Summary:
Molecular chaperones govern proteome health to support cell homeostasis. An essential eukaryotic component of the chaperone system is Hsp90. Using a chemical-biology approach we characterized the features driving the Hsp90 physical interactome. We found that Hsp90 associated with ~20% of the yeast proteome using its three domains to preferentially target intrinsically disordered regions (IDRs) of client proteins. Hsp90 selectively utilized an IDR to regulate client activity as well as maintained IDR-protein health by preventing the transition to stress granules or P-bodies at physiological temperatures. We also discovered that Hsp90 controls the fidelity of ribosome initiation that triggers a heat shock response when disrupted. Our study provides insights into how this abundant molecular chaperone supports a dynamic and healthy native protein landscape.
eTOC blurb:
The central eukaryotic molecular chaperone Hsp90 physically interacts with a large fraction of the yeast proteome by targeting select intrinsically disordered regions (IDRs) on client proteins to both regulate their cellular activities and maintain the proteostasic status of IDR-containing clients.
Graphical Abstract
Introduction
Molecular chaperones are the sentinels of the protein homeostasis (proteostasis) process that are required to safeguard polypeptides by continuously scrutinizing the conformational status of all cellular proteins1. Constant monitoring facilitates the forward flow of biological pathways including the folding of nascent chains, regulation of native proteins, and removal of damaged polypeptides. Breakdown in proteostasis, which can be triggered by stress and aging, correlates with many diseases including neurodegeneration, type II diabetes, and heart failure2. Insights into proteostasis have been primarily gained by studying protein biogenesis and triage events1–3. Yet, the proverbial birth and death of any polypeptide represent a small percentage of the protein-lifecycle. The mainstay of a protein’s time is spent functioning in pathways to support life4. Minimally, proteins adopt distinct physical forms to accomplish work or interact with cofactors, yet the mechanisms maintaining a healthy proteome or fostering transitions between different physical states are not well understood. Although it has long been suggested that molecular chaperones regulate native proteins5, how any chaperone might recognize and/or modulate the broad spectrum of mature proteins found within a cell is unknown.
Hsp90 and Hsp70 are the central hubs of eukaryotic chaperone networks3,6,7. Hsp70s comprise a highly conserved protein family that serves in protein biogenesis and degradation by recognizing short, hydrophobic motifs exposed in nonnative proteins8. Hsp90s evolved from a single nonessential factor in bacteria to an essential family in eukaryotes7. Hsp90 was first identified with metastable factors, including unactivated hormone receptors and kinases9,10, leading to the concept that Hsp90 recognizes polypeptides late in the folding process with recent efforts extending Hsp90’s reach to fully mature proteins7. Large-scale genetic interaction screens revealed that this abundant chaperone is linked to a wide variety of cellular processes11,12, nevertheless most physical interaction screens only identified a limited number of hits (e.g., 2-hybrid found 20)12,13. Likely, Hsp90 transiently interacts with its clients thereby abating detection by standard methods. A study incorporating a chemical crosslinker followed by mass spectrometry showed that Hsp90 associated with ~8% of the yeast proteome14. How Hsp90 recognizes these diverse clients was unresolved.
Hsp90 consists of an amino (N), middle (M), and carboxyl (C) terminal domain with the N-domain minimally housing ATPase activity and the C-domain mediating Hsp90 dimerization7. A high-resolution structural study showed that Hsp90 contacted the Cdk4 kinase client primarily using its M-domains15. Whether Hsp90 uses one or all three domains to commonly recognize other clients had not been determined. To better comprehend the mechanistic features driving client recognition by the Hsp90 molecular chaperone we exploited the non-natural amino acid p-benzoyl-L-phenylalanine (Bpa), which is also a photo-activatable crosslinker16, and large-scale mass spectrometry to build a domain-centric Hsp90 interaction map. Our presented findings establish a large physical interactome for Hsp90, provide insights into how Hsp90 is able to broadly associate with native protein clients, reveal a chaperone-dependent mechanism for maintaining the health of proteins with intrinsically disordered regions (IDRs), and identify a novel role for Hsp90 in regulating the fidelity of translation initiation that seemingly drives the heat shock response triggered by the inhibition of Hsp90.
Results
Constructing an Hsp90 interactome
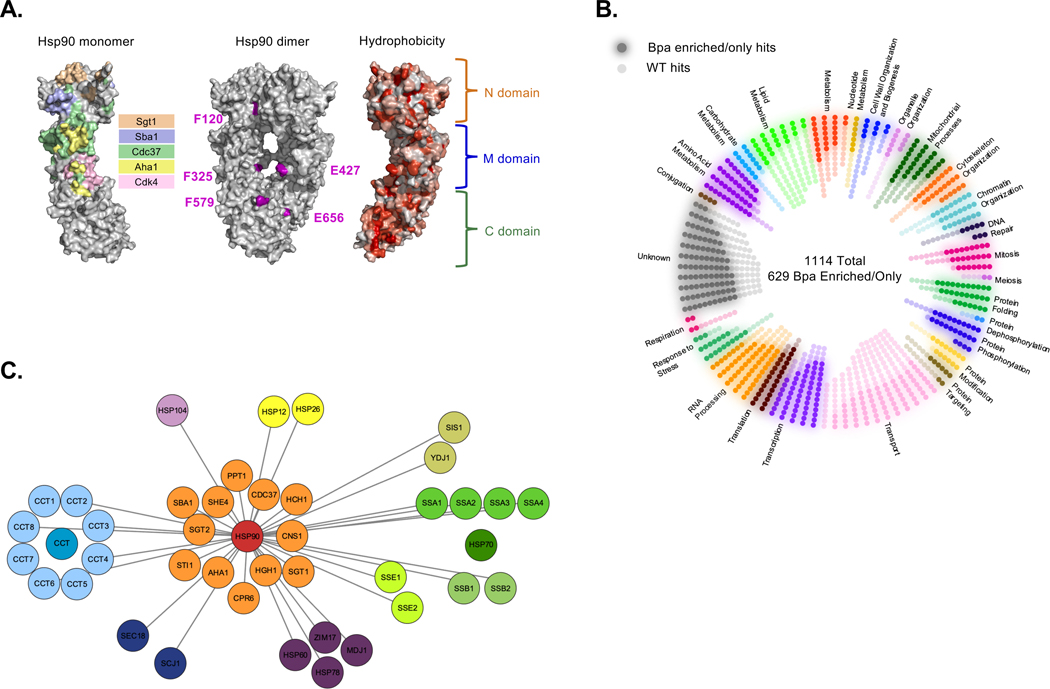
To create our Hsp90 physical interactome we engineered variants of yeast Hsp90 that express a single Bpa substitution at defined residues (Figure 1A). Our Bpa design criteria considered the structure of the 3 Hsp90 domains, known interaction sites (e.g., cochaperone partners and Cdk4 client), and the hydrophobicity topology map of Hsp90. However, despite the use of hydrophobic pockets by other chaperones to bind clients3, the hydrophobic surface areas of Hsp90 are distributed across its 3 domains and therefore did not seem insightful. We generated 5 Bpa derivatives positioned at yeast Hsp90 residues F120 (N-domain), E325 (M), E427 (M), F579 (C), and E656 (C) (Figure 1A) that supported life in a manner comparable to wild type (WT) and the maturation of the well-established Hsp90 client protein Glucocorticoid Receptor when serving as the sole source of full-length Hsp90 (Figure S1A, S1B, and S1C). WT Hsp90 and the Bpa-variants were expressed as histidine-tagged fusions to facilitate mass spectroscopic analysis of the affinity purified chaperone and associated proteins. Of note, our UV crosslinking conditions do not appear to be physiologically stressful since the conditions did not invoke a heat shock response (Figure S1D).
Figure 1. Establishing an Hsp90 physical interactome.
(A) Topology maps of Hsp90 showing the interaction areas with the indicated cochaperones or Cdk4 client (left), hydrophobicity surfaces (right) or amino acid positions that were engineered to Bpa residues (center). (B) Hsp90 interactors identified by mass spectrometry that were Bpa-dependent (dark spots) or Bpa-independent (light spot) were separated into gene ontology categories, each hit was assigned to an initial cell process and displayed. (C) The Bpa-hits demonstrate a substantial physical connection between Hsp90 and numerous other molecular chaperones and cochaperones, which we will refer to as (co)chaperone interactions. (See also Figure S1, Table S1)
We reasoned that the use of both WT and Bpa-modified Hsp90 would permit the isolation of stably- and transiently-bound interactors. A total of 1114 proteins or ~20% of the yeast proteome were identified with 629 being Bpa-dependent (Figure 1B; Table S1). Gene ontology (GO) analysis of the hits showed heightened connections to protein folding, protein targeting, and response to heat (Figure S1E). Our dataset supports the concept that Hsp90 serves as a central hub of a eukaryotic chaperone network since Hsp90 shared connections to numerous other chaperones including Hsp70s, Hsp40s, CCT, Hsp104, as well as the majority of known Hsp90 cochaperones (11 of 14) (Figure 1C; Table S1). Comparison to the known Hsp90 physical and genetic interactors (Saccharomyces Genome Database; www.yeastgenome.org) showed overlaps of ~30% for each (Figure S1F). Given that our Bpa-tactic uses only 5 sites with a crosslinker that has a ~3 Å reach, which only covers 2% of the surface area of Hsp90, the ~30% overlap with the existing physical interactome is considerable. Overall, our approach built an intricate Hsp90 interactome.
Hsp90 uses all three domains to associate with proteins
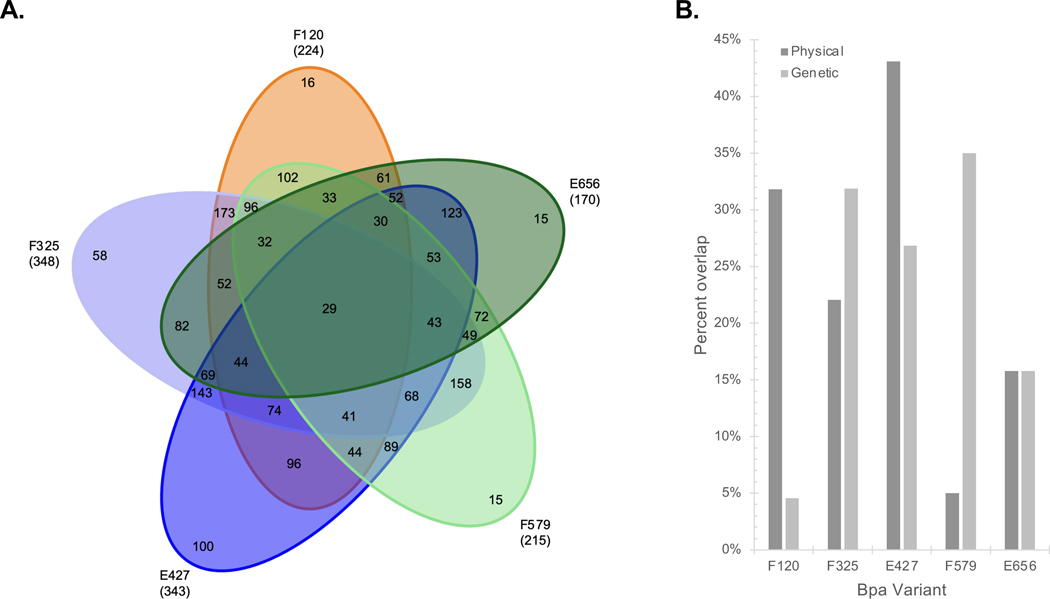
By incorporating Bpa at select sites along the chaperone’s surface, the domains used to coordinate physical interactions with targets can be resolved. Our results indicate that all 3 Hsp90 domains are used to associate other proteins since each Bpa site mediated a comparable number of contacts (Figure 2A; Table S2). Moreover, the same target proteins were often captured with more than one Bpa site suggesting that Hsp90 uses multiple contact points when interacting with other proteins. We had anticipated that established interactors would be equally distributed across our Bpa sites. Yet, our analysis showed that the N-domain residue 120 preferred known physical binders and the C-domain site 579 preferentially overlapped with genetic interactors (Figure 2B). A GO assessment revealed a differential use of the Hsp90 sites with proteins working in different pathways (Figure S2). Minimally, our domain-centric interactome supports the concept that Hsp90 binds to numerous proteins by using all 3 of its domains thereby exploiting Hsp90’s large hydrophobic surface area to foster associations with a broad array of clients and cochaperones (Figure 1B; Table S1).
