Highlights
-
•
ALKBH5 is downregulated in CRC and interrelated with poor prognosis.
-
•
ALKBH5 attenuates proliferation, migration and invasion of CRC cells.
-
•
Elevated levels of ALKBH5 are linked to enhanced CD8+ T cells recruitment to the tumor.
-
•
CD8A was significantly associated with CCL5 in CRC.
-
•
ALKBH5 promotes CD8+ T cells infiltration through NF-kB-CCL5 signaling pathway.
Keywords: Colorectal cancer, ALKBH5, NF-κB, CCL5, Tumor microenvironment, CD8+ T cells
Summary
Background
ALKBH5 belongs to the ALKB family consists of a Fe (II) and a-ketoglutarate-dependent dioxygenase. ALKBH5 directly catalyzes the oxidative demethylation of m6A-methylated adenosine. ALKBH5 involves in tumorigenesis and tumor progression, and is often dysregulated in a wide range of cancers, including colorectal cancer. Emerging evidence indicates that the expression of ALKBH5 is associated with the abundance of infiltrating immune cells in the microenvironment. However, how ALKBH5 affects immune cell infiltration in the microenvironment in colorectal cancer (CRC) has not been reported. The aim of this study was to identify how the expression of ALKBH5 affects the biological behaviors of CRC cell lines and regulates the effects on infiltrating CD8+ T cells in CRC microenvironment with its specific mechanism.
Methods
Firstly, the transcriptional expression profiles of CRC were downloaded from TCGA database and integrated via R software (4.1.2). Between CRC and normal colorectal tissues, ALKBH5 mRNA expressions were compared (Wilcoxon rank-sum). We further identified the expression levels of ALKBH5 in CRC tissues and cell lines through quantitative PCR, western blot, and immunohistochemistry. Then, how ALKBH5 affects the biological behaviors of CRC cells were confirmed by gain- and loss-of-function analysis. Furthermore, the relationship between ALKBH5 level and 22 tumor-infiltrating immune cells was examined through CIBERSORT in R software. Furthermore, we explored the correlation between ALKBH5 expression and tumor-infiltrated CD8+, CD4+ and regulatory T cells by utilizing the TIMER database. Finally, the association between chemokines and CD8+ T cells infiltration in CRC was analyzed using GEPIA online database. qRT-PCR, WB and IHC were used to further determine the effect of ALKBH5 on NF-κB-CCL5 signaling axis and CD8+ T cells infiltration.
Results
Clinically, ALKBH5 expression was downregulated in CRC and low levels of ALKBH5 expression were correlated with poor overall survival (OS). Functionally, overexpression of ALKBH5 reduced the proliferation, migration and invasion of CRC cells, and vice versa. Overexpression of ALKBH5 suppresses NF-κB pathway, thus reduces CCL5 expression and promotes CD8+ T cells infiltration in CRC microenvironment.
Conclusions
ALKBH5 is poorly expressed in CRC, and overexpression of ALKBH5 attenuates CRC malignant progression by inhibiting CRC cell proliferation, migration, invasion and promoting CD8+ T cells infiltration in the tumor microenvironment through NF-κB-CCL5 axis.
Introduction
Colorectal cancer is one of the most common cancers worldwide, ranking third for incidence and fifth for mortality rates [1]. Due to the insidious onset of CRC, approximately 25% of patients are already at an advanced stage when initially diagnosed [2]. Neoadjuvant chemotherapy is one of the standard of treatments in advanced colorectal cancer, and liver transplantation combined with chemotherapy has proven to improve overall survival in advanced colorectal cancer patients with liver metastases [3]. In recent years, new treatments, including targeted therapeutic agents and immunotherapy, have been extensively explored in the field of colorectal cancer treatment. Regorafenib (REG), an oral multikinase inhibitor, has been approved to treat colorectal cancer clinically [4]. Preclinically, immune checkpoint molecular targeted therapies have been widely used to treat tumors. However, only about 15% of colorectal cancer patients benefit from immune checkpoint blockade (ICB) therapy [5]. Recent studies have shown that immune checkpoint inhibitors combined with other treatments may prevent early mortality in some CRC patients [6]. MEK inhibitors combined with immunotherapy drug spartalizumab can improve the durability of responses in CRC patients [7]. Therefore, identifying new molecular regulating the occurrence and development of CRC which could synergize with ICB treatment, and the analysis of its underlying regulatory mechanism are of great significance to reduce the mortality and improve the prognosis of colorectal cancer patients.
N6-methyladenosine (m6A), as one of the most common site being intrinsic modified in eukaryotic mRNAs, is dynamically regulated by three categories proteins: methyltransferase as "writer," demethylase as "eraser," and specific "reader" m6A-binding protein [8,9]. To date, only fat mass and obesity-related protein (FTO) and ALKHB5 have been identified to function as m6A demethylases, which removes m6A modifications [10,11]. ALKBH5 acts as double-edged sword in different cancer types. ALKBH5 was reported to act as a tumor promoter in malignant tumors, such as gastric cancer, renal cell cancer and breast cancer [12], [13], [14]. Whereas in pancreatic cancer, bladder cancer, hepatocellular cancer [15], [16], [17] as well as CRC [18,19], ALKBH5 functions as a tumor suppressor.
Recent studies have shown that ALKBH5 may mediate tumor progression through immune-modulatory effects [20]. In intrahepatic cholangiocarcinoma, ALKBH5 upregulates PD-L1 expression in an m6A-YTHDF2-dependent manner, thereby inhibiting the cytotoxicity of T cells [21]. ALKBH5 is induced in a hypoxic environment and upregulates IL-8 expression, thereby promoting immunosuppression and tumor progression in GBM through tumor-associated macrophage (TAM) recruitment [22]. ALKBH5 promotes the accumulation of myeloid-derived suppressor cells (MDSCs) and Tregs and paralyzes the therapeutic efficacy of anti-PD-1 in melanoma [23]. Infiltration of CD8+ T cells in tumor tissues is associated with a better prognosis [24], [25], [26]. However, aberrant expression of ALKBH5 affecting CD8+ T cells infiltration into the tumor microenvironment has rarely been reported. Therefore, how expression of ALKBH5 impact immune cell infiltration in CRC TME requires further study.
To further explore the potential immunomodulatory role of ALKBH5 in CRC, we evaluated the expression of ALKBH5 in clinical CRC samples, verified the biological effect of ALKBH5 in colon cancer cells, and analyzed the mechanism of ALKBH5′s role in CD8+ T infiltration in tumor tissues.
