ABSTRACT
The metal-resistant bacterium Cupriavidus metallidurans uses its copper resistance components to survive the synergistic toxicity of copper ions and gold complexes in auriferous soils. The cup, cop, cus, and gig determinants encode as central component the Cu(I)-exporting PIB1-type ATPase CupA, the periplasmic Cu(I)-oxidase CopA, the transenvelope efflux system CusCBA, and the Gig system with unknown function, respectively. The interplay of these systems with each other and with glutathione (GSH) was analyzed. Copper resistance in single and multiple mutants up to the quintuple mutant was characterized in dose-response curves, Live/Dead-staining, and atomic copper and glutathione content of the cells. The regulation of the cus and gig determinants was studied using reporter gene fusions and in case of gig also RT-PCR studies, which verified the operon structure of gigPABT. All five systems contributed to copper resistance in the order of importance: Cup, Cop, Cus, GSH, and Gig. Only Cup was able to increase copper resistance of the Δcop Δcup Δcus Δgig ΔgshA quintuple mutant but the other systems were required to increase copper resistance of the Δcop Δcus Δgig ΔgshA quadruple mutant to the parent level. Removal of the Cop system resulted in a clear decrease of copper resistance in most strain backgrounds. Cus cooperated with and partially substituted Cop. Gig and GSH cooperated with Cop, Cus, and Cup. Copper resistance is thus the result of an interplay of many systems.
IMPORTANCE The ability of bacteria to maintain homeostasis of the essential-but-toxic “Janus”-faced element copper is important for their survival in many natural environments but also in case of pathogenic bacteria in their respective host. The most important contributors to copper homeostasis have been identified in the last decades and comprise PIB1-type ATPases, periplasmic copper- and oxygen-dependent copper oxidases, transenvelope efflux systems, and glutathione; however, it is not known how all these players interact. This publication investigates this interplay and describes copper homeostasis as a trait emerging from a network of interacting resistance systems.
KEYWORDS: Cupriavidus metallidurans, P-type ATPases, copper resistance, multicopper oxidases
INTRODUCTION
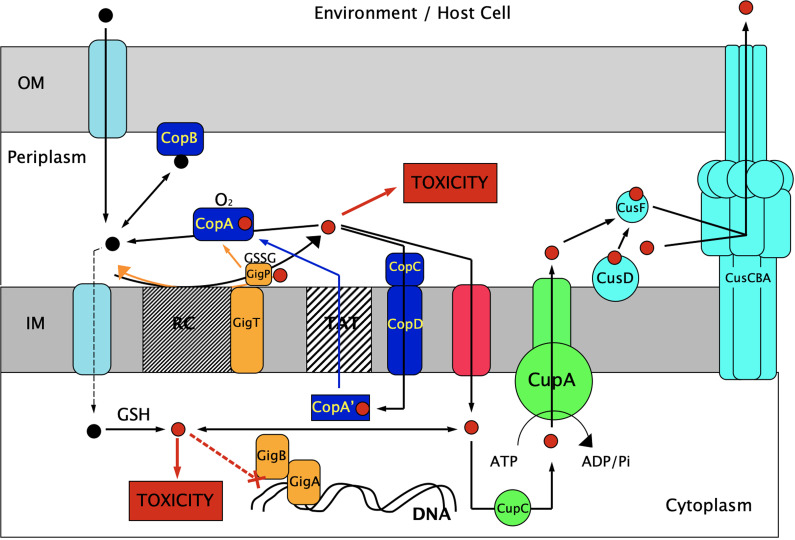
The betaproteobacterium Cupriavidus metallidurans survives in environments rich in transition metals (1–3). The necessary metal resistance determinants are located on the bacterial chromosome, a chromid and two plasmids (4). C. metallidurans also occurs in biofilms of bacterio-formed gold (5). In auriferous soils, gold complexes are rapidly accumulated within the cell but later precipitated as gold nanoparticles in the periplasm (6). Upon contact with gold complexes, defense systems against oxidative stress and copper resistance systems (Fig. 1 and Fig. S1 in the supplemental material) are upregulated (7), for instance, the yet uncharacterized gig genes (gold induced genes, Fig. S1A). Auriferous soils usually contain an elevated copper content. Copper ions and gold complexes exert synergistic toxicity because cytoplasmic gold compounds inhibit the PIB1-type ATPase CupA, which is responsible for removal of surplus cytoplasmic Cu(I) ions (Fig. 1, Fig. S1D) (8). To prevent this combined toxic effect, C. metallidurans induces the cop determinant. The periplasmic Cu(I)/Au(I) oxidase CopA (Fig. 1, Fig. S1E) oxidizes both compounds back to Au(III) and Cu(II), which are less toxic than the respective monovalent ions (6, 7, 9, 10). Au(III) complexes can be subsequently reduced to metallic Au(0) in the periplasm, leading to direct formation of gold nanoparticles in the periplasm without using the toxic Au(I) state as an intermediate (11).
FIG 1.
Model for copper homeostasis in C. metallidurans. In the plasmid-free C. metallidurans strain AE104, which does not possess the pMOL30-encoded cop and sil determinants (3), Cu(II) (black dots) is imported into the periplasm of this Gram-negative bacterium by outer membrane (OM) porins and further on with a low rate into the cytoplasm as unspecific substrate of metal cation import systems such as ZupT (light blue) (48). Periplasmic Cu(II) ions are reduced upon contact with the respiratory chain (RC) to Cu(I) (red dots) (45) and cytoplasmic Cu(II) ions via glutathione (GSH) (28), which leads to toxic effects in both compartments. Periplasmic Cu(I) is a much better substrate for import and is subsequently imported into the cytoplasm at a higher rate than Cu(II) by unspecific import (red system) (9, 10). The central component of the Cop system (dark blue) is the copper- and oxygen-dependent oxidase CopA, which oxidizes Cu(I) back to Cu(II) (69, 70, 126, 127), decreasing copper import into the cytoplasm. Cu(II) is sequestered by CopB, which is attached to the OM (75). CopA is exported to the periplasm by the twin-arginine transport system (TAT) in a partially folded, probably copper-loaded form CopA’ (74) with CopCD-mediated import or GSH-mediated reduction of Cu(II) providing the required copper ions (72, 73). The Cup system (green) is centered around the PIB1-type, Cu(I)-exporting ATPase CupA, which may receive Cu(I) from the cytoplasmic copper chaperone CupC (128–130). The Cus system (green-blue) exports periplasmic Cu(I) from the periplasm to the outside by the transmembrane efflux system CusCBA, which may receive Cu(I) from the periplasmic copper chaperone CusF (19). CusF may have previously accepted Cu(I) from CupA or the IM-attached protein CusD. GigBA (orange) may protect the DNA or DNA-operating enzymes against copper toxicity. GigT in combination with GigP and oxidized glutathione (GSSG) may oxidize periplasmic Cu(I) to Cu(II) by feeding the electron into the quinol pool and GigP may additionally assist in maturation of CopA.
This scenario highlights that gold transformation and resistance is interwoven with copper resistance and homeostasis in C. metallidurans. Gold resistance and transformation of C. metallidurans CH34 wild type and its plasmid-free derivative strain AE104 is similar (7), so that plasmid pMOL30 with its large copper resistance gene region (12, 13) does not need to be considered to understand the copper-gold interaction network. C. metallidurans possesses, in addition to CupA, three other PIB1-type, copper exporting ATPases, pMOL30-encoded CopF and chromosome-encoded CtpA1 and RdxI. CtpA1 and RdxI are “anabolic” exporters that supply copper ions to periplasmic copper binding sites of cuproproteins (14). Their genes are not upregulated by gold compounds in contrast to cupA (7). The contribution of these proteins to copper resistance has already been characterized (8).
Resistance-nodulation-cell division family of proteins (RND)-driven transenvelope efflux systems (15) such as CusCBA from E. coli export Cu(I) and Ag(I) from the periplasm to the outside (16–19), so that the orthologue CusCBA from C. metallidurans (Fig. 1, Fig. S1C) may also export Au(I), reminiscent to the RND-type GesCBA transport system in Salmonella (20, 21). The thiol-containing tripeptide glutathione (GSH; Fig. 1, Fig. S1B) is involved in copper resistance of Escherichia coli (10, 22) and present in many bacteria (23). It is synthesized by the gamma-glutamylcysteine synthetase GshA and the glutathione synthetase GshB from glutamate, cysteine, and glycine (24). If copper is imported into the cytoplasm as Cu(II), the ion should be immediately reduced by GSH to Cu(I) and GS-SG, and subsequently bound to GSH (25–28), until Cu(I) can be sequestered by copper chaperones such as CupC for further delivery to a copper-exporting P-type ATPase (29–32).
A first step into a deeper insight into the synergistic toxicity of copper and gold compounds, as well as into the synergetic gold-copper detoxification, would be an analysis of the network of copper resistance determinants in the plasmid-free C. metallidurans strain AE104 (Fig. 1). In this publication, we investigate the interplay of systems centered around the PIB1-type ATPase CupA, the periplasmic copper oxidase CopA, the RND efflux system CusCBA, or the unknown Gig components, respectively, with each other in the absence and presence of glutathione.
RESULTS
Contribution of Cup, Cop, Cus, Gig, and glutathione to copper resistance in C. metallidurans.
Single, double, triple, quadruple mutants, and the quintuple mutant in the cupC/AR, copA2B2C2D2, cusDCBAF, gigTBAP, and gshA gene regions (further referred to a Δcup, Δcop, Δcus, Δgig; Fig. S1) were constructed in the plasmid-free C. metallidurans strain AE104. While a mutant carrying a marker-free deletion of gshA could be obtained in the parent strain background, it was neither possible to construct marker-free deletions of gshA in the strains with deletions in the copper resistance determinants nor to delete copper resistance determinants in the ΔgshA mutant. Consequently, gshA was interrupted in all these strains, which also should lead to a decreased expression of gshB due to a polar effect (Fig. S1B). Tables 1, 2, and 3 show only the results from the ΔgshA mutants with the interrupted gene, and Table 4 included the comparison with the marker-free ΔgshA deletion in the parent strain AE104.
TABLE 1.
Copper resistance of strains with and without disruption of ΔgshAa
| Bacterial strain | IC50 (μM) | Bacterial strain | IC50 (μM) | Q | D |
|---|---|---|---|---|---|
| Δcus | 743 ± 62 | Δcus ΔgshA | 612 ± 32 | 1.21 | 1.40 |
| AE104 | 615 ± 68 | ΔgshA | 450 ± 6 | 1.37 | 2.22 |
| Δcus Δgig | 597 ± 26 | Δcus Δgig ΔgshA | 472 ± 49 | 1.26 | 1.68 |
| Δgig | 580 ± 30 | Δgig ΔgshA | 458 ± 43 | 1.26 | 1.67 |
| Δcop Δgig | 492 ± 50 | Δcop Δgig ΔgshA | 110 ± 9 | 4.46 | 6.40 |
| Δcop | 389 ± 43 | Δcop ΔgshA | 161 ± 24 | 2.41 | 3.37 |
| Δcop Δcus | 197 ± 21 | Δcop Δcus ΔgshA | 26.4 ± 2.9 | 7.45 | 7.18 |
| Δcop Δcus Δgig | 158 ± 16 | Δcop Δcus Δgig ΔgshA | 29.0 ± 4.4 | 5.44 | 6.41 |
| Δcup | 13.5 ± 1.2 | Δcup ΔgshA | 13.2 ± 2.5 | 1.02 | 0.07 |
| Δcup Δgig | 11.0 ± 2.1 | Δcup Δgig ΔgshA | 6.22 ± 1.03 | 1.77 | 1.52 |
| Δcup Δcus | 4.25 ± 0.89 | Δcup Δcus ΔgshA | 3.83 ± 1.37 | 1.11 | 0.19 |
| Δcop Δcup | 3.41 ± 0.62 | Δcop Δcup ΔgshA | 0.42 ± 0.10 | 8.12 | 4.15 |
| Δcup Δcus Δgig | 2.22 ± 0.14 | Δcup Δcus Δgig ΔgshA | 0.51 ± 0.12 | 4.35 | 6.58 |
| Δcop Δcup Δcus | 0.81 ± 0.23 | Δcop Δcup Δcus ΔgshA | 0.43 ± 0.09 | 1.88 | 1.19 |
| Δcop Δcup Δcus Δgig | 0.47 ± 0.10 | Δcop Δcup Δcus Δgig ΔgshA | 0.44 ± 0.16 | 1.07 | 0.12 |
| Δcop Δcup Δgig | 0.43 ± 0.21 | Δcop Δcup Δgig ΔgshA | 0.46 ± 0.07 | 0.93 | 0.11 |
Copper resistance was determined in 96-well plates at 30°C. These data are sorted in decreasing order of the IC50 values of the gshA+ strains. The Q values give the IC50 ratios plus and minus gshA. D, respective distance value. Q values in italics indicate no difference between the IC50 value (D < 1); bold-faced values indicate Q > 2. Table S1 compares the effect of other deletions. The D value is used as an easy-to-handle statistical value and is the ratio of the absolute difference of the mean value of two data sets, divided by the sum of the respective deviations. At n ≥ 3, D > 1 means a significance of 95% (33, 34). D > 1 means that the deviation bars of two mean values do not overlap or touch.
TABLE 2.
Percentage of dead cells in cultures of C. metallidurans mutant cellsa
| Bacterial strain |
% dead cells |
|||
|---|---|---|---|---|
| Glutathione | Present (GSH+) |
Absent (ΔgshA) |
||
| Added Cu(II) | No | At IC50 | No | At IC50 |
| Δcus | 2.81 ± 2.72 | 15.9 ± 16.7 | 1.32 ± 0.58 | 7.04 ± 4.36 |
| AE104 | 4.93 ± 2.40 | 9.12 ± 5.98 | 2.81 ± 1.76 | 5.54 ± 3.23 |
| Δcus Δgig | 3.41 ± 3.88 | 18.2 ± 26.4 | 2.24 ± 0.44 | 3.64 ± 2.07 |
| Δgig | 3.45 ± 0.71 | 5.73 ± 2.68 | 2.54 ± 1.91 | 4.10 ± 2.38 |
| Δcop Δgig | 2.51 ± 2.30 | 10.1 ± 10.1 | 4.50 ± 0.39 | 30.7 ± 10.0 |
| Δcop | 2.90 ± 1.58 | 9.67 ± 5.66 | 2.74 ± 3.04 | 4.69 ± 3.28 |
| Δcop Δcus | 3.57 ± 2.11 | 27.3 ± 17.1 | 2.75 ± 1.89 | 13.7 ± 5.6 |
| Δcop Δcus Δgig | 5.51 ± 3.13 | 17.1 ± 6.1 | 6.54 ± 4.06 | 26.1 ± 12.6 |
| Δcup | 2.43 ± 2.85 | 8.61 ± 6.25 | 1.24 ± 0.62 | 4.91 ± 2.79 |
| Δcup Δgig | 4.53 ± 2.20 | 4.66 ± 4.53 | 4.35 ± 1.41 | 10.6 ± 1.9 |
| Δcup Δcus | 4.35 ± 3.16 | 2.58 ± 2.65 | 4.19 ± 2.23 | 20.0 ± 13.5 |
| Δcop Δcup | 3.96 ± 2.04 | 10.5 ± 4.9 | 9.47 ± 5.69 | 9.37 ± 3.62 |
| Δcup Δcus Δgig | 5.12 ± 2.44 | 3.78 ± 2.42 | 3.95 ± 3.57 | 2.76 ± 2.84 |
| Δcop Δcup Δcus | 4.47 ± 1.42 | 6.18 ± 8.05 | 4.12 ± 2.43 | 7.34 ± 3.73 |
| Δcop Δcup Δcus Δgig | 3.86 ± 3.22 | 9.04 ± 9.46 | 5.92 ± 2.86 | 10.0 ± 2.00 |
| Δcop Δcup Δgig | 2.55 ± 0.99 | 4.85 ± 6.21 | 1.38 ± 0.64 | 7.82 ± 3.97 |
The indicated mutant strains of C. metallidurans AE104 and the ΔgshA mutant were incubated for 20 h in the presence of Cu(II) provided at the IC50 concentration (Table 1) of the respective strain, without copper (negative control) and a Live/Dead staining was applied. The percentage of the dead cells (filter 3, λEx 546 nm/λEm 590 nm) of the total cells (filter 2, λEx 450 to 490 nm/λEm 520 nm) is given. Bold-faced values are Q > 1.5 and D > 1, n = 4 without copper and n = 6 with copper. Bold-faced and italic values have higher min, median and max values of these 4 or 6 experiments in the presence of copper compared to without copper but the mean values could not be judged as different due to a large deviation. Underlined values indicate a >50% increase in the presence of copper at the IC50 concentration but not judged as different due to large deviations. Light gray fields indicates no increased copper-mediated killing in the presence of GSH. Midgray field indicates neither in the presence nor the absence of GSH.
TABLE 3.
