Abstract
Around the world, more the 700,000 individuals die by suicide every year. It is necessary to understand the mechanisms associated with suicidal behavior. Recently, an increase in gene expression studies has been in development. Through a systematic review, we aimed to find a candidate gene in gene expression studies on postmortem brains of suicide completers. Databases were systematically searched for published studies. We performed an online search using PubMed, Scopus and Web of Science databases to search studies up until May 2023. The terms included were “gene expression”, “expressed genes”, “microarray”, “qRT–PCR”, “brain samples” and “suicide”. Our systematic review included 59 studies covering the analysis of 1450 brain tissues from individuals who died by suicide. The majority of gene expression profiles were obtained of the prefrontal cortex, anterior cingulate cortex, dorsolateral prefrontal cortex, ventral prefrontal cortex and orbital frontal cortex area. The most studied mRNAs came of genes in glutamate, γ-amino-butyric acid and polyamine systems. mRNAs of genes in the brain-derived neurotrophic factor, tropomyosin-related kinase B (TrkB), HPA axis and chemokine family were also studied. On the other hand, psychiatric comorbidities indicate that suicide by violent death can alter the profile of mRNA expression.
Keywords: expression analysis, brains, suicide, systematic review
1. Introduction
Suicide is a serious global health problem and one of the primary causes of death worldwide [1]. Moreover, it is one of the most devastating outcomes of individuals with psychiatric disorders [2]. For instance, completed suicide is regarded as the deliberate act of killing oneself and succeeding; commonly, individuals who die by suicide go through a series of suicidal ideations and suicide attempts before completion [1]. There are more studies regarding the psychopathology, risk factor profiles, neurobiology, and neurochemistry of suicide completers (SC) than other traits of the suicide spectrum [3]. Additionally, many factors could be associated to exacerbate or suppress the expression of the genes, factors such as a polymorphism, environment such as childhood abuse, and exposition to trauma such as wars in general post-traumatic stress.
Therefore, several studies have aimed to identify potential suicide biomarkers via the examination of postmortem tissue [4]. Molecular markers and processes identified in postmortem designs may reflect a long-standing risk and/or a more proximal precursor of death by suicide [5]. Specifically, brain tissue has complex patterns of neurochemical and neuroplasticity alterations linked to a variety of psychiatric diseases including suicide [6]. With regards to completed suicide, several reports have proposed a potential causal impact of a differential gene expression on this complex psychiatric trait [7,8]. Therefore, examining the gene expression could help us identify functional variants that might play a more direct role in the SB predisposition [9].
Up until today, the impact of these candidate genetic variants and the risk of suicide is not completely understood [10]. Hence, it is important to analyze conceptual frameworks that enhance our understanding of death by suicide as part of the suicide–spectrum behavior, which could guide us towards supporting or performing hypotheses. Our primary aim was to perform a detailed and updated systematic review of gene expression of postmortem studies from brain tissue of individuals who died by suicide.
2. Methods
This study was performed using a predetermined protocol in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) statement (Table S1). The registry of the systematic review is CDR42021274922 (by Gonzalez Castro and Carlos Tovilla).
2.1. Search Strategy
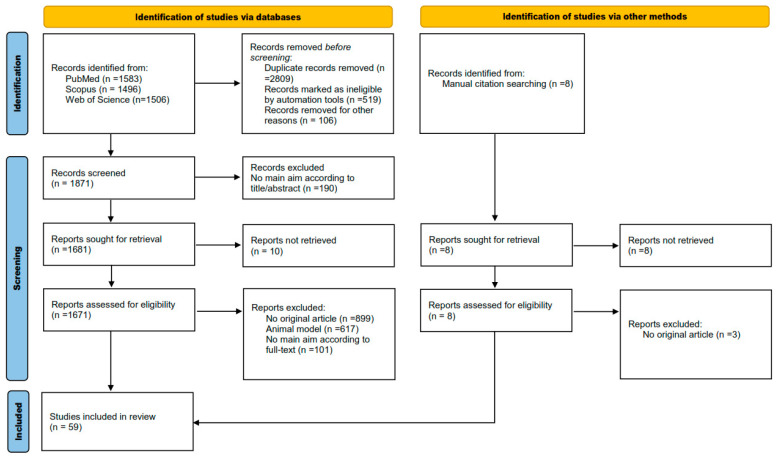
PubMed, Scopus and Web of Science databases were used to search for relevant studies published up until May 2023. The search terms were the following: (“gene expression” OR “expressed genes” OR “microarray” OR “qRT–PCR” OR “brain samples”) AND (“suicide” OR “suicidal” OR “suicidality”). References in these studies were examined to identify other possible papers that were not indexed in the databases used. (Figure 1 shows the strategy flowchart.) Search results were uploaded into EndNote X9 for a first screening; subsequently, those files were exported to Covidence for a formal screening.
Figure 1.
Flow diagram of the search criteria for the systematic review.
2.2. Inclusion and Exclusion Criteria
To be eligible, the studies had to meet the following criteria: (a) full-text articles, (b) case-control designs, (c) evaluated the association between gene expression and completed suicide, (d) included candidate genes related to suicide risk, (e) analyzed the gene expression using a microarray, next-generation sequencing or a quantitative RT-PCR, (f) published in English, (g) published in peer-reviewed journals and (h) cause of death in controls (anything except suicide).
The exclusion criteria were as follows: (a) data available of no use, (b) non-research papers or (c) duplicates.
2.3. Data Extraction
Data of each retrieved publication were independently collected in duplicate by two investigators (González-Castro and Tovilla-Zárate) following a standard procedure. Disagreements were solved through discussion until reaching a consensus. The following data were extracted: (a) first author’s name, (b) publication year, (c) country, (d) gene expression candidate, (e) laboratory methods, (f) suicide methods (cases), (g) diagnostics, (h) sample size, (i) mean age, (j) range age, (k) gender proportion in cases and controls, (l) RNA integrity, (m) postmortem interval and (n) pH of brain. These characteristics were gathered in both cases and controls.
2.4. Quality Assessment
The Newcastle–Ottawa Scale (NOS) was applied to assess the quality of the eligible articles. NOS involves three perspectives: study group selection, group comparability and whether the exposure or the outcome of interest for a case–control study is listed in the scale. Each study can receive a maximum of nine stars. Furthermore, all studies were critically appraised using the ROBINS-I tool according to the intervention bias, missing data, confounding factors, outcome bias, report, selection and overall risk bias.
3. Results
3.1. Selection of the Studies with Gene Expression Analysis
We used the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines for reporting methodology. Our search provided 4585 studies from electronic databases and 8 from other sources. After the first stage of removed records before screening, 1871 were analyzed. Then, detailed screening showed that 59 studies were eligible for qualitative synthesis in the current systematic review [5,7,8,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66]. The process of the study selection is depicted in Figure 1.
3.2. Characteristics of the Studies
The main population analyzed in the included studies was from North America: the United States of America, Canada and Mexico. Other less studied countries were Hungary, Japan, Slovenia, Spain, Singapore, Germany and Sweden. Concerning the regions and structures of the brain analyzed for gene expression in suicide completers, the most frequent were the prefrontal cortex, anterior cingulate cortex, dorsolateral pre-frontal cortex, ventral prefrontal cortex, orbital frontal cortex, hypothalamus, amygdala and hippocampus. The majority of the studies performed or validated the gene expression by quantitative RT-PCR, microarrays or a western blot analysis (Table 1).
Table 1.
Principal features of studies that searched for an association between gene expression and suicide.
