Abstract
Corethrodendron fruticosum is an endemic forage grasses in China with high ecological value. In this study, the complete chloroplast genome of C. fruticosum was sequenced using Illumina paired-end sequencing. The C. fruticosum chloroplast genome was 123,100 bp and comprised 105 genes, including 74 protein-coding genes, 4 rRNA-coding genes, and 27 tRNA-coding genes. The genome had a GC content of 34.53%, with 50 repetitive sequences and 63 simple repeat repetitive sequences that did not contain reverse repeats. The simple repeats included 45 single-nucleotide repeats, which accounted for the highest proportion and primarily comprised A/T repeats. A comparative analysis of C. fruticosum, C. multijugum, and four Hedysarum species revealed that the six genomes were highly conserved, with differentials primarily located in the conserved non-coding regions. Moreover, the accD and clpP genes in the coding regions exhibited high nucleotide variability. Accordingly, these genes may serve as molecular markers for the classification and phylogenetic analysis of Corethrodendron species. Phylogenetic analysis further revealed that C. fruticosum and C. multijugum appeared in different clades than the four Hedysarum species. The newly sequenced chloroplast genome provides further insights into the phylogenetic position of C. fruticosum, which is useful for the classification and identification of Corethrodendron.
Keywords: Corethrodendron fruticosum, chloroplast genome, codon usage, repeat analysis, phylogenetic relationship
1. Introduction
Corethrodendron fruticosum (Leguminosae) is a subshrub distributed primarily in the grassland areas of eastern Inner Mongolia and western northeast China [1]. It is suitable for planting on semi-fixed or flowing sand with good aeration and water [2]. C. fruticosum is a valuable forage grass that is unique to China that can also be employed for windbreak and sand fixation [3]. Moreover, it is resistant to drought, high temperatures, and wind erosion, with a low seed germination rate, strong asexual reproduction, and vigorous growth in the third year of planting [2]. Currently, C. fruticosum is broadly distributed in arid and semi-arid areas in northern China as an excellent forage grass and for wind and sand control [4]. In particular, mild saline stress stimulates C. fruticosum root growth at the end of the growth period [5]. Meanwhile, light sand burial accelerates the growth of C. fruticosum meristems, whereas high sand burial (at depths of 80–100% of the C. fruticosum plant height) weakens the survival and growth of C. fruticosum meristems [6]. C. fruticosum was originally classified as Hedysarum [7]; however, more recent morphological and molecular phylogenetic evidence supports that Corethrodendron is a species that is independent of Hedysarum [8,9]. The primary Corethrodendron species include C. scoparium, C. multijugum, and C. fruticosum.
Chloroplasts are plastids that are common in land plants, algae, and protists that function as semi-autonomous organelles possessing the chloroplast genome or plastome as their genetic material [10]. With the development of next-generation sequencing (NGS) technologies, chloroplast genome sequencing has becoming a research hot spot. Chloroplast genomes have crucial roles in phylogeny, species identification, and crop breeding [11]. The chloroplast genomes of most plants comprise four regions: a large single-copy region, a small single-copy region, and two inverted repeat regions (IR) that act as chloroplast spacers between the large and small single-copy regions. There are four regions: a large single-copy region, a small single-copy region, and two inverted repeat regions (IR) that act as spacers between the large and small single-copy regions in the chloroplast genomes of most species [12,13,14]. Chloroplast genomes are typically 115–160 kb in length and generally encode 110–130 genes [15]. The variation in genome size is primarily influenced by variation in the IR region length [16,17,18]. However, certain legume species, including Medicago truncatula [19], Pisum sativum [20], and Caragana microphylla [21], exhibit a loss of the IR region and are collectively designated as the IR-lacking clade (IRLC) [22,23]. In other species, such as Pinus thunbergia [24], the IR region undergoes contraction, while in others, such as Pelargonium hortorum [25], it is expanded.
The chloroplasts of Nicotiana tabacum [26] and Marchantia polymorpha [27] have been successively sequenced and annotated, and the number of sequences has rapidly increased. In fact, 11,946 chloroplast genomes from 19,388 species, including 604 Leguminosae, have been integrated and curated in the chloroplast genome information resource (CGIR, https://ngdc.cncb.ac.cn/cgir accessed on 25 May 2023) [28,29]. In particular, the chloroplast genomes of C. multijugum and four Hedysarum species (Hedysarum semenovii, Hedysarum polybotrys, Hedysarum petrovii, and Hedysarum taipeicum) have been sequenced and annotated, thereby revealing that all five species have lost the IR regions [30,31,32].
In this study, the complete chloroplast genome of C. fruticosum was sequenced and annotated; it was then compared to those of C. multijugum and the four Hedysarum species chloroplast genome sequences mentioned above. Repeat sequences, simple sequence repeats (SSRs), nucleotide diversity (Pi), and the evolution of the six species were comparatively studied to gain further insight into the chloroplast genome of C. fruticosum. Additionally, we created a phylogenetic tree based on the 30 species chloroplast genome sequences in order to study their evolutionary relationships.
2. Materials and Methods
2.1. DNA Extraction and Sequencing, Genome Assembly, and Annotation
One C. fruticosum was collected from Ordos, Inner Mongolia, China (40.42° N, 110.04° E) and stored at the National Medium Term Genebank Forage Germplasm (Hohhot, China). Total genomic DNA was extracted from fresh leaves using a TIANamp Genomic DNA Kit (Tiangen Biotech Co., Ltd., Beijing, China). NGS was performed using a MiSeq PE150 platform to generate 150 bp-paired reads. The chloroplast genome was de novo assembled using GetOrganelle [33] and annotated using the Plastid Genome Annotator tool [34]. Geneious V9.0.2 was used to manually fix incorrect annotations of initiation codons and to stop codons made by Plastid Genome Annotator [35,36]. The chloroplast genome sequences of C. multijugum (NC_069301.1), H. taipeicum (NC_046493.1), H. polybotrys (MZ322397.1), H. semenovii (NC_047344.1), and H. petrovii (MT120797.1) were obtained from GenBank.
2.2. Identification of Repetitive Sequences and SSRs
Repetitive sequences, including forward repeats, reverse repeats, complementary repeats, and palindromic repeats, were identified using REPuter [37] under the following minimal repeat lengths: 30, with a hamming distance of 3. SSRs were identified using MISA [38], with the following parameter settings: unit size (nucleotide) minimum repeat: 1–10, 2–6, 3–4, 4–-3, 5–3, and 6–3. The minimum distance between two SSRs of 100 bp.
