Abstract
Despite having a generally conserved structure, chloroplast genome data have been helpful for plant population genetics and evolution research. To mine Pueraria montana chloroplast genome variation architecture and phylogeny, we investigated the chloroplast variation architecture of 104 P. montana accessions from across China. P. montana’s chloroplast genome showed high diversity levels, with 1674 variations, including 1118 single nucleotide polymorphisms and 556 indels. The intergenic spacers, psbZ-trnS and ccsA-ndhD, are the two mutation hotspot regions in the P. montana chloroplast genome. Phylogenetic analysis based on the chloroplast genome dataset supported four P. montana clades. P. montana variations were conserved among and within clades, which showed high gene flow levels. Most P. montana clades were estimated to have diverged at 3.82–5.17 million years ago. Moreover, the East Asian summer monsoon and South Asian summer monsoon may have accelerated population divergence. Our results show that chloroplast genome sequences were highly variable and can be used as molecular markers to assess genetic variation and relationships in P. montana.
Keywords: Pueraria montana, population structure, genetic diversity, chloroplast genome, divergence time estimation
1. Introduction
Chloroplasts are maternally inherited in most angiosperms and have their own genome. The chloroplast genome is circular, ranging from 107 to 218 kb among the photosynthetic plant species [1]. Comparative genomic architecture studies have shown that chloroplast genome structures, gene numbers, and gene content organization are highly conserved among most land plants. The typical chloroplast genome contains two inverted repeats (IR) regions that separate large (LSC) and small single-copy (SSC) regions. The chloroplast genome encodes 120–130 genes that primarily participate in photosynthesis, transcription, and translation [2].
The chloroplast genome’s evolution rate is moderate compared with nuclear or mitochondrial genomes, and the ratio of mutation rates in the mitochondrial, chloroplast, and nuclear genes is about 1:3:10 in seed plants [3]. The chloroplast genome’s protein-coding gene sequences are widely used for phylogenetic analysis at high taxonomic levels, such as family or order levels [4,5,6,7,8]. Moreover, whole chloroplast genome sequences are popular molecular markers for reconstructing relationships and species identification due to their variability among interspecies levels [9,10,11]. For example, the coding regions of maturase K (matK), ribulose bisphosphate carboxylase large subunit (rbcL), and ycf1 are used as DNA barcodes for plants [12,13]. Similarly, spacer regions trnH-psbA, rpl32-trnL, rps16-trnQ, and rbcL-accD have a long history in plants evolution analysis [14,15].
Chloroplast genome sequences have been widely used as the DNA marker for genetic diversity research with the development of next-generation sequencing (NGS) methods. Previous studies have provided many genetic resources for single sequence repeats (SSRs), single nucleotide polymorphisms (SNPs), and insertion/deletion polymorphisms (indels) in chloroplast genomes at the intraspecies level. Chloroplast genetic resources have been used to examine genetic divergence within endangered or medicinal species [16,17,18], biogeographical structure [19,20,21,22], gene flow among subpopulations [23,24], and cultivar origins and domestication [25,26,27]. Maternally inherited chloroplast-genome-based evolutionary studies must sometimes be complemented by nuclear genomic information.
Pueraria Candolle includes ~20 species distributed in Southeast Asia [28]. Lee and Hymowitz (2001) were first to use molecular methods to confirm polyphyly within Pueraria, although they concluded that a “more rigorous molecular investigation” was required to definitively comprehend its evolutionary history. Egan et al. (2016) further confirmed polyphyly within Pueraria s. l., and all analyses reveal five distinct clades defined as five genera throughout the Phaseoleae tribe—P. wallichii, P. stricta, P. phaseoloides, P. peduncularis + P. yunnanensis, and the remaining species grouped into the main clade of Pueraria, called Pueraria sensu stricto (Pueraria s. str.) [29]. And Pueraria s. s. is sister to Psoraleeae. Some Pueraria species located in Pueraria s. str., especially for Pueraria montana (Lour.) Merr. (kudzu), have been used to treat medical ailments and as textile, food, and paper sources since 500 BC in China. The P. montana was estimated to have separated from its closely related species at around 4.63 million years ago (mya). It has been divided into three varieties: var. lobata (Willd.) Maesen & S.M.Almeida ex Sanjappa & Predeep, var. montana, and var. thomsonii (Benth.) M.R.Almeida. However, these three varieties are notoriously difficult to identify morphologically due to overlapping taxonomic characteristics, such as the flower size and terminal leaflet shape, which are the predominant characteristics for distinguishing them [28,30]. Recent molecular data supported the three varieties forming a monophyletic group [29]; however, the three varieties did not form as separate clades [31]. P. montana var. lobata is a noxious invasive species in North America, first introduced at the 1876 Centennial Exposition in Philadelphia [32].
Previous studies have explored P. montana’s genetic diversity using various markers, including allozymes [32,33], random amplified polymorphic DNAs (RAPDs) [34], sequence-related amplified polymorphisms [35], inter-simple sequence repeats (ISSRs) [36,37,38], and microsatellites [39,40,41]. However, few studies have examined its population structure and genetic diversity based on DNA sequence markers. With their high reproducibility, DNA sequence markers increase our likelihood and ability to detect genetic variation [42]. P. montana’s chloroplast and nuclear genomes have been sequenced [43,44,45], enabling their use for genetic and evolutionary studies.
This study sequenced the chloroplast genomes of 70 P. montana accessions from mostly wild distributions in China. The complete chloroplast genome was assembled to identify sequence variations and diversity among accessions. Phylogenetic, population structure, and principal component analyses were used to investigate genetic divergence among P. montana accessions and identify their subclades. Furthermore, we assessed genetic divergence and distance among subclades. These results provide a further case study for discovering chloroplast genome mutation rates at the intraspecies level and a genetic variation resource for breeding this ecologically and economically important species, and management of invasive species.
2. Results
2.1. Pueraria Montana Plastid Genome Assembly
This study used genome skimming methods to assemble complete chloroplast genomes for 70 P. montana accessions with high mean coverage (120×). The P. montana chloroplast genomes were circular molecules with a typical quadripartite structure and sizes of 149,452–154,595 bp, including two IRs of 23,678–25,655 bp, separated by an LSC of 84,055–85,316 bp and SSC of 17,957–18,017 bp. Their overall GC content was 35.3%–35.4%. P. montana’s chloroplast genome encodes 124 genes, including 87 protein-coding genes, 29 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. The 87 protein-coding genes are divided into self-replicated, photosynthesis-related, and other types. All annotated chloroplast genomes were deposited in GenBank (Table S1).