Figure 2. Combinatorial use of Hsp90’s 3 domains support a broad physical interactome.
(A) A Venn diagram with the total number of hits for each Hsp90 Bpa site is shown as well as the overlap between the hits linked to the various sites. (B) Bpa site-specific hits were compared to the established physical and genetic interactors of yeast Hsp90s. (See also Figure S2, Table S2)
Hsp90 client binding sites are favored within intrinsically disordered regions
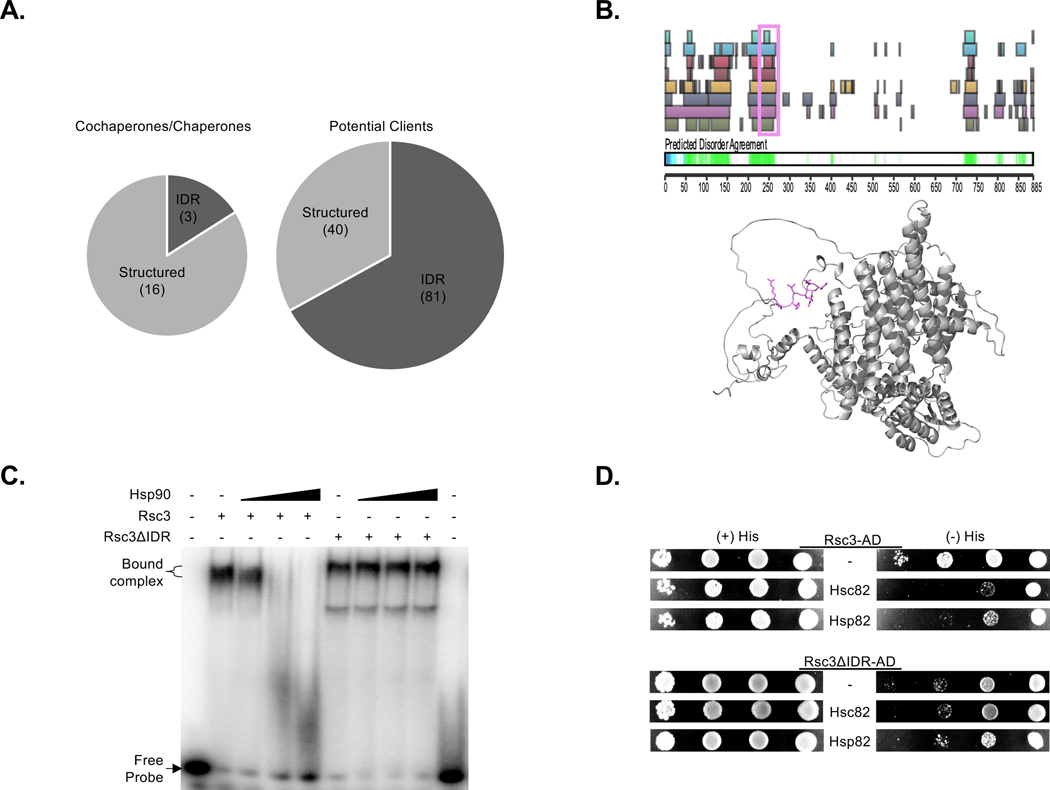
Besides mapping the Hsp90 domains used for protein interactions, we exploited the Bpa-crosslinking tactic to identify what client features are recognized by Hsp90. In brief, the Bpa-peptide serves as a unique tag in the mass spectrometry spectra since it is a non-natural amino acid. First, we identified crosslinked peptide pairs using the MeroX algorithm17 and then further filtered the pairs as described (Figure S3A; Table S3). Crosslinked products containing peptides from the Sti1 cochaperone supported the validity of our approach since the Bpa-crosslinked peptide pairs aligned in the known structure of the Hsp90-Sti1 complex (Figure S3B). Similarly, the established interactions between the Cpr6 and Sgt2 involved the carboxyl-domain of Hsp90 and the respective tetratricopeptide repeat (TPR) domains of each cochaperone (Table S3).
For the crosslinked products representing potential clients, we had anticipated finding an Hsp90 recognition motif within the identified Bpa-crosslinked peptide pairs (Table S3). However, no consensus sequence was apparent within these 152 peptides. Rather, we found that the bound client peptides were preferentially located within intrinsically disordered regions (IDRs) whereas Hsp90 interacted with structured regions of other chaperones and Hsp90 cochaperones (collectively referred to as (co)chaperones) (Figure 3A). The presence and locations of IDRs were based upon the D2P2 prediction database and further supported by visualizing their locations using AlphaFold structures (Figure 3B and S3C)18,19. A previous study had shown how Hsp90 interacts with the intrinsically disordered protein Tau using its N- and M-domains and favored certain motifs and positively charged residues20. Our findings indicate that Hsp90 uses all 3 domains to broadly target IDR peptides independent of sequence and charge (Figure S3E; Table S3 and S4). The propensity of the Hsp90-client peptides to be within certain IDRs suggests that selective pressure has acted on the nature of the Hsp90-client interaction.
Figure 3. Hsp90 associates with a specific IDR to regulate client activity.
(A) The propensity of Hsp90-bound peptides (numbers in parentheses) to be within structured (e.g., α-helix or β-sheet) or IDR regions of a target protein was determined for known (co)chaperones and potential clients, as indicated including total numbers for each, using D2P2 and AlphaFold software. (B) The D2P2 (top; the rows of the colored bars represent the prediction of disorder by the 9 different algorithms used by D2P2, each row/color corresponds to a single predictor and the bars within the row are a prediction of disorder) and AlphaFold (bottom) predictions for the Hsp90 client Rsc3 is shown as well as the position of the Hsp90-linked peptide (highlighted in violet). (C) The influence of Hsp90 (1, 4, 16 μM) on the DNA binding activity of Rsc3 and Rsc3 missing the Hsp90-associated IDR (Rsc3ΔIDR) was monitored by EMSA using purified recombinant proteins and a radiolabeled dsDNA RSC3 consensus oligonucleotide. (D) The capacity of Rsc3-AD and Rsc3ΔIDR-AD to drive the expression of a HIS3 reporter through an upstream RSC3 consensus motif was monitored in yeast transformants carrying an empty vector (−) or expressing the yeast Hsp90 homologs Hsc82 or Hsp82, as marked. (See also Figure S3, Table S3, Table S4)
Hsp90 targets select IDRs to regulate client function
To test whether a targeted IDR was relevant we interrogated the established Hsp90 client Rsc3, which is a DNA binding subunit of the RSC chromatin remodeler. In brief, chaperones disrupt the DNA binding activity of Rsc321. Guided by structural predictions with both D2P2 and AlphaFold, the Rsc3 peptide linked to the Hsp90 Bpa moiety is within an IDR (Figure 3B). Using recombinant purified proteins, including normal Rsc3 and Rsc3 deleted of the target IDR (Rsc3ΔIDR), Hsp90 was unable to dissociate the DNA bound Rsc3 protein in the absence of this IDR (Figure 3C). In a modified 2-hybrid assay where Rsc3 drives expression of the HIS3 reporter gene through a RSC3 consensus element in the promoter21, Hsp90 fused to the Gal4-DBD did not influence the activation potential of Rsc3ΔIDR-AD whereas it disrupted the activation potential of Rsc3-AD (Figure 3D). Of note, based on the prediction programs, Rsc3ΔIDR still has at least 2 IDRs (Figure 3B), which suggests Hsp90 selectively recognizes its target IDR both in vitro and in vivo. Hence, the IDRs targeted by Hsp90 may have evolved to allow chaperone-mediated client regulation.