2.3. Analysis and Comparison of Genome Structures
Relative synonymous codon usage (RSCU) was calculated for all codons using CodonW v.1.4.2 (https://codonw.sourceforge.net accessed on 16 April 2023). The number of codons of protein-coding genes in the C. fruticosum chloroplast genome was determined. Pi values and sequence polymorphisms of C. fruticosum, C. multijugum, and the four Hedysarum species were analyzed using DNAsp v.6.10 [39]. The complete chloroplast genome sequences of C. fruticosum, C. multijugum, H. taipeicum, H. polybotrys, and H. semenovii were compared using mVISTA, with H. petrovii as the reference sequence [40] using default parameters.
2.4. Phylogenetic Analysis
The chloroplast genome sequences of C. fruticosum and 30 other species retrieved from NCBI were used to construct a phylogenetic tree using Arabidopsis thaliana and Oryza sativa as outgroups (Table S1). The concatenated protein-coding genes were used for phylogenetic analysis. All sequences were aligned using MAFFT (parameter default) [41]. Trees were constructed using the maximum likelihood and Bayesian methods. The best-fitting substitution model was selected using Modeltest 3.7 [42]. The maximum likelihood tree was constructed using IQ-TREE 1.6.12 [43] with the GTR + F + R4 model, and branch support was analyzed using bootstrap analysis with 1000 replicates. The Bayesian tree was constructed using MrBayes v.3.2.6 [44] with the GTR +F + I + G4 model.
3. Results
3.1. Genomic Characteristics of the C. fruticosum Chloroplast Genome
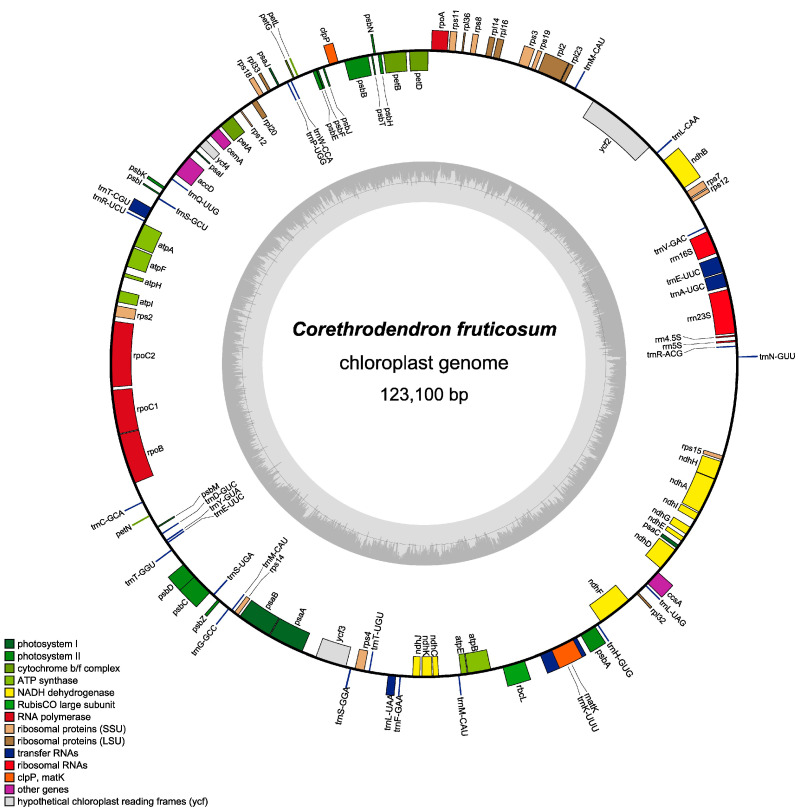
We sequenced and annotated the C. fruticosum chloroplast genome (Figure 1), which was missing a copy of the IR region; this supports the placement of C. fruticosum in the IRLC in Papilionoideae. The chloroplast genome of C. fruticosum was calculated as 123,100 bp that comprised 105 genes (Table 1), including 74 protein-coding genes, 4 rRNA-coding genes, and 27 tRNA-coding genes. Forty-three genes were associated with photosynthesis. The genes associated with transcription included 8 encoding ribosomal large subunits, 11 encoding ribosomal small subunits, and 4 encoding DNA-dependent RNA polymerases.
Figure 1.
Gene map of the C. fruticosum chloroplast genome. Genes distributed outside the outer circle are transcribed in the clockwise direction; those distributed inside the outer circle are transcribed in the counterclockwise direction. The dashed darker gray area of the inner circle indicates GC base content; the lighter gray indicates AT base content. Different functional groups genes are coded with different colors.
Table 1.
Genes encoded by the chloroplast genome of C. fruticosum.
| Gene Category | Gene Group | Gene Names |
|---|---|---|
| Photosynthesis | Subunits of Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Subunits of Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbM, psbN, psbT, psbZ | |
| NDH complex | ndhA *, ndhB *, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Subunits of cytochrome b/f complex | petA, petB *, petD *, petG, petL, petN | |
| Subunits of ATP synthase | atpA, atpB, atpE, atpF*, atpH, atpI | |
| Subunits of Rubisco | rbcL | |
| Transcription | Large subunits of ribosomes | rpl14, rpl16, rpl2 *, rpl20, rpl23, rpl32, rpl33, rpl36 |
| Small subunits of ribosomes | rps11, rps12 *, rps14, rps15, rps18, rps19, rps2, rps3, rps4, rps7, rps8 | |
| DNA-dependent RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | |
| rRNA genes | rrn16S, rrn23S, rrn4.5S, rrn5S | |
| tRNA genes | trnA-UGC *, trnC-GCA, trnD-GUC, trnE-UUC, trnE-UUC *, trnF-GAA, trnG-GCC, trnH-GUG, trnK-UUU *, trnL-CAA, trnL-UAA *, trnL-UAG, trnM-CAU(3), trnN-GUU, trnP-UGG, trnQ-UUG, trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-CGU *, trnT-GGU, trnT-UGU, trnV-GAC, trnW-CCA, trnY-GUA | |
| Other genes | C-type cytochrome synthesis genes | ccsA |
| Envelope membrane proteins | cemA | |
| Proteases | clpP | |
| Subunits of acetyl-CoA carboxylase | accD | |
| Maturases | matK | |
| Components of the translocon | ycf2 * | |
| Unknown | Conserved open reading frames | ycf3 **, ycf4 |
* Intron number; Gene (3): number of copies of multiple gene copies; ** meant ycf3 owned two introns.
