 (Clontech), pEGFP-N1 (Clontech), pEGFP-actin (Clontech), AC-TagGFP2 (AC-GFP; Chromotek), and LifeAct-TagGFP2 (Clontech). Each GFP-conjugated protein was coexpressed with mCherry to highlight neuronal morphology. After transfection (24 h), cells were either fixed or used for live cell imaging.
Jasplakinolide (HelloBio; #HB3946) was used in a final concentration of 5 μM on glass-bottom Petri dishes mounted for live cell imaging, 7–15 min before imaging, and was present throughout the imaging session.
Confocal microscopy in fixed samples
The cultures were fixed using 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min, followed by 3 × 5 min PBS washes. For membrane permeabilization, 0.1% Triton X-100 dissolved in PBS was applied for 5 min, followed by a PBS rinse. The coverslips were incubated in phalloidin-Alexa Fluor 633 conjugate (Invitrogen; #A22284) for 1.5 h to label endogenous F-actin, followed by 3 × 5 min PBS washes. Coverslips were mounted in ProLong Diamond (Invitrogen; #P36961).
Images were taken with a Zeiss LSM 800 microscope with a Plan-Apochromat 20×/0.8 dry objective for the dendritic tree analysis and GFP intensity measurement, and with a Plan-Apochromat 63×/1.4 oil objective for the dendritic spine density analysis. Z-stack images were taken in 0.7 and 0.2 μm intervals, respectively.
Live cell imaging and FRAP experiments
For FRAP experiments, the culture medium was changed to a custom-made imaging buffer containing 142 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 1 mM NaH2PO4, 0.8 mM MgSO4, 5 mM glucose, and 25 mM HEPES with a pH of 7.4. Time-lapse images were taken with a Zeiss Axio Observer microscope equipped with a CSU-X1 spinning disk module, using a Plan-Apochromat 100×/1.46 oil objective. Selective photobleaching of the GFP signal was done by a separately controlled RAPP UGA42 laser, emitting a 473-nm laser beam. Images were taken every second for 360 s. After the first 10–20 images were taken, 5–10 circular spots were bleached separately. Fluorescence recovery was recorded for at least 180 s postbleach in every case (Supplemental Video 1).
Image analysis methods
Dendritic branching was analyzed using Fiji and Ilastik softwares. Z-stack images were max-projected and a two-stage Ilastik random forest model was trained to segment the foreground (the neuronal dendritic tree) and background in the pictures, followed by manual correction. A second Ilastik model was trained to segment the binary images further, adding dendritic skeleton and endpoints. Sholl analysis was carried out on the segmented images, using the built-in Fiji function. Dendritic endpoints were counted, and image skeletons were analyzed also by the corresponding Fiji functions (Supplemental Figure 2A).
Dendritic spine densities and morphotypes were analyzed on max-projected z-stack images as well. Here, Ilastik models were trained to segment first, the dendrites and background, and second, the intraspine components: base, neck, and head. A separate third model was trained to draw a line inside the spines and the dendrite so their running length could be measured. Spine density and morphotype data were then summarized in Fiji: dendritic length was measured, spines counted, and their length, base dimeter, and head diameter were measured (Supplemental Figure 2B). Three morphotypes were distinguished: stubby spines that were shorter than 0.8 μm, thin spines that were longer than 0.8 μm and their head/neck diameter ratios were smaller than 1.5, and mushroom spines that were longer than 0.8 μm and their head/neck diameter ratios higher than 1.5. The ratios used for morphotyping were according to Morales et al. (2021).
The spine-to-shaft ratio of GFP fluorescence was measured on max-projected z-stack images that had been used for spine density measurements in fixed cells, or the first frame of live cell recordings. In both cases Ilastik models were trained to segment background, shaft, and spine. The spines were then enlarged as circles and taken as regions of interest (ROI). Spine pixels in the ROI were used as a mask for spine intensity, shaft pixels for shaft intensity, and background pixels for background intensity (Supplemental Figure 2C). Hence, the spine intensities were normalized to an adjacent dendritic segment only instead of the full branch, which had visible heterogeny in intensity, as well as in the case of background, which was also heterogeneous.
