ABSTRACT
Genetic engineering of hyperthermophilic organisms for the production of fuels and other useful chemicals is an emerging biotechnological opportunity. In particular, for volatile organic compounds such as ethanol, fermentation at high temperatures could allow for straightforward separation by direct distillation. Currently, the upper growth temperature limit for native ethanol producers is 72°C in the bacterium Thermoanaerobacter ethanolicus JW200, and the highest temperature for heterologously-engineered bioethanol production was recently demonstrated at 85°C in the archaeon Pyrococcus furiosus. Here, we describe an engineered strain of P. furiosus that synthesizes ethanol at 95°C, utilizing a homologously-expressed native alcohol dehydrogenase, termed AdhF. Ethanol biosynthesis was compared at 75°C and 95°C with various engineered strains. At lower temperatures, the acetaldehyde substrate for AdhF is most likely produced from acetate by aldehyde ferredoxin oxidoreductase (AOR). At higher temperatures, the effect of AOR on ethanol production is negligible, suggesting that acetaldehyde is produced by pyruvate ferredoxin oxidoreductase (POR) via oxidative decarboxylation of pyruvate, a reaction known to occur only at higher temperatures. Heterologous expression of a carbon monoxide dehydrogenase complex in the AdhF overexpression strain enabled it to use CO as a source of energy, leading to increased ethanol production. A genome reconstruction model for P. furiosus was developed to guide metabolic engineering strategies and understand outcomes. This work opens the door to the potential for ‘bioreactive distillation’ since fermentation can be performed well above the normal boiling point of ethanol.
IMPORTANCE Previously, the highest temperature for biological ethanol production was 85°C. Here, we have engineered ethanol production at 95°C by the hyperthermophilic archaeon Pyrococcus furiosus. Using mutant strains, we showed that ethanol production occurs by different pathways at 75°C and 95°C. In addition, by heterologous expression of a carbon monoxide dehydrogenase complex, ethanol production by this organism was driven by the oxidation of carbon monoxide. A genome reconstruction model for P. furiosus was developed to guide metabolic engineering strategies and understand outcomes.
KEYWORDS: Pyrococcus furiosus, archaea, thermophile, ethanol, genetic engineering, CO dehydrogenase, bioethanol, carbon monoxide, hyperthermophiles, metabolic engineering, metabolic modeling
INTRODUCTION
The use of extremely thermophilic microorganisms is becoming an increasingly attractive option for biological production of fuels and chemicals, and improved genetic systems in thermophiles have expanded the possibilities in recent years (1–4). In particular, organisms growing at temperatures of 70°C to 100°C have distinct advantages over mesophiles and moderate thermophiles for industrial applications: cultivation at high temperatures makes them less prone to contamination and limits phage infection, volatile fuels and chemical products can be continuously distilled from culture, and less cooling is needed for a large-scale fermenter, leading to reduced cooling costs (1, 4–6). With that said, metabolic engineering of extreme thermophiles for industrial fuels and chemicals is still in its beginning stages. However, with the expansion of molecular genetic tools, a growing number of these organisms hold promise for industrial biotechnology.
One such organism that is being developed as a platform for high-temperature metabolic engineering applications is the marine archaeon Pyrococcus furiosus, which grows optimally near 100°C. As a heterotrophic organism with a doubling time (37 min [7]) approaching that of Escherichia coli and for which a robust genetic system has been developed (7–9), it is a prime candidate for genetic engineering applications. Current engineering approaches in this organism have required the use of a temperature shift strategy for expression of heterologous pathways and synthesis of the resulting products because most pathway enzymes come from organisms which have optimum growth temperatures lower than that of the host. Engineering efforts in P. furiosus for product formation have included lactate at 72°C using a bacterial thermophilic lactate dehydrogenase gene (10), ethanol at 78°C using a bacterial alcohol dehydrogenase gene (AdhA) (10, 11), 3-hydroxypriopionate at 73°C using a five-gene archaeal pathway (12), and butanol at 60°C using a combination of genes from three bacterial sources (13). It has also been engineered for utilization of formate at temperatures up to 95°C (14) and for utilization of CO at 80°C (15) via expression of multi-subunit membrane-bound complexes from a related archaeon.
Engineered ethanol production above 70°C has been accomplished in only two organisms to date. The cellulose-degrading thermophilic bacterium Caldicellulosiruptor bescii has been engineered with a bifunctional AdhE enzyme containing both acetaldehyde dehydrogenase and an alcohol dehydrogenase domains for the conversion of acetyl-CoA to ethanol; this engineered strain, also containing a deletion of the lactate dehydrogenase gene, produced up to 2.3 mM ethanol at 75°C (16). P. furiosus has been engineered to produce ethanol, up to 78°C, via insertion of a single alcohol dehydrogenase gene from Thermoanaerobacter strain X514 (11, 17). Intriguingly, this AdhATX514 enzyme was found to act in concert with the native aldehyde ferredoxin oxidoreductase (AOR) for the production of ethanol from acetate in a redox-balanced energy-conserving pathway. The ferredoxin-dependent AOR enzyme was previously demonstrated to oxidize acetaldehyde to acetate and thought to detoxify aldehyde buildup in the cell (18, 19). In the AOR-Adh pathway, however, AOR functions in reverse, converting acetate to acetaldehyde, which is used by AdhATX514 for ethanol production. Not only could the pathway convert native acetate to ethanol, but the broad specificity of the two enzymes allowed exogenously added organic acids, such as propionate and isobutyrate, to be converted to their corresponding alcohol (17). Furthermore, engineering a CO dehydrogenase complex into the AdhA-containing strain allowed CO to supply the reductant for organic acid conversion (17). P. furiosus was also engineered to contain a primary alcohol dehydrogenase from the thermophile Caldanaerobacter subterraneus which enabled ethanol production, presumably through the AOR-Adh pathway, optimally at 80°C and at a maximum of 85°C (20).
The bifunctional AdhE enzyme catalyzes ethanol production from acetyl-CoA, and P. furiosus has also been engineered with AdhE enzymes in an aor deletion background to eliminate competition from the AOR-Adh pathway (21). However, this ethanol production was an order of magnitude lower than that of the AOR-Adh pathway, and the temperature of ethanol formation was limited to 65°C. Of the eight AdhE enzymes tested, only two, from Thermoanaerobacter species, were functional in P. furiosus.
P. furiosus produces a small amount (~1 mM) of ethanol natively during growth at lower temperatures (e.g., 65°C to 75°C) (17, 21). Another study also reported the production of ethanol at 90°C in concentrated cell suspensions, using NMR studies with 13C-labeled glucose (22). Interestingly, a strain in which the AOR gene had been deleted still produced background levels of ethanol at 65°C, indicating that AOR is not the only source for native acetaldehyde production in P. furiosus (21). The native pyruvate ferredoxin oxidoreductase enzyme (POR), which catalyzes the ferredoxin-dependent conversion of pyruvate to acetyl-CoA, non-oxidatively decarboxylates pyruvate to acetaldehyde in vitro (23, 24). Additional potential sources of acetaldehyde include other members of the tungsten-containing AOR family, such as formaldehyde ferredoxin oxidoreductase (FOR) (25), and two other partially characterized tungsten-containing oxidoreductases (WOR4 and WOR5) (26, 27).
