Abstract
Background: The outbreak of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) resulted in the global COVID-19 pandemic. The urgency for an effective SARS-CoV-2 vaccine has led to the development of the first series of vaccines at unprecedented speed. The discovery of SARS-CoV-2 spike-glycoprotein mutants, however, and consequentially the potential to escape vaccine-induced protection and increased infectivity, demonstrates the persisting importance of monitoring SARS-CoV-2 mutations to enable early detection and tracking of genomic variants of concern. Results: We developed the CoVigator tool with three components: (1) a knowledge base that collects new SARS-CoV-2 genomic data, processes it and stores its results; (2) a comprehensive variant calling pipeline; (3) an interactive dashboard highlighting the most relevant findings. The knowledge base routinely downloads and processes virus genome assemblies or raw sequencing data from the COVID-19 Data Portal (C19DP) and the European Nucleotide Archive (ENA), respectively. The results of variant calling are visualized through the dashboard in the form of tables and customizable graphs, making it a versatile tool for tracking SARS-CoV-2 variants. We put a special emphasis on the identification of intrahost mutations and make available to the community what is, to the best of our knowledge, the largest dataset on SARS-CoV-2 intrahost mutations. In the spirit of open data, all CoVigator results are available for download. The CoVigator dashboard is accessible via covigator.tron-mainz.de. Conclusions: With increasing demand worldwide in genome surveillance for tracking the spread of SARS-CoV-2, CoVigator will be a valuable resource of an up-to-date list of mutations, which can be incorporated into global efforts.
Keywords: SARS-CoV-2, dashboard, genomic variants, software, pipeline, virus genome assemblies, knowledge base, intrahost
1. Introduction
The identification, characterization and monitoring of the pathogen responsible for a novel emerging disease is crucial for the development of a timely public health response. This includes rapid and open sharing of data [1], which has been adapted in past outbreaks to advance research and improve medical support [2,3]. The outbreak of the respiratory disease COVID-19 caused by SARS-CoV-2 demonstrated the increasing value of high-throughput sequencing through enabling the publication of the complete virus genome within one month of sampling [4,5]. The identification of the SARS-CoV-2 spike protein as a valuable target for vaccine design [6,7] led to the development of vaccines at unprecedented speed [8,9] and is still fostering further developments.
Nevertheless, the discovery of SARS-CoV-2 spike-glycoprotein mutants, associated with the potential to escape vaccine-induced protection, demonstrates the importance of monitoring SARS-CoV-2 genomic sequences to enable early detection of these genomic variants of concern. In a first study, we analyzed 1,036,030 genomic assemblies from the Global Initiative on Sharing Avian Influenza Data (GISAID) [10,11,12] and 30,806 Next Generation Sequencing (NGS) datasets from the European Nucleotide Archive (ENA). We reported non-synonymous spike protein mutations and their frequencies and analyzed the effect on known T-cell epitopes [13]. Although we confirmed low mutation rates of the spike protein, we experienced an increase in the number of genomic variants over time. Therefore, we further developed our computational pipeline to tackle potential escape mutations [14].
There are multiple initiatives to monitor SARS-CoV-2 mutations based on the GISAID dataset: NextStrain [15], CoV-GLUE [16], CoV-Spectrum [17] and Coronapp [18]. A further initiative based on the ENA dataset is the Galaxy project COVID-19 [19]; other systems use regional data: CLIMB-COVID (COG-UK) [20] and CovRadar [21]. The COVID-19 Data Portal [22] is provided by EMBL-EBI and the European COVID-19 Data Platform to facilitate data sharing and accelerate research through making all the data available in the public domain and encouraging the research community to share SARS-CoV-2 data. Furthermore, there are some open-source pipelines to identify mutations on SARS-CoV-2 data, i.e., Cecret [23], nf-core viralrecon [24], ncov2019-artic-nf [25],ViralFlow [26], Havoc [27], NCBI SARS-CoV-2 Variant Calling (SC2VC) [28] and ASPICoV [29].
Each dataset has its own advantages. While genomic assemblies are easier to share and interpret, raw reads provide granular information about the mutations through access to the pileup of reads supporting each mutation, also allowing the characterization of intrahost mutations. Analyzing both datasets together may support the identification of potential false positives in the data and the confirmation of trends.
To enable monitoring of SARS-CoV-2 sequences from both sources, we have developed CoVigator, an NGS pipeline and dashboard that allows geographical and temporal navigation through SARS-CoV-2 genomic variants. We automatically download and analyze genomic assemblies from the COVID-19 Data Portal and raw reads from the European Nucleotide Archive (ENA). Furthermore, we screen the literature for studies on SARS-CoV-2 intrahost mutations [30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45] and propose a filtering strategy to obtain a high-quality set of intrahost mutations in the large and heterogeneous dataset obtained from ENA. Thus, the CoVigator platform supports the early detection of variants that can potentially serve as the basis for further research in the field of vaccines.
2. Materials and Methods
The CoVigator knowledge base is implemented in Python version 3.8 and the database for storing data is PostgreSQL (version 13.4).
The CoVigator pipeline (version 0.14.0) is implemented in the Nextflow framework version 19.10.0. All dependencies are managed within conda (version 4.9) environments [46] (see Table 1). The pipeline may receive as input either (1) a single-end FASTQ, (2) two paired-end FASTQs, (3) an assembly in FASTA format or (4) a VCF file with mutations. Adapter sequences are trimmed from FASTQs using fastp [47], alignment to the reference genome is performed with BWA mem2 [48], Base Quality Score Recalibration (BQSR) is performed with GATK [49], duplicate reads are marked with sambamba [50] and, finally, a horizontal and vertical coverage analysis is performed with samtools [51]. Variant calling on the BAM files derived from the FASTQs is performed with LoFreq [52], GATK [49], BCFtools [53] and iVar [54] (only the results from LoFreq are shown in the dashboard). Variant calling on the FASTA assemblies is performed with a custom script using Biopython’s Needleman–Wunsch global alignment [55]. Further processing of VCF files adds functional annotations with SnpEff [56], technical annotations with VAFator [14], ConsHMM conservation scores [57] and Pfam protein domains [58]. Pangolin [59] is employed to determine the lineage of every sample. The input for pangolin is either the input assembly in FASTA format or the consensus assembly derived from the clonal mutations (i.e., VAF ≥ 0.8) and the reference genome. See Table 1 for more details on the specific settings of each tool.
Table 1.
