Abstract
Meg8-DMR is the first maternal methylated DMR to be discovered in the imprinted Dlk1-Dio3 domain. The deletion of Meg8-DMR enhances the migration and invasion of MLTC-1 depending on the CTCF binding sites. However, the biological function of Meg8-DMR during mouse development remains unknown. In this study, a CRISPR/Cas9 system was used to generate 434 bp genomic deletions of Meg8-DMR in mice. High-throughput and bioinformatics profiling revealed that Meg8-DMR is involved in the regulation of microRNA: when the deletion was inherited from the mother (Mat-KO), the expression of microRNA was unchanged. However, when the deletion occurred from the father (Pat-KO) and homozygous (Homo-KO), the expression was upregulated. Then, differentially expressed microRNAs (DEGs) were identified between WT with Pat-KO, Mat-KO, and Homo-KO, respectively. Subsequently, these DEGs were subjected to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) term enrichment analysis to explore the functional roles of these genes. In total, 502, 128, and 165 DEGs were determined. GO analysis showed that these DEGs were mainly enriched in axonogenesis in Pat-KO and Home-KO, while forebrain development was enriched in Mat-KO. Finally, the methylation levels of IG-DMR, Gtl2-DMR, and Meg8-DMR, and the imprinting status of Dlk1, Gtl2, and Rian were not affected. These findings suggest that Meg8-DMR, as a secondary regulatory region, could regulate the expression of microRNAs while not affecting the normal embryonic development of mice.
Keywords: genomic imprinting, differentially methylated region, CRISPR/Cas9, Meg8-DMR, Dlk1-Dio3 domain
1. Introduction
Genomic imprinting is an epigenetic mechanism in mammals that leads to the functional inequivalence of the parental genome [1]. The imprinted genes are regulated by differentially methylated regions (DMRs), which are dependent on the parent of origin. Imprinted genes are mainly characterized by a single allelic expression [2,3,4]. The differential methylation patterns originate during spermatogenesis or after fertilization [5]. A loss of imprinting can lead to the silencing or biallelic expression of genes, which leads to various embryonic developmental defects and postnatal diseases. The imprinted Dlk1-Dio3 domain that is located on the distal arm of mouse chromosome 12 and human chromosome 14 contains three paternally expressed protein-coding genes (Dlk1, Rtl1, and Dio3), four maternally expressed long non-coding RNA genes (Gtl2, anti-Rtl1, Rian, and Mirg) [6,7,8,9,10,11,12,13], and a large number of microRNAs and snoRNAs [14,15]. The Dlk1-Dio3 imprinting region in mammals has been reported as highly conserved in humans and mice, including chromosome localization, sequence characteristics, and gene imprinting status. In addition, imprinted genes within the Dlk1-Dio3 domain have also been reported to be imprinted conserved in other mammals, such as sheep, cattle, and pigs [16,17]. These imprinted genes have been shown to have important functions in controlling prenatal development and postnatal differentiation.
The Dlk1-Dio3 imprinted domain contains three paternally methylated DMRs (IG-DMR, Gtl2-DMR, Dlk1-DMR) and a functionally unknown maternally methylated differentially methylated region, Meg8-DMR [18,19]. The Dlk1-Dio3 domain is mainly regulated by IG-DMR, which is methylated during spermatogenesis [20]. In mice, when the IG-DMR knockout occurs on paternal alleles, the offspring are born with normal phenotype and gene expression. However, when it occurs on maternal alleles, this results in perinatal lethality. The expression of Dlk1, Rtl1, and Dio3 all become biallelic, while maternally expressed genes are suppressed [21,22]. The maternally expressed genes of the Dlk1-DIo3 domain are secondarily regulated by Gtl2-DMR. In mice, when the Gtl2-DMR knockout occurs on a maternal allele, the offspring are born with a normal phenotype. However, all the offspring die within four weeks, mainly due to severe hypoplastic pulmonary alveoli and hepatocellular necrosis. When the deletion occurs on the paternal allele, the embryos show severe developmental disabilities and high lethality in the perinatal period [23]. The Meg8-DMR located inside the second intron of Rian (Rian and Meg8 are alternative names given to the same transcript) is the first maternally methylated, somatically derived, differentially methylated region and acquired differential methylation prior to E7.5 in the domain. In addition, the CTCF binds to the Meg8-DMR non-allele, specifically in vivo. Deletion of the Meg8-DMR in the mouse MLTC-1 cell line enhanced cell migration and invasion [24]. A previous study found that Meg8-DMR shows hypermethylation in TS14 patients, while hypomethylation occurred in KOS14 patients [25]. The methylation levels in the brain of Temple syndrome patients are quite interesting. The methylation levels are very low in fetal and 4-month-old patient brains [26]. However, for patients aged from 44 to 79 years, methylation levels are 26–30%. Thus, it seems that the methylation level in the brain increases with age [27]. To the best of our knowledge, Meg8-DMR will be the first reported imprinting site to obtain methylation in a tissue-specific manner with age. In addition, the methylation level may be associated with the development of intelligence in patients with Temple syndrome. However, little is known about the role of Meg8-DMR in mice.
To investigate the biological function of Meg8-DMR, the CRISPR/Cas9 system was used to generate genomic deletions in mice with paternal deletion, maternal deletion, and the parental double-deletion of Meg8-DMR. The weight, mode of inheritance, morphology, and methylation levels are not changed. The expression level of multiple microRNAs is exchanged. High-throughput analysis showed that differentially expressed genes are enriched in axonogenesis. These results imply that somatic-cell-derived Meg8-DMR could regulate the expression of microRNAs while not affecting the normal embryonic development of mice.
