Abstract
Autosomal dominant polycystic kidney disease (ADPKD) is the most common genetic cause of kidney failure in adult life. Rarely, ADPKD can be diagnosed in utero or in infancy, and the genetic mechanism underlying such severe presentation has been shown to be related to reduced gene dosage. Biallelic PKD1 variants are often identified in early onset ADPKD, with one main pathogenic variant and a modifier hypomorphic variant showing an in trans configuration. We describe two unrelated individuals with early onset cystic kidney disease and unaffected parents, where a combination of next-generation sequencing of cystic genes including PKHD1, HNF1B and PKD1 allowed the identification of biallelic PKD1 variants. Furthermore, we review the medical literature in order to report likely PKD1 hypomorphic variants reported to date and estimate a minimal allele frequency of 1/130 for this category of variants taken as a group. This figure could help to orient genetic counseling, although the interpretation and the real clinical impact of rare PKD1 missense variants, especially if previously unreported, remain challenging.
Keywords: ADPKD, hypomorphic variants, disease modifiers, biallelic inheritance, PKD1, PKD2
1. Introduction
Autosomal dominant polycystic kidney disease (ADPKD) is the most common hereditary form of renal disease and one of the most common genetic disorders in humans, with an estimated prevalence at birth of 1:1000 [1]. It is the fourth leading cause of end-stage renal disease (ESRD) worldwide, but the clinical course varies considerably among individuals, with some reaching ESRD before 40 years of age and others living a normal lifespan without requiring renal replacement therapy [2]. The two major genes involved are PKD1 (chromosome locus 16p13.3), responsible for ~78% of cases, and PKD2 (4q21), responsible for ~15% of cases [1,3]. Genotype–phenotype correlations are well established: pathogenic variants in PKD1 are associated with more severe disease with an earlier age at diagnosis than in PKD2-related ADPKD [4], and truncating PKD1 mutations lead to earlier ESRD rather than non-truncating variants [5].
Although these two genes classically show an autosomal dominant inheritance pattern and adult onset of disease, biallelic variants have been described, especially in early (childhood) or very early (prenatal) onset cases [6]. In this group of patients, earlier onset with respect to an affected parent can be caused by coinheritance of the familial inactivating PKD1 mutation plus an additional low-penetrant variant on the second allele [7,8,9,10]. Furthermore, ADPKD can be caused by a de novo variant in sporadic cases, but some patients were found to carry biallelic PKD1 variants, inherited from healthy or mildly affected parents; these variants acted as hypomorphic or reduced-penetrance alleles [11]. Occasionally, digenic inheritance has also been described, with patients carrying a trans-heterozygous pathogenic allele in PKD2 and a hypomorphic variant in PKD1 [12,13,14,15].
According to the established loss-of-function mechanism, due to a dosage/threshold model or somatic second-hit mechanism [16], biallelic full inactivation of both copies of ADPKD genes are unnecessary to cystogenesis as well as incompatible with life, while residual function of low-penetrance alleles can trigger cyst initiation below a critical threshold (approximately 20–30%) [17].
Nevertheless, the definition of low-penetrant variants is a major problem: regarding PKD1, 30–35% of pathogenic alleles are non-truncating [16], and it is challenging to define pathogenic or neutral changes and their degree of penetrance, also due to the lack of available functional tests. Moreover, some of these hypomorphic alleles are relatively frequent in the general population. For these reasons, most of them are classified as Variants of Unknown Significance (VoUS) or Likely Benign according to ACMG classification [11,18].
Here, we describe two unrelated children with early/very early onset cystic nephropathy and biallelic PKD1 hypomorphic variants. Inspired by this, we performed an extensive literature review in order to gather available information on biallelic inheritance of PKD1 hypomorphic variants and applied stringent selection criteria to evaluate their global frequency.
2. Materials and Methods
2.1. Patient Recruitment
The two probands described in this study were referred to the Paediatric Nephrology Program of the Paediatrics Unit of our hospital and afterwards to the Medical Genetics Unit. Genetic counselling was provided before and after genetic testing, and informed consent was signed by the parents. Ethical review and approval were waived for this study because, according to the local policy, informed consent is considered sufficient for reports of an observational nature concerning a limited number of patients.
2.2. Genetic Analysis
Genetic testing was performed on DNA isolated from EDTA peripheral blood using a semi-automatic Maxwell 16 instrument (Promega Corporation, Madison, WI, USA).
Considering the early age of onset in both probands, targeted NGS analysis of the coding sequence of PKHD1 (MIM#606702; HGNC:9016; RefSeq NM_138694.3), DZIP1L (MIM#617570; HGNC:26551; RefSeq NM_173543.3) and HNF1B (MIM#189907; HGNC:11630 RefSeq NM_000458.4) was performed as a first-line genetic exam. Following negative results, genetic testing was focused on PKD1 (MIM#601313 HGNC:9008, RefSeq NM_001009944.3) and PKD2 (MIM#173910; HGNC:9009; RefSeqNM_000297.4), using targeted NGS-based testing strategies validated in a diagnostic setting in order to overcome the well-known difficulties related to the molecular characteristic of PKD1 [19]. Our workflow included LR-PCR targeted re-sequencing for both PKD1 and PKD2, combined with a specific alignment pipeline for PKD1; the complete procedure is described in Mantovani et al. 2020 [12]. The raw sequencing data were transferred to the TorrentServer where Torrent SuiteTM performed alignment to a reference genome in order to generate Fastq files, a Binary Alignment Map (BAM) in conjunction with the Binary Alignment Index (BAI) and Variant Call Format (VCF) files. PKD1 reads were aligned against a modified reference genome based on chromosome 16 of Human Genome 38 (Grch38), where all the nucleotides outside the PKD1 locus were masked and replaced with “Ns.” All the VCF files were uploaded into Ion Reporter software (Thermo Fisher Scientific Inc., Waltham, MA, USA), selecting the Annotation Variant workflow in order to associate to each variant the nucleotide change in mRNA transcript, the aminoacidic change, the exons or IVSs, and the function. The BAM/BAI files, generated following alignment, were visualized using Integrative Genome Viewer (IGV) software. IGV was used to assess the depth of coverage of the sequencing reads, zygosity, quality of the sequencing reads and the mapping quality.
