Abstract
Background:
Esophagitis is prevalent in patients with esophageal dysmotility despite acid suppression, likely related to poor esophageal clearance. Esophageal atresia (EA) is a classic model of dysmotility where this observation holds true. In adult non-dysmotility populations, failure of esophagitis to respond to proton pump inhibitors (PPI) has been linked to variants in CYP2C19 that influence the activity of the encoded enzyme. It is unknown if CYP2C19 metabolizer phenotype contributes to PPI-refractory, non-allergic esophagitis in EA.
Methods:
We performed a cross-sectional study of 314 children with (N = 188) and without (N = 126) EA who were on PPI therapy at the time of endoscopy to evaluate for possible gastroesophageal reflux disease. Patients with eosinophilic esophagitis and/or fundoplication were excluded. Clinical and histology data were collected. Genomic DNA from biopsy samples was genotyped for polymorphisms in CYP2C19.
Results:
CYP2C19 metabolizer phenotypes were not associated with presence or severity of esophagitis (P = 0.994). In a multivariate logistic regression adjusted for potential confounders, EA was the strongest and only significant predictor of esophagitis (odds ratio 2.72, P = 0.023). Using negative binomial regression, we found that CYP2C19 phenotype was not a significant predictor of eosinophil count in children with PPI-refractory esophagitis.
Conclusions:
Patients with EA are significantly more likely to experience PPI-refractory, non-allergic esophagitis than controls regardless of CYP2C19 metabolizer phenotype, suggesting that factors other than CYP2C19 genetics, including dysmotility, are the primary drivers of esophagitis in EA. CYP2C19 genotype failed to predict PPI-refractory, non-allergic esophagitis in both EA and non-EA children.
Keywords: CYP2C19, dysmotility, esophageal atresia, esophagitis, proton pump inhibitor metabolism
1 |. INTRODUCTION
Esophagitis is highly prevalent among patients with esophageal atresia (EA) despite widespread use of acid-suppressive medications.1–4 Esophagitis in EA has been attributed to increased rates of gastroesophageal reflux disease (GERD), eosinophilic esophagitis, and dysmotility; however, the relative importance of each of these potential mechanisms is unclear.
In non-EA populations, failure of reflux esophagitis to respond to proton pump inhibitors (PPI) has been linked to variants in CYP2C19, which encodes the CYP2C19 enzyme that is primarily responsible for metabolizing PPI. For example, CYP2C19 rapid metabolizers (ie, with lower circulating PPI) have increased esophageal acid exposure times (pH <4 time 5.7% vs 2.7%),5 increased GERD symptom recurrence,6 and decreased response to PPIs for healing erosive reflux esophagitis in adult populations.7,8 There have been no studies of the role of CYP2C19 metabolizer phenotype in patients at highest risk for esophagitis: patients with esophageal dysmotility. One model of significant esophageal dysmotility is EA, an ideal model to determine the relative impact of PPI pharmacogenetics and dysmotility. This understanding is important to understand if genetic testing is an important test in the algorithm for persistent esophagitis in these patients.
Esophageal motility is near universally compromised in EA patients and has been associated with esophagitis and Barrett’s esophagus;2 multiple manometry and impedance studies in patients with EA have found abnormal patterns of low amplitude peristalsis and impaired bolus transit in 80%−100% of patients.9–12
The relative importance of dysmotility versus CYP2C19 metabolizer phenotype in EA is unclear. We hypothesized that dysmotility, rather than genetic variants in CYP2C19, is the primary driver of PPI-refractory non-allergic esophagitis in EA. In this case–control study of 314 patients with (N = 188) and without (N = 126) EA, we aimed to characterize CYP2C19 metabolizer phenotype to determine if they are associated with non-allergic PPI-refractory esophagitis in EA. As a secondary aim, because of the high eosinophil counts sometimes seen in EA, we examined STAT6 rs324011 genotype as a possible confounder, as homozygosity for this variant is known to associate with elevated eosinophil counts in allergic-eosinophilia.13–15
2 |. METHODS
This study was approved by the Boston Children’s Hospital institutional review board. Two patient cohorts were included in this study: 188 patients with EA and 126 control patients (Figure 1). Control patients were recruited prospectively as part of a large study to assess for risk factors for the development of esophagitis in children. Surveys of symptom frequency at the time of study enrollment and the PedsQL were collected and assessed (Data S1).16 All control patients recruited to the larger study were included in the CYP2C19 study except: (1) those with eosinophilic esophagitis (EoE; defined below); (2) those who had their endoscopy performed off of acid suppression; and (3) those with a history of prior fundoplication. All EA patients who underwent routine surveillance endoscopy with biopsy between 2016 and 2018 at Boston Children’s Hospital were included, except those who met the same exclusion criteria (1)-(3) as above. In our routine clinical practice, children with repaired EA undergo surveillance endoscopy to detect esophagitis and other long-term complications of esophageal atresia every 1–3 years, regardless of the presence or absence of symptoms.17 Medical record review was performed to identify clinical patient characteristics including demographic information; symptoms at time of endoscopy; acid-suppressive medication type, dosing, and frequency at the time of endoscopy; presence and characteristics of the esophagitis by histopathology; endoscopy reports of gross esophageal findings; and radiologic diagnostic evaluation reports. Erosive esophagitis was defined as gross erosive disease in the esophagus meeting the criteria for the Los Angeles classification grades A-D.18
FIGURE 1.
