Abstract
Background:
The number of somatic mutations detectable in circulating tumor DNA (ctDNA) is highly heterogeneous in metastatic colorectal cancer (mCRC). The optimal number of mutations required to assess disease kinetics is relevant and remains poorly understood.
Objectives:
To determine whether increasing panel breadth (the number of tracked variants in a ctDNA assay) would alter the sensitivity in detecting ctDNA in patients with mCRC.
Design:
We used archival tissue sequencing to perform an in silico assessment of the optimal number of tracked mutations to detect and monitor disease kinetics in mCRC using sequencing data from the Canadian Cancer Trials Group CO.26 trial.
Methods:
For each patient, 1, 2, 4, 8, 12, or 16 of the most clonal (highest variant allele frequency) somatic variants were selected from archival tissue-based whole-exome sequencing and assessed for the proportion of variants detected in matched ctDNA at baseline, week 8, and progression timepoints.
Results:
Data from 110 patients were analyzed. Genes most frequently encountered among the top four highest VAF variants in archival tissue were TP53 (51.9% of patients), APC (43.3%), KRAS (42.3%), and SMAD4 (9.6%). While the frequency of detecting at least one tracked variant increased when expanding beyond variant pool sizes of 1 and 2 in baseline (p = 0.0030) and progression (p = 0.0030) ctDNA samples, we observed no significant benefit to increases in variant pool size past four variants in any of the ctDNA timepoints (p < 0.05).
Conclusion:
While increasing panel breadth beyond two tracked variants improved variant re-detection in ctDNA samples from patients with treatment refractory mCRC, increases beyond four tracked variants yielded no significant improvement in variant re-detection.
Keywords: circulating tumor DNA, colorectal cancer, ctDNA panel breadth, number of variants tracked, monitoring tumor burden
Background
An increasing number of studies have demonstrated the utility of circulating tumor DNA (ctDNA) in the care of patients with colorectal cancer (CRC). However, there exists a high degree of variability in methodology and genomic coverage (both depth and breadth) of assays.1–3 Determining the number of tracked variants required for ctDNA panels remains an unmet need.
In the context of following disease kinetics or detecting minimal residual disease (MRD), most assays use a mixture of detecting single nucleotide variants (SNVs) with or without concurrent analytes, such as methylation. A subset of ctDNA assays aimed at tracking MRD use a tumor-informed approach, in which mutations identified through tumor sequencing are used to design a ctDNA panel that is tailored to an individual’s tumor. Assays from different providers have a differing number of SNVs that are followed in the blood, although the extent by which sensitivity is increased as a result of incorporating a larger number of variants on a monitoring assay remains unclear. A small number of variants may be sufficient to design a personalized tumor biomarker to follow disease kinetics in the context of metastatic disease, where ctDNA concentration is generally high. 4
Previous studies have revealed that a subset of genes are recurrent targets of somatic mutation in CRC. The concept of the ‘adenoma-carcinoma’ pathway, first described in 1990 by Fearon and Vogelstein, 5 states that the majority of non-hypermutated colorectal tumors exhibit a stepwise acquisition of mutations in APC, TP53, KRAS, or PIK3CA.6–8 From this, we hypothesized that there exists a threshold beyond which including additional variants to a monitoring assay for metastatic CRC provides little improvement in performance due to the large number of early truncal events. We performed an in silico analysis to determine whether the number of variants chosen from tissue-based whole-exome sequencing (WES) would alter the sensitivity in detecting mutations in subsequent ctDNA timepoints from samples that were available as part of the Canadian Cancer Trials Group CO.26 trial (NCT02870920).
Methods
Trial design and patient enrollment
This study is a correlative study from the CO.26 trial, a phase II trial that randomized 180 patients with refractory mCRC 2:1 to either durvalumab 1500 mg intravenously every 4 weeks + tremelimumab 75 mg intravenously every 4 weeks for the initial four cycles + best supportive care (BSC) or BSC alone. Eligible patients had refractory metastatic CRC and received all available standard systemic therapies. Full study details and the study protocol were previously published. 9 All participants provided written informed consent and the study was approved by the institutional review board of each participating center.
