Abstract
The COVID-19 pandemic caused by SARS-CoV-2 has led to hundreds of millions of infections and millions of deaths, however, human monoclonal antibodies (mAbs) can be an effective treatment. Since SARS-CoV-2 emerged, a variety of strains have acquired increasing numbers of mutations to gain increased transmissibility and escape from the immune response. Most reported neutralizing human mAbs, including all approved therapeutic ones, have been knocked down or out by these mutations. Broadly neutralizing mAbs are therefore of great value, to treat current and possible future variants. Here, we review four types of neutralizing mAbs against the spike protein with broad potency against previously and currently circulating variants. These mAbs target the receptor-binding domain, the subdomain 1, the stem helix, or the fusion peptide. Understanding how these mAbs retain potency in the face of mutational change could guide future development of therapeutic antibodies and vaccines.
Current Opinion in Virology 2023, 61:101332
This review comes from a themed issue on Virus structure and expression
Edited by Kuan-Ying Arthur Huang and CheAlex Ma
https://doi.org/10.1016/j.coviro.2023.101332
1879–6257/© 2023 The Author(s). Published by Elsevier B.V. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
Introduction
Following its emergence in late 2019, COVID-19 rapidly established a pandemic, which has caused a public health crisis and economic recession. According to data from WHO, there have been more than 660 million reported cases and more than 6.7 million deaths, as of January 2023. Vaccines have been widely and effectively used to reduce disease severity and a number of drugs have been approved for clinical use, including the small-molecule drugs Paxlovid and Veklury and several monoclonal antibodies (mAbs): bebtelovimab, bamlanivimab, etesevimab, Xevudy (sotrovimab), REGEN-COV (casirivimab and imdevimab), and Evusheld (cilgavimab and tixagevimab) 1, 2, 3.
Since the first cases were reported in China, Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has mutated rapidly and multiple variants have appeared. From the Wuhan strain to currently dominant Omicron strains ( Figure 1a, phylogenetic tree), the virus has gained increased transmissibility and immune escape. Many mAbs, including approved therapeutic ones that neutralize earlier variants, have largely or totally lost their ability to neutralize new variants 3, 4. It is therefore essential to develop broadly neutralizing antibodies for the ongoing Omicron variants and, ideally, new variants that will emerge in the future.
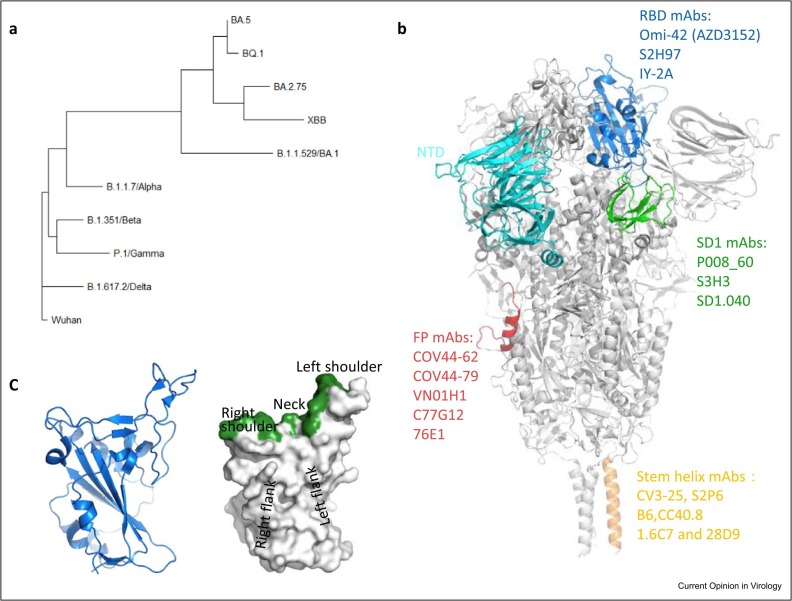
Figure 1.
(a) Phylogenetic tree of SARS-CoV-2 previous VoCs (Wuhan, Alpha, Beta, Gamma, Delta, and Omicron BA.1) and currently dominant strains (BA.5, BQ.1, BA.2.75, and XBB). The tree is based on the amino acid sequences of the spike protein. (b) Regions of SARS-CoV-2 spike protein (in gray, PDB: 6XR8) targeted by 4 types of broadly neutralizing mAbs. The RBD, NTD, SD1, fusion peptide, and stem helix are colored in blue, cyan, green, red, and orange, respectively. (c) Cartoon representation of the RBD (left) and the surface representation of the RBD showing the locations of the left shoulder, neck, right shoulder, left flank, and right flank (right), PDB: 7BEI. The binding site of ACE2 on the RBD is colored in green.
