Abstract
The large quantity of olive roots resulting from a large number of old and unfruitful trees encouraged us to look for ways of adding value to these roots. For this reason, the current research work is devoted to the valorization of olive roots by identifying active phytochemicals and assessing their biological activities, including the cytotoxicity and antiviral potential of different extracts from the Olea europaea Chemlali cultivar. The extract, obtained by ultrasonic extraction, was analyzed using the liquid chromatography-mass spectrometry technique (LC-MS). The cytotoxicity was evaluated through the use of the microculture tetrazolium assay (MTT) against VERO cells. Subsequently, the antiviral activity was determined for HHV-1 (Human Herpesvirus type 1) and CVB3 (Coxsackievirus B3) replication in the infected VERO cells. LC-MS analysis allowed the identification of 40 compounds, classified as secoiridoids (53%), organic acids (13%), iridoids (10%), lignans (8%), caffeoylphenylethanoid (5%), phenylethanoids (5%),sugars and derivatives (2%), phenolic acids (2%), and flavonoids (2%). It was found that extracts were not toxic to the VERO cells. Moreover, the extracts did not influence the appearance of HHV-1 or CVB3 cytopathic effects in the infected VERO cells and failed to decrease the viral infectious titer.
Keywords: olive roots, ultrasonic extraction, LC-MS, phenolic compounds, biological activities
1. Introduction
Tunisia is regarded as the most important olive-producing country in the southern Mediterranean region. More than 30% of the agricultural land proved to be devoted to cultivating olive trees. These trees are distributed from north to south, with very different bioclimatic conditions and a wide range of varieties. Today, Tunisia ranks the fourth country in the world for the number of cultivated olive trees, estimated at more than 86 million trees covering an area of 1,800,000 hectares [1]. In addition to oil as the basic product, the olive oil industry produces a large number of by-products, including pruning products, pomace, and margines. The valorization of these residues has become crucial in order to avoid increased contamination. Prunings display multiple uses, including animal feed [2] and compost production [3].
These residues are known for their beneficial properties for human health, referring to their richness in phenolic compounds, especially oleuropein [4]. These compounds exhibit, among others, antioxidant, anticancer, and antimicrobial potentials, which makes them very significant for the health and food industries [5,6]. The growing interest in phenolic compounds in olive oil permeates all olive products that can be consumed as food (table olives, olive paste) or that are generated as by-products of the olive industry [6].
Phenolic compounds are known for their intrinsic physiological roles in both the plant and animal kingdoms. Referring to their structure, phenolic compounds are able to bind to specific enzymes and proteins and thus modify enzymatic balances. In the plant kingdom, these substances are often involved in the defense mechanisms developed by plants against predation by insects and herbivores, infections, and numerous microbial aggressions. In addition, phenolic compounds are associated with different physiological processes, namely cell growth, differentiation, bud dormancy, and flowering [7,8]. As far as herbal medicine is concerned, the effects of various medicinal plants are attributed, in whole or in part, to the phenolic compounds existing in these plants. These substances have biological activities, making them beneficial to human health. Multiple studies emphasize that polyphenols can reduce the risk of a number of pathologies, particularly those related to aging and oxidative damage, such as cancer and cardiovascular or neurodegenerative diseases [9]. Thus far, the structural characterization of phenolic compounds has significantly improved thanks to the use of liquid chromatography coupled with mass spectrometry (LC-MS) [10,11]. Among the most prominent researchers, we mention Ammar et al. [12], who conducted a phytochemical study of olive wood by means of analyses based on mass spectrometry and its comparison with the by-products derived from leaves [12].
The current study is dedicated in its first part to the valorization of the roots of the olive tree by identifying the active phytochemical substances through the use of various chromatographic techniques. In its second part, our central focus is on the biological activities of the extracts.
2. Results and Discussion
2.1. The Quantitative Study of the Different Extracts
The extraction yield of dry residue rich in phenolic compounds from the external part of the olive roots was 31.16 g/100 g, the internal part was 10.23 g/100 g, and the root sum was 15.72 g/100 g. The percentages of extraction yields are illustrated in Table 1.
Table 1.
The extraction yield of the dry residue rich in phenolic compounds from olive tree roots.
| Organs | Yield (%) |
|---|---|
| External part | 31.61 |
| Internal part | 10.23 |
| Root sum | 15.72 |
Our results show that extract yields for all three samples are important, ranging on average from 10.23% to 31.61%. The highest yield was registered for the outer part of the olive root, with a value of 31.61%.
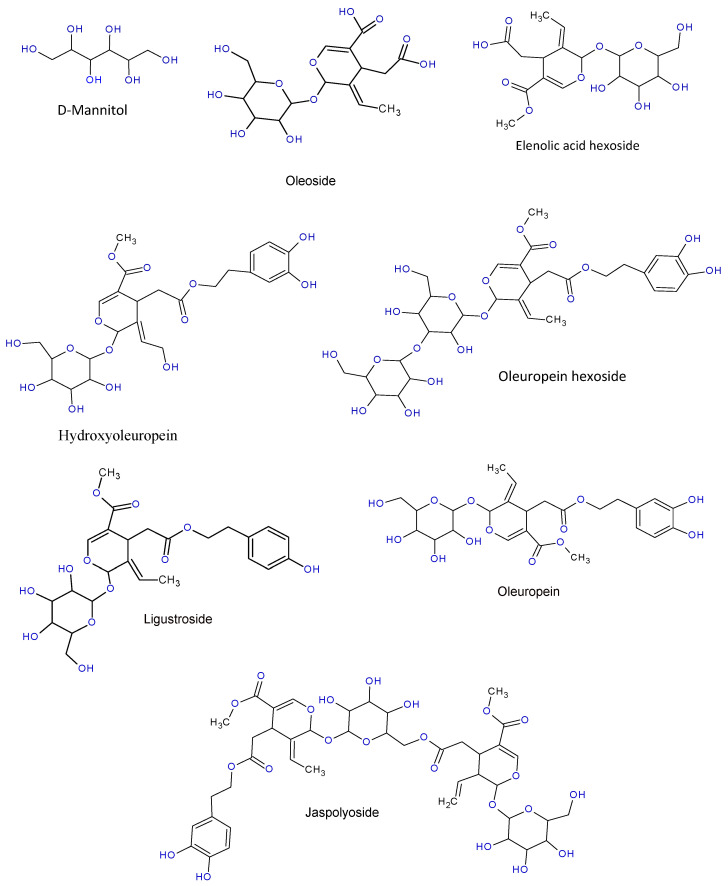
The quantitative analysis of the different phenolic compounds was undertaken by high-performance liquid chromatography (HPLC). Several compounds from different families (flavonoids, secoiridoids, lignans, iridoids, organic acids, and sugars) were quantified (Table 2), such as oleuropein, which is the major compound, whose concentration varies between 5.892 mg/g for the root bark and 10.362 mg/g for the inner part of the root; elenolic acid, with concentrations varying from 4.54 mg/g to 0.116 mg/g; oleuropein hexoside content ranges from 3.484 mg/g to 0.141 mg/g;and ligstroside, which varied from 2.112 mg/g to 5.241 mg/g. Figure 1 displays the structure of the detected main compounds.
Table 2.
Content of the major detected compound.
| N° | m/z | Compound | Concentration [mg/g] | |
|---|---|---|---|---|
| External Part | Internal Part | |||
| 1 | 181 | D-Mannitol | 0.493 ± 0.02 | 1.530 ± 0.06 |
| 2 | 389 | Oleoside | 0.317 ± 0.01 | - |
| 3 | 403 | Elenolic acid hexoside | 4.54 ± 0.19 | 0.116 ± 0.01 |
| 4 | 583 | 2″-Ethoxyoleuropein | 2.227 ± 0.10 | 3.356 ± 0.13 |
| 5 | 555 | Hydroxyoleuropein | 4.892 ± 0.19 | 3.342 ± 0.15 |
| 6 | 701 | Oleuropein hexoside | 3.484 ± 0.14 | 0.141 ± 0.01 |
| 8 | 539 | Oleuropein | 5.892 ± 0.41 | 10.362 ± 0.44 |
| 9 | 623 | verbascoside | 0.115 ± 0.01 | 3.207 ± 0.13 |
| 10 | 523 | Ligstroside | 2.112 ± 0.09 | 5.241 ± 0.22 |
| 11 | 925 | Jaspolyoside | 2.648 ± 0.12 | 0.460 ± 0.018 |
Figure 1.
Structures of the major quantified compounds.