Tools employed in the pipeline, specific versions and settings.
| Tool | Purpose | Settings | References | Version | FASTQ | FASTA |
|---|---|---|---|---|---|---|
| fastp | Adapter trimming | [47] | 0.20.1 | X | ||
| BWA mem 2 | Alignment | Default | [48] | 2.2.1 | X | |
| GATK | Variant calling and alignments preprocessing | MQ ≥ 20, BQ ≥ 20, ploidy = 1 | [49] | 4.2.0.0 | X | |
| sambamba | Read deduplication | MQ ≥ 20, BQ ≥ 20, ploidy = 1 | [50] | 0.8.2 | X | |
| samtools | Coverage analysis | [51] | 1.12 | X | ||
| LoFreq | Variant calling | MQ ≥ 20, BQ ≥ 20 | [52] | 2.1.5 | X | |
| BCFtools | Variant calling, normalization and annotation | MQ ≥ 20, BQ ≥ 20 | [53] | 1.14 | X | X |
| iVar | Variant calling | MQ ≥ 20, BQ ≥ 20 | [54] | 1.3.1 | X | |
| Biopython | Custom variant calling on assemblies sequences based on Needleman-Wunsch global alignment | aligner.mode = ‘global’ aligner.match = 2 aligner.mismatch = −1 aligner.open_gap_score = −3 aligner.extend_gap_score = −0.1 aligner.target_end_gap_score = 0.0 aligner.query_end_gap_score = 0.0 |
[55] | 1.79 | X | |
| SnpEff | Functional annotations | [56] | 5.0 | X | X | |
| VAFator | Technical annotations | MQ > 0, BQ > 0 | [14] | 1.2.5 | X | |
| Pangolin | Lineage calling | [59] | 4.1.2 | X | X | |
| ConsHMM | Conservation annotations | [57] | Not available | X | X | |
| Pfam | SARS-CoV-2 protein domains | [58] | Not available | X | X |
The CoVigator dashboard is also implemented in Python using the visualization framework Dash (version 2.1.0). The computation is distributed through a high-performance computing cluster with a library that provides advanced parallelism, Dask (version 2022.9.2).
3. Results and Discussion
3.1. System Description
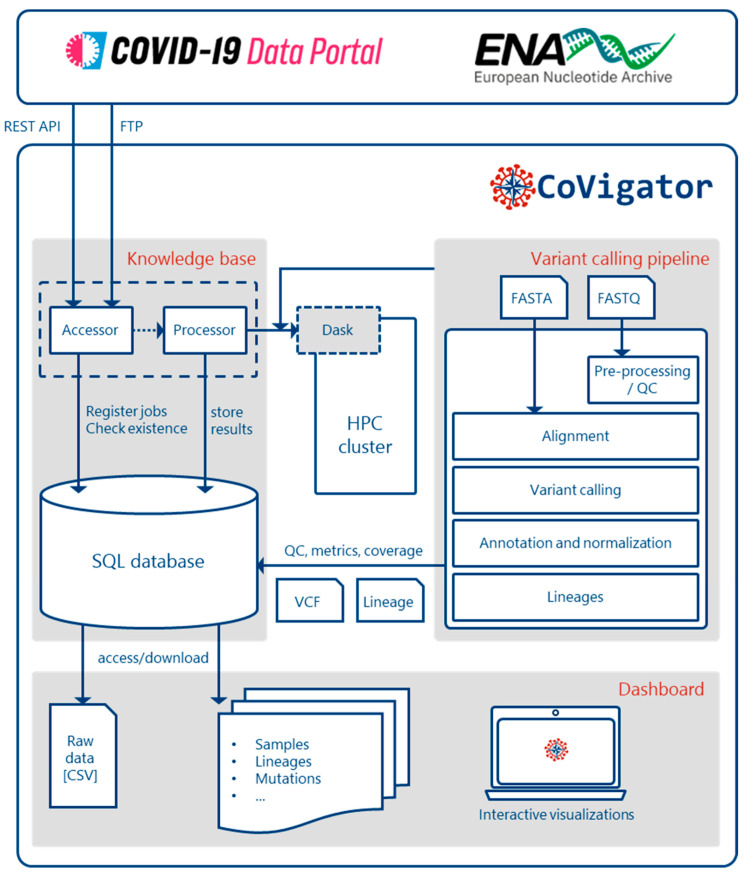
The CoVigator system (Figure 1) has three main components: (1) the knowledge base, (2) the analysis pipeline and (3) the dashboard. For every sample, the knowledge base orchestrates the metadata retrieval, raw data download and finally its analysis through the pipeline for the detection of mutations. Furthermore, it makes all necessary data available through a database (Postgre-SQL version 13). Finally, the dashboard presents the data to the end user through a set of interactive visualizations.
Figure 1.
CoVigator system components. The accessor reads external data and stores it in an SQL database. The processor reads the stored data and distributes the processing of every sample in an HPC cluster via Dask. The pipeline processes FASTA and FASTQ data and finally stores the results back in the database (See Figure S1 for a more detailed FASTA and FASTQ processing pipeline). The dashboard reads the results and displays them in a set of interactive plots. The results are also available in raw format.
CoVigator operates via interaction with external systems: a high-performance computing (HPC) cluster and the ENA and COVID-19 Data Portal Application Programming Interfaces (APIs). Samples between both original datasets (raw reads and genomic assemblies) may overlap. As recommended, some data providers might automatically upload both data formats. The results presented in the dashboard are stratified by dataset.
3.2. Knowledge Base
The CoVigator knowledge base collects data from both genomic assemblies and raw reads, orchestrates its processing through the variant calling pipeline and stores all the metadata, raw data and processed results in a relational database.
The data for both datatypes are fetched via the corresponding API hosted by the European Bioinformatics Institute [60]; the metadata are normalized, the FASTQ (raw NGS reads) and FASTA (genomic assemblies) files are downloaded and their MD5 checksums are confirmed to ensure data integrity.
Furthermore, the knowledge base iteratively builds a variant co-occurrence matrix (only for the raw read dataset) and precomputes analyses of the data (binned abundance of mutations, dN/dS ratios per gene and domain, top occurring variants, pairwise co-occurrence and counts of variants per lineage, country, sample, mutation type, length and nucleotide substitution) that ensure low-latency responses.
3.3. Analysis Pipeline
In general, the CoVigator pipeline processes FASTQ and FASTA files into annotated and normalized analysis-ready VCF files via two independent workflows (Figure 1 and Figure S1). We implemented the pipeline in the Nextflow framework [61] and managed all dependencies with Conda environments to enable seamless installation. We have embedded the SARS-CoV-2 reference genome ASM985889v3 [5]. Using a different reference, this pipeline could instantly analyze other virus sequences as well.
