Abstract
Phenotypes of athletic performance and exercise capacity are complex traits influenced by both genetic and environmental factors. This update on the panel of genetic markers (DNA polymorphisms) associated with athlete status summarises recent advances in sports genomics research, including findings from candidate gene and genome-wide association (GWAS) studies, meta-analyses, and findings involving larger-scale initiatives such as the UK Biobank. As of the end of May 2023, a total of 251 DNA polymorphisms have been associated with athlete status, of which 128 genetic markers were positively associated with athlete status in at least two studies (41 endurance-related, 45 power-related, and 42 strength-related). The most promising genetic markers include the AMPD1 rs17602729 C, CDKN1A rs236448 A, HFE rs1799945 G, MYBPC3 rs1052373 G, NFIA-AS2 rs1572312 C, PPARA rs4253778 G, and PPARGC1A rs8192678 G alleles for endurance; ACTN3 rs1815739 C, AMPD1 rs17602729 C, CDKN1A rs236448 C, CPNE5 rs3213537 G, GALNTL6 rs558129 T, IGF2 rs680 G, IGSF3 rs699785 A, NOS3 rs2070744 T, and TRHR rs7832552 T alleles for power; and ACTN3 rs1815739 C, AR ≥21 CAG repeats, LRPPRC rs10186876 A, MMS22L rs9320823 T, PHACTR1 rs6905419 C, and PPARG rs1801282 G alleles for strength. It should be appreciated, however, that elite performance still cannot be predicted well using only genetic testing.
Keywords: sports, genetics, genotype, polymorphism, genomics, physical performance, athletes, GWAS, WGS, WES
1. Introduction
Athletic success is influenced by many genetically determined factors, including transcriptomic, biochemical, histological, anthropometric, physiological, and psychological traits, as well as general health status [1,2,3,4,5,6,7,8]. On average, 66% of the variance in athlete status can be explained by genetic factors [9]. The remaining variance is due to environmental factors, such as deliberate practice, nutrition, ergogenic aids, birthplace, the availability of medical and social support, and even luck (e.g., birthdate) [6,10,11,12].
Starting in the late 1990s, research began to identify DNA polymorphisms associated with predisposition to certain types of sports and exercise-related phenotypes, with initial focus on variants of the ACE, ACTN3, AMPD1, PPARA, PPARD, and PPARGC1A genes [13,14,15,16,17,18,19,20,21,22,23]. Initially, most research was conducted using the candidate gene approach [24,25,26,27,28,29], which limited progress in the discovery of new genetic markers associated with exercise- and sport-related phenotypes [30]. In addition to the fact that this approach studies only a single genetic variant in isolation, most candidate gene studies in the field of sports genomics are limited by sample size. This is a potential source of type I error (false positive findings), underpinning why replication of positive associations in independent cohorts is essential. Conversely, the genome-wide association approach is considered the most efficient study design thus far in identifying genetic markers associated with sport-related characteristics. Indeed, the application of this approach has enabled the discovery of hundreds of single nucleotide polymorphisms (SNPs) directly or indirectly associated with exercise and sport, such as height (12,111 SNPs) [31], appendicular lean mass (1059 SNPs) [32], testosterone levels (855 SNPs) [33], handgrip strength (170 SNPs) [34,35,36], sarcopenia (78 SNPs) [37], and brisk walking (70 SNPs) [38].
It is important to consider that the discovery of 12,111 independent genome-wide (p < 5 × 10−8) significant SNPs associated with height (which in combination account for 40% of phenotypic variance) required the study of 5.4 million individuals [31]. Accordingly, such findings indicate that each DNA locus is likely to explain only a very small proportion of the reported phenotypic variance (~0.0033%). This also suggests that very large samples of athletes and non-athletic controls are needed to detect a substantial number of genetic markers associated with athlete status, with some suggesting those markers be used as part of talent identification strategies. To date, five genome-wide association (GWAS) studies [39,40,41,42,43], one whole-genome sequencing (WGS) study [44], and one exome-wide association (EWAS) study [45] involving athletic cohorts have been conducted. However, due to their limited sample sizes, these investigations and subsequent replication studies have resulted in the identification of only 13 genetic markers. Six markers were associated with endurance athlete status (CDKN1A rs236448 A, GALNTL6 rs558129 C, NFIA-AS2 rs1572312 C, MYBPC3 rs1052373 G, SIRT1 rs41299232 G, and TRPM2 rs1785440 G alleles) [39,40,42,43,45,46], five with sprinter/strength athlete status (AGRN rs4074992 C, CDKN1A rs236448 C, CPNE5 rs3213537 G, NUP210 rs2280084 A, and GALNTL6 rs558129 T alleles) [41,45,47,48], and two with reaction time in wrestlers and athlete status in combat/team sports (APC rs518013 A and LRRN3 rs80054135 T alleles, respectively) [44].
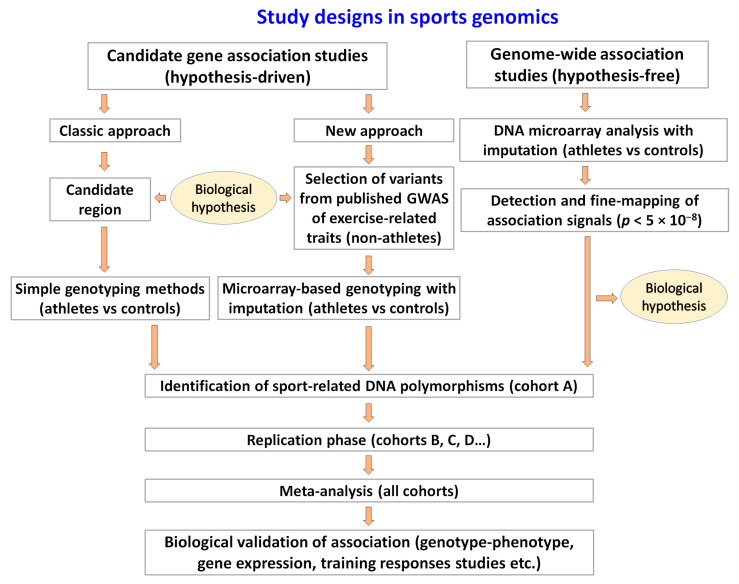
To overcome the issue of small samples of elite athletes, the use of genome-wide significant (p < 5 × 10−8) markers of exercise-related phenotypes (correlated with sport-related traits) discovered in large cohorts of untrained subjects (for example, UK Biobank, FinnGen, BioBank Japan, etc.) was proposed [49,50]. This approach may significantly decrease the risk of obtaining false-positive results in the discovery of markers associated with athlete status, which is recognized as a key limitation of sports genomics studies conducted on limited sample sizes. Accordingly, the first stage of such an investigation is to list candidate genetic markers previously identified in GWAS of exercise-related phenotypes in non-athletic populations. For example, this may include SNPs associated with phenotypes of handgrip strength, brisk walking, physical activity, lean mass, forced vital capacity, haemoglobin, or levels of hormones such as testosterone and IGF1. At the second stage, microarray analysis (with imputation; covering > 10 million SNPs) is carried out in a group of athletes to test the hypothesis that favourable alleles of exercise-related phenotypes are over-represented in those athletes compared to non-athletic controls (Figure 1). This approach may equally be applied to determine whether favourable alleles could be associated with direct measures of athletic capability, such as weightlifting performance, running times, or scores recorded during competitive events. In this case, the significance threshold for the second stage is set at p < 0.05 (to reduce the likelihood of potentially important findings being overlooked).
Figure 1.
Case-control study designs in sports genomics. In this approach, allelic frequencies are compared between athletes and controls (e.g., endurance athletes vs. untrained subjects or endurance vs. power athletes). A case-control study may be the first step followed by a genotype–phenotype study (e.g., identification of VO2max or weightlifting performance-increasing genotypes among athletes). In some cases, studies begin with a genotype–phenotype approach, and the findings are subsequently validated by a case-control study.
A recent example of such an approach being implemented is the study by Guilherme et al. [49], which investigated two correlated phenotypes: brisk walking pace (using UK Biobank participants) and sprint athlete status (using elite Russian sprinters). Brisk walkers perform more physical activity, are taller, have reduced adiposity and demonstrate greater physical performance and strength versus slower walkers, with such traits also recorded more commonly in sprinters than other athletes of other disciplines. Therefore, it was hypothesized that the alleles associated with high-speed walking (discovered in untrained subjects) would also be over-represented in elite sprinters. Accordingly, 70 genetic markers of brisk walking were identified from the literature [38], of which 15 SNPs had a significantly different allele frequency when comparing sprinters with non-athletic controls [49]. The same innovative approach later identified 23 SNPs associated with strength athlete status [51] based on genome-wide significant markers for handgrip strength in a non-athletic population using the UK Biobank [34,35]. Furthermore, using a panel of 822 testosterone-related SNPs from the UK Biobank study [33], five DNA-polymorphisms associated with muscle fibre size and weightlifting performance were identified [50].
Another approach that has proven effective in addressing the possibility of false positive results in sports genomics literature is to perform replication studies in two or more independent athletic cohorts (even with small or moderate sample sizes), followed by a meta-analysis to quantify the overall effect of a polymorphism on athlete status and/or a sport- and exercise-related trait [43,52,53,54,55,56,57,58,59,60,61,62,63,64,65]. However, in some cases, replication is not possible due to the exclusivity of a polymorphism to specific populations based on their geographic ancestry. For example, the rs671 G/A polymorphism of the aldehyde dehydrogenase 2 (ALDH2) gene was associated with strength in athletes and non-athletes from the Japanese population [66,67,68]. Interestingly, the unfavourable (associated with reduced strength) rs671 A allele is not present in Europeans or South Asians (frequency 0%), but common in Chinese, Japanese, and Vietnamese populations (15–25%). This demonstrates a notable challenge seeking to replicate genomic findings in larger samples, as increasing the study sample must also consider the geographic ancestry of participants. This also highlights the possibility that the genetic determinants of some sport- and exercise-related phenotypes are restricted to certain populations, demonstrating that increasing sample size is not as straightforward as simply recruiting participants from multiple countries and/or continents.
As well as the phenotypes of athlete status or competitive performance, several recent studies have investigated a broader range of traits which may relate directly or indirectly to athletic capability. These include flexibility, coordination, cardiorespiratory fitness, spatial ability, stress resilience, mental toughness, fat loss efficiency, and cardiovascular and metabolic responses to training, amongst others [69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84]. For example, combat athletes are more likely than untrained subjects to have the warrior (COMT rs4680 GG) genotype [85], whilst chess players demonstrate an increased frequency of an allele linked to improved memory and spatial ability (KIBRA rs17070145 T) [86]. Such discoveries demonstrate the broadening nature of sports genomics in recent times, with focus expanding from the traditional domain of investigating what makes elite performers different from the general population into other domains, such as sports nutrigenetics [87,88,89,90,91,92,93,94,95,96,97,98,99,100] and areas of sports medicine, such as genomic variants associated with soft-tissue injuries and sports-related concussion [101,102,103,104,105,106,107,108,109,110,111,112,113,114].
Technological advancement has lowered the cost of conducting genomic studies, increasing accessibility to researchers who wish to investigate the genetic underpinnings of sport and exercise phenotypes. Consequently, sports genomics is a dynamic and continually developing field, making it important to regularly appraise the contribution of recent advances to the field. Therefore, the aim of the current review was to summarise recent progress in understanding the genetic determinants of athlete status, and to detail novel DNA polymorphisms that may underpin differences between individuals in their athletic potential.
At the time of writing (end of May 2023), the total number of DNA polymorphisms associated with athletic performance since the first discovery in 1998 is 251 (Figure 2). Our search for sports genomics publications was based on journals indexed in major databases (i.e., PubMed etc.) using specific key words (e.g., athletes + polymorphism/genotype etc.). However, not all articles were included in the current review due to language limitations (articles written in languages other than English must contain at least abstracts in English). In addition, papers with very small cohort (less than 25 in athletes/controls), or articles with combined groups of athletes (for example, endurance + power without separation) were not included. Abstracts of conference proceedings were not considered. In recognition of the fact that many studies in the field of sports genomics report associations based on the investigation of small sample sizes, we stipulated that only markers where statistically significant associations have been reported in at least two studies (two case-control studies and/or one case-control plus one functional study; including those presented in one article) would be included in the present review.
Figure 2.
Sports-related genetic markers discovered between 1998 and 2023.
According to these criteria, 128 markers could be associated with athlete status (41 endurance-related, 45 power-related, and 42 strength-related) from the original 251 identified in our literature search. The most promising genetic markers (i.e., most replicated and had fewer negative or controversial findings) include AMPD1 rs17602729 C, CDKN1A rs236448 A, HFE rs1799945 G, MYBPC3 rs1052373 G, NFIA-AS2 rs1572312 C, PPARA rs4253778 G, and PPARGC1A rs8192678 G alleles for endurance; ACTN3 rs1815739 C, AMPD1 rs17602729 C, CDKN1A rs236448 C, CPNE5 rs3213537 G, GALNTL6 rs558129 T, IGF2 rs680 G, IGSF3 rs699785 A, NOS3 rs2070744 T, and TRHR rs7832552 T alleles for power; and ACTN3 rs1815739 C, AR ≥ 21 CAG repeats, LRPPRC rs10186876 A, MMS22L rs9320823 T, PHACTR1 rs6905419 C, and PPARG rs1801282 G alleles for strength. This update on the panel of genetic markers associated with athlete status covers advances in research reported in the past two years (previous online version was published in 2021 [115]). The current review also lists all known markers associated with endurance, power, or strength athlete status/performance. This article does not aim to review genetic markers associated with team (game) and combat sports, markers for which are well described elsewhere [26,61,116,117].
2. Gene Variants for Endurance Athlete Status
An individual’s endurance capacity is determined by many factors, including their muscle fibre typology, haemoglobin mass, mitochondrial biogenesis, maximal cardiac output, and maximal rate of oxygen consumption (VO2max), among others [118,119,120,121,122,123,124]. Indeed, there is evidence that these intermediate phenotypes have a substantial genetic influence, with literature indicating that genetic factors account for up to 70% of the variability in endurance-related traits [125]. Usually, genetic markers associated with endurance athlete status are determined by comparing allelic frequencies between endurance athletes (e.g., biathletes, road cyclists etc.) and controls.
To support the observed findings from endurance-related case-control studies, researchers subsequently perform functional, lab-based studies to determine the relationship between genotypes and physiological measures. Examples of measurements used to complement genomic studies include (but are not limited to) VO2max, forced expiratory volume in one second (FEV1), proportion of slow-twitch muscle fibres, recovery speed, long-distance running performance, running economy, lactate threshold, erythropoietin and haemoglobin levels, number of erythrocytes, capillary density, mitochondrial density, fat metabolism, and fatigue resistance.
Our literature search revealed that at least 41 of the 114 reported markers could be associated with endurance athlete status based on our criteria (Table 1). The most promising genetic markers for endurance athlete status include AMPD1 rs17602729 C, CDKN1A rs236448 A, HFE rs1799945 G, MYBPC3 rs1052373 G, NFIA-AS2 rs1572312 C, PPARA rs4253778 G, and PPARGC1A rs8192678 G alleles. In contrast, the other 73 markers (endurance alleles) have not passed our strict criteria: ACOXL rs13027870 G, ADRA2A 6.7kb, ADRA2A rs1800544 G, ADRB1 rs1801252 G, AGT rs699 A, BDKRB2 rs1799722 T, CAMK1D rs11257754 A, CHRNB3 rs4950 G, CLSTN2 rs2194938 A, CNDP2 rs6566810 A, COL5A1 rs71746744 AGGG, COL6A1 rs35796750 T, CPQ rs6468527 A, CYP2D6 rs3892097 G, DMT1 258 bp, EPAS1 (HIF2A) rs1867785 G, EPAS1 (HIF2A) rs11689011 T, GABPB1 rs8031031 T, GALM rs3821023 A, GNB3 rs5443 T, GRM3 rs724225 G, GSTT rs17856199 (+), IGF1R rs1464430 A, IL6 rs1800795 C, IL15RA rs2228059 A, ITPR1 rs1038639 T, ITPR1 rs2131458 T, FMNL2 rs12693407 G, KCNJ11 rs5219 C, L3MBTL4 rs17483463 T, MSTN rs11333758 D, MtDNA loci (G1, HV, L0, M*, m.11215T, m.152C, m.15518T, m.15874G, m.4343G, m.514(CA) ≤ 4, poly(C ≥ 7) stretch at m.568–573, m.16080G, m.5178C, N9, V, unfavourable: B, J2, T, L3*), NALCN-AS1 rs4772341 A, NACC2 rs4409473 C, NATD1 rs732928 G, NOS3 rs1799983 G, NOS3 (CA)n 164bp, NOS3 27bp 4B, PPARD rs2016520 C, PPARD rs1053049 T, PPARGC1A rs4697425 A, PPARGC1B rs11959820 A, PPP3CA rs3804358 C, PPP3CB rs3763679 C, SGMS1 rs884880 A, SLC2A4 rs5418 A, SOD2 rs4880 C, SPOCK1 rs1051854 T, TPK1 rs10275875 T, TTN rs10497520 T, Y-chromosome haplogroups (E*, E3*, and K*(xP); unfavourable: E3b1), and ZNF429 rs1984771 G. Most of these markers have been described in previous reviews [27,126,127] but cannot be included in our current list of endurance-associated markers until they are validated through replication by additional studies.
Table 1.