2. Materials and Methods
2.1. Generation of Meg8-DMR Mice
A sgRNA DNA temple was generated using PCR amplification with the following primers: sgRNA F-5′-TTAAGATTTGTGCCAACTCA-3′ and sgRNA R-5′-CAGGAGCCCAGGAAACCCAA-3′. PCR products were purified using Qiaquick (#28014, Qiagen, Frankfurt, Hessian, Germany) columns, and approximately 100 ng of DNA was used as a template for the in vitro transcription reaction of T7 (#AM1344, Invitrogen, Carlsbad, CA, USA). In vitro transcription and Cas9/sgRNA co-injection were performed as described previously. DBA and C57BL6/J mice were mated to produce the hybrid strain DBAB6F1. The Meg8-DMR knockout mice lines were maintained by mating heterozygous mice with ICR mice. The Pat-KO, Mat-KO, and WT mice were generated by interbreeding with WT and heterozygous mutants. The Homo-KO mutants were obtained by mating male and female heterozygous mutants. Genotype identification was carried out using PCR amplification with the following primers: Meg8-F-5′-CAACAGGATAACTCTGGCAACTTCC-3′and Meg8-R-5′-CTAACCCCTTATGACTCTCCATCGT-3′.
2.2. RNA Extraction and qRT-PCR
Total RNA was extracted from the tissue using RNAiso Plus (TaKaRa, Dalian, China). The cDNA was synthesized with reverse transcription using the PrimeScriptTM RT kit (Takara). qRT-PCR was performed on the ABI 7500 real-time fluorescent quantitative PCR system using FastStart Universal SYBR Green Master (Roche, Basel, Switzerland). The primers used for the analysis are shown in Table S1. MicroRNA reverse transcription reactions were performed with the TaqMan miRNA Reverse Transcription Kit (#4366596, Applied Biosystems, Carlsbad, CA, USA) and miRNA-specific reverse transcription primers (#4366596, Applied Biosystems, USA), and quantitative PCR was performed with TaqMan 2× Universal PCR Master Mix (#4369016, Applied Biosystems, USA) and miRNA-specific quantitative PCR primers (#4409156, Applied Biosystems, USA).
2.3. RNA-Seq Analysis
Five separated total RNAs were extracted from embryos and mixed together. RNA electrophoresis was used for quality detection, and Nanodrop was used for purity analysis. The mixed RNAs were provided to a private Bio Company for transcriptome sequencing. DEGs were defined as the level of gene expression in mutant mice relative to expression in WT mice, and absolute fold change >1 and p-value < 0.05 were used to identify DEGs. For DEGs, log2 fold change (FC) was defined as upregulated DEGs if it was positive and downregulated DEGs if it was negative. David Bioinformatics Resources (v6.8, Frederick, MD, USA) was used to annotate the GO functions of different genes.
2.4. DNA Methylation Analysis
DNA was isolated from the embryo of WT, Pat-KO, Mat-KO, and Homo-KO mutants. Sequencing was performed as described previously. The primers used were as follows: IG-DMR, forward 5′-GTATTGTAATATAGGTTAGGTG-3′, reverse 5′-CTACATAATACCATATAAACATATCTC-3′. Gtl2-DMR, forward 5′-AAATTCTGCAAGGAAAAGAATCCTCAGG-3′, reverse 5′-TTCAGAATTGCTGGTCAACATGAACCTC-3′. Meg8-DMR, forward 5′-AGTTGGGTTATTTAAGATTTGTGTT-3′, reverse 5′-AATTTCCTAAACTCCTAACCACTTC-3′.
2.5. Allelic Analysis of Gene Expression
According to the analysis of SNP sites in the imprinting region on the bioinformatics website (http://www.informatics.jax.org/strains_SNPs.shtml, accessed on 5 June 2021), SNP sites of Gtl2 exist between the ICR mouse strain and C57BL/6J mouse strain, and SNP sites of Dlk1 and Rian exist between the ICR mouse strain and DBA/2J mouse strain. Using the DNA of each pure line of mice, the correctness of SNP sites was verified. Two different hybrid lines were crossed. The pregnant mice were dissected in E12.5 and E15.5, and two groups of hybrid lines were obtained: BIF1 and IBFI and IDF1 and DIF1, respectively. The total RNA of the E12.5 embryo and E15.5 tissues were extracted, and the cDNA was obtained with reverse transcription, which was used as an experimental sample to establish the crossed cDNA library. At the same time, the DNA of the parents of each hybrid line and pure line was extracted. cDNA primers and DNA primers containing SNP sites were designed in the detected imprinted gene sequences. DNA samples of parents and hybrids were PCR and product recovery sequencing, and SNP sites of bioinformatics analysis were verified. Then, cDNA bit templates of hybrids were used for PCR product recovery sequencing.
3. Results
3.1. CRISPR/Cas9-Mediated Meg8-DMR Deletion in Mice
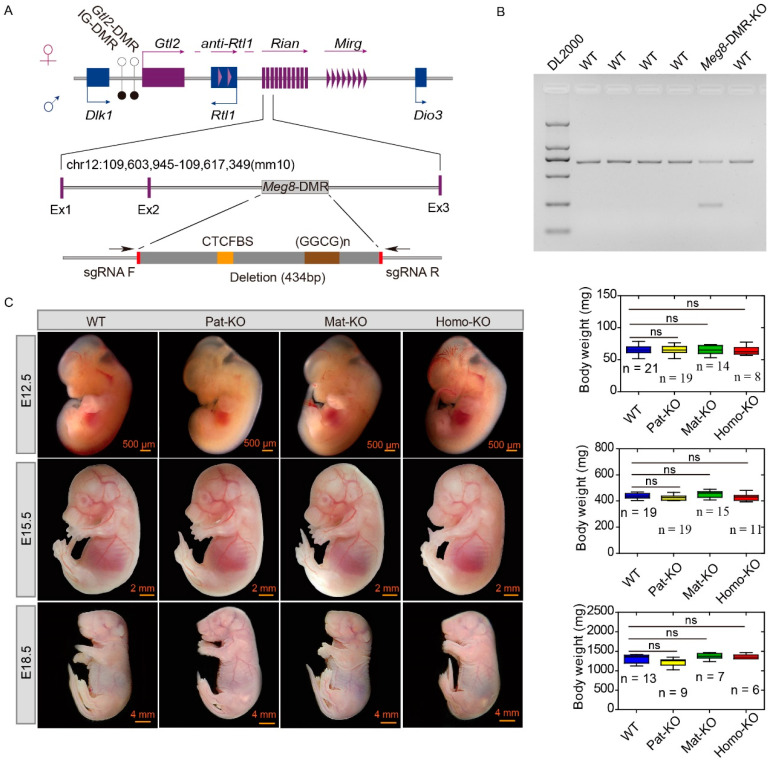
To investigate the functional role of the Meg8-DMR in mice, the CRISPR/Cas9 system was used to knockout the Meg8-DMR. The CRISPR/Cas9 system with two sgRNAs (sgRNA F and sgRNA R) was used to induce a 434 bp deletion in the Meg8-DMR. Cas9 mRNA and sgRNAs were transcribed in vitro. Then, they were injected together into blastocyst-stage embryos (Figure 1A). In total, six pups were successfully obtained from 83 transferred embryos. The primers Meg8-F and Meg8-R were used to validate the deletion of Meg8-DMR. PCR products indicated that one of the six founders harbored the deletion (Figure 1B). The PCR product was cloned into TA cloning vectors and further confirmed by sequencing. Sequencing of this pup showed that 434 bp between the two targeted sites was successfully knocked out. To investigate whether deletions could be stably inherited by pups, the founder was mated with wild-type ICRs. The genotypic identification of litters from F1 animals showed that Meg8-DMR deletion can be stably inherited by their offspring.
