ABSTRACT
Acetyl-CoA synthetase (ACS) and acetate ligase (ACD) are widespread among microorganisms, including archaea, and play an important role in their carbon metabolism, although only a few of these enzymes have been characterized. Anaerobic methanotrophs (ANMEs) have been reported to convert methane anaerobically into CO2, polyhydroxyalkanoate, and acetate. Furthermore, it has been suggested that they might be able to use acetate for anabolism or aceticlastic methanogenesis. To better understand the potential acetate metabolism of ANMEs, we characterized an ACS from ANME-2a as well as an ACS and an ACD from ANME-2d. The conversion of acetate into acetyl-CoA (Vmax of 8.4 μmol mg−1 min−1 and Km of 0.7 mM acetate) by the monomeric 73.8-kDa ACS enzyme from ANME-2a was more favorable than the formation of acetate from acetyl-CoA (Vmax of 0.4 μmol mg−1 min−1 and Km of 0.2 mM acetyl-CoA). The monomeric 73.4-kDa ACS enzyme from ANME-2d had similar Vmax values for both directions (Vmax,acetate of 0.9 μmol mg−1 min−1 versus Vmax,acetyl-CoA of 0.3 μmol mg−1 min−1). The heterotetrameric ACD enzyme from ANME-2d was active solely in the acetate-producing direction. Batch incubations of an enrichment culture dominated by ANME-2d fed with 13C2-labeled acetate produced 3 μmol of [13C]methane in 7 days, suggesting that this anaerobic methanotroph might have the potential to reverse its metabolism and perform aceticlastic methanogenesis using ACS to activate acetate albeit at low rates (2 nmol g [dry weight]−1 min−1). Together, these results show that ANMEs may have the potential to use acetate for assimilation as well as to use part of the surplus acetate for methane production.
IMPORTANCE Acetyl-CoA plays a key role in carbon metabolism and is found at the junction of many anabolic and catabolic reactions. This work describes the biochemical properties of ACS and ACD enzymes from ANME-2 archaea. This adds to our knowledge of archaeal ACS and ACD enzymes, only a few of which have been characterized to date. Furthermore, we validated the in situ activity of ACS in ANME-2d, showing the conversion of acetate into methane by an enrichment culture dominated by ANME-2d.
KEYWORDS: acetate metabolism, acetyl-CoA synthetase, acetate ligase, ANME, aceticlastic methanogenesis
INTRODUCTION
Methane is an abundant C1 compound and plays an important role in global warming (1). Two types of microorganisms contribute to the methane balance on earth: methanogens, producing methane anaerobically, and methanotrophs, consuming this methane with or without oxygen. Anaerobic methanotrophic (ANME) archaea are key drivers of the anaerobic oxidation of methane in the environment, but a mechanistic understanding of their metabolic potential is still largely missing. ANME archaea can be divided into the subgroups ANME-1, -2abcd, and -3 (2). These microorganisms convert methane into carbon dioxide using several electron acceptors such as sulfate, nitrate, and metal oxides (3–8). Other metabolic products formed by anaerobic methanotrophs (ANMEs) have not been investigated in detail so far. An analysis of the proteome of a “Candidatus Methanoperedens” ANME-2d culture pointed toward pyruvate, acetate, and formate as potential intermediates or products (9). Physiological experiments indeed confirmed the production of acetate from methane via polyhydroxyalkanoate (PHA) by a “Ca. Methanoperedens” enrichment culture (10, 11). This observation raised the question of whether ANMEs could also work in the reverse direction and, like aceticlastic methanogens, convert acetate into methane. Anaerobic methanotrophs have many physiological properties in common with methanogenic archaea, but their metabolisms differ in their directionality (2). It is already possible to reverse the methanogenesis pathway of the aceticlastic methanogen Methanosarcina acetivorans by introducing a methyl-coenzyme M reductase (MCR) gene from ANME-1 into the genome, thereby enabling M. acetivorans to convert methane via the reverse methanogenesis pathway using iron oxides as electron acceptors (12). According to genome analyses, ANMEs cannot conduct hydrogenotrophic or methylotrophic methanogenesis as ANME genomes lack hydrogenases and methyltransferases (13), which are essential for these pathways (14, 15). However, ANME-2a and -d harbor crucial genes for acetyl-coenzyme A synthesis, similar to those of the aceticlastic methanogens Methanothrix and Methanosarcina (16): acetyl-CoA synthetase (ACS) and acetate ligase (ACD). ACS-encoding genes are widely distributed over all three domains of life (17–19) and have been reported to catalyze the reversible conversion of acetate into acetyl-CoA. However, in Methanothrix, ACS is predominantly an acetate-activating enzyme and converts acetate, ATP, and coenzyme A into acetyl-CoA, AMP, and pyrophosphate (20). In contrast to Methanothrix sp., the aceticlastic methanogen Methanosarcina sp. uses a two-step process to activate acetate, converting acetate into acetyl-phosphate using acetate kinase (ACK), and subsequently, acetyl-phosphate is converted into acetyl-CoA using phosphotransacetylase (PTA) (21). These two enzymes used for acetate activation typically have different acetate affinities: whereas Methanothrix sp. uses ACS with a high substrate affinity, Methanosarcina uses ACK and PTA, which is coupled with a much lower affinity. This is consistent with the presence of Methanothrix in environments with low acetate concentrations (22). Acetyl-CoA plays a key role in carbon metabolism and is found at the junction of many anabolic and catabolic reactions. Only a few ACS enzymes from archaea have been biochemically characterized, including those from Haloarcula marismortui, Methanothermobacter thermautotrophicus, Archaeoglobus fulgidus, Methanothrix soehngenii, and Methanothrix thermoacetophila, leaving much to be explored (17, 19, 20, 23). Another enzyme that plays a crucial role in the acetate metabolism of many microorganisms is ACD; this enzyme is found mainly in the archaeal domain (24–26) and mainly converts acetyl-CoA into acetate while phosphorylating ADP into ATP. In Methanothrix sp., ACD was found to not be expressed under aceticlastic conditions, therefore suggesting that this enzyme was active only when storage compounds were converted into acetate (17). For the ACD enzyme, the kinetic parameters have been determined only for the archaea Methanocaldococcus jannaschii, Pyrococcus furiosus, and Archaeoglobus fulgidus (24, 25). A previous study on the ACS and ACD enzymes from ANME-2a focused mostly on evaluating the potential of ANMEs to perform acetogenesis from methane and did not describe the enzymatic properties of ACS and ACD in detail (27). In addition, several environmental studies explored the ability of ANMEs to reverse their pathway and produce methane from acetate (28–32).
In these studies, anoxic microbial mats dominated by ANME-1 and -2 were used in the laboratory to investigate the versatility of ANME metabolism. Only small amounts of methane were produced when acetate was used as a substrate (29). An in situ study with White Oak River estuary cores suggested that ANME-1 displayed methanogenesis activity as methane was produced at a depth of between 60 and 80 cm in sediments where ANME-1 comprised 92.8% of all of the methanomicrobiota (31, 32). Ding et al. (28) performed batch incubations with an ANME-2d-dominated enrichment culture, but they did not detect any methane production from acetate. To clarify whether “Ca. Methanoperedens” can reverse its metabolism and to obtain a better understanding of archaeal ACS and ACD enzymes, we characterized the respective enzymes with regard to their kinetic properties and performed a physiological study by providing an ANME-2d culture with 13C2-labeled acetate to evaluate whether this substrate can be used for aceticlastic methanogenesis.
RESULTS
Phylogenetic relationship of ACS enzymes.