Genetic markers for endurance athlete status.
| Gene | Full Name | Locus | Polymorphism | Endurance-Related Allele | References | |
|---|---|---|---|---|---|---|
| Studies with Positive Results | Studies with Negative or Controversial Results | |||||
| ACE | Angiotensin I converting enzyme | 17q23.3 | Alu I/D (rs4343 A/G or rs4341 C/G) | I (A or C) | [14,15,16,128,129,130,131,132,133,134,135,136,137,138,139,140,141] | [133,142,143,144,145,146,147,148,149,150,151,152,153] |
| ACTN3 | Actinin α 3 | 11q13.1 | rs1815739 C/T | T | [17,154,155,156] | [152,157,158,159,160,161,162,163,164,165,166,167,168,169,170] |
| ADRB2 | Adrenoceptor β 2 | 5q31-q32 | rs1042713 G/A | A | [160,171,172] | [173,174] |
| ADRB2 | Adrenoceptor β 2 | 5q31-q32 | rs1042714 G/C | C | [153,175] | [173,174] |
| ADRB3 | Adrenoceptor β 3 | 8p11.23 | rs4994 A/G | G | [170,173] | |
| AGTR2 | Angiotensin II receptor type 2 | Xq22-q23 | rs11091046 A/C | C | [176] | [177] |
| AQP1 | Aquaporin 1 | 7p14 | rs1049305 C/G | C | [178,179,180] | |
| AMPD1 | Adenosine monophosphate deaminase 1 | 1p13 | rs17602729 C/T | C | [19,153,181,182,183] | [184] |
| BDKRB2 | Bradykinin receptor B2 | 14q32.1-q32.2 | +9/−9 (exon 1) | –9 | [185,186] | [153,187,188,189] |
| CDKN1A | Cyclin Dependent Kinase Inhibitor 1A | 6p21.2 | rs236448 A/C | A | [43] | |
| CKM | Creatine kinase M-type | 19q13.32 | rs8111989 A/G | A | [190,191,192] | [133,193] |
| COL5A1 | Collagen type V α 1 chain | 9q34.2-q34.3 | rs12722 C/T | T | [194,195] | |
| FTO | FTO α-Ketoglutarate Dependent Dioxygenase | 16q12.2 | rs9939609 T/A | T | [196,197] | [198] |
| GABPB1 | GA binding protein transcription factor subunit β 1 | 15q21.2 | rs12594956 A/C | A | [199,200] | |
| rs7181866 A/G | G | [199,201] | [200] | |||
| GALNTL6 | Polypeptide N-acetylgalactosaminyltransferase 6 | 4q34.1 | rs558129 T/C | C | [40] | |
| GSTP1 | Glutathione S-transferase Pi 1 | 11q13.2 | rs1695 A/G | G | [202,203] | |
| HFE | Homeostatic iron regulator | 6p21.3 | rs1799945 C/G | G | [153,204,205,206,207] | |
| HIF1A | Hypoxia inducible factor 1 subunit α | 14q23.2 | rs11549465 C/T | C | [208,209] | [144,210] |
| MCT1 | Monocarboxylate transporter 1 | 1p12 | rs1049434 A/T | T | [60,211,212,213,214] | [215] |
| MtDNA loci | Mitochondrial DNA | MtDNA | MtDNA haplogroups | H | [161,216] | |
| Unfavourable: K | [161,216] | |||||
| MYBPC3 | Myosin Binding Protein C3 | 11p11.2 | rs1052373 A/G | G | [42] | |
| NFATC4 | Nuclear factor of activated T cells 4 | 14q11.2 | rs2229309 G/C | G | [144] | |
| NFIA-AS2 | NFIA antisense RNA 2 | 1p31.3 | rs1572312 C/A | C | [39,46] | |
| NOS3 | Nitric oxide synthase 3 | 7q36 | rs2070744 T/C | T | [153,217,218] | [219] |
| PPARA | Peroxisome proliferator activated receptor α | 22q13.31 | rs4253778 G/C | G | [20,220,221,222] | |
| PPARGC1A | Peroxisome proliferative activated receptor, γ, coactivator 1 α | 4p15.1 | rs8192678 G/A | G | [18,20,170,223] | [216,224,225] |
| PPARGC1B | Peroxisome proliferative activated receptor, γ, coactivator 1 β | 5q32 | rs7732671 G/C | C | [144,226] | |
| PPP3R1 | Protein phosphatase 3 regulatory subunit B, α | 2p15 | Promoter 5I/5D | 5I | [144,227] | |
| PRDM1 | PR/SET Domain 1 | 6q21 | rs10499043 C/T | T | [228,229] | |
| RBFOX1 | RNA binding fox-1 homolog 1 | 16p13.3 | rs7191721 G/A | G | [39] | |
| SIRT1 | Sirtuin 1 | 10q21.3 | rs41299232 C/G | G | [45] | |
| SPEG | Striated Muscle Enriched Protein Kinase | 2q35 | rs7564856 G/A | G | [230] | |
| TFAM | Transcription factor A, mitochondrial | 10q21 | rs1937 G/C | C | [144,231] | [216] |
| TRPM2 | Transient Receptor Potential Cation Channel Subfamily M Member 2 | 21q22.3 | rs1785440 A/G | G | [45] | |
| TSHR | Thyroid stimulating hormone receptor | 14q31 | rs7144481 T/C | C | [39] | |
| UCP2 | Uncoupling protein 2 | 11q13 | rs660339 C/T | T | [130,144,232] | |
| UCP3 | Uncoupling Protein 3 | 11q13 | rs1800849 C/T | T | [130,144] | [233] |
| VEGFA | Vascular endothelial growth factor A | 6p12 | rs2010963 G/C | C | [144,234,235] | |
| VEGFR2 | Vascular endothelial growth factor receptor 2 | 4q11-q12 | rs1870377 T/A | A | [236,237] | |
3. Gene Variants for Power Athlete Status
Several characteristics are positively associated with power performance, including circulating levels of testosterone, percentage and cross-sectional area of fast-twitch muscle fibres, muscle mass and strength, body and calcaneus height, muscle fascicle length, and reaction time, among others [3,238,239,240,241,242,243,244]. The heritability of power-related phenotypes has been reported in the literature to range from approximately 49 to 86% in a range of phenotypes, including jumping ability [245,246]. Typically, genetic markers associated with power athlete status are determined by comparing allelic frequencies between power athletes (e.g., 100 m runners, shot putters, arm wrestlers, etc.) and untrained subjects. To support findings from case-control studies, investigators perform genotype–phenotype studies by measuring sprint times, jump performance, muscle fibre size, muscle fibre typology, maximal strength, rate of force development, and circulatory levels of anabolic hormones such as testosterone. Our literature search revealed that at least 45 of the 95 markers reportedly associated with power athlete status met our new criteria (Table 2). The most promising of these genetic markers associated with power athlete status currently include ACTN3 rs1815739 C, AMPD1 rs17602729 C, CDKN1A rs236448 C, CPNE5 rs3213537 G, GALNTL6 rs558129 T, IGF2 rs680 G, IGSF3 rs699785 A, NOS3 rs2070744 T, and TRHR rs7832552 T alleles. In contrast, the remaining 50 genetic markers (power alleles) did not meet our strict criteria: ARHGEF28 rs17664695 G, CACNG1 rs1799938 A, CALCR rs17734766 G, CLSTN2 rs2194938 C, CNDP1 rs2887 A, CNDP1 rs2346061 C, CNDP2 rs3764509 G, COTL1 rs7458 T, CREM rs1531550 A, DMD rs939787 T, EPAS1 (HIF2A) rs1867785 G, EPAS1 (HIF2A) rs11689011 C, FOCAD rs17759424 C, GABRR1 rs282114 A, GALNT13 rs10196189 G, GPC5 rs852918 T, IGF1R rs1464430 C, IL1RN rs2234663 *2, IP6K3 rs6942022 C, MCT1 rs1049434 A, MED4 rs7337521 T, MPRIP rs6502557 A, MtDNA loci (favourable: F, m.204C, m.151T, m.15314A, Non-L/U6, unfavourable: m.16278T, m.5601T, m.4833G, m.5108C, m.7600A, m.9377G, m.13563G, m.14200C, m.14569A), MTR rs1805087 G, MTRR rs1801394 G, NOS3 rs1799983 G, NRG1 rs17721043 A, PPARGC1A rs8192678 A, PPARGC1B rs10060424 C, RC3H1 rs767053 G, SLC6A2 rs1805065 C, SUCLA2 rs10397 A, TPK1 rs10275875 C, UCP2 rs660339 C, VEGFR2 rs1870377 T, WAPL rs4934207 C, and ZNF423 rs11865138 C. The majority of these markers are reported in previous reviews [25,126,127] and should be validated in additional studies before they can meet the criteria to be included in our list of power-associated genetic variants.
Table 2.
Genetic markers for power athlete status.
| Gene | Full Name | Locus | Polymorphism | Power-Related Allele | References | |
|---|---|---|---|---|---|---|
| Studies with Positive Results | Studies with Negative or Controversial Results | |||||
| ACE | Angiotensin I converting enzyme | 17q23.3 | Alu I/D (rs4343 A/G or rs4341 C/G) | D (G) | [16,128,145,169,247,248,249,250] | [150,251,252,253,254] |
| ACVR1B | Activin A type IB receptor | 12q13.13 | rs2854464 A/G | A | [255,256] | [256,257] |
| ACTN3 | Actinin α 3 | 11q13.1 | rs1815739 C/T | C | [17,161,162,168,170,250,258,259,260,261,262,263,264] | [159,165,254,265,266] |
| ADAM15 | ADAM Metallopeptidase Domain 15 | 1q21.3 | rs11264302 G/A | G | [49] | |
| ADRB2 | Adrenoceptor β 2 | 5q31-q32 | rs1042713 G/A | G | [41,174] | |
| rs1042714 C/G | G | [41,174] | ||||
| AGRN | Agrin | 1p36.33 | rs4074992 C/T | C | [45] | |
| AGT | Angiotensinogen | 1q42.2 | rs699 T/C | C | [41,267,268] | |
| AGTR2 | Angiotensin II receptor type 2 | Xq22-q23 | rs11091046 A/C | A | [176,177] | [55] |
| AKAP6 | A-Kinase Anchoring Protein 6 | 14q12 | rs12883788 C/T | C | [49] | |
| AMPD1 | Adenosine monophosphate deaminase 1 | 1p13 | rs17602729 C/T | C | [184,269,270] | |
| AUTS2 | Activator of Transcription and Developmental Regulator AUTS2 | 7q11.22 | rs10452738 A/G | A | [49] | |
| BDNF | Brain derived neurotrophic factor | 11p14.1 | rs10501089 G/A | A | [271] | |
| CCT3 | Chaperonin Containing TCP1 Subunit 3 | 1q22 | rs11548200 T/C | T | [49] | |
| CDKN1A | Cyclin Dependent Kinase Inhibitor 1A | 6p21.2 | rs236448 A/C | C | [43] | |
| CKM | Creatine kinase, M-type | 19q13.32 | rs8111989 A/G | G | [65,272,273] | [274] |
| CNTFR | Ciliary neurotrophic factor receptor | 9p13.3 | rs41274853 C/T | T | [275] | |
| CPNE5 | Copine V | 6p21.2 | rs3213537 G/A | G | [41,48] | |
| CRTAC1 | Cartilage Acidic Protein 1 | 10q24.2 | rs2439823 A/G | A | [49] | |
| CRTC1 | CREB Regulated Transcription Coactivator 1 | 19p13.11 | rs11881338 T/A | A | [49] | |
| E2F3 | E2F Transcription Factor 3 | 6p22.3 | rs4134943 C/T | T | [49] | |
| FHL2 | Four and a Half LIM Domains 2 | 2q12.2 | rs55680124 C/T | C | [49] | |
| GALNTL6 | Polypeptide N-acetylgalactosaminyltransferase like 6 | 4q34.1 | rs558129 C/T | T | [47,276] | |
| GDF5 | Growth Differentiation Factor 5 | 20q11.22 | rs143384 A/G | G | [49] | |
| HIF1A | Hypoxia inducible factor 1 α subunit | 14q21-q24 | rs11549465 C/T | T | [277,278,279] | |
| HSD17B14 | Hydroxysteroid 17-β dehydrogenase 14 | 19q13.33 | rs7247312 A/G | G | [41] | |
| IGF1 | Insulin like growth factor 1 | 12q23.2 | rs35767 C/T | T | [280,281] | |
| IGF2 | Insulin like growth factor 2 | 11p15.5 | rs680 A/G | G | [41,281,282] | |
| IGSF3 | Immunoglobulin Superfamily Member 3 | 1p13.1 | rs699785 G/A | A | [49] | |
| IL6 | Interleukin 6 | 7p21 | rs1800795 C/G | G | [41,283,284] | |
| ILRUN | Inflammation and Lipid Regulator with UBA-Like and NBR1-Like Domains | 6p21.31 | rs205262 A/G | A | [49] | |
| MTHFR | Methylenetetrahydrofolate reductase | 1p36.3 | rs1801131 A/C | C | [285,286] | |
| NOS3 | Nitric oxide synthase 3 | 7q36 | rs2070744 T/C | T | [219,279,287] | |
| NRXN3 | Neurexin 3 | 14q24.3-q31.1 | rs8011870 G/A | G | [49] | |
| NUP210 | Nucleoporin 210 | 3p25.1 | rs2280084 C/A | C | [45] | |
| PIEZO1 | Piezo Type Mechanosensitive Ion Channel Component 1 | 16q24.3 | rs572934641 (TCC/-) E756del | D | [288] | |
| PPARA | Peroxisome proliferator activated receptor α | 22q13.31 | rs4253778 G/C | C | [159,220,289] | |
| PPARG | Peroxisome proliferator activated receptor γ | 3p25.2 | rs1801282 C/G | G | [279,290,291] | [292] |
| rs2920503 C/T | T | [49] | ||||
| SLC39A8 | Solute Carrier Family 39 Member 8 | 4q24 | rs13107325 C/T | C | [49] | |
| SOD2 | Superoxide dismutase 2 | 6q25.3 | rs4880 C/T | C | [293] | |
| TRHR | Thyrotropin releasing hormone receptor | 8q23.1 | rs7832552 C/T | T | [65,294,295] | |
| UBR5 | Ubiquitin Protein Ligase E3 Component N-Recognin 5 | 8q22.3 | rs10505025 G/A | A | [296] | |
| rs4734621 G/A | A | [296] | ||||
| ZNF568 | Zinc Finger Protein 568 | 19q13.12 | rs1667369 A/C | A | [49] | |
4. Gene Variants for Strength Athlete Status
Performance in strength-based sports is based on multiple factors. However, the factors considered to contribute substantially to strength phenotypes include skeletal muscle hypertrophy (muscle fibre size), hyperplasia, the predominance of fast-twitch muscle fibres, a greater muscle fascicle pennation angle, improved neurological adaptation, high glycolytic capacity, and increased circulatory testosterone [297]. Importantly, evidence exists that strength athletes exhibit vastly different transcriptomic, biochemical, anthropometric, physiological, and biomechanical characteristics compared to endurance athletes and/or controls [1,4]. These differences can be explained by the presence of both deliberate environmental (training, nutrition, etc.) and genetic factors. Indeed, studies indicate that there is a strong heritability of power- and strength-related traits, where genetic factors account for up to 85% of the variation in maximal isometric, isotonic, and isokinetic strength [246]. In a recent study investigating the genetic component of severe sarcopenia (the age-related decline in skeletal muscle mass, strength, and gait speed) [37], it was found that the alleles associated with higher risk of severe sarcopenia were closely linked to tiredness, alcohol intake, smoking, time spent watching television, and a higher self-reported consumption of salt and processed meat. In contrast, alleles associated with lower risk of severe sarcopenia were positively associated with levels of serum testosterone, IGF1, and 25-hydroxyvitamin D; height; physical activity; as well as indicators of healthier dietary habits (self-reported intake of cereal, cheese, oily fish, protein, water, fruit, and vegetables). Whilst muscle strength phenotypes in the general population may be less pronounced than in strength athletes, the latter may represent an ideal population to identify genomic variants associated with skeletal muscle capacity, potentially aiding the advancement of knowledge surrounding sarcopenia and directing strategies to reduce the negative impact of age-related declines in muscle mass. In general, genetic markers associated with strength athlete status can be determined by comparing allelic frequencies between strength athletes and controls. To support these findings, scientists perform genotype–phenotype studies by measuring handgrip and isokinetic strength, powerlifting/weightlifting performance, as well as evaluating the acute and chronic responses to resistance training.
Previously, 170 DNA polymorphisms were reported to be associated with handgrip strength in three large GWASs [34,35,36]. In a follow-up study involving elite weightlifters and powerlifters, Moreland et al. [51] tested the hypothesis that alleles associated with greater handgrip strength would be over-represented in these athletes compared to controls. Accordingly, they identified 23 DNA polymorphisms that were associated with strength athlete status. Of these SNPs, the LRPPRC rs10186876, MMS22L rs9320823, and PHACTR1 rs6905419 polymorphisms were also associated with superior competitive weightlifting performance [298].
Our literature search based on our new inclusion criteria revealed at that least 42 genetic markers could be associated with strength athlete status (Table 3). The most promising genetic markers for strength athlete status include ACTN3 rs1815739 C, AR ≥ 21 CAG repeats, LRPPRC rs10186876 A, MMS22L rs9320823 T, PHACTR1 rs6905419 C, and PPARG rs1801282 G alleles.
Table 3.
Genetic markers for strength athlete status.
| Gene | Full Name | Locus | Polymorphism | Strength-Related Allele | References | |
|---|---|---|---|---|---|---|
| Studies with Positive Results | Studies with Negative or Controversial Results | |||||
| ABHD17C | Abhydrolase domain containing 17C | 15q25.1 | rs7165759 G/A | A | [35,51] | |
| ACE | Angiotensin I converting enzyme | 17q23.3 | Alu I/D (rs4343 A/G or rs4341 C/G) |
D (G) | [299,300,301,302,303,304] | [305] |
| ACTG1 | Actin γ 1 | 17q25.3 | rs6565586 T/A | A | [34,51] | |
| ACTN3 | Actinin α 3 | 11q13.1 | rs1815739 C/T | C | [302,306,307,308] | [51,305,309,310] |
| ADCY3 | Adenylate cyclase 3 | 2p23.3 | rs10203386 T/A | T | [35,51] | |
| ADPGK | ADP dependent glucokinase | 15q24.1 | rs4776614 C/G | C | [35,51] | |
| AGT | Angiotensinogen | 1q42.2 | rs699 T/C | C | [310,311] | [51] |
| ALDH2 | Aldehyde Dehydrogenase 2 Family Member | 12q24.12 | rs671 G/A | G | [66,67,68] | |
| ANGPT2 | Angiopoietin 2 | 8p23.1 | rs890022 G/A | A | [51,312] | |
| AR | Androgen Receptor | Xq12 | (CAG)n | ≥21 | [313,314] | |
| ARPP21 | CAMP regulated phosphoprotein 21 | 3p22.3 | rs1513475 T/C | C | [35,51] | |
| BCDIN3D | Bicoid interacting 3 domain containing RNA methyltransferase | 12q13.12 | rs12367809 C/T | C | [35,51] | |
| CKM | Creatine kinase, M-type | 19q13.32 | rs8111989 A/G | G | [54,315] | [51,274] |
| CNTFR | Ciliary neurotrophic factor receptor | 9p13.3 | rs41274853 C/T | T | [275,316] | [51] |
| CRTAC1 | Cartilage acidic protein 1 | 10q24.2 | rs563296 G/A | G | [35,51] | |
| DHODH | Dihydroorotate dehydrogenase (Quinone) | 16q22.2 | rs12599952 G/A | A | [35,51] | |
| GALNTL6 | Polypeptide N-acetylgalactosaminyltransferase-like 6 | 4q34.1 | rs558129 C/T | T | [47] | |
| GBE1 | 1, 4-α-glucan branching enzyme 1 | 3p12.2 | rs9877408 A/G | A | [35,51] | |
| GBF1 | Golgi brefeldin A resistant guanine nucleotide exchange factor 1 | 10q24.32 | rs2273555 G/A | A | [34,317] | |
| GLIS3 | GLIS Family Zinc Finger 3 | 9p24.2 | rs34706136 T/TG | TG | [50] | |
| HIF1A | Hypoxia inducible factor 1 α | 14q21-q24 | rs11549465 C/T | T | [278,295,318] | [51] |
| IGF1 | Insulin-like growth factor 1 | 12q23.2 | rs35767 C/T | T | [51,280,319] | |
| IL6 | Interleukin 6 | 7p21 | rs1800795 C/G | G | [51,283] | |
| ITPR1 | Inositol 1, 4, 5-Triphosphate Receptor Type 1 | 3p26.1 | rs901850 G/T | T | [35,51] | |
| KIF1B | Kinesin family member 1B | 1p36.22 | rs11121542 G/A | G | [35,51] | |
| LRPPRC | Leucine-rich pentatricopeptide repeat cassette | 2p21 | rs10186876 A/G | A | [34,51,298] | |
| MLN | Motilin | 6p21.31 | rs12055409 A/G | G | [35,317] | |
| MMS22L | Methyl methanesulfonate-sensitivity protein 22-Like | 6q16.1 | rs9320823 T/C | T | [35,51,298] | |
| MTHFR | Methylenetetrahydrofolate reductase | 1p36.3 | rs1801131 A/C | C | [51,286,298] | |
| NPIPB6 | Nuclear pore complex interacting protein family member B6 | 16p12.1 | rs2726036 A/C | A | [35,51] | |
| PHACTR1 | Phosphate and actin regulator 1 | 6p24.1 | rs6905419 C/T | C | [35,51,298] | |
| PLEKHB1 | Pleckstrin homology domain containing B1 | 11q13.4 | rs7128512 A/G | G | [51,312] | |
| PPARA | Peroxisome proliferator activated receptor α | 22q13.31 | rs4253778 G/C | C | [220,320,321] | [51] |
| PPARG | Peroxisome proliferator activated receptor γ | 3p25.2 | rs1801282 C/G | G | [51,290,291] | |
| PPARGC1A | Peroxisome proliferative activated receptor, γ, coactivator 1 α | 4p15.2 | rs8192678 G/A | A | [51,305,308] | |
| R3HDM1 | R3H domain containing 1 | 2q21.3 | rs6759321 G/T | T | [35,51] | |
| RASGRF1 | Ras protein specific guanine nucleotide Releasing Factor 1 | 15q25.1 | rs1521624 C/A | A | [35,51] | |
| RMC1 | Regulator of MON1-CCZ1 | 18q11.2 | rs303760 C/T | C | [35,51] | |
| SLC39A8 | Solute carrier family 39 member 8 | 4q24 | rs13135092 A/G | A | [35,51] | |
| TFAP2D | Transcriptional factor AP-2 delta | 6p12.3 | rs56068671 G/T | T | [35,51] | |
| ZKSCAN5 | Zinc finger with KRAB and SCAN domains 5 | 7q22.1 | rs3843540 T/C | C | [35,51] | |
| ZNF608 | Zinc finger protein 608 | 5q23.2 | rs4626333 G/A | G | [312,317] | |
5. Conclusions
The current review demonstrates that at least 251 genetic markers are reportedly linked to sport-related traits. However, only 128 (51%) of these markers (41 endurance-related, 45 power-related, and 42 strength-related) have been associated with athlete status in two or more studies. On the other hand, of these 128 genetic markers, the significance of 29 (22.7%) DNA polymorphisms was not replicated in at least one study, raising the possibility that a number of findings may represent false positives. It is important to consider that there may be one of several reasons why the findings of a study may not be replicated by another, including disparity of sample sizes, small sample sizes in one or more of the studies, different study designs, inconsistent classification of sporting groups or types of sport (strength, power, etc.), variability in how researchers or research groups define the term “elite athletes” (some researchers define the term “elite” as performances at the international level, others if the athlete is a prize winner in international competitions), and the ethnicity/geographical ancestry of the cohorts studied, amongst others.