Hsp90 governs translation initiation by maintaining the physical state of a client
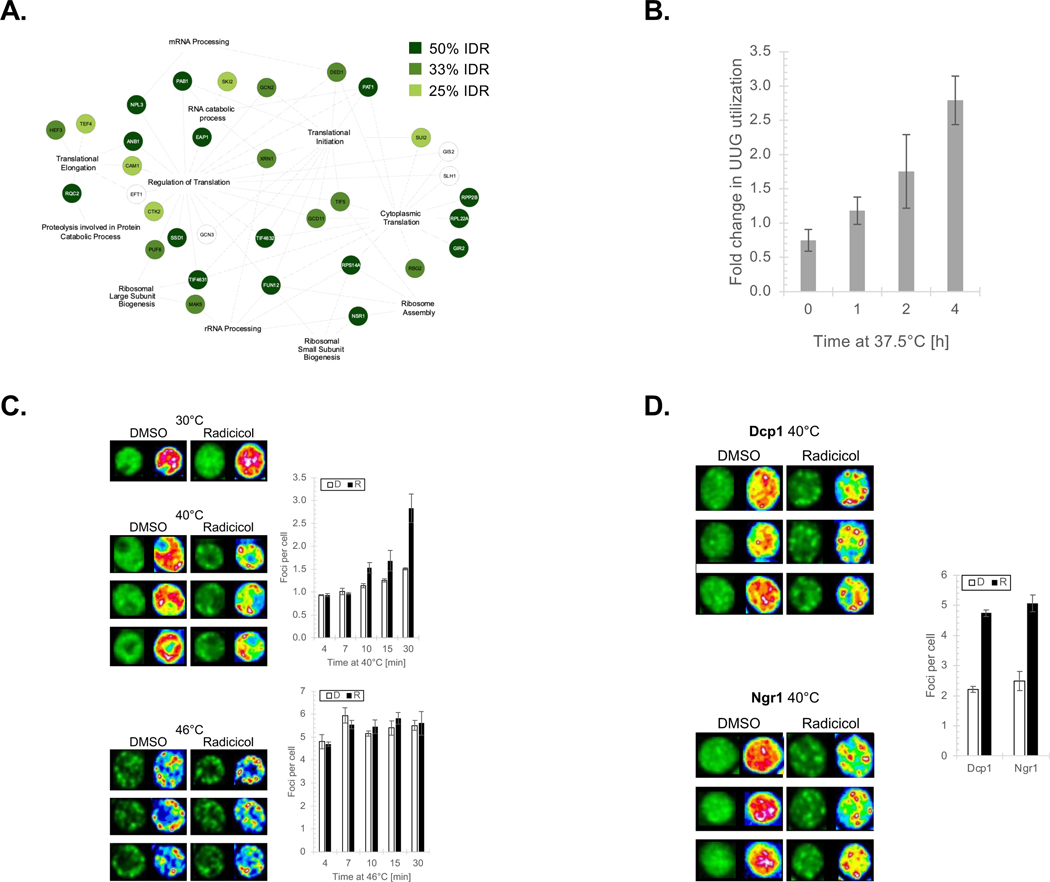
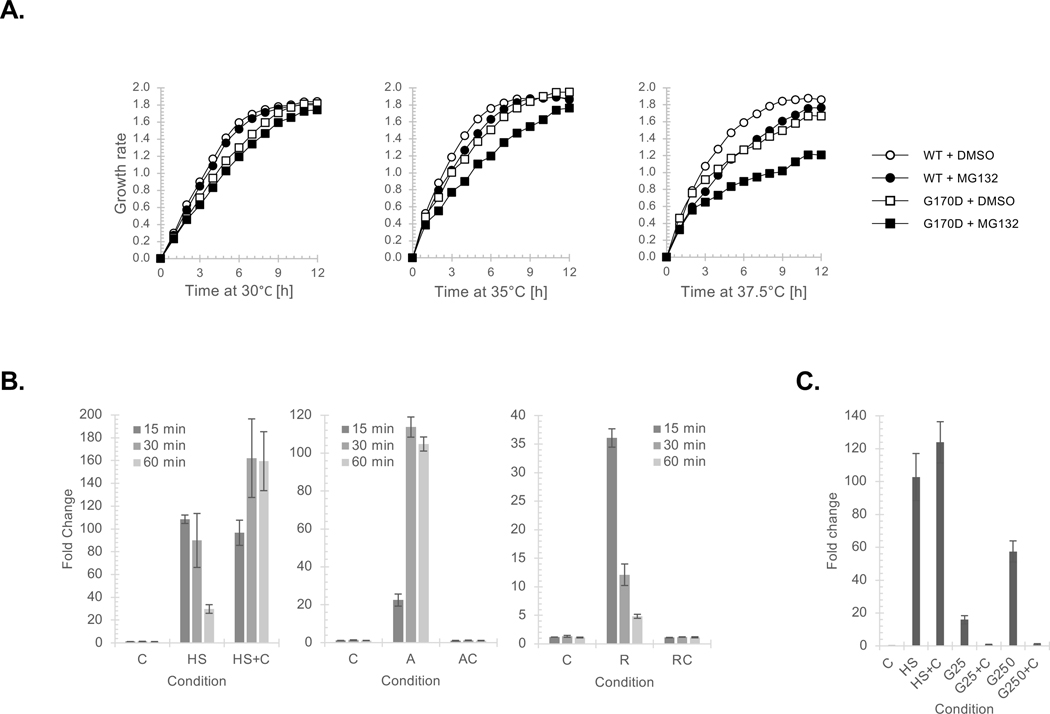
To further understand the connection between Hsp90 and IDRs we focused on an Hsp90-linked cell process that is enriched with IDR-containing factors—protein translation (Figure 1B and 4A). We used polysome fractionation to identify the translation complex associated with Hsp90 and found that it is primarily with the 40S small ribosome subunit peak whereas the nascent chain binding chaperone Hsp70 was apparent across the ribosome fractions (Figure S4A). A key function of the 40S complex is to scan the mRNA 5’-UTR for the AUG start codon22. Hence, we tested whether Hsp90 influences 40S activity by checking the fidelity of translation initiation using the ts Hsp90 allele G170D and a dual luciferase system where the firefly luciferase ORF begins with the non-conical start codon UUG and Renilla luciferase has the control AUG23. Shifting to the non-permissive temperature of 37.5°C, which fully inactivates G170D24, reduced the fidelity of translation initiation since the production of UUG-driven firefly luciferase increased (Figure 4B; all values were normalized to the WT Renilla signal). Next, we wished to identify the translation protein(s) targeted by Hsp90.
Figure 4. Hsp90 regulates translation initiation, which controls an HSR, by influencing the physical state of the Ded1 RNA helicase.
(A) Hsp90-hits functioning in various steps of the protein translation process are shown. Each protein is shaded based on its relative percent of IDR with dark green (50% IDR), medium green (33%), light green (25%), and white (<25%). (B) The fidelity of translation initiation was determined using the dual luciferase assay and yeast expressing WT or the G170D ts allele as a sole source of Hsp90. Data is presented as the mean ± SEM and the p-values are the following: 1 h 0.165, 2 h 0.147, and 4 h 0.006. Fold increase in luciferase expression of firefly (UUG noncanonical) relative to Renilla (AUG) in G170D was normalized to WT following incubation at 37.5°C. (C) Formation of Ded1 SGs were visualized in yeast expressing GFP-Ded1 that were treated with DMSO (white bars) or Radicicol (black bars) and shifted to 40° or 46°C, as marked for the indicated times. Data are presented as mean ± SEM and the p-values are the following: 40°C 4 min 0.90, 7 min 0.41, 10 min 0.08, 15 min 0.21, 30 min 0.04 and 46°C 4 min 0.75, 7 min 0.34, 10 min 0.41, 15 min 0.35, 30 min 0.86. (D) The physical status of the SG component Dcg1 and P-Body factor Ngr1 were followed using GFP-fusions in yeast treated with DMSO (white bars) or Radicicol (black bars) exposed to 40°C for 30 min. Data are presented as mean ± SEM and the p-values are the following: Dcp1 0.0001 and Ngr1 0.0037.
Classic Hsp90 clients degrade following Hsp90 inhibition7. Yet, none of the 32 translation hits displayed steady state protein changes upon loss of Hsp90 activity (Figure S4B). However, when translation factors inactivate in reaction to stress, the proteins do not necessarily degrade but rather can form stress granules (SGs), which are membraneless compartments comprised of phase-transitioned proteins25. To assess whether Hsp90 influences SG formation we followed the heat-induced transition of the SG-marker Ded126, which is also an Hsp90-hit (Table S1). Ded1 is a DEAD-box RNA helicase that promotes assembly of the 48S preinitiation complex by fostering the scanning of 5’-UTRs and forms SGs in an IDR-dependent manner27. Typically, SG formation is triggered by exposing cells to a super-physiological temperature that also results in a substantial cell death (e.g., 46°C)28. At 46°C the inhibition of Hsp90 had no apparent effect on GFP-Ded1 SG development (Figure 4C). Yet, at the physiologically and, notably, clinically relevant temperature of 40°C29, the formation of GFP-Ded1 SGs increased in the presence of the Hsp90 inhibitor Radicicol (Figure 4C). Significantly, the role of Hsp90 in maintaining the physical status of clients extends beyond Ded1 since the SG protein Ngr1 and P-Body factor Dcp1 more readily transitioned upon inhibition of Hsp90 with Radicicol (Figure 4D). Thus, Hsp90 activity is required to maintain the physical state of client proteins at physiologically relevant temperatures. Minimally, our data demonstrate that Hsp90 promotes the fidelity of translation initiation by maintaining the physical status of IDR-containing translation proteins such as the Ded1 RNA helicase.
HSR triggered by Hsp90 inhibition is translation-dependent
As would be expected for cells producing aberrant polypeptides because of mis-initiation, cells were more sensitive to the proteosome inhibitor MG132 following the progressive inactivation of the G170D allele (Figure 5A). We reasoned that the influence of Hsp90 on protein translation initiation might account for the HSR triggered by most Hsp90 inhibitors, which correlates with numerous failed Hsp90 clinical trials30. We tested whether active translation is required for a HSR triggered by different stresses (i.e., elevated temperature (42°C), the proline analog azetidine, or an Hsp90 inhibitor (Radicicol or Ganetespib)) using the translation inhibitor cycloheximide. Addition of cycloheximide exacerbated the HSR triggered by 42°C treatment, alleviated the azetidine effect, and prevented the HSR upon Hsp90 inhibitor application in either yeast (Figure 5B) or human cells (Figure 5C). Hence, comparable to azetidine, the HSR triggered by Hsp90 inhibitors is dependent upon active protein translation to create anomalous polypeptides.
Figure 5. The HSR response triggered by Hsp90 loss is translation dependent.
(A) Growth rates were determined for parental (circles) or G170D (squares) yeast grown at 30°, 35°, or 37.5°C in the absence (open) or presence (closed) of MG132 for the marked times. SEM for each data point derived from biological triplicates are included. (B) The induction of SSA4 expression in yeast exposed to cycloheximide (C), 42°C (HS), Azetidine (A), and/or Radicicol (R) was measured by qRT-PCR relative to cells maintained at 30°C. Data are presented as mean ± SEM. (C) The induction of HSP70 expression in human HeLA cells exposed to 42°C (HS) or Ganetespib (G25 – 25 μM; G250 – 250 μM) with or without cycloheximide (C) was measured by qRT-PCR relative to cells maintained at 37°C. Data are presented as mean ± SEM and the p-values are the following: HS vs C 0.0001, HS+C vs HS 0.2930, G25 vs C 0.0001, G25+C 0.1375, G250 vs C 0.0001, and G250+C 0.0523. (See also Figure S4)
Discussion
Our presented findings support a new paradigm for governing a native protein landscape in which the abundant Hsp90 molecular chaperone targets specific IDRs to both regulate client protein activity and maintain the physical health of a proteome. Traditionally, protein function has been conceptualized around the dogma that the 1° amino acid sequence defines the 2° structure (i.e., α-helices and β-sheets) that forms the basis for the 3° fold which, in turn, determines the operational capacity of a polypeptide31,32. In other words, form equals function. This framework has been useful for understanding and assigning functions to individual domains or full-length proteins as well as comprehending molecular evolution where the overall fold of distantly related polypeptides can be comparable despite differences in the 1° sequence33. While the existence of unstructured protein regions has long been acknowledged, these areas typically have been cast as linkers or as sites that adopt a structure when an appropriate partner or ligand is encountered, yet on the whole the biological significance of IDRs has remained enigmatic34. Our presented work indicates that intrinsically disordered regions might be central to an ancient mechanism of chaperone-mediated proteome control.
The Hsp90 molecular chaperone is a highly abundant protein that is conserved in bacteria and Eukarya but absent in Archaea35. The biological relevance of Hsp90 has risen in parallel with complexity, as it is nonessential in bacteria but essential in Eukarya7. Comparably, IDR presence appears to intensify going from Archaea to bacteria to Eukarya. Although in silico IDR identification is difficult, most prediction algorithms reveal the same trend with Archaea having the lowest and Eukarya having the highest level of IDR presence. For instance, estimations based on DISOPRED2 showed that Archaea proteins have 2% IDRs of 30 residues or more, bacteria 4.2%, and Eukarya 33%36. In general, prediction software suggest bacterial polypeptides are ~10% IDR and Eukarya ~30%, which equates to 135 disordered residues for the average Eukarya protein (average length 450) and 36 amino acids for the typical bacterial factor (average length 300)37. In S. cerevisiae and based on D2P2, 75% of the yeast proteome is considered to have at least 10% IDRs and 60% of the proteome is covered if 33% of each protein is considered intrinsically disordered. Despite the prevalence of Hsp90 and IDRs in Eukarya proteomes, the physiological relevance for both is still unclear.