Copper content of C. metallidurans mutant cellsa
| Bacterial strain |
1,000 Cu/cell |
Bacterial strain |
1,000 Cu/cell |
||
|---|---|---|---|---|---|
| Added Cu(II) (μM) | 0 | 25 | Added Cu(II) (μM) | 0 | 25 |
| Δcus | 7.59 ± 2.64 | 49.6 ± 12.9 | Δcus ΔgshA | 5.13 ± 0.15 | 36.3 ± 5.7 |
| AE104 | 5.97 ± 1.00 | 32.8 ± 5.6 | AE104 ΔgshA | 6.11 ± 1.41 | 23.0 ± 2.5 |
| Δgig Δcus | 7.33 ± 2.02 | 28.1 ± 3.0 | Δgig Δcus ΔgshA | 6.58 ± 0.74 | 25.3 ± 1.3 |
| Δgig | 7.01 ± 1.21 | 26.2 ± 2.7 | Δgig ΔgshA | 5.02 ± 0.47 | 24.4 ± 3.5 |
| Δcop Δgig | 5.67 ± 1.05 | 36.3 ± 7.5 | Δcop Δgig ΔgshA | 6.69 ± 0.41 | 253 ± 17 |
| Δcop | 7.83 ± 3.20 | 52.4 ± 8.4 | Δcop ΔgshA | 4.62 ± 0.67 | 125 ± 25 |
| Added Cu(II) (μM) | 0 | 1 | Added Cu(II) (μM) | 0 | 1 |
| AE104 | 6.88 ± 1.29 | 20.5 ± 3.9 | |||
| Δcop Δcus | 7.06 ± 1.33 | 26.0 ± 3.2 | Δcop Δcus ΔgshA | 5.00 ± 0.53 | 45.5 ± 20.3 |
| Δcop Δcus Δgig | 6.71 ± 2.21 | 30.6 ± 8.4 | Δcop Δcus Δgig ΔgshA | 4.71 ± 0.62 | 33.3 ± 12.4 |
| Δcup | 9.00 ± 3.41 | 19.9 ± 6.1 | Δcup ΔgshA | 5.50 ± 0.94 | 19.1 ± 2.6 |
| Δcup Δgig | 6.59 ± 2.29 | 20.6 ± 7.0 | Δcup Δgig ΔgshA | 6.02 ± 0.75 | 17.5 ± 0.8 |
| Δcup Δcus | 5.22 ± 1.16 | 20.3 ± 3.5 | Δcup Δcus ΔgshA | 5.09 ± 0.39 | 19.6 ± 1.1 |
| Added Cu(II) (μM) | 0 | 0.02 | Added Cu(II) (μM) | 0 | 0.02 |
| AE104 | 6.92 ± 2.10 | 10.8 ± 0.8 | |||
| Δcop Δcup | 6.48 ± 2.23 | 8.0 ± 1.0 | Δcop Δcup ΔgshA | 6.03 ± 0.58 | 13.6 ± 4.0 |
| Δcup Δcus Δgig | 6.11 ± 2.12 | 9.7 ± 0.3 | Δcup Δcus Δgig ΔgshA | 3.30 ± 1.03 | 8.0 ± 1.3 |
| Δcop Δcup Δcus | 8.04 ± 3.82 | 8.3 ± 3.2 | Δcop Δcup Δcus ΔgshA | 5.64 ± 0.44 | 9.8 ± 0.8 |
| Δcop Δcup Δcus Δgig | 6.32 ± 1.88 | 9.2 ± 0.3 | Δcop Δcup Δcus Δgig ΔgshA | 5.78 ± 1.10 | 12.0 ± 1.4 |
| Δcop Δcup Δgig | 5.85 ± 1.03 | 7.4 ± 2.6 | Δcop Δcup Δgig ΔgshA | 3.68 ± 0.67 | 11.5 ± 1.2 |
The cells were cultivated in medium without (negative control) or with copper at the indicated concentrations. The copper content of the Tris-buffered mineral salts medium without added copper was 38 ± 23 nM (n = 4), as determined by ICP-MS. Cells were mineralized in 67% nitric acid at 70°C for 2 h. Samples were diluted to a final concentration of 2% nitric acid. Bold indicates ([Q ≥ 1.5 OR Q ≤ 0.66] AND D > 1) in the comparison to the parent strain AE104 cultivated in the presence of the same copper concentration.
TABLE 4.
Glutathione content of C. metallidurans mutant cellsa
| Bacterial strain | nmol GSH/mg proteins |
|
|---|---|---|
| Added Cu(II) (μM) | 0 | 25 |
| Δcus | 120 ± 38d | 136 ± 42d,e |
| AE104 | 593 ± 18 | 535 ± 73 |
| AE104 ΔgshA | −14 ± 7d | ND |
| AE104 ΔgshAb | −7 ± 3d | ND |
| Δgig Δcus | 536 ± 13 | 533 ± 73 |
| Δgig | 603 ± 2 | 540 ± 63 |
| Δcop Δgig | 593 ± 158 | 418 ± 88 |
| Δcop | 668 ± 45 | 553 ± 45 |
| Added Cu(II) (μM) | 0 | 1 |
| AE104 | 593 ± 18c | 505 ± 78 |
| Δcop Δcus | 608 ± 119 | 650 ± 81 |
| Δcop Δcus Δgig | 501 ± 38 | 366 ± 19d |
| Δcup | 587 ± 14 | 576 ± 73 |
| Δcup Δgig | 422 ± 112 | 662 ± 110f |
| Δcup Δcus | 487 ± 79 | 585 ± 77 |
| Added Cu(II) (μM) | 0 | 0.02 |
| AE104 | 593 ± 18c | 671 ± 83 |
| Δcop Δcup | 654 ± 182 | 552 ± 42 |
| Δcup Δcus Δgig | 498 ± 45 | 460 ± 45 |
| Δcop Δcup Δcus | 448 ± 80 | 548 ± 60 |
| Δcop Δcup Δcus Δgig | 403 ± 71 | 544 ± 82 |
| Δcop Δcup Δgig | 557 ± 98 | 630 ± 108 |
Cells were cultivated in medium without (negative control) or with copper at the indicated concentrations and 5 mg cells were harvested, washed and disrupted by thaw-freeze cycling in liquid nitrogen. Protein content of the supernatant was determined with BSA as reference protein for a standard curve and GSH-content was determined by using the glutathione assay kit (CS0260, Sigma-Aldrich, Taufkirchen, Germany) with glutathione solution for the standard curve. ND; not determined.
Deletion of gshA without markers.
Same negative-control value for parent AE104 in all three parts of the table. Bold-faced numbers indicate differences ([Q ≥ 1.5 OR Q ≤ 0.66] AND D > 1) in comparison to: (i) AE104 without added copper; (ii) AE104 with the same concentration of added copper; or (iii) to the same strain without added copper.
AE104 without added copper.
AE104 with the same concentration of added copper.
The same strain without added copper.
Copper resistance of these mutant strains was determined in dose-response experiments (Fig. S2) and used to calculate the IC50 values (Table 1). Copper resistance of these mutant strains was also compared with each other in other combinations to evaluate the effect of each system on copper resistance in C. metallidurans mutant strains (see Table S1 in the supplemental material). The Q value was the quotient of two IC50 values and the D value was the distance of their mean values divided by the sum of both deviations, as published (7, 33, 34). At n ≥ 3, D > 1 means a > 95% significance in the Student's t test. The residual resistance level of many multiple deletion mutants was about IC50 = 0.4 μM and this resistance level was referred to a “IC50-min.” The IC50-min resistance level was reached by all mutants with Δcop Δcup (Δcus and/or Δgig) mutations independent from the presence of glutathione, and additionally in ΔgshA mutants with the genotype Δcup (Δcop or Δcus Δgig) (Table 1). Independent from the presence of glutathione, Cus or Gig were not able to mediate copper resistance above the IC50-min. In the absence of glutathione, Cop or Cus together with Gig also did not promote copper resistance. All five systems contributed to copper resistance and their functions were studied by the specific effect of each deletion in the parent and all mutant strains.
After the contribution of these systems to copper resistance has been outlined, the influence of the mutations on survival of the mutants and their metal and glutathione content was analyzed. Finally, the regulation of cus and gig was studied using a reporter gene system.
Cup.
Deletion of cup, with the PIB1-type Cu(I)-exporting ATPase CupA as a central component, decreased copper resistance of the parent strain 46-fold. Cop, Cus, Gig, and GSH together were not able to fully substitute a missing Cup system. Copper resistance of the Δcup single mutant was even lower than that of the Δcus Δgig, Δcop Δgig, and Δcop Δcus double mutants or the Δcop Δcus Δgig triple mutant (Table 1), in the presence as well as in the absence of GSH. The impact of the combined Cop, Cus, and Gig systems and of GSH was smaller than that of Cup alone. This indicated the outstanding importance of Cup and sorted the mutants with respect to their copper resistance into two groups: (i) in the presence of GSH, the Cup-containing strains exhibited a mid to high degree of copper resistance with an IC50 > 150 μM; and (ii) all combinations of Δcup mutants possessed only an IC50 < 15 μM (Table 1), a 10-fold difference in resistance. In the absence of GSH, all Δcup mutants also had an IC50 value below 15 μM but the IC50 values of all Cup-containing strains was just above 30 μM (Table 1), a 2-fold difference. In the single comparison of all mutants with and without cup, the smallest decrease in the IC50 value was 34-fold (ΔgshA ± Δcup; Table S1), while the largest of any non-Δcup deletion in all the other mutants was 31.5-fold (Δcup ΔgshA ± Δcop). Cup was by far the most important copper resistance system in C. metallidurans.
The strongest, 1,145-fold decrease in copper resistance resulted from deletion of cup in the Δcop Δgig double mutant, which still contained Cus and GSH (Table S1). It went down to the IC50-min value. The effect of a Δcup deletion in a Δcop Δcus Δgig background (which still contained GSH) was 336-fold and in a Δcop Δgig ΔgshA background (which still contained Cus) 240-fold, leading to the IC50-min value in both Δcup-containing quadruple mutant strains. Cus was not able to substitute Cup in any way, not in the presence nor the absence of GSH.
The comparison of the effect of an additional Δcup deletion in parent and single mutant strains yielded a ranking by the declining Q values of Δcus (175-fold) > Δcop (114-fold) > Δgig (53-fold) > AE104 parent (46-fold) > ΔgshA (34-fold). Cup was more important in the absence of Cus or Cop than in that of Gig or GSH. Ranking of the declining Q values resulting from a Δcup mutation in double mutants with a Δcop or a Δcus mutation led for the Δcop-containing double mutants to Δcop Δgig (1,145-fold to IC50-min) > Δcop ΔgshA (383-fold to IC50-min) > Δcop Δcus (243-fold to IC50 = 0.8 μM) > Δcop single mutant (114-fold) and for the mutants with a Δcus mutation Δcus Δgig (269-fold) > Δcop Δcus (243-fold) > Δcus single mutant (175-fold) > Δcus ΔgshA (160-fold). Cup was more important in Δcop mutants than in Δcus mutants. The IC50 of 0.8 μM the Δcop Δcup Δcus triple mutant, with a 2-fold IC50-min, was not different from the copper resistance of the quadruple Δcop Δcup Δcus Δgig mutant (Fig. S2C, open diamonds compared to open inverted triangles) and the D value was just 1.03 (Table S1). Taking this into consideration, all Δcop-containing double mutants went down to the IC50-min by an additional Δcup mutation. Cooperation of Cup and Cop was the most important contributor to copper resistance in C. metallidurans. Due to the increasing importance of Cup in double mutants with a Δcop mutation, Cus, Gig, and GSH supported this Cup-Cop cooperation in the ranking of importance Gig > GSH > Cus.
Cup was the only system characterized here that was able to increase copper resistance of the quintuple mutant from the IC50-min value 66-fold to IC50 = 29 μM (Δcop Δcus Δgig ΔgshA mutant, Table 1). The task of the Cup system was unique, had a stronger impact on copper resistance than all the other systems combined, and could not be substituted by Cus or GSH, but needed cooperative interaction with Cop.
Since the Cup system is centered around the Cu(I)-exporting PIB1-type ATPase CupA, the unique and strong contribution of Cup to copper resistance would be the export of surplus cytoplasmic Cu(I) ions to the periplasm. Nevertheless, the other three systems and glutathione were also required to increase copper resistance in the presence of Cup from IC50 = 29 μM 21-fold to the resistance level of the parent strain, IC50 = 615 μM. If full copper resistance in C. metallidurans was considered the increase in the IC50 from the IC50-min of the quintuple mutant, first Cup was needed (IC50 = 29 μM, Δcop Δcus Δgig ΔgshA), closely interacting with Cop (472 μM, Δcus Δgig ΔgshA), followed by contribution of Cus, Gig, and GSH (615 μM, parent). On the other hand, all strains with an IC50-min resistance level were Δcup mutants so that a Δcup mutation was the first condition for a complete loss of copper resistance. Removal of surplus Cu(I) ions from the cytoplasm was an essential contribution to copper resistance, but alone, on the other hand, was not sufficient to reach full copper resistance in C. metallidurans. This phenotype resulted from an interplay of many systems.
Cop.
Removal of the Cop system, centered around the periplasmic Cu(I) oxidase CopA (Fig. 1), resulted in a moderate 1.58-fold decrease in copper resistance (D = 2.04) in the parent strain background (Table 1). This effect of the cop deletion in the parent strain AE104 was well documented in the dose-response curves (Fig. S2A, closed diamonds versus circles). The other resistance systems were not fully able to substitute a missing Cop system but to 63% (1/1.58). Deletion of cop mediated a clear decrease in copper resistance in most mutants with the exception of the quintuple mutant and a very small 1.18-fold decrease (D = 1.09) in the Δgig single mutant (Table S1). Deletion of cop in all strains with a Δcup deletion except the Δcup single mutant resulted in the IC50-min, in agreement with a close cooperation of Cop and Cup. The resistance level of the Δcop Δcup mutant was IC50 = 3.4 μM, 8.5-fold of the IC50-min, so that Cus, Gig, or GSH together could mediate some copper resistance. This was also demonstrated by the 925-fold decrease in copper resistance down to the IC50-min that resulted from the Δcup deletion in the Δcus Δgig ΔgshA mutant background, which still contained Cop.
Deletion of cop had the strongest effect (31.5-fold decrease) in the Δcup ΔgshA background and led to the IC50-min level (Table S1). This was the strongest effect of all non-Δcup deletions. The decrease in resistance by deletion of cop was only 3-fold in the ΔgshA and only 4-fold in the Δcup single mutants, indicating an important contribution of GSH to copper resistance to the function of the respective remaining system in the absence of Cup or Cop. A 26-fold decrease in the IC50-level resulting from the Δcop deletion occurred in the Δcup Δgig mutant, whereas a cop deletion in the Δgig background had nearly no effect. Cup, Gig, and GSH were the most important interaction partners of Cop, but Gig only in the absence of Cup.
Cop was not able to increase copper resistance of the quintuple mutant but of all quadruple mutants. Cop in combination either with GSH, Gig, Cus, or Cup mediated a 5-fold, 9-fold, 13-fold or 16-fold increase in copper resistance, respectively. All four compounds led to an essential support to the Cop-mediated contribution to copper resistance in the ranking Cup > Cus > Gig > GSH. Cus, Gig, and GSH together, but not alone and not in single pairs, were able to mediate an IC50 = 3.4 μM. These three systems contributed to a function that could not be provided by any of the other two systems and only the resulting triple partner interaction network led to the emergence of a low degree of copper resistance. Together with Cup, these three systems were nearly able to substitute a missing Cop (63% of the IC50 of the parent strain, Table S1). Here, Cus and GSH were important, but not Gig.
Cop was second most important for copper resistance in C. metallidurans, but in contrast to Cup, Cop alone had no resistance effect; Cop needed the interaction with Cup, Cus, Gig, or GSH to mediate an increase in copper resistance in C. metallidurans above the IC50-min. Cooperation of Cup and Cop mediated 77% of the copper resistance level of the parent (Δcus Δgig ΔgshA compared with parent, D = 1.22) so that the combined contribution of Cus, Gig, and GSH to copper resistance was small (23%), but these systems together partially substituted a missing Cop. All Δcop Δcup mutants with any other deletion (Δcus, Δgig or ΔgshA) displayed a complete loss of copper resistance, so that Δcop was the second most important condition for complete loss of this phenotype. On the other hand, a Δcup Δcus Δgig ΔgshA quadruple mutant also had an IC50 = IC50-min, indicating again that Cop alone was not able to mediate any increase in copper resistance.
Cus.
Deletion of cus in the parent strain had no effect. There was even a slight increase in copper resistance, which was well documented in the dose-response curves (see Fig. S2A in the supplemental material). This increase in resistance as consequence of Δcus did not occur in the Δgig, Δcop, or Δcup single but in the ΔgshA double mutant (Table S1). A Δgig deletion in the parent or the ΔgshA mutant abolished the Δcus-mediated increase in copper resistance. Even in the presence of Cop and Cup, Gig supported copper resistance to a small degree.
Since deletion of cus in the parent strain had no effect, Cup, Cop, Gig, and GSH fully substituted a missing Cus system. Deletion of cus led to a 2-fold decrease of copper resistance in the Δcop mutant (Fig. S2A), a 3-fold decrease in the Δcup mutant, and a 4-fold decrease in the Δcup Δcop mutant, down to the IC50-min value (Table 1). Additional deletion of gig in the Δcop and Δcup single mutants led to a stronger decrease of copper resistance mediated by Δcus as additional deletion in the double compared to the single mutants (Δcop ± Δcus Q = 2 but Δcop Δgig ± Δcus Q = 3; Δcup ± Δcus Q = 3 but Δcup Δgig ± Δcus Q = 5; Table S1). Similarly, additional deletion of gshA led to a similar effect (Δcop ± Δcus Q = 2 but Δcop ΔgshA ± Δcus Q = 6; Δcup ± Δcus Q = 3 but Δcup ΔgshA ± Δcus Q = 3.5; Table S1). Gig and GSH supported the contribution of Cus to copper resistance.