In P. furiosus, native ethanol production likely comes from acetaldehyde via one or more native alcohol dehydrogenase enzymes, some of which have been biochemically characterized. The short-chain alcohol dehydrogenase termed AdhA (PF0074) was heterologously-expressed in E. coli and characterized with a substrate specificity for pyruvaldehyde (specific activity: 30.2 U mg−1) in the reductive reaction and 2-pentanol (Vmax: 41.6 U mg−1) in the oxidative reaction (28). The same study identified an iron-containing AdhB (PF0075) encoded adjacent to the AdhA gene; however, the heterologously expressed protein was not biochemically characterized due to its rapid loss of activity (28). Interestingly, a Thermotoga neapolitana protein highly homologous to AdhB (74% identity over the entire sequence) was characterized as a bifunctional Adh, leading to the possibility that P. furiosus AdhB may also be bifunctional (29). A mid-chain, zinc-containing alcohol dehydrogenase, AdhC (PF0991), heterologously expressed and purified from E. coli, was demonstrated to oxidize 2,3-butanediol to acetoin (specific activity: 5 U mg−1) (30). Another alcohol dehydrogenase homolog, termed AdhD (PF1960), was heterologously expressed in E. coli and characterized with a substrate specificity for diacetyl-acetoin (Vmax: 22.5 U mg−1) in the reductive reaction and 2,3-butanediol (Vmax: 108.3 U mg−1) in the oxidative reaction (31). Interestingly, this AdhD is transcribed immediately upstream of, and is likely in an operon with WOR4 (PF1961). There was also a report of a characterized alcohol dehydrogenase from P. furiosus before its genome was sequenced; however, it is not easily identifiable because the 14-amino acid N-terminal sequence determined for the purified protein does not correspond to any gene in the P. furiosus genome (32).
Here, we describe a previously unstudied P. furiosus Adh that is responsible for native ethanol production and suggest the mechanisms by which ethanol is produced at suboptimal versus optimal growth temperatures. Furthermore, its homologous overexpression allows for increased ethanol production at 95°C in P. furiosus. Additionally, heterologous expression of a CO dehydrogenase in this strain was shown to enable CO to serve as a source of energy for ethanol biosynthesis. Genome-scale modeling revealed an important role for redox balancing ferredoxin in controlling the mechanism of ethanol production at different temperatures and in the presence of CO dehydrogenase.
RESULTS AND DISCUSSION
P. furiosus Adh gene expression.
Since P. furiosus natively produces low concentrations (~1 mM) of ethanol at lower growth temperatures (around 75°C), we were interested in determining the expression levels of the five putative adh genes. P. furiosus COM1 was cultivated at both 95°C and 75°C, RNA was extracted, and quantitative RT-PCR was performed for adhA, adhB, adhC, and adhD. PF0608 was also included because it is another annotated, although uncharacterized, iron-containing alcohol dehydrogenase (Fig. 1). Expression levels of adhA and adhB transcripts were significantly lower at 75°C, approaching 10-fold lower than that of the control gene, while the expression of adhC, adhD, and PF0608 was similar to that of the control gene. Since AdhC and AdhD have characterized activities with 2,3-butanediol and acetoin, respectively (30, 31), we selected PF0608 to pursue as a potential Adh with ethanol-producing activity. Here, we refer to the PF0608 gene product as AdhF to keep to the naming sequence of previous P. furiosus Adh-type enzymes and avoid confusion with the well-characterized bacterial bifunctional AdhE.
FIG 1.
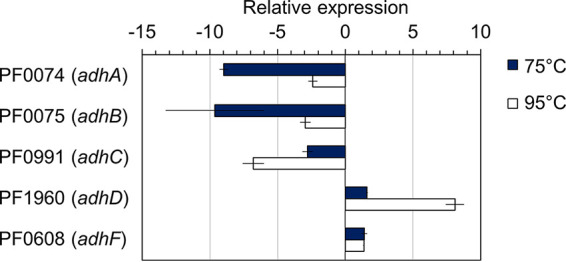
Expression levels of Pyrococcus furiosus Adh genes. P. furiosus control strain (MW004) was grown in minimal maltose medium at 75°C and 95°C to mid-log phase. Expression of the constitutive gene PF0983 encoding the DNA polymerase sliding clamp was used as an internal standard to calculate relative expression. Error bars represent standard deviation (SD), n = 2 biological replicates.
Deletion of adhF abolishes ethanol production.
To determine whether AdhF plays a role in ethanol production in P. furiosus, we generated adhF deletions in both the COM1 and Δaor backgrounds; here, these strains are referred to as ΔAdhF and ΔAOR ΔAdhF, respectively (Fig. 2 and Table 1). Previous work has demonstrated native production of up to ~1 mM ethanol by P. furiosus at 65°C and 75°C (17, 21). A Δaor strain was also shown to produce similar amounts of ethanol at 65°C (21). The ΔAdhF and ΔAOR ΔAdhF strains were cultured at 75°C and 95°C in minimal medium containing 5 g L−1 maltose and 0.5 g L−1 yeast extract and compared to the control and ΔAOR strains (Fig. 3). At 95°C, the growth of all strains was comparable, and at 75°C, the ΔAOR ΔAdhF strain appeared to perform better than the other strains. At 75°C, ethanol production by the ΔAdhF strains after 96 h of growth was <0.2 mM. Also, ΔAOR produced slightly more ethanol than COM1 (0.4 versus 0.2 mM, respectively), consistent with previous observations at 65°C (21). Interestingly, at 95°C, both the control and ΔAOR strains produced more ethanol, particularly after the cultures reached stationary phase, with the ΔAOR strain again producing more ethanol than the control (0.8 versus 0.6 mM, respectively). At 95°C, neither of the ΔAdhF strains produced ethanol, confirming the role of AdhF in native ethanol production in P. furiosus. The observation that the ΔAOR strain produced more ethanol than the control suggests that AOR is not involved in native ethanol production, as also previously proposed (21). In the hybrid synthetic AOR-Adh pathway developed previously, AOR drives ethanol production via a heterologously expressed Adh operating at 75°C (17).
FIG 2.
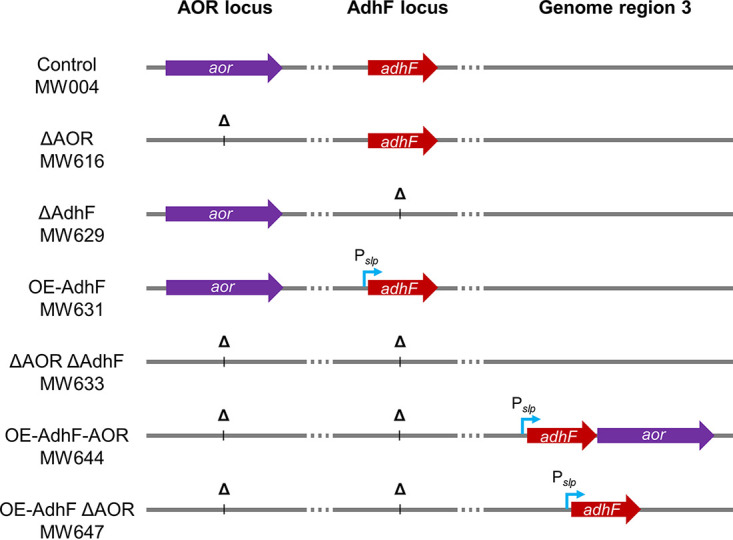
Strain diagrams. Strains are listed on the left with loci of interest labeled at the top. aor and adhF genes are indicated by purple and red arrows, respectively. Overexpression promoter Pslp is represented by a bent blue arrow. Deleted loci are represented by Δ.
TABLE 1.
