Highlights
-
•
12 interleukin genes were identified from Takifugu obscurus.
-
•
Most ToIL genes were broadly expressed in all the examined tissues.
-
•
12 ToIL genes increased significantly following bacterial infection.
-
•
12 ToILs were involved in antibacterial immune response of T. obscurus.
Keywords: Interleukins, Expression pattern analysis, Antibacterial immunity, Takifugu obscurus
Abstract
Interleukins (ILs) are a subgroup of secreted cytokines, which are molecules involved in the intercellular regulation of the immune system. In this study, 12 IL homologs were cloned and functionally identified from obscure puffer Takifugu obscurus, and they were termed as ToIL-1β, ToIL-1, ToIL-6, ToIL-10, ToIL-11, ToIL-12, ToIL-17, ToIL-18, ToIL-20, ToIL-24, ToIL-27, and ToIL-34. Multiple alignment results showed that except for ToIL-24 and ToIL-27, other deduced ToIL proteins shared typical characteristics and structure with other known fish ILs. Phylogenetic analysis revealed that 12 ToILs were evolutionarily closely related to their counterparts in other selected vertebrates. Tissue distribution assay demonstrated that the mRNA transcripts of most ToIL genes were constitutively expressed in all tissues examined, with relatively high expression in immune tissues. Following Vibrio harveyi and Staphylococcus aureus infection, the expression levels of 12 ToILs in the spleen and liver were significantly upregulated, and their response over time varied. Taken together, these data were discussed accordingly with the ToIL expression and the immune response under the different situations tested. The results suggest that the 12 ToIL genes are involved in the antibacterial immune response in T. obscurus.
1. Introduction
Cytokines are small glycoproteins that act as major regulators of innate and adaptive immunity that enable cells of the immune system to communicate over short distances [1]. Interleukins (ILs) are a subgroup of secreted cytokines, which are molecules involved in the intercellular regulation of the immune system. The term interleukin was first coined in 1979 to refer to molecules that signal between different leucocyte types, although this communication is not exclusively restricted to leucocyte communication [2]. To date, dozens of ILs are produced by various cell types, and many of them are synthesized by immune cells such as T lymphocytes, B lymphocytes, and macrophages/monocytes cells [3,4]. Most ILs are conserved from fish to mammals and play essential roles in physiological inflammatory and immunological processes [5,6].
Fish comprises the first group in the animal evolution that possess both innate and adaptive immunity [7]. In the past two decades, considerable progress has been made in the cloning, sequencing, and functional analysis of fish IL genes [5]. In fish, the first IL-1β cDNA was identified from rainbow trout Oncorhynchus mykiss by cDNA library screening [8]. Subsequently, IL-1βs have been found in several teleost and cartilaginous fish species, such as catshark Scyliorhinus canicula [9], channel catfish Ictalurus punctatus [10], and Nile tilapia Oreochromis niloticus [11], and fish IL-1β proteins are involved in regulating the inflammatory response to bacterial or parasitic infection [12]. In comparison with IL-1β, fish IL-18 s are under-investigated and have been reported only in a few fish species, including rainbow trout [13], seabream Sparus aurata [14], turbot Scophthalmus maximus [15], and elephant shark Callorhinchus milii [16]. Studies have suggested that IL-18 may play a role in mucosal immunity [14]. In teleosts such as Japanese pufferfish Fugu rubripes [17], gilthead seabream S. aurata [18], zebrafish Danio rerio [19], and Siberian sturgeon Acipenser baeri [20], IL-6 s play vital roles in inflammatory response, driving lymphocyte differentiation and the induction of antimicrobial peptides. IL-10 s have been identified in model fish, such as zebrafish D. rerio [21], and economically important species, such as grass carp Ctenopharyngodon idella [22], common carp Cyprinus carpio [23], and golden pompano Trachinotus ovatus [24]. IL-10 in common carp exerts prototypical anti-inflammatory activities on macrophages and neutrophils; it down regulates oxygen and nitrogen radical production and cytokine synthesis by inhibiting pro-inflammatory gene expression [23]. IL-17 family genes play crucial roles in fish immune responses by stimulating the release of immune factors in different ways. In large yellow croaker Larimichthys crocea, IL-17C can strongly stimulate the expression of chemokines, pro-inflammatory factors, and antibacterial peptide and may promote the inflammatory response and host defense by activating the NF-κB signaling [25]. The members of IL-20 subfamily have been described in green pufferfish Tetraodon nigroviridis, zebrafish, and rainbow trout and play important roles in homeostasis, cell proliferation, and host immune defenses [26]. IL-34 in rainbow trout [27], large yellow croaker [28], and grouper Epinephelus coioides [29] responds to stimulation by pathogens and certain pathogen-associated molecular pattern molecules. Grass carp IL-34 can induce the expression of pro-inflammatory cytokines in primary head kidney macrophages [30]. The increasing knowledge based on the regulation of immune responses in fish by ILs and their potential use as good immunopotentiator candidate for disease prevention or treatment in fish make fish IL biology an attractive and rapidly expanding field.
Takifugu obscurus, commonly known as obscure puffer, is an anadromous species that migrates to freshwater from the sea for spawning [31]. In recent years, the immune system of T. obscurus has attracted widespread attention because of its important evolutionary status and great economic value. T. obscurus is among the useful models for studying the immune system of fish. Thus, the systematic study of different types of cytokines in T. obscurus is expected to lay the foundation for in-depth studies of the regulatory mechanism of fish immunity. In the present study, the structural and functional features of ILs in T. obscurus were investigated. Based on the data from a previously constructed transcriptome, 12 IL genes were identified from T. obscurus, namely, ToIL-1β, ToIL-1, ToIL-6, ToIL-10, ToIL-11, ToIL-12, ToIL-17, ToIL-18, ToIL-20, ToIL-24, ToIL-27, and ToIL-34. The sequence alignment, phylogenetic relationship, and mRNA tissue distribution of the 12 T. obscurus ILs were comparatively analyzed. Furthermore, the inducible expression of these ToIL genes was detected in different tissues following exposure to Staphylococcus and Vibrio. Overall, this study may provide evidence for the 12 ToIL genes in T. obscurus with characterization of their gene expression and function.
2. Materials and methods
2.1. Experimental fish
Healthy T. obscurus (approximately 50 g in weight) were obtained from a breeding farm in Yangzhong City, Jiangsu Province, China. All fish samples brought to the laboratory were used during the assay. They were cultured in a recirculating aquaculture system at 23–25 °C for 2 weeks until the start of the experiment. During this period, the fish samples were kept under standard rearing conditions and fed with commercial pellets regularly.
2.2. Bacterial challenge and sample collection
Staphylococcus aureus ATCC 25,923 was purchased from China Center of Industrial Culture Collection. Vibrio harveyi E385 was kept in our laboratory. The tissue distribution of 12 ToIL genes was investigated by obtaining three samples of heart, liver, kidney, spleen, gills, and intestine tissues from normal T. obscurus. Each tissue sample was pooled from three individual fish. For immune challenge, fish samples were intraperitoneally injected with 100 μL of S. aureus (3.0 × 107 CFU/mL) and V. harveyi (3.0 × 107 CFU/mL). The spleen and liver were sampled. Three samples of spleen and liver (each pooled from three fish) were sampled at 0, 12, 24, 36, 48 and 72 h post-injection in each group. Fish injected with phosphate-buffered saline (PBS,140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) were sampled as controls.
2.3. RNA extraction and cDNA synthesis
From all samples, total RNA was extracted using the TRIzol reagent kit (Invitrogen, USA) in accordance with the manufacturer's instructions. The extracted RNA was treated with RNase-Free DNase (Takara, Japan) to remove contaminating DNA. RNA concentration was assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, USA), and RNA integrity was confirmed by 1.5% agarose gel electrophoresis. Total RNA (1 μg) was reverse-transcribed into first-strand cDNA by using the PrimeScript™ II 1st strand cDNA synthesis kit (Takara, Japan) following the manufacturer's instructions. The resulting cDNA was diluted six times and used as template for gene cloning.