Deletion of cus decreased copper resistance in half of the mutant backgrounds (Table S1), in all mutants that carried a Δcop, Δcup, or Δcup Δcop mutation. Cus was partially able to support Cup in the absence of Cop as well as Cop in the absence of Cup, providing a back-up function for both systems. Cus was not able to raise copper resistance of the quintuple mutant or the quadruple mutants still containing Gig or GSH, but in the quadruple mutants possessing Cop or Cup. On the one hand, Cop and Cup fully substituted a missing Cus, on the other hand, Cus could also cooperate with either Cop or Cup to mount a moderate degree of copper resistance. The strongest decrease in copper resistance as result of a Δcus deletion occurred in the Δcup Δgig ΔgshA mutant, 12-fold down to the IC50-min, with a smaller effect in the Δcup Δgig (5-fold) and the Δcup ΔgshA (6-fold) but none in the Δgig ΔgshA background.
Cooperation of Cup, centered around the PIB1-type ATPase CupA, and of Cop, centered around the periplasmic Cu(I) oxidase CopA, was central to copper resistance in C. metallidurans. Cus, centered around the transenvelope efflux complex CusCBA, was not required when Cup and Cop were present but was required when either system was absent. The absence of copper resistance above the IC50-min value in the quadruple mutant with Cus as sole component clearly demonstrated that CusCBA was not able to substitute the function of CupA as efflux pump for cytoplasmic Cu(I) ions. Instead, substitution of CopA gave evidence that CusCBA removed periplasmic Cu(I) ions by export to the outside while CopA oxidized these to Cu(II) ions. Either way resulted in decreased uptake of Cu(I) into the cytoplasm and consequently relief the necessity of CupA (Fig. 1).
Gig.
Deletion of gig had no effect in the parent, so the other systems were fully able to substitute a missing Gig system. The effect of a gig deletion in all strains with a functional Cup system was small (1 > Q > 2, D > 1, mutants Δcus ± ΔgshA, Δcop ΔgshA, Δcop Δcus) or not existing (D < 1, ΔgshA, Δcop Δcus ΔgshA, Table S1), indicating a minor contribution of Gig in Cup+ strain with additional possession of Cop, Cus without GSH, and GSH without Cus but not with Cus and GSH. There was no effect of the Δgig deletion in the Δcup mutant but in the Δcup mutants with an additional ΔgshA ± Δcus or Δcop deletion. The strongest decrease in resistance (7-fold to 8-fold down to the IC50-min) occurred in the Δcop Δcup double and in the Δcup Δcus ΔgshA triple mutant. Contribution of Gig to copper resistance was visible in a Δcup background with additional absence of Cop or of Cus plus GSH.
Gig was not able to increase copper resistance of the quintuple mutant, not in quadruple mutants still containing Cus, GSH, or Cup but in the quadruple mutant still containing Cop. Gig cooperated with Cop to increase copper resistance of the quintuple mutant to a moderate degree of IC50 = 3.8 μM, with Cus plus GSH to a similar level (IC50 = 3.4 μM), and doubled copper resistance in cells having either GSH and Cop, or Cus, and Cop.
The contribution of Gig to copper resistance was visible in cells with impaired removal of excess cytoplasmic (Cup) and of periplasmic (Cop, Cus) Cu(I) ions. In multiple mutants, Gig was required for full function of Cop and of Cus plus GSH.
GSH.
Absence of GSH had a small (1 < Q < 2, D > 1) effect in the parent strain, the Δcus, Δgig, or Δcus Δgig mutants (Table 1). Contribution of GSH to copper resistance was small in cells with Cup and Cop systems, which together largely substituted missing GSH. Copper resistance decreased as result of a ΔgshA mutation in Δcop ± Δcus ± Δgig strains and in Δcup mutants with an additional deletion of Δgig (moderate effect), Δcop, Δcus Δgig, or Δcop Δcus (moderate effect) but not in the Δcup single mutant. GSH was not able to substitute a missing Cup system but was needed when Cop, Gig, or Cus were also absent in the Δcup mutant. The strongest decrease in copper resistance mediated by ΔgshA occurred in the Δcop Δcus (7-fold) and the Δcop Δcup (8-fold) mutants. In Δcop Δcup, any additional deletion yielded the IC50-min. As judged by the increase in the Q value following deletion of gshA, importance of GSH increased from Δcop (2.4-fold) to Δcop Δgig (4.5-fold), Δcop Δcus (7.4-fold) with a decrease of 5.4-fold when gshA was deleted in the Δcop Δcus Δgig mutant, indicating a contribution of GSH to Gig- and Cus-mediated substitution of Cop.
Presence of GSH was not able to increase copper resistance of the quintuple mutant, quadruple mutants still containing Cus, or Gig, but those with a remaining Cup or Cop. GSH alone was not able to mediate copper resistance but GSH increased Cup-mediated copper resistance 5-fold, was essential for a Cop-mediated 4-fold-, and a Cus-Gig-mediated 8-fold increase above the IC50-min. GSH could not increase copper resistance when Cop and Gig or Cop, Gig, and Cus were present, but to a small part (Q = 1.77, D = 1.52) when only Cop and Cus were present but Gig was absent.
The absence of GSH in the Δcup mutant, which lacked the major efflux system CupA for cytoplasmic Cu(I) ions but possessed Cop, Cus, and Gig, indicated that a Cu(I)-buffering activity of GSH in the cytoplasm did not contribute to copper resistance in C. metallidurans. Instead, GSH became important when Cop was missing in the parent or Δcup background. GSH supported copper resistance based on the CopA-mediated oxidation of Cu(I) in the periplasm. Lack of Cus and Gig increased importance of GSH for this process. Moreover, GSH even allowed a 4-fold increase of copper resistance above the IC50-min by an interplay of GSH with Cop, and an 8-fold increase by an interplay with Cus and Gig.
Interaction of the five systems.
Cup, centered around the PIB1-type ATPase CupA, which exports excess Cu(I) from the cytoplasm, was by far the most important copper resistance system in C. metallidurans. Nevertheless, full copper resistance required cooperation of Cup with Cop. Cus, Gig, and GSH, forming a triple-partner interaction network, supported the Cup-Cop cooperation. The Cop system, centered around the periplasmic Cu(I)-oxidase CopA, needed the interaction with Cup and with Cus-Gig-GSH. Any decline of the oxygen tension may impair Cop and enhance the importance of Cus, Gig, and GSH as substitute for the interaction with Cup. Cop and Cup fully substituted a missing Cus, but Cus could also cooperate with either Cop or Cup to mount a moderate degree of copper resistance. Cus alone was not able to substitute Cup in any way, not in the presence nor the absence of GSH, so the transenvelope efflux complex CusCBA should not be able to export cytoplasmic Cu(I) ions. The contribution of Gig to copper resistance was visible in cells with impaired removal of excess cytoplasmic (Δcup) as well as of periplasmic Cu(I) ions by CopA or CusCBA. Gig supported the function of Cop and of Cus plus GSH. A potential Cu(I)-buffering activity of GSH in the cytoplasm did not contribute to copper resistance. GSH increased copper resistance by an interplay of Cup, Cop, with Cus plus Gig, or to a moderate degree when Cop and Cus were present, but Gig was absent. Full copper resistance in C. metallidurans results from an interplay of these five systems.
Copper kills.
Live/Dead staining followed by microscopy was used to investigate whether the copper has a more bactericidal or bacteriostatic effect on multiple deletion strains (Table 2, Fig. S3 and S4). The cells were incubated in the presence and absence of Cu(II) in the growth medium. The IC50 value of the individual strain was used as copper concentration (Table 1). This allowed comparison of strains with IC50 values of copper between 743 μM and the IC50-min of about 0.4 μM. These concentrations should have the same physiological impact on the compared strains. A bactericidal effect should increase the percentage of dead cells in the culture compared to the control without copper, and the deviation of the results since the difference in turbidity of individual cultures in biological repeats of dose-response experiments was always larger close to the IC50 value than at lower or higher copper concentrations (Fig. S2). Intact cells displayed a green fluorescence when filter 2 was used while membrane-damaged cells fluoresced in red (filter 3; an example is shown in Fig. S3). From the images, the percentage of dead cells was calculated. Isopropanol-treated cells served as negative control and were all dead (Table S2). Not only the mean values of the percentage of dead cells plus its deviation was calculated but also the minimum, median and maximum value from the biological repeats (Table S3). These values served also to judge the impact of copper at the IC50-concentration on the cells (Table 2).
In GshA+ cells (Table 2), the mean value of the percentage of dead cells in the absence of copper ions was 3.77 ± 0.98%. About 4% of the cells in a C. metallidurans culture were always dead. Mutations in the copper resistance systems did not change this value in the absence of copper ions. When the cells were incubated with copper added at the concentration of their individual IC50 for copper, large differences between the individual cultures led to strong deviations of percentage of dead cells (Table S2), leading to low D values (Table 2), as expected. The increase of the percentage of dead cells was also counted as “different” compared to the control without copper when the minimum, median, and maximum values were all three higher in copper-treated cells compared to the control (Table S3, bold-faced and italic values in Table 2) or at least the mean value was 50% higher (underlined values in Table 2). In the parent strain and most mutants, treatment with copper at the IC50 concentration increased the percentage of dead cells so that copper ions indeed killed a larger part of the population than in control cells. This effect could also be observed with only the IC50-min concentration of 0.4 μM Cu(II) and the Δcop Δcup Δcus Δgig quadruple mutant, albeit with huge deviations of the measurements between the individual biological repeats. Even very low copper concentrations were able to kill bacteria that did not possess copper resistance systems.
In ΔgshA cells (Table 2), this pattern was not different. Between 1.2% and 6.5% of the cells were dead even in the absence of copper ions, and these values increased when copper was added. The highest percentage of killed cells (31%) were reached in the Δcop Δgig ΔgshA mutant, indicating that Cop, Gig, and GSH were important contributors with respect to protection of the cells against copper-mediated killing.
Several mutant strains were not killed by copper at their respective IC50 values. In the Δcup Δgig and the Δcup Δcus mutants, presence of copper did not increase the percentage of dead cells in GSH+ cells but did increase in the corresponding ΔgshA mutants (Table 2, light gray field). In the Δcup Δcus Δgig triple mutant, presence of copper did not lead to increased death ratios, even in the absence of GSH (Table 2, medium-gray field). The respective Δcup Δcus Δgig ΔgshA quadruple mutant contained the Cop system as the sole remaining system, and had an IC50 equal to the IC50-min. This copper concentration did not increase killing of this quadruple mutant but clearly increased killing of the quintuple and several other mutants at the same copper concentration. Cop alone protected the cells against copper-mediated increased killing in the Δcup Δcus Δgig ΔgshA mutant at the IC50-min. The additional presence of GSH (Δcup Δcus Δgig mutant) allowed an IC50 of 2 μM and Cop was still able to protect the cells. GSH allowed the cells to survive and grow at a 4.35-fold higher concentration than the IC50-min by protecting them against copper-mediated killing in cooperation with Cop, which defines a role of GSH in copper resistance.
When, in the next step, presence either of Cus or of Gig mediated an IC50 between 4 μM and 11 μM, protection by Cop required the presence of GSH. Cus and Gig were able to support Cop and GSH in their respective protecting action in the Δcup Δgig or Δcup Δcus double mutants. When Gig was added to the Δcup Δgig mutant or Cus to the Δcup Δcus mutant, this resulted in the Δcup single mutant with an IC50 of 13.5 μM, which was killed by addition of this copper concentration, although there was no difference between the copper resistance levels of the Δcup single and the Δcup Δgig double mutant. When Cup was added to the Δcup Δgig or the Δcup Δcus mutants, parent resistance levels were reached, and the cells were inactivated by copper at the respective IC50 value.
The Cop system, centered around the periplasmic Cu(I) oxidase CopA, contributed to copper resistance in C. metallidurans by protecting the cells against copper-mediated killing at the IC50-min, although the Cop system did not increase copper resistance above the IC50-min in the respective Δcup Δcus Δgig ΔgshA quadruple mutant. GSH alone did not protect the cells against copper-mediated killing at the IC50-min in the Δcop Δcup Δcus Δgig quadruple mutant but Cop plus GSH protected and increased the IC50 level 4-fold in the Δcup Δcus Δgig triple mutant. Further addition of Gig (Δcup Δcus double mutant) or Cus (Δcup Δgig double mutant) increased the resistance level of the protected cells a second time (IC50 = 4 μM and 11 μM, respectively) but only in the presence of GSH. Cop-GSH mediated a first layer of protection against copper-mediated killing and Gig or Cus built upon this. Addition of Cus and Gig to the Δcup Δcus Δgig triple mutant increased the IC50 5-fold to 13 μM but the cells were not protected any more against killing at this copper concentration so that the effects of Gig and Cus were additive with some overlap with respect to resistance but counteracting with respect to Cop-GSH-mediated protection against killing. Similarly, any addition of Cup strongly increased the copper concentration that could be tolerated, which nevertheless increased the number of cells that were killed by copper.
Copper content of the mutant cells.
In general, the cellular copper and metal content of the mutant cells was determined in the presence and absence of copper ions added to the growth medium. Since dead cells hyperaccumulate copper ions (35) and most copper-treated mutant cell populations contained an increased number of mutant cells (Table 2), the copper concentrations used in these experiments were well below the IC50 values (Table 1). For cells with and without gshA with an IC50 > 100 μM, a concentration of 25 μM added Cu(II) was used, for cells with and without gshA with an 100 μM > IC50 > 1 μM, a concentration of 1 μM added Cu(II) was used, and for the remaining cells, a concentration of 0.02 μM (Table 3). This careful approach not only eliminated artifacts resulting from unspecific binding of copper to dead cells but also led to a high resolution of the effects of the single copper resistance systems on copper resistance and accumulation at low copper concentrations, e.g., those that allowed Cop-GSH to protect the cells against copper-mediated killing.
All cells cultivated without added copper contained about 6,000 atoms of Cu per cell (Table 3), caused by the 38 ± 23 nM Cu(II) in the growth medium. Only two quadruple mutants cultivated in medium without added extra copper exhibited a lower cellular copper concentration of about 3,500 Cu per cell. Since one of these strains contained 4,710 ± 620 Cu per cell when it served as negative control for cells cultivated in the presence of 1 μM added copper instead of 20 nM added copper (Table S4), both low values were irrelevant fluctuations and not further considered.
The AE104 parent cells increased their copper content in the presence of 25 μM copper to about 33,000 Cu atoms per cell, in the presence of 1 μM Cu to 20,000 Cu atoms per cell, and in the presence of 20 nM Cu to 10,000 Cu atoms per cell, showing a saturation curve of the amount of cell-bound copper instead of a linear relationship. None of the multiple mutant strains incubated with 20 nM added copper showed an increased cellular copper content compared to the parent cell, although all these mutants carried a Δcup deletion. This included the Δcup Δcus Δgig and Δcup Δcus Δgig ΔgshA mutants, which were protected by Cop against copper-mediated killing at the IC50-min and in the presence of GSH at an IC50 = 2 μM. At 1 μM added copper, none of the Δcup strains displayed an increased cellular copper content. This included the Δcup Δgig and the Δcup Δcus double mutants, which were protected by Cop-GSH against killing at their IC50 of 11 μM and 4 μM, respectively. This indicated that cup with the PIB1-type ATPase CupA as the central component was not required to adjust the cellular copper content at 1 μM added copper. Second, the protecting effect of Cop, GSH, Cus, or Gig was not accompanied by a decreased cellular copper content at low copper concentrations.
At 25 μM copper, the copper content of the ΔgshA single mutant was slightly below that of the parent cell (23,000 Cu per cell versus 33,000 Cu per cell, Q = 0.70, D = 1.21), so that presence of GSH mediated a small increase in the cellular copper content. The presence of GSH alone did not increase copper resistance of the quintuple mutant (Table 1). The copper content of the Δcup Δgig and Δcup Δcus mutants plus or minus GSH was not different (Table 3) although GSH was essential for the Cop-mediated protection against copper-caused enhanced killing (Table 2) at 2 μM added copper. Since GSH, as the main cellular thiol compound, is required for repair of oxidative damage, this all means that a GSH-mediated increase in copper resistance by interaction of GSH with the other resistance systems was not based on complexation of copper by GSH but rather by repair of copper-mediated oxidative damage.
While the cellular copper content of the parent cells at 25 μM added copper was 33,000 Cu per cell, the Δcop mutant contained 52,400 Cu atoms per cell (Table 3). This demonstrated that Cop, which was no longer able to protect the cells at this concentration against copper-mediated killing, nevertheless contributed at these copper concentrations to copper resistance by causing decreased accumulation of copper, even in the presence of Cup. Consequently, Cop interacted with GSH and subsequently Gig and Cus to prevent copper-mediated killing at low concentrations, and with Cup to prevent accumulation of copper at high concentrations.