Figure 1.
CRISPR/Cas9-mediated deletion of Meg8-DMR and the growth of the mutants. (A) Regional physical map of the Dlk1-Dio3 domain and deletion locus of Meg8-DMR on mouse chromosome 12. The positions of imprinted genes are indicated by squares, vertical bars (snoRNAs), or triangles (microRNA). Maternally expressed genes are indicated by purple, paternally expressed genes are indicated by blue, CTCFBS is indicated by yellow and (GGCG)n is indicated by brown. The sgRNA primers are indicated by red. The direction of sgRNA primers are indicated by the arrows. IG-DMR and Gtl2-DMR are indicated by circles (filled circles, hypermethylation; open circles, hypomethylation). (B) Demonstration of 434bp deletion in the Meg8-DMR in founder animals. One of the six founders showed a 434bp deletion in the Meg8-DMR. (C) Gross phenotype of E12.5, E15.5, and E18.5 Meg8-DMR embryos. Comparison of WT with Pat-KO, Mat-KO, and Homo-KO mutants.
3.2. The Mode of Inheritance of Meg8-DMR Mutants
To investigate the epigenetic mode of Meg8-DMR knockout mice in the embryos, we generated four genotypes of E12.5 embryos, WT, Homo-KO, Pat-KO, or Mat-KO by crossing male Meg8-DMR heterozygous with female heterozygous mutants. However, it was impossible to distinguish whether the deletion was from the paternal or maternal allele. By crossing male wild-type mice with female heterozygotes mice, we generated Mat-KO embryos, and by crossing male heterozygotes with female wild-type mice, we generated Pat-KO embryos. Compared with the wild-type, Pat-KO, Mat-KO, and Homo-KO mutant embryos had normal physical characteristics in terms of body weight and length (Figure 1C). In order to eliminate the interference caused by the cohabitation scheme, we tried to combine the mating scheme. The results were consistent: the number of pups and proportion of each genotype were close to the Mendelian inheritance proportion in the Meg8-DMR mutants (Table 1). To investigate whether Meg8-DMR knockout results in perinatal lethality, E15.5 embryos and E18.5 embryos were generated in the same crossing. The mode of inheritance and phenotype were the same as the E12.5 mutant embryos. Furthermore, all the mutants survived at the embryonic stage and developed into normal adults with reproductive abilities.
Table 1.
Number of pups per litter in the Meg8-DMR mutants.
| Mating Pattern | No. of Litters | No. of Pups/Total (%) | ||||
|---|---|---|---|---|---|---|
| Total | WT | Pat-KO | Mat-KO | Homo-KO | ||
| WT (♀) × WT (♂) | 3 | 33 | 33 ± 0.67 (100) | — | — | — |
| WT (♀) × Pat-KO (♂) | 4 | 36 | 18 ± 0.75 (50) | 18 ± 1.0 (50) | — | — |
| WT (♀) × Mat-KO (♂) | 3 | 30 | 14 ± 0.83 (46.7) | 16 ± 0.63 (53.3) | — | — |
| WT (♀) × Homo-KO (♂) | 3 | 39 | — | 39 ± 0.66 (100) | — | — |
| Homo (♀) × WT (♂) | 3 | 37 | — | — | 37 ± 1.06 (100) | — |
| Homo (♀) × Pat-KO (♂) | 3 | 34 | — | — | 15 ± 1.3 (44.1) | 19 ± 0.76 (55.9) |
| Homo (♀) × Mat-KO (♂) | 3 | 30 | — | — | 14 ± 0.5 (46.7) | 16 ± 1.1 (53.3) |
| Pat-KO (♀) × Pat-KO (♂) | 3 | 24 | 5 ± 0.5 (20.8) | 13 ± 0.66 (54.2) | 6 ± 0.66 (25) | |
| Pat-KO (♀) × Mat-KO (♂) | 5 | 64 | 18 ± 1.12 (28.1) | 30 ± 1.2 (46.9) | 16 ± 1.04 (25) | |
| Pat-KO (♀) × Homo-KO (♂) | 3 | 36 | — | 17 ± 0.46 (47.2) | — | 19 ± 0.9 (52.8) |