The major antigens of SARS-CoV-2 are the nucleoprotein and the trimeric spike glycoprotein, and numerous spike-binding antibodies have been characterized as potent neutralizers 5, 6. Spike consists of the S1 and S2 subunits that are linked by a furin protease cleavage site. S1 mediates binding with the receptor angiotensin-converting enzyme 2 (ACE2) [7] and S2 facilitates membrane fusion with the host cell. The S1 subunit has a string of domains with the N-terminal domain (NTD), subdomain 1 (SD1), and receptor-binding domain (RBD) all characterized as binding neutralizing antibodies (Figure 1b). The RBD harbors the ACE2-binding site, which lies across the top of the RBD, spanning the neck and shoulders (Figure 1c). The RBD adopts a range of configurations on the spike, from ‘up’ to ‘down’, and only the up conformation can interact with ACE2 [8]. Both up and down conformations are observed in published spike structures 6, 8•, 9, 10. Previously, we introduced a naming convention for the RBD to describe the epitopes of RBD-specific neutralizing mAbs, which can be grouped in several clusters: left shoulder, neck, right shoulder, left flank, and right flank [6] (Figure 1c). Most potent neutralizing mAbs induced by vaccination or natural infection target the RBD and usually interfere with ACE2 binding 5, 6, 9, 10, 11••. In line with this, many mutations in variants occur on the edge of (or in some cases more central to) the ACE-binding site, presumably allowing escape from ACE2-blocking antibodies without significantly impairing ACE2 binding ( Figure 2a–e). To date, a little over three years since the virus emerged, the observed variants already account for changes at more than 30 of the ∼200 residues in the RBD. Although neutralizing antibodies have been found that bind a single, so-called supersite within the NTD, this epitope is not well conserved between variants and so activity is generally narrow 12, 13, 14, 15. Although much less common and usually less potent than the best ACE2 blockers, neutralizing antibodies have also been found against SD1, the stem-helix region, and the fusion peptide 16•, 17, 18. Here, we mainly review broadly neutralizing mAbs directed against highly conserved epitopes in portions of the RBD and SD1 on S1 and the stem-helix region and the fusion peptide in S2 (Figure 1b).
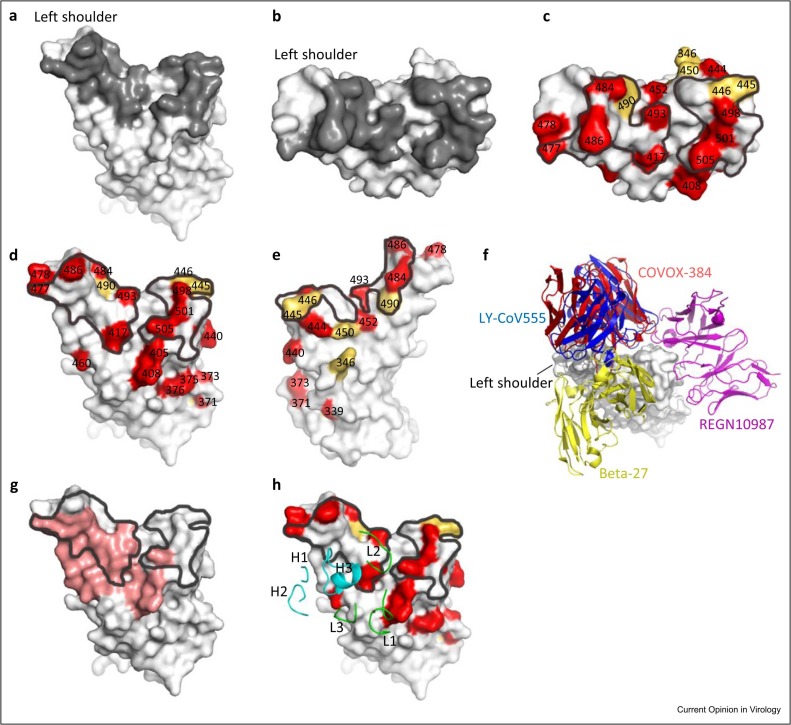
Figure 2.
(a and b) ACE2 footprint on the RBD. The RBD is colored in light gray and the ACE2 footprint in dark gray. (c–e) Mutations of SARS-CoV-2 variants in the RBD. Mutations from strain BQ.1 are colored in red and mutations from other strains that are not present in BQ.1 are colored in yellow. The ACE2 footprint is marked by black lines. (f) Different binding modes of four mAbs (LY-CoV555: blue, PDB: 7KMG; COVOX-384: red, PDB: 7BEP; REGN10987: magenta, PDB: 6XDG; Beta-27: yellow, PDB: 8BH5) with the RBD. (g) Binding region of Omi-42 (red) on the RBD, which is partially overlapped with the ACE2 footprint (marked by black lines). (h) Binding of the CDRs of Omi-42 with the RBD (CDRs of the heavy chain are colored in cyan and light chain in green), PDB: 7ZR7.