Large rearrangements were excluded by multiplex-ligation probe amplification (MLPA). In both families, an EDTA peripheral blood sample from each parent was taken in order to perform segregation studies.
Variant filtering based on population frequency was performed using population databases ExAC, gnomAD [20], 1000Genomes [21] and dbSNP to include only alleles with a minor allele frequency (MAF) ≤0.01. The variants were then annotated according to the guidelines published by the Human Genome Variation Society [22] and classified into five categories, according to American College of Medical Genetics and Genomics (ACMG) standards [18]. To achieve this, we made use of public variant databases (ADPKD Variant Database [23], ClinVar [24] and LOVD [25]) and online tools such as VarSome Premium [26] and Franklin [27].
2.3. Literature Review
In order to assess prevalence in the general population and the alleged role in a clinical setting, we conducted a literature review and collected the hypomorphic PKD1 variants published so far. The inclusion criteria were (1) a confirmed “in trans” configuration of the PKD1 variants and (2) a known clinical and family history of disease that allowed us to assess the clinical course as undoubtedly more severe in the proband than in the affected parent (or both parents with no renal disease).
Reports in which these data were unavailable were discarded. In order not to overestimate the true frequency of hypomorphic variants, we also excluded patients with a de novo PKD1 variant in which the phase was not known. Some studies also reported biallelic PKD2 variants or digenic PKD1/PKD2 inheritance [12,13,14]: for the sake of this article, we decided to exclude these patients, focusing on PKD1 variants only.
3. Results
3.1. Family F1
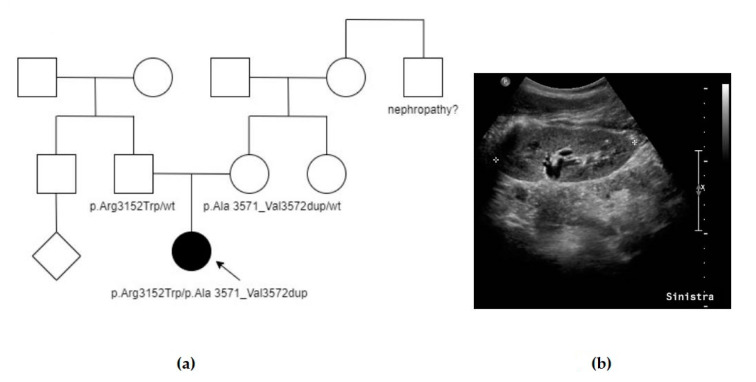
The proband (F1, III-2, Figure 1a) was the only child of non-consanguineous healthy parents. During her pregnancy, increased nuchal translucency was identified, and cytogenetic analysis on amniocytes revealed a normal female karyotype. Subsequent ultrasound (US) scans did not detect any other malformations. A few days after birth, abdominal US revealed enlarged and hyperechogenic kidneys with no corticomedullary differentiation, without any liver involvement. She was therefore addressed to our pediatric nephrology service and received periodical clinical and instrumental follow-up. No extrarenal manifestations were detected at the time of our report. At 12 years old (last clinical evaluation), she was normotensive and had normal estimated renal function (GFR 187 mL/min/1.73 m2) and urinalysis; her kidneys showed bilateral diffuse hyperechogenicity with multiple small cysts (Figure 1b). Family history was unremarkable except for a maternal uncle affected with an unspecified nephropathy. Her mother and father underwent an abdominal US study, at the ages of 44 and 41 years, respectively, and no renal cysts or other structural anomalies were detected. Further imaging (abdominal MRI or CT scans) was not performed in the parents. As informed consent was obtained, a blood sample was collected from the proband. Sequencing and MLPA analysis of PKHD1 and HNF1B were performed as a first basis but resulted negative. Afterwards, analysis of ADPKD genes revealed two previously unreported PKD1 heterozygous variants: p.Ala3571_Val3572dup and p.Arg3152Trp, whose configuration “in trans” was subsequently confirmed by parental segregation analysis (Table 1). The first variant is an extremely rare in-frame insertion located in a mutational hotspot, classified as likely pathogenic in VarSome and as VoUs in Franklin. The second one is a rare missense variant, an alternative to another likely pathogenic variant in the same codon (p.Arg3152Ser), classified as likely pathogenic in VarSome and VoUS in Franklin. No further genes were tested.
Figure 1.
Summary of case F1: (a) Pedigree of F1, showing negative family history, except for an uncle with unspecified nephropathy. Affected individuals are colored in black, the arrow indicates the proband. (b) Abdominal US scan taken at 12 years old, showing hyperechogenicity with multiple small cysts in the left kidney. Similar signs were seen also in the right kidney (bilateral involvement).
Table 1.
Genetic variants in PKD1 gene identified in F1 and F2.
| Pt | PKD1 Variant | Total MAF 1 | ACMG Criteria | ACMG Class 2 | Reports 3 |
|---|---|---|---|---|---|
| F1 | c.10710_10715dup (p.Ala3571_Val3572dup) | 0.000005 | PM1, PM2, PM4 | 4 | none |
| c.9454C > T (p.Arg3152Trp) | 0.00001 | PM1, PM2, PM5, PP3 | 4 | none | |
| F2 | c.6124G > A (p.Ala2042Thr) | 0.000008 | PM1, PM2, PM5, PP3 | 4 | none |
| c.2356C > A (p.Pro786Thr) | 0 | PM2-BP4 | 2 | none |
1 Minor allele frequency as reported in GnomAD. 2 According to VarSome Premium. 3 Reports in literature, ClinVar or ADPKD Variants Database (Mayo).