Genes associated with transcription were also identified, and they comprised four genes encoding ribosomal RNAs: rrn16S, rrn23S, rrn4.5S, and rrn5S. What is more, six other genes and two unknown genes, ycf3, and ycf4, were identified. The gene ycf2 was also catalogued to an unknown gene before, but it was identified as a component of the translocon recently [45]. Of the 15 genes in the introns, all except ycf3 contained one intron, while ycf3 contained two. Additionally, trnK-UUU contained the longest amount of introns (2451 bp) and was discovered to be the longest intron (Table 2).
Table 2.
Size of introns and exons in split genes in the C. fruticosum complete chloroplast genome.
| Gene | Strand | Start | End | Exon I (bp) | Intron I (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) |
|---|---|---|---|---|---|---|---|---|
| atpF | − | 54,104 | 55,196 | 172 | 685 | 407 | ||
| ndhA | − | 113,391 | 115,680 | 553 | 1198 | 539 | ||
| ndhB | + | 11,627 | 13,789 | 721 | 678 | 764 | ||
| petD | − | 30,512 | 31,706 | 8 | 712 | 475 | ||
| petB | − | 31,901 | 33,335 | 6 | 787 | 642 | ||
| rpl2 | + | 21,974 | 23,497 | 397 | 696 | 431 | ||
| rpoC1 | − | 63,215 | 66,006 | 430 | 737 | 1625 | ||
| rps12 | − | 10,591 | 43,117 | 114 | 258 | |||
| ycf2 | − | 14,983 | 19,560 | 2711 | 30 | 1837 | ||
| ycf3 | − | 85,395 | 87,263 | 124 | 714 | 236 | 787 | 132 |
| trnT-CGU | + | 51,384 | 52,138 | 35 | 677 | 43 | ||
| trnL-UAA | + | 90,065 | 90,571 | 35 | 422 | 50 | ||
| trnK-UUU | + | 99,750 | 102,272 | 37 | 2451 | 35 | ||
| trnE-UUC | − | 5824 | 6855 | 32 | 960 | 40 | ||
| trnA-UGC | − | 4918 | 5759 | 37 | 805 | 36 |
3.2. Codon Usage Analysis of Protein-Coding Genes
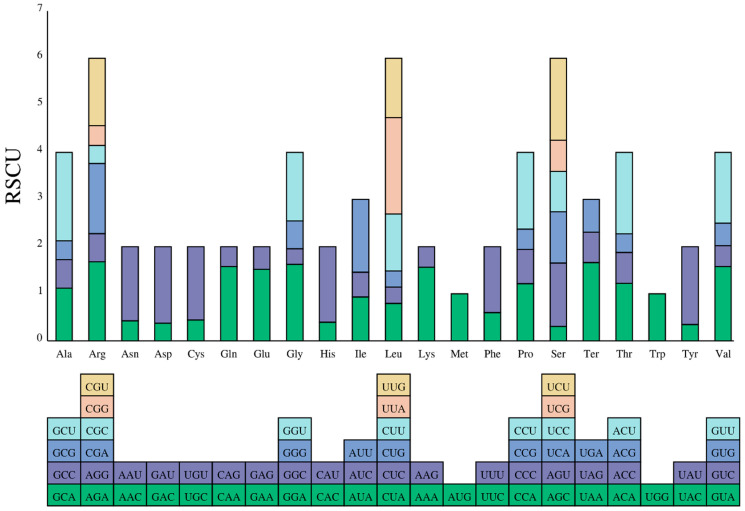
The chloroplast genome of C. fruticosum was found to contain 19,798 codons. Arg was the most common amino acid, whereas Trpwas the least common (Table 3). Even if the termination codon is counted in, the most common codon was ATT, which appeared 911 times and encoded Ile, whereas the least common codon was TGA encoding Ter, which appeared only 17 times. The RSCU reflects the ratio of the actual codon usage frequency compared to the expected frequency (Figure 2) [46]. In the C. fruticosum chloroplast genome, most codons with a high RSCU end in A/T bases, while the codon TTG occurred in Leu (RSCU > 1). Met and Trp were encoded by only one codon and had no codon preference.
Table 3.
Codon counts in the C. fruticosum chloroplast genome.
| Codon | Count | Codon | Count | Codon | Count | Codon | Count |
|---|---|---|---|---|---|---|---|
| TAA | 41 | GGC | 119 | ATG | 470 | AGT | 325 |
| TAG | 16 | GGG | 212 | AAC | 192 | TCA | 263 |
| TGA | 17 | GGT | 525 | AAT | 702 | TCC | 207 |
| GCA | 332 | CAC | 94 | CCA | 252 | TCG | 160 |
| GCC | 179 | CAT | 378 | CCC | 151 | TCT | 420 |
| GCG | 117 | ATA | 549 | CCG | 90 | ACA | 313 |
| GCT | 555 | ATC | 312 | CCT | 338 | ACC | 168 |
| TGC | 52 | ATT | 911 | CAA | 558 | ACG | 100 |
| TGT | 182 | AAA | 762 | CAG | 150 | ACT | 443 |
| GAC | 140 | AAG | 212 | AGA | 308 | GTA | 443 |
| GAT | 607 | CTA | 281 | AGG | 111 | GTC | 124 |
| GAA | 755 | CTC | 122 | CGA | 273 | GTG | 134 |
| GAG | 237 | CTG | 121 | CGC | 71 | GTT | 422 |
| TTC | 339 | CTT | 426 | CGG | 77 | TGG | 342 |
| TTT | 794 | TTA | 722 | CGT | 263 | TAC | 124 |
| GGA | 587 | TTG | 446 | AGC | 74 | TAT | 588 |
Figure 2.
Codon usage (RSCU) in protein-coding genes in the C. fruticosum chloroplast genome.
3.3. Repeat Analysis
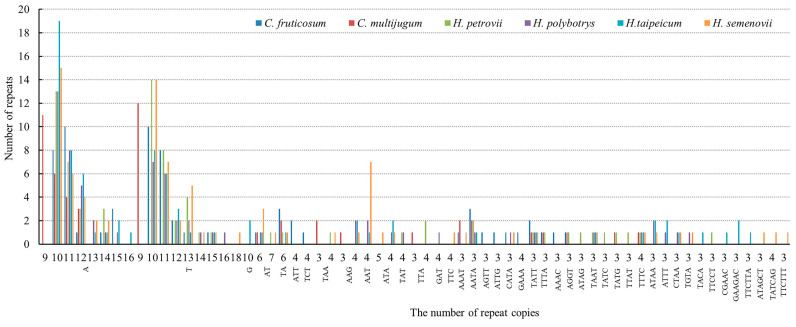
In the C. fruticosum chloroplast genome, 50 repetitive sequences (Figure 3) were identified, including forward, complementary, and palindromic repeats. No reverse repeat sequences were detected. Forward repeats (58%) accounted for the largest proportion of repetitive sequences, followed by palindromic (40%) and complementary (2%) repeats. Meanwhile, the C. multijugum chloroplast genome lacked complementary repeats; however, it contained similar numbers of forward and palindromic repeats as C. fruticosum. The chloroplast genomes of H. polybotrys, H. taipeicum, and H. semenovii only contained forward and palindromic repeats, with the former found to be the most abundant, accounting for 94%, 94.2%, and 90.9%, of the repetitive sequences, respectively. In contrast, four types of repetitive sequences were identified in the chloroplast genome of H. petrovii: forward (48%), reverse (8%), complementary (4%), and palindromic (40%) sequences. The genes containing the most repetitive sequences in C. fruticosum were rps15 and trnN-GUU, which contained palindromic (2) and forward (12) sequences (Table S3).