As discussed previously, height remains not only the most studied exercise-related phenotype at the genetic level, but also the most studied human trait, with 12,111 associated SNPs [31]. It is estimated that the final number of height-related SNPs may reach 25,000 (with a minor allele frequency of ≥1%), but the sample size needs to be increased to approximately 100 million individuals of the same ethnicity. These values should be noteworthy and serve as a benchmark for the direction of future research in the field of sports genomics, where the current number of 251 genetic markers must be increased by a considerable magnitude in order to fully comprehend the genomic underpinnings of exercise performance, and thus to be considered as potential predictors of talent in sport. Given that effective talent identification remains a challenging task despite decades of research and strategy [322,323,324], it remains possible that the development of predictive genetic performance tests in future may be able to contribute to the advancement of this field. However, the literature currently available does not support the use of genetic testing for these purposes [325,326,327].
Whilst genomics is the among the most established molecular sub-disciplines of sport and exercise research, sport- and exercise-related DNA polymorphisms do not fully explain the heritability of athlete status. Consequently, other forms of variation, such as rare mutations [328,329] and epigenetic markers (i.e., stable and heritable changes in gene expression) [330], must be considered. Newly emerging high-throughput technologies enable the design of multi-omics approaches integrating various -omics levels (metabolomics, transcriptomics, proteomics, epigenomics, etc.) with the aim of determining how each level contributes to the biological mechanisms underpinning physical performance. For example, transcriptomic analyses have revealed the roles of both genomic and epigenomic mechanisms in modulating the transcription of genes regulated by exercise [2,331,332]. Incorporating multi-omics approaches has the potential to drastically advance the understanding of how the acute response to exercise is regulated, and consequently how chronic adaptations to exercise are mediated in the context of elite performance and/or health and wellbeing. Accordingly, future research, including collaborative multicentre GWASs and whole-genome sequencing of large athlete cohorts with further validation and replication, as well as the use of large purpose-built Biobanks, should focus on identifying genetic and other -omics markers of sport-related phenotypes and their underlying biology.
Our review does have limitations. First, we have not provided information regarding genetic markers associated with team (game) and combat sports, flexibility, coordination, personality, cognitive abilities, muscle fibre composition, skeletal muscle hypertrophy, injuries, and responses to training/supplements. These markers are well described elsewhere [4,24,26,28,37,61,74,79,115,116,117,333,334]. Second, we have not described all studies in detail (ethnicity, specific sporting disciplines, sample size, p-values etc.) given word limit. Third, some genetic markers (out of the 128 most significant) were selected based on data obtained in case-control studies only, without confirmation of functional significance (genotype–phenotype studies are therefore warranted).
In conclusion, our literature search revealed at least 251 DNA polymorphisms that could be associated with endurance, power, and strength athlete statuses. Most of these genetic markers have been discovered in studies involving Australian, Brazilian, British, Canadian, Chinese, Croatian, Czech, Ethiopian, Finnish, French, German, Greek, Hungarian, Indian, Iranian, Israeli, Italian, Jamaican, Japanese, Kenyan, Korean, Lithuanian, Polish, Qatari, Russian, Slovenian, South African, Spanish, Taiwanese, Tatar, Tunisian, Turkish, Ukrainian, and US athletes.
Author Contributions
Conceptualization, I.I.A.; formal analysis, E.A.S. and I.I.A.; writing—original draft preparation, E.A.S. and I.I.A.; writing—review and editing, E.C.R.H. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Stepto N.K., Coffey V.G., Carey A.L., Ponnampalam A.P., Canny B.J., Powell D., Hawley J.A. Global Gene Expression in Skeletal Muscle from Well-Trained Strength and Endurance Athletes. Med. Sci. Sport. Exerc. 2009;41:546–565. doi: 10.1249/MSS.0b013e31818c6be9. [DOI] [PubMed] [Google Scholar]
- 2.Zhelankin A.V., Iulmetova L.N., Ahmetov I.I., Generozov E.V., Sharova E.I. Diversity and Differential Expression of MicroRNAs in the Human Skeletal Muscle with Distinct Fiber Type Composition. Life. 2023;13:659. doi: 10.3390/life13030659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmetov I.I., Stepanova A.A., Biktagirova E.M., Semenova E.A., Shchuplova I.S., Bets L.V., Andryushchenko L.B., Borisov O.V., Andryushchenko O.N., Generozov E.V., et al. Is testosterone responsible for athletic success in female athletes? J. Sport. Med. Phys. Fit. 2020;60:1377–1382. doi: 10.23736/S0022-4707.20.10171-3. [DOI] [PubMed] [Google Scholar]
- 4.Fuku N., Kumagai H., Ahmetov I.I. Genetics of muscle fiber composition. In: Barh D., Ahmetov I., editors. Sports, Exercise, and Nutritional Genomics: Current Status and Future Directions. Academic Press; Cambridge, MA, USA: 2019. pp. 295–314. [DOI] [Google Scholar]
- 5.Hall E.C.R., Semenova E.A., Borisov O.V., Andryushchenko O.N., Andryushchenko L.B., Zmijewski P., Generozov E.V., Ahmetov I.I. Association of muscle fiber composition with health and exercise-related traits in athletes and untrained subjects. Biol. Sport. 2021;38:659–666. doi: 10.5114/biolsport.2021.102923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rees T., Hardy L., Güllich A., Abernethy B., Côté J., Woodman T., Montgomery H., Laing S., Warr C. The Great British Medalists Project: A Review of Current Knowledge on the Development of the World’s Best Sporting Talent. Sport. Med. 2016;46:1041–1058. doi: 10.1007/s40279-016-0476-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blume K., Wolfarth B. Identification of Potential Performance-Related Predictors in Young Competitive Athletes. Front. Physiol. 2019;10:1394. doi: 10.3389/fphys.2019.01394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dines H.R., Nixon J., Lockey S.J., Herbert A.J., Kipps C., Pedlar C.R., Day S.H., Heffernan S.M., Antrobus M.R., Brazier J., et al. Collagen Gene Polymorphisms Previously Associated with Resistance to Soft-Tissue Injury Are More Common in Competitive Runners Than Nonathletes. J. Strength Cond. Res. 2023;37:799–805. doi: 10.1519/JSC.0000000000004291. [DOI] [PubMed] [Google Scholar]
- 9.De Moor M.H.M., Spector T.D., Cherkas L.F., Falchi M., Hottenga J.J., Boomsma D.I., de Geus E.J.C. Genome-wide linkage scan for athlete status in 700 British female DZ twin pairs. Twin Res. Hum. Genet. 2007;10:812–820. doi: 10.1375/twin.10.6.812. [DOI] [PubMed] [Google Scholar]
- 10.Macnamara B.N., Hambrick D.Z., Oswald F.L. Deliberate practice and performance in music, games, sports, education, and professions: A meta-analysis. Psychol. Sci. 2014;25:1608–1618. doi: 10.1177/0956797614535810. [DOI] [PubMed] [Google Scholar]
- 11.Beck K.L., Thomson J.S., Swift R.J., von Hurst P.R. Role of nutrition in performance enhancement and postexercise recovery. Open Access J. Sports Med. 2015;6:259–267. doi: 10.2147/OAJSM.S33605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bezuglov E., Morgans R., Butovskiy M., Emanov A., Shagiakhmetova L., Pirmakhanov B., Waśkiewicz Z., Lazarev A. The relative age effect is widespread among European adult professional soccer players but does not affect their market value. PLoS ONE. 2023;18:e0283390. doi: 10.1371/journal.pone.0283390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rivera M.A., Dionne F.T., Wolfarth B., Chagnon M., Simoneau J.A., Pérusse L., Boulay M.R., Gagnon J., Song T.M., Keul J., et al. Muscle-specific creatine kinase gene polymorphisms in elite endurance athletes and sedentary controls. Med. Sci. Sport. Exerc. 1997;29:1444–1447. doi: 10.1097/00005768-199711000-00009. [DOI] [PubMed] [Google Scholar]
- 14.Montgomery H.E., Marshall R., Hemingway H., Myerson S., Clarkson P., Dollery C., Hayward M., Holliman D.E., World M., Thomas E.L., et al. Human gene for physical performance. Nature. 1998;393:221–222. doi: 10.1038/30374. [DOI] [PubMed] [Google Scholar]
- 15.Gayagay G., Yu B., Hambly B., Boston T., Hahn A., Celermajer D.S., Trent R.J. Elite endurance athletes and the ACE I allele--the role of genes in athletic performance. Hum. Genet. 1998;103:48–50. doi: 10.1007/s004390050781. [DOI] [PubMed] [Google Scholar]
- 16.Nazarov I.B., Woods D.R., Montgomery H.E., Shneider O.V., Kazakov V.I., Tomilin N.V., Rogozkin V.A. The angiotensin converting enzyme I/D polymorphism in Russian athletes. Eur. J. Hum. Genet. 2001;9:797–801. doi: 10.1038/sj.ejhg.5200711. [DOI] [PubMed] [Google Scholar]
- 17.Yang N., MacArthur D.G., Gulbin J.P., Hahn A.G., Beggs A.H., Easteal S., North K. ACTN3 genotype is associated with human elite athletic performance. Am. J. Hum. Genet. 2003;73:627–631. doi: 10.1086/377590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lucia A., Gómez-Gallego F., Barroso I., Rabadán M., Bandrés F., San Juan A.F., Chicharro J.L., Ekelund U., Brage S., Earnest C.P., et al. PPARGC1A genotype (Gly482Ser) predicts exceptional endurance capacity in European men. J. Appl. Physiol. 2005;99:344–348. doi: 10.1152/japplphysiol.00037.2005. [DOI] [PubMed] [Google Scholar]
- 19.Rubio J.C., Martín M.A., Rabadán M., Gómez-Gallego F., San Juan A.F., Alonso J.M., Chicharro J.L., Pérez M., Arenas J., Lucia A. Frequency of the C34T mutation of the AMPD1 gene in world-class endurance athletes: Does this mutation impair performance? J. Appl. Physiol. 2005;98:2108–2112. doi: 10.1152/japplphysiol.01371.2004. [DOI] [PubMed] [Google Scholar]
- 20.Akhmetov I.I., Popov D.V., Mozhaĭskaia I.A., Missina S.S., Astratenkova I.V., Vinogradova O.L., Rogozkin V.A. Association of regulatory genes polymorphisms with aerobic and anaerobic performance of athletes. Ross. Fiziol. Zh. Im. I.M. Sechenova. 2007;93:837–843. [PubMed] [Google Scholar]
- 21.Akhmetov I.I., Astranenkova I.V., Rogozkin V.A. Association of PPARD gene polymorphism with human physical performance. Mol. Biol. 2007;41:852–857. [PubMed] [Google Scholar]
- 22.Bray M.S., Hagberg J.M., Pérusse L., Rankinen T., Roth S.M., Wolfarth B., Bouchard C. The human gene map for performance and health-related fitness phenotypes: The 2006–2007 update. Med. Sci. Sports Exerc. 2009;41:35–73. doi: 10.1249/MSS.0b013e3181844179. [DOI] [PubMed] [Google Scholar]
- 23.Gineviciene V., Utkus A., Pranckevičienė E., Semenova E.A., Hall E.C.R., Ahmetov I.I. Perspectives in Sports Genomics. Biomedicines. 2022;10:298. doi: 10.3390/biomedicines10020298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cagnin S., Chemello F., Ahmetov I.I. Genes and response to aerobic training. In: Barh D., Ahmetov I., editors. Sports, Exercise, and Nutritional Genomics: Current Status and Future Directions. Academic Press; Cambridge, MA, USA: 2019. pp. 169–188. [DOI] [Google Scholar]
- 25.Maciejewska-Skrendo A., Sawczuk M., Cięszczyk P., Ahmetov I.I. Genes and power athlete status. In: Barh D., Ahmetov I., editors. Sports, Exercise, and Nutritional Genomics: Current Status and Future Directions. Academic Press; Cambridge, MA, USA: 2019. pp. 41–72. [DOI] [Google Scholar]
- 26.Massidda M., Calò C.M., Cięszczyk P., Kikuchi N., Ahmetov I.I., Williams A.G. Genetics of Team Sports. In: Barh D., Ahmetov I., editors. Sports, Exercise, and Nutritional Genomics: Current Status and Future Directions. Academic Press; Cambridge, MA, USA: 2019. pp. 105–128. [DOI] [Google Scholar]
- 27.Semenova E.A., Fuku N., Ahmetov I.I. Genetic profile of elite endurance athletes. In: Barh D., Ahmetov I., editors. Sports, Exercise, and Nutritional Genomics: Current Status and Future Directions. Academic Press; Cambridge, MA, USA: 2019. pp. 73–104. [DOI] [Google Scholar]
- 28.Valeeva E.V., Ahmetov I.I., Rees T. Psychogenetics and sport. In: Barh D., Ahmetov I., editors. Sports, Exercise, and Nutritional Genomics: Current Status and Future Directions. Academic Press; Cambridge, MA, USA: 2019. pp. 147–165. [DOI] [Google Scholar]
- 29.Varillas-Delgado D., Del Coso J., Gutiérrez-Hellín J., Aguilar-Navarro M., Muñoz A., Maestro A., Morencos E. Genetics and sports performance: The present and future in the identification of talent for sports based on DNA testing. Eur. J. Appl. Physiol. 2022;122:1811–1830. doi: 10.1007/s00421-022-04945-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang G., Tanaka M., Eynon N., North K.N., Williams A.G., Collins M., Moran C.N., Britton S.L., Fuku N., Ashley E.A., et al. The Future of Genomic Research in Athletic Performance and Adaptation to Training. Genet. Sport. 2016;61:55–67. doi: 10.1159/000445241. [DOI] [PubMed] [Google Scholar]
- 31.Yengo L., Vedantam S., Marouli E., Sidorenko J., Bartell E., Sakaue S., Graff M., Eliasen A.U., Jiang Y., Raghavan S., et al. A saturated map of common genetic variants associated with human height. Nature. 2022;610:704–712. doi: 10.1038/s41586-022-05275-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pei Y.F., Liu Y.Z., Yang X.L., Zhang H., Feng G.J., Wei X.T., Zhang L. The genetic architecture of appendicular lean mass characterized by association analysis in the UK Biobank study. Commun. Biol. 2020;3:608. doi: 10.1038/s42003-020-01334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruth K.S., Day F.R., Tyrrell J., Thompson D.J., Wood A.R., Mahajan A., Beaumont R.N., Wittemans L., Martin S., Busch A.S., et al. Using human genetics to understand the disease impacts of testosterone in men and women. Nat. Med. 2020;26:252–258. doi: 10.1038/s41591-020-0751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willems S.M., Wright D.J., Day F.R., Trajanoska K., Joshi P.K., Morris J.A., Matteini A.M., Garton F.C., Grarup N., Oskolkov N., et al. Large-scale GWAS identifies multiple loci for hand grip strength providing biological insights into muscular fitness. Nat. Commun. 2017;8:16015. doi: 10.1038/ncomms16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tikkanen E., Gustafsson S., Amar D., Shcherbina A., Waggott D., Ashley E.A., Ingelsson E. Biological Insights into Mus-cular Strength: Genetic Findings in the UK Biobank. Sci. Rep. 2018;8:6451. doi: 10.1038/s41598-018-24735-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones G., Trajanoska K., Santanasto A.J., Stringa N., Kuo C.L., Atkins J.L., Lewis J.R., Duong T., Hong S., Biggs M.L., et al. Genome-wide meta-analysis of muscle weakness identifies 15 susceptibility loci in older men and women. Nat. Commun. 2021;12:654. doi: 10.1038/s41467-021-20918-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Semenova E.A., Pranckevičienė E., Bondareva E.A., Gabdrakhmanova L.J., Ahmetov I.I. Identification and Characterization of Genomic Predictors of Sarcopenia and Sarcopenic Obesity Using UK Biobank Data. Nutrients. 2023;15:758. doi: 10.3390/nu15030758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timmins I.R., Zaccardi F., Nelson C.P., Franks P.W., Yates T., Dudbridge F. Genome-wide association study of self-reported walking pace suggests beneficial effects of brisk walking on health and survival. Commun. Biol. 2020;3:634. doi: 10.1038/s42003-020-01357-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmetov I., Kulemin N., Popov D., Naumov V., Akimov E., Bravy Y., Egorova E., Galeeva A., Generozov E., Kostryukova E., et al. Genome-wide association study identifies three novel genetic markers associated with elite endurance performance. Biol. Sport. 2015;32:3–9. doi: 10.5604/20831862.1124568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rankinen T., Fuku N., Wolfarth B., Wang G., Sarzynski M.A., Alexeev D.G., Ahmetov I.I., Boulay M.R., Cieszczyk P., Eynon N., et al. No evidence of a common DNA variant profile specific to world class endurance athletes. PLoS ONE. 2016;11:e0147330. doi: 10.1371/journal.pone.0147330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pickering C., Suraci B., Semenova E.A., Boulygina E.A., Kostryukova E.S., Kulemin N.A., Borisov O.V., Khabibova S.A., Larin A.K., Pavlenko A.V., et al. A genome-wide association study of sprint performance in elite youth football players. J. Strength Cond. Res. 2019;33:2344–2351. doi: 10.1519/JSC.0000000000003259. [DOI] [PubMed] [Google Scholar]
- 42.Al-Khelaifi F., Yousri N.A., Diboun I., Semenova E.A., Kostryukova E.S., Kulemin N.A., Borisov O.V., Andryushchenko L.B., Larin A.K., Generozov E.V., et al. Genome-Wide Association Study Reveals a Novel Association Between MYBPC3 Gene Polymorphism, Endurance Athlete Status, Aerobic Capacity and Steroid Metabolism. Front. Genet. 2020;11:595. doi: 10.3389/fgene.2020.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Semenova E.A., Zempo H., Miyamoto-Mikami E., Kumagai H., Larin A.K., Sultanov R.I., Babalyan K.A., Zhelankin A.V., Tobina T., Shiose K., et al. Genome-Wide Association Study Identifies CDKN1A as a Novel Locus Associated with Muscle Fiber Composition. Cells. 2022;11:3910. doi: 10.3390/cells11233910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boulygina E.A., Borisov O.V., Valeeva E.V., Semenova E.A., Kostryukova E.S., Kulemin N.A., Larin A.K., Nabiullina R.M., Mavliev F.A., Akhatov A.M., et al. Whole genome sequencing of elite athletes. Biol. Sport. 2020;37:295–304. doi: 10.5114/biolsport.2020.96272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bulgay C., Kasakolu A., Kazan H.H., Mijaica R., Zorba E., Akman O., Bayraktar I., Ekmekci R., Koncagul S., Ulucan K., et al. Exome-Wide Association Study of Competitive Performance in Elite Athletes. Genes. 2023;14:660. doi: 10.3390/genes14030660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Malczewska-Lenczowska J., Orysiak J., Majorczyk E., Sitkowski D., Starczewski M., Zmijewski P. HIF-1α and NFIA-AS2 Polymorphisms as Potential Determinants of Total Hemoglobin Mass in Endurance Athletes. J. Strength Cond. Res. 2022;36:1596–1604. doi: 10.1519/JSC.0000000000003686. [DOI] [PubMed] [Google Scholar]
