Abstract
The ribosome is one of the largest complexes in the cell. Adding to its complexity are more than 200 RNA modification sites present on ribosomal RNAs (rRNAs) in a single human ribosome. These modifications occur in functionally important regions of the rRNA molecule, and they are vital for ribosome function and proper gene expression. Until recent technological advancements, the study of rRNA modifications and their profiles has been extremely laborious, leaving many questions unanswered. Small nucleolar RNAs (snoRNAs) are non-coding RNAs that facilitate and dictate the specificity of rRNA modification deposition, making them an attractive target for ribosome modulation. Here, we propose that through the mapping of rRNA modification profiles, we can identify cell-specific modifications with high therapeutic potential. We also describe the challenges of achieving the targeting specificity needed to implement snoRNAs as therapeutic targets in cancers.
Keywords: snoRNAs, 2′-O-methylation, ribosome, therapy
1. Introduction
DNA and histone modifications (epigenomics) have an established role in regulating gene expression and disease onset [1,2,3]. However, the role of RNA modifications (the epitranscriptome) in these processes has only been recently delineated [4,5,6].
With 80 proteins and 4 RNA molecules, the ribosome is one of the largest complexes in the cell [7]. Adding to its complexity are more than 200 RNA modification sites present on ribosomal RNAs in a single human ribosome. rRNA modifications occur in evolutionary conserved and functionally important regions of the rRNA molecule. Moreover, rRNA modifications are essential for the processing and secondary structure of rRNA, and hence they are vital for ribosome function and proper gene expression [8,9,10]. The two most common post-transcriptional modifications include ribose 2′-OH hydroxyl methylation (2´OMe) and the conversion of uridine to pseudouridine (pseudouridylation, or Ψ). Studies have shown that changes to 2′OMe and Ψ are linked to the translational capacity of ribosomes [11,12], cellular properties, such as antibiotic sensitivity and immune function, as well as the maintenance of cancer cells [10,13,14,15,16,17,18,19]. We have previously demonstrated that aberrant 2′OMe is the etiological basis for bone marrow failure in dyskeratosis congenita patients [20]. Hence, aberrant rRNA modifications play a crucial role in human physiology and disease.
Together, 2′OMe and Ψ comprise more than 95% of modified rRNA nucleotides, with more than 110 2′OMe and ~100 Ψ modified residues in a single ribosome [19,21,22,23,24]. 2′OMe and Ψ are mostly introduced by two classes of small nucleolar RNAs, box C/D and box H/ACA, respectively. These two classes of snoRNAs are defined by their structure and sequence motifs, which are required for their association within snoRNPs and guidance of the complex to the nucleotide intended for modification. Box C/D snoRNA are associated with snoRNPs containing methyltransferase fibrillarin (FBL), and box H/ACA snoRNA with pseudouridine synthase dyskerin (DKC1). As 2′OMe and Ψ installation is based on extensive base-pairing between a snoRNA and an rRNA, many snoRNAs facilitate the modification of a single site [21,22]. This exclusive interaction between a snorRNA and a modification site enables precise targeting of specific rRNA modification sites through perturbations of the related snoRNA.
Even though many RNA modifications have been known since the 1950s, due to technological limitations, their investigation has thus far been extremely laborious and mostly confined to a single modified site at a time. This hindered our understanding of their function. Moreover, the lack of genetic tools for research in vertebrates and/or mammalian models has restricted our capacity to explore their physiological function and relevance to human disease. In the past decade, we have witnessed a boom in technological advancements, making it possible to detect RNA modifications transcriptome-wide and with single nucleotide resolution. This technological leap enables us to revisit the contribution of rRNA 2′OMe and Ψ in human disease to reassess the therapeutic potential of snoRNAs.
Indeed, in recent years, aberrant snoRNA expression has been described in various malignancies. The description of this body of work has been extensively reviewed by others [25,26,27]. However, it is important to note that increased ribogenesis and high rRNA modification levels are a feature of malignancies. A possible explanation for this is the malignant cell’s need for the fast accumulation of biomass in order to facilitate division and a high proliferative rate. This dependence on high modification levels might serve as a liability of the cancer cell as we can use it to specifically target those cells. Due to their pivotal role in the installment of rRNA modifications, ribogenesis, and gene expression, snoRNAs have great therapeutic potential.