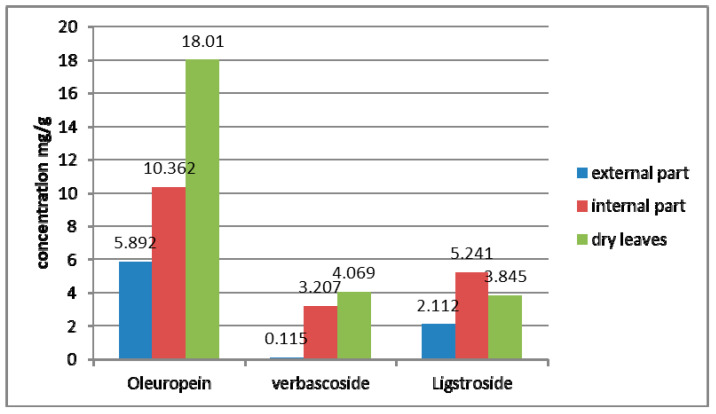
Figure 2 illustrates a comparison between the content of selected compounds in roots and dry leaves studied by Talhaoui et al. [13]. The oleuropein content in olive roots varies from 5.892 to 10.362 mg/g, while that in leaves is around 18.01 mg/g. The verbascoside concentration in dry leaves is around 4 mg/g, while in the inner part of olive roots it amounts to 3.207 mg/g. Moreover, the ligesroside content of olive roots (5.241 mg/g) is higher than that of olive leaves (3.845 mg/g).
Figure 2.
Comparison of the content of selected compounds in roots and leaves.
2.2. Qualitative Profiling via HPLC-DAD and LC-ESIMS/MS and Their Relative MS/MS Data
The identified phenolic compounds marked by HPLC-DAD and LC-ESI- MS/MS in negative ionization mode are exhibited in Table 3. Each compound is related to its retention time (RT), its molecular monoisotopic neutral mass, its experimental m/z, its molecular formula, its mass error (in ppm), and its main MS/MS fragments.
Table 3.
Identified compounds in olive roots using the negative ionization mode.
| N° | RT (min) | Molecular Mass | [M-H]- | Molecular Formula | Mass Error (ppm) | Main Fragments via MS/MS | Proposed Compound | External Part | Internal Part | Total of the Root |
|---|---|---|---|---|---|---|---|---|---|---|
| sugar and derivates | ||||||||||
| 1 | 1.49 | 182.079 | 181.0708 | C6H14O6 | 5.28 | 89.0244, 71.0141, 163.0600, 119.0343 | D-Mannitol | + | + | + |
| organic acids | ||||||||||
| 2 | 1.657 | 196.0583 | 195.0500 | C6H12O7 | −5.23 | 177.0418, 159.0282, 75.0095, 129.0192, 99.0107 | Gluconic acid | + | + | + |
| 3 | 1.875 | 136.0372 | 135.0299 | C4H8O5 | −5.86 | 75.0095, 117.0167, 89.0242 | Threonic acid | + | - | + |
| 4 | 1.958 | 134.0215 | 133.0142 | C4H6O5 | −5.57 | 115.0038, 71.0142 | Malic acid | + | + | + |
| 5 | 2.410 | 192.027 | 191.0177 | C6H8O7 | 0.55 | 111.0085, 87.0084, 129.0197 173.0060 | Citric acid | + | + | + |
| 6 | 5.221 | 168.0423 | 167.0354 | C8H8O4 | −6.44 | 123.0429, 149.0199, 109.0304 93.0356 | 3,4-dihydroxyphenylacetic acid | + | - | + |
| iridoids | ||||||||||
| 7 | 3.748 | 376.1369 | 375.1285 | C16H24O10 | 1.41 | 213.0763, 341.1061 | Loganic acid | + | + | + |
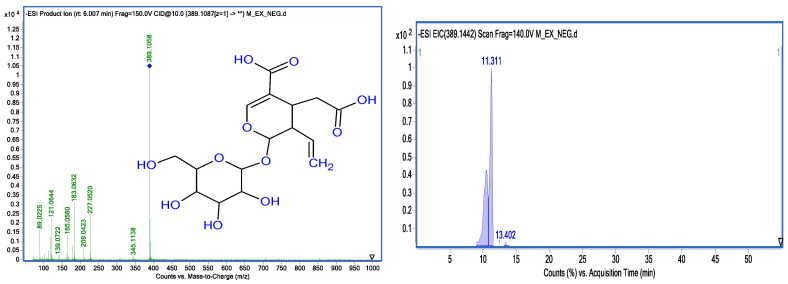
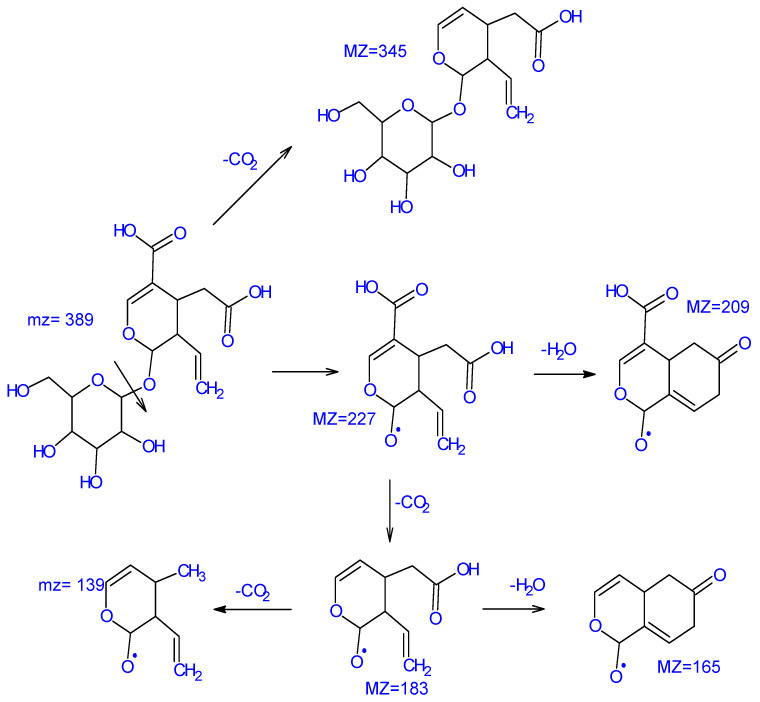
| 8 | 6.007 | 390.1162 | 389.1087 | C16H22O11 | 0.68 | 345.1138, 183.0676, 121.0673 227.0571, 89.0225, 165.0560 | Secologanoside | + | + | + |
| 9 | 11.244 | 390.1526 | 389.1442 | C17H26O10 | 1.74 | 345.1559, 115.0395, 301.1637 151.0773, 101.0252, 83.0132 | Loganin | + | - | + |
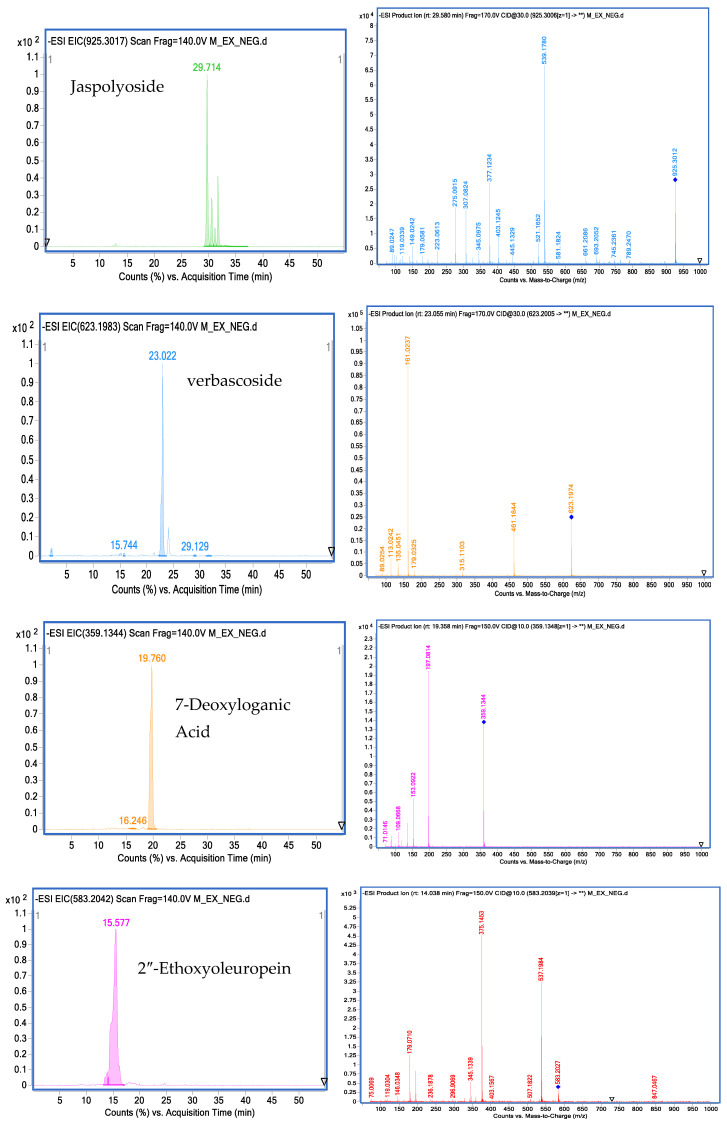
| 10 | 19.358 | 360.142 | 359.1344 | C16H24O9 | −0.99 | 197.0814, 153.0922, 135.0815, 109.0668 | 7-Deoxyloganic acid | + | + | + |
| Phenylethanoids | ||||||||||
| 11 | 4.384 | 316.1158 | 315.1085 | C14H20O8 | −3.93 | 153.0539, 135.0447, 101.0248 | Hydroxytyrosol-glucoside | + | + | + |
| 12 | 4.669 | 154.063 | 153.0546 | C8H10O3 | −7.26 | 123.0447 | Hydroxytyrosol | + | + | + |
| Caffeoyl phenylethanoid derivatives | ||||||||||
| 13 | 23.055 | 624.2054 | 623.1983 | C29H36O15 | −1.19 | 461.1662, 161.0238, 113.0242 135.0451, 315.1103 |
verbascoside | + | + | + |
| 14 | 23.089 | 624.2054 | 623.1983 | C29H36O15 | −1.19 | 461.1662, 161.0238, 113.0242 135.0451, 315.1103 |
Isoverbascoside | + | + | + |
| Phenolic acids | ||||||||||
| 15 | 7.814 | 448.1581 | 447.1508 | C19 H28 O12 | 3.06 | 153.0547, 285.0980, 363.1466 89.0221, 112.9829 |
Dihydroxybenzoic acid hexoside pentoside | + | - | + |
| Flavonoids | ||||||||||
| 16 | 46.227 | 286.1846 | 285.1866 | C15H10O6 | 2.18 | 201.0876, 270.1543, 255.1314, 131.9020, 114.9548, 135.0262 | Luteolin | + | - | + |
| Secoiridoids and derivatives | ||||||||||
| 17 | 4.970 | 390.1162 | 389.1087 | C16H22O11 | 0.6 | 183.0681, 227.0553, 121.0674 209.0414, 165.0562 |
Oleoside | + | - | + |