Figure 3.
Numbers of repetitive sequences in the complete chloroplast genomes of C. fruticosum, C. multijugum, and four Hedysarum species.
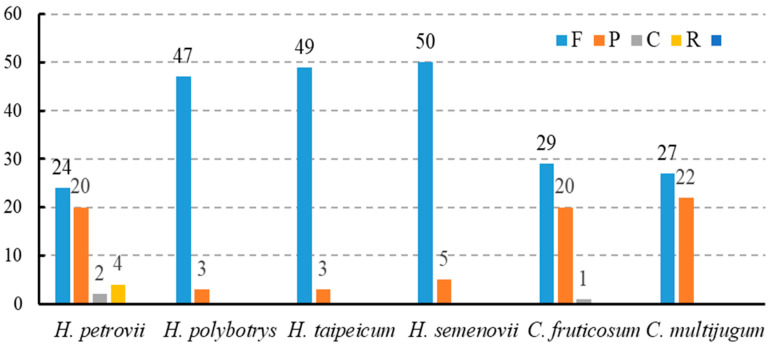
Sixty-three SSRs (Table 4) were ascertained in the chloroplast genome of C. fruticosum, with single-nucleotide repeats comprising 10–15 of repeat units, dinucleotide repeats comprising 6 repeat units, trinucleotide repeats comprising 4 repeat units, and tetranucleotide repeats comprising 3 repeat units (Figure 4). There were markedly more mononucleotide repeats in these six species than in the compound SSRs (Table S2). Of the six Fabaceae species, C. multijugum had the fewest compound SSRs (7), while H. taipeicum and H. semenovii had the most (16). Mononucleotide repeats included only the A/T in C. fruticosum, and C. multijugum, while the G/C in H. taipeicum, C. fruticosum and H. polybotrys had a single-nucleotide repeat. C. fruticosum, C. multijugum, and H. polybotrys had no pentanucleotide or hexanucleotide repeats, whereas H. petrovii had one hexanucleotide repeat (AAAGG/CCTTT). Regarding C. fruticosum, mononucleotide repeats (45) were the most abundant, followed by tetranucleotide repeats (12). H. petrovii and H. taipeicum each carried one pentanucleotide repeat, whereas H. taipeicum and H. semenovii had two and three hexanucleotide repeats, respectively. The chloroplast genome of H. taipeicum had one type of tetranucleotide repeat (AATC/ATTG) that was not found in the other Hedysarum species. The number and variety of SSRs in the rps15 and trnN-GUU was also the highest in C. fruticosum (Table S4).
Table 4.
Number of SSRs of four Hedysarum species, C. fruticosum, and C. multijugum.
| Species | Total SSRs | Compounds SSRs | Type | |||||
|---|---|---|---|---|---|---|---|---|
| Mono- | Di- | Tri- | Tetra- | Penta- | Hexa- | |||
| C. fruticosum | 63 | 8 | 45 | 3 | 3 | 12 | 0 | 0 |
| C. multijugum | 59 | 7 | 43 | 3 | 4 | 9 | 0 | 0 |
| H. semenovii | 68 | 10 | 50 | 1 | 6 | 11 | 0 | 0 |
| H. taipeicum | 80 | 16 | 59 | 2 | 4 | 11 | 1 | 3 |
| H. semenovii | 88 | 16 | 60 | 5 | 12 | 8 | 0 | 3 |
| H. petrovii | 76 | 14 | 56 | 1 | 6 | 12 | 1 | 0 |
Figure 4.
Numbers of SSRs of C. fruticosum, C. multijugum, and four Hedysarum species.
3.4. Comparative Analysis of the C. fruticosum Chloroplast Genome
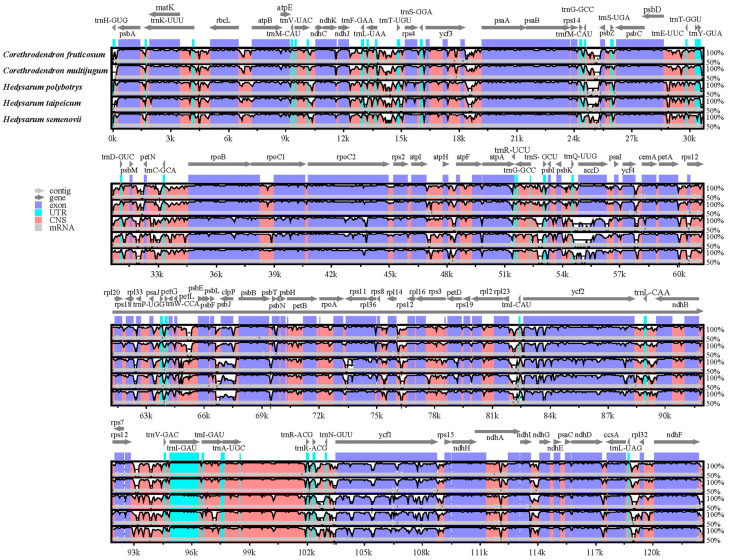
A comparison of the overall sequence variation in the chloroplast genomes using mVISTA revealed that the six chloroplast genomes were highly conserved (Figure 5). The gene intergenic regions of ycf3-psaA, trnG-GCC-psbZ, trnT-GGU-psbD, ndhC-trnV-UAC, psbE-petL, rpl16-rpl14, trnI-CAU-rpl123, trnR-ACC-trnN-GUU, rps12-trnV-GAC, ndhI-ndhG, ndhF-rpl32, and rpl32-trnL-UAG exhibited high variation and were located in the conserved non-coding regions (CNS) of the chloroplast genomes of these six species. In the exonic region, rpoB, rpoC2, ycf1, ycf2, and clpP exhibited significant differences. However, in all the studied chloroplast genomes, the regions with evident differences were primarily observed in the CNS.
