Abstract
Abstract
There is currently an urgent need to identify factors predictive of immunogenicity in colorectal cancer (CRC). Mucinous CRC is a distinct histological subtype of CRC, associated with a poor response to chemotherapy. Recent evidence suggests the commensal facultative anaerobe Fusobacterium may be especially prevalent in mucinous CRC. The objectives of this study were to assess the association of Fusobacterium abundance with immune cell composition and prognosis in mucinous CRC. Our study included two independent colorectal cancer patient cohorts, The Cancer Genome Atlas (TCGA) cohort, and a cohort of rectal cancers from the Beaumont RCSI Cancer Centre (BRCC). Multiplexed immunofluorescence staining of a tumour microarray (TMA) from the BRCC cohort was undertaken using Cell DIVE technology. Our cohorts included 87 cases (13.3%) of mucinous and 565 cases (86.7%) of non-mucinous CRC. Mucinous CRC in the TCGA dataset was associated with an increased proportion of CD8 + lymphocytes (p = 0.018), regulatory T-cells (p = 0.001) and M2 macrophages (p = 0.001). In the BRCC cohort, mucinous RC was associated with enhanced CD8 + lymphocyte (p = 0.022), regulatory T-cell (p = 0.047), and B-cell (p = 0.025) counts. High Fusobacterium abundance was associated with an increased proportion of CD4 + lymphocytes (p = 0.031) and M1 macrophages (p = 0.006), whilst M2 macrophages (p = 0.043) were under-represented in this cohort. Patients with increased Fusobacterium relative abundance in our mucinous CRC TCGA cohort tended to have better clinical outcomes (DSS: likelihood ratio p = 0.04, logrank p = 0.052). Fusobacterium abundance may be associated with improved outcomes in mucinous CRC, possibly due to a modulatory effect on the host immune response.
Key messages
• Increased Fusobacterium relative abundance was not found to be associated with microsatellite instability in mucinous CRC.
• Increased Fusobacterium relative abundance was associated with an M2/M1 macrophage switch, which is especially significant in mucinous CRC, where M2 macrophages are overexpressed.
• Increased Fusobacterium relative abundance was associated with a significant improvement in disease specific survival in mucinous CRC.
• Our findings were validated at a protein level within our own in house mucinous and non-mucinous rectal cancer cohorts.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00109-023-02324-5.
Keywords: Mucinous colorectal cancer, Fusobacterium, Microsatellite instability
Introduction
Mucinous colorectal cancer (CRC) is a histological subtype of colorectal adenocarcinoma, which accounts for approximately 5–15% of all colorectal tumours [1]. These tumours are characterised by an abundance of extracellular mucin, which constitutes more than 50% of the tumour volume [2, 3]. When compared with non-mucinous rectal cancer (RC), mucinous rectal adenocarcinoma is associated with reduced rates of pathological complete response (pCR) and tumour downstaging following neoadjuvant chemoradiotherapy. As a consequence, patients with this disease have an increased likelihood of having a positive resection margin and are associated with poorer definitive outcomes [4]. Mucinous adenocarcinoma of the colon is associated with an increased risk of metastasis, and this cohort has also been shown to be associated with resistance to oxaliplatin and irinotecan-based chemotherapy [5]. Our group have previously demonstrated that mucinous CRCs are more likely to harbour KRAS and BRAF mutations, and are more likely to demonstrate microsatellite instability (MSI), and be of the CPG island methylator phenotype as compared to non-mucinous colorectal tumours [6].
Fusobacterium are a genus of gram-negative facultative anaerobes, commonly encountered in gastrointestinal tract pathologies such as inflammatory bowel disease and cancer [7, 8]. The species of Fusobacterium most commonly associated with colorectal cancer is Fusobacterium nucleatum (F. nucleatum) [7]. F. nucleatum has been shown to be more abundant in colorectal tumour tissue compared with matched adjacent normal mucosa, which has led to the suggestion there may be a potential causative relationship [9, 10]. F. nucleatum promotes a pro-inflammatory state [11], and has been shown in pre-clinical studies to modulate the T-cell-mediated immune response in CRC [12]. F. nucleatum is also known to be more abundant in MSI-high tumours [13]. This is pertinent; given the emergence of evidence demonstrating an association between an increased abundance of various members of the gut microbiome with improved rates of responsivity to immunotherapy in cancer [14, 15].
The prognostic impact of F. nucleatum has been evaluated in a number of cohort studies [16]. Though some studies managed to demonstrate correlation between F. nucleatum abundance and poor prognosis [17, 18], this association was not observed in other studies [16, 19]. A recent publication by our group, premised on the hypothesis that the impact of F. nucleatum/Fusobacteriales may differ according to underlying tumour biology, demonstrated a correlation between increased Fusobacterium abundance and poor prognosis in mesenchymal-type tumours only [11]. A previous whole genome sequencing study, again undertaken by our group, examined 10 mucinous rectal adenocarcinomas, and found F. nucleatum to be significantly more abundant within mucinous tumour tissue [20]. F. nucleatum has previously been shown to promote MUC2, TNF-α and mucin production in colonic cells [21]. F. nucleatum has also been implicated in exacerbations of chronic obstructive pulmonary disease, where it has been shown to contribute towards inflammation and mucin production [22].
In light of this evidence, it was hypothesised there may be a relationship between Fusobacterium abundance and outcomes in mucinous CRC. The aims of this study were to compare Fusobacterium relative abundance at a genus rank level between mucinous and non-mucinous CRC cohorts within the Cancer Genome Atlas (TCGA) dataset, and examine whether an association exists between Fusobacterium abundance and immune cell composition and prognosis in mucinous CRC. We then sought to validate TCGA findings at a protein level, by hyperplexing tumour microarrays (TMAs) which included our own mucinous and non-mucinous cohort from the Beaumont RCSI Cancer Centre (BRCC) with a pan-fusobacterium antisera alongside an array of immune markers.
Materials and methods
TCGA gene expression analysis
We performed a search of TCGA to identify cases of CRC eligible for inclusion. Institutional approval was not required for these open-access data. The inclusion criteria specified stages 1 to 4; mucinous and non-mucinous colorectal adenocarcinoma. Patients of both genders and all age groups and ethnicities were eligible for inclusion in the analysis. The demographic, pathologic, and clinical data for each eligible patient were collated and harmonised from the GDC Legacy Archive and the TCGA-Clinical Data Resource publication [23]. This study focused on the impact of Fusobacterium in mucinous adenocarcinoma, thus we restricted the analysis to patients of the TCGA-Colorectal Adenocarcinoma (COAD)-Rectal Adenocarcinoma (READ) cohort that have both clinical information and Fusobacterium data (n = 594 of 631 candidate cases (94%)). The pathologic variables recorded included TNM stage, lymphovascular invasion (LVI), extramural vascular invasion (EMVI) and MSI status. Patients were also categorised according to consensus molecular subtype [24]. Regarding survival, three end-points were considered; disease-free survival (DFS), disease-specific survival (DSS), and overall survival (OS).