To add another layer to this interaction network, Gig, Cus, and GSH influenced the Cop-Cup-mediated decrease in cell-bound copper (Table 3). In the presence of Cus and at 25 μM added copper, the Δcop ΔgshA cells contained the 2.5-fold amount of cell-bound copper compared to that of the Δcop single mutant. Additional deletion of gig doubled the number of cell-bound copper again, while this number was not increased compared to that of the parent strain AE104 in the Δcop Δgig mutant. GSH quenched copper accumulation in Δcop Cus+ Cup+ cells by allowing Cus or Cup to export copper ions. Gig was able to substitute GSH to some extent in these cells. Since the IC50 values of the Δcup, the Δcup ΔgshA, and the Δcup Δgig strains were not different (Table 1), Cup did not need GSH or Gig for function so that the underlying interaction network is the Cus-Gig-GSH triangle, which also caused a small increase in copper resistance compared to the quintuple mutant. In the presence of GSH, Gig seemed to stimulate copper accumulation in Δcop cells but this effect was close to the significance threshold when the copper contents of Δcop Δgig and Δcop cells were directly compared with each other (Table 3, Q = 1.44, D = 1.01), whereas the comparison of the copper contents of the Δcop ΔgshA ± gig cells yielded a significant result (Table 3, Q = 2.02, D = 3.05). GSH quenched copper accumulation in Δcop cells by interacting with Cus. Gig could substitute GSH here to some part.
At 1 μM added copper, the copper content of the Δcop Δcus Δgig and Δcop Δcus cells was not different from the parent (Table 3). There was some increase in the Δcop Δcus ΔgshA mutant compared to the parent but this comparison possessed only a D value close to the significance threshold due to a large deviation of one value (Q = 2.22, D = 1.03). No difference could be observed between the copper contents of the Δcop Δcus ± ΔgshA cells (Q = 1.75, D < 1) and of the Δcop Δcus ΔgshA ± Δgig cells (Q = 1.37, D < 1). At 1 μM added copper, neither Cup nor Cop were required to limit accumulation of copper by the cells and consequently, Cus-Gig-GSH were not needed to substitute a missing Cop.
Glutathione.
Marker-free deletion or interruption of the gshA gene in C. metallidurans resulted in a decrease of the measured cytoplasmic GSH in wild-type cells from 600 nmol GSH per mg protein to GSH values that could no longer be detected (Table 4, negative values). This cellular GSH concentration corresponds to 42 million GSH molecules per cell (Table 4), so that the slightly increased copper content of 10,000 atoms per cell represents only a neglectable portion of the total GSH pool that was occupied by copper ions.
The GSH content of the cells with the deletions of copper resistance determinants was not different from that of the parent strain AE104 in cells incubated without added copper (Table 4) with exception of the Δcus mutant. This strain exhibited only 20% of the GSH content of the parent. Any additional deletion (Δgig Δcus, Δcop Δcus, Δcup Δcus) increased the GSH content back to the parent level. The decreased GSH content in the Δcus strain depends on the presence of Gig, Cop, or Cup. In the presence of copper, added again at the same concentration used for the determination of the cellular copper content, the GSH level of the Δcus strain was also decreased.
The GSH content of mutant strains incubated in the presence of 1 μM or of 20 nM added copper was not different from that of the parent or the same mutant cultivated without copper (Table 4), with two exceptions. The GSH content of copper-treated cells was decreased in the Δcop Δcus Δgig triple and increased in the Δcup Δgig double mutant but not in the respective Gig+ strains, which were also incubated with 1 μM copper. Gig prevented a decrease in the GSH level in the Δcop Δcus but an increase in the Δcup strain. Gig seems to influence the GSH level in C. metallidurans.
Regulation of the cus determinant.
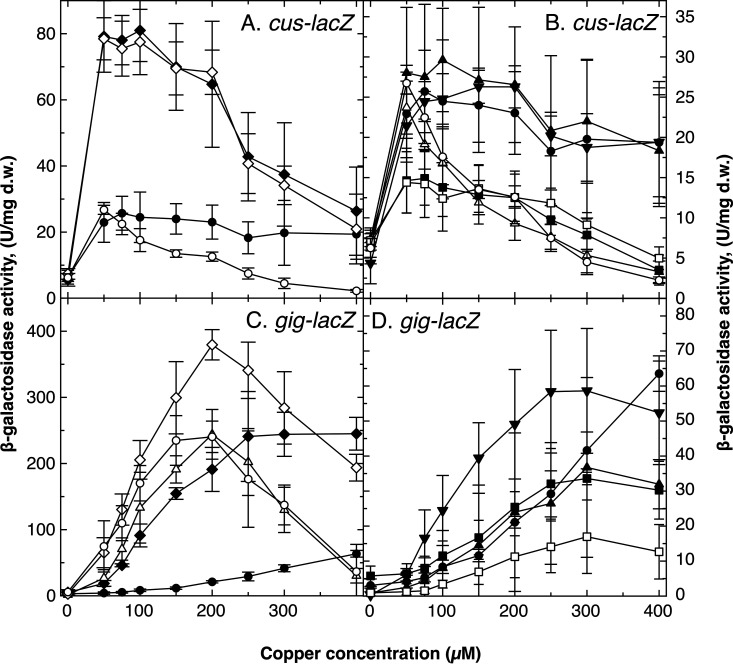
A lacZ reporter gene was inserted downstream of the cusDCBAF genes on the chromid of a variety of C. metallidurans AE104 mutants. The cells were incubated in the presence of increasing copper concentrations and the reporter activity was measured (Fig. 2A and B). In the parent strain AE104, cus-lacZ was upregulated by 50 μM copper to a specific activity of about 23 ± 8 U/mg dry mass and remained on this level with increasing copper concentrations. In the Δcop and Δcop Δgig mutants, the activity reached the 4-fold specific activity at 50 μM copper and decreased with increasing copper concentrations to the level of the AE104 parent cells of 20 U/mg at 400 μM Cu(II). This copper concentration was in the same range as the IC50 values of both strains.
FIG 2.
Regulation of cus and gig. Reporter gene fusions with the lacZ gene were constructed with the cus (A and B) and gig operons (C and D) in various mutant backgrounds. The strains were incubated in the presence of increasing copper concentrations and the beta-galactosidase activity was determined. A to D: strain AE104 (closed circles, ●), Δcop (closed diamonds, ◆), ΔgshA (closed inverted triangles, ▾), Δcop Δcup (open triangles, △), Δcup (closed squared, ◼). A and B: Δgig (closed triangles, ▴), Δcop Δgig (open diamonds, ◇), Δcop Δcup Δgig (open circles, ○), Δcup Δgig (open squares, ☐). C and D: Δcus (closed triangles, ▴), Δcop Δcus (open diamonds, ◇), Δcop Δcup Δcus (open circles, ○), Δcup Δcus (open squares, ☐). Deviations shown (n ≥ 3).
In total, four different modes of copper-dependent cus-activation were evident: first, the strong activation by copper in the Δcop ± Δgig mutants (Fig. 2A); second, activation similar to the parent cells in Δgig and ΔgshA cells; third, activation similar to the parent up to 50 μM added Cu(II) but a rapid decline at higher concentration in Δcop Δcup ± Δgig cells; and last, a lower level of activation at 50 μM Cu(II) followed by declining expression levels in Δcup ± Δgig cells. Since the IC50 values of all Δcup cells were ≤ 13.5 μM, this decline was probably the result of copper-mediated inhibition of the cells. Gig and GSH were not involved in regulation of cus. The strong activation in all Δcop mutants with lacking periplasmic Cu(I) oxidase CopA indicated that periplasmic Cu(I) ions were most likely the regulator of cus expression.
Regulation of the gig determinant.
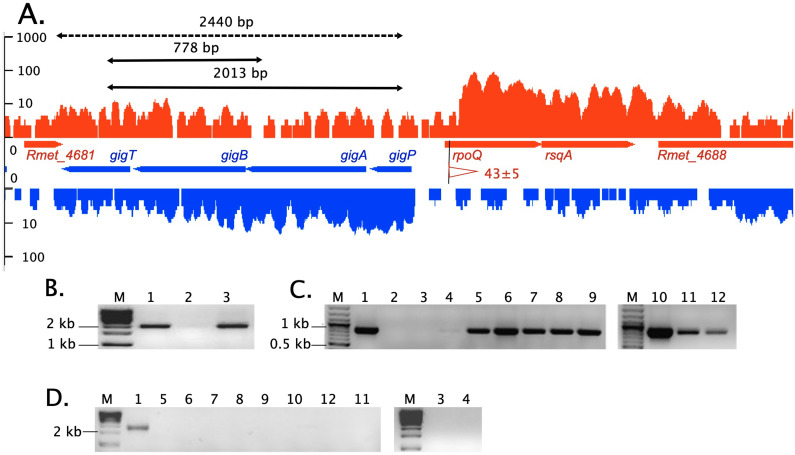
RT-PCR experiments verified the operon structure and regulation of gig transcription (Fig. 3). The gigPABT genes formed an operon from upstream of gigP to downstream of gigT but not beyond. These experiments also demonstrated that copper induced transcription of the gig (gold induced) genes as well as gold complexes.
FIG 3.
The gigPABT-rpoQ-rsqA gene region on the chromid of C. metallidurans. (A) As part of Fig. S1A, the open reading frames for the genes Rmet_4681, gigPABT, rpoQ, rsqA, and Rmet_4688 and the corresponding transcriptional activity in C. metallidurans wild-type cells as nucleotide activities per kilobase of exon model per million mapped reads (NPKM) values are shown in red (plus strand) or blue (minus strand) on a logarithmic scale (100). This region corresponds to base pairs 1.313.350 to 1.317.350 on the chromid (accession number CP000353). A transcriptional start site with a low TSS score (<50) is shown as a flag with the red color indicating dependence on RpoD (103). No transcriptional start site was found upstream of gigP under nonchallenging conditions. The two-sided arrows on the top show the result of an RT-PCR experiment with positive (solid line) or negative (dashed line) outcome. The RT-PCR results are shown in panels B to D with the marker (M) always shown on the left hand. (B) (2,031-bp fragment, primers gigT>gigP): 1, positive-control DNA; 2, negative-control water; 3, RNA from cells incubated in the presence of 25 μM Au(III). (C) (827 bp, primers gigT>gigA). (D) (2,458 bp, primers Rmet_4681>gigP): 1; positive and 2 negative control (not shown in panel D). Lanes 4 to 12 RT-PCR with RNA from cells incubated in the presence of various conditions: 3, no metal; 4 to 8 Au(III) (4, 30 min 10 μM; 5, 10 min 25 μM; 6, 30 min 25 μM; 7, 10 min 50 μM; 8, 30 min 50 μM); 9 to 12 Cu(II) (9; 10 min 100 μM; 10, 30 min 100 μM; 11, 30 min 500 μM; 12, 10 min 500 μM). Please note that the lanes in panel D, which correspond to that in panel C, are in a different order. The PCR results clearly prove the existence of the predicted gigPABT operon Op1355r_1.
A lacZ gene was inserted downstream of gigT and the activity of the gig-lacZ fusion was determined at increasing copper concentrations (Fig. 2C and D). In parent strain AE104, gig-lacZ activity increased with increasing copper concentrations to a specific activity of 67 U/mg. In a Δcop strain (Fig. 2C, closed diamonds), activity reached 250 U/mg at 250 μM copper and remained there up to a copper concentration of 400 μM (Fig. 2C). There was no decline of the reporter activity in the Δcop mutant with an IC50 of around 400 μM at this concentration, so the decline of the cus-lacZ fusion (Fig. 2A) was not the result of copper-mediated inhibition of translation but of a decreased upregulation of cus. The strong expression in Δcop cells indicated that gig was also regulated by periplasmic Cu(I) ions. Moreover, Gig was needed at higher copper concentrations than Cus.
Expression of gig-lacZ in the Δcop Δcus double mutant was even stronger than in the Δcop mutant and peaked at 200 μM added Cu(II). Since gig was regulated by periplasmic Cu(I) ions and its expression increased following deletion of cus in Δcop mutants, this was evidence that Cus indeed decreased the periplasmic Cu(I) level by CusCBA-mediated copper efflux to the outside.
Expression of gig-lacZ in Δcop Δcup ± Δcus cells also increased at copper concentrations up to 200 μM added Cu(II) and decreased thereafter, probably due to the lower copper tolerance of the Δcup deletion strains. Single mutants with a Δcup or a Δcus deletion regulated gig-lacZ similar to the parent (Fig. 2D) but decreased expression at 400 μM Cu(II). In the presence of Cop, a Δcus deletion no longer resulted in an upregulation of gig. Absence of GSH in the ΔgshA strain led to an upregulation of gig-lacZ.
DISCUSSION
The cellular stage for copper homeostasis.
Homeostasis of copper as an “essential-but-toxic” element is an important cellular process in many organisms. Consequently, copper resistance systems are widespread in cultivated bacteria and in complex communities in many ecosystems. Here, most important are Cup- and Cus-like systems (36). Copper tolerance is a virulence factor (37). It is not only involved in the interaction of pathogenic bacteria with a human host but also for survival of bacteria within amoeba (38), indicating an ancient origin of the strategy to use copper ions as “war heads” against intruding bacteria. This all points out the necessity to understand the full picture of copper toxicity, homeostasis, and resistance. These processes involve multiple pathways (39) and our publication describes that copper resistance is an emergent feature based on the interaction of several resistance and tolerance mechanisms, some of them acting in the periplasm of Gram-negative bacteria.
For cells growing under oxic conditions, copper is usually present as Cu(II) ion. The redox potential of the Cu(II)/Cu(I) at pH 7 is Eo’ = −267 mV (40). Cu(II) may pass the outer membrane of C. metallidurans and other Gram-negative bacteria by facilitated diffusion across unspecific porins or by TonB-dependent active transport across this membrane (1, 41–44). In the periplasm, Cu(II) can be reduced by contact with respiratory chain components (45) or by reducing equivalents provided by a cysteine-cystine shuttle (46), probably by electrons stemming from NADH (Eo’ = −320 mV) (47). Several pathways allow import of copper ions from the periplasm into the cytoplasm. Cu(II) can be imported as an unspecific substrate, for instance by the Zn(II)-importer ZupT of the ZIT/IRT (ZIP) transporter family (48). Subsequently, incoming Cu(II) should be immediately reduced to Cu(I) by a rapid, thiol-mediated process (28).
Copper is more toxic to E. coli under anaerobic conditions compared to aerobic conditions (10). Export of cytoplasmic copper in E. coli depends on a CupA-orthologue. Expression of the respective gene is under the control of the MerR-type regulator CueR with its zeptomolar Cu(I) affinity (49). The time-dependent profiles of the CueR-dependent expression of its target-genes clearly indicate that much more Cu(I) arrives in the cytoplasm under anaerobic conditions than under aerobic conditions, leading to increased anoxic copper sensitivity (10). An even lower resistance level occurs in E. coli cells growing in the presence of substrates that are imported by Na(I)-dependent import systems. This indicates that a third, unspecific Cu(I) import system exists in bacteria that may use Na(I)-dependent importers such as MelB. Na(I) binds to copper-binding proteins and copper ions inhibit Na(I) transport across membranes (50). C. metallidurans can survive anoxic conditions by using nitrate respiration (51). Mutants of C. metallidurans carrying multiple deletions in the genes for metal uptake systems nevertheless import metal ions by an unknown high-rate importer, which also transports Cu(I) and even gold ions into the cytoplasm (8, 34, 52, 53). Cu(I) is a better substrate for import into the cytoplasm than Cu(II) and unspecific high-rate uptake systems, e.g., by Na(I)-dependent transporters, may be responsible for Cu(I) import into the cytoplasm.
In Gram-negative bacteria such as E. coli, DNA is not damaged by Cu(I)-catalyzed oxidation in the presence of GSH since most of the hydrogen peroxide-oxidizable copper is located in the periplasm. Most of the copper-mediated hydroxyl radical formation occurs in this compartment (54), which affects processes here such as the assembly of c-type cytochromes (55). In the cytoplasm, one of the Cu(I) targets are FeS-clusters. FeS-dependent dehydratases are inhibited (56) and FeS-cluster assembly is interrupted (57–59), e.g., by binding of Cu(I) to IscA (60). Due to the higher reduction rate of Cu(II) to Cu(I) and the resulting higher import rates of copper ions, this leads to strong copper-mediated inhibition under anoxic conditions. The lower iron content of cells with deleted copper resistance determinants plus a ΔgshA mutation, especially in the presence of copper ions, indicates that iron homeostasis of C. metallidurans is also affected by copper and that GSH protects to some part against this damage (Table S4). Moreover, and reminiscent to Cd(II), Cu(I) strongly binds to thiol residues, leading to damaged proteins (61, 62) and upregulation of the small heat shock proteins IpoA/B (63), similar to Cd(II), which also damages FeS-clusters (22, 64).
In summary, Cu(II) is reduced to Cu(I) in the periplasm, under anoxic conditions more rapidly than under oxic conditions, and Cu(I) is a better substrate for import into the cytoplasm than Cu(II). Copper damages by formation of hydroxyl radicals in the periplasm, and in the cytoplasm by acting on FeS cluster or preventing their assembly, and by binding to and unfolding proteins. Consequently, cytoplasmic and periplasmic functions are needed to protect either compartment against copper-mediated damage, and the cytoplasmic membrane between them. Tolerance functions should decrease the availability of Cu(I), protect or repair targets that are sensitive to Cu(I)-mediated damage.
The strongest one is not strong enough alone: Cup needs partners.