| Pat-KO (♀) × WT (♂) | 5 | 51 | 28 ± 1.2 (54.9) | — | 23 ± 1.5 (45.1) | — |
| Mat-KO (♀) × Pat-KO (♂) | 4 | 56 | 16 ± 1.0 (28.6) | 30 ± 1.0 (53.5) | 10 ± 1.0 (17.9) | |
| Mat-KO (♀) × Mat-KO (♂) | 4 | 51 | 12 ± 2.0 (23.5) | 28 ± 1.5 (54.9) | 11 ± 0.9 (21.6) | |
| Mat-KO (♀) × Homo-KO (♂) | 3 | 39 | — | 21 ± 1.3 (53.8) | — | 18 ± 1.3 (46.2) |
| Mat-KO (♀) × WT (♂) | 3 | 27 | 15 ± 0.67 (55.5) | — | 12 ± 1.3 (44.5) | — |
3.3. Analysis of DEGs in WT and CRISPR/Cas9-Mediated Meg8-DMR Knockout Mice
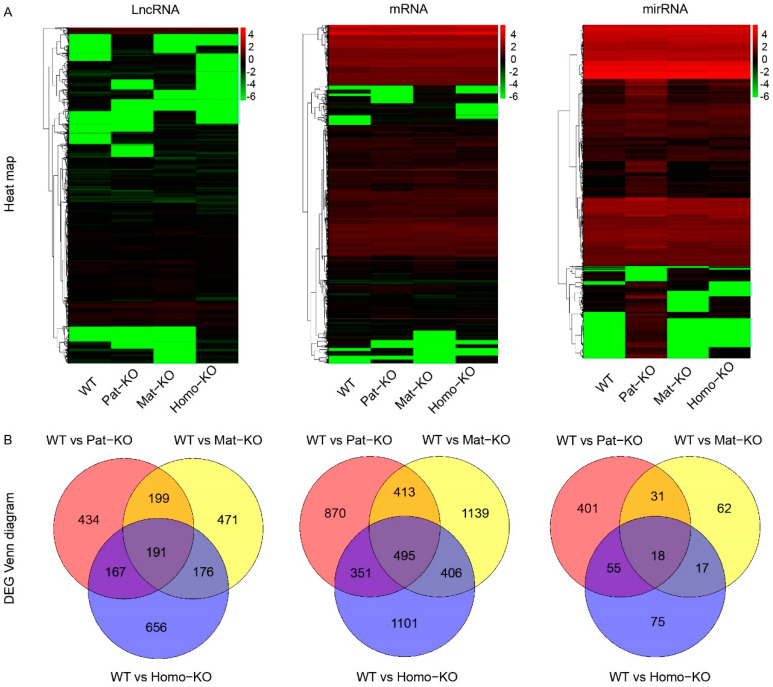
To further investigate the molecular mechanism underlying Meg8-DMR regulation, RNA-seq was performed to analyze E12.5 embryos. We performed a differentially expressed genes’ (DEGs’) heat map analysis of lncRNA, mRNAs, and microRNAs in total embryonic RNA from each of the four genotypes (Figure 2A). The results showed that microRNA expression profiles were significantly different in Pat-KO, Mat-KO, and Homo-KO compared with WT. To our surprise, microRNAs, following Meg8-DMR knockout, had the greatest impact on Pat-KO, followed by Homo-KO and Mat-KO. This result was consistent with the pattern of imprinting regulation. A subsequent Venn diagram analysis of microRNA expression profiles was conducted; the threshold was ∣log2 fold change (FC)∣ > 1 and adjusted p value < 0.05. The results showed 505, 128, and 165 DEGs for Pat-KO, Mat-KO, and Homo-KO, respectively, compared with WT (Figure 2B). Overall, Meg8-DMR knockout had broad effects on microRNA expression profiles.
Figure 2.
Analysis of differentially expressed genes (DEGs) in WT and CRISPR/Cas9-mediated Meg8-DMR knockout mice. (A) Heat map of LncRNA, mRNA, and miRNA DEGs in WT and Meg8-DMR knockout mice. From left to right: lncRNA, mRNA, and microRNA; The horizontal coordinate represents the sample name and the clustering result of the sample, and the vertical coordinate represents the differential gene and the clustering result of the gene. Different columns in the diagram represent different samples, and different rows represent different genes. The color represents the level of gene expression in the sample log10 (gene + 0.000001). (B) Venn diagram of DEGs in Pat-KO (red), Mat-KO (yellow), and Homo-KO (blue) mutants compared to the wild-type. From left to right: lncRNA, mRNA, and microRNA.
3.4. Detection of miRNA DEGs in WT and Meg8-DMR Knockout Mice
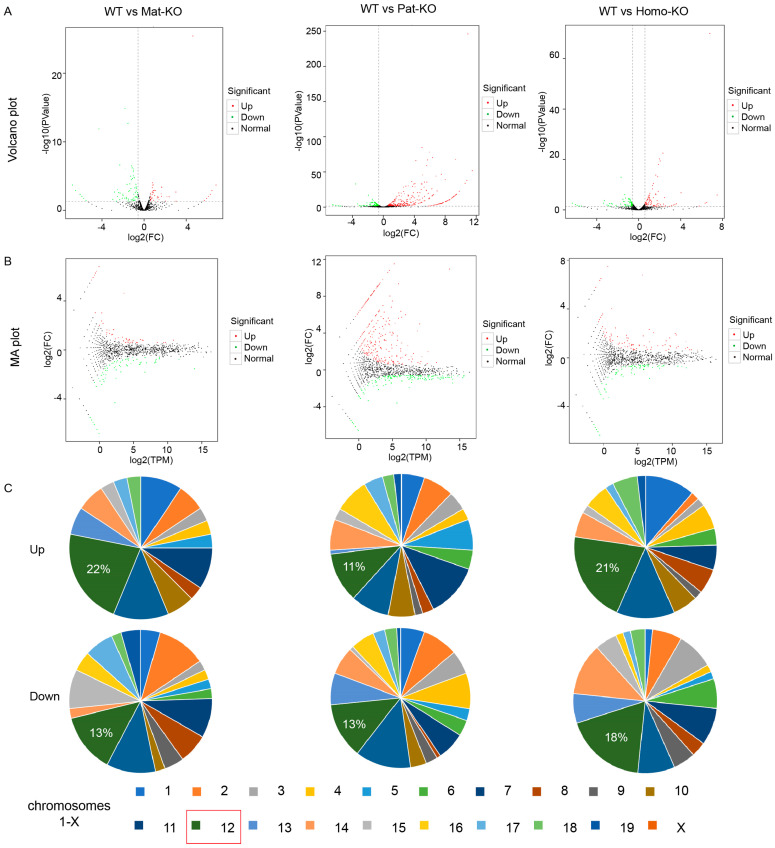
To learn more about the involvement of Meg8-DMR in microRNA regulation, we obtained the DEGs of Pat-KO, Mat-KO, and Homo-KO and compared them with WT. The DEGs are presented in a volcano plot (Figure 3A), and the chromosome distribution of the DEGs are presented in the pie charts (Figure 3B). The red dots represented upregulated DEGs, and the green dots represented downregulated DEGs. In total, 152 upregulated and 353 downregulated DEGs were identified in WT vs. Pat-KO, 77 upregulated and 51 downregulated DEGs were identified in WT vs. Mat-KO, and 79 upregulated and 86 downregulated DEGs were identified in WT vs. Homo-KO. In comparison to the WT, the majority of upregulated and downregulated DEGs were found on chromosome 12, where the Meg8-DMR was located.