| First Author | Year of Publication | Location | Brodmann Area | GE Lab Method | Some Genes Analyzed | NOS |
|---|---|---|---|---|---|---|
| USA | ||||||
| Hiroi N [11] | 2001 | AMY, THAL, PG, HPC, CB, SN | NA | qRT–PCR | CRH-R1, CRH-R2 | 7 |
| Dwivedi Y [12] | 2001 | PFC, HPC, CB | 8, 9, 10 | qRT–PCR | MKP2, ERK1/2 | 8 |
| Dwivedi Y [13] | 2003 | PFC, HPC | 9 | qRT–PCR | BNDF, TrkB | 9 |
| Sibille E [14] | 2004 | PFC | 9, 47 | Microarray | BDNF, TrkB, CREB, HTR1A, HTR2C, ADRA1A, ADRA2B | 9 |
| Choudary PV [16] | 2005 | ACC, DLPFC | 24, 9, 46 | Microarray | GABAAα1, GABAAß3 | 8 |
| Kim S [21] | 2007 | PFC | 46/10 | Microarray | PLSCR4, EMX2 | 6 |
| Garbett K [25] | 2008 | PFC | NA | Microarray | TOB1, NFIA, TLOC, AL119182, HTR2A | 9 |
| Pandey GN [28] | 2009 | PFC, HPC | 9 | qRT–PCR | GSK-3b | 8 |
| Simmons M [33] | 2010 | DLPFC | NA | qRT–PCR | ADAR1 | 7 |
| Choi K [36] | 2011 | PFC, HPC | 46 | Microarray | CAMK2B, CDK5, MAPK9, PRKCI | 7 |
| Sequeira A [40] | 2012 | DLPFC, ACC, NACC | NA | Microarray | 5-HT2A, MT1E, MT1F, MT1G, MT1H, MT1X, MT2A | 8 |
| Galfalvy H [42] | 2013 | DLPFC, ACC | 9, 24 | Microarray | CYP19A1, MBNL2, KTBBD2, FOXN3, DSC2, CD300LB | 8 |
| Ren X [43] | 2013 | PFC, HPC | 9 | qRT–PCR | GSK-3b, b-catenin | 8 |
| Pandey GN [45] | 2013 | PFC, HPC, AMY | 9 | qRT–PCR | GR-α, GR | 7 |
| Gray AL [51] | 2015 | DLPFC | NA | qRT–PCR | GRIN2B, GRIK3, GRM2 | 9 |
| Fuchsova B [52] | 2015 | PFC | 9 | qRT–PCR | GPM6A, GPM6B | 9 |
| Gray AL [51] | 2015 | DLPFC | NA | qRT–PCR | GRIN2B, GRIK3, GRM2 | 9 |
| Fuchsova B [52] | 2015 | PFC | 9 | qRT–PCR | GPM6A, GPM6B | 9 |
| Zhao J [53] | 2015 | DLPFC, ACC | 24, 9 | qRT–PCR | CRH, NIDD | 9 |
| Yin H [54] | 2016 | DLPFC | 9 | Microarray | NR3C1 | 9 |
| Pandey GN [55] | 2016 | PFC | 9 | qRT–PCR | SKA2 | 8 |
| Pantazatos SP [57] | 2017 | DLPFC | 9 | NGS | MTRNR2L8, SERPINH1 | 7 |
| Zhang L [61] | 2020 | DLPFC, ACC | 46, 24 | qRT–PCR | P2RY12 | 8 |
| Zhang Lb [62] | 2020 | DLPFC, ACC | 46, 24 | qRT–PCR | TREM2, P2RY12 | 8 |
| Yoshino Y [5] | 2020 | DLPFC | 9 | Microarray | GRP78, GRP94, ATF4C | 8 |
| Pandey GN [7] | 2021 | PFC | 9 | qRT–PCR | CXCL1, CXCL2, CXCL3, CCL2 | 8 |
| Canada | ||||||
| De Luca V [18] | 2006 | DLPFC | 46 | qRT–PCR | TPH2 | 8 |
| Sequeira A [19] | 2006 | OC, DLPFC, MC | 4, 8/9, 11 | Microarray | SSAT | 7 |
| Sequeira A [20] | 2007 | AMY, HPC, ACG, PCG | 24, 29 | Microarray | ADCY8, APLP2 | 7 |
| Feldcamp LA [24] | 2008 | DLPFC | 46 | qRT–PCR | DARPP-32 | 9 |
| McGowan PO [26] | 2009 | HPC | NA | qRT–PCR | NR3C1 | 7 |
| Ernst C [27] | 2009 | FC, CB | 4, 6, 10, 11, 44, 45, 46, 47, 8/9 | Microarray | TrkB.T1 | 6 |
| Klempan TA [29] | 2009 | OFC, IFG | 4, 6, 8/9, 10, 11, 20, 21, 24, 29, 38, 44, 45, 46, 47 | Microarray | QKI | 9 |
| Sequeira A [30] | 2009 | ACC | NA | Microarray | GABARAPL1, GABARA4, GABARB1, GRIA3, GRIA4, GRIA1 | 9 |
| Lalovic A [31] | 2010 | DLPFC, OFC, VPFC | 8/9, 11, 47 | Microarray | FADS1, LEPR, PIK3C2A, SCD | 8 |
| Fiori LM [37] | 2011 | AMY, CB, HPC, HPT, NACC, THAL | 4, 6, 8/9, 10, 11, 20, 21, 24, 29, 38, 44, 45, 46, 47 | Microarray | SAT1, ALDH3A2, AMD1, ARG2 | 7 |
| Smalheiser N [39] | 2012 | PFC | 9 | RT-PCR | DMNT3b, BCL2 | 9 |
| Labonté B [41] | 2013 | HPC, DG | NA | Microarray | NR2E1, GRM7 | 8 |
| Gross J [46] | 2013 | 44 | Microarray | OAZ1, OAZ2, AMD1, ARG2 | 7 | |
| Lopez JP [47] | 2014 | PFC | 44 | qRT–PCR | SAT1, SMOX | 7 |
| Nagy C [56] | 2017 | MDTHAL, CN, CBTX, CTX | 4, 17 | qRT–PCR | CX30, CX43 | 8 |
| Postolache TT [64] | 2020 | DLPFC, ACC | 46, 24, 32, 33 | qRT–PCR | CRAMP | 9 |
| Squassina A [65] | 2020 | ACG | 24 | qRT–PCR | PRKAB2, CREB1, PTEN, PRKAG1, PTPN11, INSR | 9 |
| Mexico | ||||||
| Cabrera B [60] | 2019 | PFC | 9 | Microarray | ARL16, KLHL28, SUCLA2, ATP6V0C, TRAK2, CDK19, FNBP1 | 8 |
| Cabrera-Mendoza Bb [63] | 2020 | DLPFC | 9 | Microarray | BBS4, NKX6-2, AXL, CTNND1, MBP, PAOX | 8 |
| Romero-Pimentel A [66] | 2021 | DLPFC | 9 | Microarray | ADCY9, CRH, NFATC4, ABCC8, HMGA1, KAT2A, EPHA2, TRRAP | 9 |
| Asian countries | ||||||
| Yanagi M [17] | 2005 | AMY | NA | Microarray | 14-3-3 ε | 8 |
| Tochigi M [22] | 2008 | PFC | 10 | Microarray | CAD, ATP1A3 | 9 |
| Sherrin T [15] | 2004 | CB, CG, PFC | NA | qRT–PCR | CCKB | 8 |
| European countries | ||||||
| Thalmeier A [23] | 2008 | OFC | 11 | Microarray | AMPH, CDH12, CDH22, CHGB, MYR8, PENK, PTPRR, SCN2B | 9 |
| Perroud N [32] | 2010 | VPFC | 11 | qRT–PCR | TPH2 | 7 |
| Keller S [34] | 2010 | Wernicke | NA | Microarray | BDNF | 7 |
| Keller S [35] | 2011 | Wernicke | 8, 9 | Microarray | TrkB | 7 |
| Zhurov V [38] | 2012 | FPC | 10 | Microarray | MEF2D, TFE3, PLAGL1, C1D, XRCC5, EP300, FMR1, VTA1 | 8 |
| Pérez-Ortiz JM [44] | 2013 | AMY | NA | qRT–PCR | FKBP5, GR | 7 |
| Du L [49] | 2014 | FPC, OFC | 10, 11, 12, 45, 47 | qRT–PCR | COMT | 8 |
| Monsalve E [50] | 2014 | DLPFC, AMY | NA | qRT–PCR | NOTCH2, NOTCH1, NOTCH3, NOTCH4, DLL4, JAGGED1 | 9 |
| García-Gutiérrez MS [58] | 2018 | DLPFC | 9 | qRT–PCR | CB2r, GPR55 | 8 |
| Kouter K [59] | 2019 | PFC, HPC | 9 | NGS | NRIP3, ZNF714 | 8 |
| Mixed populations | ||||||
| Di Narzo AF [48] | 2014 | OFC, ACC | 11, 25 | Microarray | 5 HT2CR | 7 |
| Cabrera-Mendoza B [8] | 2020 | DLPFC | 9 | Microarray | GRM3, GRM8, GRIA2, GRIN2A, GRIN2C | 8 |
PFC: prefrontal cortex; DLPFC: dorsolateral prefrontal cortex; ACC: anterior cingulate cortex; ACG: anterior cingulate gyrus; HPC: hippocampus; HPT: hypothalamus; AMY: amygdala; THAL: thalamus; PG: pituitary; CB: cerebellum; SN: substantia nigra; CG: cingulate gyrus; PCG: posterior cingulate gyrus; OC: orbital cortex; MC: motor cortex; OFC: orbitofrontal cortex; FC: frontal cortex; IFG: inferior frontal gyrus; VPFC: ventral prefrontal cortex; NACC: nucleus accumbens; DG: dentate gyrus; MDTHAL: mediodorsal thalamus; CN: caudate nucleus; CBTX: cerebellar cortex; CTX: cerebral cortex; qRT–PCR: real-time quantitative reverse transcription PCR; NGS: next-generation sequencing; NA: not available. USA: United States of America; NOS: Newcastle-Ottawa Scale.
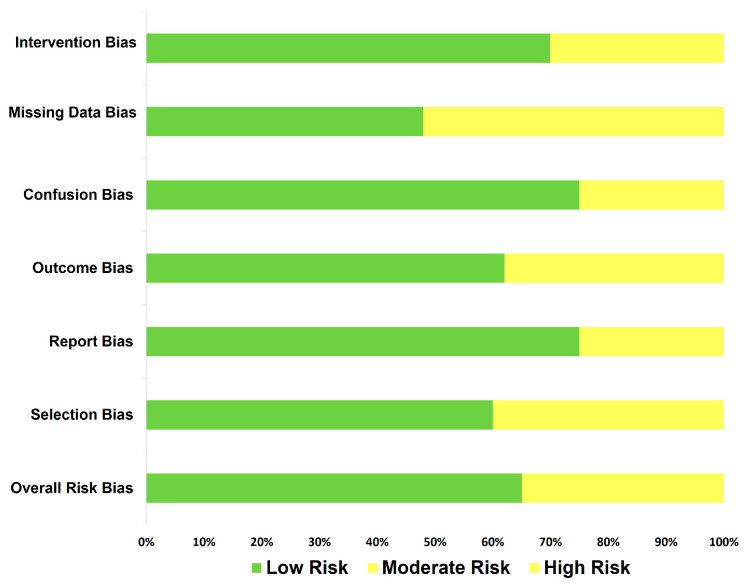
On the other hand, when the quality measurements were applied, we could observe that studies differed in methodological standardization (e.g., type of method applied in gene expression, percent of male/female, among others). Nevertheless, the quality level of the studies did not report an important evidence of bias. However, any results should be taken with caution (Figure 2).
Figure 2.
Risk of bias summary: a review of authors’ judgments about risk of bias.