Receptor-binding domain
MAbs binding near the neck and shoulders of the RBD have either footprint or spatial overlap with ACE2, enabling them to block ACE2 binding and thus have the strongest neutralizing abilities against SARS-CoV-2. Mabs with epitopes far from this area, lower on the flanks of the RBD, for example, EY6A [19], S304 [20], and S309 [21], are usually much less potent.
The succession of mutations from earlier-circulating variants of concern (VoCs) B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), and B.1.617.2 (Delta) to B.1.1.529 (Omicron BA.1) and the currently worldwide-dominant Omicron strains (with respective prevalence: BQ.1 (33.4%), sublineages of BA.5 (7.3%), BA.2.75 (12.4%) and sublineages of XBB (31.6%), in epidemiological week 4 of 2023 (23rd–29th January), WHO data), have accumulated mutations in the RBD (Figures 1a and 2c–e). While some of these increase the binding affinity for ACE2, increasing viral transmissibility 6, 22, many more impair the potency of neutralizing antibodies, facilitating reinfection of the recovered or vaccinated population 22, 23, 24.
Many potent neutralizing mAbs, including all current commercial therapeutic mAbs, have been rendered wholly or largely ineffective by the emerging RBD mutations. MAbs COVOX-384 and LY-CoV555 (bamlanivimab), two potent antibodies against the Wuhan strain targeting the RBD at the left shoulder, were knocked out by the E484K substitution of Beta and Gamma variants 23, 25. REGN10987 (imdevimab), another commercial mAb that binds at the right shoulder of the RBD, was knocked out by the N440K and G446S substitutions of the Omicron BA.1 strain [23]. Beta-27, a potent mAb that binds at the neck and back of the left shoulder and broadly neutralizes Alpha, Beta, Gamma, and Delta variants, was impaired by Q493R and Y505H of Omicron BA.1 [23] (Figure 2f). In fact, the BA.1 strain marked a step change in divergence with no less than 15 substitutions in the RBD, causing widespread escape from most existing, early pandemic generated, neutralizing antibodies 4, 23, 24. BA.2 took over from BA.1 in March 2022 as the dominant variant and globally the virus has since fragmented into a plethora of strains derived from BA.2, many showing convergent mutations around the ACE2-binding site that confer considerable escape from responses against earlier versions of Omicron [26] .
Despite the loss of activity of most neutralizing mAbs, a few have been reported recently that retain potency against Alpha, Beta, Gamma, Delta, BA.1, BA.2, and BA.4/5 variants, and in some cases BA.1.1 or BA.2.12.1. Based on their footprints on the RBD, these mAbs can be roughly divided into two groups: those targeting the back of the left-shoulder (F61 [27] and Omi-42 [28]) and right-shoulder binders (LY-CoV1404 (bebtelovimab) [29], 2–7 [30], XGv265 [31], XG005 [32], AZD1061 [1], MB.02 [33], SP1–77 [34], and 002-S21F2) [35]. However, it is likely that many of these mAbs will lose potency against the most recently emerging Omicron sublineages (BA.2.3.20, BA.2.75, BA.4.6, BA.5, BJ.1, BQ.1, and XBB) with one or several of five mutations: R346X, K444X, V445X, N450D, and N460X. Specifically, MAbs mentioned above binding in the front of the right shoulder of the RBD may be knocked out by R346T/S, K444T/R, V445P, G446S, and N450D substitutions (Figure 2e). For example, the potency of LY-CoV1404 is severely impaired by the V445P substitution in BJ.1 and XBB [28]. AZD1061 is knocked out by R346T in BA.2.75.2 and is ineffective against BA.2.3.20, possibly due to the K444R and N450D mutations [28]. MAb F61, which targets the back of the left shoulder, may be impaired by the N460K substitution found in a series of BA.5 sublineages or G476S mutation in BS.1 (BA.2.3.2.1). MAb Omi-42 is very unusual in retaining potent neutralizing ability against all VoCs and recent emerging Omicron sublineages [11].
Omi-42 is a highly potent antibody isolated from a patient infected by BA.1. A cryo-EM structure (PDB: 7ZR7) shows Omi-42 to bind at the back of the left shoulder [11] (Figure 2g, h). Omi-42 and ACE2 have substantially overlapping binding footprints on the RBD (Figure 2g), so mutations in the overlap region that compromise ACE2 binding would not be allowed. The mutations that are on the edge of the ACE-binding site and may affect binding of Omi-42 are K417N, S477N, Q493R, and Y505H. Structural analysis [11] proves that these four amino acid residues all interact with Omi-42 but Omi-42 retains its potency both in a live-virus neutralization assay in vitro and a hamster challenge study, despite the mutations at these sites [36]. A slightly modified version of Omi-42, AZD3152, has been combined with cilgavimab as AZD5156 that is in Phase-I/-III clinical trial (https://clinicaltrials.gov/ct2/show/NCT05648110) for pre-exposure prophylaxis of immunocompromised patients.