3.2. Family F2
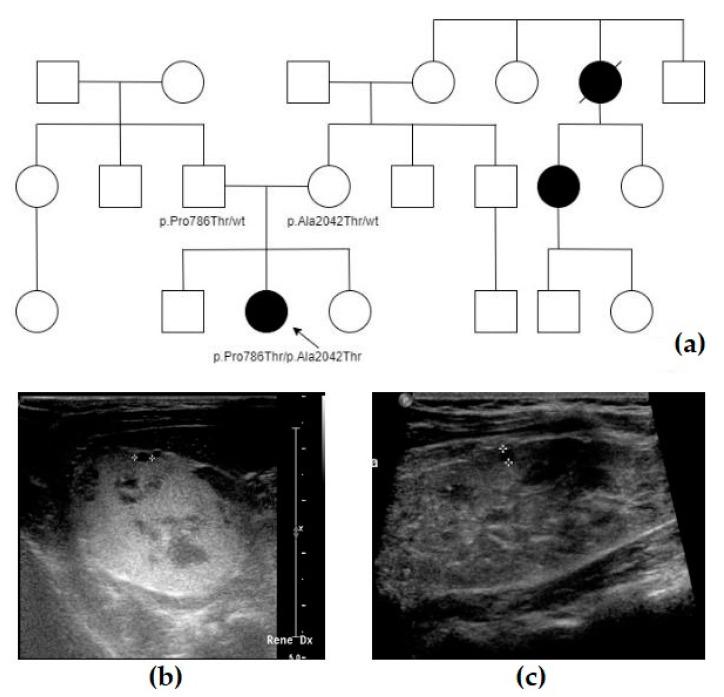
The proband (F2, IV-3, Figure 2a) was the second child of non-consanguineous healthy parents. During her pregnancy, her mother was referred to our Genetic Clinic because a moderate risk for trisomy 21 was identified on first trimester non-invasive maternal screening: a normal fetal female karyotype was detected on a chorionic villus sample. She came back to our attention after a 23-week US scan showed bilateral dysplastic hyperechogenic kidneys, with normal amniotic fluid volume and without any additional malformations. No further tests were performed. The amniotic fluid index remained normal throughout pregnancy. She was born at term by uncomplicated eutocic delivery. Soon after birth, abdomen ultrasound revealed enlarged and hyperechogenic kidneys with multiple cystic spots and no hepatic structural anomalies (Figure 2b); serum creatinine and urinalysis were normal. From neonatal age on, she has been followed up at the pediatric nephrology service of our hospital and received clinical and instrumental periodic evaluations. She had no extra renal anomalies except for segmental facial vitiligo. The most recent abdominal ultrasound, performed at the age of 7 years, showed renal cortical hyperechogenicity with multiple cystic lesions ranging from a point-like size to 3–4 mm in diameter, mild right pelvic dilatation (4–5 mm) and normal hepatic echogenicity (Figure 2c). Blood pressure, serum creatinine and urinalysis were within the normal range according to her age.
Figure 2.
Summary of case F2: (a) Pedigree of F2, showing the presence of some relatives on the maternal side with ADPKD. Affected individuals are colored in black, the arrow indicates the proband. (b) Abdominal US scan taken right after birth, showing an hyperechogenic right kidney with small multiple cysts. (c) Most recent abdominal US scan, taken at 7 years old, showing renal cortical hyperechogenicity with multiple cystic lesions ranging from point-like size to 3–4 mm in diameter.
After postnatal genetic counselling, informed consent was obtained and a blood sample was collected from the proband. Sequencing and MLPA analysis of PKHD1 and HNF1B were performed as a first basis but resulted negative. Afterwards, analysis of ADPKD genes revealed two previously unreported PKD1 heterozygous variants: p.Ala2042Thr, inherited from her mother, and p.Pro786Thr, inherited from her father. The first missense variant is extremely rare, an alternative to another likely pathogenic variant in the same codon (p.Ala2042Pro), classified as likely pathogenic in VarSome and as VoUS in Franklin. The second one, though absent in general population databases, is poorly conserved evolutionarily and has weak in silico prediction scores, so it is classified as likely benign in VarSome and VoUS in Franklin. In this case, sequencing and MLPA analysis of NPHP1 was also performed, together with the sequence analysis of other rarer genes related to nephronophthisis (INVS, NPHP3, NPHP4, IQCB1): no pathogenic variant was found in these genes.
As soon as a fetal kidney dysplasia diagnosis was made, her parents underwent abdominal US studies that did not reveal any anomalies. Further imaging (abdominal MRI or CT scans) was not performed in the parents. The mother reported to have relatives affected with ADPKD (see pedigree), but it was not known if genetic tests were ever performed. The proband had two siblings, aged 8 and 3 years old at the time of the report, who were in good general health but did not receive a renal US study or genetic tests (as a parental choice).