- 47.Díaz Ramírez J., Álvarez-Herms J., Castañeda-Babarro A., Larruskain J., Ramírez de la Piscina X., Borisov O.V., Semenova E.A., Kostryukova E.S., Kulemin N.A., Andryushchenko O.N., et al. The GALNTL6 Gene rs558129 Polymorphism is Associated with Power Performance. J. Strength Cond. Res. 2020;34:3031–3036. doi: 10.1519/JSC.0000000000003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guilherme J.P.L.F., Semenova E.A., Zempo H., Martins G.L., Lancha-Junior A.H., Miyamoto-Mikami E., Kumagai H., Tobina T., Shiose K., Kakigi R., et al. Are Genome-Wide Association Study Identified Single-Nucleotide Polymorphisms Associated with Sprint Athletic Status? A Replication Study With 3 Different Cohorts. Int. J. Sport. Physiol. Perform. 2021;16:489–495. doi: 10.1123/ijspp.2019-1032. [DOI] [PubMed] [Google Scholar]
- 49.Guilherme J.P.L.F., Semenova E.A., Larin A.K., Yusupov R.A., Generozov E.V., Ahmetov I.I. Genomic Predictors of Brisk Walking Are Associated with Elite Sprinter Status. Genes. 2022;13:1710. doi: 10.3390/genes13101710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guilherme J.P.L., Semenova E.A., Borisov O.V., Larin A.K., Moreland E., Generozov E.V., Ahmetov I.I. Genomic predictors of testosterone levels are associated with muscle fiber size and strength. Eur. J. Appl. Physiol. 2022;122:415–423. doi: 10.1007/s00421-021-04851-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moreland E., Borisov O.V., Semenova E.A., Larin A.K., Andryushchenko O.N., Andryushchenko L.B., Generozov E.V., Williams A.G., Ahmetov I.I. Polygenic Profile of Elite Strength Athletes. J. Strength Cond. Res. 2022;36:2509–2514. doi: 10.1519/JSC.0000000000003901. [DOI] [PubMed] [Google Scholar]
- 52.Ma F., Yang Y., Li X., Zhou F., Gao C., Li M., Gao L. The association of sport performance with ACE and ACTN3 genetic polymorphisms: A systematic review and meta-analysis. PLoS ONE. 2013;8:e54685. doi: 10.1371/journal.pone.0054685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopez-Leon S., Tuvblad C., Forero D.A. Sports genetics: The PPARA gene and athletes’ high ability in endurance sports. A systematic review and meta-analysis. Biol. Sport. 2016;33:3–6. doi: 10.5604/20831862.1180170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen C., Sun Y., Liang H., Yu D., Hu S. A meta-analysis of the association of CKM gene rs8111989 polymorphism with sport performance. Biol. Sport. 2017;34:323–330. doi: 10.5114/biolsport.2017.69819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yvert T.P., Zempo H., Gabdrakhmanova L.J., Kikuchi N., Miyamoto-Mikami E., Murakami H., Naito H., Cieszczyk P., Leznicka K., Kostryukova E.S., et al. AGTR2 and sprint/power performance: A case-control replication study for rs11091046 polymorphism in two ethnicities. Biol. Sport. 2018;35:105–109. doi: 10.5114/biolsport.2018.71599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weyerstraß J., Stewart K., Wesselius A., Zeegers M. Nine genetic polymorphisms associated with power athlete status—A Meta-Analysis. J. Sci. Med. Sport. 2018;21:213–220. doi: 10.1016/j.jsams.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 57.Chen Y., Wang D., Yan P., Yan S., Chang Q., Cheng Z. Meta-analyses of the association between the PPARGC1A Gly482Ser polymorphism and athletic performance. Biol. Sport. 2019;36:301–309. doi: 10.5114/biolsport.2019.88752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tharabenjasin P., Pabalan N., Jarjanazi H. Association of the ACTN3 R577X (rs1815739) polymorphism with elite power sports: A meta-analysis. PLoS ONE. 2019;14:e0217390. doi: 10.1371/journal.pone.0217390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tharabenjasin P., Pabalan N., Jarjanazi H. Association of PPARGC1A Gly428Ser (rs8192678) polymorphism with potential for athletic ability and sports performance: A meta-analysis. PLoS ONE. 2019;14:e0200967. doi: 10.1371/journal.pone.0200967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guilherme J.P.L.F., Bosnyák E., Semenova E.A., Szmodis M., Griff A., Móra Á., Almási G., Trájer E., Udvardy A., Kostryukova E.S., et al. The MCT1 gene Glu490Asp polymorphism (rs1049434) is associated with endurance athlete status, lower blood lactate accumulation and higher maximum oxygen uptake. Biol. Sport. 2021;38:465–474. doi: 10.5114/biolsport.2021.101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McAuley A.B.T., Hughes D.C., Tsaprouni L.G., Varley I., Suraci B., Roos T.R., Herbert A.J., Kelly A.L. The association of the ACTN3 R577X and ACE I/D polymorphisms with athlete status in football: A systematic review and meta-analysis. J. Sport. Sci. 2021;39:200–211. doi: 10.1080/02640414.2020.1812195. [DOI] [PubMed] [Google Scholar]
- 62.Ipekoglu G., Bulbul A., Cakir H.I. A meta-analysis on the association of ACE and PPARA gene variants and endurance athletic status. J. Sport. Med. Phys. Fitness. 2022;62:795–802. doi: 10.23736/S0022-4707.21.12417-X. [DOI] [PubMed] [Google Scholar]
- 63.Konopka M.J., van den Bunder J.C.M.L., Rietjens G., Sperlich B., Zeegers M.P. Genetics of long-distance runners and road cyclists-A systematic review with meta-analysis. Scand. J. Med. Sci. Sport. 2022;32:1414–1429. doi: 10.1111/sms.14212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saito M., Ginszt M., Semenova E.A., Massidda M., Huminska-Lisowska K., Michałowska-Sawczyn M., Homma H., Cięszczyk P., Okamoto T., Larin A.K., et al. Is COL1A1 Gene rs1107946 Polymorphism Associated with Sport Climbing Status and Flexibility? Genes. 2022;13:403. doi: 10.3390/genes13030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saito M., Ginszt M., Semenova E.A., Massidda M., Huminska-Lisowska K., Michałowska-Sawczyn M., Homma H., Cięszczyk P., Okamoto T., Larin A.K., et al. Genetic profile of sports climbing athletes from three different ethnicities. Biol. Sport. 2022;39:913–919. doi: 10.5114/biolsport.2022.109958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kikuchi N., Tajima T., Tamura Y., Yamanaka Y., Menuki K., Okamoto T., Sakamaki-Sunaga M., Sakai A., Hiranuma K., Nakazato K. The ALDH2 rs671 polymorphism is associated with athletic status and muscle strength in a Japanese population. Biol. Sport. 2022;39:429–434. doi: 10.5114/biolsport.2022.106151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Almeida K.Y., Saito M., Homma H., Mochizuki Y., Saito A., Deguchi M., Kozuma A., Okamoto T., Nakazato K., Kikuchi N. ALDH2 gene polymorphism is associated with fitness in the elderly Japanese population. J. Physiol. Anthropol. 2022;41:38. doi: 10.1186/s40101-022-00312-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saito A., Saito M., Almeida K.Y., Homma H., Deguchi M., Kozuma A., Kobatake N., Okamoto T., Nakazato K., Kikuchi N. The Association between the ALDH2 rs671 Polymorphism and Athletic Performance in Japanese Power and Strength Athletes. Genes. 2022;13:1735. doi: 10.3390/genes13101735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ahmetov I.I., Donnikov A.E., Trofimov D.Y. Actn3 genotype is associated with testosterone levels of athletes. Biol. Sport. 2014;31:105–108. doi: 10.5604/20831862.1096046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baranova T.I., Berlov D.N., Glotov O.S., Korf E.A., Minigalin A.D., Mitrofanova A.V., Ahmetov I.I., Glotov A.S. Genetic determination of the vascular reactions in humans in response to the diving reflex. Am. J. Physiol. Heart Circ. Physiol. 2017;312:H622–H631. doi: 10.1152/ajpheart.00080.2016. [DOI] [PubMed] [Google Scholar]
- 71.Leońska-Duniec A., Ahmetov I.I., Zmijewski P. Genetic variants influencing effectiveness of exercise training programmes in obesity—an overview of human studies. Biol. Sport. 2016;33:207–214. doi: 10.5604/20831862.1201052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leońska-Duniec A., Jastrzębski Z., Jażdżewska A., Moska W., Lulińska-Kuklik E., Sawczuk M., Gubaydullina S.I., Shakirova A.T., Cięszczyk P., Maszczyk A., et al. Individual Responsiveness to Exercise-Induced Fat Loss and Improvement of Metabolic Profile in Young Women is Associated with Polymorphisms of Adrenergic Receptor Genes. J. Sport. Sci. Med. 2018;17:134–144. [PMC free article] [PubMed] [Google Scholar]
- 73.Leonska-Duniec A., Cięszczyk P., Ahmetov I.I. Genes and individual responsiveness to exercise-induced fat loss. In: Barh D., Ahmetov I., editors. Sports, Exercise, and Nutritional Genomics: Current Status and Future Directions. Academic Press; Cambridge, MA, USA: 2019. pp. 231–247. [DOI] [Google Scholar]
- 74.Massidda M., Miyamoto N., Beckley S., Kikuchi N., Fuku N. Genetics of flexibility. In: Barh D., Ahmetov I., editors. Sports, Exercise, and Nutritional Genomics: Current Status and Future Directions. Academic Press; Cambridge, MA, USA: 2019. pp. 273–293. [DOI] [Google Scholar]
- 75.Peplonska B., Safranow K., Adamczyk J., Boguszewski D., Szymański K., Soltyszewski I., Barczak A., Siewierski M., Ploski R., Sozanski H., et al. Association of serotoninergic pathway gene variants with elite athletic status in the Polish population. J. Sport. Sci. 2019;37:1655–1662. doi: 10.1080/02640414.2019.1583156. [DOI] [PubMed] [Google Scholar]
- 76.Yusof H.A., Aziz A.R., Muhamed A.M.C. The influence of angiotensin I-converting enzyme (ACE) I/D gene polymorphism on cardiovascular and muscular adaptations following 8 weeks of isometric handgrip training (IHG) in untrained normotensive males. Biol. Sport. 2019;36:81–94. doi: 10.5114/biolsport.2019.79975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mazur I.I., Drozdovska S., Andrieieva O., Vinnichuk Y., Polishchuk A., Dosenko V., Andreev I., Pickering C., Ahmetov I.I. PPARGC1A gene polymorphism is associated with exercise-induced fat loss. Mol. Biol. Rep. 2020;47:7451–7457. doi: 10.1007/s11033-020-05801-z. [DOI] [PubMed] [Google Scholar]
- 78.Zhao Y., Huang G., Chen Z., Fan X., Huang T., Liu J., Zhang Q., Shen J., Li Z., Shi Y. Four Loci Are Associated with Cardiorespiratory Fitness and Endurance Performance in Young Chinese Females. Sci. Rep. 2020;10:10117. doi: 10.1038/s41598-020-67045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mountford H.S., Hill A., Barnett A.L., Newbury D.F. Genome-Wide Association Study of Motor Coordination. Front. Hum. Neurosci. 2021;15:669902. doi: 10.3389/fnhum.2021.669902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Williams C.J., Li Z., Harvey N., Lea R.A., Gurd B.J., Bonafiglia J.T., Papadimitriou I., Jacques M., Croci I., Stensvold D., et al. Genome wide association study of response to interval and continuous exercise training: The Predict-HIIT study. J. Biomed. Sci. 2021;28:37. doi: 10.1186/s12929-021-00733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klevjer M., Nordeidet A.N., Hansen A.F., Madssen E., Wisløff U., Brumpton B.M., Bye A. Genome-Wide Association Study Identifies New Genetic Determinants of Cardiorespiratory Fitness: The Trøndelag Health Study. Med. Sci. Sport. Exerc. 2022;54:1534–1545. doi: 10.1249/MSS.0000000000002951. [DOI] [PubMed] [Google Scholar]
- 82.Bojarczuk A., Boulygina E.A., Dzitkowska-Zabielska M., Łubkowska B., Leońska-Duniec A., Egorova E.S., Semenova E.A., Andryushchenko L.B., Larin A.K., Generozov E.V., et al. Genome-Wide Association Study of Exercise-Induced Fat Loss Efficiency. Genes. 2022;13:1975. doi: 10.3390/genes13111975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McAuley A.B.T., Hughes D.C., Tsaprouni L.G., Varley I., Suraci B., Baker J., Herbert A.J., Kelly A.L. Genetic associations with personality and mental toughness profiles of English academy football players: An exploratory study. Psychol. Sport Exerc. 2022;61:102209. doi: 10.1016/j.psychsport.2022.102209. [DOI] [Google Scholar]
- 84.McAuley A.B., Hughes D.C., Tsaprouni L.G., Varley I., Suraci B., Baker J., Herbert A.J., Kelly A.L. Genetic associations with technical capabilities in English academy football players: A preliminary study. J. Sport. Med. Phys. Fitness. 2023;63:230–240. doi: 10.23736/S0022-4707.22.13945-9. [DOI] [PubMed] [Google Scholar]
- 85.Tartar J.L., Cabrera D., Knafo S., Thomas J.D., Antonio J., Peacock C.A. The “Warrior” COMT Val/Met Genotype Occurs in Greater Frequencies in Mixed Martial Arts Fighters Relative to Controls. J. Sport. Sci. Med. 2020;19:38–42. [PMC free article] [PubMed] [Google Scholar]
- 86.Ahmetov I.I., Valeeva E.V., Yerdenova M.B., Datkhabayeva G.K., Bouzid A., Bhamidimarri P.M., Sharafetdinova L.M., Egorova E.S., Semenova E.A., Gabdrakhmanova L.J., et al. KIBRA Gene Variant Is Associated with Ability in Chess and Science. Genes. 2023;14:204. doi: 10.3390/genes14010204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Womack C.J., Saunders M.J., Bechtel M.K., Bolton D.J., Martin M., Luden N.D., Dunham W., Hancock M. The influence of a CYP1A2 polymorphism on the ergogenic effects of caffeine. J. Int. Soc. Sport. Nutr. 2012;9:7. doi: 10.1186/1550-2783-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guest N.S., Horne J., Vanderhout S.M., El-Sohemy A. Sport Nutrigenomics: Personalized Nutrition for Athletic Performance. Front. Nutr. 2019;6:8. doi: 10.3389/fnut.2019.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guest N., Corey P., Vescovi J., El-Sohemy A. Caffeine, CYP1A2 Genotype, and Endurance Performance in Athletes. Med. Sci. Sport. Exerc. 2018;50:1570–1578. doi: 10.1249/MSS.0000000000001596. [DOI] [PubMed] [Google Scholar]
- 90.Guest N.S., Corey P., Tyrrell P.N., El-Sohemy A. Effect of Caffeine on Endurance Performance in Athletes May Depend on HTR2A and CYP1A2 Genotypes. J. Strength Cond. Res. 2022;36:2486–2492. doi: 10.1519/JSC.0000000000003665. [DOI] [PubMed] [Google Scholar]
- 91.Muñoz A., López-Samanes Á., Aguilar-Navarro M., Varillas-Delgado D., Rivilla-García J., Moreno-Pérez V., Del Coso J. Effects of CYP1A2 and ADORA2A Genotypes on the Ergogenic Response to Caffeine in Professional Handball Players. Genes. 2020;11:933. doi: 10.3390/genes11080933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wong O., Marshall K., Sicova M., Guest N.S., García-Bailo B., El-Sohemy A. CYP1A2 Genotype Modifies the Effects of Caffeine Compared With Placebo on Muscle Strength in Competitive Male Athletes. Int. J. Sport Nutr. Exerc. Metab. 2021;31:420–426. doi: 10.1123/ijsnem.2020-0395. [DOI] [PubMed] [Google Scholar]
- 93.Sicova M., Guest N.S., Tyrrell P.N., El-Sohemy A. Caffeine, genetic variation and anaerobic performance in male athletes: A randomized controlled trial. Eur. J. Appl. Physiol. 2021;121:3499–3513. doi: 10.1007/s00421-021-04799-x. [DOI] [PubMed] [Google Scholar]
- 94.Hall E.C.R., Semenova E.A., Bondareva E.A., Andryushchenko L.B., Larin A.K., Cięszczyk P., Generozov E.V., Ahmetov I.I. Association of Genetically Predicted BCAA Levels with Muscle Fiber Size in Athletes Consuming Protein. Genes. 2022;13:397. doi: 10.3390/genes13030397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rahimi R. The effect of CYP1A2 genotype on the ergogenic properties of caffeine during resistance exercise: A randomized, double-blind, placebo-controlled, crossover study. Ir. J. Med. Sci. 2019;188:337–345. doi: 10.1007/s11845-018-1780-7. [DOI] [PubMed] [Google Scholar]
- 96.Loy B.D., O’Connor P.J., Lindheimer J.B., Covert S.F. Caffeine is ergogenic for adenosine A2A receptor gene (ADORA2A) T allele homozygotes: A pilot study. J. Caffeine Res. 2015;5:73–81. doi: 10.1089/jcr.2014.0035. [DOI] [Google Scholar]
- 97.Pickering C., Kiely J. Are the current guidelines on caffeine use in sport optimal for everyone? Inter-individual variation in caffeine ergogenicity, and a move towards personalised sports nutrition. Sport. Med. 2018;48:7–16. doi: 10.1007/s40279-017-0776-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thomas R., Algrain H., Ryan E., Popojas A., Carrigan P., Abdulrahman A., Carrillo A. Influence of a CYP1A2 polymorphism on post-exercise heart rate variability in response to caffeine intake: A double-blind, placebo-controlled trial. Ir. J. Med. Sci. 2017;186:285–291. doi: 10.1007/s11845-016-1478-7. [DOI] [PubMed] [Google Scholar]
- 99.Rahimi M.R., Semenova E.A., Larin A.K., Kulemin N.A., Generozov E.V., Łubkowska B., Ahmetov I.I., Golpasandi H. The ADORA2A TT Genotype Is Associated with Anti-Inflammatory Effects of Caffeine in Response to Resistance Exercise and Habitual Coffee Intake. Nutrients. 2023;15:1634. doi: 10.3390/nu15071634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guntoro A.Y., Melita S., Dijaya R., Subali D., Kartawidjajaputra F., Suwanto A. Evaluation of Caffeine Ingested Timing on Endurance Performance based on CYP1A2 rs762551 Profiling in Healthy Sedentary Young Adults. Rep. Biochem. Mol. Biol. 2023;11:663–671. doi: 10.52547/rbmb.11.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rahim M., Collins M., September A. Genes and Musculoskeletal Soft-Tissue Injuries. Med. Sport Sci. 2016;61:68–91. doi: 10.1159/000445243. [DOI] [PubMed] [Google Scholar]