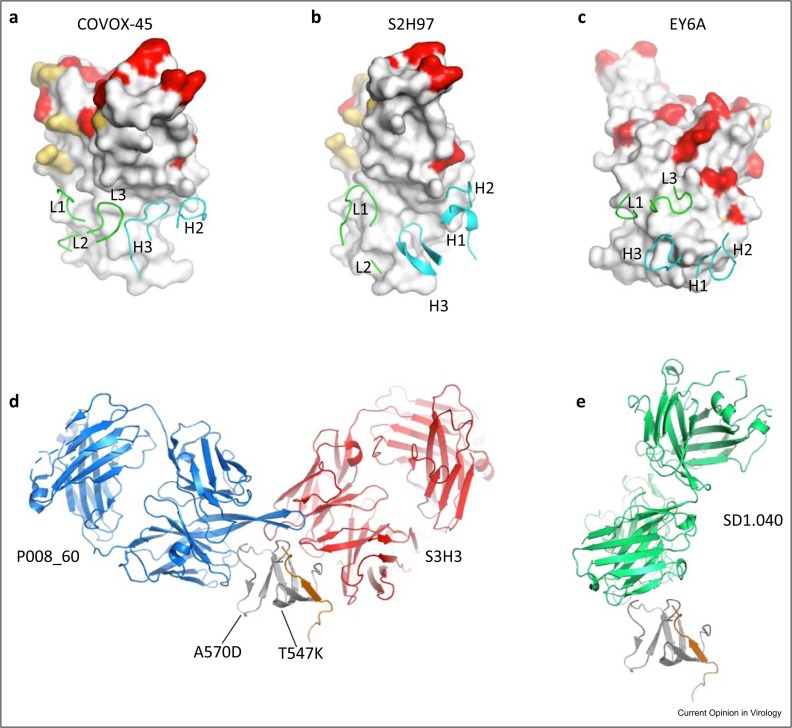
In addition to the highly potent mAbs, there are also more weakly neutralizing anti-RBD mAbs with potential broad neutralizing ability 6, 19, 37, 38. These weakly neutralizing mAbs have little or no effect on ACE2 binding but they bind RBD in two different conserved regions and utilize different neutralization mechanisms. These two large continuous conserved regions on the RBD surface are: one starts from the front of the left shoulder and runs all the way down to the left flank ( Figures 2e and 3a, b); another one is at the bottom part of the back of the RBD, just below the Omi-42-binding site (Figures 2d and 3c). Representative neutralizing mAbs binding in the vicinity of the left-flank conserved region include COVOX-45 [6] and S2H97 [37]. S2H97 binds RBD exactly at the left flank, while COVOX-45 binds somewhat more toward the front (Figure 3a, b). It was reported that S2H97 protected hamsters infected by SARS-CoV-2 Wuhan strain and could neutralize BA.1 variant. S2H97 binds a panel of 45 RBDs of SARS-related coronaviruses (sarbecoviruses) with high affinity and is a promising pan-sarbecovirus-neutralizing antibody 37, 39. COVOX-45 recognizes a similar epitope to S2H97 but the breadth of its potency has not been checked. S2H97 induces receptor-independent conversion of spike protein to the post-fusion state, thereby inhibiting ACE2-dependent cell entry [37]. COVOX-45 may use the same neutralization mechanism since it recognizes a similar epitope to S2H97. S309 (sotrovimab) is another therapeutic antibody, which binds the right flank. S309 was an effective therapeutic broadly neutralizing antibody against earlier variants, but its potency was impaired by BA.2 sublineage, possibly due to the S371F mutation [40], so it is no longer in widespread therapeutic use.
Figure 3.
(a–c) Binding of the CDRs of COVOX-45, S2H97, and EY6A with the more conserved regions of the RBD (COVOX-45, PDB: 7PRY; S2H97, PDB: 7M7W; EY6A, PDB: 6ZCZ). (d and e) Different binding patterns of P008_60 (blue, PDB: 7ZBU), S3H3 (red, PDB: 7WD8), and SD1.040 (green, PDB: 8D48) with the SD1 (gray). The mutations T547K and A570D of the SD1 are labeled. The peptide 320–330 of the RBD that interacts with S3H3 is shown as an orange cartoon.