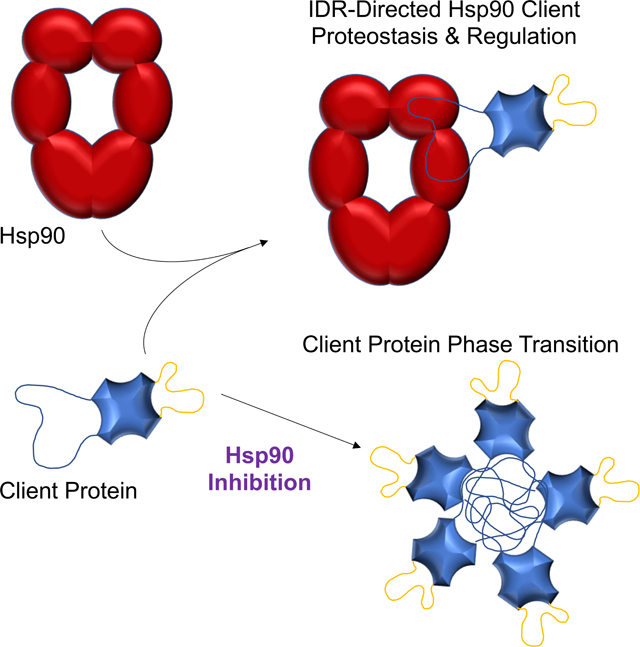
The biological purpose for Hsp90 generally has been relegated to maintaining the steady-state levels of metastable signaling proteins including kinases and steroid hormone receptors9,10. By developing the Bpa chemical-biology tactic into a high-throughput approach, we mapped which Hsp90 domains associated with 1114 different yeast proteins as well as identified 181 direct contact sites (Figure 1, Tables S1 and S3). Our data supports a model in which Hsp90 broadly associates with diverse protein clientele by targeting specific IDRs utilizing its large hydrophobic surface area. We suggest that by distributing the interaction areas across the 3 domains of Hsp90, the chaperone gains a combinatorial binding mechanism in which different permutations of the interaction surfaces are coordinated to associate with the wider variety of targets (Figure 6). The specificity and timing of the Hsp90-client complexes might be dictated by various structural forms of the chaperone.
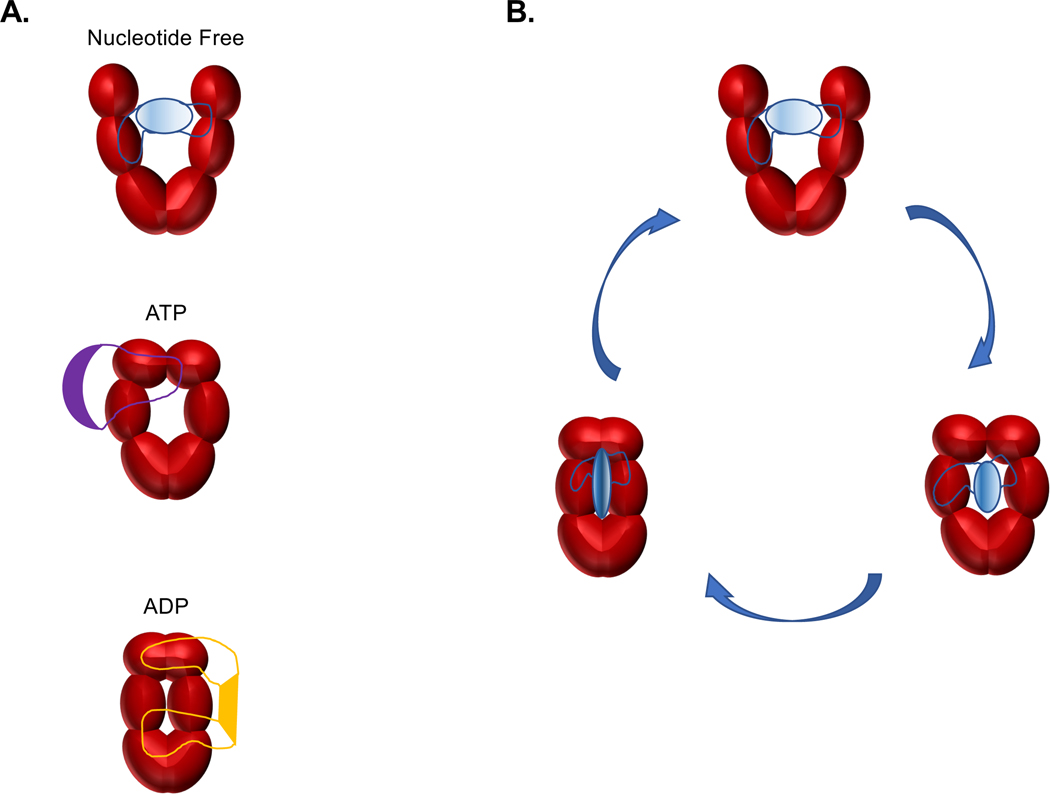
Figure 6. Client protein recognition and regulation by the Hsp90 chaperone.
The well-established ATP-dependent conformations of Hsp907 further add to the client recognition capacity of Hsp90 by providing distinct binding platforms for different target proteins (A) and mediating a physical manipulation of a bound client polypeptide as Hsp90 progresses through its ATPase cycle (B).
It is well-established that Hsp90 itself undergoes drastic conformational changes while progressing through its ATPase cycle7 and that interactions with a client also can modulate the conformational dynamics of Hsp9038. Minimally, Hsp90 has a nucleotide-free open state, an ATP client loading configuration, and an ADP client maturation form (Figure 6). As the angles and distances between the 3 domains change during the ATP cycle, we propose that these Hsp90 states create different binding platforms to accommodate diverse clients (Figure 6A). Additionally, once a target protein is bound, the ATP-dependent changes in the structure of the Hsp90 dimer can manipulate the conformation of an engaged client (Figure 6B). Hence, by exploiting numerous weak binding areas across the highly flexible 6 domains of the Hsp90 dimer, diverse targets can both be recognized and manipulated to regulate client activity (Figure 3) and maintain polypeptide health (Figure 4). Overall, our model opens new possibilities into the molecular mechanisms by which Hsp90 chaperones client proteins.
In addition to understanding the chaperone attributes contributing to Hsp90-client complexes, our data uncovered a central client feature recognized by Hsp90—Hsp90 relied on a specific IDR to control client function (Figure 3). The selectivity of the IDR targeting suggests that Hsp90 has coevolved with Rsc3 IDRs to recognize and/or control this client, although additional work is needed to confirm this concept. Beyond modulating target function, Hsp90 also maintained the physical state of IDR-containing proteins since inhibition of Hsp90 prompted a transition of targets into SGs or P-bodies at physiological temperatures (Figure 4). Notably, Hsp90’s influence on stability of the Ded1 translation initiation factor led to the discovery that the HSR linked to Hsp90 inhibition/loss is dependent on active protein translation. Does this finding afford an opportunity to hone Hsp90 therapeutic trials (e.g., cancer and pathogen infections) to avoid triggering an HSR? Minimally, our study provides insights into the workings of an essential eukaryotic molecular chaperone and reveals new pathways governed by the Hsp90 molecular chaperone. Overall, our discoveries suggest that the highly conserved Hsp90 chaperone has coevolved with IDRs as means to allow molecular chaperone-mediated regulation across the native protein landscape that facilitates a dynamic and healthy proteome.
Limitations of the Study
One intention of our study was to resolve the client physical features recognized by the Hsp90 molecular chaperone. While our approach captured numerous proteins associated with Hsp90, we were limited in our ability to identify the crosslinked peptides formed between the Hsp90 Bpa-sites and the bound clients. Hence, our dataset was insufficient to discover whether Hsp90 has a select client recognition motif such as the one used by Hsp708. Nevertheless, we were able to deduce that Hsp90 preferentially recognizes intrinsically disordered regions of client proteins especially relative to the cochaperone interaction sites. A caveat of this finding is the basis for what constitutes an IDR, as these sites were primarily defined using prediction methods including D2P2 and AlphaFold. Furthermore, we suggested that the Hsp90-IDR interaction might have coevolved, yet our hypothesis is based on our singular studies between Hsp90 and the Rsc3 client and therefore the concept should be treated with appropriate caution.
STAR Methods
Resource Availability
Data and code availability
All mass spectrometry (PRIDE) and imaging data (Mendeley Data) generated in this study have been deposited and are publicly available as of the date of publication. Accession numbers and DOI are listed in the key resources table.