Fusobacterium composition was derived from RNASeq experiments as previously described by Salvucci et al. [11]. Briefly, composition was computationally inferred using PathSeq from RNA experiments by aligning reads not mapped to the host to microbial taxonomic references [25]. We reported Fusobacterium relative abundance (in %) at the genus and species taxonomic ranks. Sub-species/strains reported by PathSeq were remapped to their parent species following sequence blasting, as described in Salvucci et al. [11]. Level 4 transcriptomic data were retrieved from TCGA pancancer release and subset to include only measurements from primary tumours of patients diagnosed with COAD and READ. Cell type composition was computed using the quanTIseq package, as previously described [11].
Immunofluorescence staining of tumour microarrays
Formalin-fixed, paraffin-embedded primary tumour tissue sections were obtained from patients with stage I-III rectal cancer following tumour resection at the BRCC. Tissue was provided from the Beaumont Hospital Colorectal Biobank with written consent provided by all patients. Institutional ethical approval was granted by the Beaumont Ethics (Medical Research) Committee (Reference 21/98). Mucinous tumours were defined by a consultant histopathologist as those with greater than 50% of the tumour composed of mucin. To construct the TMA 1 mm punches were taken from different regions within the centre of the tumour. Multiplexed immunofluorescence staining of the tumour micro-array with relevant immune markers (CD3, CD4, CD8, CD20 and forkhead box P3 (FOXP3)) were performed using Cell DIVE™ technology (Leica Microsystems, Issaquah, USA). This involves multiple rounds of antibody staining performed on the same tissue section with mild dye oxidation between successive rounds of staining and imaging [26]. Epithelial cells were segmented using stains against DAPI, and antibodies for pan-cytokeratin (CK-26), ribosomal protein S6 (S6) and Na+K+ATPase and stromal cells were segmented using DAPI. Antibodies were acquired commercially and underwent a multi-step process of validation and conjugation (as previously described by Gerdes et al. [26]). Detailed description of the image analysis workflow was published in a larger analysis of 373 tumour cores by our research group [27]. To summarise, immune cells were classified according to cell-level expression and were quantified at tissue core (tumour and stroma) patient level [27]. Immune cell composition in the tumour cores varied significantly, with some cores showing predominantly cancerous/epithelial cells in the absence of immune cell infiltration, and others showing very high levels (up to 55%) of immune cells. A bootstrap analysis using randomly sampled pairings; found cell type composition in cores from the same patient, to be more similar to each other compared to random pairings, suggesting that cell type composition was a biological feature of individual tumours.
Pan-fusobacterium outer membrane antisera
A pan-fusobacterium outer membrane antisera was produced and made available by Professor Slade’s group (Department of Biochemistry, Virginia Polytechnic Institute and State University, Blacksburg, VA 24,061, USA) [28]. Staining of the TMAs with the pan-fusobacterium membrane antisera was conducted separately using the GeoMx imaging platform (Nanostring, Seattle, WA, USA). Separate tissue sections from the corresponding TMAs underwent standard deparaffinisation, ahead of antigen target retrieval using 1X Tris EDTA (PH 9.0) (Abcam). Microarrays were then fixed using 10% neutral buffered formalin. Slides were next blocked with Buffer W (Nanostring) for 30 min at 37 °C. This was followed by the addition of 1:300 dilution of the antisera for 1 h at 37 °C. Slides were next washed twice for 2 min in 2 × SSC wash buffer and incubated for 1 h with Alexa Fluor 594 goat anti-rabbit antibody (Abcam) diluted in buffer W (Nanostring). After 3 further washes with 2 × SSC, TMAs were finally incubated for 1 h with Syto 13 conjugated to Alexa Flour 488 (Nuclear stain) (Nanostring) and pancytokeratin conjugated to Alexa Flour 532 (Nanostring). Exposure time was set to 300 ms for the 594 and 532 channel and 100 ms for the 488 channel. Images were analysed using FIJI imaging software [29]. Images were thresholded and area of staining intensity quantified [30]. To verify the validity of the antisera, sections from 8 tumours which had previously undergone whole genome sequencing, were stained with the antisera. Fusobacterium burden was quantified and levels were compared with Fusobacterium relative abundance as interpreted previously from existing sequencing data [20].
Statistical analysis
For the statistical analysis reported in Fig. 1A, we used Fusobacterium relative abundance as a continuous variable. For all other analyses, we grouped patients into high versus low Fusobacterium relative abundance. We defined high versus low Fusobacterium relative abundance using the 75th percentile as a cut-off. This cut-off was determined from the data and is in agreement with cut-offs used previously in the literature [11].
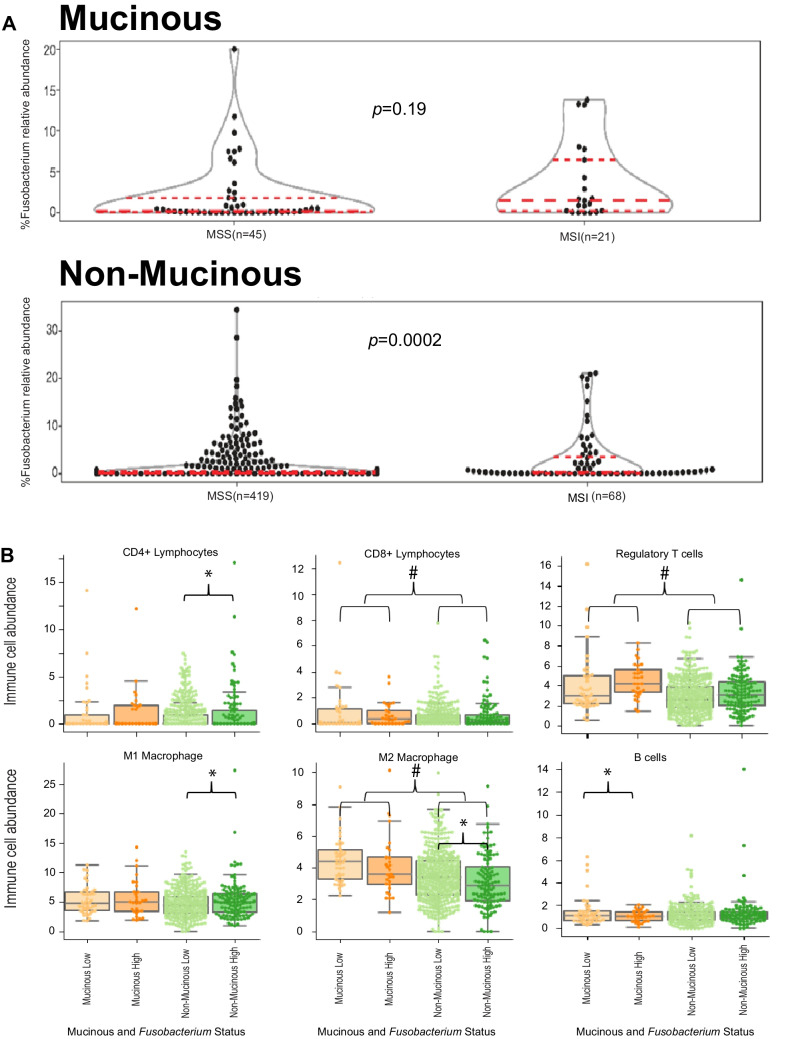
Fig. 1.