C. metallidurans CH34 wild type contains four PIB1-type ATPases able to export Cu(I) from the cytoplasm to the periplasm. CupA and the plasmid pMOL30-encoded CopF are part of copper resistance determinants and only cupA is present in the plasmid-free strain AE104 that was used in this study. The cupA gene is followed by the gene for the metal-binding MerR-type and CueR-orthologous regulator cupR (49, 65) downstream of cupA. The gene cupC for a copper chaperone (29, 30) is located on the other DNA strand (Fig. S1C). CtpA and RdxI are “anabolic” PIB1-type ATPases involved in the assembly of periplasmic copper sites of copper-dependent proteins (4, 13, 66). The contribution of all four proteins to copper homeostasis of C. metallidurans CH34 wild type and its plasmid-free derivative AE104 has been characterized (8); however, the IC50 values published there and in this study cannot be directly compared because the Tris-buffered mineral salts medium has been improved in the meantime to reveal effects of metal starvation conditions at higher resolution. This has been accomplished by using mineral salts of higher purity. Especially, the sodium sulfate source previously used contained a larger amount of iron and zinc contaminations. This protected the cells so that the IC50 value of AE104 was 956 μM (8) compared to 615 μM published here (Table 1), and the IC50 of the ΔcupA mutant of strain AE104 was 400 μM (8) compared to 13.5 μM (Table 1), respectively. In strain AE104 cultivated in the previously used version of TMM, additional deletion of rdxI or of ctpA in the Δcup strain decreased the IC50 value by half and deletion of both down to 25% of the IC50 of the Δcup mutant (8). This would be an effect like a deletion of cus or cop in the Δcup mutant (Table 1).
CupA is the only PIB1-type ATPase in strain AE104 that is upregulated by excess cytoplasmic Cu(I) ions via the CueR-orthologue CupR and accepts its substrate from the cytoplasmic copper chaperone CupC (29, 30, 49, 65). Deletion of the cupC/AR determinant (Fig. S1D) results in a strong decrease of copper resistance in all mutant backgrounds (Table 1, Table S1). The Δcup deletion decreased the resistance level of strain AE104 45.6-fold to a resistance level half of that of a Cup+ quadruple mutant deleted in all the other copper resistance determinants (Δcop Δcus Δgig ΔgshA, 29.0 μM versus 13.5 μM, Table 1). Consequently, the Cup system is the strongest contributor to copper resistance in C. metallidurans because it mediates efflux of excess cytoplasmic Cu(I) to the periplasm, but the other four factors are needed for an additional 21-fold increase in copper resistance. Efflux and influx systems adjust the concentration of their substrate in a cellular compartment via a kinetic flow equilibrium of the export and import rates (67, 68). This explains the necessity of interaction partners for CupA, which influence import of copper into the cytoplasm.
The kinetic flow equilibrium of Cop and Cup controls the cytoplasmic copper levels.
The chromid-encoded cop determinant of the plasmid-free strain AE104 (Fig. S1E) contains genes for a periplasmic Cu(I) and Au(I) oxidase CopA (11), which is an orthologue of CueO from E. coli (16, 17, 69, 70). Additional cop products are the possible outer membrane-attached protein CopB, the periplasmic protein CopC, and the inner membrane protein CopD (71). Expression of cop in nonamended TMM medium is below the threshold of nucleotide activities per kilobase of exon model per million mapped reads (NPKM) = 10 (Fig. S1E) but expression can be upregulated about 10-fold by metal stress. The two-component regulatory system CopRS, which is encoded adjacent to copABCD on the other DNA strand, should be responsible for this upregulation.
CopC-like proteins usually contain distinct Cu(I) and Cu(II) binding sites and they are frequently fused as domain to a CopD-like domain (72) as in Bacillus subtilis (73) to form another copper uptake system, which is independent of the three import routes outlined above. Since CueO and its relatives are copper-dependent proteins, they are transported by the twin-arginine transport (TAT) system in a partially folded state (74). Cytoplasmic copper ions are needed to fold CopA/PcoA/CueO-like proteins and export them into the periplasm (71). It would be a disaster to the cells if a CopCD copper uptake system was produced in response to high periplasmic copper concentrations and would subsequently increase copper import into the cytoplasm and increase copper-mediated damage. Copper imported by CopCD should be exclusively used to fold CopA and not be released into the cytoplasm.
The outer membrane-attached and metal-binding (75) CopB protein contains at least two metal-binding motifs, HXHXCHXXH and EHXXXHXXDEH, which resemble His-rich Cu(II)-binding sites of CopC/PcoC-like proteins (76). CopA oxidizes Cu(I) back to Cu(II) which has two effects. First, CopA protects against copper-mediated killing (Table 2). Copper ions released from solid copper surfaces kill by damaging the cytoplasmic membrane (77–79). The cells are killed by the production of superoxide radicals by Cu(I)-mediated reduction of molecular oxygen, which is also responsible for the copper-mediated hydroxyl radical formation in the periplasm (54). Reoxidation of Cu(I) to Cu(II) in the periplasm by CopA prevents damage of the cytoplasmic membrane by reactive oxygen species and subsequent killing of the cells. CopB binds the resulting Cu(II) ions to prevent anew reduction. Second, since Cu(I) is a better substrate for copper uptake than Cu(II), oxidation of Cu(I) to Cu(II) by CopA decreases copper accumulation (Table 3). Acting together on the kinetic flow equilibrium that determines the cytoplasmic copper concentration, Cop and Cup increase the IC50 value of the respective Cop+ Cup+ triple deletion mutant Δcus Δgig ΔgshA to 472 μM, 77% of the IC50 of the parent. On the other hand, Cop alone was not able to mediate a resistance level above the IC50-min unless in cooperation with Cup, or Cus, GSH, or Gig (Table 1, Table S1). This indicates that Cus, Gig, and GSH should decrease the cytoplasmic or periplasmic Cu(I) concentration, repair Cu(I)-mediated membrane damage, or support Cop in another way.
Cus exports periplasmic but not many cytoplasmic Cu(I) ions to the outside.
Reminiscent to cop, the cusDCBAF determinant on the chromid (Fig. S1C) is not expressed in nonamended mineral salts medium but upregulated under metal stress conditions. Only the cusD gene is expressed in nonamended medium. There is no two-component regulatory system able to sense copper encoded in the vicinity of cus so that CopRS might be responsible for cus expression like CusRS from E. coli, which also controls expression of pcoE (80). CusD is a periplasmic lipoprotein with a TAT leader as indicated by a signalP prediction (81) (Fig. S5) with several metal binding motifs such as MXMXXMDEHXXMEXXMXCXDM, CMXHC, HXDH, and HXXXHCC (68), indicating the potential to sequester Cu(I). Periplasmic CusF-like proteins were first found in E. coli (19) and bind Cu(I) to a WXHD-MXM site (82, 83). CusF can accept copper ions from the CupA-orthologue of E. coli (84) and deliver them to the CusCBA transenvelope efflux complex (85, 86). The CusF orthologue from C. metallidurans is only distantly related to the E. coli protein although both belong to the CusF-Ec protein superfamily. The copper binding site of the E. coli protein is not present in the C. metallidurans protein but Met-rich stretches that might represent three triple-Met Cu(I) binding sites, which would allow CusF from C. metallidurans to accept Cu(I) from CusD.
The CusCBA transenvelope efflux complex was first identified in E. coli (18, 19, 80). Because CusCBA can substitute a missing periplasmic CueO/CopA/PcoA-type Cu(I) oxidase (16, 70) and the periplasmic CusF delivers copper ions directly to this protein complex (85, 86), CusCBA exports periplasmic Cu(I) to the outside. In C. metallidurans, cus expression is strongly upregulated in a cop strain (Fig. 2A), which is unable to oxidize periplasmic Cu(I), so that periplasmic Cu(I) regulates cus expression. This may be accomplished by the CopRS two-component regulatory system, which is still present in Δcop mutant cells (Fig. S1E). The gig determinant is also upregulated in Δcop mutants (Fig. 2C), so that periplasmic Cu(I) also serves as inducer for gig expression. Metal-sensing histidine kinases such as CopS and CusS indeed sense periplasmic copper ions (9) so that regulation of cus expression in C. metallidurans by periplasmic copper ions is highly probable. Since the gig operon was more strongly upregulated in a Δcus Δcop double mutant than in a Δcop single mutant (Fig. 2C), presence of Cus clearly decreases the periplasmic Cu(I) concentration in C. metallidurans.
C. metallidurans can survive under anaerobic conditions by nitrate respiration (51) but the oxygen-dependent CopA protein cannot oxidize Cu(I) here; CusCBA would be important to remove periplasmic Cu(I) especially under anaerobic conditions, as has been shown for E. coli (17, 87). This makes Cus- together with Cup-like systems into the most frequently occurring copper resistance systems in gamma proteobacteria (88) and in natural communities (36).
As previously shown for another metal-exporting efflux system (89), CusA can export in vitro metal ions across a membrane that would resemble the cytoplasmic membrane in vivo (90, 91). Presence of an open periplasmic and a cytoplasmic copper binding site and a transport channel between them, however, would result in a copper uniport from the periplasm to the cytoplasm. The CusA-mediated transport of Cu(I) from the cytoplasm across the membrane has either no relevance in vivo or a regulatory function, for instance, CusCBA functions only when sufficient cytoplasmic copper ions are present. In C. metallidurans, Cus alone was not able to mount any copper resistance above the IC50-min value (Table 1, Table S1). Presence of Cus increased copper resistance only in the presence of Cup, Cop, or Gig plus GSH. Export of excess cytoplasmic Cu(I) by CupA to the periplasm and successively by CusF and CusCBA to the outside would sufficiently explain the interaction of Cup and Cus, without assuming an export of cytoplasmic Cu(I) by Cus. Periplasmic Cu(I) ions oxidized by CopA back to Cu(II) may result in a new reduction of the ions not sequestered by CopB, which would be a futile cycle and export of periplasmic Cu(I) by Cus could interrupt this cycle. This function of Cus also fully explains the fact that Cus can substitute a missing Cop system in Cup+ strains. On the other hand, together with Cop, Gig, and GSH, Cus mediates the copper resistance level of the Δcup mutant of IC50 = 13.5 μM. This IC50 is 3.17-fold decreased by a Δcus deletion (Table S1). Cus was also able to substitute missing Cup in Cop+ strains, and Cop as well as Cup were able to substitute a missing Cus, despite the different functions of Cup and Cop.
Together with Gig and GSH, Cus was able to increase copper resistance of C. metallidurans 4.2-fold above the IC50-min value. The Cop system needs either Gig or GSH to mediate an 8.9- or 4.7-fold increase, respectively, above the IC50-min value but Cus requires them both to substitute 1/3 of the performance of Cop. Gig and GSH perform an overlapping function with respect to Cop but an essential, additive function with respect to Cus. Possible contributions of the Gig and GSH to copper tolerance could be (i) repair of copper-mediated damage, (ii) oxidation of periplasmic Cu(I) to substitute Cop, (iii) sequestration of cytoplasmic or periplasmic copper ions, or (iv) reduction of Cu(II) to Cu(I) to feed the resulting ion into Cus-mediated efflux from the periplasm or the cytoplasm. A missing Cop system resulted in increased accumulation of copper by the cells even in the presence of Cup because more periplasmic Cu(I) as the substrate for import into the cytoplasm was available and the kinetic flow equilibrium of uptake and CupA-mediated export reactions reached a new, higher concentration level. GSH quenched copper accumulation in Δcop cells by interacting with Cus. Gig could substitute GSH here to some part (Table 3). Repair of copper-mediated damage by Gig or GSH would not explain this fact. Sequestration of copper by Gig or GSH would increase the copper content of Δcop cells independent of Cus, leaving a redox change of copper ions in the periplasm or cytoplasm as possible functions of Gig and GSH.
The redox potential of Cu(II)/Cu(I) of E0’ = −267 mV (40) is close to the redox potential of GSSG/GSH of E0’ = −240 mV under standard conditions and E0’ = −260 mV in vivo (92, 93), leading to a ratio of GSH to GSSG in the cytoplasm of 1,000:1, according to the Nernst equation (22). Taking the low concentration of copper in the cytoplasm and the high ratio GSH to GSSG into account, oxidation of Cu(I) by GSSG is not a favorable reaction but instead Cu(II) is rapidly reduced by GSH (26–28, 94, 95). The resulting Cu(I) is promptly bound by CupC and exported by CupA. In the periplasm and in the Δcop strain with its upregulated cus expression, a reduction of Cu(II) to Cu(I) for the purpose of exporting the resulting Cu(I) by CusCBA so that Cu(I) cannot be imported into the cytoplasm would require an efficient Cu(I) removal by CusCBA. The increased copper sensitivity of E. coli under anaerobic conditions (10) would not agree with such an efficient Cu(I) removal by Cus, at least not in E. coli.
In the periplasm, the DsbA/DsbB-, thioredoxin/DsbB-, or glutaredoxin/GSSG-mediated higher redox level (96) could indeed oxidate Cu(I) back to Cu(II) as an alternative, CopA-independent pathway of Cu(I) oxidation, which cooperates with removal of periplasmic Cu(I) by Cus. Adding Gig plus GSH to a Cus+-only quadruple mutant increased its copper resistance 33% compared to the increase resulting from the addition of Cop to this quadruple mutant (Table S1), so that the efficiency of a possible Gig/GSSG electron shuttle should be 33% of that of CopA, and both Gig and GSSG are needed for this little increase in resistance. The decrease of the GSH content of the C. metallidurans Δcus mutant (Table 4) would agree to this fact because more exported, periplasmic GSSG would be needed if Cus no longer exports periplasmic Cu(I) ions.
On the other hand, Gig or GSH are required for CopA to mediate any increase in the resistance level. Since CopA is 3-times more efficient than Gig/GSSG, both are not needed for Cu(I) oxidation when CopA is present. But CopA is exported in a partially folded form (74). GSH may be required for efficient loading of copper into nascent CopA in the cytoplasm and Gig in the periplasm. This would explain the 4.72-fold increase above the IC50-min value resulting from addition of Cop to the Gig+-only and the 9.81-fold increase by addition of Cop to the GSH-only quadruple mutants, respectively (Table S1).
These contributions of GSH and Gig to copper tolerance also indicate that the main physiological function of Cus is export of periplasmic Cu(I) to the outside. If Cus also exports cytoplasmic Cu(I) ions, the export rate should be in the range of Cu(I) export by the anabolic PIB1-type ATPases RdxI and CtpA and much lower than that mediated by CupA. Cus interacts with Cup to form an export route of excess cytoplasmic Cu(I) by CupA-CusF-CusCBA, with Cop to remove periplasmic Cu(I) by oxidation, and with Gig plus GSH to the same end.
Hints concerning a possible function of Gig.
The gigPABT operon (Fig. 3) is in a divergon situation with the gene for the extracytoplasmic function (ECF) sigma factor (97–102) gene rpoQ and a putative gene for an antisigma factor. In nonamended growth medium, gig is expressed on a low level around the threshold of NPKM = 10 (Fig. S1A). Due to the low expression level, no promoter upstream of gig has been identified (103). The rpoQ-rsqA region for the sigma factor and its predicted membrane-bound antisigma factor RsqA depends on the main housekeeping sigma factor RpoD (103). No evidence for initiation of expression of gig from a RpoQ-dependent RNA polymerase has been obtained since deletion of rpoQ results in an upregulation of gig expression in metal-treated mutant cells (100) due to a compensatory mechanism of the sigma factor network of C. metallidurans. Since gig was clearly upregulated in response to copper ions in Δcop cells more than in parent cells and in Δcop Δcus cells more than in Δcop cells (Fig. 2B), gig is under the control of periplasmic signals. Regulation by RpoQ and its antisigma factor can be assumed despite the currently lacking further experimental evidence. This would also hint that Gig may protect against stress originating in the periplasm (98, 102).
The first Gig protein is GigP (Rmet_4685), a small putative periplasmic protein without an outstanding metal binding motif. The overall size is 94 aa in length of the preprotein, minus 22 aa for the leader as determined by SignalP 5.0. The remaining 72-aa peptide contains 4 Cys and 5 Met residues not very close to each other. The low hydrophobicity of the 72 aa argues for a periplasmic rather than an integral membrane protein. GigA (Rmet_4684) is a metal-binding protein with a few His-containing putative metal-binding motifs, which resemble trinuclear zinc-binding sites involved in phosphodiester cleavage in endonuclease IV involved in base excision repair (104). GigB (Rmet_4683) is a predicted cytoplasmic protein with a thioredoxin fold, His- and Cys-containing metal-binding motifs. GigB contains a DUF2063 domain at the amino terminus, which occurs in the DNA-binding protein NGO1945 from Neisseria gonorrhoeae (105). GigT (Rmet_4682) has three or four predicted transmembrane spans and is loosely related to DoxX (COG2259), a subunit of a terminal quinol oxidase in an archaeon. While GigA and GigB might be associated with the DNA, GigP and GigT may have a function in the cellular envelope, cytoplasmic membrane, or periplasm.
The distant relationship of GigT with a component of a quinol oxidase indicates the potential of GigT and GigP to oxidize periplasmic Cu(I)-GSSG and channel the resulting electron into the quinol pool, which would explain why Gig plus glutathione enable Cus to mediate some increase in copper resistance above the IC50-min (Table 1, Table S1). Repair or protection of the DNA by GigB and GigA is also in agreement with the importance of Gig in the mutants Δcup ΔgshA, Δcup Δcus ΔgshA, and Δcop Δcup, which are devoid of Cup-mediated removal of cytoplasmic Cu(I) ions in combination with increased import and decreased removal of Cu(I) bound to proteins by GSH, including zinc-binding enzymes involved in DNA repair.
