Abstract
Bone morphogenetic proteins (BMPs) play important roles in a lot of biological processes, such as bone development, cell proliferation, cell differentiation, growth, etc. However, the functions of abalone BMP genes are still unknown. This study aimed to better understand the characterization and biological function of BMP7 of Haliotis discus hannai (hdh-BMP7) via cloning and sequencing analysis. The coding sequence (CDS) length of hdh-BMP7 is 1251 bp, which encodes 416 amino acids including a signal peptide (1–28 aa), a transforming growth factor-β (TGF-β) propeptide (38–272 aa), and a mature TGF-β peptide (314–416 aa). The analysis of expression showed that hdh-BMP7 mRNA was widely expressed in all the examined tissues of H. discus hannai. Four SNPs were related to growth traits. The results of RNA interference (RNAi) showed that the mRNA expression levels of hdh-BMPR I, hdh-BMPR II, hdh-smad1, and hdh-MHC declined after hdh-BMP7 was silenced. After RNAi experiment for 30 days, the shell length, shell width, and total weight were found to be reduced in H. discus hannai (p < 0.05). The results of real-time quantitative reverse transcription PCR revealed that the hdh-BMP7 mRNA was lower in abalone of the S-DD-group than in the L-DD-group. Based on these data, we hypothesized that BMP7 gene has a positive role in the growth of H. discus hannai.
Keywords: BMP7, Haliotis discus hannai, growth, RNAi, SNP
1. Introduction
The transforming growth factor β (TGF-β) family is a multifunctional group of secretory signal transducers that are involved in cell proliferation, differentiation, apoptosis, embryonic development, growth, bone formation and reconstruction, and other important biological processes [1,2,3,4,5]. The TGF-β superfamily includes TGF-βs, growth differentiation factors (GDFs), bone morphogenetic proteins (BMPs), Inhibins/Activins, and Nodal. All members of the TGF-β superfamily release mature polypeptides by division of precursors at specific sites, and the activity relies on the formation of dimers from the same or different subunits [6,7]. Although the TGF-β family contains many diverse factors, they all signal via conserved signaling pathways. The TGF-β family ligands bind to receptors and then mediate the phosphorylation of Smad proteins. The interaction between Smads and other signaling proteins mediates regulatory signals, controlling the expression of target genes, the translation of mRNA translation, and the regulation of cytoplasmic protein [8,9,10,11]. In recent years, the TGF-β superfamily has been found to play a critical role in the growth and development of shellfish [7,12,13,14].
BMPs, as the largest secreted cytokines in the TGF-β superfamily, are originally named for their ability in bone formation [15]. BMPs are widely expressed during the development of mammals, with many biological activities, such as development, proliferation, etc. [7,16]. BMPs can distinguish between serine and threonine kinase receptors and bind to them, and the subsequent signaling is mediated by Smad-dependent and -independent pathways [7,17,18]. BMP7, as a member of the BMPs, plays important roles in various biological functions including cell proliferation, differentiation, and apoptosis [19,20,21,22,23,24]. BMP7 has the classical TGF-β domains [7,25]. In vertebrates, BMP7 is closely related to bone development, organogenesis, and embryo development [20,22,23,26]. Recombinant human BMP7 protein can promote bone regeneration [7,27,28,29]. In mice, BMP7 was related to the proliferation of sertoli cells, and BMP7 knockout mice were infertile [7,30,31]. BMP7 was showed to be associated with growth in cattle and chickens, and growth-related SNPs have been founded [7,32,33,34]. Overexpression of BMP7 promotes muscle growth and prevents muscle atrophy [35,36]. In invertebrates, so far, the BMP7 gene has been reported in several bivalve species, including Tegillarca granosa [37], Pinctada martensii [38,39], and Hyriopsis cumingii [40]. The interference experiments of PmBMP7 showed that BMP7 played an important role in the formation of nymphal and columnar layers of shells in the P. martensii [38]. The mRNA expression of TG-BMP7 in the mantle and gill of T. granosa is high, and the expression level in the d-larva stage is high during embryo development, suggesting that BMP7 may be related to shell formation and regulation of embryo growth and development [7].
The H. discus hannai is the most popular cultivated abalone throughout in China [41,42]. The abalone growth cycle generally lasts for 2 years, which restricts the development of abalone aquaculture. At present, the molecular genetics and function of most genes closely associated with growth in H. discus hannai are not clear, which limits the genetic improvement of related growth traits. Hybridization is an effective method to introduce improved traits [43,44,45]. Lvpan abalone (H. discus hannai♀× H. fulgens♂), a hybrid with fast growth and large size, has become a new abalone variety in Fujian Province [46]. Lvpan abalone may be a good material for proving the mechanism related to abalone growth. BMP7 is identified to be involved in animal growth and development, but there are few studies conducted on BMP7 in marine mollusks, and there are no reports on whether BMP7 is involved in the growth of abalone. In this work, we analyzed the gene structure of BMP7 and studied its role in the H. discus hannai growth. Our results in the paper would lay a foundation for further study on molecular mechanisms of growth regulation in abalone.
2. Materials and Methods
2.1. Sample Collection
All abalones (H. discus hannai and Lvpan abalone) used in this study were obtained from Fuda Aquaculture and were consistent with the previous study [43]. The adductor muscles were collected in different stages of development of H. discus hannai, that is, at 1, 4, 7, 10, 12, 14, 16, and 18 months. All experimental samples used to analyze the expression pattern in tissues were consistent with the previous study [43]. The adductor muscle tissue samples of the larger H. discus hannai (“L-DD-group”), the smaller H. discus hannai (“S-DD-group”), the larger Lvpan abalone (“L-DF-group”), and the smaller Lvpan abalone (“S-DF-group”) were also consistent with the previous study [43]. All the samples were stored at −80 °C.
2.2. Cloning and Sequence Analysis
The isolation of total RNA and the synthesis of cDNA followed the methods in the previous study by Huang et al. (2023) [43]. The primers (hdh-BMP7-F and hdh-BMP7-R), used to amplify the hdh-BMP7 open reading frame (ORF), were designed using the software of Primer3Plus (Table 1). The ORF sequences of hdh-BMP7 were uploaded to GenBank (accession numbers: OP856631). Lasergene software tools were used to determine the amino acid sequence of hdh-BMP7. SingalP 5.0 Server was used for signal peptide prediction; Prop 1.0 server was used for proteolytic processing site analysis; SOPMA was used for secondary structure analysis; CDD was used for protein domains; the ClustalW2 program was used for aligning the sequence of BMP7, and the MEGA program with the neighbor-joining algorithm was used for constructing the phylogenetic tree.
Table 1.