Figure 3.
Detection of miRNA DEGs in WT and Meg8-DMR knockout mice. (A) Volcano plot corresponding to the WT, Pat-KO, Mat-KO, and Homo-KO. From left to right: WT vs. Pat-KO, WT vs. Mat-KO, and WT vs. Homo-KO. (B) Meandiff plot for the WT compared with Pat-KO, Mat-KO, and Homo-KO. (C) Distribution of upregulated and downregulated miRNAs in Pat-KO, Mat-KO, and Homo-KO on chromosomes.
3.5. GO and KEGG Pathway Enrichment of DEGs
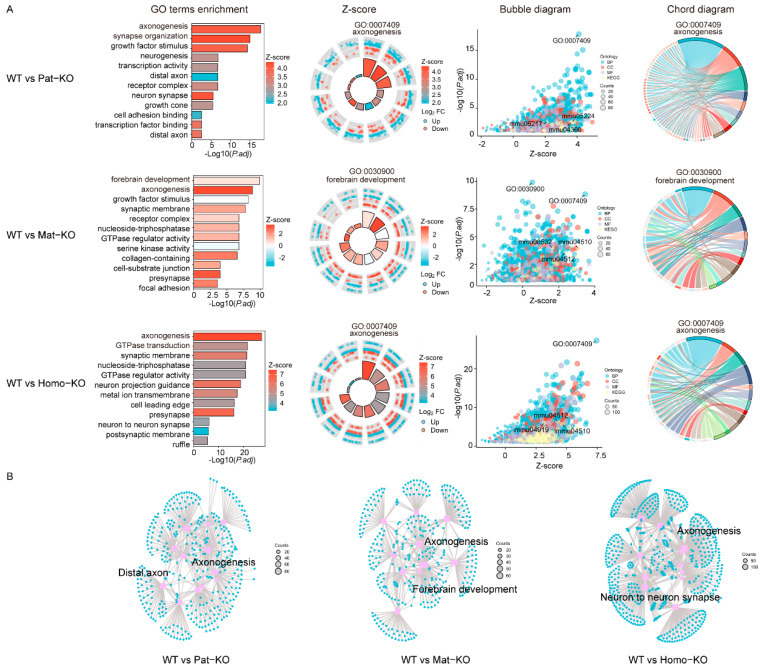
To further understand the effects of Meg8-DMR at the embryonic stage, we compared Pat-KO, Mat-KO, and Homo-KO mutants with WT using GO and KEGG pathway enrichment analysis. We used the top 12 GO terms for comparative analysis. The results of microRNA-associated DEGs analysis in Pat-KO and Homo-KO suggest that the biological processes of these genes were mainly involved in axonogenesis and forebrain development in Mat-KO (Figure 4A). The network diagram and relationships between DEG and the top 12 GO enrichment results showed that axonogenesis was located at the central node of the network (Figure 4B). Taken together, the GO and KEGG pathway enrichment analysis of DEGs in mutations revealed that Meg8-DMR played a role in the axonogenesis and forebrain development of mice.
Figure 4.
Go and KEGG pathway enrichment of DEGs. (A) GO terms and KEGG pathway enrichment results. From left to right: the top 12 GO enrichment results and Z-score results; bubble diagram and relationships between DEGs and the KEGG pathway. (B) Network diagram and relationships between DEGs and the top 12 GO enrichment results.
3.6. Expression of the Imprinted Genes in the Dlk1-Dio3 Domain
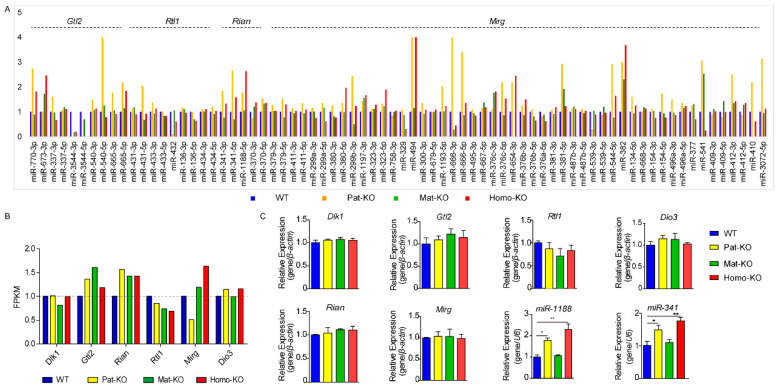
The Dlk1-Dio3 domain contains a number of microRNAs. We also analyzed the effect on microRNA on the cluster. RNA-seq showed that Meg8-DMR knockout resulted in altered expression of the microRNA. To our surprise, microRNA expression was upregulated in Pat-KO and Homo-KO mutants, whereas microRNA in Mat-KO mutants showed little change compared with WT (Figure 5A). The maternally expressed Gtl2 and Rian were upregulated and paternally expressed; Rtl1 was downregulated. The expression of Dlk1, Mirg, and Dio3 did not differ from that of WT (Figure 5B). QRT-PCR analysis showed that the expression of Gtl2 was increased by 8% in Pat-KO, 14% in Mat-KO, and 12.4% in Homo-KO compared to WT. The expression of Rian was increased by 3.7% in Pat-KO, 10% in Mat-KO, and 9.8% in Homo-KO. The expression of Rtl1 was decreased by 8.6% in Pat-KO, 7.1% in Mat-KO, and 8.3% in Homo-KO. The expression of Dlk1, Mirg, and Dio3 was not altered. QRT-PCR analysis of miR-1188 and miR-341 showed that the expression of miR-1188 was increased by 77% in Pat-KO, 6% in Mat-KO, and 120% in Homo-KO compared with WT. The expression of miR-341 was increased by 68% in Pat-KO, 5% in Mat-KO, and 90% in Homo-KO (Figure 5C), the primer sequences were shown in Table S1. In summary, the paternal expression gene Rtl1 was downregulated, and maternal expression genes Gtl2 and Rian were upregulated in the Dlk1-Dio3 domain; the expression of microRNA was upregulated in Pat-KO and Homo-KO mutations; little changed in Mat-KO.