2.2. Chloroplast Genome Sequence Variation in Pueraria Montana
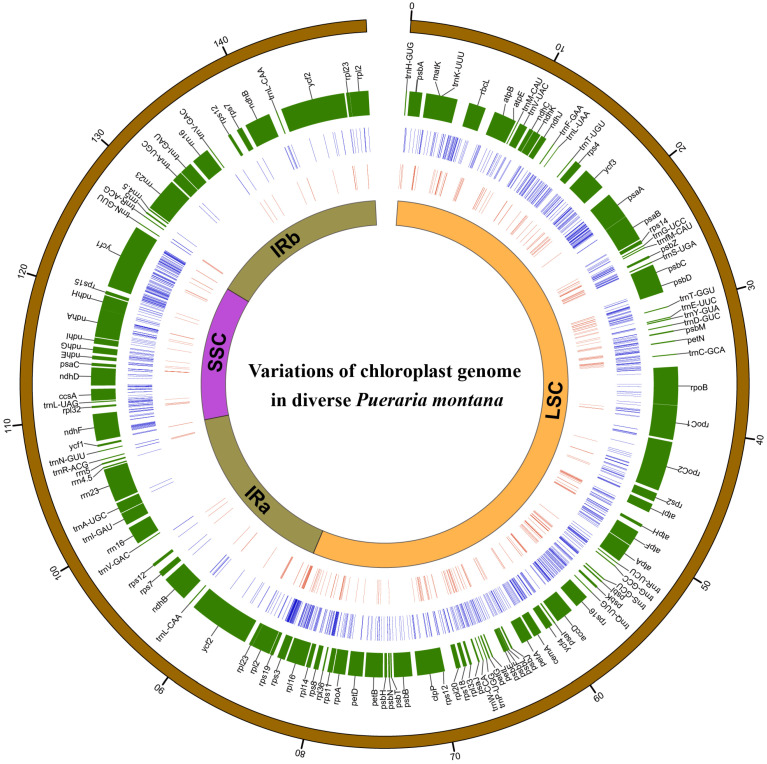
Combining these P. montana chloroplast genomes with those from GenBank, there were 104 accessions in total (Table S1). The alignment length was 158,274 bp, and 1674 variations, including 1118 SNPs and 556 indels, were identified in the P. montana chloroplast genomes (Figure 1). The 1118 SNPs included 615 singleton and 503 parsimony-informative sites. The average SNP density was 10.46/kb across the entire chloroplast genome, 9.26/kb in the LSC, 13.28/kb in the SSC, and 1.7/kb in the IR region. The overall π was 0.00055. The IR region had a lower SNP-based genetic diversity than the LSC and SSC regions. When π was averaged over 800 bp windows, two intergenic spacers (psbZ-trnS and ccsA-ndhD) had the highest sequence divergence (Figure S1). We identified 556 indels in the chloroplast genomes of the 104 P. montana accessions, most located in noncoding regions.
Figure 1.
The overall distribution of SNPs and indels across the P. montana chloroplast genome. A circular map showing the chloroplast genome structure with the outer distance shown in kb. All genes (including protein coding, tRNAs, and rRNAs) and their locations are shown in the second circle. The third and fourth circles show SNPs and indels identified across all 104 accessions, respectively.
2.3. Pueraria Montana Phylogeny based on the Chloroplast Genome
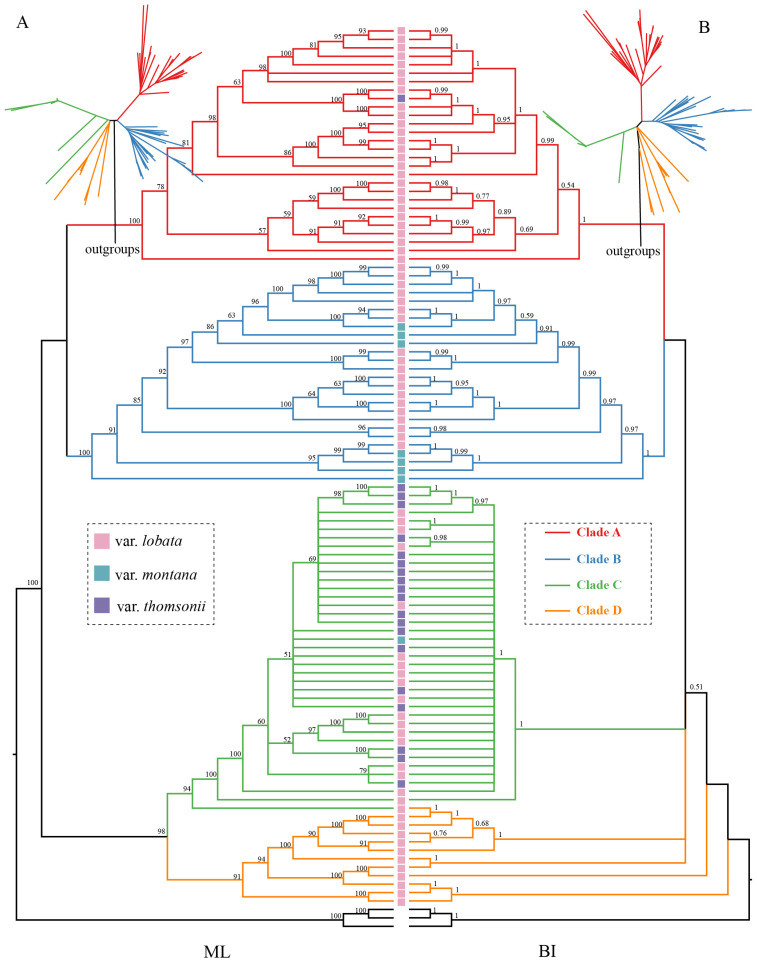
We used maximum likelihood (ML) and Bayesian inference (BI) methods to construct phylogenetic trees based on entire chloroplast genome sequences (Figure 2). A tanglegram was used to compare ML and BI trees, showing that the overall phylogenetic structure and clustering of the accessions in the two trees were nearly identical (the same accession in two trees can connect with itself at the same location in the clusters). Both results supported the separation of the 104 accessions into 4 clades (Figure 2). The resulting network with 4 subclades is shown in Figure S2.
Figure 2.
A tanglegram phylogenetic analysis using (A) ML and (B) BI trees to illustrate the relationships among P. montana accessions. Different colored squares represent different variants.
Twenty-eight accessions from Anhui, Chongqing, Gansu, Guangdong, Henan, Hubei, Jiangsu, Jilin, Liaoning, Shaanxi, Shandong, Shanxi, and Yunnan formed Clade A, including 27 haplotypes. Clade B contained 26 samples, including 24 haplotypes. Clade A was sister to Clade B with high support values in both ML and BI trees. Clade C included the most samples with 39 accessions, containing 26 samples from GenBank, including 2 varieties of var. thomsonii and var. lobata. The ML tree supported Clade C being a sister to Clade D with 98% support. However, the BI tree did not support Clade D as a separate clade. Clade D included 11 samples from Gansu, Shandong, Shanxi, Sichuan, Jiangsu, Zhejiang, Shaanxi, Hubei, and Hebei. Clades C and D had 10 and 11 distinct haplotypes, respectively.