Sequences of the primer pairs.
| Primer Name | Sequence Information (5′-3′) |
|---|---|
| hdh-BMP7-F | CACAGAAGGGGATCGAGTCA |
| hdh-BMP7-R | AGGCTACGAAAAGGTCACGT |
| hdh-BMP7-dsF | TAATACGACTCACTATAGGGGCCACCAAGACTTCCACATT |
| hdh-BMP7-dsR | TAATACGACTCACTATAGGGGATCGCTATCCATCCGAAAA |
| EGFP-dsF | TAATACGACTCACTATAGGGGTGCCCATCCTGGTCGAGCT |
| EGFP-dsR | TAATACGACTCACTATAGGGTGCACGCTGCCGTCCTCGAT |
| hdh-BMP7-qF | GCGGTCCCATCTTCGTCA |
| hdh-BMP7-qR | TGCACGTTCTTTGTACAGCC |
| hdh-BMPR I-qF | GACCTCATAGAACAGTCCC |
| hdh-BMPR I-qR | ACCATAACGCCCCTTGCCT |
| hdh-BMPR II-qF | GGCGGGGAGAGATGTAATG |
| hdh-BMPR II-qR | TAAGTTGGGTCGGGTGTAG |
| hdh-Smad1-qF | GGACTCCTCTCCAACGTCAA |
| hdh-Smad1-qR | ACATTCCGCAAACACCTCAC |
| hdh-MHC-qF | GACCCCAACGACCCTGATAT |
| hdh-MHC-qR | TCTTCTCCCTTGGTGCTCTG |
| β-actin-qF | GGTATCCTCACCCTCAAGT |
| β-actin-qR | GGGTCATCTTTTCACGGTTG |
2.3. Single Nucleotide Polymorphism Analysis
To explore the SNPs of hdh-BMP7, the abalones, the foot muscle samples, the method of DNA extraction, the growth-related traits, and the method of hdh-BMP7 ORF sequence genotype were consistent with the previous study [43]. The POPGENE 1.32 software was used to evaluate the PIC of the SNPs according to the manufacturer’s instructions.
2.4. RNA Interference of hdh-BMP7
A pair of specific primers, dsBMP7-F and dsBMP7-R, were designed to amplify a 303 bp fragment of the hdh-BMP7. EGFP acted as a control gene, amplifying a 497 bp fragment. The information of specific primers is shown in Table 1. The dsRNA synthesis and purity were performed as previously described [47]. For the hdh-BMP7 RNAi assays, approximately 80 H. discus hannai (~4–5 g) were randomly divided into two groups: the hdh-BMP7 RNAi group and the EGFP control group. The abalones were intramuscularly injected. The RNAi experimental methods were consistent with the previous study [43].
2.5. Real-Time Quantitative Reverse Transcription PCR
The qRT-PCR was used to detect the gene expression levels of hdh-BMP7 (gene ID: evm.model.scaffold76053.25.1), hdh-BMPR I (gene ID: evm.model.scaffold168485.18), hdh-BMPR II (gene ID: evm.model.scaffold36941.4), hdh-Smad1 (gene ID: evm.model.scaffold148433.104_evm.model.scaffold148433.105), and MHC (gene ID: evm.model.scaffold19291.11). These primers (Table 1) were designed using the software of Primer3Plus. The data processing used the 2−∆∆CT method. The qRT-PCR runs and the cycling parameters used were performed using similar conditions as described previously [48]. The PCR amplification was performed in 20 μL reaction mixtures containing the following components: 10 μL FastStart Universal SYBR Green Master (ROX), 1 μL forward and reverse primers (10 μM each), 5 μL of cDNA (100-times diluted), and 4 μL distilled water [29].
2.6. Statistical Analysis
All experimental data in this study were analyzed using one-way ANOVAs (ANOVA) in SPSS 19.0 (IBM, Tulsa, OK, USA). The significance level for these analyses was specified at p < 0.05.
3. Results
3.1. Characterization of Hdh-BMP7
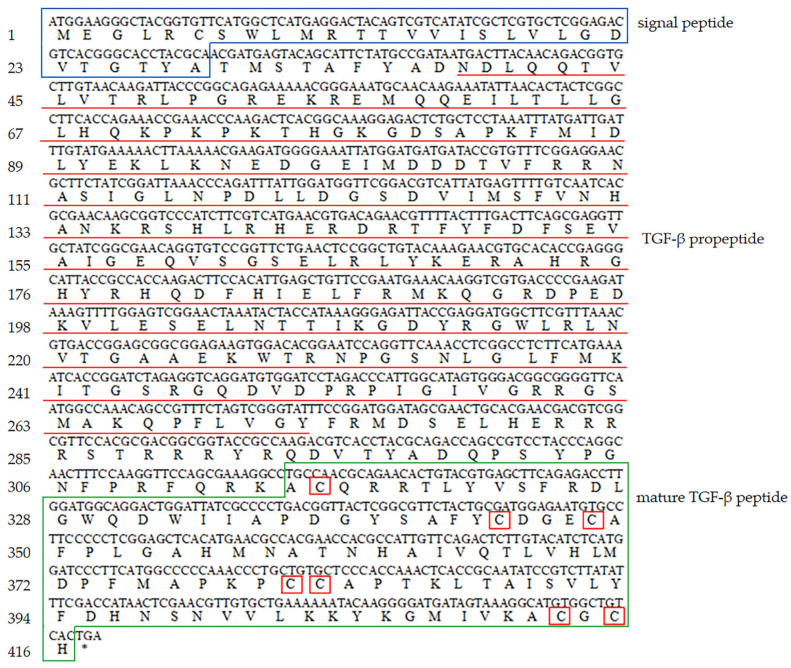
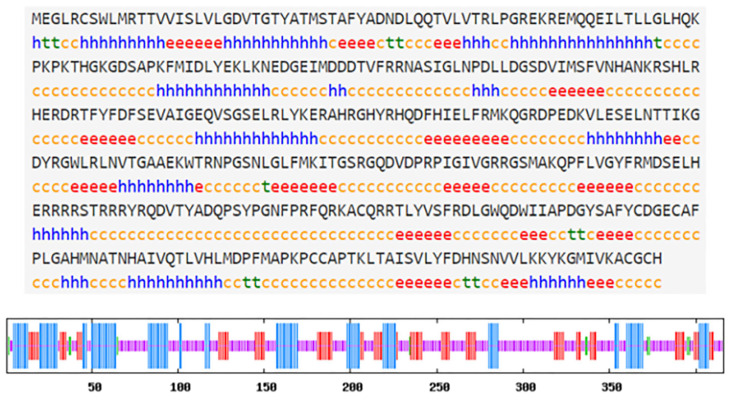