2. The Challenges
2.1. Specificity
SnoRNAs are ubiquitous and highly abundant RNAs [21,28]. So, how can one target the malignant cell with little to no harm to healthy cells?
Recent publications have demonstrated that some rRNA modifications occur in sub-stoichiometric levels [10,29]. This finding has overturned the notion that all rRNA modifications are constitutively present in ribosomes [23]. This also highlights nucleotide modifications as an important source of ribosomal heterogeneity, or cell-type-specific ribosomes. Together with the identification of high snoRNA levels in malignancies, this suggests that it is possible to identify cancer-specific rRNA modification patterns (Figure 1).
Figure 1.
2′OMe rRNA profiles provide specificity to target malignant cells. Cell-type-specific rRNA 2′OMe profiles in healthy (yellow), malignant (pink), and leukemia stem cells (LSC; brown). Methylation pattern shifts in cancer, with specific sites highly methylated in leukemia blasts generally and in leukemia stem cells specifically, exemplified here by Um116 and Gm1447, respectively (depicted in cyan). This figure was created by BioRender.com.
These cancer-specific modification profiles, once identified, will lead to the subsequent snoRNAs, which are crucial for the malignant cell. Those snoRNAs would have the highest potential as candidates for therapeutic targeting due to their ability to provide cancer specificity, and so their targeting will mostly affect cancer cells and not healthy, by standard, cells.
Recent works have already uncovered a few such snoRNA-dependent modification sites crucial for the maintenance of cancers. Zhou et al. revealed that rRNA 2′OMe modulates protein translation, affecting leukemia stem cell (LSC) phenotypes and propagation [30]. Their work shows a distinct methylation pattern of patient-derived acute myeloid leukemia (AML) cells compared to healthy hematopoietic cells. Furthermore, they identified a cluster of 2′OMe sites in which methylation correlated with the expression of LSC genes. LSCs are a small subset of self-renewing AML stem cells crucial for cancer’s propagation. Zhou et al. demonstrate that methylation level at these sites directs the leukemogenicity of AML cells, likely through the translation regulation of amino acid transporters and heightened amino acid metabolism needed for cell stemness. The authors then focus on a single modification site of the aforementioned cluster, 18S-Gm1447. By genetic manipulations of the snoRNA, which guides the site’s methylation, SNORD127, the authors demonstrate its role in the maintenance of LSC properties and leukemogenesis.
Additionally, Pauli and Liu et al. uncovered another single methylation site important for the proliferation of AML cell lines [15]. They performed a snoRNA screen to identify snoRNAs essential for leukemia cells’ proliferation. One of the top hits was SNORD42A, suggesting that this snoRNA is essential for the growth of leukemic cells. Importantly, SNORD42A was also found to be highly expressed in primary AML blasts. The knockout of SNORD42A reduces the methylation at 18S-Um116 while hindering the proliferation and colony formation ability of cells. It seems that a reduction in U116 methylation in SNORD42A knockout cells decreases the overall translation in the cells, as well as a decreased translation of specific proteins, most prominently ribosomal proteins.
Herein, we focus on leukemia; however, snoRNAs have been found to be implicated in additional cancers [31,32]. For example, it was found that in non-small cell lung carcinoma, the knockout of SNORA7A, SNORA7B, and SNORA65 and subsequent reduction in pseudouridylation resulted in the inhibition of the cancer cell’s proliferation, invasion, and migration [33]. Additionally, snoRNA U50, which mediates the methylation of C2848 in 28 S rRNA, appears to be downregulated in several cancers including breast, prostate, and colorectal. U50 inhibits cell colony formation and is suggested to be a tumor-suppressor-like gene [34,35,36].
The above-mentioned examples demonstrate the central role of even a single modification site on rRNAs in regulating malignancies. The need of the malignant cell to tailor its ribosome and achieve a high proliferative rate becomes a liability as it generates specificity. Thus, the identification of tumor-specific modification profiles may expose new therapeutic targets and pose as a promising avenue for cancer therapy.
2.2. Targeting SnoRNAs
Several techniques have been developed to target cellular RNAs. Here, we discuss two.