2.4. Population Structure and PCA of Pueraria Montana
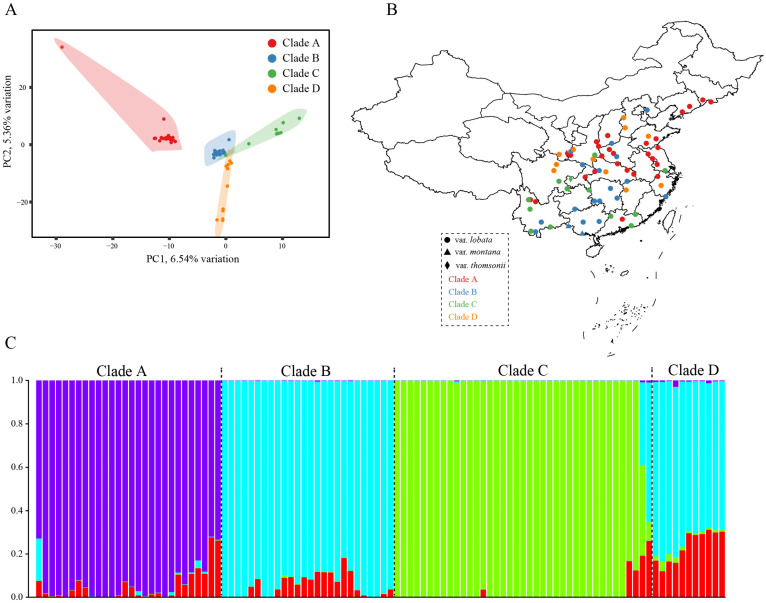
A PCA based on the entire chloroplast genome sequences showed four significant principal components (PCs; Figure 3A). The first two PCs only explain >11.9 % of the total variance. The first PC significantly distinguished Clade A from Clade C, and the second PC distinguished Clade A from some accessions in Clade D. These findings indicate that population genetics research might use analyses based on the chloroplast genome.
Figure 3.
Population structure and PCA of all 104 accessions. (A) PCA of all accessions. (B) The position of 70 collected P. montana accessions (C) Population structure clustering with the optimal K (4).
STRUCTURE, which calculates individual ancestry and admixture proportions assuming K populations, was used to examine population structure among the 104 accessions based on the entire chloroplast genome. We examined population structures with values of K between 2 and 10, with 10 iterations performed for each. The most suitable “blood lineages” of P. montana accessions were determined using the DeltaK, and the largest value was K = 4 (Figure 3B). The population structures with K = 3, 5, and 6 are shown in Figure S4.
2.5. Pueraria Montana Divergence Time
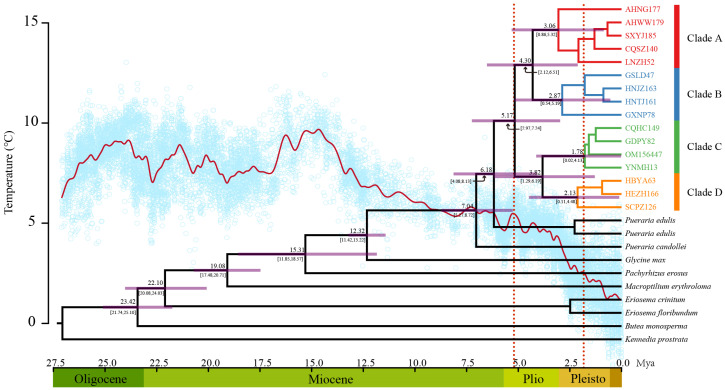
The divergence time estimates for the stem and crown nodes of Pueraria s. str. were 12.32 million years ago (Ma) (95% highest posterior density [HPD]: 11.42–13.22 Ma) in the middle Miocene and 7.04 Ma (95% HPD: 5.72–8.72 Ma) in the late Miocene, respectively (Figure 4). Chloroplast genome-based phylogenetic inferences subdivided the 104 accessions into four clades. Molecular dating analyses suggested that they first diverged during the early Pliocene, around 5.17 Ma (95% HPD: 2.97–7.24 Ma). The crown age of Clades A and B was 4.3 Ma (95% HPD: 2.12–6.51 Ma) in the middle Pliocene. The divergence time between Clades C and D was 3.82 Ma in the middle Pliocene. The crown ages of Clades A, B, C, and D were 3.06, 2.87, 1.78, and 2.13 Ma, respectively, indicating that most genetic variants arose during the Pleistocene. The divergence time of the four clades coincided with global cooling during the Pliocene (Figure 4).
Figure 4.
Glycininae divergence time. Numbers at nodes refer to their mean age (Ma). The purple bars correspond to the 95% HPD. A global average δ18O curve derived from benthic foraminifera, which mirrors the major global temperature trends about 28 MYA (red line), is shown below the tree [46,47].
2.6. Genetic Evolution in Different Groups
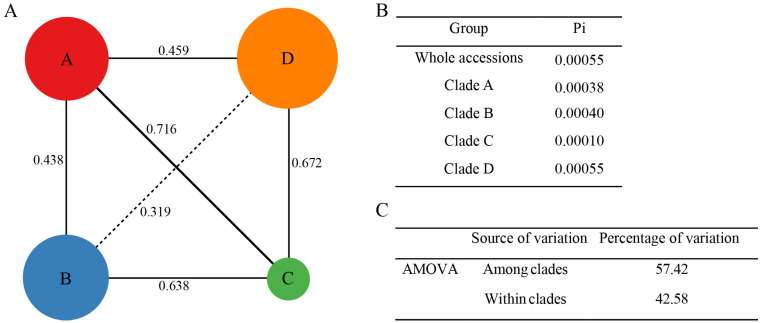
The four subclades identified in the phylogeny and PCA analyses were used to compare intra-clade divergence. The mean π of the four subclades were calculated as 0.00038, 0.00040, 0.00010, and 0.00055, respectively (Figure 5A,B). Clade A, comprising 464 SNPs and 190 indels, had the highest diversity, and Clade C, comprising 221 SNPs and 104 indels, had the lowest (Table 1).
Figure 5.
Chloroplast genome nucleotide diversity (π) and genetic distance (Fst). (A) The π (circle size) of the four clades (circles) and Fst among them. (B) Average π for the entire collection and each clade. (C) The AMOVA results among and within clades.
Table 1.
Summary of the total variations (SNPs/indels) detected in the whole collection and each clade.
| Group | Accessions | All Variations | |||
|---|---|---|---|---|---|
| SNPs | Indels | Total | Density/kb | ||
| Clade A | 28 | 464 | 190 | 654 | 4.09 |
| Clade B | 26 | 422 | 225 | 647 | 4.04 |
| Clade C | 39 | 221 | 104 | 325 | 2.03 |
| Clade D | 11 | 232 | 151 | 383 | 2.39 |
| Total | 104 | 1118 | 556 | 1674 | 10.46 |
Genetic distances (Fst) between the four subclades were 0.319–0.716, indicating high genetic differences among them (Figure 5A). Clade C had the highest genetic difference with Clades A and D. Clade C mainly included the var. thomsonii accession from GenBank. Four clades and the entire collection showed relatively high divergences. An analysis of molecular variance (AMOVA) suggested that both variations contributed to inter- and intra-clade differences (Figure 5C).