Strains used and constructed in this study
| Strain | Alias | Parent | Genotype/description | Source |
|---|---|---|---|---|
| MW002 | COM1 | DSM 3638 | ΔpyrF | 9 |
| MW004 | Control | MW002 | ΔpyrF::pyrF | 43 |
| MW616 | ΔAOR | MW002 | ΔpyrF Δaor::Pgdh-pyrF | 21 |
| MW252 | - | MW616 | ΔpyrF Δaor | 21 |
| MW629 | ΔAdhF | MW002 | ΔpyrF ΔadhF::Pgdh-pyrF | This work |
| MW631 | OE-AdhF | MW002 | ΔpyrF PslpadhF::Pgdh-pyrF | This work |
| MW633 | ΔAOR ΔAdhF | MW252 | ΔpyrF Δaor ΔadhF::Pgdh-pyrF | This work |
| MW641 | - | MW633 | ΔpyrF Δaor ΔadhF | This work |
| MW644 | OE-AdhF-AOR | MW641 | ΔpyrF Δaor ΔadhF Pslp-adhF-aor::Pgdh-pyrF | This work |
| MW647 | OE-AdhF ΔAOR | MW641 | ΔpyrF Δaor ΔadhF Pslp-adhF::Pgdh-pyrF | This work |
| MW191 | CODH | MW002 | ΔpyrF Pmbh-TonCODH::Pgdh-pyrF | 15 |
| MW304 | - | MW191 | ΔpyrF Pmbh-TonCODH | This work |
| MW650 | CODH-OE-AdhF | MW304 | ΔpyrF Pmbh-TonCODH Pslp-adhF::Pgdh-pyrF | This work |
FIG 3.
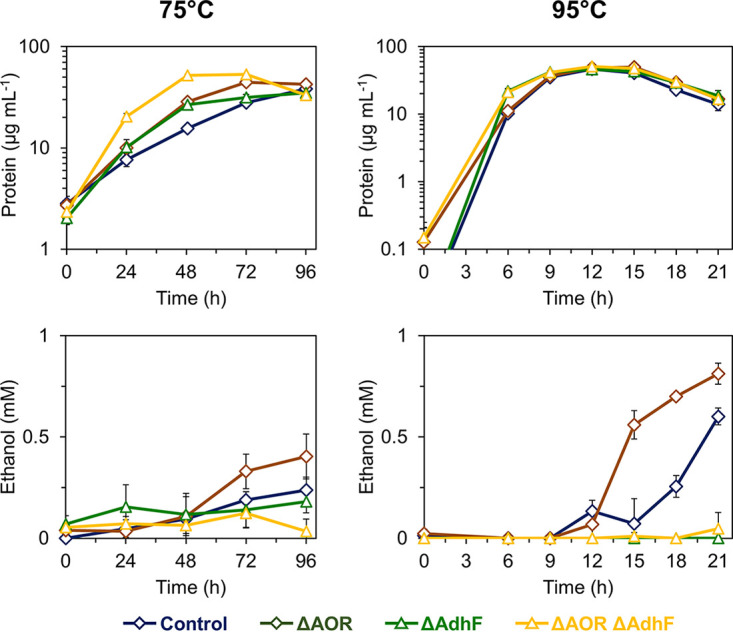
Effect of adhF deletion on native ethanol production at 75°C and 95°C. Growth (top panels) and ethanol production (bottom panels) of control (dark blue, diamonds), ΔAOR (brown, diamonds), ΔAdhF (green, triangles) and ΔAOR ΔAdhF (yellow, triangles) strains grown in minimal medium (containing 5 g L−1 maltose and 0.5 g L−1 yeast extract) without shaking at 75°C (left panels) and 95°C (right panels). Error bars represent SD, n = 3 biological replicates.
Overexpression of AdhF increases ethanol production at 75°C and 95°C.
To dissect the role of AdhF in ethanol production in P. furiosus, we constructed adhF overexpression strains (Fig. 2 and Table 1). For adhF overexpression, the high-level slp promoter was inserted in front of the gene at its native location (strain OE-AdhF). Two additional adhF overexpression strains were generated in a ΔadhF Δaor background, one which overexpressed adhF only (strain OE-AdhF ΔAOR) and a second which overexpressed both adhF and aor (strain OE-AdhF-AOR), both at a separate genome region from the ΔadhF locus. These strains were cultured in minimal medium at both 75°C and 95°C to monitor growth and metabolite production (Fig. 4). While the endpoint growth of these strains at both temperatures was comparable to the control (shown in Fig. 3), each of the three strains had a significant increase in ethanol production. At 95°C, all three strains produced ethanol at similar levels, with a maximum of around 2 mM. At 75°C, the OE-AdhF-AOR strain also produced ~2 mM ethanol after 96 h of growth, while the OE-AdhF and OE-AdhF ΔAOR strains produced a maximum of ~0.75 mM. These results indicate a clear difference in how ethanol is produced at low and high temperatures. At 75°C, overexpression of AOR along with AdhF appears to enhance ethanol production. However, at 95°C, AOR appears to play no significant role in ethanol production, since the same levels of ethanol are produced in its presence and absence. Correspondingly, ethanol:acetate ratios were highest for OE-AdhF-AOR at 75°C, approaching 0.6 at 96 h of growth, while at 95°C, the ratios for the three overexpression strains are similar (0.3 to 0.4) (Fig. S2 in the supplemental material). At 95°C, the yield of ethanol was approximately 0.26 mol per mol of maltose consumed (Fig. S3). Therefore, ethanol production in P. furiosus follows different metabolic routes depending on the temperature.
FIG 4.
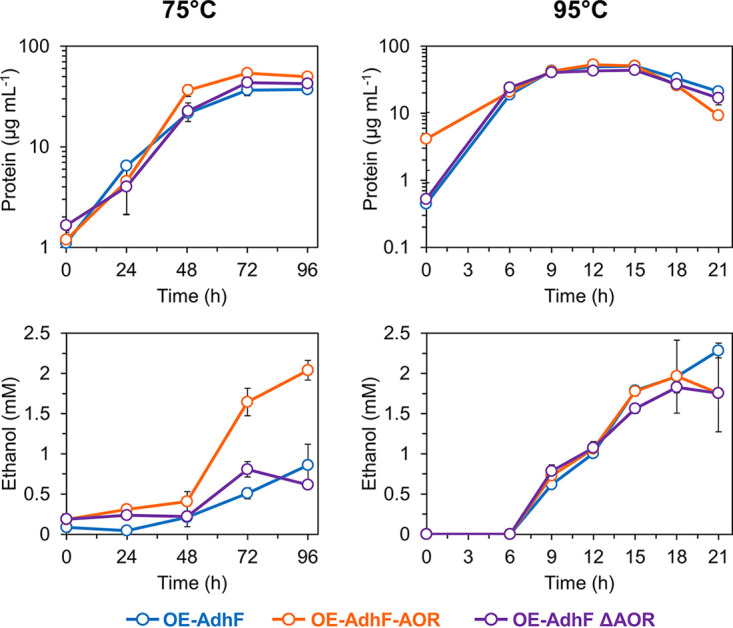
Effect of adhF overexpression on ethanol production at 75°C and 95°C. Growth (top panels) and ethanol production (bottom panels) of OE-AdhF (light blue, circles), OE-AdhF-AOR (orange, circles), and OE-AdhF ΔAOR (purple, circles) strains grown in minimal medium (containing 5 g L−1 maltose and 0.5 g L−1 yeast extract) without shaking at 75°C (left panels) and 95°C (right panels). Error bars represent SD, n = 3 biological replicates.