2.2.1. Antisense Oligonucleotides (ASOs)
ASOs are single-stranded deoxy-oligonucleotides, usually ~20 nt in length, which are designed to bind specifically to the target RNA and modulate its abundance. ASOs act through several mechanisms of action. Regarding the regulation of snoRNA abundance, we bring forth two such mechanisms.
RNase H1-Mediated Degradation
Ribonuclease H1, or RNase H1, resides in nuclei [37,38] and plays a role in maintaining chromosomal stability by removing RNA-DNA hybrids [39]. ASOs target RNA, forming ASO (DNA)-RNA heteroduplexes, which act as substrates for RNase enzymes [40]. A specific design of ASOs, in which the central region is made of unmodified nucleotides, named Gapmers, is used to this end. In Gapmers, the incorporation of flanking modified nucleotides increases the ASO/Gapmer binding affinity, and the unmodified nucleotides promote RNase H1 activity. Many FDA-approved ASO drugs act through the activation of RNases [41,42,43]. Using this mode of action ASO treatment will lead to reduced snoRNA abundance and low rRNA modification of the specific site.
Modulation of Splice Sites
Most snoRNAs are encoded from the introns of host genes [21]. The host genes are transcribed by RNA polymerase II, and after splicing occurs, the excised intron undergoes additional processing steps to produce the mature snoRNA [44]. Thus, the modulation of splicing constitutes a natural tactic to regulate snoRNA abundance. ASOs can also exert their function by redirecting splicing, leading to exon-inclusion or exclusion [45,46,47,48]. If ASOs are directed against a snoRNA host gene, they will affect snoRNA abundance. An example of the benefits of this mode of action by ASOs would be the targeting of a mutated beta-globin pre-mRNA to produce splice variant mRNA that restored hemoglobin production [45,48].
2.2.2. Small Molecules Interacting with RNA
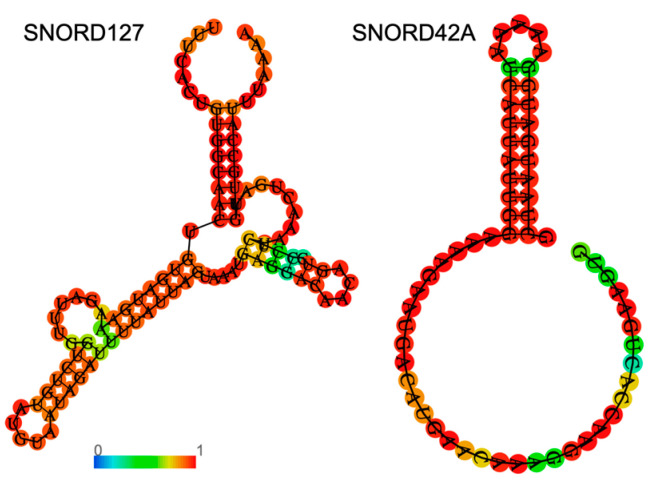
An interesting and promising alternative to sequence-specific targeting is structure-specific small molecules that bind to structures in the RNA molecule. Small molecules can be designed to lead to the degradation of their target, thus modulating their targets’ function. Small molecules are attractive therapeutic agents as they have low molecular weight and are less charged. These traits facilitate improved delivery efficiency compared with ASOs. Importantly, small molecules have been shown to be bioactive and tolerated in multiple models [49,50]. Importantly, while snoRNAs from the same family, either C/D or H/ACA box, share consensus sequences, snoRNAs can differ significantly in their secondary structure (Figure 2). These differences enable specific targeting of the desired snoRNA by custom-designed small molecules.
Figure 2.
Examples of specific secondary structures of C/D box snoRNAs. Shown in the figure are predictions for the secondary structures of human SNORD127 (left) and SNORD42A (right), which were previously discussed. An RNA fold web server was used for structure prediction. The color of the nucleotides represents base-pair probability according to the color bar.