Figure 5.
Global alignment of six chloroplast genomes using the H. petrovii genome as a reference sequence. The horizontal axis indicates the coordinates in the chloroplast genome. The vertical scale represents the average percentage of sequence similarity in the aligned regions, which ranged between 50% and 100%.
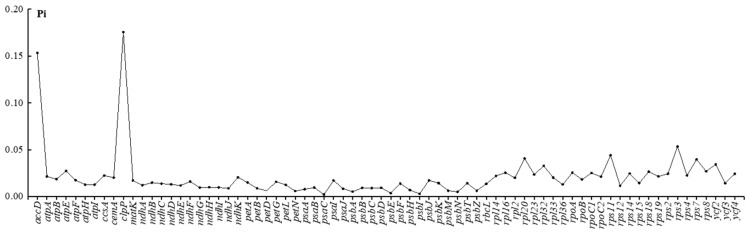
We calculated the Pi values of the six Fabaceae species to further clarify the variation in the coding regions (Figure 6). Although most sequences were relatively conserved (Pi < 0.01), accD and clpP—encoding an acetyl-CoA carboxylase subunit and protease, respectively—had high Pi values. Moreover, we identified four hotspot regions with Pi > 0.04 (rps3, rps11, rps7, and rpl20), all of which were related to ribosome subunit formation during transcription in plants.
Figure 6.
Line graph showing the gene nucleotide diversity (Pi) value of six Fabaceae species. X-axis: gene names. Y-axis: Pi value.
3.5. Phylogenetic Analysis
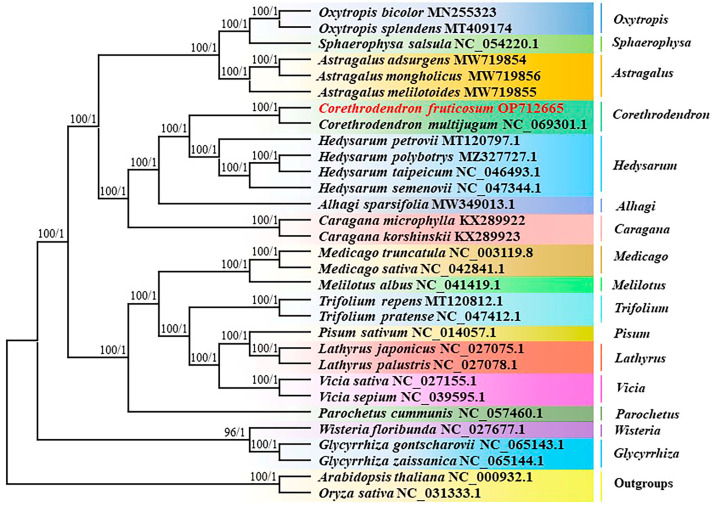
The topology of the phylogenetic tree comprising 16 genera and 31 species of Papilionoideae, and the taxonomic agreement of O. sativa and A. thaliana as outgroups of Papilionoideae, had strong bootstrap support (Figure 7). Corethrodendron was independent of Hedysarum, whereas C. fruticosum and C. multijugum formed a high-support branch with the four Hedysarum species. However, among Hedysarum species, the closest relatives were H. polybotrys and H. taipeicum, which formed a branch with H. semenovii with high support. Moreover, besides Hedysarum species, Corethrodendron was more closely related to Alhagi and Caragana compared with the other Papilionoideae genura.
Figure 7.
Phylogenetic tree of C. fruticosum and 30 other representative Fabaceae species based on 69 concatenated protein-coding genes. Arabidopsis thaliana and Oryza sativa were used as outgroups. Numbers associated with branches represent maximum likelihood values/Bayesian inference values (posterior probability).
4. Discussion
4.1. Sequence Variation in C. fruticosum
In the present research, five previously published complete chloroplast genomes were compared to that of C. fruticosum. No significant structural rearrangements were found in the genome of C. fruticosum, except for the deletion of the IR region. The gene contents and sequences of Corethrodendron and Hedysarum were highly conserved. We found that the codons in the C. fruticosum chloroplast genome exhibited a preference for A/T bases, which is often found in higher plants [47,48,49]. Accordingly, the GC content in C. fruticosum, as with C. multijugum and four Hedysarum species, was low [30,31,32].
The results of mVISTA analysis indicated that the length and gene order of the chloroplast genomes in these six plant were highly uniform; however, the CNS exhibited greater variation than other regions. As is consistent with previous studies, certain gene intergenic regions can be used as DNA barcodes for plant classification and identification [50,51]. More specifically, the divergent CNS regions in C. fruticosum, namely, psbE-petL and ndhF-rpl32, might prove effective when developed as DNA barcodes. Similarly, the chloroplast matK, trnL-trnF, and psbA-trnH sequences can be used as a basis for Hedysarum taxon delimitation [8,9]. The two genes with the highest Pi values in the coding region in the C. fruticosum chloroplast genome were accD and clpP. ClpP encodes a protease that is involved in chloroplast protein homeostasis and gene expression regulation [52]. AccD encodes the β-carboxyltransferase subunit of acetyl-CoA carboxylase [53]. Acetyl-CoA carboxylase is the rate-limiting enzyme in fatty acid biosynthesis, and its expression is induced by light [54,55,56]. Hence, these two genes may be responsible for the superior ability of C. fruticosum to grow on sand compared with the other four Hedysarum species [57]. As such, they have potential applications in C. fruticosum related to high light efficiency and stress tolerance breeding.
4.2. Repeat Sequences
The main source of duplication, rearrangement, and deletion events occurring in the chloroplast genome are repetitive sequences [58]. C. fruticosum had a low variety and few repetitive sequences. H. petrovii was the most similar to C. fruticosum in terms of the types and numbers of repetitive sequences, which may reflect the degree of relatedness between these species. Chloroplast SSRs are primarily located in non-coding regions and have the advantages of being highly conserved, endowing with uniparental inheritance, and having relative evolutionary independence [59,60,61]. Many plant species harbor chloroplast SSR markers [62]. The chloroplast SSRs in C. fruticosum, C. multijugum, and the four Hedysarum species primarily included poly-A/T and multi-base repeats, which is a consistent result with those found in other species in the IRLC clade [63]. Indeed, the chloroplast SSRs of C. fruticosum, C. multijugum, and the four Hedysarum species were highly variable, particularly for the composite SSRs. Hence, these SSRs can be used as molecular markers to differentiate C. fruticosum with other species and can provide a basis for studying the phylogeny and population of C. fruticosum.