3.3. Gene Expression Associated with Completed Suicide
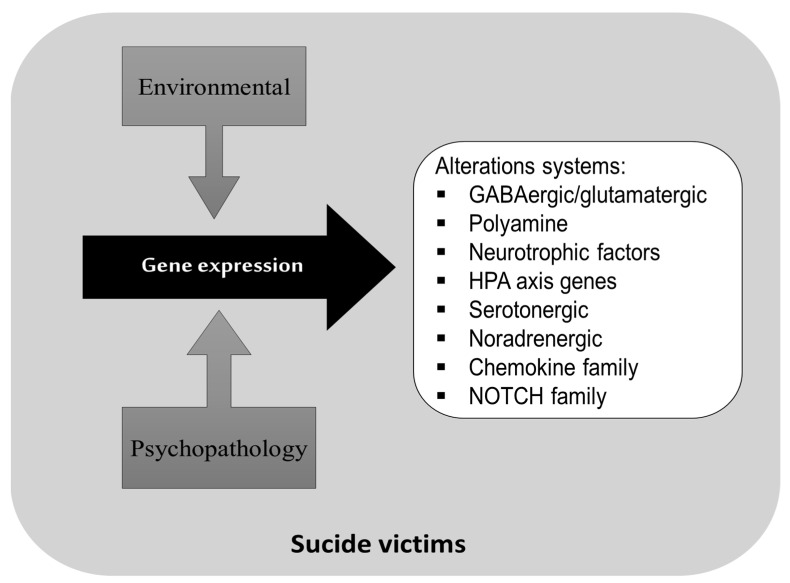
The included results reported several pathways that have been implicated as risk factors of suicide; the glutamate pathway and the γ-amino-butyric acid and polyamine systems have been the most studied. One study of brain expression in suicide completers with and without major depression reported global changes in the synaptic transmission of GABAergic (inhibitory) and glutamatergic (excitatory) systems [30]. In another study of Canadian individuals who died by suicide, the polyamine biosynthetic gene expression was analyzed and it was observed that the AMD1 and ARG2 genes correlated with a decreased methylation of specific CpGs in the promoter region of these genes [46] (Figure 3).
Figure 3.
Principal neurobiological systems involved in postmortem studies on brains of suicide completers.
Other pathways commonly associated with completed suicide are the neurotrophic factor systems. The brain-derived neurotrophic factor and tropomyosin-related kinase B (TrkB), even a truncated variant (TrkB.T1), have been found in the frontal cortex and Wernicke area of a completed suicide subpopulation [27,34,35]. The HPA axis genes have been investigated as possible candidate genes for suicide behavior markers, including the corticotropin-releasing hormone receptor genes [11], the FKBP5 and glucocorticoid receptor gene [44]. Other genes commonly investigated are those related to glia or astrocyte cell functioning or proliferation; for instance, we found that the mRNA expression of the chemokines CXCL1, CXCL2, CXCL3 and CCL2 was significantly decreased in the prefrontal cortex (PFC) of suicide completers who had depression when compared with non-suicide individuals [7].
Additionally, a high astrocytic CX gene expression has been found in individuals with depression who died by suicide [56].
Finally, some studies addressed the gene expression of serotonergic and noradrenergic pathways, which are thought to be etiologically relevant to suicide risk and other psychiatric disorders [18,25,32,48,49].
3.4. Description of Cases Group (Brains of Suicide Completers)
In this review, we evaluated 1450 individuals who died by suicide; the mean age was 39, ranging from 12 to 94 years old. The majority of suicide completers were men (n = 1058). Regarding the characteristics of brain tissue, they showed a mean pH of 6.5, and the postmortem interval was around 24 h. RNA integrity was approximately 7. The main characteristics of each study are described in Table 2.
Table 2.
Summary of main characteristics of suicide victims and brain samples of the studies included.
| First Author | Diagnostic | N | Mean Age | Range Age | M | F | RIN | pH | PMI |
|---|---|---|---|---|---|---|---|---|---|
| Hiroi N 2001 [11] | - | 9 | 3.8 | 17–72 | 3 | 6 | - | - | - |
| Dwivedi Y 2001 [12] | DD | 11 | 36.2 | 21–53 | 5 | 6 | - | - | 17.8 |
| Dwivedi Y 2003 [13] | Psychiatric | 27 | 41 | 21–87 | 19 | 8 | - | 6.1 | 19.2 |
| Sibille E 2004 [14] | DD | 19 | 44.6 | - | 14 | 5 | - | - | 16.5 |
| Sherrin T 2004 [15] | Psychiatric | 10 | 37.2 | 16–54 | 9 | 1 | - | - | 16.6 |
| Choudary PV 2005 [16] | DD | 9 | - | - | 7 | 2 | - | - | - |
| BD | 6 | - | - | 5 | 1 | - | - | - | |
| Yanagi M 2005 [17] | Psychiatric | 14 | 43.9 | 26–65 | 5 | 9 | - | - | 18.2 |
| De Luca V 2006 [18] | SCHZ, BD | 23 | 42.6 | - | 11 | 12 | - | 6.47 | 38.5 |
| Sequeira A 2006 [19] | With DD | 16 | 34 | 18–53 | 16 | 0 | - | 6.49 | 22.34 |
| Without DD | 8 | 35.12 | 21–51 | 8 | 0 | - | 6.3 | 24.25 | |
| Sequeira A 2007 [20] | Without DD | 8 | 35.1 | 21–51 | 8 | 0 | - | 6.3 | 24.3 |
| With DD | 18 | 36.5 | 19–53 | 18 | 0 | - | 6.5 | 24.1 | |
| Kim S 2007 [21] | BD | 22 | 44.3 | - | 11 | 11 | - | 6.4 | 36.8 |
| SCHZ | 10 | 34.5 | - | 6 | 4 | - | 6.4 | 35.3 | |
| Tochigi M 2008 [22] | DP | 11 | 46 | - | 6 | 5 | - | - | 27 |
| BD | 11 | 39 | - | 8 | 3 | - | - | 32 | |
| SCHZ | 13 | 44 | - | 8 | 5 | - | - | 33 | |
| Thalmeier A 2008 [23] | Psychiatric | 11 | 55.4 | 33–81 | 8 | 3 | - | 6.72 | 59.7 |
| Feldcamp LA 2008 [24] | SCHZ | 6 | 36.5 | - | 3 | 3 | - | 6.6 | 36.8 |
| BD | 16 | 45.1 | - | 8 | 8 | - | 6.4 | 39.34 | |
| Garbett K 2008 [25] | SCHZ | 6 | 38 | 25–50 | 3 | 3 | - | 6.9 | 19.4 |
| McGowan PO 2009 [26] | With CA | 12 | 34.2 | 12 | 0 | 6.3 | 24.6 | ||
| Without CA | 12 | 22.8 | 12 | 0 | 6.5 | 39.0 | |||
| Ernst C 2009 [27] | Psychiatric | 28 | 39 | 18–72 | 28 | 0 | - | 6.5 | 26 |
| Pandey GN 2009 [28] | Psychiatric | 29 | 16.17 | 13–20 | 17 | 12 | - | 6.17 | 18.41 |
| Psychiatric | 27 | 42.7 | 22–87 | 17 | 10 | - | 6.12 | 19.52 | |
| Klempan TA 2009 [29] | DD | 16 | 36.5 | 18–53 | 16 | 0 | >6 | - | 24.6 |
| Sequeira A 2009 [30] | Psychiatric | 10 | 34 | 21–51 | 10 | 0 | 7.14 | 6.32 | 29 |
| DD | 16 | 37 | 18–53 | 16 | 0 | 7.14 | 6.55 | 25 | |
| Lalovic A 2010 [31] | With DD | 15 | 34.5 | 19–53 | 15 | 0 | 6.5 | 6.6 | 25 |
| Without DD | 7 | 32.4 | 21–51 | 7 | 0 | 6.5 | 6.4 | 25.9 | |
| Perroud N 2010 [32] | Psychiatric | 39 | 47.36 | 15–94 | 26 | 13 | - | 6.84 | 37.38 |
| Simmons M 2010 [33] | DD, BD, SCHZ | 15 | - | - | 9 | 6 | - | 6.2 | 31.2 |
| Keller S 2010 [34] | Psychiatric | 44 | - | 15–79 | 21 | 23 | - | - | - |
| Keller S 2011 [35] | Psychiatric | 19 | - | 14–59 | 10 | 9 | - | 6.6 | <24 |
| Choi K 2011 [36] | Psychiatric | 45 | 41.7 | - | 25 | 20 | >7 | 6.5 | 32.9 |
| Fiori LM 2011 [37] | DD, BD | 29 | 39.8 | - | 29 | 0 | - | 6.6 | 27 |
| Zhurov V 2012 [38] | DD | 10 | 52.5 | - | 10 | 0 | 7.4 | 6.63 | 5.3 |
| Smalheiser NR 2012 [39] | DD | 18 | 40 | 19–65 | 16 | 2 | 8.98 | 6.5 | 10.7 |
| Sequeira A 2012 [40] | DD, BD | 15 | 44 | 24–77 | 12 | 3 | 7.8 | 6.82 | 24.64 |
| DD, BD | 9 | 43.3 | 34–56 | 6 | 3 | 8.03 | 6.91 | 23.52 | |
| DD, BD | 13 | 42.72 | 29–58 | 10 | 3 | 8.1 | 6.8 | 24.03 | |
| Labonté B 2013 [41] | Psychiatric | 13 | 30.9 | - | 13 | 0 | 6.23 | 6.6 | 23.2 |
| Galfalvy H 2013 [42] | DD | 18 | 55.8 | - | 8 | 10 | >7 | - | - |
| Ren X 2013 [43] | Psychiatric | 24 | 15.92 | 12–20 | 14 | 10 | 7.2 | 6.21 | 19 |
| Pérez-Ortiz JM 2013 [44] | Psychiatric | 13 | 40 | 18–66 | 13 | 0 | 6.15 | - | 17 |
| Pandey GN 2013 [45] | Psychiatric | 24 | 15.92 | 12–20 | 14 | 10 | 7.15 | 6.21 | 19 |
| Gross JA 2013 [46] | SCHZ, DD, BD | 34 | 38.6 | - | 34 | 0 | 6.7 | 6.6 | 33.9 |
| Lopez JP 2014 [47] | DD | 15 | 37.9 | - | 15 | 0 | 6.4 | 6.6 | 29.3 |
| Di Narzo AF 2014 [48] | Psychiatric | 22 | 32.18 | 16–47 | 16 | 6 | 7.64 | 6.8 | 24 |
| DD | 10 | 47.7 | 26–72 | 2 | 8 | 8.13 | 6.61 | 16.1 | |
| Du L 2014 [49] | DD | 49 | 48.91 | - | 35 | 14 | - | 6.57 | 4.9 |
| Monsalve EM 2014 [50] | Psychiatric | 13 | 40 | 18–66 | 13 | 0 | 6.15 | - | 17 |
| Gray AL 2015 [51] | DD | 34 | 41.1 | 16–83 | 16 | 18 | 8.36 | 6.3 | 34.6 |
| Fuchsova B 2015 [52] | DD | 25 | 41.92 | 22–74 | 12 | 13 | 6.6 | 7 | 20.16 |
| Zhao J 2015 [53] | DD | 17 | 40 | 24–63 | 10 | 7 | - | 6.67 | 29.6 |
| Yin H 2016 [54] | DD | 21 | 52.1 | - | 13 | 8 | - | - | - |
| Pandey GN 2016 [55] | DD | 24 | 38.96 | 18–74 | 14 | 10 | >6.6 | 6.96 | 18.92 |
| SCHZ | 16 | 36.31 | 20–54 | 13 | 3 | >6.6 | 6.68 | 16.69 | |