- 102.Kim S.K., Roche M.D., Fredericson M., Dragoo J.L., Horton B.H., Avins A.L., Belanger H.G., Ioannidis J.P.A., Abrams G.D. A Genome-wide Association Study for Concussion Risk. Med. Sci. Sport. Exerc. 2021;53:704–711. doi: 10.1249/MSS.0000000000002529. [DOI] [PubMed] [Google Scholar]
- 103.Lim T., Santiago C., Pareja-Galeano H., Iturriaga T., Sosa-Pedreschi A., Fuku N., Pérez-Ruiz M., Yvert T. Genetic variations associated with non-contact muscle injuries in sport: A systematic review. Scand. J. Med. Sci. Sport. 2021;31:2014–2032. doi: 10.1111/sms.14020. [DOI] [PubMed] [Google Scholar]
- 104.Antrobus M.R., Brazier J., Stebbings G.K., Day S.H., Heffernan S.M., Kilduff L.P., Erskine R.M., Williams A.G. Genetic Factors That Could Affect Concussion Risk in Elite Rugby. Sports. 2021;9:19. doi: 10.3390/sports9020019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brazier J., Antrobus M.R., Herbert A.J., Callus P.C., Khanal P., Stebbings G.K., Day S.H., Heffernan S.M., Kilduff L.P., Bennett M.A., et al. Gene variants previously associated with reduced soft-tissue injury risk: Part 2—Polygenic associations with elite status in Rugby. Eur. J. Sport Sci. 2022;26:1–10. doi: 10.1080/17461391.2022.2155877. [DOI] [PubMed] [Google Scholar]
- 106.Brazier J., Antrobus M.R., Herbert A.J., Callus P.C., Stebbings G.K., Day S.H., Heffernan S.M., Kilduff L.P., Bennett M.A., Erskine R.M., et al. Gene variants previously associated with reduced soft tissue injury risk: Part 1—independent associations with elite status in rugby. Eur. J. Sport Sci. 2022;29:1–10. doi: 10.1080/17461391.2022.2155877. [DOI] [PubMed] [Google Scholar]
- 107.de Almeida K.Y., Cetolin T., Marrero A.R., Aguiar Junior A.S., Mohr P., Kikuchi N. A Pilot Study on the Prediction of Non-Contact Muscle Injuries Based on ACTN3 R577X and ACE I/D Polymorphisms in Professional Soccer Athletes. Genes. 2022;13:2009. doi: 10.3390/genes13112009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ginevičienė V., Urnikytė A. Association of COL12A1 rs970547 Polymorphism with Elite Athlete Status. Biomedicines. 2022;10:2495. doi: 10.3390/biomedicines10102495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jacob Y., Anderton R.S., Cochrane Wilkie J.L., Rogalski B., Laws S.M., Jones A., Spiteri T., Hince D., Hart N.H. Genetic Variants within NOGGIN, COL1A1, COL5A1, and IGF2 are Associated with Musculoskeletal Injuries in Elite Male Australian Football League Players: A Preliminary Study. Sport. Med. Open. 2022;8:126. doi: 10.1186/s40798-022-00522-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Antrobus M.R., Brazier J., Callus P.C., Herbert A.J., Stebbings G.K., Khanal P., Day S.H., Kilduff L.P., Bennett M.A., Erskine R.M., et al. Concussion-Associated Polygenic Profiles of Elite Male Rugby Athletes. Genes. 2022;13:820. doi: 10.3390/genes13050820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hall E.C.R., Baumert P., Larruskain J., Gil S.M., Lekue J.A., Rienzi E., Moreno S., Tannure M., Murtagh C.F., Ade J.D., et al. The genetic association with injury risk in male academy soccer players depends on maturity status. Scand. J. Med. Sci. Sport. 2022;32:338–350. doi: 10.1111/sms.14077. [DOI] [PubMed] [Google Scholar]
- 112.Kumagai H., Miyamoto-Mikami E., Kikuchi N., Kamiya N., Zempo H., Fuku N. A rs936306 C/T Polymorphism in the CYP19A1 Is Associated with Stress Fractures. J. Strength Cond. Res. 2022;36:2322–2325. doi: 10.1519/JSC.0000000000003825. [DOI] [PubMed] [Google Scholar]
- 113.Varillas-Delgado D., Morencos E., Gutierrez-Hellín J., Aguilar-Navarro M., Maestro A., Perucho T., Coso J.D. Association of the CKM rs8111989 Polymorphism with Injury Epidemiology in Football Players. Int. J. Sport. Med. 2023;44:145–152. doi: 10.1055/a-1945-8982. [DOI] [PubMed] [Google Scholar]
- 114.Varillas-Delgado D., Gutierrez-Hellín J., Maestro A. Genetic Profile in Genes Associated with Sports Injuries in Elite Endurance Athletes. Int. J. Sport. Med. 2023;44:64–71. doi: 10.1055/a-1917-9212. [DOI] [PubMed] [Google Scholar]
- 115.Ahmetov I.I., Hall E.C.R., Semenova E.A., Pranckevičienė E., Ginevičienė V. Advances in sports genomics. Adv. Clin. Chem. 2022;107:215–263. doi: 10.1016/bs.acc.2021.07.004. [DOI] [PubMed] [Google Scholar]
- 116.Egorova E.S., Borisova A.V., Mustafina L.J., Arkhipova A.A., Gabbasov R.T., Druzhevskaya A.M., Astratenkova I.V., Ahmetov I.I. The polygenic profile of Russian football players. J. Sport. Sci. 2014;32:1286–1293. doi: 10.1080/02640414.2014.898853. [DOI] [PubMed] [Google Scholar]
- 117.Youn B.Y., Ko S.G., Kim J.Y. Genetic basis of elite combat sports athletes: A systematic review. Biol. Sport. 2021;38:667–675. doi: 10.5114/biolsport.2022.102864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Abernethy P.J., Thayer R., Taylor A.W. Acute and chronic responses of skeletal muscle to endurance and sprint exercise. A review. Sports Med. 1990;10:365–389. doi: 10.2165/00007256-199010060-00004. [DOI] [PubMed] [Google Scholar]
- 119.Simoneau J.A., Bouchard C. Genetic determinism of fiber type proportion in human skeletal muscle. FASEB J. 1995;9:1091–1095. doi: 10.1096/fasebj.9.11.7649409. [DOI] [PubMed] [Google Scholar]
- 120.Bassett D.R., Jr., Howley E.T. Limiting factors for maximum oxygen uptake and determinants of endurance performance. Med. Sci. Sport. Exerc. 2000;32:70–84. doi: 10.1097/00005768-200001000-00012. [DOI] [PubMed] [Google Scholar]
- 121.Ahmetov I.I., Vinogradova O.L., Williams A.G. Gene polymorphisms and fiber-type composition of human skeletal muscle. Int. J. Sport Nutr. Exerc. Metab. 2012;22:292–303. doi: 10.1123/ijsnem.22.4.292. [DOI] [PubMed] [Google Scholar]
- 122.Malczewska-Lenczowska J., Orysiak J., Majorczyk E., Zdanowicz R., Szczepańska B., Starczewski M., Kaczmarski J., Dybek T., Pokrywka A., Ahmetov I.I., et al. Total Hemoglobin Mass, Aerobic Capacity, and HBB Gene in Polish Road Cyclists. J. Strength Cond. Res. 2016;30:3512–3519. doi: 10.1519/JSC.0000000000001435. [DOI] [PubMed] [Google Scholar]
- 123.Semenova E.A., Khabibova S.A., Borisov O.V., Generozov E.V., Ahmetov I.I. The Variability of DNA Structure and Muscle-Fiber Composition. Hum. Physiol. 2019;45:pp. 225–232. doi: 10.1134/S0362119719010122. [DOI] [Google Scholar]
- 124.Konopka M.J., Zeegers M.P., Solberg P.A., Delhaije L., Meeusen R., Ruigrok G., Rietjens G., Sperlich B. Factors associated with high-level endurance performance: An expert consensus derived via the Delphi technique. PLoS ONE. 2022;17:e0279492. doi: 10.1371/journal.pone.0279492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Miyamoto-Mikami E., Zempo H., Fuku N., Kikuchi N., Miyachi M., Murakami H. Heritability estimates of endurance-related phenotypes: A systematic review and meta-analysis. Scand. J. Med. Sci. Sport. 2018;28:834–845. doi: 10.1111/sms.12958. [DOI] [PubMed] [Google Scholar]
- 126.Ahmetov I.I., Fedotovskaya O.N. Current Progress in Sports Genomics. Adv. Clin. Chem. 2015;70:247–314. doi: 10.1016/bs.acc.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 127.Ahmetov I.I., Egorova E.S., Gabdrakhmanova L.J., Fedotovskaya O.N. Genes and Athletic Performance: An Update. Med. Sport Sci. 2016;61:41–54. doi: 10.1159/000445240. [DOI] [PubMed] [Google Scholar]
- 128.Myerson S., Hemingway H., Budget R., Martin J., Humphries S., Montgomery H. Human angiotensin I-converting enzyme gene and endurance performance. J. Appl. Physiol. 1999;87:1313–1316. doi: 10.1152/jappl.1999.87.4.1313. [DOI] [PubMed] [Google Scholar]
- 129.Jelakovic B., Kuzmanic D., Milicic D. Influence of angiotensin converting enzyme (ACE) gene polymorphism and circadian blood pressure (BP) changes on left ventricle (LV) mass in competitive oarsmen. Am. J. Hypertens. 2000;13:182A. doi: 10.1016/S0895-7061(00)01168-7. [DOI] [Google Scholar]
- 130.Ahmetov I.I., Popov D.V., Astratenkova I.V., Druzhevskaia A.M., Missina S.S., Vinogradova O.L., Rogozkin V.A. The use of molecular genetic methods for prognosis of aerobic and anaerobic performance in athletes. Hum. Physiol. 2008;34:338–342. doi: 10.1134/S0362119708030110. [DOI] [PubMed] [Google Scholar]
- 131.Alvarez R., Terrados N., Ortolano R., Iglesias-Cubero G., Reguero J.R., Batalla A., Cortina A., Fernández-García B., Rodríguez C., Braga S., et al. Genetic variation in the renin-angiotensin system and athletic performance. Eur. J. Appl. Physiol. 2000;82:117–120. doi: 10.1007/s004210050660. [DOI] [PubMed] [Google Scholar]
- 132.Collins M., Xenophontos S.L., Cariolou M.A., Mokone G.G., Hudson D.E., Anastasiades L., Noakes T.D. The ACE gene and endurance performance during the South African Ironman Triathlons. Med. Sci. Sport. Exerc. 2004;36:1314–1320. doi: 10.1249/01.MSS.0000135779.41475.42. [DOI] [PubMed] [Google Scholar]
- 133.Lucía A., Gómez-Gallego F., Chicharro J.L., Hoyos J., Celaya K., Córdova A., Villa G., Alonso J.M., Barriopedro M., Pérez M., et al. Is there an association between ACE and CKMM polymorphisms and cycling performance status during 3-week races? Int. J. Sport. Med. 2005;26:442–447. doi: 10.1055/s-2004-821108. [DOI] [PubMed] [Google Scholar]
- 134.Hruskovicová H., Dzurenková D., Selingerová M., Bohus B., Timkanicová B., Kovács L. The angiotensin converting enzyme I/D polymorphism in long distance runners. J. Sports Med. Phys. Fitness. 2006;46:509–513. [PubMed] [Google Scholar]
- 135.Scanavini D., Bernardi F., Castoldi E., Conconi F., Mazzoni G. Increased frequency of the homozygous II ACE genotype in Italian Olympic endurance athletes. Eur. J. Hum. Genet. 2002;10:576–577. doi: 10.1038/sj.ejhg.5200852. [DOI] [PubMed] [Google Scholar]
- 136.Turgut G., Turgut S., Genc O., Atalay A., Atalay E.O. The angiotensin converting enzyme I/D polymorphism in Turkish athletes and sedentary controls. Acta Med. 2004;47:133–136. doi: 10.14712/18059694.2018.79. [DOI] [PubMed] [Google Scholar]
- 137.Tsianos G., Sanders J., Dhamrait S., Humphries S., Grant S., Montgomery H. The ACE gene insertion/deletion polymorphism and elite endurance swimming. Eur. J. Appl. Physiol. 2004;92:360–362. doi: 10.1007/s00421-004-1120-7. [DOI] [PubMed] [Google Scholar]
- 138.Cieszczyk P., Krupecki K., Maciejewska A., Sawczuk M. The angiotensin converting enzyme gene I/D polymorphism in Polish rowers. Int. J. Sport. Med. 2009;30:624–627. doi: 10.1055/s-0029-1202825. [DOI] [PubMed] [Google Scholar]
- 139.Min S.K., Takahashi K., Ishigami H., Hiranuma K., Mizuno M., Ishii T., Kim C.S., Nakazato K. Is there a gender difference between ACE gene and race distance? Appl. Physiol. Nutr. Metab. 2009;34:926–932. doi: 10.1139/H09-097. [DOI] [PubMed] [Google Scholar]
- 140.Shenoy S., Tandon S., Sandhu J., Bhanwer A.S. Association of Angiotensin Converting Enzyme gene Polymorphism and Indian Army Triathletes Performance. Asian J. Sport. Med. 2010;1:143–150. doi: 10.5812/asjsm.34855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Znazen H., Mejri A., Touhami I., Chtara M., Siala H., LE Gallais D., Ahmetov I.I., Messaoud T., Chamari K., Soussi N. Genetic advantageous predisposition of angiotensin converting enzyme id polymorphism in Tunisian athletes. J. Sport. Med. Phys. Fit. 2016;56:724–730. [PubMed] [Google Scholar]
- 142.Ash G.I., Scott R.A., Deason M., Dawson T.A., Wolde B., Bekele Z., Teka S., Pitsiladis Y.P. No association between ACE gene variation and endurance athlete status in Ethiopians. Med. Sci. Sport. Exerc. 2011;43:590–597. doi: 10.1249/MSS.0b013e3181f70bd6. [DOI] [PubMed] [Google Scholar]
- 143.Tobina T., Michishita R., Yamasawa F., Zhang B., Sasaki H., Tanaka H., Saku K., Kiyonaga A. Association between the angiotensin I-converting enzyme gene insertion/deletion polymorphism and endurance running speed in Japanese runners. J. Physiol. Sci. 2010;60:325–330. doi: 10.1007/s12576-010-0100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ahmetov I.I., Williams A.G., Popov D.V., Lyubaeva E.V., Hakimullina A.M., Fedotovskaya O.N., Mozhayskaya I.A., Vinogradova O.L., Astratenkova I.V., Montgomery H.E., et al. The combined impact of metabolic gene polymorphisms on elite endurance athlete status and related phenotypes. Hum. Genet. 2009;126:751–761. doi: 10.1007/s00439-009-0728-4. [DOI] [PubMed] [Google Scholar]
- 145.Papadimitriou I.D., Papadopoulos C., Kouvatsi A., Triantaphyllidis C. The ACE I/D polymorphism in elite Greek track and field athletes. J. Sport. Med. Phys. Fit. 2009;49:459–463. [PubMed] [Google Scholar]
- 146.Scott R.A., Moran C., Wilson R.H., Onywera V., Boit M.K., Goodwin W.H., Gohlke P., Payne J., Montgomery H., Pitsiladis Y.P. No association between Angiotensin Converting Enzyme (ACE) gene variation and endurance athlete status in Kenyans. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2005;141:169–175. doi: 10.1016/j.cbpb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 147.Rankinen T., Wolfarth B., Simoneau J.A., Maier-Lenz D., Rauramaa R., Rivera M.A., Boulay M.R., Chagnon Y.C., Pérusse L., Keul J., et al. No association between the angiotensin-converting enzyme ID polymorphism and elite endurance athlete status. J. Appl. Physiol. 2000;88:1571–1575. doi: 10.1152/jappl.2000.88.5.1571. [DOI] [PubMed] [Google Scholar]
- 148.Taylor R.R., Mamotte C.D., Fallon K., van Bockxmeer F.M. Elite athletes and the gene for angiotensin-converting enzyme. J. Appl. Physiol. 1999;87:1035–1037. doi: 10.1152/jappl.1999.87.3.1035. [DOI] [PubMed] [Google Scholar]
- 149.Orysiak J., Zmijewski P., Klusiewicz A., Kaliszewski P., Malczewska-Lenczowska J., Gajewski J., Pokrywka A. The association between ace gene variation and aerobic capacity in winter endurance disciplines. Biol. Sport. 2013;30:249–253. doi: 10.5604/20831862.1077549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ginevičienė V., Pranculis A., Jakaitienė A., Milašius K., Kučinskas V. Genetic variation of the human ACE and ACTN3 genes and their association with functional muscle properties in Lithuanian elite athletes. Medicina. 2011;47:40. doi: 10.3390/medicina47050040. [DOI] [PubMed] [Google Scholar]
- 151.Muniesa C.A., González-Freire M., Santiago C., Lao J.I., Buxens A., Rubio J.C., Martín M.A., Arenas J., Gomez-Gallego F., Lucia A. World-class performance in lightweight rowing: Is it genetically influenced? A comparison with cyclists, runners and non-athletes. Br. J. Sport. Med. 2010;44:898–901. doi: 10.1136/bjsm.2008.051680. [DOI] [PubMed] [Google Scholar]
- 152.Papadimitriou I.D., Lockey S.J., Voisin S., Herbert A.J., Garton F., Houweling P.J., Cieszczyk P., Maciejewska-Skrendo A., Sawczuk M., Massidda M., et al. No association between ACTN3 R577X and ACE I/D polymorphisms and endurance running times in 698 Caucasian athletes. BMC Genomics. 2018;19:13. doi: 10.1186/s12864-017-4412-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Varillas-Delgado D., Morencos E., Gutiérrez-Hellín J., Aguilar-Navarro M., Muñoz A., Mendoza Láiz N., Perucho T., Maestro A., Tellería-Orriols J.J. Genetic profiles to identify talents in elite endurance athletes and professional football players. PLoS ONE. 2022;17:e0274880. doi: 10.1371/journal.pone.0274880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shang X., Huang C., Chang Q., Zhang L., Huang T. Association between the ACTN3 R577X polymorphism and female endurance athletes in China. Int. J. Sport. Med. 2010;31:913–916. doi: 10.1055/s-0030-1265176. [DOI] [PubMed] [Google Scholar]
- 155.Gasser B., Flück M., Frey W.O., Valdivieso P., Spörri J. Association of Gene Variants for Mechanical and Metabolic Muscle Quality with Cardiorespiratory and Muscular Variables Related to Performance in Skiing Athletes. Genes. 2022;13:1798. doi: 10.3390/genes13101798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Orysiak J., Sitkowski D., Zmijewski P., Malczewska-Lenczowska J., Cieszczyk P., Zembron-Lacny A., Pokrywka A. Overrepresentation of the ACTN3 XX genotype in elite canoe and kayak paddlers. J. Strength Cond. Res. 2015;29:1107–1112. doi: 10.1519/JSC.0000000000000717. [DOI] [PubMed] [Google Scholar]
- 157.Ahmetov I.I., Druzhevskaya A.M., Astratenkova I.V., Popov D.V., Vinogradova O.L., Rogozkin V.A. The ACTN3 R577X polymorphism in Russian endurance athletes. Br. J. Sport. Med. 2010;44:649–652. doi: 10.1136/bjsm.2008.051540. [DOI] [PubMed] [Google Scholar]
- 158.Döring F.E., Onur S., Geisen U., Boulay M.R., Pérusse L., Rankinen T., Rauramaa R., Wolfahrt B., Bouchard C. ACTN3 R577X and other polymorphisms are not associated with elite endurance athlete status in the Genathlete study. J. Sport. Sci. 2010;28:1355–1359. doi: 10.1080/02640414.2010.507675. [DOI] [PubMed] [Google Scholar]
- 159.Ginevičienė V., Pranckevičienė E., Milašius K., Kučinskas V. Relating fitness phenotypes to genotypes in Lithuanian elite athletes. Acta Med. Litu. 2010;17:1–10. doi: 10.2478/v10140-010-0001-0. [DOI] [Google Scholar]
- 160.Tsianos G.I., Evangelou E., Boot A., Zillikens M.C., van Meurs J.B., Uitterlinden A.G., Ioannidis J.P. Associations of polymorphisms of eight muscle- or metabolism-related genes with performance in Mount Olympus marathon runners. J. Appl. Physiol. 2010;108:567–574. doi: 10.1152/japplphysiol.00780.2009. [DOI] [PubMed] [Google Scholar]