3.3. Literature Review
Following the criteria mentioned in the Materials and Methods, we selected a total of 50 patients described in the literature: altogether, they harbored 43 distinct hypomorphic variants. The complete list is reported in Table 2 and Table 3. Table 2 lists 14 patients with biallelic hypomorphic variants, with parents described as unaffected or having mild manifestations of disease (e.g., a few kidney cysts or liver cysts only). Table 3 reports 36 individuals who carry a pathogenic and hypomorphic variant in trans configuration: for each selected patient, we first reported the pathogenic PKD1 variant considered as “main”, together with the parental allele in which it was identified. For each hypomorphic variant, we reported the parental origin, global minor allele frequency (from GnomAD) and ACMG current classification according to the online tools cited above. Finally, we reported the presence of family history related to the disease, including mild cases, the reported age of onset in the proband and main clinical features in the proband. The global list is not exhaustive, and several putative hypomorphic variants from the referenced papers were excluded. The main reasons for exclusions were uncertainty on the phase of the variants (e.g., one of the variants was de novo) and insufficient clinical data. For example, the recent paper from Durkie et al. [11] reported 21 cases of early onset/very early onset ADPKD with possible biallelic inheritance: three cases were discarded since at least one of the variants was in PKD2, another three because it was impossible to determine the phase of the variants and, finally, one for insufficient information about family history and inheritance, leaving only 14 cases.
Some variants were reported in more than one study: p.Arg3277Cys (eight reports), p.Asp1332Asn (four reports), p.Arg4154Cys (three reports) and Ile3167Phe (two reports). The most frequent hypomorphic variants in the general population were p.Leu2696Arg, p.Ile3167Phe and p.Arg4154Cys, all with an MAF of nearly 0.001. Nine variants were absent in the general population. Globally, by summing up the MAF of all the putative hypomorphic variants that we could identify, we estimated a minimal allele frequency of 0.0076 (1/130) for PKD1 hypomorphic variants taken as a group.
In 12 cases (24%), there was no family history of kidney cystic disease, with another 2 cases with very few signs (single cysts) or incomplete penetrance in the family. We also included two patients with de novo variants, since they were reported “in trans” in the respective paper [11].
Variant p.Phe2132Cys is actually described in other two articles, and some authors hypothesized a correlation with Congenital Hepatic Fibrosis [28,29]; they were excluded since they did not meet the selection criteria (e.g., insufficient information).
Table 2.
Results of the literature review for biallelic hypomorphic PKD1 variants. Recurrent hypomorphic variants are in gray boxes. For selection criteria, see “Materials and Methods”.
| Reference | Paternal Allele | ACMG Class | H-MAF (GnomAD) |
Maternal Allele | ACMG Class |
H-MAF (GnomAD) |
Family History | Clinical Data of the Proband |
|---|---|---|---|---|---|---|---|---|
| Mantovani 2020 [12] | R459P | 3 | 0 | G1185D | 3 | 0 | Some cysts in the father | Prenatal US diagnosis, termination of pregnancy |
| Gulati 2023 [30] |
V1971M | 3 | 0.00007 | T2250M | 2 | 0.002 | Mild cystic disease in the mother | Perinatal onset (1 y) + stillborn |
| Al-Hamed 2019 [31] | G2713R | 3 | 0 | G2713R | 3 | 0 | None | Prenatal US diagnosis |
| Bergmann 2011 [8] | V1274M | 3 | 0.000004 | V1274M G2906S |
32 | 0.000004 0.0001 |
None | Three affected children, early onset (neonatal, 7 years and 17 months) |
| Durkie 2021 [11] |
S3037L | 3 | 0.000006 | S3037L | 3 | 0.000006 | None | Prenatal bilateral multicystic kidneys and hydronephrosis. Postnatal: PKD |
| Durkie 2021 [11] |
G960S | 4H | 0.000009 | N3074K | 3 | 0.00003 | None | 18 months, bilateral multicystic kidneys, atypical, no renal failure |
| Durkie 2021 [11] |
N3188S | 4H | 0 | N3188S | 4 | 0 | None, consanguineous parents | Neonatal diagnosis (bilateral), hypertension |
| Mantovani 2020 [12] | R3277C | 3 | 0.0002 | R3277C | 3 | 0.0002 | None | 22 years, typical ADPKD |
| Gilbert 2017 [32] |
C2495R | 4H | 0 | R3277C | 3 | 0.0002 | None | Neonatal onset |
| Rossetti 2009 [6] |
R3277C | 3 | 0.0002 | R3277C | 3 | 0.0002 | None | 62 years, late-onset ADPKD |
| Durkie 2021 [11] |
E3121K | 4H | 0 | R3277C | 3 | 0.0002 | None | Prenatal US diagnosis, severe oligohydramnios. Termination of pregnancy |
| Al-Hamed 2019 [31] | R3938W | 3 | 0.00002 | R3938W | 3 | 0.00002 | None | Prenatal US diagnosis, oligohydramnios, bilateral polycystic kidneys, hypertension |
| Durkie 2021 [11] |
R3892H | 3 | 0.0005 | A3959V | 3 | 0 | None | Prenatal US diagnosis, hypertension at birth, enlarged polycystic kidneys |
| Izzi 2022 [13] |
R4154C | 2 | 0.001 | R4154C | 2 | 0.001 | None | Atypical ADPKD, hypertension, CKD IV at 65 years |
Table 3.