Examples of broadly neutralizing antibodies that target the conserved region at the back of the RBD are S304, IY-2A [38], and EY6A (Figure 3c). They all recognize a cryptic epitope buried inside the RBD-down trimeric spike and neutralize SARS-COV-2 Wuhan strain by disrupting the spike trimer 19, 20, 38. This epitope is conserved across all established and emerging variants. S304, IY-2A, and EY6A all retain their strong binding affinity for Omicron BA.1 RBD, however, S304 completely lost its neutralizing ability against Omicron sublineages BA.1, BA.1.1, BA.2, and BA.3, the potency of EY6A against Omicron BA.2 and BA.5 was impaired while IY-2A is still strongly active 38, 41. Structural studies show that Wuhan spike usually adopts the two- and three-RBD-up conformations after binding ACE2, while Omicron spike packs more tightly and maintains the preferential one-RBD-up conformation, both before and after ACE2 binding [42]. S304 cannot bind to the up-RBD in the one-RBD-up conformation of the Omicron spike because it will clash with adjacent RBD that has a ‘down’ conformation 41, 42. This may be why S304 can neutralize previous VoCs but not the Omicron sublineage, indicating that an antibody can lose its potency when its conserved epitope becomes inaccessible.
In contrast to Omi-42, broadly neutralizing mAbs binding at the conserved regions in the left flank or back of the RBD usually neutralize more weakly, possibly because they have no or little blocking effect on ACE2 binding.
N-terminal domain
Although there are also neutralizing antibodies that recognize the NTD, this spike domain is less immunogenic than the RBD, possibly due to its extensive glycan shielding 20, 43, 44. Neutralizing mAbs targeting the NTD appear to mainly recognize an extended single epitope called the NTD supersite, which comprises the N1 (residues 14–26), N3 (residues 141–156), and N5 (residues 246–260) loops 5, 44, 45. This restricted mode of binding, together with the high level of variation seen in the NTD, caused not only by point substitutions (as in the RBD), but also by multiple deletions/insertions, facilitates antibody escape. The currently dominant BA.5 strain has nine substitutions/deletions in the NTD, with nearly half of these in the supersite. Thus, NTD-specific mAbs are generally not broadly neutralizing 24, 44, 45.
Subdomain 1
The SD1 domain is highly conserved across all VoCs and currently circulating strains. The only reported mutations in SD1 of these strains are T547K and A570D, but these two amino acids are buried inside the spike trimer, so they generally cannot be recognized by neutralizing mAbs [8]. Representative neutralizing mAbs targeting SD1 include S3H3 [16], P008_60 [46], and sd1.040 [47]. Pseudovirus or live-virus neutralization assays showed that these antibodies neutralize a panel of variants but recognize different epitopes 16•, 46, 47. While all interact with SD1, S3H3 also interacts with the N-terminal region and sd1.040 interacts with the C-terminal region of the RBD (Figure 3d, e). S3H3 binds trimeric spike but P008_60 and sd1.040 dissociate trimeric spike and only complexes of S1 with Fab could be observed, due to clashes of Fabs with the adjacent NTD in the trimeric spike. Given the ability of P008_60 and sd1.040 to neutralize the virus, they may recognize an uncharacterized rare conformational state of the spike in which its epitope on SD1 is accessible. P008–60 and sd1.040 therefore neutralize the virus by destabilizing the spike trimer 46, 47. S3H3 may neutralize SARS-CoV-2 by blocking the release of S1 from the spike to inhibit the virus entry [48]. However, to date, these SD1 binders neutralize relatively weakly, compared with potent ACE2-blocking RBD-binding mAbs 16•, 46.
Stem-helix region
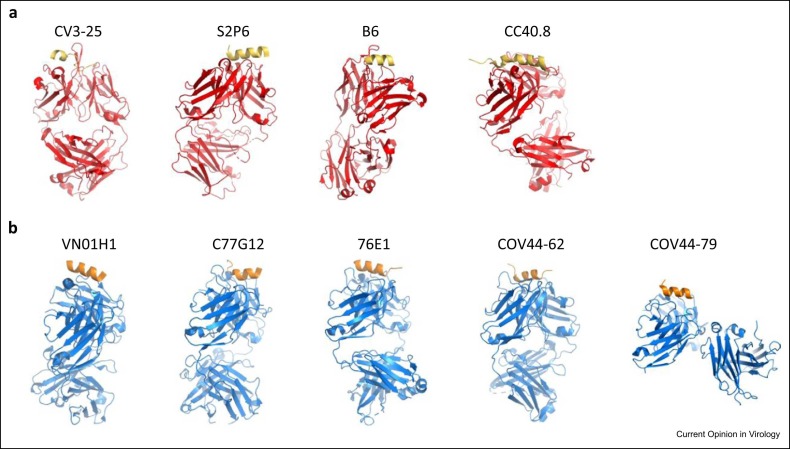
The stem-helix region (residues 1144–1158 of Wuhan strain) is part of the viral fusion machinery and is highly conserved among betacoronaviruses [49]. Several neutralizing mAbs target this region, including CV3–25 [17], S2P6 [50], B6 [51], CC40.8 [52], 1.6C7, and 28D9 [53]. They all have the ability to inhibit membrane fusion but recognize different epitopes in the stem-helix region. CV3–25 interacts with the C terminus of the stem helix (residues 1153–1158) as well as the hinge (residues 1159–1166), while the other five mAbs mainly recognize the N terminus of the stem helix (residues 1144–1154) ( Figure 4a). High-resolution crystal structures showed that S2P6, B6, and CC40.8 bind to the hydrophobic face of the stem helix, which is mostly buried in the native trimeric prefusion spike, indicating that binding of these mAbs induces significant conformational changes in the prefusion spike helix bundle. These conformational changes were demonstrated by structures of prefusion spike–Fab complexes 50•, 51, 52. In contrast, CV3–25 binds to the hydrophilic face of the helix, however, the CV3–25-bound stem helix (seen in a crystal structure) adopts a different conformation to that in the native prefusion spike visualized by cryo-EM structure [17]. All these antibodies were broadly neutralized in pseudovirus neutralization assay or animal models, but their potency was weak 17, 50•, 51, 52, 53.