3.4. Dashboard
The dashboard is the user interface to CoVigator. There are two separate views for the raw reads and genomic assembly datasets. Each view provides a set of tabs that allows the user to explore different aspects of the data held in the database. Each tab provides some interactive visualizations, described below. When applicable, the tabs provide a set of filters on the left side. These have been excluded from the screenshots for the purpose of clarity.
The most relevant tabs are described below, and some notable findings are highlighted. The data shown here include 137,025 samples downloaded from ENA on 21 October 2022 and 6,165,681 samples downloaded from the COVID-19 Data Portal on 18 November 2022.
3.5. Samples
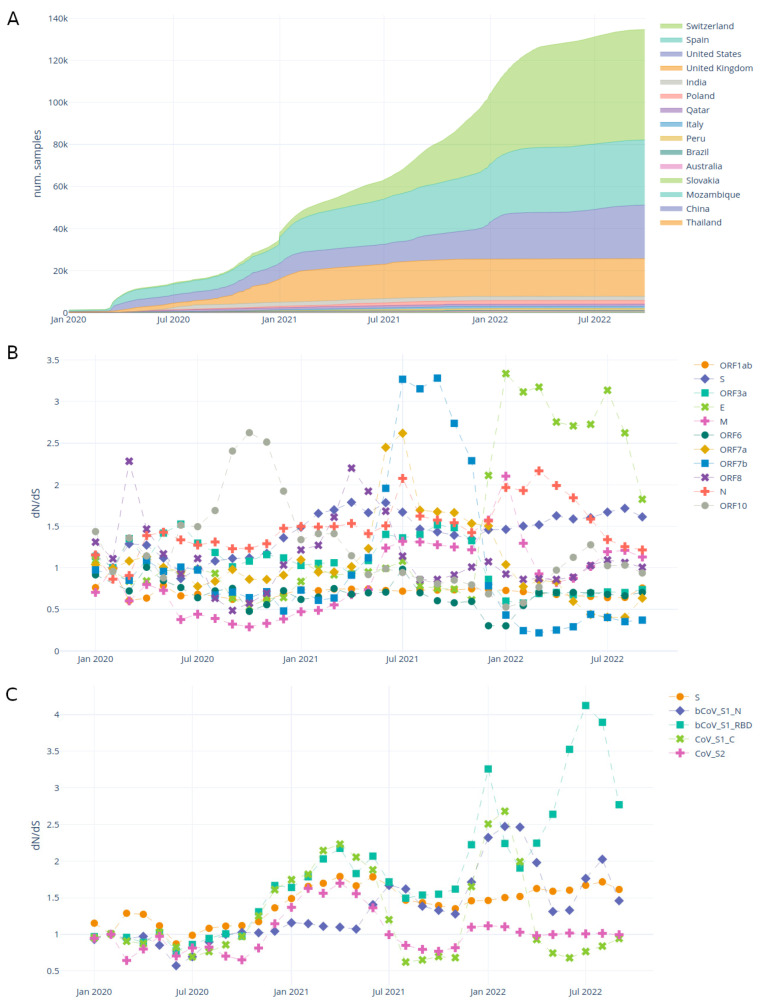
The samples tab (Figure 2) enables the user to explore the accumulation of samples through time and the evolution of the dN/dS ratio in different genomic regions.
Figure 2.
Samples by country tab plots for raw read dataset. (A) accumulation of samples through time by country; (B) dN/dS ratio through time on each SARS-CoV-2 protein; (C) dN/dS ratio through time in the domains of the spike protein. See Figure S2 for a screenshot including the filters.
Figure 2A shows the accumulation of samples in each country. The dashboard allows the user to select specific countries and/or lineages.
Figure 2B,C show the dynamics of the dN/dS ratio through time, over genes and protein domains. The dN/dS ratio aims to estimate the evolutionary pressure on SARS-CoV-2 proteins and domains. This metric, although originally developed for assessing diverging species, is an imperfect but simple estimation of the evolutionary pressure within the same species [62,63], in this case, SARS-CoV-2. There have been recent efforts to develop better alternatives for estimating the evolutionary pressure on SARS-CoV-2 [64]. The traditional interpretation of dN/dS is as follows: dN/dS < 0 indicates purifying selection, dN/dS = 1 indicates neutral evolution and dN/dS > 1 indicates positive selection.
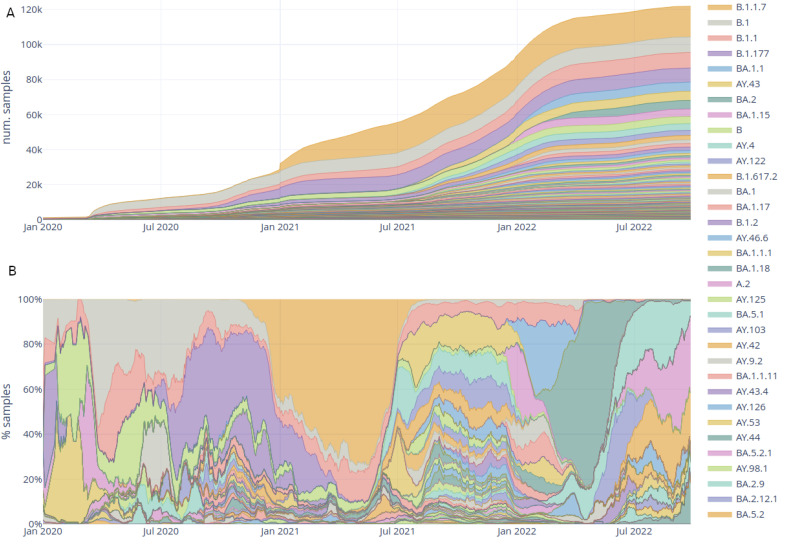
3.6. Lineages
The lineages tab enables the user to explore the different lineages through time and geography (Figure 3). Both the accumulation of samples in every lineage worldwide (Figure 3A) and the dominant lineage through time (Figure 3B) can be viewed. In the screenshot, the displacement of B.1 by Alpha (B.1.1.7), the subsequently displacement by multiple Delta lineages (AY.*) and finally displacement by the three Omicron lineages (BA.1, BA.2 and BA.3) can be seen.
Figure 3.
Interactive plots in the lineages tab for the raw read dataset. (A) Accumulation of samples in each lineage through time; (B) dominant lineages through time. See Figure S3 for a screenshot including the filters.