Results
ALKBH5 is downregulated in CRC and interrelated with poor prognosis
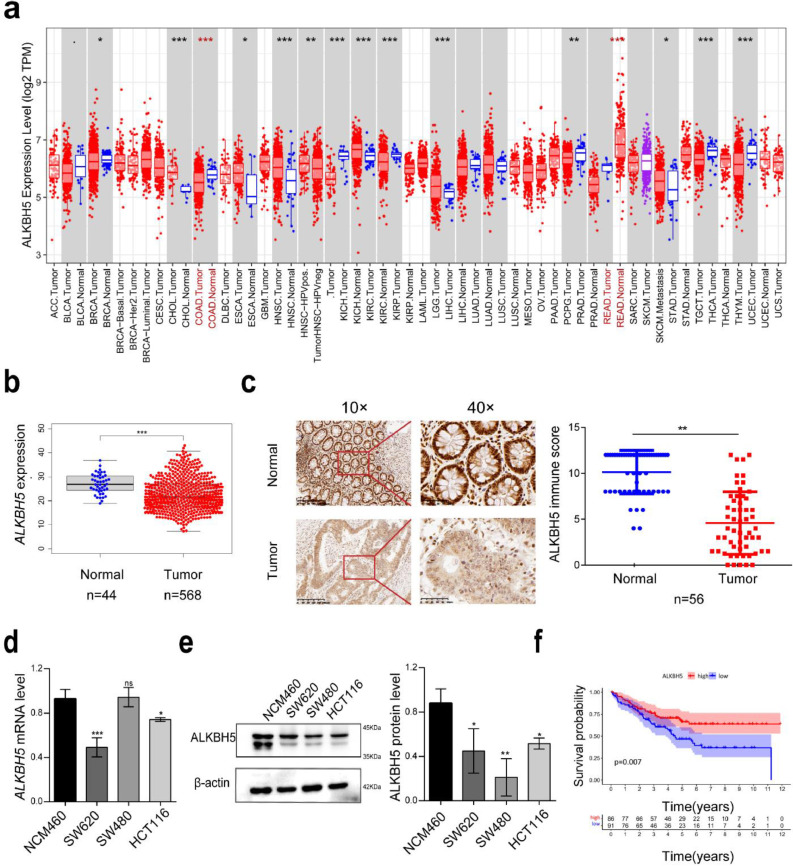
TIMER database results showed lower expression of ALKBH5 in CRC than in paired paraneoplastic tissues (Fig. 1a). To further determine the level of ALKBH5 expression in CRC, we analyze a cohort of CRC patients enrolled in TCGA. It exhibited that the expression of ALKBH5 was significantly down-regulated in CRC (Fig. 1b). Moreover, ALKBH5 expression level was decreased in CRC tissue microarray, which was in line with the results from TCGA database (Fig. 1c). Besides, ALKBH5 expression were also lower in CRC cell lines than the normal colonic epithelial cell line NCM460 at the mRNA and protein levels (Fig. 1d and e). Importantly, Kaplan-Meier analysis demonstrated that CRC patients with low ALKBH5 expression had lower overall survival (OS) rate indicating the potential prognostic role of ALKBH5 in CRC patients (Fig. 1f).
Fig. 1.
ALKBH5 expression is downregulated in CRC and strongly correlates with patient prognoses. a Pan-cancer analysis of ALKBH5 expression levels in TIMER database. b Expression of ALKBH5 in TCGA CRC cohort. c IHC showing representative images of ALKBH5 protein staining in tissue microarrays constructed from 56 CRC tissues and paired adjacent paraneoplastic tissues and statistical analysis. d Detection of mRNA levels of ALKBH5 in human normal colonic mucosal erithelial cells (NCM460) and CRC cancer cells (HCT116, SW620, SW480) by qRT-PCR. e Protein expression levels of ALKBH5 in normal NCM460 cells and three CRC cell lines. f Kaplan-Meier survival analysis shows the relationship between expression of ALKBH5 and overall patient survival time.
ALKBH5 attenuates proliferation, migration and invasion of CRC cells
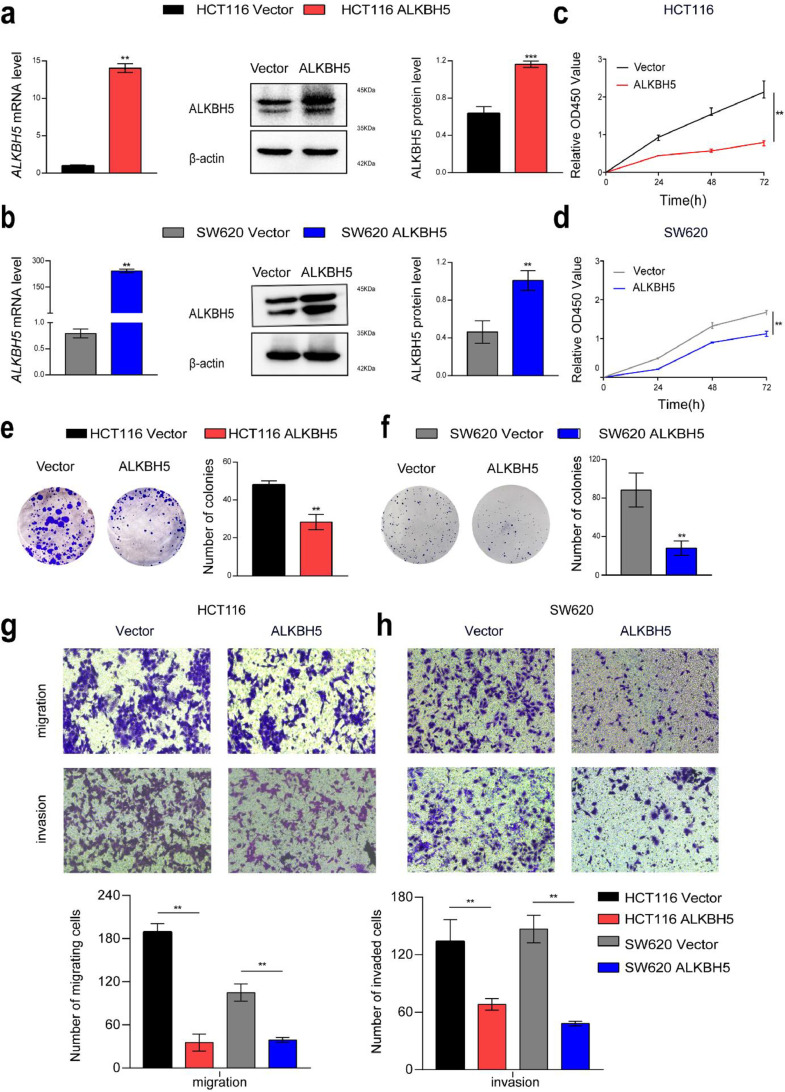
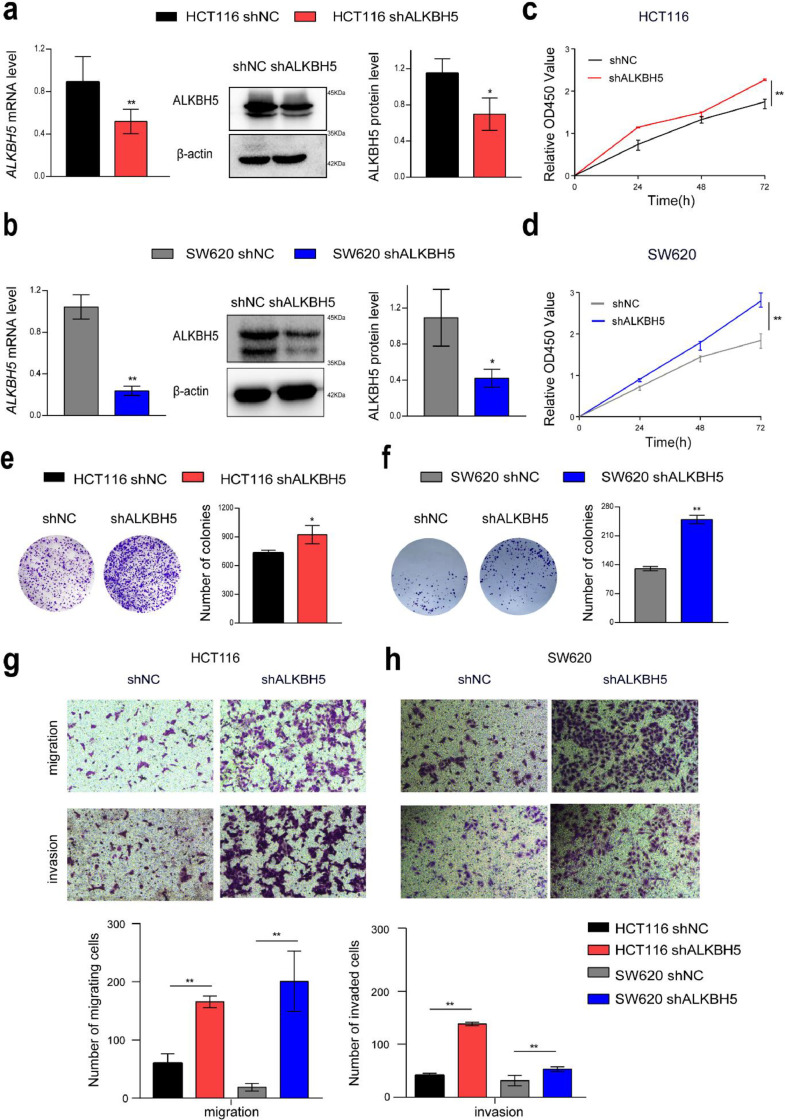
To explore the effects of ALKBH5 on the biological characteristics of CRC cells, we overexpressed or knocked down ALKBH5 in HCT116 and SW620 cells (Figs. 2a and b, and 3a and b). Overexpression of ALKBH5 functionally suppressed cell proliferation and viability, and cellular growth and viability was enhanced after knockdown of ALKBH5, determined by the cell counting kit-8 (CCK-8) and colony formation assay (Figs. 2c–f, and 3c–f). Transwell migration and invasion assay revealed that forced expression of ALKBH5 apparently impeded the invasiveness and migratory ability of HCT116 and SW620 cells, and attenuation of ALKBH5 expression significantly elevated the invasion and migration speed of HCT116 and SW620 cells (Figs. 2g and h, and 3g and h).