Figure 4.
(a) Binding patterns of Fabs (red) with stem-helix peptide (yellow). (b) Different Fabs (blue) binding with fusion peptide (orange).
Fusion peptide
The fusion peptide is a hydrophobic segment of ∼20 residues, in the S2 domain of the spike protein, which is immediately downstream of the S2′ cleavage site and responsible for initiation of the fusion of the virus and host cell membranes (residues 816–837, Figure 1b). This peptide is highly conserved across all SARS-CoV-2 variants. Sequence alignment of more than 7.9 million deposited SARS-CoV-2 sequences indicates residues in this region have identities> 99.7% [54]. Indeed, this peptide is conserved among all coronaviruses [55]. Several neutralizing mAbs target this region, including COV44–62, COV44–79 [18], VN01H1, C77G12 [54], and 76E1 [56] (Figure 4b). These mAbs broadly neutralize all alpha- and betacoronaviruses, including the currently dominant SARS-CoV-2 variants. Experiments on Syrian hamster or mouse models showed they all limited disease caused and reduced viral load 18, 54•, 56. Structural studies showed the epitope recognized by all these antibodies is inaccessible in the prefusion spike trimer and it appears the epitope becomes exposed upon conformational changes induced by binding of ACE2. Once again, compared with anti-RBD antibodies, these antibodies have weak potency against SARS-CoV-2 18, 54•, 56.
Conclusion
The rapid evolution of antibody-binding epitopes enables viruses to escape host immune response. The BA.1 strain identified in November 2021 showed widespread escape from antibody responses to vaccination or infection by early pandemic virus [23] and in turn antibody responses from BA.1 infection led to responses with significantly poorer neutralization of BA.4/5, compared with BA.1 and BA.2 [4]. Indeed, the majority of potent anti-RBD mAbs were knocked out by BA.4/5. Since BA.4/5 SARS-COV-2 has continued to evolve to gain increased immune escape, leading to the proliferation of a large number of variants, often arriving, via convergent evolution of common amino acid substitutions at positions around the ACE2 footprint that abrogate the binding of groups of antibodies. Thus, BQ.1 and XBB, for instance, show substantial escape [57]. Very few anti-RBD antibodies survive this onslaught of variation, although Omi-42, that acts by blocking ACE2 binding, remains highly active against all variants to date. It seems likely that antibodies such as Omi-42 have survived with neutralization intact because they contribute little to the overall neutralization titer of polyclonal responses and have been fortunate not to have been affected by bystander effects from mutations selected because of their value in abrogating more dominant neutralizers, but it also seems likely that such antibodies will eventually also lose potency as SARS-CoV-2 continues to evolve.
Away from the ACE2-binding site, neutralizing antibodies have been identified that bind at either the left flank or back of the RBD, two large areas that are highly conserved across all SARS-CoV-2 variants. In addition, antibodies have been found against the SD1 domain in S1 and the stem helix and the fusion peptide in S2, all of which are functionally essential and highly conserved. MAbs targeting these regions are potentially very broadly neutralizing, although they are usually not very potent, and so may not be very effective when used alone. To reduce the likelihood of antibody escape of SARS-CoV-2, in principle, broadly neutralizing mAbs targeting different epitopes can be used as cocktails and these weak mAbs can be combined with potent anti-RBD mAbs. Bispecific antibodies can also be developed, with two different broadly neutralizing Fabs in the two arms of one IgG molecule. Some well-designed bispecific antibodies showed higher binding and neutralization activities against SARS-CoV-2, with their two arms interacting simultaneously with a single-spike protein 47, 58, 59.
Declaration of Competing Interest
Oxford University holds intellectual property related to SARS-CoV-2 mAbs discovered in Gavin R Screaton’s laboratory and DIS consults for AstraZeneca
Acknowledgements
D.Z. is supported by the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Science (CIFMS), China (grant number: 2018-I2M-2-002). J.R. is funded by the UK Wellcome Trust (101122/Z/13/Z), and D.I.S. and E.E.F. by the UK Medical Research Council (MR/N00065X/1).