3.7. Mutation Statistics
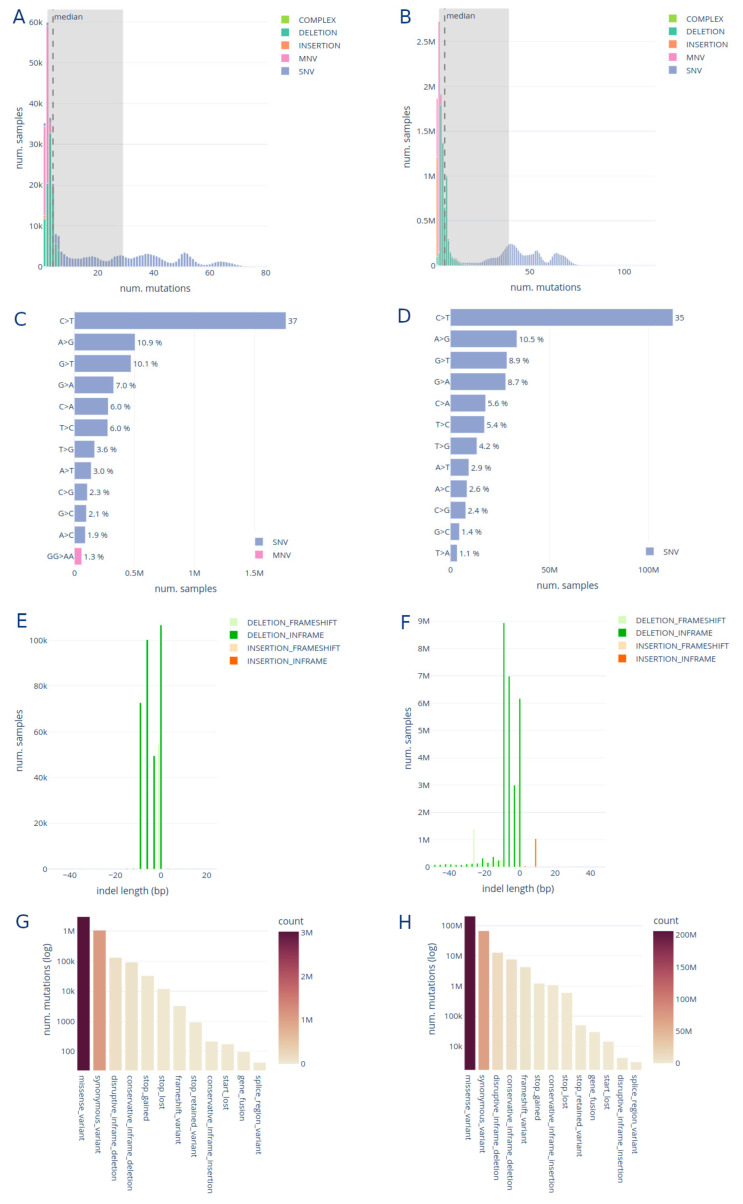
The mutation statistics tab provides insights into the variant calling results on the different datasets and genomic regions (Figure 4). Expected trends in the data can be confirmed in these visualizations.
Figure 4.
Interactive plots on the mutation statistics tab showing results for raw reads and genomic assembly datasets. (A) ENA distribution of the number of mutations per sample; (B) C19DP distribution of the number of mutations per sample; (C) ENA frequency of base substitutions, (D) C19DP frequency of base substitutions; (E) ENA indel length distribution; (F) C19DP indel length distribution; (G) ENA frequency of mutation effect on the protein; (H) C19DP frequency of mutation effect on the protein. See Figure S4 for a screenshot including the filters.
The median number of SNVs per sample in the raw read dataset is 32, with an interquartile range (IQR) of 30 (Figure 4A). Additionally, the median number of MNVs is two with an IQR of one. The number of deletions is lower (median: 3, IQR: 2) than the number of SNVs, and the number of insertions is even lower, with few samples having just one insertion. For the genomic assemblies, the numbers are slightly different, with median SNVs = 44 (IQR: 19), MNVs = 2 (IQR: 0), deletions = 4 (IQR: 3) and again just one insertion in a few samples (Figure 4B).
We observe that the base substitution C > T is by and large the most frequent, followed by G > T and A > G; the deletion TA > T and the MNV GG > AA is the most frequent in both datasets (Figure 4C,D).
In Figure 4E,F, we confirm that deletions are more frequent than insertions with an insertion-to-deletion ratio of 0.002 and 0.032 for raw reads and genomic assemblies, respectively. We also confirm two previous findings: (1) shorter deletions and insertions are more common than longer ones [65,66] and (2) the deletions and insertions not causing a frameshift are overrepresented as their impact in the resulting protein are more subtle [67]. In the genomic assemblies, we observe a long tail of deletions longer than 8 bp, which is not observed in the raw read results. We suspect this is a technical artefact introduced via our variant calling method. Finally, as shown in Figure 4G,H we observe that the most frequent mutation effect is a missense variant, followed by a synonymous variant. This is coherent between both datasets.
3.8. Recurrent Mutations
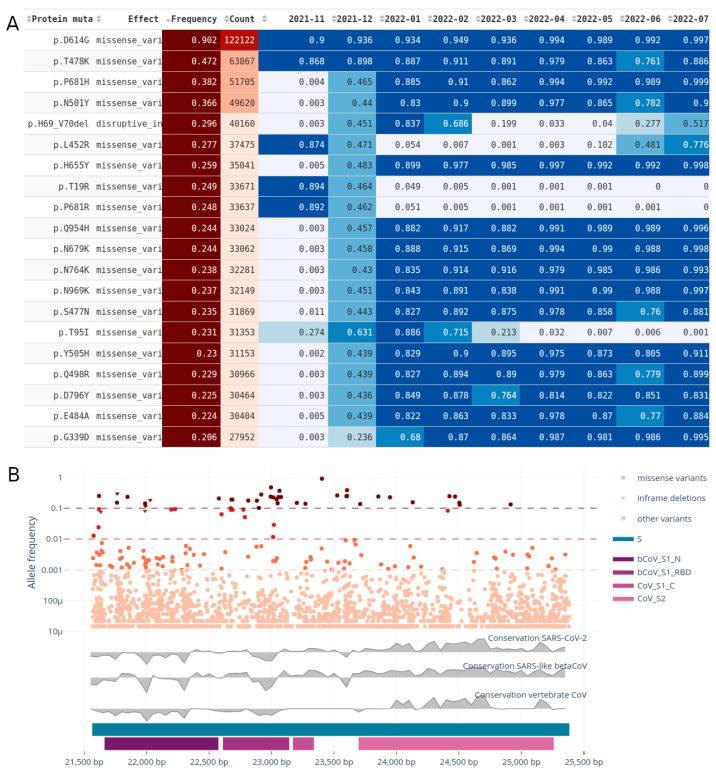
The recurrent mutations tab allows the user to explore the most recurrent mutations by the total count of observations through time within their genomic context (Figure 5). In Figure 5A, the top recurrent mutations and their frequency and counts through time are shown. The size of the table can be parametrized for up to 100 mutations. For instance, the user can explore the most recurrent mutations in the whole genome, a given gene or a given protein domain. Furthermore, the period in which the monthly counts are shown can be parameterized. The gene viewer (Figure 5B) has multiple tracks: (i) a scatter plot with the relevant mutations and their frequencies in the virus population, (ii) ConsHMM conservation tracks and (iii) gene and Pfam protein domains. The table in Figure S5 provides the decline and rise of the Alpha and Delta lineages, respectively, in the counts of mutations between April and July 2021.