Fig. 2.
ALKBH5 attenuated proliferation, migration, and invasion of CRC cells. a-b qRT-PCR and western blot analysis of ALKBH5 mRNA and protein levels after overexpression ALKBH5 in HCT116 and SW620. c-d CCK8 was performed to detect the proliferation of CRC cells transfected with ALKBH5 plasmid compared with vector. e-f The colony formation assay provides representative images and quantification of ALKBH5 or vector-transfected HCT116 and SW620 cells. g-h The migratory and invasive capacities of ALKBH5 overexpressed HCT116 and SW620 cells were assessed using transwell migration and invasion assays. All data were indicative of three separate studies and shown as mean ± SD. Student's t-test. The Student's t-test was used to determine P values.
Fig. 3.
Knockdown of ALKBH5 promoted proliferation, migration, and invasion of CRC cells in vitro. a-b Knockdown of ALKBH5 was confirmed by real-time PCR and western blot (upper panel) in HCT116 and SW620 cells. c–h Transwell assay, colony formation, and cell counting assay in wild-type and ALKBH5 knocked-down cells.
Elevated levels of ALKBH5 are linked to enhanced CD8+ T cells recruitment to the tumor
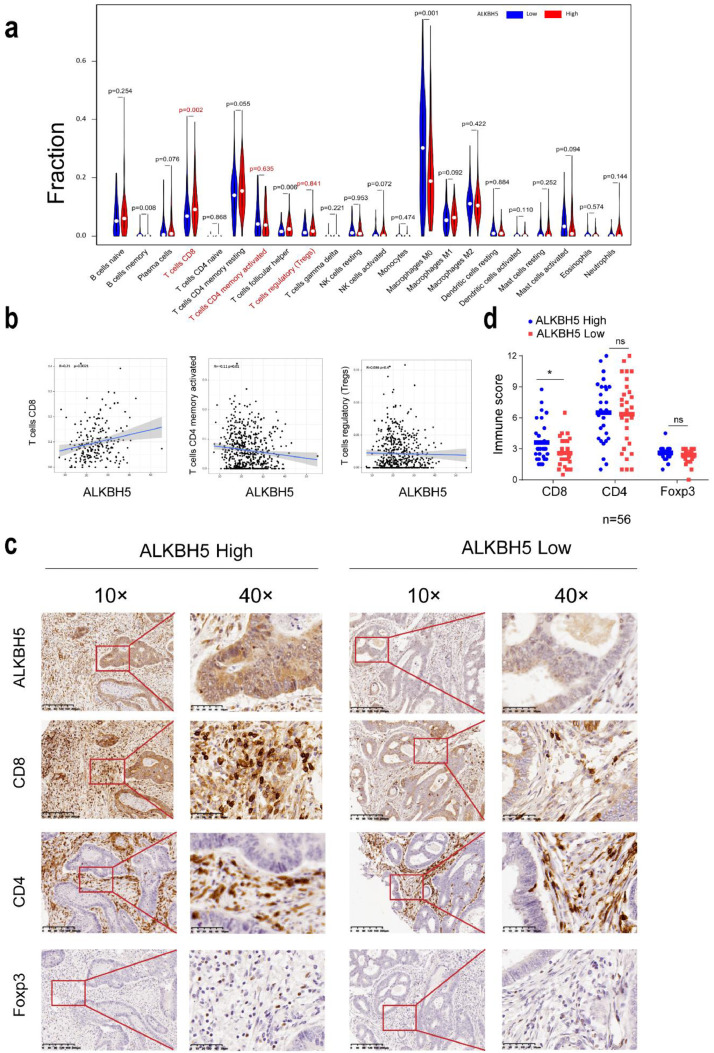
We continue to investigate if ALKBH5 was involved in regulating immune response to tumors. We performed CIBERSORT to identify the correlation between the infiltration of 22 types of immune cell and the expression level of ALKBH5 in CRC. It was found that ALKBH5 expression is positively correlated to elevated CD8+ T cells and activated memory CD4+ T infiltration, where no significant correlation to Treg infiltration (Fig. 4a and b). To further verify the results of bioinformatics analysis, we assessed the immune score of T-cell infiltration versus ALKBH5 expression via IHC staining of tissue microarray. Consistently, we found that the abundance of CD8+ T cells was significantly reduced in ALKBH5 low expression group. However, there was no significant correlations found in the infiltration of total CD4+ T cells and Foxp3+ Treg cells between two groups (Fig. 4c and d). According to these findings, ALKBH5 could be crucial to promote CD8+ T cells infiltration into the tumor site in CRC patients.
Fig. 4.