Colocalization of the expressed fluorescent proteins and phalloidin was measured on images consisting of multiple optical slices. The previously described segmentation was used to highlight the dendrites and spines. Manders’ coefficient was calculated between the GFP and phalloidin signals using the Coloc2 plugin in Fiji.
Two-channel FRAP recordings were analyzed with a custom-made Fiji plugin called FRAP2ch in the following way: in the bleached spines, mCherry signal was used as a mask to select the ROI in the GFP channel, which was then measured, in every time frame. Every spine intensity was normalized to the intensity of a nearby dendritic segment, which were both background-corrected. Then, spine intensities were normalized to the mean of the last 10 prebleach time points. A one-phase decay equation was then fitted to the postbleach values, according to Koskinen and Hotulainen (2014). t-half and plateau values were calculated for each spine individually, using GraphPad Prism.
To measure dendritic spine motility, another custom-made Fiji plugin, called DFMA was used (Tárnok et al., 2015). Here, time-lapse images were segmented with Ilastik, to provide a standard quality input for the DFMA plugin, which is known to depend on signal quality. The segmented images of filopodial protrusions were used to locate their center of mass on every time point. Then, cumulative displacement curves were calculated, and the 60-s points were compared and statistically analyzed (Supplemental Video 2).
All used Fiji macro codes and Ilastik models can be accessed on the GitHub account of A.I.
Statistical analysis
Statistical analysis of the acquired data was carried out using GraphPad Prism software 6. First, a Shapiro-Wilk test was used to determine the normality of distribution for each dataset. Then, a comparison was done depending on the normality: for datasets showing Gaussian distribution one-way ANOVA tests were carried out and a Kruskal-Wallis test for those showing nonparametric distribution.
Supplementary Material
Acknowledgments
This work was supported by a grant from the German Research Foundation (DFG Grant no. HA-357/11-3) to A.H. and a travel exchange program by German Academic Exchange Service (DAAD; PPP Hungary 57392635 and 57602951; 449419 and 169036 by Tempus Foundation [TKA]) to A.H. and K.S., respectively, as well as by the Hungarian Brain Research Program (2017-1.2.1-NKP-2017-00002) and by the VEKOP-2.3.3-15-2016-00007 infrastructural grants. A.I. and D.N-H. are grateful for the support of the ÚNKP-21-3 New National Excellence Program of the Ministry for Innovation and Technology from the source of the National Research, Development and Innovation Fund, and National Talent Program NTP-NFTÖ-21-B-0250, respectively.
Abbreviations used:
- AC-GFP
Actin-Chromobody-TagGFP2
- DIV
days in vitro
- FRAP
fluorescence recovery after photobleaching
- LifeAct-GFP
LifeAct-TagGFP2
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-08-0331) on March 29, 2023.