2.4. Cloning of T. obscurus ILs
Transcriptome data from T. obscurus liver (not shown) were previously obtained in our laboratory. These data were searched for IL homologs by using Takifugu rubripes IL sequences as the query by local blast. Twelve pairs of primers were designed to amplify the complete open reading frames (ORFs) of 12 ToIL genes (ToIL-1β-F and ToIL-1β-R, ToIL-1-F and ToIL-1-R, ToIL-6-F and ToIL-6-R, ToIL-10-F and ToIL-10-R, ToIL-11-F and ToIL-11-R, ToIL-12-F and ToIL-12-R, ToIL-17-F and ToIL-17-R, ToIL-18-F and ToIL-18-R, ToIL-20-F and ToIL-20-R, ToIL-24-F and ToIL-24-R, ToIL-27-F and ToIL-27-R, and ToIL-34-F and ToIL-34-R, Table 1). PCR amplification was carried out in a 20 μL reaction mix containing 2 μL of 10 × Ex Taq buffer, 1.6 μL of dNTP mix (2.50 mM), 0.4 μL of each primer (10 mM), 0.2 μL of Ex Taq polymerase (Takara, Japan), 1 μL of template cDNA, and 14.4 μL of nuclease-free water. The cycling conditions were as follows: initial denaturation at 95 °C for 5 min; 30 times of main amplification cycle: 30 s of denaturation at 95 °C, 30 s of annealing at 56 °C and 2 min of extension at 72 °C; and 5 min of final extension at 72 °C. The PCR products were electrophoresed on 1.2% agarose gel to determine successful amplification. The PCR amplified fragments were purified using a DNA gel extraction kit (Generay, China), then cloned into the pEASY-T3 vector (TransGen, China), and transformed into Escherichia coli DH5α (TransGen Biotech, China). The positive clones were picked and confirmed by sequencing (Sangon Biotech, China).
Table 1.
Sequences of the primers used in the study.
| Primers name | Sequences (5′−3′) | Tm ( °C) |
|---|---|---|
| ToIL-1β-F | ATGGAATCTCAGATGAAATCCAACG | 63.6 |
| ToIL-1β-R | TTACATCTCTCCCTCACACGTGGGCG | 72.1 |
| ToIL-1-F | ATGGACCTGAAGGAATCAGAGGTA | 61.1 |
| ToIL-1-R | TCATTTCTTAATGACAAAGAGGAAGG | 60.8 |
| ToIL-6-F | ATGGCTTCCATCAGTTACCTGCTCGC | 70.7 |
| ToIL-6-R | TCACGATCTCTTCTTTGGGTGGTAG | 63.8 |
| ToIL-10-F | ATGACTCCCGGCTCTCTGCTCTCCG | 72.8 |
| ToIL-10-R | TCAGGCCACGTTGCGGCGCCTCTTT | 79.3 |
| ToIL-11-F | ATGAAATTGCTCCTCGACTCCTCCTC | 67.5 |
| ToIL-11-R | TCATTGCACCGCGGCATTGACCTTGG | 78.5 |
| ToIL-12-F | ATGGCCTCGCTCCACCTCTGTGAGTA | 71.2 |
| ToIL-12-R | TCAGCTGCTGGGCTGAGCATCGGCAC | 78.6 |
| ToIL-17-F | ATGCAGGTGGTGTCGGGAACGCTGCT | 77.0 |
| ToIL-17-R | GGCCCACTGTTATACCCCAACAGTGA | 69.2 |
| ToIL-18-F | ATGGCAGCTAACAATGGCAATTTTG | 66.6 |
| ToIL-18-R | CTAAGACATGGTGACGTAGCAGGTC | 62.2 |
| ToIL-20-F | ATGAAGACTCTGTCATCCCTCTGCCT | 65.9 |
| ToIL-20-R | TCAGTTCGGACTTTTCTGCTCGGAG | 68.1 |
| ToIL-24-F | ATGCTCGCCTCCACACTCCGCTGCTC | 77.0 |
| ToIL-24-R | TCAGGCCACGGTAACGTTGGTCTGG | 72.8 |
| ToIL-27-F | ATGATTGGGTGGACCACTCTGGCTCT | 70.1 |
| ToIL-27-R | CTACAGATTGCTTTTGGGTAAAGTGAT | 62.1 |
| ToIL-34-F | ATGTGCACGCCCCTGAAGACCATCAA | 74.1 |
| ToIL-34-R | TCACACACGTAAGGTGGGCGCCGCGT | 79.8 |
| ToIL-1β-qF | ATGAGCATGCAGGAGTACAG | 53.1 |
| ToIL-1β-qR | GGTTCACTTTGCGACATTCC | 57.8 |
| ToIL-1-qF | AACACGTGGCCTCTCATTAC | 54.9 |
| ToIL-1-qR | CTCCTCCTCATTTGCCTTGTAG | 58.5 |
| ToIL-6-qF | CGACTTTCTGGACCGTTACAA | 57.9 |
| ToIL-6-qR | CCGCTTGAGAAGAACTGAGTAG | 56.5 |
| ToIL-10-qF | TTCTACCTGGACTCGGTTCT | 53.7 |
| ToIL-10-qR | TGGTCGAAGATCTGCTGAATG | 58.5 |
| ToIL-11-qF | GTCGGCCCATAGTCTCAATAATC | 59.8 |
| ToIL-11-qR | CCTGTTCAGCCACTCAAAGT | 54.9 |
| ToIL-12-qF | GAGGTGGAAGAAGAGAAGGTTG | 57.4 |
| ToIL-12-qR | ACAGCTCTGTTGATGGTGATG | 55.4 |
| ToIL-17-qF | TACTATCCTACGCTGGTCCTC | 54.0 |
| ToIL-17-qR | GTCCTCAGCTGGAACTCATATT | 55.4 |
| ToIL-18-qF | GAGAATCACAGAAGGGTGCTATC | 57.5 |
| ToIL-18-qR | GGATGACTTTGTTCAGCGTTTG | 59.9 |
| ToIL-20-qF | ACGTAGAGAGAGTGTTCAGGAG | 52.9 |
| ToIL-20-qR | CGCAGAGGCAGTGACATTTA | 57.1 |
| ToIL-24-qF | GCCCAACATCAAGCCAAATC | 60.2 |
| ToIL-24-qR | CAGCACGTTGTTCAGGTAGA | 54.2 |
| ToIL-27-qF | TGCTGGTGCGTCAGTTATC | 54.7 |
| ToIL-27-qR | CAGACTCTGTGTGTCTCTCTCT | 50.2 |
| ToIL-34-qF | TATCCAGGAGACTTGGGATCA | 56.8 |
| ToIL-34-qR | ATGGTGTAGCACAGGTTGTC | 52.6 |
| β-actin-qF | GCATTGTCACCAACTGGGATG | 60.8 |
| β-actin-qR | GCAGGACTGGATGCTCCTCT | 58.9 |
2.5. Bioinformatic analysis
The ORF finder program (https://www.ncbi.nlm.nih.gov/orffinder/) and ExPASy (http://web.expasy.org/translate/) were used to deduce the amino acid sequences of ToILs. The BLASTX and BLASTP programs (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were used to analyze the nucleotide and deduced protein sequences, respectively. Conserved domain footprints were analyzed using Conserved Domain Database (www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and SMART (http://smart.embl-heidelberg.de/). Three-dimensional (3D) models were constructed using the SWISS-MODEL (https://swissmodel.expasy.org/) and PyMOL program. The theoretical isoelectric points (pI) and molecular weight (Mw) of ToILs were predicted using the ExPASy compute pI/Mw tool (http://web.expasy.org/compute_pi/). The Clustal Omega program (https://www.ebi.ac.uk/Tools/msa/clustalo/) and GENEDOC software were used to create multiple sequence alignment by using the full ORF sequences of ToILs. Phylogenetic tree based on Neighbor-Joining (NJ) method was constructed using MEGA 7.0 software [32], with 1000 bootstrap samples.
2.6. Gene expression analysis by RT-qPCR
After tissue collection, total RNA extraction, and cDNA synthesis, gene expression analysis via quantitative real-time PCR (RT-qPCR) was carried out using the SYBR Premix Ex Taq™ II kit (Perfect Real Time, Takara, Japan) on LightCycler® 96 (Roche, USA). The amplification reaction was performed using a MicroAmp Optical 96-well reaction plate (Applied Biosystems, USA), with a total reaction volume of 10 μL, consisting of 5 μL of 2 × SYBR Premix Ex Taq™ II, 1 μL of the cDNA template, 0.4 μL each of the sense and reverse primers (10 μM), and 3.2 μL of ddH2O. The cycling parameters were as follows: 95 °C for 60 s, 95 °C for 5 s, 60 °C for 30 s, then go to step 2 and repeated for 40 cycles. After each RT-qPCR reaction, dissociation analysis was performed to confirm the detection of only one product. β-actin was amplified as the reference gene. All primers (ToIL-1β-qF and ToIL-1β-qR, ToIL-1-qF and ToIL-1-qR, ToIL-6-qF and ToIL-6-qR, ToIL-10-qF and ToIL-10-qR, ToIL-11-qF and ToIL-11-qR, ToIL-12-qF and ToIL-12-qR, ToIL-17-qF and ToIL-17-qR, ToIL-18-qF and ToIL-18-qR, ToIL-20-qF and ToIL-20-qR, ToIL-24-qF and ToIL-24-qR, ToIL-27-qF and ToIL-27-qR, ToIL-34-qF and ToIL-34-qR, and β-actin-qF and β-actin-qR) used in this study were designed using IDT's PrimerQuest™ tool (https://sg.idtdna.com/PrimerQuest/Home/Index) and are listed in Table 1.