3. Discussion
3.1. Highly Variable Pueraria Montana Chloroplast Genome Sequences
Compared with nuclear DNA, the chloroplast genome had a lower substitution rate, occurring mostly at the species level or above. Therefore, detecting useful intraspecies polymorphisms is difficult in practice [23,48,49]. However, with the advent of NGS methods, many species have had more than two individuals sequenced, significantly improving our ability to assess mutation rates or detect intraspecies variations. Previous studies have assessed intraspecies mutations in chloroplast genome sequences, such as the medicine plants Arnebia euchroma and Arnebia guttata [50], the model grass plant Brachypodium distachyon [51], the endangered species Bretschneidera sinensis [16], the tertiary relict tree Ginkgo biloba [52], and cultivated rice [27]. These results indicate that the chloroplast genome sequences contain many mutations for evolutionary research.
This study analyzed the chloroplast genomes of 104 P. montana accessions, identifying 1118 SNPs and 556 indel variations with high intraspecies variability (Table 1). P. montana chloroplast genome showed higher variability compared with other published plant species, such as Ginkgo biloba (135 SNPs, 71 accessions) [52], Brachypodium distachyon (298 SNPs, 53 accessions) [51], soybean (44 SNPs, 2580 accessions) [53], Coptis chinensis (111 SNPs, 227 accessions) [19], and Ulmus laevis (32 SNPs, 54 accessions). Mutation heterogeneity across different plant lineages or species has been previously examined. There are two hypotheses invoking the mutation rates. The first hypothesis is that mutation rates are negatively correlated with generation times. This hypothesis suggests that long-lived plants will have lower mutation rates than short-lived species, such as herbaceous plants, due to their longer generation time. P. montana’s high intraspecific mutation rate may be partially explained by generation time since it is a perennial, semi-woody, climbing legume species with a middle to long generation time.
The second hypothesis is that long divergence times enable more mutations to accumulate. Estimated divergence times showed that P. montana evolved very early, around 5.17 Ma in the early Pliocene (Figure 4). Therefore, the persistence of genetically distinct populations through periods of historical climate variation during its long evolutionary history is the likely cause of its relatively high genetic diversity. The Northeastern and Southeastern Tibet Plateau’s uplift from the Pliocene to the early Pleistocene impacted Eastern China [54], with the East Asian summer monsoon [55] and South Asian summer monsoon [56] intensifying during that period.
Another possible reason for P. montana’s high mutation rate may be related to its type of chloroplast inheritance. Approximately 20% of angiosperm species have biparental plastid inheritance [57,58], which is associated with chloroplast genome rearrangement events and accelerated mutation rates. Some species in the Papilionoideae group have shown that the type of chloroplast inheritance is biparental [57,58], and its chloroplast genome structure has a large inversion (~56 kb) and gene and intron loss. In addition, the chloroplast genome has a high mutation rate [59,60,61]. Aberrant DNA repair, recombination, and replication with biparental inheritance may underlie increased substitution rates in the chloroplast genome [60,62].
Mutation heterogeneity also occurs in different regions of the chloroplast genome, such as mutation hotspots, with higher mutation rates. The IR regions are known to be more conserved than LSC and SSC regions. Mutations in the P. montana chloroplast genomes were found mostly in the LSC and SSC regions. The spacers psbZ-trnS and ccsA-ndhD had the highest marker variability in P. montana (Figure S1), and these markers are suited for investigating genetic diversity and population or subpopulation structure.
3.2. Pueraria Montana Genetic Divergence and Diversity
The P. montana complex includes three species (P. montana, Pueraria lobata, and Pueraria thomsonii) or three varieties (var. montana, lobata, and thomsonii). Phylogenetic evidence indicates that the P. montana complex forms a monophyletic group [29]. It is challenging to morphologically distinguish between species or varieties in the P. montana complex because characteristics used to differentiate taxa in dichotomous keys appear continuous and overlapping across its range, particularly for vars. lobata and thomsonii [28,63]. In this study, the monophyletic group of each species or variety was also not supported using the chloroplast genome data (Figure 2). The phylogeny of rDNA ITS data showed a lack of resolution (Figure S5) but still could be clustered into three clades by three well supported nodes (Table S1). In general, the varieties of Pueraria appear polyphyletic based on the results. Alternatively, the samples obtained from public databases could be by named incorrectly because of small differences between varieties. A previous study also supported a close relationship in nuclear sequence data between P. montana var. lobata and P. montana var. thomsonii, which were intermixed, suggesting their possible hybridization [36]. Nuclear SNP data has also indicated a higher potential for hybridization or introgression among the three varieties [30]. Hybridization or introgression may blur species boundaries and influence population structure.
P. montana genetic diversity has been assessed with various molecular markers. For example, an analysis of 13 allozymes across 1000 US accessions concluded that P. montana var. lobata possessed considerable genetic variation and lacked geographic structuring [33]. Analyses using RAPD [34], ISSR [36,37,38], and SSR [39,40] markers discovered higher genetic diversity in P. montana populations. Chloroplast genome dataset discovered P. montana with high diversity levels and population structure analyses identified four P. montana clades (Figure 2). P. montana is widely distributed in north-central, south-central, and southeast of China. The samples of each four clades did not reflect geographical distribution, indicating that geographic distance does not explain the genetic structure patterns.
In the context of its high ornamental, horticultural, agricultural, and ecological interest, P. montana was likely to migrate from multiple sources multiple times [36,39,41]. P. montana genetic studies have repeatedly revealed a high degree of variation among introduced populations, such as those found in the United States [32,34,36,64]. High genetic diversity levels in introduced populations may indicate multiple introductions [65]. Bringing such genetic diversity into proximity with untried environments that enable gene flow may also allow for novel adaptations, the consequences of which may have played important roles in P. montana’s invasiveness. However, the long-lived P. montana with high genetic diversity may reduce the likelihood of adaptive evolution due to its relatively small effective number of generations [30].
4. Materials and Methods
4.1. Samples and Chloroplast Genome Sequencing
We collected 70 P. montana samples from China, covering most of its wild distribution in China (Table S1). The 70 samples (accessions) were collected from more than 19 provinces, including Anhui, Chongqing, Gansu, Guangdong, Guangxi, Guizhou, Hebei, Henan, Hubei, Hunan, Jiangsu, Jilin, Liaoning, Shaanxi, Shandong, Shanxi, Sichuan, Yunnan, and Zhejiang (Table S1). Owing to the overlapping taxonomic characteristics, we did not distinguish the three varieties in this study. For each accession, young leaves were sampled and dried with silica gel. Voucher specimens were deposited in the herbarium of the National Resource Center for Chinese Materia Medica at the China Academy of Chinese Medical Sciences. Total genomic DNA was extracted using the modified CTAB (mCTAB) method, and DNA quality and quantity were assessed by agarose gel electrophoresis (1% w/w).