Fusobacterium relative abundance was found to impact immunogenicity, prognosis and MSI status in mucinous and non-mucinous CRC in the TCGA-COAD-READ cohort. A Violin plots depicting Fusobacterium relative abundance within the TCGA-COAD-READ cohort according to mucinous status and MSI status. Median, lower (25th) and upper (75th) percentiles are indicated by dashed lines. Statistical significance was evaluated using Kruskal–Wallis tests and p-values are reported. B Box and whisker plots depicting specific immune cell counts; according to mucinous status and Fusobacterium relative abundance (high and low) within the TCGA-COAD-READ cohort. Statistical significance was evaluated using Kruskal–Wallis tests. * indicates a statistically significant difference between Fusobacterium high and Fusobacterium low cohorts. # indicates a statistically significant difference between mucinous and non-mucinous cohorts
Continuous variables were reported as medians with interquartile ranges (IQRs). Group comparisons of continuous variables were determined by Kruskall-Wallis test. Categorical variables were reported as numbers with percentages, group comparisons of categorical variables were determined by Fisher’s exact tests. Differences between Fusobacterium relative abundance by group were determined using a non-negative binomial test (function GLM.NB from the MASS R package) (Fig. 1A). Association between expression of cell types with mucinous status and Fusobacterium relative abundance was assessed by fitting a linear regression model with cell type abundance (continuous, in %) as the response variable and the mucinous status (mucinous vs. non-mucinous), Fusobacterium relative abundance (binary, low vs. high) and the interaction between mucinous status and Fusobacterium relative abundance as predictor terms. We report effect sizes, 95% confidence intervals and likelihood ratio p-values, (Table 2). To avoid overfitting in the BRCC cohort, we conducted variance analyses, and reported p-values only (Table 4). Differences in survival according to Fusobacterium relative abundance were assessed by logrank tests (pLR) and univariate Cox proportional hazard models for which we reported hazard ratio, 95% CI and likelihood ratio test p-value (pLRT) and concordance index (ci), (Fig. 2). Cox regression models were fitted on relative abundance of species from the Fusobacterium genus (binary, low vs. high) by clinical endpoint. The low subgroup was used as reference when reporting the hazard ratios (HRs) estimated from the Cox regression models. Univariate Cox regression models were fitted when evaluating the association between species subgroups in the whole unselected patient population. Cox regression models with an interaction term between species subgroup (binary, low vs. high) and mucinous status (mucinous vs. non-mucinous) were fitted to evaluate differential effect of species relative abundance on clinical outcome by mucinous status (Sup. Fig. 1). A p-value of < 0.05 was defined as the cut off for statistical significance unless otherwise stated. Data pre-processing and analysis was performed in Python (version 3.8.10, Python Software Foundation, Wilmington, DE, USA), unless otherwise stated.
Table 2.
Immune cell expression in the TCGA cohort according to mucinous status and fusobacterium relative abundance as computed using the quanTIseq package
| Cell Type | Level | Coefficient | Lower 95% CI | Upper 95% CI | p-value |
|---|---|---|---|---|---|
| Epithelium/stroma |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium(high) |
− 3.70 − 1.30 1.8 |
− 5.80 − 2.60 − 1.60 |
1.70 0.06 5.10 |
< 0.001 0.061 0.297 |
| CD4 + lymphocytes |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium(high) |
0.40 0.42 − 0.30 |
− 0.19 0.04 − 1.20 |
0.98 0.79 0.65 |
0.182 0.032 0.538 |
| CD8 + lymphocytes |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium (high) |
0.42 0.14 − 0.36 |
0.07 − 0.08 − 0.92 |
0.76 0.37 0.20 |
0.018 0.207 0.204 |
| T Regs |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium (high) |
1.1 0.39 0.12 |
0.46 < − 0.01 − 0.87 |
1.70 0.78 1.10 |
< 0.001 0.053 0.807 |
| Dendritic cells |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium (high) |
− 0.02 0.03 − 0.04 |
− 0.15 − 0.05 − 0.25 |
0.11 0.12 0.17 |
0.744 0.458 0.713 |
| B cells |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium (high) |
0.28 0.14 − 0.54 |
− 0.03 − 0.06 − 1.00 |
0.59 0.35 − 0.04 |
0.078 0.159 0.035 |
| M1 macrophage |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium (high) |
0.63 0.73 − 0.39 |
− 0.18 0.2 − 1.7 |
1.40 1.30 0.93 |
0.129 0.007 0.565 |
| M2 macrophage |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium (high) |
0.96 − 0.35 < − 0.01 |
0.44 − 0.69 − 0.85 |
1.5 − 0.01 0.85 |
< 0.001 0.040 0.990 |
| Natural killer cells |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium (high) |
− 0.03 − 0.09 0.01 |
− 0.41 − 0.34 − 0.62 |
0.36 0.16 0.63 |
0.899 0.484 0.976 |
| Neutrophils |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium (high) |
0.31 − 0.16 − 0.25 |
− 0.96 − 0.80 − 1.90 |
1.00 0.49 1.40 |
0.951 0.635 0.756 |
Table 4.
