SUMMARY
To determine which transcripts should reach the cytoplasm for translation, eukaryotic cells have established mechanisms to regulate selective mRNA export through the nuclear pore complex (NPC). The nuclear basket, a substructure of the NPC protruding into the nucleoplasm, is thought to function as a stable platform where mRNA-protein complexes (mRNPs) are rearranged and undergo quality control prior to export, ensuring that only mature mRNAs reach the cytoplasm. Here, we use proteomic, genetic, live-cell, and single-molecule resolution microscopy approaches in budding yeast to demonstrate that basket formation is dependent on RNA polymerase II transcription and subsequent mRNP processing. We further show that while all NPCs can bind Mlp1, baskets assemble only on a subset of nucleoplasmic NPCs, and these basket-containing NPCs associate a distinct protein and RNA interactome. Taken together, our data points towards NPC heterogeneity and an RNA-dependent mechanism for functionalization of NPCs in budding yeast through nuclear basket assembly.
Keywords: mRNA export, nuclear pore complex, nuclear basket, nuclear compartmentalization, Mlp1, Nucleolus, NPC heterogeneity, basket accessory interactome, poly(A) transcripts
INTRODUCTION
Exchange of macromolecules between nucleus and cytoplasm occurs through the NPC, a large multi-protein complex assembled by 30 different proteins (nucleoporins/nups), which forms a transport channel that spans the nuclear envelope (Kim et al., 2018; Lin and Hoelz, 2019). Cargo access and release from the NPC is modulated by asymmetrically distributed subcomplexes of the NPC. On the nuclear face, this is accomplished by a large basket-like structure protruding ~100nm into the nucleoplasm, the nuclear basket (Bensidoun et al., 2021).
The basket’s main scaffold is assembled by the filamentous protein TPR (Translocated Promoter Region) in humans and the two paralogues Mlp1 and Mlp2 (Myosin-like protein) in budding yeast (Frosst et al., 2002; Galy et al., 2004; Zhao et al., 2004), large proteins with predicted coiled-coil regions and an intrinsically disordered C-terminal domain that are thought to form the spokes of the basket (Krull et al., 2004; Niepel et al., 2013). Yet unlike for the central framework and cytoplasmic face of the NPC, high resolution structures of the nuclear basket have not yet been obtained (Lin and Hoelz, 2019). Moreover, even determining the exact stoichiometry of these proteins at individual NPCs using either ensemble (Kim et al., 2018; Ori et al., 2013; Rajoo et al., 2018) or single molecule data (Mi et al., 2015) has been challenging and not conclusive across studies.
The nuclear basket was shown to contribute to a range of nuclear activities, including DNA repair, epigenetic regulation, anchoring of genes at the nuclear periphery, and release of imported cargoes; however, the main role of the basket in gene regulation is thought to be mediating the access of mRNPs to the NPC (Björk and Wieslander, 2017; Brickner and Walter, 2004; Pascual-Garcia and Capelson, 2019; Raices and D’Angelo, 2012; Stewart, 2007; Taddei et al., 2006; Wing et al., 2022). Various studies have shown mRNP reorganization at the nuclear basket, and single molecule studies further confirmed the involvement of the basket in a rate-limiting step at the NPC as part of mRNP export (Bretes et al., 2014; Grünwald and Singer, 2010; Iglesias et al., 2010; Mor et al., 2010; Saroufim et al., 2015; Björk and Wieslander, 2015). After association with the nuclear side of the NPC, mRNPs are translocated with the help of export receptors and released from the cytoplasmic face of the NPC through RNP-remodelling (Xie and Ren, 2019).
Surprisingly however, Mlp1/2 and TPR are not required for mRNA export per se, as their loss only leads to a mild or partial export defect, respectively, indicating that the basket may facilitate export of only some transcripts, or has additional function (Aksenova et al., 2020; Lee et al., 2020; Powrie et al., 2011; Soucek et al., 2016; Zander et al., 2016). One such proposed function is that of establishing a quality control platform that ensures that only mature mRNPs are exported, as deletion of Mlp1 was shown to result in leakage of intron-containing mRNAs to the cytoplasm (Bonnet et al., 2015; Galy et al., 2004). While the mechanism of this process is not yet fully understood, it might involve RNA-binding proteins (RBPs) associating with mRNPs that serve as signals for export or retention, possibly modulating the ability of mRNPs to interact with baskets (Hackmann et al., 2014). Consistent with such a model, in yeast, various proteins show pre-mRNA leakage phenotypes when mutated including Gbp1, Hrb1, the Pre-mRNA Leakage proteins Pml1 and Pml39, the nuclear envelope protein Esc1, and the basket protein Nup60 (Bonnet et al., 2015; Dziembowski et al., 2004; Hackmann et al., 2014; Lewis et al., 2007; Palancade et al., 2005). Moreover, different RBPs required for mRNA export were shown to interact with Mlp1, including the nuclear poly(A)-binding protein (PABP) Nab2, which directly interacts with the C-terminus of Mlp1, further pointing towards the basket as a site of late mRNA maturation and/or initial contact point between mRNPs and NPCs to regulate export (Green et al., 2003; Saroufim et al., 2015; Soucek et al., 2016; Zander et al., 2016).
While one might expect of such a platform to be stably anchored to the rest of the NPC, various studies have shown that exchange of Mlp1 at NPCs is faster when compared to scaffold-forming nups (Hakhverdyan et al., 2020; Niepel et al., 2005; Niño et al., 2016). Moreover, Mlp1/2 dissociate from NPCs during heat-shock at 42°C and assemble into intra-nuclear granules together with RBPs, including Nab2 and Yra1, and poly(A) mRNAs (Carmody et al., 2010; Zander et al., 2016). The reasons for the dynamic association of Mlp1/2 with the NPC are still unknown, and the mechanisms leading to the formation of Mlp1 granules as well as their function are poorly understood. Moreover, in S. cerevisiae, NPCs adjacent to the nucleolus, which occupies about a third of the nuclear volume, are devoid of baskets (Galy et al., 2004; Niepel et al., 2013). Yet how cells establish these Mlp/basket-less NPCs, and whether they represent specialized NPCs with functions differing from nucleoplasmic, Mlp/basket-containing ones is not known.
Here, we show that, unlike previously thought, basket-less NPCs are not a specialized state of NPCs. Rather, our data demonstrate that basket-less NPCs are the default assembly state formed throughout the nuclear envelope and baskets assemble dynamically onto a subset of nucleoplasmic NPCs in an mRNA/mRNP-dependent manner. Specifically, inhibition of RNA polymerase II (Pol II) transcription results in the abrogation of basket assembly at nucleoplasmic NPCs, and interference with 3’end processing and polyadenylation leads to the loss of nuclear baskets, linking specific steps of mRNA maturation to basket assembly. Moreover, these disruptions result either in the formation of nuclear Mlp1-containing granules or re-location of baskets to the nucleolar periphery, the latter inducing a concomitant internalization of the nucleolus away from the periphery, suggesting an incompatibility between NPC basket assembly and the nucleolus. Proteomic, microscopy and RNA sequencing experiments further show that basket-containing NPCs assemble a unique protein and RNA interactome and that, while mRNAs associate with both types of NPCs which are likely export-competent, Mlp1 preferentially associates with longer mRNAs, but not short and/or intron-containing mRNAs. Taken together, our data identifies an RNA-dependent mechanism for functionalization of NPCs in budding yeast through nuclear basket assembly.
RESULTS
Mlp1 can traverse the nucleolus and form rare ectopic baskets at the nucleolar periphery
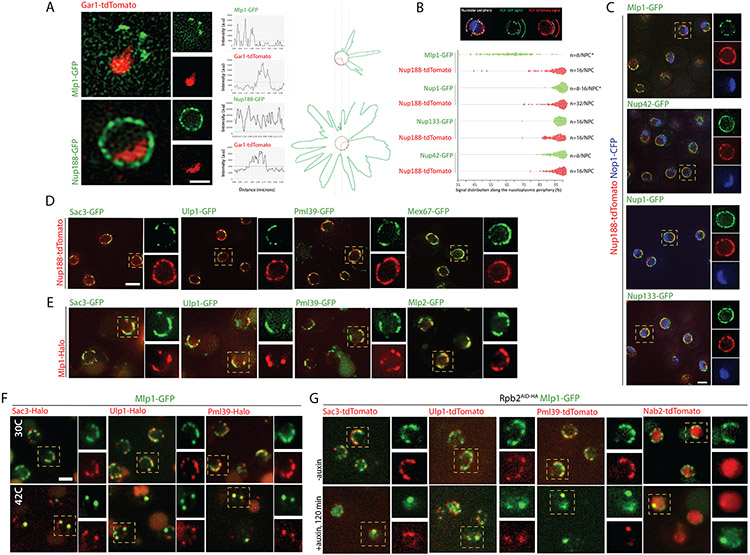
In S. cerevisiae, while NPCs are evenly distributed along the nuclear periphery, the nuclear basket proteins Mlp1/2 are excluded from NPCs next to the nucleolus, a crescent-shaped membrane-less compartment positioned adjacent to the nuclear membrane. Significant variability in nucleolar size and shape between cells in non-synchronous cultures occurs in response to stress, cell age, cell-cycle stage and overall metabolic activity (Neumüller et al., 2013; Sirri et al., 2008), suggesting that the number of NPCs bound by Mlp1/2 may also vary. While Mlp1 and Mlp2 are both considered part of the basket scaffold, Mlp2 localization at NPCs requires Mlp1, but not vice versa (Palancade et al., 2005); we therefore focussed our study on Mlp1. Comparing Mlp1-GFP distribution to a nucleolar marker showed a negative correlation between nucleolar and Mlp1-occupied territories (Figures 1A, B), suggesting that variations in nucleolar size negatively affect the number of NPCs available to bind Mlp1 and, moreover, that the number of basket-containing NPCs may vary from cell to cell and over time.
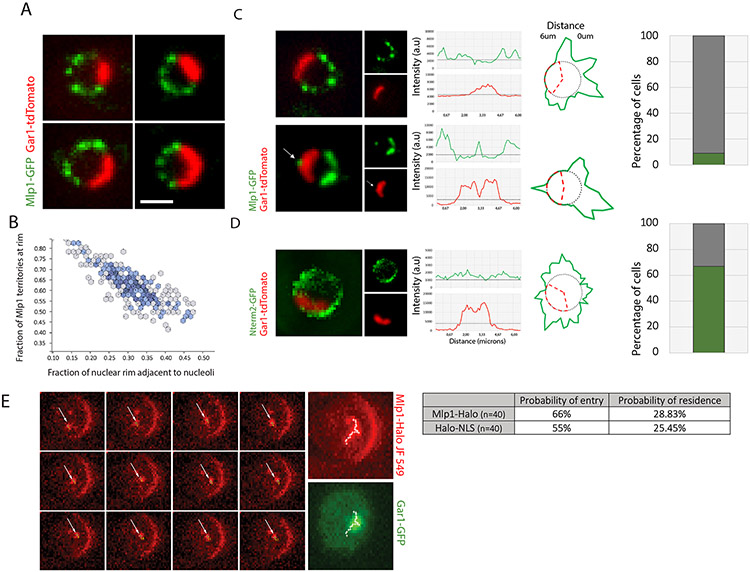
Figure 1. Mlp1 can access the nucleolus and assemble baskets at nucleolar NPCs.
A. Mlp1-GFP and Gar1-tdTomato distribution. B. Quantification of nuclear periphery occupied by Mlp1-GFP versus nucleolus (n=200). C. Mlp1-GFP assembling an ectopic basket (arrow), correlating with a Gar1-tdTomato signal loss (Gar1 intensity plot, arrow). Histogram quantification of cells with ectopic baskets (n=150). D. Mlp1-Nterm2-GFP fragment distribution along the nuclear periphery in Δmlp1/2 and quantification of cells with Mlp1-Nterm2-GFP signal at periphery adjacent to nucleolus (n=100). In C, D, signal intensity distribution shown in line plots and circular diagrams where red dashes delimit nucleolar region and average background signal shown as grey circles; Mlp1 distribution in green. E. Live-cell tracking of single Mlp1-Halo-JF549 molecules. Individual frames acquired at 20ms intervals are shown. White arrows indicate Mlp1-Halo-JF549; MAX, maximum intensity projection of all frames; track shown as white crosses. Quantification of observed probability of entry and residence for Mlp1 and control (n=40). Scale bar = 2μm.
Although Mlp1 is generally excluded from the nucleolus, a small percentage of cells show an Mlp1-GFP focus along the nucleolar-occupied territory, here referred to as an ectopic basket, which correlated with an invagination in the nucleolar signal surrounding the Mlp1 focus (Figure 1C, arrow; S1C), indicating that baskets can assemble along the nucleolar periphery but that these territories are mutually exclusive. To determine if Mlp1 can associate with nucleolar NPCs, an N-terminal fragment of Mlp1 (N-term 2), previously shown to be sufficient to associate with NPCs, was tested for NPC binding along the nucleolar periphery in an MLP1 deletion background (Niepel et al., 2013). Indeed, the Mlp1 N-term 2 region was able to bind NPCs both in the nucleoplasm and nucleolus (Figure 1D), suggesting that all NPCs along the nucleolar periphery are, in principle, competent to bind Mlp1.