These results show that native ethanol production at 95°C is most likely mediated by AdhF, and it does not depend on the presence of AOR (Fig. 3 and 4). In fact, the absence of AOR leads to a slight increase in ethanol production since acetaldehyde is not being detoxified by AOR (Fig. 3). The acetaldehyde intermediate most likely comes from a side reaction of POR via oxidative decarboxylation of pyruvate (23, 24), as illustrated in Fig. 5. Conversely, overexpression of adhF along with aor changes the dynamics of this metabolism. At 75°C, when AOR is overexpressed, it becomes the major player in supplying acetaldehyde, presumably because the lower specific activity of POR at that temperature cannot keep up with the demand. POR has an optimal temperature of ≥90°C in vitro, but at 75°C, its specific activity drops to approximately 25% of that at 95°C (23). Also, labeling experiments performed with a P. furiosus strain expressing AdhATX514 at 72°C showed that 13C-labeled acetate was converted to 13C-labeled ethanol, confirming the role of AOR in engineered ethanol production at suboptimal growth temperatures (17). At 95°C, however, the absence of AOR has little if any effect on ethanol production by AdhF (Fig. 4). Therefore, the acetaldehyde intermediate used by AdhF at 95°C is most likely supplied by POR. In addition, the RNA expression level of porA at 95°C is at least 30-fold higher than that of aor and other genes representing oxidoreductase enzymes with the potential to supply acetaldehyde (vorA, iorA, kgor, for, wor4, and wor5; see Fig. S4). The putative bifunctional AdhB enzyme may also provide acetaldehyde, given its high homology to a recently characterized bifunctional alcohol dehydrogenase in Thermotoga neapolitana (29).
FIG 5.
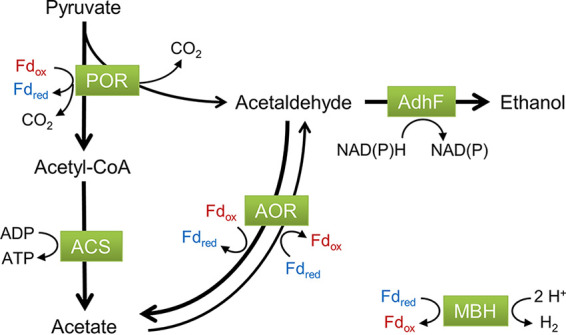
Model for native ethanol production in P. furiosus. Native ethanol is produced predominantly through two routes in P. furiosus, depending on the growth temperature. At 95°C, the acetaldehyde intermediate is generated by pyruvate ferredoxin oxidoreductase (POR) from pyruvate via an oxidative decarboxylation side-reaction, and at 75°C, it is generated predominantly by AOR from acetate. ACS, acetyl-CoA synthetase I; AOR, aldehyde ferredoxin oxidoreductase; AdhF, alcohol dehydrogenase F; MBH, membrane-bound hydrogenase; Fd, ferredoxin.
Improving growth and ethanol production at 95°C.
The growth experiments described above were performed using a minimal medium; therefore, to improve ethanol yields, we used a rich medium containing 5 g L−1 maltose and 5 g L−1 yeast extract. In closed bottle cultures at 95°C, the use of rich medium increased ethanol production to 5 to 6 mM for the three AdhF overexpression strains (Fig. S5). There was still little difference in ethanol production among these, with OE-AdhF, OE-AdhF-AOR, and OE-AdhF ΔAOR forming 5.8, 5.5, and 5.0 mM ethanol, respectively, after 21 h of growth. The ΔAdhF strain still produced no ethanol after 21 h. However, the ΔAOR ΔAdhF strain produced almost 1 mM, indicating that one or more of the other alcohol dehydrogenases must also contribute to ethanol production when sufficient acetaldehyde is available (i.e., in the absence of AOR).
Ethanol production was further increased when the OE-AdhF strain was grown in rich medium in a custom 20-L stainless steel fermenter with gas flow and pH control (Fig. 6). Using this setup, ethanol production reached 12 mM at 21 h, with an ethanol:acetate ratio of 0.9. It is worth noting that some ethanol is not accounted for in these measurements from the growth medium because the high growth temperature and continuous sparging of the vessel likely led to the loss of ethanol by evaporation. Growth began to decline after 15 h due to acetate accumulation, as shown by the decline in the concentration of soluble protein (Fig. 6).
FIG 6.
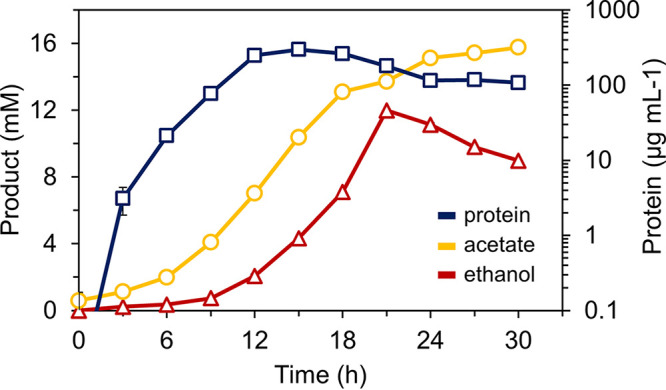
Fermenter growth of strain OE-AdhF at 95°C. Growth of the OE-AdhF strain in rich medium (containing 5 g L−1 maltose and 5 g L−1 yeast extract) in a 20-L custom fermenter with pH control, sparged with N2:CO2 (80:20) showing ethanol (red triangles) and acetate (yellow circles) production on the left axis. Growth as measured by total protein (blue squares) is plotted on the right y axis.
Effect of CODH on growth and ethanol production by AdhF.
Pyruvate via POR is thought to supply most, if not all, of the acetaldehyde for ethanol production at 95°C. Thus, ethanol production from this point after glycolysis will result in a net loss of ATP. For each ethanol molecule produced, one less molecule of acetyl-CoA is converted to acetate by acetyl-CoA synthetase, and consequently one less ATP is generated from ADP (Fig. 5). Heterologous expression of the CO-utilizing CODH (CO dehydrogenase) complex from the carboxydotrophic thermophile Thermococcus onnurineus enables P. furiosus to utilize CO as an energy source for growth (15). The CODH complex is encoded by a 16-gene cluster and includes a CO dehydrogenase, a membrane-bound [NiFe] hydrogenase, and a Na+/H+ antiporter. This complex oxidizes CO to CO2 coupled to H2 production, while also generating a sodium ion gradient that can drive ATP synthesis via the native sodium-dependent ATP synthase. When added to a P. furiosus strain with the engineered AOR-AdhA pathway, this CODH complex increased alcohol production from organic acids (17).
We explored whether addition of the CODH complex with overexpression of adhF would increase ethanol production. The AdhF expression cassette was inserted into a strain expressing the T. onnurineus CODH, here referred as strain CODH OE-AdhF (Table 1 and Fig. S6). Growth analyses of the CODH OE-AdhF and OE-AdhF strains were performed in serum bottles at 90°C (due to the stability of T. onnurineus CODH) (15) using rich medium in the presence or absence of 100% CO in the headspace, with agitation to disperse CO into the culture. Under these growth conditions, the CODH-containing strain showed improved growth and ethanol production in the presence of CO. With CO, the CODH OE-AdhF strain reached a maximum of 5.8 mM ethanol after 15 h of growth, compared to 3.5 mM in strain OE-AdhF (Fig. 7). Ethanol begins to accumulate near the beginning of stationary-phase growth (9 h, see Fig. S7), likely due to excess pyruvate production. At 15 h, the CODH-containing strain had consumed all the CO and produced twice as much H2 compared to the OE-AdhF strain (67 mM versus 28 mM, Fig. S7). Previous work suggests that the CODH in P. furiosus produces CO2 in amounts equivalent to the CO that is utilized (15). Pressure buildup from excess H2 and CO2 could have contributed to the decrease in both ethanol and H2 during headspace sampling for time points after 15 h. Also, depending on the balance of redox carriers within the cell, ethanol consumption could also account for the slow decrease of ethanol during the stationary phase. The activity of CODH with CO not only provided extra energy for growth, but also increased the ethanol:acetate ratio: up to 1.4 in the CODH OE-AdhF strain compared to <0.5 for the OE-AdhF strain (Fig. S7).