2.7. Statistical analysis
Relative gene expression was calculated using the 2−ΔΔCt method [33] and then normalized using the reference gene β-actin. Gene expression data were log-transformed (log base 2) before being subjected to one-way ANOVA. All analyses were implemented in GraphPad Prism v5 (San Diego, CA, USA). The quantitative expression data are presented as the means ± standard deviation (SD, N = 3). Statistical significance was determined using Student's t-tests. Significance was defined at *P < 0.05, ** P < 0.01, and *** P < 0.001.
3. Result
3.1. Sequence information of ToIL genes
A total of 12 ToIL genes were identified from T. obscurus (Supplementary Fig. S1). Based on the results of BLASTP and sequence analysis, these 12 ToIL genes were named ToIL-1β, ToIL-1, ToIL-6, ToIL-10, ToIL-11, ToIL-12, ToIL-17, ToIL-18, ToIL-20, ToIL-24, ToIL-27, and ToIL-34. The complete ORF sequences of these ToIL genes had lengths of 771, 1050, 684, 684, 606, 633, 477, 570, 531, 549, 759, and 786 bp, respectively. Twelve ToIL genes encoded proteins of 256, 349, 227, 227, 201, 210, 158, 189, 176, 182, 252, and 261 amino acids. The sequences of ToILs were submitted to NCBI GenBank. Detailed information is summarized in Table 2.
Table 2.
Overall identity and pI values for the 12 ILs from T. obscurus.
| Gene name | Number of amino acids | Molecular weight (kDa) | Isoelectric point (pI) | GenBank accession no. |
|---|---|---|---|---|
| ToIL-1β | 256 | 29.05 | 4.98 | OP351710 |
| ToIL-1 | 349 | 39.55 | 5.05 | OP351711 |
| ToIL-6 | 227 | 25.61 | 7.02 | OP351712 |
| ToIL-10 | 227 | 25.53 | 8.23 | OP351713 |
| ToIL-11 | 201 | 23.27 | 9.12 | OP351714 |
| ToIL-12 | 210 | 23.33 | 6.16 | OP351715 |
| ToIL-17 | 158 | 17.65 | 10.20 | OP351716 |
| ToIL-18 | 189 | 22.10 | 5.08 | OP351717 |
| ToIL-20 | 176 | 20.24 | 8.07 | OP351718 |
| ToIL-24 | 182 | 20.75 | 5.96 | OP351719 |
| ToIL-27 | 252 | 27.78 | 8.96 | OP351720 |
| ToIL-34 | 261 | 29.80 | 8.96 | OP351721 |
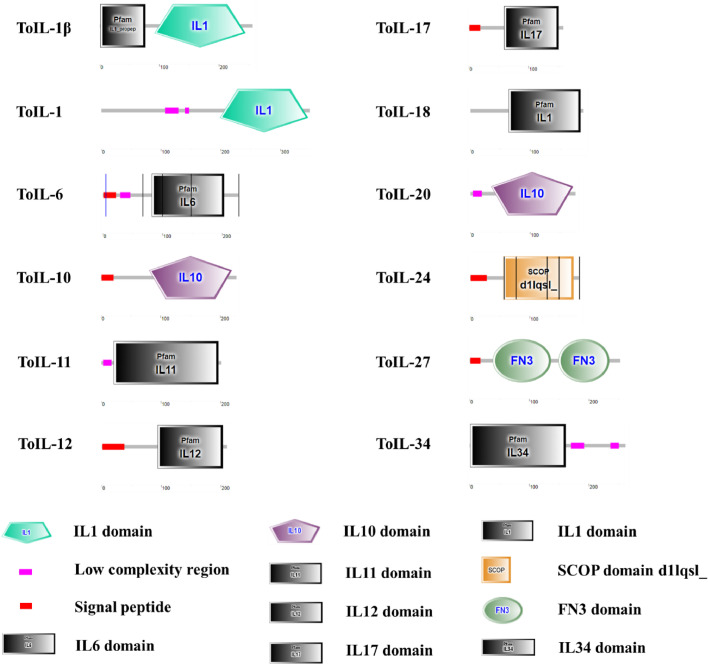
Based on functional domain prediction analysis by using the SMART program, the predicted ToIL proteins contained different domains (Fig. 1). ToIL-1β contained an IL1_propep domain (position 1–76) and an IL1 domain (position 92–244). ToIL-1 contained two low-complexity regions (positions 107–129 and 140–147) and an IL1 domain (position 199–346). ToIL-6 contained a signal peptide (position 1–22), a low-complexity region (position 29–46), and an IL6 domain (position 82–203). ToIL-10 contained a signal peptide (position 1–21) and an IL10 domain (position 81–219). ToIL-11 contained a low-complexity region (position 3–17) and an IL11 domain (position 21–196). ToIL-12 contained a signal peptide (position 1–38) and a IL12 domain (position 94–203). ToIL-17 contained a signal peptide (position 1–19) and an IL17 domain (position 59–150). ToIL-18 contained an IL1 domain (position 64–184). ToIL-20 contained a low-complexity region (position 4–19) and an IL10 domain (position 35–172). ToIL-24 contained a signal peptide (position 1–28) and a SCOP domain d1lqsl_ (position 56–173). ToIL-27 contained a signal peptide (position 1–18) and two fibronectin type 3 (FN3) domains (positions 39–136 and 149–234). ToIL-34 contained an IL34 domain (position 1–161) and two low-complexity regions (position 170–192 and 236–250). Except for ToIL-24 and ToIL-27, all other ToIL proteins contained a typical IL domain.
Fig. 1.
Diagram of the domain architectures of 12 IL proteins from T. obscurus. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. Tertiary structures of ToILs
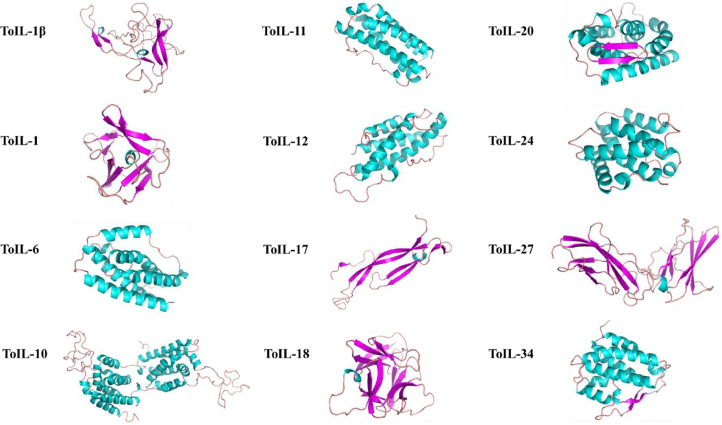
Protein function is defined by structure. Thus, homology modeling of 12 ToILs tertiary structures was performed using the SWISS-MODEL algorithm (Fig. 2). The template structures used are shown in Table 3. ToIL-1β, ToIL-1, ToIL-6, ToIL-10, ToIL-11, ToIL-12, ToIL-17, ToIL-18, ToIL-20, ToIL-24, ToIL-27, and ToIL-34 were built up by using one, two, six, fourteen, six, five, one, one, six, seven, one, and nine α helices, and eight, five, zero, zero, zero, zero, seven, thirteen, two, zero, thirteen, and two β-sheets, respectively. The results indicated that these ToIL proteins were structurally similar to their corresponding IL homologs, implying the possible functional resemblance between the ILs of T. obscurus and other species.
Fig. 2.
Tertiary structures of 12 ToIL proteins from T. obscurus. Three-dimensional models of 12 ToILs were generated using SWISS-MODEL and PyMOL. The figure displays of the animated structure models, where the α-helices are colored blue, the β-sheets are colored purple, and the random coils are colored brown. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 3.
