Abstract
Introduction
P21-activated kinase 1 (PAK1) is known to be overexpressed in several human tumour types, including breast cancer (BC). It is located on chromosome 11 (11q13.5-q14.1) and plays a significant role in proliferation in BC. In this study we aimed to assess PAK1 gene copy number (CN) in primary breast tumours and their corresponding lymph node metastases, and associations between PAK1 CN and proliferation status, molecular subtype, and prognosis. In addition, we aimed to study associations between CNs of PAK1 and CCND1. Both genes are located on the long arm of chromosome 11 (11q13).
Methods
Fluorescence in situ hybridization for PAK1 and Chromosome enumeration probe (CEP)11 were used on tissue microarray sections from a series of 512 BC cases. Copy numbers were estimated by counting the number of fluorescent signals for PAK1 and CEP11 in 20 tumour cell nuclei. Pearson’s x2 test was performed to assess associations between PAK1 CN and tumour features, and between PAK1 and CCND1 CNs. Cumulative risk of death from BC and hazard ratios were estimated in analysis of prognosis.
Results
We found mean PAK1 CN ≥4<6 in 26 (5.1%) tumours, and CN ≥ 6 in 22 (4.3%) tumours. The proportion of cases with copy number increase (mean CN ≥4) was highest among HER2 type and Luminal B (HER2-) tumours. We found an association between PAK1 CN increase, and high proliferation, and high histological grade, but not prognosis. Of cases with PAK1 CN ≥ 6, 30% also had CCND1 CN ≥ 6.
Conclusions
PAK1 copy number increase is associated with high proliferation and high histological grade, but not with prognosis. PAK1 CN increase was most frequent in the HER2 type and Luminal B (HER2-) subtype. PAK1 CN increase is associated with CN increase of CCND1.
Introduction
P21-activated kinases (PAK) are a family of serine/threonine protein kinases comprising six isoforms (PAK1–6). They are overexpressed in several human tumours, such as breast cancer (BC), colon cancer and lung cancer, and in neurofibromatosis [1]. The six PAK isoforms are subdivided in PAK1-3 (group I) and PAK4-6 (group II) [2, 3]. PAKs play a significant role in proliferation, cytoskeletal dynamics, and cell survival [1, 4]. Their roles in these cell processes make them potential therapeutic targets. More is known of the functions of PAK1 and PAK4, than of the other isoforms [5, 6].
PAK1 is located on chromosome 11 (q13.5-q14.1). Amplification of PAK1 and high PAK1 protein levels are found in several human cancers, including BC [7–9], and are linked to aggressive tumour types, chemotherapy resistance and poor prognosis [4, 10–14]. In 2000, Mira et al. first discovered that PAK1 had an important role in proliferation in BC cell lines [15]. Since then, PAK1 has been found to be involved in many stages of the BC process and is known to regulate several signaling pathways. [4, 16–21]. PAK1 amplification has recently been found to be significantly associated with reduced relapse-free survival of ER-positive BC patients [19]. PAK1 is localized in the same chromosomal region as CCND1, 11q13 [22, 23]. Cyclin D1 (CCND1) has been found to be overexpressed in breast cancer, and PAK1 is shown to regulate the expression of CCND1 in BC [8, 23].
In this study we aimed to assess PAK1 gene copy number (CN) in a well-characterized series of primary BCs and their corresponding axillary lymph node metastases. We studied associations between PAK1 CN and proliferation, molecular subtypes, and prognosis. In addition, we examined associations between CN of CCND1, assessed in an earlier study by our group [24], and PAK1 CN.
Materials and methods
Study population
A population-based survey for the early detection of BC was conducted in the county of Nord-Trøndelag, Norway, between 1956 and 1959. The study included 25,727 women born 1886–1928 [25]. These women were followed for BC occurrence, through linkage with data from the Cancer Registry of Norway. During the follow-up years, between 1961 and 2008, 1379 new BCs were registered. Of these, 909 cases were included in the study population and were first reclassified into molecular subtypes in a previous published by our group in 2013 (Table 1) [26]. All patients were followed from time of diagnosis until death or December 31st, 2015.
Table 1. Reclassification of breast cancers into molecular subtypes [26].
| Molecular subtype | Classified by |
|---|---|
| Luminal A | ER+ and/or PR+, HER2-, Ki-67<15% |
| Luminal B (HER2-) | ER+ and/or PR+, HER2-, Ki-67≥15% |
| Luminal B (HER2+) | ER+ and/or PR+, HER2+ |
| HER2 type | ER-, PR-, HER2+ |
| Basal-like | ER-, PR-, HER2-, CK5+ and/or EGFR+ |
| 5-negative phenotype | ER-, PR-, HER2-, CK5-, EGFR- |
*ER = Oestrogen receptor, PR = Progesterone receptor, HER2 = Human epidermal growth factor receptor 2, CK5 = Cytokeratin 5, EGFR = Epidermal growth factor receptor 1
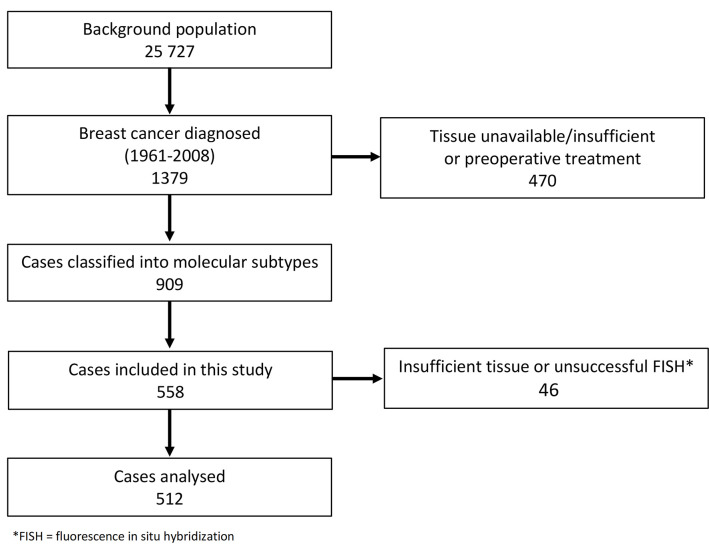
For the present study, we performed fluorescence in situ hybridization (FISH) on tissue specimens from cases mainly diagnosed after 1985 (n = 558). Of these, 46 were excluded due to missing or insufficient tumour tissue (n = 25), or due to unsuccessful FISH (n = 21). Thus, 512 cases were suitable for assessment of PAK1 and chromosome enumeration probe 11 (CEP11) CN in primary tumours (Fig 1). Of the 512 cases, 172 had lymph node metastases, and tissue from lymph node metastases was available for 143 cases. Cases with unsuccessful FISH (n = 9) or insufficient amounts of tumour tissue (n = 11) were excluded. Hence, lymph node metastases from 123 cases were included in the analyses.