Figure 5.
Expression of the imprinted genes in the Dlk1-Dio3 domain. (A) TPM analyses of the microRNA expression in the Dlk1-Dio3 domain. (B) FPKM analyses of the lncRNA and mRNA expression in the Dlk1-Dio3 domain. (C) The expression level of the six imprinted genes Dlk1, Gtl2, Rtl1, Rian, Mirg, Dio3, and two microRNA mir-1188; mir-341 was measured at E12.5. Error bars, mean ± SD. n = 4. p values were calculated using two-way ANOVA. ** p < 0.01, * p < 0.05.
3.7. Methylation Status and Allelic Analysis of Gene Expression in the Meg8-DMR Mutants
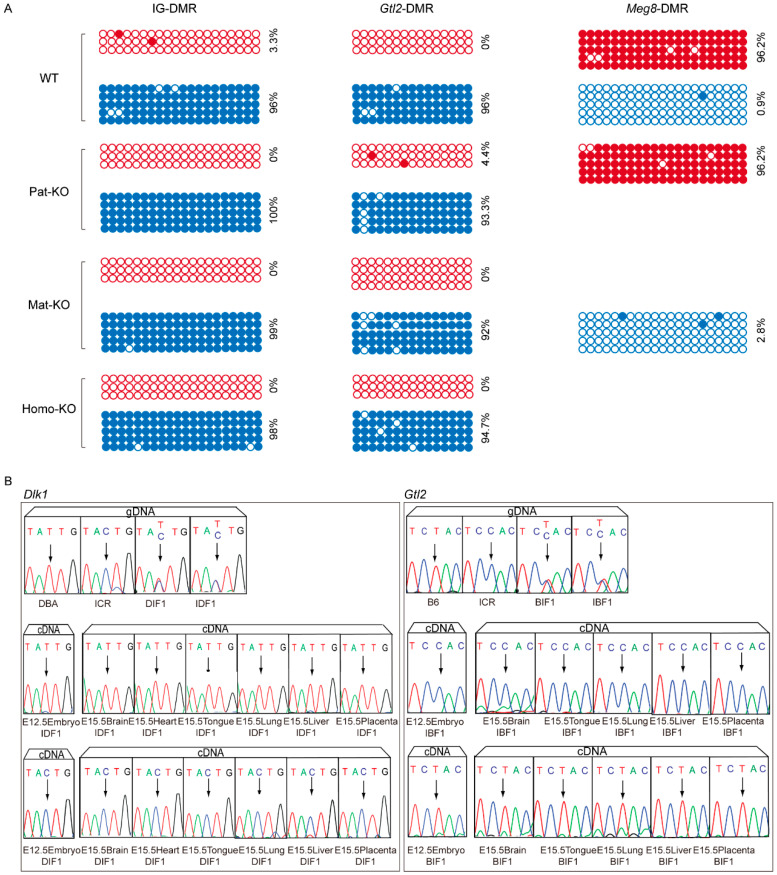
The Dlk1-Dio3 domain was mainly regulated by IG-DMR and Gtl2-DMR. We examined whether Meg8-DMR knockout changed the methylation status. We found that the paternal methylation levels of IG-DMR and maternal methylation levels of Gtl2-DMR were not changed. The methylation status of Pat-KO, Mat-KO, and Homo-KO mutants was consistent with that of WT. Then, we investigated the methylation level of Meg8-DMR in WT, Pat-Ko, and Homo-KO mutants. The results showed the hypermethylation level of the maternal allele in Pat-KO mutants, while the unmethylation level of paternal alleles was shown in Mat-KO mutants (Figure 6A). These results indicated that the deletion of Meg8-DMR does not change the methylation status of IG-DMR, Gtl2-DMR, and Meg8-DMR.
Figure 6.
Methylation status and allelic analysis of gene expression in the Meg8-DMR mutants. (A) IG-DMR, Gtl2-DMR, and Meg8-DMR methylation status in the Meg8-DMR mutants. Maternal allele and paternal allele are represented by red and blue circles, respectively. Methylated and unmethylated CpG sites are represented by filled and open circles. (B) Sequence analysis of the PCR products of Dlk1, Gtl2, and Rian. The samples were obtained from genomic DNA (Dlk1 was obtained from DBA/2J and ICR; Gtl2 was obtained from C57BL/6J and ICR) and cDNA. The polymorphic bases in the sequences are arrowed.
To further understand the expression status of imprinting genes in Meg8-DMR knockout mutants, we analyzed the imprinting status of Dlk1 and Gtl2 in transgenic mice. The imprinting patterns in E12.5.5 whole embryo and E15.5 brain, tongue, lung, liver, and placenta tissues were detected. The SNP sites of Dlk1 (T in DBA/2J strain, C in ICR strain) and Gtl2 (T in C57BL/6, C in ICR strain) marked the parents of hybrid mice, respectively, and the DNA of embryos and tissues of IDF1 and DIF1 were detected. The peak pattern at the selected SNP sites showed double peaks. The expression of paternally expressed gene Dlk1 was only expressed by the paternal allele, while the expression of the maternally expressed gene Gtl2 was only expressed by the maternal allele (Figure 6B). These results indicated that the imprinting status of Dlk1 and Gtl2 in the whole embryo and tissues was not changed.
4. Discussion
Meg8-DMR, the first discovered maternally methylated DMRs within the Dlk1-Dio3 imprinting domain, has not been well-studied. In this study, we generated Meg8-DMR knockout mouse models using the CRISPR/Cas9 system and obtained WT, Pat-KO, Mat-KO, and Homo-KO mutants using genotypic hybridization. The characterization of these mice suggested that the deletion of Meg8-DMR caused the expression of microRNA to be upregulated in Pat-KO and Homo-KO mutations, while little changed in Mat-KO. However, the effect is not sufficient to have a significant impact on morphological characteristics or embryonic development. This provides new insight into the role of Meg8-DMR in embryonic development.