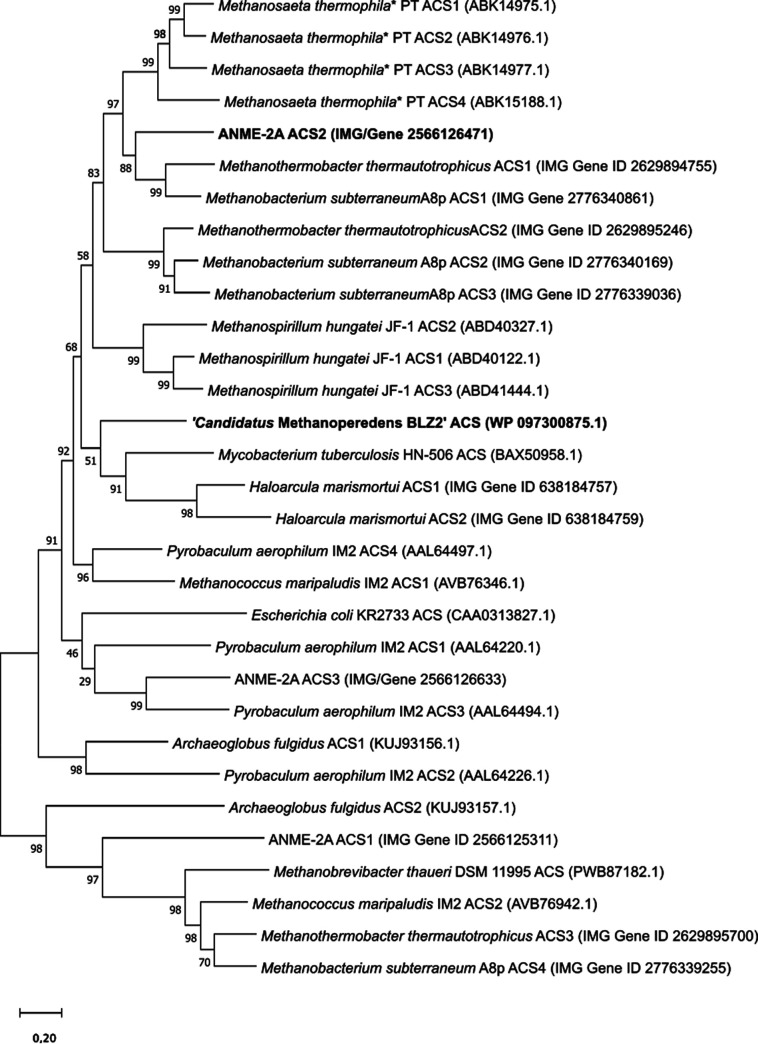
To compare the ACS enzymes from anaerobic methanotrophs with previously characterized enzymes, we constructed a phylogenetic tree based on ACS amino acid sequences (Fig. 1). The ACS enzyme from ANME-2a is most closely related to the ACS enzyme from the aceticlastic methanogen Methanothrix thermoacetophila and two ACS enzymes from the hydrogenotrophic methanogens Mt. thermautotrophicus and Methanobacterium subterraneum, while the ACS enzyme from ANME-2d is most closely related to the ACS enzymes from Mycobacterium tuberculosis and H. marismortui. Most microorganisms have multiple ACS enzymes encoded in their genomes, which may have different properties and therefore may serve different functions; bootstrap support values of <70 can be removed, while different ACS enzymes from the same microorganism do not cluster. Methanothrix thermoacetophila encodes four different ACS enzymes that are closely related to each other, but only one of them (Methanosaeta thermophila PT ACS1) is highly expressed in the exponential growth phase (17). The three ACS enzymes encoded by ANME-2a may also serve different functions, while ANME-2d encodes only one ACS enzyme.
FIG 1.
Phylogenetic tree based on the protein sequences of acetyl-CoA synthetases (ACSs), for which the characteristics are summarized in Table 1. The tree was generated with MegaX using the maximum likelihood method and the Jones-Taylor-Thornton (JTT) matrix-based model (1,000 ultrafast bootstraps). *, Methanosaeta thermophila is currently known as Methanothrix thermoacetophila.
Characterization of ACS enzymes from ANME-2a and ANME-2d and ACD from ANME-2d.
Analysis of the metagenome-assembled genome (MAG) from the in-house “Ca. Methanoperedens BLZ2” strain revealed the presence of one putative ACS-encoding gene and one ACD-encoding gene (16, 33). For a MAG from ANME-2a, three putative ACS-encoding genes were found. ACS1, -2, and -3 from ANME-2a and ACS from ANME-2d share identities of 30%, 41%, and 50%, respectively, based on the protein sequences (see Fig. S6 in the supplemental material). ACS2 (IMG/N gene identifier 2566126471) (1,965 bp) was most closely related to ACS enzymes from aceticlastic methanogens and therefore was selected for further investigation. Transcriptome analysis of the “Ca. Methanoperedens BLZ2” biomass grown in bioreactors fed with nitrate and methane (33) showed that ACS and ACD of ANME-2d were expressed at 20 and 40 transcripts per million (TPM), respectively, which are about one-half the values of the nitrate reductase genes narA (79 TPM) and narB (109 TPM) as well as the F420-dependent methylene-tetrahydromethanopterin (H4MPT) reductase gene mer (75 TPM) and the methenyl-H4MPT cyclohydrolase gene mch (65 TPM), which encode enzymes that are required for reverse methanogenesis. ACS and ACD of ANME-2d were also previously detected in the proteome of ANME-2d (9). The ACS and ACD enzymes from ANME-2d and the ACS2 enzyme from ANME-2a were successfully purified after heterologous expression in Escherichia coli. The molecular masses of the proteins on SDS-polyacrylamide gel electrophoresis (PAGE) gels matched the predicted sizes of these proteins quite well (Fig. S1). ACD from ANME-2d consists of two subunits, α and β, with molecular masses of 22.9 kDa and 49.6 kDa, respectively. The ACS enzymes consist of only one subunit, with molecular masses of 73.8 kDa for ACS2 from ANME-2a and 73.4 kDa for ACS from ANME-2d. Blue native (BN) PAGE showed that ACD forms a heterotetramer with a putative α2β2 stoichiometry, while the ACS enzymes from ANME-2a and ANME-2d are monomeric proteins (Fig. S2). The kinetic properties of all enzymes were determined after the optimization of the enzyme assay. The optimal temperatures and pHs (Fig. S3 and S4) appeared to be 50°C and pH 7.5 to 8.0 for ACS from ANME-2a, 55°C and pH 7.5 for ACS from ANME-2d, and 60°C and pH 7.5 for ACD from ANME-2d. Changes in temperature largely impacted activity, whereas only small differences in activity were observed over the tested pH range.
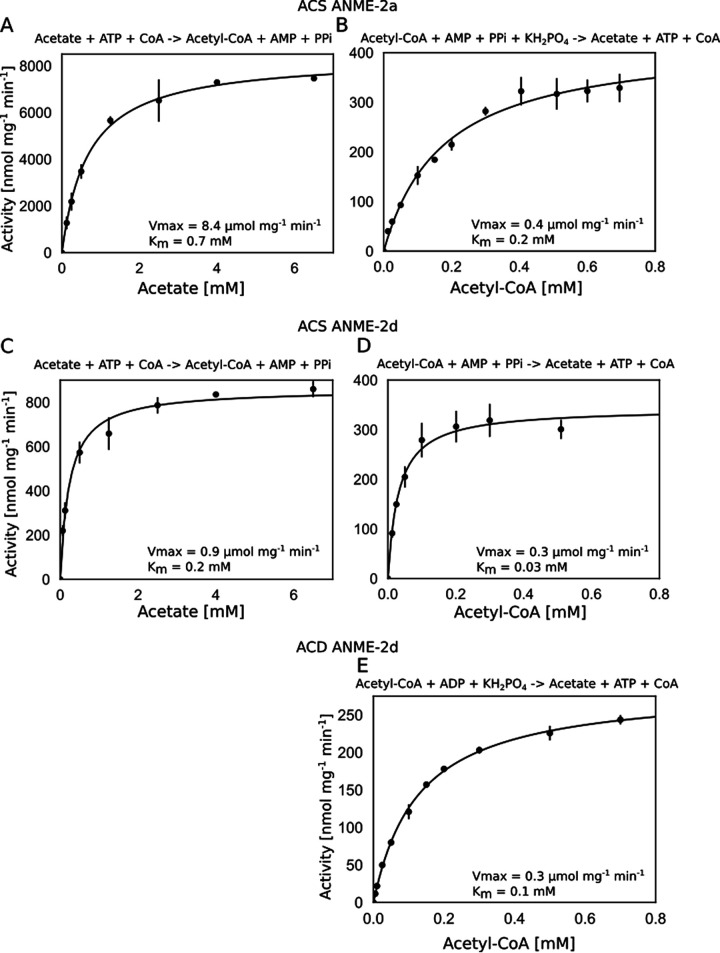
The reaction rates at different acetate and acetyl-CoA concentrations followed Michaelis-Menten kinetics, as displayed in Fig. 2. The conversion of acetate into acetyl-CoA by ACD was not detectable (using 200 μg enzyme, 6.5 mM acetate, and 10 mM ATP), as was also observed previously for other ACD enzymes (25). Furthermore, the specificity for different adenosine phosphates was tested, which confirmed that ACS was AMP dependent and that ACD was ADP dependent in the acetate-producing direction. For ACS from ANME-2a, the activity was eight times lower when ADP and KH2PO4 were used than when AMP, pyrophosphate, and KH2PO4 were used. The ACS enzyme from ANME-2d was a hundred times less active. For ACD from ANME-2d, the enzyme was three times less active when AMP and pyrophosphate were used than when ADP and KH2PO4 were used. In addition, ACS from ANME-2a was more active when KH2PO4 was added, while this was not the case for ACS from ANME-2d.