Fig 1. Overview of study population and cases included in this study.
Specimen characteristics
The primary tumours were previously reclassified into histological type and grade according to present-day guidelines [26–28]. Tissue microarray (TMA) blocks were made using the TissueArrayer Minicore with TMA Designer2 software (Alphelys). Three 1-mm in diameter tissue cylinders were extracted from the periphery of the primary tumour, and from lymph node metastases and transferred to TMA recipient blocks. Using sections from the TMAs, primary tumours were then reclassified into molecular subtypes using immunohistochemistry (IHC) and chromogenic in situ hybridization (CISH) as previously described (Table 1). Briefly, Oestrogen Receptor (ER), Progesterone Receptor (PR), the proliferation marker Ki-67, Cytokeratin 5 (CK5) and Epidermal Growth Factor Receptor 1 (EGFR) were assessed using IHC, and Human Epidermal Growth Factor Receptor 2 (HER2) was assessed using both CISH and IHC [26] (Table 2). In a previous study of CCND1 CN, FISH was used to target CCND1 and CEP11, using Dako Histology FISH Accessory Kit K 579911 probes for CCND1 (3 μL, Empire Genomics) and CEP11 (1 μL, Abbott/VYSIS) [24].
Table 2. Sources and dilutions of primary antibodies used for molecular subtyping [26].
| Antibody | Clone | Manufacturer | Concentration of antibody | Dilution |
|---|---|---|---|---|
| ER | SP1 | Cell Marque | 33 mg/ml | 1:100 |
| PR | 16 | Novocastra | 360 mg/l | 1:400 |
| HER2 | CB11 | Novocastra | 3.9 g/l | 1:640 |
| Ki-67 | MIB1 | Dako | 35 mg/l | 1:100 |
| CK5 | XM26 | Novocastra | 50 mg/l | 1:100 |
| EGFR | 2-18C9 | Dako | Ready to use | No dilution |
Fluorescence in situ hybridization
For the present study of PAK1 and CEP11 CN, FISH was done using DAKO Histology FISH Accessory Kit K 579911 according to the manufacturer’s instructions. TMA sections were preheated at 60°C for 1–2 h, then de-waxed and rehydrated. The slides were then boiled in a microwave oven for 10 min. in pretreatment solution and washed in DAKO wash buffer (2x3min.) after cooling (15 min.), followed by protein digestion in pepsin solution (37°C, 25 min.). After protein digestion, the slides were washed in DAKO wash buffer (2x3 min.), dehydrated (2 min. in 70%, 85% and 95% ethanol), then air-dried for 15 min. at room temperature.
PAK1 (3 μL, PAK1-20-RE, SpectrumRed fluorochrome Empire Genomics) and CEP11 (3 μL, CEP11 [D11Z19], SpectrumGreen fluorochrome, VYSIS) probes were mixed with hybridizing buffer (9 μL, Empire Genomics) and applied to TMA slides according to the manufacturer’s instructions. Coverslips were then applied to the slides, sealed with DAKO coverslip sealant, and the slides were dried for 20 min. After drying, denaturation was performed at 83°C for 3 min., followed by hybridization at 37°C overnight using DAKO hybridizer. Post-hybridization washes were done in 0.4 X SSC/ 0.3% NP-40 stringent wash buffer at 72°C (2 min.) and 2 X SSC/ 0.1% NP-40 wash buffer at room temperature (1 min.). Slides were then dried at 37°C for 15 min., DAPI II VYSIS (15 μl, no 06J50-001) was applied. The slides were then coverslipped and stored at −20°C.
Scoring and reporting
A fluorescence microscope (Nikon Eclipse 90i) was used for counting PAK1 and CEP11 CN. For each case, all available tissue spots were examined and the number of fluorescent signals for PAK1 and CEP11 were counted in 20 well-preserved, non-overlapping tumour cell nuclei. Mean PAK1 and CEP11 CNs was calculated for tumours and lymph node metastases and were first categorized as <4 and ≥4. In addition, to distinguish between low-level CN gain and high-level gain or gene amplification, we also subdivided CN into three categories: <4; ≥4<6; and ≥6 according to guidelines for categorizing HER2 CNs [29], a strategy which has been used in previous studies of other genes by our group [24, 30–32]. The Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) were followed [33].
Statistical analyses
Pearson’s chi square test was used to compare tumour characteristics across categories of PAK1 mean CN. Cumulative incidence of death from breast cancer was estimated, and Gray’s test was used to compare equality between cumulative incidence curves. Cox proportional hazard analyses were used to estimate hazard ratios (HR) of breast cancer death with 95% confidence intervals (CI). The analyses were adjusted for age (≤ 49, 50–59, 60–64, 65–69, 70–74, ≥ 75), stage (I–IV), histological grade (1–3), and Ki67 status (</≥ 15%). Adjustments were made for each variable separately, and for age, grade, and stage combined. No clear violations of proportionality were observed in log minus-log plots. All statistical tests were two-sided and statistical significance was assessed at the 5% level. We used Stata 16 (Stata corp., College station, TX, USA) in the statistical analyses.
Ethics statement
This study was granted approval including dispensation from the general requirement of informed consent, by the Regional Committee for Medical and Health Research Ethics, Midt-Norge (REK 836/2009). All methods were carried out in accordance with relevant guidelines and regulations (The Declaration of Helsinki and national regulations (ACT 2008-06-20 no. 44: Act on medical and health research (the Health Research Act)).
Results
Patient and tumour characteristics for the 512 patients included in the present study are given in Table 3. The mean age at diagnosis was 75.4 years (range 41–96) and the mean follow-up after diagnosis was 9.1 years (SD = 7.2). At end of follow-up, 35.4% of patients had died from BC and 54.3% had died from other causes.
Table 3. Patient and tumour characteristics according to PAK1 copy number.