Data from patients who received both tissue-based WES and subsequent ctDNA sequencing were used for analysis. Tissue-based sequencing (hereafter referred to as ‘archival’) was performed using archival tissue from diagnostic biopsies or surgical resection specimens, while ctDNA timepoints were defined relative to when patients received treatment as part of the CO.26 trial of tremelimumab and durvalumab versus BSC. ctDNA sampling timepoints included the following: prior to the start of trial treatment (baseline), 8 week post-treatment initiation (week 8), and upon disease progression (progression). For each patient, we selected 1, 2, 4, 8, 12, and 16 of the most clonal [highest variant allele frequency (VAF)] somatic variants from the archival tissue and assessed the proportion of such variants detected at each patient’s matched ctDNA timepoint.
Tissue and ctDNA sequencing
Archival formalin fixed and paraffin-embedded (FFPE) tumor tissue and leukocytes from peripheral blood were utilized for tumor/matched normal WES. Tissue was sequenced with an average coverage before de-duplication of 100× for normal and 400× for tumor samples. For ctDNA sequencing, plasma was isolated from blood collected in Streck tubes prior to treatment on the CO.26 trial (baseline), 8 weeks after initiation of therapy (week 8), and on disease progression (progression). DNA extraction and next-generation sequencing were subsequently performed at Guardant Health using the GuardantOMNI 2.15 Mb, 500 gene panel, which has been previously validated and demonstrated 98.7% accuracy to detect SNVs with 95% limits of detection of 0.24–0.6% VAF. 10
Tissue-based variant calling
Somatic variants were called on tumor-normal pairs through a combination of Manta v1.5.0 and Strelka v2.9.10 (Illumina Inc., CA, USA), using default parameters and genome build GRCh37/hg19. 11 Variants were annotated using SnpEff v4.3 with parameters -v GRCh37.75 -canon -no-downstream -no-upstream -noLog -noStats -no-intergenic. 12 VCF files were converted to MAF format using vcf2maf v1.6.18 with default parameters. Variants were filtered to exclude those located outside of exons (using consensus exon regions GRCh37.p13; GCF_000001405.25, downloaded 19 June 2020) and any common variants identified by ExAC (ExAC_nonTCGA.r0.3.1.sites.vep.vcf.gz). 13 Variants were further filtered based on established guidelines, including VAF ⩾ 0.05, tumor depth ⩾ 25, and alternate allele count ⩾ 3.14,15 Only missense, nonsense, and in-frame/frameshift variants were included for analysis. Relative variant allele fraction (rVAF) was calculated as the allele frequency of a variant divided by the maximum detected allele frequency of any somatic variant in the sample. Variants were filtered for those included in the GuardantOMNI ctDNA assay (n = 500 genes). Tumor content values for FFPE samples were calculated using Facets v0.6.0 with default parameters. 16
Variant detectability analysis
For each sample, archival tissue variants were ordered by decreasing rVAF and the top x variants were chosen for ctDNA tracking, where iterations of x = {1, 2, 4, 8, 12, 16}. If a sample had less than x archival tissue variants to choose from, it was omitted from that iteration. For each sample, presence/absence of the x variants was then assessed in each ctDNA timepoint (baseline, 8 week, and progression) available for that sample.
Statistics
One-way ANOVA tests were used to assess differences in a continuous variable across discrete groupings. Spearman correlation tests were used to assess differences between two continuous variables. Fisher’s exact tests were used to assess differences in frequency across discrete groupings. All p values were subjected to Benjamini–Hochberg multiple test correction. Analyses were performed using R v3.6.3 (Posit Software, MA, USA).