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-TAP | ThermoFisher | Cat#CAB1001 |
| Anti-His | ThermoFisher | Cat#MA1–21315 |
| Bacterial and virus strains | ||
| Rosetta(DE3) | Novagen | Cat#70954–3 |
| Chemicals, peptides, and recombinant proteins | ||
| All chemicals unless otherwise noted | ThermoFisher | N/A |
| Yeast Media Components | DIFCO | N/A |
| LB Media Components | DIFCO | N/A |
| p-Benzoyl-L-phenylalanine | ThermoFisher (Bachem) | Cat#NC0169631 |
| Radicicol | AG Scientific | Cat#R-1130 |
| Ganetespib | Selleckchem | Cat#S1159 |
| L-Azetidine-2-carboxylic Acid | TCI | Cat#A1043 |
| Cycloheximide | GoldBio | Cat#C-930–10 |
| D-Luciferin, Potassium salt | GoldBio | Cat#LUCK-100 |
| Coelenterazine | GoldBio | Cat#CZ2.5 |
| CPRG | Roche | Cat#99792–50-4 |
| ATP [γ−32P] | Perkin Elmer | Cat#BLU002Z250UC |
| SYPRO Ruby Protein Gel Stain | ThermoFisher | Cat#S12000 |
| TALON Metal Affinity Resin | Takara Bio | Cat#635503 |
| Restriction Enzymes | New England BioLabs | N/A |
| Critical commercial assays | ||
| Pierce BCA Assay Kit | Fisher Scientific | Cat#PI23223 |
| Power SYBR Green RNA-to-CT 1-Step kit | Applied Biosystems | Cat#4388869 |
| In-Fusion Snap Assembly kit | Takara Bio | Cat#638946 |
| Q5 Site-Directed Mutagenesis Kit | New England BioLabs | Cat#E0554S |
| Deposited data | ||
| Imaging Data | Mendeley Data, V1 | doi:10.17632/8rnk4fggd8.1 |
| Mass Spectrometry Data | PRIDE | PXD040875 |
| Experimental models: Cell lines | ||
| HeLa Human Epithelial cells | ATCC | Cat#CRM-CCL-2 |
| Experimental models: Organisms/strains | ||
| Saccharomyces cerevisiae | Strain Table S2 | N/A |
| TAP tag strain library | Ghaemmaghami et al.42 | N/A |
| Mat a Knockout library | Open Biosystems | Cat#YSC1053 |
| GFP Library | Yofe et al.43 | N/A |
| JJ117 | Wang et al.44 | N/A |
| Oligonucleotides | ||
| SSA4 qRT FP | GCTCAACGTGTTCAAGCTAA | N/A |
| SSA4 qRT RP | ACCCACCTTCTCCTTGAAGT | N/A |
| TAF10 qRT FP | TACGAATATTCCAGGATCAGG | N/A |
| TAF10 qRT RP | CAATAGCTGCCTAGCTCTC | N/A |
| Hsp70 qRT FP | CAGAGCGGAGCCGACAGAG | N/A |
| Hsp70 qRT RP | CCACCTTGCCGTGTTGGAAC | N/A |
| GAPDH qRT FP | ACACCCACTCCTCCACCTTTGAC | N/A |
| GAPDH qRT RP | ACCACCCTGTTGCTGTAGCCA | N/A |
| pGBK Hsc82 N FP | GAATTCCCGGGGATCATGGCTGGTGAAACTTTTGAATTTC | N/A |
| pGBK Hsc82 C RP | GCAGGTCGACGGATCCTTAATCAACTTCTTCCATCTCGGT | N/A |
| Hsc82 N FP | TTGTTGTCTAGAATGGCTGGTGAAACTTTTGAATTTC | N/A |
| Hsc82 N RP | TTGTTGCTGCAGTTACTTAGTCTTGTTCAACTCTTCTAATTC | N/A |
| Hsc82 M FP | TTGTTGTCTAGAGTTCCAATTCCAGAAGAAGAAAAG | N/A |
| Hsc82 M RP | TTGTTGCTGCAGTTATTCTTCCAATTCGAAATCTTTAGT | N/A |
| Hsc82 C FP | TTGTTGTCTAGAACTAAAGATTTCGAATTGGAAGAA | N/A |
| Hsc82 C RP | TTGTTGCTGCAGTTAATCAACTTCTTCCATCTCGG | N/A |
| pGBK Rsc3 FP | CATGGAGGCCGAATTCATGGATATTCGTGGTAGAAAAATG | N/A |
| pGBK Rsc3 RP | GGATCCCCGGGAATTTTAGCTACGAATAGCCTCAAAAAC | N/A |
| Rsc3ΔIDR FP | ATTCAGTATTTGATTGAACG | N/A |
| Rsc3ΔIDR RP | TGTAATTTCTTGCAATTCC | N/A |
| F120-Bpa FP | GCCGATGTATCCATGATTGGTCAATAGGGTGTTGGTTTTTACTCTTTATTC | N/A |
| F120-Bpa RP | GAATAAAGAGTAAAAACCAACACCCTATTGACCAATCATGGATACATCGGC | N/A |
| F325-Bpa FP | TTGTTCATTCCAAAGAGAGCACCATAGGACTTATTTGAGAGTAAGAAGAAG | N/A |
| F325-Bpa RP | CTTCTTCTTACTCTCAAATAAGTCCTATGGTGCTCTCTTTGGAATGAACAA | N/A |
| E427-Bpa FP | AAGAACATTAAGCTGGGTGTACATTAGGACACTCAAAACAGAGCTGCTTTA | N/A |
| E427-Bpa RP | TAAAGCAGCTCTGTTTTGAGTGTCCTAATGTACACCCAGCTTAATGTTCTT | N/A |
| F579-Bpa FP | CATCAGAACTGGTCAATAGGGCTGGTCTGCTAACAT | N/A |
| F579-Bpa RP | ATGTTAGCAGACCAGCCCTATTGACCAGTTCTGATG | N/A |
| E656-Bpa FP | TTGACTTCTGGTTTCAGTTTGGAATAGCCAACTTCTTTTGCATCAAGAATA | N/A |
| E656-Bpa RP | TATTCTTGATGCAAAAGAAGTTGGCTATTCCAAACTGAAACCAGAAGTCAA | N/A |
| ACC1_TIM23 F | ACGCGCGCGCGGCCGGGCCA | N/A |
| ACC1_TIM23 R | TGGCCCGGCCGCGCGCGCGT | N/A |
| Recombinant DNA | ||
| pTGPD-HSP82 | S. Lindquist (Whitehead Institute) | N/A |
| pTGPD-hsp82 G170D | S. Lindquist (Whitehead Institute) | N/A |
| pSNR-tRNA-pBpaRS | A. Mapp (University of Michigan) | N/A |
| pRS313GPD His-Hsc82 | E. Craig (University of Wisconsin, Madison) | N/A |
| pRS313GPD His-Hsc82 F120-Bpa | This manuscript | N/A |
| pRS313GPD His-Hsc82 F325-Bpa | This manuscript | N/A |
| pRS313GPD His-Hsc82 E427-Bpa | This manuscript | N/A |
| pRS313GPD His-Hsc82 F579-Bpa | This manuscript | N/A |
| pRS313GPD His-Hsc82 E656-Bpa | This manuscript | N/A |
| pFJZ1052 (AUG) | J. Lorsch (Division of Intramural Research, NIH) | N/A |
| pFJZ1054 (UUG) | J. Lorsch (Division of Intramural Research, NIH) | N/A |
| pGADT7 | Clontech Matchmaker | Cat#630442 |
| pGBKT7 | Clontech Matchmaker | Cat#630489 |
| pGBKT7 Hsp82 | Echtenkamp et al.21 | N/A |
| pGADT7 Rsc3 | Echtenkamp et al.21 | N/A |
| pGBKT7 Hsc82 | This manuscript | N/A |
| pGADT7 Rsc3ΔIDR | This manuscript | N/A |
| pETSUMO-Rsc3 | Echtenkamp et al.21 | N/A |
| pETSUMO-Rsc3ΔIDR | This manuscript | N/A |
| Software and algorithms | ||
| artMS | Jimenez-Morales et al.40 | http://artms.org/ |
| MeroX | Götze et al.17 | http://stavrox.com/ |
Experimental Model and Study Participant Details
Saccharomyces cerevisiae cultures were grown in complete (YPD) or minimal media supplemented with the required amino acids at 30°C unless otherwise noted. The main yeast strain (JJ117) used in this study has the endogenous Hsp90 genes disrupted (hsc82Δhsp82Δ) and is maintained by plasmid (Yep24-HSP82-URA) borne expression of Hsp82 protein. The Bpa Hsc82 derivatives were generated by introducing Amber stop codons (UAG) into an Hsc82 expression plasmid (pRS313GPD His-Hsc82) at the positions encoding for the indicated amino acid codons of Hsc82 (F120, F325, E427, F579, and E656) by one-step overlap extension PCR. Plasmids to express wild type Hsc82 (WT) or each Bpa variant were introduced in parental yeast (JJ117) transformed with pSNR-BPA16 and YEp24-HSP82-URA. Plasmid shuffling with 5-FOA selection was used to eventually drop the YEp24-HSP82 plasmid. All yeast used in the described experiments express a variant of Hsc82, which is referred to as Hsp90.
Method Details
Growth media and conditions
Bpa containing liquid media was prepared with sterile, Synthetic Dextrose (SD) media supplemented with the required amino acids or complete (YPD) media. Bpa stock solution (200 mM; prepared by dissolving 53.8 mg of Bpa in 1 mL of 1 M NaOH) was added by slowly dripping the solution into the liquid media prior to inoculation to a final concentration of 2 mM. The pH was neutralized with equal volume of 1 M HCl. Bpa containing solid media was prepared with autoclaved SD media with 2% agar, cooled to 55°C, required amino acid drop out mix was added while stirring with a magnetic stir bar, Bpa stock solution was slowly dripped into the media to a final concentration of 2 mM, and pH was neutralized with equal volume of 1 M HCl.
For cell culture experiments, human HeLa cervical adenocarcinoma cells grown in DMEM supplemented with 10% FBS to 70%−80% confluency in 6 well plates. Ganetespib (25 nM or 250 nM) and/or cycloheximide (1 ng/mL) were added in the indicated combinations to each well for the specified durations. For heat stress, the 6 well plates were wrapped in parafilm 2x and submerged in a 42°C water bath for the specified durations. Cells were lysed directly in the well using TRIzol and manufacturer instructions were followed for RNA isolation. RNA (100 ng) was used for qRT PCR with HSP70 and GAPDH qRT primers using Power SYBR Green RNA-to-CT 1-Step kit according to manufacturer’s instructions.
Bpa variant production and growth tests
To check the ability of each Hsp90 derivative to support cell growth, yeast expressing either Hsp90 WT or a Bpa variant were grown to exponential log phase, collected by centrifugation, and resuspended to OD595 1.0. Each sample was serially diluted 10-fold and 5 μL of each dilution was spotted onto selective SD media plates supplement with or without Bpa. Plates were incubated at 30°C or 37°C and imaged after two days of growth.
To assess the production length of each Hsp90 derivative, each variant was grown overnight to saturation in selective SD media with and without Bpa. Collected cell pellets were used to generate whole cell extracts by glass bead beating for 37 min with a 1 min ON/2 min OFF cycle. Lysates were clarified by centrifugation at 14000 rpm for 15 min. Aliquots of each extract (100 μg) was resolved by SDS PAGE. The Hsp90 proteins were detected by immunoblot analysis using anti-His antibody.
Bpa crosslinking
Yeast transformants expressing either wild type His-tagged Hsp90 (WT) or a Bpa variant were maintained on 2 mM Bpa supplemented selective SD solid media as described. Overnight cultures in selective SD media supplemented with Bpa (2 mM) were used to seed ~325 mL YPD media supplemented with Bpa (since each variant produced full length Hsp90-Bpa only in the presence of Bpa, there is an inherent selective pressure to retain the Hsp90-Bpa variant plasmid, thus, YPD media was used for growth of crosslinking cultures as cells were empirically determined to grow better in the YPD media supplemented with Bpa when compared to selective SD media supplemented with Bpa) to a final OD595 of 0.3. Cells were harvested at OD595 0.6–0.7, collected by centrifugation, and resuspended in 25 mL YPD without Bpa. Aliquots (5 mL) were spread onto five 10 cm2 plates each. (~0.8 OD595 units of cells per cm2 surface area to achieve a 0.1 cm thickness of cell suspension). To maintain temperature during the UV crosslinking step, a Styrofoam box was prepared by adding crushed dry ice to fill the bottom third of the bottom of the box, followed by wet ice packed over the top and a metal plate was placed over the ice for levelling and support. The petri plate containing cells was placed on top of the metal plate and the box was placed in the crosslinker with paper towels added under it to raise the box till the petri plate was ~2 cm from the UV bulbs. Crosslinking was carried out with default energy settings for 3 hours. After UV exposure, cells were collected by centrifugation, washed once with water, and pellet was flash frozen in liquid nitrogen.
Cultures expressing His-tagged WT Hsc82 were used to detect stable Hsp90 interactors, while Hsp90-Bpa variants were used to generate an Hsp90-Bpa dependent interactor list. The crosslinking procedure was also carried out with W303 yeast grown with an untagged Hsp90 without selective pressure but in YPD media to serve as a negative control for analysis of the mass spectrometry data.