Immune cell expression in the BRCC cohort according to mucinous status and fusobacterium relative abundance
| Cell type | Level | p-value |
|---|---|---|
| Epithelium/stroma |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium(high) |
0.914 0.384 0.491 |
| CD4 + lymphocytes |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium(high) |
0.373 0.202 0.950 |
| CD8 + lymphocytes |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium (high) |
0.022 0.097 0.491 |
| T regs |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium (high) |
0.047 0.454 0.754 |
| B cells |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium (high) |
0.025 0.029 0.228 |
| Other immune cells |
Mucinous status(mucinous) Fusobacterium (high) Mucinous status(mucinous): Fusobacterium (high) |
0.103 0.006 0.662 |
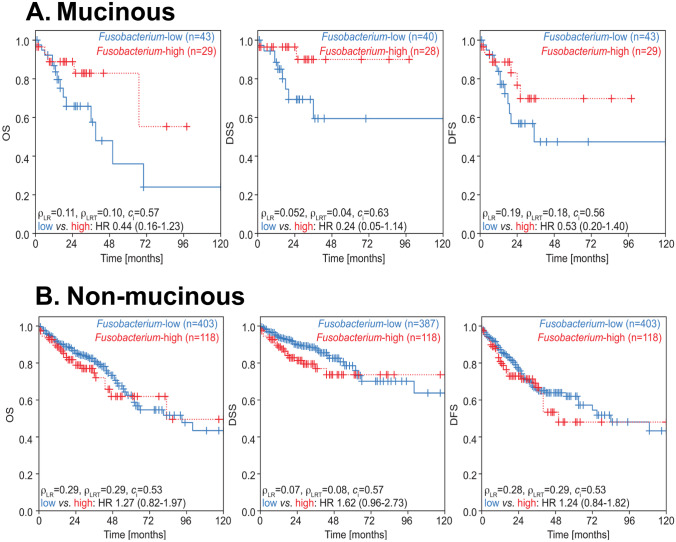
Fig. 2.
Kaplan Meier Curves depicting survival differences between patients grouped by mucinous status and by Fusobacterium relative abundance. Differences in survival outcomes were assessed by logrank tests (p_LR) and univariate Cox proportional hazard models (hazard ratio, 95% CI and likelihood ratio test p-value (p_LRT) and concordance index (c_i))
Results
Increased Fusobacterium relative abundance was not associated with MSI status in mucinous CRC in the TCGA dataset
From the TCGA dataset, we included 72 cases (12%) of mucinous and 522 cases (88%) of non-mucinous CRC. The clinical and pathologic characteristics of the included patients are summarised in Table 1. We found mucinous tumours were more likely to be proximal (p < 0.001) and had a higher incidence of MSI compared to the non-mucinous CRC group (p < 0.001). The two cohorts also demonstrated significant differences when categorised by consensus molecular subtype (p < 0.001). Mucinous tumours were over-represented in CMS1 (immune) (26.4% vs 11.7%) and CMS4 (mesenchymal) (31.9% vs 23%) categories and under-represented in the CMS2 (canonical) group. Following an investigation into whether the relative abundance (RA) of Fusobacterium at the genus taxonomic rank differed between mucinous and non-mucinous CRC, we found a trend, albeit not statistically significant, whereby patients with mucinous CRC trended to have higher Fusobacterium relative abundance compared to patients with non-mucinous CRC (p = 0.07).
Table 1.
Clinical characteristics of the patients of the TCGA-COAD-READ cohort. CRC, colorectal cancer; AJCC, American Joint Committee on Cancer; CMS, consensus molecular subtypes; IQR, inter-quartile range. Categorical data reported as n (%). Continuous data reported as median (IQR)
|
Mucinous CRC (n = 72) |
Non-mucinous CRC (n = 522) |
p-value | ||
|---|---|---|---|---|
| Age | 67 (52–77.5) | 68 (58–76) | 0.611 | |
| Sex | Male | 37 (51.4%) | 241 (46.2%) | 0.48 |
| Female | 35 (48.6%) | 281 (53.8%) | ||
| Tumor location (n = 579)a | Colon | 58 (81.7%) | 366 (72%) | 0.115 |
| Rectum | 13 (18.3%) | 142 (28%) | ||
| Stage (n = 574)a | AJCC 1 | 10 (14.1%) | 90 (17.9%) | 0.571 |
| AJCC 2 | 28 (39.4%) | 189 (37.6%) | ||
| AJCC 3 | 25 (35.2%) | 147 (29.2) | ||
| AJCC 4 | 8 (11.3%) | 77 (15.3%) | ||
| T Stage (n = 593)a | T1 | 2 (2.8%) | 18 (3.5%) | 0.528 |
| T2 | 9 (12.5%) | 90 (17.3%) | ||
| T3 | 50 (69.4%) | 358 (68.7%) | ||
| T4 | 11 (15.3%) | 55 (10.6%) | ||
| N Stage (n = 591)a | N0 | 39 (54.2%) | 297 (57.2%) | 0.528 |
| N1 | 16 (22.2%) | 128 (24.7%) | ||
| N2 | 17 (23.6%) | 94 (18.1%) | ||
| M Stage (n = 525)a | M0 | 51 (86.4%) | 389 (83.5%) | 0.693 |
| M1 | 8 (13.6%) | 77 (16.5%) | ||
| Resection margin (n = 438)a | R0 | 51 (96.2%) | 381 (99%) | 0.157 |
| R1/R2 | 2 (3.8%) | 4 (1%) | ||
| Lymphovascular invasion (n = 535)a | Yes | 22 (33.8%) | 197 (41.9%) | 0.269 |
| No | 43 (66.2%) | 273 (58.1%) | ||
| Perineural invasion (n = 230)a | Yes | 6 (22.2%) | 54 (26.6%) | 0.800 |
| No | 21 (77.8%) | 149 (73.4%) | ||
| Vascular invasion (n = 514)a | Yes | 12 (20%) | 115 (25.3%) | 0.459 |
| No | 48 (80%) | 339 (74.7%) | ||
| Microsatellite status (n = 553)a | MSI | 21 (31.8%) | 68 (14%) | < 0.001 |
| MSS | 45 (68.2%) | 419 (86%) | ||
| CMS (n = 536)a | CMS1 | 19 (26.4%) | 61 (11.7%) | < 0.001 |
| CMS2 | 4 (5.6%) | 234 (44.8%) | ||
| CMS3 | 20 (27.8%) | 55 (10.5%) | ||
| CMS4 | 23 (31.9%) | 120 (23%) |
aData not available in full cohort: n in parentheses = number with data available
Next, we sought to investigate the relationship between Fusobacterium abundance and MSI status. In keeping with pre-existing evidence, when we restricted our analysis to non-mucinous CRC cases, we found MSI tumours to be strongly associated with an increased abundance of Fusobacterium (p < 0.001) (Fig. 1A). No statistically significant association was evident between Fusobacterium relative abundance and MSI status in the mucinous cohort (p = 0.19, Fig. 1A).
Mucinous status and elevated Fusobacterium relative abundance were both independently found to impact composition of immune cells in the TCGA CRC cohort
Overall, mucinous tumours were associated with a significantly greater ratio of total immune cells to epithelial/stromal cells in the TCGA dataset (p < 0.001) (Table 2, Fig. 1B). Specifically, the mucinous cohort were associated with significantly greater proportions of CD8 + lymphocytes (p = 0.018), regulatory T-cells (p < 0.001), and M2 macrophages (p = 0.003) (Table 2, Fig. 1B).