Data Availability
Data will be made available on request.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
-
•
of special interest
-
••
of outstanding interest
- 1.Dong J., et al. Genetic and structural basis for SARS-CoV-2 variant neutralization by a two-antibody cocktail. Nat Microbiol. 2021;6:1233–1244. doi: 10.1038/s41564-021-00972-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim C., et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat Commun. 2021;12:1–10. doi: 10.1038/s41467-020-20602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumari M., et al. A critical overview of current progress for COVID-19: development of vaccines, antiviral drugs, and therapeutic antibodies. J Biomed Sci. 2022;29:1–36. doi: 10.1186/s12929-022-00852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuekprakhon A., et al. Antibody escape of SARS-CoV-2 Omicron BA. 4 and BA.5 from vaccine and BA. 1 serum. Cell. 2022;185 doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 6.Dejnirattisai W., et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184 doi: 10.1016/j.cell.2021.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. cell. 2020;181 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8•.Wrapp D., et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]; This was the first paper reported the structure of the SARS-CoV-2 spike protein.
- 9.Barnes C.O., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu C., et al. The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants. Cell Host Microbe. 2022;30 doi: 10.1016/j.chom.2021.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Nutalai R., et al. Potent cross-reactive antibodies following Omicron breakthrough in vaccinees". Cell. 2022;185:2116–2131. doi: 10.1016/j.cell.2022.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; A potent broadly neutralizing mAb (Omi-42) against all previously and currently circulating SARS-CoV-2 strains was reported in this paper.
- 12.Hastie K.M., et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: a global consortium study. Science. 2021;374:472–478. doi: 10.1126/science.abh2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang P., et al. Antibody resistance of SARS-CoV-2 variants B. 1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 14.Graham C., et al. Neutralization potency of monoclonal antibodies recognizing dominant and subdominant epitopes on SARS-CoV-2 Spike is impacted by the B.1.1.7 variant. Immunity. 2021;54 doi: 10.1016/j.immuni.2021.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andreano E., et al. Anatomy of Omicron BA. 1 and BA. 2 neutralizing antibodies in COVID-19 mRNA vaccinees. Nat Commun. 2022;13:1–8. doi: 10.1038/s41467-022-31115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16•.Xu S., et al. Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein. Emerg Microbes Infect. 2022;11:351–367. doi: 10.1080/22221751.2021.2024455. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reported a relatively strong broadly neutralizing mAb targeting the SD1 region of SARS-CoV-2 spike.
- 17.Hurlburt N.K., et al. Structural definition of a pan-sarbecovirus neutralizing epitope on the spike S2 subunit. Commun Biol. 2022;5:1–13. doi: 10.1038/s42003-022-03262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dacon C., et al. Broadly neutralizing antibodies target the coronavirus fusion peptide. Science. 2022;377:728–735. doi: 10.1126/science.abq3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou D., et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat Struct Mol Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- 20.Piccoli L., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183 doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinto D., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 22.Zhou D., et al. Evidence of escape of SARS-CoV-2 variant B. 1.351 from natural and vaccine-induced sera. Cell. 2021;184 doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dejnirattisai W., et al. SARS-CoV-2 Omicron-B. 1.1. 529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185 doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao Y., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Starr T.N., Greaney A.J., Dingens A.S., Bloom J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep Med. 2021;2 doi: 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ai J., et al. Antibody evasion of SARS-CoV-2 Omicron BA. 1, BA. 1.1, BA. 2, and BA. 3 sub-lineages. Cell Host Microbe. 2022;30 doi: 10.1016/j.chom.2022.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X., et al. Structural basis of a two-antibody cocktail exhibiting highly potent and broadly neutralizing activities against SARS-CoV-2 variants including diverse Omicron sublineages. Cell Discov. 2022;8.1 doi: 10.1038/s41421-022-00449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dijokaite-Guraliuc A., et al. Rapid escape of new SARS-CoV-2 Omicron variants from BA. 2-directed antibody responses. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.112271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westendorf K., et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerutti G., et al. Structural basis for accommodation of emerging B. 1.351 and B.1.1. 7 variants by two potent SARS-CoV-2 neutralizing antibodies. Structure. 2021;29 doi: 10.1016/j.str.2021.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang K., et al. Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants. Nature. 2022;603:919–925. doi: 10.1038/s41586-022-04466-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z., et al. An ultrapotent pan-β-coronavirus lineage B (β-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope. Protein Cell. 2022;13:655–675. doi: 10.1007/s13238-021-00871-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ren, P. et al.: Function and Cryo-EM structures of broadly potent bispecific antibodies against multiple SARS-CoV-2 Omicron sublineages. bioRxiv. 2022. [DOI] [PMC free article] [PubMed]
- 34.Luo S., et al. An antibody from single human VH-rearranging mouse neutralizes all SARS-CoV-2 variants through BA. 5 by inhibiting membrane fusion. Sci Immunol. 2022;7 doi: 10.1126/sciimmunol.add5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar S., et al. Structural insights for neutralization of Omicron variants BA. 1, BA. 2, BA. 4, and BA. 5 by a broadly neutralizing SARS-CoV-2 antibody. Sci Adv. 2022;8 doi: 10.1126/sciadv.add2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Francica J. et al.: The SARS-CoV-2 monoclonal antibody AZD3152 potently neutralises historical and currently circulating variants. Poster presented at: 33rd European Congress of Clinical Microbiology & Infectious Diseases (ECCMID); Copenhagen, Denmark; 17 April 2023.