A hallmark of liquid-phase separated compartments, such as nucleoli, is their ability to concentrate or exclude specific factors depending on their biochemical properties (Feric et al., 2016; Miné-Hattab et al., 2019; Sirri et al., 2008). Hence, the nucleolus could represent a diffusion barrier for full-length Mlp1. FRAP experiments have shown that Mlp1 associates dynamically with the NPC and exists in two pools within the nucleus: an NPC-bound fraction and a free pool that in theory could also assemble baskets on NPCs in the nucleolus (Niepel et al., 2013; Niño et al., 2016). We thus analyzed diffusion patterns of free Mlp1 using single-protein tracking of Halo-tagged Mlp1 and Halo-NLS by HILO illumination microscopy (Movies S1, S2). While most single Mlp1 molecules were bound to the periphery and static, nuclear diffusing molecules were observed to enter the nucleolus, similar to a Halo-NLS control, with 29% of the tracks overlapping with the nucleolar area (Figures 1E, S1A, B). These data suggest that Mlp1 can access the nucleolus, and NPCs along the nucleolar periphery can bind Mlp1; however, formation of baskets at nucleolar NPCs is rare and correlates with an invagination of the nucleolus.
Basket assembly requires mRNA production
The main differentiating feature between the nucleoplasmic and nucleolar region is the synthesis of two distinct types of RNA: RNA Pol I synthesizing ribosomal RNA (rRNA) in the nucleolus, and RNA Pol II producing mRNA in the nucleoplasm. We considered the possibility that basket assembly is linked to events solely occurring in the nucleoplasm and tested whether this requires processes linked to mRNA metabolism. To that end, we constructed strains in which the large subunit of either RNA Pol I (RpbA135) or RNA Pol II (Rpb2) were tagged with an Auxin Inducible Degron cassette (AID-HA) to enable their depletion upon auxin addition (Figure 2A). Depletion of RNA Pol II, but not RNA Pol I, resulted in the loss of Mlp1-GFP at the nuclear periphery and its redistribution into the nucleoplasm after 120 min (Figure 2B). A similar phenotype was observed upon cytoplasmic sequestration of Rbp1 using the anchor away system 40 min after rapamycin addition (Figure S2A)(Geisberg et al., 2014; Jeronimo and Robert, 2014) and in rpb1-1 but not WT cells at non-permissive temperature (37°C) after 10 min (Figure 2C). Moreover, in all three strains, a large fraction of cells (~80%) exhibited formation of a singular nuclear Mlp1 granule (Figures 2D and S2B) similar to what has previously been observed upon heat-shock at 42°C (Carmody et al., 2010), where Mlp1 dissociates from NPCs and forms intranuclear foci that contain several mRNA maturation factors, as well as upon deletion of NUP60, which is required for the anchoring of Mlp1 at the NPC (Mészáros et al., 2015; Niepel et al., 2013). These observations link basket formation, which is restricted to the nucleoplasm, to RNA Pol II activity and hence nuclear mRNA metabolism. However, these experiments do not discriminate whether loss of transcriptional activity per se or the consequential absence of mRNA maturation and/or mRNA export is responsible for the observed phenotype.
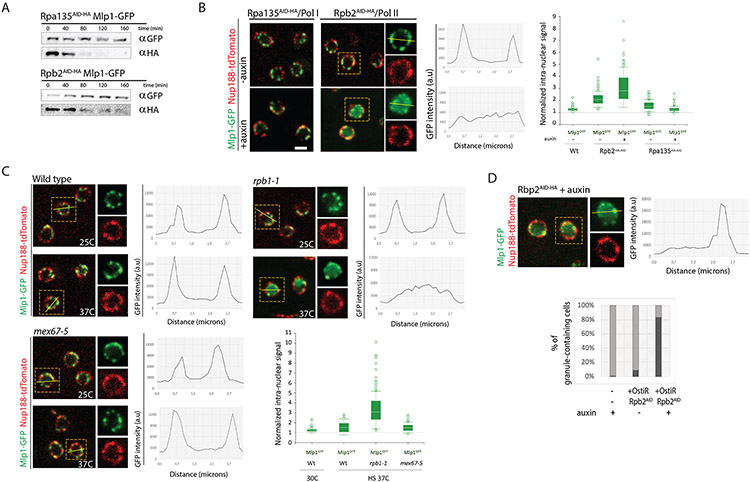
Figure 2. Inhibition of RNA polymerase II transcription, but not mRNA export, affects basket assembly.
A. Western blot of total cell lysates from Rpb2AID-HA and Rpa135AID-HA cells upon auxin addition. B. Mlp1-GFP localization pre- and post-addition of auxin (500 μM, 120mins) in Rpb2AID-HA and Rpa135AID-HA cells. Nup188-tdTomato outlines nuclear periphery. For Rpb2AID-HA cells, line scan intensity plot shows Mlp1-GFP signal distribution in a single cell pre/post-auxin. Bar graph shows quantification of Mlp1-GFP intranuclear signal (n=100). C. Mlp1-GFP distribution in wt, rpb1-1 and mex67-5 cells at 25°C and 37°C. Line scan intensity plots show Mlp1-GFP signal distribution, bar graphs quantification of average Mlp1-GFP intranuclear signal intensities in the center of nuclei over background. D. Mlp1-GFP granule formation in Rpb2AID-HA cells 120 min post-auxin. Histogram quantification of % cells with Mlp1-GFP granules post-Rbp2 depletion. Scale bar = 2μm. Data are represented as mean ± SEM.
To discriminate between these possibilities, we next blocked mRNA export using a temperature-sensitive mutant of the main mRNA export receptor Mex67, mex67-5 (Santos-Rosa et al., 1998). Upon shift to non-permissive temperature (37°C), no Mlp1 redistribution (Figure 2C) was seen, suggesting that blocking RNA Pol II activity, and possibly downstream events, but not mRNA export affects basket formation. Alternatively, mRNPs reaching the periphery could be required for basket assembly.
Rerouting of mRNPs induces basket formation at nucleolar NPCs
Similar to the compartmentalization of RNA Pol I and II, mRNA and the mRNA maturation machinery are generally restricted to the nucleoplasm and absent from the nucleolus. However, mutations in some RNA maturation/surveillance factors such as Dis3, Csl4, and the ribosome biogenesis factor Enp1 were shown to accumulate different types of polyadenylated RNAs (rRNAs, snoRNAs, and mRNAs) and mRNP components in the nucleolus (Aguilar et al., 2020; Paul and Montpetit, 2016). We therefore asked how a redistribution of mRBPs and mRNPs to the nucleolus affects basket formation at NPCs. Enp1 was depleted using a Enp1AID-HA strain and basket assembly monitored using Mlp1-GFP. 120 min after auxin addition, Mlp1 was mostly lost from nucleoplasmic NPCs and redistributed to the nucleolar periphery (Figures 3A, S4A). Relocalization was specific for Mlp1 and not observed for Nup188, indicating that Enp1 depletion does not lead to NPC clustering along the nucleolar territory (Figure S3A). A similar Mlp1 redistribution was observed in Csl4AID-HA cells (Figure S3B), suggesting that NPCs adjacent to nucleoli are able to assemble baskets upon redistribution of mRNPs to the nucleolus. Moreover, under these conditions, nucleoli were often spherical, fragmented, and internalized with little or no overlap between Mlp1 and Gar1 signals (Figure S3B), reminiscent of the nucleolar invaginations caused by ectopic baskets (Figure 1C; S1C). To test that redistribution of Mlp1/nuclear baskets to the nucleolar periphery is not due to nucleolar fragmentation per se, we disrupted nucleolar organization by either depleting Sgs1, a nucleolar DNA helicase, or treatment of cells with 1,6-hexanediol, both causing nucleolar fragmentation (Abraham et al., 2020; Kroschwald et al., 2015; Sinclair et al., 1997). While number and circularity of nucleolar domains was similar under all conditions, Mlp1 localization along nucleolar territories was only observed upon Enp1 depletion (Figure S3C, D).
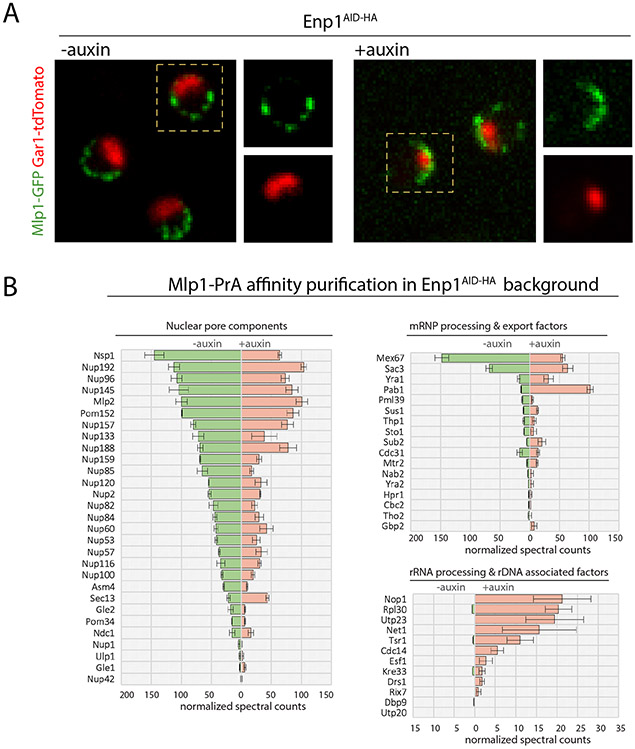
Figure 3. Nucleolar basket assembly upon depletion of the ribosome biogenesis factor Enp1.
A. Distribution of Mlp1-GFP along the nuclear and nucleolar peripheries in Enp1AID-HA cells pre- and post-auxin (120 min). Nucleolus labeled by Gar1-tdTomato. B. Normalized median MS spectral counts of proteins co-purified with Mlp1-PrA from Enp1AID-HA cells pre- and post-auxin treatment (120 min). Data are represented as mean ± SEM (n=3).
Lastly, to determine that Mlp1 assembles into bona fide baskets upon Enp1 depletion, we performed affinity purification (AP) of Mlp1-PrA and analyzed its interactome by semi-quantitative mass spectrometry (MS) in the presence and absence of auxin. In both conditions, Mlp1-associated interactomes were similar in terms of co-isolated NPC components (Figure 3B). Furthermore, consistent with an accumulation of mRNPs in the nucleolus, we identified mRNA maturation and export factors upon Enp1 depletion, in addition to low levels of nucleolar proteins. While the overall levels of mRNA maturation factors were comparable in both conditions, the poly(A)-binding protein Pab1 was ~4-fold enriched upon Enp1 depletion (Figure 3B; S6B). These data suggest that basket assembly requires elements or events of the mRNA maturation pathway that under normal conditions are restricted to the nucleoplasm, but which upon their sequestration in the nucleolus can lead to basket assembly along the nucleolar periphery. However, the assembly of nucleolar baskets appears to be incompatible with the cooccurrence of a nucleolus.
Basket assembly requires 3’end processed and polyadenylated mRNA
To further dissect the processes along the mRNA maturation pathway required for basket assembly, we carried out a targeted AID screen, depleting selected mRNA maturation factors and monitoring Mlp1 distribution upon auxin treatment. This included factors involved in co-transcriptional mRNP assembly, i.e., THO/TREX/TREX-2; 5’cap-binding; splicing; 3’end cleavage, polyadenylation, and poly(A)-binding; the TRAMP complex; mRNA export; and proteins linked to pre-mRNP surveillance (Wende et al., 2019). Phenotypes were compared to Nup60 depletion, shown to be required for Mlp1 binding to NPCs, as well as Rpb2AID-HA and Enp1AID-HA (Figure 4A; Table S4) (Lewis et al., 2007; Mészáros et al., 2015).
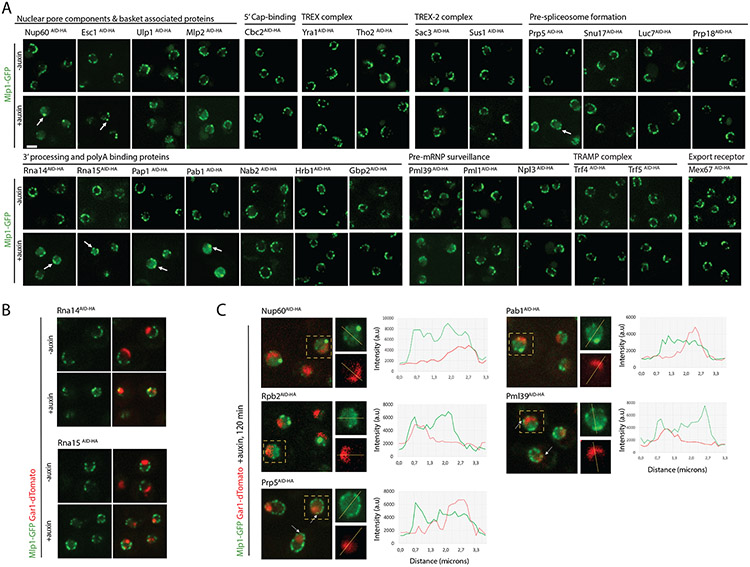
Figure 4. Depletion of mRNP maturation factors affecting basket assembly and Mlp1 localization.
A. Depletion screen for changes in Mlp1 localization (120 min post-auxin); changes indicated by arrows. B. Distribution of Mlp1-GFP in Rna14AID-HA and Rna15AID-HA cells. C. Mlp1-GFP distribution with respect to nucleolus in strains with altered perinuclear Mlp1-GFP signal. Line scan intensity plot show comparative Mlp1-GFP and Gar1-tdTomato distribution; arrows indicate Mlp1 signal at nucleolar periphery. Scale bar =2μm.