4.3. Phylogeny of C. fruticosum
Our phylogenetic understanding of Corethrodendron is incomplete. Corethrodendron was originally classified as Hedysarum [7], and early phylogenetic studies of Hedysarum species used the sequence of the chloroplast gene matK for all Leguminosae [64,65]. Later, phylogenetic trees of Hedysarum were constructed using nuclear, gene intergenic regions and sequences of multiple chloroplast loci, including matK, trnL-trnF, and psbA-trnH [8,9]. During this period, a study used morphological data for the reclassification of Hedysarum [66]; the results were included in the Flora of China [1]. Collectively, these results highlight the independence of Corethrodendron from Hedysarum, which is supported by our findings. One study revealed that, among the species classified as Leguminosae IRLC, Hedysarum is more closely related to Astragalus than is Medicago [31], which is also supported by our findings. This may be due to the fact that the IR region of Astragalus is completely missing in Hedysarum and Corethrodendron [67], whereas the IR region in Medicago species, such as Medicago truncatula, is partially deleted [19]. The results of the present study indicate that Corethrodendron is more closely related to Alhagi and Caragana than to Astragalus. This may be due to most Astragalus species being herbaceous, whereas most others are subshrubs.
5. Conclusions
In this study, we sequenced and assembled the complete chloroplast genome of C. fruticosum and compared it to those of C. multijugum and four Hedysarum species, all of which belong to Papilionoideae. These species all belong to the IRLC. Their chloroplast genomes were found to be rich in repetitive sequences and SSRs, some of which can be used as molecular markers in genetic diversity analysis and Corethrodendron species identification. The marked differences in the CNS region can be used as novel DNA barcodes. The chloroplast genome of C. fruticosum had distinctly differentiated coding regions compared to the Hedysarum species. This further supports the independence of Corethrodendron from Hedysarum. However, the specific evolutionary relationship between Corethrodendron and Hedysarum remain unclear, and the few studies on other Corethrodendron are scarce. Collectively, this study provides useful information for the phylogenetic analysis and species identification of Corethrodendron.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14061289/s1, Table S1: GenBank accession numbers of chloroplast genomes; Table S2: SSR motif numbers in the C. fruticosum, C. multijugum, and four Hedysarum species chloroplast genomes; Table S3: Repeat sequences in the Corethrodendron fruticosum chloroplast genome; Table S4: SSR locations in the Corethrodendron fruticosum chloroplast genomes.
Author Contributions
T.N. carried out the analyses and wrote the first manuscript. C.T. collected the plant materials and was in charge of manuscript revision. Y.Y. had a certain contribution to the revision of the manuscript. Q.L., L.L., and Q.T. helped with the data analysis. Z.L. conceived the experiments, and Z.W. designed the experiment, carried out the analyses, and revised the manuscript, as well as provided funding. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The complete chloroplast genome sequences of the C. fruticosum we sequenced were deposited in the NCBI; the GenBank accession number is the following: OP712665.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The research was funded by the Central Public Interest Scientific Institution Basal Research Fund (No. Y2023PT02), the Key Projects in Science and Technology of Inner Mongolia (2021ZD0031), and the Inner Mongolia Science and Technology Plan (2020GG0127 and 2022YFHH0140).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Xu L.R., Choi B.H. Hedysarum L. In: Wu Z.Y., Raven P.H., Hong D.Y., editors. Flora of Chinal. Volume 10. Science Press; St. Louis, MO, USA: Botanical Garden Press; Beijing, China: 2010. pp. 514–525. [Google Scholar]
- 2.Yan G., Huang Z. A Study of Biological Character for Seeds and Seedling of Hedysarum laeve and H. scoparium. Grassl. China. 1996;3:37–41. [Google Scholar]
- 3.Li S., Li Q., Cui Z. Determination of Optimum Condition for Seed Germination Testing of Hedysarum Leave. Grassl. China. 1995;4:52–55. [Google Scholar]
- 4.Huang J., Zhang Y. Main Wind-proof Sand-fixing Plants and Their Application Value. Mod. Agric. Res. 2020;26:145–146. [Google Scholar]
- 5.Chen X., Gao Y., Zhao N., Li W., Zhang C., Zhai B., Wang Y. Adaptation evaluation of saline-alkali condition on roots of Hedysarum laeve in Mu Us sandy lande. J. Northwest A&F Univ. (Nat. Sci. Ed.) 2017;45:89–94. [Google Scholar]
- 6.Liu F., Ye X., Yu F., Dong M. Clonal Integration Modifies Responses of Hedysarum laeve to LOCAL Sand Burial in MU US Sandland. J. Plant Ecol. 2006;30:278–285. [Google Scholar]
- 7.Ferguson I.K., Skvarla J.J. Advances in Legumes Systematics 1. Royal Botanic Gardens, Kew; Richmond, UK: 1981. Pollen morphology of the subfamily Papilionoideae (Leguminosae) [Google Scholar]
- 8.Amirahmadi A., Kazempour Osaloo S., Moein F., Kaveh A., Maassoumi A.A. Molecular systematics of the tribe Hedysareae (Fabaceae) based on nrDNA ITS and plastid trnL-F and matK sequences. Plant Syst. Evol. 2014;300:729–747. doi: 10.1007/s00606-013-0916-5. [DOI] [Google Scholar]
- 9.Duan L., Wen J., Yang X., Liu P.L., Arslan E., Ertuğrul K., Chang Z.Y. Phylogeny of Hedysarum and tribe Hedysareae (Leguminosae: Papilionoideae) inferred from sequence data of ITS, matK, trnL-F and Psba-Trnh. TAXON. 2015;64:49–64. doi: 10.12705/641.26. [DOI] [Google Scholar]
- 10.Ravi V., Khurana J.P., Tyagi A.K., Khurana P. An update on chloroplast genomes. Plant Syst. Evol. 2008;271:101–122. doi: 10.1007/s00606-007-0608-0. [DOI] [Google Scholar]
- 11.Xing S., Liu C.J. Progress in Chloroplast Genome Analysis. Prog. Biochem. Biophys. 2008;35:21–28. [Google Scholar]
- 12.Sugiura M. The chloroplast genome. Plant Mol. Biol. 1992;19:149–168. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- 13.Sugiura M., Hirose T., Sugita M. Evolution and mechanism of translation in chloroplasts. Annu. Rev. Genet. 1998;32:437–459. doi: 10.1146/annurev.genet.32.1.437. [DOI] [PubMed] [Google Scholar]