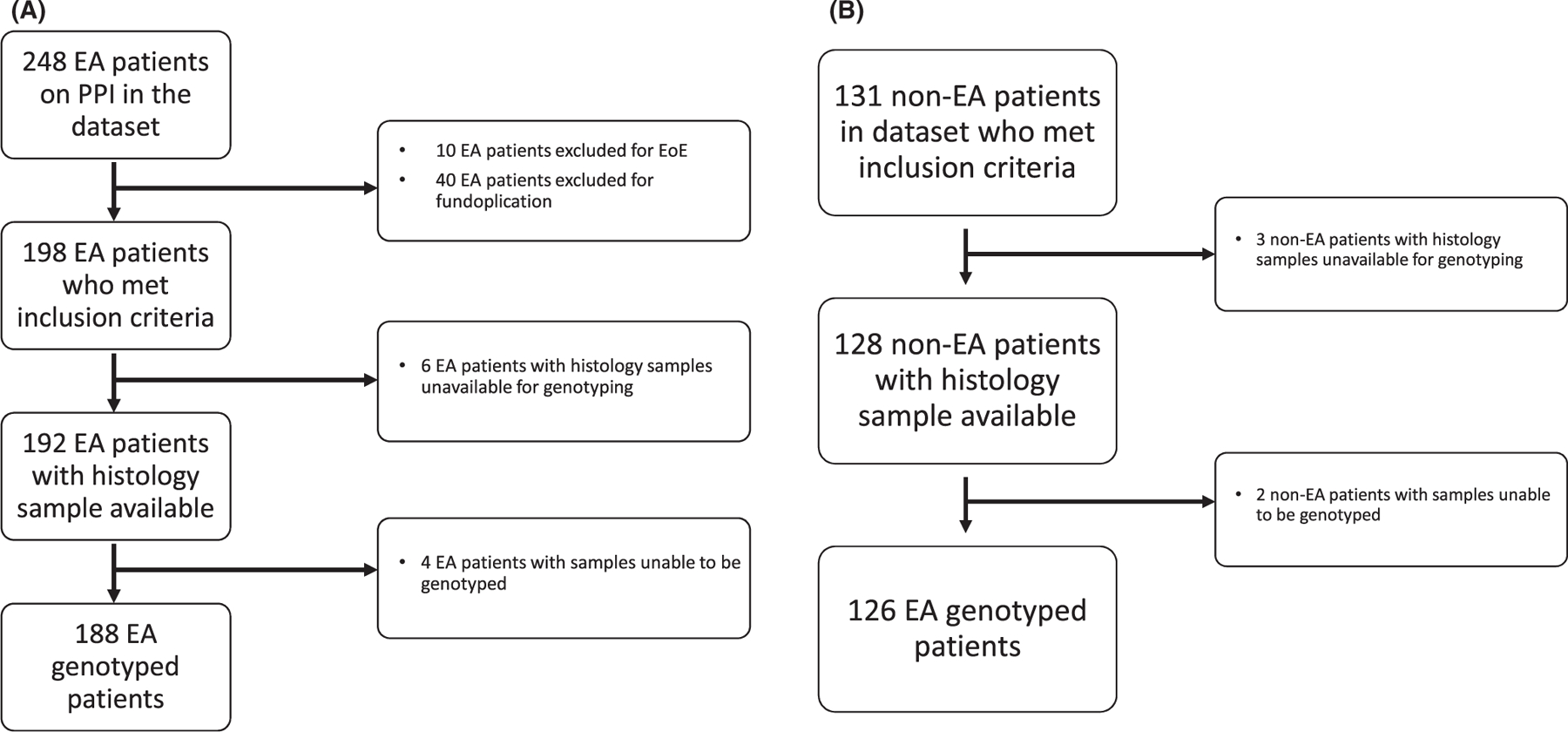
Diagram detailing datasets and excluded patients for EA cases (A) and non-EA controls (B)
Patients with a diagnosis of allergic EoE and/or fundoplication at the time of endoscopy with biopsy were excluded from both the EA cases and non-EA controls. To define which patients were excluded, we defined EoE as peak eosinophil count ≥ 15 eosinophils/hpf accompanied by symptoms of esophageal dysfunction, plus either suggestive gross endoscopic findings (furrows, rings, exudates) and/or suggestive atopic or family history that resulted in EoE directed treatment and response. Having eosinophil counts greater than 15 per hpf alone was not an automatic exclusion, as severe reflux-associated esophagitis may produce this histologic finding.19 In addition, we genotyped the STAT6 rs324011 variant as a possible confounder, as this variant is known to associate with PPI-refractory EoE.20 The primary outcome measure was the difference in distribution of CYP2C19 metabolizer phenotypes between EA and non-EA groups, stratified by the degree of inflammation (0 eosinophils, 1–15 eosinophils, and >15 eosinophils per hpf). Secondary outcomes were the impact of PPI genetics on erosive esophagitis and esophageal stricture formation.
2.1 |. Genotyping
Genomic DNA was isolated from formalin-fixed paraffin-embedded (FFPE) sections of esophageal biopsy tissue.5 SNPs were interrogated by TaqMan fluorescent assay using a ViiA7 real-time polymerase chain reaction instrument (Applied Biosystems, Foster City, CA). Genotypes were called automatically by the ViiA7 software and were confirmed manually. The SNPs interrogated and assays used were as follows: CYP2C19: loss of function variant *2, rs4244285, C_25986767_70; loss of function variant *3, rs4986893, C_27861809_10; gain of function variant *17 rs12248560, C__469857_10; STAT6: rs324011, C__620399_10. The STAT6 rs324011 SNP was sequenced as this variant has been most strongly tied to EoE.20 Genotypes for all SNPs were confirmed to be in Hardy–Weinberg equilibrium within the complete study population using the R statistical package Hardy–Weinberg.21
CYP2C19 diplotypes were assigned to predicted metabolizer phenotypes as follows: ultra-rapid metabolizer (UM; *17/*17), rapid metabolizer (RM; *1/*17), normal metabolizer (NM; *1/*1), intermediate metabolizer (IM; *1/*2, *1/*3), or poor PPI metabolizer (PM; *2/*2, *2/*3, *3/*3).22 As discussed in depth elsewhere, rapid and ultra-rapid metabolizers generally achieve lower PPI blood levels, clear PPI faster, and experience shorter periods of acid suppression compared to normal metabolizers; intermediate and poor metabolizers achieve higher PPI blood levels and longer periods of acid suppression compared to normal metabolizers.22
2.2 |. Statistical analysis
Continuous data are presented as median and interquartile range (IQR) and categorical data are presented as frequency and percentage. Univariate comparisons were performed using Fisher’s exact test or the Wilcoxon rank-sum test. Multivariable logistic regression analysis was used to determine adjusted associations between outcome and predictor variables, with results reported using adjusted odds ratios (OR) with corresponding 95% confidence intervals and P values. Collinear terms were identified among PPI variables to avoid multicollinearity in regression modeling. Statistical analyses were performed using Stata (version 16.0, StataCorp LLC., College Station, Texas). A two-tailed statistical significance threshold of P<0.05 was implemented.