The current understanding of DMRs within the Dlk1-Dio3 domain was mainly based on IG-DMR and Gtl2-DMR. However, the genetic pattern, characterization, and molecular performance of Meg8-DMR at the embryonic stages were significantly different from those of IG-DMR and Gtl2-DMR. Meg8-DMR did not affect the development of mouse embryos, as Meg8-DMR was a somatic cell-derived DMR. The expression of lncRNA, mRNA, microRNA, and snoRNAs in the imprinted domain changed when IG-DMR or Gtl2-DMR were knocked out. However, the expression of microRNAs changed most significantly after Meg8-DMR deletion, indicating that the regulatory ability of Meg8-DMR was the weakest. In addition, the methylation status of DMRs in Dlk1-Dio3 was not altered; it seems that the transcriptional regulation altered the expression of microRNAs via the cis and trans effects [28].
In this study, the expression of Gtl2, Rtl1, and Rian changed in all Meg8-DMR mutants, although the dosage was less than 10%. This suggested that both chromosomes derived from this parent of Meg8-DMR may have biological functions. This bears some resemblance to the unique phenotype of Gtl2 knockout mice. A previous study found that the effect on mice was lethal during the embryonic period when Gtl2 deletion occurred in Pat-KO. When Gtl2 deletion occurred in Mat-KO, all mice died within 4 weeks of birth. It is likely that this regulatory relationship requires the process of transcription or the transcript itself. We hypothesized that the changes induced by Meg8-DMR knockout in the embryo would continue to accumulate until the mice were born or grew into adult mice.
Our previous experiments showed that the same gene expression pattern could be observed in MLTC-1 cells by knocking out only the CTCF binding site or all the Meg8-DMR [24]. In particular, the paternally expressed genes Dlk1 and Rtl1 were significantly downregulated. The invasion and migration abilities of MLTC-1 cells were enhanced. However, the changes were only insignificant when GGCG repeats were knocked out. This indicates that the GGCG repeats are required for the function of Meg8-DMR. In MLTC-1, we knocked out 644bp, including the GGCG repeats, but only knocked out 434bp when the knockout occurred in mice. We suspect that the deletion of the 210bp nucleotide sequence may be the main cause of the phenotypic difference.
In this study, GO and KEGG pathway enrichment showed that axonogenesis and forebrain development were enhanced, although this phenotype was not present in the embryonic development period. Axonogenesis and forebrain development led to a change in neural connections and early hippocampal development. This is essential for neural conduction, spatial learning, and memory formation in later cognitive functioning [29,30]. High-throughput sequencing analysis revealed that genes associated with hippocampal development, such as NFAT and Ndufb1, were upregulated. Moreover, the intraventricular injection of adenovirus and overexpression of Rian can improve sevoflurane-induced neuronal injury and apoptosis, and the activity of the hippocampal neurons is enhanced in this process. Therefore, Rian has a neuroprotective effect on cognitive dysfunction [31]. In this study, the expression of Rian tended to be upregulated at the embryonic stage. This suggests that Meg8-DMR may be associated with the development of intelligence in adult mice. This study identified Meg8-DMR as a potential target for improving cognitive performance in adult mice.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14061264/s1. Table S1. Primer sequences. References [6,19] are cited in the supplementary materials.
Author Contributions
Q.W. conceived this project and designed the experiment. L.Z. performed the experiments, wrote the paper, and analyzed the data for Figures. Z.H. bred the Meg8-DMR knockout mice. H.H. edited the paper. X.Z. performed the methylation analysis. M.Z. analyzed the RNA-seq data. B.L. prepared materials for the experiment. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Animal experiments were carried out in strict accordance with the Guide for the Care and Use of Laboratory Animals from the Harbin Institute of Technology (HIT) and approved by the Institutional Animal Care and Use Committee (IACUC-2023013) or Animal Experimental Ethics Committee of HIT.
Informed Consent Statement
Not applicable.
Data Availability Statement
RNA-seq sequencing data have been deposited in GEO under accession number.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (No. 3177090206).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Takahashi N., Kobayashi R., Kono T. Restoration of Dlk1 and Rtl1 Is Necessary but Insufficient to Rescue Lethality in Intergenic Differentially Methylated Region (IG-DMR)-deficient Mice. J. Biol. Chem. 2010;285:26121–26125. doi: 10.1074/jbc.M109.075325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Ferguson-Smith A.C., Surani M.A. Imprinting and the epigenetic asymmetry between parental genomes. Science. 2001;293:1086–1089. doi: 10.1126/science.1064020. [DOI] [PubMed] [Google Scholar]
- 4.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 5.Barlow D.P., Bartolomei M.S. Genomic Imprinting in Mammals. CSH Perspect. Biol. 2014;6:a018382. doi: 10.1101/cshperspect.a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rocha S.T.D., Edwards C.A., Ito M., Ogata T., Ferguson-Smith A.C. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Hagan J.P., O’Neill B.L., Stewart C.L., Kozlov S.V., Croce C.M. At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1. PLoS ONE. 2009;4:e4352. doi: 10.1371/journal.pone.0004352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tierling S., Dalbert S., Schoppenhorst S., Tsai C.E., Oliger S., Ferguson-Smith A.C., Paulsen M., Walter J. High-resolution map and imprinting analysis of the Gtl2-Dnchc1 domain on mouse chromosome 12. Genomics. 2006;87:225–235. doi: 10.1016/j.ygeno.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 9.Da R.S., Tevendale M., Knowles E., Takada S., Watkins M., Ferguson-Smith A.C. Restricted co-expression of Dlk1 and the reciprocally imprinted non-coding RNA, Gtl2: Implications for cis-acting control. Dev. Biol. 2007;306:810–823. doi: 10.1016/j.ydbio.2007.02.043. [DOI] [PubMed] [Google Scholar]