Phenotypes varied considerably among strains; however, shared phenotypes were observed among proteins implicated in specific stages of the pathway. Overall, depletion of early mRNP maturation factors did not affect basket formation, as neither depletion of Cbc2 nor of THO/TREX complex components changed Mlp1 localization (Figure 4A). Interfering with different stages of splicing did also not induce Mlp1 relocalization with the exception of Prp5, an ATPase required for pre-spliceosome assembly (Figure 4A) (Jurica et al., 2003). Prp5 depletion resulted in an increased nucleoplasmic Mlp1 signal, albeit less pronounced than in Rbp2AID-HA cells (Figure 2, 4C), and Mlp1 redistribution along the entire nuclear periphery. In addition to its role in splicing, prp5 mutants were shown to affect RNA Pol II transcription suggesting that this phenotype might be linked to transcription rather than splicing (Shao et al., 2020).
Depletion of 3’end processing and polyadenylation factors, however, induced strong Mlp1 relocalization phenotypes (Figure 4A). Specifically, depletion of 3’end cleavage factors (Rna14, Rna15), the RNA poly(A) polymerase Pap1 and poly(A)-binding protein Pab1 showed altered Mlp1 distribution suggesting that these steps are significant for basket formation. Upon Rna14 and Rna15 depletion, Mlp1 was redistributed to the nucleolar periphery, similar to Enp1AID-HA cells (Figure 4B and 3A) and frequently exhibited fragmented nucleoli, a phenotype previously been described for rna14-1 and rna15-2 at non-permissive temperature (37°C) (Carneiro et al., 2007). Accumulation of poly(A) RNA in the nucleolus has also been observed in rna14 and rna15 mutants due to disruption of 3’end cleavage, polyadenylation, and export (Brodsky and Silver, 2000; Casolari et al., 2004; Dunn et al., 2005). Similar to csl4 and enp1 mutants, where nucleolar poly(A) RNA accumulation results in nucleolar sequestration of mRNPs, redistribution of mRNPs to nucleoli might be responsible for basket formation along the nucleolar periphery in Rna14AID-HA and Rna15AID-HA cells (Aguilar et al., 2020). While Pap1 and Pab1 depletions also led to Mlp1 redistribution, an increase in nucleoplasmic Mlp1 levels and decrease of Mlp1 signal along the nuclear periphery was observed with concomitant formation of larger Mlp1 foci, reminiscent of a Rbp2 depletion phenotype (Figure 4A, C). No change in Mlp1 pattern was observed upon depletion of the Mlp1 interactor and poly(A) binding protein Nab2, nor upon that of Gbp2 or Hrb1, two poly(A)-binding proteins linked to the maturation of intron-containing mRNAs, or Npl3 (Figure 4A).
We also tested proteins linked to nuclear basket function for a role in Mlp1 localization. Of the two pre-mRNA surveillance factors only Pml39 displayed a phenotype, where Mlp1 signal was decreased along the nuclear periphery with an increase in nucleoplasmic Mlp1 (Figure 4A, C). TREX-2 components showed no phenotype. We furthermore determined Mlp1 localization upon depletion of NPC- and nuclear basket-associated factors Esc1, Ulp1, and Mlp1-paralog Mlp2. Esc1AID-HA cells exhibited formation of large Mlp1 granules along the nuclear rim and simultaneous loss of evenly distributed Mlp1 rim staining, while Ulp1 and Mlp2 depletion showed a moderate decrease in peri-nuclear and increased nucleoplasmic Mlp1 signal (Figure 4A).
We further asked whether Mlp1 loss at the nuclear periphery upon RNA Pol II or specific mRBP depletion could be due to altered levels of nucleoporins required for Mlp1 binding to NPCs, in particular Nup60. However, neither Rpb2, Pab1 or Pap1 resulted in altered localization of Nup60 at the periphery (Figure S4B).
Taken together, our results suggest that basket assembly at NPCs is linked to specific steps of nuclear mRNA maturation and requires the presence of poly(A) transcripts as we observed a significant loss of Mlp1 along the nuclear periphery and its redistribution into the nucleoplasm upon depletion of Rbp2, Prp5, Pap1, Pab1, and Pml39. Yet not all poly(A)-binding proteins appeared to be required for basket formation as neither depletion of Gbp2, Hrb1 nor Nab2, which has been linked to Mlp1 in the context of mRNA export and shown to directly interact with the Mlp1 C-terminal region (Green et al., 2003), affected nuclear basket formation.
Not all nucleoplasmic NPCs contain baskets
The requirement of an active mRNP maturation pathway for basket formation might suggest that baskets assemble randomly at individual NPCs, and that not all NPCs assemble baskets at all times or with the same frequency, which could result in a functional heterogeneity at nucleoplasmic NPCs. Consistent with such a model, Mlp1-GFP distribution at the nuclear periphery, imaged using spinning disk confocal microscopy, often showed a discontinuous staining pattern (Figure 1A). To investigate possible NPC heterogeneity at nucleoplasmic NPCs, we analyzed distribution and co-localization of different NPC components using Structured Illumination microscopy (SIM) (Figure 5, S5). Signal distribution of Nup188, a component of the inner ring, showed an overall continuous staining along the nuclear periphery whereas Mlp1 staining was discontinuous, suggesting that several regions of Mlp1/basket-less NPCs occupy the nuclear periphery (Figure 5A, S5A). Quantification of the distribution of stoichiometrically different nups along the nucleoplasmic periphery showed that only Mlp1 exhibited a discontinuous signal (Figure 5B).
Figure 5. Baskets assemble on a subset of NPCs in the nucleoplasm and co-localize with select proteins at the nuclear periphery.
A. SIM of Nup188-GFP or Mlp1-GFP and Gar1-tdTomato; signal intensity distribution shown in line plots and circular diagrams where red dashes delimit nucleolar region; average background signal shown as grey circles and GFP distribution in green. B. Signal distribution analysis of selected GFP-tagged Nups or Mlp1-GFP compared to Nup188-tdTomato along the nucleoplasmic periphery; each dot represents the occupied proportion/cell (in %; n=100). C. SIM co-localization analysis of Nop1-CFP, Nup188-tdTomato, and Nup42-, Nup1-, Nup133- or Mlp1-GFP. D. SIM co-localization analysis of Nup188-tdTomato and Sac3-, Ulp1-, Pml39- and Mex67-GFP. E. SIM co-localization analysis of Mlp1-Halo and Sac3-, Ulp1-, Pml39- and Mlp2-GFP. F. SIM co-localization analysis of Mlp1-GFP and Sac3-, Ulp1-, and Pml39-Halo at 30°C or 42°C. G. SIM co-localization analysis of Mlp1-GFP and Sac3-, Ulp1-, Pml39-, and Nab2-Halo in Rbp2AID-HA cells pre- and 120 min post-auxin. Scale bar =2μm.
Moreover, co-staining of the nuclear asymmetric nups Nup60, Nup1, and Nup2, usually also considered basket nups, with Nup-188 and the nucleolar marker Nop1 showed association with all NPCs, in line with previous reports, and suggests a different behavior of Mlp1 compared to other so-called basket nups (Figure 5C, S5B) (Galy et al., 2004). These observations suggest that nuclear asymmetric nups should not necessarily be referred to as basket nups in the context of the basket as a structure, as they appear not to be part of the physical basket structure, which only assembles in the presence of Mlp1 (Krull et al., 2010; Niepel et al., 2013). Mlp2, however, showed a localization pattern similar to that of Mlp1 and co-localized with Mlp1 (Figure 5E), in line with the requirement of Mlp1 for Mlp2 perinuclear localization (Palancade et al., 2005). Together, our data show that NPC heterogeneity extends beyond the nucleolus and implies that basket formation is not a default state of nucleoplasmic NPCs.
Specific nuclear mRNA maturation factors are enriched at basket-containing NPCs
Various NPC-associated proteins generally not considered bona fide nucleoporins have been linked to the nuclear basket (e.g., TREX-2, Pml39, Ulp1) and were previously shown to be excluded from the nucleolar periphery (Bonnet et al., 2015; Zhao et al., 2004). Distribution of the TREX-2 main scaffold protein Sac3, Ulp1 and Pml39 relative to Nup188 displayed an uneven pattern along the periphery, similar to Mlp1 (Figure 5D) and co-localized with Mlp1-Halo, suggesting that these proteins associate preferentially with basket-containing nucleoplasmic NPCs (Figure 5E, S5C). These proteins also colocalized in Mlp1 granules that formed upon heat-shock at 42°C or Rpb2 depletion (Figure 5F, G).
Localization of Mex67 showed a distribution along the entire nuclear periphery (Figure 5D), consistent with recent models suggesting Mex67 as a bona fide NPC component associating with NPCs independent of its RNP association (Derrer et al., 2019). Moreover, it may also indicate that all NPCs are, in principle, able to export mRNPs using the Mex67-dependent mRNA export pathway, but that some are further functionalized by the presence of a basket and associated factors implicated in mRNA metabolism.
To further determine whether the presence of baskets on NPCs correlates with a specific basket protein interactome, we characterized the interactomes of NPCs with and without nuclear baskets by AP-MS. First, to analyze the general NPC interactome (‘All NPCs’), we carried out single-step affinity purifications (ssAP) of NPCs followed by MS from an Mlp1-PrA/Nup133-GFP double-tagged yeast strain, using Nup133-GFP as bait (Figure S6A)(Trahan and Oeffinger, 2022). To ensure the capture of dynamic interactors such as Mlp1 (Hakhverdyan et al., 2020), we stabilized NPCs and associated proteins using a short in-lysate glutaraldehyde fixation prior to affinity purification (Subbotin and Chait, 2014). To compare the interactomes of NPCs with and without nuclear baskets, we applied a differential affinity purification approach (dAP, STAR Methods) that enabled separation of the two types of NPCs from the same lysate via two consecutive APs: first, isolation of Mlp1-PrA associated complexes, including basket-containing NPCs (‘Basketplus’), followed by purification of the remaining basket-less NPCs (‘Basketminus’) from the flow-through via Nup133-GFP (Figure S6A). To identify proteins associated with NPCs in an Mlp1 - and/or poly(A) RNA-dependent manner, we also isolated NPCs and their interactome from a Δmlp1/2/Nup133-GFP cells and Enp1AID-HA cells upon auxin treatment using Nup133-GFP and Mlp1-PrA, respectively (Tables S5, S6). While for our analysis Mlp1-PrA complexes were considered NPC-associated, we cannot rule out that some proteins identified with Mlp1 interact only transiently with the nuclear periphery or with free nucleoplasmic Mlp1.
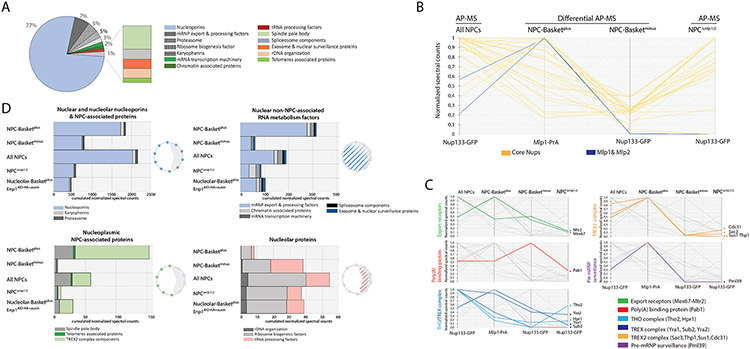
AP-MS data for ‘All NPCs’ (Nup133-GFP) revealed an enrichment for nucleoporins, which constituted the majority of the NPC interactome (77%), mRNA maturation/export factors (7%), and proteins implicated in various nuclear processes previously linked to NPCs (Figure 6A). Efficient separation of basket-containing (‘Basketplus’, Mlp1-PrA) and basket-less NPCs (‘Basketminus’, Nup133-GFP) was confirmed by a significant decrease of Mlp1/Mlp2 levels in ‘Basketminus’ samples compared to ‘All NPCs’ (Figure 6B).
Figure 6. Dissection of differential NPC interactomes.
A. Overview NPC interactome from the total pore AP. Each protein categories represent the sum of the normalized spectral counts. B, C. Parallel coordinate plots representing the sum of normalized spectral count values for NPC and basket proteins (B) and nuclear mRNA metabolism proteins (C) affinity purified via Nup133-GFP (‘All NPC’); differentially affinity purified via Mlp1-PrA (Basketplus) and Nup133-GFP (Basketminus), or Δmlp1/2/ Nup133-GFP cells. Data has been compressed for visualization and comparison purposes; 1 represents maximal value of ESC median between experiments (n=3). Lines represent the relative abundance of proteins between different APs. D. Histograms representing the sum of normalized spectral counts of proteins co-purified with either ‘All NPCs’, Mlp1-PrA (Basketplus), and Nup133-GFP (Basketminus) from Mlp1-PrA/Nup133-GFP, Δmlp1/2/ Nup133-GFP cells and Mlp1-PrA/Nup133-GFP/Enp1AID-HA cells upon auxin treatment and analyzed according to their function in RNA metabolism across different subnuclear compartments. Cartoons: yeast nuclei, nucleolus outlined as grey crescent; spheres represent associating proteins/group.