Results of the literature review for hypomorphic PKD1 variants in trans with pathogenic PKD1 variants. Rows are sorted by the codon of the hypomorphic variant, in order to underline the recurrence of the same variant in more than one article (gray boxes). For selection criteria, see “Materials and Methods”.
| Reference | Pathogenic Variant | Origin | Hypomorphic Allele | Origin | ACMG Class |
H-MAF (GnomAD) |
Family History | Clinical Data of the Proband |
|---|---|---|---|---|---|---|---|---|
| Audrézet 2016 [9] |
T2183fs * | Paternal | D1332N | Maternal | 3 | 0.0002 | Father, typical ADPKD | Prenatal US diagnosis (kidneys +3 SD) |
| Izzi 2022 [13] |
Q4231 * | Paternal | D1332N | Maternal | 3 | 0.0002 | Father, ESRD 46 years | Enlarged hyperechogenic kidneys in utero, enlarged palpable kidneys at birth, ESRD at 35 years |
| Gulati 2023 [30] |
T2192fs * | Maternal | D1332N | Paternal | 3 | 0.0002 | Mother, typical ADPKD | Prenatal US diagnosis, perinatal demise |
| Gulati 2023 [30] |
H526fs * | Paternal | D1332N | Maternal | 3 | 0.0002 | Father, typical ADPKD | 3 years old, focal cystic disease, hypertension |
| Gulati 2023 [30] |
R2767H | Paternal | V1611I | Maternal | 3 | 0.00003 | Some liver cysts in the mother, father unknown but probably ADPKD | 20 years old, CKD2, hypertension |
| Audrézet 2016 [9] |
S4169fs * | Maternal | V1611I | Paternal | 3 | 0.00003 | Mother, typical ADPKD | Prenatal US diagnosis (kidneys +3 SD, hyperechogenicity) |
| Durkie 2021 [11] |
A1961_Q1962del | Maternal | E1929K | Paternal | 3 | 0.000009 | Some maternal relatives affected | Prenatal US diagnosis |
| Audrézet 2016 [9] |
W1839C | Maternal | G1944R | Paternal | 3 | 0.0002 | Mother, typical ADPKD | Prenatal US diagnosis (kidneys +2 SD, hyperechogenicity) |
| Durkie 2021 [11] |
S788fs * | Maternal | V1950M | Paternal | 3 | 0.00001 | Mother, typical ADPKD | Prenatal US diagnosis, neonatal death |
| Mantovani 2020 [12] |
Y1599 * | Maternal | S2000C | Paternal | 3 | 0 | Mother, typical ADPKD | 3 years old, focal cystic disease |
| Audrézet 2016 [9] |
N3188D | Paternal | Q2058R | Maternal | 3 | 0.000008 | Father, typical ADPKD | At birth, kidneys +14 SD, hyperechogenicity |
| Gulati 2023 [30] |
R3750Q | Paternal | F2132C (1) | Maternal | 2 | 0.0003 | Father, typical ADPKD | 2 years, ESRD, associated congenital hepatic fibrosis (CHF) and bile duct proliferation |
| Durkie 2021 [11] |
R2266fs * | DN (Pat) | R2162W | Maternal | 2 | 0.0001 | No (de novo variant confirmed in trans) | Prenatal echogenic kidneys. Postnatally—multiple bilateral renal cysts identified |
| Bergmann 2011 [8] |
Y2753 * | Paternal | R2255C | Maternal | 3 | 0.00003 | Father, typical ADPKD | Earlier onset and more severe disease |
| Pandita 2019 [33] |
c.529 + 3G > C | Maternal | V2267G | Paternal | 3 | 0 | Mother affected but with normal renal function | 15 years, bilateral polycystosis, dialysis |
| Bergmann 2011 [8] |
R1351fs * | Maternal | L2696R | Paternal | 2 | 0.001 | Mother, typical ADPKD | Neonatal diagnosis |
| Audrézet 2016 [9] | L339fs * | Paternal | T2710N | Maternal | 3 | 0.0004 | Father, typical ADPKD | Prenatal US diagnosis (kidneys +3 SD, hyperechogenicity) |
| Carrera 2016 [34] |
V4038fs * | Maternal | V2897delins | Paternal | 3 | 0 | Mother, typical ADPKD | Young age (earlier onset) |
| Durkie 2021 [11] |
M1247V | DN (Pat) | R3000C | Maternal | 3 | 0.00002 | No (de novo variant confirmed in trans) | Prenatal US diagnosis, postnatal poor renal function, renal transplant aged 7 |
| Durkie 2021 [11] |
E2780 * | Paternal | G3150S | Maternal | 3 | 0.000008 | Father, typical ADPKD | Prenatal US diagnosis, confirmed bilateral polycystosis on postnatal scan |
| Mantovani 2020 [12] | L1479fs * | Paternal | I3167F | Maternal | 3 | 0.001 | Father, typical ADPKD | Prenatal US diagnosis, two interrupted pregnancies |
| Durkie 2021 [11] |
P252fs * | Paternal | I3167F | Maternal | 3 | 0.001 | Father, typical ADPKD | Prenatal US diagnosis reduced amniotic fluid. Postnatal renal impairment and hypertension |
| Audrézet 2016 [9] | E3872 * | Paternal | R3183Q | Maternal | 3 | 0.0003 | Father, typical ADPKD | Prenatal US diagnosis (kidneys +6 SD, hyperechogenicity) |
| Durkie 2021 [11] |
Q1828 * | Maternal | S3238S | Paternal | 2 | 0.00003 | Mother, typical ADPKD | 2 months, bilateral involvement |
| Durkie 2021 [11] |
R2163 * | Maternal | R3269Q | Paternal | 3 | 0.000006 | Mother, typical ADPKD | Prenatal US diagnosis, neonatal demise |
| Al-Hamed 2019 [31] | L2046P | Paternal | G3227W | Maternal | 3 | 0 | Father, typical ADPKD. Single cyst in the mother | Prenatal US diagnosis, postnatal hypertension, enlarged polycystic kidneys |
| Audrézet 2016 [9] | Gross deletion | Maternal | R3277C | Paternal | 3 | 0.0002 | Mother, typical ADPKD | Prenatal US diagnosis, termination of pregnancy |
| Rossetti 2009 [6] |
Q2158 * | Paternal | R3277C | Maternal | 3 | 0.0002 | Father, typical ADPKD | Prenatal US diagnosis |
| Vujic 2010 [7] |
R2220W | Maternal | R3277C | Paternal | 3 | 0.0002 | Mother, typical ADPKD | Prenatal US diagnosis |
| Durkie 2021 [11] |
C2495R | Maternal | R3277C | Paternal | 3 | 0.0002 | Mother, typical ADPKD | Prenatal US diagnosis, severe neonatal PKD, multiple bilateral cysts, hypertension |
| Audrézet 2016 [9] | W3411 * | Paternal | N3295S | Maternal | 3 | 0.00005 | Father, typical ADPKD | Prenatal US diagnosis, hyperechogenicity |