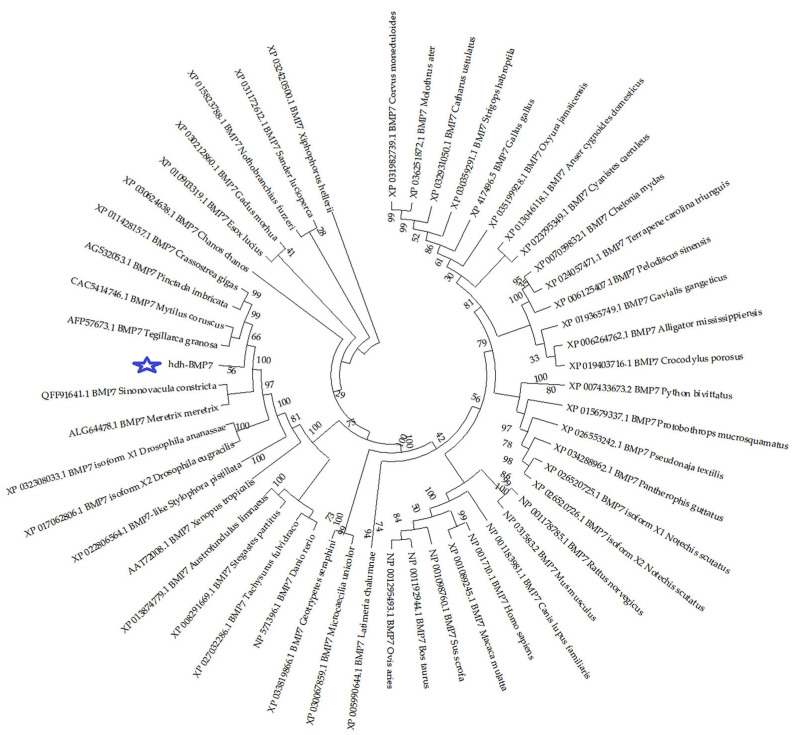
The hdh-BMP7 protein contained 416 amino acid residues and was encoded by 1251 nucleotides. The estimated molecular weight of the mature hdh-BMP7 was 47.80 kDa, and the pl was 9.15. The hdh-BMP7 protein contained a signal peptide (1–28 aa), a TGF-β propeptide (38–272 aa), and a mature TGF-β peptide (314–416 aa). The mature protein has two putative proteolytic sites of Arg-X-X-Arg (52–55 aa and 282–285 aa) with seven conserved cysteine residues (Figure 1). The secondary structure of hdh-BMP7 was predicted to possess 26.44% α-helix, 20.43% extended strand, 2.88% β-turns, and 50.24% random coils (Figure 2). The phylogenetic tree demonstrated a close relationship between H. discus hannai, T. granosa, Mytilus coruscus, Pinctada imbricata, and Crassostrea gigas, which clustered together and formed a branch (Figure 3). This suggests that the BMP7 protein is evolutionarily conserved in sequence and structure in shellfish.
Figure 1.
Gene information of hdh-BMP7. The cysteine residues are shown in the red boxes. The signal peptides are shown in the blue boxes. The TGF-β propeptide is shown in red lines. The mature TGF-β peptides are shown in the green boxes.
Figure 2.
The information of hdh-BMP7 in secondary structure. Hh expresses the α-helix. Tt expresses the β-turn. Ee expresses the extended strand. Cc expresses the random coil.
Figure 3.
Phylogenetic tree of BMP7 from vertebrates and invertebrates. Hdh-BMP7 is shown with a pentagram.
3.2. Expression Detection of Hdh-BMP7
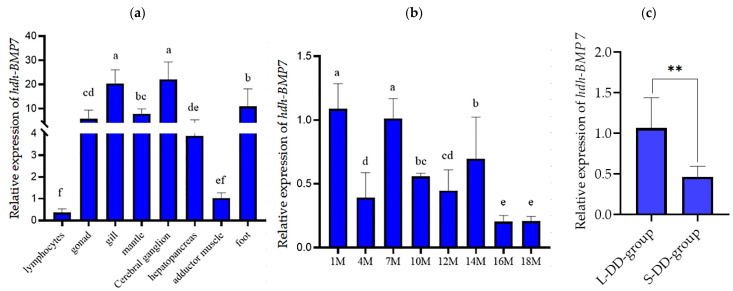
The expression levels of hdh-BMP7 in tissue and developmental stage-specific abalones were determined by qRT-PCR. The highest expression levels of hdh-BMP7 were found in the gills and cerebral ganglion (p < 0.05, Figure 4a). Hdh-BMP7 mRNA was widely expressed at different developmental stages of H. discus hannai (Figure 4b). The expression of hdh-BMP7 mRNA was lower in the S-DD-group than in the L-DD-group (p < 0.01, Figure 4c).
Figure 4.
The expression pattern of hdh-BMP7 mRNA in (a) various tissues (n = 6), (b) development stages (n = 4), and (c) the L-DD-group (n = 12) and S-DD-group (n = 12). Different letters and ** indicate p < 0.05 and p < 0.01, respectively.
3.3. SNPs in Hdh-BMP7
Sequence comparisons detected eight single nucleotide polymorphisms (SNPs) in hdh-BMP7 (Table 2). The average Polymorphic information content (PIC) was 0.147, which is considered a low polymorphism level (PIC < 0.25). Four SNPs (744 A > G, 819 A > G, 834 T > A, and 852 C > G) were related to growth traits in H. discus hannai. The MW/TW of AA was higher than that of abalones with the genotype of GG at the 744 A > G locus (p < 0.05). The MW/TW of AA was higher than that of abalones with the GG and GA genotype at the 819 A > G locus (p < 0.05). The MW/TW of TT was lower than that of abalones with the AT genotype at the 834 T > A locus (p < 0.05). The MW/TW of CC was higher than that of abalones with the genotype of GG at the 852 C > G locus (p < 0.05).
Table 2.
Growth traits of H. discus hannai with different genotypes.
