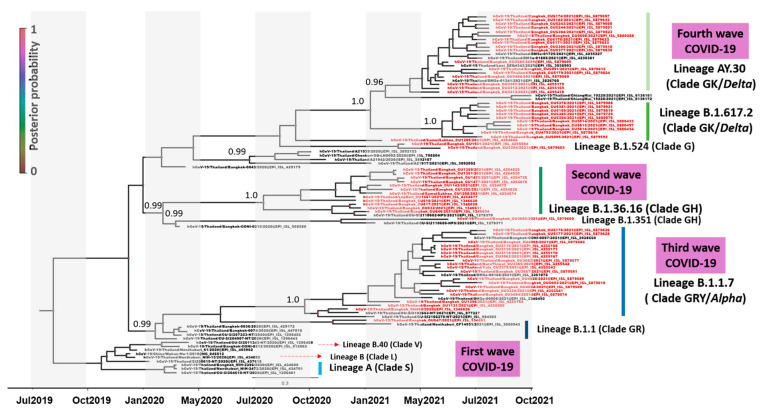
Figure 1.
Time-scaled phylogenetic tree of 96 complete SARS-CoV-2 genomes (nt positions 56–29,739, 29,684 bp) detected before the B.1.1.529 omicron variant predominated. Shown is a maximum clade credibility tree constructed from 10,000 trees sampled from the posterior distribution with mean node ages. Clades described in GISAID are identified (S, L, V, G, GH, GR, GRY, and GK). Several lineages predominantly represent outbreaks in Thailand, and posterior probability support is given.