| Audrézet 2016 [9] | L727P | Paternal | T3945M | Maternal | 3 | 0 | Father, typical ADPKD | Prenatal US diagnosis, extremely enlarged kidney, hyperechogenicity, termination of pregnancy |
| Audrézet 2016 [9] | W861 * | Paternal | E4025G | Maternal | 3 | 0.000004 | Father, typical ADPKD | Prenatal US diagnosis, extremely enlarged kidneys, ESRD 2 at 4 years |
| Bergmann 2011 [8] | L1400fs * | Maternal | R4138H | Paternal | 3 | 0.000004 | Mother, typical ADPKD | Two children (neonatal onset and prenatal onset) |
| Audrézet 2016 [9] | C2370S | Paternal | R4154C | Maternal | 2 | 0.001 | Father, typical ADPKD | Prenatal US diagnosis (kidneys +3 SD, hyperechogenicity) |
| Audrézet 2016 [9] | W1958 * | Maternal | R4154C | Paternal | 2 | 0.001 | Mother, typical ADPKD | Prenatal US diagnosis (kidneys +3 SD, hyperechogenicity) |
4. Discussion
We described two patients who came to our attention for early/very early onset cystic nephropathy and biallelic PKD1 hypomorphic variants. Both girls had enlarged and echogenic kidneys with onset in utero or soon after birth and unaffected parents. In the absence of a positive family history in either of the parents, these clinical pictures could be misdiagnosed at first for an alternative congenital cystic nephropathy such as Autosomal Recessive Polycystic Kidney Disease (ARPKD), presenting with enlarged echogenic kidneys, hypertension and varying degrees of renal dysfunction due to PKHD1 or DZIP1L biallelic mutations [35], or as HNF1B-related disease, where bilateral hyperechogenic kidneys can be detected on prenatal ultrasound [36]. Therefore, these genes were tested in the two probands, ruling out their involvement in both cases. Subsequently, non-truncating, previously unreported biallelic PKD1 variants were identified, but heterozygous parents did not have renal cysts on ultrasound. It would be interesting to look for minor signs of kidney/liver involvement through abdominal MRI or CT scans, especially in the mother of patient 2, who reported some affected relatives.
These variants were extremely rare in the general population and were classified as Likely Benign, VoUS or Likely Pathogenic according to ACMG standards. Some authors [11] suggested that a different approach could be used while applying ACMG criteria to variants for which a hypomorphic status has been speculated in the literature, for example using the PM3 criteria (recessive in trans) or adding more value to PP5 (reputable source data). As a matter of fact, applying these proposed parameters to the four variants that we reported here contributes to strengthening their pathogenicity. If a diagnosis of bi-allelic ADPKD can be confirmed in these two families, it still leaves many questions unanswered, especially in terms of long-term prognosis and expected age of ESRD, possibility and usefulness of Tolvaptan therapy, and risks related to living kidney donors if the candidate carries a hypomorphic variant.
In general, the biological effect of hypomorphic PKD1 alleles is difficult to ascertain, thus making allele-specific clinical case reporting especially valuable [28]. To date, the only variant with unequivocal proven ‘reduced’ function in a genetically engineered mouse mutant (Pkd1 RC mouse) is p.Arg3277Cys, which is also the most frequently reported variant in our literature review. Cellular studies have shown that this amino acid change leads to PC1 misfolding, resulting in increased ER retention and reduced surface expression [37]. Other hypomorphic variants were tested through generated mutant structures in silico by Durkie et al. [11]. In general, the most reliable hypomorphic variants are those reported in multiple affected individuals, such as p.Asp1332Asn, p.Arg4154Cys and again p.Arg3277Cys. Unfortunately, functional studies for PKD1 variants are cumbersome at the moment, so it is crucial to gather families, obtain long-term follow-up of early onset cases, perform segregation analysis of putative hypomorphic variants and collect population data on variant frequencies. With all the possible limitations, ADPKD serves as a good model for the study of hypomorphic variants, because it is a relatively frequent condition with limited genetic heterogeneity.
The prevalence of hypomorphic variants also has important implications for reproductive genetic counseling. In families where both parents are heterozygous carriers, there will be a 25% recurrence risk in future pregnancies of biallelic ADPKD. In families with typical ADPKD, the risk of bearing a child with early/very early onset of disease can be estimated: the global allele frequency of the hypomorphic variants reported in Table 2 adds up to 1/130; this figure is likely underestimated but would translate into a population frequency of heterozygous carriers of 1/65. The risk is thus relatively low but justifies a warning and consideration of molecular tests in partners with a history of benign cysts.
5. Conclusions
PKD1 hypomorphic variants emerge clearly as modifiers of ADPKD, giving a significant contribution to early onset cases. Allele-specific clinical case reporting is extremely valuable in order to increase the recognition of these variants and bring new data related to disease progression, inheritance and response to therapy.
Acknowledgments
We thank all of the family members for their collaboration.