Total DNA was fragmented into 350 bp fragments by sonication. Illumina paired-end DNA libraries were constructed using NEB Next Ultra DNA Library Prep Kit for Illumina (NEB, USA). Each sample was barcoded with a unique index, and libraries were pooled for sequencing on an Illumina HiSeq X-Ten platform at Novogene (Tianjin, China). Each accession yielded ~10 Gb of 150-bp paired-end reads.
The raw data was subject to quality control using Trimmomatic 0.36 [66] with the following parameters: LEADING, 20; TRAILING, 20; SLIDING WINDOW, 4:15; MIN LEN, 36; and AVG QUAL, 20. The cleaned data were used to assemble the chloroplast genome and rDNA ITS using GetOrganelle [67] with 85, 95, and 105 kmer lengths. Where GetOrganelle failed to assemble the complete chloroplast genome, we assembled it following the methods of Dong et al. [5]. The Perl script Plann [68] was used to annotate the chloroplast genome using the published P. montana var. lobata genome (GenBank accession number: MT818508) as the reference sequence. The annotated chloroplast genome sequences are deposited in the GenBank, accession number OP963859- OP963928.
4.2. Variation Identification and Statistics
The chloroplast genomes of 70 P. montana accessions and 44 downloaded from GenBank were aligned using MAFFT version 7.490 [69]. Se-Al version 2.0 [70] was used to manually correct alignment errors, such as polymeric repeat structures and small inversions. SNPs and indels were identified over the entire chloroplast genome, and genetic distances were calculated among accessions. The numbers and distributions of SNPs and indels were summarized using MEGA version 7.0 [71] and DnaSP version 6 [72].
4.3. Population Structure and Principal Component Analysis
The accessions’ population structures were investigated using STRUCTURE [73]. The optimal cluster number (K) was determined by running the K-means clustering algorithm from K = 2 to K = 10 with ten runs for each K. The STRUCTURE workflow was according to the previous study (Wang et al., 2022c). A principal component analysis (PCA) was performed using PLINK [74] to evaluate the genetic structure. The R v4.1 statistical software’s ggbiplot package was used to plot the graphs [75].
The complete chloroplast genomes of 104 P. montana accessions were aligned with those for Pueraria edulis and Pueraria candollei var. mirifica as the outgroups using MAFFT version 7 [69]. Two methods, maximum likelihood (ML) and Bayesian inference (BI), were used to reconstruct the phylogenetic tree for the P. montana accessions. The best-fit model for the ML analyses was selected by ModelFinder [76] based on the Akaike Information Criterion and BI analyses were based on the Bayesian information criterion. The ML analysis was performed in RAxML-NG [77], and support values were assessed by 1000 bootstrap replicates.
BI analysis was performed in Mrbayes v3.2 [78]. Four Markov chain Monte Carlo (MCMC) simulations were run from random trees for 10 million generations, with trees sampled every 1000 generations. The stationary phase was examined using Tracer 1.6 [79], and the first 25% of the sampled trees were discarded as “burn-in.” The majority-rule consensus tree was created using the remaining trees and estimated posterior probabilities. All trees were visualized using FigTree version 1.4 (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 21 April 2023).
4.4. Genetic Diversity in P. montana Subclades
Nucleotide diversity (π) and subclade divergence (fixation index [Fst]) were calculated using the whole chloroplast genome dataset to assess genetic diversity among the different clades. The network of all samples was constructed. Haplotype data were analyzed with DnaSP version 6 [72], and a median-joining network was constructed using PopArt version 1.7 [80,81].
4.5. Divergence Time Estimation and Profiling
We used the chloroplast genome to estimate the divergence times of the different subclades. This dataset included 16 P. montana accessions from different subclades and nine Glycininae species as the outgroups. According to the average values of Egan et al. [29] in a calibrated analysis, we used four secondary priors for this study: (1) the crown age of the core Phaseoleae was set to 23.52 million years ago (Ma); (2) the Phaseolinae stem age was set to 19.26 Ma; (3) the crown node of the Pueraria s. str. and Glycine was 12.71 Ma; and (4) the Pueraria s. str. crown age was 7.85 Ma. All four secondary priors were placed under normal distribution with a standard deviation of 1.
The divergence time analyses were performed using BEAST 2 [82]. We chose a general time reversible model and a relaxed molecular clock model with an uncorrelated lognormal distribution. We ran 500,000,000 generations of the MCMC algorithm, with samples taken every 50,000 generations. Effective sampling sizes > 200 were used to measure convergence using Tracer version 1.6 [79]. A maximum clade credibility tree with mean heights was built in TreeAnnotator after discarding the first 10% of the trees as burn-in.
5. Conclusions
This study used whole chloroplast genomes to examine P. montana’s genetic diversity, identifying genome-wide variability and contributing to a better understanding of the divergence history among its populations. The phylogenetic, PCA, and structure analyses showed genetic differentiation and divergence among P. montana populations. Chloroplast genome sequences showed high variability and provided novel insights into evolutionary history.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12122231/s1, Figure S1. Nucleotide diversity (π) in P. montana’s chloroplast genome based on a window size of 800 bp and a step size of 100 bp. Figure S2. Network of all 104 P. montana accessions. Figure S3. The delta K results of STRUCTURE analysis. Figure S4. Population structure of all 104 accessions with K = 3, 5, and 6. Figure S5. The ITS phylogenetic tree of 70 P. montana collected accessions. Table S1. Detail information of 107 sequenced or downloaded chloroplast genomes.