ALKBH5 expression promotes CD8 + T cells infiltration in human CRC tissues. a Infiltration level of 22 immune cells in the ALKBH5 high and low expression groups were examined using CIBERSORT. b Scatterplots show the correlation among activated memory CD4+T cells, CD8+T cells, Tregs and the ALKBH5 expression level. c-d Correlation and quantitative analyses of ALKBH5 expression with CD4, CD8 and Foxp3 in human CRC tissues using immunohistochemistry, the data represented mean ± SD (n=56).
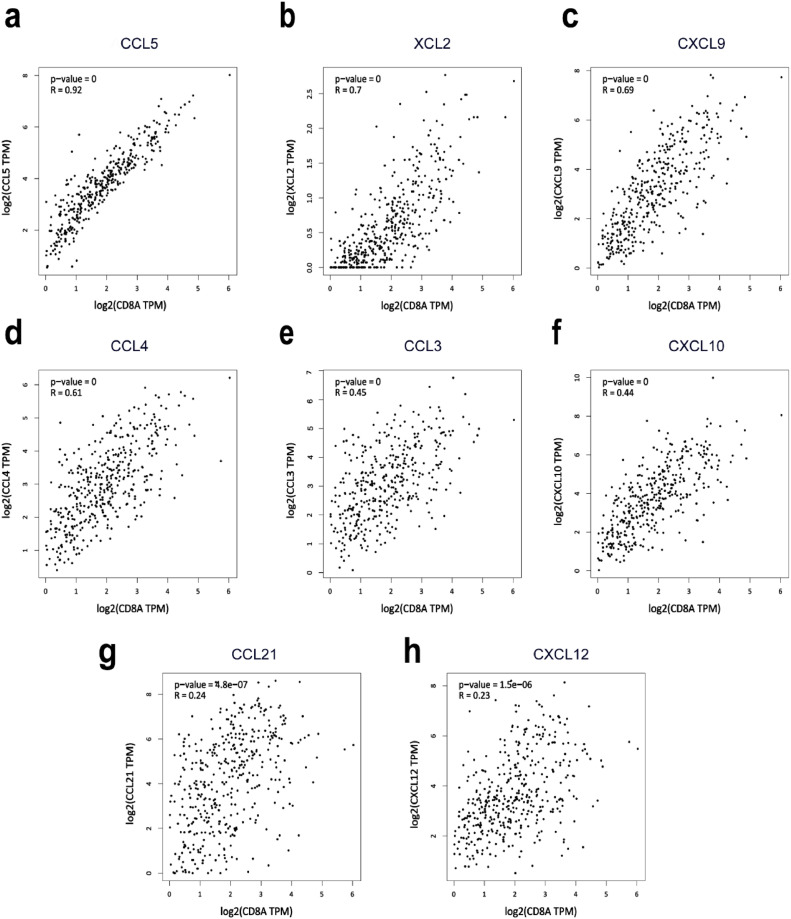
CD8A was significantly associated with CCL5 in CRC
Next, we sought to examine the underlying mechanism of how ALKBH5 expression regulates CD8+ T cells recruitment. Chemokines have been identified to contribute to the enrollment of effector T cells to the irradiated tumor. We then asked whether increased CD8+ T cells infiltration in high ALKBH5 group is associated with chemokine expression. We selected CD8A as a genetic marker for measuring CD8+ T cells infiltration [27]. GEPIA online database was utilized to analyze the association between multiple chemokines and CD8A in colorectal cancer, and CCL5 positively correlates to CD8A expression the most. Given the results above, we postulated here that ALKBH5 may impact CD8+ T cells recruitment by regulating CCL5 expression (Fig. 5).
Fig. 5.
Analysis of the correlations between chemokines and CD8A expression level. a–h CCL5, XCL2, CXCL9, CCL4, CCL3, CXCL10, CCL21 and CXCL12, respectively.
ALKBH5 promotes CD8+ T cells infiltration through NF-κB-CCL5 signaling pathway
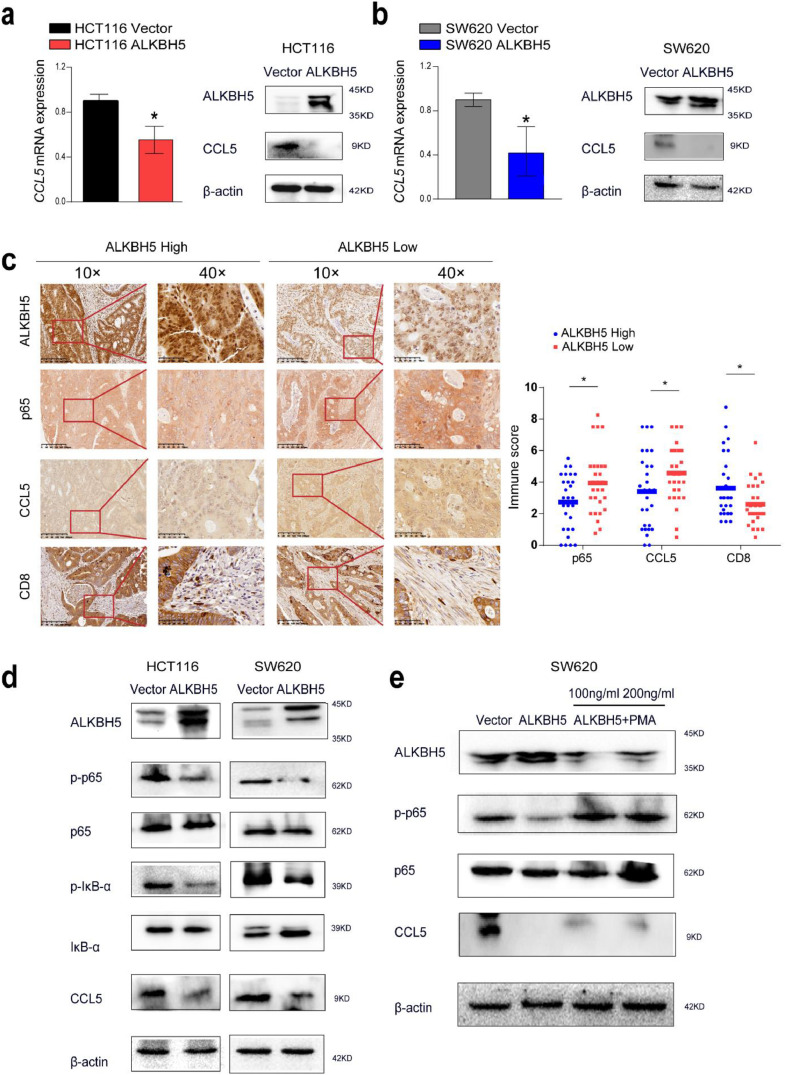
To further confirm the above hypothesis, we first evaluated whether ALKBH5 expression affected the expression of CCL5. Real-time PCR and western blot showed that CCL5 mRNA and protein abundance was significantly decreased in the ALKBH5-overexpressed cell lines compared with the control (Fig. 6a and b). It was reported that the promoter region of CCL5 has the binding site of nuclear transcription factor κappa- B (NF-κB) [28]. To confirm whether the NF-κB pathway is involved in the reduction of CCL5 expression regulated by ALKBH5, Immunohistochemical results confirmed that ALKBH5 high expression group suppressed the nuclear localization of p65 and CCL5 expression and promoted CD8+ T cells infiltrations (Fig. 6c). To confirm whether the NF-κB pathway is involved in the reduction of CCL5 expression regulated by ALKBH5, we performed western blotting analysis to examine the phosphorylation status of p65 and IκB-α. It was found that the phosphorylation levels of p65 and IκB-α were significantly reduced in the ALKBH5 overexpressed cell lines compared with the control cell lines (Fig. 6d). NF-κB is known to be instantaneously activated by PMA in a variety of mammalian cell types [29,30]. Thus, we treated SW620 cells with PMA and observed that PMA enhanced the phosphorylation of p65 and partially restore CCL5 expression compared with ALKBH5 alone (Fig. 6e). These data suggest that ALKBH5 promotes CD8+T cells infiltration via inhibiting the NF-κB-CCL5 axis.