| Total study population | Mean PAK1 copy number, three categories | Mean PAK1 copy number, two categories | ||||||
|---|---|---|---|---|---|---|---|---|
| <4 | ≥4 to <6 | ≥6 | p value (χ2) | <4 | ≥4 | p value (χ2) | ||
| N (%) | 512 | 464 (90.6) | 26 (5.1) | 22 (4.3) | 464 (90.6) | 48 (9.4) | ||
| Mean age at diagnosis, years (SD) | 75.4(41–96) (8.2) | 75.5 (8.1) | 75.2 (7.3) | 74.3 (10.0) | 75.5 (8.1) | 74.8 (8.6) | ||
| Mean follow-up, years (SD) | 9.1 (7.2) | 9.0 (7.0) | 9.6 (6.5) | 9.0 (7.5) | 9.0 (7.0) | 9.3 (6.9) | ||
| Deaths from breast cancer (%) | 181 (35.4) | 161 (34.7) | 9 (34.6) | 11 (50.0) | 161 (34.7) | 20 (41.7) | ||
| Deaths from other causes (%) | 278 (54.3) | 255 (55.0) | 15 (57.7) | 8 (36.4) | 255 (55.0) | 23 (47.9) | ||
| Histological grade (%) | ||||||||
| I | 56 (10.9) | 55 (11.9) | 0 (0) | 1 (4.6) | 0.082 | 55 (11.9) | 1(2.1) | 0.037 |
| II | 287 (56.1) | 262 (56.5) | 12 (46.2) | 13 (59.1) | 262 (56.5) | 25 (52.1) | ||
| III | 169 (33.0) | 147 (31.7) | 14 (53.9) | 8 (36.4) | 147 (31.7) | 22 (45.8) | ||
| Lymph node metastasis (%) | ||||||||
| Yes | 172 (33.6) | 153 (33.0) | 13 (50.0) | 6 (27.3) | 0.272 | 153 (33.0) | 19 (39.6) | 0.360 |
| No | 228 (44.5) | 209 (45.0) | 9 (34.6) | 10 (45.5) | 209 (45.0) | 19 (39.6) | ||
| Unknown histology | 112 (21.9) | 102 (22.0) | 4 (15.4) | 6 (27.3) | 102 (22.0) | 10 (20.8) | ||
| Tumor size (%) | ||||||||
| ≤2 cm | 245 (47.9) | 217 (46.8) | 16 (61.5) | 12 (54.6) | 0.327 | 217 (46.8) | 28 (58.3) | 0.516 |
| >2 cm, ≤ 5 cm | 95 (18.6) | 88 (19.0) | 4 (15.4) | 3 (13.6) | 88 (19.0) | 7 (14.6) | ||
| >5 cm | 10 (2.0) | 9 (1.9) | 1 (3.9) | 0 (0) | 9 (1.9) | 1(2.1) | ||
| Uncertain, but >2 cm | 63 (12.3) | 60 (12.9) | 3 (11.5) | 0 (0) | 60 (12.9) | 3 (6.3) | ||
| Uncertain | 99 (19.3) | 90 (19.4) | 2 (7.7) | 7 (31.8) | 90 (19.4) | 9 (18.8) | ||
| Stage (%) | ||||||||
| I | 242 (47.3) | 221 (47.6) | 9 (34.6) | 12 (54.6) | 0.027 | 221 (47.6) | 21 (43.8) | 0.117 |
| II | 218 (42.6) | 198 (42.7) | 14 (53.9) | 6 (27.3) | 198 (42.7) | 20 (41.7) | ||
| III | 27 (5.3) | 22 (4.7) | 3 (11.5) | 2 (9.1) | 22 (4.7) | 5 (10.4) | ||
| IV | 23 (4.5) | 22 (4.7) | 0 (0) | 1 (4.6) | 22 (4.7) | 1 (2.1) | ||
| Unknown | 2 (0.4) | 1 (0.2) | 0 (0) | 1 (4.6) | 1 (0.2) | 1 (2.1) | ||
| Molecular subtype (%) | ||||||||
| Luminal A | 272 (53.1) | 251 (54.1) | 11 (42.3) | 10 (45.5) | 0.649 | 251 (54.1) | 21 (43.8) | 0.375 |
| Luminal B (HER2-) | 121 (23.6) | 105 (22.6) | 8 (30.8) | 8 (36.4) | 105 (22.6) | 16 (33.3) | ||
| Luminal B (HER2+) | 42 (8.2) | 39 (8.4) | 1 (3.9) | 2 (9.1) | 39 (8.4) | 3 (6.3) | ||
| HER2 type | 27 (5.3) | 23 (5.0) | 3 (11.5) | 1 (4.6) | 23 (5.0) | 4 (8.3) | ||
| 5NP | 11 (2.2) | 11 (2.4) | 0 (0) | 0 (0) | 11 (2.4) | 0 (0) | ||
| BP | 39 (7.6) | 35 (7.5) | 3 (11.5) | 1 (4.6) | 35 (7.5) | 4 (8.3) | ||
| Histological type (%) | ||||||||
| Invasive carcinoma NOS | 353 (69.0) | 318 (68.5) | 19 (73.1) | 16 (72.7) | 0.593 | 318 (68.5) | 35 (69.0) | 0.273 |
| Lobular carcinoma | 66 (12.9) | 61 (13.2) | 2 (7.7) | 3 (13.6) | 61 (13.2) | 5 (10.4) | ||
| Tubular carcinoma | 1 (0.2) | 1 (0.2) | 0 (0) | 0 (0) | 1 (0.2) | 0 (0) | ||
| Mucinous carcinoma | 24 (4.7) | 23 (5.0) | 1 (3.9) | 0 (0) | 23 (5.0) | 1 (2.1) | ||
| Medullary carcinoma | 14 (2.7) | 10 (2.2) | 3 (11.5) | 1 (4.6) | 10 (2.2) | 4 (8.3) | ||
| Papillary carcinoma | 25 (4.9) | 23 (5.0) | 1 (3.9) | 1 (4.6) | 23 (5.0) | 2 (4.2) | ||
| Metaplastic | 8 (1.6) | 8 (1.7) | 0 (0) | 0 (0) | 8 (1.7) | 0 (0) | ||
| Other | 21 (4.1) | 20 (4.3) | 0 (0) | 1 (4.6) | 20 (4.3) | 1 (2.1) | ||
| Ki67 high/low (%) | ||||||||
| Ki67 <15% | 308 (60.2) | 286 (61.6) | 12 (46.2) | 10 (45.5) | 0.104 | 286 (61.6) | 22 (45.8) | 0.033 |
| Ki67 ≥15% | 204 (39.8) | 178 (38.4) | 14 (53.9) | 12 (54.6) | 178 (38.4) | 26 (54.2) | ||
| Mitoses/10 HPF, median (IQR p25, p75) | 5 (1, 12) | 5 (1, 11) | 9 (3,20) | 6 (2, 12) | 5 (1, 12) | 8 (2.5, 16.5) | ||
| Mitoses/10 HPF, quartiles (%) | ||||||||
| ≤1 | 136 (26.6) | 128 (27.6) | 6 (23.1) | 2 (9.1) | 0.025 | 128 (27.6) | 8 (16.7) | 0.162 |
| >1 ≤5 | 133 (26.0) | 123 (26.5) | 1 (3.9) | 9 (40.9) | 123 (26.5) | 10 (20.8) | ||
| >5 ≤12 | 123 (24.0) | 107 (23.1) | 9 (34.6) | 7 (31.8) | 107 (23.1) | 16 (33.3) | ||
| >12 | 120 (23.4) | 106 (22.8) | 10 (38.5) | 4 (18.2) | 106 (22.8) | 14 (29.2) | ||
Abbreviations: SD = standard deviation, HER2 = human epidermal growth factor receptor 2, 5NP = 5 negative phenotype, BP = basal phenotype, HPF = high power fields
PAK1 and CEP11 copy number, and histological grade and proliferation
PAK1 CN ≥4 was found in 48 (9.4%) tumours (Table 3, Fig 2). Of these, 26 (5.1%) cases had mean CN ≥4<6, and 22 (4.3%) had mean CN ≥6. While 147/464 (31.7%) cases with CN <4 were grade 3, 22/48 (45.8%) cases with CN ≥4 were grade 3 (p = 0.037). We found no significant associations between PAK1 CN increase and high histological grade using three categories of mean PAK1 CN (Table 3).