We then analyzed relative abundance of RBPs and factors required for mRNA maturation, export and surveillance by comparing normalized spectral counts of ESC ≥10 across different samples as well as log2 fold change relative to ‘All NPCs’ (Figure 6C, D; Figure S6B, C). Mex67 and Mtr2 were found with both Basketplus and Basketminus NPCs (Figure S6B, C), consistent with Mex67 localization (Figure 5C), supporting the notion that Mex67 is systematically present at NPCs and, moreover, that RNP export may occur through both types of NPCs. In Δmlp1/2 cells, both proteins were decreased which may reflect, at least in part, a lower residence time of Mex67-bound mRNPs at NPCs lacking baskets, as suggested previously (Saroufim et al., 2015). Yra1, other THO/TREX components and Pab1 were also purified with both Basketplus and Basketminus and Δmlp1/2 NPCs (Figure 6C; Figure S6B) suggesting that mRNPs containing these factors can interact with NPCs independent of a basket or Mlp1, consistent with previous observations of an only moderate mRNA export defect in Δmlp1/2 cells (Galy et al., 2004; Green et al., 2003). These proteins, however, were all enriched with Mlp1-PrA in Enp1AID-HA cells upon auxin treatment (Figure S6B) indicating that while THO/TREX components are not required for basket formation they are tightly linked to nuclear poly(A) mRNP metabolism (Aguilar et al., 2020). Nab2 and Gbp2 behaved similarly, yet their overall spectral counts were too low to compare their distribution across samples (Figure S6B, C).
TREX-2 components (Sac3, Sus1, Thp1, Cdc31) were enriched with Basketplus and nucleolar over ‘All NPCs’ and significantly decreased in Basketminus NPCs and Δmlp1/2 cells (Figure 6C; S6B, C). These results are consistent with our previous observation that Sac3 colocalizes with Mlp1 along the nuclear periphery and the suggestions that TREX-2 associates preferentially with basket-containing nucleoplasmic NPCs. Nevertheless, Sac3 was found at the nuclear periphery in Δmlp1/2 cells at very low levels compared to Nup1 whose distribution did not change, indicating potential alternative, or additional, associations with the nuclear periphery in the absence of the basket (Figure S5D). While Ulp1 levels were too low to compare its distribution across samples, Pml39 was enriched with Basketplus NPCs (Figure 6C; S6B, C), again in agreement with our observation that Pml39 localizes only with Mlp1 and Basketplus NPCs (Figure 5D, E; S5C). Pml39 was significantly decreased in Δmlp1/2 cells, while it was enriched with Mlp1 at nucleolar NPCs in Enp1AID-HA cells (Figure S6B), consistent with previous data that its NPC association is dependent on the presence of a basket, i.e., Mlp1 (Palancade et al., 2005).
To further analyze the differences between basket-containing and basket-less NPC interactomes, we binned proteins in four groups based on their nuclear sub-localization. Nuclear/nucleolar nups and NPC-associated proteins (i), and nuclear non-periphery-associated RNA metabolism factors (ii) were identified across all samples, consistent with their relative abundance compared to ‘All NPCs’ (Figure S6B). While many mRNA maturation and export factors (ii) were enriched with Mlp1/Basketplus NPCs, their presence with Basketminus and NPCs in Δmlp1/2 cells indicates that these interactions are not basket-dependent, yet their significantly lower numbers in Δmlp1/2 suggest that the basket may facilitate these interactions.
Nucleoplasmic NPC-associated proteins (iii) (e.g., TREX-2, SPB proteins) were significantly enriched with Mlp1/Basketplus NPCs in Wt and Enp1AID-HA cells and mostly absent from Basketminus. Conversely, nucleolar proteins (iv) were significantly under-represented with nucleoplasmic Basketplus compared to Basketminus NPCs and ‘All NPCs’ (Figure 6D). This category was also slightly increased with nucleolar Mlp1/Basketplus NPCs in Enp1-depleted cells, however, as no ribosome export factors were identified, and depletion of Enp1 is a terminal phenotype, it is unlikely that these NPCs are actively exporting pre-ribosomes or mRNPs (Aguilar et al., 2020); alternatively, these factors, except for Mex67, may be more transiently associated with NPCs.
Taken together, our AP-MS data show that basket-less, and basket-containing NPCs associate with a shared set of mRNA maturation and export factors, suggesting that mRNPs can bind, and likely be exported, through both types of NPCs, in accordance with the observation that mRNA export does not require Mlp1/2 or a nuclear basket (Powrie et al., 2011). However, several factors (i.e., TREX-2, Pml39) were enriched with Mlp1/Basketplus NPCs suggesting that these NPCs may represent a differentially regulated export route for at least a subset of mRNPs. Lastly, these AP-MS data link basket formation to overall poly(A)-RNA metabolism as many mRNA maturation factors associated with nucleolar Mlp1 and basket-containing NPCs upon poly(A) transcript sequestration to the nucleolus.
A distinct Mlp1 mRNA interactome suggests differential export via basket and basket-less NPCs
Our NPC interactome dissections show that while mRNP-associated factors are found with all NPCs, there is a distinct subset of mRNP export and processing factors enriched with Mlp1, suggesting that some mRNAs may be preferentially exported via basket-containing NPCs. To determine if subsets of nuclear mRNAs are differentially associated with nucleoplasmic Basketplus or Basketminus NPCs, AP-RNA-seq was performed on oligo-dT-purified RNA samples isolated with either ‘All NPCs’ (Nup133-GFP), or Basketplus and Basketminus’ NPCs via differential APs as above. Following background removal, RNAs were compared against a total poly(A)-RNA library to identify transcripts differentially associated with NPCs, using a log2 fold change (FC) cut-off of greater than (>) 1.
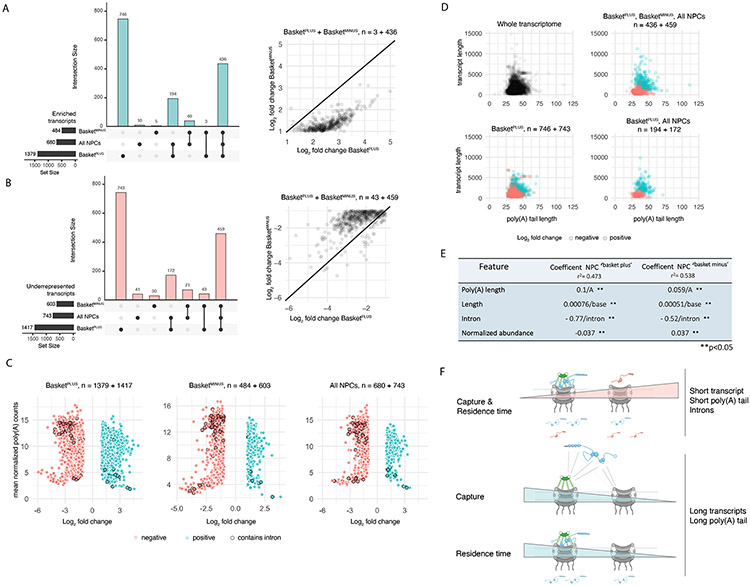
680 transcripts were enriched with ‘All NPCs’, 1379 with Basketplus, and 484 with Basketminus NPCs (Figure 7A). Among these, 746 transcripts were exclusively enriched with Basketplus, yet only five with Basketminus NPCs (Figure 7A), suggesting a preferential association of a subset of transcripts with basket-containing NPCs. Consistent with this, comparing log2(FC) for the 439 mRNAs enriched in both Basketplus and Basketminus samples, values were consistently higher in Basketplus (paired t-test, estimate = 1.22, p < 0.001) (Figure 7A) indicating that their enrichment is greater in basket-containing NPCs, in line with a preferential association of transcripts with basket-containing NPCs, or the association of Mlp1 with transcripts in the nucleoplasm prior to their NPC association.
Figure 7. A differential Mlp1 RNA interactome suggests selective transport.
A. Upset plot. Bar graph, bottom left: transcripts identified with ‘All NPCs’, and differential ‘Basketplus’ and ‘Basketminus’ NPC APs with a log2(FC) > 1 over poly(A) libraries. Bar graph, top: exclusively enriched transcripts and intersection sizes; dots indicate shared transcript sets. Scatter plot, right: transcripts enriched in both ‘Basketplus’ and ‘Basketminus’ with higher log2(FC) values in ‘Basketplus’ compared to ‘Basketminus’. B. Upset plot. Bar graph, left: transcripts significantly underrepresented with a log2(FC)< 1 over poly(A) libraries across samples. Bar graph, top: exclusively underrepresented transcripts and intersection sizes; dots indicate shared transcript sets. Scatter plot, right: transcripts depleted in both ‘Basketplus’ and ‘Basketminus’ with lower log2(FC) values in ‘Basketplus’ compared to ‘Basketminus’. C. Scatter plots of log2(FC) values against mean normalized transcript expression in the poly(A) library. D. Scatter plots of poly(A) tail length vs. transcript length. E. Linear regression models built from poly(A) tail length, transcript length, intron presence/absence and mean poly(A) normalized transcript counts for ‘Basketplus’ and ‘Basketminus’ samples. All coefficents were significant (p<0.05). F. Cartoons illustrating models of mRNP capture and residence time at ‘Basketplus’ and ‘Basketminus’ NPCs based on select features.
Conversely, 743 transcripts were underrepresented with ‘All NPCs’, 1417 transcripts with Basketplus, and 603 with Basketminus (Figure 7B). Among these, 743 mRNAs were specifically depleted from Basketplus samples and only 30 from Basketminus (Figure 7B). Considering log2(FC) values, the vast majority of the 502 mRNAs underrepresented across Basketplus and Basketminus samples had lower log2(FC) values in basket-less compared to basket-containing NPCs (paired t-test, estimate = −0.85, p < 0.001) (Figure 7B). Overall, this suggests that these transcripts may either form unstable interactions with basket-containing NPCs, have shorter residence times at the NPC, are exported more rapidly, or preferentially associate with basket-less NPCs.
Gene ontology (GO) enrichment analyses did not reveal any significant bias in the functional classification of transcripts co-purified in Basketminus and Basketplus. We also did not observe a strong correlation between cellular transcript abundance and relative enrichment at NPCs (Figure 7C; S7A). However, transcripts generated from intron-containing genes, which are among the most highly expressed transcripts, were underrepresented across all three samples (Figure 7C; S7A), suggesting that mRNA interaction with NPCs may vary depending on specific features. Moreover, no differential enrichment of unspliced pre-mRNAs was detected across samples (Figure S7B; Table S7). We also identified pervasive and non-coding transcripts most of which were predominately underrepresented across samples, particularly with Basketplus NPCs (Figure S7C).
To identify other features that may influence the relative enrichment of transcripts with NPCs, we further considered transcript- and poly(A) tail length, using published poly(A) sequencing data (Tudek et al., 2021). Transcripts enriched across all samples (except controls) tended to be longer and had longer poly(A) tails compared to underrepresented transcripts (Figure 7D; S7B, C, D, E), suggesting that mRNA and poly(A) tail length may influence mRNA-NPC interactions.
To further examine the impact of transcript length, poly(A) tail length, presence of an intron, and transcript abundance on the association of mRNAs with Basketplus and Basketminus NPCs, we utilized linear regression to model the relationship between these variables and log2(FC) (enrichment) in the dAPs. The resulting models show that presence of introns negatively influenced the enrichment of transcripts with NPCs (Figure 7E) and the penalty for the presence of an intron was greater for mRNAs associating with Basketplus than Basketminus NPCs. The model further confirmed that transcript length and poly(A) tail length had a positive influence on transcript enrichment for both Basketminus and Basketplus NPCs, which was larger for mRNAs found associated with basket-containing NPCs (Figure 7E). This suggests that longer transcript and poly(A) tail lengths favor mRNA-NPC association at different types of NPCs, while the presence of an intron makes it less likely to observe an interaction. Interestingly, the log2(FC) increase per base was higher for the poly(A) tail length than for the total transcript length, indicating increased poly(A) tail length to be more significant than overall transcript length.
Taken together, our results suggest a preferential association of mRNAs with basket-containing and basket-less NPCs based on features such as poly(A) tail length, transcript length and the initial presence of introns. This may be reflective of differential export dynamics for mRNAs/mRNPs. Considering that, in yeast, intron-containing transcripts are generally short (<1000nt), their short size and/or presence of an intron may lead to differential interactions with NPCs and faster export kinetics as suggested by their general under-representation across all NPCs (Figure 7F, i). Conversely, longer mRNAs were enriched across all NPCs, suggesting slower export kinetics or longer residence times, possibly due to necessary mRNP rearrangements prior to export (Figure 7F, ii). Notably, longer transcript and poly(A) tail lengths favored basket-containing NPCs/Mlp1, suggesting a potential role for the basket in directing the export of a subset of mRNPs with distinct features.
DISCUSSION
A functionally specialized Mlp1 and basket interactome
The observation that not all NPCs assemble a basket raises the question of what initiates the assembly at only some NPCs, beyond the release of correctly processed and polyadenylated mRNAs from their sites of transcription. As basket assembly requires mRNA and a subset of nuclear RBPs, it is possible that only a specific subset of mRNPs triggers basket formation and preferential transit through basket-containing NPCs. Pml39 and the TREX-2 complex were highly enriched with Mlp1/basket-containing NPCs. Mlp1/the nuclear basket and Pml39 have been linked to surveillance and retention of intron-containing transcripts at the NPC. TREX-2 has been implicated in the recruitment of transcribing genes to the NPC in connection with SAGA, a general co-factor for RNA Pol II transcription (Baptista et al., 2017; Jani et al., 2009, 2014); however, the observation that a specific Mlp1/basket interactome, including TREX-2, is still found at nucleolar NPCs upon Enp1 depletion might suggest that basket assembly is not linked to an association of actively transcribing genes at the periphery as it is improbable that chromatin will move to the nucleolus under these conditions.