| Locus | Genotype | Sample Number | Shell Length (mm) | Shell Width (mm) | Total Weight (g) | Muscle Weight (g) | MW/TW |
|---|---|---|---|---|---|---|---|
| 570 C > T | CT | 111 | 73.76 ± 9.82 a | 49.64 ± 6.27 a | 42.07 ± 16.40 a | 17.48 ± 7.92 a | 0.4064 ± 0.0458 a |
| TT | 104 | 73.99 ± 9.61 a | 49.86 ± 6.21 a | 41.25 ± 15.29 a | 16.73 ± 7.15 a | 0.3981 ± 0.0407 a | |
| CC | 4 | 71.19 ± 7.99 a | 48.03 ± 4.85 a | 36.63 ± 15.55 a | 15.30 ± 8.35 a | 0.4071 ± 0.0501 a | |
| 606 C > T | CC | 111 | 74.47 ± 9.60 a | 50.11 ± 6.13 a | 42.10 ± 15.57 a | 17.06 ± 7.30 a | 0.3975 ± 0.0416 a |
| TC | 106 | 73.21 ± 9.48 a | 49.30 ± 6.14 a | 41.05 ± 15.75 a | 17.08 ± 7.68 a | 0.4066 ± 0.0451 a | |
| 744 A > G | AA | 139 | 73.67 ± 9.72 a | 49.66 ± 6.18 a | 42.55 ± 16.36 a | 17.62 ± 7.83 a | 0.4060 ± 0.0439 a |
| GA | 74 | 74.30 ± 9.20 a | 49.88 ± 6.08 a | 40.15 ± 14.59 a | 16.33 ± 6.95 a | 0.3985 ± 0.0422 ab | |
| GG | 4 | 69.57 ± 16.74 a | 47.47 ± 10.31 a | 32.95 ± 15.75 a | 12.13 ± 6.51 a | 0.3595 ± 0.0385 b | |
| 805 A > G | AG | 107 | 73.44 ± 9.29 a | 49.50 ± 6.08 a | 41.53 ± 16.17 a | 17.20 ± 7.77 a | 0.4056 ± 0.0448 a |
| GG | 106 | 74.07 ± 9.96 a | 49.88 ± 6.26 a | 41.47 ± 15.45 a | 16.91 ± 7.33 a | 0.3994 ± 0.0426 a | |
| AA | 5 | 75.21 ± 12.42 a | 49.81 ± 8.41 a | 41.84 ± 17.68 a | 16.80 ± 8.31 a | 0.3929 ± 0.0403 a | |
| 819 A > G | AA | 86 | 73.47 ± 9.41 a | 49.56 ± 5.95 a | 41.88 ± 16.24 a | 17.58 ± 7.83 a | 0.4115 ± 0.0433 a |
| GA | 97 | 74.32 ± 9.69 a | 50.09 ± 6.39 a | 42.40 ± 15.87 a | 17.27 ± 7.56 a | 0.3987 ± 0.0448 bc | |
| GG | 35 | 73.10 ± 10.30 a | 48.94 ± 6.30 a | 38.14 ± 14.31 a | 15.14 ± 6.59 a | 0.3895 ± 0.0370 c | |
| 834 T > A | TT | 174 | 73.14 ± 9.74 a | 49.20 ± 6.24 a | 40.48 ± 15.74 a | 16.49 ± 7.47 a | 0.3988 ± 0.0445 a |
| AT | 42 | 75.74 ± 8.59 a | 51.24 ± 5.50 a | 44.51 ± 14.95 a | 18.70 ± 7.15 a | 0.4144 ± 0.0367 b | |
| 852 C > G | CC | 133 | 72.93 ± 9.47 a | 49.33 ± 6.09 a | 41.33 ± 15.97 a | 17.12 ± 7.66 a | 0.4058 ± 0.0440 a |
| GC | 80 | 75.28 ± 9.43 a | 50.29 ± 6.11 a | 41.97 ± 15.47 a | 17.06 ± 7.34 a | 0.3982 ± 0.0421 ab | |
| GG | 4 | 69.57 ± 16.74 a | 47.47 ± 10.31 a | 32.95 ± 15.75 a | 12.13 ± 6.51 a | 0.3595 ± 0.0385 b | |
| 903 G > C | GG | 124 | 73.08 ± 9.88 a | 49.36 ± 6.30 a | 41.80 ± 16.26 a | 17.39 ± 7.84 a | 0.4070 ± 0.0445 a |
| CG | 86 | 74.83 ± 8.86 a | 50.17 ± 5.85 a | 40.82 ± 14.64 a | 16.53 ± 6.88 a | 0.3979 ± 0.0407 a | |
| CC | 7 | 72.16 ± 14.87 a | 48.48 ± 9.01 a | 42.29 ± 22.71 a | 16.69 ± 10.60 a | 0.3781 ± 0.0506 a |
Note: Different little letters within a column are p < 0.05. MW means muscle weight; TW means total weight.
3.4. Effects of RNA Interference
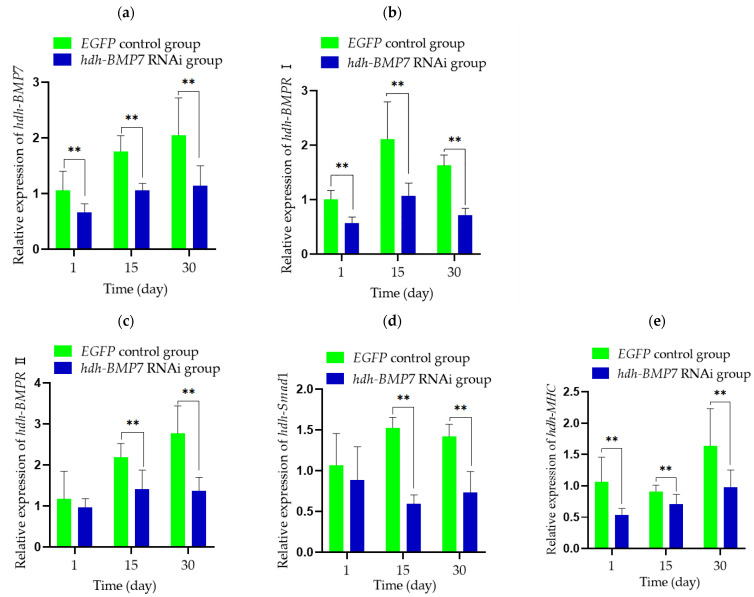
The expression levels of hdh-BMP7 in the adductor muscle decreased about 40–50% after RNAi experiment at 1 day, 15 days, and 30 days (p < 0.01, Figure 5a) compared to the EGFP control group. After RNAi experiment for 1 day, hdh-BMPR I and myosin heavy chain (hdh-MHC) mRNA levels were significantly lower in the hdh-BMP7 RNAi group than in the EGFP control group (Figure 5b,e). After RNAi experiment for 15 and 30 days, hdh-BMPR I, hdh-BMPR II, hdh-Smad1, and hdh-MHC mRNA levels were lower in the hdh-BMP7 RNAi group compared with the EGFP control group (p < 0.01, Figure 5).
Figure 5.
Expression analysis of RT-qPCR. Expression analysis of (a) hdh-BMP7, (b) hdh-BMPR I, (c) hdh-BMPR II, (d) hdh-Smad1, and (e) hdh-MHC after RNAi in adductor muscle (n = 4). ** indicate p < 0.05.
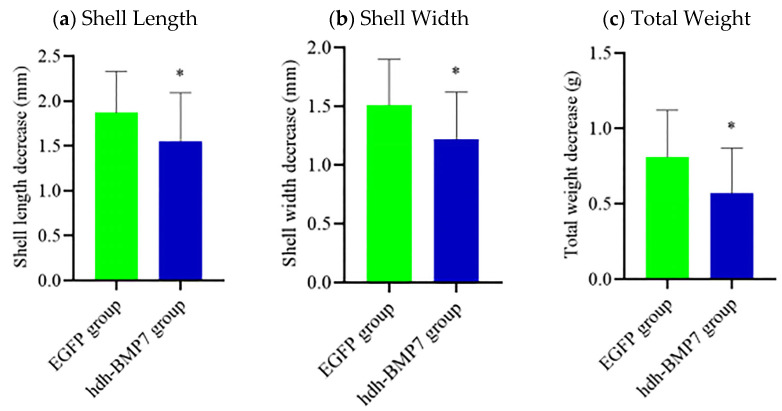
After RNAi experiment for 30 days, the decrease in shell length, shell width, and total weight in the hdh-BMP7 RNAi group was significantly higher than in the EGFP control group (p < 0.05, Figure 6a–c).