Information on 3D model templates used in the study.
| Protein name | Template name | SMTL ID | Sequence identity (%) | Oligo-State | GMQE | Citation |
|---|---|---|---|---|---|---|
| ToIL-1β | Interleukin-1 beta | 3ltq.1.A | 35.00 | Monomer | 0.31 | [34] |
| ToIL-1 | IL-1 beta | 4 × 37.1.A | 23.48 | Monomer | 0.17 | unpublished |
| ToIL-6 | Interleukin-6 | 7nxz.1.A | 22.09 | Monomer | 0.45 | [35] |
| ToIL-10 | Interleukin-10 | 1ilk.1.A | 29.33 | Homo-dimer | 0.46 | [36] |
| ToIL-11 | Interleukin-11 | 6o4o.1.A | 28.75 | Monomer | 0.63 | [37] |
| ToIL-12 | Interleukin-12 subunit alpha | 3hmx.1.B | 28.48 | Monomer | 0.53 | [38] |
| ToIL-17 | Interleukin-17F | 6hgo.2.A | 33.04 | Homo-dimer | 0.45 | [39] |
| ToIL-18 | Interleukin-18 | 7al7.1.B | 22.29 | Monomer | 0.54 | [40] |
| ToIL-20 | Interleukin-20 | 4doh.1.A | 38.78 | Monomer | 0.69 | [41] |
| ToIL-24 | Interleukin 22 | 4o6k.3.A | 31.11 | Monomer | 0.55 | [42] |
| ToIL-27 | Interleukin-27 subunit beta | 7u7n.1.C | 32.83 | Monomer | 0.54 | [43] |
| ToIL-34 | Interleukin-34 | 4exn.1.A | 31.76 | Homo-dimer | 0.43 | [44] |
3.3. Phylogenetic analysis
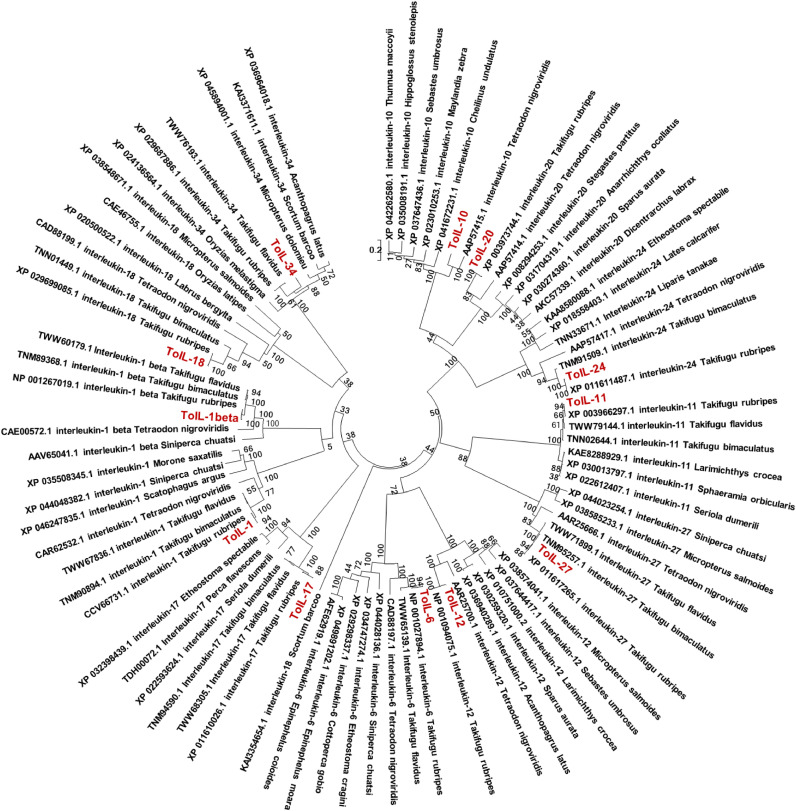
Based on the multiple alignment analysis, these 12 ILs from T. obscurus are lowly conserved. A phylogenetic tree was constructed to evaluate the molecular evolution relationship of 12 ToIL proteins of T. obscurus and other IL homologs from different species (Fig. 3). In the phylogenetic tree, 12 ToILs were clustered into different groups. ToIL-1β, together with three IL-1βs from T. rubripes (100.00% identity), Takifugu bimaculatus (95.70% identity), and Takifugu flavidus (95.31% identity), belong to one cluster. ToIL-1 is closely related to three IL-1 s from T. rubripes (99.71% identity), T. bimaculatus (97.43% identity), and T. flavidus (97.14% identity). ToIL-6 and two IL-6 s from T. rubripes (100.00% identity) and T. flavidus (97.16% identity) belong to one cluster. ToIL-10 and IL-10 from T. nigroviridis (61.08% identity) belong to one group. ToIL-11 and three IL-11 s from T. rubripes (100.00% identity), T. flavidus (98.91% identity), and T. bimaculatus (98.91% identity) belong to one clade. ToIL-12 and IL-12 from T. rubripes (100.00% identity) belong to one group. ToIL-17 and three IL-17 s from T. rubripes (100.00% identity), T. flavidus (95.57% identity), and T. bimaculatus (95.57% identity) belong to one clade. ToIL-18 and four IL-18 s from T. rubripes (100.00% identity), T. bimaculatus (90.00% identity), T. nigroviridis (65.22% identity), and Labrus bergylta (45.55% identity) were clustered into one group. ToIL-20 and IL-20 from T. rubripes (100.00% identity) belong to one cluster. ToIL-24 and two IL-24 s from T. rubripes (100.00% identity) and T. bimaculatus (99.21% identity) belong to one group. ToIL-27 and three IL-27 s from T. rubripes (100.00% identity), T. bimaculatus (99.21% identity), and T. flavidus (98.41% identity) belong to one cluster. ToIL-34 and two IL-34 s from T. rubripes (97.04% identity) and T. flavidus (83.25% identity) belong to one group.
Fig. 3.
Phylogenetic tree of ILs in T. obscurus and other species. The tree was constructed using the NJ method based on the amino acid alignments. Numbers were bootstrap values for 1000 trials. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Tissue distributions of ToIL genes
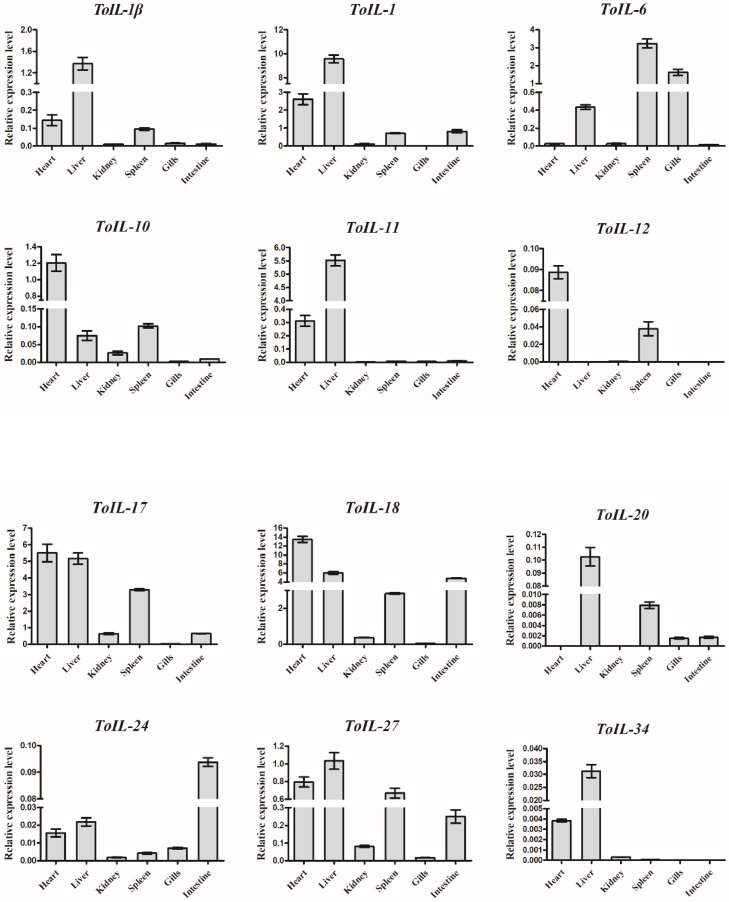
The mRNA expression of 12 ToIL genes was determined in six tissues of T. obscurus by using RT-qPCR. The results show that most ToILs were widely expressed in the heart, liver, kidney, spleen, gills, and intestine (Fig. 4). However, the expression levels of different ToILs differed. The transcripts of ToIL-1β, ToIL-1, ToIL-11, ToIL-17, ToIL-18, ToIL-27, and ToIL-34 were detected mainly in the liver and heart. ToIL-10 and ToIL-12 were primarily expressed in the heart and spleen. ToIL-20 was mainly expressed in the liver and spleen. Other ToILs such as ToIL-6 and ToIL-24 showed the highest expression level in the spleen and intestine, respectively.
Fig. 4.