FIG 7.
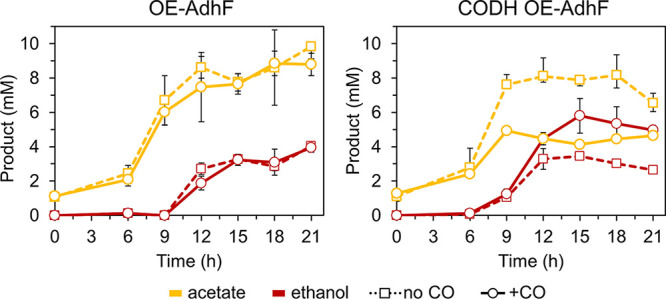
Addition of CO dehydrogenase (CODH) with CO improves ethanol production by AdhF in shaking cultures. Ethanol (red) and acetate (yellow) production of OE-AdhF (left) and CODH OE-AdhF (right) strains grown in rich medium (containing 5 g L−1 maltose and 5 g L−1 yeast extract) with shaking at 90°C, with CO (solid lines, circles) or without CO (dashed lines, squares) in the headspace. Error bars represent SD, n = 3 biological replicates.
Ethanol production in P. furiosus is linked to the production and uptake of H2. P. furiosus contains a membrane-bound hydrogenase (MBH), which generates H2 and recycles reduced ferredoxin generated during glycolysis (33). In static, closed cultures, the H2 produced by MBH diffuses slowly to the gas phase, but agitation of the culture during growth causes faster mass transfer of the evolved H2, allowing more efficient recycling of reduced ferredoxin by MBH. A steady supply of oxidized ferredoxin should favor the ferredoxin-dependent activity of POR over the oxidative decarboxylation side-reaction and also push AOR in the direction of acetaldehyde detoxification (Fig. 5). Therefore, agitation of the culture effectively decreases ethanol output by keeping the pool of oxidized ferredoxin high. Evidence of this effect can be seen when comparing the OE-AdhF strain grown statically (Fig. S5A) to that grown in agitation (Fig. 7), where the strain reached a maximum of 5.8 mM ethanol compared to 4.0 mM, respectively. When the CODH complex was added to the strain, however, H2 output doubled, increasing hydrogen partial pressure and reducing the efficiency of MBH at recycling ferredoxin. An insufficient pool of oxidized ferredoxin should increase the side-reaction of POR to produce acetaldehyde, resulting in improved ethanol production. Therefore, addition of the CODH with CO in the OE-AdhF strain prevents ethanol production from diminishing due to culture agitation. This is an important factor to consider because agitation or stirring is a necessity when scaling up from bottles to a fermentor.
P. furiosus also has two soluble hydrogenases (SHI and SHII), one of which is thought to be involved in H2 uptake for NADPH biosynthesis (33). If AdhF is NADPH-dependent, then increased H2 supplied via CODH could also increase NADPH availability for ethanol synthesis. It is not known whether CODH or its associated MBH can utilize P. furiosus ferredoxin, as has been suggested (17); however, if so, it could contribute to not only the balance of the intracellular NADP+/NADPH pools but also those of reduced and oxidized ferredoxin.
Simulation of redox-driven responses to temperature shift and CODH expression.
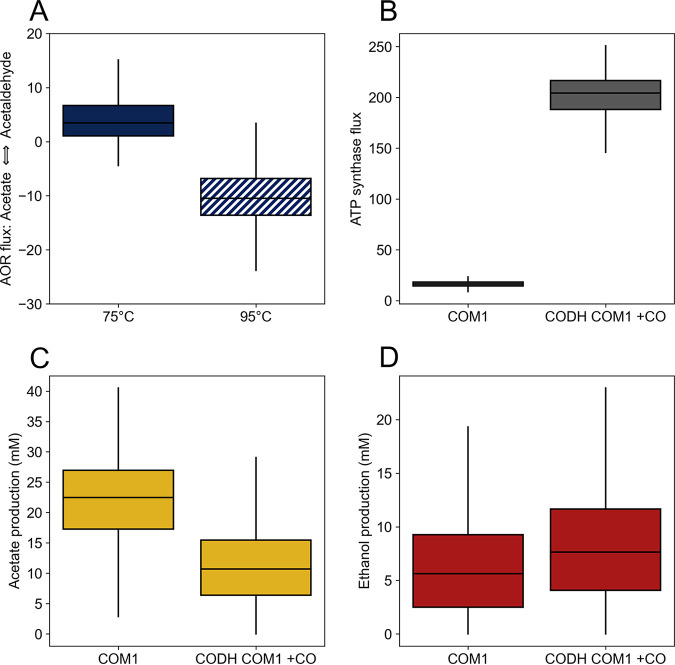
The newly developed genome-scale model of P. furiosus (34) was used to investigate redox balance in response to temperature shifts and CODH expression. We hypothesized that the temperature-dependent activity of POR would cause a shift in the average direction of AOR from acetaldehyde-producing at 75°C to acetaldehyde-consuming at 95°C. POR is most active above 90°C, with the acetaldehyde-producing decarboxylation reaction generally producing less than the oxidative acetyl-CoA-producing reaction but becoming proportionally more active at higher temperatures (24). Accordingly, we approximated the two temperature conditions by constraining the acetaldehyde-producing reaction to be less than one-third of the flux of the oxidative reaction at 75°C, and greater than or equal to the flux of the oxidative reaction at 95°C. A flux sampling approach was used to measure the effect of these constraints on the overall distribution of fluxes in the model. We observed a median AOR flux of 3.26 (acetaldehyde-producing) in the 75°C condition and −10.50 (acetaldehyde-consuming) in the 95°C condition, with a significant difference between the two AOR flux distributions (P < 1e–10, effect size = −2.59) (Fig. 8A). Despite differences in the AOR flux distribution, the overall ferredoxin balance remained similar between the two temperatures. The flux of energy-conserving MBH, the major enzyme that oxidizes reduced ferredoxin, showed no significant change between the 75°C and 95°C conditions (P = 0.14). Therefore, it appears that AOR responds to changes in POR flux as a way to maintain a supply of reduced ferredoxin; when more flux is directed through the decarboxylation reaction of POR than the oxidative (oxidized ferredoxin-using) reaction, AOR is driven to consume acetaldehyde and produce acetate and reduced ferredoxin.
FIG 8.
Simulated flux distributions for key reactions. (A) AOR flux at 75°C and 95°C. (B to D) ATP synthase flux (B), acetate production (C), and ethanol production (D) with and without CODH and CO.