Author Contributions
Conceptualization, project administration W.D., Y.Z., J.S. and L.H.; Data curation, investigation, conceptualization, and resources, J.S., Y.W., P.Q., L.Z., E.L., Y.Z. and W.D.; Funding acquisition, J.S., Y.Z. and L.H.; Writing-review and editing, visualization, and supervision, J.S., Y.Z. and W.D. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
The data is contained within the manuscript and Supplementary Materials. The chloroplast genome sequences under this study are deposited in the GenBank, accession number OP963859- OP963928.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was financially supported by the National Key Research and Development Program of China (Grant No. 2017YFC1702901), and Science & Technology Fundamental Resources Investigation Program (Grant No.2022FY101000).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Daniell H., Lin C.-S., Yu M., Chang W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17:134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wicke S., Schneeweiss G.M., Depamphilis C.W., Muller K.F., Quandt D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011;76:273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drouin G., Daoud H., Xia J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 2008;49:827–831. doi: 10.1016/j.ympev.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Zhao F., Chen Y.-P., Salmaki Y., Drew B.T., Wilson T.C., Scheen A.-C., Celep F., Bräuchler C., Bendiksby M., Wang Q., et al. An updated tribal classification of Lamiaceae based on plastome phylogenomics. BMC Biol. 2021;19:2. doi: 10.1186/s12915-020-00931-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong W., Li E., Liu Y., Xu C., Wang Y., Liu K., Cui X., Sun J., Suo Z., Zhang Z., et al. Phylogenomic approaches untangle early divergences and complex diversifications of the olive plant family. BMC Biol. 2022;20:92. doi: 10.1186/s12915-022-01297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Wang S., Liu Y., Yuan Q., Sun J., Guo L. Chloroplast genome variation and phylogenetic relationships of Atractylodes species. BMC Genom. 2021;22:103. doi: 10.1186/s12864-021-07394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li E., Liu K., Deng R., Gao Y., Liu X., Dong W., Zhang Z. Insights into the phylogeny and chloroplast genome evolution of Eriocaulon (Eriocaulaceae) BMC Plant Biol. 2023;23:32. doi: 10.1186/s12870-023-04034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Y., Liu K., Li E., Wang Y., Xu C., Zhao L., Dong W. Dynamic evolution of the plastome in the Elm family (Ulmaceae) Planta. 2023;257:14. doi: 10.1007/s00425-022-04045-4. [DOI] [PubMed] [Google Scholar]
- 9.Dong W., Sun J., Liu Y., Xu C., Wang Y., Suo Z., Zhou S., Zhang Z., Wen J. Phylogenomic relationships and species identification of the olive genus Olea (Oleaceae) J. Syst. Evol. 2022;60:1263–1280. doi: 10.1111/jse.12802. [DOI] [Google Scholar]
- 10.Dong W., Liu Y., Li E., Xu C., Sun J., Li W., Zhou S., Zhang Z., Suo Z. Phylogenomics and biogeography of Catalpa (Bignoniaceae) reveal incomplete lineage sorting and three dispersal events. Mol. Phylogenet. Evol. 2022;166:107330. doi: 10.1016/j.ympev.2021.107330. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y., Wang J., Garran T., Liu H., Lin H., Luo J., Yuan Q., Sun J., Dong W., Guo L. Genetic diversity and population divergence of Leonurus japonicus and its distribution dynamic changes from the last interglacial to the present in China. BMC Plant Biol. 2023;23:276. doi: 10.1186/s12870-023-04284-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong W., Xu C., Li C., Sun J., Zuo Y., Shi S., Cheng T., Guo J., Zhou S. ycf1, the most promising plastid DNA barcode of land plants. Sci. Rep. 2015;5:8348. doi: 10.1038/srep08348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.China Plant BOLG. Li D.Z., Gao L.M., Li H.T., Wang H., Ge X.J., Liu J.Q., Chen Z.D., Zhou S.L., Chen S.L., et al. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Nat. Acad. Sci. USA. 2011;108:19641–19646. doi: 10.1073/pnas.1104551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong W., Liu J., Yu J., Wang L., Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE. 2012;7:e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaw J., Lickey E.B., Beck J.T., Farmer S.B., Liu W.S., Miller J., Siripun K.C., Winder C.T., Schilling E.E., Small R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005;92:142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- 16.Shang C., Li E., Yu Z., Lian M., Chen Z., Liu K., Xu L., Tong Z., Wang M., Dong W. Chloroplast Genomic Resources and Genetic Divergence of Endangered Species Bretschneidera sinensis (Bretschneideraceae) Front. Ecol. Evol. 2022;10:873100. doi: 10.3389/fevo.2022.873100. [DOI] [Google Scholar]
- 17.Torre S., Sebastiani F., Burbui G., Pecori F., Pepori A.L., Passeri I., Ghelardini L., Selvaggi A., Santini A. Novel Insights Into Refugia at the Southern Margin of the Distribution Range of the Endangered Species Ulmus laevis. Front. Plant Sci. 2022;13:826158. doi: 10.3389/fpls.2022.826158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Sun J., Wang J., Mao Q., Dong W., Yuan Q., Guo L., Huang L. Coptis huanjiangensis, a new species of Ranunculaceae from Guangxi, China. PhytoKeys. 2022;213:131–141. doi: 10.3897/phytokeys.213.96546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y., Sun J., Zhao Z., Xu C., Qiao P., Wang S., Wang M., Xu Z., Yuan Q., Guo L., et al. Multiplexed Massively Parallel Sequencing of Plastomes Provides Insights Into the Genetic Diversity, Population Structure, and Phylogeography of Wild and Cultivated Coptis chinensis. Front. Plant Sci. 2022;13:923600. doi: 10.3389/fpls.2022.923600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perdereau A., Klaas M., Barth S., Hodkinson T.R. Plastid genome sequencing reveals biogeographical structure and extensive population genetic variation in wild populations of Phalaris arundinacea L. in north-western Europe. GCB Bioenergy. 2017;9:46–56. doi: 10.1111/gcbb.12362. [DOI] [Google Scholar]
- 21.Xue C., Geng F.D., Li J.J., Zhang D.Q., Gao F., Huang L., Zhang X.H., Kang J.Q., Zhang J.Q., Ren Y. Divergence in the Aquilegia ecalcarata complex is correlated with geography and climate oscillations: Evidence from plastid genome data. Mol. Ecol. 2021;30:5796–5813. doi: 10.1111/mec.16151. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Sun J., Qiao P., Wang J., Wang M., Du Y., Xiong F., Luo J., Yuan Q., Dong W., et al. Evolutionary history of genus Coptis and its dynamic changes in the potential suitable distribution area. Front. Plant Sci. 2022;13:1003368. doi: 10.3389/fpls.2022.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang D.I., Hefer C.A., Kolosova N., Douglas C.J., Cronk Q.C. Whole plastome sequencing reveals deep plastid divergence and cytonuclear discordance between closely related balsam poplars, Populus balsamifera and P. trichocarpa (Salicaceae) New Phytol. 2014;204:693–703. doi: 10.1111/nph.12956. [DOI] [PubMed] [Google Scholar]