REFERENCES
- Amtul Z, Atta-Ur-Rahman (2015). Neural plasticity and memory: molecular mechanism. Rev Neurosci 26, 253–268. [DOI] [PubMed] [Google Scholar]
- Arellano JI, Benavides-piccione R, Defelipe J, Yuste R (2007). Ultrastructure of dendritic spines: correlation between synaptic and spine morphologies. Front Neurosci 1, 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belin BJ, Goins LM, Mullins RD (2014). Comparative analysis of tools for live cell imaging of actin network architecture. Bioarchitecture 4, 189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyy A, Merino F, Sitsel O, Raunser S (2020). Structure of the Lifeact–F-actin complex. PloS Biol 18, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg S, Kutra D, Kroeger T, Straehle CN, Kausler BX, Haubold C, Schiegg M, Ales J, Beier T, Rudy M, et al. (2019). Ilastik: interactive machine learning for image analysis. Nat Methods 16, 1226–1232. [DOI] [PubMed] [Google Scholar]
- Bosch M, Hayashi Y (2015). Structural plasticity of dendritic spines. Curr Opin Neurobiol 22, 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne JN, Harris KM (2008). Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci 31, 47–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD (2010). A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol 11, 237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Nag S, Pollard TD (2012). Formins filter modified actin subunits during processive elongation. J Struct Biol 177, 32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtemanche N, Pollard T, Chen Q (2013). Avoiding artefacts when counting polymerized actin in live cells with LifeAct fused to fluorescent proteins. Nat Cell Biol 18, 676–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti CG, Sullivan CA, Banker GA (1988). The establishment of polarity by hippocampal neurons in culture. J Neurosci 8, 1454–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores LR, Keeling MC, Zhang X, Sliogeryte K, Gavara N (2019). Lifeact-TagGFP2 alters F-actin organization, cellular morphology and biophysical behaviour. Sci Rep 91, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlushchenko I, Koskinen M, Hotulainen P (2016). Dendritic spine actin dynamics in neuronal maturation and synaptic plasticity. Cytoskeleton 73, 435–441. [DOI] [PubMed] [Google Scholar]
- Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GCR, Kasai H (2008). The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron 57, 719–729. [DOI] [PubMed] [Google Scholar]
- Hotulainen P, Hoogenraad CC (2010). Actin in dendritic spines: connecting dynamics to function. J Cell Biol 189, 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P, Llano O, Smirnov S, Tanhuanpää K, Faix J, Rivera C, Lappalainen P (2009). Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol 185, 323–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensuu M, Lanoue V, Hotulainen P (2018). Dendritic spine actin cytoskeleton in autism spectrum disorder. Prog Neuro-Psychopharmacol Biol Psychiatry 84, 362–381. [DOI] [PubMed] [Google Scholar]
- Johnson HW, Schell MJ (2009). Neuronal IP3 3-kinase is an F-actin-bundling protein: role in dendritic targeting and regulation of spine morphology. Mol Biol Cell 20, 5166–5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H, Fukuda M, Watanabe S, Hayashi-Takagi A, Noguchi J (2010). Structural dynamics of dendritic spines in memory and cognition. Trends Neurosci 33, 121–129. [DOI] [PubMed] [Google Scholar]
- Kayser MS, Nolt MJ, Dalva MB (2008). EphB receptors couple dendritic filopodia motility to synapse formation. Neuron 59, 56–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konietzny A, Bär J, Mikhaylova M (2017). Dendritic actin cytoskeleton: structure, functions, and regulations. Front Cell Neurosci 11, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Svitkina TM (2010). Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell 21, 165–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskinen M, Bertling E, Hotulainen P (2012). Methods to measure actin treadmilling rate in dendritic spines. Methods Enzymol 505, 47–58. [DOI] [PubMed] [Google Scholar]