| 18 | 6.559 | 408.1632 | 407.1559 | C17H28O11 | −3.88 | 389.1447, 357.1187, 89.0239, | Acyclodihydroelenolic acid hexoside I | + | - | + |
| 19 | 9.738 | 422.1788 | 421.1697 | C18H30O11 | −0.79 | 403.1614, 359.1688, 115.0389, 101.0235, 73.0283, 379.1549 151.0737 |
Oleoside methyl ester derivative |
+ | + | + |
| 20 | 11.528 | 404.1319 | 403.1243 | C17H24O11 | −0.71 | 89.0251, 223.0607, 119.0338, 179.0564, 71.0146, 101.0236 |
Elenolic acid hexoside | + | + | + |
| 21 | 13.837 | 392.1682 | 391.1602 | C17H28O10 | −0.18 | 225.1114, 183.1017, 255.1255, 285.1284, 167.0704, 113.0238, 89.0228 |
Methyl oleuropein aglycone | + | + | + |
| 22 | 14.038 | 584.2105 | 583.2032 | C27H36O14 | 1.15 | 537.1984, 375.1453, 179.0710, 195.0667, 345.1339 |
2″-Ethoxyoleuropein | + | + | + |
| 23 | 18.187 | 462.1373 | 461.1301 | C19H26O13 | 0.08 | 403.1209,75.0099, 223.0617 149.0428 |
Elenolic acid hexoside Derivative |
+ | + | + |
| 24 | 21.399 | 556.1792 | 555.1759 | C25H32O14 | 1.75 | 151.0396, 509.2231, 537.1625 403.1237, 223.0607 |
Hydroxyoleuropein | + | + | + |
| 25 | 21.282 | 526.1686 | 525.1618 | C24H30O13 | 0.83 | 195.0662, 389.1059, 345.0980 319.1235, 209.0476 |
Demethyloleuropein | + | + | + |
| 26 | 21.449 | 702.2371 | 701.2319 | C31H42O18 | 2.65 | 315.1071, 539.1808, 469.1349 437.1439 |
Oleuropein hexoside | + | + | + |
| 27 | 21.7 | 702.2371 | 701.2304 | C31H42O18 | 0.8 | 315.1054, 539.1741, 469.1353 437.1428 |
Oleuropein hexoside | + | + | + |
| 28 | 23.708 | 686.2422 | 685.2371 | C31H42O17 | −1.05 | 523.1801, 453.1276, 421.1484 299.1130 |
Nuezhenide | + | - | + |
| 29 | 25.498 | 570.1949 | 569.498 | C26H34O14 | 0.74 | 537.1619, 403.1237, 223.0612 151.0396, 179.0555, 119.0350 89.0250, 337.0904, 305.1026 |
Methoxyoleuropein | + | + | + |
| 30 | 26.358 | 544.2215 | 543.2316 | C25H36O13 | 1.18 | 377.1364, 197.0780, 153.0949 183.1068, 109.1033 |
Dihydro oleuropein | + | + | + |
| 31 | 26.586 | 540.1843 | 539.1781 | C25H32O13 | 2.01 | 403.1217, 377.1217, 307.0792 275.0898, 223.0594, 179.0533 149.0228, 119.0331, 89.0240 |
Oleuropein | + | + | + |
| 32 | 27.054 | 584.1741 | 583.1732 | C27H36O14 | 1.29 | 537.1590, 403.1224, 223.0573 151.0403, 179.0521 | Lucidumoside C | + | - | + |
| 33 | 28.058 | 558.2312 | 557.2225 | C26H38O13 | −2.62 | 513.2315, 227.1269, 371.0920 121.0659, 183.0636, 165.0614 |
60-O-[(2E)-2.6-Dimethyl-8- hydroxy-2-octenoyloxy]- secologanoside |
+ | - | + |
| 34 | 28.426 | 524.1894 | 523.1822 | C25H32O12 | 0.19 | 361.1297, 291.0869, 259.0975 101.0252, 127.0393, 179.0593 |
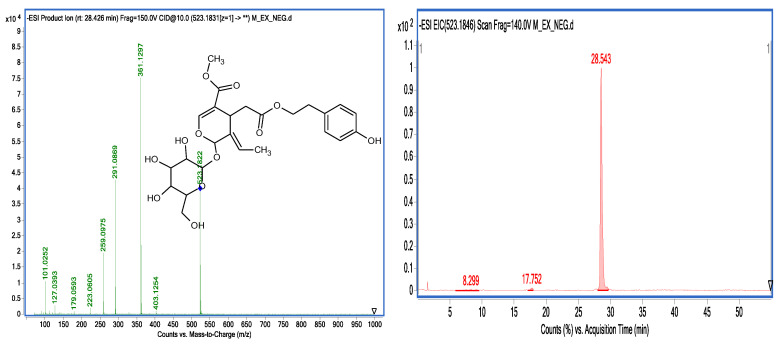
Ligustroside | + | + | + |
| 35 | 28.644 | 362.1366 | 361.1193 | C19H22O7 | 1.17 | 291.0882, 259.0991, 127.0401 101.0245, 171.0286 |
Ligstroside aglycone | + | + | + |
| 36 | 29.580 | 926.3056 | 925.3006 | C42H54O23 | 2.47 | 789.2470, 745.2361, 661.2086 539.1780, 521.1652, 403.1245 377.1234, 307.0824, 275.0915 223.0613, 149.0242, 119.0339 89.0247 |
Jaspolyoside I | + | + | + |
| 37 | 31.354 | 394.1264 | 393.1194 | C19H22O9 | 0.75 | 317.1027, 349.1270, 289.1046 181.0490, 137.0590, 153.0540 | Hydroxy oleuropein aglycone | + | + | + |
| 38 | 32,19 | 706.2837 | 705.2774 | C35H46O15 | 1.42 | 539.1748, 521.1635, 377.1211 327.2138, 307.0799, 275.0896 223.0596, 149.0223 |
Oleuropein derivate | + | + | + |
| Lignan | ||||||||||
| 39 | 15.376 | 538.205 | 537.1990 | C26H34O12 | 0.84 | 375.1448, 179.0703, 195.0666 345.1343, 327.1202 |
Olivil 4-O-β-D-glucopyranoside | + | + | + |
| 40 | 15.510 | 376.1522 | 375.1476 | C20H24O7 | 2.85 | 360.1440, 345.1361, 327.1244 195.0672, 179.0725, 161.0561 146.0376, 122.0408 |
Olivil | + | + | + |
| 41 | 16.096 | 416.1682 | 415.1581 | C19H28O10 | −0.17 | 149.0447, 89.0233, 191.0577 251.0803, 131.0396 |
1-acetoxy-pinoresinol | + | - | + |
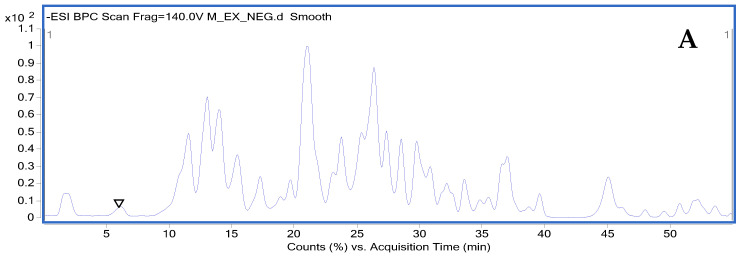
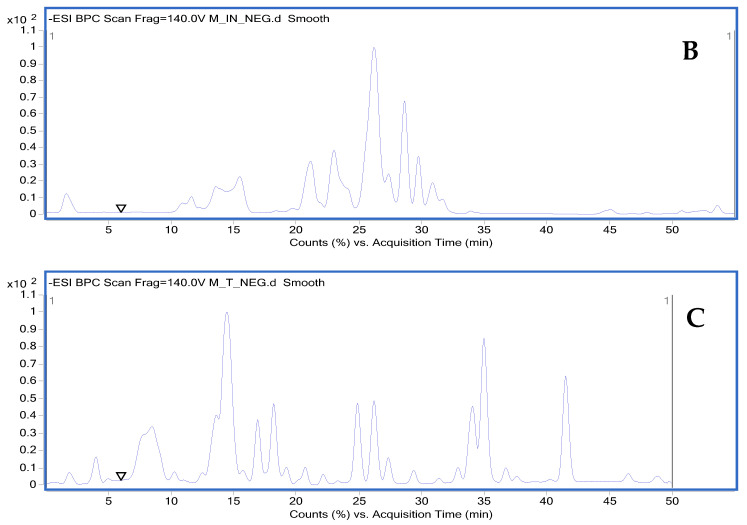
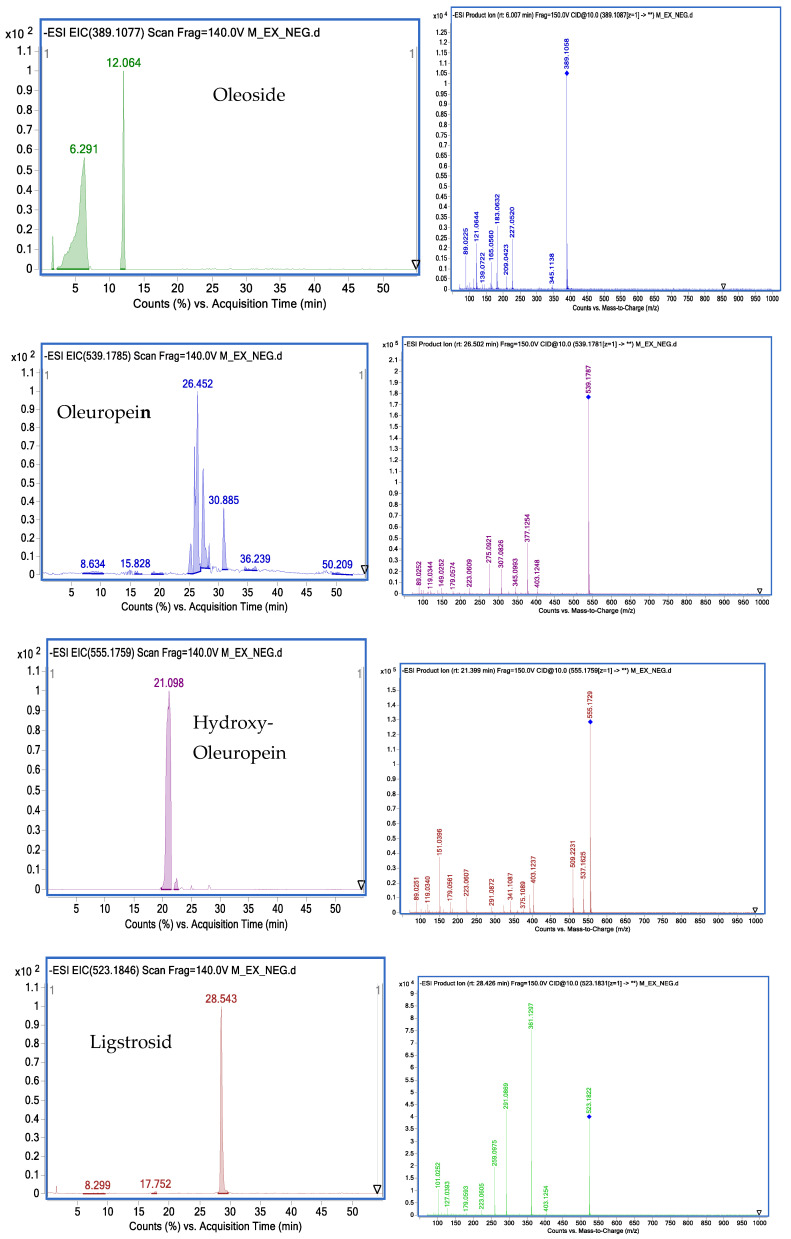