Results
Clonal variants in TP53, APC, and KRAS are highly recurrent in archival tissue
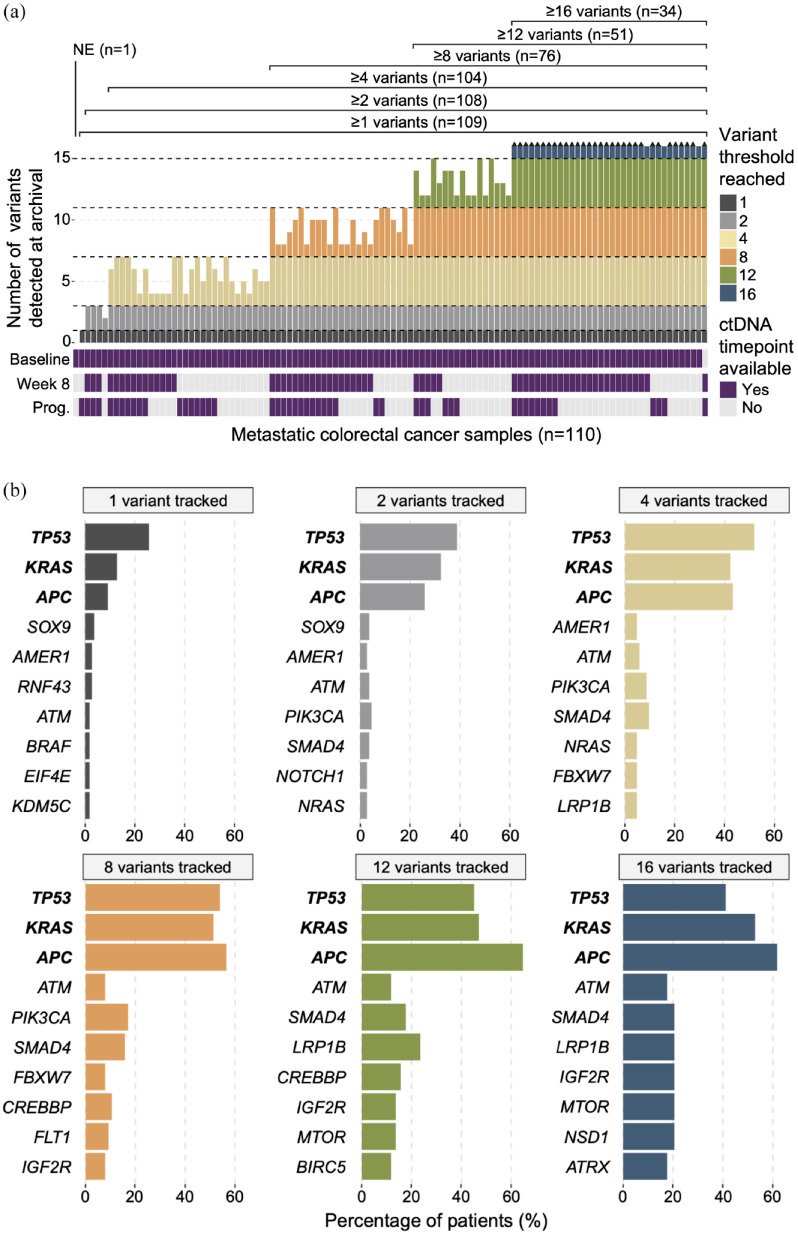
To assess the detectability of clonal mutations across ctDNA timepoints, data from 110 patients that received both tissue-based WES (hereafter referred to as ‘archival’) and at least one of three subsequent ctDNA sequencing timepoints (baseline, week 8, and progression) were selected for bioinformatic analysis. Somatic variant calling was applied to each archival patient sample, and variants were filtered for those affecting genes included in the Guardant OMNI ctDNA panel (n = 500 genes). Samples were grouped according to whether they had at least 1 (n = 109 patients), 2 (n = 108), 4 (n = 104), 8 (n = 76), 12 (n = 51), or 16 (n = 34) archival tissue variants. 17 One patient had no mutation detected and was therefore non-evaluable (NE) and excluded from downstream analysis (Figure 1(a)). For each variant pool, samples meeting the variant number threshold had their top 1, 2, 4, 8, 12, or 16 most clonal VAF variants selected for downstream analysis. We then assessed the frequency at which specific genes were affected by the top clonal variants across the variant pool groupings. For the 1, 2, 4, 8, 12, and 16 variant pool groups, TP53 (25.7%, 38.9%, 51.9%, 53.9%, 45.1%, and 41.2%, respectively), APC (9.2%, 25.9%, 43.3%, 56.6%, 64.7%, and 61.8%, respectively), and KRAS (12.8%, 32.4%, 42.3%, 51.3%, 47.1%, and 52.9%, respectively) were consistently the top three most frequent clonal events across patients (Figure 1(b)). Across all evaluable archival tissue samples combined (n = 109), the distribution of relative VAF (rVAF) levels for variants affecting these genes were as follows: TP53: min. 9.1%, max 100%, median 68.0%; APC: min. 7.4%, max 100%, median 41.6%; and KRAS: min. 19.3%, max 100%, median 54.2%. Mutation in TP53, KRAS, or APC was found in 88.1% of archival tissue samples, and 79.8% had at least one of KRAS, TP53, or APC mutations among their top four top clonal variants. These results indicate a recurrent pattern in clonal mutation events across patients, aligning to the notion that CRC development involves stepwise, truncal mutation acquisition in a small subset of key driver genes. 5
Figure 1.