- 14.Palmer J.D. Isolation and structural analysis of chloroplast DNA. Methods Enzymol. 1986;118:167–186. [Google Scholar]
- 15.Cheng H., Li J., Zhang H., Cai B., Mi L. The complete chloroplast genome sequence of strawberry (Fragaria × ananassa Duch.) and comparison with related species of Rosaceae. PeerJ. 2017;5:e3919. doi: 10.7717/peerj.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asaf S., Khan A.L., Khan M.A., Imran Q.M., Lee I.J. Comparative analysis of complete plastid genomes from wild soybean (Glycine soja) and nine other Glycine species. PLoS ONE. 2017;12:e0182281. doi: 10.1371/journal.pone.0182281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheon K.S., Kim K.A., Han J.S., Yoo K.O. The complete chloroplast genome sequence of Codonopsis minima (Campanulaceae), an endemic to Korea. Conserv. Genet. Resour. 2017;9:541–543. doi: 10.1007/s12686-017-0718-0. [DOI] [Google Scholar]
- 18.Yin D., Wang Y., Zhang X., Ma X., He X., Zhang J. Development of chloroplast genome resources for peanut (Arachis hypogaea L.) and other species of Arachis. Sci. Rep. 2017;7:11649. doi: 10.1038/s41598-017-12026-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin G., Rousseau-Gueutin M., Cordonnier S., Lima O., Michon-Coudouel S., Naquin D., de Carvalho J., Aïnouche M., Salmon A., Aïnouche A. The first complete chloroplast genome of the Genistoid legume Lupinus luteus: Evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Ann. Bot. Lond. Oup Acad. Press Oxf. Univ. Press. 2014;113:1197–1210. doi: 10.1093/aob/mcu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiong Y., Xiong Y., He J., Yu Q., Zhao J., Lei X., Dong Z., Yang J., Peng Y., Zhang X., et al. The Complete Chloroplast Genome of Two Important Annual Clover Species, Trifolium alexandrinum and T. resupinatum: Genome Structure, Comparative Analyses and Phylogenetic Relationships with Relatives in Leguminosae. Plants. 2020;9:478. doi: 10.3390/plants9040478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saski C., Lee S.B., Daniell H., Wood T.C., Tomkins J., Kim H.G., Jansen R.K. Complete Chloroplast Genome Sequence of Glycine max and Comparative Analyses with other Legume Genomes. Plant Mol. Biol. 2005;59:309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- 22.Magee A.M., Aspinall S., Rice D.W., Cusack B.P., Semon M., Perry A.S., Stefanovic S., Milbourne D., Barth S., Palmer J.D. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 2010;20:1700–1710. doi: 10.1101/gr.111955.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B., Duan N., Zhang H., Liu S., Shi J., Chai B. Characterization of the whole chloroplast genome of Caragana microphylla Lam (Fabaceae) Conserv. Genet. Resour. 2016;8:371–373. doi: 10.1007/s12686-016-0561-8. [DOI] [Google Scholar]
- 24.Wakasugi T., Tsudzuki J., Ito S., Nakashima K., Tsudzuki T. Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii. Proc. Natl. Acad. Sci. USA. 1994;91:9794–9798. doi: 10.1073/pnas.91.21.9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chumley T.W., Palmer J.D., Mower J.P., Matthew F.H., Calie P.J., Boore J.L., Jansen R.K. The Complete Chloroplast Genome Sequence of Pelargonium × hortorum: Organization and Evolution of the Largest and Most Highly Rearranged Chloroplast Genome of Land Plants. Mol. Biol. Evol. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- 26.Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohyama K., Fukuzawa H., Kohchi T., Shirai H., Sano T., Sano S., Umesono K., Shiki Y., Takeuchi M., Chang Z. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322:572–574. doi: 10.1038/322572a0. [DOI] [Google Scholar]
- 28.Hua Z., Tian D., Jiang C., Song S., Chen Z., Zhao Y., Jin Y., Huang L., Zhang Z., Yuan Y. Towards comprehensive integration and curation of chloroplast genomes. Plant Biotechnol. J. 2022;20:2239–2241. doi: 10.1111/pbi.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Memberspartners C.N., Gao F. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2021;50:D27–D38. doi: 10.1093/nar/gkab951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.She R.X., Li W.Q., Xie X.M., Gao X.X., Zhao P. The complete chloroplast genome sequence of a threatened perennial herb species Taibai sweetvetch (Hedysarum taipeicum K.T. Fu) Mitochondrial DNA Part B. 2019;4:1439–1440. doi: 10.1080/23802359.2019.1598817. [DOI] [Google Scholar]
- 31.Zhang R., Wang Y.H., Jin J., Stull G.W., Anne B., Domingos C., Paganucci D.Q.L., Moore M.J., Zhang S.D., Chen S.Y. Exploration of Plastid Phylogenomic Conflict Yields New Insights into the Deep Relationships of Leguminosae. Syst. Biol. 2020;69:613–622. doi: 10.1093/sysbio/syaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao J., Han C., Yang Y. Characterization of the complete chloroplast genome of Hedysarum polybotrys var. alaschanicum (Fabaceae) and its phylogeny. Mitochondrial DNA Part B Resour. 2021;6:3312–3313. doi: 10.1080/23802359.2021.1994900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin J.J., Yu W.B., Yang J.B., Song Y., Li D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qu X.J., Moore M.J., Li D.Z., Yi T.S. PGA: A software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 2019;15:50. doi: 10.1186/s13007-019-0435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohse M., Drechsel O., Bock R. OrganellarGenomeDRAW (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 36.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurtz S., Schleiermacher C. REPuter: Fast computation of maximal repeats in complete genomes. Bioinformatics. 1999;15:426–427. doi: 10.1093/bioinformatics/15.5.426. [DOI] [PubMed] [Google Scholar]