Fig. 6.
ALKBH5 disrupting NF-κB-CCL5 signaling. a-b Relative expression of CCL5 in ALKBH5-overexpressed HCT116 and SW620 cells. c Immunohistochemistry was used to detect the expression of ALKBH5, p65, CCL5, and CD8 in human colorectal cancer tissues, the data represented mean ± SD (n=56). d CCL5 expression and the total and phosphorylation level of p65 and IκB-α after ALKBH5 overexpression were examined by western blot analysis. e SW620 were pretreated with 100 and 200 ng/ml of specific NF-κB activator PMA, and western blot analysis was to assess the expression of CCL5 and p-p65.
Materials and methods
Bioinformatics analysis
Using the TIMER database (https://cistrome.shinyapps.io/timer/), the expression profile of ALKBH5 in all types of cancer was investigated. The log2 TPM measurements are used to represent the gene expression levels. RNA-seq data for 568 CRC and 44 normal tissue samples from the TCGA database were downloaded. The Wilcoxon rank sum test was performed to examine the differences in ALKBH5 mRNA expression between CRC and normal tissues. CIBERSORT was employed in order to evaluate immune cell infiltration in CRC samples from the UCSC Xena database.
Clinical sample and immunohistochemistry
From January 4 to March 30, 2015, Shanxi Cancer Hospital collected 56 pairs of CRC and adjacent paraneoplastic paraffin specimens. Each patient gave informed permission prior to surgery. The expression of ALKBH5 and the infiltration levels of T cells (CD8+, CD4+ and Treg) in microenvironment were detected by immunohistochemistry. The antibodies used in this study included CD8 (1:1000, ab209775) (Abcam, Cambridge, UK), CD4 (1:1000, ab133636) (Cell Signaling Technology, MA, USA), Foxp3 (1:1000, ab20034) (Abcam, Cambridge, UK).
Paraffin-embedded tissues were sliced to a thickness of 2–4 μm using a slicer, dewaxed and hydrated in xylene and graded alcohol, after which the tissues were submerged in sodium citrate repair solution to achieve antigen repair, and then the sections were immersed in 3% hydrogen peroxide to block the activity of endogenous peroxidase. Afterwards, they were closed in a 5% bovine serum albumin closure solution. Then ALKBH5, CD8, CD4, Foxp3 and CCL5 antibodies were incubated on slides overnight and then exposed to anti-mouse/rabbit IgG polymer that has been HRP-labeled and complexes of diaminobenzidine.
Two pathologists evaluated expression in a double-blind fashion. The results were determined by the staining positive rate score which was defined as 1 (<25%), 2 (25%−50%), 3 (51%−75%), 4 (76%−100%) and the staining intensity score which was designated as 0 (negative), 1 (weak positive), 2 (medium positive) and 3 (strong positive). The staining positive rate score and the staining intensity score were multiplied to generate the overall score. The patients were split into groups with high and low ALKBH5 expression based on the median value of the ALKBH5 total score.
CRC cell lines and cell culture
The human CRC cell lines HCT116, SW480, and SW620 in this study were purchased from the Chinese Academy of Sciences' Cell Bank of Typical Cultures Preservation Committee. Human normal colonic epithelial cell NCM460 was bought from Shanxi Zheyuan Scientific Instrument Co. LTD. L15 medium was used for culturing SW480 and SW620 cells, and DMEM medium was used for NCM460 and HCT116 cells. Cells were grown in a medium that also contained 10% FBS and 100 U/mL of a penicillin-streptomycin mixture. The cells were kept at 37 °C in a humidified environment that contained 95% air and 5% CO2.
Construction of ALKBH5 overexpression and knockdown plasmid
Full-length ALKBH5 cDNA was synthesized from NCM460 by RT-PCR and subcloned into the pEGFP-C1-3×Flag vector to construct pEGFP-ALKBH5 overexpression plasmid. Short hairpin RNA (shRNA) sequences targeting ALKBH5 were synthesized by mature annealing and then cloned into plvx-zsGreen-scramble.
The sequences of hairpin sequence utilized in this article were listed below:
ALKBH5 forward primer:5′-CCGGAATTCGATGGCGGCCGCATCTTC-3′
reverse primer:5′-CGGGGTACCGTGCCGCCGCATCTTC-3′.
shALKBH5: 5′-GATGAAATCACTCACTGCATA-3′.
Plasmid transfection
Plasmids were transfected to tumor cells via Lipofectamine 8000 (Shanghai Biyuntian, China). After 48–72 h, its efficiency of overexpression or knockdown was detected by western blotting and qRT-PCR. Cancer cells with stable transfection were obtained by Geneticin (G418) screening for four weeks. All transfectants were periodically tested by western blot to confirm the efficiency of overexpression or knockdown.
Cell viability assay
The impact of ALKBH5 on the proliferation of colorectal cancer cells was measuerd by using CCK-8 assay. 5 × 103 per well HCT116 and SW620 cells were placed in 96-well plates and kept there for 24, 48 or 72 h. After treating the cells with 10 μl of CCK-8, the absorbance at 450 nm was measured after 1 to 2 h.
Colony formation assay
A colony formation experiment was used to investigate the ALKBH5 impacts CRC cells' ability to form colonies. Overexpression or knockdown plasmids stably expressing ALKBH5 were seeded at a density of 5000 cells per well in 6-well plates and maintaining their viability for two weeks in medium containing 10% fetal bovine serum were both done. After being preserved with methanol, the resulting colonies were then colored using crystal violet (Solarbio). ImageJ software was used to count the colonies by counting how many were stained.
Transwell assay
Transwell method was used for detecting the invasion and migration ability changes of CRC cells after the exogenous intervention. 8 × 104 cells (8.0 μm pore size, Corning, USA) were placed in 200 μl serum-free medium, also 600 μl of medium having 20% FBS, and was seeded in the bottom chamber as an inductance. Cells in the upper portion of the chamber that had not invaded the lower part of the chamber after 24 h were taken out. After that, they were dyed with crystal violet and captured on camera.
Transwell invasion assays were performed using 24-well transwell inserts of 60 μl matrix (ABW Matrigengel) in the top chamber (8.0 μm pore size, Corning, USA) to mimic the basement membrane, followed by overnight incubation of the 37 °C gel.