Figure 5.
Gene view for the spike protein on the raw read dataset. (A) Table of the top 20 recurrent mutations with the frequency segregated by month between November 2021 and July 2022; (B) gene view showing mutations (synonymous and unique mutations excluded) in the spike protein and their frequencies in the virus population, the ConsHMM conservation tracks in grey and the Pfam protein domains in tones of purple. See Figure S4 for a screenshot including the filters.
Additionally, the mutation statistics tab provides a co-occurrence analysis that points to clusters of co-occurring mutations and their correspondence with virus lineages; or in the case of mutations shared between lineages, these clusters may contain a mixture of different but related lineages. Due to performance limitations, this analysis is only available in the raw read dataset and at the gene level. In Figure S6, we show the Jaccard index co-occurrence matrix in the spike protein and its clustering results annotated with SARS-CoV-2 lineages in Table S1.
3.9. Clonal and Intrahost Mutations in the Raw Read Dataset
The FASTQ files provide the pile-up of reads across the genome, and this gives detailed information into the called variants. In particular, we can count the number of reads supporting each variant, and this allows us to identify subclonal variants supported using only a fraction of the reads. These variants likely emerged within the host and are referred to as intrahost variants. The identification of intrahost variants is not possible on the genomic assemblies.
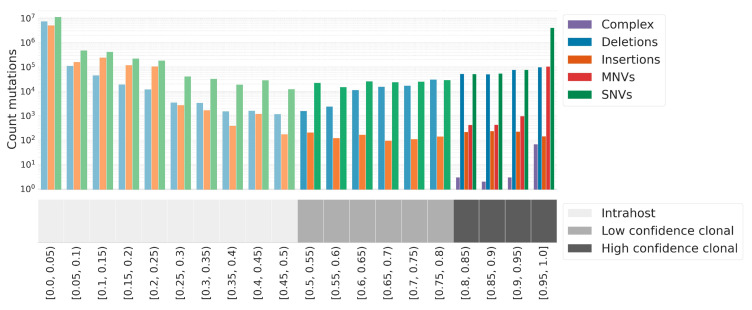
We consider high-quality clonal mutations as those with a VAF greater than or equal to 80%, and those with a VAF greater than or equal to 50% and lower than 80% as low-confidence clonal mutations. Only high-confidence clonal mutations are used to determine a consensus sequence and assign a SARS-CoV-2 lineage (Figure 6).
Figure 6.
Distribution of VAF across all mutation calls (4,665,192 with VAF ≥ 0.8; 222,297 with VAF ≥ 0.5 and < 0.8; 26,231,409 with VAF < 0.5) in 135,347 samples. High-confidence clonal mutations overlapping the same amino acid are merged into MNVs or complex variants. See Figure S7 for a screenshot of intrahost mutations tab including the filters.
The remaining dataset of mutations poses a different technical challenge due to the difficulty of separating true low VAF mutations from noise. We first determine those mutations with a VAF below 50% as raw candidate intrahost mutations.
We observed a large number of low-frequency mutations among SARS-CoV-2 genomes. In order to establish a high-quality set of intrahost mutations for studying viral evolution, we screened and compared the literature on SARS-CoV-2 intrahost mutations for different filtering approaches and implemented a conservative approach (Table 2).
Table 2.
Published and implemented filtering approaches for intrahost variants.
| Approach | Sample Filters | Variant Filters |
|---|---|---|
| Valesano-like [44] | ≥50,000 mapped reads ≥29,000 bp horizontal coverage |
VAF ≥ 2%, VAF < 50% DP ≥ 100 ≥10 supporting reads |
| Sapoval-like [39] | ≥20,000 mapped reads | VAF ≥ 2%, VAF < 50% DP ≥ 10 Mask extremes of genome + homoplasmic positions [68] |
| Tonkin-Hill-like [38] | Excessive number iSNVs (99.9th percentile) Outlier number of iSNVs with mid-VAFs, between 40% and 80% |
VAF ≥ 5%, VAF < 50% DP ≥ 100 ≥5 supporting reads |
| CoVigator approach | ≥50,000 mapped reads ≥29,000 bp horizontal coverage Excessive number iSNVs (99.9th percentile) Outlier number of iSNVs with mid-VAFs, between 40% and 80% |
VAF ≥ 2%, VAF < 50% DP ≥ 100 ≥10 supporting reads Mask extremes of genome + homoplasmic positions [68] from indels ≤ 10 bp |
4. Conclusions
The persistently increasing amount of publicly available SARS-CoV-2 sequencing data calls for robust platforms that allow constant monitoring of genomic SARS-CoV-2 variants in heterogeneous data sets. Our CoVigator pipeline covers the essential steps of preparing the data and calling variants from SARS-CoV-2 raw sequencing data from ENA and genome assemblies from the COVID-19 Data Portal. The pipeline is integrated within the CoVigator knowledge base that orchestrates the download, processing and storage of the underlying samples and results. The CoVigator dashboard provides different visualizations and features for selecting clonal variants across all genes from the SARS-CoV-2 genome in a selected period. The dashboard also provides a comprehensive analysis of intrahost variants observed across detected mutations in the raw read dataset. To this end, we propose a conservative filtering approach based on filtering samples and mutations. The dataset of intrahost mutations derived from public data that we make available through CoVigator is, to the best of our knowledge, the largest published dataset of SARS-CoV-2 intrahost mutations. The main strength of CoVigator is the combination of a software pipeline with a dashboard, which ensures both processing of the data and its interpretation. Uniquely, CoVigator processes genome assemblies and raw sequencing data types, making it open-data-friendly and allowing it to be adopted to other SARS-CoV-2 data sources. A brief comparison of the important features of CoVigator with other pipelines is tabulated in Table S2.