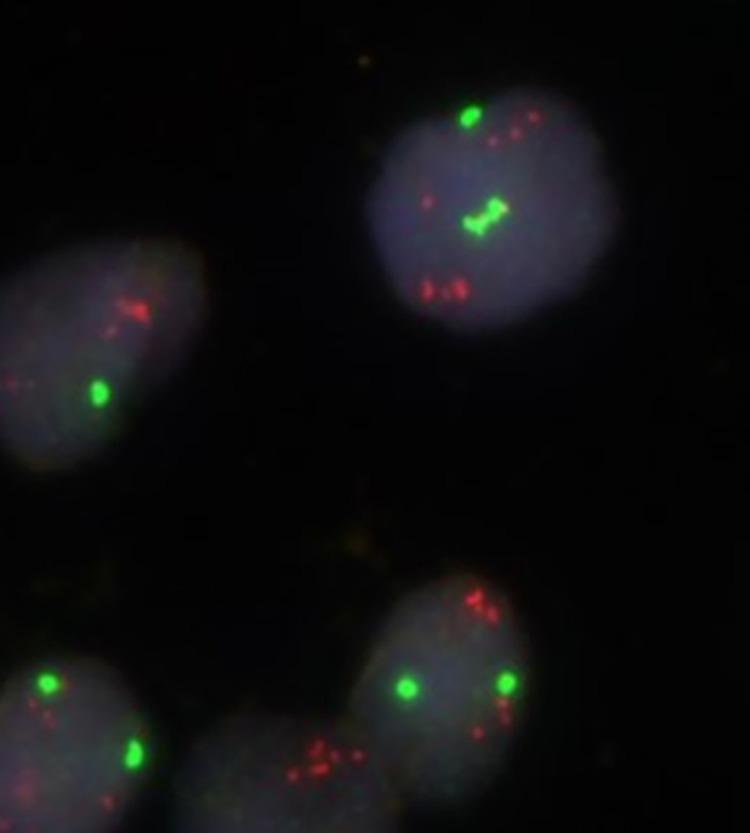
Fig 2. Fluorescence in situ hybridization using probes for CEP11 (fluorochrome SpectrumGreen) and PAK1 (fluorochrome SpectrumRed).
Fig 2 showing 2–3 copies of CEP11 and 6–8 copies of PAK1 in each tumour cell nucleus.
PAK1 CN ≥4 was associated with high Ki-67 (≥15%). Of cases with PAK1 CN <4, 178/464 (38.4%) had Ki-67 ≥15%, compared to 26/48 (54.2%) among those with PAK1 CN ≥4 (p = 0.033). No association between PAK1 CN increase and Ki-67 status was found when PAK1 CN was subdivided into three categories. The median mitotic count was higher in cases with mean PAK1 CN ≥4, compared to cases with mean CN <4 (8 mitoses/10 high power fields [HPF] and 5 mitoses/10 HPF, respectively). The proportion of cases with mitotic counts in the upper quartile was also higher for cases with mean PAK1 CN ≥4, compared to those with mean CN <4 (106/464 [22.8%] and 14/48 (29.2%), respectively (p = 0.162)) (Table 3). Only seven cases showed CEP11 CN increase. Five of these were in cases with PAK1 CN <4. Of the 26 cases with PAK1 CN ≥4<6, only two were accompanied by CEP11 CN increase (≥4<6). Of the 22 cases with PAK1 CN ≥6, none had concurrent CN increase of CEP11.
PAK1 copy number and molecular subtypes
Copy number increase of PAK1 was found in all molecular subtypes, except the 5-negative phenotype (5NP). The highest proportion of cases with PAK1 CN ≥4 was found in the HER2 type, followed by Luminal B (HER2-). Of a total of 27 cases of the HER2 type, four (14.7%) had PAK1 CN ≥4, one of which (3.7%) had PAK1 CN ≥6. In Luminal B (HER2-), 16/121 (13.2%) had PAK1 CN ≥4, and of these, 8/121 (6.6%) had PAK1 CN ≥6. Among Luminal B (HER2+) cases, 3/42 (7.1%) showed PAK1 CN ≥4 (Table 3).
PAK1 and prognosis
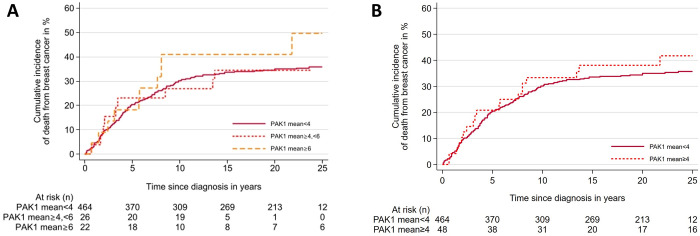
The cumulative risk of death from BC during the first 5 years after diagnosis was 20.3% (95%CI 16.9–24.2) for cases with mean PAK1 CN <4, 23.1% (95% CI 11.1–44.3) for cases with CN ≥4<6, and 18.2% (95% CI 7.2–41.5) for cases with CN ≥6 (Fig 3, Table 4). During the first 10 years after diagnosis, the cumulative risk of death from BC was 30.1% (95% CI 26.1–34.5) for cases with mean PAK1 CN <4, 26.9% (95% CI 13.9–48.3) for cases with CN ≥4<6, and 40.9% (95% CI 23.8–63.9) for cases with CN ≥6. In the Cox regression analyses using mean PAK1 CN <4 as the reference, no significantdifference was observed in the rate of death from breast cancer for cases with PAK1 CN increase (HR 1.4 [95% CI 0.8–2.7]) for cases with mean PAK1 copy number ≥6). Fourteen of the 123 cases for which lymph node metastases were available had PAK1 CN ≥4 in the primary tumour. Of these, 8 also had PAK1 CN ≥4 in the corresponding lymph node metastasis. Of the five cases with PAK1 CN ≥6 in the primary tumour, 3 also had PAK1 CN ≥6 in the corresponding lymph node metastasis (Table 5).
Fig 3. Cumulative incidence of death from breast cancer according to mean PAK1 copy number in primary breast cancer tumours.
Cumulative incidence curves show no significant association between PAK1 copy number and risk of death. A) Mean PAK1 copy number <4, ≥4<6 and ≥6. p = 0.39. B) Mean PAK1 copy number <4 and ≥4. p = 0.42.