RNA isolation and reverse transcription (RT)-PCR assay
Total RNA was extracted using an RNA extraction kit (Cwbio, Beijing, China). After applying the Reverse transcription kit, total RNA was reverse transcribed into complimentary cDNA following the removal of genomic DNA with the gDNA eraser (Cwbio, Beijing, China). The SYBR Green PCR Master Mix was used to conduct cDNA amplification (Qiagen, Hilden, Germany). Following the reaction, the proportional gene expression was determined using the formula 2−ΔΔCt. The internal control was GAPDH.
The primers sequences are as follows:
ALKBH5: sense: 5′- CGGCGAAGGCTACACTTACG-3′,
antisense: 5′- CCACCAGCTTTTGGATCACCA-3′
CCL5: sense: 5′- CAGCAGTCGTCCACAGGTCAAG-3′,
antisense: 5′- TTTCTTCTCTGGGTTGGCACACAC-3′
GAPDH: sense: 5′-CAGGAGGCATTGCTGATGAT-3′,
antisense: 5′-GAAGGCTGGGGCTCATTT-3′
Western blot
The cells were washed with PBS and dissolved with RIPA buffer. To eliminate cell debris, the lysates were then ultrasound at 14,000 g for 5 min at 4 °C while being ultrasounded on ice. Next, the supernatant was decomposed in 10% SDS-PAGE (25 μg/ lane) and transferred to a 0.45 μm polyvinylidene fluoride (PVDF) membrane (GE Healthcare, Milwaukee, USA). Cell membranes were implemented at 4 °C with appropriate antibodies. After three washes, let the secondary antibody that has been HRP-labeled on film sit at room temperature for one hours. Enhanced chemiluminescence was used to show protein rings (ECL). In this research, antibodies were used that included ALKBH5(1:1000, ab195377, Abcam, Cambridge, UK), β-Actin (1:4000, TransGen Biotech, Beijing, China), CCL5 (1:500, AF5151, Affinity Biosciences, Cincinnati, OH, USA), NF-κB /p65 (1:1000, #8242), p-NF-κB /p-p65 (1:1000, #3033), IκB-α (1:1000, #4814), p-IκB-α (1:1000, #2859) (Cell Signaling Technology, MA, USA).
Statistical analysis
GraphPad Prism 8 was used to perform all statistical analyses, the Wilcoxon signed-rank test and the student t test were applied. The survival curves were measured using the Kaplan-Meier method, and the differences were evaluated using the log-rank test. The Pearson correlation analysis was used to evaluate correlations. Statistical significance was indicated by P values lower than 0.05. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
In our present study, we identified that the downregulated ALKBH5 expression correlates to worse CRC prognosis. Based on the transient ALKBH5 knockdown or forced expression cells, we then functionally delineated that ALKBH5 attenuates CRC growth and invasion in vitro. Furthermore, ALKBH5 expression was proved to promote CD8+ T cells recruitment by regulating CCL5 expression.
ALKBH5 is aberrantly expressed in a variety of tumors and significantly correlated with the prognosis of cancer patients [31]. Altered ALKBH5 expression could both promote or inhibit carcinogenesis, depending on the kind of cancer [31]. Ectopic upregulation of ALKBH5 occurs in NSCLC which is directly related to decreased patient survival. However, pancreatic cancer patients with high levels of ALKBH5 experienced an improved prognosis [32,33]. According to our research preclinically and clinically, down-regulation of ALKBH5 expression strongly correlates to worse OS in CRC patients. The demethylation of the lncRNA KCNK15-AS1 by ALKBH5 is shown to have tumor-suppressive effects, regulating KCNK15-AS1-dependent cell invasion and migration in PC [34]. In a m6A-YTHDF2-dependent manner, ALKBH5 post-transcriptionally upregulates PER1 expression, activating ATMCHK2-P53/CDC25C signaling and inhibiting PC cell growth [35]. Functional experiments in the present study further supported the finding that ALKBH5 prevents CRC cells proliferation, migration and invasion. These results demonstrated the cancer suppressor role of ALKBH5 in CRC.
There have been some reports that m6A methylation treatment of genes affects CD8+ T cells infiltration. Loss of the m6A methylase METTL14 in C1q+ macrophages leads to decreased m6A modification on Ebi3 transcripts and increased Ebi3 expression in macrophages, which results in lower and less potent of CD8+ T cells in TME [36]. METTL3 or METTL14 deletion in mouse melanoma and CRC cells enhances CD8+ T cell infiltration by modulating cytokine and chemokine production in the tumor microenvironment, thereby enhancing immune responses to anti-PD-1 therapy [37]. In- YTHDF1 knockout mice, lysosomal protein expression is reduced in dendritic cells and the antigen is not catabolized but rather enhances CD8+ T cell and natural killer (NK) cell infiltration [38]. RNA demethylase FTO can promote tumor cell glycolytic gene expression and glycolytic process, thus reducing the tumor infiltration and cell killing ability of CD8+T cells [39]. However, the relationship between ALKBH5 and CD8+ T cell infiltration in CRC remain unclear. In our study, we used bioinformatics to discover a positive correlation between ALKBH5 and CD8+ T cells infiltration that was statistically significant, and we achieved consistent results when study clinical tissue samples. Results suggested that lowering ALKBH5 expression would facilitate immune evasion by preventing CD8+ T cells recruitment, which could be further explored as a new therapeutic target in CRC patients.
Chemokines are the primary factors affecting CD8+ T cells trafficking to tumor locations [40,41]. Targeting KDM4C has been demonstrated to improve CD8+ T cells ability to fight tumors by triggering the transcription of the chemokine CXCL10 in lung cancer [42]. Chemokines CCL5 and CXCL9 are closely related to CD8+ T cells infiltration in various solid tumors [41].. By interfering with STAT1-CXCL10 signaling, TRIB3 prevents CD8+T cell invasion in CRC [43]. Recent research demonstrates that CCL5 deficiency in colorectal cancer boosts CD8+ T cells infiltration, the specific mechanism is that CCL5 absence leads in altered CD11b(hi)F4/80(low) tumor-associated macrophage (TAM) metabolism and decreased S100a9 expression [44]. In our study, to identify specific chemokines for ALKBH5-induced CD8+ T cells recruitment, we performed bioinformatics analysis and the data revealed eight chemokines with significantly positive correlation and CD8+ T cells infiltration. The discordance between literature report and bioinformatics analysis in the role of CXCL5 affecting CD8+ T cells trafficking to tumor locations attracted our attention. So we selected CCL5 which had the most notable correlation with CD8A. To interpret the disparity between the literature data and the mentioned bioinformatics, we confirmed that ALKBH5 overexpression could decrease the level of CCL5 both in mRNA and protein level, and highly expressed ALKBH5 encourages CD8+ T cells infiltration by lowering CCL5 expression in our immunohistochemical studies. These results are consistent with those reported in the literature [44]. The reason for this discrepancy may be attributed to that CD8, as a disulfide-linked dimeric glycoprotein on the cell surface, consists of two chains, CD8α and CD8β [45,46]. T cell surface glycoprotein CD8α chain (CD8A) alone has limitations as a marker of CD8+ T cells infiltration.