Cop needs Cup, Cus, GSH, or Gig to mediate some increase in copper resistance above the IC50-min level. Cup is required as Cu(I)-exporting counterpart for excess cytoplasmic copper ions while Cop oxidizes periplasmic Cu(I) back to Cu(II) to decrease copper uptake into the cytoplasm. Cop and Cus remove periplasmic Cu(I) alternatively by oxidation or efflux, respectively. GSH may be required to load copper imported by CopCD in the cytoplasm into the pre-CopA apoprotein ahead of its TAT-mediated export to the periplasm. GigPT could also be involved of an efficient activation of apo-CopA, e.g., by mobilization of periplasmic Cu(I) for subsequent import by CopCD. In summary, Gig contributes to copper tolerance, but its contribution is small, only visible in a few mutant backgrounds and the biochemical mechanisms behind its function have not been revealed yet (Fig. 1). This may change when gold complexes are added to the picture.
GSH does not form a copper-complexing pool in the cytoplasm.
In Streptococcus pyogenes, glutathione buffers excess intracellular copper (106). The higher copper content of GSH+ cells of C. metallidurans compared to ΔgshA cells indicates that presence of GSH also leads to increased cellular copper binding in this bacterium (Table 3) but presence of GSH did not increase the copper resistance of the Cup+-only mutant (Δcop Δcus Δgig ΔgshA) or Δcup mutant at all (Table S1). GSH was not required for function of Cup and did not substitute a missing Cup system.
In the presence of copper ions, copper-catalyzed oxidation of GSH to GSSG produces hydrogen peroxide and superoxide radicals (27, 95). Cu(II) is immediately reduced by the high intracellular GSH content to Cu(I)-GSH2 complexes, which react with molecular oxygen to Cu(II)-GSSG and superoxide radicals (95). GSH is able to reduce Cu(II) bound to GSSG again and this reaction is quantitative and complete due to the high intracellular concentrations of GSH (26). This is an exceptional reaction only catalyzed by GSH while other thiols bind Cu(II) and convert subsequently to time-stable Cu(I)-thiol complexes. Consequently, GSH cannot protect the cytoplasm against copper-mediated oxidative damage but is the source of it in aerobic bacteria. Cytoplasmic copper chaperones such as CupC in C. metallidurans or its orthologue from E. coli (107, 108) are needed to keep Cu(I) away from GSH and to deliver the ion to CupA-like proteins for export to the outside. One function of GSH is to prevent interaction of copper with cytoplasmic proteins, which leads to unfolding reactions (109), a second to repair oxidative damage caused by its own interaction with Cu(I).
GSH in C. metallidurans was not required for the function of CupA but nearly all mutants with a Δcop mutation now needed GSH, except those with a Δcop Δgig double mutation. Both, Cu(I) and Cu(II) bind to GSSG, the oxidized form of GSH that is the predominant form in the periplasm (26, 95, 96, 110). In the cytoplasm, only GSH is able to reduce Cu(II) bound to GSSG (26). In the periplasm, a Gig-mediated oxidation of Cu(I)-GSSG would explain why GSH is especially required in Δcop mutants (Table 1) for removal of periplasmic Cu(I) ions as outlined above. GSH can be exported into the periplasm by a CydDC-type bacterial glutathione exporter (111). Four proteins in C. metallidurans are related to CydD: Rmet_0391 or AtmA (29% identity in 428 aa), Rmet_0705 (27% identity in 515 aa), Rmet_2516 (30% identity in 426 aa), and Rmet_0757 (24% identity in 493 aa). Three proteins are related over a long range to CydC: Rmet_2516 (37% in 289 aa), Rmet_0705 (28% in 493 aa), and Rmet_0757 (26% in 508 aa) again. Rmet_0757 is probably the lipid A exporter MsbA but AtmA, Rmet_0705, and Rmet_2516 could export GSH or GSH complexes, and AtmA is involved in nickel tolerance in C. metallidurans (112).
Conclusion.
Copper resistance in the plasmid-free C. metallidurans strain AE104 is accomplished by the interaction of many systems and not predominantly by the function of a single factor (Fig. 1). The Cup system is under the control of the cytoplasmic Cu(I) content via the MerR-type regulator CupR, binds these ions to CupC, and exports them to the periplasm by the PIB1-type ATPase CupA. In analogy to other bacteria, Cu(I) may be forwarded to CusCBA for further export to the outside by CusF. While the CusCBA transenvelope efflux system clearly affects the periplasmic copper concentration, no evidence argues for an efficient CusCBA-mediated efflux of cytoplasmic Cu(I) ions in vivo. In contrast to E. coli, the Cus system from C. metallidurans additionally contains the periplasmic TAT-exported lipoprotein CusD, which contains a copper binding site, may sequester periplasmic Cu(I) close to the cytoplasmic membrane, and forward the ions via CusF to CusCBA for export. While Cus removes excess periplasmic Cu(I) by export to the outside, the Cop system oxidizes Cu(I) to Cu(II), which is inferior to Cu(I) as the substrate for copper import, leading to a Cop-mediated decreased copper accumulation. Moreover, Cop also protects against copper-mediated oxidative damage and subsequent killing of the cells. CopA is a copper-containing oxygen-dependent Cu(I) oxidase. CopC and CopD may import copper into the cytoplasm for loading of copper into CopA and subsequent TAT-mediated export into the periplasm. Glutathione could stimulate this process. CopB is attached to the outer membrane and seems to bind Cu(II) to prevent reduction of Cu(II) to Cu(I) by respiratory chain components in the cytoplasmic membrane. Glutathione seems not to be a cytoplasmic Cu(I) buffer because interaction of GSH with copper ions leads to production of superoxide radicals. Instead, it is required for full function of Cop, maybe by assisting insertion of copper into apo-CopA, and periplasmic GSSG may interact with the Gig system to mediate a CopA-independent oxidation of periplasmic Cu(I). Moreover, Gig, which is under the control of periplasmic copper ions and a minor contributor to copper tolerance, contains two proteins that may protect the DNA against copper-mediated damage in the absence of CupA and GSH, or repair this damage, or circumvent DNA-interacting zinc-dependent proteins inactivated by copper. In comparison with the plasmid-free C. metallidurans strain AE104, the wild type additionally contains a huge copper resistance region on plasmid pMOL30, which adds another level of sophistication to copper homeostasis of this bacterium.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains used for experiments were derivatives of the plasmid-free derivative AE104 of C. metallidurans CH34 (3) and are listed in Table S5 in the supplemental material. Tris-buffered mineral salts medium (3) containing 2 g sodium gluconate/l (TMM) was used to cultivate these strains aerobically with shaking at 30°C. The medium had been improved compared to the previously published medium by choosing mineral salts with a higher purity. Solid Tris-buffered media contained 20 g/L agar. Strains were routinely transferred to fresh TMM plates every 2 weeks and taken from the −80°C stock culture twice a year.
Dose-response growth curves in 96-well plates were conducted in TMM.
A preculture was incubated at 30°C, 200 rpm up to early stationary phase, then diluted 1:20 into fresh medium and incubated for 24 h at 30°C and 200 rpm. Overnight cultures were used to inoculate parallel cultures with increasing metal concentrations in 96-well plates (Greiner). Cells were cultivated for 20 h at 30°C and 1,300 rpm in a neoLab Shaker DTS-2 (neoLab, Heidelberg, Germany) and the optical density was determined at 600 nm as indicated in a TECAN Infinite 200 PRO reader (Tecan Group Ltd., Männedorf, Switzerland). To calculate the IC50 values (inhibitory concentration: metal concentration that led to turbidity reduction by half) and the corresponding b value (measure of the slope of the sigmoidal dose-response curve), the data were adapted to the formula OD(c) = OD0/{1 + exp([c − IC50]/b)}, which is a simplified version of a Hill-type equation, as introduced by Pace and Scholtz (1997) (113, 114). OD(c) is the turbidity at a given metal concentration, OD0 that had no added metal and c, the metal concentration.
β-galactosidase assay.
C. metallidurans cells with a lacZ reporter gene fusion were cultivated as a preculture in TMM containing 1.5 g/L−1 kanamycin at 30°C, 250 rpm for 30 h, diluted 20-fold into fresh medium with 1 g/L−1 kanamycin, incubated with shaking at 30°C for 24 h, diluted 50-fold into fresh medium, and incubated with shaking at 30°C until a cell density of 100 Klett units was reached. This culture was distributed into sterile 96-well plates (Greiner Bio-One, Frickenhausen, Germany). After addition of metal salts, incubation in the 96-well plates was continued for 3 h at 30°C in a neoLab Shaker DTS-2 (neoLab Migge Laborbedarf, Heidelberg, Germany). The turbidity at 600 nm was determined in a TECAN Infinite 200 Pro reader (TECAN, Männedorf, Switzerland) and the cells sedimented by centrifugation at 4°C for 30 min at 4,500 × g. The supernatant was discarded and the cell pellets were frozen at −20°C. For the enzyme assay, the pellet was suspended in 190 μL Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.5 M beta-mercaptoethanol) and 10 μL permeabilization buffer was added (6.9 mM CTAB, cetyl-trimethyl-ammonium bromide, 12 mM sodium deoxycholate). The suspension was incubated with shaking at 30°C and 20 μL ONPG solution (13.3 mM ortho-nitrophenyl-beta-d-galactopyranoside in Z-buffer without beta-mercaptoethanol) were added. Incubation was continued with shaking in a neoLab Shaker DTS-2 at 30°C until the yellow color of o-nitrophenol was clearly visible and stopped by addition of 50 μL 1 M Na2CO3. The extinction at 420 nm and 550 nm was measured in a TECAN Infinite 200 Pro reader. The activity was determined as published (115) with a factor of 315.8 μM calculated from the path length of the 96-well plate and the extinction coefficient of o-nitrophenol:
specific activity: activity divided by the cellular dry mass as published (115).
Genetic techniques.
Standard molecular genetic techniques were used (116, 117). For conjugative gene transfer, overnight cultures of donor strain E. coli S17/1 (118) and of the C. metallidurans recipient strains grown at 30°C in Tris- buffered medium were mixed (1:1) and plated onto nutrient broth agar. After 2 d, the bacteria were suspended in TMM, diluted, and plated onto selective media as previously described (116). Primer sequences are provided in Table S6.
Gene deletions.
Primer sequences are also provided in Table S6. Plasmid pECD1002, a derivate of plasmid pCM184 (119), was used to construct deletion mutants. These plasmids harbor a kanamycin resistance cassette flanked by loxP recognition sites. Plasmid pECD1002 additionally carries alterations of 5 bp at each loxP-site. Using these mutant lox sequences, multiple gene deletions within the same genome are possible without interferences by secondary recombination events (120, 121). Fragments of 300 bp upstream and downstream of the target gene were amplified by PCR, cloned into vector pGEM T-Easy (Promega), sequenced, and further cloned into plasmid pECD1002. The resulting plasmids were used in a double-crossover recombination in C. metallidurans strains to replace the respective target gene by the kanamycin-resistance cassette, which was subsequently also deleted by transient introduction of cre expression plasmid pCM157 (119). Cre recombinase is a site-specific recombinase from the phage P1 that catalyzes the in vivo excision of the kanamycin resistance cassette at the loxP recognition sites. The correct deletions of the respective transporter genes were verified by Southern DNA-DNA hybridization. For construction of multiple deletion strains, these steps were repeated. The resulting mutants carried a small open reading frame instead of the wild-type gene to prevent polar effects.
Gene insertions and disruptions.
For reporter operon fusions, lacZ was inserted downstream of several targets. This was done without interrupting any open reading frame downstream of the target genes to prevent polar effects. The 300- to 400-bp 3′ ends of the respective target genes were amplified by PCR from total DNA of strain AE104 and the resulting fragments cloned into plasmid pECD794 (pLO2-lacZ) (122). The respective operon fusion vectors (pECD1386 for cusF-lacZ, pECD1667 for gigT-lacZ) were inserted into the open reading frame of the target gene by single crossover recombination. Using the 300-bp 5′ part of the gshA gene, this procedure was also used to interrupt the gshA gene with pECD1668 in strain AE104 parent strain and its mutant derivatives.
PCR.
DNA was amplified by PCR with 0.2 μM each primer and 1.5 U of Taq polymerase (Roche Diagnostics GmbH, Mannheim, Germany). All primer pairs used are listed in Table S6. The resulting amplified DNA was separated on agarose gels stained with ethidium bromide (117).
RNA isolation.
C. metallidurans cells were cultivated as described above. At a cell turbidity of 150 Klett, metal salts were added. After a 10- or 30-min incubation at 30°C, the cells were rapidly harvested and stored at −80°C. Total RNA was isolated as published (123) DNase treatment was performed. RNA concentration was determined photometrically, and RNA quality was checked on formamide gels (117).
RT-PCR.
For the RT reaction, 1 μg of total RNA and 0.1 μg hexamer primers were incubated at 65°C for 5 min and snap-cooled on ice. After addition of 0.5 mM (each) dATP, dGTP, dTTP, and dCTP, 20 mM DTT, and 100 U of reverse transcriptase (superscript II) in reaction buffer (Thermo Fisher Scientific, Germany), reverse transcription proceeded for 10 min at room temperature, followed by 1 h at 42°C. After finishing the RT reaction, the enzyme was inactivated at 70°C for 10 min. Then, 1 μL of the resulting cDNA was amplified by PCR with 0.2 μM each primer and 1 U of Taq-Polymerase (Roche Diagnostics GmbH, Mannheim, Germany). A no-template control and a no-RT (negative) control were performed under identical conditions as for the target genes.
Live/Dead staining of surface-exposed cells.
Live/Dead staining (Live/Dead BacLight bacterial viability kit, LifeTechnologies, Darmstadt, Germany) differentiates between undamaged and damaged bacterial membranes by employing two fluorescence dyes, which intercalate into DNA. Live, undamaged bacteria fluoresce green, while membrane-damaged bacteria fluoresce red. This is because the green stain SYTO 9 penetrates the membrane of both undamaged and damaged cells, while the red stain propidium iodide can only penetrate damaged membranes, and after intercalating into the bacterial DNA, reduces the green fluorescence of SYTO 9.
A preculture incubated at 30°C, 200 rpm for 18 h in TMM (with kanamycin for mutant strains carrying a gene disruption) was diluted into fresh medium at 5% and incubated at 30°C shaking, 200 rpm for 24 h. Cells were diluted 10-fold into fresh medium and continue shaking at 200 rpm for 20 h. Cultures contained no copper or copper resembling the IC50 concentration (calculated from dose response experiments) unique for each strain. After incubation cells were harvested and washed two times in saline (0.85% NaCl solution). Pellets were resuspended in 80 μL saline and divided in two samples each containing 40 μL of suspension. One sample was diluted with 800 μL of saline (Live sample), the other with 70% isopropanol (Dead sample) and incubated for 1 h at room temperature. Cells were harvested, washed two times in saline and resuspended in 400 μL saline. Cell suspensions were mixed 1:1 in a 1:1 mixture of Syto9 and propidium iodide solution (L13152 Live/Dead BacLight bacterial viability kit, Thermo Fisher Scientific, Dreiech), incubated for 10 to 15 min and examined in a confocal fluorescence microscope (Carl Zeiss MicroImagine, Jena, Germany) with λEx 450 to 490/546 nm, λEm 520/590 nm.
Inductively coupled plasma mass spectrometry.
Cells were incubated in TMM for 20 h at 30°C shaking at 200 rpm, diluted 20-fold into fresh TMM medium, and shaking continued at 30°C for 24 h. Cells were diluted 50-fold into fresh medium containing added copper or not, and continued shaking at 30°C at 200 rpm until 150 Klett were reached (midexponential phase of growth). Then, 10 mL of the cells were harvested by centrifugation, washed twice with 50 mM TrisHCl buffer (pH 7.0) containing 50 mM EDTA at 0°C and suspended in 50 mM TrisHCl buffer (pH 7.0). For inductively coupled plasma mass spectrometry (ICP-MS) analysis, HNO3 (trace metal grade; Normatom/PROLABO) was added to the samples to a final concentration of 67% (wt/vol) and the mixture mineralized at 70°C for 2 h. Samples were diluted to a final concentration of 2% (wt/vol) nitric acid. Indium and germanium were added as internal standards at a final concentration of 10 ppb each. Elemental analysis was performed via ICP-MS using Cetac ASX-560 sampler (Teledyne, Cetac Technologies, Omaha, Nebraska), a MicroFlow PFA-200 nebulizer (Elemental Scientific, Mainz, Germany) and an ICAP-TQ ICP-MS instrument (Thermo Fisher Scientific, Bremen) operating with a collision cell and flow rates of 4.8 mL × min−1 of He/H2 (93%/7% [124]), with an Ar carrier flow rate of 0.76 L × min−1 and an Ar make-up flow rate at 15 L × min−1. An external calibration curve was recorded with ICP-multi-element standard solution XVI (Merck) in 2% (vol/vol) nitric acid. The sample was introduced via a peristaltic pump and analyzed for its metal content. For blank measurement and quality/quantity thresholds, calculations based on DIN32645 TMM were used. The results were calculated from the ppb data as atoms per cell as described (125).