- 38.Sebastian B., Thomas T., Thomas M., Uwe S., Martin M. MISA-web: A web server for microsatellite prediction. Bioinformatics. 2017;33:2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Julio R., Albert F.M., Carlos S.D.J., Sara G.R., Pablo L., Ramos-Onsins S.E., Alejandro S.G. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017;34:12. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 40.Stephan G., Pascal L., Ralph B. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: Expanded toolkit for the graphical visualization of organellar genomes. Nuclc Acids Res. 2019;47:W59–W64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh K., Standley D. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:722–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Posada D., Crandall K.A. MODELTEST: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 43.Lam-Tung N., Schmidt H.A., Arndt V.H., Quang M.B. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ronquist F., Teslenko M., Mark P., Ayres D.L., Darling A., Hhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kikuchi S., Imai M., Nakahira Y., Kotani Y., Hashiguchi Y., Nakai Y., Takafuji K., Bédard J., Hirabayashi-Ishioka Y., Mori H., et al. A Ycf2-FtsHi Heteromeric AAA-ATPase Complex Is Required for Chloroplast Protein Import. Plant Cell. 2018;30:2677–2703. doi: 10.1105/tpc.18.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nie L., Cui Y., Wu L., Zhou J., Yao H. Gene Losses and Variations in Chloroplast Genome of Parasitic Plant Macrosolen and Phylogenetic Relationships within Santalales. Int. J. Mol. Sci. 2019;20:5812. doi: 10.3390/ijms20225812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang S., Ge X., Cano A., Salazar B., Deng Y. Comparative analysis of chloroplast genomes for five Diclipteraspecies (Acanthaceae): Molecular structure, phylogenetic relationships, and adaptive evolution. PeerJ. 2020;8:e8450. doi: 10.7717/peerj.8450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei F., Tang D., Wei K., Qin F., Li L., Lin Y., Zhu Y., Khan A., Kashif M.H., Miao J. The complete chloroplast genome sequence of the medicinal plant Sophora Tonkinensis. Sci. Rep. 2020;10:12473. doi: 10.1038/s41598-020-69549-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hohmann N., Wolf E.M., Lysak M.A., Koch M.A. A Time-Calibrated Road Map of Brassicaceae Species Radiation and Evolutionary History. Plant Cell. 2015;27:2770–2784. doi: 10.1105/tpc.15.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong W., Liu J., Jing Y., Wang L., Zhou S. Highly Variable Chloroplast Markers for Evaluating Plant Phylogeny at Low Taxonomic Levels and for DNA Barcoding. PLoS ONE. 2012;7:e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui N., Liao B.S., Liang C.L., Shi-Feng L.I., Zhang H., Jiang X.U., Xi-Wen L.I., Chen S.L. Complete chloroplast genome of Salvia plebeia: Organization, specific barcode and phylogenetic analysis. Chin. J. Nat. Med. 2020;18:563–572. doi: 10.1016/S1875-5364(20)30068-6. [DOI] [PubMed] [Google Scholar]
- 52.Sun Y., Li J., Zhang L., Lin R. Regulation of chloroplast protein degradation. J. Genet. Genom. 2023 doi: 10.1016/j.jgg.2023.02.010. [DOI] [PubMed] [Google Scholar]
- 53.Sasaki Y. The compartmentation of acetyl-coenzyme A carboxylase in plants. Plant Physiol. 1995;108:445–449. doi: 10.1104/pp.108.2.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kozaki A., Kamada K., Nagano Y., Iguchi H., Sasaki Y. Recombinant Carboxyltransferase Responsive to Redox of Pea Plastidic Acetyl-CoA Carboxylase. J. Biol. Chem. 2000;275:10702–10708. doi: 10.1074/jbc.275.14.10702. [DOI] [PubMed] [Google Scholar]
- 55.Kozaki A., Mayumi K., Sasaki Y. Thiol-Disulfide Exchange between Nuclear-encoded and Chloroplast-encoded Subunits of Pea Acetyl-CoA Carboxylase. J. Biol. Chem. 2001;276:39919–39925. doi: 10.1074/jbc.M103525200. [DOI] [PubMed] [Google Scholar]
- 56.Logemann E., Tavernaro A., Schulzt W.G. UV light selectively coinduces supply pathways from primary metabolism and flavonoid secondary product formation in parsley. Proc. Natl. Acad. Sci. USA. 2000;97:1903–1907. doi: 10.1073/pnas.97.4.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang C., Yang C., Dong M. The clonal integration of photosynthates in the rhizomatous half-shrub Hedysarum laeve. Acta Ecol. Sin. 2001;21:1986–1993. [Google Scholar]
- 58.Li B., Zheng Y. Dynamic evolution and phylogenomic analysis of the chloroplast genome in Schisandraceae. Sci. Rep. 2018;8:9285. doi: 10.1038/s41598-018-27453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang S., Gao M., Zaitlin D. Molecular Linkage Mapping and Marker-Trait Associations with NlRPT, a Downy Mildew Resistance Gene in Nicotiana Langsdorffii. Front. Plant Sci. 2012;3:185. doi: 10.3389/fpls.2012.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Breidenbach N., Gailing O., Krutovsky K.V. Genetic structure of coast redwood (Sequoia sempervirens [D Don] Endl) populations in and outside of the natural distribution range based on nuclear and chloroplast microsatellite markers. PLoS ONE. 2020;15:e0243556. doi: 10.1371/journal.pone.0243556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gichira A.W., Avoga S., Li Z., Hu G., Wang Q., Chen J. Comparative genomics of 11 complete chloroplast genomes of Senecioneae (Asteraceae) species: DNA barcodes and phylogenetics. Bot. Stud. 2019;60:17. doi: 10.1186/s40529-019-0265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang L., Zhao H., Peng Z., Dong L., Gao Z. Development and Application of SSR Molecular Markers from the Chloroplast Genome of Bamboo. J. Trop. Subtrop. Bot. 2014;22:263–269. [Google Scholar]
- 63.Lei W., Ni D., Wang Y., Shao J., Chang L. Intraspecific and heteroplasmic variations, gene losses and inversions in the chloroplast genome of Astragalus membranaceus. Sci. Rep. 2016;6:21669. doi: 10.1038/srep21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wojciechowski M.F., Lavin M., Sanderson M.J. A phylogeny of Legumes (leguminosae) based on analysis of the plastid matk gene resolves many well-supported subclades within the family. Am. J. Bot. 2004;91:1846–1862. doi: 10.3732/ajb.91.11.1846. [DOI] [PubMed] [Google Scholar]
- 65.Wojciechowski M.F., Sanderson M.J., Steele K.P., Liston A. Molecular phylogeny of the “temperate herbaceous tribes” of papilionoid legumes: A supertree approach. Adv. Legume Syst. 2000;9:277–298. [Google Scholar]
- 66.Choi B.H., Ohashi H. Generic criteria and an infrageneric system for Hedysarum and related genera (Papilionoideae-Leguminosae) TAXON. 2003;52:567–576. doi: 10.2307/3647455. [DOI] [Google Scholar]
- 67.Tian C., Li X., Wu Z., Li Z., Hou X., Li F.Y. Characterization and Comparative Analysis of Complete Chloroplast Genomes of Three Species from the Genus Astragalus (Leguminosae) Front. Genet. 2021;12:705482. doi: 10.3389/fgene.2021.705482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete chloroplast genome sequences of the C. fruticosum we sequenced were deposited in the NCBI; the GenBank accession number is the following: OP712665.