| Psychiatric | 12 | 40.08 | 19–87 | 11 | 1 | >6.6 | 6.67 | 22.58 | |
| Nagy C 2017 [56] | DD | 22 | 39.7 | - | 22 | 0 | 6.7 | 17.5 | |
| Pantazatos SP 2017 [57] | DD | 21 | 52 | - | 13 | 8 | 6.6 | 6.4 | 16.1 |
| García-Gutiérrez MS 2018 [58] | Psychiatric | 18 | 43 | 18–78 | 18 | 0 | 6.26 | - | 17 |
| Kouter K 2019 [59] | Psychiatric | 9 | 50.56 | - | 9 | 0 | - | - | 21.33 |
| Cabrera B 2019 [60] | With SUD | 23 | 31.95 | - | 21 | 2 | >7 | - | 14.91 |
| Without SUD | 20 | 32.8 | - | 12 | 8 | >7 | - | 15.03 | |
| Zhang L 2020 [61] | SCHZ | 35 | 43 | 19–59 | 26 | 9 | >7 | 6.5 | 31.3 |
| Zhang L 2020b [62] | BD | 13 | 44.5 | 29–59 | 7 | 6 | 8.7 | 6.5 | 39.7 |
| Cabrera-Mendoza B 2020 [63] | DD, BD | 48 | 31 | - | 38 | 10 | >6 | - | 15.8 |
| Cabrera-Mendoza B 2020b [63] | DD, PD | 21 | 28.4 | - | 21 | 0 | >6 | - | 14.3 |
| Postolache TT 2020 [64] | DD | 15 | 39 | - | 13 | 2 | 7.9 | 6.6 | 35 |
| Yoshino Y 2020 [65] | DD | 43 | 50.3 | - | 26 | 17 | 8 | 6.8 | 17.9 |
| Squassina A 2020 [66] | BD | 7 | 40.6 | 4 | 3 | ||||
| Pandey GN 2021 [67] | DD | 24 | 38.95 | 19–74 | 14 | 10 | ≈7 | 6.95 | 18.91 |
| Romero-Pimentel AL 2021 [68] | Psychiatric | 35 | 33.11 | - | 35 | 0 | 7.4 | - | 11.2 |
| Overall | 1450 | 39 | 12–94 | 1058 | 392 | 7 | 6.5 | 24 |
BD: Bipolar disorder; DD: Depression disorder; SCHZ: Schizophrenia; CA: Child abuse; SUD: Substance-use disorder; N: Sample size; M: Male; F: Female; RIN: RNA integrity numbers; postmortem interval.
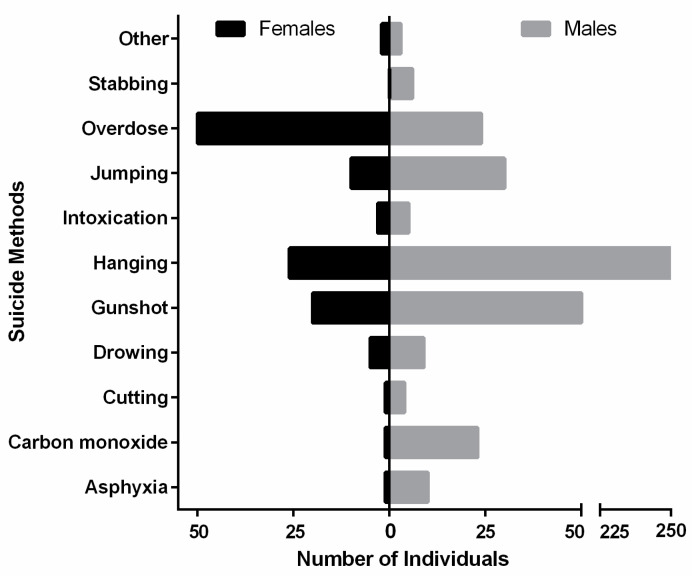
Regarding the suicide method selection, the most common one in females was drug overdose; while in males, hanging was the most frequently selected. Males showed a strong tendency to use more violent suicide methods (i.e., hanging, a gunshot and jumping) (Figure 4).
Figure 4.
Distribution of suicide method selection by gender.
3.5. Description of the Comparison Group (Brains of Non-Suicidal Individuals)
The comparison group consisted of 1314 individuals with a mean age of 43 years; they died by any other causes (Table 3). The majority of individuals in this group were also men (n = 972). The brain samples in the comparison group had a mean pH of 7 and a postmortem interval of 23 h.
Table 3.
Summary of main characteristics of controls and brain samples of the studies included.
| First Author | Diagnostic | N | Mean Age | Range Age | M | F | RIN | pH | PMI |
|---|---|---|---|---|---|---|---|---|---|
| Hiroi N 2001 [11] | - | 7 | 48.7 | - | 4 | 3 | - | - | - |
| Dwivedi Y 2001 [12] | Non-psychiatric | 11 | 37.8 | 22–46 | 8 | 3 | - | - | 15.7 |
| Dwivedi Y 2003 [13] | Non-psychiatric | 21 | 49.2 | 22–83 | 17 | 4 | - | 6.1 | 18.7 |
| Sibille E 2004 [14] | Non-psychiatric | 19 | 44.5 | 14 | 5 | - | - | 18.5 | |
| Sherrin T 2004 [15] | Non-psychiatric | 10 | 37.6 | 20–56 | 9 | 1 | - | - | 18.7 |
| Choudary PV 2005 [16] | Non-psychiatric | 7 | - | - | 6 | 1 | - | - | - |
| Yanagi M 2005 [17] | Non-psychiatric | 14 | 54.6 | 28–75 | 5 | 9 | - | - | 12.8 |
| De Luca V 2006 [18] | SCHZ, BD | 23 | 44 | - | 11 | 12 | - | 6.53 | 32 |
| Sequeira A 2006 [19] | Non-psychiatric | 12 | 35.58 | 19–55 | 12 | 0 | - | 6.44 | 25.91 |
| Sequeira A 2007 [20] | Non-psychiatric | 13 | 35.3 | 19–55 | 13 | 0 | - | 6.5 | 23.7 |
| Kim S 2007 [21] | BD | 23 | 45.4 | - | 12 | 11 | - | 6.4 | 36.8 |
| SCHZ | 35 | 44.3 | - | 28 | 7 | - | 6.4 | 31.4 | |
| Tochigi M 2008 [22] | Non-psychiatric | 15 | 48 | - | 9 | 6 | - | - | 24 |
| Thalmeier A 2008 [23] | Non-psychiatric | 10 | 64.1 | 48–83 | 7 | 3 | - | 6.71 | 69.5 |
| Feldcamp LA 2008 [24] | SCHZ | 29 | 44 | - | 23 | 6 | - | 6.5 | 30.14 |
| BD | 18 | 46.2 | - | 9 | 9 | - | 6.4 | 34.3 | |
| Garbett K 2008 [25] | Non-psychiatric | 6 | 39 | 19–52 | 4 | 2 | - | 6.8 | 18.2 |
| Ernst C 2009 [26] | Non-psychiatric | 11 | 39 | 28–58 | 11 | 0 | - | 6.5 | 22 |
| McGowan PO 2009 [27] | Non-psychiatric | 12 | 35.8 | 12 | 0 | 6.5 | 23.5 | ||
| Pandey GN 2009 [28] | Non-psychiatric | 26 | 16.46 | 13–19 | 18 | 8 | - | 6.19 | 18.41 |
| Non-psychiatric | 20 | 43.55 | 22–83 | 16 | 4 | - | 6.1 | 18.45 | |
| Klempan TA 2009 [29] | Non-psychiatric | 13 | 35.3 | 19–55 | 13 | 0 | >6 | - | 23.7 |
| Sequeira A 2009 [30] | Psychiatric | 13 | 35 | 19–55 | 13 | 0 | 7.14 | 6.44 | 24 |
| Lalovic A 2010 [31] | Non-psychiatric | 13 | 37 | 19–55 | 13 | 0 | 6.5 | 6.4 | 22.5 |
| Perroud N 2010 [32] | Mostly non-psychiatric | 40 | 51.13 | 16–97 | 27 | 13 | - | 6.79 | 39.58 |
| Simmons M 2010 [33] | Non-psychiatric | 15 | - | - | 9 | 6 | - | 6.2 | 23.7 |
| Keller S 2010 [34] | Non-psychiatric | 33 | - | 13–76 | 16 | 17 | - | - | - |
| Keller S 2011 [35] | Non-psychiatric | 18 | - | 13–70 | 7 | 11 | - | 6.8 | <24 |
| Choi K 2011 [36] | Psychiatric | 38 | 47.2 | - | 21 | 17 | >7 | 6.4 | 33 |
| Fiori LM 2011 [37] | Non-psychiatric | 16 | 39.8 | - | 16 | 0 | - | 6.6 | 27 |
| Zhurov V 2012 [38] | Non-psychiatric | 9 | 59.4 | - | 9 | 0 | 6.5 | 6.5 | 3.7 |
| Smalheiser NR 2012 [39] | Non-psychiatric | 17 | 35.5 | 19–63 | 17 | 0 | 9 | 6.5 | 26.9 |
| Sequeira A 2012 [40] | Psychiatric | 6 | 59 | 19–59 | 4 | 2 | 8.5 | 6.8 | 25.5 |
| Psychiatric | 6 | 53.3 | 44–59 | 3 | 3 | 8 | 6.76 | 15.9 | |
| Psychiatric | 8 | 52.3 | 44–66 | 6 | 2 | 8.1 | 6.6 | 27.7 | |
| Labonté B 2013 [41] | Non-psychiatric | 9 | 37.4 | - | 9 | 0 | 6.48 | 6.7 | 27.8 |
| Galfalvy H 2013 [42] | Non-psychiatric | 21 | 51.8 | - | 14 | 7 | >7 | - | - |
| Ren X 2013 [43] | Non-psychiatric | 24 | 16.29 | 13–19 | 17 | 7 | 7.2 | 6.15 | 18.13 |
| Pérez-Ortiz JM 2013 [44] | Non-psychiatric | 13 | 46 | 19–64 | 13 | 0 | 6.71 | 16 | |
| Pandey GN 2013 [45] | Non-psychiatric | 24 | 16.29 | 13–19 | 17 | 7 | 7.21 | 6.15 | 18.13 |
| Gross JA 2013 [46] | Non-psychiatric | 34 | 43.6 | - | 34 | 0 | 6.4 | 6.5 | 45.2 |
| Lopez JP 2014 [47] | Non-psychiatric | 16 | 39.8 | - | 16 | 0 | 6.4 | 6.6 | 23.8 |
| Di Narzo AF 2014 [48] | Non-psychiatric | 29 | 37.2 | 19–65 | 24 | 5 | 7.5 | 6.72 | 21.4 |
| DD | 24 | 49.7 | 16–74 | 8 | 16 | 8.2 | 6.55 | 17.7 | |
| Du L 2014 [49] | Non-psychiatric | 72 | 64.47 | - | 46 | 26 | 5.96 | 6.67 | 4.07 |
| Monsalve EM 2014 [50] | Non-psychiatric | 13 | 46 | 19–64 | 13 | 0 | 6.71 | - | 16 |
| Gray AL 2015 [51] | Non-psychiatric | 32 | 39.2 | 16–65 | 19 | 13 | 8.2 | 6.5 | 29 |
| Fuchsova B 2015 [52] | Non-psychiatric | 25 | 42.8 | 22–72 | 18 | 7 | 6.6 | 7.01 | 17.72 |
| Zhao J 2015 [53] | Non-psychiatric | 12 | 47 | 24–63 | 8 | 4 | - | 6.64 | 25.3 |
| Yin H 2016 [54] | Mostly non-psychiatric | 38 | 46.9 | - | 29 | 9 | - | - | - |
| Pandey GN 2016 [55] | DD | 12 | 49.5 | 14–74 | 7 | 5 | >6.6 | 6.8 | 17.92 |
| SCHZ | 15 | 50.6 | 24–83 | 10 | 5 | >6.6 | 6.61 | 15.33 | |
| Non-psychiatric | 24 | 42.08 | 19–83 | 20 | 4 | >6.6 | 7.02 | 16.54 | |
| Nagy C 2017 [56] | Non-psychiatric | 22 | 41.6 | - | 22 | 0 | - | 6.5 | 21.5 |
| Pantazatos SP 2017 [57] | Non-psychiatric | 29 | 43.5 | - | 23 | 6 | 7.1 | 6.5 | 13.2 |
| García-Gutiérrez MS 2018 [58] | Non-psychiatric | 15 | 46 | 19–64 | 15 | 0 | 6.35 | - | 15 |
| Kouter K 2019 [59] | Non-psychiatric | 9 | 53.11 | - | 9 | 0 | - | - | 24.22 |
| Cabrera B 2019 [60] | With SUD | 9 | 30.88 | - | 8 | 1 | >7 | - | 17.76 |