Table 4. Absolute and relative risk of death from breast cancer according to mean PAK1 copy number/tumour cell nucleus in primary tumours.
| Mean PAK1 copy number | |||
|---|---|---|---|
| <4 | ≥4<6 | ≥6 | |
| Cumulative risk after 5 years (%) (95% CI) | 20.3(16.9–24.2) | 23.1 (11.1–44.3) | 18.2 (7.2–41.5) |
| Cumulative risk after 10 years (%) (95% CI) | 30.1 (26.1–34.5) | 26.9 (13.9–48.3) | 40.9 (23.8–63.9) |
| HR unadjusted (95% CI) | 1.0 | 0.9 (0.5–1.8) | 1.4 (0.8–2.7) |
| HR adjusted for age (95% CI) | 1.0 | 0.9 (0.5–1.8) | 1.5 (0.8–2.7) |
| HR adjusted for stage (95% CI) | 1.0 | 0.8 (0.4–1.6) | 1.7 (0.9–3.2) |
| HR adjusted for grade (95% CI) | 1.0 | 0.8 (0.4–1.6) | 1.4 (0.8–2.6) |
| HR adjusted for Ki-67 (95% CI) | 1.0 | 0.8 (0.4–1.7) | 1.3 (0.7–2.3) |
| HR adjusted for age, stage, and grade (95% CI) | 1.0 | 0.8 (0.4–1.5) | 1.7 (0.9–3.2) |
Abbreviations: HR = Hazard ratio, CI = confidence interval
Table 5. PAK1 copy number in primary tumours and corresponding axillary lymph node metastases.
| Mean PAK1 copy number in primary tumours (%) | ||||
|---|---|---|---|---|
| Mean PAK1 copy number in lymph node metastases (%) | <4 | ≥4<6 | ≥6 | Total |
|
<4
≥4<6 ≥6 Total |
103 (94.5) | 6 (66.7) | 0 | 109 |
| 5 (4.6) | 3 (33.3) | 2 (40) | 10 | |
| 1 (0.9) | 0 | 3 (60) | 4 | |
| 109 | 9 | 5 | 123 | |
| Mean PAK1 copy number in primary tumours (%) | ||||
| Mean PAK1 copy number in lymph node metastases (%) | <4 | ≥4 | Total | |
|
<4
≥4 Total |
103 (94.5) | 6 (42.9) | 109 | |
| 6 (5.5) | 8 (57.1) | 14 | ||
| 109 | 14 | 123 | ||
PAK1 and CCND1
Among the 512 cases included in this study, CCND1 CN status was available for 504 cases [24]. A total of 84/504 cases showed CCND1 CN ≥4 and 40 of these had ≥6 copies of CCND1/nucleus (Table 6). Of the 22 patients with PAK1 CN ≥6, 12 (54.6%) cases also had CCND1 CN ≥6. Of the 48 cases with PAK1 CN ≥4, 30 (62.5%) cases also had CCND1 CN ≥4. However, 54 cases had CCND1 CN ≥4 without a corresponding increase in PAK1 CN and 18 cases showed CN increase ≥4 for PAK1 without CN increase of CCND1 (Table 6).
Table 6. PAK1 and CCND1 copy numbers in primary tumours.
| Mean PAK1 CN in primary tumours (%) | |||||
|---|---|---|---|---|---|
| Mean CCND1 CN | <4 | ≥4<6 | ≥6 | Total | |
| <4 | 402 (88.2) | 11 (42.3) | 7 (31.8) | 420 | p<0.001 |
| ≥4<6 | 31 (6.8) | 10 (38.5) | 3 (13.6) | 44 | |
| ≥6 | 23 (5.0) | 5 (19.2) | 12 (54.6) | 40 | |
| Total | 456 | 26 | 22 | 504 | |
| Mean PAK1 CN in primary tumours (%) | |||||
| Mean CCND1 CN | <4 | ≥4 | Total | ||
| <4 | 402 (88.2) | 18 (37.5) | 420 | p<0.001 | |
| ≥4 | 54 (11.8) | 30 (62.5) | 84 | ||
| Total | 456 | 48 | 504 | ||
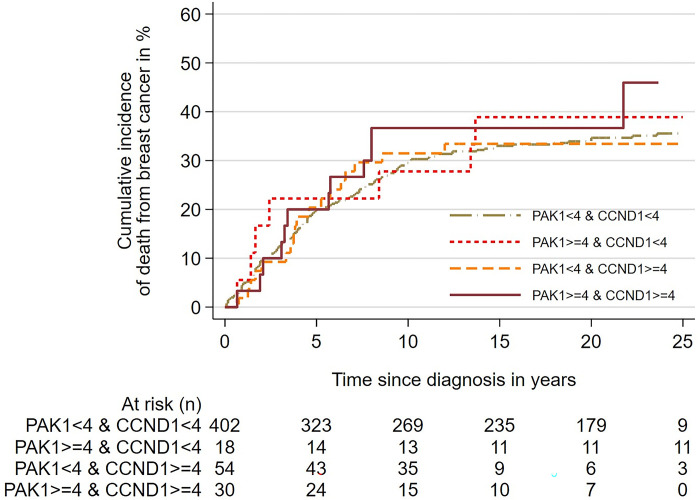
We found no significant difference in the cumulative risk of death from BC between cases with CN ≥4 of PAK1 alone, CN ≥4 CCND1 alone, and cases with CN ≥4 for both PAK1 and CCND1 combined (Fig 4). Similarly, The Cox regression analysis using combined PAK1 CN <4 and CCND CN <4 as the reference value, showed no significant difference in the rate of death from BC between the three groups of patients with copy number increase (Table 7).
Fig 4. Cumulative incidence of death from breast cancer according to copy numbers of PAK1 and CCND1, and co-amplification of PAK1 and CCND1.
Cumulative incidence curves show no significant association between PAK1 copy number, CCND1 copy number, and co-amplification of PAK1 and CCND1, and risk of death. p = 0,81.
Table 7. Relative risk of death from breast cancer according to copy numbers of PAK1 and CCND1, and co-amplification of PAK1 and CCND1.
| Copy number of PAK1 and CCND1 | Hazard ratio | ||
|---|---|---|---|
| HR | CI | p-value | |
| PAK1 CN<4 & CCND1 CN<4 (reference value) | 1.0 | 0.872 | |
| PAK1 CN≥4 & CCND1 CN<4 | 1.3 | 0.6–2.6 | |
| PAK1 CN<4 & CCND1 CN≥4 | 0.9 | 0.6–1.5 | |
| PAK1 CN≥4& CCND1 CN≥4 | 1.1 | 0.6–2.0 | |
Hazard ratio = HR, Confidence interval = CI
Discussion
In this study of 512 primary BC tumours, we found PAK1 CN ≥4 in 48 (9.4%) cases, of which 22 cases showed high grade CN increase of PAK1 CN ≥6. We found an association between PAK1 CN ≥4, and high Ki-67 (≥15%) and high histological grade. The highest proportion of cases with increased CN of PAK1 (≥4) was found in the HER2 type and Luminal B (HER2-) breast cancer subtype. Concurrent CN increase (≥4) of PAK1 and CCND1 was observed in 30 cases. Of the 123 cases with available lymph node metastases, only three cases had PAK1 CN ≥6 in both the primary tumour and the corresponding lymph node metastases.