Given the importance of producing reduced ferredoxin and maintaining a high MBH flux in the model to maximize energy conservation, we hypothesized that expression of energy-conserving CODH would reduce acetate production by allowing MBH to maintain a lower flux. Sampling the solution space with and without the CODH reaction showed notable shifts in the product distributions and overall energy balance of the cell. When CODH was active, acetate production was greatly reduced from a median of 22.48 to 10.69 mM, while ethanol production saw a significant median increase from 5.62 to 7.64 mM (P < 1e–10, effect size = 0.33), consistent with the experimental results (Fig. 8C and D). CODH activity allowed the model to take up slightly less carbon on average while achieving a higher median biomass (Table S1). Additionally, the median flux of MBH was reduced from 79.25 to 67.49, and the fluxes of GAPOR and POR saw similar reductions (Table S1). When CODH was active, ATP synthase carried an extremely high median flux of 204.22 compared to 16.36 in the absence of CO oxidation (Fig. 8B).
Taken together, these results highlight the importance of MBH and ferredoxin redox balancing to ATP generation in P. furiosus. Without CODH, MBH drives the production of a sodium gradient and the recycling of reduced ferredox in that is generated through glycolysis (via GAPOR) and acetate production (via AOR and POR). AOR is also important for maintaining ferredoxin redox balance by MBH, and this partially explains the shift from AOR acting as the main acetaldehyde source at 75°C to POR becoming the acetaldehyde source at 95°C. When CODH is expressed, it becomes an additional source of energy conservation by generating a sodium gradient; this reduces dependence on both MBH activity and acetate production for ATP generation, allowing greater amounts of ethanol to be produced.
Conclusions.
There are many advantages to high-temperature fermentation, including reduced risk of contamination, lower fermenter cooling costs, and the potential to separate volatile products based on temperature (4). Currently, native ethanol producers are more readily metabolically engineered for high-yield ethanol production (35). P. furiosus natively produces a very small amount of ethanol (<1 mM), and we were able to identify the primary alcohol dehydrogenase responsible for ethanol production, AdhF. Overexpression of this gene resulted in an order-of-magnitude increase in ethanol production of up to 12 mM. While this is still low by industry standards, further modifications to the metabolism, such as overexpression of POR or deletion of genes encoding acetyl-CoA synthase (ACS), could potentially improve yield. Addition of the CODH complex to the strain provided an additional pathway to boost ATP synthesis and influence redox pools, allowing more pyruvate to be diverted for ethanol production (Fig. 5). This resulted in a 3-fold increase in the ethanol:acetate ratio compared to a strain without the CODH complex. The ease of genetic manipulation of P. furiosus coupled with a robust understanding of its metabolism makes it a useful model for metabolic engineering of high-temperature pathways, particularly for producing volatile products such as ethanol as demonstrated here.
MATERIALS AND METHODS
PCR product and plasmid construction.
For construction of PF0608 (AAL80732.1, here referred to as adhF) deletion strains, 0.5-kb flanking regions upstream and downstream of adhF were joined on either side of the Pgdh-pyrF pop-out marker cassette (12) by splice overlap extension (SOE) PCR (36). To insert the slp promoter (Pslp, consisting of 184 b of the promoter region immediately upstream of the S-layer protein gene, PF1399) in front of adhF at its locus, 0.5-kb flanking regions upstream and downstream of the first base of the adhF gene were joined on either side of the Pgdh-pyrF pop-out marker cassette, together with Pslp, by SOE PCR.
Expression constructs for insertion into Genome Region 3 were constructed by standard cloning. The slp promoter and T1 terminator sequence (5′-aatcttttttagcactttt, 19 b from the 3′ untranslated region of PF1722) were combined with either adhF alone or adhF and aor via SOE PCR and cloned into pGL020 (a plasmid essentially identical to pGL007 [12], but lacking a single point mutation at the SphI and AscI sites) to generate pGL117 and pGL118, respectively (Fig. S1). Resulting plasmids were confirmed by sequencing.
Strain construction.
Transformation of P. furiosus was performed essentially as previously described, using linearized plasmids or PCR products (9). The COM1 strain (MW002) was transformed with the adhF deletion PCR construct and the Pslp insertion PCR construct to generate strains ΔAdhF (MW629) and OE-AdhF (MW631), respectively. The MW252 strain (21) harboring a deletion of the aor gene was transformed with the AdhF deletion PCR construct to generate the double deletion strain ΔAOR ΔAdhF (MW633). To generate a markerless version of the ΔAOR ΔAdhF strain, loss of the Pgdh-pyrF marker cassette was selected for using 5-fluoroorotic acid (5-FOA), as previously described (9, 12). The resulting strain, MW641, was transformed with linearized pGL117 and pGL118 to generate strains OE-AdhF-AOR (MW644) and OE-AdhF ΔAOR (MW647), respectively. To generate an AdhF expression strain also containing T. onnurineus CODH complex, a markerless version of the CODH strain MW191 was obtained as described previously for MW641, and the resulting strain, MW304, was transformed with a PCR construct for insertion of Pslp at the adhF locus to generate strain CODH OE-AdhF (MW650). For strains constructed from PCR products, the altered loci were verified by sequencing. Strains constructed in this work are summarized in Table 1.
Growth of P. furiosus.
P. furiosus strains were cultured anaerobically in an artificial seawater-based medium containing, per L, 1× base salts (37), 1× trace minerals (37), 5 g maltose, 1× vitamins (9), 10 μM sodium tungstate, 0.25 μg resazurin, 0.5 g cysteine, 1 g sodium bicarbonate, and 1 mM potassium phosphate buffer, with pH adjusted to 6.8 prior to bottling. Yeast extract was added at a concentration of 0.5 or 5 g L−1 to make ‘minimal’ and ‘rich’ media, respectively. Media were aliquoted into serum bottles, and the headspace was replaced with argon after three cycles of vacuum and argon. For experiments with CODH-containing strains, sodium tungstate was reduced to 1 μM and headspace was replaced with 100% CO as indicated. Cultures were incubated at 95°C or 75°C with or without shaking at 150 rpm, as indicated. When necessary, cell counts were performed using a Petroff-Hausser counting chamber.
RNA extraction and quantitative RT-PCR.
Cultures of control strain MW004 were grown in duplicate at 95°C and 75°C in minimal maltose medium, and cells were harvested from 25 mL culture samples. RNA was extracted using a phenol:chloroform extraction method as previously described (38). Contaminating genomic DNA was digested using TURBO DNase (Ambion) after which RNA was purified by phenol:chloroform extraction and ethanol precipitation. The Affinity Script QPCR cDNA synthesis kit (Agilent) was used for cDNA synthesis with 1 μg purified RNA. The Brilliant III Ultra-Fast SYBR QPCR Master Mix (Agilent) was used for quantitative reverse transcription-PCR (RT-PCR) experiments with primers designed to amplify an ~150-b product within the target genes. The constitutively expressed PF0983 gene encoding the DNA polymerase sliding clamp was used as a reference.
Cell protein and metabolite analyses.
During growth, 1-mL samples were collected in 96-well deep-well plates using a syringe. Plates were centrifuged at 12°C for 25 min at 3,500 × g. A multichannel pipette was used to transfer 200 μL of medium supernatant to a 96-well polypropylene storage plate. The remaining medium was decanted and cell pellets were used for cell protein quantitation using the Bradford protein assay kit (Bio-Rad) as previously described (14). For metabolite analyses, 10 μL 88% formic acid was pipetted into each 200-μL spent medium sample in the 96-well plates. Acidified samples were analyzed for acetate and ethanol using GC-FID (Agilent 7890A Gas Chromatograph with a Flame Ionization Detector fitted with a Carbowax 20M column) and for maltose using high-performance liquid chromatography (Waters Alliance 2690 fitted with a Bio-Rad Aminex HPX-87H column and a Waters 410 RI detector).