The identification of mutations over such heterogeneous datasets obtained with different sequencing protocols is challenging. With CoVigator, we observed VAF dilution on mutations identified via targeted amplicon sequencing with overlapping primers, genome edge effects and read edge effects. We aim to address these challenges in the future, e.g., through inferring the primers used in an arbitrary sample. Additionally, we implemented a simplistic phasing of clonal mutations occurring in the same amino acid to ensure their correct annotation. However, we identified the need for a phasing method for low-VAF mutations that existing germline phasing tools do not cover. CoVigator is currently limited to processing Illumina sequencing data, while the majority of SARS-CoV-2 sequencing projects (i.e., Artic network) and pipelines use Oxford Nanopore sequencing. SARS-CoV-2 Nanopore data processing will be implemented in subsequent releases of CoVigator.
Future versions of CoVigator can be broadened to other use cases, such as other infectious organisms or co-existing infections during the pandemic (see supplementary methods for further details [69,70]). Additionally, we envision the annotation of all possible mutations before their observation to potentially improve preparation for future variants of concern.
Acknowledgments
We thank Franziska Lang, Özlem Muslu, Jonas Ibn-Salem, Jos de Graaf and Rudolf Koopmann for critical discussions. We thank Karen Chu and Paul Kerbs for reviewing, editing and proofreading this article. We gratefully acknowledge the authors from the originating laboratories responsible for obtaining the specimens, as well as the submitting laboratories where the sequence data were generated and shared via the European Nucleotide Archive and the COVID-19 Data Portal.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v15061391/s1, Figure S1. Workflow of CoVigator pipeline; Figure S2. screenshot of the lineages tab in the ENA dataset; Figure S3. screenshot of mutation statistics tab in the ENA dataset; Figure S4. recurrent mutations tab for the spike protein in the ENA dataset; Figure S5. top 30 mutations in the spike protein from COVID-19 Data Portal; Figure S6. screenshot of intrahost mutations tab; Figure S7. Jaccard index co-occurrence matrix on the spike protein; Table S1. co-occurrence clusters on the spike protein with its matching lineage information; Table S2. Comparison of SARS-CoV-2 data processing pipelines with CoVigator. Supplementary methods for co-occurrence analysis and for CoVigator extension to other viruses.
Author Contributions
U.S., M.L., B.S. and T.B. were involved in conceptualization. T.B., P.R.-F., P.S. and J.H. participated in implementation of the dashboard, developing the pipeline and hosting the webserver. P.R.-F. and P.S. were involved in developing documentation and releasing the pipeline. T.B., P.R.-F., P.S., R.G., T.R. and J.H. were involved in design of the dashboard and pipeline. R.G. and P.R.-F. performed additional analysis to show the performance of the pipeline. R.G., P.R.-F. and T.B. prepared the original draft of the manuscript. B.S., U.S. and M.L. were involved in writing, critical review and editing the final draft of the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The CoVigator dashboard is accessible via covigator.tron-mainz.de and can be installed via https://github.com/TRON-bioinformatics/covigator. A standalone version of the CoVigator pipeline with nextflow is available at https://github.com/TRON-Bioinformatics/covigator-ngs-pipeline. CoVigator documentation is available at https://covigator.readthedocs.io. All links accessed on 16 June 2023.
Conflicts of Interest
Author U.S. is co-founder, shareholder and CEO at BioNTech SE. The remaining authors declare no conflict of interest.
Funding Statement
BioNTech SE: Mainz, Germany, supports the study. The funder provided support in the form of a salary for author U.S., but did not have any additional role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. The specific roles of this author are articulated in the ‘Author Contributions’ section. In addition, the other authors are employees of the non-profit company TRON gGmbH and are supported in the form of salaries. TRON gGmbH did not have any additional role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. Intel is committed to accelerating access to technology that can combat the current pandemic and enabling scientific discovery that better prepares our world for future crises. Funding for this solution was funded in part by Intel’s Pandemic Response Technology Initiative. For more information about healthcare solutions from Intel, visit intel.com/healthcare. For more information about Intel’s COVID-19 response, visit https://www.intel.com/content/www/us/en/corporate-responsibility/covid-19-response.html, accessed on 16 June 2023.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Moorthy V.S., Karam G., Vannice K.S., Kieny M.-P. Rationale for WHO’s new position calling for prompt reporting and public disclosure of interventional clinical trial results. PLoS Med. 2015;12:e1001819. doi: 10.1371/journal.pmed.1001819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Ventura C.V., Maia M., Bravo-Filho V., Góis A.L., Belfort R. Zika virus in Brazil and macular atrophy in a child with microcephaly. Lancet. 2016;387:228. doi: 10.1016/S0140-6736(16)00006-4. [DOI] [PubMed] [Google Scholar]
- 4.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shang W., Yang Y., Rao Y., Rao X. The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines. NPJ Vaccines. 2020;5:18. doi: 10.1038/s41541-020-0170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tai W., He L., Zhang X., Pu J., Voronin D., Jiang S., Zhou Y., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: Implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khare S., Gurry C., Freitas L., Schultz M.B., Bach G., Diallo A., Akite N., Ho J., Lee R.T., Yeo W., et al. GISAID’s Role in Pandemic Response. China CDC Wkly. 2021;3:1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data—From vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrörs B., Riesgo-Ferreiro P., Sorn P., Gudimella R., Bukur T., Rösler T., Löwer M., Sahin U. Large-scale analysis of SARS-CoV-2 spike-glycoprotein mutants demonstrates the need for continuous screening of virus isolates. PLoS ONE. 2021;16:e0249254. doi: 10.1371/journal.pone.0249254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riesgo-Ferreiro P. VAFator. [(accessed on 16 June 2023)]. Available online: https://github.com/TRON-Bioinformatics/vafator.git.