- 161.Niemi A.K., Majamaa K. Mitochondrial DNA and ACTN3 genotypes in Finnish elite endurance and sprint athletes. Eur. J. Hum. Genet. 2005;13:965–969. doi: 10.1038/sj.ejhg.5201438. [DOI] [PubMed] [Google Scholar]
- 162.Papadimitriou I.D., Papadopoulos C., Kouvatsi A., Triantaphyllidis C. The ACTN3 gene in elite Greek track and field athletes. Int. J. Sport. Med. 2008;29:352–355. doi: 10.1055/s-2007-965339. [DOI] [PubMed] [Google Scholar]
- 163.Paparini A., Ripani M., Giordano G.D., Santoni D., Pigozzi F., Romano-Spica V. ACTN3 genotyping by real-time PCR in the Italian population and athletes. Med. Sci. Sport. Exerc. 2007;39:810–815. doi: 10.1097/mss.0b013e3180317491. [DOI] [PubMed] [Google Scholar]
- 164.Saunders C.J., September A.V., Xenophontos S.L., Cariolou M.A., Anastassiades L.C., Noakes T.D., Collins M. No association of the ACTN3 gene R577X polymorphism with endurance performance in Ironman Triathlons. Ann. Hum. Genet. 2007;71:777–781. doi: 10.1111/j.1469-1809.2006.00385.x. [DOI] [PubMed] [Google Scholar]
- 165.Yang N., MacArthur D.G., Wolde B., Onywera V.O., Boit M.K., Lau S.Y., Wilson R.H., Scott R.A., Pitsiladis Y.P., North K. The ACTN3 R577X polymorphism in East and West African athletes. Med. Sci. Sports Exerc. 2007;39:1985–1988. doi: 10.1249/mss.0b013e31814844c9. [DOI] [PubMed] [Google Scholar]
- 166.Lucia A., Gómez-Gallego F., Santiago C., Bandrés F., Earnest C., Rabadán M., Alonso J.M., Hoyos J., Córdova A., Villa G., et al. ACTN3 genotype in professional endurance cyclists. Int. J. Sports Med. 2006;27:880–884. doi: 10.1055/s-2006-923862. [DOI] [PubMed] [Google Scholar]
- 167.Grealy R., Smith C.L., Chen T., Hiller D., Haseler L.J., Griffiths L.R. The genetics of endurance: Frequency of the ACTN3 R577X variant in Ironman World Championship athletes. J. Sci. Med. Sport. 2013;16:365–371. doi: 10.1016/j.jsams.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 168.Mikami E., Fuku N., Murakami H., Tsuchie H., Takahashi H., Ohiwa N., Tanaka H., Pitsiladis Y.P., Higuchi M., Miyachi M., et al. ACTN3 R577X genotype is associated with sprinting in elite Japanese athletes. Int. J. Sport. Med. 2014;35:172–177. doi: 10.1055/s-0033-1347171. [DOI] [PubMed] [Google Scholar]
- 169.Wang G., Mikami E., Chiu L.L., DE Perini A., Deason M., Fuku N., Miyachi M., Kaneoka K., Murakami H., Tanaka M., et al. Association analysis of ACE and ACTN3 in elite Caucasian and East Asian swimmers. Med. Sci. Sport. Exerc. 2013;45:892–900. doi: 10.1249/MSS.0b013e31827c501f. [DOI] [PubMed] [Google Scholar]
- 170.Yang R., Jin F., Wang L., Shen X., Guo Q., Song H., Hu J., Zhao Q., Wan J., Cai M. Prediction and Identification of Power Performance Using Polygenic Models of Three Single-Nucleotide Polymorphisms in Chinese Elite Athletes. Front. Genet. 2021;12:726552. doi: 10.3389/fgene.2021.726552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wagoner L.E., Craft L.L., Singh B., Suresh D.P., Zengel P.W., McGuire N., Abraham W.T., Chenier T.C., Dorn G.W., 2nd, Liggett S.B. Polymorphisms of the β(2)-adrenergic receptor determine exercise capacity in patients with heart failure. Circ. Res. 2000;86:834–840. doi: 10.1161/01.RES.86.8.834. [DOI] [PubMed] [Google Scholar]
- 172.Wolfarth B., Rankinen T., Mühlbauer S., Scherr J., Boulay M.R., Pérusse L., Rauramaa R., Bouchard C. Association between a beta2-adrenergic receptor polymorphism and elite endurance performance. Metabolism. 2007;56:1649–1651. doi: 10.1016/j.metabol.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 173.Santiago C., Ruiz J.R., Buxens A., Artieda M., Arteta D., González-Freire M., Rodríguez-Romo G., Altmäe S., Lao J.I., Gómez-Gallego F., et al. Trp64Arg polymorphism in ADRB3 gene is associated with elite endurance performance. Br. J. Sport. Med. 2011;45:147–149. doi: 10.1136/bjsm.2009.061366. [DOI] [PubMed] [Google Scholar]
- 174.Sawczuk M., Maciejewska-Karlowska A., Cieszczyk P., Skotarczak B., Ficek K. Association of the ADRB2 Gly16Arg and Glu27Gln polymorphisms with athlete status. J. Sports Sci. 2013;31:1535–1544. doi: 10.1080/02640414.2013.786184. [DOI] [PubMed] [Google Scholar]
- 175.Moore G.E., Shuldiner A.R., Zmuda J.M., Ferrell R.E., McCole S.D., Hagberg J.M. Obesity gene variant and elite endurance performance. Metabolism. 2001;50:1391–1392. doi: 10.1053/meta.2001.28140. [DOI] [PubMed] [Google Scholar]
- 176.Mustafina L.J., Naumov V.A., Cieszczyk P., Popov D.V., Lyubaeva E.V., Kostryukova E.S., Fedotovskaya O.N., Druzhevskaya A.M., Astratenkova I.V., Glotov A.S., et al. AGTR2 gene polymorphism is associated with muscle fibre composition, athletic status and aerobic performance. Exp. Physiol. 2014;99:1042–1052. doi: 10.1113/expphysiol.2014.079335. [DOI] [PubMed] [Google Scholar]
- 177.Guilherme J.P.L.F., Silva M.S., Bertuzzi R., Lancha Junior A.H. The AGTR2 rs11091046 (A>C) polymorphism and power athletic status in top-level Brazilian athletes. J. Sport. Sci. 2018;36:2327–2332. doi: 10.1080/02640414.2018.1455260. [DOI] [PubMed] [Google Scholar]
- 178.Martínez J.L., Carrión A., Florián M.E., Martín J.A., López-Taylor J.R., Fahey T.D., Rivera M.A. Aquaporin-1 gene DNA variation predicts performance in Hispanic marathon runners. Med. Sport. 2009;13:251–255. doi: 10.2478/v10036-009-0039-9. [DOI] [Google Scholar]
- 179.Rivera M.A., Martínez J.L., Carrion A., Fahey T.D. AQP-1 association with body fluid loss in 10-km runners. Int. J. Sport. Med. 2011;32:229–233. doi: 10.1055/s-0030-1268489. [DOI] [PubMed] [Google Scholar]
- 180.Saunders C.J., Posthumus M., O’Connell K., September A.V., Collins M. A variant within the AQP1 3′-untranslated region is associated with running performance, but not weight changes, during an Ironman Triathlon. J. Sports Sci. 2015;33:1342–1348. doi: 10.1080/02640414.2014.989535. [DOI] [PubMed] [Google Scholar]
- 181.Rico-Sanz J., Rankinen T., Joanisse D.R., Leon A.S., Skinner J.S., Wilmore J.H., Rao D.C., Bouchard C., HERITAGE Family study Associations between cardiorespiratory responses to exercise and the C34T AMPD1 gene polymorphism in the HERITAGE Family Study. Physiol. Genom. 2003;14:161–166. doi: 10.1152/physiolgenomics.00165.2002. [DOI] [PubMed] [Google Scholar]
- 182.Thomaes T., Thomis M., Onkelinx S., Fagard R., Matthijs G., Buys R., Schepers D., Cornelissen V., Vanhees L. A genetic predisposition score for muscular endophenotypes predicts the increase in aerobic power after training: The CAREGENE study. BMC Genet. 2011;12:84. doi: 10.1186/1471-2156-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ciȩszczyk P., Eider J., Ostanek M., Leońska-Duniec A., Ficek K., Kotarska K., Girdauskas G. Is the C34T polymorphism of the AMPD1 gene associated with athlete performance in rowing? Int. J. Sport. Med. 2011;32:987–991. doi: 10.1055/s-0031-1283186. [DOI] [PubMed] [Google Scholar]
- 184.Ginevičienė V., Jakaitienė A., Pranculis A., Milašius K., Tubelis L., Utkus A. AMPD1 rs17602729 is associated with physical performance of sprint and power in elite Lithuanian athletes. BMC Genet. 2014;15:58. doi: 10.1186/1471-2156-15-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Williams A.G., Dhamrait S.S., Wootton P.T., Day S.H., Hawe E., Payne J.R., Myerson S.G., World M., Budgett R., Humphries S.E., et al. Bradykinin receptor gene variant and human physical performance. J. Appl. Physiol. 2004;96:938–942. doi: 10.1152/japplphysiol.00865.2003. [DOI] [PubMed] [Google Scholar]
- 186.Saunders C.J., Xenophontos S.L., Cariolou M.A., Anastassiades L.C., Noakes T.D., Collins M. The bradykinin β 2 receptor (BDKRB2) and endothelial nitric oxide synthase 3 (NOS3) genes and endurance performance during Ironman Triathlons. Hum. Mol. Genet. 2006;15:979–987. doi: 10.1093/hmg/ddl014. [DOI] [PubMed] [Google Scholar]
- 187.Sawczuk M., Timshina Y.I., Astratenkova I.V., Maciejewska-Karlowska A., Leońska-Duniec A., Ficek K., Mustafina L.J., Cięszczyk P., Klocek T., Ahmetov I.I. The −9/+9 polymorphism of the bradykinin receptor β 2 gene and athlete status: A study involving two European cohorts. Hum. Biol. 2013;85:741–756. doi: 10.3378/027.085.0511. [DOI] [PubMed] [Google Scholar]
- 188.Grenda A., Leońska-Duniec A., Cięszczyk P., Zmijewski P. Bdkrb2 gene -9/+9 polymorphism and swimming performance. Biol. Sport. 2014;31:109–113. doi: 10.5604/20831862.1096047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zmijewski P., Grenda A., Leońska-Duniec A., Ahmetov I., Orysiak J., Cięszczyk P. Effect of BDKRB2 Gene -9/+9 Polymorphism on Training Improvements in Competitive Swimmers. J. Strength Cond. Res. 2016;30:665–671. doi: 10.1519/JSC.0000000000001145. [DOI] [PubMed] [Google Scholar]
- 190.Rivera M.A., Dionne F.T., Simoneau J.A., Pérusse L., Chagnon M., Chagnon Y., Gagnon J., Leon A.S., Rao D.C., Skinner J.S., et al. Muscle-specific creatine kinase gene polymorphism and VO2max in the HERITAGE Family Study. Med. Sci. Sport. Exerc. 1997;29:1311–1317. doi: 10.1097/00005768-199710000-00006. [DOI] [PubMed] [Google Scholar]
- 191.Rivera M.A., Pérusse L., Simoneau J.A., Gagnon J., Dionne F.T., Leon A.S., Skinner J.S., Wilmore J.H., Province M., Rao D.C., et al. Linkage between a muscle-specific CK gene marker and VO2max in the HERITAGE Family Study. Med. Sci. Sport. Exerc. 1999;31:698–701. doi: 10.1097/00005768-199905000-00012. [DOI] [PubMed] [Google Scholar]
- 192.Fedotovskaya O.N., Popov D.V., Vinogradova O.L., Akhmetov I.I. Association of muscle-specific creatine kinase (CKMM) gene polymorphism with physical performance of athletes. Hum. Physiol. 2012;38:89–93. doi: 10.1134/S0362119712010082. [DOI] [PubMed] [Google Scholar]
- 193.Martínez J.L., Khorsandi S., Sojo R., Martínez C., Martín J.A., López-Taylor J.R., Fahey T.D., Rivera M.A. Lack of an Association Between CKMM Genotype and Endurance Performance Level in Hispanic Marathon Runners. Med. Sport. 2009;13:219–223. doi: 10.2478/v10036-009-0034-1. [DOI] [Google Scholar]
- 194.Posthumus M., Schwellnus M.P., Collins M. The COL5A1 gene: A novel marker of endurance running performance. Med. Sci. Sport. Exerc. 2011;43:584–589. doi: 10.1249/MSS.0b013e3181f34f4d. [DOI] [PubMed] [Google Scholar]
- 195.Brown J.C., Miller C.J., Posthumus M., Schwellnus M.P., Collins M. The COL5A1 gene, ultra-marathon running performance, and range of motion. Int. J. Sport. Physiol. Perform. 2011;6:485–496. doi: 10.1123/ijspp.6.4.485. [DOI] [PubMed] [Google Scholar]
- 196.Guilherme J., Egorova E.S., Semenova E.A., Kostryukova E.S., Kulemin N.A., Borisov O.V., Khabibova S.A., Larin A.K., Ospanova E.A., Pavlenko A.V., et al. The A-allele of the FTO gene rs9939609 polymorphism is associated with decreased proportion of slow oxidative muscle fibers and over-represented in heavier athletes. J. Strength Cond. Res. 2019;33:691–700. doi: 10.1519/JSC.0000000000003032. [DOI] [PubMed] [Google Scholar]
- 197.Zmijewski P., Leońska-Duniec A. Association between the FTO A/T Polymorphism and Elite Athlete Status in Caucasian Swimmers. Genes. 2021;12:715. doi: 10.3390/genes12050715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Eynon N., Nasibulina E.S., Banting L.K., Cieszczyk P., Maciejewska-Karlowska A., Sawczuk M., Bondareva E.A., Shagimardanova R.R., Raz M., Sharon Y., et al. The FTO A/T polymorphism and elite athletic performance: A study involving three groups of European athletes. PLoS ONE. 2013;8:e60570. doi: 10.1371/journal.pone.0060570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.He Z., Hu Y., Feng L., Lu Y., Liu G., Xi Y., Wen L., McNaughton L.R. NRF2 genotype improves endurance capacity in response to training. Int. J. Sport. Med. 2007;28:717–721. doi: 10.1055/s-2007-964913. [DOI] [PubMed] [Google Scholar]
- 200.Eynon N., Ruiz J.R., Bishop D.J., Santiago C., Gómez-Gallego F., Lucia A., Birk R. The rs12594956 polymorphism in the NRF-2 gene is associated with top-level Spanish athlete’s performance status. J. Sci. Med. Sport. 2013;16:135–139. doi: 10.1016/j.jsams.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 201.Maciejewska-Karłowska A., Leońska-Duniec A., Cięszczyk P., Sawczuk M., Eider J., Ficek K., Sawczyn S. The GABPB1 gene A/G polymorphism in Polish rowers. J. Hum. Kinet. 2012;31:115–120. doi: 10.2478/v10078-012-0012-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zarebska A., Jastrzebski Z., Kaczmarczyk M., Ficek K., Maciejewska-Karlowska A., Sawczuk M., Leońska-Duniec A., Krol P., Cieszczyk P., Zmijewski P., et al. The GSTP1 c.313A>G polymorphism modulates the cardiorespiratory response to aerobic training. Biol. Sport. 2014;31:261–266. doi: 10.5604/20831862.1120932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zarebska A., Jastrzebski Z., Ahmetov I.I., Zmijewski P., Cieszczyk P., Leonska-Duniec A., Sawczuk M., Leznicka K., Trybek G., Semenova E.A., et al. GSTP1 c.313A>G polymorphism in Russian and Polish athletes. Physiol. Genom. 2017;49:127–131. doi: 10.1152/physiolgenomics.00014.2016. [DOI] [PubMed] [Google Scholar]
- 204.Deugnier Y., Loréal O., Carré F., Duvallet A., Zoulim F., Vinel J.P., Paris J.C., Blaison D., Moirand R., Turlin B., et al. Increased body iron stores in elite road cyclists. Med. Sci. Sport. Exerc. 2002;34:876–880. doi: 10.1097/00005768-200205000-00023. [DOI] [PubMed] [Google Scholar]
- 205.Chicharro J.L., Hoyos J., Gómez-Gallego F., Villa J.G., Bandrés F., Celaya P., Jiménez F., Alonso J.M., Córdova A., Lucia A. Mutations in the hereditary haemochromatosis gene HFE in professional endurance athletes. Br. J. Sport. Med. 2004;38:418–421. doi: 10.1136/bjsm.2002.003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hermine O., Dine G., Genty V., Marquet L.A., Fumagalli G., Tafflet M., Guillem F., Van Lierde F., Rousseaux-Blanchi M.P., Palierne C., et al. Eighty percent of French sport winners in Olympic, World and Europeans competitions have mutations in the hemochromatosis HFE gene. Biochimie. 2015;119:1–5. doi: 10.1016/j.biochi.2015.09.028. [DOI] [PubMed] [Google Scholar]
- 207.Semenova E.A., Miyamoto-Mikami E., Akimov E.B., Al-Khelaifi F., Murakami H., Zempo H., Kostryukova E.S., Kulemin N.A., Larin A.K., Borisov O.V., et al. The association of HFE gene H63D polymorphism with endurance athlete status and aerobic capacity: Novel findings and a meta-analysis. Eur. J. Appl. Physiol. 2020;120:665–673. doi: 10.1007/s00421-020-04306-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Prior S.J., Hagberg J.M., Phares D.A., Brown M.D., Fairfull L., Ferrell R.E., Roth S.M. Sequence variation in hypoxia-inducible factor 1alpha (HIF1A): Association with maximal oxygen consumption. Physiol. Genom. 2003;15:20–26. doi: 10.1152/physiolgenomics.00061.2003. [DOI] [PubMed] [Google Scholar]
- 209.Döring F., Onur S., Fischer A., Boulay M.R., Pérusse L., Rankinen T., Rauramaa R., Wolfarth B., Bouchard C. A common haplotype and the Pro582Ser polymorphism of the hypoxia-inducible factor-1alpha (HIF1A) gene in elite endurance athletes. J. Appl. Physiol. 2010;108:1497–1500. doi: 10.1152/japplphysiol.01165.2009. [DOI] [PubMed] [Google Scholar]
- 210.Bosnyák E., Trájer E., Alszászi G., Móra Á., Györe I., Udvardy A., Tóth M., Szmodis M. Lack of association between the GNB3 rs5443, HIF1A rs11549465 polymorphisms, physiological and functional characteristics. Ann. Hum. Genet. 2020;84:393–399. doi: 10.1111/ahg.12387. [DOI] [PubMed] [Google Scholar]
- 211.Merezhinskaya N., Fishbein W.N., Davis J.I., Foellmer J.W. Mutations in MCT1 cDNA in patients with symptomatic deficiency in lactate transport. Muscle Nerve. 2000;23:90–97. doi: 10.1002/(SICI)1097-4598(200001)23:1<90::AID-MUS12>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 212.Cupeiro R., Benito P.J., Maffulli N., Calderón F.J., González-Lamuño D. MCT1 genetic polymorphism influence in high intensity circuit training: A pilot study. J. Sci. Med. Sport. 2010;13:526–530. doi: 10.1016/j.jsams.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 213.Fedotovskaya O.N., Mustafina L.J., Popov D.V., Vinogradova O.L., Ahmetov I.I. A common polymorphism of the MCT1 gene and athletic performance. Int. J. Sport. Physiol. Perform. 2014;9:173–180. doi: 10.1123/ijspp.2013-0026. [DOI] [PubMed] [Google Scholar]
- 214.Ramírez de la Piscina-Viúdez X., Álvarez-Herms J., Bonilla D.A., Castañeda-Babarro A., Larruskain J., Díaz-Ramírez J., Ahmetov I.I., Martínez-Ascensión A., Kreider R.B., Odriozola-Martínez A. Putative Role of MCT1 rs1049434 Polymorphism in High-Intensity Endurance Performance: Concept and Basis to Understand Possible Individualization Stimulus. Sports. 2021;9:143. doi: 10.3390/sports9100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sawczuk M., Banting L.K., Cięszczyk P., Maciejewska-Karłowska A., Zarębska A., Leońska-Duniec A., Jastrzębski Z., Bishop D.J., Eynon N. MCT1 A1470T: A novel polymorphism for sprint performance? J. Sci. Med. Sport. 2015;18:114–118. doi: 10.1016/j.jsams.2013.12.008. [DOI] [PubMed] [Google Scholar]
- 216.Maruszak A., Adamczyk J.G., Siewierski M., Sozański H., Gajewski A., Żekanowski C. Mitochondrial DNA variation is associated with elite athletic status in the Polish population. Scand. J. Med. Sci. Sport. 2014;24:311–318. doi: 10.1111/sms.12012. [DOI] [PubMed] [Google Scholar]
- 217.Drozdovska S.B., Dosenko V.E., Ilyin V.N., Filippov M.M., Kuzmina L.M. Allelic Polymorphism of Endothelial No-Synthase (eNOS) Association with Exercise-Induced Hypoxia Adaptation. Baltic. J. Health Phys. Activ. 2009;1:13–19. doi: 10.2478/v10131-009-0001-1. [DOI] [Google Scholar]