FIG 2.
(A to D) Michaelis-Menten kinetics for acetyl-CoA synthetase (ACS) purified from ANME-2a at the optimal temperature of 50°C for acetate (A) and acetyl-CoA (B) as well as for ACS from ANME-2d at the optimal temperature of 55°C for acetate (C) and acetyl-CoA (D). (E) Michaelis-Menten curve for acetate ligase (ACD) from ANME-2d at the optimal temperature of 60°C for acetyl-CoA conversion. For the reverse reaction using acetate as a substrate, the activity was not detectable using up to 200 μg mL−1 enzyme. The data are shown as averages, with error bars depicting the distributions from two biological replicates. For ACS from ANME-2d, 100 μg enzyme was used; for all other enzymes, 50 μg was used. A curve was fitted through the data using Michaelis-Menten kinetics {V = Vmax × [S]/(Km + [S])} in Excel, from which the Km and Vmax values were calculated (Km,A = 0.68 mM, Vmax,A = 8.4 μmol mg−1 min−1, Km,B = 0.2 mM, Vmax,B = 0.4 μmol mg−1 min−1, Km,C = 0.2 mM, Vmax,C = 0.9 μmol mg−1 min−1, Km,D = 0.03 mM, Vmax,D = 0.3 μmol mg−1 min−1, Km,E = 0.1 mM, and Vmax,E = 0.3 μmol mg−1 min−1).
Substrate spectrum of ACS enzymes.
Besides acetate, ACS enzymes have been reported to activate propionate, butyrate, and formate (19, 23, 34). Therefore, we tested the substrate spectrum of the purified ACS enzymes (Table 1). ACS from ANME-2a converted propionate with approximately one-third of the activity that was measured for acetate, while butyrate and formate were not used. For ACS from ANME-2d, propionate conversion was approximately one-half as low as the activity that was measured for acetate, while butyrate could also be converted by the enzyme, with an activity that was approximately 10 times lower than the activity with acetate. Formate was not converted.
TABLE 1.
Average activities of ACS enzymes from ANME-2a and -2d with different substratesa
| Substrate | Avg activity of ACS from ANME-2a (μmol mg−1 min−1) | Activities of ACS from ANME-2d (μmol mg−1 min−1) |
|---|---|---|
| Acetate | 7.46 ± 0.031 | 0.84 and 0.88 |
| Propionate | 2.47 ± 0.27 | 0.45 and 0.49 |
| Butyrate | ND | 0.098 and 0.061 |
| Formate | ND | ND |
Data are shown as measured values obtained from two biological replicates for ANME-2d and average values obtained from three biological replicates for ANME-2a. These activities were measured with 6.5 mM substrate at pH 7.5 at 55°C (ACS of ANME-2a) or at pH 8.0 at 50°C (ACS of ANME-2d), using 100 μg enzyme (ACS of ANME-2d) or 50 μg enzyme (ACS of ANME-2a). ND, not detected.
A “Ca. Methanoperedens” enrichment culture converted acetate into methane.
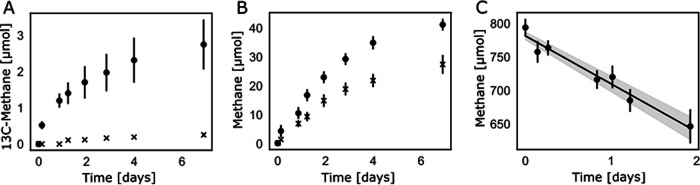
ACS is widely used by microorganisms for assimilation and dissimilation. Hydrogenotrophic methanogens encode ACS enzymes and use them for the assimilation of acetate into biomass rather than for aceticlastic methanogenesis (35, 36). The ACS enzymes of the investigated ANMEs are most likely acetate activating; thus, we wondered whether these ANMEs could also perform aceticlastic methanogenesis. To this end, we performed batch incubations without acetate (negative control) and with 13C2-labeled acetate using our available “Ca. Methanoperedens” enrichment culture. 13C-labeled methane (Fig. 3A) was produced only when 13C-labeled acetate was fed to the culture. Unlabeled methane was also produced with acetate (40 μmol in 7 days) and without acetate (28 μmol in 7 days) supplied during the incubations (Fig. 3B). 16S rRNA gene sequencing was performed, which showed that “Ca. Methanoperedens” was the most highly abundant (>98%) archaeon in the inoculum and the incubation mixtures after a week of feeding 13C-labeled acetate (Table 2). Methanobacteriaceae were the only other archaeal family present in the samples, at <0.16% of the reads. The remaining reads arose from the unspecific binding of the archaeal primers.
FIG 3.
Batch incubations of an enrichment culture dominated by “Ca. Methanoperedens.” (A) 13CH4 produced under two conditions, with 13C-labeled acetate added to the medium (circles) and a negative control without the substrate added (crosses). (B) Total methane concentrations (in micromoles) produced during the experiment shown in panel A. Circles, acetate added; crosses, no acetate added. (C) Total methane (in micromoles) consumed by a “Ca. Methanoperedens” enrichment culture supplied with methane and nitrate as a positive control to validate the activity of the methanotrophic culture.
TABLE 2.
Relative abundances of archaea in the batch incubation experimentsa
| Incubation condition | Relative abundance |
||
|---|---|---|---|
| “Ca. Methanoperedens” | Methanobacteriaceae | Remainder | |
| T 0 | 98.1 | 0.01 | 1.8 |
| With acetate | 99.1 | 0.16 | 0.7 |
| Without acetate | 99.2 | 0.03 | 0.8 |
See Fig. 2. The abundances were calculated for the inoculum (time zero [T0]), for the experiment with acetate supplied, and for the experiment without acetate supplied. These numbers were obtained by amplicon sequencing using general archaeal primers that target the archaeal 16S rRNA gene. The remainder is bacteria that were the result of the unspecific binding of the primers to the bacterial 16S rRNA gene.
DISCUSSION
ACD from ANME-2d produces acetate from acetyl-CoA.