Figure 6.
The growth characteristics after hdh-BMP7 RNAi in H. discus hannai. (a) The decrease in shell length after hdh-BMP7 RNAi. (b) The decrease in shell width after hdh-BMP7 RNAi. (c) The decrease in total weight after hdh-BMP7 RNAi. * represents p < 0.05.
3.5. Verification of the Lvpan Abalone
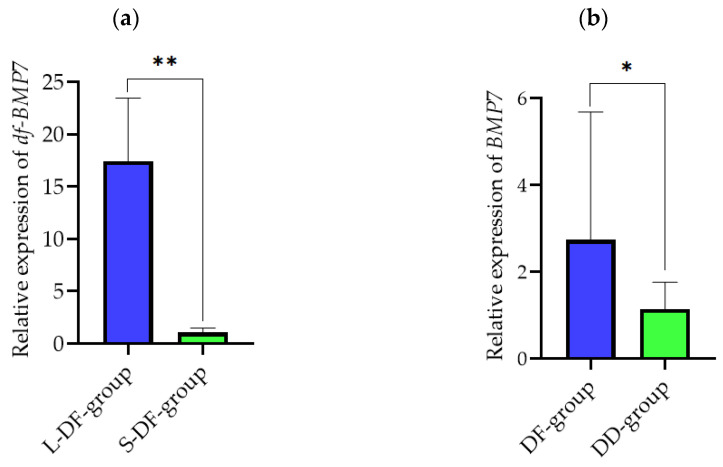
In order to further verify the function of BMP7 in abalone, we compared the expression level of BMP7 mRNA between H. discus hannai (DD) and Lvpan abalone (DF). The expression of df-BMP7 mRNA in Lvpan abalone was lower in the S-DF-group than in the L-DF-group (p < 0.05, Figure 7). In addition, the expression of BMP7 mRNA in the H. discus hannai (DD-group) was lower than in the Lvpan abalone (DF-group) (p < 0.05).
Figure 7.
Expression of BMP7 mRNA in Lvpan abalone. (a) Expression of hdh-BMP7 in adductor muscle between L-DF-group (n = 12) and S-DF-group (n = 12). (b) Expression of BMP7 in adductor muscle between DD-group (n = 24) and DF-group (n = 24). * and ** indicate p < 0.05 and p < 0.01, respectively.
4. Discussion
BMP7 is one of the members of the TGF-β superfamily. In this paper, we cloned the ORF frame of the BMP7 from H. discus hannai. We found that the hdh-BMP7 protein contained 416 amino acid residues encoded by 1251 nucleotides. Our results also demonstrate that hdh-BMP7 has a TGF-β propeptide. The hdh-BMP7 protein has the same TGF-β functional domain as other BMP members, which is required for the BMP ligand to be functional [49]. Hdh-BMP7 has seven conserved cysteine residues, which is consistent with the BMP7 in Meretrix meretrix [50]. However, not all mollusks have seven highly conservative cysteine residues, such as Sinonovacula constricta [7] and T. granosa [37]. Highly conserved cysteine residues are the structural characteristics that are typical of the TGF-β superfamily. Therefore, the data indicate that hdh-BMP7 is a member of the BMP family [51]. The result of the phylogenetic tree suggests that H. discus hannai is most closely related to these shellfish of T. granosa, M. coruscus, P. imbricata, and C. gigas.
BMPs are widely expressed during the development of mammals, with many biological activities [7,16]. BMP7 is a member of the BMPs. To explore the potential roles of BMP7 in H. discus hannai, we analyzed the expression of hdh-BMP7 mRNA in various tissues of individuals in different development stages. The qRT-PCR results revealed that hdh-BMP7 was widely expressed in the cerebral ganglion, gills, mantle, and other tissues, which is consistent with results previously found in zebrafish [52], M. meretrix [50] and T. granosa [37]. Previous studies have suggested that the BMP system plays a role in pituitary function in vertebrates [53], and BMP7 is mainly involved in shell growth and nacre formation [7,37,50]. Thus, our results of tissues distribution suggest that hdh-BMP7 may be related to shell growth and shell formation and play an important role in the nervous system. In addition, the hdh-BMP7 mRNA was widely expressed in different developmental stages of abalone, and its expression level was higher in the L-DD-group than in the S-DD-group. Thus, we speculate that hdh-BMP7 may be related to the growth and development of H. discus hannai.
SNP is an important molecular marker, widely divided in the genome of organisms. The SNPs in candidate genes were analyzed to find the loci related to the target trait, which helps to effectively screen individuals. For example, in Haliotis diversicolor supertexta, a total of nine SNP sites were identified in the MSTN gene. The association analysis of SNP sites and growth traits showed that the SNP site g909C > T in the coding region of the MSTN gene in H. diversicolor supertexta was significantly correlated with the shell length, shell height, and body weight [54]. In the myostatin gene of H. discus hannai, nine SNPs were significantly associated with growth traits [43]. In previous studies, BMP7 has been identified as a potential growth-related gene in SNP studies in bovines [33] and chickens [34] but not in abalone. In order to explore the SNPs and growth correlation of the BMP7 gene of H. discus hannai, hdh-BMP7 was experimentally measured by direct sequencing of PCR products, and the association between hdh-BMP7 SNPs and five growth-related traits (shell length, shell width, total weight, muscle weight, and muscle weight/wet weight ratio) was analyzed. These data identified eight SNPs in the CDS region of hdh-BMP7, four of which were significantly associated with growth traits, which suggests that hdh-BMP7 is closely correlated with growth traits of abalone. Previous research has shown that BMP signaling is the dominant pathway controlling muscle mass and the inhibition of BMP signaling causes muscle atrophy [35]. Overexpression of BMP7 promotes muscle growth and prevents muscle atrophy [36]. In our study, the higher ratio of MW/TW means that the adductor muscle contributes to a large part of the total weight, which suggests that hdh-BMP7 also may be related to the abalone muscle growth. There is a strong correlation between BMP7 and the growth of shellfish such as S. constricta [7], M. meretrix [50], and T. granosa [37]. Therefore, we speculate hdh-BMP7 may play a crucial role in the process of growth regulation of H. discus hannai. In this study, only four SNP sites related to growth traits were found in the CDS region. In order to provide more useful molecular markers for breeding, we plan to screen SNP in the intron and promoter regions next. Meanwhile, we will further expand the abalone samples to verify the accuracy of SNP.