Based on our prior studies, we found the incidence of poor metabolizers was 3% in patients requiring antireflux surgery versus 21% in patients who did not require surgery. Assuming this same difference—that patients with persistent esophagitis (a population similar to those requiring antireflux surgery) would have a 3% incidence of PM genotype compared to 21% of patients without esophagitis (a population similar to those who did not require antireflux surgery)—we calculated that we would need a sample size of 48 patients in each group to provide 80% power, assuming a 5% type I error rate.
3 |. RESULTS
Baseline clinical characteristics and presenting symptoms at time of endoscopy of EA cases and non-EA controls are provided in Table 1. Symptoms prompting endoscopy in control patients are additionally enumerated in Data S1 and are reflected by mean PedsQL total score of 79.2 ± 16.0, which is well above previously published cutoffs for impaired quality of life.23 Similarly, most EA patients (77%) were symptomatic at time of endoscopy (Table 1). Presenting symptoms at time of endoscopy differed significantly between the groups with EA cases more likely to have respiratory symptoms or frequent respiratory infections, likely due to the high prevalence of concurrent tracheomalacia and aspiration in EA.
TABLE 1.
Cohort characteristics
| EA (N=188) | % or IQR | Non-EA (N=126) | % or IQR | P value | |
|---|---|---|---|---|---|
| Gender = Female | 92 | 48.9% | 63 | 49.2% | 0.909 |
| Age at EGD, months | 32.5 | (14–71) | 108 | (63–171) | <0.001* |
| Long gap EA | 57 | 30.3 | — | — | — |
| Neurological impairment and/or genetic syndromea | 17 | 9.0% | 15 | 11.9% | 0.449 |
| Symptoms at EGDb | |||||
| Reflux | 61 | 32.4% | 102 | 81.0% | <0.001* |
| Dysphagia | 52 | 27.7% | 34 | 27.0% | 0.999 |
| Feeding issues | 77 | 41.0% | 9 | 7.1% | <0.001* |
| Respiratory sx | 72 | 38.3% | 3 | 2.4% | <0.001* |
| Freq. resp. infections | 45 | 23.9% | 0 | 0% | <0.001* |
| PPI | 0.062 | ||||
| Omeprazole (1st gen) | 112 | 59.6% | 64 | 50.8% | |
| lansoprazole(1st gen) | 51 | 27.1% | 47 | 37.3% | |
| Esomeprazole (2nd gen) | 17 | 9.0% | 5 | 4.0% | |
| Pantoprazole (1st gen) | 7 | 3.7% | 9 | 7.2% | |
| Dexlansoprazole (2nd gen) | 1 | 0.5% | 1 | 0.8% | |
| PPI dose frequency | 0.0002* | ||||
| QD | 58 | 30.9% | 66 | 51.6% | |
| BID | 129 | 68.6% | 60 | 46.9% | |
| PPI dose (mg/kg/d) | 1.54 | (1.01–1.9) | 0.97 | (0.65–1.49) | <0.001* |
| Length of PPI usage (months) | 15.5 | (8.3–30.5) | 3.25 | (1.5–8) | <0.001* |
| H2RA use | 62 | 33.0% | 7 | 5.5% | <0.001* |
| Erosive Esophagitis | 13 | 6.9% | 0 | 0% | <0.001* |
| Histologic Esophageal Eosinophilia | 0.059 | ||||
| ≥ 15 eos/hpf | 32 | 17.0% | 10 | 7.9% | |
| 1–14 eos/hpf | 67 | 35.6% | 46 | 36.5% | |
| 0 eos/hpf | 89 | 47.3% | 70 | 55.6% | |
| CYP2C19 metabolizer | 0.258 | ||||
| UM | 11 | 5.9% | 3 | 2.4% | |
| RM | 36 | 19.2% | 30 | 23.8% | |
| NM | 93 | 49.5% | 53 | 42.1% | |
| IM | 42 | 22.3% | 37 | 29.4% | |
| PM | 6 | 3.2% | 3 | 2.4% | |
| Copies of STAT6 rs324011 variant | 0.719 | ||||
| 0/1 copies | 165 | 87.8% | 113 | 89.7% | |
| 2 copies | 23 | 12.2% | 13 | 10.3% |
Abbreviations: and PM, poor metabolizer; EGD, esophagogastroduodenoscopy; IM, intermediate metabolizer; NM, normal metabolizer; RM, rapid metabolizer; UM, ultra-rapid metabolizer.
Syndromes were trisomy 21, CHARGE syndrome, trisomy 18, and various duplication or deletion syndromes with associated neurological impairment.
Multiple symptoms may be present in the same patient and are not mutually exclusive. Reflux symptoms included heartburn, chest pain, abdominal pain, vomiting, regurgitation. Feeding issues included gastrostomy or gastrojejunostomy dependence and oral aversion. Respiratory symptoms included persistent cough, exercise intolerance. Statistically significant results denoted by bold face type and asterisk (*).
3.1 |. Esophagitis characteristics
Rates of esophagitis are shown in Table 1. Among patients with ≥ 15 eosinophils/hpf, median worst eosinophil counts in the esophagus were 36.5 eosinophils/hpf (IQR 20–50) in the EA group versus 25 eosinophils/hpf (IQR 20–35.3) in the control group (P = 0.441). To support that our chosen outcome measure of histologic esophagitis ≥ 15 eosinophils/hpf was related to GERD rather than EoE, we performed STAT6 rs324011 variant genotyping. While homozygosity for STAT6 rs324011 is known to associate with allergy-mediated eosinophilia in EoE,15 we found no association between peak eosinophil counts ≥ 15 eosinophils/hpf and STAT6 rs324011 genotype (P = 0.260) suggesting that allergy-mediated eosinophilia is not a significant confounder in this cohort. Furthermore, the geographic distribution of esophageal eosinophil counts was evaluated and was significantly higher in distal compared to proximal biopsy specimens supporting our hypothesis of significant reflux in this population (Figure S2, P < 0.001).