The cohort of Norwegian BC patients from which the cases of this study are derived is well-described, with mean follow-up of nine years. Since recurrence and death from BC may occur many years after the primary diagnosis, long-term follow-up is important in studies of prognostic markers. While recurrence data was unavailable to us, long-term survival data is complete, enabling us to assess the influence of biomarkers on prognosis. Histological typing and grading of all cases in this cohort were revised by experienced pathologists according to current guidelines. All biomarkers were stained at the same laboratory, and the same antibodies, cut-off levels and algorithm for molecular subtyping were used for all cases in the cohort [26].
In this study we used FISH on TMAs. TMAs provide the opportunity to efficiently study biomarkers in a large number of samples simultaneously under similar laboratory conditions at a relatively low cost. FISH is a method available in most laboratories, contrary to multigene assays. It enables us to assess the morphology of the section and ensure that only invasive tumour cell nuclei are assessed. Despite this, FISH applied to tissue sections may lead to an underestimation of CN compared to analysis of whole nuclei, due to nuclear truncation [34]. This would be of particular importance in cases with low CN increase. Preanalytical conditions will have varied considering that the cases included in the present study were diagnosed over decades. This could have affected the cases suitable for FISH analysis. However, few cases were discarded due to unsuccessful FISH. There are no established guidelines for cut-off levels in the assessment of PAK1 CN. We chose to follow HER2 guidelines for categorizing CN, as in previous studies by our group [24, 29–32]. While we also registered CN of CEP 11, we did not calculate the ratio between CNs of PAK1 and CEP11 as this would have masked the true gene CN increase. Furthermore, we found that CEP11 CN increase was observed in only seven cases, of which only two were accompanied by CN increase of PAK1.
Tamoxifen is an established hormonal therapy used in ER positive BC. Five years of tamoxifen therapy nearly halves the risk of BC recurrence among ER positive patients [35]. Phosphorylation of ER by PAK1 may induce tamoxifen-resistance in ER positive tumours and tamoxifen itself may also increase nuclear PAK1 and PAK1 kinase activity [14, 23]. Patients with PAK1 amplification have reduced benefit from tamoxifen and PAK1 CN may therefore be a predictor of tamoxifen resistance [23]. Thus, PAK1-inhibitors may be useful in ER positive tumours, to improve the effect of tamoxifen in these cases [36].
Both PAK1 and CCND1 encode proteins shown to activate ER [23, 36]. Both are located on 11q13 and are thought to be frequently co-amplified. In this study, of the 504 patients analyzed for both CCND1 and PAK1, 84 cases had CN ≥4 for CCND1 and 48 with PAK1 CN ≥4. A total of 30 (62.5%) cases had CN increase of both genes. These results are in accordance with the findings of others [23]. In the present study, co-amplification of PAK1 and CCND1 was not associated with prognosis.
The proportion of cases with increased PAK1 CN in this study was lower compared to the results of previous studies [7, 8]. However, the mean age at diagnosis in our study was 75.4 years, which is high compared to other studies and higher than the mean age for diagnosis of breast cancer in Norway which is 62 years of age [37]. Fumagalli et al found CN increase in 11% of cases in a selected series of ER+, metastatic breast cancer cases. In our series of cases, PAK1 CN increase was found among Luminal B HER2- and the HER2 type [38]. High proliferation rate and poor prognosis are found to be associated in BC [39, 40], and the prognostic effect of proliferation has been shown to vary with age, exerting a greater effect on prognossis among younger BC patients [41]. This may, in part, explain the discrepant results compared to other studies of PAK1 and further studies including a wider age range are warranted. Furthermore, the choice of method may also have contributed to these results. Tissue microarrays include only small tissue cylinders from the tumour and may not be representative of the whole tumour, particularly in cases with tumour heterogeneity [42, 43]. In the TMAs used in our study, tissue cylinders were extracted from the tumour periphery and are therefore not necessarily representative of other areas of the tumour. However, we considered the tumour periphery to be the region of greatest interest in the tumour given its greater proliferative activity [44] and its proximity to surrounding breast tissue. Furthermore, selecting tissue for TMA form the same region of all tumours contributes to a certain standardization of the material examined in the study.
Despite associations between PAK1 CN increase and high histological grade and high proliferation, we failed to demonstrate a statistically significant association between increased PAK1 CN and prognosis. It would be interesting to study prognosis according to PAK1 CN for each of the molecular subtypes separately. However, in the present study the number of cases in some of the molecular subtypes was too low to warrant further analyses of subgroups. The numbers of cases showing PAK1 CN increase in primary tumours only, lymph node metastases only, or both were too low to give reliable prognostic information. The frequency of PAK1 CN change in this study was lower than the expression of established biomarkers, such as ER, PR and HER2 in BC. However, in an era of personalized medicine, its known influence on the effect of tamoxifen in BC makes it an interesting biomarker and potential target for treatment.
Conclusion
PAK1 CN increase is found in all molecular subtypes, except the 5-negative phenotype (5NP), and most frequently in the HER2 and Luminal B (HER2-) subtypes. It is associated with aggressive tumour characteristics such as high histological grade and high Ki-67 protein expression, but not with prognosis. It is co-amplified with CCND1 in a proportion of cases. Few cases showed PAK1 CN increase in both the primary tumour and the corresponding lymph node metastases.
Acknowledgments
The authors thank the Department of Pathology, St. Olav´s Hospital, Trondheim University Hospital for making the diagnostic archives available for the study, and the Cancer Registry of Norway for supplying the patient data.
Data Availability
The datasets generated in the current study are not publicly available due to ethical and legal restrictions imposed by General Data Protection Regulations (GDPR), National health research legislation and the conditions for approval by the Regional Committee for Medical and Health Research Ethics, Midt-Norge (REK 836/2009), but may be available from the corresponding author on reasonable request and/or the Institutional Research Officer, Department of Clinical and Molecular Medicine, Faculty of Medicine, NTNU at postmottak@mh.ntnu.no.