| Without SUD | 14 | 31.78 | - | 8 | 6 | >7 | - | 16.84 | |
| Zhang L 2020 [61] | Non-psychiatric | 34 | 45 | 31–60 | 25 | 9 | >7 | 6.69 | 28.5 |
| Zhang L 2020b [62] | Non-psychiatric | 34 | 45 | 31–60 | 25 | 9 | 8.4 | 6.69 | 28.5 |
| Cabrera-Mendoza B 2020 [63] | Non-psychiatric | 27 | 35 | - | 20 | 7 | >6 | - | 19 |
| Cabrera-Mendoza B 2020b [63] | Non-psychiatric | 6 | 29.33 | - | 6 | 0 | - | - | 15.53 |
| Postolache TT 2020 [64] | Non-psychiatric | 15 | 36.6 | - | 15 | 0 | 7.9 | 6.5 | 34.6 |
| Yoshino Y 2020 [65] | Non-psychiatric | 27 | 48.4 | - | 16 | 11 | 7.9 | 6.6 | 18.7 |
| Squassina A 2020 [66] | Non-psychiatric | 12 | 38 | 3 | 9 | ||||
| Pandey GN 2021 [67] | Non-psychiatric | 24 | 42.08 | 19–62 | 20 | 4 | ≈7 | 7.01 | 16.54 |
| Romero-Pimentel AL 2021 [68] | Non-psychiatric | 13 | 32.4 | - | 13 | 0 | 7.2 | - | 11.6 |
| Overall | 1314 | 43 | 13–97 | 972 | 342 | 7 | 6.5 | 23 |
SCHZ: Schizophrenia; BD: Bipolar disorder; DD: Depression disorder; SUD: Substance-use disorder; N: Sample size; M: Male; F: Female; RIN: RNA integrity numbers; postmortem interval.
4. Discussion
The use of postmortem brain samples of individuals who died by suicide leads to new opportunities to study molecular mechanisms underlying suicide behavior [67]. Research focusing on the identification of candidate genetic factors could increase our knowledge of different neurobiological elements responsible for this pathology [68]. Hence, in the present work, we aimed to perform a systematic review of the gene expression association studies on postmortem brain samples from completed suicide.
4.1. GABAergic/Glutamatergic Systems
Gamma-amino butyric acid (GABA) and glutamate are the major inhibitory and excitatory neurotransmitters in the mammalian central nervous system (CNS), respectively, and they are thereby involved directly or indirectly in several mental aspects, such as learning, memory, cognition and mood regulation, among other normal functions [69]. Some studies have reported an association of the GABAergic/glutamatergic gene expression with suicide behavior. One study found that the expression of both GABA and glutamate genes is increased in the anterior cingulate cortex of individuals with depression who died by suicide when compared with individuals with depression who died by other causes [70]. Specifically, one study found that the subunit of GABAA receptors, GABRG2, had a higher gene expression in the dorsolateral prefrontal cortex of individuals with major depression [16], while another study found lower GABRG2 brain expression in suicide behavior individuals who died by suicide [71]. We have found strong evidence that the mRNA expression of glutamatergic and GABAergic proteins is similarly altered in completed suicide. Then, is there a disturbed balance between neuronal/brain excitation and inhibition in individuals who died by suicide? It is plausible to assume that brains of suicide completers started with variations in neuronal/brain excitation followed by proportional changes in neuronal/brain inhibition [47,66].
4.2. Polyamine System
The cellular roles of polyamines (putrescine, spermidine, spermine and agmatine) include the modulation of synaptic activity and ion channels that participate in the excitability of the neuronal network, as well as the regulation of gene transcription and post-transcriptional modifications [72]. One of the most common genes studied of the polyamine system is the spermine/spermidine N1-acetyltransferase (SSAT) gene. In a Canadian population, for instance, a study found a significant downregulation of SSAT1 in suicide completers [73]. A subsequent study hypothesized that the dysregulation of the SSAT1 gene in suicide completers could be influenced by miRNA post-transcriptional regulation. In fact, this group also found that several miRNAs showed a significant up-regulation in suicide completers when compared with non-psychiatric controls [47]. This altered gene expression of SSAT1 would be expected when there is homeostatic disruption, causing increases in spermine levels, spermidine levels, or both. Additionally, an analysis of transcriptomic and DNA methylation profiles of 21 individuals who died by suicide showed that polyamine oxidase (PAOX) gene expression is up-regulated, which provides a better explanation of the altered levels of polyamines in the brain of completed suicide [63]. Together, these findings suggest that changes in polyamine levels may have deep effects, due to the multiple processes that polyamines contribute to in the CNS.
4.3. Neurotrophic Factor Systems
Neurotrophic factors, also known as the neurotrophin signaling system, are a wide variety of polypeptides essential for the development and survival of neurons in the central and peripheral nervous systems [74]. The hypothesis of the relation between neurotrophic factors and behavior is widely proposed by some investigators; in suicide behavior, it has been suggested that alterations in gene expression of neurotrophic factors partly underlie changes in plasticity observed in the brains of suicide completers [75]. One of the most important neurotrophins is the brain-derived neurotrophic factor (BDNF); after binding and activating the receptor tyrosine kinase B (TrkB), it is directly involved in the functioning of neurons and the synapsis [76]. For instance, one study reported that mRNA levels of BDNF and TrkB were reduced in the PFC and hippocampus of suicide completers when compared with individuals who died of other causes [13]. Another report indicates that the BDNF promoter is hypermethylated in the brain of suicide completers, which could have contributed to the downregulation of BDNF expression in suicide completers [34]. Overall, the findings suggest that BDNF and TrkB are promising markers of suicide behavior. Gene expression in postmortem studies underly the dysregulation of specific regions of the brain and neuronal plasticity.
4.4. HPA Axis Genes
Our research indicates a dysregulation of the hypothalamic–pituitary–adrenal (HPA) axis stress response activity, as well as an impairment of other typical neurotransmitter systems [77], in the diathesis of suicide behavior. In suicide completers, the receptors of the corticotropin-releasing hormone (CRH), specifically CRH-R1 and CRH-R2, were highly expressed in the brain, particularly in the pituitary [11]. A study found decreased gene expression levels of the neuron-specific glucocorticoid receptor in suicide completers with a history of child abuse [26]. Additionally, investigations have found that the gene expression of co-chaperone FK506-binding protein 51 (FKBP5) was significantly reduced in the amygdala of individuals who died by suicide when compared with controls [44]. It is well known that in situations where our body does not have a successful control of stressors, the outcome effects can be negative; therefore, we could assume that alteration of any member of the HPA axis could induce an endocrine response affecting the brain and peripheral tissues [44].