Although there are some reports on functional genes related to abalone growth, most of these studies use high-throughput sequencing technology to mine relevant genes and pathways and lack further functional verification, or the functional verification only stays at the level of cloning and mRNA expression, and the in-depth study on the mechanism of abalone functional gene is still very lacking. The technology of RNAi refers to the phenomenon of the gene silencing by foreign dsRNAs at the mRNA level. At present, researchers often use RNAi to reveal the genetic function of aquatic animals, such as H. discus hannai [43], P. martensii [38], etc. To further investigate the interaction between the BMP system and growth in abalone, we successfully inhibited the expression of hdh-BMP7 by RNAi, which resulted in a significant decrease in shell length growth gain, shell width growth gain, and total weight gain compared with the EGFP control group. This further demonstrates that hdh-BMP7 can promote abalone growth. In vertebrates, BMPs can act as regulators of myoblasts and facilitate the development and regeneration of myoblasts after muscle injury [55,56]. Overexpression of BMP7 or ALK3 promotes muscle growth and prevents muscle atrophy [35,36]. A previous study [57] suggested that BMP signaling is a positive regulator of muscle mass in animals. In this study, the ratio of MW/TW was significantly different in several SNP loci. Muscle is the main edible part of abalone, accounting for about half of the body weight. The growth of muscle directly affects the overall growth. Some studies have investigated the mechanism of BMP7 in growth regulation on shellfish [7,37,50], although no reports have been made about abalone. In a future study, we will conduct more long-term RNAi experiments and detect the morphological change of abalone muscle fibers, so as to further reveal the regulatory function of BMP7 on regulating abalone muscle growth.
As we all know, BMPs initially bind to two BMP receptors, BMPR I and BMPR II, which activate Smads and target genes [58]. The interaction between Smads and other proteins mediates signals, controlling the expression of target genes, the translation of mRNA translation, and the regulation of cytoplasmic protein [8,9,10,11]. Previous studies have reported that TGF-β signaling was transduced from the extra-cellular space to the cell nucleus via Smad proteins to activate downstream pathways and regulate general life processes [59,60]. In Scylla paramamosain, BMP7 also obeys this signaling mechanism [15]. In this study, when hdh-BMP7 expression was inhibited, the expression levels of hdh-BMPR I and hdh-BMPR II changed in response, which indicates that hdh-BMPR I and hdh-BMPR II are the receptors of hdh-BMP7 in abalone. In addition, the expression levels of hdh-smad1 and hdh-MHC were significantly decreased (p < 0.01) compared to the EGFP control group at 15 days and 30 days, suggesting that hdh-BMP7 may promote muscle growth by hdh-Smad1 signaling in H. discus hannai.
In the end, we verified the role of BMP7 in Lvpan abalone. Lvpan abalone, a hybrid with fast growth, has become a good material for proving the mechanism related to abalone growth. In this research, we compared the expression level of BMP7 mRNA between H. discus hannai (DD) and Lvpan abalone (DF). We found that the expression in L-group DD and DF at the same age was higher than that in S-group equivalents. The BMP7 expression level in the DF was higher than that in DD at the same age, which further indicates that BMP7 plays a positive role in regulating abalone growth. Ultimately, our data indicate that hdh-BMP7 potentially impacts the growth of H. discus hannai.
5. Conclusions
In the study, we cloned the ORF sequence and described the structure of hdh-BMP7 from H. discus hannai. The expression level of hdh-BMP7 mRNA was higher in the fast-growing group (p < 0.05) than in the slow-growing group, suggesting that it may be correlated with the growth of abalone. Further results from RNAi indicated that the hdh-BMP7 gene plays a vital role in promoting growth of H. discus hannai. Association analysis identified four SNPs that were significantly associated with growth traits, suggesting that hdh-BMP7 is closely involved in abalone growth. The results of this paper would lay a foundation for further study on the function of hdh-BMP7 in abalone growth and contribute to molecular marker assisted breeding in H. discus hannai.
Author Contributions
Designing research, J.H., W.Y. and C.K.; Performing experiments, J.H. and M.Z.; Writing original draft, J.H.; Revising manuscript, X.L. and C.K. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The research was approved by the Laboratory Animal Ethics Committee of Xiamen University (1 January 2019).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The study was funded by the Seed Industry Innovation and Industrialization in Fujian Province (No.2021FJSCZY02), Special Fund for Ocean and Fisheries of Xiamen (No. 21CZY018HJ01), Key S & T Program of Fujian Province (No. 2020NZ08003), Earmarked Fund for CARS (No. CARS-49). Fujian Provincial S & T Project (2020N5001).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Feng X., Derynck R. Specificity and versatility in TGF-β signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- 2.Massague J. TGF-β in cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramesh S., Wildey G., Howe P. Transforming growth factor β (TGFβ)-induced apoptosis: The rise and fall of Bim. Cell Cycle. 2009;8:11–17. doi: 10.4161/cc.8.1.7291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y., Ma B.W., Wang X.C., Zha X.J., Sheng C.J., Yang P., Qu S. Potential Functions of the BMP Family in Bone, Obesity, and Glucose Metabolism. J. Diabetes Res. 2021;2021:6707464. doi: 10.1155/2021/6707464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan C.Y., Wong J.C., Chan P.S., Tan H., Liao D., Chen W.M., Tan L.W., Ackers-Johnson M., Wakimoto H., Seidman J.G., et al. Yin Yang 1 Suppresses Dilated Cardiomyopathy and Cardiac Fibrosis through Regulation of Bmp7 and Ctgf. Circ. Res. 2019;125:834–846. doi: 10.1161/CIRCRESAHA.119.314794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kingsley D. The TGF-β superfamily: New members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994;8:133–146. doi: 10.1101/gad.8.2.133. [DOI] [PubMed] [Google Scholar]