Purification of crosslinked Hsp90 complexes
Frozen pellets were resuspended in TALON Binding Buffer (TBB; 50 mM Sodium phosphate pH 7.2, 300 mM NaCl) with 0.75% Triton-X and lysed using glass bead beating with a 1 min ON and 2 min OFF cycle for 37 min at 1400 rpm. Lysates were collected and clarified by centrifugation at 14000 rpm for 15 minutes. TALON Cobalt beads prewashed with TBB were added and the mixture was nutated for 2 hours at 4°C. Beads were collected by centrifugation at 3000 rpm for 3 min and the lysate was discarded. The beads were washed 4x with 10x bead volume of TBB supplemented with 20 mM Imidazole. Samples were eluted with a 1x bead volume of TBB containing 150 mM Imidazole for 15 min at 4°C and pooled with a second round of elution. 325 mL of crosslinked culture was purified using 200 μL of wet bead pellet and yielded ~500 μL of combined eluant which was concentrated down to ~200 μL. An aliquot (10 μL) of the samples was analyzed by SDS-PAGE and visualized by Coomassie blue or SYPRO Ruby Protein gel staining for crosslinking products. The remaining sample was flash frozen in Dry Ice/Methanol and stored at −20°C until mass spectrometry analysis.
Mass spectrometry analysis
Multidimensional Protein Identification Technology mass spectrometry was performed on the purified samples by the Sanford Burnham Prebys Proteomics facility. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE39 partner repository with the dataset identifier PXD040875. The Bioconductor package artMS was used for analysis of the mass spectrometry data40. Default settings were used for relative quantification of identified proteins using triplicates of samples generated for Hsp90-Bpa variants (Hsp90-Bpa), wild type His-Hsc82 (WT) and untagged Hsc/p82 (W303). Relative quantification tables were generated by providing a contrast text file comparing WT and Hsp90-Bpa with W303 and Hsp90-Bpa with full length. Hsp90 interacting proteins (stable interactors) were identified as proteins with a 1.5-fold or higher enrichment and adjusted p value of 0.05 or lower in Hsp90-Bpa and/or WT when compared to W303. Crosslinked proteins (transient interactors) were determined as proteins with a 1.5-fold or higher enrichment and adjusted p value of 0.05 or lower in Hsp90-Bpa when compared to WT. To identify the Bpa crosslinked peptide pairs, files for each Hsp90 variant were generated in accordance with the MeroX software’s guidelines. Briefly, the site of Bpa incorporation in the Hsc82 fasta sequence was replaced with an ‘x’ and fasta sequences of the variant specific hits were combined into the same file (due to many variants specific hits, multiple files were generated for each variant with a subset of the hits to significantly reduce computational time of the software). After crosslinked peptide list was generated, peptides were filtered to select those with more than two b and/or y ions from both peptides as well as more than 5% relative intensity for at least one ion that included the crosslink from both peptides.
Yeast two-hybrid
Two-hybrid interaction analysis was carried out using Matchmaker Gal4 2-hybrid system (Clontech). Cloning in pGAD-T7 and pGBK-T7 was done with InFusion cloning kit (Takara Bio). IDR deletion in Rsc3 was created with Q5 Site-Directed Mutagenesis protocol (New England BioLabs). For the Rsc3 yeast two hybrid (Y2H) analysis to assess the effects of Hsp90 on Rsc3 or Rsc3ΔIDR DNA binding activity, spot test assay was carried out to test the relative activation of the HIS3 reporter. Single colonies from each Y2H combination were patched onto fresh plates and inoculated from the patches into selective SD media for 2 days to saturation. 10-fold serial dilution was carried out from saturated cultures and 5 μL of each dilution was spotted onto selective solid SD media with and without Histidine. Plates were imaged every two days for 6 days in total.
Rsc3 electrophoretic mobility shift assay
IDR deletion (Rsc3ΔIDR) in Rsc3 was generated using the Q5 Site directed mutagenesis protocol with pETSUMO-Rsc3 as a template. Recombinant Rsc3 or Rsc3ΔIDR (region deleted was G197-Q303) proteins were purified as His6-SUMO fusion proteins following expression in Rosetta(DE3) transformants. The DNA binding assay used a radiolabeled ACC1_TIM23 probe in Binding Buffer (10 mM Tris pH 7.4, 40 mM NaCl, 1 mM DTT, 4% glycerol, 50 mg/mL BSA). Where indicated, reactions were supplemented with Hsp90 (1, 4, 16 μM) or BSA to balance total protein levels (16 μM total). Reactions were resolved on 4% native polyacrylamide 1x GTG, gels were dried, and the products were visualized using a PhosphoImager (Molecular Dynamics).
Polysome fractionation
To determine the locations of the yeast Hsp90 and Hsp70 molecular chaperones relative to established ribosome complexes, we used a standard polysome fractionation procedure41 with some modification. In brief, an overnight culture of wild type yeast (BY4741) was used to inoculate 100 mL of YPD to OD595 0.25 and allowed to grow till OD595 0.7. Cycloheximide (CHX) was added to the culture to a final concentration of 100 μg/mL for 15 minutes followed by collection of cells by centrifugation at 4°C, 4000 rpm for 1 min. Cells were resuspended in chilled polysome gradient buffer (20 mM Tris pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 100 μg/mL CHX). Resuspended cells were dripped into liquid N2 to create frozen cell pellets. Lysates were prepared by grinding the frozen cells in liquid N2 using a mortar and pestle and thawing the frozen powder at 4°C. Absorbance (A260) of the lysates was measured and lysate corresponding to 200 μg of RNA was layered onto a premade 10%−50% sucrose gradient. Fractionation in sucrose density gradient was carried out using the SW41Ti rotor and ultra-centrifugation at 32000 rpm for 3 hours. AKTA fraction collector set up with a UV reader was flushed with 50% sucrose solution and then used for sample fractionation with a fraction volume of 1 mL. Fractions were collected corresponding to the crude extract, 40S, 60S, monosome and polysome fractions based on A260 readings. Fractions were TCA precipitated with 25% TCA and washed twice with acetone. Pellets were dried on a 95°C heat block and resuspended in 40 μL 2xSDS PAGE loading dye. Aliquots (20 μL) of each fraction were resolved by SDS-PAGE. The positions of yeast Hsp90 and Hsp70 were visualized by immunoblot analysis using anti-Ssa1/2 or anti-Hsc/p82 antibodies.
Heat shock response assays
For yeast studies, wild type cells were grown in YPD from OD595 0.25 to OD595 0.5–0.6. Cells were concentrated 4-fold. Radicicol (10 μM), L-azetidine 2-carboxylic acid (5 mM), and/or CHX (100 μg/mL) were added in the required combinations. Cultures were then kept at 30°C or 42°C for heat stress for the specified duration. Cells were collected by centrifugation and total RNA was extracted by hot acidic phenol method. Briefly, cell pellet was resuspend in 400 μL TES buffer (10 mM Tris-Cl pH 7.5, 10 mM EDTA, 0.5% SDS) and 400 μL of acid phenol preheated to 65°C. Samples were incubated at 65°C for 60 minutes with brief vortexing for 10 sec every 15 min. Phases were separated by centrifugation at 14000 rpm at 4°C for 5 min. Aqueous phase was transferred to a clean microcentrifuge tube and 400 μL of chloroform was added followed by vigorous vortexing and centrifugation at 14000 rpm, 4°C for 5 min. 300 μL of aqueous phase was transferred to a new tube and 30 μL 3 M sodium acetate, pH 5.3 and 750 μL of 100% ethanol was added. Samples were incubated overnight at −80°C and precipitated by centrifugation at 14000 rpm at 4°C for 5 min. RNA pellet was washed in ice-cold 70% ethanol and dried for 5 min. RNA pellet was resuspended in 50 μL of H2O and concentration was measured by nanodrop. RNA (100 ng) was used for qRT PCR with SSA4 and TAF10 qRT primers using Power SYBR Green RNA-to-CT 1-Step kit according to manufacturer’s instructions with tm at 58°C.
For mammalian cell work, HeLa cells were cultured to 70%−80% confluency in 6 well plates. Ganetespib (25 nM or 250 nM) and/or CHX (1 ng/mL) were added in the required combinations to each well for the specified duration. For heat stress, an entire 6 well plate was parafilm wrapped twice and submerged in a 42°C water bath for the specified duration. Cells were lysed directly in the well using TRIzol and manufacturer instructions were followed for RNA isolation. 100 ng RNA was used for qRT PCR with HSP70 and GAPDH qRT primers using Power SYBR Green RNA-to-CT 1-Step kit according to manufacturer’s instructions with tm at 58°C.
Dual luciferase assay
To determine if Hsp90 influences the fidelity of translation initiation, we used a standard dual luciferase expression system23. The dual luciferase expression plasmids were kind gifts of Dr. Jon Lorsch (NIH). WT and G170D expressing yeast were transformed with each plasmid and four single colonies were assayed for dual luciferase activity. Overnight cultures for each transformant were inoculated to OD595 0.2 in a 96 well plate and incubated at 30°C for 4–6 hours to OD595 0.6–0.8. 50 μL of cultures were moved to a new 96 well plate and 50 μL of SD Ura- media prewarmed to 45°C was added to bring the temperature to 37.5°C. Cultures were allowed to grow for another 4–6 hours at 37.5°C. Aliquots (2 μL) of the cultures (30°C and 37.5°C) were added to 50 μL 1x Passive lysis buffer and incubated at room temperature with shaking for 50 min. Firefly luciferase activity was measured in the TECAN Evolution plate reader at 560 nm by the addition of 50 μL luciferin in substrate buffer (15 mM Tris pH 8, 25 mM glycylglycine, 4 mM EGTA, 15 mM MgSO4, 1 mM DTT, 2 mM ATP, 0.1 mM CoA, 75 μM Luciferin).
Renilla luciferase activity was subsequently measured at 440 nm by the addition of 50 μL coelenterazine substrate (0.22 M citric acid-sodium citrate pH 5, 1.1 M NaCl, 2.2 mM EDTA, 1.3 mM NaN3, 0.44 mg/mL BSA, 1.43 μM Coelenterazine) to the same wells. To calculate UUG utilization, UUG and AUG Firefly luciferase values were first normalized to their respective AUG Renilla luciferase values (from the same plasmid as the Firefly luciferase) to determine the and values. was then normalized to for the final UUG utilization value. Since the activity levels of Renilla/Firefly proteins are controlled by normalization of signal to yeast expressing the same luciferases under the canonical AUG start codon, differences in Hsp90 dependence of Renilla/Firefly luciferase can be discounted.
To determine fold change, the G170D UUG utilization value was compared to the WT UUG utilization value.