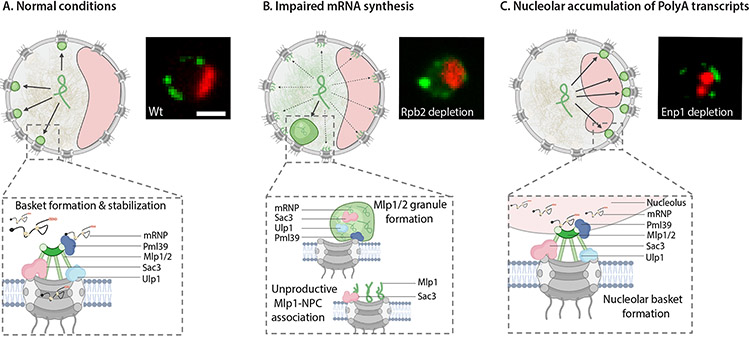
Overall, our observations suggest that basket formation at NPCs depends on a balance of Mlp1 concentration, mRNA production and release, and specific RBPs (Figure 8A) rather than subnuclear compartments; this, as well as the assembly of a basket at only some NPCs, may point towards basket assembly as a stochastic process. Interestingly, upon Pml39, Prp5, Pap1, and Pab1 depletion and, to some extent, RNA Pol II shutdown, a weak Mlp1 signal is still observed at the nuclear periphery, often continuous along the nuclear rim including the nucleolar periphery. This may be due to Mlp1 molecules binding to NPCs below a required stoichiometry for fully formed baskets (Figure 8B) and suggests that self-assembly of Mlp1 at the NPC is not enough to form a functional basket but instead requires other processes along the mRNA maturation pathway. One such process could be pre-association of Mlp1 with mRNPs in the nucleoplasm as Mlp1 has a free-diffusing nucleoplasmic pool and has been found co-isolating with mRNPs (Oeffinger et al., 2007). Reaching the NPC as part of an mRNP could then contribute to basket assembly, either via stabilization or recruitment of more Mlp1 molecules, a notion in line with assembly of nucleolar NPCs in Enp1 and Csl4 mutants (Figure 8C).
Figure 8. mRNA metabolism drives basket assembly.
Cartoons of yeast nuclei illustrating different models of Mlp1-assemblies. A. Assembly of productive nuclear baskets, B. unproductive Mlp1-NPC binding and Mlp1 granules, and C. nucleolar baskets. Nucleoli, red crescents; Mlp1, green ampersands. Fluorescent images representing the three different types of Mlp1 assemblies shown on the right.
Alternatively, Mlp1 may need a critical concentration at the NPC to form a basket. Our observations show that Mlp1 is an aggregation-prone protein as inhibition of RNA Pol II transcription and Pap1 and Pab1 depletion cause granule formation at the nuclear periphery similar to Nup60 depletion. As nucleoplasmic Mlp1 levels are increased under these conditions, and Mlp1-granule formation appears to scale with the severity of the loss of Mlp1 from the periphery, basket formation may depend on a balance of Mlp1 concentration and mRNAs/mRNPs in flux towards the periphery. Mlp1 granule formation, or aggregation, would then be prevented by its contact with mRNPs, yet this capacity for aggregation may have been retained by cells either to rapidly assemble granules for sequestration of specific mRNPs and/or to remove baskets and basket-mediated processes from the NPC under stress conditions.
The role of the basket in selective transport
Previous data suggested that the basket might serve as a quality control platform ensuring that partially or unspliced mRNAs do not exit the nucleus. However, literature concerning the role of the basket in the retention of intron-containing mRNAs is conflicting. In general, the majority of intron-containing mRNAs in yeast are short, while observations of leakage and retention of intron-containing transcripts are based on an inefficiently spliced ~3kb reporter transcript (Bonnet et al., 2015; Galy et al., 2004; Palancade et al., 2005). We observed that while most mRNAs preferentially associated with Mlp1/basket-containing NPCs, there was a clear underrepresentation of short mRNAs, which include the majority of intron-containing transcripts. Moreover, recent data in human cells saw no increase in the leakage of endogenous pre-mRNAs upon TPR depletion (Aksenova et al., 2020; Lee et al., 2020; Zuckerman et al., 2020), unlike previous work using inefficiently spliced reporter constructs (Rajanala and Nandicoori, 2012), and transcripts whose export depends on TPR—and human TREX-2—were in general short, intron-poor or intron-less. However, it is important to note that while one of these features is low number of introns, it is not—as previously thought—the mere presence or absence of introns, suggesting the basket may not be primarily a designated gatekeeper for unspliced pre-mRNAs. Yet it is possible that the function of a basket in pre-mRNA retention becomes more pronounced when splicing is inefficient.
Overall, Mlp1/TPR-dependent transcripts may have common features, which would point towards a role for the basket in selective transport. However, while our data also points towards such a selectivity model, it is, at least in budding yeast, unclear what this selectivity involves. The basket might mediate an increased residence of longer mRNAs at the NPC prior to export, leading to a kinetic delay for larger mRNPs that require rearrangements and/or quality control. Such model would be in line with the enrichment of longer mRNAs observed with Mlp1/basket-containing NPCs in this study, including those containing introns, and would further explain the dependence of the 3kb-intron-leakage/retention reporter on Mlp1/2.
While the nuclear basket is not a requirement for nuclear mRNA export per se, it nevertheless appears to serve as a facilitator. Either model for the nuclear basket as mediator of selective export kinetics, whether it is to enhance or slow access to the central NPC channel for translocation, would be compatible with different models of basket formation and function. In addition to mRNP-dependent assembly, the previously shown slow binding of Mlp1 to new NPCs may in part also contribute to basket-related NPC heterogeneity (Onischenko et al., 2020). Independent of which of the proposed models for assembly and function of the nuclear basket will prevail, our study shows that in budding yeast, its role in mRNP transport and metabolism is dedicated to a select subset of NPCs. Whether a similar heterogeneous functionalization of NPCs exists in other organisms and whether such specialization extends to other NPC functions is an exciting topic for future research.
LIMITATIONS OF THIS STUDY
While SIM resolution allowed us to reveal the existence of basket related NPC heterogeneity along the nuclear periphery, it was not sufficient to determine whether basket-containing and basket-less NPCs form in distinct regions along the nucleoplasmic periphery, nor whether all baskets contain a defined number of Mlp1 molecules, or how many molecules would be sufficient to form a functional basket. Moreover, while differential affinity purification enables us to dissect the interactomes of basket-containing and -less NPCs, a limitation of the approach is that we cannot distinguish whether mRNPs can bind free nucleoplasmic Mlp1 in addition to Mlp1 at the periphery, i.e., at the basket. Lastly, while the affinity purification of NPC-associated RNAs was only performed in unperturbed conditions, investigating the RNA interactome upon perturbation of RNA processing will allow further dissection of the role of the basket in quality control and export.
STAR METHODS
RESOURCES AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed and will be fulfilled by the lead contact, Marlene Oeffinger (Marlene.Oeffinger@ircm.qc.ca).
Materials availability
All unique reagents generated in this study are available from the lead contact with a completed material transfer agreement.
Data and code availability
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot and raw images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE |
SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-HA | Sigma-Aldrich | 11583816001 |
| Mouse monoclonal anti-GFP | Sigma-Aldrich | 11814460001 |
| Rabbit IgG | Sigma-Aldrich | I5006 |
| Anti-mouse HRP | Abcam | ab6728 |
| Nanobody Anti-GFP | Rothbauer et al., 2008 | GBP |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Rapamycin | Bio Basic | R706203 |
| Indole-3-acetic acid (IAA/Auxin) | Sigma-Aldrich | I3750 |
| 1-6 Hexanediol | Sigma-Aldrich | H11807 |
| Dynabeads M-270 Epoxy | Invitrogen | 14302D |
| RNAsin | Promega | N2111 |
| Pepstatin A | Sigma-Aldrich | P5318 |
| PMSF | Sigma-Aldrich | P7626 |
| antifoam | Sigma-Aldrich | A5633 |
| Glutaraldehyde | Sigma-Aldrich | G6257 |
| Trypsin Protease | Promega | V511A |
| Phusion DNA polymerase | ThermoFischer | F630S |
| Q5 DNA polymerase | NEB | M0491L |
| YPD | Biobasic | SD7022 |
| Yeast nitrogen base w/o amino acids | Biobasic | S505 |
| Concanavalin A | Sigma-Aldrich | C5275 |
| Halo-ligand JF-549 | Luke Lavis, Janelia Research Campus | Grimm et al., 2015 |
| Trizol | Invitrogen | 15596026 |
| Ultra-pure water | Invitrogen | 10977023 |
| Critical Commercial Assays | ||
| QIAquick PCR purification kit | Biobasic | BS664 |
| Direct-zol Miniprep Kit | Zymo research | R2050 |
| Deposited data | ||
| NPC affinity purification and Mass spectrometry | This manuscript. | Project Name: Nuclear pore complexes interactome dissection PRIDE Project DOI: 10.6019/PXD027872 |
| NPC-AP-RNAseq | This manuscript. | GEO: GSM5492330 |
| RNA poly(A) tail sequencing | Tudek et al., 2021 | https://doi.org/10.17632/v5vm3dmm8y.1. |
| Raw data files | This study | Mendeley, DOI: 10.17632/krwndnhw87.1 |
| Experimental model: Cell lines | ||
| Saccharomyces cerevisiae: W303-1A and W303-1B | Laboratory of Rodney Rothstein | ATCC:208352 ATCC:201238 |
| Yeast strains used | This manuscript. | Supplemental Table S1 |
| Oligonucleotides | ||
| Oligonucleotides used | This manuscript. | Supplemental Table S3 |
| Recombinant DNA | ||
| Plasmids used | This manuscript. | Supplemental Table S2 |
| Software and Algorithms | ||
| FIJI – Open source edition | ImageJ | http://fiji.sc; RRID:SCR_002285 |
| ZEN Microscopy software | Zeiss | Version ZEN 2012 SP5 |
| Scaffold (version 4.8.4) | Proteome Software, Inc., Portland, OR | https://www.proteomesoftware.com/ |
| RNA-seq analysis code | This manuscript. | Zenodo, DOI: 10.5281/zenodo.7063132 |
| Figures | This manuscript. | BioRender.com |
| RNA-seq analysis | Trimmomatic (version 0.38.0) | https://toolshed.g2.bx.psu.edu/repository?repository_id=ef9e620e9ac844b3 |
| RNA-seq analysis | STAR (version 2.5.2a) | https://toolshed.g2.bx.psu.edu/repository?repository_id=b8c12e1910b4735a |
| RNA-seq analysis | Htseq-count (version 0.11.3) | https://toolshed.g2.bx.psu.edu/repository?repository_id=2df7e24ce6c1f224 |
All original code has been deposited at Zenodo and is publicly available as of the date of publication. The DOI has been listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast (S. cerevisiae) strains used in this study are described in Table S1. Epitope tagging was performed by standard DNA recombination methods using the plasmids listed in Table S2. Each construction was validated by sequencing, PCR, immunoblotting, and/or microscopy.
Method Details
Yeast tagging and growth.
Yeast strains and plasmids used in this study are listed in Tables S1 and S2. Yeast strains are all derived from W303, and epitope-tagged proteins have been C-terminally tagged. Yeast strains were constructed by homologous recombination using a recombination cassette generated by PCR (for primers, see Table S3) with 50 bp homology arms as described in (Bensidoun et al., 2016). Cells were grown in YPD or synthetic complete media lacking the appropriate amino acid to maintain plasmids when appropriate. Unless noted otherwise, cells for RNA-based or protein-based analyzes were isolated, rapidly frozen in liquid nitrogen, and cryo-lysis was performed by solid-phase milling in a planetary ball mill (Retsch) producing a fine cell grindate (Oeffinger et al., 2007). All grindates were stored at −80°C until processed either for affinity purification or RNA extractions.
Mlp1 N-terminal fragment expression.
All expression plasmids encoding yEGFP3-NLS-Mlp1 truncations have been generously provided by C. Strambio-De-Castillia (Niepel et al., 2013). Cells transformed with these expression plasmids were grown in the presence of 150 mg/l methionine for live-cell microscopy to reduce the expression level of the GFP-NLS–tagged Mlp1p fragments in target yeast cells.
Auxin depletion.
Yeast cells were grown at 30°C in YPD to an OD600 ~0.3. Indole-3-acetic acid (auxin; Sigma-Aldrich) was added to a final concentration of 500 μM (Morawska et al, 2008). After 120 min, cells were prepared for live-cell imaging in SD with 2% glucose supplemented with 500 μM of auxin. For western blotting, 20 ml of culture was harvested at an OD600 of ~0.6 with or without 500 μm auxin, and protein extraction and western blotting were carried out as described above.
Rapamycin and 1-6 Hexanediol treatment.
Yeast cells were grown at 30°C in YPD to an OD600 ~0.3. Rapamycin was added to a final concentration of 10% for 40min then cells were prepared for live-cell imaging in SD with 2% glucose supplemented with 10% rapamycin. 1-6 Hexanediol was added to a final concentration of 10% for 15 min then cells were prepared for live-cell imaging in SD with 2% glucose supplemented with 10% hexanediol.
Protein extraction and western blot analysis.