- Koskinen M, Hotulainen P (2014). Measuring F-actin properties in dendritic spines. Front Neuroanat 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A, Kesarwani S, Javoor MG, Vinothkumar RK, Sirajuddin M (2020). Structural insights into actin filament recognition by commonly used cellular actin markers. EMBO J 39, e104006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman J, Yasuda R, Raghavachari S (2012). Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci 13, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukinavičius G, Reymond L, D’Este E, Masharina A, Göttfert F, Ta H, Güther A, Fournier M, Rizzo S, Waldmann H, et al. (2014). Fluorogenic probes for live-cell imaging of the cytoskeleton. Nat Methods 11, 731–733. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Bear MF (2004). LTP and LTD: an embarrassment of riches. Neuron 44, 5–21. [DOI] [PubMed] [Google Scholar]
- Melak M, Plessner M, Grosse R (2017). Actin visualization at a glance. J Cell Sci 130, 525–530. [DOI] [PubMed] [Google Scholar]
- Morales CO, Ignácz A, Bencsik N, Sziber Z, Rátkai AE, Lieb WS, Eisler SA, Szűcs A, Schlett K, Hausser A (2021). Protein kinase D promotes activity-dependent AMPA receptor endocytosis in hippocampal neurons. Traffic 22, 454–470. [DOI] [PubMed] [Google Scholar]
- Munsie LN, Caron N, Desmond CR, Truant R (2009). Lifeact cannot visualize some forms of stress-induced twisted f-actin. Nat Methods 6, 317. [DOI] [PubMed] [Google Scholar]
- Nägerl UV, Eberhorn N, Cambridge SB, Bonhoeffer T (2004). Bidirectional activity-dependent morphological plasticity in hippocampal neurons. Neuron 44, 759–767. [DOI] [PubMed] [Google Scholar]
- Nishiyama J, Yasuda R (2015). Biochemical computation for spine structural plasticity. Neuron 87, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan AS (2017). Filopodia: a rapid structural plasticity substrate for fast learning. Front Synaptic Neurosci 9, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panza P, Maier J, Schmees C, Rothbauer U, Söllner C (2015). Live imaging of endogenous protein dynamics in zebrafish using chromobodies. Development 142, 1879–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Fok SYY, Stefen H, Tomanić T, Parić E, Herold R, Brettle M, Djordjevic A, Fath T (2017). Functional characterisation of filamentous actin probe expression in neuronal cells. PLoS One 12, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelucchi S, Stringhi R, Marcello E (2020). Dendritic spines in Alzheimer’s disease: how the actin cytoskeleton contributes to synaptic failure. Int J Mol Sci 21, 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A, Kaiserman-Abflamof IR (1970). The small pyramidal neuron of the rat cerebral cortex. The perikaryon, dendrites and spines. Am J Anat 127, 321–355. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Web Z, et al. (2008). Lifeact: A versatile marker to visualize F-actin. Nat Methods 5, 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochetti A, Hawes C, Kriechbaumer V (2014). Fluorescent labelling of the actin cytoskeleton in plants using a cameloid antibody. Plant Methods 10, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochefort NL, Konnerth A (2012). Dendritic spines: from structure to in vivo function. EMBO Rep 13, 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy JW (2015). Actin dynamics and the evolution of the memory trace. Brain Res 1621, 17–28. [DOI] [PubMed] [Google Scholar]
- Schubert V, Dotti CG (2007). Transmitting on actin: synaptic control of dendritic architecture. J Cell Sci 120, 205–212. [DOI] [PubMed] [Google Scholar]
- Star EN, Kwiatkowski DJ, Murthy VN (2002). Rapid turnover of actin in dendritic spines and its regulation by activity. Nat Neurosci 5, 239–246. [DOI] [PubMed] [Google Scholar]
- Tárnok K, Gulyás M, Bencsik N, Ferenc K, Pfizenmaier K, Hausser A, Schlett K (2015). A new tool for the quantitative analysis of dendritic filopodial motility. Cytometry, Part A 87, 89–96. [DOI] [PubMed] [Google Scholar]
- Traenkle B, Rothbauer U (2017). Under the microscope: single-domain antibodies for live-cell imaging and super-resolution microscopy. Front Immunol 8, 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner W, Ilgen P, Gregor C, Van Dort J, Mott AC, Steffens H, Willig KI (2017). In vivo mouse and live cell STED microscopy of neuronal actin plasticity using far-red emitting fluorescent proteins. Sci Rep 7, 11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Du S (2021). Overexpression of Lifeact-GFP disrupts F-actin organization in cardiomyocytes and impairs cardiac function. Front Cell Dev Biol 9. 746818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R (2010). Dendritic Spines. Cambridge, MA: MIT Press Scolarship Online. [Google Scholar]
- Yuste R, Bonhoeffer T (2004). Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat Rev Neurosci 5, 24–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.