Tissue distribution of 12 ILs in healthy T. obscurus. The transcripts of ToIL-1β, ToIL-1, ToIL-6, ToIL-10, ToIL-11, ToIL-12, ToIL-17, ToIL-18, ToIL-20, ToIL-24, ToIL-27, and ToIL-34 were measured by RT-qPCR. β-actin was used as an internal control. The tissues, including heart, liver, kidney, spleen, gills, and intestine were collected from three individuals. Three biological repeats were set in each group. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.5. ToIL gene expression after bacterial infection
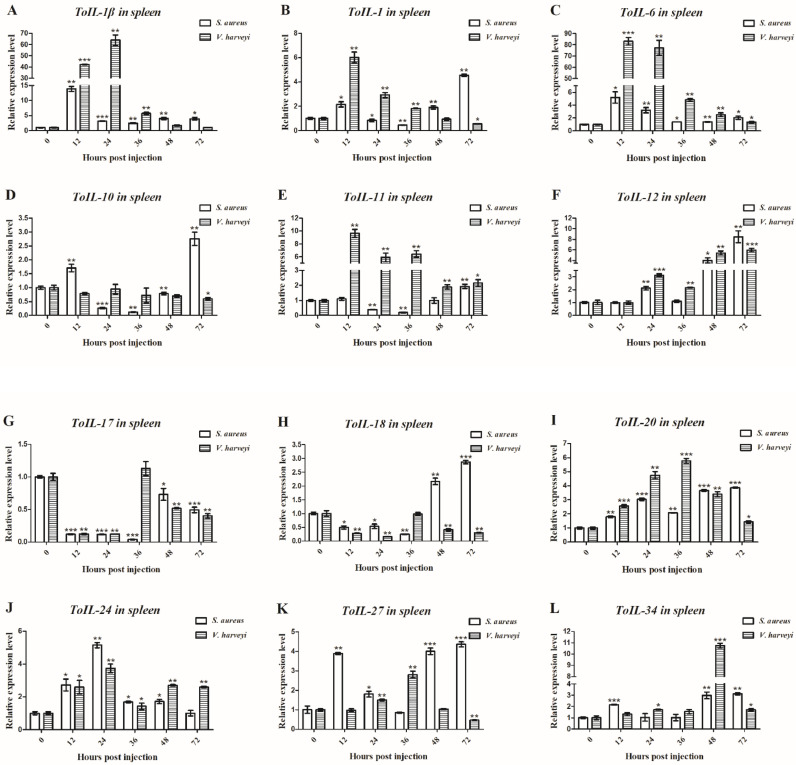
The roles of 12 ToIL genes in antibacterial innate immunity were investigated by observing the temporal mRNA expression of ToILs in the spleen and liver of T. obscurus upon challenge with S. aureus and V. harveyi by using RT-qPCR. The results showed that S. aureus and V. harveyi induced the expression of all ToIL genes in time-dependent manners in the tested tissues (Fig. 5, Fig. 6). However, the maximum induction was observed at different times. In spleen, the ToIL-1β transcript reached the highest level at 12 h post S. aureus challenge (13.91-fold, P < 0.01) and at 24 h following V. harbeyi challenge (63.82-fold, P < 0.01, Fig. 5A). The expression of ToIL-1 reached the peak at 72 h after stimulation by S. aureus (4.54-fold, P < 0.01) and at 12 h after V. harbeyi challenge (6.02-fold, P < 0.01, Fig. 5B). ToIL-6 mRNA was quickly upregulated at 12 h post-injection of S. aureus (5.22-fold, P < 0.05) and V. harveyi (83.14-fold, P < 0.001, Fig. 5C). ToIL-10 expression significantly increased at 72 h following S. aureus challenge (2.76-fold, P < 0.01) but lower after V. harveyi infection compared with uninfected controls, and the differences were not significant (Fig. 5D). The ToIL-11 transcript reached the highest level at 72 h post S. aureus challenge (1.94-fold, P < 0.01) and at 12 h post V. harbeyi challenge (9.64-fold, P < 0.01, Fig. 5E). The ToIL-12 expression was significantly upregulated at 72 h following S. aureus (8.49-fold, P < 0.01) and V. harveyi (5.96-fold, P < 0.001) challenges (Fig. 5F). ToIL-17 expression was downregulated from 12 h to 72 h after S. aureus and V. harveyi infection (Fig. 5G). ToIL-18 mRNA level was upregulated at 72 h after S. aureus infection (2.87-fold, P < 0.001) and then downregulated from 12 h to 72 h following V. harbeyi challenge (Fig. 5H). The ToIL-20 transcript reached the highest level at 72 h post S. aureus challenge (3.86-fold, P < 0.001) and at 36 h post V. harbeyi challenge (5.77-fold, P < 0.001, Fig. 5I). The ToIL-24 expression reached the peak at 24 h following S. aureus (5.15-fold, P < 0.01) and V. harveyi (3.73-fold, P < 0.01) challenge (Fig. 5J). The relative gene expression of ToIL-27 significantly increased at 72 h upon S. aureus challenge (4.37-fold, P < 0.001) and at 36 h after V. harveyi challenge (2.81-fold, P < 0.01, Fig. 5K). The ToIL-34 expression reached the peak at 72 h following S. aureus challenge (3.13-fold, P < 0.01) and at 48 h post V. harveyi challenge (10.74-fold, P < 0.001, Fig. 5L).
Fig. 5.
Relative expression levels of 12 ILs in the spleen of T. obscurus at different time points (0, 12, 24, 36, 48, and 72 h) as measured with RT-qPCR after S. aureus and V. harveyi infection. β-actin was employed as an internal control. Each experiment was performed in triplicate. Data are shown as mean ± S.D. Asterisks indicate significant differences (*P < 0.05, **P < 0.01, ***P < 0.001). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
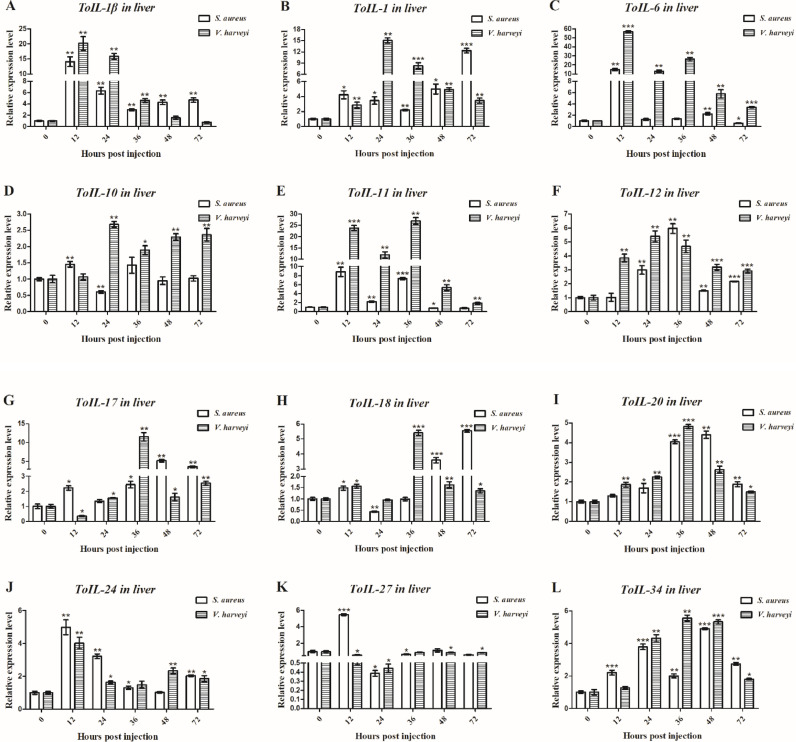
Fig. 6.
Relative expression levels of 12 ILs in the liver of T. obscurus at different time points (0, 12, 24, 36, 48, and 72 h) as measured with RT-qPCR after S. aureus and V. harveyi infection. β-actin was employed as an internal control. Each experiment was performed in triplicate. Data are shown as mean ± S.D. Asterisks indicate significant differences (*P < 0.05, **P < 0.01, ***P < 0.001). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In liver, both ToIL-1β and ToIL-6 expression reached the highest level at 12 h following S. aureus (14.09- and 14.91-fold, P < 0.01) and V. harveyi (20.19- and 57.03-fold, P < 0.01) challenge (Fig. 6A and C). The gene expression level of ToIL-1 significantly increased at 72 h following S. aureus challenge (12.31-fold, P < 0.001) and at 24 h post V. harveyi challenge (15.01-fold, P < 0.01, Fig. 6B). The ToIL-10 expression reached the peak at 12 h following S. aureus challenge (1.56-fold, P < 0.01) and at 24 h post V. harveyi challenge (2.68-fold, P < 0.01, Fig. 6D). The ToIL-11 transcript reached the highest level at 12 h post S. aureus challenge (8.83-fold, P < 0.01) and at 36 h post V. harbeyi challenge (26.94-fold, P < 0.01, Fig. 6E). The transcript of ToIL-12 was significantly upregulated at 36 h following S. aureus challenge (5.96-fold, P < 0.01) and at 24 h post V. harveyi challenge (5.41-fold, P < 0.01, Fig. 6F). ToIL-17, ToIL-20, and ToIL-34 expression reached the highest level at 48 h upon S. aureus challenge (5.19-, 4.40-, and 4.91-fold, P < 0.01) and at 36 h after V. harveyi challenge (11.53-, 4.82-, and 5.56-fold, P < 0.01, Fig. 6G, I and L). The ToIL-18 transcript reached the pick at 72 h post S. aureus challenge (5.55-fold, P < 0.001) and at 36 h following V. harbeyi challenge (5.42-fold, P < 0.001, Fig. 6H). The maximum ToIL-24 expression was reached at 12 h following S. aureus (4.98-fold, P < 0.01) and V. harveyi (4.01-fold, P < 0.01) challenge (Fig. 6J). The ToIL-27 mRNA expression level decreased in the V. harveyi infection group but remarkably increased at 12 h after S. aureus infection (5.47-fold, P < 0.01, Fig. 6K). In addition, the transcripts of all ToIL genes in the spleen and liver did not change evidently after PBS challenge from 0 h to 72 h (data not shown).