4.5. Chemokine Family
Chemokines are small proteins with several implications in neuroendocrine regulation, blood barrier permeability control, pre- and post-synaptic modulation and other essential activities for the normal functioning of the relation between the central nervous system and immune system (Nakagawa & Chiba, 2015; Stuart & Baune, 2014). For that reason, it is not surprising that chemokines such as CXCL1 and CCL2, among others, have been implicated in a number of neurological diseases. In fact, some papers suggest that the disruption of the previously mentioned chemokine functions in neurodevelopmental periods or in later life contribute to the pathophysiology of psychiatric traits (e.g., suicide) (Nakagawa & Chiba, 2015). Namely, CXCL proteins are involved in the inhibition of glutamatergic activity in hippocampal neurons and regulation processes of neuroplasticity (Rogers et al., 2011; Tokac et al., 2016).
4.6. Considerations and Limitations
We want to highlight that gene expression is controlled by a variety of factors that should be considered when drawing conclusions. There is evidence that epigenetic changes such as post-translational histone modifications could alter gene expression without altering the DNA sequence [78]. Different diagnoses are another important factor. We recognize that it was difficult to determine the changes in gene expression associated with suicide behavior, particularly when analyzing genes associated with other psychiatric disorders such as depression. However, having more detailed information regarding the expression of candidate genes could impact the understanding of the pathophysiology in major depression and the diathesis for suicide, including differences and similitudes. We recognize that in future studies, it will be necessary to take into consideration gender analyses, analyses of other suicide features and analyses of the suicide method selected. For instance, we observed that men usually complete suicide using a violent method, while for women, the most frequent methods used are non-violent. In this sense, several studies have suggested that the courage to carry out self-harm with the aim of death is partially heritable [79]. In particular, the serotonin transporter gene consists of one short (S) and one long (L) allelic variant; the S allele is associated with lower gene expression and is repeatedly associated with violent suicide methods [80].
To sum up, even with the limitations mentioned above, we consider that the findings obtained in the present review are of great value and will help to focus future research on relevant pathways that will help improve our knowledge of suicide behavior. Postmortem studies provide a deep insight of brain neurobiology and how it changes in suicide.
5. Conclusions
Our results show that mRNA expression in postmortem brains of suicide completers could be increased or decreased depending on the area, axis and/or pathway studied. On the other hand, psychiatric comorbidities indicate that suicide by violent death can alter the profile of mRNA expression. Therefore, more studies are necessary to determine the role of mRNA expression profiles in order to understand the molecular changes in brains of suicide completers.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/brainsci13060906/s1. Table S1: PRISMA 2020 Checklist.
Author Contributions
Conceptualization, T.B.G.-C. and C.A.T.-Z.; Software, A.D.G.-M. and T.B.G.-C.; Validation, H.N. and M.A.R.-M.; Formal analysis, I.E.J.-R. and M.L.L.-N.; Investigation, M.A.R.-M. and C.A.T.-Z.; Resources, T.B.G.-C. and H.N.; Data curation, C.A.T.-Z. and T.B.G.-C.; Writing—original draft preparation, T.B.G.-C.; Writing—original draft preparation, C.A.T.-Z., A.D.G.-M. and H.N.; Visualization, M.L.L.-N. and I.E.J.-R.; Supervision, I.E.J.-R. and A.D.G.-M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.O’Rourke M.C., Jamil R.T., Siddiqui W. StatPearls. StatPearls Publishing LLC.; Treasure Island, FL, USA: 2021. Suicide Screening and Prevention. StatPearls Publishing Copyright © 2021. [PubMed] [Google Scholar]
- 2.Ludwig B., Dwivedi Y. The concept of violent suicide, its underlying trait and neurobiology: A critical perspective. Eur. Neuropsychopharmacol. 2018;28:243–251. doi: 10.1016/j.euroneuro.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lengvenyte A., Conejero I., Courtet P., Olié E. Biological bases of suicidal behaviours: A narrative review. Eur. J. Neurosci. 2021;53:330–351. doi: 10.1111/ejn.14635. [DOI] [PubMed] [Google Scholar]
- 4.Gifuni A.J., Chakravarty M.M., Lepage M., Ho T.C., Geoffroy M.C., Lacourse E., Gotlib I.H., Turecki G., Renaud J., Jollant F. Brain cortical and subcortical morphology in adolescents with depression and a history of suicide attempt. J. Psychiatry Neurosci. JPN. 2021;46:E347–E357. doi: 10.1503/jpn.200198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshino Y., Dwivedi Y. Elevated expression of unfolded protein response genes in the prefrontal cortex of depressed subjects: Effect of suicide. J. Affect. Disord. 2020;262:229–236. doi: 10.1016/j.jad.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodríguez-López M.L., Martínez-Magaña J.J., Cabrera-Mendoza B., Genis-Mendoza A.D., García-Dolores F., López-Armenta M., Flores G., Vázquez-Roque R.A., Nicolini H. Exploratory analysis of genetic variants influencing molecular traits in cerebral cortex of suicide completers. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet. 2020;183:26–37. doi: 10.1002/ajmg.b.32752. [DOI] [PubMed] [Google Scholar]
- 7.Pandey G.N., Rizavi H.S., Bhaumik R., Zhang H. Chemokines gene expression in the prefrontal cortex of depressed suicide victims and normal control subjects. Brain Behav. Immun. 2021;94:266–273. doi: 10.1016/j.bbi.2021.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabrera-Mendoza B., Fresno C., Monroy-Jaramillo N., Fries G.R., Walss-Bass C., Glahn D.C., Ostrosky-Wegman P., Mendoza-Morales R.C., García-Dolores F., Díaz-Otañez C.E., et al. Sex differences in brain gene expression among suicide completers. J. Affect. Disord. 2020;267:67–77. doi: 10.1016/j.jad.2020.01.167. [DOI] [PubMed] [Google Scholar]
- 9.Cabrera-Mendoza B., Fresno C., Monroy-Jaramillo N., Fries G.R., Walss-Bass C., Glahn D.C., Ostrosky-Wegman P., Genis-Mendoza A.D., Martínez-Magaña J.J., Romero-Pimentel A.L., et al. Brain Gene Expression Profiling of Individuals with Dual Diagnosis Who Died by Suicide. J. Dual Diagn. 2020;16:177–190. doi: 10.1080/15504263.2019.1692160. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y., Lutz P.E., Ibrahim E.C., Courtet P., Tzavara E., Turecki G., Belzeaux R. Suicide and suicide behaviors: A review of transcriptomics and multiomics studies in psychiatric disorders. J. Neurosci. Res. 2020;98:601–615. doi: 10.1002/jnr.24367. [DOI] [PubMed] [Google Scholar]
- 11.Hiroi N., Wong M.L., Licinio J., Park C., Young M., Gold P.W., Chrousos G.P., Bornstein S.R. Expression of corticotropin releasing hormone receptors type I and type II mRNA in suicide victims and controls. Mol. Psychiatry. 2001;6:540–546. doi: 10.1038/sj.mp.4000908. [DOI] [PubMed] [Google Scholar]
- 12.Dwivedi Y., Rizavi H.S., Roberts R.C., Conley R.C., Tamminga C.A., Pandey G.N. Reduced activation and expression of ERK1/2 MAP kinase in the post-mortem brain of depressed suicide subjects. J. Neurochem. 2001;77:916–928. doi: 10.1046/j.1471-4159.2001.00300.x. [DOI] [PubMed] [Google Scholar]
- 13.Dwivedi Y., Rizavi H.S., Conley R.R., Roberts R.C., Tamminga C.A., Pandey G.N. Altered gene expression of brain-derived neurotrophic factor and receptor tyrosine kinase B in postmortem brain of suicide subjects. Arch. Gen. Psychiatry. 2003;60:804–815. doi: 10.1001/archpsyc.60.8.804. [DOI] [PubMed] [Google Scholar]
- 14.Sibille E., Arango V., Galfalvy H.C., Pavlidis P., Erraji-Benchekroun L., Ellis S.P., John Mann J. Gene expression profiling of depression and suicide in human prefrontal cortex. Neuropsychopharmacology. 2004;29:351–361. doi: 10.1038/sj.npp.1300335. [DOI] [PubMed] [Google Scholar]
- 15.Sherrin T., Heng K.Y., Zhu Y.Z., Tang Y.M., Lau G., Tan C.H. Cholecystokinin-B receptor gene expression in cerebellum, pre-frontal cortex and cingulate gyrus and its association with suicide. Neurosci. Lett. 2004;357:107–110. doi: 10.1016/j.neulet.2003.11.072. [DOI] [PubMed] [Google Scholar]
- 16.Choudary P.V., Molnar M., Evans S.J., Tomita H., Li J.Z., Vawter M.P., Myers R.M., Bunney W.E., Jr., Akil H., Watson S.J., et al. Altered cortical glutamatergic and GABAergic signal transmission with glial involvement in depression. Proc. Natl. Acad. Sci. USA. 2005;102:15653–15658. doi: 10.1073/pnas.0507901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yanagi M., Shirakawa O., Kitamura N., Okamura K., Sakurai K., Nishiguchi N., Hashimoto T., Nushida H., Ueno Y., Kanbe D., et al. Association of 14-3-3 epsilon gene haplotype with completed suicide in Japanese. J. Hum. Genet. 2005;50:210–216. doi: 10.1007/s10038-005-0241-0. [DOI] [PubMed] [Google Scholar]
- 18.De Luca V., Likhodi O., Van Tol H.H., Kennedy J.L., Wong A.H. Gene expression of tryptophan hydroxylase 2 in post-mortem brain of suicide subjects. Int. J. Neuropsychopharmacol. 2006;9:21–25. doi: 10.1017/S1461145705005572. [DOI] [PubMed] [Google Scholar]