To fully assess the phenolic composition of olive roots, we have chosen to study each part on its own in order to compare them with each other. The first part corresponds to the outer part or bark as represented by the chromatogram (A); the second part corresponds to the inner part of the root as represented by the chromatogram (B); and the third part corresponds to the sum of the roots as represented by the chromatogram (C). The base-peak chromatograms (BPC) of olive roots are plotted in Figure 3, indicating their richness in phenolic compounds, as 40 phenolic compounds were identified for the roots. Olive metabolites were classified into eight structural classes: sugars and derivatives, organic acids, phenolic acids (simple), flavonoids, caffeoyl phenylethanoid derivatives, iridoids and derivatives, secoiridoids and derivatives, and lignans.
Figure 3.
Base peak chromatogram (BPC) of the olive root methanol extract obtained by HPLC-DAD-ESI-MS in the negative ionization mode ((A)—chromatogram of the extract from the outer part of the olive tree root; (B)—chromatogram of the extract from the inner part of the olive tree root; (C)—chromatogram of the extract from the olive tree whole roots).
2.2.1. Sugars and Derivatives
Low-molecular-weight sugars share multiple chemical properties. They are optically active aliphatic polyhydroxy compounds that are generally highly soluble in water. The presence of sugars and their derivatives was denoted with a peak at 1.49 min, representing the monosaccharide D-mannitol (C6H14O6) (181.07 m/z) with its fragment (163.06 m/z) (C6H12O5), corresponding to the loss of water molecules (-H2O), (89.0244 m/z), (71.0141 m/z), and (119.0343 m/z). This is in good agreement with previous data addressing olive wood from Spanish cultivars [14,15].
2.2.2. Organic Acids
Organic acids are ubiquitous in nature, as they are found in animal, vegetable, and microbial sources. They contain one or more carboxylic acid groups that can be covalently bonded with such groups as amides, esters, and peptides. Five compounds were observed in olive root extracts, including: gluconic acid (C6H12O7) (195.05 m/z) at 1.657 min with their two fragments (177.0418 m/z) (C6H10O6) and (159.0282 m/z) (C6H8O5) corresponding to a successive dehydration (-2H2O), and another fragment of molecular mass (129.0192 m/z) (C5H5O4) ascribed to the loss of the group (-CH2OH). Threonic acid (135.02 m/z) (C4H8O5) is represented by a peak at 1.87 min, with a fragment (117.0167 m/z) assigned to the elimination of a water molecule (H2O). Another fragment (89.0242 m/z) corresponded to the loss of the group (HCOOH) from threonic acid. Malic acid (C4H6O5) was identified at 1.95 min with (133.01 m/z) along with the main fragments: (115.0038 m/z) and (71.0142 m/z). Citric acid (191.0177 m/z) and 3,4-dihydroxyphenylacetic acid (167.03 m/z). The fragmentation pattern of the olive root extracts displayed similar results to those reported in the literature [16,17,18].
2.2.3. Flavonoids
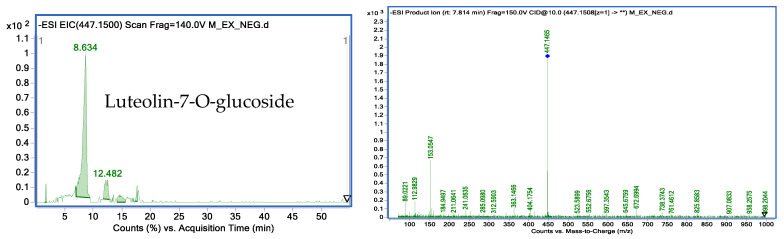
Luteolin (C15H9O6) (285.18 m/z) at 7.4 min with the main fragments: (201.0876 m/z), (270.1543 m/z), (255.1314 m/z), (131.9020 m/z), (114.9548 m/z), and (135.0262 m/z). The identification was carried out by comparing their ECs and fragmentation pathways to those reported in the literature and databases [18,19].
2.2.4. Caffeoylphenylethanoid Derivatives
Verbascoside and isoverbascoside isomers were identified at (623.1983 m/z) (C29H36O15). The main fragment at (461.16 m/z) was obtained from the loss of the glucose (C6H11O5) residue from the deprotonated molecular ion (623.1983 m/z). Other fragments were present, such as (161.02 m/z), (113.02 m/z), (135.04 m/z), and (315.11 m/z). Their retention pattern, MS, and MS/MS spectra are in good accordance with the available data [10,13,18].