Patients with EA received higher doses of PPIs compared to controls (Table 1, 1.54 vs 0.97 mg/kg/day, P < 0.001), were more likely to receive twice-daily dosing compared to once-daily dosing (69% vs 48%, P < 0.001), and were more likely to be treated with concurrent H2 receptor antagonist (H2RA; 33% vs 5%; P < 0.001). Despite more aggressive acid suppression, children with EA were significantly more likely to have erosive esophagitis (6.9% vs 0%, P < 0.001) and more likely to have significant histologic esophagitis (≥15 eos/hpf, 17% vs 7.8%, P = 0.027) than controls.
3.2 |. CYP2C19 metabolizer phenotypes
To understand the mechanism behind differential rates of esophagitis, we then compared the distribution of CYP2C19 metabolizer phenotypes between EA cases and non-EA controls (Table 1; P = 0.258). Distributions of CYP2C19 metabolizer phenotypes and STAT6 genotypes were similar among patients with varying levels of histologic inflammation (Figure 2a, P = 0.994; Figure 3a, P = 0.490, respectively).
FIGURE 2.
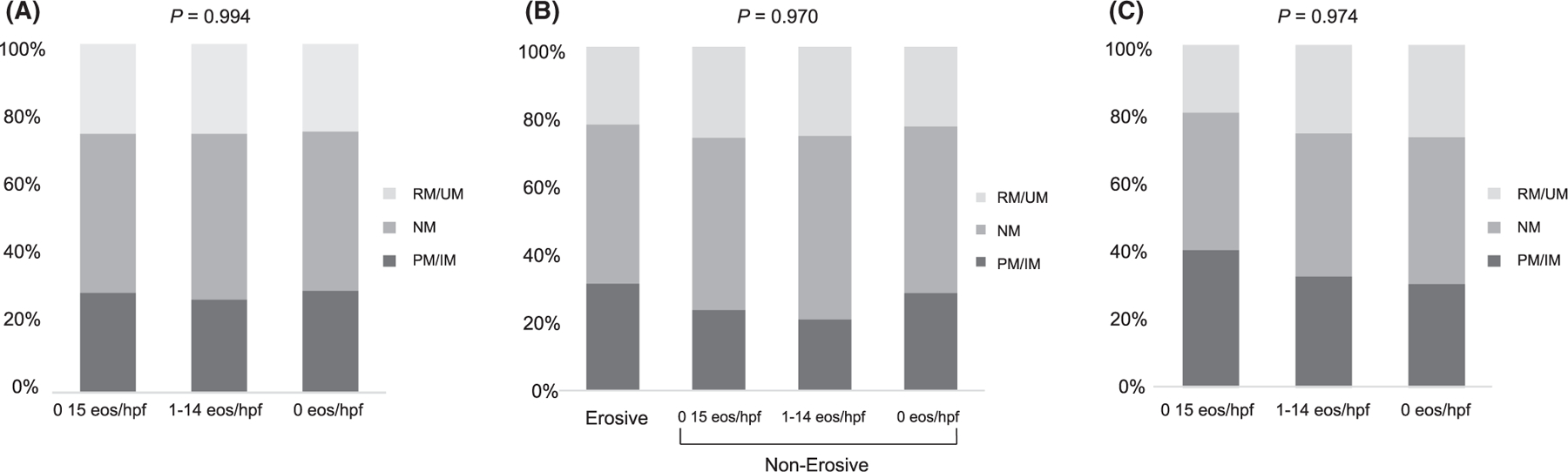
Distributions of CYP2C19 metabolizer phenotypes among sub-populations with and without esophagitis expressed as a percentage of the total at-risk patients. A. Entire cohort. B. EA subgroup. C. non-EA subgroup
FIGURE 3.
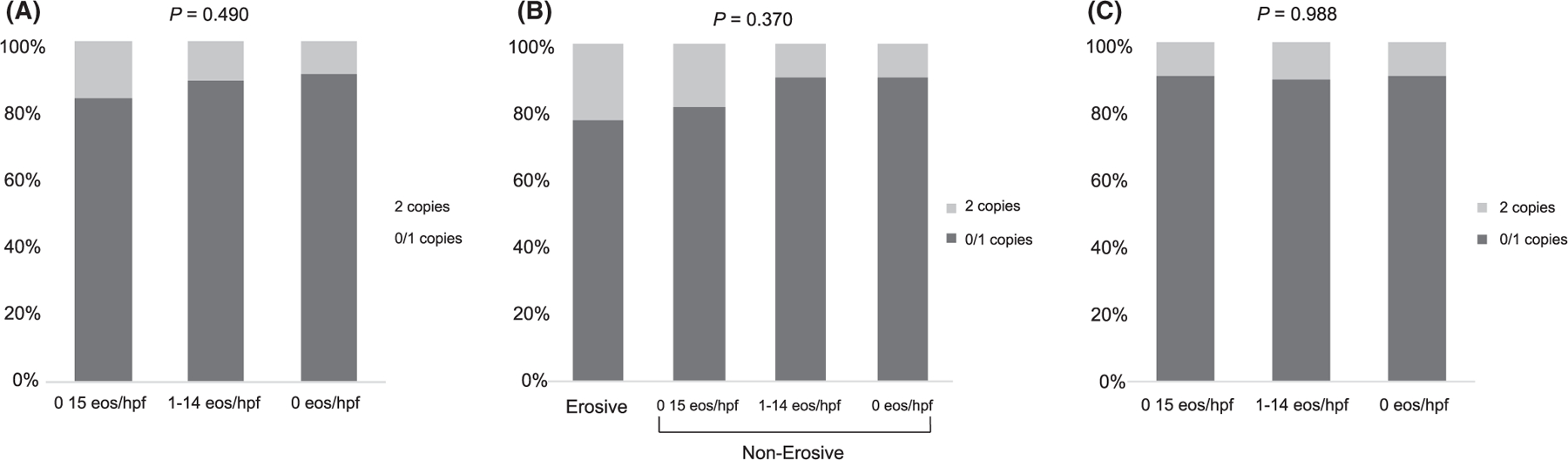
Distributions of STAT6 rs324011 genotypes among sub-populations with and without esophagitis expressed as a percentage of the total at-risk patients. A. Entire cohort. B. EA subgroup. C. non-EA subgroup
When performing a subgroup analysis of CYP2C19 metabolizer phenotypes in just the EA patients (N = 188), eosinophil/hpf counts did not significantly differ between CYP2C19 metabolizer phenotypes or STAT6 variant genotype by Kruskal–Wallis testing (P = 0.930 and P = 0.714, respectively). There was no significant difference in the distribution of CYP2C19 metabolizer phenotypes between EA patients with any type of esophagitis and those with no inflammation (Figure 2b; P = 0.970). Similarly, STAT6 variant homozygosity was not significantly more common in EA patients with erosive disease or histologic inflammation compared to no inflammation (Figure 3b; P = 0.370). There was no association between stricture formation and CYP2C19 metabolizer phenotype (P > 0.49).