- 37.Starr T.N., et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102. doi: 10.1038/s41586-021-03807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang K.-Y.A., et al. Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2. Nat Commun. 2023;14 doi: 10.1038/s41467-023-35949-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L., et al. An antibody class with a common CDRH3 motif broadly neutralizes sarbecoviruses. Sci Transl Med. 2022;14 doi: 10.1126/scitranslmed.abn6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iketani S., et al. Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature. 2022;604:553–556. doi: 10.1038/s41586-022-04594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang M., et al. Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA. 3. Immunity. 2022;55 doi: 10.1016/j.immuni.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Z., et al. Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape. Nat Commun. 2022;13:1–12. doi: 10.1038/s41467-022-32665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walls A.C., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCallum M., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184 doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suryadevara N., et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184 doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seow J., et al. A neutralizing epitope on the SD1 domain of SARS-CoV-2 spike targeted following infection and vaccination. Cell Rep. 2022;40 doi: 10.1016/j.celrep.2022.111276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bianchini F., et al. Human neutralizing antibodies to cold linear epitopes and subdomain 1 of the SARS-CoV-2 spike glycoprotein. Sci Immunol. 2023;8 doi: 10.1126/sciimmunol.ade0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong Q., et al. Molecular basis of receptor binding and antibody neutralization of Omicron. Nature. 2022;604:546–552. doi: 10.1038/s41586-022-04581-9. [DOI] [PubMed] [Google Scholar]
- 49.Li W., et al. Structural basis and mode of action for two broadly neutralizing antibodies against SARS-CoV-2 emerging variants of concern. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2021.110210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50•.Pinto D., et al. Broad betacoronavirus neutralization by a stem helix–specific human antibody. Science. 2021;373:1109–1116. doi: 10.1126/science.abj3321. [DOI] [PMC free article] [PubMed] [Google Scholar]; A neutralizing mAb recognizing the stem helix was reported and its neutralizing mechanism was proposed in this paper.
- 51.Sauer M.M., et al. Structural basis for broad coronavirus neutralization. Nat Struct Mol Biol. 2021;28:478–486. doi: 10.1038/s41594-021-00596-4. [DOI] [PubMed] [Google Scholar]
- 52.Zhou P., et al. A human antibody reveals a conserved site on beta-coronavirus spike proteins and confers protection against SARS-CoV-2 infection. Sci Transl Med. 2022;14 doi: 10.1126/scitranslmed.abi9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C., et al. A conserved immunogenic and vulnerable site on the coronavirus spike protein delineated by cross-reactive monoclonal antibodies. Nat Commun. 2021;12:1–15. doi: 10.1038/s41467-021-21968-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54•.Low J.S., et al. ACE2-binding exposes the SARS-CoV-2 fusion peptide to broadly neutralizing coronavirus antibodies. Science. 2022;377:735–742. doi: 10.1126/science.abq2679. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reported neutralizing mAbs targeting the fusion peptide and proved epitope on the fusion peptide becomes exposed upon conformational changes induced by binding of ACE2.
- 55.Madu I.G., Roth S.L., Belouzard S., Whittaker G.R. Characterization of a highly conserved domain within the severe acute respiratory syndrome coronavirus spike protein S2 domain with characteristics of a viral fusion peptide. J Virol. 2009;83:7411–7421. doi: 10.1128/JVI.00079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun X., et al. Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2. Nat Microbiol. 2022;7:1063–1074. doi: 10.1038/s41564-022-01155-3. [DOI] [PubMed] [Google Scholar]
- 57.Wang Q., et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell. 2022;186:279–286.e8. doi: 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y., et al. Inactivated vaccine-elicited potent antibodies can broadly neutralize SARS-CoV-2 circulating variants. Nat Commun. 2023;14 doi: 10.1038/s41467-023-37926-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y., et al. Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA. 1-BA. 5. Cell Discov. 2023;9 doi: 10.1038/s41421-022-00509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.