Translation factor steady-state protein levels
TAP-tagged strains of the required translation factors were grown from OD595 0.25 to OD595 0.5–0.6. Cells were concentrated 5-fold and an aliquot was immediately collected by centrifugation and the pellet was frozen as time 0. Radicicol was added to a final concentration of 25 μM to the remaining culture and incubated at 30°C, with aliquots being taken at 15 min and 60 min. Frozen pellets were lysed by bead beating in TENG (Tris pH 7.4, EDTA, NaCl, 10% Glycerol) buffer and lysates were clarified. BCA assay was used to determine the total protein concentration of the lysate. Proteins were TCA precipitated and resuspended in 1x SDS loading buffer to a final concentration of 25 μg/μL. Protein expression levels based on placement in the TAP-tag library were used to determine the amount of whole cell extract (WCE) resolved42. For abundant TAP-tagged proteins (library plates S1) 100 μg WCE was used, for mid-expressed proteins (library plates S2 and S3) 250 μg WCE was used, and for low expression factors (library plates S4) 500 μg of WCE was resolved on 10% SDS polyacrylamide gel and analyzed by immunoblotting using αTAP and αSBA1 antibodies.
MG132 growth curves
Overnight cultures of parental and G170D yeast were inoculated to an OD595 0.25 and grown to OD595 0.6–0.7. Cultures were diluted down to OD595 0.5 and added 1:1 to tubes pre-aliquoted with media containing MG132 (100 μM) or equivalent amounts of DMSO. Culture tubes were shifted to 30°C/35°C/37.5°C water baths and absorbance was measured every hour for 12 hours. Growth rate was calculated as log (Xt/X0) with Xt being the absorbance at time t and X0 being the initial absorbance of the cultures.
Stress granule analysis
GFP-Ded1 yeast from the N-terminal GFP library kindly provided by Dr. Maya Schuldiner (Weizmann Institute of Science), was grown to OD595 0.5. Required volume of culture was aliquoted into conical tubes and DMSO or Radicicol (200 μM) was added to each. DMSO/Radicicol containing cultures were then aliquoted (1 mL) into test tubes and moved to the indicated water bath (40°/46°C). Samples were collected at 4/7/10/15/30 min and added to formaldehyde to a final formaldehyde concentration of 3.7%. Cells were fixed on ice for 10 min followed by 2x washes in KPO4 buffer (pH 6.5). Cells were resuspended in KPO4 buffer and mounted onto polylysine coated slides using VectaShield. GFP-Ded1 was visualized using DeltaVision OMX with the FITC channel, 30 s exposure, and 50% transmission. Z-stacks (2 μM) were generated and deconvoluted. A custom Fiji script provided by Dr. Titus Franzmann (TU Dresden) was used for identification and counting of stress granules and cell numbers. In brief, the GFP signal (GFP-Ded1, GFP-Dcp1, or GFP-Ngr1, as indicated) compiled z-stacked images were pseudocolorized into a 16-color topology map where the formation of SGs (Ded1 and Dcp1) or P-bodies (Ngr1) correlated with the coalescence of GFP signal into high intensity puncta (pink/white) that was accompanied with a reduction in diffuse GFP signal (i.e., increase in blue/green). The high intensity puncta (pink/white) were counted as SGs or P-bodies and the average number per cell was calculated for a minimum of 50 cells for each experimental condition.
Quantification and statistical analysis
All experiments were performed in 3 biological replicates. Additionally, in the microscopy experiments, at least 50 cells were counted in each condition. The error bars represent the SEM of the 3 biological replicates. Student’s t-test (unpaired or paired when appropriate) was used to compare with WT or DMSO controls to determine significance. Further details for each experiment including the exact number of replicates (cells) and p values are provided in the figure legends.
Supplementary Material
Table S1. (Related to Figure 1): Bpa-Dependent and -Independent Hsp90 Interactors
Table S2. (Related to Figure 2): Bpa site specific capture of Hsp90 interactors
Table S3. (Related to Figure 3): Peptide sequences of Bpa crosslinked targets.
Highlights:
Hsp90 associates with ~20% of the yeast proteome using all 3 of its domains
Hsp90 targets specific intrinsically disordered regions to regulate client activity
Hsp90 maintains the physical status of IDR-proteins at physiological temperatures
The HSR triggered by Hsp90 inhibition is dependent upon active protein translation
Acknowledgments:
We are grateful to Anna Mankovich (UIUC), Dr. Matthias Mayer (U. Heidleberg) and Dr. Titus Franzmann (T.U. Dresden) for their technical help with experimental approaches. We thank Dr. Maya Schuldiner (Weizmann Institute of Science) for her kind gift of the amino-terminal GFP fusion library, Dr. Jon Lorsch (NIH) for his generous gift of the dual luciferase system, and Dr. Alex Campos (Sanford Burnham Prebys) with the mass spectrometry analysis.
Funding:
NIH R35 GM136660
Footnotes
Declaration of Interests: No competing interests
Inclusion and Diversity Statement: We support inclusive, diverse, and equitable conduct of research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jayaraj GG, Hipp MS, and Hartl FU (2019) Functional Modules of the Proteostasis Network. Cold Spring Harbor Perspective in Biology. 10.1101/cshperspect.a033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Labbadia J, and Morimoto RI (2015) The Biology of Proteostasis in Aging and Disease. Annu. Rev. Biochem. 84, 435–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahiya V., and Buchner J. (2019) Functional principles and regulation of molecular chaperones. Adv. Protein Chem. Struct. Biol. 114, 1–60. [DOI] [PubMed] [Google Scholar]
- 4.DeZwaan DC, and Freeman BC (2008) Hsp90: the Rosetta stone for cellular protein dynamics? Cell Cycle 7, 1006–1012. [DOI] [PubMed] [Google Scholar]
- 5.Ellis J (1987) Proteins as molecular chaperones. Nature 328, 378–379. [DOI] [PubMed] [Google Scholar]
- 6.Rosenzweig R, Nillegoda NB, Mayer MP, and Bukau B (2019) The Hsp70 chaperone network. Nat. Rev. Mol. Cell. Biol. 20, 665–680. [DOI] [PubMed] [Google Scholar]
- 7.Schopf FH, Biebl MM, and Buchner J (2017) The HSP90 chaperone machinery. Nat. Rev. Mol. Cell. Biol. 18, 345–360. [DOI] [PubMed] [Google Scholar]
- 8.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, and Hendrickson WA (1996) Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brugge JS (1981) The specific interaction of the Rous sarcoma virus transforming protein, pp60src, with two cellular proteins. Cell 25, 363–372. [DOI] [PubMed] [Google Scholar]
- 10.Joab I, Radanyi C, Renoir M, Buchou T, Catelli M, Binart N, Mester J, and Baulieu E (1984) Common non-hormone binding component in non-transformed chick oviduct receptors of four steroid hormones. Nature 308, 850–853. [DOI] [PubMed] [Google Scholar]
- 11.McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, and Frydman J (2007) Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell 131, 121–135. [DOI] [PubMed] [Google Scholar]
- 12.Zhao R, Davey M, Hsu YC, Kalplaek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J et al. (2005) Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120, 715–727. [DOI] [PubMed] [Google Scholar]
- 13.Millson SH., Truman AW., Wolfram F., King V., Panaretou B., Prodromou C., Pearl LH., and Piper PW. (2004) Investigating the protein-protein interactions of the yeast Hsp90 chaperone system by two-hybrid analysis: potential uses and limitations of this approach. Cell Stress Chaperones 9, 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gristmair H, Tippel F, Lopez A, Tych K, Stein F, Haberkant P, Schmid PWN, Helm D, Rief M, Sattler M, and Buchner J (2019) The Hsp90 isoforms from S. cerevisiae differ in structure, function, and client range. Nat Commun. 10, 3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verba KA, Wang RY, Arakawa A, Liu Y, Shirouzu M, Yokoyama S, and Agard DA (2016) Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science 352, 1542–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin JW, and Schultz PG (2002) In vivo photocrosslinking with unnatural amino acid mutagenesis. Chembiochem. 3, 1135–1137. [DOI] [PubMed] [Google Scholar]
- 17.Götze M, Pettelkau J, Fritzsche R, Ihlinh CH, Schӓfer M, and Sinz A (2015) Automated assignment of MS/MS cleavable cross-links in protein 3D-Structure analysis. J. Am. Soc. Mass Spectrom. 26, 83–97. [DOI] [PubMed] [Google Scholar]
- 18.Oates ME, Romero P, Ishida T, Ghalwash M, Mizianty MJ, Xue B, Dosztányi S, Uversky VN, Obradovic Z, Kurgan L et al. (2013) D2P2: Database of disordered protein predictions. Nucleic Acids Research 41, 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A et al. (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karagöz GE., Duarte AMS., Akoury E., Ippel H., Biernat J., Morán Luengo T., Radli M., Didenko T., Nordhues BA., Veprintsev DB. et al. (2014) Hsp90-Tau complex reveals molecular basis for specificity in chaperone action. Cell 156, 963–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Echtenkamp FJ, Gvozdenov Z, Adkins NL, Zhang Y, Day ML, Watanabe S, Peterson CL, and Freeman BC (2016) Hsp90 and p23 molecular chaperones control chromatin architecture by maintaining the functional pool of RSC chromatin remodeler. Mol. Cell 64, 888–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hershey JWB, Sonenberg N, and Mathews MB (2019) Principles of translation control. Cold Spring Harbor Perspective Biology 10.1101/cshperspect.a032607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takacs JE, Neary TB, Ingolia NT, Saini AK, Martin-Marcos P, Pelletier J, Hinnebusch AG, and Lorsch JR (2011) Identification of compounds that decrease the fidelity of start codon recognition by the eukaryotic translation machinery. RNA 17, 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nathan DF, and Lindquist S (1995) Mutational analysis of Hsp90 function: interactions with a steroid receptor and a protein kinase. Mol. Cell. Biol. 15, 3917–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Protter DSW, and Parker R (2016) Principles and properties of Stress Granules. Trends Cell Biology 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iserman C, Altamirano CD, Jegers C, Friedrich U, Zarin T, Fritsch AW, Mittasch M, Domingues A, Hersemann L, Jahnel M et al. (2020) Condensation of Ded1p promotes a translational switch from housekeeping to stress protein production. Cell 181, 818–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guenther UP., Weinberg DE., Zubradt MM., Tedschi FA., Stawicki BN., Zagore LL., Brar GA., Licatalosi DD., Bartel DP., Weissman JS., and Jankowsky E. (2018) The helicase Ded1p controls use of near-cognate translation initiation codons in 5’ UTRs. Nature 559, 130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindquist S, and Craig EA (1998) The heat-shock proteins. Ann. Rev. Genetics 22, 631–677. [DOI] [PubMed] [Google Scholar]
- 29.Liti G (2015) The fascinating and secret wild life of the budding yeast S. cerevisiae. eLife 4, e05835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitesell L, and Lindquist SL (2005) Hs90 and the chaperoning of cancer. Nature Reviews Cancer 5, 761–772. [DOI] [PubMed] [Google Scholar]
- 31.Murzin AG, Brenner SE, Hubbard T, and Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 32.Sillitoes I, Lewis TE, Cuff A, Das S, Ashford P, Dawson NL, Furnham N, Laskowski RA, Lee D, Lees JG, Lehtinen S, Studer RA, Thornton J, and Orengo CA (2015) CATH: comprehensive structural and functional annotations for genome sequences. Nucleic Acids Res. 43, D376–D381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski RA, and Thornton JM (2008) Understanding the molecular machinery of genetics through 3D structures. Nat. Rev. Genet. 9, 141–151. [DOI] [PubMed] [Google Scholar]
- 34.Oldfield CJ, and Dunker AK (2014) Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 83, 553–584. [DOI] [PubMed] [Google Scholar]
- 35.Chen B, Zhong D, and Monteiro A (2006) Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. BMC Genomics. 7, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward JJ., McGuffin LJ., Bryson K., Buxton BF., and Jones DT. (2004) The DISOPRED server for the prediction of protein disorder. Bioinformatics 20, 2138–2139. [DOI] [PubMed] [Google Scholar]
- 37.Basile W, Salvatore M, Bassot C, and Elofsson A (2019) Why do eukaryotic proteins contain more intrinsically disordered regions? PLoS Comput. Bio. 15, e1007186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lopez A, Dahiya V, Delhommel F, Freiburger L, Stehle R, Asami S, Rutz D, Blair L, Buchner J, and Sattler M (2021) Client binding shifts the populations of dynamic Hsp90 conformations through an allosteric network. Sci Adv. 7, eabl7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perez-Riverol Y, Bai J, Bandla C, Garcia-Seisdedos S, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M, Walzer M, Wang S, Brazma A, and Vizaino JA (2022) The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jimenez-Morales D, Rosa AC, Dollen JV, Krogan N, and Swaney D (2022) artMS: Analytical R tools for Mass Spectrometry. R package version 1.14.0. Bioconductor 10.18129/B9.bioc.artMS. [DOI] [Google Scholar]
- 41.Panasenko OO (2012) Ribosome Fractionation in Yeast. Bio-Protocols 10.21769/BioProtoc.251. [DOI] [Google Scholar]
- 42.Ghaemmaghami S., Huh W., Bower K., Howson RW., Belle A., Dephoure N., O’Shea EK., and Weissman JS. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741. [DOI] [PubMed] [Google Scholar]
- 43.Yofe I, Weill U, Meurer M, Chuartzman S, Zalckvar E, Goldman O, Ben-Dor S, Schütze C, Wiedemann N, Knop M, Khmelinskii A, and Schuldiner M (2016). One library to make them all: streamlining the creation of yeast libraries via a SWAp-Tag strategy. Nature Methods 13, 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang RYR, Noddings CM, Kirschke E, Myasnikov G, Johnson JL, and Agard DA (2022). Structure of Hsp90-Hsp70-Hop-GR reveals the Hsp90 client-loading mechanism. Nature 601, 460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. (Related to Figure 1): Bpa-Dependent and -Independent Hsp90 Interactors
Table S2. (Related to Figure 2): Bpa site specific capture of Hsp90 interactors
Table S3. (Related to Figure 3): Peptide sequences of Bpa crosslinked targets.