- 15.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singer J., Gifford R., Cotten M., Robertson D. CoV-GLUE: A Web Application for Tracking SARS-CoV-2 Genomic Variation. Preprints.org. 2020 doi: 10.20944/preprints202006.0225.v1. [DOI] [Google Scholar]
- 17.Chen C., Nadeau S., Yared M., Voinov P., Xie N., Roemer C., Stadler T. CoV-Spectrum: Analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics. 2022;38:1735–1737. doi: 10.1093/bioinformatics/btab856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mercatelli D., Triboli L., Fornasari E., Ray F., Giorgi F.M. Coronapp: A web application to annotate and monitor SARS-CoV-2 mutations. J. Med. Virol. 2021;93:3238–3245. doi: 10.1002/jmv.26678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maier W., Bray S., van den Beek M., Bouvier D., Coraor N., Miladi M., Singh B., de Argila J.R., Baker D., Roach N., et al. Ready-to-use public infrastructure for global SARS-CoV-2 monitoring. Nat. Biotechnol. 2021;39:1178–1179. doi: 10.1038/s41587-021-01069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholls S.M., Poplawski R., Bull M.J., Underwood A., Chapman M., Abu-Dahab K., Taylor B., Colquhoun R.M., Rowe W.P.M., Jackson B., et al. CLIMB-COVID: Continuous integration supporting decentralised sequencing for SARS-CoV-2 genomic surveillance. Genome Biol. 2021;22:196. doi: 10.1186/s13059-021-02395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wittig A., Miranda F., Hölzer M., Altenburg T., Bartoszewicz J.M., Beyvers S., Dieckmann M.A., Genske U., Giese S.H., Nowicka M., et al. CovRadar: Continuously tracking and filtering SARS-CoV-2 mutations for genomic surveillance. Bioinformatics. 2022;38:4223–4225. doi: 10.1093/bioinformatics/btac411. [DOI] [PubMed] [Google Scholar]
- 22.Harrison P.W., Lopez R., Rahman N., Allen S.G., Aslam R., Buso N., Cummins C., Fathy Y., Felix E., Glont M., et al. The COVID-19 Data Portal: Accelerating SARS-CoV-2 and COVID-19 research through rapid open access data sharing. Nucleic Acids Res. 2021;49:W619–W623. doi: 10.1093/nar/gkab417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cecret. [(accessed on 13 October 2021)]. Available online: https://github.com/UPHL-BioNGS/Cecret.
- 24.Harshil P., Sarai V., Sara M., Jose E.-C., Michael L.H., Gisela G., nf-core bot. Phil E., Miguel J., Stephen K., et al. Nf-Core/Viralrecon. Zenodo. 2021 doi: 10.5281/zenodo.5146252. [DOI] [Google Scholar]
- 25.Connor-Lab Ncov2019-Artic-Nf.: GitHub. 2022. [(accessed on 15 June 2023)]. Available online: https://github.com/connor-lab/ncov2019-artic-nf.
- 26.Dezordi F.Z., Neto A.M.d.S., Campos T.d.L., Jeronimo P.M.C., Aksenen C.F., Almeida S.P., Wallau G.L., Fiocruz COVID-19 Genomic Surveillance Network ViralFlow: A Versatile Automated Workflow for SARS-CoV-2 Genome Assembly, Lineage Assignment, Mutations and Intrahost Variant Detection. Viruses. 2022;14:217. doi: 10.3390/v14020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Truong Nguyen P.T., Plyusnin I., Sironen T., Vapalahti O., Kant R., Smura T. HAVoC, a bioinformatic pipeline for reference-based consensus assembly and lineage assignment for SARS-CoV-2 sequences. BMC Bioinform. 2021;22:373. doi: 10.1186/s12859-021-04294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.NCBI SARS-CoV-2 Variant Calling (SC2VC) Pipeline. [(accessed on 12 June 2023)]. Available online: https://github.com/ncbi/sars2variantcalling.
- 29.Tilloy V., Cuzin P., Leroi L., Guérin E., Durand P., Alain S. ASPICov: An automated pipeline for identification of SARS-Cov2 nucleotidic variants. PLoS ONE. 2022;17:e0262953. doi: 10.1371/journal.pone.0262953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Al Khatib H.A., Benslimane F.M., Elbashir I.E., Coyle P.V., Al Maslamani M.A., Al-Khal A., Al Thani A.A., Yassine H.M. Within-Host Diversity of SARS-CoV-2 in COVID-19 Patients With Variable Disease Severities. Front. Cell. Infect. Microbiol. 2020;10:575613. doi: 10.3389/fcimb.2020.575613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armero A., Berthet N., Avarre J.-C. Intra-Host Diversity of SARS-Cov-2 Should Not Be Neglected: Case of the State of Victoria, Australia. Viruses. 2021;13:133. doi: 10.3390/v13010133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karamitros T., Papadopoulou G., Bousali M., Mexias A., Tsiodras S., Mentis A. SARS-CoV-2 exhibits intra-host genomic plasticity and low-frequency polymorphic quasispecies. J. Clin. Virol. 2020;131:104585. doi: 10.1016/j.jcv.2020.104585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lythgoe K.A., Hall M., Ferretti L., de Cesare M., MacIntyre-Cockett G., Trebes A., Andersson M., Otecko N., Wise E.L., Moore N., et al. SARS-CoV-2 within-host diversity and transmission. Science. 2021;372:eabg0821. doi: 10.1126/science.abg0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moreno G., Katarina M.B., Peter J.H., Trent M.P., Kasen K.R., Amelia K.H., Joseph L., Kelsey R.F., Yoshihiro K., Thomas C.F., et al. Limited SARS-CoV-2 diversity within hosts and following passage in cell culture. bioRxiv. 2020 doi: 10.1101/2020.04.20.051011. [DOI] [Google Scholar]
- 35.Popa A., Genger J.-W., Nicholson M.D., Penz T., Schmid D., Aberle S.W., Agerer B., Lercher A., Endler L., Colaço H., et al. Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2. Sci. Transl. Med. 2020;12:eabe2555. doi: 10.1126/scitranslmed.abe2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose R., Nolan D.J., Moot S., Feehan A., Cross S., Garcia-Diaz J., Lamers S.L. Intra-Host Site-Specific Polymorphisms of SARS-CoV-2 Is Consistent across Multiple Samples and Methodologies. MedRxiv. 2020 doi: 10.1101/2020.04.24.20078691. [DOI] [Google Scholar]
- 37.Siqueira J.D., Goes L.R., Alves B.M., de Carvalho P.S., Cicala C., Arthos J., Viola J.P.B., de Melo A.C., Soares M.A. SARS-CoV-2 genomic and quasispecies analyses in cancer patients reveal relaxed intrahost virus evolution. bioRxiv. 2020 doi: 10.1101/2020.08.26.267831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tonkin-Hill G., Martincorena I., Amato R., Lawson A.R.J., Gerstung M., Johnston I., Jackson D.K., Park N.R., Lensing S.V., Quail M.A., et al. Patterns of within-host genetic diversity in SARS-CoV-2. Elife. 2020;10:e66857. doi: 10.7554/eLife.66857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sapoval N., Mahmoud M., Jochum M.D., Liu Y., Elworth R.A.L., Wang Q., Albin D., Ogilvie H.A., Lee M.D., Villapol S., et al. SARS-CoV-2 genomic diversity and the implications for qRT-PCR diagnostics and transmission. Genome Res. 2021;31:635–644. doi: 10.1101/gr.268961.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Z.-Y., Liu H., Zhang Y.-D., Wu Y.-Q., Peng M.-S., Li A., Irwin D.M., Li H., Lu J., Bao Y., et al. Worldwide tracing of mutations and the evolutionary dynamics of SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.08.07.242263. [DOI] [Google Scholar]