For western blot analysis, proteins were extracted from yeast cells was performed as previously described in (Kushnirov et al., 2000) and separated on 10% or 4–12% Tris/Glycine SDS-PAGE gels. Proteins were transfer to PVDF membranes and detected using either monoclonal anti-GFP (1:1000; Sigma, 11814460001), anti-HA 12CA5 (1:1000; Sigma, 11583816001), or anti-mouse HRP (1:5000; Abcam, ab6728) antibodies. Images were acquired using a ChemiDoc MP Imaging System (Biorad).
Preparing cells for live-cell imaging.
Yeast cells were grown at 30°C in SD with 2% glucose to an OD600 ~ 0.4–0.6. For imaging, 100-μl cell suspension was added to a 96-well glass-bottom plate (MGB096-1-2-LG-L; Brooks Life Science Systems) previously coated with concanavalin A (Con A) and concentrated on the bottom of the well by centrifugation. Wells were coated by adding 100 μl of 1 mg/ml Con A (Sigma-Aldrich) for 10 min before unbound Con A is removed and the Con A activated by adding 100 μl of 50 mM CaCl2/50 mM MnSO4 for 10 min. The solution was then removed, washed once with 100 μl ddH2O, and air-dried. To minimize cell motion for SIM imaging, cells were briefly fixed with 70% EtOH for 5 min chilled at −20C, wash with cold 1x PBS before being added to glass-bottom plates coated with ConA.
Cell labeling with HaloTag ligands.
Cells were grown in YPD to an OD600 ~0.15 in log phase before incubating with 100 nM of Halo-ligand JF-549 (generously provided by Luke Lavis, Janelia Research Campus) (Grimm et al., 2015) for 90 min, followed by 3 quick YPD washes and a 30min wash in YPD with agitation. Finally, cells were washed 3 times in SD with 2% glucose before imaging. For SIM experiments, cells were fixed after the last wash as described above.
Image acquisition.
Unless mentioned otherwise in the text, images were acquired on a spinning disk confocal microscope (Observer SD; Carl Zeiss) using a 100×/1.43 NA objective (Carl Zeiss), 488-nm (100 mW), and 561-nm (40 mW) excitation laser lines, and Semrock single bandpass filters for GFP (525 nm/50 nm) and RFPs (617 nm/73 nm). Images were captured using an electron-multiplying charge-coupled device camera (Evolve 512; Photometrics) using Zen blue software. For heat-shock experiments, the stage was preheated to 42°C using a Zeiss incubation chamber. Cells were incubated in the chamber for 1h at 42C in SD with 2% glucose.
Structured illumination microscopy (SIM).
SIM images were acquired with a 63x NA 1.46 oil objective on a Zeiss Elyra PS.1 system equipped with an Andor EMCCD iXon3 DU-885CSO VP461 camera (1004x1002 pixels), and with the following lasers: 50 mW405 nm HR diode, 100 mW 488 nm HR diode, 100 mW 561 nm HR DPSS, 150 mW 642 nm HR diode. Each image was acquired using 3 rotations and a grid size of 42mm for all channels.
Single-molecule imaging using Highly Inclined Laminated Optical (HILO) illumination.
Cells were concentrated and mounted for imaging as described above. Images were acquired using Zeiss Elyra PS.1 microscope setup equipped with a 63x NA 1.46 oil objective, an Andor EMCCD iXon3 DU-885CSO VP461 camera (1004x1002 pixels), and with the following lasers: 50 mW405 nm HR diode, 100 mW 488 nm HR diode, 100 mW 561 nm HR DPSS, 150 mW 642 nm HR diode. To minimize the out-of-focus light, time-lapse movies were acquired using Highly Inclined Laminated Optical (HILO) sheet illumination. Image size was cropped to 512x512 pixels, and images were acquired at 50 Hz (frames per second). To quantify single-molecule diffusion in the nucleoplasm and the nucleolus, images were acquired until most fluorophores were bleached and single molecules became visible to allow single-particle tracking. Tracking was done using the Trackmate plugin in ImageJ (Tinevez et al., 2017). To measure the fraction of tracks from single diffusing particles overlapping with the nucleolus, the nucleolar areas were first manually delimited using the ROI manager tool in ImageJ, followed by counting the fraction of time individual single particles overlapped with the nucleolus. 40 tracks for Mlp1-Halo and Halo-NLS, varying from 10 to 40 frames were analyzed.
Affinity purification and mass spectrometry.
Affinity purification.
All affinity purifications (AP) were performed in triplicate per conditions as previously described (Oeffinger et al., 2007b; Trahan and Oeffinger, 2022). In brief, cells were grown to late log phase, frozen by immersion in liquid nitrogen, and mechanically ground using a planetary ball mill (Retsch). For each AP, 1 g of cell powder was thawed in 9 ml of extraction buffer (1X tributyltin, 50 mM NaCl, 1 mM DTT, 0.5 % Triton X-100, 1X protease inhibitor cocktail [4 mg/mL pepstatin A (Sigma), 180 mg/mL PMSF (Sigma)], antifoam B (Sigma, 1:5000), and 40U/mL RNAsin (Promega), homogenized with a Polytron for 25 s, and cleared by centrifugation at 4,000 g for 5 min. 10mM glutaraldehyde was added for 5 min and samples were gently agitated on ice before the reaction was quenched with Tris-HCl pH8 to a final concentration of 100mM. Lysates were incubated with either IgG (anti-rabbit IgG, Sigma) or GFP-nanobody (expressed from pDZ580-pET28a-GBP and purified as described in-conjugated magnetic beads (Dynabeads M-280) for 30 min (Rothbauer et al., 2008). After removal of the supernatant, beads were extensively washed in extraction buffer, then detergents were removed by washing the bead-bound complexes in 0.1 M NH4OAc/0.1 mM MgCl2 before a final was and resuspension in 50μl of 20-mM Tris-HCl, pH 8.0. Isolated proteins were digested on-bead at 37°C with 1 μg trypsin (Pierce Trypsin Protease, MS Grade) for 16 h (Trahan and Oeffinger, 2022). The digestion was stopped by adding formic acid to a final concentration of 2%.
For differential APs, the flow-through was incubated with GFP-nanobody-conjugated magnetic beads for 30 min and the beads were then treated as described above.
Peptide preparation for injection into the mass spectrometer.
Tryptic peptides were cleaned using C18 ZipTips as per supplier recommendations (Milli-pore). Samples were injected to near saturation of the signal, while an equivalent volume of their respective negative controls was injected. Liquid chromatography was performed using a PicoFrit fused silica capillary column (15 cm × 75 μm i.d; New Objective, Woburn, MA, USA), self-packed with C-18 reverse-phase resin (Jupiter 5 um particles, 300 Å pore size; Phenomenex, Torrance, CA, USA) using a high-pressure packing cell on the Easy-nLC II system (Proxeon Biosystems, Odense, Denmark) and coupled to an Orbitrap Fusion™ Tribrid™ Mass Spectrometer equipped with a Proxeon nanoelectrospray Flex ion source. 0.2% formic acid (Solvent A) and 100% acetonitrile/0.2% formic acid (Solvent B) were used for chromatography and peptides were loaded on-column at a flowrate of 600 ml/min and eluted with a three-slope gradient at a flowrate of 250 nl/min. Solvent B was first increased from 2 to 25% over 20 min, then from 25 to 45% over 40 min, and finally from 45 to 80% B over 10 min.
Protein identification.
The peak list files were generated with Proteome Discoverer the following as described in (Aguilar et al, 2020). Protein database searching was performed with Mascot 2.5 (Matrix Science) against the NCBI - S. cerevisiae protein database (20160802). The mass tolerances for precursor and fragment ions were set to 10 ppm and 0.6 Da, respectively. Trypsin was used as the enzyme allowing for up to 1 missed cleavage. Cysteine carbamidomethylation was specified as a fixed modification and methionine oxidation as variable modification. Data analysis was performed using Scaffold (version 4.8.4).
Mass spectrometry data analysis.
Protein and peptide identification thresholds in Scaffold™ were set to 95% which resulted in a decoy false discovery rate of 6%. Exclusive spectrum counts (ESC) were used for semi-quantification of protein preys, and mass spectrometry results were analyzed as previously described in (Scott et al., 2017). Briefly, only Exclusive Spectral Counts (ESCs) above background detected in controls were retained, and of those only ESCs ≥10 used for quantitive analyses across samples. In silico digestion using MS digest (http://prospector.ucsf.edu) was performed for each protein to take protein size and predicted cleavage sites into account. Values were normalized against the average values of proteins associated with the bait proteins in the different APs: Mlp2 for Mlp1-PrA and Y complex nucleoporins (Nup84, Nup85, Nup120 Nup145C) for Nup133-GFP. This allowed normalization of the data sets against proteins with a similar size, stoichiometry, and segregation behavior. Data are available via ProteomeXchange with identifier PXD027872. Project Name: Nuclear pore complexes interactome dissection, Project DOI: 10.6019/PXD027872.
Affinity purification RNA-seq
Poly(A)-RNA preparation and sequencing.
RNA Affinity purification was performed using 1 g of cell powder per triplicate as described above but without crosslinking. Lysates were incubated with either IgG- or GFP-nanobody-conjugated magnetic beads for 30 min. Beads were washed extensively (8 times) in extraction buffer before being resuspended in 1 ml of Trizol (Invitrogen, 15596026) and vortexed vigorously. Control experiments were carried out using strains expressing Protein A or GFP alone, and RNAs identified in these samples were considered background and filtered from those identified with Mlp1-PrA and Nup133-GFP, respectively. The total poly(A) library was generated by RNA extraction using 100 mg of cryo-ground cell powder thawed into Trizol in triplicates. RNA was then extracted using the Direct-zol Miniprep Kit (Zymo Research, R2050) and Dnase treatment was performed on-column according to the manufacturer’s instructions. Samples were resuspended in 30 μl ultra-pure water (Invitrogen, 10977023), and the quality of RNA was assessed by Qbit and Bioanalyzer chip. RNA extracts were Poly(A)-RNA enriched via a Poly(A) mRNA magnetic Isolation Module oligo-dT (NEBNext), and cDNA libraries were prepared using the Kapa RNA HyperPrep Kit (96 rxns, Roche) and TruSeq DNA UDI 96 indexes (Illumina). RNA-sequencing was performed using Nocaseq6000 flowcell S2 PE50.
RNA sequencing analysis.
RNA-seq reads were trimmed using Trimmomatic (version 0.38.0) to quality trim and remove adapters. Trimmed reads were mapped to the S. cerevisiae genome (GCF_000146045.2_R64) using STAR (version 2.5.2a) using default parameters. Genes were counted using htseq-count (version 0.11.3) with parameters -m intersection-nonempty -s yes -r pos.
Differential enrichment analysis was performed using DESeq2 (version 1.28.1) using the Wald test. All five dAPs (Basketplus, Basketminus, ‘All pores’, IgG, and GFP alone) were compared against the total poly(A) library representative of the total transcriptome to generate log2 fold change values for each transcript. Transcripts with an absolute log2 fold change value > 1 and p < 0.05 were considered significantly differentially enriched. Transcripts that were significantly enriched in the IgG and GFP libraries were removed from further analysis from the Basketplus (IgG), Basketminus (GFP), and ‘All pores’ (GFP) results according to purification type. The R library UpSetR was used to generate upset plots and determined overlapping sets of enriched and depleted genes across libraries. The R library clusterProfiler was used to perform gene set enrichment analysis using Gene Ontology terms for each dAP library. To determine whether enriched transcripts correlated with other expression features, we parsed the S. cerevisiae GTF gene annotation file (GCF_000146045.2_R64) to determine the presence of introns and transcript length, and used poly(A) tail lengths as determined previously (Tudek et al., 2021). All analysis code is available at Zenodo, DOI: 10.5281/zenodo.7063132
QUANTIFICATION AND STATISTICAL ANALYSIS
Image quantitation
For image analysis, composite images including all channels were generated in Fiji. To measure the signal intensities across the nucleus or along its periphery, straight lines or circular ROIs respectively were manually selected and intensities values along the ROIs were obtained using the plot profile analysis tool in ImageJ. The shape and sizes of nucleoli were estimated by measuring two parameters (“solidity” and “circularity”) using the shape descriptors available in ImageJ analysis tools after manual segmentation of the nucleolar areas. Averaged intranuclear signals for Mlp1-GFP and Nup60-tdTomato in Wt, Rpb2AID-HA, Pab1AID-HA, Pap1AID-HA, and Esc1AID-HA were calculated from squared intra-nuclear ROIs and normalized by their respective backgrounds. Channel-specific backgrounds have been estimated using averaged values measured along lines or within sections of the fields without cells.
For signal distribution analysis in Figure 5B, the periphery adjacent to the nucleolus was segmented and excluded prior to measuring signal intensities of Nup188-tdTomato and selected GFP-tagged Nups or Mlp1-GFP. Each dot in graph represents the proportion/cell (in %) of the nucleoplasmic periphery (non-nucleolar) occupied by tdTomato and GFP signals (n=100 for each strain).
Peptide quantitation – MS
Protein and peptide identification thresholds in Scaffold™ were set to 95% which resulted in a decoy false discovery rate of 6%. Exclusive spectrum counts (ESC) were used for semi-quantification of protein preys, and peptides were quantified as follows: only Exclusive Spectral Counts (ESCs) above background detected in controls were retained, and of those only ESCs ≥10 used for quantitive analyses across samples. In silico digestion using MS digest (http://prospector.ucsf.edu) was performed for each protein to take protein size and predicted cleavage sites into account. These values were normalized against the average values of proteins associated with the bait proteins in the different APs: Mlp2 for Mlp1-PrA, and Y complex nucleoporins (Nup84, Nup85, Nup120 Nup145C) for Nup133-GFP. This allowed normalization of the data sets against proteins with a similar size, stoichiometry, and segregation behavior. For comparison in Figure 6B and C, data has been compressed so that 1 represents maximal value of ESC median between experiments (n=3) and lines represent the relative abundance of proteins between different affinity purifications. Relative abundance of isolated RBPs and factors required for mRNA maturation, export and surveillance was further quantified by comparing normalized spectral counts of ESC≥10 across all samples (Supplemental Table 6C) as well as log2 fold change (FC) relative to ‘All NPCs’ (Supplemental Table 6B).