Similar to the EA patients, when performing a subgroup analysis of CYP2C19 metabolizer phenotypes in just the control patients (N=126), eosinophil/hpf count did not significantly differ between CYP2C19 metabolizer phenotypes or STAT6 genotypes (P = 0.765 and P = 0.316, respectively). Distributions of CYP2C19 phenotypes (Figure 2c; P = 0.974) and STAT6 variant homozygosity (Figure 3c, P = 0.988) were similar between control patients with histologic esophagitis versus control patients with no inflammation. No patients in the control group had stricture that required dilation or erosive esophagitis.
3.3 |. Gastric dysmotility
Gastric factors such as delayed gastric emptying may contribute to esophageal reflux burden. Nine of 126 control children (7%) and 18 of 188 EA patients (9.6%) underwent gastric emptying studies within 6 months of their included study EGD. Of these children with gastric emptying studies, 2/9 control patients (22%) and 6/18 EA patients (33%) had delayed gastric emptying. There was no significant difference in rates of delayed gastric emptying between the control and EA cohorts (P = 0.6758); there was a trend toward but no statistically significant association between delayed gastric emptying and esophagitis >15 eosinophils/hpf (P = 0.0646).
3.4 |. Multivariate predictors of esophagitis
In a multivariate logistic regression, EA was the strongest and only statistically significant predictor of esophagitis (Table 2; odds ratio (OR) 2.72, P = 0.023). Additional analysis was performed with a negative binomial regression using peak eosinophil count as a continuous outcome, and similarly found that EA was the strongest predictor of esophagitis (coefficient 1.22, P < 0.001) while pharmacogenetics remained not significant (Data S3). Regardless of how the metabolizers were grouped (PM vs IM vs NM vs RM vs UM; IM/PM vs NM/RM/UM; IM/PM vs NM vs RM/UM; NM/IM/PM vs RM/UM), there was no difference in metabolizer phenotype and the presence of esophagitis (P > 0.40). A subgroup analysis of individuals who were not homozygous for the STAT6 rs324011 variant (N = 277) found that EA remained the strongest predictor of esophagitis, achieving strong statistical significance in the negative binomial regression model (Data S4; coefficient 1.23, P = 0.001).
TABLE 2.
Multivariable logistic regression of esophagitis (≥ 15 eosinophils/hpf N = 42; < 15 eos/hpf N = 272)
| Odds ratio | 95% Confidence interval (CI) | P value | |
|---|---|---|---|
| EA | 2.72 | 1.14–6.47 | 0.023* |
| Gender (ref = female) | 1.49 | 0.75–2.98 | 0.259 |
| Age | 1.00 | 1.00–1.01 | 0.599 |
| Total PPI (mg/kg/day) | 1.47 | 0.80–2.72 | 0.215 |
| PPI Generation (ref = 1st gen) | 1.16 | 0.36–3.73 | 0.802 |
| H2RA | 0.77 | 0.33–1.76 | 0.531 |
| CYP2C19 phenotype | |||
| PM/IM | 1.22 | 0.54–2.72 | 0.631 |
| NM | ref | — | — |
| RM/UM | 1.17 | 0.51–2.67 | 0.704 |
| STAT6 rs324011 2 copies | 1.63 | 0.64–4.13 | 0.302 |
Statistically significant results denoted by bold face type and asterisk (*).
When performing multivariate logistic regression within the subgroup of patients with EA, neither intensity of acid suppression, type of acid suppression, CYP2C19 phenotype nor STAT6 genotype was significantly associated with histologic esophagitis, erosive esophagitis, or presence of stricture requiring dilation at the time of endoscopy with biopsies by multivariate logistic regression analyses (Data S5).
When performing multivariate logistic regression within control patients, there was no significant relationship between CYP2C19 metabolizer phenotype and histologic esophagitis in multivariate analyses (Data S6).
4 |. DISCUSSION
Despite the importance of CYP2C19 genotype in achieving sufficient PPI levels to effectively treat GERD and reflux esophagitis in adult patients, we failed to observe a significant impact of CYP2C19 metabolizer phenotype on microscopic or erosive esophagitis in this study in children. Instead, esophageal atresia was the strongest and only statistically significant predictor of esophagitis in this large pediatric cross-sectional, case–control study of esophageal biopsies obtained on PPI therapy.
Given that EoE was an exclusion criteria, we hypothesized that dysmotility with poor clearance is a major contributor to this persistent, PPI-refractory esophagitis. There are two disease models of dysmotility similar to EA which support this hypothesis: scleroderma and endoscopically treated achalasia. In both of these diseases, high dose acid suppression is frequently needed for symptom control, and while microscopic esophagitis is not reported in adult studies, gross esophagitis often persists.24 For example, one study of 53 adult patients with scleroderma found that erosive esophagitis was present in 60% and did not correlate with acid suppression treatment; all patients with erosive esophagitis had absent distal esophageal peristalsis by manometry, while no patients with normal manometry had erosive esophagitis.25 Similarly, of 43 peroral endoscopy myotomy (POEM)-treated achalasia patients with erosive esophagitis, persistent erosive esophagitis was seen in 18.6% of patients on PPI.26 Another study of 231 post-POEM achalasia patients found erosive esophagitis in 74% of patients taking PPIs at the time of endoscopy.27 While it is clear that esophagitis is highly prevalent in these populations, and in our EA population, this is the first study to look at PPI metabolizer status in any dysmotility population, pediatric, or adult.