Glutathione determination.
Cells were cultivated as described above for ICP-MS. Next, 5 mg cells were harvested (15 min, 4,500 × g, 4°C) and washed twice with TMM. Cell pellets were resuspended in 100 μL 5% sulfosalicylic acid (SSA) solution and disrupted by three freeze-thawing cycles (liquid nitrogen, water bath at 37°C, 2 min per treatment). Cell debris was removed by centrifugation (10 min, 15,300 × g, 4°C). The supernatant was used to determine the protein concentration with the QuantiPro bicinchoninic acid assay (BCA assay) kit (Sigma-Aldrich, Taufkirchen, Germany) using bovine serum albumin as a standard and to measure the GSH content by the glutathione assay kit (CS0260, Sigma-Aldrich, Taufkirchen, Germany) according to manufacturer’s instructions. The enzymatic determination of the total amount of glutathione (GSH and GSSG) after deproteinization with SSA were measured photometrically at 412 nm by increasing amounts of TNB [5,5=dithiobis(2-nitrobenzoic acid)] using a kinetic assay.
Statistics.
Student’s t test was used but in most cases the distance (D) value has been used several times previously for such analyses (7, 33, 34). It is a simple, more useful value than Student's t test because nonintersecting deviation bars of two values (D > 1) for three repeats always means a statistically relevant (≥95%) difference provided the deviations are within a similar range. At n = 4, significance is ≥97.5%, at n = 5 ≥ 99% (significant), and at n = 8 ≥ 99.9% (highly significant).
ACKNOWLEDGMENTS
We thank Grit Schleuder for skillful technical assistance and for keeping our strain collection for nearly 30 years. This paper was initiated by Josè Argüello and his tale of the spider at the copper conference 2018 in Sorrento. The work was funded by the Deutsche Forschungsgemeinschaft as Ni262/20-1.
Footnotes
Supplemental material is available online only.
Contributor Information
Dietrich H. Nies, Email: d.nies@mikrobiologie.uni-halle.de.
John R. Spear, Colorado School of Mines
REFERENCES
- 1.Nies DH. 2016. The biological chemistry of the transition metal “transportome” of Cupriavidus metallidurans. Metallomics 8:481–507. doi: 10.1039/c5mt00320b. [DOI] [PubMed] [Google Scholar]
- 2.Mergeay M, Houba C, Gerits J. 1978. Extrachromosomal inheritance controlling resistance to cadmium, cobalt and zinc ions: evidence for curing a Pseudomonas. Arch Int Physiol Biochim 86:440–441. [PubMed] [Google Scholar]
- 3.Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, van Gijsegem F. 1985. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol 162:328–334. doi: 10.1128/jb.162.1.328-334.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, Michaux A, Benotmane MA, Leys N, Vallaeys T, Lapidus A, Monchy S, Medigue C, Taghavi S, McCorkle S, Dunn J, van der Lelie D, Mergeay M. 2010. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS One 5:e10433. doi: 10.1371/journal.pone.0010433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reith F, Rogers SL, McPhail DC, Webb D. 2006. Biomineralization of gold: biofilms on bacterioform gold. Science 313:233–236. doi: 10.1126/science.1125878. [DOI] [PubMed] [Google Scholar]
- 6.Reith F, Etschmann B, Grosse C, Moors H, Benotmane MA, Monsieurs P, Grass G, Doonan C, Vogt S, Lai B, Martinez-Criado G, George GN, Nies DH, Mergeay M, Pring A, Southam G, Brugger J. 2009. Mechanisms of gold biomineralization in the bacterium Cupriavidus metallidurans. Proc Natl Acad Sci USA 106:17757–17762. doi: 10.1073/pnas.0904583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiesemann N, Mohr J, Grosse C, Herzberg M, Hause G, Reith F, Nies DH. 2013. Influence of copper resistance determinants on gold transformation by Cupriavidus metallidurans strain CH34. J Bacteriol 195:2298–2308. doi: 10.1128/JB.01951-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wiesemann N, Bütof L, Herzberg M, Hause G, Berthold L, Etschmann B, Brugger J, Martinez-Criado G, Dobritzsch D, Baginsky S, Reith F, Nies DH. 2017. Synergistic toxicity of copper and gold compounds in Cupriavidus metallidurans. Appl Environ Microbiol 83:e01679-17. doi: 10.1128/AEM.01679-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rismondo J, Große C, Nies DH. 2023. The sensory histidine kinase CusS of Escherichia coli senses periplasmic copper ions. Microbiol Spectr 11. doi: 10.1128/spectrum.00291-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Große C, Schleuder G, Schmole C, Nies DH. 2014. Survival of Escherichia coli cells on solid copper surfaces is increased by glutathione. Appl Environ Microbiol 80:7071–7078. doi: 10.1128/AEM.02842-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bütof L, Wiesemann N, Herzberg M, Altzschner M, Holleitner A, Reith F, Nies DH. 2018. Synergetic gold-copper detoxification at the core of gold biomineralisation in Cupriavidus metallidurans. Metallomics 10:278–286. doi: 10.1039/c7mt00312a. [DOI] [PubMed] [Google Scholar]
- 12.Monchy S, Benotmane MA, Janssen P, Vallaeys T, Taghavi S, van der Lelie D, Mergeay M. 2007. Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J Bacteriol 189:7417–7425. doi: 10.1128/JB.00375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monchy S, Benotmane MA, Wattiez R, van Aelst S, Auquier V, Borremans B, Mergeay M, Taghavi S, van der Lelie D, Vallaeys T. 2006. Transcriptomics and proteomic analysis of the pMOL30-encoded copper resistance in Cupriavidus metallidurans strain CH34. Microbiology (Reading) 152:1765–1776. doi: 10.1099/mic.0.28593-0. [DOI] [PubMed] [Google Scholar]
- 14.Raimunda D, Gonzalez-Guerrero M, Leeber BW, Argüello JM. 2011. The transport mechanism of bacterial Cu+-ATPases: distinct efflux rates adapted to different function. Biometals 24:467–475. doi: 10.1007/s10534-010-9404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tseng T-T, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MHJ. 1999. The RND superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol 1:107–125. [PubMed] [Google Scholar]
- 16.Grass G, Rensing C. 2001. Genes involved in copper homeostasis in Escherichia coli. J Bacteriol 183:2145–2147. doi: 10.1128/JB.183.6.2145-2147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Outten FW, Huffman DL, Hale JA, O'Halloran TV. 2001. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. J Biol Chem 276:30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 18.Franke S, Grass G, Nies DH. 2001. The product of the ybdE gene of the Escherichia coli chromosome is involved in detoxification of silver ions. Microbiology (Reading) 147:965–972. doi: 10.1099/00221287-147-4-965. [DOI] [PubMed] [Google Scholar]
- 19.Franke S, Grass G, Rensing C, Nies DH. 2003. Molecular analysis of the copper-transporting CusCFBA efflux system from Escherichia coli. J Bacteriol 185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pontel LB, Audero ME, Espariz M, Checa SK, Soncini FC. 2007. GolS controls the response to gold by the hierarchical induction of Salmonella-specific genes that include a CBA efflux-coding operon. Mol Microbiol 66:814–825. doi: 10.1111/j.1365-2958.2007.05963.x. [DOI] [PubMed] [Google Scholar]
- 21.Cerminati S, Giri GF, Mendoza JI, Soncini FC, Checa SK. 2017. The CpxR/CpxA system contributes to Salmonella gold-resistance by controlling the GolS-dependent gesABC transcription. Environ Microbiol 19:4035–4044. doi: 10.1111/1462-2920.13837. [DOI] [PubMed] [Google Scholar]
- 22.Helbig K, Bleuel C, Krauss GJ, Nies DH. 2008. Glutathione and transition metal homeostasis in Escherichia coli. J Bacteriol 190:5431–5438. doi: 10.1128/JB.00271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fahey RC, Brown WC, Adams WB, Worsham MB. 1978. Occurrence of glutathione in bacteria. J Bacteriol 133:1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apontoweil P, Berends W. 1975. Glutathione biosynthesis in Escherichia coli K 12. Properties of the enzymes and regulation. Biochim Biophys Acta 399:1–9. doi: 10.1016/0304-4165(75)90205-6. [DOI] [PubMed] [Google Scholar]
- 25.Corazza A, Harvey I, Sadler PJ. 1996. 1H,13C-NMR and X-ray absorption studies of copper(I) glutathione complexes. Eur J Biochem 236:697–705. doi: 10.1111/j.1432-1033.1996.0697d.x. [DOI] [PubMed] [Google Scholar]
- 26.Aliaga ME, Lopez-Alarcon C, Garcia-Rio L, Martin-Pastor M, Speisky H. 2012. Redox-changes associated with the glutathione-dependent ability of the Cu(II)-GSSG complex to generate superoxide. Bioorg Med Chem 20:2869–2876. doi: 10.1016/j.bmc.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 27.Kachur AV, Koch CJ, Biaglow JE. 1998. Mechanism of copper-catalyzed oxidation of glutathione. Free Radic Res 28:259–269. doi: 10.3109/10715769809069278. [DOI] [PubMed] [Google Scholar]
- 28.Rigo A, Corazza A, di Paolo ML, Rossetto M, Ugolini R, Scarpa M. 2004. Interaction of copper with cysteine: stability of cuprous complexes and catalytic role of cupric ions in anaerobic thiol oxidation. J Inorg Biochem 98:1495–1501. doi: 10.1016/j.jinorgbio.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Zammit CM, Weiland F, Brugger J, Wade B, Winderbaum LJ, Nies DH, Southam G, Hoffmann P, Reith F. 2016. Proteomic responses to gold(III)-toxicity in the bacterium Cupriavidus metallidurans CH34. Metallomics 8:1204–1216. doi: 10.1039/c6mt00142d. [DOI] [PubMed] [Google Scholar]
- 30.O'Halloran TV, Culotta VC. 2000. Metallochaperones, an intracellular shuttle service for metal ions. J Biol Chem 275:25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 31.Banci L, Bertini I, Cantini F, Felli IC, Gonnelli L, Hadjiliadis N, Pierattelli R, Rosato A, Voulgaris P. 2006. The Atx1-Ccc2 complex is a metal-mediated protein-protein interaction. Nat Chem Biol 2:367–368. doi: 10.1038/nchembio797. [DOI] [PubMed] [Google Scholar]
- 32.Banci L, Bertini I, Ciofi-Baffoni S, Kozyreva T, Zovo K, Palumaa P. 2010. Affinity gradients drive copper to cellular destinations. Nature 465:645–648. doi: 10.1038/nature09018. [DOI] [PubMed] [Google Scholar]
- 33.Herzberg M, Schüttau M, Reimers M, Große C, Nies DH, Hans-Günther-Schlegel. 2015. Synthesis of nickel-iron hydrogenase in Cupriavidus metallidurans is controlled by metal-dependent silencing and un-silencing of genomic islands. Metallomics 7:632–649. doi: 10.1039/c4mt00297k. [DOI] [PubMed] [Google Scholar]
- 34.Große C, Herzberg M, Schüttau M, Wiesemann N, Hause G, Nies DH. 2016. Characterization of the Δ7 mutant of Cupriavidus metallidurans with deletions of seven secondary metal uptake systems. mSystems 1:e00004-16. doi: 10.1128/mSystems.00004-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bleichert P, Bütof L, Rückert C, Herzberg M, Francisco R, Morais P, Grass G, Kalinowski J, Nies DH. 2020. Mutant strains of Escherichia coli and methicillin-resistant Staphylococcus aureus obtained by laboratory selection to survive on metallic copper surfaces. Appl Environ Microbiol 87:e01788-20. doi: 10.1128/AEM.01788-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillan DC. 2016. Metal resistance systems in cultivated bacteria: are they found in complex communities? Curr Opin Biotechnol 38:123–130. doi: 10.1016/j.copbio.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Ladomersky E, Petris MJ. 2015. Copper tolerance and virulence in bacteria. Metallomics 7:957–964. doi: 10.1039/c4mt00327f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hao XL, Luthje FL, Qin Y, McDevitt SF, Lutay N, Hobman JL, Asiani K, Soncini FC, German N, Zhang SY, Zhu YG, Rensing C. 2015. Survival in amoeba-a major selection pressure on the presence of bacterial copper and zinc resistance determinants? Identification of a “copper pathogenicity island.” Appl Microbiol Biotechnol 99:5817–5824. doi: 10.1007/s00253-015-6749-0. [DOI] [PubMed] [Google Scholar]
- 39.Giachino A, Waldron KJ. 2020. Copper tolerance in bacteria requires the activation of multiple accessory pathways. Mol Microbiol 114:377–390. doi: 10.1111/mmi.14522. [DOI] [PubMed] [Google Scholar]
- 40.Weast RC. 1984. CRC handbook of chemistry and physics, 64 ed CRC Press, Inc., Boca Raton, Florida, USA. [Google Scholar]
- 41.Nies DH. 2022. Chemical constraints for transition metal cation allocation, p 21–52. In Hurst CJ (ed), Microbial metabolism of metals and metalloids, vol 10. Springer, Heidelberg. [Google Scholar]
- 42.Egler M, Grosse C, Grass G, Nies DH. 2005. Role of ECF sigma factor RpoE in heavy metal resistance of Escherichia coli. J Bacteriol 187:2297–2307. doi: 10.1128/JB.187.7.2297-2307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhamidimarri SP, Young T, Shanmugam M, Soderholm S, Basle A, Bumann D, van den Berg B. 2021. Acquisition of ionic copper by the bacterial outer membrane protein OprC through a novel binding site. PLoS Biol 19:e3001446. doi: 10.1371/journal.pbio.3001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gonzalez-Sanchez A, Cubillas CA, Miranda F, Davalos A, Garcia-de los Santos A. 2018. The ropAe gene encodes a porin-like protein involved in copper transit in Rhizobium etli CFN42. MicrobiologyOpen 7:e00573. doi: 10.1002/mbo3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Volentini SI, Farias RN, Rodriguez-Montelongo L, Rapisarda VA. 2011. Cu(II)-reduction by Escherichia coli cells is dependent on respiratory chain components. Biometals 24:827–835. doi: 10.1007/s10534-011-9436-3. [DOI] [PubMed] [Google Scholar]
- 46.Ohtsu I, Wiriyathanawudhiwong N, Morigasaki S, Nakatani T, Kadokura H, Takagi H. 2010. The L-cysteine/L-cystine shuttle system provides reducing equivalents to the periplasm in Escherichia coli. J Biol Chem 285:17479–17487. doi: 10.1074/jbc.M109.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Housecroft CE, Constable EC. 2006. Chemistry (3rd edition). Pearson Education Limited, Essex, England. [Google Scholar]
- 48.Grass G, Franke S, Taudte N, Nies DH, Kucharski LM, Maguire ME, Rensing C. 2005. The metal permease ZupT from Escherichia coli is a transporter with a broad substrate spectrum. J Bacteriol 187:1604–1611. doi: 10.1128/JB.187.5.1604-1611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Changela A, Chen K, Xue Y, Holschen J, Outten CE, O'Halloran TV, Mondragon A. 2003. Molecular basis of metal-ion selectivity and zeptomolar sensitivity by CueR. Science 301:1383–1387. doi: 10.1126/science.1085950. [DOI] [PubMed] [Google Scholar]
- 50.Suwalsky M, Ungerer B, Quevedo L, Aguilar F, Sotomayor CP. 1998. Cu2+ ions interact with cell membranes. J Inorg Biochem 70:233–238. doi: 10.1016/s0162-0134(98)10021-1. [DOI] [PubMed] [Google Scholar]
- 51.Goris J, De Vos P, Coenye T, Hoste B, Janssens D, Brim H, Diels L, Mergeay M, Kersters K, Vandamme P. 2001. Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis Steinle et al. 1998 emend. Int J Syst Evol Microbiol 51:1773–1782. doi: 10.1099/00207713-51-5-1773. [DOI] [PubMed] [Google Scholar]
- 52.Herzberg M, Bauer L, Kirsten A, Nies DH. 2016. Interplay between seven secondary metal transport systems is required for full metal resistance of Cupriavidus metallidurans. Metallomics 8:313–326. doi: 10.1039/c5mt00295h. [DOI] [PubMed] [Google Scholar]
- 53.Koch D, Nies DH, Grass G. 2007. The RcnRA (YohLM) system of Escherichia coli: a connection between nickel, cobalt and iron homeostasis. Biometals 20:759–771. doi: 10.1007/s10534-006-9039-6. [DOI] [PubMed] [Google Scholar]