2.2.5. Phenylethanoids
Two peaks are present at 4.38 and 4.66 min with precursor ion values equal to (315.1085 m/z) and (153.0546 m/z), corresponding to hydroxytyrosol-glucoside (C14H20O8) and hydroxytyrosol (C8H10O3), respectively. The MS-MS fragmentation spectrum of hydroxytyrosol exhibits a main fragment at (123.0447 m/z) obtained through the loss of the (CH2OH) group at(123.0447 m/z).
2.2.6. Iridoids and Derivatives
Iridoids are known to offer a wide range of health benefits, including resisting a wide assortment of physical, biological, and chemical stressors. Multiple scientific studies have highlighted that iridoids possess antioxidant, antibacterial, anti-cancer, and anti-viral properties. Iridoids have been identified in olive tree root extracts. Among the most important ones, we state loganic acid (C16H24O10) (375.12 m/z) with main fragments at (213.0763 m/z), (341.1061 m/z); secologanosid (389.1087 m/z) (C16H22O11) (Figure 4); and main fragments at (345.1138 m/z), (183.0676 m/z), (121.0673 m/z), and (227.0571 m/z) related to the loss of (C6H11O5), (89.0225 m/z), and (165.0560 m/z). Relying on the spectrophotometric data, Figure 5 illustrates a proposed mechanism for the fragmentation of secologanosid. The appearing peak has a retention time of 19.35 min (359.13 m/z), and the fragments (197.0814 m/z), (153.0922 m/z), (135.0815 m/z), and (109.0668 m/z) have been identified as 7-deoxyloganic acid (C16H24O9). The peak with a retention time of 11.24 min (389.14 m/z) and the fragments (345.1559 m/z), (115.0395 m/z), (301.1637 m/z), (151.0773 m/z), (101.0252 m/z), and (83.0132 m/z) were identified as loganin.
Figure 4.
Ion chromatogram and MS-MS spectra of secologanosid.
Figure 5.
Proposed mechanism of secologanosid fragmentation based on MS/MS data.
This type of monoterpene is widely distributed in the Oleaceae (Obied et al., 2008) [20]. It is to be noted that their fragmentation patterns are consistent with previous works [10,14,21].
2.2.7. Secoiridoids and Derivatives
Secoiridoids are the major class of secondary metabolites identified in olives. Among them, oleuropein corresponds to the major compound in leaf, stem, and root extracts. Several secoiridoids were detected in the extracts, and all were identified by comparing their ECs and fragmentation pathways to those found in the literature and databases [10,16].
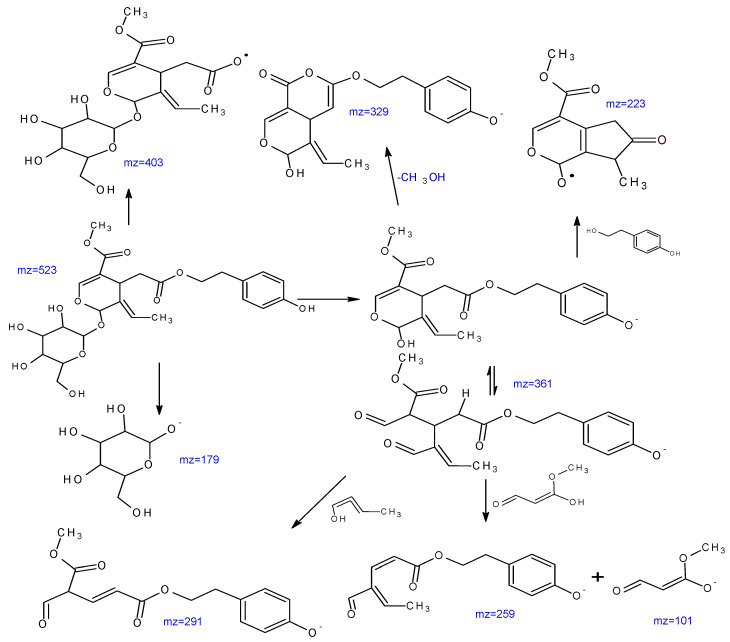
A total of 22 known secoiridoids have been tentatively identified in olive roots. They notably correspond to oleoside (C16H22O11) (389.1087 m/z), acyclodihydroelenolic acid hexoside I (C17H28O11) (407, 1559 m/z), oleoside methyl ester derivative (C18H30O11) (421.1697 m/z),elenolic acid hexoside(C17H24O11) (403.1243 m/z) methyl oleuropeinaglycone (C17H28O10) (391.1602 m/z), 2″-ethoxyoleuropein (C27H36O14) (583.2032 m/z), hydroxyoleuropein (C25H32O14) (555.1759 m/z), demethyloleuropein (C24 H30 O13) (525.1618 m/z), oleuropein hexoside (C31H42O18) (701.2319 m/z),oleuropein hexoside (C31H42O18) (701.2304 m/z), methoxyoleuropein (C26H34O14) (569.498 m/z), dihydrooleuropein (543.2316 m/z), oleuropein (C25H32O13) (539.1781 m/z), lucidumoside C (C27H36O14) (583.1732 m/z), 60-O-[(2E)-2.6-dimethyl-8-hydroxy-2-octenoyloxy]-secologanoside (C26H38O13) (557.2225 m/z), ligustroside (C25H32O12) (523.1822 m/z), ligstrosideaglycone (C19H22O7) (361.1193 m/z), jaspolyoside (C42H54O23) (925.3006 m/z), hydroxyoleuropeinaglycone (C19H22O9) (393.1194 m/z) and oleuropein derivate (C35H46O15) (705.2774 m/z), elenolic acid hexoside derivative (C19H26O13) (461.1301 m/z) and nuezhenide (C31H42O17) (685.2371 m/z). We opted to characterize two compounds by examining their MS and MS/MS spectra. Oleuropein at 25.586 min is present at (539.1811 m/z) (C25H31O12). Two main fragments at (403.1217 m/z) and (377.1217 m/z) are obtained by the loss of the (C8H9O2) moiety as well as the loss of the glucose residue group of the deprotonated molecular ion (539.1811 m/z). Other characteristic fragments can be detected, such as (275.0898 m/z), which is characteristic of the loss of (C4H6O3) from (377.1217 m/z), and (307.0792 m/z), which is characteristic of the loss of (C4H6O) from (377.1217 m/z). Ligstroside at 28.42 min, another well-known compound of the Oleaceae family, is present with 523.1811 m/z(C25H31O12) (Figure 6). The main fragment (361.1277 m/z) is obtained by the loss of a glucose residue from the deprotonated molecular ion (523.1811 m/z). Other characteristic fragments can be observed, such as (291.0865 m/z), obtained through the loss of (C4H6O) from (361.1297 m/z), and (259.0975 m/z), characteristic of the loss of (CH3OH) from (291.0865 m/z). Figure 7 depicts a proposed mechanism for the fragmentation of ligstroside. The fragmentation model was checked through comparison with previous studies [22].
Figure 6.
Ion chromatogram and MS-MS spectra of ligstroside.
Figure 7.
Proposed mechanism of ligestrosid fragmentation based on MS/MS data.
2.2.8. Lignans
Another equally significant group of compounds present in the olive tree, namely the lignans, characterizes mainly the stem and the roots. After careful interpretation of their MS and MS/MS spectra, the identified compounds are: olivil (C20H24O7) (375.1476 m/z), and their main fragments are: (195.0672 m/z), (179.0725 m/z), (161.0561 m/z), and (122.0408 m/z). This is in good conformity with the opening and cleavage of the tetrahydrofuran ring, the additional loss of methyl groups, and the loss of water molecules, as reported by Sanz et al. [23]. Olivil 4-O-β-D-glucopyranoside (C26H34O12) was characterized by a peak appearing at 15.76 min with (537.1990 m/z) and the fragments: (375.1448 m/z), (179.0703 m/z), (195.0666 m/z), (345.1343 m/z), and (327.1202 m/z). Another peak emerged at 15.44 min with (415.1581 m/z) and the fragments: (149.0447 m/z), (89.0233 m/z), (191.0577 m/z), (251.0803 m/z), and (131.0396 m/z) were identified as 1-acetoxy-pinoresinol (C19H28O10). All aforementioned lignans were previously reported in olive wood [14,15].
Figure 8 illustrates the extracted ion chromatogram (EIC) profiles and MS-MS spectra of selected phenolic compounds identified in methanolic extracts from Chemlali olive roots.
Figure 8.
Profiles of extracted ion chromatograms (EIC) and their MS-MS spectra obtained by HPLC-DAD-MS of some phenolic compounds identified in methanolic extracts of Chemlali olive tree roots.