- 41.James S.E., Ngcapu S., Kanzi A.M., Tegally H., Fonseca V., Giandhari J., Wilkinson E., Chimukangara B., Pillay S., Singh L., et al. High Resolution analysis of Transmission Dynamics of Sars-Cov-2 in Two Major Hospital Outbreaks in South Africa Leveraging Intrahost Diversity. MedRxiv. 2020 doi: 10.1101/2020.11.15.20231993. [DOI] [Google Scholar]
- 42.Sashittal P., Luo Y., Peng J., El-Kebir M. Characterization of SARS-CoV-2 viral diversity within and across hosts. bioRxiv. 2020 doi: 10.1101/2020.05.07.083410. [DOI] [Google Scholar]
- 43.Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., Zhou Z., Yang J., Zhong J., Yang D., et al. Genomic Diversity of Severe Acute Respiratory Syndrome-Coronavirus 2 in Patients With Coronavirus Disease 2019. Clin. Infect. Dis. 2020;71:713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valesano A.L., Rumfelt K.E., Dimcheff D.E., Blair C.N., Fitzsimmons W.J., Petrie J.G., Martin E.T., Lauring A.S. Temporal dynamics of SARS-CoV-2 mutation accumulation within and across infected hosts. PLoS Pathog. 2021;17:e1009499. doi: 10.1371/journal.ppat.1009499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Wang D., Zhang L., Sun W., Zhang Z., Chen W., Zhu A., Huang Y., Xiao F., Yao J., et al. Intra-host variation and evolutionary dynamics of SARS-CoV-2 populations in COVID-19 patients. Genome Med. 2021;13:30. doi: 10.1186/s13073-021-00847-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conda Anaconda Software Distribution. [(accessed on 7 February 2022)]. Available online: https://docs.conda.io/
- 47.Chen S., Zhou Y., Chen Y., Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vasimuddin M., Misra S., Li H., Aluru S. Efficient Architecture-Aware Acceleration of BWA-MEM for Multicore Systems; Proceedings of the 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS); Rio de Janeiro, Brazil. 20–24 May 2019; pp. 314–324. [Google Scholar]
- 49.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tarasov A., Vilella A.J., Cuppen E., Nijman I.J., Prins P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilm A., Aw P.P.K., Bertrand D., Yeo G.H.T., Ong S.H., Wong C.H., Khor C.C., Petric R., Hibberd M.L., Nagarajan N. LoFreq: A sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danecek P., McCarthy S.A. BCFtools/csq: Haplotype-aware variant consequences. Bioinformatics. 2017;33:2037–2039. doi: 10.1093/bioinformatics/btx100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grubaugh N.D., Gangavarapu K., Quick J., Matteson N.L., de Jesus J.G., Main B.J., Tan A.L., Paul L.M., Brackney D.E., Grewal S., et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20:8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cingolani P., Platts A., Le Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwon S.B., Ernst J. Single-nucleotide conservation state annotation of the SARS-CoV-2 genome. Commun. Biol. 2021;4:698. doi: 10.1038/s42003-021-02231-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ensembl Annotations SARS-CoV-2. [(accessed on 7 May 2021)]. Available online: ftp://ftp.ensemblgenomes.org/pub/viruses/json/sars_cov_2/sars_cov_2.json.
- 59.O’Toole Á., Scher E., Underwood A., Jackson B., Hill V., McCrone J.T., Colquhoun R., Ruis C., Abu-Dahab K., Taylor B., et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7:veab064. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D., et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Di Tommaso P., Chatzou M., Floden E.W., Barja P.P., Palumbo E., Notredame C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017;35:316–319. doi: 10.1038/nbt.3820. [DOI] [PubMed] [Google Scholar]
- 62.Kryazhimskiy S., Plotkin J.B. The population genetics of dN/dS. PLoS Genet. 2008;4:e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spielman S.J., Wilke C.O. The relationship between dN/dS and scaled selection coefficients. Mol. Biol. Evol. 2015;32:1097–1108. doi: 10.1093/molbev/msv003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kistler K., Huddleston J., Bedford T. Rapid and parallel adaptive mutations in spike S1 drive clade success in SARS-CoV-2. bioRxiv. 2022 doi: 10.2139/ssrn.3932602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rogozin I.B., Saura A., Bykova A., Brover V., Yurchenko V. Deletions across the SARS-CoV-2 Genome: Molecular Mechanisms and Putative Functional Consequences of Deletions in Accessory Genes. Microorganisms. 2023;11:229. doi: 10.3390/microorganisms11010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garushyants S.K., Rogozin I.B., Koonin E.V. Insertions in SARS-CoV-2 genome caused by template switch and duplications give rise to new variants that merit monitoring. bioRxiv. 2021 doi: 10.1101/2021.04.23.441209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Montgomery S.B., Goode D.L., Kvikstad E., Albers C.A., Zhang Z.D., Mu X.J., Ananda G., Howie B., Karczewski K.J., Smith K.S., et al. The origin, evolution, and functional impact of short insertion-deletion variants identified in 179 human genomes. Genome Res. 2013;23:749–761. doi: 10.1101/gr.148718.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Maio N., Walker C., Borges R., Weilguny L., Slodkowicz G., Goldman N. Issues with SARS-CoV-2 Sequencing Data. [(accessed on 8 April 2021)]. Available online: https://virological.org/t/issues-with-sars-cov-2-sequencing-data/473.
- 69.Ankerst M., Breunig M.M., Kriegel H.-P., Sander J. Optics. SIGMOD Rec. 1999;28:49–60. doi: 10.1145/304181.304187. [DOI] [Google Scholar]
- 70.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The CoVigator dashboard is accessible via covigator.tron-mainz.de and can be installed via https://github.com/TRON-bioinformatics/covigator. A standalone version of the CoVigator pipeline with nextflow is available at https://github.com/TRON-Bioinformatics/covigator-ngs-pipeline. CoVigator documentation is available at https://covigator.readthedocs.io. All links accessed on 16 June 2023.