Tumours with high Fusobacterium relative abundance were found to be associated with significantly greater proportions of CD4 + lymphocytes (p = 0.031) and M1 macrophages (p = 0.006), whilst M2 macrophages (p = 0.043) were under-represented across this group (Table 2, Fig. 1B). Evaluation of the mucinous cohort in isolation, found a significant reduction in the proportion of B-cells (p = 0.035) in patients with elevated Fusobacterium relative abundance (Table 2, Fig. 1B).
Elevated Fusobacterium prevalence is associated with better outcomes in mucinous CRC in the TCGA dataset
Existing evidence has linked Fusobacterium abundance with prognostic outcomes in CRC. To examine its precise impact with regards to mucinous tumours, we compared outcomes between patients with high and low Fusobacterium relative abundance, in both mucinous and non-mucinous cohorts in isolation (Fig. 2A, B). When we restricted our analysis to non-mucinous CRC patients, we found high Fusobacterium relative abundance did not appear to significantly impact DFS (HR 1.24, 95% CI 0.84 to 1.82, likelihood ratio test p = 0.29, logrank p = 0.28, Fig. 2B), DSS (HR 1.62, 95% CI 0.96 to 2.73, likelihood ratio test p = 0.08, logrank p = 0.07, Fig. 2B) or OS (HR 1.27, 95% CI 0.82 to 1.97, likelihood ratio test p = 0.29, logrank p = 0.29, Fig. 2B). However, univariate Cox regression models demonstrated how mucinous CRC patients with elevated Fusobacterium relative abundance trended to have more favourable clinical outcomes, specifically with reference to DSS (HR 0.24, 95% CI 0.05 to 1.14, likelihood ratio test p = 0.04, logrank p = 0.052, Fig. 2A).
We investigated the association between Fusobacterium relative abundance at higher resolution, namely at the species level, with mucinous status and clinical outcome (Sup. Fig. 1). In line with previous literature reports, we observed that F. nucleatum was the most abundant species (average 1.17%, 95% CI 0.00 to 10.60%), both across the whole unselected patient population and by mucinous status (mucinous: average 1.67%, 95% CI 0.00 to 11.46% vs. non-mucinous: average 1.10%, 95% CI 0.00 to 10.14%), (Sup. Fig. 1A-B). Fusobacterium periodonticum (average 0.15%, 95% CI 0.00% to 1.11%), Fusobacterium necrophorum (average 0.07%, 95% CI 0.00 to 0.22%), Fusobacterium gonidiaformans (average %, 95% CI 0.00 to 0.37%) and Fusobacterium mortiferum (average 0.04%, 95% CI 0.00 to 0.20%) were amongst the species with the highest mean relative abundance (Sup. Fig. 1A). When restricting the analysis to mucinous CRC patients, we observed an enrichment for Fusobacterium necrophorum (mucinous: average 0.18%, 95% CI 0.00 to 1.30% vs. non-mucinous: average 0.06%, 95% CI 0.00 to 0.21%) species (Sup. Fig. 1A).
Next, we sought to investigate the association between the relative abundance of species from the Fusobacterium genus with clinical outcome (OS, DSS, DFS, Sup. Fig. 1C). Univariate Cox regression models fitted on the whole unselected patients population revealed that patients with high F. nucleatum relative abundance have worse OS (HR 1.57, 95% CI 1.05–2.36, p = 0.03) and DSS (HR 1.87, 95% CI 1.16–3.03, p = 0.01), (grey-shaded panels, Sup. Fig. 1C). Furthermore, Cox regression models fitted with an interaction term capturing the differential impact of species abundance by mucinous status confirmed the findings at the genus taxonomic rank (light-red-shaded panels, Sup. Fig. 1C). High species relative abundance is associated with more favourable clinical outcomes in the mucinous subpopulation. In contrast, the reverse is observed in the non-mucinous subpopulation whereby high species relative abundance is associated with worse clinical outcomes.
Fusobacterium abundance in rectal cancer tumour microarray validation cohort
The BRCC cohort included 15 cases (26%) of mucinous and 43 cases (74%) of non-mucinous rectal cancer, with 66% of the cohort having underwent neoadjuvant chemoradiotherapy. 14% (n = 2) of the mucinous cohort were MSI-high compared to 2% (n = 1) of the non-mucinous group. Further clinical and pathologic characteristics of the included patients are summarised in Table 3.
Table 3.
Clinical characteristics of the patients of the BRCC cohort. RC, rectal cancer; AJCC, American Joint Committee on Cancer; Categorical data reported as n (%). Continuous data reported as median (IQR)
| Mucinous RC (N = 15) | Non-Mucinous RC (N = 43) | p-value | ||
|---|---|---|---|---|
| Male | 53.3% (8) | 58.1% (25) | 0.75 | |
| Age | Median (IQR) | 71 (29–81) | 70(42–89) | 0.10 |
| Stage | AJCC 1 | 13.3% (2) | 11.6% (5) | 0.17 |
| AJCC 2 | 60.0% (9) | 34.8% (15) | ||
| AJCC 3 | 26.0% (4) | 53.4%(23) | ||
| AJCC4 | 0.0% (0) | 0.0%(0) | ||
| T stage | Tis-T1-T-2 | 20.0% (3) | 25.5% (11) | 0.28 |
| T3 | 60.0% (9) | 65.1% (28) | ||
| T4 | 20.0% (3) | 9.3% (4) | ||
| N stage | N0 | 73.3% (11) | 48.8% (21) | 0.52 |
| N1 | 6.7% (1) | 41.8%(18) | ||
| N2 | 20.0% (3) | 9.3%(4) | ||
| M stage | M0 | 100% (15) | 100% (43) | NA |
| M1 | 0.0% (0) | 0.0% (0) | ||
| Neoadjuvant CRT(n = 56)a | 53.3% (8) | 69.8% (30) | 0.32 | |
| Adjuvant CRT (n = 52)a | 53.3% (8) | 39.5% (17) | 0.26 | |
| MSI (n = 55)a | 14.3%(2) | 2.4% (1) | 0.09 | |
| KRAS (n = 55)a | Mutant | 35.7%(5) | 17.1%(7) | 0.15 |
| BRAF (n = 54)a | Mutant | 7.7%(1) | 2.4% (1) | 0.38 |
| LVI (n = 57)a | 6.7%(1) | 19%(8) | 0.26 | |
| Perineural invasion (n = 56)a | 14.3%(2) | 11.9%(5) | 0.82 | |