2.3. Cytotoxicity and Antiviral Activity
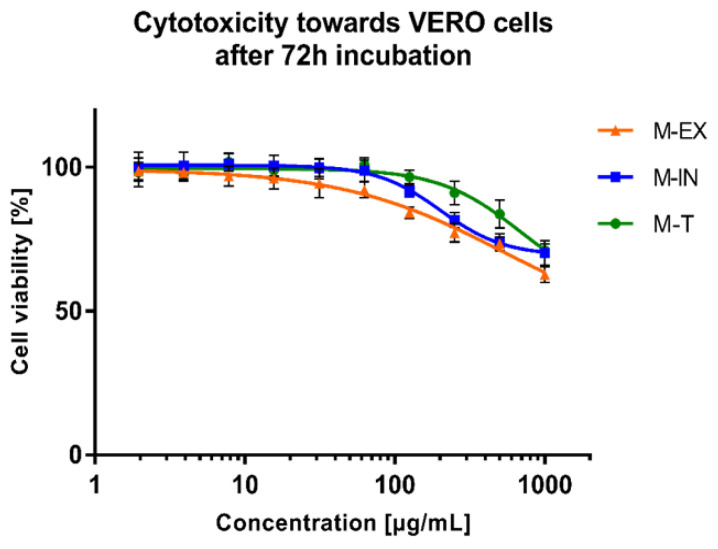
The dose-response effect of the extracts (external part M-EX, internal part M-IN, and all the roots M-T) on the viability of VERO cells is portrayed in Figure 9. The exact CC50 values (concentrations of the tested extract decreasing the cellular viability by 50%) could not be assessed but were above 500 µg/mL for all tested extracts. According to a previously published work [24], no significant cytotoxicity of plant extract is recorded when the CC50 is above 500 µg/mL.
Figure 9.
The cytotoxicity of extracts. (M-EX—external part; M-IN—internal part; M-T—whole roots).
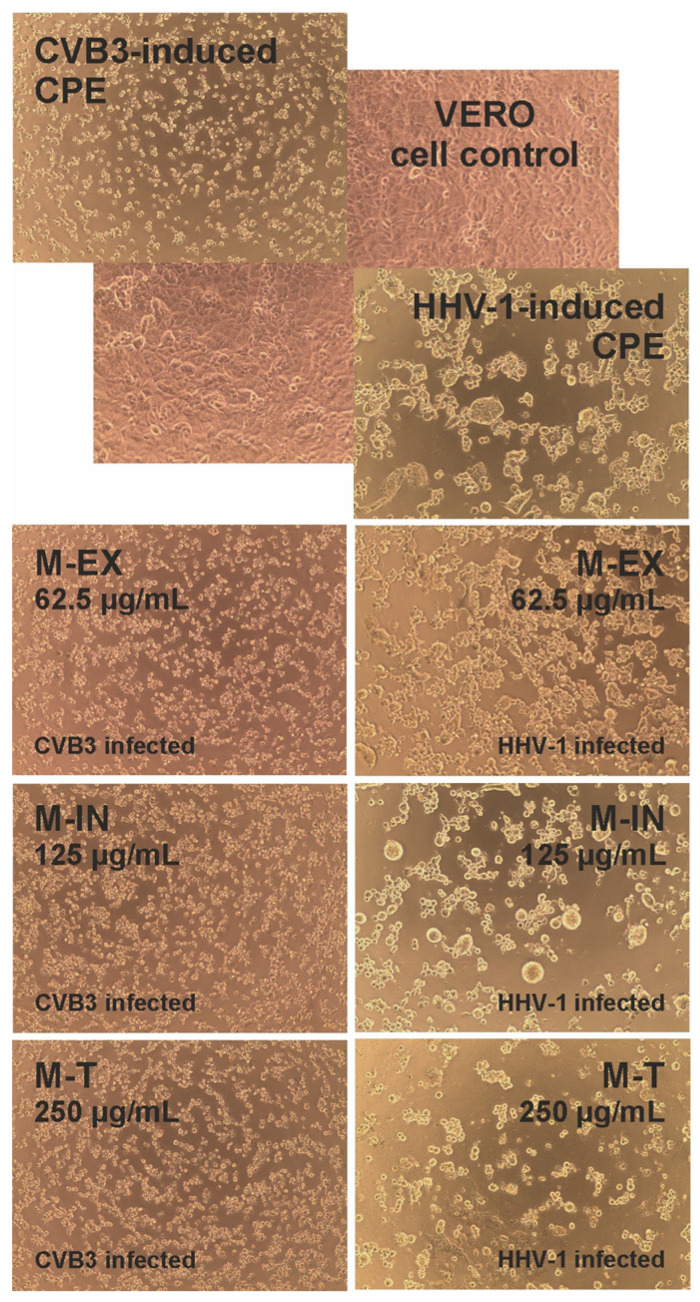
None of the tested extracts displayed any noticeable effect on the formation of virus-induced cytopathic effects(CPEs) in the infected VERO cells. Subsequent end-point dilution assays revealed that the infectious titers of both viruses in the extract-treated samples were comparable to the titers in the appropriate virus control samples.
At this stage of analysis, it is noteworthy that the extracts obtained from the underground parts of the olive tree Chemlalai cultivar exhibit no significant cytotoxicity towards non-cancerous VERO cells. Interestingly, methanolic extracts from the twigs of Chemlali and Chétoui cultivars presented moderate cytotoxicity towards VERO cells with CC50 values of 155 and 151 μg/mL [25]. Aqueous extracts from the leaves of two olive tree varieties (Meski and Chemlali) proved to increase the viability of murine oligodendrocytes (158N) in the concentration range of 5–200 μg/mL, while at 400 μg/mL, the viability of 158N was reduced [26]. The olive leaf ethanolic extracts also suppressed cytotoxicity induced by oxidative stress on the mouse embryonic fibroblast (MEF) [27]. Hydroethanolic extract from the leaves of the Chemlali cultivar proved to inhibit the proliferation of a cell line derived from human chronic myeloid leukemia (K562) and to exert G0/G1 cell cycle arrest on the 1st and 2nd days of incubation, and then also at G2/M phase (3rd and 4th days). Additionally, the extract induced apoptosis and differentiation of K562 cells towards the monocyte lineage [28]. Essafi Rhouma et al. [29] extracted the flowers of the Chemlali cultivar with 80% (v/v) ethanol and tested their cytotoxicity towards breast cancer cells (MCF-7) and human mammary epithelial cells (MCF-10A). The extracts in the concentration range between 31.25 and 250 μg/mL decreased the viability of MCF-7 cells without displaying a detrimental effect on MCF-10A. Moreover, the western blot analysis revealed the potential pro-apoptotic activity of flower extracts in MCF-7 cells [29].
None of the tested extracts demonstrated antiviral activity against the CVB3 or HHV-1 viruses replicating in the VERO cells. Leila et al. [25] investigated the cytotoxicity and antiviral activity against CVB3 or HHV-2 (Human Herpesvirus type 2) of twig extracts obtained using hexane, dichloromethane, ethyl acetate, and methanol from two Tunisian olive cultivars (Chemlali and Chétoui). Extracts from both cultivars displayed no effect on the replication of HHV-2. However, hexane extracts from Chemlali and Chétoui cultivars exhibited antiviral effects towards CVB-3 with IC50 (50% inhibitory concentration) of 20.41 and 23.28 μg/mL, respectively, and a selectivity index (SI) > 5 [25]. The hydroethanolic (water:ethanol; 50:50 (v/v)) extracts from the leaves of O. europaea var. sativa and O. europaea var. sylvestris were found to inhibit the replication of HHV-1 in the HeLa cells, reducing the infectious titer (plaque-forming assay) as well as the viral load (real-time PCR) [30]. Despite the literature data describing olive twigs or leaves as potential sources of antiviral compounds, we have not observed any antiviral activity for the extracts from underground parts of the olive tree (Chemlalai cultivar). In the case of hexane extracts from twigs of Chemlali and Chétoui cultivars, antiviral activity against CVB-3 was observed, but no detailed phytochemical analysis was performed. It was suggested that 2,4-di-tert-butylphenol was the bioactive compound responsible for this activity, but this was only shown through bio-guided assays using thin-layer chromatography [25]. This compound was not present in the root extracts we studied. The O. europaea var. sativa and O. europaea var. sylvestris extracts showed potent anti-HHV-1 activity, and this activity was probably due to the presence of oleuropein [30]. We have also identified oleuropein in the extracts from underground parts. Still, the content of this compound is the highest in leaves, and this may be the reason for the lack of antiviral activity of root extracts. Unfortunately, the cytotoxicity of root extracts did not allow us to test the antiviral activity using higher concentrations than those presented in Figure 10.
Figure 10.
The influence of extracts on viral-induced cytopathic effects (CVB3—Coxsackievirus B3; CPE—cytopathic effect; HHV-1—Human Herpesvirus type 1; M-EX—external part; M-IN—internal part; M-T—whole roots).
3. Materials and Methods
3.1. Sample Extraction
The olive root (2 kg) was collected from Chemlali olive trees grown in the Sfax region (Tunisia). The olive trees were about 45 years old. Olive root samples were collected in early January 2021. The samples were dried for 90 days at room temperature in a dark and airy room, and then the root was scraped at a local sawmill to separate the bark from the root wood. All samples were then ground to a fine powder using a domestic mill before being extracted. The roots of the olive tree have been separated into three parts, namely the external part, the internal part, and the total roots. Each part of the olive roots was placed separately in amber glass bottles and homogenized in a solution of methanol/water 80:20 (v/v) using an ultrasonic bath for 30 min. After filtration with Whatman filter paper No. 42 (125 mm), the extracts were put in a rotary evaporator under vacuum at 40 °C. Eventually, the extracts were kept at −20 °C until future analysis. After extraction and obtaining dry residues, the extraction yield was computed.