- 218.Zmijewski P., Cięszczyk P., Ahmetov I.I., Gronek P., Lulińska-Kuklik E., Dornowski M., Rzeszutko A., Chycki J., Moska W., Sawczuk M. The NOS3 G894T (rs1799983) and -786T/C (rs2070744) polymorphisms are associated with elite swimmer status. Biol. Sport. 2018;35:313–319. doi: 10.5114/biolsport.2018.76528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gómez-Gallego F., Ruiz J.R., Buxens A., Artieda M., Arteta D., Santiago C., Rodríguez-Romo G., Lao J.I., Lucia A. The -786 T/C polymorphism of the NOS3 gene is associated with elite performance in power sports. Eur. J. Appl. Physiol. 2009;107:565–569. doi: 10.1007/s00421-009-1166-7. [DOI] [PubMed] [Google Scholar]
- 220.Ahmetov I.I., Mozhayskaya I.A., Flavell D.M., Astratenkova I.V., Komkova A.I., Lyubaeva E.V., Tarakin P.P., Shenkman B.S., Vdovina A.B., Netreba A.I., et al. PPARalpha gene variation and physical performance in Russian athletes. Eur. J. Appl. Physiol. 2006;97:103–108. doi: 10.1007/s00421-006-0154-4. [DOI] [PubMed] [Google Scholar]
- 221.Maciejewska A., Sawczuk M., Cięszczyk P. Variation in the PPARα gene in Polish rowers. J. Sci. Med. Sport. 2011;14:58–64. doi: 10.1016/j.jsams.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 222.Tural E., Kara N., Agaoglu S.A., Elbistan M., Tasmektepligil M.Y., Imamoglu O. PPAR-α and PPARGC1A gene variants have strong effects on aerobic performance of Turkish elite endurance athletes. Mol. Biol. Rep. 2014;41:5799–5804. doi: 10.1007/s11033-014-3453-6. [DOI] [PubMed] [Google Scholar]
- 223.Maciejewska A., Sawczuk M., Cieszczyk P., Mozhayskaya I.A., Ahmetov I.I. The PPARGC1A gene Gly482Ser in Polish and Russian athletes. J. Sport. Sci. 2012;30:101–113. doi: 10.1080/02640414.2011.623709. [DOI] [PubMed] [Google Scholar]
- 224.He Z.H., Hu Y., Li Y.C., Gong L.J., Cieszczyk P., Maciejewska-Karlowska A., Leonska-Duniec A., Muniesa C.A., Marín-Peiro M., Santiago C., et al. PGC-related gene variants and elite endurance athletic status in a Chinese cohort: A functional study. Scand. J. Med. Sci. Sport. 2015;25:184–195. doi: 10.1111/sms.12188. [DOI] [PubMed] [Google Scholar]
- 225.Hall E.C.R., Lockey S.J., Heffernan S.M., Herbert A.J., Stebbings G.K., Day S.H., Collins M., Pitsiladis Y.P., Erskine R.M., Williams A.G. The PPARGC1A Gly482Ser polymorphism is associated with elite long-distance running performance. J. Sport. Sci. 2023;41:56–62. doi: 10.1080/02640414.2023.2195737. [DOI] [PubMed] [Google Scholar]
- 226.Akhmetov I.I., Popov D.V., Missina S.S., Vinogradova O.L., Rogozkin V.A. The analysis of PPARGC1B gene polymorphism in athletes. Ross. Fiziol. Zh. Im. I.M. Sechenova. 2009;95:1247–1253. [PubMed] [Google Scholar]
- 227.Akhmetov I.I., Linde E.V., Shikhova I.V., Popov D.V., Missina S.S., Vinogradoba O.L., Rogozkin V.A. The influence of calcineurin gene polymorphism on morphofunctional characteristics of cardiovascular system of athletes. Ross. Fiziol. Zh. Im. I.M. Sechenova. 2008;94:915–922. [PubMed] [Google Scholar]
- 228.Bouchard C., Sarzynski M.A., Rice T.K., Kraus W.E., Church T.S., Sung Y.J., Rao D.C., Rankinen T. Genomic predictors of the maximal O₂ uptake response to standardized exercise training programs. J. Appl. Physiol. 2011;110:1160–1170. doi: 10.1152/japplphysiol.00973.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Hall E.C.R., Almeida S.S., Heffernan S.M., Lockey S.J., Herbert A.J., Callus P., Day S.H., Pedlar C.R., Kipps C., Collins M., et al. Genetic Polymorphisms Related to VO2max Adaptation Are Associated with Elite Rugby Union Status and Competitive Marathon Performance. Int. J. Sport. Physiol. Perform. 2021;16:1858–1864. doi: 10.1123/ijspp.2020-0856. [DOI] [PubMed] [Google Scholar]
- 230.Kusić D., Connolly J., Kainulainen H., Semenova E.A., Borisov O.V., Larin A.K., Popov D.V., Generozov E.V., Ahmetov I.I., Britton S.L., et al. Striated muscle-specific serine/threonine-protein kinase β segregates with high versus low responsiveness to endurance exercise training. Physiol. Genom. 2020;52:35–46. doi: 10.1152/physiolgenomics.00103.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ahmetov I.I., Popov D.V., Missina S.S., Vinogradova O.L., Rogozkin V.A. Association of mitochondrial transcription factor (TFAM) gene polymorphism with physical performance in athletes. Hum. Physiol. 2010;36:229–233. doi: 10.1134/S0362119710020155. [DOI] [PubMed] [Google Scholar]
- 232.Gronek P., Gronek J., Lulińska-Kuklik E., Spieszny M., Niewczas M., Kaczmarczyk M., Petr M., Fischerova P., Ahmetov I.I., Żmijewski P. Polygenic Study of Endurance-Associated Genetic Markers NOS3 (Glu298Asp), BDKRB2 (-9/+9), UCP2 (Ala55Val), AMPD1 (Gln45Ter) and ACE (I/D) in Polish Male Half Marathoners. J. Hum. Kinet. 2018;64:87–98. doi: 10.1515/hukin-2017-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Hudson D.E., Mokone G.G., Noakes T.D., Collins M. The -55 C/T polymorphism within the UCP3 gene and performance during the South African Ironman Triathlon. Int. J. Sport. Med. 2004;25:427–432. doi: 10.1055/s-2004-815850. [DOI] [PubMed] [Google Scholar]
- 234.Prior S.J., Hagberg J.M., Paton C.M., Douglass L.W., Brown M.D., McLenithan J.C., Roth S.M. DNA sequence variation in the promoter region of the VEGF gene impacts VEGF gene expression and maximal oxygen consumption. Am. J. Physiol. Heart Circ. Physiol. 2006;290:H1848–H1855. doi: 10.1152/ajpheart.01033.2005. [DOI] [PubMed] [Google Scholar]
- 235.Ahmetov I.I., Khakimullina A.M., Popov D.V., Missina S.S., Vinogradova O.L., Rogozkin V.A. Polymorphism of the vascular endothelial growth factor gene (VEGF) and aerobic performance in athletes. Hum. Physiol. 2008;34:477–481. doi: 10.1134/S0362119708040129. [DOI] [PubMed] [Google Scholar]
- 236.Ahmetov I.I., Hakimullina A.M., Popov D.V., Lyubaeva E.V., Missina S.S., Vinogradova O.L., Williams A.G., Rogozkin V.A. Association of the VEGFR2 gene His472Gln polymorphism with endurance-related phenotypes. Eur. J. Appl. Physiol. 2009;107:95–103. doi: 10.1007/s00421-009-1105-7. [DOI] [PubMed] [Google Scholar]
- 237.Eider J., Leonska-Duniec A., Maciejewska-Karlowska A., Sawczuk M., Ficek K., Sawczyn S. The VEGFR2 gene His472Gln polymorphism in Polish endurance athletes. Int. SportMed. J. 2013;14:29–35. [Google Scholar]
- 238.Kumagai K., Abe T., Brechue W.F., Ryushi T., Takano S., Mizuno M. Sprint performance is related to muscle fascicle length in male 100-m sprinters. J. Appl. Physiol. 2000;88:811–816. doi: 10.1152/jappl.2000.88.3.811. [DOI] [PubMed] [Google Scholar]
- 239.Weyand P.G., Davis J.A. Running performance has a structural basis. J. Exp. Biol. 2005;208:2625–2631. doi: 10.1242/jeb.01609. [DOI] [PubMed] [Google Scholar]
- 240.Uth N. Anthropometric comparison of world-class sprinters and normal populations. J. Sport. Sci. Med. 2005;4:608–616. [PMC free article] [PubMed] [Google Scholar]
- 241.Tønnessen E., Haugen T., Shalfawi S.A. Reaction time aspects of elite sprinters in athletic world championships. J. Strength Cond. Res. 2013;27:885–892. doi: 10.1519/JSC.0b013e31826520c3. [DOI] [PubMed] [Google Scholar]
- 242.Haugen T., Seiler S., Sandbakk Ø., Tønnessen E. The Training and Development of Elite Sprint Performance: An Integration of Scientific and Best Practice Literature. Sport. Med. Open. 2019;5:44. doi: 10.1186/s40798-019-0221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Suga T., Terada M., Tanaka T., Miyake Y., Ueno H., Otsuka M., Nagano A., Isaka T. Calcaneus height is a key morphological factor of sprint performance in sprinters. Sci. Rep. 2020;10:15425. doi: 10.1038/s41598-020-72388-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hall E.C.R., Lysenko E.A., Semenova E.A., Borisov O.V., Andryushchenko O.N., Andryushchenko L.B., Vepkhvadze T.F., Lednev E.M., Zmijewski P., Popov D.V., et al. Prediction of muscle fiber composition using multiple repetition testing. Biol. Sport. 2021;38:277–283. doi: 10.5114/biolsport.2021.99705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Hughes D.C., Day S.H., Ahmetov I.I., Williams A.G. Genetics of muscle strength and power: Polygenic profile similarity limits skeletal muscle performance. J. Sport. Sci. 2011;29:1425–1434. doi: 10.1080/02640414.2011.597773. [DOI] [PubMed] [Google Scholar]
- 246.Zempo H., Miyamoto-Mikami E., Kikuchi N., Fuku N., Miyachi M., Murakami H. Heritability estimates of muscle strength-related phenotypes: A systematic review and meta-analysis. Scand. J. Med. Sci. Sport. 2017;27:1537–1546. doi: 10.1111/sms.12804. [DOI] [PubMed] [Google Scholar]
- 247.Woods D., Hickman M., Jamshidi Y., Brull D., Vassiliou V., Jones A., Humphries S., Montgomery H. Elite swimmers and the D allele of the ACE I/D polymorphism. Hum. Genet. 2001;108:230–232. doi: 10.1007/s004390100466. [DOI] [PubMed] [Google Scholar]
- 248.Costa A.M., Silva A.J., Garrido N.D., Louro H., de Oliveira R.J., Breitenfeld L. Association between ACE D allele and elite short distance swimming. Eur. J. Appl. Physiol. 2009;106:785–790. doi: 10.1007/s00421-009-1080-z. [DOI] [PubMed] [Google Scholar]
- 249.Boraita A., de la Rosa A., Heras M.E., de la Torre A.I., Canda A., Rabadán M., Díaz A.E., González C., López M., Hernández M. Cardiovascular adaptation, functional capacity and Angiotensin-converting enzyme I/D polymorphism in elite athletes. Rev. Esp. Cardiol. 2010;63:810–819. doi: 10.1016/S0300-8932(10)70184-6. [DOI] [PubMed] [Google Scholar]
- 250.Papadimitriou I.D., Lucia A., Pitsiladis Y.P., Pushkarev V.P., Dyatlov D.A., Orekhov E.F., Artioli G.G., Guilherme J.P., Lancha A.H., Jr., Ginevičienė V., et al. ACTN3 R577X and ACE I/D gene variants influence performance in elite sprinters: A multi-cohort study. BMC Genom. 2016;17:285. doi: 10.1186/s12864-016-2462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Amir O., Amir R., Yamin C., Attias E., Eynon N., Sagiv M., Sagiv M., Meckel Y. The ACE deletion allele is associated with Israeli elite endurance athletes. Exp. Physiol. 2007;92:881–886. doi: 10.1113/expphysiol.2007.038711. [DOI] [PubMed] [Google Scholar]
- 252.Kim C.H., Cho J.Y., Jeon J.Y., Koh Y.G., Kim Y.M., Kim H.J., Park M., Um H.S., Kim C. ACE DD genotype is unfavorable to Korean short-term muscle power athletes. Int. J. Sport. Med. 2010;31:65–71. doi: 10.1055/s-0029-1239523. [DOI] [PubMed] [Google Scholar]
- 253.Shahmoradi S., Ahmadalipour A., Salehi M. Evaluation of ACE gene I/D polymorphism in Iranian elite athletes. Adv. Biomed. Res. 2014;3:207. doi: 10.4103/2277-9175.143242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Scott R.A., Irving R., Irwin L., Morrison E., Charlton V., Austin K., Tladi D., Deason M., Headley S.A., Kolkhorst F.W., et al. ACTN3 and ACE genotypes in elite Jamaican and US sprinters. Med. Sci. Sport. Exerc. 2010;42:107–112. doi: 10.1249/MSS.0b013e3181ae2bc0. [DOI] [PubMed] [Google Scholar]
- 255.Windelinckx A., De Mars G., Huygens W., Peeters M.W., Vincent B., Wijmenga C., Lambrechts D., Delecluse C., Roth S.M., Metter E.J., et al. Comprehensive fine mapping of chr12q12-14 and follow-up replication identify activin receptor 1B (ACVR1B) as a muscle strength gene. Eur. J. Hum. Genet. 2011;19:208–215. doi: 10.1038/ejhg.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Voisin S., Guilherme J.P., Yan X., Pushkarev V.P., Cieszczyk P., Massidda M., Calò C.M., Dyatlov D.A., Kolupaev V.A., Pushkareva Y.E., et al. ACVR1B rs2854464 Is Associated with Sprint/Power Athletic Status in a Large Cohort of Europeans but Not Brazilians. PLoS ONE. 2016;11:e0156316. doi: 10.1371/journal.pone.0156316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Venckunas T., Degens H. Genetic polymorphisms of muscular fitness in young healthy men. PLoS ONE. 2022;17:e0275179. doi: 10.1371/journal.pone.0275179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Druzhevskaya A.M., Ahmetov I.I., Astratenkova I.V., Rogozkin V.A. Association of the ACTN3 R577X polymorphism with power athlete status in Russians. Eur. J. Appl. Physiol. 2008;103:631–634. doi: 10.1007/s00421-008-0763-1. [DOI] [PubMed] [Google Scholar]
- 259.Massidda M., Vona G., Calò C.M. Association between the ACTN3 R577X polymorphism and artistic gymnastic performance in Italy. Genet. Test. Mol. Biomark. 2009;13:377–380. doi: 10.1089/gtmb.2008.0157. [DOI] [PubMed] [Google Scholar]
- 260.Chiu L.L., Wu Y.F., Tang M.T., Yu H.C., Hsieh L.L., Hsieh S.S. ACTN3 genotype and swimming performance in Taiwan. Int. J. Sport. Med. 2011;32:476–480. doi: 10.1055/s-0030-1263115. [DOI] [PubMed] [Google Scholar]
- 261.Ahmetov I.I., Druzhevskaya A.M., Lyubaeva E.V., Popov D.V., Vinogradova O.L., Williams A.G. The dependence of preferred competitive racing distance on muscle fibre type composition and ACTN3 genotype in speed skaters. Exp. Physiol. 2011;96:1302–1310. doi: 10.1113/expphysiol.2011.060293. [DOI] [PubMed] [Google Scholar]
- 262.Cięszczyk P., Eider J., Ostanek M., Arczewska A., Leońska-Duniec A., Sawczyn S., Ficek K., Krupecki K. Association of the ACTN3 R577X Polymorphism in Polish Power-Orientated Athletes. J. Hum. Kinet. 2011;28:55–61. doi: 10.2478/v10078-011-0022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Yang R., Shen X., Wang Y., Voisin S., Cai G., Fu Y., Xu W., Eynon N., Bishop D.J., Yan X. ACTN3 R577X Gene Variant Is Associated With Muscle-Related Phenotypes in Elite Chinese Sprint/Power Athletes. J. Strength Cond. Res. 2017;31:1107–1115. doi: 10.1519/JSC.0000000000001558. [DOI] [PubMed] [Google Scholar]
- 264.Akazawa N., Ohiwa N., Shimizu K., Suzuki N., Kumagai H., Fuku N., Suzuki Y. The association of ACTN3 R577X polymorphism with sports specificity in Japanese elite athletes. Biol. Sport. 2022;39:905–911. doi: 10.5114/biolsport.2022.108704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Koku F.E., Karamızrak S.O., Çiftçi A.S., Taşlıdere H., Durmaz B., Çoğulu Ö. The relationship between ACTN3 R577X gene polymorphism and physical performance in amateur soccer players and sedentary individuals. Biol. Sport. 2019;36:9–16. doi: 10.5114/biolsport.2018.78900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Zaccagni L., Lunghi B., Barbieri D., Rinaldo N., Missoni S., Šaric T., Šarac J., Babic V., Rakovac M., Bernardi F., et al. Performance prediction models based on anthropometric, genetic and psychological traits of Croatian sprinters. Biol. Sport. 2019;36:17–23. doi: 10.5114/biolsport.2018.78901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Gomez-Gallego F., Santiago C., González-Freire M., Yvert T., Muniesa C.A., Serratosa L., Altmäe S., Ruiz J.R., Lucia A. The C allele of the AGT Met235Thr polymorphism is associated with power sports performance. Appl. Physiol. Nutr. Metab. 2009;34:1108–1111. doi: 10.1139/H09-108. [DOI] [PubMed] [Google Scholar]
- 268.Zarębska A., Sawczyn S., Kaczmarczyk M., Ficek K., Maciejewska-Karłowska A., Sawczuk M., Leońska-Duniec A., Eider J., Grenda A., Cięszczyk P. Association of rs699 (M235T) polymorphism in the AGT gene with power but not endurance athlete status. J. Strength Cond. Res. 2013;27:2898–2903. doi: 10.1519/JSC.0b013e31828155b5. [DOI] [PubMed] [Google Scholar]
- 269.Cieszczyk P., Ostanek M., Leońska-Duniec A., Sawczuk M., Maciejewska A., Eider J., Ficek K., Sygit K., Kotarska K. Distribution of the AMPD1 C34T polymorphism in Polish power-oriented athletes. J. Sport. Sci. 2012;30:31–35. doi: 10.1080/02640414.2011.623710. [DOI] [PubMed] [Google Scholar]
- 270.Fedotovskaya O.N., Danilova A.A., Akhmetov I.I. Effect of AMPD1 gene polymorphism on muscle activity in humans. Bull. Exp. Biol. Med. 2013;154:489–491. doi: 10.1007/s10517-013-1984-9. [DOI] [PubMed] [Google Scholar]
- 271.Guilherme J.P.L.F., Semenova E.A., Borisov O.V., Kostryukova E.S., Vepkhvadze T.F., Lysenko E.A., Andryushchenko O.N., Andryushchenko L.B., Lednev E.M., Larin A.K., et al. The BDNF-Increasing Allele is Associated with Increased Proportion of Fast-Twitch Muscle Fibers, Handgrip Strength, and Power Athlete Status. J. Strength Cond. Res. 2022;36:1884–1889. doi: 10.1519/JSC.0000000000003756. [DOI] [PubMed] [Google Scholar]
- 272.Fedotovskaya O., Eider J., Cięszczyk P., Ahmetov I., Moska W., Sawczyn S., Leońska–Duniec A., Maciejewska-Karłowska A., Sawczuk M., Czubek Z., et al. Association of muscle-specific creatine kinase (CKM) gene polymorphism with combat athlete status in Polish and Russian cohorts. Arch. Budo. 2013;9:233–237. [Google Scholar]
- 273.He E.P., Li Y.H., Qian J.D., Yan H.W. Association of CKMM gene A/G polymorphism and athletic performance of uyghurnationality. Zhongguo Ying Yong Sheng Li Xue Za Zhi. 2016;32:82–86. [PubMed] [Google Scholar]
- 274.Ginevičienė V., Jakaitienė A., Utkus A., Hall E.C.R., Semenova E.A., Andryushchenko L.B., Larin A.K., Moreland E., Generozov E.V., Ahmetov I.I. CKM Gene rs8111989 Polymorphism and Power Athlete Status. Genes. 2021;12:1499. doi: 10.3390/genes12101499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Miyamoto-Mikami E., Fujita Y., Murakami H., Ito M., Miyachi M., Kawahara T., Fuku N. CNTFR Genotype and Sprint/power Performance: Case-control Association and Functional Studies. Int. J. Sport. Med. 2016;37:411–417. doi: 10.1055/s-0035-1564257. [DOI] [PubMed] [Google Scholar]
- 276.Zmijewski P., Trybek G., Czarny W., Leońska-Duniec A. GALNTL6 Rs558129: A Novel Polymorphism for Swimming Performance? J. Hum. Kinet. 2021;80:199–205. doi: 10.2478/hukin-2021-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Cieszczyk P., Eider J., Arczewska A., Ostanek M., Leońska-Duniec A., Sawczyn S., Ficek K., Jascaniene N., Kotarska K., Sygit K. The HIF1A gene Pro582Ser polymorphism in polish power-orientated athletes. Biol. Sport. 2011;28:111–114. doi: 10.5604/945117. [DOI] [Google Scholar]