| Extramural invasion (n = 56)a | 7.1%(1) | 19.0%(8) | 0.29 |
aData not available in full cohort: n in parentheses = number with data available
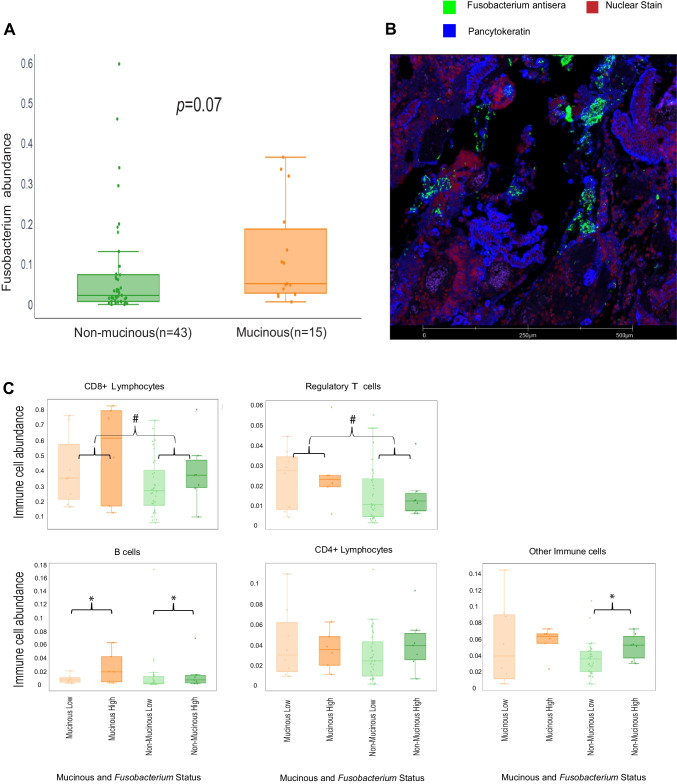
Fusobacterium abundance was quantified at a patient level and compared between patients with mucinous and non-mucinous RC. We again observed a trend whereby, Fusobacterium was more abundant in mucinous as opposed to non-mucinous RC; however, this trend fell short of statistical significance (p = 0.070) (Fig. 3A).
Fig. 3.
Fusobacterium abundance and mucinous status were found to impact immune cell expression in RC in our BRCC cohort. A Box and whisker blots depicting Fusobacterium abundance according to mucinous status. Statistical significance was evaluated using a Kruskal–Wallis test and the p-value is reported. B Image derived from the GeoMx platform of a mucinous core depicting pancytokeratin (Blue), Fusobacterium (Green) and Syto 13 (Red). C Box and whisker plots depicting specific immune cell counts; according to mucinous status and Fusobacterium relative abundance (high and low) within the BRCC RC cohort. Statistical significance was evaluated using Kruskal–Wallis tests. * indicates a statistically significant difference between Fusobacterium high and Fusobacterium low cohorts. # indicates a statistically significant difference between mucinous and non-mucinous cohorts
Mucinous status and elevated Fusobacterium abundance are associated with increased proportions of immune cells in our rectal cancer tumour microarray validation cohort
Next, we looked to determine an association between mucinous status and Fusobacterium abundance with immune cell populations in our BRCC rectal cancer cohort. Mucinous rectal tumours were associated with significantly greater CD8 + lymphocyte (p = 0.022), regulatory T-cell (p = 0.047), and B-cell (p = 0.025) counts (Table 4, Fig. 3C).
Tumours with high Fusobacterium abundance were found to be associated with a significantly greater proportion of B cells (p = 0.031) (Table 4, Fig. 3C).
Discussion
The previously determined association between high Fusobacterium relative abundance and MSI status in CRC, was not found to extend to mucinous CRC in our analysis [13]. Both mucinous status and high Fusobacterium relative abundance were independently found to influence immune cell composition in both the TCGA CRC and BRCC RC cohorts. However, some differences existed amongst the cell types affected across our two datasets. High Fusobacterium relative abundance tended to be associated with improved outcomes in mucinous CRC, suggesting Fusobacterium may have a protective function in this specific histological subtype of colorectal adenocarcinoma.
Increased Fusobacterium abundance within tumour tissue has not previously been associated with positive outcomes in CRC [16]. However, findings from our group’s recent publication suggest the prognostic impact of Fusobacterium may differ significantly according to underlying tumour biology [11]. In this previous analysis, increased Fusobacterium colonisation was only associated with poor prognosis in mesenchymal-type tumours (CMS group 4) [11]. Our mucinous TCGA cohort was over-represented in CMS group 1, and had a far higher incidence of MSI compared to the non-mucinous group. We initially hypothesised that Fusobacterium may play a causative role in this context, inducing MSI thus resulting in improved outcomes in those patients with higher Fusobacterium relative abundance. However, our analysis demonstrated no significant association between Fusobacterium relative abundance and MSI status in mucinous CRC. Though this finding may simply be a reflection of the relatively smaller size of our mucinous cohort, it raises the question could the improved outcomes observed in patients with mucinous CRC with elevated Fusobacterium abundance, be due to factors beyond MSI status? Further analysis involving larger datasets is required to validate our preliminary findings.
The immune characteristics of mucinous CRC have been examined to a limited extent in the literature. Tozawa et al. demonstrated reduced peri-tumoural lymphocyte infiltration in mucinous CRC compared to non-mucinous CRC in a cohort of 152 patients [31]. Meanwhile, Nazemalhosseini-Mojarad et al. found no difference in the distribution of stromal CD8 + lymphocytes or tumour CD8 + lymphocytes in mucinous compared to non-mucinous CRC [32]. Our analysis of the TCGA dataset found immune cell proportions to be significantly greater within our mucinous CRC cohort compared to the non-mucinous group. In particular, CD8 + lymphocytes, regulatory T cells, and M2 macrophages were all found in significantly greater proportion in the mucinous cohort. Similarly, in our BRCC cohort, mucinous rectal cancer was associated with greater numbers of CD8 + lymphocytes and regulatory T cells compared to the non-mucinous group. These finding are of increased significance in the context of recently published clinical trial results, which demonstrated very encouraging outcomes for patients with MSI-high locally advanced RC, treated with immunotherapy in the neoadjuvant setting [33]. Our findings that mucinous tumours are highly immunogenic, offers hope that immunotherapy may have an important future role to play in the management of this cohort, known to demonstrate resistance to traditional adjuvant chemotherapy agents [4, 5].