- 10.Labialle S., Croteau S., Belanger V., McMurray E.N., Ruan X., Moussette S., Jonnaert M., Schmidt J.V., Cermakian N., Naumova A.K. Novel imprinted transcripts from the Dlk1-Gtl2 intergenic region, Mico1 and Mico1os, show circadian oscillations. Epigenetics. 2008;3:322–329. doi: 10.4161/epi.3.6.7109. [DOI] [PubMed] [Google Scholar]
- 11.Georgiades P., Watkins M., Surani M.A., Ferguson-Smith A.C. Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development. 2000;127:4719–4728. doi: 10.1242/dev.127.21.4719. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt J.V., Matteson P.G., Jones B.K., Guan X.J., Tilghman S.M. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 2000;14:1997–2002. doi: 10.1101/gad.14.16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatada I., Morita S., Obata Y., Sotomaru Y., Shimoda M., Kono T. Identification of a new imprinted gene, Rian, on mouse chromosome 12 by fluorescent differential display screening. J. Biochem. 2001;130:187–190. doi: 10.1093/oxfordjournals.jbchem.a002971. [DOI] [PubMed] [Google Scholar]
- 14.Seitz H., Royo H., Bortolin M.L., Lin S.P., Ferguson-Smith A.C., Cavaille J. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavaille J., Seitz H., Paulsen M., Ferguson-Smith A.C., Bachellerie J.P. Identification of tandemly-repeated C/D snoRNA genes at the imprinted human 14q32 domain reminiscent of those at the Prader-Willi/Angelman syndrome region. Hum. Mol. Genet. 2002;11:1527–1538. doi: 10.1093/hmg/11.13.1527. [DOI] [PubMed] [Google Scholar]
- 16.Fahrenkrug S.C., Freking B.A., Smith T.P. Genomic organization and genetic mapping of the bovine PREF-1 gene. Biochem. Biophys. Res. Commun. 1999;264:662–667. doi: 10.1006/bbrc.1999.1558. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z.L., Cheng H.C., Xia Q.Y., Jiang C.D., Deng C.Y., Li Y.M. Imprinting analysis of RTLI and DIO3 genes and their association with carcass traits in pigs. Agric. Sci. China. 2009;8:101–105. doi: 10.1016/S1671-2927(08)60253-9. [DOI] [Google Scholar]
- 18.Takada S., Paulsen M., Tevendale M., Tsai C.E., Kelsey G., Cattanach B.M., Ferguson-Smith A.C. Epigenetic analysis of the Dlk1-Gtl2 imprinted domain on mouse chromosome 12: Implications for imprinting control from comparison with Igf2-H19. Hum. Mol. Genet. 2002;11:77–86. doi: 10.1093/hmg/11.1.77. [DOI] [PubMed] [Google Scholar]
- 19.Zeng T.B., He H.J., Han Z.B., Zhang F.W., Huang Z.J., Liu Q., Cui W., Wu Q. DNA methylation dynamics of a maternally methylated DMR in the mouse Dlk1-Dio3 domain. FEBS Lett. 2014;588:4665–4671. doi: 10.1016/j.febslet.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 20.Hiura H., Komiyama J., Shirai M., Obata Y., Ogawa H., Kono T. DNA methylation imprints on the IG-DMR of the Dlk1-Gtl2 domain in mouse male germline. FEBS Lett. 2007;581:1255–1260. doi: 10.1016/j.febslet.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 21.Lin S.P., Coan P., Da R.S., Seitz H., Cavaille J., Teng P.W., Takada S., Ferguson-Smith A.C. Differential regulation of imprinting in the murine embryo and placenta by the Dlk1-Dio3 imprinting control region. Development. 2007;134:417–426. doi: 10.1242/dev.02726. [DOI] [PubMed] [Google Scholar]
- 22.Lin S.P., Youngson N., Takada S., Seitz H., Reik W., Paulsen M., Cavaille J., Ferguson-Smith A.C. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 23.Takahashi N., Okamoto A., Kobayashi R., Shirai M., Obata Y., Ogawa H., Sotomaru Y., Kono T. Deletion of Gtl2, imprinted non-coding RNA, with its differentially methylated region induces lethal parent-origin-dependent defects in mice. Hum. Mol. Genet. 2009;18:1879–1888. doi: 10.1093/hmg/ddp108. [DOI] [PubMed] [Google Scholar]
- 24.Han X., He H., Shao L., Cui S., Yu H., Zhang X., Wu Q. Deletion of Meg8-DMR Enhances Migration and Invasion of MLTC-1 Depending on the CTCF Binding Sites. Int. J. Mol. Sci. 2022;23:8828. doi: 10.3390/ijms23158828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beygo J., Kuchler A., Gillessen-Kaesbach G., Albrecht B., Eckle J., Eggermann T., Gellhaus A., Kanber D., Kordass U., Ludecke H.J., et al. New insights into the imprinted MEG8-DMR in 14q32 and clinical and molecular description of novel patients with Temple syndrome. Eur. J. Hum. Genet. 2017;25:935–945. doi: 10.1038/ejhg.2017.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Court F., Tayama C., Romanelli V., Martin-Trujillo A., Iglesias-Platas I., Okamura K., Sugahara N., Simon C., Moore H., Harness J.V., et al. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014;24:554–569. doi: 10.1101/gr.164913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lokk K., Modhukur V., Rajashekar B., Martens K., Magi R., Kolde R., Koltsina M., Nilsson T.K., Vilo J., Salumets A., et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome Biol. 2014;15:3248. doi: 10.1186/gb-2014-15-4-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demars J., Gicquel C. Epigenetic and genetic disturbance of the imprinted 11p15 region in Beckwith-Wiedemann and Silver-Russell syndromes. Clin. Genet. 2012;81:350–361. doi: 10.1111/j.1399-0004.2011.01822.x. [DOI] [PubMed] [Google Scholar]
- 29.Murai K., Qu Q., Sun G., Ye P., Li W., Asuelime G., Sun E., Tsai G.E., Shi Y. Nuclear receptor TLX stimulates hippocampal neurogenesis and enhances learning and memory in a transgenic mouse model. Proc. Natl. Acad. Sci. USA. 2014;111:9115–9120. doi: 10.1073/pnas.1406779111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Squire L.R. Memory and the hippocampus: A synthesis from findings with rats, monkeys, and humans. Psychol. Rev. 1992;99:195–231. doi: 10.1037/0033-295X.99.2.195. [DOI] [PubMed] [Google Scholar]
- 31.Yu Y., Zhang W., Zhu D., Wang H., Shao H., Zhang Y. LncRNA Rian ameliorates sevoflurane anesthesia-induced cognitive dysfunction through regulation of miR-143-3p/LIMK1 axis. Hum. Cell. 2021;34:808–818. doi: 10.1007/s13577-021-00502-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq sequencing data have been deposited in GEO under accession number.