A meta-analysis of 19 adult studies (N = 1112 patients) identified that odds of PPI-refractory reflux esophagitis were significantly greater among rapid metabolizers compared to poor metabolizers (OR 1.661, 95% CI 1.023–2.659, P = 0.040).8 While there have been no prior studies of CYP2C19 genotype and reflux esophagitis in children, a study of 74 children who underwent pH-impedance studies showed that CYP2C19 rapid metabolizers had significantly longer acid exposure times in the esophagus (pH < 4 time: 5.7% vs 2.7%).5 In another retrospective case–control study among 34 children with medically refractory GERD defined by pH probe despite PPI therapy, carriage of CYP2C19*17 allele corresponding to the UM and RM phenotypes was associated with antireflux surgery compared to controls (OR 9.78, CI 1.25–76.55, P < 0.03).28 Our study is the first study of CYP2C19 genotype in children examining reflux esophagitis, and the only study in patients with dysmotility. Despite our large study size, we failed to find a significant relationship between CYP2C19 metabolizer phenotype and esophagitis in EA as well as non-EA controls. Rapid PPI metabolizers were not more common among patients with esophagitis in this large pediatric study. While it is possible we may be underpowered to detect such an association, it is also possible that the CYP2C19 genotype is less relevant for reflux esophagitis outcomes in pediatric patients in the setting of more frequent (eg, BID) and/or higher PPI dosing (1–2 mg/kg/day) routinely used in children compared to adults.7
The widespread practice of long-term acid suppression of children with EA stems in part from the assumption that stasis of acid is primarily responsible for esophagitis and its complications like Barrett’s esophagus, and that treatment with PPI should be expected to prevent and treat these issues. We have shown in a separate study that acid suppression is associated with less esophagitis in EA,1 but we have previously shown and again demonstrate here that acid suppression in standard doses (1–3 mg/kg/day) does not uniformly prevent esophagitis. While higher dosing or divided dosing of PPI has been shown to successfully achieve sufficient plasma levels of PPI to effectively suppress acid and treat acid-related disorders in patients without esophageal dysmotility,7 higher dosing and greater frequency was not linked to reduced esophagitis in EA suggesting that different therapies to address stasis and prolonged esophageal exposure such as motility medications or coating agents may need to be added. Rational reflux management strategies are particularly important in EA and dysmotility patients who may be at greater risk for pulmonary complications, independently of the risk of PPI-associated respiratory infections. 17,29
To better understand the relative importance of acid suppression and dysmotility in esophagitis, we intentionally excluded children with diagnoses of EoE from this study. The development of EoE and its response to PPI treatment is linked to STAT6, a transcription factor which has been extensively linked to a variety of allergic disorders13 including eosinophilic esophagitis.14,30–33 STAT6 drives allergic eosinophilic inflammation in the esophagus14,31–33 and has been tied to likelihood of PPI response in EoE20 as PPIs have been shown to block STAT6-mediated recruitment of eosinophils to the esophagus.14,33 Furthermore, STAT6-mediated eotaxin-3 levels differentiate between GERD and EoE.30 We genotyped our study cohort for STAT6 rs324011 to support that our lack of CYP2C19 association with esophagitis was not because PPI-treated patients with EoE were erroneously classified as non-EoE. We found that pro-allergic STAT6 variant homozygosity was not enriched among patients with high eosinophil counts, supporting our belief that we generally accurately excluded EoE patients. Even excluding those few included patients with STAT6 variant homozygosity, EA remained the strongest predictor of esophagitis. We found no association between CYP2C19 or STAT6 genotype and esophagitis in this large non-allergic pediatric cohort. Our study again raises the idea that patients with dysmotility may need to have a different standard for the diagnosis of EoE and that many patients have peptic disease with high eosinophil counts. To rationally treat EA-related esophagitis, additional studies are needed to define peptic disease in patients with dysmotility.
While this is a large study of CPY2C19 pharmacogenetics, it is possible that the study was underpowered to find difference in metabolizer status. However, if there is an effect, it is likely to be small given the sample size. Another possible limitation is physician practice such that PPI dosing may not be representative of other centers. However, we think this is also unlikely given that the dosing range used in this study was the same as that reported in the NASPGHAN GERD guidelines.34 While histologic esophagitis defined solely by eosinophil count does not capture additional described histologic markers of esophagitis such as basal cell hyperplasia and papillary elongation, our institutional pathology practice is to not report these measures in clinical pathology reports, making these findings impossible for us to report in this study. These features are not routinely reported by our pathology group because they depend heavily on esophageal specimen orientation35–37 and have been described to have lower interobserver reproducibility than eosinophil count, neutrophil count, and erosions (64–74% versus 83–97%).38
Presence of hiatal hernias was unfortunately not recorded reliably for the control group, limiting our ability to make any adjustments for this potential confounder. A final limitation is the lack of correlated pH-impedance or formal motility testing in these patients to confirm the severity of dysmotility. Again, however, we know from numerous studies from EA patients that dysmotility is universally present making this less relevant.2,9–12,39–41
In summary, while CYP2C19 genotype has explained variability in PPI response in adult non-dysmotility acid-related disorders, EA-related esophagitis cannot be explained by variations in PPI metabolizer status and occurs despite aggressive acid suppression. PPI therapy alone may be insufficient to treat EA-associated esophagitis, as dysmotility is likely a critical factor in the development of esophagitis in EA. More broadly, while PPI metabolizer status has been linked to esophagitis outcomes in adults, CYP2C19 genotype failed to explain PPI-refractory esophagitis in children.
Supplementary Material
ACKNOWLEDGMENTS
The authors declare no conflicts of interest. The authors have no competing interests.