Existing evidence from pre-clinical studies has linked F. nucleatum with recruitment of myeloid derived suppressor cells [7] and inhibition of Natural Killer cell activity [34] in CRC. Immune cell proportions varied considerably in our analysis according to the degree of tumour Fusobacterium abundance. Within our TCGA cohort, high Fusobacterium relative abundance was associated with significantly increased proportions of CD4 + lymphocytes and M1 macrophages, whilst M2 macrophages were significantly under-represented in this group. The association with macrophages in TCGA dataset could not be assessed in our BRCC cohort. M1 macrophages play an integral role in the anti-tumour immune response, via identification and direct cytotoxic effects against tumour cells [35]. Increased CD4 + lymphocyte infiltration has also been found to be associated with improved survival in mismatch repair proficient colorectal tumours [36]. Findings from a previous meta-analysis demonstrated an association between M1 macrophage and M2 macrophage infiltration and mucinous CRC, this corresponds with our analysis where M2 macrophages were over-represented in mucinous CRC [37]. High-density M2 macrophage infiltration is associated with poor survival in solid-organ tumours [37]. These cells have been implicated in tumour migration, invasion, and have been found to induce an attenuated anti-tumour immune response [37]. In the context of mucinous CRC, the finding that Fusobacterium are associated with a significant decrease in M2 macrophage infiltration is pertinent and may further explain how Fusobacterium influences outcomes positively in mucinous CRC.
Though our findings regarding the impact of Fusobacterium are important, there are a number of limitations to our study. Firstly, our findings are limited by the significantly smaller proportion of mucinous tumours as compared to non-mucinous across our cohorts. Preliminary findings pertaining to mucinous CRC will require further validation in a larger dataset. It is also important to note the differences in immune cell expression according to Fusobacterium abundance in our BRCC and TCGA cohorts. Fusobacterium was positively associated with B cell proportions in our BRCC cohort whilst there was no such association observed in the TCGA group. Similarly CD4 + lymphocytes were not found in greater proportion in tumours with high Fusobacterium abundance in our BRCC group.
Mucinous CRC is a molecularly distinct subtype of colorectal adenocarcinoma, which appears to have a unique relationship with Fusobacterium. Fusobacterium abundance may be associated with positive outcomes in mucinous CRC, this is likely through modulation of immune moderators.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The results included here are in part based on data generated by the TCGA Research Network (https:// www cancer gov/tcga). We wish to acknowledge the Information Technology department at the Royal College of Surgeons in Ireland and the Irish Centre for High End Computing (ICHEC) for the provision of computational facilities and support. We acknowledge the technical support provided by Ina Woods, Éanna B. Ryan and Sanghee Cho.
Author contribution
WP Duggan: formal analysis, investigation, methodology, visualisation, writing original draught, writing-review and -editing. M Salvucci: formal analysis, methodology, software, visualisation, writing-review and -editing. B Kisakol: methodology, software, formal analysis, validation writing-review and -editing. AU Lindner: methodology, software, formal analysis, writing-review and -editing. IS Reynolds: concept design, visualisation, writing-review and -editing. H Dussmann: methodology, writing-review and -editing. J Fay: methodology, writing-review and -editing. T O’Grady: methodology, resources, writing-review and -editing. DB Longley: conceptualization, resources, writing-review and -editing. F Ginty: conceptualization, funding acquisition, resources, supervision, writing-review and -editing. E McDonough: data curation, resources, writing-review and -editing. DJ Slade: conceptualization, resources, writing-review and –editing. JP Burke: conceptualization, funding acquisition, resources, supervision, writing-review and -editing. JHM Prehn: conceptualization, funding acquisition, resources, supervision, writing-review and -editing.
Funding
Open Access funding provided by the IReL Consortium. This work was funded by a US-Northern Ireland-Ireland Tripartite grant from Science Foundation Ireland and the Health Research Board to JHMP (16/US/3301) and the National Cancer Institute of the National Institutes of Health under award number R01CA208179 supporting (FG, EMcD) and HSCNI, STL/5715/15 (DBL). WPD is supported by an RCSI/ Bon Secours Hospital MD StAR fellowship. BK is supported by Science Foundation Ireland through the SFI Centre for Research Training in Genomics Data Science under Grant number 18/CRT/6214, EU’s Horizon 2020 research, and innovation programme under the Marie Sklodowska-Curie grant H2020-MSCA-COFUND-2019–945385.
Availability of data and materials
TCGA data is readily available via the TCGA repository at the National Cancer Institute (NCI) (www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). Other data that support the findings of this study are available from the corresponding author upon request.
Declarations
Ethical approval and consent to participate
Tissue was provided from the Beaumont Hospital Colorectal Biobank with written consent provided by all patients. Institutional ethical approval was granted by the Beaumont Ethics (Medical Research) Committee (Reference 21/98).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
William P. Duggan and Manuela Salvucci contributed equally as first authors on this manuscript.
References
- 1.Kang H, O’Connell JB, Maggard MA, Sack J, Ko CY. A 10-year outcomes evaluation of mucinous and signet-ring cell carcinoma of the colon and rectum. Dis Colon Rectum. 2005;48(6):1161–1168. doi: 10.1007/s10350-004-0932-1. [DOI] [PubMed] [Google Scholar]
- 2.Bosman FT, Carneiro F, Hruban RH, Theise ND (2010) WHO classification of tumours of the digestive system: World Health Organization
- 3.Symonds DA, Vickery AL. Mucinous carcinoma of the colon and rectum. Cancer. 1976;37(4):1891–1900. doi: 10.1002/1097-0142(197604)37:4<1891::AID-CNCR2820370439>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 4.McCawley N, Clancy C, O’Neill BD, Deasy J, McNamara DA, Burke JP. Mucinous rectal adenocarcinoma is associated with a poor response to neoadjuvant chemoradiotherapy: a systematic review and meta-analysis. Dis Colon Rectum. 2016;59(12):1200–1208. doi: 10.1097/DCR.0000000000000635. [DOI] [PubMed] [Google Scholar]
- 5.Debunne H, Ceelen W. Mucinous differentiation in colorectal cancer: molecular, histological and clinical aspects. Acta Chir Belg. 2013;113(6):385–390. doi: 10.1080/00015458.2013.11680951. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds IS, Furney SJ, Kay EW, McNamara DA, Prehn JHM, Burke JP. Meta-analysis of the molecular associations of mucinous colorectal cancer. Br J Surg. 2019;106(6):682–691. doi: 10.1002/bjs.11142. [DOI] [PubMed] [Google Scholar]