4. Discussion
Fish innate immune systems have become an important barrier to the invasion of defense pathogens. ILs constitute an emerging family of cytokines and appear to be a distinct and potent signaling system involved in the control of immune response in fish [5]. In recent years, the functions of fish IL molecules have been extensively studied. The present study reports the identification and molecular characterization of the whole coding sequences of 12 ILs from T. obscurus, a teleost fish. The structural analysis of the 12 ToILs shared certain similar features with their mammalian orthologs. In addition to ToIL-24 and ToIL-27, the 10 other deduced ToIL proteins possessed a typical IL superfamily domain, which is consistent with the protein characteristics of ILs from other teleost fish. ToIL-27 contained two repeat units of FN3, which is a small autonomous folding unit present in many animal proteins involved in ligand binding. ToIL-6, ToIL-10, ToIL-12, ToIL-17, ToIL-24, and ToIL-27 proteins contained a signal peptide sequence, indicating that these molecules may be secreted through the classical pathway involving the golgi/endoplasmic reticulum route. ToIL-1, ToIL-6, ToIL-11, ToIL-20, and ToIL-34 also contained one or two low-complexity regions, which are protein sequences formed by a set of compositionally biased residues. Low-complexity regions are extremely abundant in cellular proteins and have also been reported in viruses, where they may partake in the evasion of the host immune system [45]. The conservative structures of ToILs implied their biological functions in innate immunity. In addition, the predicted ToIL proteins shared strong identities of 80%–100% with their counterparts in other selected vertebrates, and the phylogenetic tree and analysis of protein 3D structure both demonstrated that ToILs are evolutionarily conserved genes in animals and clear homologs of mammalian and fish ILs.
In general, the analysis of tissue distribution for genes provides a good understanding of their physiological roles. In T. obscurus, the expression of most ToIL molecules occurs constitutively in multiple tissues. The broad expression profiles of ToILs suggest that they are pleiotropic cytokines that may be required for tissue homeostasis. In fish, the expression profiles of ILs quite differ among species. The highest expression of ToIL-1β, ToIL-1, ToIL-11, ToIL-20, ToIL-27, and ToIL-34 was detected in the liver, which consists of tissues with multifaceted functions in innate immunity, metabolism, and growth. In roughskin sculpin Trachidermus fasciatus, rsIL-1β gene was expressed most abundantly in the skin and moderately in the blood, heart, gills, and intestine [46]. CsIL-11 from tongue sole Cynoglossus semilaevis was highly expressed in the blood, muscle, and heart [47]. In snakehead Channa argus, shIL-20 was widely expressed, with the highest transcription in the liver and a relatively high transcription in the mucosal immune organs (gut and skin) [48]. In Japanese flounder Paralichthys olivaceus, PoIL-34 expression is relatively high in the intestine and heart [49], and a relatively high ToIL-6 expression was observed in the spleen. In A. baeri, a notably high level of Abil-6 expression was reported in the spleen [20]. Spleen is regarded as the major primary lymphoid organ in fish, where monocytes/macrophages and lymphocytes reside, differentiate, and interact. The maximum expression of ToIL-24 gene was detected in the intestine, which is a multifunctional organ central to both nutrient uptake, pathogen recognition, and regulation of the intestinal microbiome, suggesting a potential role in the mucosal immunity. A novel and unexpected finding in this study was that ToIL-10, ToIL-12, ToIL-17, and ToIL-18 expression levels were very high in the heart, which possesses an extensive lymphatic network. IL-10 from mandarin fish Siniperca chuatsi exhibited relatively high mRNA levels in the spleen, trunk kidney, head kidney, and intestine [50]. In rainbow trout, IL-18A and IL-18B were highly expressed in the gut, heart, and kidney [13]. The abundance of IL transcripts in different tissues differs across species possibly because of the difference in the genetics and behavior and the culturing conditions of different fish samples.
As key members of cytokines, ILs play essential roles in infections and inflammatory diseases [6]. In the present study, the involvement of 12 ToIL genes in the immune response of T. obscurus was investigated by examining their expression in the liver and spleen of fish challenged with S. aureus (a Gram-positive bacterium) and V. harveyi (a Gram-negative bacterium). S. aureus, a metabolically flexible pathogen that causes infections in diverse settings, is increasingly being detected in various food-producing animals, including fish [51]. V. harveyi, which belongs to family Vibrionaceae of class Gammaproteobacteria, is one of the most serious pathogenic Vibrio in aquaculture [52]. The expression levels of all ToIL genes in the liver and spleen were significantly upregulated during infection with both pathogens, and their response varies over time. Similarly, the increase in IL expression was observed in vivo in other fish species treated with bacteria. In yellow catfish Pelteobagrus fulvidraco, the expression level of Pf_IL-1β was upregulated in the trunk kidney, head kidney, blood, spleen, heart, and liver after Edwardsiella ictaluri challenge [53]. Ayu Plecoglossus altivelis PaIL-6 mRNA was significantly induced in the head kidney, liver, gills, and spleen upon Vibrio anguillarum infection [54], and gilthead seabream sbIL-6 expression was remarkably upregulated in the head kidney by V. anguillarum challenge [18]. In golden pompano T. ovatus, TOIL-10 and TOIL-22 were rapidly activated after Streptococcus agalactiae SAΔphoB immunization and significantly increased to peak levels at 12 h and 4 days in the kidney and spleen, respectively, following challenge [24]. In crucian carp (Carassius auratus gibelio), the IL-11 expression was enhanced in the kidney at early time points upon Aeromonas hydrophila infection [55]. After V. anguillarum stimulation, the mRNA expression of three L. crocea ILs (lcIL12A, lcIL16 and lcIL34) was upregulated in the liver, spleen, and kidney [28]. In large yellow croaker, the expression of LcIL-17C and LcIL-17D significantly increased in the gills, head kidney, and spleen after A. hydrophila challenge [25]. In snakehead C. argus, injection with Aeromonas schubertii and Nocardia seriolae increased the shIL-20 expression in the spleen and head kidney [48]. Grass carp CiIL-34 expression was upregulated in the spleen and head kidney upon infection with Flavobacterium columnare [30]. The discrepancies in IL expression profiles in different species may have resulted from multiple factors such as the challenge routes (injection and bath), pathogen types, fish species, and age. Collectively, our results provided evidence that 12 ToIL genes are tightly associated with bacterial challenges and have great importance in host defense in T. obscurus.
In conclusion, the characterization of 12 ToIL genes from T. obscurus was described herein. ToILs possessed the characteristic structure features present in their respective homologs. Most ToILs were preferentially expressed in immune tissues such as liver, spleen, and intestine and increased significantly upon bacterial challenge. These data provide more detailed insights into the functional activities of the obscure puffer IL family. However, further in-depth studies are still needed to ascertain the functional role of fish ILs and their implication in immunity.
Ethics statement
The protocol was approved by the Ethics Committee of Experimental Animals at Hohai University.
Consent for publication
Manuscript is approved by all authors for publication.
Funding
The current study was supported by the Project for Seed Industry Vitalization of Jiangsu Province (JBGS[2021]133), the Guangdong Provincial Key Laboratory of Applied Marine Biology (LAMB20221006), the Fundamental Research Funds for the Central Universities (B220202048), and the National Natural Science Foundation of China (32002423).
CRediT authorship contribution statement
Ying Huang: Data curation, Formal analysis, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. Peng Luo: Validation, Resources, Writing – review & editing. Fu-Hui Jiang: Methodology, Formal analysis, Software, Writing – original draft. Hui-Ze Gao: Formal analysis, Data curation, Software. Li-Fan Cui: Methodology, Software, Validation. Zhe Zhao: Conceptualization, Project administration, Resources, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.fsirep.2023.100103.