- 24.Cui H., Ding Z., Zhu Q., Wu Y., Gao P. Population structure and genetic diversity of watermelon (Citrullus lanatus) based on SNP of chloroplast genome. 3 Biotech. 2020;10:374. doi: 10.1007/s13205-020-02372-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y., Liao B., Ostevik K.L., Zhou H., Wang F., Wang B., Xia H. The Maternal Donor of Chrysanthemum Cultivars Revealed by Comparative Analysis of the Chloroplast Genome. Front. Plant Sci. 2022;13:923442. doi: 10.3389/fpls.2022.923442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nock C.J., Hardner C.M., Montenegro J.D., Ahmad Termizi A.A., Hayashi S., Playford J., Edwards D., Batley J. Wild Origins of Macadamia Domestication Identified Through Intraspecific Chloroplast Genome Sequencing. Front. Plant Sci. 2019;10:334. doi: 10.3389/fpls.2019.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong W., Kim T.-S., Park Y.-J. Rice Chloroplast Genome Variation Architecture and Phylogenetic Dissection in Diverse Oryza Species Assessed by Whole-Genome Resequencing. Rice. 2016;9:57. doi: 10.1186/s12284-016-0129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van der Maesen L. Revision of the Genus Pueraria DC with Some Notes on Teyleria Backer (Leguminosae) Taylor & Francis; London, UK: 1985. [Google Scholar]
- 29.Egan A.N., Vatanparast M., Cagle W. Parsing polyphyletic Pueraria: Delimiting distinct evolutionary lineages through phylogeny. Mol. Phylogenet. Evol. 2016;104:44–59. doi: 10.1016/j.ympev.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Haynsen M.S. Population Genetics of Pueraria montana var. Lobata. The George Washington University; Washington, DC, USA: 2018. [Google Scholar]
- 31.Sun Y., Shaw P.-C., Fung K.-P. Molecular Authentication of Radix Puerariae Lobatae and Radix Puerariae Thomsonii by ITS and 5S rRNA Spacer Sequencing. Biol. Pharm. Bull. 2007;30:173–175. doi: 10.1248/bpb.30.173. [DOI] [PubMed] [Google Scholar]
- 32.Kartzinel T.R., Hamrick J.L., Wang C., Bowsher A.W., Quigley B.G.P. Heterogeneity of clonal patterns among patches of kudzu, Pueraria montana var. lobata, an invasive plant. Ann. Bot. 2015;116:739–750. doi: 10.1093/aob/mcv117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pappert R.A., Hamrick J.L., Donovan L.A. Genetic variation in Pueraria lobata (Fabaceae), an introduced, clonal, invasive plant of the southeastern United States. Am. J. Bot. 2000;87:1240–1245. doi: 10.2307/2656716. [DOI] [PubMed] [Google Scholar]
- 34.Heider B., Fischer E., Berndl T., Schultze-Kraft R. Analysis of Genetic Variation Among Accessions of Pueraria montana (Lour.) Merr. var. lobata and Pueraria phaseoloides (Roxb.) Benth. based on RAPD Markers. Genet. Resour. Crop. Evol. 2007;54:529–542. doi: 10.1007/s10722-006-0009-1. [DOI] [Google Scholar]
- 35.Chen D., Peng R., Li L., Zhang X., Wang Y. Analysis of genetic relationships of Pueraria thomsonii based on SRAP markers. Zhongguo Zhong Yao Za Zhi China J. Chin. Mater. Med. 2011;36:538–541. [PubMed] [Google Scholar]
- 36.Sun J.H., Li Z.C., Jewett D.K., Britton K.O., Ye W.H., Ge X.J. Genetic diversity of Pueraria lobata (kudzu) and closely related taxa as revealed by inter-simple sequence repeat analysis. Weed Res. 2005;45:255–260. doi: 10.1111/j.1365-3180.2005.00462.x. [DOI] [Google Scholar]
- 37.Chen J., Tan L., Zhang Q., Guan Q., Yang Y., Tian S., Yang Z., Zhu Z., Xu L. Genetic diversity of Pueraria lobata varieties and differences of puerarin analysis in Three Gorges Reservoir Area. Southwest China J. Agric. Sci. 2015;28:2334–2336. [Google Scholar]
- 38.Tsia P., Ho S., Hou C., Lin J., Li T. The analysis of genetic diversity of Pueraria montana in Taiwan revealed by ISSR method. J. Taiwan Livest. Res. 2017;50:294–302. [Google Scholar]
- 39.Hoffberg S.L., Bentley K.E., Lee J.B., Myhre K.E., Iwao K., Glenn T.C., Mauricio R. Characterization of 15 microsatellite loci in kudzu (Pueraria montana var. lobata) from the native and introduced ranges. Conserv. Genet. Resour. 2015;7:403–405. doi: 10.1007/s12686-014-0381-7. [DOI] [Google Scholar]
- 40.Bentley K.E., Mauricio R. High degree of clonal reproduction and lack of large-scale geographic patterning mark the introduced range of the invasive vine, kudzu (Pueraria montana var. lobata), in North America. Am. J. Bot. 2016;103:1499–1507. doi: 10.3732/ajb.1500434. [DOI] [PubMed] [Google Scholar]
- 41.Haynsen M.S., Vatanparast M., Mahadwar G., Zhu D., Moger-Reischer R.Z., Doyle J.J., Crandall K.A., Egan A.N. De novo transcriptome assembly of Pueraria montana var. lobata and Neustanthus phaseoloides for the development of eSSR and SNP markers: Narrowing the US origin(s) of the invasive kudzu. BMC Genom. 2018;19:439. doi: 10.1186/s12864-018-4798-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar P., Gupta V.K., Misra A.K., Modi D.R., Pandey B.K. Potential of Molecular Markers in Plant Biotechnology. Plant Omics. 2009;2:141–162. [Google Scholar]
- 43.Li J., Yang M., Li Y., Jiang M., Liu C., He M., Wu B. Chloroplast genomes of two Pueraria DC. species: Sequencing, comparative analysis and molecular marker development. FEBS Open Bio. 2022;12:349–361. doi: 10.1002/2211-5463.13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mo C., Wu Z., Shang X., Shi P., Wei M., Wang H., Xiao L., Cao S., Lu L., Zeng W., et al. Chromosome-level and graphic genomes provide insights into metabolism of bioactive metabolites and cold-adaption of Pueraria lobata var. montana. DNA Res. 2022;29:dsac030. doi: 10.1093/dnares/dsac030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shang X., Yi X., Xiao L., Zhang Y., Huang D., Xia Z., Ou K., Ming R., Zeng W., Wu D., et al. Chromosomal-level genome and multi-omics dataset of Pueraria lobata var. thomsonii provide new insights into legume family and the isoflavone and puerarin biosynthesis pathways. Hortic. Res. 2022;9:uhab035. doi: 10.1093/hr/uhab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zachos J.C., Dickens G.R., Zeebe R.E. An early Cenozoic perspective on greenhouse warming and carbon-cycle dynamics. Nature. 2008;451:279–283. doi: 10.1038/nature06588. [DOI] [PubMed] [Google Scholar]