To identify potential small molecules in a high-throughput approach, RNA motif libraries are used to screen for small molecules that bind to a certain motif with high specificity. As these libraries are modular, they enable diverse experimental designs where one can either screen for the binding of many motifs or, alternatively, focus on specific motif families. Various mechanisms of action have been demonstrated by small molecules; for example, direct binding of the target RNA and its neutralization. Other small molecules have been designed to recruit endogenous nucleases, leading to the degradation of the target RNA [51,52]. The selectivity of small molecules can be even further increased by designing them to bind multiple sites on the RNA molecule [52,53]. To this end, dimeric compounds have been developed to simultaneously target two adjacent motifs within the RNA target with a defined distance between the motifs, increasing specificity. With respect to snoRNAs, this approach especially holds high potential, as the function of snoRNAs is dependent on their strong secondary structure. Thus, taking advantage of their molecular properties by targeting snoRNAs with small molecules serves as an interesting new therapeutic avenue.
3. Perspective
Thanks to technological developments and the rise of the non-coding RNA field, snoRNAs are now appreciated as facilitators of many molecular processes. These include ribogenesis, translation regulation by rRNA modifications or miRNAs, splicing, and more [31,54]. Because of their multi-functional nature, it is not surprising to find aberrant snoRNA expression in many malignancies, making them attractive therapeutic targets.
The growing appreciation of cell-type-specific rRNA modification profiles [10,15,29] enables the identification of snoRNAs that will facilitate high specificity for targeted therapy of the cancer cell’s ribosome. With the improved mapping of cancer-specific rRNA modification patterns, more cancer-specific snoRNAs can be identified, and therapeutic agents may be designed to promote the targeting of cancer cells.
Author Contributions
Conceptualization, O.R. and D.N., writing O.R. and D.N. Funding acquisition, D.N. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
D.N. is supported by the Israel Science Foundation personal research grant 1193/22, The Binational Science Foundation grant 2021127, the Abisch-Frenkel Faculty Development Lectureship, and the Alon Fellowship from the Israeli Council for Higher Education.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Arechederra M., Recalde M., Garate-Rascon M., Fernandez-Barrena M.G., Avila M.A., Berasain C. Epigenetic Biomarkers for the Diagnosis and Treatment of Liver Disease. Cancers. 2021;13:1265. doi: 10.3390/cancers13061265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavalli G., Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571:489–499. doi: 10.1038/s41586-019-1411-0. [DOI] [PubMed] [Google Scholar]
- 3.Hanna C.W., Demond H., Kelsey G. Epigenetic regulation in development: Is the mouse a good model for the human? Hum. Reprod. Update. 2018;24:556–576. doi: 10.1093/humupd/dmy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui L., Ma R., Cai J., Guo C., Chen Z., Yao L., Wang Y., Fan R., Wang X., Shi Y. RNA modifications: Importance in immune cell biology and related diseases. Signal Transduct. Target. Ther. 2022;7:334. doi: 10.1038/s41392-022-01175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiapaer Z., Su D., Hua L., Lehmann H.I., Gokulnath P., Vulugundam G., Song S., Zhang L., Gong Y., Li G. Regulation and roles of RNA modifications in aging-related diseases. Aging Cell. 2022;21:e13657. doi: 10.1111/acel.13657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jonkhout N., Tran J., Smith M.A., Schonrock N., Mattick J.S., Novoa E.M. The RNA modification landscape in human disease. RNA. 2017;23:1754–1769. doi: 10.1261/rna.063503.