Top clonal variants show recurrent patterns across mCRC samples. (a) Upper bar plot depicts the number of variants detected in each archival tissue sample, for variants affecting genes assayed by the subsequent ctDNA panel (n = 500 genes). Black arrowheads above bars indicate samples with more than 16 archival variants detected. Lower heatmap shows availability of subsequent ctDNA timepoints for each sample. (b) Bar plots illustrating genes most frequently affected by the top 1, 2, 4, 8, 12, and 16 variant pools (left to right) across all samples.
Archival tissue variant pool size does not impact downstream ctDNA sensitivity
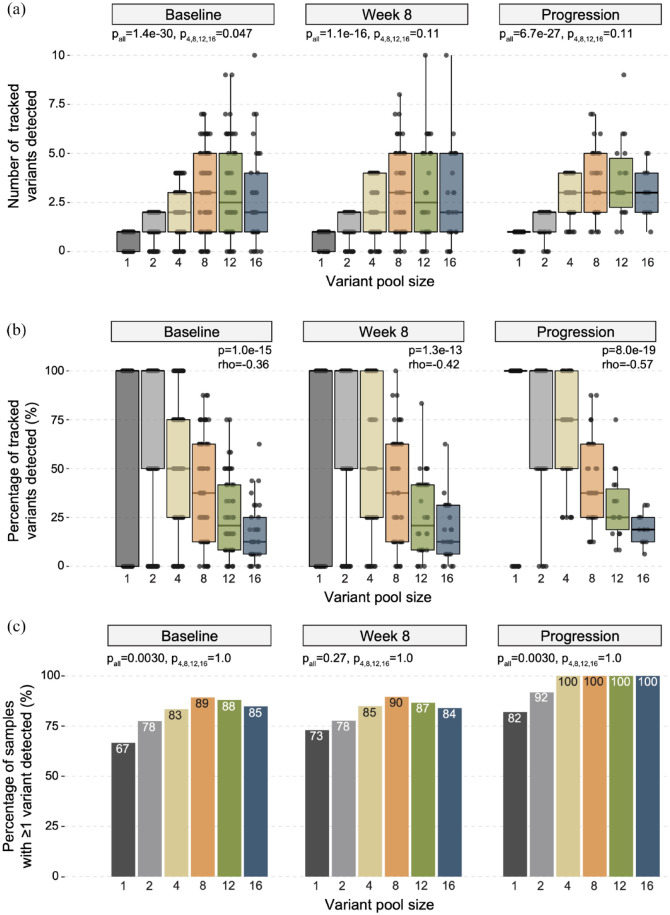
We next assessed the number of variants detected (VAF > 0) at subsequent ctDNA timepoints for each sample, for each of the variant pool groupings (n = 1, 2, 4, 8, 12, and 16 variants tracked) defined in the archival tissue dataset. In this analysis, both genomic coordinate and nucleotide alteration were used to define each variant, such that the exact same variant location and type was required to be considered ‘re-detected’ at a later ctDNA timepoint. In baseline ctDNA samples, the number of re-detected variants varied across all variant pool sizes, with a median of 1.0 (n = 108 patient samples), 2.0 (n = 107), 2.0 (n = 103), 3.0 (n = 75), 2.5 (n = 50), and 2.0 (n = 33) archival variants re-detected for variant pool sizes 1, 2, 4, 8, 12, and 16 (respectively; pall ⩽ 0.0001), though the impact of increasing variant pool size on the number of re-detected variants was diminished when considering only variant pool sizes of four or greater (p4,8,12,16 = 0.047; Figure 2(a)). Similarly, the median number of re-detected variants also varied across all variant pool sizes in week 8 [median 1.0 (n = 63), 2.0 (n = 63), 2.0 (n = 60), 3.0 (n = 48), 2.5 (n = 30), and 2.0 (n = 25) re-detected variants for variant pool sizes 1, 2, 4, 8, 12, and 16, respectively; pall < 0.0001] and progression [median 1.0 (n = 50), 2.0 (n = 49), 3.0 (n = 46), 3.0 (n = 32), 3.0 (n = 18), and 3.0 (n = 12) re-detected variants for variant pool sizes 1, 2, 4, 8, 12, and 16, respectively; pall < 0.0001] ctDNA timepoints. For both week 8 and progression ctDNA timepoints, the impact of variant pool size on variant re-detection was no longer significant when only variant pool sizes greater than or equal to 4 were considered (p4,8,12,16 = 0.11 and 0.11, respectively), highlighting a pattern of diminishing returns, and thereby limited benefit, to increasing panel size past four tracked variants.