Data Availability Statement
All mass spectrometry (PRIDE) and imaging data (Mendeley Data) generated in this study have been deposited and are publicly available as of the date of publication. Accession numbers and DOI are listed in the key resources table.
This paper does not report any original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-TAP | ThermoFisher | Cat#CAB1001 |
| Anti-His | ThermoFisher | Cat#MA1–21315 |
| Bacterial and virus strains | ||
| Rosetta(DE3) | Novagen | Cat#70954–3 |
| Chemicals, peptides, and recombinant proteins | ||
| All chemicals unless otherwise noted | ThermoFisher | N/A |
| Yeast Media Components | DIFCO | N/A |
| LB Media Components | DIFCO | N/A |
| p-Benzoyl-L-phenylalanine | ThermoFisher (Bachem) | Cat#NC0169631 |
| Radicicol | AG Scientific | Cat#R-1130 |
| Ganetespib | Selleckchem | Cat#S1159 |
| L-Azetidine-2-carboxylic Acid | TCI | Cat#A1043 |
| Cycloheximide | GoldBio | Cat#C-930–10 |
| D-Luciferin, Potassium salt | GoldBio | Cat#LUCK-100 |
| Coelenterazine | GoldBio | Cat#CZ2.5 |
| CPRG | Roche | Cat#99792–50-4 |
| ATP [γ−32P] | Perkin Elmer | Cat#BLU002Z250UC |
| SYPRO Ruby Protein Gel Stain | ThermoFisher | Cat#S12000 |
| TALON Metal Affinity Resin | Takara Bio | Cat#635503 |
| Restriction Enzymes | New England BioLabs | N/A |
| Critical commercial assays | ||
| Pierce BCA Assay Kit | Fisher Scientific | Cat#PI23223 |
| Power SYBR Green RNA-to-CT 1-Step kit | Applied Biosystems | Cat#4388869 |
| In-Fusion Snap Assembly kit | Takara Bio | Cat#638946 |
| Q5 Site-Directed Mutagenesis Kit | New England BioLabs | Cat#E0554S |
| Deposited data | ||
| Imaging Data | Mendeley Data, V1 | doi:10.17632/8rnk4fggd8.1 |
| Mass Spectrometry Data | PRIDE | PXD040875 |
| Experimental models: Cell lines | ||
| HeLa Human Epithelial cells | ATCC | Cat#CRM-CCL-2 |
| Experimental models: Organisms/strains | ||
| Saccharomyces cerevisiae | Strain Table S2 | N/A |
| TAP tag strain library | Ghaemmaghami et al.42 | N/A |
| Mat a Knockout library | Open Biosystems | Cat#YSC1053 |
| GFP Library | Yofe et al.43 | N/A |
| JJ117 | Wang et al.44 | N/A |
| Oligonucleotides | ||
| SSA4 qRT FP | GCTCAACGTGTTCAAGCTAA | N/A |
| SSA4 qRT RP | ACCCACCTTCTCCTTGAAGT | N/A |
| TAF10 qRT FP | TACGAATATTCCAGGATCAGG | N/A |
| TAF10 qRT RP | CAATAGCTGCCTAGCTCTC | N/A |
| Hsp70 qRT FP | CAGAGCGGAGCCGACAGAG | N/A |
| Hsp70 qRT RP | CCACCTTGCCGTGTTGGAAC | N/A |
| GAPDH qRT FP | ACACCCACTCCTCCACCTTTGAC | N/A |
| GAPDH qRT RP | ACCACCCTGTTGCTGTAGCCA | N/A |
| pGBK Hsc82 N FP | GAATTCCCGGGGATCATGGCTGGTGAAACTTTTGAATTTC | N/A |
| pGBK Hsc82 C RP | GCAGGTCGACGGATCCTTAATCAACTTCTTCCATCTCGGT | N/A |
| Hsc82 N FP | TTGTTGTCTAGAATGGCTGGTGAAACTTTTGAATTTC | N/A |
| Hsc82 N RP | TTGTTGCTGCAGTTACTTAGTCTTGTTCAACTCTTCTAATTC | N/A |
| Hsc82 M FP | TTGTTGTCTAGAGTTCCAATTCCAGAAGAAGAAAAG | N/A |
| Hsc82 M RP | TTGTTGCTGCAGTTATTCTTCCAATTCGAAATCTTTAGT | N/A |
| Hsc82 C FP | TTGTTGTCTAGAACTAAAGATTTCGAATTGGAAGAA | N/A |
| Hsc82 C RP | TTGTTGCTGCAGTTAATCAACTTCTTCCATCTCGG | N/A |
| pGBK Rsc3 FP | CATGGAGGCCGAATTCATGGATATTCGTGGTAGAAAAATG | N/A |
| pGBK Rsc3 RP | GGATCCCCGGGAATTTTAGCTACGAATAGCCTCAAAAAC | N/A |
| Rsc3ΔIDR FP | ATTCAGTATTTGATTGAACG | N/A |
| Rsc3ΔIDR RP | TGTAATTTCTTGCAATTCC | N/A |
| F120-Bpa FP | GCCGATGTATCCATGATTGGTCAATAGGGTGTTGGTTTTTACTCTTTATTC | N/A |
| F120-Bpa RP | GAATAAAGAGTAAAAACCAACACCCTATTGACCAATCATGGATACATCGGC | N/A |
| F325-Bpa FP | TTGTTCATTCCAAAGAGAGCACCATAGGACTTATTTGAGAGTAAGAAGAAG | N/A |
| F325-Bpa RP | CTTCTTCTTACTCTCAAATAAGTCCTATGGTGCTCTCTTTGGAATGAACAA | N/A |
| E427-Bpa FP | AAGAACATTAAGCTGGGTGTACATTAGGACACTCAAAACAGAGCTGCTTTA | N/A |
| E427-Bpa RP | TAAAGCAGCTCTGTTTTGAGTGTCCTAATGTACACCCAGCTTAATGTTCTT | N/A |
| F579-Bpa FP | CATCAGAACTGGTCAATAGGGCTGGTCTGCTAACAT | N/A |
| F579-Bpa RP | ATGTTAGCAGACCAGCCCTATTGACCAGTTCTGATG | N/A |
| E656-Bpa FP | TTGACTTCTGGTTTCAGTTTGGAATAGCCAACTTCTTTTGCATCAAGAATA | N/A |
| E656-Bpa RP | TATTCTTGATGCAAAAGAAGTTGGCTATTCCAAACTGAAACCAGAAGTCAA | N/A |
| ACC1_TIM23 F | ACGCGCGCGCGGCCGGGCCA | N/A |
| ACC1_TIM23 R | TGGCCCGGCCGCGCGCGCGT | N/A |
| Recombinant DNA | ||
| pTGPD-HSP82 | S. Lindquist (Whitehead Institute) | N/A |
| pTGPD-hsp82 G170D | S. Lindquist (Whitehead Institute) | N/A |
| pSNR-tRNA-pBpaRS | A. Mapp (University of Michigan) | N/A |
| pRS313GPD His-Hsc82 | E. Craig (University of Wisconsin, Madison) | N/A |
| pRS313GPD His-Hsc82 F120-Bpa | This manuscript | N/A |
| pRS313GPD His-Hsc82 F325-Bpa | This manuscript | N/A |
| pRS313GPD His-Hsc82 E427-Bpa | This manuscript | N/A |
| pRS313GPD His-Hsc82 F579-Bpa | This manuscript | N/A |
| pRS313GPD His-Hsc82 E656-Bpa | This manuscript | N/A |
| pFJZ1052 (AUG) | J. Lorsch (Division of Intramural Research, NIH) | N/A |
| pFJZ1054 (UUG) | J. Lorsch (Division of Intramural Research, NIH) | N/A |
| pGADT7 | Clontech Matchmaker | Cat#630442 |
| pGBKT7 | Clontech Matchmaker | Cat#630489 |
| pGBKT7 Hsp82 | Echtenkamp et al.21 | N/A |
| pGADT7 Rsc3 | Echtenkamp et al.21 | N/A |
| pGBKT7 Hsc82 | This manuscript | N/A |
| pGADT7 Rsc3ΔIDR | This manuscript | N/A |
| pETSUMO-Rsc3 | Echtenkamp et al.21 | N/A |
| pETSUMO-Rsc3ΔIDR | This manuscript | N/A |
| Software and algorithms | ||
| artMS | Jimenez-Morales et al.40 | http://artms.org/ |
| MeroX | Götze et al.17 | http://stavrox.com/ |