While ACD plays an important role in acetate metabolism, only four archaeal ACD enzymes have been characterized so far (24–27). To find out more about the properties and functions of ACD enzymes in anaerobic methanotrophs (ANMEs), we characterized an ACD enzyme from ANME-2d. The kinetic parameters of the ACD enzyme from ANME-2d obtained in this study were compared to those of previously characterized enzymes (Table 3). ACD from ANME-2d was active only in the acetate-forming direction. This is similar to the ACD enzyme from the hydrogenotrophic methanogen Methanococcus jannaschii (25) and the ACD enzyme from ANME-2a (27), while all other characterized ACD enzymes have been reported to be reversible (see Table S1 in the supplemental material). ACD encoded by “Ca. Methanoperedens” has a Vmax value that is 1 to 2 orders of magnitude lower than the Vmax values reported for other microorganisms such as Archaeoglobus fulgidus (hyperthermophilic archaeon) (25), Chloroflexus aurantiacus (thermophilic photosynthetic bacterium) (26), Pyrococcus furiosus (heterotrophic, thermophilic archaeon) (24), and Methanococcus jannaschii (hydrogenotrophic methanogen) (25). On the other hand, the ACD enzyme from “Ca. Methanoperedens” is 20 times more active than the ACD enzyme from ANME-2a. The Km for acetyl-CoA is 1 order of magnitude higher than those for all other microorganisms shown in Table 3. The pH optima are similar for all ACD enzymes, while the optimum temperature is much higher for ACD from Chloroflexus aurantiacus.
TABLE 3.
Kinetic parameters of acetate ligases from ANME-2d and ANME-2a obtained in this study compared to these parameters for the same enzymes obtained in previous studies for acetyl-CoA conversion
| Microorganism | Km of acetyl-CoA (mM) | Vmax of acetyl-CoA (μmol mg−1 min−1) | Mean Kcat/Km ratio (s−1 mM−1) ± SD | Optimum temp (°C) | Optimum pH | Subunit composition |
|---|---|---|---|---|---|---|
| Archaeoglobus fulgidus a | 0.01 | 75 (at 55°C) | 7.0 | Homodimer | ||
| Methanococcus jannaschii a | 0.04 | 3–6 (at 55°C) | 18.6 ± 0.5 | 55 | 7.5 | Homodimer |
| Chloroflexus aurantiacus b | 0.04 | 51 | 90 | 7.0 | Homotetramer | |
| Pyrococcus furiosus c | 0.02 | 18 | Heterodimer | |||
| ANME-2ad | 0.03 | 0.01 | ||||
| ANME-2d BLZ2e | 0.13 | 0.29 | 0.01 | 60 | 7.5 | Heterotetramer |
Based on these properties, we assume that “Ca. Methanoperedens” most likely uses the ACD enzyme for substrate-level phosphorylation by converting some of its cellular acetyl-CoA, ADP, and phosphate into acetate and ATP. This reaction might take place if there is an excess of acetyl-CoA present in the cell that is not used for anabolism, most likely during the breakdown of polyhydroxyalkanoates (PHAs) (10) when acetate production needs to fulfill “Ca. Methanoperedens” energy requirements. For Haloarcula marismortui, it has been shown that the ACD enzyme is produced in the exponential growth phase with glucose as a substrate when acetyl-CoA is in excess. Later during the stationary phase, when glucose is limited, acetate is taken up again and activated by ACS (37). In vitro, ACD from ANME-2d is active only in the acetate-forming direction and thus may have a role similar to that of ACD from Haloarcula marismortui, producing acetate and ATP when acetyl-CoA is present in excess, such as during PHA degradation.
The ACD enzyme of ANME-2d had a heterotetrameric composition, while other archaeal ACDs seem to be either homodimeric, heterodimeric, or homotetrameric (Table 3).
ACS enzymes from ANME-2d and -2a interconvert acetate into acetyl-CoA and vary regarding their Vmax and Km values.
For ACS enzymes, there is still much to be explored as only a few archaeal ACSs have been characterized so far. The kinetic parameters of the ACS enzymes from ANME-2a and -2d obtained in this study were compared to those of previously characterized enzymes (Table 4 and Table S2). The affinity of the ACS enzymes from ANME-2a and -2d align quite well with the majority of the reported kinetic values, although the Vmax values are in the lower range. The slow conversion of acetate by ACS enzymes in ANMEs may indicate that anaerobic methanotrophs most likely are not dependent on acetate as a carbon or energy source, in contrast to aceticlastic methanogens such as Methanothrix sp. Surprisingly, the Vmax of ACS from ANME-2a reported previously by Yang et al. (27) is 1 order of magnitude lower than the value reported in our study. This may be because we determined the Vmax at the optimum temperature of 50°C, whereas Yang et al. measured activities at room temperature. The ACS enzyme from ANME-2a is apparently more active in the direction of acetyl-CoA production, with a Vmax that is 20 times higher in this direction than during acetate production. The ACS enzyme of ANME-2d has comparable rates for both directions. There is some variation in the Km and Vmax values of the ANMEs. The Km for acetate, however, is in the same low-nanomolar range as that of Methanothrix ACS, making it likely that ANMEs can scavenge acetate for anabolism (Table 4).
TABLE 4.
Kinetic parameters of acetyl-CoA synthetases from ANME-2d and ANME-2a obtained in this study compared to these parameters for the same enzymes obtained in previous studies for acetate conversion
| Microorganism | Km of acetate (mM) | Vmax of acetate (μmol mg−1 min−1) | Kcat/Km ratio (s−1 mM−1) | Optimum temp (°C) | Optimum pH | Subunit composition |
|---|---|---|---|---|---|---|
| Methanothrix soehngenii a | 0.8 | 55 | 35 | 8.5 | Dimer | |
| Methanothrix thermophila b | 0.4 | 21.7–28 | 55 | |||
| ANME-2ac | 0.5 | 0.1 | ||||
| ANME-2ad | 0.7 | 8.4 | 0.03 | 50 | 7.5 | Monomer |
| ANME-2d BLZ2d | 0.2 | 0.9 | 0.02 | 55 | 8 | Monomer |
| E. coli e | 0.2 | |||||
| Mycobacterium tuberculosis f | 1.2 | |||||
| Methanothermobacter thermautotrophicus g | 3.5 | 18.6 | 65–70 | Dimer | ||
| Archaeoglobus fulgidus g | 1.7 | 21.2 | 65–70 | Trimer | ||
| Dunaliella tertiolecta h | 4.7 | 40 | 8 | |||
| Haloarcula marismortui i | 0.2 | 26.5 | 41 | 7.5 | Monomer | |
| Pyrobaculum aerophilum j | 0.003 | 37 | >97 | Octamer |
The subunit composition of ACS enzymes in different species is not uniform, varying from monomeric in H. marismortui and the ANMEs in this study to octameric in Pyrobaculum aerophilum (Table 4).
The optimum temperatures of the ACS enzymes from ANME-2a and “Ca. Methanoperedens” are much higher than the temperatures in sediments where these microorganisms normally reside and may be a remnant of their thermophilic ancestry.
ACS enzymes from ANMEs can convert other substrates besides acetate.
Some ACS enzymes have been reported to convert a variety of substrates. We found that ANME-2a ACS converts propionate with 33% of the activity obtained with acetate, while butyrate and formate could not be converted, similar to ACS from Haloarcula marismortui (19). ACS from “Ca. Methanoperedens” converts propionate with 49% of the activity and butyrate with 9.2% of the activity obtained with acetate as the substrate, and the conversion of formate was not detected. Archaeoglobus fulgidus ACS activates acetate, propionate, butyrate, and isobutyrate (23), showing a weak (2-fold) preference for acetate over propionate, similar to ACS from ANME-2d. The potential to use a variety of substrates may give ANME-2d a competitive advantage in situ without the need to express and produce a different enzyme, which gives it the option to adapt quickly to changing environmental conditions. Microorganisms with a high substrate specificity for ACS, such as Methanothrix sp., have a high affinity for acetate and therefore might outcompete other organisms with lower substrate specificities for acetate. The high substrate affinity might prevent the enzyme from having a broader substrate specificity. ACS from ANME-2d has a substrate range and specificity similar to those of A. fulgidus; however, these enzymes are phylogenetically not related. Instead, ANME-2d ACS is more closely related to ACS from H. marismortui, with a narrow substrate range. On the other hand, ACS from H. marismortui and ACS from ANME-2d resemble each other in their monomeric protein configurations. The fact that enzymes with similar biochemical characteristics do not cluster in phylogenetic trees emphasizes the need to characterize enzymes biochemically rather than inferring enzymological properties from amino acid identity only.