Appendix. Supplementary materials
References
- 1.Conlon K.C., Miljkovic M.D., Waldmann T.A. Cytokines in the Treatment of Cancer. J. Interferon. Cytokine Res. 2019;39(1):6–21. doi: 10.1089/jir.2018.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dinarello C.A., Mier J.W. Interleukins. Annu. Rev. Med. 1986;37:173–178. doi: 10.1146/annurev.me.37.020186.001133. [DOI] [PubMed] [Google Scholar]
- 3.Garlanda C., Dinarello C.A., Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39(6):1003–1018. doi: 10.1016/j.immuni.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isailovic N., Daigo K., Mantovani A., Selmi C. Interleukin-17 and innate immunity in infections and chronic inflammation. J. Autoimmun. 2015;60:1–11. doi: 10.1016/j.jaut.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Secombes C.J., Wang T., Bird S. The interleukins of fish. Dev. Comp. Immunol. 2011;35(12):1336–1345. doi: 10.1016/j.dci.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Zou J., Secombes C.J. The function of fish cytokines. Biology (Basel) 2016;5(2):23. doi: 10.3390/biology5020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.González-Fernández C., Chaves-Pozo E., Cuesta A. Identification and Regulation of Interleukin-17 (IL-17) Family Ligands in the Teleost Fish European Sea Bass. Int. J. Mol. Sci. 2020;21(7):2439. doi: 10.3390/ijms21072439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zou J., Grabowski P.S., Cunningham C., Secombes C.J. Molecular cloning of interleukin 1beta from rainbow trout Oncorhynchus mykiss reveals no evidence of an ice cut site. Cytokine. 1999;11(8):552–560. doi: 10.1006/cyto.1998.0470. [DOI] [PubMed] [Google Scholar]
- 9.Bird S., Wang T., Zou J., Cunningham C., Secombes C.J. The first cytokine sequence within cartilaginous fish: IL-1 beta in the small spotted catshark (Scyliorhinus canicula) J. Immunol. 2002;168(7):3329–3340. doi: 10.4049/jimmunol.168.7.3329. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y., Wang Q., Baoprasertkul P., Peatman E., Liu Z. Genomic organization, gene duplication, and expression analysis of interleukin-1beta in channel catfish (Ictalurus punctatus) Mol. Immunol. 2006;43(10):1653–1664. doi: 10.1016/j.molimm.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 11.Lee D.S., Hong S.H., Lee H.J., Jun L.J., Chung J.K., Kim K.H., Jeong H.D. Molecular cDNA cloning and analysis of the organization and expression of the IL-1beta gene in the Nile tilapia, Oreochromis niloticus. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2006;143(3):307–314. doi: 10.1016/j.cbpa.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 12.Hong S., Peddie S., Campos-Pérez J.J., Zou J., Secombes C.J. The effect of intraperitoneally administered recombinant IL-1beta on immune parameters and resistance to Aeromonas salmonicida in the rainbow trout (Oncorhynchus mykiss) Dev. Comp. Immunol. 2003;27(9):801–812. doi: 10.1016/s0145-305x(03)00056-9. [DOI] [PubMed] [Google Scholar]
- 13.Zou J., Bird S., Truckle J., Bols N., Horne M., Secombes C. Identification and expression analysis of an IL-18 homologue and its alternatively spliced form in rainbow trout (Oncorhynchus mykiss) Eur. J. Biochem. 2004;271(10):1913–1923. doi: 10.1111/j.1432-1033.2004.04101.x. [DOI] [PubMed] [Google Scholar]
- 14.Pérez-Cordón G., Estensoro I., Benedito-Palos L., Calduch-Giner J.A., Sitjà-Bobadilla A., Pérez-Sánchez J. Interleukin gene expression is strongly modulated at the local level in a fish-parasite model. Fish Shellfish Immunol. 2014;37(2):201–208. doi: 10.1016/j.fsi.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 15.Pereiro P., Balseiro P., Romero A., Dios S., Forn-Cuni G., Fuste B., Planas J.V., Beltran S., Novoa B., Figueras A. High-throughput sequence analysis of turbot (Scophthalmus maximus) transcriptome using 454-pyrosequencing for the discovery of antiviral immune genes. PLoS ONE. 2012;7(5):e35369. doi: 10.1371/journal.pone.0035369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkatesh B., Lee A.P., Ravi V., Maurya A.K., Lian M.M., Swann J.B., Ohta Y., Flajnik M.F., Sutoh Y., Kasahara M., Hoon S., Gangu V., Roy S.W., Irimia M., Korzh V., Kondrychyn I., Lim Z.W., Tay B.H., Tohari S., Kong K.W., Ho S., Lorente-Galdos B., Quilez J., Marques-Bonet T., Raney B.J., Ingham P.W., Tay A., Hillier L.W., Minx P., Boehm T., Wilson R.K., Brenner S., Warren W.C. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505(7482):174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bird S., Zou J., Savan R., Kono T., Sakai M., Woo J., Secombes C. Characterisation and expression analysis of an interleukin 6 homologue in the Japanese pufferfish, Fugu rubripes. Dev. Comp. Immunol. 2005;29(9):775–789. doi: 10.1016/j.dci.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Castellana B., Iliev D.B., Sepulcre M.P., MacKenzie S., Goetz F.W., Mulero V., Planas J.V. Molecular characterization of interleukin-6 in the gilthead seabream (Sparus aurata) Mol. Immunol. 2008;45(12):3363–3370. doi: 10.1016/j.molimm.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 19.Varela M., Dios S., Novoa B., Figueras A. Characterisation, expression and ontogeny of interleukin-6 and its receptors in zebrafish (Danio rerio) Dev. Comp. Immunol. 2012;37(1):97–106. doi: 10.1016/j.dci.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Wang X., Chen J., Zhang R., Liu L., Ma G., Zhu H. Interleukin-6 in Siberian sturgeon (Acipenser baeri): molecular characterization and immune functional activity. Fish Shellfish Immunol. 2020;102:296–306. doi: 10.1016/j.fsi.2020.03.023. [DOI] [PubMed] [Google Scholar]
- 21.Zhang D.C., Shao Y.Q., Huang Y.Q., Jiang S.G. Cloning, characterization and expression analysis of interleukin-10 from the zebrafish (Danio rerion) J. Biochem. Mol. Biol. 2005;38(5):571–576. doi: 10.5483/bmbrep.2005.38.5.571. [DOI] [PubMed] [Google Scholar]
- 22.Wei H., Yang M., Zhao T., Wang X., Zhou H. Functional expression and characterization of grass carp IL-10: an essential mediator of TGF-β1 immune regulation in peripheral blood lymphocytes. Mol. Immunol. 2013;53(4):313–320. doi: 10.1016/j.molimm.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 23.Piazzon M.C., Savelkoul H.S., Pietretti D., Wiegertjes G.F., Forlenza M. Carp Il10 has anti-inflammatory activities on phagocytes, promotes proliferation of memory T cells, and regulates B cell differentiation and antibody secretion. J. Immunol. 2015;194(1):187–199. doi: 10.4049/jimmunol.1402093. [DOI] [PubMed] [Google Scholar]
- 24.Peng Y., Cai X., Zhang G., Wang J., Li Y., Wang Z., Wang B., Xiong X., Wu Z., Jian J. Molecular characterization and expression of interleukin-10 and interleukin-22 in golden pompano (Trachinotus ovatus) in response to Streptococcus agalactiae stimulus. Fish Shellfish Immunol. 2017;65:244–255. doi: 10.1016/j.fsi.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 25.Ding Y., Ao J., Chen X. Comparative study of interleukin-17C (IL-17C) and IL-17D in large yellow croaker Larimichthys crocea reveals their similar but differential functional activity. Dev. Comp. Immunol. 2017;76:34–44. doi: 10.1016/j.dci.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 26.Wang T., Díaz-Rosales P., Martin S.A., Secombes C.J. Cloning of a novel interleukin (IL)-20-like gene in rainbow trout Oncorhynchus mykiss gives an insight into the evolution of the IL-10 family. Dev. Comp. Immunol. 2010;34(2):158–167. doi: 10.1016/j.dci.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Wang T., Kono T., Monte M.M., Kuse H., Costa M.M., Korenaga H., Maehr T., Husain M., Sakai M., Secombes C.J. Identification of IL-34 in teleost fish: differential expression of rainbow trout IL-34, MCSF1 and MCSF2, ligands of the MCSF receptor. Mol. Immunol. 2013;53(4):398–409. doi: 10.1016/j.molimm.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Wang L., Jiang L., Wu C., Lou B. Molecular characterization and expression analysis of large yellow croaker (Larimichthys crocea) interleukin-12A, 16 and 34 after poly I:c and Vibrio anguillarum challenge. Fish Shellfish Immunol. 2018;74:84–93. doi: 10.1016/j.fsi.2017.12.041. [DOI] [PubMed] [Google Scholar]