Funding Information
Financial Support provided by Boston Children’s Hospital Translational Research Program Senior Investigator Award (NIH R01 DK097112)
Abbreviations:
- CYP2C19 PPI
metabolizer phenotypes
- EA
Esophageal atresia
- EoE
eosinophilic esophagitis
- GERD
gastroesophageal reflux disease
- H2RA
histamine-2 receptor antagonist
- Hpf
high powered field
- IM
intermediate metabolizer
- NM
normal metabolizer
- PM
poor metabolizer
- PPI
proton pump inhibitor
- RM
rapid metabolizer
- UM
ultra-rapid metabolizer
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
DATA AVAILABILITY STATEMENT
Data not available due to ethical restrictions. Participants of this study did not agree for their data to be shared publicly, so supporting data are not available.
REFERENCES
- 1.Yasuda JL, Clark SJ, Staffa SJ, et al. Esophagitis in pediatric esophageal atresia: acid may not always be the issue. J Pediatr Gastroenterol Nutr 2019;69(2):163–170. [DOI] [PubMed] [Google Scholar]
- 2.Sistonen SJ, Koivusalo A, Nieminen U, et al. Esophageal morbidity and function in adults with repaired esophageal atresia with tracheoesophageal fistula: A population-based long-term follow-up. Ann Surg 2010;251(6):1167–1173. [DOI] [PubMed] [Google Scholar]
- 3.Deurloo JA, Ekkelkamp S, Bartelsman JFWM, et al. Gastroesophageal reflux: prevalence in adults older than 28 years after correction of esophageal atresia. Ann Surg 2003;238(5):686–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deurloo JA, Ekkelkamp S, Taminiau JAJM, et al. Esophagitis and Barrett esophagus after correction of esophageal atresia. J Pediatr Surg 2005;40(8):1227–1231. [DOI] [PubMed] [Google Scholar]
- 5.Franciosi JP, Mougey EB, Williams A, et al. Association between CYP2C19*17 Alleles and pH probe testing outcomes in children with symptomatic gastroesophageal reflux. J Clin Pharmacol 2018;58(1):89–96. [DOI] [PubMed] [Google Scholar]
- 6.Saitoh T, Otsuka H, Kawasaki T, et al. Influences of CYP2C19 polymorphism on recurrence of reflux esophagitis during proton pump inhibitor maintenance therapy. Hepatogastroenterology 2009;56(91–92):703–706. [PubMed] [Google Scholar]
- 7.Furuta T, Sugimoto M, Shirai N. Individualized therapy for gastroesophageal reflux disease: potential impact of pharmacogenetic testing based on CYP2C19. Mol Diagn Ther 2012;16 (4):223–234. [DOI] [PubMed] [Google Scholar]
- 8.Ichikawa H, Sugimoto M, Sugimoto K, Andoh A, Furuta T. Rapid metabolizer genotype of CYP2C19 is a risk factor of being refractory to proton pump inhibitor therapy for reflux esophagitis. J Gastroenterol Hepatol 2016;31(4):716–726. [DOI] [PubMed] [Google Scholar]
- 9.Tong S, Mallitt KA, Krishnan U. Evaluation of gastroesophageal reflux by combined multichannel intraluminal impedance and pH monitoring and esophageal motility patterns in children with esophageal atresia. Eur J Pediatr Surg 2016;26(4):322–331. [DOI] [PubMed] [Google Scholar]
- 10.Pedersen RN, Markøw S, Kruse-Andersen S, et al. Esophageal atresia: Gastroesophageal functional follow-up in 5–15-year-old children. J Pediatr Surg 2013;48(12):2487–2495. [DOI] [PubMed] [Google Scholar]
- 11.Tovar JA, Diez Pardo JA, Murcia J, Prieto G, Molina M, Polanco I. Ambulatory 24-hour manometric and pH metric evidence of permanent impairment of clearance capacity in patients with esophageal atresia. J Pediatr Surg 1995;30(8):1224–1231. [DOI] [PubMed] [Google Scholar]
- 12.Fröhlich T, Otto S, Weber P, et al. Combined esophageal multichannel intraluminal impedance and pH monitoring after repair of esophageal atresia. J Pediatr Gastroenterol Nutr 2008;47(4):443–449. [DOI] [PubMed] [Google Scholar]
- 13.Krishnamurthy P, Kaplan MH. STAT6 and PARP family members in the development of T cell-dependent allergic inflammation. Immune Netw 2016;16(4):201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park JY, Zhang X, Nguyen N, Souza RF, Spechler SJ, Cheng E. Proton pump inhibitors decrease eotaxin-3 expression in the proximal esophagus of children with esophageal eosinophilia. PLoS ONE 2014;9(7):e101391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mougey EB, Williams A, Coyne AJK, et al. CYP2C19 and STAT6 variants influence the outcome of proton pump inhibitor therapy in pediatric eosinophilic esophagitis. J Pediatr Gastroenterol Nutr 2019;69(5):581–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varni JW, Bendo CB, Denham J, et al. PedsQL Gastrointestinal Symptoms Module. J Pediatr Gastroenterol Nutr 2014;59(3):347–355. [DOI] [PubMed] [Google Scholar]
- 17.Krishnan U, Mousa H, Dall’Oglio L, et al. ESPGHAN-NASPGHAN guidelines for the evaluation and treatment of gastrointestinal and nutritional complications in children with esophageal atresia-tracheoesophageal fistula. J Pediatr Gastroenterol Nutr 2016;63(5):550–570. [DOI] [PubMed] [Google Scholar]
- 18.Armstrong D, Bennett JR, Blum AL, et al. The endoscopic assessment of esophagitis: a progress report on observer agreement. Gastroenterology 1996;111(1):85–92. [DOI] [PubMed] [Google Scholar]
- 19.Dellon ES, Liacouras CA, Molina-Infante J, et al. Updated International Consensus Diagnostic Criteria for Eosinophilic Esophagitis: Proceedings of the AGREE Conference. Gastroenterology 2018;155(4):1022–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mougey EB, Williams A, Coyne AJK, et al. CYP2C19 and STAT6 Variants influence the outcome of proton pump inhibitor therapy in pediatric eosinophilic esophagitis. J Pediatr Gastroenterol Nutr 2019;69(5):581–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graffelman J, Chang C, Puig X. Package “HardyWeinberg” Type Package Title Statistical Tests and Graphics for Hardy-Weinberg Equilibrium Published online 2019:Version 1.6.3. doi: 10.1126/science.28.706.49 [DOI]