- 19.Sequeira A., Gwadry F.G., Ffrench-Mullen J.M., Canetti L., Gingras Y., Casero R.A., Jr., Rouleau G., Benkelfat C., Turecki G. Implication of SSAT by gene expression and genetic variation in suicide and major depression. Arch. Gen. Psychiatry. 2006;63:35–48. doi: 10.1001/archpsyc.63.1.35. [DOI] [PubMed] [Google Scholar]
- 20.Sequeira A., Klempan T., Canetti L., ffrench-Mullen J., Benkelfat C., Rouleau G.A., Turecki G. Patterns of gene expression in the limbic system of suicides with and without major depression. Mol. Psychiatry. 2007;12:640–655. doi: 10.1038/sj.mp.4001969. [DOI] [PubMed] [Google Scholar]
- 21.Kim S., Choi K.H., Baykiz A.F., Gershenfeld H.K. Suicide candidate genes associated with bipolar disorder and schizophrenia: An exploratory gene expression profiling analysis of post-mortem prefrontal cortex. BMC Genom. 2007;8:413. doi: 10.1186/1471-2164-8-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tochigi M., Iwamoto K., Bundo M., Sasaki T., Kato N., Kato T. Gene expression profiling of major depression and suicide in the prefrontal cortex of postmortem brains. Neurosci. Res. 2008;60:184–191. doi: 10.1016/j.neures.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Thalmeier A., Dickmann M., Giegling I., Schneider B., Hartmann A.M., Maurer K., Schnabel A., Kauert G., Möller H.J., Rujescu D. Gene expression profiling of post-mortem orbitofrontal cortex in violent suicide victims. Int. J. Neuropsychopharmacol. 2008;11:217–228. doi: 10.1017/S1461145707007894. [DOI] [PubMed] [Google Scholar]
- 24.Feldcamp L.A., Souza R.P., Romano-Silva M., Kennedy J.L., Wong A.H. Reduced prefrontal cortex DARPP-32 mRNA in completed suicide victims with schizophrenia. Schizophr. Res. 2008;103:192–200. doi: 10.1016/j.schres.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 25.Garbett K., Gal-Chis R., Gaszner G., Lewis D.A., Mirnics K. Transcriptome alterations in the prefrontal cortex of subjects with schizophrenia who committed suicide. Neuropsychopharmacologia Hungarica Magyar Pszichofarmakologiai Egyesulet lapja = Off. J. Hung. Assoc. Psychopharmacol. 2008;10:9–14. [PubMed] [Google Scholar]
- 26.McGowan P.O., Sasaki A., D’Alessio A.C., Dymov S., Labonté B., Szyf M., Turecki G., Meaney M.J. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ernst C., Deleva V., Deng X., Sequeira A., Pomarenski A., Klempan T., Ernst N., Quirion R., Gratton A., Szyf M., et al. Alternative splicing, methylation state, and expression profile of tropomyosin-related kinase B in the frontal cortex of suicide completers. Arch. Gen. Psychiatry. 2009;66:22–32. doi: 10.1001/archpsyc.66.1.22. [DOI] [PubMed] [Google Scholar]
- 28.Pandey G.N., Dwivedi Y., Rizavi H.S., Teppen T., Gaszner G.L., Roberts R.C., Conley R.R. GSK-3beta gene expression in human postmortem brain: Regional distribution, effects of age and suicide. Neurochem. Res. 2009;34:274–285. doi: 10.1007/s11064-008-9770-1. [DOI] [PubMed] [Google Scholar]
- 29.Klempan T.A., Ernst C., Deleva V., Labonte B., Turecki G. Characterization of QKI gene expression, genetics, and epigenetics in suicide victims with major depressive disorder. Biol. Psychiatry. 2009;66:824–831. doi: 10.1016/j.biopsych.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Sequeira A., Mamdani F., Ernst C., Vawter M.P., Bunney W.E., Lebel V., Rehal S., Klempan T., Gratton A., Benkelfat C., et al. Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PLoS ONE. 2009;4:e6585. doi: 10.1371/journal.pone.0006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lalovic A., Klempan T., Sequeira A., Luheshi G., Turecki G. Altered expression of lipid metabolism and immune response genes in the frontal cortex of suicide completers. J. Affect. Disord. 2010;120:24–31. doi: 10.1016/j.jad.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Perroud N., Neidhart E., Petit B., Vessaz M., Laforge T., Relecom C., La Harpe R., Malafosse A., Guipponi M. Simultaneous analysis of serotonin transporter, tryptophan hydroxylase 1 and 2 gene expression in the ventral prefrontal cortex of suicide victims. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet. 2010;153B:909–918. doi: 10.1002/ajmg.b.31059. [DOI] [PubMed] [Google Scholar]
- 33.Simmons M., Meador-Woodruff J.H., Sodhi M.S. Increased cortical expression of an RNA editing enzyme occurs in major depressive suicide victims. Neuroreport. 2010;21:993–997. doi: 10.1097/WNR.0b013e32833f11c3. [DOI] [PubMed] [Google Scholar]
- 34.Keller S., Sarchiapone M., Zarrilli F., Videtic A., Ferraro A., Carli V., Sacchetti S., Lembo F., Angiolillo A., Jovanovic N., et al. Increased BDNF promoter methylation in the Wernicke area of suicide subjects. Arch. Gen. Psychiatry. 2010;67:258–267. doi: 10.1001/archgenpsychiatry.2010.9. [DOI] [PubMed] [Google Scholar]
- 35.Keller S., Sarchiapone M., Zarrilli F., Tomaiuolo R., Carli V., Angrisano T., Videtic A., Amato F., Pero R., di Giannantonio M., et al. TrkB gene expression and DNA methylation state in Wernicke area does not associate with suicidal behavior. J. Affect. Disord. 2011;135:400–404. doi: 10.1016/j.jad.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Choi K., Le T., Xing G., Johnson L.R., Ursano R.J. Analysis of kinase gene expression in the frontal cortex of suicide victims: Implications of fear and stress. Front. Behav. Neurosci. 2011;5:46. doi: 10.3389/fnbeh.2011.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiori L.M., Bureau A., Labbe A., Croteau J., Noël S., Mérette C., Turecki G. Global gene expression profiling of the polyamine system in suicide completers. Int. J. Neuropsychopharmacol. 2011;14:595–605. doi: 10.1017/S1461145710001574. [DOI] [PubMed] [Google Scholar]
- 38.Zhurov V., Stead J.D., Merali Z., Palkovits M., Faludi G., Schild-Poulter C., Anisman H., Poulter M.O. Molecular pathway reconstruction and analysis of disturbed gene expression in depressed individuals who died by suicide. PLoS ONE. 2012;7:e47581. doi: 10.1371/journal.pone.0047581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smalheiser N.R., Lugli G., Rizavi H.S., Torvik V.I., Turecki G., Dwivedi Y. MicroRNA expression is down-regulated and reorganized in prefrontal cortex of depressed suicide subjects. PLoS ONE. 2012;7:e33201. doi: 10.1371/journal.pone.0033201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sequeira A., Morgan L., Walsh D.M., Cartagena P.M., Choudary P., Li J., Schatzberg A.F., Watson S.J., Akil H., Myers R.M., et al. Gene expression changes in the prefrontal cortex, anterior cingulate cortex and nucleus accumbens of mood disorders subjects that committed suicide. PLoS ONE. 2012;7:e35367. doi: 10.1371/journal.pone.0035367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Labonté B., Suderman M., Maussion G., Lopez J.P., Navarro-Sánchez L., Yerko V., Mechawar N., Szyf M., Meaney M.J., Turecki G. Genome-wide methylation changes in the brains of suicide completers. Am. J. Psychiatry. 2013;170:511–520. doi: 10.1176/appi.ajp.2012.12050627. [DOI] [PubMed] [Google Scholar]
- 42.Galfalvy H., Zalsman G., Huang Y.Y., Murphy L., Rosoklija G., Dwork A.J., Haghighi F., Arango V., Mann J.J. A pilot genome wide association and gene expression array study of suicide with and without major depression. World J. Biol. Psychiatry Off. J. World Fed. Soc. Biol. Psychiatry. 2013;14:574–582. doi: 10.3109/15622975.2011.597875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren X., Rizavi H.S., Khan M.A., Dwivedi Y., Pandey G.N. Altered Wnt signalling in the teenage suicide brain: Focus on glycogen synthase kinase-3β and β-catenin. Int. J. Neuropsychopharmacol. 2013;16:945–955. doi: 10.1017/S1461145712001010. [DOI] [PubMed] [Google Scholar]
- 44.Pérez-Ortiz J.M., García-Gutiérrez M.S., Navarrete F., Giner S., Manzanares J. Gene and protein alterations of FKBP5 and glucocorticoid receptor in the amygdala of suicide victims. Psychoneuroendocrinology. 2013;38:1251–1258. doi: 10.1016/j.psyneuen.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 45.Pandey G.N., Rizavi H.S., Ren X., Dwivedi Y., Palkovits M. Region-specific alterations in glucocorticoid receptor expression in the postmortem brain of teenage suicide victims. Psychoneuroendocrinology. 2013;38:2628–2639. doi: 10.1016/j.psyneuen.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gross J.A., Fiori L.M., Labonté B., Lopez J.P., Turecki G. Effects of promoter methylation on increased expression of polyamine biosynthetic genes in suicide. J. Psychiatr. Res. 2013;47:513–519. doi: 10.1016/j.jpsychires.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez J.P., Fiori L.M., Gross J.A., Labonte B., Yerko V., Mechawar N., Turecki G. Regulatory role of miRNAs in polyamine gene expression in the prefrontal cortex of depressed suicide completers. Int. J. Neuropsychopharmacol. 2014;17:23–32. doi: 10.1017/S1461145713000941. [DOI] [PubMed] [Google Scholar]
- 48.Di Narzo A.F., Kozlenkov A., Roussos P., Hao K., Hurd Y., Lewis D.A., Sibille E., Siever L.J., Koonin E., Dracheva S. A unique gene expression signature associated with serotonin 2C receptor RNA editing in the prefrontal cortex and altered in suicide. Hum. Mol. Genet. 2014;23:4801–4813. doi: 10.1093/hmg/ddu195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du L., Merali Z., Poulter M.O., Palkovits M., Faludi G., Anisman H. Catechol-O-methyltransferase Val158Met polymorphism and altered COMT gene expression in the prefrontal cortex of suicide brains. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2014;50:178–183. doi: 10.1016/j.pnpbp.2013.12.016. [DOI] [PubMed] [Google Scholar]