3.2. HPLC Analysis
In order to characterize the phenolic compounds by HPLC (Agilent 1200 series), the mobile phases relied on a combination of solvent A (water) and solvent B (methanol). The following multi-step gradient was applied: 0 min, 50% B; 5 min, 60% B; 20 min, 80% B; 25 min, 100% B; 26 min, 50% B; 35 min, 50% B. The flow rate was set at 1 mL/min throughout the gradient. The separation was undertaken with a Gemini 5 mcm NX-C18 110A column (250 × 4.6 mm) at room temperature. UV spectra were recorded from 190 to 600 nm. The injection volume was 10 uL. The identification of each compound was based on a comparison of their retention times with those of the reference compounds and recording the UV spectra in the 190–400 nm range. The quantitative determination was performed using calibration curves for phenolic standards. The content of the phenolic compounds was expressed in mg/g of dry olive roots.
3.3. MS-MS Analysis
The system was coupled to a 6540 Agilent Ultra-High-Definition Accurate-Mass QTOF, equipped with an ESI source (Agilent Dual Jet Stream) (Agilent Technologies, Palo Alto, CA, USA). The operating conditions in negative ionization mode were as follows: gas temperature, 350 °C; drying gas, nitrogen at 12 L/min; nebulizer pressure, 40 psig; sheath gas temperature, 400 °C; sheath gas flow, nitrogen at 12 L/min; capillary voltage, 4000 V; skimmer, 645 V; octopol radiofrequency voltage, 750 V, with the corresponding polarity automatically set. The spectra were acquired over a mass range from 100 to 1000 m/z, and for MS/MS experiments, from 100 to 1000 m/z. The reference mass correction of each sample was carried out with a continuous infusion of Agilent API TOF reference mixture (61,969–85,001). The data analysis was conducted on a Mass Hunter Qualitative Analysis B.10.00 (Agilent Technologies).
3.4. Cytotoxicity and Antiviral Activity
The VERO cells were cultured using Dulbecco’s Modified Eagle’s Medium (DMEM, Corning, Tewksbury, MA, USA) supplemented with fetal bovine serum (FBS; Capricorn Scientific, Ebsdorfergrund, Germany) and antibiotics (Penicillin-Streptomycin Solution, Corning). The dimethyl sulfoxide and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were obtained from Sigma-Aldrich (St. Louis, MO, USA), whereas phosphate-buffered saline (PBS) and trypsin were purchased from Corning. Incubation was undertaken at 37 °C in a 5% CO2 atmosphere (CO2 incubator, Panasonic Healthcare Co., Ltd., Tokyo, Japan).
HHV-1 (ATCC, No. VR-260) and CVB3 (ATCC, No. VR-30) were propagated in VERO cells, and the infectious titers (CCID50—50% cell culture infectious dose) were assessed using an end-point dilution assay.
3.4.1. Cytotoxicity Testing
Cytotoxicity was tested according to the previously described methodology [31]. The monolayer of VERO cells in 96-well plates was treated with serial dilutions of extracts in cell media for 72 h. Afterwards, the media were removed. After washing with PBS, the MTT solution in DMEM was added, and the incubation continued for 4 h. Eventually, the formazan crystals were dissolved using SDS/DMF/PBS solvent. Following the overnight incubation, the absorbance (540 and 620 nm) was determined using the Synergy H1 Multi-Mode Microplate Reader (BioTek Instruments, Inc., Winooski, VT, USA). Collected data were exported from Gen5 software (ver. 3.09.07; BioTek Instruments, Inc., Winooski, VT, USA) to GraphPad Prism (v7.0.4) for further analysis.
3.4.2. Antiviral Activity
The antiviral activity was tested, as previously described [31]. The VERO cells seeded in 48-well plates were infected with HHV-1 or CVB3 at a 100-fold CCID50 infectious titer. After 1 h of pre-incubation allowing virus attachment, the cell monolayer was washed with PBS and treated with extracts in non-toxic concentrations. The non-toxic doses were selected based on dose-response curves obtained during cytotoxicity studies. They were defined as concentrations of selected extracts that did not reduce cellular viability by more than 10%. Incubation continued until a typical CPE was observed in the virus control (infected, non-treated cells). Next, the sample-treated infected cells were also observed for the occurrence of CPE using an inverted microscope (CKX41, Olympus Corporation, Tokyo, Japan) equipped with a camera (Moticam 3+, Motic, Hong Kong), and the results were recorded (Motic Images Plus 2.0, Motic, Hong Kong). Afterwards, the plates were thrice frozen (−72 °C) and thawed, and the samples were collected and kept frozen at −72 °C until the assessment of the viral infectious titer was carried out.
Virus titrations were conducted using an end-point dilution assay [31]. Briefly, monolayers of VERO in 96-well plates were treated with tenfold dilutions of samples from the above-described antiviral assays. After 72 h, the viral titers were calculated using an MTT assay. The antiviral activity was specified, referring to the difference in viral titer between the extract-treated samples and the virus control. A significant antiviral activity needs at least a 3 log reduction of the viral infectious titer.
4. Conclusions
Overall, olive roots may be considered a source of polyphenolic compounds. Spectrometric analysis applied through the use of the liquid chromatography-mass spectrometry technique proved to be a powerful tool for the characterization of several compounds in olive roots. 41 molecules of different families were identified, including secoiridoids, iridoids, lignans, and organic acids. Moreover, according to the quantification by UHPLC-DAD, oleuropein is the major compound detected in olive roots, with a concentration of 10.36 mg/g. Ligestroside has also been detected, and its amount is higher than that in leaves. These results provide a deeper and better insight into the bioactive compounds in the less-considered organs of the olive tree. Several compounds remained unidentified, revealing the chemical diversity of Olea. The extracts proved to be non-toxic to VERO cells and did not exert any noticeable antiviral effect against HHV-1 or CVB3 replicating in VERO cells. Indeed, in future works, we may tackle the identified compounds, attempt to isolate and investigate their characteristics, and fully understand how significant they are.
Author Contributions
Conceptualization, M.B.; methodology, K.S.-W.; validation, K.T., Ł.Ś. and A.B.; formal analysis, K.S.-W.; data curation, K.T.; writing—original draft preparation, K.T. and Ł.Ś.; writing—review and editing, K.T. and M.B.; supervision, M.B. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are available from the authors.