- 29.Mo Z.Q., Li Y.W., Zhou L., Li A.X., Luo X.C., Dan X.M. Grouper (Epinephelus coioides) IL-34/MCSF2 and MCSFR1/MCSFR2 were involved in mononuclear phagocytes activation against Cryptocaryon irritans infection. Fish Shellfish Immunol. 2015;43(1):142–149. doi: 10.1016/j.fsi.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 30.Xue Y., Jiang X., Gao J., Li X., Xu J., Wang J., Gao Q., Zou J. Functional characterization of interleukin 34 in grass carp Ctenopharyngodon idella. Fish Shellfish Immunol. 2019;92:91–100. doi: 10.1016/j.fsi.2019.05.059. [DOI] [PubMed] [Google Scholar]
- 31.Kang S., Kim J.H., Jo E., Lee S.J., Jung J., Kim B.M., Lee J.H., Oh T.J., Yum S., Rhee J.S., Park H. Chromosomal-level assembly of Takifugu obscurus (Abe, 1949) genome using third-generation DNA sequencing and Hi-C analysis. Mol. Ecol. Resour. 2020;20(2):520–530. doi: 10.1111/1755-0998.13132. [DOI] [PubMed] [Google Scholar]
- 32.Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Barthelmes K., Reynolds A.M., Peisach E., Jonker H.R., DeNunzio N.J., KN Allen, Imperiali B., Schwalbe H. Engineering encodable lanthanide-binding tags into loop regions of proteins. J. Am. Chem. Soc. 2011;133(4):808–819. doi: 10.1021/ja104983t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reif A., Lam K., Weidler S., Lott M., Boos I., Lokau J., Bretscher C., Mönnich M., Perkams L., Schmälzlein M., Graf C., Fischer J.P., Lechner C., Hallstein K., Becker S., Weyand M., Steegborn C., Schultheiss G., Rose-John S., Garbers C., Unverzagt C. Natural glycoforms of human interleukin 6 show atypical plasma clearance. Angew. Chem. Int. Ed. Engl. 2021;60(24):13380–13387. doi: 10.1002/anie.202101496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zdanov A., Schalk-Hihi C., Gustchina A., Tsang M., Weatherbee J., Wlodawer A. Crystal structure of interleukin-10 reveals the functional dimer with an unexpected topological similarity to interferon gamma. Structure. 1995;3(6):591–601. doi: 10.1016/s0969-2126(01)00193-9. [DOI] [PubMed] [Google Scholar]
- 37.Metcalfe R.D., Aizel K., Zlatic C.O., Nguyen P.M., Morton C.J., Lio D.S., Cheng H.C., Dobson R.C.J., Parker M.W., Gooley P.R., Putoczki T.L., Griffin M.D.W. The structure of the extracellular domains of human interleukin 11α receptor reveals mechanisms of cytokine engagement. J. Biol. Chem. 2020;295(24):8285–8301. doi: 10.1074/jbc.RA119.012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo J., Wu S.J., Lacy E.R., Orlovsky Y., Baker A., Teplyakov A., Obmolova G., Heavner G.A., Richter H.T., Benson J. Structural basis for the dual recognition of IL-12 and IL-23 by ustekinumab. J. Mol. Biol. 2010;402(5):797–812. doi: 10.1016/j.jmb.2010.07.046. [DOI] [PubMed] [Google Scholar]
- 39.Goepfert A., Lehmann S., Blank J., Kolbinger F., Rondeau J.M. Structural Analysis Reveals that the Cytokine IL-17F Forms a Homodimeric Complex with Receptor IL-17RC to Drive IL-17RA-Independent Signaling. Immunity. 2020;52(3):499–512. doi: 10.1016/j.immuni.2020.02.004. .e5. [DOI] [PubMed] [Google Scholar]
- 40.Detry S., Andries J., Bloch Y., Gabay C., Clancy D.M., Savvides S.N. Structural basis of human IL-18 sequestration by the decoy receptor IL-18 binding protein in inflammation and tumor immunity. J. Biol. Chem. 2022;298(5) doi: 10.1016/j.jbc.2022.101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Logsdon N.J., Deshpande A., Harris B.D., Rajashankar K.R., Walter M.R. Structural basis for receptor sharing and activation by interleukin-20 receptor-2 (IL-20R2) binding cytokines. Proc. Natl. Acad. Sci. U. S. A. 2012;109(31):12704–12709. doi: 10.1073/pnas.1117551109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siupka P., Hamming O.J., Frétaud M., Luftalla G., Levraud J.P., Hartmann R. The crystal structure of zebrafish IL-22 reveals an evolutionary, conserved structure highly similar to that of human IL-22. Genes. Immun. 2014;15(5):293–302. doi: 10.1038/gene.2014.18. [DOI] [PubMed] [Google Scholar]
- 43.Caveney N.A., Glassman C.R., Jude K.M., Tsutsumi N., Garcia K.C. Structure of the IL-27 quaternary receptor signaling complex. Elife. 2022;11:e78463. doi: 10.7554/eLife.78463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y., Yu S., Chai Y., Zhang Q., Yang H., Zhu Q. Lipopolysaccharide-induced gene expression of interleukin-1 receptor-associated kinase 4 and interleukin-1β in roughskin sculpin (Trachidermus fasciatus) Fish Shellfish Immunol. 2012;33(4):690–698. doi: 10.1016/j.fsi.2012.05.035. [DOI] [PubMed] [Google Scholar]
- 45.Becerra A., Muñoz-Velasco I., Aguilar-Cámara A., Cottom-Salas W., Cruz-González A., Vázquez-Salazar A., Hernández-Morales R., Jácome R., Campillo-Balderas J.A., Lazcano A. Two short low complexity regions (LCRs) are hallmark sequences of the Delta SARS-CoV-2 variant spike protein. Sci. Rep. 2022;12(1):936. doi: 10.1038/s41598-022-04976-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu H., Leo C., Chen X., Wong B.R., Williams L.T., Lin H., He X. The mechanism of shared but distinct CSF-1R signaling by the non-homologous cytokines IL-34 and CSF-1. Biochim. Biophys. Acta. 2012;1824(7):938–945. doi: 10.1016/j.bbapap.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X.P., Chen G.Y., Jin Q., Lou F.R., Liu B.J., Zhang J., Feng J.X., Chen T.T. CsIL-11, a teleost interleukin-11, is involved in promoting phagocytosis and antibacterial immune defense. Int. J. Biol. Macromol. 2021;192:1021–1028. doi: 10.1016/j.ijbiomac.2021.10.080. [DOI] [PubMed] [Google Scholar]
- 48.Cui Z., Zhu X., Zhao F., Li D., Deng Y., Tan A., Lai Y., Huang Z., Gong H. Molecular identification and functional exploration of interleukin-20 in snakehead (Channa argus) involved in bacterial invasion and the proliferation of head kidney leukocytes. Fish Shellfish Immunol. 2022;127:623–632. doi: 10.1016/j.fsi.2022.07.005. [DOI] [PubMed] [Google Scholar]
- 49.Yu C., Zhang P., Zhang T.F., Sun L. IL-34 regulates the inflammatory response and anti-bacterial immune defense of Japanese flounder Paralichthys olivaceus. Fish Shellfish Immunol. 2020;104:228–236. doi: 10.1016/j.fsi.2020.05.073. [DOI] [PubMed] [Google Scholar]
- 50.Huo H.J., Chen S.N., Li L., Nie P. Functional characterization of IL-10 and its receptor subunits in a perciform fish, the mandarin fish, Siniperca chuatsi. Dev. Comp. Immunol. 2019;97:64–75. doi: 10.1016/j.dci.2019.03.017. [DOI] [PubMed] [Google Scholar]
- 51.Prince A., Wong Fok Lung T. Consequences of Metabolic Interactions during Staphylococcus aureus Infection. Toxins (Basel) 2020;12(9):581. doi: 10.3390/toxins12090581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X.H., He X., Austin B. Vibrio harveyi: a serious pathogen of fish and invertebrates in mariculture. Mar. Life Sci. Technol. 2020;2(3):231–245. doi: 10.1007/s42995-020-00037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liao C.L., Zhang G.R., Zhu D.M., Ji W., Shi Z.C., Jiang R., Fan Q.X., Wei K.J. Molecular cloning and expression analysis of interleukin-1β and interleukin-1 receptor type I genes in yellow catfish (Pelteobagrus fulvidraco): responses to challenge of Edwardsiella ictaluri. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2018;223:1–15. doi: 10.1016/j.cbpb.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 54.Zhu K., Lu X.J., Lu J.-F., Chen J. The interleukin-6 regulates the function of monocytes/macrophages (MO/MФ) via the interleukin-6 receptor β in ayu (Plecoglossus altivelis) Fish Shellfish Immunol. 2019;93:191–199. doi: 10.1016/j.fsi.2019.07.049. [DOI] [PubMed] [Google Scholar]
- 55.Podok P., Xu L., Xu D., Lu L. Different expression profiles of Interleukin 11 (IL-11), Intelectin (ITLN) and Purine nucleoside phosphorylase 5a (PNP 5a) in crucian carp (Carassius auratus gibelio) in response to Cyprinid herpesvirus 2 and Aeromonas hydrophila. Fish Shellfish Immunol. 2014;38(1):65–73. doi: 10.1016/j.fsi.2014.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.