Headspace gas analyses.
Headspace samples were collected using a pressure-lock syringe. H2 and CO were quantified using GC (Shimadzu GC8A with TCD detector, oven 70°C, injector/detector 120°C, Alltech Molecular Sieve column 5A 80/100). Samples from bottles of uninoculated medium bottles containing known amounts of H2 and CO were used as standards.
Metabolic modeling.
Metabolic fluxes were simulated using the genome-scale model of P. furiosus COM1 (34). The model was represented in YAML format following the PSAMM software package guidelines (39, 40). A medium file was defined based on the minimal maltose medium used in this study, with 5 g L−1 maltose as the carbon source and 0.5 g L−1 yeast extract added. The model was exported to sbml format for use with the COBRApy package (41) using the “sbmlexport” function in PSAMM. Flux samples were collected using the optGpSampler method (42) as implemented by the “sample” function in COBRApy. Whenever samples were collected, the total sample size was 2,500, split between 10 independent sampling runs of size 250. Additionally, constraints were applied to restrict the flux of GAPDH (glyceraldehyde-3-phosphate dehydrogenase; R01063 and R01061) to zero to remove unrealistic looping behavior. To emulate the temperature conditions, constraints were set as follows:
75°C: POR_acetaldehyde ≤ (1/3) × POR_acetyl-CoA
95°C: POR_acetaldehyde ≥ POR _acetyl-CoA
The added CODH reaction in the model was formulated as the following:
where [c] denotes compounds within the cytoplasm and [e] denotes extracellular compounds. When CODH was added, the model was allowed to freely take up carbon monoxide. CODH expression was simulated without any temperature-dependent constraints. The temperature constraints were applied to simulate AOR directionality. The CODH reaction was used to examine ethanol, acetate, and ATP synthase usage.
ACKNOWLEDGMENTS
We thank Gerrit Schut for helpful discussions and Farris Poole for technical assistance.
This material is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomic Science Program under award no. DE-SC0022191 and DE-SC0022192.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/AEM.00563-23.
Supplemental material is available online only.
Contributor Information
Michael W. W. Adams, Email: adamsm@uga.edu.
Isaac Cann, University of Illinois Urbana-Champaign.
REFERENCES
- 1.Crosby JR, Laemthong T, Lewis AM, Straub CT, Adams MWW, Kelly RM. 2019. Extreme thermophiles as emerging metabolic engineering platforms. Curr Opin Biotechnol 59:55–64. doi: 10.1016/j.copbio.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Straub CT, Counts JA, Nguyen DMN, Wu CH, Zeldes BM, Crosby JR, Conway JM, Otten JK, Lipscomb GL, Schut GJ, Adams MWW, Kelly RM. 2018. Biotechnology of extremely thermophilic archaea. FEMS Microbiol Rev 42:543–578. doi: 10.1093/femsre/fuy012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor MP, van Zyl L, Tuffin IM, Leak DJ, Cowan DA. 2011. Genetic tool development underpins recent advances in thermophilic whole-cell biocatalysts. Microb Biotechnol 4:438–448. doi: 10.1111/j.1751-7915.2010.00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeldes BM, Keller MW, Loder AJ, Straub CT, Adams MW, Kelly RM. 2015. Extremely thermophilic microorganisms as metabolic engineering platforms for production of fuels and industrial chemicals. Front Microbiol 6:1209. doi: 10.3389/fmicb.2015.01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keller M, Loder A, Basen M, Izquierdo J, Kelly RM, Adams MWW. 2014. Production of lignofuels and electrofuels by extremely thermophilic microbes. Biofuels 5:499–515. doi: 10.1080/17597269.2014.996729. [DOI] [Google Scholar]
- 6.Yun SH, Kwon SO, Park GW, Kim JY, Kang SG, Lee JH, Chung YH, Kim S, Choi JS, Kim SI. 2011. Proteome analysis of Thermococcus onnurineus NA1 reveals the expression of hydrogen gene cluster under carboxydotrophic growth. J Proteomics 74:1926–1933. doi: 10.1016/j.jprot.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Fiala G, Stetter KO. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch Microbiol 145:56–61. doi: 10.1007/BF00413027. [DOI] [Google Scholar]
- 8.Farkas J, Stirrett K, Lipscomb GL, Nixon W, Scott RA, Adams MW, Westpheling J. 2012. Recombinogenic properties of Pyrococcus furiosus strain COM1 enable rapid selection of targeted mutants. Appl Environ Microbiol 78:4669–4676. doi: 10.1128/AEM.00936-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipscomb GL, Stirrett K, Schut GJ, Yang F, Jenney FE Jr, Scott RA, Adams MW, Westpheling J. 2011. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl Environ Microbiol 77:2232–2238. doi: 10.1128/AEM.02624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basen M, Sun J, Adams MW. 2012. Engineering a hyperthermophilic archaeon for temperature-dependent product formation. mBio 3:e00053-12. doi: 10.1128/mBio.00053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen DM, Lipscomb GL, Schut GJ, Vaccaro BJ, Basen M, Kelly RM, Adams MW. 2016. Temperature-dependent acetoin production by Pyrococcus furiosus is catalyzed by a biosynthetic acetolactate synthase and its deletion improves ethanol production. Metab Eng 34:71–79. doi: 10.1016/j.ymben.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Keller MW, Schut GJ, Lipscomb GL, Menon AL, Iwuchukwu IJ, Leuko TT, Thorgersen MP, Nixon WJ, Hawkins AS, Kelly RM, Adams MW. 2013. Exploiting microbial hyperthermophilicity to produce an industrial chemical, using hydrogen and carbon dioxide. Proc Natl Acad Sci USA 110:5840–5845. doi: 10.1073/pnas.1222607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keller MW, Lipscomb GL, Loder AJ, Schut GJ, Kelly RM, Adams MW. 2015. A hybrid synthetic pathway for butanol production by a hyperthermophilic microbe. Metab Eng 27:101–106. doi: 10.1016/j.ymben.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Lipscomb GL, Schut GJ, Thorgersen MP, Nixon WJ, Kelly RM, Adams MW. 2014. Engineering hydrogen gas production from formate in a hyperthermophile by heterologous production of an 18-subunit membrane-bound complex. J Biol Chem 289:2873–2879. doi: 10.1074/jbc.M113.530725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schut GJ, Lipscomb GL, Nguyen DM, Kelly RM, Adams MW. 2016. Heterologous production of an energy-conserving carbon monoxide dehydrogenase complex in the hyperthermophile Pyrococcus furiosus. Front Microbiol 7:29. doi: 10.3389/fmicb.2016.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung D, Cha M, Snyder EN, Elkins JG, Guss AM, Westpheling J. 2015. Cellulosic ethanol production via consolidated bioprocessing at 75°C by engineered Caldicellulosiruptor bescii. Biotechnol Biofuels 8:163. doi: 10.1186/s13068-015-0346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basen M, Schut GJ, Nguyen DM, Lipscomb GL, Benn RA, Prybol CJ, Vaccaro BJ, Poole FL 2nd, Kelly RM, Adams MW. 2014. Single gene insertion drives bioalcohol production by a thermophilic archaeon. Proc Natl Acad Sci USA 111:17618–17623. doi: 10.1073/pnas.1413789111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heider J, Ma K, Adams MW. 1995. Purification, characterization, and metabolic function of tungsten-containing aldehyde ferredoxin oxidoreductase from the hyperthermophilic and proteolytic archaeon Thermococcus strain ES-1. J Bacteriol 177:4757–4764. doi: 10.1128/jb.177.16.4757-4764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukund S, Adams MW. 1991. The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an aldehyde ferredoxin oxidoreductase. Evidence for its participation in a unique glycolytic pathway. J Biol Chem 266:14208–14216. doi: 10.1016/S0021-9258(18)98669-2. [DOI] [PubMed] [Google Scholar]