- 278.Gabbasov R.T., Arkhipova A.A., Borisova A.V., Hakimullina A.M., Kuznetsova A.V., Williams A.G., Day S.H., Ahmetov I.I. The HIF1A Gene Pro582Ser Polymorphism in Russian Strength Athletes. J. Strength Cond. Res. 2013;27:2055–2058. doi: 10.1519/JSC.0b013e31827f06ae. [DOI] [PubMed] [Google Scholar]
- 279.Drozdovska S.B., Dosenko V.E., Ahmetov I.I., Ilyin V.N. The association of gene polymorphisms with athlete status in ukrainians. Biol. Sport. 2013;30:163–167. doi: 10.5604/20831862.1059168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Ben-Zaken S., Meckel Y., Nemet D., Eliakim A. Can IGF-I polymorphism affect power and endurance athletic performance? Growth Horm. IGF Res. 2013;23:175–178. doi: 10.1016/j.ghir.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 281.Ben-Zaken S., Meckel Y., Remmel L., Nemet D., Jürimäe J., Eliakim A. The prevalence of IGF-I axis genetic polymorphisms among decathlon athletes. Growth Horm. IGF Res. 2022;64:101468. doi: 10.1016/j.ghir.2022.101468. [DOI] [PubMed] [Google Scholar]
- 282.Itaka T., Agemizu K., Aruga S., Machida S. G Allele of the IGF2 ApaI Polymorphism Is Associated with Judo Status. J. Strength Cond. Res. 2016;30:2043–2048. doi: 10.1519/JSC.0000000000001300. [DOI] [PubMed] [Google Scholar]
- 283.Ruiz J.R., Buxens A., Artieda M., Arteta D., Santiago C., Rodríguez-Romo G., Lao J.I., Gómez-Gallego F., Lucia A. The -174 G/C polymorphism of the IL6 gene is associated with elite power performance. J. Sci. Med. Sport. 2010;13:549–553. doi: 10.1016/j.jsams.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 284.Eider J., Cieszczyk P., Leońska-Duniec A., Maciejewska A., Sawczuk M., Ficek K., Kotarska K. Association of the 174 G/C polymorphism of the IL6 gene in Polish power-orientated athletes. J. Sport. Med. Phys. Fitness. 2013;53:88–92. [PubMed] [Google Scholar]
- 285.Terruzzi I., Senesi P., Montesano A., La Torre A., Alberti G., Benedini S., Caumo A., Fermo I., Luzi L. Genetic polymorphisms of the enzymes involved in DNA methylation and synthesis in elite athletes. Physiol. Genom. 2011;43:965–973. doi: 10.1152/physiolgenomics.00040.2010. [DOI] [PubMed] [Google Scholar]
- 286.Zarebska A., Ahmetov I., Sawczyn S., Weiner A.S., Kaczmarczyk M., Ficek K., Maciejewska-Karlowska A., Sawczuk M., Leonska-Duniec A., Klocek T., et al. Association of the MTHFR 1298A>C (rs1801131) polymorphism with speed and strength sports in Russian and Polish athletes. J. Sport. Sci. 2013;32:375–382. doi: 10.1080/02640414.2013.825731. [DOI] [PubMed] [Google Scholar]
- 287.Murtagh C.F., Brownlee T.E., Rienzi E., Roquero S., Moreno S., Huertas G., Lugioratto G., Baumert P., Turner D.C., Lee D., et al. The genetic profile of elite youth soccer players and its association with power and speed depends on maturity status. PLoS ONE. 2020;15:e0234458. doi: 10.1371/journal.pone.0234458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Nakamichi R., Ma S., Nonoyama T., Chiba T., Kurimoto R., Ohzono H., Olmer M., Shukunami C., Fuku N., Wang G., et al. The mechanosensitive ion channel PIEZO1 is expressed in tendons and regulates physical performance. Sci. Transl. Med. 2022;14:eabj5557. doi: 10.1126/scitranslmed.abj5557. [DOI] [PubMed] [Google Scholar]
- 289.Maciejewska-Skrendo A., Mieszkowski J., Kochanowicz A., Niespodziński B., Cieszczyk P., Leźnicka K., Leońska-Duniec A., Kolbowicz M., Kaczmarczyk M., Piskorska E., et al. Does the PPARA Intron 7 Gene Variant (rs4253778) Influence Performance in Power/Strength-Oriented Athletes? A Case-Control Replication Study in Three Cohorts of European Gymnasts. J. Hum. Kinet. 2021;79:77–85. doi: 10.2478/hukin-2020-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Ahmetov I.I., Mozhayskaya I.A., Lyubaeva E.V., Vinogradova O.L., Rogozkin V.A. PPARG Gene polymorphism and locomotor activity in humans. Bull. Exp. Biol. Med. 2008;146:630–632. doi: 10.1007/s10517-009-0364-y. [DOI] [PubMed] [Google Scholar]
- 291.Maciejewska-Karlowska A., Sawczuk M., Cieszczyk P., Zarebska A., Sawczyn S. Association between the Pro12Ala polymorphism of the peroxisome proliferator-activated receptor γ gene and strength athlete status. PLoS ONE. 2013;8:e67172. doi: 10.1371/journal.pone.0067172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Guilherme J.P.L.F., Bertuzzi R., Lima-Silva A.E., Pereira A.D.C., Lancha Junior A.H. Analysis of sports-relevant polymorphisms in a large Brazilian cohort of top-level athletes. Ann. Hum. Genet. 2018;82:254–264. doi: 10.1111/ahg.12248. [DOI] [PubMed] [Google Scholar]
- 293.Ahmetov I.I., Naumov V.A., Donnikov A.E., Maciejewska-Karłowska A., Kostryukova E.S., Larin A.K., Maykova E.V., Alexeev D.G., Fedotovskaya O.N., Generozov E.V., et al. SOD2 gene polymorphism and muscle damage markers in elite athletes. Free Radic. Res. 2014;48:948–955. doi: 10.3109/10715762.2014.928410. [DOI] [PubMed] [Google Scholar]
- 294.Miyamoto-Mikami E., Murakami H., Tsuchie H., Takahashi H., Ohiwa N., Miyachi M., Kawahara T., Fuku N. Lack of association between genotype score and sprint/power performance in the Japanese population. J. Sci. Med. Sport. 2017;20:98–103. doi: 10.1016/j.jsams.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 295.Khanal P., He L., Herbert A.J., Stebbings G.K., Onambele-Pearson G.L., Degens H., Morse C.I., Thomis M., Williams A.G. The Association of Multiple Gene Variants with Ageing Skeletal Muscle Phenotypes in Elderly Women. Genes. 2020;11:1459. doi: 10.3390/genes11121459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Seaborne R.A., Hughes D.C., Turner D.C., Owens D.J., Baehr L.M., Gorski P., Semenova E.A., Borisov O.V., Larin A.K., Popov D.V., et al. UBR5 is a novel E3 ubiquitin ligase involved in skeletal muscle hypertrophy and recovery from atrophy. J. Physiol. 2019;597:3727–3749. doi: 10.1113/JP278073. [DOI] [PubMed] [Google Scholar]
- 297.Kuschel L.B., Sonnenburg D., Engel T. Factors of Muscle Quality and Determinants of Muscle Strength: A Systematic Literature Review. Healthcare. 2022;10:1937. doi: 10.3390/healthcare10101937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Kikuchi N., Moreland E., Homma H., Semenova E.A., Saito M., Larin A.K., Kobatake N., Yusupov R.A., Okamoto T., Nakazato K., et al. Genes and Weightlifting Performance. Genes. 2022;13:25. doi: 10.3390/genes13010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Colakoglu M., Cam F.S., Kayitken B., Cetinoz F., Colakoglu S., Turkmen M., Sayin M. ACE genotype may have an effect on single versus multiple set preferences in strength training. Eur. J. Appl. Physiol. 2005;95:20–26. doi: 10.1007/s00421-005-1335-2. [DOI] [PubMed] [Google Scholar]
- 300.Giaccaglia V., Nicklas B., Kritchevsky S., Mychalecky J., Messier S., Bleecker E., Pahor M. Interaction between angiotensin converting enzyme insertion/deletion genotype and exercise training on knee extensor strength in older individuals. Int. J. Sport. Med. 2008;29:40–44. doi: 10.1055/s-2007-964842. [DOI] [PubMed] [Google Scholar]
- 301.Costa A.M., Silva A.J., Garrido N., Louro H., Marinho D.A., Cardoso Marques M., Breitenfeld L. Angiotensin-converting enzyme genotype affects skeletal muscle strength in elite athletes. J. Sport. Sci. Med. 2009;8:410–418. [PMC free article] [PubMed] [Google Scholar]
- 302.Pimjan L., Ongvarrasopone C., Chantratita W., Polpramool C., Cherdrungsi P., Bangrak P., Yimlamai T. A Study on ACE, ACTN3, and VDR Genes Polymorphism in Thai Weightlifters. Walailak J. Sci. Technol. (WJST) 2017;15:609–626. doi: 10.48048/wjst.2018.3525. [DOI] [Google Scholar]
- 303.Melián Ortiz A., Laguarta-Val S., Varillas-Delgado D. Muscle Work and Its Relationship with ACE and ACTN3 Polymorphisms Are Associated with the Improvement of Explosive Strength. Genes. 2021;12:1177. doi: 10.3390/genes12081177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Khanal P., Morse C.I., He L., Herbert A.J., Onambélé-Pearson G.L., Degens H., Thomis M., Williams A.G., Stebbings G.K. Polygenic Models Partially Predict Muscle Size and Strength but Not Low Muscle Mass in Older Women. Genes. 2022;13:982. doi: 10.3390/genes13060982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Gineviciene V., Jakaitiene A., Aksenov M.O., Aksenova A.V., Astratenkova A.D., Egorova E.S., Gabdrakhmanova L.J., Tubelis L., Kucinskas V., Utkus A. Association analysis of ACE, ACTN3 and PPARGC1A gene polymorphisms in two cohorts of European strength and power athletes. Biol. Sport. 2016;33:199. doi: 10.5604/20831862.1201051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Roth S.M., Walsh S., Liu D., Metter E.J., Ferrucci L., Hurley B.F. The ACTN3 R577X nonsense allele is under-represented in elite-level strength athletes. Eur. J. Hum. Genet. 2007;16:391–394. doi: 10.1038/sj.ejhg.5201964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Erskine R.M., Williams A.G., Jones D.A., Stewart C.E., Degens H. The individual and combined influence of ACE and ACTN3 genotypes on muscle phenotypes before and after strength training. Scand. J. Med. Sci Sport. 2014;24:642–648. doi: 10.1111/sms.12055. [DOI] [PubMed] [Google Scholar]
- 308.Homma H., Saito M., Saito A., Kozuma A., Matsumoto R., Matsumoto S., Kobatake N., Okamoto T., Nakazato K., Nishiyama T., et al. The Association between Total Genotype Score and Athletic Performance in Weightlifters. Genes. 2022;13:2091. doi: 10.3390/genes13112091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Orysiak J., Mazur-Różycka J., Busko K., Gajewski J., Szczepanska B., Malczewska-Lenczowska J. Individual and Combined Influence of ACE and ACTN3 Genes on Muscle Phenotypes in Polish Athletes. J. Strength Cond. Res. 2018;32:2776–2782. doi: 10.1519/JSC.0000000000001839. [DOI] [PubMed] [Google Scholar]
- 310.Ben-Zaken S., Eliakim A., Nemet D., Meckel Y. Genetic Variability among Power Athletes: The Stronger vs. Faster. J. Strength Cond. Res. 2019;33:1505–1511. doi: 10.1519/JSC.0000000000001356. [DOI] [PubMed] [Google Scholar]
- 311.Aleksandra Z., Zbigniew J., Waldemar M., Agata L.D., Mariusz K., Marek S., Agnieszka M.S., Piotr Ż., Krzysztof F., Grzegorz T., et al. The AGT Gene M235T Polymorphism and Response of Power-Related Variables to Aerobic Training. J. Sport. Sci. Med. 2016;15:616–624. [PMC free article] [PubMed] [Google Scholar]
- 312.Matteini A.M., Tanaka T., Karasik D., Atzmon G., Chou W.C., Eicher J.D., Johnson A.D., Arnold A.M., Callisaya M.L., Davies G., et al. GWAS analysis of handgrip and lower body strength in older adults in the CHARGE consortium. Aging Cell. 2016;15:792–800. doi: 10.1111/acel.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Walsh S., Zmuda J.M., Cauley J.A., Shea P.R., Metter E.J., Hurley B.F., Ferrell R.E., Roth S.M. Androgen receptor CAG repeat polymorphism is associated with fat-free mass in men. J. Appl. Physiol. 2005;98:132–137. doi: 10.1152/japplphysiol.00537.2004. [DOI] [PubMed] [Google Scholar]
- 314.Guilherme J.P.L., Shikhova Y.V., Dondukovskaya R.R., Topanova A.A., Semenova E.A., Astratenkova I.V., Ahmetov I.I. Androgen receptor gene microsatellite polymorphism is associated with muscle mass and strength in bodybuilders and power athlete status. Ann. Hum. Biol. 2021;48:142–149. doi: 10.1080/03014460.2021.1919204. [DOI] [PubMed] [Google Scholar]
- 315.Fedotovskaia O.N., Popov D.V., Vinogradova O.L., Akhmetov I.I. Association of the muscle-specific creatine kinase (CKMM) gene polymorphism with physical performance of athletes. Fiziol. Cheloveka. 2012;38:105–109. doi: 10.1134/S0362119712010082. [DOI] [PubMed] [Google Scholar]
- 316.Homma H., Kobatake N., Sekimoto Y., Saito M., Mochizuki Y., Okamoto T., Nakazato K., Nishiyama T., Kikuchi N. Ciliary Neurotrophic Factor Receptor rs41274853 Polymorphism Is Associated with Weightlifting Performance in Japanese Weightlifters. J. Strength Cond. Res. 2020;34:3037–3041. doi: 10.1519/JSC.0000000000003750. [DOI] [PubMed] [Google Scholar]
- 317.Grishina E.E., Zmijewski P., Semenova E.A., Cięszczyk P., Humińska-Lisowska K., Michałowska-Sawczyn M., Maculewicz E., Crewther B., Orysiak J., Kostryukova E.S., et al. Three DNA Polymorphisms Previously Identified as Markers for Handgrip Strength Are Associated with Strength in Weightlifters and Muscle Fiber Hypertrophy. J. Strength Cond. Res. 2019;33:2602–2607. doi: 10.1519/JSC.0000000000003304. [DOI] [PubMed] [Google Scholar]
- 318.Ahmetov I.I., Hakimullina A.M., Lyubaeva E.V., Vinogradova O.L., Rogozkin V.A. Effect of HIF1A gene polymorphism on human muscle performance. Bull. Exp. Biol. Med. 2008;146:351–353. doi: 10.1007/s10517-008-0291-3. [DOI] [PubMed] [Google Scholar]
- 319.Kostek M.C., Devaney J.M., Gordish-Dressman H., Harris T.B., Thompson P.D., Clarkson P.M., Angelopoulos T.J., Gordon P.M., Moyna N.M., Pescatello L.S., et al. A polymorphism near IGF1 is associated with body composition and muscle function in women from the Health, Aging, and Body Composition Study. Eur. J. Appl. Physiol. 2010;110:315–324. doi: 10.1007/s00421-010-1500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Ahmetov I.I., Gavrilov D.N., Astratenkova I.V., Druzhevskaya A.M., Malinin A.V., Romanova E.E., Rogozkin V.A. The association of ACE, ACTN3 and PPARA gene variants with strength phenotypes in middle school-age children. J. Physiol. Sci. 2013;63:79–85. doi: 10.1007/s12576-012-0233-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Petr M., Stastny P., Pecha O., Šteffl M., Šeda O., Kohlíková E. PPARA intron polymorphism associated with power performance in 30-s anaerobic Wingate Test. PLoS ONE. 2014;9:e107171. doi: 10.1371/journal.pone.0107171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Mann D.L., Dehghansai N., Baker J. Searching for the elusive gift: Advances in talent identification in sport. Curr. Opin. Psychol. 2017;16:128–133. doi: 10.1016/j.copsyc.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 323.Johnston K., Wattie N., Schorer J., Baker J. Talent Identification in Sport: A Systematic Review. Sport. Med. 2018;48:97–109. doi: 10.1007/s40279-017-0803-2. [DOI] [PubMed] [Google Scholar]
- 324.Baker J., Wilson S., Johnston K., Dehghansai N., Koenigsberg A., de Vegt S., Wattie N. Talent Research in Sport 1990-2018: A Scoping Review. Front. Psychol. 2020;11:607710. doi: 10.3389/fpsyg.2020.607710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Wang G., Padmanabhan S., Wolfarth B., Fuku N., Lucia A., Ahmetov I.I., Cieszczyk P., Collins M., Eynon N., Klissouras V., et al. Genomics of elite sporting performance: What little we know and necessary advances. Adv. Genet. 2013;84:123–149. doi: 10.1016/B978-0-12-407703-4.00004-9. [DOI] [PubMed] [Google Scholar]
- 326.Webborn N., Williams A., McNamee M., Bouchard C., Pitsiladis Y., Ahmetov I., Ashley E., Byrne N., Camporesi S., Collins M., et al. Direct-to-consumer genetic testing for predicting sports performance and talent identification: Consensus statement. Br. J. Sport. Med. 2015;49:1486–1491. doi: 10.1136/bjsports-2015-095343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Pitsiladis Y.P., Tanaka M., Eynon N., Bouchard C., North K.N., Williams A.G., Collins M., Moran C.N., Britton S.L., Fuku N., et al. Athlome Project Consortium: A concerted effort to discover genomic and other “omic” markers of athletic performance. Physiol. Genom. 2016;48:183–190. doi: 10.1152/physiolgenomics.00105.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.de la Chapelle A., Sistonen P., Lehväslaiho H., Ikkala E., Juvonen E. Familial erythrocytosis genetically linked to erythropoietin receptor gene. Lancet. 1993;341:82–84. doi: 10.1016/0140-6736(93)92558-B. [DOI] [PubMed] [Google Scholar]
- 329.Schuelke M., Wagner K.R., Stolz L.E., Hübner C., Riebel T., Kömen W., Braun T., Tobin J.F., Lee S.J. Myostatin mutation associated with gross muscle hypertrophy in a child. N. Engl. J. Med. 2004;350:2682–2688. doi: 10.1056/NEJMoa040933. [DOI] [PubMed] [Google Scholar]
- 330.Turner D.C., Gorski P.P., Maasar M.F., Seaborne R.A., Baumert P., Brown A.D., Kitchen M.O., Erskine R.M., Dos-Remedios I., Voisin S., et al. DNA methylation across the genome in aged human skeletal muscle tissue and muscle-derived cells: The role of HOX genes and physical activity. Sci. Rep. 2020;10:15360. doi: 10.1038/s41598-020-72730-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Seaborne R.A., Strauss J., Cocks M., Shepherd S., O’Brien T.D., van Someren K.A., Bell P.G., Murgatroyd C., Morton J.P., Stewart C.E., et al. Human Skeletal Muscle Possesses an Epigenetic Memory of Hypertrophy. Sci. Rep. 2018;8:1898. doi: 10.1038/s41598-018-20287-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Turner D.C., Seaborne R.A., Sharples A.P. Comparative Transcriptome and Methylome Analysis in Human Skeletal Muscle Anabolism, Hypertrophy and Epigenetic Memory. Sci. Rep. 2019;9:4251. doi: 10.1038/s41598-019-40787-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Demirci B., Bulgay C., Ceylan H.İ., Öztürk M.E., Öztürk D., Kazan H.H., Ergun M.A., Cerit M., Semenova E.A., Larin A.K., et al. Association of ACTN3 R577X Polymorphism with Elite Basketball Player Status and Training Responses. Genes. 2023;14:1190. doi: 10.3390/genes14061190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Murtagh C.F., Hall E.C., Brownlee T.E., Drust B., Williams A.G., Erskine R.M. The genetic association with athlete status, physical performance and injury risk in soccer. Int. J. Sport. Med. 2023 doi: 10.1055/a-2103-0165. in press . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.