- 7.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi Y, Wu HM, Yang Z, Zhou YF, Jin L, Yang MF, et al. New insights into the role of oral microbiota dysbiosis in the pathogenesis of inflammatory bowel disease. Dig Dis Sci. 2022;67(1):42–55. doi: 10.1007/s10620-021-06837-2. [DOI] [PubMed] [Google Scholar]
- 9.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. European J Clin Microbiol Infect Dis Official Public European Soc Clin Microbiol. 2014;33(8):1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 10.Brennan CA, Garrett WS. Fusobacterium nucleatum — symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17(3):156–166. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salvucci M, Crawford N, Stott K, Bullman S, Longley DB, Prehn JHM (2021) Patients with mesenchymal tumours and high Fusobacteriales prevalence have worse prognosis in colorectal cancer (CRC). Gut [DOI] [PMC free article] [PubMed]
- 12.Borowsky J, Haruki K, Lau MC, Dias Costa A, Väyrynen JP, Ugai T, et al. Association of Fusobacterium nucleatum with specific T-cell subsets in the colorectal carcinoma microenvironment. Clinical Cancer Res: An Official J Am Assoc Cancer Res. 2021;27(10):2816–2826. doi: 10.1158/1078-0432.CCR-20-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee DW, Han SW, Kang JK, Bae JM, Kim HP, Won JK, et al. Association Between Fusobacterium nucleatum, pathway mutation, and patient prognosis in colorectal cancer. Ann Surg Oncol. 2018;25(11):3389–3395. doi: 10.1245/s10434-018-6681-5. [DOI] [PubMed] [Google Scholar]
- 14.Sun JY, Yin TL, Zhou J, Xu J, Lu XJ. Gut microbiome and cancer immunotherapy. J Cell Physiol. 2020;235(5):4082–4088. doi: 10.1002/jcp.29359. [DOI] [PubMed] [Google Scholar]
- 15.Nosho K, Sukawa Y, Adachi Y, Ito M, Mitsuhashi K, Kurihara H, et al. Association of Fusobacterium nucleatum with immunity and molecular alterations in colorectal cancer. World J Gastroenterol. 2016;22(2):557–566. doi: 10.3748/wjg.v22.i2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gethings-Behncke C, Coleman HG, Jordao HWT, Longley DB, Crawford N, Murray LJ, et al. Fusobacterium nucleatum in the colorectum and its association with cancer risk and survival: a systematic review and meta-analysis. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2020;29(3):539–548. doi: 10.1158/1055-9965.EPI-18-1295. [DOI] [PubMed] [Google Scholar]
- 17.Yamaoka Y, Suehiro Y, Hashimoto S, Hoshida T, Fujimoto M, Watanabe M, et al. Fusobacterium nucleatum as a prognostic marker of colorectal cancer in a Japanese population. J Gastroenterol. 2018;53(4):517–524. doi: 10.1007/s00535-017-1382-6. [DOI] [PubMed] [Google Scholar]
- 18.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33(8):1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 19.Oh HJ, Kim JH, Bae JM, Kim HJ, Cho N-Y, Kang GH. Prognostic impact of fusobacterium nucleatum depends on combined tumor location and microsatellite instability status in stage II/III colorectal cancers treated with adjuvant chemotherapy. J Pathol Transl Med. 2019;53(1):40–49. doi: 10.4132/jptm.2018.11.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynolds IS, Thomas V, O’Connell E, Fichtner M, McNamara DA, Kay EW, et al. Mucinous adenocarcinoma of the rectum: a whole genome sequencing study. Front Oncol. 2020;10:1682. doi: 10.3389/fonc.2020.01682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dharmani P, Strauss J, Ambrose C, Allen-Vercoe E, Chadee K. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infect Immun. 2011;79(7):2597–2607. doi: 10.1128/IAI.05118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki R, Kamio N, Kaneko T, Yonehara Y, Imai K (2022) Fusobacterium nucleatum exacerbates chronic obstructive pulmonary disease in elastase-induced emphysematous mice. FEBS open bio [DOI] [PMC free article] [PubMed]
- 23.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA Pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–16.e11. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker MA, Pedamallu CS, Ojesina AI, Bullman S, Sharpe T, Whelan CW, et al. GATK PathSeq: a customizable computational tool for the discovery and identification of microbial sequences in libraries from eukaryotic hosts. Bioinformatics (Oxford, England) 2018;34(24):4287–4289. doi: 10.1093/bioinformatics/bty501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc Natl Acad Sci USA. 2013;110(29):11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindner AU, Salvucci M, McDonough E, Cho S, Stachtea X, O'Connell EP et al (2021) An atlas of inter- and intra-tumor heterogeneity of apoptosis competency in colorectal cancer tissue at single-cell resolution. Cell Death Different [DOI] [PMC free article] [PubMed]
- 28.Casasanta MA, Yoo CC, Udayasuryan B, Sanders BE, Umaña A, Zhang Y et al (2020) Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci Signal 13(641) [DOI] [PMC free article] [PubMed]
- 29.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern. 1979;9(1):62–66. doi: 10.1109/TSMC.1979.4310076. [DOI] [Google Scholar]
- 31.Tozawa E, Ajioka Y, Watanabe H, Nishikura K, Mukai G, Suda T, et al. Mucin expression, p53 overexpression, and peritumoral lymphocytic infiltration of advanced colorectal carcinoma with mucus component: is mucinous carcinoma a distinct histological entity? Pathol Res Pract. 2007;203(8):567–574. doi: 10.1016/j.prp.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 32.Nazemalhosseini-Mojarad E, Mohammadpour S, Torshizi Esafahani A, Gharib E, Larki P, Moradi A, et al. Intratumoral infiltrating lymphocytes correlate with improved survival in colorectal cancer patients: independent of oncogenetic features. J Cell Physiol. 2019;234(4):4768–4777. doi: 10.1002/jcp.27273. [DOI] [PubMed] [Google Scholar]
- 33.Cercek A, Lumish M, Sinopoli J, Weiss J, Shia J, Lamendola-Essel M, et al. PD-1 blockade in mismatch repair-deficient, locally advanced rectal cancer. N Engl J Med. 2022;386(25):2363–2376. doi: 10.1056/NEJMoa2201445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan Y, Yu Y, Wang X, Zhang T (2020) Tumor-associated macrophages in tumor immunity. 11 [DOI] [PMC free article] [PubMed]
- 36.Qi J, Liu X, Yan P, He S, Lin Y, Huang Z, et al. Analysis of immune landscape reveals prognostic significance of cytotoxic CD4(+) T cells in the central region of pMMR CRC. Front Oncol. 2021;11:724232. doi: 10.3389/fonc.2021.724232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Y, Ge X, Xu X, Yu S, Wang J, Sun L. Prognostic value and clinicopathological roles of phenotypes of tumour-associated macrophages in colorectal cancer. J Cancer Res Clin Oncol. 2019;145(12):3005–3019. doi: 10.1007/s00432-019-03041-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TCGA data is readily available via the TCGA repository at the National Cancer Institute (NCI) (www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). Other data that support the findings of this study are available from the corresponding author upon request.