- 20.Straub CT, Schut G, Otten JK, Keller LM, Adams MWW, Kelly RM. 2020. Modification of the glycolytic pathway in Pyrococcus furiosus and the implications for metabolic engineering. Extremophiles 24:511–518. doi: 10.1007/s00792-020-01172-2. [DOI] [PubMed] [Google Scholar]
- 21.Keller MW, Lipscomb GL, Nguyen DM, Crowley AT, Schut GJ, Scott I, Kelly RM, Adams MW. 2017. Ethanol production by the hyperthermophilic archaeon Pyrococcus furiosus by expression of bacterial bifunctional alcohol dehydrogenases. Microb Biotechnol 10:1535–1545. doi: 10.1111/1751-7915.12486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kengen SW, de Bok FA, van Loo ND, Dijkema C, Stams AJ, de Vos WM. 1994. Evidence for the operation of a novel Embden-Meyerhof pathway that involves ADP-dependent kinases during sugar fermentation by Pyrococcus furiosus. J Biol Chem 269:17537–17541. doi: 10.1016/S0021-9258(17)32474-2. [DOI] [PubMed] [Google Scholar]
- 23.Blamey JM, Adams MW. 1993. Purification and characterization of pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim Biophys Acta 1161:19–27. doi: 10.1016/0167-4838(93)90190-3. [DOI] [PubMed] [Google Scholar]
- 24.Ma K, Hutchins A, Sung SJ, Adams MW. 1997. Pyruvate ferredoxin oxidoreductase from the hyperthermophilic archaeon, Pyrococcus furiosus, functions as a CoA-dependent pyruvate decarboxylase. Proc Natl Acad Sci USA 94:9608–9613. doi: 10.1073/pnas.94.18.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy R, Mukund S, Schut GJ, Dunn DM, Weiss R, Adams MW. 1999. Purification and molecular characterization of the tungsten-containing formaldehyde ferredoxin oxidoreductase from the hyperthermophilic archaeon Pyrococcus furiosus: the third of a putative five-member tungstoenzyme family. J Bacteriol 181:1171–1180. doi: 10.1128/JB.181.4.1171-1180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bevers LE, Bol E, Hagedoorn PL, Hagen WR. 2005. WOR5, a novel tungsten-containing aldehyde oxidoreductase from Pyrococcus furiosus with a broad substrate specificity. J Bacteriol 187:7056–7061. doi: 10.1128/JB.187.20.7056-7061.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy R, Adams MW. 2002. Characterization of a fourth tungsten-containing enzyme from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol 184:6952–6956. doi: 10.1128/JB.184.24.6952-6956.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Oost J, Voorhorst WG, Kengen SW, Geerling AC, Wittenhorst V, Gueguen Y, de Vos WM. 2001. Genetic and biochemical characterization of a short-chain alcohol dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. Eur J Biochem 268:3062–3068. doi: 10.1046/j.1432-1327.2001.02201.x. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, Sha C, Wang H, Ma K, Wiegle J, Abomohra AE, Shao W. 2021. A novel bifunctional aldehyde/alcohol dehydrogenase catalyzing reduction of acetyl-CoA to ethanol at temperatures up to 95°C. Sci Rep 11:1050. doi: 10.1038/s41598-020-80159-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kube J, Brokamp C, Machielsen R, van der Oost J, Markl H. 2006. Influence of temperature on the production of an archaeal thermoactive alcohol dehydrogenase from Pyrococcus furiosus with recombinant Escherichia coli. Extremophiles 10:221–227. doi: 10.1007/s00792-005-0490-z. [DOI] [PubMed] [Google Scholar]
- 31.Machielsen R, Uria AR, Kengen SW, van der Oost J. 2006. Production and characterization of a thermostable alcohol dehydrogenase that belongs to the aldo-keto reductase superfamily. Appl Environ Microbiol 72:233–238. doi: 10.1128/AEM.72.1.233-238.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma K, Adams MW. 1999. An unusual oxygen-sensitive, iron- and zinc-containing alcohol dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol 181:1163–1170. doi: 10.1128/JB.181.4.1163-1170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schut GJ, Nixon WJ, Lipscomb GL, Scott RA, Adams MW. 2012. Mutational analyses of the enzymes involved in the metabolism of hydrogen by the hyperthermophilic archaeon Pyrococcus furiosus. Front Microbiol 3:163. doi: 10.3389/fmicb.2012.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vailionis JZW, Zhang K, Rodionov D, Lipscomb GL, Tanwee TNN, O’Quinn HC, Kelly RM, Adams MWW, Zhang Y. 2023. Optimizing strategies for bio-based ethanol production using genome-scale metabolic model of the hyperthermophilic archaeon, Pyrococcus furiosus. Appl Environ Microbiol. doi: 10.1016/j.copbio.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olson DG, Sparling R, Lynd LR. 2015. Ethanol production by engineered thermophiles. Curr Opin Biotechnol 33:130–141. doi: 10.1016/j.copbio.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 37.Adams MW, Holden JF, Menon AL, Schut GJ, Grunden AM, Hou C, Hutchins AM, Jenney FE Jr, Kim C, Ma K, Pan G, Roy R, Sapra R, Story SV, Verhagen MF. 2001. Key role for sulfur in peptide metabolism and in regulation of three hydrogenases in the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol 183:716–724. doi: 10.1128/JB.183.2.716-724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipscomb GL, Hahn EM, Crowley AT, Adams MWW. 2017. Reverse gyrase is essential for microbial growth at 95°C. Extremophiles 21:603–608. doi: 10.1007/s00792-017-0929-z. [DOI] [PubMed] [Google Scholar]
- 39.Steffensen JL, Dufault-Thompson K, Zhang Y. 2016. PSAMM: a Portable System for the Analysis of Metabolic Models. PLoS Comput Biol 12:e1004732. doi: 10.1371/journal.pcbi.1004732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dufault-Thompson K, Steffensen JL, Zhang Y. 2018. Using PSAMM for the Curation and Analysis of Genome-Scale Metabolic Models. Methods Mol Biol 1716:131–150. doi: 10.1007/978-1-4939-7528-0_6. [DOI] [PubMed] [Google Scholar]
- 41.Ebrahim A, Lerman JA, Palsson BO, Hyduke DR. 2013. COBRApy: COnstraints-Based Reconstruction and Analysis for Python. BMC Syst Biol 7:74. doi: 10.1186/1752-0509-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Megchelenbrink W, Huynen M, Marchiori E. 2014. optGpSampler: an improved tool for uniformly sampling the solution-space of genome-scale metabolic networks. PLoS One 9:e86587. doi: 10.1371/journal.pone.0086587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thorgersen MP, Stirrett K, Scott RA, Adams MW. 2012. Mechanism of oxygen detoxification by the surprisingly oxygen-tolerant hyperthermophilic archaeon, Pyrococcus furiosus. Proc Natl Acad Sci USA 109:18547–18552. doi: 10.1073/pnas.1208605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.00012-23-s0001.pdf, PDF file, 0.6 MB (625.6KB, pdf)