NF-κB has been well established as a transcription factor for CCL5 [47,48]. Furthermore, the effect of ALKBH5 on CCL5 expression was associated with inhibition of NF-κB pathway. To further analyze whether ALKBH5 down-regulates the expression of CCL5 by NF-κB in CRC, we used PMA to activate NF-κB, and the results showed that NF-κB activator could partially reverse the down-regulated expression of CCL5 by ALKBH5. Notably, NF-κB activation inhibited the expression of ALKBH5, suggesting that a negative feedback pathway may exist between ALKBH5 and NF-κB. Negative feedback control of NF-κB signaling has been well studied. It is reported that Zinc-Finger-protein A20 can regulate TRAF6 and RIP of NF-κB pathway through ubiquitination, which in turn NF-κB can negatively affect A20 [49]. Therefore, we hypothesize that ALKBH5 and NF-κB constitute a feedback loop to regulate each other's expression, however the specific mechanism of action deserves further investigation.
Our study was not free of limitations: first of all, we, did not investigate the target RNAs regulated by ALKBH5, as well as the specific m6A-binding protein which influence the translation and degradation of target RNAs. Secondly, the data dimension of this article contains tissue and cellular, but we still need further validation of the mechanism at animal levels. These unresolved limitations need to be addressed in future studies.
Funding
Supported by the Science and Technology Innovation Team of Shanxi Province [grant numbers 202204051002030]; Central leading local science and Technology Development Fund Project [grant numbers YDZJSX2022A057]; Basic Research Program of Shanxi Province [grant numbers 202103021223226 and 20210302124415].
Data availability
The data can be obtained upon a reasonable request from the corresponding author.
Ethics approval and consent to participate
The experiments were approved by the Ethics Committee of Shanxi Cancer Hospital. The processing of clinical tissue samples is in strict compliance with the ethical standards of the Declaration of Helsinki. Each patient gave informed permission prior to surgery.
Consent for publication
Consent for publication was obtained from the participants.
CRediT authorship contribution statement
Jing Ge: Data curation, Formal analysis, Writing – original draft. Sheng-Lu Liu: Methodology, Data curation, Resources. Jing-Xiu Zheng: Funding acquisition, Writing – review & editing. Yu Shi: Conceptualization, Validation. Ying Shao: Formal analysis, Funding acquisition. Yu-Jing Duan: Validation, Visualization. Rui Huang: Investigation, Project administration. Li-Jun Yang: Supervision, Project administration, Validation. Tao Yang: Supervision, Funding acquisition, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no competing interests.
References
- 1.Siegel R.L., Miller K.D., Goding S.A., et al. Colorectal cancer statistics, 2020 [J] CA Cancer J. Clin. 2020;70(3):145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 2.He S.Y., Li Y.C., Wang Y., et al. Fecal gene detection based on next generation sequencing for colorectal cancer diagnosis [J] World J. Gastroenterol. 2022;28(25):2920–2936. doi: 10.3748/wjg.v28.i25.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandi G., Ricci A.D., Rizzo A., et al. Is post-transplant chemotherapy feasible in liver transplantation for colorectal cancer liver metastases? [J] Cancer Commun. (Lond.) 2020;40(9):461–464. doi: 10.1002/cac2.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizzo A., Nannini M., Novelli M., et al. Dose reduction and discontinuation of standard-dose regorafenib associated with adverse drug events in cancer patients: a systematic review and meta-analysis [J] Ther. Adv. Med. Oncol. 2020;12 doi: 10.1177/1758835920936932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Man S.M., Jenkins B.J. Context-dependent functions of pattern recognition receptors in cancer [J] Nat. Rev. Cancer. 2022;22(7):397–413. doi: 10.1038/s41568-022-00462-5. [DOI] [PubMed] [Google Scholar]
- 6.Viscardi G., Tralongo A.C., MASSARI F., et al. Comparative assessment of early mortality risk upon immune checkpoint inhibitors alone or in combination with other agents across solid malignancies: a systematic review and meta-analysis [J] Eur. J. Cancer. 2022;177:175–185. doi: 10.1016/j.ejca.2022.09.031. [DOI] [PubMed] [Google Scholar]
- 7.Tuca A., Gallego R., Ghanem I., et al. Chemotherapy and targeted agents in the treatment of elderly patients with metastatic colorectal cancer [J] J. Clin. Med. 2020;9(12) doi: 10.3390/jcm9124015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao G.C., Li H.B., Yin Z.N., et al. Recent advances in dynamic m(6)A RNA modification [J] Open Biol. 2016;6(4) doi: 10.1098/rsob.160003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong J.Y., Flavell R.A., Li H.B. RNA m(6)A modification and its function in diseases [J] Front. Med. 2018;12(4):481–489. doi: 10.1007/s11684-018-0654-8. [DOI] [PubMed] [Google Scholar]
- 10.Zhang S.C., Zhao B.S., Zhou A.D., et al. m(6)A Demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program [J] Cancer Cell. 2017;31(4):591. doi: 10.1016/j.ccell.2017.02.013. -+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng G.Q., Dahl J.A., Niu Y.M., et al. ALKBH5 Is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility [J] Mol. Cell. 2013;49(1):18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J., Guo S., Piao H.Y., et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1 [J] J. Physiol. Biochem. 2019;75(3):379–389. doi: 10.1007/s13105-019-00690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X., Wang F., Wang Z., et al. ALKBH5 promotes the proliferation of renal cell carcinoma by regulating AURKB expression in an m(6)A-dependent manner [J] Ann. Transl. Med. 2020;8(10):646. doi: 10.21037/atm-20-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang C.Z., Samanta D., Lu H.Q., et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA [J] Proc. Natl. Acad. Sci. USA. 2016;113(14):E2047–E2E56. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang B., Yang Y.H., Kang M., et al. m(6)A demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF-1 RNA methylation and mediating Wnt signaling [J] Mol. Cancer. 2020;19(1) doi: 10.1186/s12943-019-1128-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Yu H., Yang X., Tang J.Y., et al. ALKBH5 inhibited cell proliferation and sensitized bladder cancer cells to cisplatin by m6A-CK2 alpha-mediated glycolysis [J] Mol. Ther. Nucleic Acids. 2021;23:27–41. doi: 10.1016/j.omtn.2020.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y., Zhao Y., Chen J., et al. ALKBH5 suppresses malignancy of hepatocellular carcinoma via m(6)A-guided epigenetic inhibition of LYPD1 [J] Mol. Cancer. 2020;19(1):123. doi: 10.1186/s12943-020-01239-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu X.B., Dai M.Z., Li J.L., et al. m(6)A demethylase ALKBH5 inhibits cell proliferation and the metastasis of colorectal cancer by regulating the FOXO3/miR-21/SPRY2 axis [J] Am. J. Transl. Res. 2021;13(10):11209–11222. [PMC free article] [PubMed] [Google Scholar]
- 19.Yang P., Wang q., Liu A. ALKBH5 holds prognostic values and inhibits the metastasis of colon cancer [J] Pathol. Oncol. Res. 2020;26(3):1615–1623. doi: 10.1007/s12253-019-00737-7. [DOI] [PubMed] [Google Scholar]