Author Contributions
Conceptualization, E.A., F.M. and C.G.; methodology, C.P.C. and C.G.; investigation, C.P.C., I.C., A.P. and C.L.S.; writing—original draft preparation, E.A. and C.G. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki. Ethical review and approval were waived for this study because, according to the local policy, informed consent is considered sufficient for reports of an observational nature concerning a limited number of patients.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patients’ parents to publish this paper.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Harris P.C., Torres V.E. In: Polycystic Kidney Disease, Autosomal Dominant. Adam M.P., Mirzaa G.M., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., Amemiya A., editors. University of Washington; Seattle, WA, USA: 1993–2023. [(accessed on 20 April 2023)]. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1246/ [Google Scholar]
- 2.Cornec-Le Gall E., Blais J.D., Irazabal M.V., Devuyst O., Gansevoort R.T., Perrone R.D., Chapman A.B., Czerwiec F.S., Ouyang J., Heyer C.M., et al. Can we further enrich autosomal dominant polycystic kidney disease clinical trials for rapidly progressive patients? Application of the PROPKD score in the TEMPO trial. Nephrol. Dial. Transpl. 2018;33:645–652. doi: 10.1093/ndt/gfx188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossetti S., Consugar M.B., Chapman A.B., Torres V.E., Guay-Woodford L.M., Grantham J.J., Bennett W.M., Meyers C.M., Walker D.L., Bae K., et al. Comprehensive molecular diagnostics in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2007;18:2143–2160. doi: 10.1681/ASN.2006121387. [DOI] [PubMed] [Google Scholar]
- 4.Torres V.E., Harris P.C., Pirson Y. Autosomal dominant polycystic kidney disease. Lancet. 2007;369:1287–1301. doi: 10.1016/S0140-6736(07)60601-1. [DOI] [PubMed] [Google Scholar]
- 5.Cornec-Le Gall E., Audrézet M.-P., Chen J.-M., Hourmant M., Morin M.-P., Perrichot R., Charasse C., Whebe B., Renaudineau E., Jousset P., et al. Type of PKD1 mutation influences renal outcome in ADPKD. J. Am. Soc. Nephrol. 2013;24:1006–1013. doi: 10.1681/ASN.2012070650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossetti S., Kubly V.J., Consugar M.B., Hopp K., Roy S., Horsley S.W., Chauveau D., Rees L., Barratt T.M., van’t Hoff W.G., et al. Incompletely penetrant PKD1 alleles suggest a role for gene dosage in cyst initiation in polycystic kidney disease. Kidney Int. 2009;75:848–855. doi: 10.1038/ki.2008.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vujic M., Heyer C.M., Ars E., Hopp K., Markoff A., Orndal C., Rudenhed B., Nasr S.H., Torres V.E., Torra R., et al. Incompletely penetrant PKD1 alleles mimic the renal manifestations of ARPKD. J. Am. Soc. Nephrol. 2010;21:1097–1102. doi: 10.1681/ASN.2009101070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergmann C., von Bothmer J., Ortiz Brüchle N., Venghaus A., Frank V., Fehrenbach H., Hampel T., Pape L., Buske A., Jonsson J., et al. Mutations in multiple PKD genes may explain early and severe polycystic kidney disease. J. Am. Soc. Nephrol. 2011;22:2047–2056. doi: 10.1681/ASN.2010101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Audrézet M.P., Corbiere C., Lebbah S., Morinière V., Broux F., Louillet F., Fischbach M., Zaloszyc A., Cloarec S., Merieau E., et al. Comprehensive PKD1 and PKD2 Mutation Analysis in Prenatal Autosomal Dominant Polycystic Kidney Disease. J. Am. Soc. Nephrol. 2016;27:722–729. doi: 10.1681/ASN.2014101051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heyer C.M., Sundsbak J.L., Abebe K.Z., Chapman A.B., Torres V.E., Grantham J.J., Bae K.T., Schrier R.W., Perrone R.D., Braun W.E., et al. Predicted mutation strength of nontruncating PKD1 mutations aids genotype-phenotype correlations in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2016;27:2872–2884. doi: 10.1681/ASN.2015050583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durkie M., Chong J., Valluru M.K., Harris P.C., Ong A.C.M. Biallelic inheritance of hypomorphic PKD1 variants is highly prevalent in very early onset polycystic kidney disease. Genet. Med. 2021;23:689–697. doi: 10.1038/s41436-020-01026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mantovani V., Bin S., Graziano C., Capelli I., Minardi R., Aiello V., Ambrosini E., Cristalli C.P., Mattiaccio A., Pariali M., et al. Gene Panel Analysis in a Large Cohort of Patients with Autosomal Dominant Polycystic Kidney Disease Allows the Identification of 80 Potentially Causative Novel Variants and the Characterization of a Complex Genetic Architecture in a Subset of Families. Front. Genet. 2020;7:11. doi: 10.3389/fgene.2020.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izzi C., Dordoni C., Delbarba E., Mazza C., Savoldi G., Econimo L., Cortinovis R., Zeni L., Martin E., Alberici F., et al. Lessons From the Clinic: ADPKD Genetic Test Unraveling Severe Phenotype, Intrafamilial Variability, and New, Rare Causing Genotype. Kidney Int. Rep. 2022;7:895–898. doi: 10.1016/j.ekir.2021.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arora V., Bijarnia-Mahay S., Tiwari V., Bansal S., Gupta P., Setia N., Puri R.D., Verma I.C. Co-inheritance of pathogenic variants in PKD1 and PKD2 genes presenting as severe antenatal phenotype of autosomal dominant polycystic kidney disease. Eur. J. Med. Genet. 2020;63:103734. doi: 10.1016/j.ejmg.2019.103734. [DOI] [PubMed] [Google Scholar]
- 15.Pei Y., Paterson A.D., Wang K.R., He N., Hefferton D., Watnick T., Germino G.G., Parfrey P., Somlo S., George-Hyslop P.S. Bilineal Disease and Trans-Heterozygotes in Autosomal Dominant Polycystic Kidney Disease. Am. J. Hum. Genet. 2001;68:355–363. doi: 10.1086/318188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornec-Le Gall E., Torres V.E., Harris P.C. Genetic Complexity of Autosomal Dominant Polycystic Kidney and Liver Diseases. J. Am. Soc. Nephrol. 2018;29:13–23. doi: 10.1681/ASN.2017050483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lanktree M.B., Haghighi A., di Bari I., Song X., Pei Y. Insights into Autosomal Dominant Polycystic Kidney Disease from Genetic Studies. Clin. J. Am. Soc. Nephrol. 2021;16:790–799. doi: 10.2215/CJN.02320220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards C.S., Bale S., Bellissimo D.B., Das S., Grody W.W., Hegde M.R., Lyon E., Ward B.E. Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 19.Rossetti S., Hopp K., Sikkink R.A., Sundsbak J.L., Lee Y.K., Kubly V., Eckloff B.W., Ward C.J., Winearls C.G., Torres V.E., et al. Identification of gene mutations in autosomal dominant polycystic kidney disease through targeted resequencing. J. Am. Soc. Nephrol. 2012;23:915–933. doi: 10.1681/ASN.2011101032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Den Dunnen J.T., Antonarakis S.E. Mutation nomenclature extensions and suggestions to describe complex mutations: A discussion. Hum. Mutat. 2000;15:7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 23.PKD Foundation Variant Database. [(accessed on 29 April 2023)]. Available online: https://pkdb.mayo.edu/variants.