Protein annotations
Protein database searching was performed with Mascot 2.5 (Matrix Science) against the NCBI - S. cerevisiae protein database (20160802). Functional protein annotations in Figure 6, Supplemental Figure S6, and Supplemental Tables S4 and S5 were sourced from UniProt columns named “Subcellular location [CC]” or “Protein names” (UniProt Consortium, 2019) and PANTHER 17.0 (Thomas et al., 2022). For Figure 6D, protein localization was strictly parsed using annotations from Saccharomyces Genome Database (SGD) (Cherry et al., 2012), LoQAtE (Cohen et al., 2011) YeastGFP (Huh et al., 2003), yeastRC Public Images (Riffle and Davis, 2010), and YeastRGB (Dubreuil et al., 2019).
RNA-seq statistical analysis and T-tests
To normalize counts to compare average transcript expression across libraries, the variance stabilized transformation as implemented in the DESeq2 function vst() was used. T-tests were performed using the t.test() function, wilcoxon rank sum tests were performed using the wilcox.test() function, and linear models were generated using the lm() function.
Linear regression model
Linear regression was performed using the lm() function in R. The prediction of log2 fold change was based on the length of the gene in base pairs, the presence or absence of an intron in the gene, mean length of the polyA tail from https://data.mendeley.com/public-files/datasets/v5vm3dmm8y/files/aceb7ee8-5ba0-4cc5-be02-bdab3dc94cb3/file_downloaded, and the mean variance-stabilized transformed expression in the oligo DT libraries. The mean variance stabilized transformed value was calculated using the DESeq2 vst() function. Genes which had an undetermined polyA tail length were removed from the analysis.
Supplementary Material
Supplemental Table S1. Yeast strains used in this study, Related to STAR Methods
Supplemental Table S4. List of AID screen candidates and phenotypes, Related to Figure 4
Supplemental Table S5. Raw AP-MS data, Related to Figure 6
Supplemental Table S6. Processed AP-MS data, Related to Figure 6
Supplemental Table S7. Splice Efficiency Pairwise t-test calculations, Related to Figure 7
Supplemental Movie S2. Single-protein tracking movie of Halo-NLS, Related to Figure 1
Supplemental Movie S1. Single-protein tracking movie of Mlp1-Halo, Related to Figure 1
ACKNOWLEDGEMENTS
We thank S. Adivarahan, P. Raymond, LC. Aguilar and C. Trahan for discussions and help. C. Strambio-De-Castillia for plasmids encoding yEGFP3-NLS-Mlp1 truncations (Niepel et al., 2013); L. Lavis for HaloTag ligands; M. Swaffer, J. Skotheim, and R. Reyes-Lamothe for the yeast strain expressing Halo-NLS. Thanks to D. Faubert (IRCM Proteomics Platform), the IRCM Molecular Biology Platform, and N. Stifani (BMM microscopy platform). This work was supported by Natural Sciences and Engineering Research Council (RGPIN-2015-05922 to D.Z.; RGPIN-2015-06568 to M.O.), Canadian Institute for Health Research (PJT-425798 to D.Z. and M.O.), Canadian Foundation for Innovation (D.Z. and M.O.), and the National Institutes of Health (R01-GM124120 to B.M.). D.Z. holds an FRQ-S Chercheur Boursier Senior.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abraham KJ, Khosraviani N, Chan JNY, Gorthi A, Samman A, Zhao DY, Wang M, Bokros M, Vidya E, Ostrowski LA, et al. (2020). Nucleolar RNA polymerase II drives ribosome biogenesis. Nature 585, 298–302. 10.1038/s41586-020-2497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar L-C, Paul B, Reiter T, Gendron L, Rajan AAN, Montpetit R, Trahan C, Pechmann S, Oeffinger M, and Montpetit B (2020). Altered rRNA processing disrupts nuclear RNA homeostasis via competition for the poly(A)-binding protein Nab2. Nucleic Acids Res 48, 11675–11694. 10.1093/nar/gkaa964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksenova V, Smith A, Lee H, Bhat P, Esnault C, Chen S, Iben J, Kaufhold R, Yau KC, Echeverria C, et al. (2020). Nucleoporin TPR is an integral component of the TREX-2 mRNA export pathway. Nat Commun 11, 4577. 10.1038/s41467-020-18266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista T, Grünberg S, Minoungou N, Koster MJE, Timmers HTM, Hahn S, Devys D, and Tora L (2017). SAGA Is a General Cofactor for RNA Polymerase II Transcription. Mol Cell 68, 130–143.e5. 10.1016/j.molcel.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensidoun P, Zenklusen D, and Oeffinger M (2021). Choosing the right exit: How functional plasticity of the nuclear pore drives selective and efficient mRNA export. Wiley Interdiscip Rev Rna e1660. 10.1002/wrna.1660. [DOI] [PubMed] [Google Scholar]
- Björk P, and Wieslander L (2015). The Balbiani Ring Story: Synthesis, Assembly, Processing, and Transport of Specific Messenger RNA–Protein Complexes. Annu Rev Biochem 84, 65–92. 10.1146/annurev-biochem-060614-034150. [DOI] [PubMed] [Google Scholar]
- Björk P, and Wieslander L (2017). Integration of mRNP formation and export. Cell Mol Life Sci 74, 2875–2897. 10.1007/s00018-017-2503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet A, Bretes H, and Palancade B (2015). Nuclear pore components affect distinct stages of intron-containing gene expression. Nucleic Acids Res 43, 4249–4261. 10.1093/nar/gkv280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretes H, Rouviere JO, Leger T, Oeffinger M, Devaux F, Doye V, and Palancade B (2014). Sumoylation of the THO complex regulates the biogenesis of a subset of mRNPs. Nucleic Acids Res 42, 5043–5058. 10.1093/nar/gku124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner JH, and Walter P (2004). Gene Recruitment of the Activated INO1 Locus to the Nuclear Membrane. Plos Biol 2, e342. 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky AS, and Silver PA (2000). Pre-mRNA processing factors are required for nuclear export. Rna New York N Y 6, 1737–1749. 10.1017/s1355838200001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody SR, Tran EJ, Apponi LH, Corbett AH, and Wente SR (2010). The Mitogen-Activated Protein Kinase Slt2 Regulates Nuclear Retention of Non-Heat Shock mRNAs during Heat Shock-Induced Stress ▿†. Mol Cell Biol 30, 5168–5179. 10.1128/mcb.00735-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro T, Carvalho C, Braga J, Rino J, Milligan L, Tollervey D, and Carmo-Fonseca M (2007). Depletion of the Yeast Nuclear Exosome Subunit Rrp6 Results in Accumulation of Polyadenylated RNAs in a Discrete Domain within the Nucleolus ▿. Mol Cell Biol 27, 4157–4165. 10.1128/mcb.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, and Silver PA (2004). Genome-Wide Localization of the Nuclear Transport Machinery Couples Transcriptional Status and Nuclear Organization. Cell 117, 427–439. 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED (2012). Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40; D700–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y, Schuldiner M (2011). Advanced methods for high-throughput microscopy screening of genetically modified yeast libraries. Methods of Molecular Biology 781;127–59. DOI: 10.1007/978-1-61779-276-2_8 [DOI] [PubMed] [Google Scholar]
- Derrer CP, Mancini R, Vallotton P, Huet S, Weis K, and Dultz E (2019). The RNA export factor Mex67 functions as a mobile nucleoporin. J Cell Biol 218, 3967–3976. 10.1083/jcb.201909028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil B, Sass E, Nadav Y, Heidenreich M, Georgeson JM, Weill U, Duan Y, Meurer M, Schuldiner M, Knop M, Levy ED (2019). YeastRGB: comparing the abundance and localization of yeast proteins across cells and libraries. Nucleic Acids Res. 47(D1); D1245–D1249. doi: 10.1093/nar/gky941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EF, Hammell CM, Hodge CA, and Cole CN (2005). Yeast poly(A)-binding protein, Pab1, and PAN, a poly(A) nuclease complex recruited by Pab1, connect mRNA biogenesis to export. Gene Dev 19, 90–103. 10.1101/gad.1267005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Ventura A, Rutz B, Caspary F, Faux C, Halgand F, Laprévote O, and Séraphin B (2004). Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. Embo J 23, 4847–4856. 10.1038/sj.emboj.7600482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosst P, Guan T, Subauste C, Hahn K, and Gerace L (2002). Tpr is localized within the nuclear basket of the pore complex and has a role in nuclear protein export. J Cell Biology 156, 617–630. 10.1083/jcb.200106046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, and Nehrbass U (2004). Nuclear Retention of Unspliced mRNAs in Yeast Is Mediated by Perinuclear Mlp1. Cell 116, 63–73. 10.1016/s0092-8674(03)01026-2. [DOI] [PubMed] [Google Scholar]
- Geisberg JV, Moqtaderi Z, Fan X, Ozsolak F, and Struhl K (2014). Global Analysis of mRNA Isoform Half-Lives Reveals Stabilizing and Destabilizing Elements in Yeast. Cell 156, 812–824. 10.1016/j.cell.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DM, Johnson CP, Hagan H, and Corbett AH (2003). The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc National Acad Sci 100, 1010–1015. 10.1073/pnas.0336594100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, Lionnet T, Lavis LD (2015). A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods 12(3), 244–50. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünwald D, and Singer RH (2010). In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature 467, 604. 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackmann A, Wu H, Schneider U-M, Meyer K, Jung K, and Krebber H (2014). Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1. Nat Commun 5, 3123. 10.1038/ncomms4123. [DOI] [PubMed] [Google Scholar]
- Hakhverdyan Z, Molloy KR, Keegan S, Herricks T, Lepore DM, Munson M, Subbotin RI, Fenyö D, Aitchison JD, Fernandez-Martinez J, et al. (2020). Dissecting the Structural Dynamics of the Nuclear Pore Complex. Mol Cell 10.1016/j.molcel.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003). Global analysis of protein localization in budding yeast. Nature 425(6959);686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Iglesias N, Tutucci E, Gwizdek C, Vinciguerra P, Dach EV, Corbett AH, Dargemont C, and Stutz F (2010). Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Gene Dev 24, 1927–1938. 10.1101/gad.583310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani D, Lutz S, Marshall NJ, Fischer T, Köhler A, Ellisdon AM, Hurt E, and Stewart M (2009). Sus1, Cdc31, and the Sac3 CID Region Form a Conserved Interaction Platform that Promotes Nuclear Pore Association and mRNA Export. Mol Cell 33, 727–737. 10.1016/j.molcel.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani D, Valkov E, and Stewart M (2014). Structural basis for binding the TREX2 complex to nuclear pores, GAL1 localisation and mRNA export. Nucleic Acids Res 42, 6686–6697. 10.1093/nar/gku252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, and Robert F (2014). Kin28 regulates the transient association of Mediator with core promoters. Nat Struct Mol Biol 21, 449–455. 10.1038/nsmb.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Fernandez-Martinez J, Nudelman I, Shi Y, Zhang W, Raveh B, Herricks T, Slaughter BD, Hogan JA, Upla P, et al. (2018). Integrative structure and functional anatomy of a nuclear pore complex. Nature 555, 475. 10.1038/nature26003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nüske E, Poser I, Richter D, and Alberti S (2015). Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. Elife 4, e06807. 10.7554/elife.06807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S, Thyberg J, Björkroth B, Rackwitz H-R, and Cordes VC (2004). Nucleoporins as Components of the Nuclear Pore Complex Core Structure and Tpr as the Architectural Element of the Nuclear Basket. Mol Biol Cell 15, 4261–4277. 10.1091/mbc.e04-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S, Dörries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, and Cordes VC (2010). Protein Tpr is required for establishing nuclear pore-associated zones of heterochromatin exclusion. Embo J 29, 1659–1673. 10.1038/emboj.2010.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov VV (2000). Rapid and reliable protein extraction from yeast. Yeast 16, 857–860. [DOI] [PubMed] [Google Scholar]
- Lee ES, Wolf EJ, Ihn SSJ, Smith HW, Emili A, and Palazzo AF (2020). TPR is required for the efficient nuclear export of mRNAs and lncRNAs from short and intron-poor genes. Nucleic Acids Res 10.1093/nar/gkaa919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Felberbaum R, and Hochstrasser M (2007). A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J Cell Biology 178, 813–827. 10.1083/jcb.200702154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DH, and Hoelz A (2019). The Structure of the Nuclear Pore Complex (An Update). Annu Rev Biochem 88, 1–59. 10.1146/annurev-biochem-062917-011901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mészáros N, Cibulka J, Mendiburo MJ, Romanauska A, Schneider M, and Köhler A (2015). Nuclear Pore Basket Proteins Are Tethered to the Nuclear Envelope and Can Regulate Membrane Curvature. Dev Cell 33, 285–298. 10.1016/j.devcel.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi L, Goryaynov A, Lindquist A, Rexach M, and Yang W (2015). Quantifying Nucleoporin Stoichiometry Inside Single Nuclear Pore Complexes In vivo. Sci Rep-Uk 5, 9372. 10.1038/srep09372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor A, Suliman S, Ben-Yishay R, Yunger S, Brody Y, and Shav-Tal Y (2010). Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. [DOI] [PubMed] [Google Scholar]