- 20.Maimaiti A., Tuersunniyazi A., Meng X.H., et al. N6-methyladenosine RNA methylation regulator-related alternative splicing gene signature as prognostic predictor and in immune microenvironment characterization of patients with low-grade glioma [J] Front. Genet. 2022;13 doi: 10.3389/fgene.2022.872186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qiu X., Yang S., Wang S., et al. M(6)A demethylase ALKBH5 regulates PD-L1 expression and tumor immunoenvironment in intrahepatic cholangiocarcinoma [J] Cancer Res. 2021;81(18):4778–4793. doi: 10.1158/0008-5472.Can-21-0468. [DOI] [PubMed] [Google Scholar]
- 22.Dong F., Qin X., Wang B., et al. ALKBH5 facilitates hypoxia-induced paraspeckle assembly and IL8 secretion to generate an immunosuppressive tumor microenvironment [J] Cancer Res. 2021;81(23):5876–5888. doi: 10.1158/0008-5472.Can-21-1456. [DOI] [PubMed] [Google Scholar]
- 23.Li N., Kang Y., Wang L., et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment [J] Proc. Natl. Acad. Sci. USA. 2020;117(33):20159–20170. doi: 10.1073/pnas.1918986117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu L., Liu H., Guo H., et al. Circulating and tumor-infiltrating myeloid-derived suppressor cells in cervical carcinoma patients [J] Oncol. Lett. 2018;15(6):9507–9515. doi: 10.3892/ol.2018.8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oberg A., Samii S., Stenling R., et al. Different occurrence of CD8+, CD45R0+, and CD68+ immune cells in regional lymph node metastases from colorectal cancer as potential prognostic predictors [J] Int. J. Colorectal Dis. 2002;17(1):25–29. doi: 10.1007/s003840100337. [DOI] [PubMed] [Google Scholar]
- 26.Pagès F., Berger A., Camus M., et al. Effector memory T cells, early metastasis, and survival in colorectal cancer [J] N. Engl. J. Med. 2005;353(25):2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 27.Dangaj D., Bruand M., Grimm A.J., et al. Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors [J] Cancer Cell. 2019;35(6):885–900. doi: 10.1016/j.ccell.2019.05.004. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moriuchi H., Moriuchi M., Fauci A.S. Nuclear factor-kappa B potently up-regulates the promoter activity of RANTES, a chemokine that blocks HIV infection [J] J. Immunol. 1997;158(7):3483–3491. [PubMed] [Google Scholar]
- 29.Lallena M.J., Diaz-Meco M.T., Bren G., et al. Activation of IkappaB kinase beta by protein kinase C isoforms [J] Mol. Cell Biol. 1999;19(3):2180–2188. doi: 10.1128/mcb.19.3.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peters R.T., Liao S.M., Maniatis T. IKKepsilon is part of a novel PMA-inducible IkappaB kinase complex [J] Mol Cell. 2000;5(3):513–522. doi: 10.1016/s1097-2765(00)80445-1. [DOI] [PubMed] [Google Scholar]
- 31.Qu J., Yan H., Hou Y., et al. RNA demethylase ALKBH5 in cancer: from mechanisms to therapeutic potential [J] J. Hematol. Oncol. 2022;15(1):8. doi: 10.1186/s13045-022-01224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho S.H., Ha M., Cho Y.H., et al. ALKBH5 gene is a novel biomarker that predicts the prognosis of pancreatic cancer: a retrospective multicohort study [J] Ann. Hepatobiliary Pancreat. Surg. 2018;22(4):305–309. doi: 10.14701/ahbps.2018.22.4.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geng Y., Guan R., Hong W., et al. Identification of m6A-related genes and m6A RNA methylation regulators in pancreatic cancer and their association with survival [J] Ann. Transl. Med. 2020;8(6):387. doi: 10.21037/atm.2020.03.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Y., Hu H., Wang Y., et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation [J] Cell Physiol. Biochem. 2018;48(2):838–846. doi: 10.1159/000491915. [DOI] [PubMed] [Google Scholar]
- 35.Guo X., Li K., Jiang W., et al. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner [J] Mol. Cancer. 2020;19(1):91. doi: 10.1186/s12943-020-01158-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong L., Chen C., Zhang Y. The loss of RNA N(6)-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8(+) T cell dysfunction and tumor growth [J] Cancer Cell. 2021;39(7):945–957. doi: 10.1016/j.ccell.2021.04.016. e10. [DOI] [PubMed] [Google Scholar]
- 37.Wang L., Hui H., Agrawal K., et al. m(6) A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy [J] EMBO J. 2020;39(20) doi: 10.15252/embj.2020104514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han D., Liu J., Chen C. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells [J] Nature. 2019;566(7743):270–274. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., Liang G., Xu H., et al. Tumors exploit FTO-mediated regulation of glycolytic metabolism to evade immune surveillance [J] Cell Metab. 2021;33(6):1221–1233. doi: 10.1016/j.cmet.2021.04.001. e11. [DOI] [PubMed] [Google Scholar]
- 40.Nagarsheth N., Wicha M.S., Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy [J] Nat. Rev. Immunol. 2017;17(9):559–572. doi: 10.1038/nri.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dangaj D., Bruand M., Grimm A.J., et al. Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors [J] Cancer Cell. 2019;35(6):885. doi: 10.1016/j.ccell.2019.05.004. -+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jie X., Chen Y., Zhao Y., et al. Targeting KDM4C enhances CD8(+) T cell mediated antitumor immunity by activating chemokine CXCL10 transcription in lung cancer [J] J. Immunother. Cancer. 2022;10(2) doi: 10.1136/jitc-2021-003716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shang S., Yang Y.W., Chen F., et al. TRIB3 reduces CD8(+) T cell infiltration and induces immune evasion by repressing the STAT1-CXCL10 axis in colorectal cancer [J] Sci. Transl. Med. 2022;14(626) doi: 10.1126/scitranslmed.abf0992. [DOI] [PubMed] [Google Scholar]
- 44.Zhang S., Zhong M., Wang C., et al. CCL5-deficiency enhances intratumoral infiltration of CD8(+) T cells in colorectal cancer [J] Cell Death Dis. 2018;9(7):766. doi: 10.1038/s41419-018-0796-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang R., Natarajan K., Margulies D.H. Structural basis of the CD8 alpha beta/MHC class I interaction: focused recognition orients CD8 beta to a T cell proximal position [J] J. Immunol. 2009;183(4):2554–2564. doi: 10.4049/jimmunol.0901276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devine L., Kieffer L.J., Aitken V. Human CD8 beta, but not mouse CD8 beta, can be expressed in the absence of CD8 alpha as a beta beta homodimer [J] J. Immunol. 2000;164(2):833–838. doi: 10.4049/jimmunol.164.2.833. [DOI] [PubMed] [Google Scholar]
- 47.Gu L., Sang Y., Nan X. circCYP24A1 facilitates esophageal squamous cell carcinoma progression through binding PKM2 to regulate NF-κB-induced CCL5 secretion [J] Mol. Cancer. 2022;21(1):217. doi: 10.1186/s12943-022-01686-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang B., Qin Y., Wu Q. mTOR signaling pathway regulates the release of proinflammatory molecule CCL5 implicated in the pathogenesis of autism spectrum disorder [J] Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.818518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martens A., Hertens P., Priem D. A20 controls RANK-dependent osteoclast formation and bone physiology [J] EMBO Rep. 2022;23(12):e55233. doi: 10.15252/embr.202255233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data can be obtained upon a reasonable request from the corresponding author.