Figure 2.
Variant re-detection across increasing tracked variant pool sizes in each ctDNA timepoint. (a) Boxplots indicating the number of variants re-detected in ctDNA data when variant pool sizes 1, 2, 4, 8, 12, and 16 were used in baseline, week 8, and progression timepoints (left to right). Box-and-whiskers indicate median, 25 and 75% interquartile ranges are shown, along with one-way ANOVA p values for all variant pool sizes (pall) and variant pool sizes greater than or equal to 4 (p4,8,12,16). (b) Boxplots depicting the percentage of tracked variants re-detected across variant pool sizes for each ctDNA timepoint. Box-and-whiskers indicate median, 25 and 75% interquartile ranges and Spearman correlation p values and correlation coefficients (rho) are shown. (c) Bar plots showing the percentage of samples with at least one tracked variant re-detected for each variant pool size at each ctDNA timepoint, with percentage values shown at the top of each bar. Fisher’s exact test p values are shown for all variant pool sizes (pall) and variant pool sizes greater than or equal to 4 (p4,8,12,16).
The percentage of variants successfully re-detected was negatively correlated with variant pool size at each ctDNA timepoint, with a median of 100%, 100%, 50.0%, 37.5%, 20.8%, and 12.5% of variants re-detected for variant pool sizes 1, 2, 4, 8, 12, and 16 (respectively) in baseline ctDNA (p < 0.0001, rho = −0.36), 100%, 100%, 50.0%, 37.5%, 20.8%, and 12.5% of variants re-detected in week 8 ctDNA (p < 0.0001, rho = −0.42) and 100%, 100%, 75.0%, 37.5%, 25.0%, and 18.8% of variants re-detected in progression ctDNA (p < 0.0001, rho = −0.57; Figure 2(b)). We next assessed the degree of correlation between archival sample tumor content and the percentage of variants detected across baseline, week 8, and progression timepoints. While archival sample tumor content was correlated with variant re-detection for variant pool size of 2 (p = 0.005, rho = 0.19), such correlation was not observed for variant pool sizes 1 (p = 0.24, rho = 0.08), 4 (p = 0.11, rho = 0.11), 8 (p = 0.90, rho = −0.01), 12 (p = 0.16, rho = −0.14) nor 16 (p = 0.75, rho = 0.04), indicating that tumor content was unlikely to be a confounding factor in our analysis.
In the context of MRD, re-detection of at least one tracked variant may be considered sufficient evidence for disease persistence or recurrence.2,18 To investigate the relationship between single variant re-detection sensitivity and variant pool size, we calculated the percentage of samples with at least one tracked variant re-detected for each variant pool size at each ctDNA timepoint (Figure 2(c)). When considering all variant pool sizes, sensitivity was significantly variable for baseline (66.7%, 77.6%, 83.5%, 89.3%, 88.0%, and 84.8% of samples having at least one variant detected for variant pool sizes of 1, 2, 4, 8, 12, and 16, respectively; pall = 0.003) and progression (82.0%, 91.8%, 100%, 100%, 100%, and 100% of samples having at least one variant detected for variant pool sizes of 1, 2, 4, 8, 12, and 16, respectively; pall = 0.003) samples. Sensitivity was not significantly variable across all variant pool sizes in week 8 samples (73.0%, 77.8%, 85.0%, 89.6%, 86.7%, and 84.0% of samples having at least one variant detected for variant pool sizes of 1, 2, 4, 8, 12, and 16, respectively; pall = 0.27). Meanwhile, when only variant pool sizes greater than or equal to 4 were considered, differences in sensitivity across variant pool sizes were not significant for baseline (p4,8,12,16 = 1.0), week 8 (p4,8,12,16 = 1.0) nor progression (p4,8,12,16 = 1.0) timepoints, further indicating a lack of benefit when expanding panel sizes beyond four tracked variants.