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fox G.E. Origin and evolution of the ribosome. Cold Spring Harb. Perspect. Biol. 2010;2:a003483. doi: 10.1101/cshperspect.a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chawla M., Oliva R., Bujnicki J.M., Cavallo L. An atlas of RNA base pairs involving modified nucleobases with optimal geometries and accurate energies. Nucleic Acids Res. 2015;43:9573. doi: 10.1093/nar/gkv925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Polikanov Y.S., Melnikov S.V., Soll D., Steitz T.A. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat. Struct. Mol. Biol. 2015;22:342–344. doi: 10.1038/nsmb.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma S., Marchand V., Motorin Y., Lafontaine D.L.J. Identification of sites of 2′-O-methylation vulnerability in human ribosomal RNAs by systematic mapping. Sci. Rep. 2017;7:11490. doi: 10.1038/s41598-017-09734-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khoshnevis S., Dreggors-Walker R.E., Marchand V., Motorin Y., Ghalei H. Ribosomal RNA 2′-O-methylations regulate translation by impacting ribosome dynamics. Proc. Natl. Acad. Sci. USA. 2022;119:e2117334119. doi: 10.1073/pnas.2117334119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoon A., Peng G., Brandenburger Y., Zollo O., Xu W., Rego E., Ruggero D. Impaired control of IRES-mediated translation in X-linked dyskeratosis congenita. Science. 2006;312:902–906. doi: 10.1126/science.1123835. [DOI] [PubMed] [Google Scholar]
- 13.Ayadi L., Galvanin A., Pichot F., Marchand V., Motorin Y. RNA ribose methylation (2′-O-methylation): Occurrence, biosynthesis and biological functions. Biochim. Biophys. Acta Gene Regul. Mech. 2019;1862:253–269. doi: 10.1016/j.bbagrm.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Cundliffe E., Thompson J. Ribose methylation and resistance to thiostrepton. Nature. 1979;278:859–861. doi: 10.1038/278859a0. [DOI] [PubMed] [Google Scholar]
- 15.Pauli C., Liu Y., Rohde C., Cui C., Fijalkowska D., Gerloff D., Walter C., Krijgsveld J., Dugas M., Edemir B., et al. Site-specific methylation of 18S ribosomal RNA by SNORD42A is required for acute myeloid leukemia cell proliferation. Blood. 2020;135:2059–2070. doi: 10.1182/blood.2019004121. [DOI] [PubMed] [Google Scholar]
- 16.Ringeard M., Marchand V., Decroly E., Motorin Y., Bennasser Y. FTSJ3 is an RNA 2′-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature. 2019;565:500–504. doi: 10.1038/s41586-018-0841-4. [DOI] [PubMed] [Google Scholar]
- 17.Ruggero D., Grisendi S., Piazza F., Rego E., Mari F., Rao P.H., Cordon-Cardo C., Pandolfi P.P. Dyskeratosis congenita and cancer in mice deficient in ribosomal RNA modification. Science. 2003;299:259–262. doi: 10.1126/science.1079447. [DOI] [PubMed] [Google Scholar]
- 18.Zhou F., Liu Y., Rohde C., Pauli C., Gerloff D., Kohn M., Misiak D., Baumer N., Cui C., Gollner S., et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat. Cell Biol. 2017;19:844–855. doi: 10.1038/ncb3563. [DOI] [PubMed] [Google Scholar]
- 19.Borchardt E.K., Martinez N.M., Gilbert W.V. Regulation and Function of RNA Pseudouridylation in Human Cells. Annu. Rev. Genet. 2020;54:309–336. doi: 10.1146/annurev-genet-112618-043830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nachmani D., Bothmer A.H., Grisendi S., Mele A., Bothmer D., Lee J.D., Monteleone E., Cheng K., Zhang Y., Bester A.C., et al. Germline NPM1 mutations lead to altered rRNA 2′-O-methylation and cause dyskeratosis congenita. Nat. Genet. 2019;51:1518–1529. doi: 10.1038/s41588-019-0502-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouchard-Bourelle P., Desjardins-Henri C., Mathurin-St-Pierre D., Deschamps-Francoeur G., Fafard-Couture E., Garant J.M., Elela S.A., Scott M.S. snoDB: An interactive database of human snoRNA sequences, abundance and interactions. Nucleic Acids Res. 2020;48:D220–D225. doi: 10.1093/nar/gkz884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jorjani H., Kehr S., Jedlinski D.J., Gumienny R., Hertel J., Stadler P.F., Zavolan M., Gruber A.R. An updated human snoRNAome. Nucleic Acids Res. 2016;44:5068–5082. doi: 10.1093/nar/gkw386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maden B.E. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 24.Taoka M., Nobe Y., Yamaki Y., Sato K., Ishikawa H., Izumikawa K., Yamauchi Y., Hirota K., Nakayama H., Takahashi N., et al. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res. 2018;46:9289–9298. doi: 10.1093/nar/gky811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pauli C., Kienhofer M., Gollner S., Muller-Tidow C. Epitranscriptomic modifications in acute myeloid leukemia: M(6)A and 2′-O-methylation as targets for novel therapeutic strategies. Biol. Chem. 2021;402:1531–1546. doi: 10.1515/hsz-2021-0286. [DOI] [PubMed] [Google Scholar]