- 47.Zachos J., Pagani M., Sloan L., Thomas E., Billups K. Trends, Rhythms, and Aberrations in Global Climate 65 Ma to Present. Science. 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 48.Mohamoud Y.A., Mathew L.S., Torres M.F., Younuskunju S., Krueger R., Suhre K., Malek J.A. Novel subpopulations in date palm (Phoenix dactylifera) identified by population-wide organellar genome sequencing. BMC Genom. 2019;20:498. doi: 10.1186/s12864-019-5834-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mariotti R., Cultrera N.G.M., Díez C.M., Baldoni L., Rubini A. Identification of new polymorphic regions and differentiation of cultivated olives (Olea europaea L.) through plastome sequence comparison. BMC Plant Biol. 2010;10:211. doi: 10.1186/1471-2229-10-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun J., Wang S., Wang Y., Wang R., Liu K., Li E., Qiao P., Shi L., Dong W., Huang L., et al. Phylogenomics and Genetic Diversity of Arnebiae Radix and Its Allies (Arnebia, Boraginaceae) in China. Front. Plant Sci. 2022;13:920826. doi: 10.3389/fpls.2022.920826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sancho R., Cantalapiedra C.P., Lopez-Alvarez D., Gordon S.P., Vogel J.P., Catalan P., Contreras-Moreira B. Comparative plastome genomics and phylogenomics of Brachypodium: Flowering time signatures, introgression and recombination in recently diverged ecotypes. New Phytol. 2018;218:1631–1644. doi: 10.1111/nph.14926. [DOI] [PubMed] [Google Scholar]
- 52.Hohmann N., Wolf E.M., Rigault P., Zhou W., Kiefer M., Zhao Y., Fu C.-X., Koch M.A. Ginkgo biloba’s footprint of dynamic Pleistocene history dates back only 390,000 years ago. BMC Genom. 2018;19:299. doi: 10.1186/s12864-018-4673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yue Y., Li J., Sun X., Li Z., Jiang B. Polymorphism analysis of the chloroplast and mitochondrial genomes in soybean. BMC Plant Biol. 2023;23:15. doi: 10.1186/s12870-022-04028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.An Z., Zhang P., Wang E., Wang S., Qiang X., Li L., Song Y., Chang H., Liu X., Zhou W. Changes of the monsoon-arid environment in China and growth of the Tibetan Plateau since the Miocene. Quat. Sci. 2006;26:678–693. [Google Scholar]
- 55.An Z. Late Cenozoic Climate Change in Asia: Loess, Monsoon and Monsoon-Arid Environment Evolution. Springer; Berlin/Heidelberg, Germany: 2014. [Google Scholar]
- 56.Chang Z., Xiao J., Lü L., Yao H. Abrupt shifts in the Indian monsoon during the Pliocene marked by high-resolution terrestrial records from the Yuanmou Basin in southwest China. J. Asian Earth Sci. 2010;37:166–175. doi: 10.1016/j.jseaes.2009.08.005. [DOI] [Google Scholar]
- 57.Corriveau J.L., Coleman A.W. Rapid screening method to detect potential biparental inheritance of plastid DNA and results for over 200 angiosperm species. Am. J. Bot. 1988;75:1443–1458. doi: 10.1002/j.1537-2197.1988.tb11219.x. [DOI] [Google Scholar]
- 58.Zhang Q., Liu Y.S. Examination of the cytoplasmic DNA in male reproductive cells to determine the potential for cytoplasmic inheritance in 295 angiosperm species. Plant Cell Physiol. 2003;44:941–951. doi: 10.1093/pcp/pcg121. [DOI] [PubMed] [Google Scholar]
- 59.Schwarz E.N., Ruhlman T.A., Weng M.-L., Khiyami M.A., Sabir J.S.M., Hajarah N.H., Alharbi N.S., Rabah S.O., Jansen R.K. Plastome-wide nucleotide substitution rates reveal accelerated rates in Papilionoideae and correlations with genome features across legume subfamilies. J. Mol. Evol. 2017;84:187–203. doi: 10.1007/s00239-017-9792-x. [DOI] [PubMed] [Google Scholar]
- 60.Guisinger M.M., Kuehl J.N.V., Boore J.L., Jansen R.K. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc. Nat. Acad. Sci. USA. 2008;105:18424–18429. doi: 10.1073/pnas.0806759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weng M.-L., Blazier J.C., Govindu M., Jansen R.K. Reconstruction of the Ancestral Plastid Genome in Geraniaceae Reveals a Correlation between Genome Rearrangements, Repeats, and Nucleotide Substitution Rates. Mol. Biol. Evol. 2014;31:645–659. doi: 10.1093/molbev/mst257. [DOI] [PubMed] [Google Scholar]
- 62.Barnard-Kubow K.B., Sloan D.B., Galloway L.F. Correlation between sequence divergence and polymorphism reveals similar evolutionary mechanisms acting across multiple timescales in a rapidly evolving plastid genome. BMC Evol. Biol. 2014;14:268. doi: 10.1186/s12862-014-0268-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keung W.M. Pueraria: The genus Pueraria. CRC Press; Boca Raton, FL, USA: 2002. [Google Scholar]
- 64.Callen S.T., Miller A.J. Signatures of niche conservatism and niche shift in the North American kudzu (Pueraria montana) invasion. Divers. Distrib. 2015;21:853–863. doi: 10.1111/ddi.12341. [DOI] [Google Scholar]
- 65.Dlugosch K.M., Parker I.M. Founding events in species invasions: Genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 2008;17:431–449. doi: 10.1111/j.1365-294X.2007.03538.x. [DOI] [PubMed] [Google Scholar]
- 66.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jin J.-J., Yu W.-B., Yang J.-B., Song Y., dePamphilis C.W., Yi T.-S., Li D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020;21:241. doi: 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang D.I., Cronk Q.C.B. Plann: A command-line application for annotating plastome sequences. Appl. Plant Sci. 2015;3:1500026. doi: 10.3732/apps.1500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Se-Al: Sequence Alignment Editor. University of Oxford; Oxford, UK: 1996. Version 2.0. [Google Scholar]
- 71.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rozas J., Ferrer-Mata A., Sanchez-DelBarrio J.C., Guirao-Rico S., Librado P., Ramos-Onsins S.E., Sanchez-Gracia A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 73.Pritchard J.K., Stephens M., Donnelly P. Inference of Population Structure Using Multilocus Genotype Data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vu V.Q. ggbiplot: A ggplot2 Based biplot. R Package Version 0.55. 2011. [(accessed on 17 November 2022)]. Available online: https://github.com/vqv/ggbiplot.
- 76.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., Von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Hohna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rambaut A., Suchard M., Xie D., Drummond A. Tracer v1. 6. 2014. [(accessed on 6 December 2022)]. Available online: http://beast.bio.ed.ac.uk/Tracer.
- 80.Clement M., Snell Q., Walker P., Posada D., Crandall K. TCS: Estimating gene genealogies. Parallel Distrib. Process. Symp. Int. Proc. 2002;2:184. [Google Scholar]
- 81.Leigh J.W., Bryant D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015;6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- 82.Bouckaert R., Heled J., Kuhnert D., Vaughan T., Wu C.H., Xie D., Suchard M.A., Rambaut A., Drummond A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comp. Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data is contained within the manuscript and Supplementary Materials. The chloroplast genome sequences under this study are deposited in the GenBank, accession number OP963859- OP963928.