“Ca. Methanoperedens” produces methane from acetate.
ANMEs and methanogens are phylogenetically related and employ similar enzymes and pathways; however, it is unclear whether ANMEs can perform aceticlastic methanogenesis by reversing their metabolism. In the aceticlastic methanogen Methanothrix sp., an ACS enzyme converts acetate into acetyl-CoA, after which the acetyl group from acetyl-CoA is transferred to H4MPT, resulting in methyl-H4MPT (21). The methyl group is ultimately converted into methane by methyl-coenzyme M reductase (MCR) after the transfer of the methyl group to coenzyme M via the membrane-bound methyltransferase MTR. ANME-2d displays all of the enzymes necessary for this pathway but lacks the membrane-bound energy-conserving modules to oxidize the reduced ferredoxin that is produced during the oxidation of the acetyl-derived carbonyl group. Therefore, it is questionable whether these microorganisms would be able to conserve energy during aceticlastic growth. We reported that ANME ACS preferably converts acetate to acetyl-CoA, so this enzyme might be used for acetate activation. We were wondering whether acetyl-CoA could be used for anabolism only or also for methanogenesis; to investigate whether “Ca. Methanoperedens” may be capable of reversing its metabolism and converting some of the acetate into methane, we incubated an enrichment culture dominated by “Ca. Methanoperedens” (>98%) with 13C-labeled acetate as a carbon source. Three micromoles of 13C-labeled methane was indeed produced in the culture fed with 13C-labeled acetate over the course of 1 week. This corresponds to a methane production rate of 2.1 nmol CH4 g (dry weight)−1 min−1. This rate is about 102 times lower than the methane oxidation rate of “Ca. Methanoperedens” but 105 times lower than the aceticlastic methanogenesis rate of Methanothrix soehngenii (methane consumption rate of 200 μmol CH4 g [dry weight]−1 min−1) and 104 times lower than that of Methanosarcina acetivorans (methane consumption rate of 25 μmol CH4 g [dry weight]−1 min−1) (38). This may be explained by the fact that ANMEs possibly have a very low or no energy gain from converting acetate into methane. In addition, acs might have much lower expression levels in the cells than in aceticlastic methanogens such as Methanothrix sp., which depends on acetate as the substrate. As Methanosarcina sp. and Methanothrix sp. were undetectable by amplicon sequencing at the end of the incubation period, we think that “Ca. Methanoperedens” is the most likely candidate for the methane production. The methane oxidation rate of the culture using nitrate as an electron acceptor was 289 nmol CH4 g (dry weight)−1 min−1, indicating that the reversal of metabolism was operational at only 0.7% of the capacity of the methanogenic/methanotrophic pathway, confirming that methanogenesis is probably not connected to energy conservation.
The production of unlabeled methane, which was detected for the batch incubations with (40 μmol in 7 days) and without (28 μmol in 7 days) labeled acetate, may originate either from the anaerobic degradation of organic matter released from lysing cells and subsequent methane production by “Ca. Methanoperedens” or from PHA conversion (10). Cai et al. (10) demonstrated previously that in the absence of an electron acceptor, which was the case with our incubations, intracellular PHA is converted to acetate, which is released into the medium. “Ca. Methanoperedens” might then convert acetate to methane at a low but measurable rate. We performed amplicon sequencing targeting the 16S rRNA gene of archaea to determine if canonical methanogens were present in the culture. A member of the Methanobacteriaceae family was found but at a very low relative abundance of 0.16% of all archaea. Members of this family are hydrogenotrophic methanogens that are not known to perform aceticlastic methanogenesis. Hydrogenotrophic methanogens encode ACS enzymes that might be used for assimilation (36). Methanothermobacter marburgensis has been reported to assimilate acetate in the presence of CO2 and H2 with up to 65% of the cell carbon derived from acetate (35). However, we cannot exclude that 13C-labeled acetate may have been converted into 13C-labeled CO2 by heterotrophic bacteria, after which it could be converted into 13C-labeled methane by hydrogenotrophic methanogens. The required cosubstrate hydrogen was not added to the culture. However, we cannot exclude that hydrogen is produced by fermenting, H2-producing bacteria such as Ignavibacterium that are present at low abundances in the ANME-2d enrichment culture.
Conclusion.
Overall, we provide insights into the acetate and acetyl-CoA metabolism of ANME-2 anaerobic methanotrophs. We found that ANME-2d ACD is active only in the direction of acetate formation and might enable ANMEs to generate ATP from excess acetyl-CoA while producing acetate, e.g., during the turnover of PHA. The ACS enzymes from ANME-2a and -2d catalyze a reversible reaction but are more active in the direction of acetate conversion. Both ACS enzymes are thus likely to be used for the conversion of acetate to acetyl-CoA, but the use of acetate can most likely be used only for anabolism and not for energy conservation by ANMEs. Furthermore, the batch cultivation experiment with 13C-labeled acetate indicated that “Ca. Methanoperedens” may be able to convert acetate into methane. The ACS enzyme and the culture showed a low rate of acetate turnover, suggesting that in situ methane formation from acetate by ANME-2d is not ecologically relevant and rather is a side effect of anabolic acetate assimilation.
MATERIALS AND METHODS
Phylogenetic analysis of ACS enzymes.
A multiple-sequence alignment of acetyl-CoA synthetases was performed using MegaX and the built-in Muscle algorithm. The tree was generated with MegaX (39) using the maximum likelihood method and the Jones-Taylor-Thornton (JTT) matrix-based model (1,000 ultrafast bootstraps). The amino acid sequences were retrieved from the NCBI. For some microorganisms, sequences were not available in the NCBI database; for these microorganisms, the sequences were retrieved from the DOE Joint Research Institute (JGI)-IMG. For the sequences obtained from the JGI, the gene identifier is shown in the tree; these sequences were translated into amino acid sequences by using protparam (https://web.expasy.org/protparam/).
Cloning.
Analysis of the metagenome-assembled genomes (MAGs) from ANME-2a and ANME-2d revealed the presence of putative ACS- and ACD-encoding genes (16, 27, 33, 40). The ANME-2a MAG was obtained from enrichment cultures from the Captain Aryutinov Mud Volcano (Spain) (40), and the ANME-2d MAG was obtained from an in-house culture seeded with Ooijpolder (Netherlands) sediments (16, 41, 42). Two acetyl-CoA synthetase genes (acs) and one acetate ligase gene (acd) were heterologously expressed in E. coli. acs genes from ANME-2d BLZ2 (MPEBLZ_01103) (1,959 bp) and ANME-2a (taxon identifier 2565956544; IMG/N gene identifier 2566126471) (1,965 bp) archaea were codon optimized, synthesized, and delivered in pUC57Kan plasmids (Baseclear, Leiden, Netherlands), with BsaI restriction endonuclease sites inserted at each end of the gene, enabling the cloning of the inserts into pASK-IBA3+ expression vectors (IBA GmbH, Gottingen, Germany). The recombinant plasmids pACS-ANME2a and pACS-ANME2d were transformed into E. coli BL21 and stored as glycerol stocks at −80°C. In the case of ACD, the genes of the two ACD subunits from ANME-2d BLZ2 (NCBI RefSeq accession numbers WP_097298476.1 and WP_097298477.1) were amplified by using DNA that was extracted from an enrichment culture dominated by “Ca. Methanoperedens BLZ2” (41). The DNA fragment consisting of the β and α subunits was amplified by using forward primer 5′-CGCATATGAATGTAATGCTGTCTTCTATATTCGAACC-3′ and reverse primer 5′-TTGCGGCCGCTTATTTTTCGAACTGCGGGTGGCTCCAGCTAGCTTCTTCCACCATCGCTCTTGC-3′ so that the restriction sites (NdeI and NotI) and a C-terminal Strep-tag were added. The PCR product was purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany). The FastDigest restriction enzymes NdeI and NotI were used according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). Ligation of the restricted PCR product into the pET30a plasmid was performed overnight at 16°C using T4 DNA ligase (Thermo Fisher Scientific, Waltham, MA, USA), resulting in plasmid pACD-ANME2d, and the ligation product was transformed into E. coli DH5α cells. For the transformation of the plasmid, the ligation product and DH5α cells were incubated on ice for 1 h, after which the mixture was incubated at 42°C for 90 s and cooled on ice for 3 min. Subsequently, LB medium containing 10 g tryptone, 5 g yeast extract, and 10 g NaCl per L was used to allow the cells to recover at 37°C for 1 h at 200 rpm. The transformed cells were grown on plates using the same LB medium supplemented with kanamycin (50 mg L−1). Colonies were transferred into LB medium supplemented with kanamycin (50 mg L−1). After incubation of the cultures overnight at 37°C, the plasmids were extracted using the GeneJET plasmid miniprep kit (Thermo Fisher Scientific, Waltham, MA, USA) and sequenced to verify the correctness of the cloned DNA sequence. For heterologous expression, the produced plasmids pACD-ANME2d, pACS-ANME2a, and pACS-ANME2d were transformed into E. coli BL21(DE3) cells as described above.