- Neumüller RA, Gross T, Samsonova AA, Vinayagam A, Buckner M, Founk K, Hu Y, Sharifpoor S, Rosebrock AP, Andrews B, et al. (2013). Conserved Regulators of Nucleolar Size Revealed by Global Phenotypic Analyses. Sci Signal 6, ra70. 10.1126/scisignal.2004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M, Strambio-de-Castillia C, Fasolo J, Chait BT, and Rout MP (2005). The nuclear pore complex–associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J Cell Biology 170, 225–235. 10.1083/jcb.200504140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M, Molloy KR, Williams R, Farr JC, Meinema AC, Vecchietti N, Cristea IM, Chait BT, Rout MP, and Strambio-De-Castillia C (2013). The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol Biol Cell 24, 3920–3938. 10.1091/mbc.e13-07-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niño CA, Guet D, Gay A, Brutus S, Jourquin F, Mendiratta S, Salamero J, Géli V, and Dargemont C (2016). Posttranslational marks control architectural and functional plasticity of the nuclear pore complex basket. J Cell Biology 212, 167–180. 10.1083/jcb.201506130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, and Rout MP (2007). Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods 4, 951–956. 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- Onischenko E, Noor E, Fischer JS, Gillet L, Wojtynek M, Vallotton P, and Weis K (2020). Maturation Kinetics of a Multiprotein Complex Revealed by Metabolic Labeling. Cell 183, 1785–1800.e26. 10.1016/j.cell.2020.11.001. [DOI] [PubMed] [Google Scholar]
- Ori A, Banterle N, Iskar M, Andrés-Pons A, Escher C, Bui HK, Sparks L, Solis-Mezarino V, Rinner O, Bork P, et al. (2013). Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol Syst Biol 9, 648. 10.1038/msb.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B, Zuccolo M, Loeillet S, Nicolas A, and Doye V (2005). Pml39, a Novel Protein of the Nuclear Periphery Required for Nuclear Retention of Improper Messenger Ribonucleoparticles. Mol Biol Cell 16, 5258–5268. 10.1091/mbc.e05-06-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Garcia P, and Capelson M (2019). Nuclear pores in genome architecture and enhancer function. Curr Opin Cell Biol 58, 126–133. 10.1016/j.ceb.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul B, and Montpetit B (2016). Altered RNA processing and export lead to retention of mRNAs near transcription sites and nuclear pore complexes or within the nucleolus. Mol Biol Cell 27, 2742–2756. 10.1091/mbc.e16-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powrie EA, Zenklusen D, and Singer RH (2011). A nucleoporin, Nup60p, affects the nuclear and cytoplasmic localization of ASH1 mRNA in S. cerevisiae. Rna 17, 134–144. 10.1261/rna.1210411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raices M, and D’Angelo MA (2012). Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nat Rev Mol Cell Bio 13, 687. 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- Rajanala K, and Nandicoori VK (2012). Localization of Nucleoporin Tpr to the Nuclear Pore Complex Is Essential for Tpr Mediated Regulation of the Export of Unspliced RNA. Plos One 7, e29921. 10.1371/journal.pone.0029921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajoo S, Vallotton P, Onischenko E, and Weis K (2018). Stoichiometry and compositional plasticity of the yeast nuclear pore complex revealed by quantitative fluorescence microscopy. Proc National Acad Sci 115, 201719398. 10.1073/pnas.1719398115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riffle M, Davis TN (2010). The Yeast Resource Center Public Image Repository: A large database of fluorescence microscopy images. BMC Bioinformatics 11(1);263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H (2008). A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics 7(2),282–9. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Moreno H, Simos G, Segref A, Fahrenkrog B, Panté N, and Hurt E (1998). Nuclear mRNA Export Requires Complex Formation between Mex67p and Mtr2p at the Nuclear Pores. Mol Cell Biol 18, 6826–6838. 10.1128/mcb.18.11.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saroufim M-A, Bensidoun P, Raymond P, Rahman S, Krause MR, Oeffinger M, and Zenklusen D (2015). The nuclear basket mediates perinuclear mRNA scanning in budding yeast. J Cell Biology 211, 1131–1140. 10.1083/jcb.201503070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DD, Trahan C, Zindy PJ, Aguilar LC, and Delubac MY, Van Nostrand EL, Adivarahan S, Wei KE, and Yeo GW, Zenklusen D and Oeffinger M (2017) Nol12 is a multifunctional RNA binding protein at the nexus of RNA and DNA metabolism. Nucleic Acids Res 45, 12509–12528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W, Ding Z, Zheng Z-Z, Shen J-J, Shen Y-X, Pu J, Fan Y-J, Query CC, and Xu Y-Z (2020). Prp5–Spt8/Spt3 interaction mediates a reciprocal coupling between splicing and transcription. Nucleic Acids Res 48, 5799–5813. 10.1093/nar/gkaa311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Mills K, and Guarente L (1997). Accelerated Aging and Nucleolar Fragmentation in Yeast sgs1 Mutants. Science 277, 1313–1316. 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- Sirri V, Urcuqui-Inchima S, Roussel P, and Hernandez-Verdun D (2008). Nucleolus: the fascinating nuclear body. Histochem Cell Biol 129, 13–31. 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek S, Zeng Y, Bellur DL, Bergkessel M, Morris KJ, Deng Q, Duong D, Seyfried NT, Guthrie C, Staley JP, et al. (2016). Evolutionarily Conserved Polyadenosine RNA Binding Protein Nab2 Cooperates with Splicing Machinery To Regulate the Fate of Pre-mRNA. Mol Cell Biol 36, 2697–2714. 10.1128/mcb.00402-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg G, Schuster M, Theisen U, Kilaru S, Forge A, and Martin-Urdiroz M (2012). Motor-driven motility of fungal nuclear pores organizes chromosomes and fosters nucleocytoplasmic transport. J Cell Biology 198, 343–355. 10.1083/jcb.201201087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M (2007). Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biology 8, 195–208. 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Subbotin RI, and Chait BT (2014). A Pipeline for Determining Protein–Protein Interactions and Proximities in the Cellular Milieu. Mol Cell Proteomics 13, 2824–2835. 10.1074/mcp.m114.041095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Houwe GV, Hediger F, Kalck V, Cubizolles F, Schober H, and Gasser SM (2006). Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441, 774. 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Tinevez J-Y, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL, and Eliceiri KW (2017). TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90. 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- Thomas PD, Ebert D, Muruganujan A, Mushayahama T, Albou LP, and Huaiyu Mi. (2022). PANTHER: Making genome-scale phylogenetics accessible to all. Protein Society 31(1); 8–22. doi: 10.1002/pro.4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trahan C, and Oeffinger M (2022). Yeast Functional Genomics, Methods and Protocols. Methods Mol Biology 2477, 195–223. 10.1007/978-1-0716-2257-5_12. [DOI] [PubMed] [Google Scholar]
- Tudek A, Krawczyk PS, Mroczek S, Tomecki R, Turtola M, Matylla-Kulińska K, Jensen TH, and Dziembowski A (2021). Global view on the metabolism of RNA poly(A) tails in yeast Saccharomyces cerevisiae. Nat Commun 12, 4951. 10.1038/s41467-021-25251-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. (2019). UniProt: a worldwide hub of protein knowledge. Nucleic Acids 47; D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wende W, Friedhoff P, and Sträßer K (2019). The Biology of mRNA: Structure and Function. Adv Exp Med Biol 1203, 1–31. 10.1007/978-3-030-31434-7_1. [DOI] [PubMed] [Google Scholar]
- Winey M, Yarar D, Giddings TH, and Mastronarde DN (1997). Nuclear Pore Complex Number and Distribution throughout the Saccharomyces cerevisiae Cell Cycle by Three-Dimensional Reconstruction from Electron Micrographs of Nuclear Envelopes. Mol Biol Cell 8, 2119–2132. 10.1091/mbc.8.11.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wing CE, Fung HYJ, and Chook YM (2022). Karyopherin-mediated nucleocytoplasmic transport. Nat Rev Mol Cell Bio 1–22. 10.1038/s41580-021-00446-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, and Ren Y (2019). Mechanisms of nuclear mRNA export: A structural perspective. Traffic 10.1111/tra.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander G, Hackmann A, Bender L, Becker D, Lingner T, Salinas G, and Krebber H (2016). mRNA quality control is bypassed for immediate export of stress-responsive transcripts. Nature 540, 593. 10.1038/nature20572. [DOI] [PubMed] [Google Scholar]
- Zhao X, Wu C-Y, and Blobel G (2004). Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J Cell Biology 167, 605–611. 10.1083/jcb.200405168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerman B, Ron M, Mikl M, Segal E, and Ulitsky I (2020). Gene Architecture and Sequence Composition Underpin Selective Dependency of Nuclear Export of Long RNAs on NXF1 and the TREX Complex. Mol Cell 10.1016/j.molcel.2020.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1. Yeast strains used in this study, Related to STAR Methods
Supplemental Table S4. List of AID screen candidates and phenotypes, Related to Figure 4
Supplemental Table S5. Raw AP-MS data, Related to Figure 6
Supplemental Table S6. Processed AP-MS data, Related to Figure 6
Supplemental Table S7. Splice Efficiency Pairwise t-test calculations, Related to Figure 7
Supplemental Movie S2. Single-protein tracking movie of Halo-NLS, Related to Figure 1
Supplemental Movie S1. Single-protein tracking movie of Mlp1-Halo, Related to Figure 1
Data Availability Statement
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot and raw images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE |
SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-HA | Sigma-Aldrich | 11583816001 |
| Mouse monoclonal anti-GFP | Sigma-Aldrich | 11814460001 |
| Rabbit IgG | Sigma-Aldrich | I5006 |
| Anti-mouse HRP | Abcam | ab6728 |
| Nanobody Anti-GFP | Rothbauer et al., 2008 | GBP |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Rapamycin | Bio Basic | R706203 |
| Indole-3-acetic acid (IAA/Auxin) | Sigma-Aldrich | I3750 |
| 1-6 Hexanediol | Sigma-Aldrich | H11807 |
| Dynabeads M-270 Epoxy | Invitrogen | 14302D |
| RNAsin | Promega | N2111 |
| Pepstatin A | Sigma-Aldrich | P5318 |
| PMSF | Sigma-Aldrich | P7626 |
| antifoam | Sigma-Aldrich | A5633 |
| Glutaraldehyde | Sigma-Aldrich | G6257 |
| Trypsin Protease | Promega | V511A |
| Phusion DNA polymerase | ThermoFischer | F630S |
| Q5 DNA polymerase | NEB | M0491L |
| YPD | Biobasic | SD7022 |
| Yeast nitrogen base w/o amino acids | Biobasic | S505 |
| Concanavalin A | Sigma-Aldrich | C5275 |
| Halo-ligand JF-549 | Luke Lavis, Janelia Research Campus | Grimm et al., 2015 |
| Trizol | Invitrogen | 15596026 |
| Ultra-pure water | Invitrogen | 10977023 |
| Critical Commercial Assays | ||
| QIAquick PCR purification kit | Biobasic | BS664 |
| Direct-zol Miniprep Kit | Zymo research | R2050 |
| Deposited data | ||
| NPC affinity purification and Mass spectrometry | This manuscript. | Project Name: Nuclear pore complexes interactome dissection PRIDE Project DOI: 10.6019/PXD027872 |
| NPC-AP-RNAseq | This manuscript. | GEO: GSM5492330 |
| RNA poly(A) tail sequencing | Tudek et al., 2021 | https://doi.org/10.17632/v5vm3dmm8y.1. |
| Raw data files | This study | Mendeley, DOI: 10.17632/krwndnhw87.1 |
| Experimental model: Cell lines | ||
| Saccharomyces cerevisiae: W303-1A and W303-1B | Laboratory of Rodney Rothstein | ATCC:208352 ATCC:201238 |
| Yeast strains used | This manuscript. | Supplemental Table S1 |
| Oligonucleotides | ||
| Oligonucleotides used | This manuscript. | Supplemental Table S3 |
| Recombinant DNA | ||
| Plasmids used | This manuscript. | Supplemental Table S2 |
| Software and Algorithms | ||
| FIJI – Open source edition | ImageJ | http://fiji.sc; RRID:SCR_002285 |
| ZEN Microscopy software | Zeiss | Version ZEN 2012 SP5 |
| Scaffold (version 4.8.4) | Proteome Software, Inc., Portland, OR | https://www.proteomesoftware.com/ |
| RNA-seq analysis code | This manuscript. | Zenodo, DOI: 10.5281/zenodo.7063132 |
| Figures | This manuscript. | BioRender.com |
| RNA-seq analysis | Trimmomatic (version 0.38.0) | https://toolshed.g2.bx.psu.edu/repository?repository_id=ef9e620e9ac844b3 |
| RNA-seq analysis | STAR (version 2.5.2a) | https://toolshed.g2.bx.psu.edu/repository?repository_id=b8c12e1910b4735a |
| RNA-seq analysis | Htseq-count (version 0.11.3) | https://toolshed.g2.bx.psu.edu/repository?repository_id=2df7e24ce6c1f224 |
All original code has been deposited at Zenodo and is publicly available as of the date of publication. The DOI has been listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.