- 26.Dong J., Wang H., Zhang Z., Yang L., Qian X., Qian W., Han Y., Huang H., Qian P. Small but strong: Pivotal roles and potential applications of snoRNAs in hematopoietic malignancies. Front. Oncol. 2022;12:939465. doi: 10.3389/fonc.2022.939465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D., Zhou J., Gao J., Wu R.Y., Huang Y.L., Jin Q.W., Chen J.S., Tang W.Z., Yan L.H. Targeting snoRNAs as an emerging method of therapeutic development for cancer. Am. J. Cancer Res. 2019;9:1504–1516. [PMC free article] [PubMed] [Google Scholar]
- 28.Fafard-Couture E., Bergeron D., Couture S., Abou-Elela S., Scott M.S. Annotation of snoRNA abundance across human tissues reveals complex snoRNA-host gene relationships. Genome Biol. 2021;22:172. doi: 10.1186/s13059-021-02391-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchand V., Pichot F., Neybecker P., Ayadi L., Bourguignon-Igel V., Wacheul L., Lafontaine D.L.J., Pinzano A., Helm M., Motorin Y. HydraPsiSeq: A method for systematic and quantitative mapping of pseudouridines in RNA. Nucleic Acids Res. 2020;48:e110. doi: 10.1093/nar/gkaa769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou F., Aroua N., Liu Y., Rohde C., Cheng J., Wirth A.K., Fijalkowska D., Gollner S., Lotze M., Yun H., et al. A Dynamic rRNA Ribomethylome Drives Stemness in Acute Myeloid Leukemia. Cancer Discov. 2023;13:332–347. doi: 10.1158/2159-8290.CD-22-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Z.H., Du Y.P., Wen J.T., Lu B.F., Zhao Y. snoRNAs: Functions and mechanisms in biological processes, and roles in tumor pathophysiology. Cell Death Discov. 2022;8:259. doi: 10.1038/s41420-022-01056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mamontova V., Trifault B., Boten L., Burger K. Commuting to Work: Nucleolar Long Non-Coding RNA Control Ribosome Biogenesis from Near and Far. Noncoding RNA. 2021;7:42. doi: 10.3390/ncrna7030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui C., Liu Y., Gerloff D., Rohde C., Pauli C., Kohn M., Misiak D., Oellerich T., Schwartz S., Schmidt L.H., et al. NOP10 predicts lung cancer prognosis and its associated small nucleolar RNAs drive proliferation and migration. Oncogene. 2021;40:909–921. doi: 10.1038/s41388-020-01570-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong X.Y., Guo P., Boyd J., Sun X., Li Q., Zhou W., Dong J.T. Implication of snoRNA U50 in human breast cancer. J. Genet. Genom. 2009;36:447–454. doi: 10.1016/S1673-8527(08)60134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong X.Y., Rodriguez C., Guo P., Sun X.D., Talbot J.T., Zhou W., Petros J., Li Q., Vessella R.L., Kibel A.S., et al. SnoRNA U50 is a candidate tumor-suppressor gene at 6q14.3 with a mutation associated with clinically significant prostate cancer. Hum. Mol. Genet. 2008;17:1031–1042. doi: 10.1093/hmg/ddm375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pacilli A., Ceccarelli C., Trere D., Montanaro L. SnoRNA U50 levels are regulated by cell proliferation and rRNA transcription. Int. J. Mol. Sci. 2013;14:14923–14935. doi: 10.3390/ijms140714923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen W., Sun H., De Hoyos C.L., Bailey J.K., Liang X.H., Crooke S.T. Dynamic nucleoplasmic and nucleolar localization of mammalian RNase H1 in response to RNAP I transcriptional R-loops. Nucleic Acids Res. 2017;45:10672–10692. doi: 10.1093/nar/gkx710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki Y., Holmes J.B., Cerritelli S.M., Sakhuja K., Minczuk M., Holt I.J., Crouch R.J. An upstream open reading frame and the context of the two AUG codons affect the abundance of mitochondrial and nuclear RNase H1. Mol. Cell. Biol. 2010;30:5123–5134. doi: 10.1128/MCB.00619-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tannous E., Kanaya E., Kanaya S. Role of RNase H1 in DNA repair: Removal of single ribonucleotide misincorporated into DNA in collaboration with RNase H2. Sci. Rep. 2015;5:9969. doi: 10.1038/srep09969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H., Lima W.F., Zhang H., Fan A., Sun H., Crooke S.T. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 2004;279:17181–17189. doi: 10.1074/jbc.M311683200. [DOI] [PubMed] [Google Scholar]
- 41.Yin W., Rogge M. Targeting RNA: A Transformative Therapeutic Strategy. Clin. Transl. Sci. 2019;12:98–112. doi: 10.1111/cts.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crooke S.T. Molecular mechanisms of action of antisense drugs. Biochim. Biophys. Acta. 1999;1489:31–44. doi: 10.1016/S0167-4781(99)00148-7. [DOI] [PubMed] [Google Scholar]
- 43.Migliorati J.M., Liu S., Liu A., Gogate A., Nair S., Bahal R., Rasmussen T.P., Manautou J.E., Zhong X.B. Absorption, Distribution, Metabolism, and Excretion of US Food and Drug Administration-Approved Antisense Oligonucleotide Drugs. Drug Metab. Dispos. 2022;50:888–897. doi: 10.1124/dmd.121.000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kufel J., Grzechnik P. Small Nucleolar RNAs Tell a Different Tale. Trends Genet. 2019;35:104–117. doi: 10.1016/j.tig.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 45.Dominski Z., Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl. Acad. Sci. USA. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu Q.L., Rabinowitz A., Chen Y.C., Yokota T., Yin H., Alter J., Jadoon A., Bou-Gharios G., Partridge T. Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc. Natl. Acad. Sci. USA. 2005;102:198–203. doi: 10.1073/pnas.0406700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Passini M.A., Bu J., Richards A.M., Kinnecom C., Sardi S.P., Stanek L.M., Hua Y., Rigo F., Matson J., Hung G., et al. Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci. Transl. Med. 2011;3:72ra18. doi: 10.1126/scitranslmed.3001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Svasti S., Suwanmanee T., Fucharoen S., Moulton H.M., Nelson M.H., Maeda N., Smithies O., Kole R. RNA repair restores hemoglobin expression in IVS2-654 thalassemic mice. Proc. Natl. Acad. Sci. USA. 2009;106:1205–1210. doi: 10.1073/pnas.0812436106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bush J.A., Williams C.C., Meyer S.M., Tong Y., Haniff H.S., Childs-Disney J.L., Disney M.D. Systematically Studying the Effect of Small Molecules Interacting with RNA in Cellular and Preclinical Models. ACS Chem. Biol. 2021;16:1111–1127. doi: 10.1021/acschembio.1c00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stelzer A.C., Frank A.T., Kratz J.D., Swanson M.D., Gonzalez-Hernandez M.J., Lee J., Andricioaei I., Markovitz D.M., Al-Hashimi H.M. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat. Chem. Biol. 2011;7:553–559. doi: 10.1038/nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang P., Liu X., Abegg D., Tanaka T., Tong Y., Benhamou R.I., Baisden J., Crynen G., Meyer S.M., Cameron M.D., et al. Reprogramming of Protein-Targeted Small-Molecule Medicines to RNA by Ribonuclease Recruitment. J. Am. Chem. Soc. 2021;143:13044–13055. doi: 10.1021/jacs.1c02248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Costales M.G., Matsumoto Y., Velagapudi S.P., Disney M.D. Small Molecule Targeted Recruitment of a Nuclease to RNA. J. Am. Chem. Soc. 2018;140:6741–6744. doi: 10.1021/jacs.8b01233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dey S.K., Jaffrey S.R. RIBOTACs: Small Molecules Target RNA for Degradation. Cell Chem. Biol. 2019;26:1047–1049. doi: 10.1016/j.chembiol.2019.07.015. [DOI] [PubMed] [Google Scholar]
- 54.Bratkovic T., Bozic J., Rogelj B. Functional diversity of small nucleolar RNAs. Nucleic Acids Res. 2020;48:1627–1651. doi: 10.1093/nar/gkz1140. [DOI] [PMC free article] [PubMed] [Google Scholar]