Discussion
Our results provide insight into the use of ctDNA for monitoring disease response in metastatic CRC, focusing on the impact of breadth of genomic coverage on assay sensitivity. Using ctDNA as a personalized dynamic biomarker could offer several advantages over serum carcinoembryonic antigen measurements and radiographic imaging for disease monitoring. For patients with refractory metastatic CRC in particular, early response assessment would identify patients who truly benefit from targeted therapies or from agents with otherwise limited benefit and potential toxicities.19,20 Currently, the use of ctDNA in this setting is limited by cost, turnaround time, and lack of standardization, among others. 2
In this study, we found increased assay performance when tracking four or more variants, but modulation of panel breadth beyond four tracked variants had insignificant impact on clonal variant detection and did not yield meaningful increases in the number of detected variants nor the percentage of samples with detected ctDNA in patients with treatment refractory metastatic CRC. Our results are a proof of concept that assays used in the metastatic setting for tracking response may only require a limited number of tracked variants, potentially reducing assay cost and complexity to facilitate routine clinical use.
Our findings also raise the question whether tracking additional variants beyond truncal mutations would improve sensitivity for MRD detection, where the stochastic distribution of ctDNA remains a significant barrier to overcome. Many assays in development are using an increasingly large number of followed variants as a means to improve assay performance. With recent advances in sequencing technologies allowing for ultra-deep sequencing, the limit of detection of an assay is now determined by the plasma DNA content of a standard 10 mL blood sample, which contains approximately 10,000–12,000 DNA molecules, setting the theoretical limit of ctDNA detection at a VAF of approximately 0.01%. 2 At such small ctDNA concentration, mathematical models predict decreased sensitivity below 16 variants tracked with little improvement beyond 25 variants,17,21 although this has not been clinically validated. In our study, we showed that the percentage of samples with at least one re-detected variant decreased when tracking only one or two variants. We also observed a slightly smaller number of variants detected of borderline statistical significance in the four-variant pool at baseline but not at week 8 or at progression. This supports the notion that increasing the number of tracked variants is more beneficial in earlier stage and less beneficial as disease becomes more advanced, and the minimal number of tracked variants required to re-detect at least one variant likely depends on the relative circulating tumor fraction.
In this study, we used archival tissue sequencing to select variants incorporated into in silico ctDNA panels. Tissue-informed assays require a longer turnaround time and delays can be a barrier to clinical implementation both in the adjuvant and metastatic setting.2,22 We found a high proportion of tumor and ctDNA samples had mutations in APC, TP53, or KRAS, which is consistent with previous reports.6–8,23,24 Given this relative homogeneity in driver alterations in CRC, tissue-informed assays may not be required, especially in the metastatic setting.
Our study has limitations that need to be considered when interpreting the results. This is a retrospective, in silico analysis with inherent bias associated with this methodology. ctDNA timepoints were chosen relative to the start of treatment in CO.26, but time between the archival tissue sampling, baseline ctDNA sampling, and progression ctDNA sampling is variable. Similarly, not all patients received the same targeted therapies (38% were previously treated with anti-EGFR, 79% with bevacizumab, and 26% with regorafenib in CO.26), 9 all of which may have altered the mutational landscape differently. Most importantly, our findings involve patients with late-stage refractory metastatic CRC and results may not apply to the MRD setting where ctDNA currently is under more intensive investigation than following response to therapy in metastatic disease. Extrapolations to the MRD setting should be regarded as exploratory or hypothesis generating only. Our findings also may not apply to a hypermutator phenotype. Our primary hypothesis was based on the model by Fearon and Vogelstein 5 ; however, hypermutators may display such tumor heterogeneity that founder or truncal mutations sometimes cannot be identified.25,26 Panels with limited genomic coverage may not be sufficient to reliably track disease kinetics in this setting. Finally, the current analysis explores the impact of genomic coverage for monitoring disease burden based on tracking truncal mutations known to persist throughout a patient’s course, this study was not designed to address resistance mechanisms or potential treatment targets. Incorporating known resistance mutations into an assay is technically feasible but adds to complexity and cost, such assays are currently less suited for routine disease monitoring.
In conclusion, our study is compatible with the notion that ctDNA panels with a relatively limited number of genes are sufficient for serial variant detection and tracking in metastatic CRC, potentially improving the feasibility of clinical implementation by decreasing the cost and complexity.
Acknowledgments
We gratefully acknowledge the participation of patients and their families.
Footnotes
ORCID iDs: Mélina Boutin  https://orcid.org/0000-0002-0799-1551
https://orcid.org/0000-0002-0799-1551
Anthony Lott  https://orcid.org/0000-0003-3563-3142
https://orcid.org/0000-0003-3563-3142
Contributor Information
Mélina Boutin, Division of Medical Oncology, BC Cancer, Vancouver, BC, Canada Centre Intégré de Cancérologie de la Montérégie, Université de Sherbrooke, QC, Canada.