- 22.El Rouby N, Lima JJ, Johnson JA. Proton pump inhibitors: from CYP2C19 pharmacogenetics to precision medicine. Expert Opin Drug Metab Toxicol 2018;14(4):447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varni JW, Limbers CA. The pediatric quality of life inventory: measuring pediatric health-related quality of life from the perspective of children and their parents. Pediatr Clin North Am 2009;56(4):843–863. [DOI] [PubMed] [Google Scholar]
- 24.Ntoumazios SK, Voulgari PV, Potsis K, Koutis E, Tsifetaki N, Assimakopoulos DA. Esophageal involvement in scleroderma: gastroesophageal reflux, the common problem. Semin Arthritis Rheum 2006;36(3):173–181. [DOI] [PubMed] [Google Scholar]
- 25.Zamost BJ, Hirschberg J, Ippoliti AF, Furst DE, Clements PJ, Weinstein WM. Esophagitis in scleroderma. Gastroenterology 1987;92(2):421–428. [DOI] [PubMed] [Google Scholar]
- 26.Nabi Z, Ramchandani M, Kotla R, et al. Gastroesophageal reflux disease after peroral endoscopic myotomy is unpredictable, but responsive to proton pump inhibitor therapy: a large, single-center study. Endoscopy 2020;52(08):643–651. [DOI] [PubMed] [Google Scholar]
- 27.Kumbhari V, Familiari P, Bjerregaard N, et al. Gastroesophageal reflux after peroral endoscopic myotomy: a multicenter case–control study. Endoscopy 2017;49(07):634–642. [DOI] [PubMed] [Google Scholar]
- 28.Franciosi JP, Mougey EB, Williams A, et al. Association between CYP2C19 extensive metabolizer phenotype and childhood anti-reflux surgery following failed proton pump inhibitor medication treatment. Eur J Pediatr 2018;177(1):69–77. [DOI] [PubMed] [Google Scholar]
- 29.Gupta M, Ghoshal UC, Jindal S, Misra A, Nath A, Saraswat VA. Respiratory dysfunction is common in patients with achalasia and improves after pneumatic dilation. Dig Dis Sci 2014;59(4):744–752. [DOI] [PubMed] [Google Scholar]
- 30.Bhattacharya B, Carlsten J, Sabo E, et al. Increased expression of eotaxin-3 distinguishes between eosinophilic esophagitis and gastroesophageal reflux disease. Hum Pathol 2007;38(12):1744–1753. [DOI] [PubMed] [Google Scholar]
- 31.Mishra A, Rothenberg ME. Intratracheal IL-13 induces eosinophilic esophagitis by an IL-5, eotaxin-1, and STAT6-dependent mechanism. Gastroenterology 2003;125(5):1419–1427. [DOI] [PubMed] [Google Scholar]
- 32.Niranjan R, Rayapudi M, Mishra A, Dutt P, Dynda S, Mishra A. Pathogenesis of allergen-induced eosinophilic esophagitis is independent of interleukin (IL)-13. Immunol Cell Biol 2013;91(6):408–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, Cheng E, Huo X, et al. Omeprazole blocks STAT6 binding to the Eotaxin-3 promoter in eosinophilic esophagitis cells. PLoS ONE 2012;7(11):e50037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosen R, Vandenplas Y, Singendonk M, et al. Pediatric Gastroesophageal Reflux Clinical Practice Guidelines: Joint Recommendations of the North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition and the European Society for Pediatric Gastroenterology, Hepatology, and Nutritio. J Pediatr Gastroenterol Nutr 2018;66(3):516–554. 10.1097/MPG.0000000000001889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown LF, Goldman H, Antonioli DA. Intraepithelial eosinophils in endoscopic biopsies of adults with reflux esophagitis. Am J Surg Pathol 1984;8(12):899–905. [DOI] [PubMed] [Google Scholar]
- 36.Frierson HF. Histology in the diagnosis of reflux esophagitis. Gastroenterol Clin North Am 1990;19(3):631–644. [PubMed] [Google Scholar]
- 37.Esposito S, Valente G, Zavallone A, Guidali P, Rapa A, Oderda G. Histological score for cells with irregular nuclear contours for the diagnosis of reflux esophagitis in children. Hum Pathol 2004;35(1):96–101. [DOI] [PubMed] [Google Scholar]
- 38.Yerian L, Fiocca R, Mastracci L, et al. Refinement and reproducibility of histologic criteria for the assessment of microscopic lesions in patients with gastroesophageal reflux disease: The esohisto project. Dig Dis Sci 2011;56(9):2656–2665. [DOI] [PubMed] [Google Scholar]
- 39.Kawahara H, Kubota A, Hasegawa T, et al. Lack of distal esophageal contractions is a key determinant of gastroesophageal reflux disease after repair of esophageal atresia. J Pediatr Surg 2007;42(12):2017–2021. [DOI] [PubMed] [Google Scholar]
- 40.Lemoine C, Aspirot A, Le Henaff G, Piloquet H, Lévesque D, Faure C. Characterization of esophageal motility following esophageal atresia repair using high-resolution esophageal manometry. J Pediatr Gastroenterol Nutr 2013;56(6):609–614. [DOI] [PubMed] [Google Scholar]
- 41.Courbette O, Omari T, Aspirot A, Faure C. Characterization of Esophageal Motility in Children with Operated Esophageal Atresia Using High-Resolution Impedance Manometry and Pressure Flow Analysis. J Pediatr Gastroenterol Nutr Published online June 11, 2020. 10.1097/MPG.0000000000002806 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data not available due to ethical restrictions. Participants of this study did not agree for their data to be shared publicly, so supporting data are not available.