Protein production and purification.
The recombinant proteins were purified by Strep-tag affinity chromatography. Six 1-L shake flasks containing 600 mL LB medium supplemented with kanamycin (50 mg L−1) were inoculated with the expression strain (E. coli BL21 with pACD-ANME2d). These cultures were grown overnight for 17 h, after which the biomass was harvested by centrifugation for 10 min at 8,000 rpm at 4°C. For the other two expression strains, precultures supplemented with ampicillin (100 mg L−1) were grown overnight; next, for each expression strain (pACS-ANME2a and pACS-ANME2d), five 1-L shake flasks containing 600 mL LB medium supplemented with ampicillin (100 mg L−1) were inoculated. The cultures were grown until an optical density at 600 nm (OD600) of 0.4 was reached, and protein production was induced by the addition of anhydrotetracycline (200 ng mL−1) and incubation for another 4 h. The biomass was harvested by centrifugation for 10 min at 8,000 rpm at 4°C. Buffer W (100 mM Tris-HCl, 150 mM NaCl [pH 8.0]) was added to the pellet, and the cells were lysed by sonication with 1-s pulses at an amplitude of 50%, with a 5-s break and a total run time of 5 min. The lysed cells were centrifuged for 30 to 45 min at 20,000 × g, and the supernatant was filtered using 0.2-μm syringe filters (Whatman, Maidstone, UK) and used for purification with a Strep-Tactin XT Superflow column (IBA Life Sciences, Göttingen, Germany). The Strep-Tactin column was washed with buffer W, and the protein extract was then loaded onto the column, after which the column was again washed with buffer W and the protein was eluted using buffer W supplemented with biotin (50 mM) and EDTA (1 mM) in different fractions of 0.5 mL, 1.5 mL, and 1 mL. All enzymes were purified aerobically. ACS from ANME-2a was purified anaerobically once, but the enzyme did not show higher activity for converting acetate into acetyl-CoA. SDS-polyacrylamide gel electrophoresis (PAGE) was performed according to the method described previously by Laemmli (43), with a 15% resolving gel and a 4% stacking gel. SDS (4×) loading buffer (250 mM Tris-Cl [pH 6.8], 8% SDS, 40% glycerol, 20% β-mercaptoethanol, 0.02% bromophenol blue) was used to dilute the protein samples. Prior to loading, the samples were heated at 95°C for 5 min. A total of 7.5 μg was added to each lane. The molecular masses of the proteins were determined by comparison with a PageRuler prestained protein ladder (Thermo Fisher, Waltham, MA, USA). The proteins were visualized by Coomassie staining. A Bradford assay (44) was used to determine the concentration of the purified enzyme using Bio-Rad (Hercules, CA, USA) protein assay dye. Blue native (BN) PAGE was performed according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA), using a 2 to 12% BN gradient gel (4% stacking gel) (Thermo Fisher Scientific, Waltham, MA, USA). Ten micrograms of protein was added to each lane after the addition of glycerol (10% final concentration) and 1 μL of sample additive per 10 μL (750 mM 6-aminocaproic acid, 5% Coomassie brilliant blue G-250). The molecular masses of the proteins were determined by comparison with a NativeMark unstained protein standard (Thermo Fisher Scientific, Waltham, MA, USA). Part of the same samples was incubated with 4× SDS loading buffer at 96°C for 5 min to denature the proteins as a comparison. Gels were run at 100 V. In the first stage, 1× NativePAGE cathode buffer (900 mL deionized water, 50 mL 20× NativePAGE running buffer, and 50 mL 20× NativePAGE cathode additive) was added to the upper buffer chamber, while 1× NativePAGE anode buffer (950 mL deionized water, 50 mL 20× NativePAGE running buffer) was added to the lower buffer chamber. When the dye front reached the middle of the gel, the cathode buffer was replaced by anode buffer. The proteins were visualized by Coomassie staining.
Enzymatic assays.
Enzymatic assays were performed for the conversion of acetate into acetyl-CoA and the reverse reaction. The formation of acetyl-CoA from acetate was determined using a discontinuous colorimetric assay adapted from methods described previously by Jones and Lipmann and Brown et al. (45, 46). The assay mixture for each reaction contained 50 mM Tris-HCl at various pH values, 100 mM hydroxylamine hydrochloride (pH 7.5), 5 mM MgCl2, various concentrations of sodium acetate, 10 mM ATP, 1 mM acetyl-CoA trilithium salt, 5 mM glutathione, and the purified enzyme in an assay mixture volume of 1 mL. For ACS from ANME-2a and ACD from ANME-2d, 50 μg enzyme was used, while for ACS from ANME-2d, 100 μg enzyme was used as the enzyme activity was lower. Samples (110 μL) were taken at different time points, added directly to 110 μL 10% trichloroacetic acid (TCA), and incubated for 5 min, after which 110 μL 2.5% (wt/vol) FeCl3 · 6H2O was added. The absorbance was measured at 520 nm using a Cary 60 spectrophotometer (Agilent, Santa Clara, CA, USA). For ACS, the optimal temperature and pH for acetyl-CoA formation were determined; these were 50°C and pH 7.5 to 8.0 for ACS from ANME-2a and 55°C and pH 7.5 for ACS from ANME-2d (see Fig. S3 and S4 in the supplemental material). For ANME-2d ACD, these parameters were obtained using the reverse reaction, namely, 60°C and pH 7.5 (Fig. S3 and S4). The optimal temperature and pH were used to obtain the Km and Vmax values for all three enzymes. The formation of acetate from acetyl-CoA was determined using Ellman’s thiol reagent [5′5-dithiobis(2-nitrobenzoic acid) (DTNB)] and measured at 412-nm wavelength (ε412 = 13.6 mM−1 cm−1) according to methods described previously by Bräsen and Schönheit (19), with some adaptations. The assay mixture contained 50 mM Tris-HCl, 5 mM MgCl2, 0.1 mM DTNB (2 mM stock solution dissolved in 100 mM Tris-HCl), various concentrations of acetyl-CoA, 1 mM ADP or AMP, 1 mM pyrophosphate (for assays with AMP), 40 μg enzyme, and 5 mM KH2PO4 (for assays with ADP and for the assay for ACS from ANME-2a). An assay mixture volume of 400 μL was used.
Feeding an ANME-2d culture with 13C-labeled acetate.