James T. Topham, Pancreas Center BC, Vancouver, BC, Canada.
Harriet Feilotter, Canadian Cancer Trials Group, Queen’s University, Kingston, ON, Canada.
Hagen F. Kennecke, Providence Cancer Institute, Portland, OR, USA
Félix Couture, CHU de Québec-Université Laval, QC, Canada.
Mohammed Harb, Moncton Hospital, Moncton, NB, Canada.
Peter Kavan, Segal Cancer Centre, Montreal, QC, Canada.
Scott Berry, Department of Oncology, Queen’s University, Kingston, ON, Canada.
Howard J. Lim, Division of Medical Oncology, BC Cancer, Vancouver, BC, Canada
John R. Goffin, Juravinski Cancer Centre, Hamilton, ON, Canada
Chaudhary Ahmad, Dr. H. Bliss Murphy Cancer Centre, St. John’s, NL, Canada.
Anthony Lott, Sunnybrook Health Sciences, Toronto, ON, Canada.
Daniel J. Renouf, Division of Medical Oncology, BC Cancer, Vancouver, BC, Canada Pancreas Center BC, Vancouver, BC, Canada
Derek J. Jonker, The Ottawa Hospital, University of Ottawa, Ottawa, ON, Canada
Dongsheng Tu, Canadian Cancer Trials Group, Queen’s University, Kingston, ON, Canada.
Chris J. O’Callaghan, Canadian Cancer Trials Group, Queen’s University, Kingston, ON, Canada
Eric X. Chen, Princess Margaret Cancer Centre, Toronto, ON, Canada
Jonathan M. Loree, Division of Medical Oncology, BC Cancer, 600 West 10th Avenue, Vancouver, BC V5Z 4E6, Canada.
Declarations
Ethics approval and consent to participate: This study is a correlative study from the CO.26 trial (NCT02870920). The study was approved by the institutional review board of each participating center and conducted according to the principles of the Declaration of Helsinki. All patients provided written informed consent.
Consent for publication: Not applicable.
Author contribution(s): Mélina Boutin: Data curation; Writing – original draft; Writing – review & editing.
James T. Topham: Data curation; Software; Validation; Writing – original draft; Writing – review & editing.
Harriet Feilotter: Data curation; Writing – review & editing.
Hagen F. Kennecke: Data curation; Writing – review & editing.
Félix Couture: Data curation; Writing – review & editing.
Mohammed Harb: Data curation; Writing – review & editing.
Peter Kavan: Data curation; Writing – review & editing.
Scott Berry: Data curation; Writing – review & editing.
Howard J. Lim: Data curation; Writing – review & editing.
John R. Goffin: Data curation; Writing – review & editing.
Chaudhary Ahmad: Data curation; Writing – review & editing.
Anthony Lott: Data curation; Writing – review & editing.
Daniel J. Renouf: Data curation; Writing – review & editing.
Derek J. Jonker: Data curation; Writing – review & editing.
Dongsheng Tu: Conceptualization; Data curation; Methodology; Writing – review & editing.
Chris J. O’Callaghan: Conceptualization; Data curation; Methodology; Writing – review & editing.
Eric X. Chen: Data curation; Writing – review & editing.
Jonathan M. Loree: Conceptualization; Data curation; Funding acquisition; Methodology; Project administration; Writing – original draft; Writing – review & editing.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This trial was sponsored by the Canadian Cancer Trials Group (CCTG), which is funded by the Canadian Cancer Society. Funding for the trial conduct and experimental drugs were provided by Astra Zeneca. Dr. Jonathan Loree and Dr. Daniel Renouf have received Michael Smith Health Professional Investigator Awards. Funding from BC Cancer Foundation helped support this analysis.
The authors declare that there is no conflict of interest.
Availability of data and materials: The Canadian Cancer Trials Group (CCTG) has an established request procedure and interested investigators should submit a brief proposal using the Request for Data Proposal Form available at http://www.ctg.queensu.ca/. Upon approval, de-identified individual participant data and variable dictionary will be made available.