Funding Statement
The research leading to these results received funding from The Liaison Committee between the Central Norway Regional Health Authority and the Norwegian University of Science and Technology (NTNU), The Joint Research Committee between St. Olav’s Hospital and the Faculty of Medicine and Health Sciences, NTNU (FFU), and the Department of Clinical and Molecular Medicine, NTNU. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ye D. Z. and Field J., "PAK signaling in cancer," (in eng), Cell Logist, vol. 2, no. 2, pp. 105–116, Apr 1 2012, doi: 10.4161/cl.21882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ong C. C. et al. , "Targeting p21-activated kinase 1 (PAK1) to induce apoptosis of tumor cells," Proceedings of the National Academy of Sciences, vol. 108, no. 17, pp. 7177–7182, 2011, doi: 10.1073/pnas.1103350108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arias-Romero L. E. and Chernoff J., "A tale of two Paks," (in eng), Biol Cell, vol. 100, no. 2, pp. 97–108, Feb 2008, doi: 10.1042/bc20070109 [DOI] [PubMed] [Google Scholar]
- 4.Radu M., Semenova G., Kosoff R., and Chernoff J., "PAK signalling during the development and progression of cancer," Nat Rev Cancer, vol. 14, no. 1, pp. 13–25, Jan 2014, doi: 10.1038/nrc3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rane C. K. and Minden A., "P21 activated kinase signaling in cancer," (in eng), Semin Cancer Biol, vol. 54, pp. 40–49, Feb 2019, doi: 10.1016/j.semcancer.2018.01.006 [DOI] [PubMed] [Google Scholar]
- 6.Kumar R., Sanawar R., Li X., and Li F., "Structure, biochemistry, and biology of PAK kinases," (in eng), Gene, vol. 605, pp. 20–31, Mar 20 2017, doi: 10.1016/j.gene.2016.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shrestha Y. et al. , "PAK1 is a breast cancer oncogene that coordinately activates MAPK and MET signaling," Oncogene, vol. 31, no. 29, pp. 3397–408, Jul 19 2012, doi: 10.1038/onc.2011.515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balasenthil S. et al. , "p21-activated kinase-1 signaling mediates cyclin D1 expression in mammary epithelial and cancer cells," J Biol Chem, vol. 279, no. 2, pp. 1422–8, Jan 9 2004, doi: 10.1074/jbc.M309937200 [DOI] [PubMed] [Google Scholar]
- 9.Dang Y. et al. , "Systemic analysis of the expression and prognostic significance of PAKs in breast cancer," Genomics, vol. 112, no. 3, pp. 2433–2444, 2020/05/01/ 2020, doi: 10.1016/j.ygeno.2020.01.016 [DOI] [PubMed] [Google Scholar]
- 10.Song P., Song B., Liu J., Wang X., Nan X., and Wang J., "Blockage of PAK1 alleviates the proliferation and invasion of NSCLC cells via inhibiting ERK and AKT signaling activity," Clinical and Translational Oncology, vol. 23, no. 4, pp. 892–901, 2021/04/01 2021, doi: 10.1007/s12094-020-02486-5 [DOI] [PubMed] [Google Scholar]
- 11.Wang R. A., Zhang H., Balasenthil S., Medina D., and Kumar R., "PAK1 hyperactivation is sufficient for mammary gland tumor formation," (in eng), Oncogene, vol. 25, no. 20, pp. 2931–6, May 11 2006, doi: 10.1038/sj.onc.1209309 [DOI] [PubMed] [Google Scholar]
- 12.Park J. et al. , "Association of p21-activated kinase-1 activity with aggressive tumor behavior and poor prognosis of head and neck cancer," (in eng), Head Neck, vol. 37, no. 7, pp. 953–63, Jul 2015, doi: 10.1002/hed.23695 [DOI] [PubMed] [Google Scholar]
- 13.Siu M. K. et al. , "Differential expression and phosphorylation of Pak1 and Pak2 in ovarian cancer: effects on prognosis and cell invasion," (in eng), Int J Cancer, vol. 127, no. 1, pp. 21–31, Jul 1 2010, doi: 10.1002/ijc.25005 [DOI] [PubMed] [Google Scholar]
- 14.Holm C., Rayala S., Jirstrom K., Stal O., Kumar R., and Landberg G., "Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients," J Natl Cancer Inst, vol. 98, no. 10, pp. 671–80, May 17 2006, doi: 10.1093/jnci/djj185 [DOI] [PubMed] [Google Scholar]
- 15.Mira J. P., Benard V., Groffen J., Sanders L. C., and Knaus U. G., "Endogenous, hyperactive Rac3 controls proliferation of breast cancer cells by a p21-activated kinase-dependent pathway," (in eng), Proc Natl Acad Sci U S A, vol. 97, no. 1, pp. 185–9, Jan 4 2000, doi: 10.1073/pnas.97.1.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanumuri R., Saravanan R., Pavithra V., Sundaram S., Rayala S. K., and Venkatraman G., "Current trends and opportunities in targeting p21 activated kinase-1(PAK1) for therapeutic management of breast cancers," Gene, vol. 760, p. 144991, Nov 15 2020, doi: 10.1016/j.gene.2020.144991 [DOI] [PubMed] [Google Scholar]
- 17.Semenova G. and Chernoff J., "Targeting PAK1," (in eng), Biochem Soc Trans, vol. 45, no. 1, pp. 79–88, Feb 8 2017, doi: 10.1042/BST20160134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pérez-Yépez E. A., Saldívar-Cerón H. I., Villamar-Cruz O., Pérez-Plasencia C., and Arias-Romero L. E., "p21 Activated kinase 1: Nuclear activity and its role during DNA damage repair," (in eng), DNA Repair (Amst), vol. 65, pp. 42–46, May 2018, doi: 10.1016/j.dnarep.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 19.Rajendran S. et al. , "p21 activated kinase-1 and tamoxifen—A deadly nexus impacting breast cancer outcomes," (in eng), Biochim Biophys Acta Rev Cancer, vol. 1877, no. 1, p. 188668, Jan 2022, doi: 10.1016/j.bbcan.2021.188668 [DOI] [PubMed] [Google Scholar]
- 20.Agarwal S. and Kashaw S. K., "Potential target identification for breast cancer and screening of small molecule inhibitors: A bioinformatics approach," (in eng), J Biomol Struct Dyn, vol. 39, no. 6, pp. 1975–1989, Apr 2021, doi: 10.1080/07391102.2020.1743757 [DOI] [PubMed] [Google Scholar]
- 21.Saldivar-Cerón H. I. et al. , "p21-Activated Kinase 1 Promotes Breast Tumorigenesis via Phosphorylation and Activation of the Calcium/Calmodulin-Dependent Protein Kinase II," (in eng), Front Cell Dev Biol, vol. 9, p. 759259, 2021, doi: 10.3389/fcell.2021.759259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wigerup C., Rayala S., Jirström K., Stål O., Kumar R., and Landberg G., "Holm C, Rayala S, Jirstrom K, Stal O, Kumar R, Landberg G Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients. J Natl Cancer Inst 98: 671–680," Journal of the National Cancer Institute, vol. 98, pp. 671–80, 06/01 2006, doi: 10.1093/jnci/djj185 [DOI] [PubMed] [Google Scholar]