- 24.ClinVar NCBI. [(accessed on 29 April 2023)]; Available online: https://www.ncbi.nlm.nih.gov/clinvar/
- 25.Leiden Open Variant Database. [(accessed on 29 April 2023)]. Available online: https://www.lovd.nl/
- 26.VarSome Premium. [(accessed on 29 April 2023)]. Available online: https://varsome.com/
- 27.Franklin Genoox. [(accessed on 29 April 2023)]. Available online: https://franklin.genoox.com/clinical-db/home.
- 28.O’Brien K., Font-Montgomery E., Lukose L., Bryant J., Piwnica-Worms K., Edwards H., Riney L., Garcia A., Daryanani K., Choyke P., et al. Congenital hepatic fibrosis and portal hypertension in autosomal dominant polycystic kidney disease. J. Pediatr. Gastroenterol. Nutr. 2012;54:83–89. doi: 10.1097/MPG.0b013e318228330c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanktree M.B., Guiard E., Li W., Akbari P., Haghighi A., Iliuta I.A., Shi B., Chen C., He N., Song X., et al. Intrafamilial Variability of ADPKD. Kidney Int. Rep. 2019;4:995–1003. doi: 10.1016/j.ekir.2019.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gulati A., Dahl N.K., Hartung E.A., Clark S.L., Moudgil A., Goodwin J., Somlo S. Hypomorphic PKD1 Alleles Impact Disease Variability in Autosomal Dominant Polycystic Kidney Disease. Kidney360. 2023;4:387–392. doi: 10.34067/KID.0000000000000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Hamed M.H., Alsahan N., Rice S.J., Edwards N., Nooreddeen E., Alotaibi M., Kurdi W., Alnemer M., Altaleb N., Ali W., et al. Bialleleic PKD1 mutations underlie early-onset autosomal dominant polycystic kidney disease in Saudi Arabian families. Pediatr. Nephrol. 2019;34:1615–1623. doi: 10.1007/s00467-019-04267-x. [DOI] [PubMed] [Google Scholar]
- 32.Gilbert R.D., Evans H., Olalekan K., Nagra A., Haq M.R., Griffiths M. Tolvaptan treatment for severe neonatal autosomal-dominant polycystic kidney disease. Pediatr. Nephrol. 2017;32:893–896. doi: 10.1007/s00467-017-3584-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pandita S., Ramachandran V., Balakrishnan P., Rolfs A., Brandau O., Eichler S., Bhalla A.K., Khullar D., Amitabh V., Ramanarayanan S., et al. Identification of PKD1 and PKD2 gene variants in a cohort of 125 Asian Indian patients of ADPKD. J. Hum. Genet. 2019;64:409–419. doi: 10.1038/s10038-019-0582-8. [DOI] [PubMed] [Google Scholar]
- 34.Carrera P., Calzavara S., Magistroni R., den Dunnen J.T., Rigo F., Stenirri S., Testa F., Messa P., Cerutti R., Scolari F., et al. Deciphering Variability of PKD1 and PKD2 in an Italian Cohort of 643 Patients with Autosomal Dominant Polycystic Kidney Disease (ADPKD) Sci. Rep. 2016;6:30850. doi: 10.1038/srep30850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sweeney W.E., Avner E.D. In: Polycystic Kidney Disease, Autosomal Recessive. Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., Amemiya A., editors. University of Washington; Seattle, WA, USA: 1993–2021. [(accessed on 20 April 2023)]. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1326/ [Google Scholar]
- 36.Naylor R., Johnson A.K., del Gaudio D. In: Maturity-Onset Diabetes of the Young Overview. Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., Amemiya A., editors. University of Washington; Seattle, WA, USA: 1993–2021. [(accessed on 20 April 2023)]. Available online: https://www.ncbi.nlm.nih.gov/books/NBK500456/ [PubMed] [Google Scholar]
- 37.Hopp K., Ward C.J., Hommerding C.J., Nasr S.H., Tuan H.F., Gainullin V.G., Rossetti S., Torres V.E., Harris P.C. Functional polycystin-1 dosage governs autosomal dominant polycystic kidney disease severity. J. Clin. Investig. 2012;122:4257–4273. doi: 10.1172/JCI64313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.