References
- 1.Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American society of clinical oncology and college of American pathologists joint review. J Clin Oncol 2018; 36: 1631–1641. [DOI] [PubMed] [Google Scholar]
- 2.Dasari A, Morris VK, Allegra CJ, et al. CtDNA applications and integration in colorectal cancer: an NCI Colon and Rectal–Anal Task Forces whitepaper. Nat Rev Clin Oncol 2020; 17: 757-–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol 2015; 26: 1715–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arisi MF, Dotan E, Fernandez SV. Circulating tumor DNA in precision oncology and its applications in colorectal cancer. Int J Mol Sci 2022; 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell 1990; 61: 759–767. [DOI] [PubMed] [Google Scholar]
- 6.Lilleberg SL, Durocher J, Sanders C, et al. High sensitivity scanning of colorectal tumors and matched plasma DNA for mutations in APC, TP53, K-RAS, and BRAF genes with a novel DHPLC fluorescence detection platform. Ann N Y Acad Sci 2004; 1022: 250–256. [DOI] [PubMed] [Google Scholar]
- 7.Muzny DM, Bainbridge MN, Chang K, et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buikhuisen JY, Torang A, Medema JP. Exploring and modelling colon cancer inter-tumour heterogeneity: opportunities and challenges. Oncogenesis. 2020; 9: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen EX, Jonker DJ, Kennecke HF, et al. CCTG CO.26 trial: A phase II randomized study of durvalumab (D) plus tremelimumab (T) and best supportive care (BSC) versus BSC alone in patients (pts) with advanced refractory colorectal carcinoma (rCRC). J Clin Oncol 2019; 37(4_suppl): 481.30620669 [Google Scholar]
- 10.Helman E, Artieri C, Vowles JV, et al. Analytical validation of a comprehensive 500-gene ctDNA panel designed for immuno-oncology and DNA damage research. Cancer Res 2018; 78(13_Suppl.): 5603. [Google Scholar]
- 11.Kim S, Scheffler K, Halpern AL, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods. 2018; 15: 591–594. [DOI] [PubMed] [Google Scholar]
- 12.Cingolani P, Platts A, Wang LL, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 2012; 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellrott K, Bailey MH, Saksena G, et al. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst 2018; 6: 271–281.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merino DM, McShane LM, Fabrizio D, et al. Establishing guidelines to harmonize tumor mutational burden (TMB): in silico assessment of variation in TMB quantification across diagnostic platforms: phase I of the Friends of Cancer Research TMB Harmonization Project. J Immunother Cancer 2020; 8: e000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 2016; 44: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Natera Inc. Natera Website, https://www.natera.com/wp-content/uploads/2020/11/Oncology-Clinical-A-personalized-tumor-informed-approach-to-detect-molecular-residual-disease-SGN_SR_WP.pdf, Natera In (accessed 6 January 2023).
- 18.Solar Vasconcelos JP, Boutin M, Loree JM. Circulating tumor DNA in early-stage colon cancer: ready for prime time or needing refinement? Ther Adv Med Oncol 2022; 14: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong ALA, Lim JSJ, Sinha A, et al. Tumour pharmacodynamics and circulating cell free DNA in patients with refractory colorectal carcinoma treated with regorafenib. J Transl Med 2015; 13: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandeputte C, Kehagias P, El Housni H, et al. Circulating tumor DNA in early response assessment and monitoring of advanced colorectal cancer treated with a multikinase inhibitor. Oncotarget 2018; 9: 17756–17769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avanzini S, Kurtz DM, Chabon JJ, et al. A mathematical model of ctDNA shedding predicts tumor detection size. Sci Adv 2020; 6: 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao P, zhang Huang X, xi Song Y, et al. Impact of timing of adjuvant chemotherapy on survival in stage III colon cancer: a population-based study. BMC Cancer 2018; 18: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burns MC, Carroll K, Jones R, et al. Molecular profiling of early-stage colorectal cancer (CRC) using targeted next generation sequencing (NGS) to predict signatures of recurrence. J Clin Oncol 2020;38(4_suppl): 242. [Google Scholar]
- 24.Ye J, Lin M, Zhang C, et al. Tissue gene mutation profiles in patients with colorectal cancer and their clinical implications. Biomed Rep 2020; 13: 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaegashi M, Iwaya T, Sasaki N, et al. Frequent post-operative monitoring of colorectal cancer using individualised ctDNA validated by multiregional molecular profiling. Br J Cancer 2021; 124: 1556–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaidi SH, Harrison TA, Phipps AI, et al. Landscape of somatic single nucleotide variants and indels in colorectal cancer and impact on survival. Nat Commun 2020; 11: 3644. [DOI] [PMC free article] [PubMed] [Google Scholar]