- 23.Bostner J., Ahnström Waltersson M., Fornander T., Skoog L., Nordenskjöld B., and Stål O., "Amplification of CCND1 and PAK1 as predictors of recurrence and tamoxifen resistance in postmenopausal breast cancer," Oncogene, vol. 26, no. 49, pp. 6997–7005, 2007/10/01 2007, doi: 10.1038/sj.onc.1210506 [DOI] [PubMed] [Google Scholar]
- 24.Valla M., Klæstad E., Ytterhus B., and Bofin A. M., "CCND1 Amplification in Breast Cancer -associations With Proliferation, Histopathological Grade, Molecular Subtype and Prognosis," (in eng), J Mammary Gland Biol Neoplasia, vol. 27, no. 1, pp. 67–77, Mar 2022, doi: 10.1007/s10911-022-09516-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.KVÂLE G., HEUCH I., and EIDE G. E., "A PROSPECTIVE STUDY OF REPRODUCTIVE FACTORS AND BREAST CANCER: I. PARITY," American Journal of Epidemiology, vol. 126, no. 5, pp. 831–841, 1987, doi: 10.1093/oxfordjournals.aje.a114720 [DOI] [PubMed] [Google Scholar]
- 26.Engstrom M. J. et al. , "Molecular subtypes, histopathological grade and survival in a historic cohort of breast cancer patients," Breast Cancer Res Treat, vol. 140, no. 3, pp. 463–73, Aug 2013, doi: 10.1007/s10549-013-2647-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elston C. W. and Ellis I. O., "Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up," Histopathology, vol. 19, no. 5, pp. 403–10, Nov 1991, doi: 10.1111/j.1365-2559.1991.tb00229.x [DOI] [PubMed] [Google Scholar]
- 28.Lakhani S. R., WHO classification of tumours of the breast, 4th ed. (World Health Organization Classification of Tumours). Lyon, France: International Agency for Research on Cancer (in English), 2012. [Google Scholar]
- 29.Wolff A. C. et al. , "Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Focused Update," (in eng), J Clin Oncol, vol. 36, no. 20, pp. 2105–2122, Jul 10 2018, doi: 10.1200/jco.2018.77.8738 [DOI] [PubMed] [Google Scholar]
- 30.Bofin A. M., Ytterhus B., Klæstad E., and Valla M., "FGFR1 copy number in breast cancer: associations with proliferation, histopathological grade and molecular subtypes," (in eng), J Clin Pathol, Mar 22 2021, doi: 10.1136/jclinpath-2021-207456 [DOI] [PubMed] [Google Scholar]
- 31.Valla M., Opdahl S., Ytterhus B., and Bofin A. M., "DTX3 copy number increase in breast cancer: a study of associations to molecular subtype, proliferation and prognosis," (in eng), Breast Cancer Res Treat, vol. 187, no. 1, pp. 57–67, May 2021, doi: 10.1007/s10549-021-06138-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klæstad E., Sawicka J. E., Engstrøm M. J., Ytterhus B., Valla M., and Bofin A. M., "ZNF703 gene copy number and protein expression in breast cancer; associations with proliferation, prognosis and luminal subtypes," (in eng), Breast Cancer Res Treat, vol. 186, no. 1, pp. 65–77, Feb 2021, doi: 10.1007/s10549-020-06035-0 [DOI] [PubMed] [Google Scholar]
- 33.Sauerbrei W., Taube S. E., McShane L. M., Cavenagh M. M., and Altman D. G., "Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): An Abridged Explanation and Elaboration," J Natl Cancer Inst, vol. 110, no. 8, pp. 803–811, Aug 1 2018, doi: 10.1093/jnci/djy088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshimoto M. et al. , "Correction to: Use of multicolor fluorescence in situ hybridization to detect deletions in clinical tissue sections," Laboratory Investigation, vol. 98, no. 6, pp. 839–839, 2018/06/01 2018, doi: 10.1038/s41374-018-0037-4 [DOI] [PubMed] [Google Scholar]
- 35.Davies C. et al. , "Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials," (in eng), Lancet, vol. 378, no. 9793, pp. 771–84, Aug 27 2011, doi: 10.1016/s0140-6736(11)60993-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh A., Awasthi S., Peterson J. R., and Hamburger A. W., "Regulation of tamoxifen sensitivity by a PAK1–EBP1 signalling pathway in breast cancer," British Journal of Cancer, vol. 108, no. 3, pp. 557–563, 2013/02/01 2013, doi: 10.1038/bjc.2013.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Norway C. R. o., "Cancer in Norway 2021—Cancer incidence, mortality, survival and prevalence in Norway.," Oslo, 2022. [Online]. Available: https://www.kreftregisteret.no/globalassets/cancer-in-norway/2021/cin_report.pdf [Google Scholar]
- 38.Fumagalli D. et al. , "Somatic mutation, copy number and transcriptomic profiles of primary and matched metastatic estrogen receptor-positive breast cancers," Ann Oncol, vol. 27, no. 10, pp. 1860–6, Oct 2016, doi: 10.1093/annonc/mdw286 [DOI] [PubMed] [Google Scholar]
- 39.Cheang M. C. et al. , "Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer," (in eng), J Natl Cancer Inst, vol. 101, no. 10, pp. 736–50, May 20 2009, doi: 10.1093/jnci/djp082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wirapati P. et al. , "Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures," (in eng), Breast Cancer Res, vol. 10, no. 4, p. R65, 2008, doi: 10.1186/bcr2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baak J. P. et al. , "The prognostic value of proliferation in lymph-node-negative breast cancer patients is age dependent," (in eng), Eur J Cancer, vol. 43, no. 3, pp. 527–35, Feb 2007, doi: 10.1016/j.ejca.2006.10.001 [DOI] [PubMed] [Google Scholar]
- 42.Pinder S. E. et al. , "The manufacture and assessment of tissue microarrays: suggestions and criteria for analysis, with breast cancer as an example," J Clin Pathol, vol. 66, no. 3, pp. 169–77, Mar 2013, doi: 10.1136/jclinpath-2012-201091 [DOI] [PubMed] [Google Scholar]
- 43.Torhorst J. et al. , "Tissue microarrays for rapid linking of molecular changes to clinical endpoints," Am J Pathol, vol. 159, no. 6, pp. 2249–56, Dec 2001, doi: 10.1016/S0002-9440(10)63075-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jimenez-Sanchez J. et al. , "Evolutionary dynamics at the tumor edge reveal metabolic imaging biomarkers," (in English), P Natl Acad Sci USA, vol. 118, no. 6, Feb 9 2021, ARTN e2018110118 doi: 10.1073/pnas.2018110118 [DOI] [PMC free article] [PubMed] [Google Scholar]