- 50.Monsalve E.M., García-Gutiérrez M.S., Navarrete F., Giner S., Laborda J., Manzanares J. Abnormal expression pattern of Notch receptors, ligands, and downstream effectors in the dorsolateral prefrontal cortex and amygdala of suicidal victims. Mol. Neurobiol. 2014;49:957–965. doi: 10.1007/s12035-013-8570-z. [DOI] [PubMed] [Google Scholar]
- 51.Gray A.L., Hyde T.M., Deep-Soboslay A., Kleinman J.E., Sodhi M.S. Sex differences in glutamate receptor gene expression in major depression and suicide. Mol. Psychiatry. 2015;20:1057–1068. doi: 10.1038/mp.2015.91. [DOI] [PubMed] [Google Scholar]
- 52.Fuchsova B., Alvarez Juliá A., Rizavi H.S., Frasch A.C., Pandey G.N. Altered expression of neuroplasticity-related genes in the brain of depressed suicides. Neuroscience. 2015;299:1–17. doi: 10.1016/j.neuroscience.2015.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao J., Qi X.R., Gao S.F., Lu J., van Wamelen D.J., Kamphuis W., Bao A.M., Swaab D.F. Different stress-related gene expression in depression and suicide. J. Psychiatr. Res. 2015;68:176–185. doi: 10.1016/j.jpsychires.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 54.Yin H., Galfalvy H., Pantazatos S.P., Huang Y.Y., Rosoklija G.B., Dwork A.J., Burke A., Arango V., Oquendo M.A., Mann J.J. Glucocorticoid receptor-related genes: Genotype and brain gene expression relationships to suicide and major depressive disorder. Depress. Anxiety. 2016;33:531–540. doi: 10.1002/da.22499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pandey G.N., Rizavi H.S., Zhang H., Bhaumik R., Ren X. The Expression of the Suicide-Associated Gene SKA2 Is Decreased in the Prefrontal Cortex of Suicide Victims but Not of Nonsuicidal Patients. Int. J. Neuropsychopharmacol. 2016;19:pyw015. doi: 10.1093/ijnp/pyw015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nagy C., Torres-Platas S.G., Mechawar N., Turecki G. Repression of Astrocytic Connexins in Cortical and Subcortical Brain Regions and Prefrontal Enrichment of H3K9me3 in Depression and Suicide. Int. J. Neuropsychopharmacol. 2017;20:50–57. doi: 10.1093/ijnp/pyw071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pantazatos S.P., Huang Y.Y., Rosoklija G.B., Dwork A.J., Arango V., Mann J.J. Whole-transcriptome brain expression and exon-usage profiling in major depression and suicide: Evidence for altered glial, endothelial and ATPase activity. Mol. Psychiatry. 2017;22:760–773. doi: 10.1038/mp.2016.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.García-Gutiérrez M.S., Navarrete F., Navarro G., Reyes-Resina I., Franco R., Lanciego J.L., Giner S., Manzanares J. Alterations in Gene and Protein Expression of Cannabinoid CB(2) and GPR55 Receptors in the Dorsolateral Prefrontal Cortex of Suicide Victims. Neurother. J. Am. Soc. Exp. NeuroTherapeutics. 2018;15:796–806. doi: 10.1007/s13311-018-0610-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kouter K., Zupanc T., Videtič Paska A. Genome-wide DNA methylation in suicide victims revealing impact on gene expression. J. Affect. Disord. 2019;253:419–425. doi: 10.1016/j.jad.2019.04.077. [DOI] [PubMed] [Google Scholar]
- 60.Cabrera B., Monroy-Jaramillo N., Fries G.R., Mendoza-Morales R.C., García-Dolores F., Mendoza-Larios A., Diaz-Otañez C., Walss-Bass C., Glahn D.C., Ostrosky-Wegman P., et al. Brain Gene Expression Pattern of Subjects with Completed Suicide and Comorbid Substance Use Disorder. Mol. Neuropsychiatry. 2019;5:60–73. doi: 10.1159/000493940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang L., Verwer R.W.H., Lucassen P.J., Huitinga I., Swaab D.F. Prefrontal cortex alterations in glia gene expression in schizophrenia with and without suicide. J. Psychiatr. Res. 2020;121:31–38. doi: 10.1016/j.jpsychires.2019.11.002. [DOI] [PubMed] [Google Scholar]
- 62.Zhang L., Verwer R.W.H., Lucassen P.J., Huitinga I., Swaab D.F. Sex difference in glia gene expression in the dorsolateral prefrontal cortex in bipolar disorder: Relation to psychotic features. J. Psychiatr. Res. 2020;125:66–74. doi: 10.1016/j.jpsychires.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 63.Cabrera-Mendoza B., Martínez-Magaña J.J., Genis-Mendoza A.D., Monroy-Jaramillo N., Walss-Bass C., Fries G.R., García-Dolores F., López-Armenta M., Flores G., Vázquez-Roque R.A., et al. Brain Gene Expression-DNA Methylation Correlation in Suicide Completers: Preliminary Results. Rev. Investig. Clin. Organo Del Hosp. Enferm. Nutr. 2020;72:283–292. doi: 10.24875/RIC.19003250. [DOI] [PubMed] [Google Scholar]
- 64.Postolache T.T., Akram F., Lee E.E., Lowry C.A., Stiller J.W., Brenner L.A., Streeten E.A., Turecki G., Dwivedi Y. Increased brain vitamin D receptor expression and decreased expression of cathelicidin antimicrobial peptide in individuals who died by suicide. J. Psychiatr. Res. 2020;125:75–84. doi: 10.1016/j.jpsychires.2020.02.027. [DOI] [PubMed] [Google Scholar]
- 65.Squassina A., Niola P., Lopez J.P., Cruceanu C., Pisanu C., Congiu D., Severino G., Ardau R., Chillotti C., Alda M., et al. MicroRNA expression profiling of lymphoblasts from bipolar disorder patients who died by suicide, pathway analysis and integration with postmortem brain findings. Eur. Neuropsychopharmacol. 2020;34:39–49. doi: 10.1016/j.euroneuro.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 66.Romero-Pimentel A.L., Almeida D., Muñoz-Montero S., Rangel C., Mendoza-Morales R., Gonzalez-Saenz E.E., Nagy C., Chen G., Aouabed Z., Theroux J.F., et al. Integrative DNA Methylation and Gene Expression Analysis in the Prefrontal Cortex of Mexicans who died by Suicide. Int. J. Neuropsychopharmacol. 2021;24:935–947. doi: 10.1093/ijnp/pyab042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Furczyk K., Schutová B., Michel T.M., Thome J., Büttner A. The neurobiology of suicide—A Review of post-mortem studies. J. Mol. Psychiatry. 2013;1:2. doi: 10.1186/2049-9256-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Heeringen K., Mann J.J. The neurobiology of suicide. Lancet Psychiatry. 2014;1:63–72. doi: 10.1016/S2215-0366(14)70220-2. [DOI] [PubMed] [Google Scholar]
- 69.Yin H., Pantazatos S.P., Galfalvy H., Huang Y.Y., Rosoklija G.B., Dwork A.J., Burke A., Arango V., Oquendo M.A., Mann J.J. A pilot integrative genomics study of GABA and glutamate neurotransmitter systems in suicide, suicidal behavior, and major depressive disorder. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet. 2016;171B:414–426. doi: 10.1002/ajmg.b.32423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao J., Verwer R.W.H., Gao S.F., Qi X.R., Lucassen P.J., Kessels H.W., Swaab D.F. Prefrontal alterations in GABAergic and glutamatergic gene expression in relation to depression and suicide. J. Psychiatr. Res. 2018;102:261–274. doi: 10.1016/j.jpsychires.2018.04.020. [DOI] [PubMed] [Google Scholar]
- 71.Klempan T.A., Sequeira A., Canetti L., Lalovic A., Ernst C., ffrench-Mullen J., Turecki G. Altered expression of genes involved in ATP biosynthesis and GABAergic neurotransmission in the ventral prefrontal cortex of suicides with and without major depression. Mol. Psychiatry. 2009;14:175–189. doi: 10.1038/sj.mp.4002110. [DOI] [PubMed] [Google Scholar]
- 72.Limon A., Mamdani F., Hjelm B.E., Vawter M.P., Sequeira A. Targets of polyamine dysregulation in major depression and suicide: Activity-dependent feedback, excitability, and neurotransmission. Neurosci. Biobehav. Rev. 2016;66:80–91. doi: 10.1016/j.neubiorev.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Almond B. Drug use and abuse: The ethical issues. Ciba Found. Symp. 1992;166:277–283. doi: 10.1002/9780470514245.ch16. discussion 284–293. [DOI] [PubMed] [Google Scholar]
- 74.Liu S. Neurotrophic factors in enteric physiology and pathophysiology. Neurogastroenterol. Motil. Off. J. Eur. Gastrointest. Motil. Soc. 2018;30:e13446. doi: 10.1111/nmo.13446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Costanza A., D’Orta I., Perroud N., Burkhardt S., Malafosse A., Mangin P., La Harpe R. Neurobiology of suicide: Do biomarkers exist? Int. J. Leg. Med. 2014;128:73–82. doi: 10.1007/s00414-013-0835-6. [DOI] [PubMed] [Google Scholar]
- 76.Dwivedi Y. Brain-derived neurotrophic factor: Role in depression and suicide. Neuropsychiatr. Dis. Treat. 2009;5:433–449. doi: 10.2147/NDT.S5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O’Connor D.B., Gartland N., O’Connor R.C. Stress, cortisol and suicide risk. Int. Rev. Neurobiol. 2020;152:101–130. doi: 10.1016/bs.irn.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 78.Nestler E.J. Epigenetic mechanisms in psychiatry. Biol. Psychiatry. 2009;65:189–190. doi: 10.1016/j.biopsych.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 79.Smith P.N., Cukrowicz K.C. Capable of suicide: A functional model of the acquired capability component of the Interpersonal-Psychological Theory of Suicide. Suicide Life-Threat. Behav. 2010;40:266–275. doi: 10.1521/suli.2010.40.3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wannemueller A., Forkmann T., Glaesmer H., Juckel G., Paashaus L., Rath D., Schönfelder A., Moser D., Kumsta R., Teismann T. The role of the 5-HTTLPR polymorphism in acquired capability for suicide. Suicide Life-Threat. Behav. 2020;50:1121–1126. doi: 10.1111/sltb.12660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.