Funding Statement
This research was funded by the Ministry of Higher Education and Scientific Research (Laboratory LR14ES08), Tunisia.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Clodoveo M.L., Yangui A., Fendri M., Giordano S., Crupi P., Corbo F. Protected Geographical Indications for EVOO in Tunisia: Towards Environmental, Social, and Economic Sustainable Development. Sustainability. 2021;13:11201. doi: 10.3390/su132011201. [DOI] [Google Scholar]
- 2.Alibes X., Berge P.H. Valorización de los Subproductos del Olivar Como Alimentos Para los Rumiantes en España. FAO; Rome, Italy: 1983. Division de la Production Animale. [Google Scholar]
- 3.Madejon E., Galli E., Tomati U. Composting of wastes produced by low water consuming olive mill technology. Agrochimica. 1998;42:135–146. [Google Scholar]
- 4.Ortega-García F., Peragón J. HPLC analysis of oleuropein, hydroxytyrosol, and tyrosol in stems and roots of Olea europaea L. cv. Picual during ripening. J. Sci. Food Agric. 2010;90:2295–2300. doi: 10.1002/jsfa.4085. [DOI] [PubMed] [Google Scholar]
- 5.Gorzynik-Debicka M., Przychodzen P., Cappello F., Kuban-Jankowska A., Marino Gammazza A., Knap N., Wozniak M., Gorska-Ponikowska M. Potential Health Benefits of Olive Oil and Plant Polyphenols. Int. J. Mol. Sci. 2018;19:686. doi: 10.3390/ijms19030686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romani A., Ieri F., Urciuoli S., Noce A., Marrone G., Nediani C., Bernini R. Health Effects of Phenolic Compounds Found in Extra-Virgin Olive Oil, By-Products, and Leaf of Olea europaea L. Nutrients. 2019;11:1776. doi: 10.3390/nu11081776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozkaya M.T., Celik M. The effects of various treatments on endogenous carbohydrate content of cuttings in easy-to-root and hard-to-root olive cultivars. Acta Hortic. 1999;474:51–54. doi: 10.17660/ActaHortic.1999.474.5. [DOI] [Google Scholar]
- 8.Malik N.S., Bradford J.M. Changes in oleuropein levels during differentiation and development of floral buds in ‘Arbequina’ olives. Sci. Hortic. 2006;110:274–278. doi: 10.1016/j.scienta.2006.07.016. [DOI] [Google Scholar]
- 9.Leong L., Shui G. An investigation of antioxidant capacity of fruits in Singapore markets. Food Chem. 2002;76:69–75. doi: 10.1016/S0308-8146(01)00251-5. [DOI] [Google Scholar]
- 10.Michel T., Khlif I., Kanakis P., Termentzi A., Allouche N., Halabalaki M., Skaltsounis A.-L. UHPLC-DAD-FLD and UHPLC-HRMS/MS based metabolic profiling and characterization of different Olea europaea organs of Koroneiki and Chetoui varieties. Phytochem. Lett. 2015;11:424–439. doi: 10.1016/j.phytol.2014.12.020. [DOI] [Google Scholar]
- 11.Luque-Muñoz A., Tapia R., Haidour A., Justicia J., Cuerva J.M. Direct determination of phenolic secoiridoids in olive oil by ultra-high performance liquid chromatography-triple quadruple mass spectrometry analysis. Sci. Rep. 2019;9:15545. doi: 10.1038/s41598-019-52060-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ammar S., Contreras M.D.M., Gargouri B., Segura-Carretero A., Bouaziz M. RP-HPLC-DAD-ESI-QTOF-MS based metabolic profiling of the potential Olea europaea by-product “wood” and its comparison with leaf counterpart. Phytochem. Anal. 2017;28:217–229. doi: 10.1002/pca.2664. [DOI] [PubMed] [Google Scholar]
- 13.Talhaoui N., Gómez-Caravaca A.M., León L., De la Rosa R., Segura-Carretero A., Fernández-Gutiérrez A. Determination of phenolic compounds of ‘Sikitita’ olive leaves by HPLC-DAD-TOF-MS. Comparison with its parents ‘Arbequina’ and ‘Picual’ olive leaves. LWT-Food Sci. Technol. 2014;58:28–34. doi: 10.1016/j.lwt.2014.03.014. [DOI] [Google Scholar]
- 14.Pérez-Bonilla M., Salido S., van Beek T.A., Altarejos J. Radical-Scavenging Compounds from Olive Tree (Olea europaea L.) Wood. J. Agric. Food Chem. 2014;62:144–151. doi: 10.1021/jf403998t. [DOI] [PubMed] [Google Scholar]
- 15.Salido S., Pérez-Bonilla M., Adams R.P., Altarejos J. Phenolic Components and Antioxidant Activity of Wood Extracts from 10 Main Spanish Olive Cultivars. J. Agric. Food Chem. 2015;63:6493–6500. doi: 10.1021/acs.jafc.5b02979. [DOI] [PubMed] [Google Scholar]
- 16.Abu-Reidah I., Contreras M., Arráez-Román D., Segura-Carretero A., Fernández-Gutiérrez A. Reversed-phase ultra-high-performance liquid chromatography coupled to electrospray ionization-quadrupole-time-of-flight mass spectrometry as a powerful tool for metabolic profiling of vegetables: Lactuca sativa as an example of its application. J. Chromatogr. A. 2013;1313:212–227. doi: 10.1016/j.chroma.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 17.Ammar S., Contreras M.D.M., Belguith-Hadrich O., Segura-Carretero A., Bouaziz M. Assessment of the distribution of phenolic compounds and contribution to the antioxidant activity in Tunisian fig leaves, fruits, skins and pulps using mass spectrometry-based analysis. Food Funct. 2015;6:3663–3677. doi: 10.1039/C5FO00837A. [DOI] [PubMed] [Google Scholar]
- 18.Quirantes-Piné R., Lozano-Sánchez J., Herrero M., Ibanez E., Segura-Carretero A., Gutierrez A.F. HPLC-ESI-QTOF-MS as a Powerful Analytical Tool for Characterising Phenolic Compounds in Olive-leaf Extracts. Phytochem. Anal. 2013;24:213–223. doi: 10.1002/pca.2401. [DOI] [PubMed] [Google Scholar]
- 19.Taamalli A., Abaza L., Román D.A., Carretero A.S., Gutiérrez A.F., Zarrouk M., Ben Youssef N. Characterisation of Phenolic Compounds by HPLC-TOF/IT/MS in Buds and Open Flowers of ‘Chemlali’ Olive Cultivar. Phytochem. Anal. 2013;24:504–512. doi: 10.1002/pca.2450. [DOI] [PubMed] [Google Scholar]
- 20.Obied H.K., Prenzler P.D., Ryan D., Servili M., Taticchi A., Esposto S., Robards K. Biosynthesis and biotransformations of phenol-conjugated oleosidic secoiridoids from Olea europaea L. Nat. Prod. Rep. 2008;25:1167–1179. doi: 10.1039/b719736e. [DOI] [PubMed] [Google Scholar]
- 21.Peralbo-Molina Á., Priego-Capote F., de Castro M.D.L. Tentative Identification of Phenolic Compounds in Olive Pomace Extracts Using Liquid Chromatography–Tandem Mass Spectrometry with a Quadrupole–Quadrupole-Time-of-Flight Mass Detector. J. Agric. Food Chem. 2012;60:11542–11550. doi: 10.1021/jf302896m. [DOI] [PubMed] [Google Scholar]
- 22.Kanakis P., Termentzi A., Michel T., Gikas E., Halabalaki M., Skaltsounis A.-L. From Olive Drupes to Olive Oil. An HPLC-Orbitrap-based Qualitative and Quantitative Exploration of Olive Key Metabolites. Planta Med. 2013;79:1576–1587. doi: 10.1055/s-0033-1350823. [DOI] [PubMed] [Google Scholar]
- 23.Sanz M., de Simón B.F., Cadahía E., Esteruelas E., Muñoz A.M., Hernández T., Estrella I., Pinto E. LC-DAD/ESI-MS/MS study of phenolic compounds in ash (Fraxinus excelsior L. and F. americana L.) heartwood. Effect of toasting intensity at cooperage. J. Mass Spectrom. 2012;47:905–918. doi: 10.1002/jms.3040. [DOI] [PubMed] [Google Scholar]
- 24.Zengin G., Mahomoodally M.F., Rocchetti G., Lucini L., Sieniawska E., Świątek Ł., Rajtar B., Polz-Dacewicz M., Senkardes I., Aktumsek A., et al. Chemical Characterization and Bioactive Properties of Different Extracts from Fibigia clypeata, an Unexplored Plant Food. Foods. 2020;9:705. doi: 10.3390/foods9060705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leila A., Lamjed B., Roudaina B., Najla T., Taamalli A., Jellouli S., Mokhtar Z. Isolation of an antiviral compound from Tunisian olive twig cultivars. Microb. Pathog. 2019;128:245–249. doi: 10.1016/j.micpath.2019.01.012. [DOI] [PubMed] [Google Scholar]
- 26.Zaïri A., Nouir S., Zarrouk A., Haddad H., Khélifa A., Achour L. Phytochemical profile, cytotoxic, antioxidant, and allelopathic potentials of aqueous leaf extracts of Olea europaea. Food Sci. Nutr. 2020;8:4805–4813. doi: 10.1002/fsn3.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ha J.Y., Goo S.Y., Sung J.S., Shin H.S. Antioxidant and cytoprotective activity of the olive leaf (Olea europaea L. var. Kalamata) extracts on the mouse embryonic fibroblast cell. Food Sci. Biotechnol. 2009;18:965–970. [Google Scholar]
- 28.Samet I., Han J., Jlaiel L., Sayadi S., Isoda H. Olive (Olea europaea) Leaf Extract Induces Apoptosis and Monocyte/Macrophage Differentiation in Human Chronic Myelogenous Leukemia K562 Cells: Insight into the Underlying Mechanism. Oxid. Med. Cell. Longev. 2014;2014:927619. doi: 10.1155/2014/927619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhouma H.E., Trabelsi N., Chimento A., Benincasa C., Tamaalli A., Perri E., Zarrouk M., Pezzi V. Olea europaea L. Flowers as a new promising anticancer natural product: Phenolic composition, antiproliferative activity and apoptosis induction. Nat. Prod. Res. 2019;35:1836–1839. doi: 10.1080/14786419.2019.1637867. [DOI] [PubMed] [Google Scholar]
- 30.Pennisi R., Ben Amor I., Gargouri B., Attia H., Zaabi R., Ben Chira A., Saoudi M., Piperno A., Trischitta P., Tamburello M.P., et al. Analysis of Antioxidant and Antiviral Effects of Olive (Olea europaea L.) Leaf Extracts and Pure Compound Using Cancer Cell Model. Biomolecules. 2023;13:238. doi: 10.3390/biom13020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Świątek Ł., Sieniawska E., Mahomoodally M.F., Sadeer N.B., Wojtanowski K.K., Rajtar B., Polz-Dacewicz M., Paksoy M.Y., Zengin G. Phytochemical Profile and Biological Activities of the Extracts from Two Oenanthe Species (O. aquatica and O. silaifolia) Pharmaceuticals. 2022;15:50. doi: 10.3390/ph15010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.