- 7.Zhao J., Cui B., Yao H., Lin Z., Dong Y. A Potential Role of Bone Morphogenetic Protein 7 in Shell Formation and Growth in the Razor Clam Sinonovacula constricta. Front. Physiol. 2020;11:1059. doi: 10.3389/fphys.2020.01059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groppe J., Hinck C.S., Samavarchi-Tehrani P., Zubieta C., Schuermann J.P., Taylor A.B., Schwarz P.M., Wrana J.L., Hinck A.P. Cooperative assembly of TGF-β superfamily signaling complexes is mediated by two disparate mechanisms and distinct modes of receptor binding. Mol. Cell. 2008;29:157–168. doi: 10.1016/j.molcel.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 9.Tzavlaki K., Moustakas A. TGF-β Signaling. Biomolecules. 2020;10:487. doi: 10.3390/biom10030487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derynck R., Budi E.H. Specificity, versatility, and control of TGF-β family signaling. Sci. Signal. 2019;12:eaav5183. doi: 10.1126/scisignal.aav5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S.N., Wu J.F. TGF-β/SMAD signaling regulation of mesenchymal stem cells in adipocyte commitment. Stem Cell Res. Ther. 2020;11:41. doi: 10.1186/s13287-020-1552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X., Meng X., Song B., Qiu X., Liu H. SNPs in the myostatin gene of the mollusk Chlamys farreri: Association with growth traits. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2010;155:327–330. doi: 10.1016/j.cbpb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Li B., Zhou Y.L., Gu W.B., Wang L.Z., Xu Y.P., Cheng Y.X., Chen D.Y., Li B.W., Xiao Y., Dong W.R., et al. Identification and functional analysis of transforming growth factor-β type III receptor (TβR3) from Scylla paramamosain: The first evidence of TβR3 involved in development and innate immunity in invertebrates. Fish Shellfish Immunol. 2020;105:41–52. doi: 10.1016/j.fsi.2020.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Niu D.H., Wang L., Bai Z.Y., Xie S.M., Zhao H.G., Li J.L. Identification and expression characterization of the myostatin (MSTN) gene and association analysis with growth traits in the razor clam Sinonovacula constricta. Gene. 2015;555:297–304. doi: 10.1016/j.gene.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 15.Shu L., Yang Y., Huang H., Ye H. A bone morphogenetic protein ligand and receptors in mud crab: A potential role in the ovarian development. Mol. Cell. Endocrinol. 2016;434:99–107. doi: 10.1016/j.mce.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 16.Salazar V., Gamer L., Rosen V. BMP signaling in skeletal development, disease and repair. Nat. Rev. Endocrinol. 2016;12:203. doi: 10.1038/nrendo.2016.12. [DOI] [PubMed] [Google Scholar]
- 17.Sánchez-Duffhues G., Hiepen C., Knaus P., ten Dijke P. Bone morphogenetic protein signaling in bone homeostasis. Bone. 2015;80:43–59. doi: 10.1016/j.bone.2015.05.025. [DOI] [PubMed] [Google Scholar]
- 18.Wordinger R., Clark A. Bone morphogenetic proteins and their receptors in the eye. Exp. Biol. Med. 2007;232:979–992. doi: 10.3181/0510-MR-345. [DOI] [PubMed] [Google Scholar]
- 19.Cook S.D., Rueger D.C. Osteogenic protein-1: Biology and applications. Clin. Orthop. Relat. Res. 1996;324:29. doi: 10.1097/00003086-199603000-00005. [DOI] [PubMed] [Google Scholar]
- 20.Panagiotou O.A., Evangelou E., Ioannidis J.P. Genome-wide significant associations for variants with minor allele frequency of 5% or less—An overview: A HuGE review. Am. J. Epidemiol. 2010;172:869–889. doi: 10.1093/aje/kwq234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyons K.M., Hogan B.L., Robertson E.J. Colocalization of BMP7 and BMP2 RNAs suggests that these factors cooperatively mediate tissue interactions during murine development. Mech. Dev. 1995;50:71–83. doi: 10.1016/0925-4773(94)00326-I. [DOI] [PubMed] [Google Scholar]
- 22.Dudley A.T., Lyons K.M., Robertson E.J. A requirement for bone morphogenetic protein-7 during development of the mammalian kidney and eye. Genes Dev. 1995;15:2795–2807. doi: 10.1101/gad.9.22.2795. [DOI] [PubMed] [Google Scholar]
- 23.Dudley A.T., Robertson E.J. Overlapping expression domains of bone morphogenetic protein family members potentially account for limited tissue defects in BMP7 deficient embryos. Dev. Dyn. 1997;208:349–362. doi: 10.1002/(SICI)1097-0177(199703)208:3<349::AID-AJA6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 24.Luo G., Hofmann C., Bronckers A.L., Sohocki M., Bradley A., Karsenty G. BMP-7 is an inducer of nephrogenesis, and is also required for eye development and skeletal patterning. Genes Dev. 1995;9:2808–2820. doi: 10.1101/gad.9.22.2808. [DOI] [PubMed] [Google Scholar]
- 25.Xu Y., Wang G., Zhou Y., Yang W. The characterization and potential roles of bone morphogenetic protein 7 during spermatogenesis in Chinese mitten crab Eriocheir sinensis. Gene. 2018;673:119–129. doi: 10.1016/j.gene.2018.06.020. [DOI] [PubMed] [Google Scholar]
- 26.Schmal H., Mehlhorn A.T., Pilz I.H., Dovi-Akue D., Kirchhoff C., Südkamp N.P., Gerlach U., Lohrmann C., Niemeyer P. Immunohistological localization of BMP-2, BMP-7, and their receptors in knee joints with focal cartilage lesions. Sci. World J. 2012;2012:467892. doi: 10.1100/2012/467892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asahina I., Sampath T., Hauschka P. Human osteogenic protein-1 induces chondroblastic, osteoblastic, and/or adipocytic differentiation of clonal murine target cells. Exp. Cell Res. 1996;222:38–47. doi: 10.1006/excr.1996.0005. [DOI] [PubMed] [Google Scholar]
- 28.Han Q., Gou S., Wang L. In vivo bone morphogenetic protein 7 gene transfection mediated by polyethyleneimine for femoral fracture healing in old rats. J. Clin. Rehabil. Tissue Eng. Res. 2008;12:1205–1208. [Google Scholar]
- 29.Hurtig M., Chubinskaya S., Dickey J., Rueger D. BMP7 protects against progression of cartilage degeneration after impact injury. J. Orthop. Res. 2009;27:602–611. doi: 10.1002/jor.20787. [DOI] [PubMed] [Google Scholar]
- 30.Puglisi R., Montanari M., Chiarella P., Stefanini M., Boitani C. Regulatory role of BMP2 and BMP7 in spermatogonia and Sertoli cell proliferation in the immature mouse. Eur. J. Endocrinol. 2004;151:511–520. doi: 10.1530/eje.0.1510511. [DOI] [PubMed] [Google Scholar]
- 31.Monsivais D., Clementi C., Peng J., Fullerton P., Jr., Prunskaite-Hyyryläinen R. BMP7 induces uterine receptivity and blastocyst attachment. Endocrinology. 2017;158:979–992. doi: 10.1210/en.2016-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen B., Li L., Shouzhi W., Xi C., Hui L. Association between polymorphism of BMP7 gene and growth and body composition traits in broiler chickens. China Poult. 2013;35:6–10. [Google Scholar]