Batch incubations with acetate as a carbon source were performed to investigate whether “Ca. Methanoperedens” is capable of aceticlastic methanogenesis. A biomass from an enrichment culture originating from Ooijpolder, the Netherlands, was used and was previously described by Ettwig et al. (42). Metagenomic sequencing reads from this bioreactor from April 2021 are available under NCBI BioProject accession number PRJNA850006 and BioSample accession number SAMN31357808. These data show that “Ca. Methanoperedens” was 33% enriched. Three different conditions were tested in batch incubations with 170 mg of the biomass in a final volume of 40 mL per experiment and in triplicate in 120-mL serum bottles closed with butyl stoppers and crimp sealed: (i) labeled acetate was used as a carbon source to test the aceticlastic methanogenesis potential of “Ca. Methanoperedens,” (ii) no carbon source was added (negative control), or (iii) methane was used as a carbon source and nitrate was used as an electron acceptor (positive control to validate that the enrichment culture was active at the start of the experiment). The medium contained 0.07 g MgSO4 · 7H2O, 0.1 g CaCl2 · 2H2O, 0.05 g KH2PO4, 2.38 g HEPES, 0.0267 g NH4Cl, 10 mM sodium acetate (13C2-labeled) (only for the conditions with acetate), and 1 mM NaNO3 (only for the positive control) (pH 7.30 [±0.1]) per L and was made anoxic by gassing for 10 min with argon. Samples from the enrichment culture were handled in an anaerobic chamber to keep the cultures anoxic and washed three times using anaerobic medium without acetate. In our laboratory, “Ca. Methanoperedens” enrichment cultures are successfully maintained without the addition of reducing agents. Fifty-microliter gas samples were taken to measure 13C-labeled methane and total methane. Labeled methane was measured using an Agilent 6890 series gas chromatograph coupled to an Agilent 5975C mass spectrometer equipped with a Porapak Q column heated at 80°C (Agilent 5975 inert MSD). Total methane was measured using a gas chromatograph with a flame ionization detector and a Porapak Q100 column (catalog number 5890; Hewlett Packard, Palo Alto, CA, USA). At the end of the experiment (8 days after inoculation), samples were taken for DNA extraction using the PowerSoil DNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions except for an extra bead-beating step at 50 s−1 for 10 min. 16S rRNA gene amplicon sequencing was performed by Macrogen (Amsterdam, Netherlands) using the Illumina MiSeq next-generation sequencing platform. Paired-end libraries were constructed using the Illumina (Eindhoven, Netherlands) Herculase II fusion DNA polymerase Nextera XT index kit V2. General primers Arch349F (5′-GYGCASCAGKCGMGAAW-3′) and Arch806R (5′-GGACTACVSGGGTATCTAAT-3′) were used to target the archaeal 16S rRNA gene (47).
Data availability.
The raw reads obtained from 16S rRNA amplicon sequencing were deposited in the NCBI database under BioProject accession number PRJNA924223.
ACKNOWLEDGMENTS
This work was supported by the Netherlands Organization for Scientific Research (NWO) through grant ALWOP.293 and by the Soehngen Institute of Anaerobic Microbiology Gravitation Program through grant NWO/OCW 024.002.002. M.S.M.J. was further supported by ERC Synergy project MARIX 8540088. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Wouter Versantvoort for experimental support.
This study was designed by J.M.K., C.U.W., H.T.O., M.S.M.J., H.J.M.O.D.C., S.B., and A.A. All experiments were performed by H.T.O., except for cloning of the plasmids pACS-ANME2a and pACS-ANME2d, which was conducted by A.A. and J.G.S.S. The data were interpreted by H.T.O., C.U.W., M.S.M.J., and J.M.K. The paper was written by H.T.O., M.S.M.J., C.U.W., and J.M.K. All authors contributed to and approved the final manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Julia M. Kurth, Email: julia.kurth@uni-marburg.de.
Nicole R. Buan, University of Nebraska—Lincoln
REFERENCES
- 1.Dean JF, Middelburg JJ, Röckmann T, Aerts R, Blauw LG, Egger M, Jetten MSM, de Jong AEE, Meisel OH, Rasigraf O, Slomp CP, in’t Zandt MH, Dolman AJ. 2018. Methane feedbacks to the global climate system in a warmer world. Rev Geophys 56:207–250. doi: 10.1002/2017RG000559. [DOI] [Google Scholar]
- 2.Knittel K, Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol 63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- 3.Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJP, Ettwig KF, Rijpstra WIC, Schouten S, Sinninghe Damsté JS, Op den Camp HJM, Jetten MSM, Strous M. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- 4.Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. 2013. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 5.Ettwig KF, Zhu B, Speth D, Keltjens JT, Jetten MSM, Kartal B. 2016. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc Natl Acad Sci USA 113:12792–12796. doi: 10.1073/pnas.1609534113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leu AO, Cai C, McIlroy SJ, Southam G, Orphan VJ, Yuan Z, Hu S, Tyson GW. 2020. Anaerobic methane oxidation coupled to manganese reduction by members of the Methanoperedenaceae. ISME J 14:1030–1041. doi: 10.1038/s41396-020-0590-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai C, Leu AO, Xie G-J, Guo J, Feng Y, Zhao J-X, Tyson GW, Yuan Z, Hu S. 2018. A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction. ISME J 12:1929–1939. doi: 10.1038/s41396-018-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glodowska M, Welte CU, Kurth JM. 2022. Metabolic potential of anaerobic methane oxidizing archaea for a broad spectrum of electron acceptors. Adv Microb Physiol 80:157–201. doi: 10.1016/bs.ampbs.2022.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Berger S, Cabrera-Orefice A, Jetten MSM, Brandt U, Welte CU. 2021. Investigation of central energy metabolism-related protein complexes of ANME-2d methanotrophic archaea by complexome profiling. Biochim Biophys Acta 1862:148308. doi: 10.1016/j.bbabio.2020.148308. [DOI] [PubMed] [Google Scholar]
- 10.Cai C, Shi Y, Guo J, Tyson GW, Hu S, Yuan Z. 2019. Acetate production from anaerobic oxidation of methane via intracellular storage compounds. Environ Sci Technol 53:7371–7379. doi: 10.1021/acs.est.9b00077. [DOI] [PubMed] [Google Scholar]
- 11.Zhang X, McIlroy SJ, Vassilev I, Rabiee H, Plan M, Cai C, Virdis B, Tyson GW, Yuan Z, Hu S. 2022. Polyhydroxyalkanoate-driven current generation via acetate by an anaerobic methanotrophic consortium. Water Res 221:118743. doi: 10.1016/j.watres.2022.118743. [DOI] [PubMed] [Google Scholar]
- 12.Soo VWC, McAnulty MJ, Tripathi A, Zhu F, Zhang L, Hatzakis E, Smith PB, Agrawal S, Nazem-Bokaee H, Gopalakrishnan S, Salis HM, Ferry JG, Maranas CD, Patterson AD, Wood TK. 2016. Reversing methanogenesis to capture methane for liquid biofuel precursors. Microb Cell Fact 15:11. doi: 10.1186/s12934-015-0397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Timmers PHA, Welte CU, Koehorst JJ, Plugge CM, Jetten MSM, Stams AJM. 2017. Reverse methanogenesis and respiration in methanotrophic archaea. Archaea 2017:1654237. doi: 10.1155/2017/1654237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krzycki JA. 2004. Function of genetically encoded pyrrolysine in corrinoid-dependent methylamine methyltransferases. Curr Opin Chem Biol 8:484–491. doi: 10.1016/j.cbpa.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Thauer RK, Kaster A-K, Goenrich M, Schick M, Hiromoto T, Shima S. 2010. Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage. Annu Rev Biochem 79:507–536. doi: 10.1146/annurev.biochem.030508.152103. [DOI] [PubMed] [Google Scholar]