- 33.Huang Y.Z., Wang X.L., He H., Lan X.Y., Lei C.Z., Zhang C.L., Chen H. Identification and genetic effect of haplotype in the bovine BMP7 gene. Gene. 2013;532:281–287. doi: 10.1016/j.gene.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y., Guo F., Qu H., Luo C., Wang J., Shu D. Associations between variants of bone morphogenetic protein 7 gene and growth traits in chickens. Br. Poult. Sci. 2018;59:264–269. doi: 10.1080/00071668.2018.1454586. [DOI] [PubMed] [Google Scholar]
- 35.Sartori R., Schirwis E., Blaauw B., Bortolanza S., Zhao J.H., Enzo E., Stantzou A., Mouisel E., Toniolo L., Ferry A., et al. BMP signaling controls muscle mass. Nat. Genet. 2013;45:1309–1318. doi: 10.1038/ng.2772. [DOI] [PubMed] [Google Scholar]
- 36.Winbanks C.E., Chen J.L., Qian H.W., Liu Y.Y., Bernardo B.C., Beyer C., Watt K.I., Thomson R.E., Connor T., Turner B.J., et al. The bone morphogenetic protein axis is a positive regulator of skeletal muscle mass. J. Cell Biol. 2013;203:345–357. doi: 10.1083/jcb.201211134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong Y. Ph.D. Thesis. Ocean University of China; Qingdao, China: 2012. Transcriptome Analysis Using 454 Pyrosequencing and Cloning and Expression of Growth-Related Genes for the Blood Clam Tegillarca granosa (Linnaeus, 1758) [Google Scholar]
- 38.Yan F., Luo S., Jiao Y., Deng Y.W., Du X.D., Huang R.L., Wang Q.H., Chen W.Y. Molecular characterization of the BMP7 gene and its potential role in shell formation in Pinctada martensii. Int. J. Mol. Sci. 2014;15:21215. doi: 10.3390/ijms151121215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan S., Zhou D., Liu B., Deng Z., Guo Y., Yu D. Molecular cloning and expression analysis of BMP 7b from Pinctada fucata. South China Fish. Sci. 2018;14:121–126. [Google Scholar]
- 40.Lin J. Master’s Thesis. Shanghai Ocean University; Shanghai, China: 2014. Cloning and Expression Analysis of Genes Involved in the Pearl Formation of Hyriopsis cumingii. [Google Scholar]
- 41.Guo X.M., Ford S.E., Zhang F.S. Molluscan aquaculture in China. J. Shellfish Res. 1999;18:19–31. [Google Scholar]
- 42.Huang J.F., You W.W., Luo X., Ke C.H. iTRAQ-Based Identification of Proteins Related to Muscle Growth in the Pacific Abalone, Haliotis discus hannai. Int. J. Mol. Sci. 2017;18:2237. doi: 10.3390/ijms18112237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang J., Zhou M., Chen J., Ke C. A Potential Negative Regulatory Function of Myostatin in the Growth of the Pacific Abalone, Haliotis discus hannai. Biology. 2023;12:14. doi: 10.3390/biology12010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Naipil C.C., Muñoz V.V., Valdés J.A., Molina A., Escárate C.G. RNA interference in Haliotis rufescens myostatin evidences upregulation of insulin signaling pathway. Agric. Gene. 2016;1:93–99. doi: 10.1016/j.aggene.2016.07.004. [DOI] [Google Scholar]
- 45.Elliott N.G. Genetic improvement programmes in abalone: What is the future? Aquac. Res. 2000;31:51–59. doi: 10.1046/j.1365-2109.2000.00386.x. [DOI] [Google Scholar]
- 46.You W.W., Guo Q., Fan F., Ren P., Luo X., Ke C.H. Experimental hybridization and genetic identifification of Pacifific abalone Haliotis discus hannai and green abalone. Aquaculture. 2015;448:243–249. doi: 10.1016/j.aquaculture.2015.05.043. [DOI] [Google Scholar]
- 47.Wu Y., He J., Yao G., Liang H., Huang X. Molecular cloning, characterization, and expression of two TNFRs from the pearl oyster Pinctada fucata martensii. Fish Shellfish Immunol. 2020;98:147–159. doi: 10.1016/j.fsi.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 48.Huang J.F., Luo X., Huang M.Q., Liu G.M., You W.W., Ke C.H. Identification and characteristics of muscle growth-related microRNA in the Pacific abalone, Haliotis discus hannai. BMC Genom. 2018;19:915. doi: 10.1186/s12864-018-5347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marques C.L., Fernandez I., Viegas M.N., Cox C.J., Martel P., Rosa J., Cancela M.L., Laize V. Comparative analysis of zebrafish bone morphogenetic proteins 2, 4 and 16: Molecular and evolutionary perspectives. Cell. Mol. Life. Sci. 2016;73:841–857. doi: 10.1007/s00018-015-2024-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan S.Y. Master’s Thesis. Ocean University of Shanghai; Shanghai, China: 2019. Association between the Polymorphisms of BMPs, TGF-β RΙ and Growth Traits in Meretrix meretrix. [Google Scholar]
- 51.Xiao Y.T., Xiang L., Shao J.Z. Bone morphogenetic protein. Biochem. Biophys. Res. Commun. 2007;362:550–553. doi: 10.1016/j.bbrc.2007.08.045. [DOI] [PubMed] [Google Scholar]
- 52.Shawi M., Serluca F.C. Identification of a BMP7 homolog in zebrafish expressed in developing organ systems. Gene Expr. Patterns. 2008;8:369–375. doi: 10.1016/j.gep.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 53.Otsuka F. Multifunctional bone morphogenetic protein system in endocrinology. Acta Med. Okayama. 2013;67:75–86. doi: 10.18926/AMO/49665. [DOI] [PubMed] [Google Scholar]
- 54.Ai J.L., Li Z.M., Liu J.Y., Shen Y.C. Single nucleotide polymorphisms of myostatin gene and its association with growth traits in Haliotis diversicolor supertexta. J. Trop. Oceanogr. 2019;38:78–85. [Google Scholar]
- 55.Ono Y., Calhabeu F., Morgan J.E., Katagiri T., Amthor H., Zammit P.S. BMP signalling permits population expansion by preventing premature myogenic differentiation in muscle satellite cells. Cell Death Differ. 2011;18:222–234. doi: 10.1038/cdd.2010.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aoyama K., Yamane A., Suga T., Suzuki E., Fukui T., Nakamura Y. Bone morphogenetic protein-2 functions as a negative regulator in the differentiation of myoblasts, but not as an inducer for the formations of cartilage and bone in mouse embryonic tongue. BMC Dev. Biol. 2011;11:44. doi: 10.1186/1471-213X-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sartori R., Gregorevic P., Sandri M. TGFβ and BMP signaling in skeletal muscle: Potential significance for muscle-related disease. Trends. Endocrinol. Metab. 2014;25:464–471. doi: 10.1016/j.tem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 58.Zhao Y.Y., Li X.X., Wang W., Chen X., Yu P., Wang J.J., Xu Y.X. Effect of BMPR IB gene silencing by siRNA on apoptosis and steroidogenesis of porcine granulosa cells. Genet. Mol. Res. 2014;13:9964. doi: 10.4238/2014.November.28.1. [DOI] [PubMed] [Google Scholar]
- 59.Heldin C.H., Miyazono K., Dijke P.T. TGF-β signaling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 60.Miyazono K. TGF-β signaling by Smad proteins. Cytokine Growth Factor Rev. 2000;11:15–22. doi: 10.1016/S1359-6101(99)00025-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.