- 16.Arshad A, Speth DR, de Graaf RM, Op den Camp HJM, Jetten MSM, Welte CU. 2015. A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like archaea. Front Microbiol 6:1423. doi: 10.3389/fmicb.2015.01423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berger S, Welte C, Deppenmeier U. 2012. Acetate activation in Methanosaeta thermophila: characterization of the key enzymes pyrophosphatase and acetyl-CoA synthetase. Archaea 2012:315153. doi: 10.1155/2012/315153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang M-H, Qv X-Y, Jin H-H, Jiang J-G. 2016. Characterization and expression of AMP-forming acetyl-CoA synthetase from Dunaliella tertiolecta and its response to nitrogen starvation stress. Sci Rep 6:23445. doi: 10.1038/srep23445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bräsen C, Schönheit P. 2005. AMP-forming acetyl-CoA synthetase from the extremely halophilic archaeon Haloarcula marismortui: purification, identification and expression of the encoding gene, and phylogenetic affiliation. Extremophiles 9:355–365. doi: 10.1007/s00792-005-0449-0. [DOI] [PubMed] [Google Scholar]
- 20.Jetten MSM, Stams AJM, Zehnder AJB. 1989. Isolation and characterization of acetyl-coenzyme A synthetase from Methanothrix soehngenii. J Bacteriol 171:5430–5435. doi: 10.1128/jb.171.10.5430-5435.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welte C, Deppenmeier U. 2014. Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim Biophys Acta 1837:1130–1147. doi: 10.1016/j.bbabio.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Jetten MSM, Stams AJM, Zehnder AJB. 1992. Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol Rev 88:181–197. doi: 10.1111/j.1574-6968.1992.tb04987.x. [DOI] [Google Scholar]
- 23.Ingram-Smith C, Smith KS. 2007. AMP-forming acetyl-CoA synthetases in Archaea show unexpected diversity in substrate utilization. Archaea 2:95–107. doi: 10.1155/2006/738517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glasemacher J, Bock A, Schmid R, Schonheit P. 1997. Purification and properties of acetyl-CoA synthetase (ADP-forming), an archaeal enzyme of acetate formation and ATP synthesis, from the hyperthermophile Pyrococcus furiosus. Eur J Biochem 244:561–567. doi: 10.1111/j.1432-1033.1997.00561.x. [DOI] [PubMed] [Google Scholar]
- 25.Musfeldt M, Schönheit P. 2002. Novel type of ADP-forming acetyl coenzyme A synthetase in hyperthermophilic archaea: heterologous expression and characterization of isoenzymes from the sulfate reducer Archaeoglobus fulgidus and the methanogen Methanococcus jannaschii. J Bacteriol 184:636–644. doi: 10.1128/JB.184.3.636-644.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt M, Schönheit P. 2013. Acetate formation in the photoheterotrophic bacterium Chloroflexus aurantiacus involves an archaeal type ADP-forming acetyl-CoA synthetase isoenzyme I. FEMS Microbiol Lett 349:171–179. doi: 10.1111/1574-6968.12312. [DOI] [PubMed] [Google Scholar]
- 27.Yang S, Lv Y, Liu X, Wang Y, Wang F, Xiao X, Zhang Y, Fan Q, Yang Z, Boon N, Wang F, Xiao X, Zhang Y. 2020. Genomic and enzymatic evidence of acetogenesis by anaerobic methanotrophic archaea. Nat Commun 11:3941. doi: 10.1038/s41467-020-17860-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding J, Fu L, Ding Z-W, Lu Y-Z, Cheng SH, Zeng RJ. 2016. Experimental evaluation of the metabolic reversibility of ANME-2d between anaerobic methane oxidation and methanogenesis. Appl Microbiol Biotechnol 100:6481–6490. doi: 10.1007/s00253-016-7475-y. [DOI] [PubMed] [Google Scholar]
- 29.Bertram S, Blumenberg M, Michaelis W, Siegert M, Krüger M, Seifert R. 2013. Methanogenic capabilities of ANME-archaea deduced from 13 C-labelling approaches. Environ Microbiol 15:2384–2393. doi: 10.1111/1462-2920.12112. [DOI] [PubMed] [Google Scholar]
- 30.Treude T, Orphan V, Knittel K, Gieseke A, House CH, Boetius A. 2007. Consumption of methane and CO2 by methanotrophic microbial mats from gas seeps of the anoxic Black Sea. Appl Environ Microbiol 73:2271–2283. doi: 10.1128/AEM.02685-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lloyd KG, Alperin MJ, Teske A. 2011. Environmental evidence for net methane production and oxidation in putative anaerobic methanotrophic (ANME) archaea. Environ Microbiol 13:2548–2564. doi: 10.1111/j.1462-2920.2011.02526.x. [DOI] [PubMed] [Google Scholar]
- 32.Kevorkian RT, Callahan S, Winstead R, Lloyd KG. 2021. ANME-1 archaea may drive methane accumulation and removal in estuarine sediments. Environ Microbiol Rep 13:185–194. doi: 10.1111/1758-2229.12926. [DOI] [PubMed] [Google Scholar]
- 33.Schoelmerich MC, Ouboter HT, Sachdeva R, Penev PI, Amano Y, West-Roberts J, Welte CU, Banfield JF. 2022. A widespread group of large plasmids in methanotrophic Methanoperedens archaea. Nat Commun 13:7085. doi: 10.1038/s41467-022-34588-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bräsen C, Urbanke C, Schönheit P. 2005. A novel octameric AMP-forming acetyl-CoA synthetase from the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. FEBS Lett 579:477–482. doi: 10.1016/j.febslet.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 35.Fuchs G, Stupperich E, Thauer RK. 1978. Acetate assimilation and the synthesis of alanine, aspartate and glutamate in Methanobacterium thermoautotrophicum. Arch Microbiol 117:61–66. doi: 10.1007/BF00689352. [DOI] [PubMed] [Google Scholar]
- 36.Jetten MSM, Stams AJM, Zehnder AJB. 1990. Acetate threshold values and acetate activating enzymes in methanogenic bacteria. FEMS Microbiol Ecol 73:339–344. doi: 10.1111/j.1574-6968.1990.tb03958.x. [DOI] [Google Scholar]
- 37.Bräsen C, Schönheit P. 2004. Regulation of acetate and acetyl-CoA converting enzymes during growth on acetate and/or glucose in the halophilic archaeon Haloarcula marismortui. FEMS Microbiol Lett 241:21–26. doi: 10.1016/j.femsle.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 38.Dolfing J, Mulder JW. 1985. Comparison of methane production rate and coenzyme F420 content of methanogenic consortia in anaerobic granular sludge. Appl Environ Microbiol 49:1142–1145. doi: 10.1128/aem.49.5.1142-1145.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang F-P, Zhang Y, Chen Y, He Y, Qi J, Hinrichs K-U, Zhang X-X, Xiao X, Boon N. 2014. Methanotrophic archaea possessing diverging methane-oxidizing and electron-transporting pathways. ISME J 8:1069–1078. doi: 10.1038/ismej.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berger S, Frank J, Dalcin Martins P, Jetten MSM, Welte CU. 2017. High-quality draft genome sequence of “Candidatus Methanoperedens sp.” strain BLZ2, a nitrate-reducing anaerobic methane-oxidizing archaeon enriched in an anoxic bioreactor. Genome Announc 5:e01159-17. doi: 10.1128/genomeA.01159-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJCT, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJM, Janssen-Megens EM, Francoijs K-J, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 43.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 44.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 45.Jones ME, Lipmann F. 1955. Aceto-CoA-kinase. Methods Enzymol 1:585–595. doi: 10.1016/0076-6879(55)01101-4. [DOI] [Google Scholar]
- 46.Brown TDK, Jones-Mortimer MC, Kornberg HL. 1977. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J Gen Microbiol 102:327–336. doi: 10.1099/00221287-102-2-327. [DOI] [PubMed] [Google Scholar]
- 47.Takai K, Horikoshi K. 2000. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol 66:5066–5072. doi: 10.1128/AEM.66.11.5066-5072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumari S, Tishel R, Eisenbach M, Wolfe AJ. 1995. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol 177:2878–2886. doi: 10.1128/jb.177.10.2878-2886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li R, Gu J, Chen P, Zhang Z, Deng J, Zhang X. 2011. Purification and characterization of the acetyl-CoA synthetase from Mycobacterium tuberculosis. Acta Biochim Biophys Sin (Shanghai) 43:891–899. doi: 10.1093/abbs/gmr076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.00367-23-s0001.docx, DOCX file, 0.8 MB (850.1KB, docx)
Data Availability Statement
The raw reads obtained from 16S rRNA amplicon sequencing were deposited in the NCBI database under BioProject accession number PRJNA924223.