- 54.Macomber L, Rensing C, Imlay JA. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol 189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Durand A, Azzouzi A, Bourbon ML, Steunou AS, Liotenberg S, Maeshima A, Astier C, Argentini M, Saito S, Ouchane S. 2015. c-type cytochrome assembly is a key target of copper toxicity within the bacterial periplasm. mBio 6. doi: 10.1128/mBio.01007-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Macomber L, Imlay JA. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc Natl Acad Sci USA 106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chillappagari S, Seubert A, Trip H, Kuipers OP, Marahiel MA, Miethke M. 2010. Copper stress affects iron homeostasis by destabilizing iron-sulfur cluster formation in Bacillus subtilis. J Bacteriol 192:2512–2524. doi: 10.1128/JB.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vallieres C, Holland SL, Avery SV. 2017. Mitochondrial ferredoxin determines vulnerability of cells to copper excess. Cell Chem Biol 24:1228–1237.e3. doi: 10.1016/j.chembiol.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan G, Yang J, Li T, Zhao J, Sun S, Li X, Lin C, Li J, Zhou H, Lyu J, Ding H. 2017. Anaerobic copper toxicity and iron-sulfur cluster biogenesis in Escherichia coli. Appl Environ Microbiol 83. doi: 10.1128/AEM.00867-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tan GQ, Cheng ZS, Pang YL, Landry AP, Li JH, Lu JX, Ding HG. 2014. Copper binding in IscA inhibits iron-sulphur cluster assembly in Escherichia coli. Mol Microbiol 93:629–644. doi: 10.1111/mmi.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiebelhaus N, Zaengle-Barone JM, Hwang KK, Franz KJ, Fitzgerald MC. 2021. Protein folding stability changes across the proteome reveal targets of Cu toxicity in E. coli. ACS Chem Biol 16:214–224. doi: 10.1021/acschembio.0c00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ansari MY, Batra SD, Ojha H, Dhiman K, Ganguly A, Tyagi JS, Mande SC. 2020. A novel function of Mycobacterium tuberculosis chaperonin paralog GroEL1 in copper homeostasis. FEBS Lett 594:3305–3323. doi: 10.1002/1873-3468.13906. [DOI] [PubMed] [Google Scholar]
- 63.Matuszewska E, Kwiatkowska J, Kuczyńska-Wiśnik D, Laskowska E. 2008. Escherichia coli heat-shock proteins IbpA/B are involved in resistance to oxidative stress induced by copper. Microbiology (Reading) 154:1739–1747. doi: 10.1099/mic.0.2007/014696-0. [DOI] [PubMed] [Google Scholar]
- 64.Helbig K, Grosse C, Nies DH. 2008. Cadmium toxicity in glutathione mutants of Escherichia coli. J Bacteriol 190:5439–5454. doi: 10.1128/JB.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jian X, Wasinger EC, Lockard JV, Chen LX, He C. 2009. Highly sensitive and selective gold(I) recognition by a metalloregulator in Ralstonia metallidurans. J Am Chem Soc 131:10869–10871. doi: 10.1021/ja904279n. [DOI] [PubMed] [Google Scholar]
- 66.Gonzalez-Guerrero M, Raimunda D, Cheng X, Argüello JM. 2010. Distinct functional roles of homologous Cu plus efflux ATPases in Pseudomonas aeruginosa. Mol Microbiol 78:1246–1258. doi: 10.1111/j.1365-2958.2010.07402.x. [DOI] [PubMed] [Google Scholar]
- 67.Legatzki A, Franke S, Lucke S, Hoffmann T, Anton A, Neumann D, Nies DH. 2003. First step towards a quantitative model describing Czc-mediated heavy metal resistance in Ralstonia metallidurans. Biodegradation 14:153–168. doi: 10.1023/a:1024043306888. [DOI] [PubMed] [Google Scholar]
- 68.Nies DH. 2022. How is a zinc ion correctly allocated to a zinc-dependent protein?, p 579–659. In Hurst CJ (ed), Microbial metabolism of metals and metalloids, vol 10. Springer, Heidelberg. [Google Scholar]
- 69.Singh SK, Grass G, Rensing C, Montfort WR. 2004. Cuprous oxidase activity of CueO from Escherichia coli. J Bacteriol 186:7815–7817. doi: 10.1128/JB.186.22.7815-7817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grass G, Rensing C. 2001. CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem Biophys Res Commun 286:902–908. doi: 10.1006/bbrc.2001.5474. [DOI] [PubMed] [Google Scholar]
- 71.Magnani D, Solioz M. 2007. How bacteria handle copper, p 259–285. In Nies DH, Silver S (ed), Molecular microbiology of heavy metals, vol 6. Springer-Verlag, Berlin. [Google Scholar]
- 72.Lawton TJ, Kenney GE, Hurley JD, Rosenzweig AC. 2016. The CopC family: structural and bioinformatic insights into a diverse group of periplasmic copper binding proteins. Biochemistry 55:2278–2290. doi: 10.1021/acs.biochem.6b00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Damle MS, Singh AN, Peters SC, Szalai VA, Fisher OS. 2021. The YcnI protein from Bacillus subtilis contains a copper-binding domain. J Biol Chem 297. doi: 10.1016/j.jbc.2021.101078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stolle P, Hou B, Brüser T. 2016. The Tat substrate CueO is transported in an incomplete folding state. J Biol Chem 291:13520–13528. doi: 10.1074/jbc.M116.729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hennaux L, Kohchtali A, Balon H, Matroule JY, Michaux C, Perpete EA. 2022. Refolding and biophysical characterization of the Caulobacter crescentus copper resistance protein, PcoB: an outer membrane protein containing an intrinsically disordered domain. Biochim Biophys Acta-Biomembranes 1864. doi: 10.1016/j.bbamem.2022.184038. [DOI] [PubMed] [Google Scholar]
- 76.Drew SC, Djoko KY, Zhang LY, Koay M, Boas JF, Pilbrow JR, Xiao ZG, Barnham KJ, Wedd AG. 2008. Electron paramagnetic resonance characterization of the copper-resistance protein PcoC from Escherichia coli. J Biol Inorg Chem 13:899–907. doi: 10.1007/s00775-008-0377-4. [DOI] [PubMed] [Google Scholar]
- 77.Espirito Santo C, Quaranta D, Grass G. 2012. Antimicrobial metallic copper surfaces kill Staphylococcus haemolyticus via membrane damage. Microbiologyopen 1:46–52. doi: 10.1002/mbo3.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bleichert P, Espirito Santo C, Hanczaruk M, Meyer H, Grass G. 2014. Inactivation of bacterial and viral biothreat agents on metallic copper surfaces. Biometals 27:1179–1189. doi: 10.1007/s10534-014-9781-0. [DOI] [PubMed] [Google Scholar]
- 79.Warnes SL, Caves V, Keevil CW. 2012. Mechanism of copper surface toxicity in Escherichia coli O157:H7 and Salmonella involves immediate membrane depolarization followed by slower rate of DNA destruction which differs from that observed for Gram-positive bacteria. Environ Microbiol 14:1730–1743. doi: 10.1111/j.1462-2920.2011.02677.x. [DOI] [PubMed] [Google Scholar]
- 80.Munson GP, Lam DL, Outten FW, O'Halloran TV. 2000. Identification of a copper-responsive two-component system on the chromosome of Escherichia coli K-12. J Bacteriol 182:5864–5871. doi: 10.1128/JB.182.20.5864-5871.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. doi: 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 82.Loftin IR, Franke S, Roberts SA, Weichsel A, Heroux A, Montfort WR, Rensing C, McEvoy MM. 2005. A novel copper-binding fold for the periplasmic copper resistance protein CusF. Biochemistry 44:10533–10540. doi: 10.1021/bi050827b. [DOI] [PubMed] [Google Scholar]
- 83.Chakravorty DK, Li PF, Tran TT, Bayse CA, Merz KM. 2016. Metal ion capture mechanism of a copper metallochaperone. Biochemistry 55:501–509. doi: 10.1021/acs.biochem.5b01217. [DOI] [PubMed] [Google Scholar]
- 84.Padilla-Benavides T, Thompson AMG, McEvoy MM, Argüello JM. 2014. Mechanism of ATPase-mediated Cu+ export and delivery to periplasmic chaperones: the interaction of Escherichia coli CopA and CusF. J Biol Chem 289:20492–20501. doi: 10.1074/jbc.M114.577668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bagai I, Rensing C, Blackburn NJ, McEvoy MM. 2008. Direct metal transfer between periplasmic proteins identifies a bacterial copper chaperone. Biochemistry 47:11408–11414. doi: 10.1021/bi801638m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ucisik MN, Chakravorty DK, Merz KM. 2015. Models for the metal transfer complex of the N-terminal region of CusB and CusF. Biochemistry 54:4226–4235. doi: 10.1021/acs.biochem.5b00195. [DOI] [PubMed] [Google Scholar]
- 87.Fung DKC, Lau WY, Chan WT, Yan AX. 2013. Copper efflux is induced during anaerobic amino acid limitation in Escherichia coli to protect iron-sulfur cluster enzymes and biogenesis. J Bacteriol 195:4556–4568. doi: 10.1128/JB.00543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hernandez-Montes G, Arguello JM, Valderrama B. 2012. Evolution and diversity of periplasmic proteins involved in copper homeostasis in gamma proteobacteria. BMC Microbiol 12. doi: 10.1186/1471-2180-12-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Goldberg M, Pribyl T, Juhnke S, Nies DH. 1999. Energetics and topology of CzcA, a cation/proton antiporter of the RND protein family. J Biol Chem 274:26065–26070. doi: 10.1074/jbc.274.37.26065. [DOI] [PubMed] [Google Scholar]
- 90.Long F, Su CC, Zimmermann MT, Boyken SE, Rajashankar KR, Jernigan RL, Yu EW. 2010. Crystal structures of the CusA efflux pump suggest methionine-mediated metal transport. Nature 467:484–488. doi: 10.1038/nature09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Su CC, Long F, Zimmermann MT, Rajashankar KR, Jernigan RL, Yu EW. 2011. Crystal structure of the CusBA heavy-metal efflux complex of Escherichia coli. Nature 470:558–562. doi: 10.1038/nature09743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rost J, Rapoport S. 1964. Reduction-potential of gutathione. Nature 201:185–187. doi: 10.1038/201185a0. [DOI] [PubMed] [Google Scholar]
- 93.Kirlin WG, Cai J, Thompson SA, Diaz D, Kavanagh TJ, Jones DP. 1999. Glutathione redox potential in response to differentiation and enzyme inducers. Free Radic Biol Med 27:1208–1218. doi: 10.1016/s0891-5849(99)00145-8. [DOI] [PubMed] [Google Scholar]
- 94.Freedman JH, Ciriolo MR, Peisach J. 1989. The role of glutathione in copper metabolism and toxicity. J Biol Chem 264:5598–5605. doi: 10.1016/S0021-9258(18)83589-X. [DOI] [PubMed] [Google Scholar]
- 95.Speisky H, Gomez M, Burgos-Bravo F, Lopez-Alarcon C, Jullian C, Olea-Azar C, Aliaga ME. 2009. Generation of superoxide radicals by copper-glutathione complexes: redox-consequences associated with their interaction with reduced glutathione. Bioorg Med Chem 17:1803–1810. doi: 10.1016/j.bmc.2009.01.069. [DOI] [PubMed] [Google Scholar]
- 96.Eser M, Masip L, Kadokura H, Georgiou G, Beckwith J. 2009. Disulfide bond formation by exported glutaredoxin indicates glutathione's presence in the E. coli periplasm. Proc Natl Acad Sci USA 106:1572–1577. doi: 10.1073/pnas.0812596106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lonetto MA, Brown KL, Rudd KE, Buttner MJ. 1994. Analysis of the Streptomyces coelicolor sigF gene reveals the existence of a subfamily of eubacterial RNA polymerase σ factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA 91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Helmann JD. 2002. The extracytoplasmic function (ECF) sigma factors. Adv Microb Phys 46:47–110. doi: 10.1016/S0065-2911(02)46002-X. [DOI] [PubMed] [Google Scholar]
- 99.Paget MSB, Helmann JD. 2003. Protein family review - The sigma(70) family of sigma factors. Genome Biol 4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Große C, Poehlein A, Blank K, Schwarzenberger C, Schleuder G, Herzberg M, Nies DH. 2019. The third pillar of metal homeostasis in Cupriavidus metallidurans CH34: preferences are controlled by extracytoplasmic functions sigma factors. Metallomics 11:291–316. doi: 10.1039/c8mt00299a. [DOI] [PubMed] [Google Scholar]
- 101.Grosse C, Friedrich S, Nies DH. 2007. Contribution of extracytoplasmic function sigma factors to transition metal homeostasis in Cupriavidus metallidurans strain CH34. J Mol Microbiol Biotechnol 12:227–240. doi: 10.1159/000099644. [DOI] [PubMed] [Google Scholar]
- 102.Nies DH. 2004. Incidence and function of sigma factors in Ralstonia metallidurans and other bacteria. Arch Microbiol 181:255–268. doi: 10.1007/s00203-004-0658-4. [DOI] [PubMed] [Google Scholar]
- 103.Große C, Grau J, Große I, Nies DH. 2022. Importance of RpoD- and non-RpoD-dependent expression of horizontally acquired genes in Cupriavidus metallidurans. Microbiol Spectr 10. doi: 10.1128/spectrum.00121-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mol CD, Hosfield DJ, Tainer JA. 2000. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3' ends justify the means. Mutat Res 460:211–229. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 105.Das D, Grishin NV, Kumar A, Carlton D, Bakolitsa C, Miller MD, Abdubek P, Astakhova T, Axelrod HL, Burra P, Chen C, Chiu HJ, Chiu M, Clayton T, Deller MC, Duan L, Ellrott K, Ernst D, Farr CL, Feuerhelm J, Grzechnik A, Grzechnik SK, Grant JC, Han GW, Jaroszewski L, Jin KK, Johnson HA, Klock HE, Knuth MW, Kozbial P, Krishna SS, Marciano D, McMullan D, Morse AT, Nigoghossian E, Nopakun A, Okach L, Oommachen S, Paulsen J, Puckett C, Reyes R, Rife CL, Sefcovic N, Tien HJ, Trame CB, van den Bedem H, Weekes D, Wooten T, Xu Q, Hodgson KO, et al. 2010. The structure of the first representative of Pfam family PF09836 reveals a two-domain organization and suggests involvement in transcriptional regulation. Acta Crystallogr Sect F Struct Biol Cryst Commun 66:1174–1181. doi: 10.1107/S1744309109022672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stewart LJ, Ong CLY, Zhang MM, Brouwer S, McIntyre L, Davies MR, Walker MJ, McEwan AG, Waldron KJ, Djoko KY. 2020. Role of glutathione in buffering excess intracellular copper in Streptococcus pyogenes. mBio 11. doi: 10.1128/mBio.02804-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Meydan S, Klepacki D, Karthikeyan S, Margus T, Thomas P, Jones JE, Khan Y, Briggs J, Dinman JD, Vazquez-Laslop N, Mankin AS. 2017. Programmed ribosomal frameshifting generates a copper transporter and a copper chaperone from the same gene. Mol Cell 65:207–219. doi: 10.1016/j.molcel.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Drees SL, Beyer DF, Lenders-Lomscher C, Lübben M. 2015. Distinct functions of serial metal-binding domains in the Escherichia coli P1B-ATPase CopA. Mol Microbiol 97:423–438. doi: 10.1111/mmi.13038. [DOI] [PubMed] [Google Scholar]
- 109.Saporito-Magrina CM, Musacco-Sebio RN, Andrieux G, Kook L, Orrego MT, Tuttolomondo MV, Desimone MF, Boerries M, Borner C, Repetto MG. 2018. Copper-induced cell death and the protective role of glutathione: the implication of impaired protein folding rather than oxidative stress. Metallomics 10:1743–1754. doi: 10.1039/C8MT00182K. [DOI] [PubMed] [Google Scholar]
- 110.Krezel A, Bal W. 1999. Coordination chemistry of glutathione. Acta Biochim Pol 46:567–580. doi: 10.18388/abp.1999_4129. [DOI] [PubMed] [Google Scholar]
- 111.Pittman MS, Robinson HC, Poole RK. 2005. A Bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J Biol Chem 280:32254–32261. doi: 10.1074/jbc.M503075200. [DOI] [PubMed] [Google Scholar]
- 112.Mikolay A, Nies DH. 2009. The ABC-transporter AtmA is involved in nickel and cobalt resistance of Cupriavidus metallidurans strain CH34. Antonie Van Leeuwenhoek 96:183–191. doi: 10.1007/s10482-008-9303-6. [DOI] [PubMed] [Google Scholar]
- 113.Pace CN, Scholtz MJ. 1997. Measuring the conformational stability of a protein, p 299–322. In Creighton T (ed), Protein structure: a practical approach. IRL press, Oxford, UK. [Google Scholar]
- 114.Anton A, Weltrowski A, Haney JH, Franke S, Grass G, Rensing C, Nies DH. 2004. Characteristics of zinc transport by two bacterial cation diffusion facilitators from Ralstonia metallidurans and Escherichia coli. J Bacteriol 186:7499–7507. doi: 10.1128/JB.186.22.7499-7507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nies DH. 1992. CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc and cadmium (czc system) in Alcaligenes eutrophus. J Bacteriol 174:8102–8110. doi: 10.1128/jb.174.24.8102-8110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nies D, Mergeay M, Friedrich B, Schlegel HG. 1987. Cloning of plasmid genes encoding resistance to cadmium, zinc, and cobalt in Alcaligenes eutrophus CH34. J Bacteriol 169:4865–4868. doi: 10.1128/jb.169.10.4865-4868.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning, a laboratory manual, 2nd. Ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 118.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 119.Marx CJ, Lidstrom ME. 2002. Broad-host-range cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33:1062–1067. doi: 10.2144/02335rr01. [DOI] [PubMed] [Google Scholar]
- 120.Suzuki N, Nonaka H, Tsuge Y, Inui M, Yukawa H. 2005. New multiple-deletion method for the Corynebacterium glutamicum genome, using a mutant lox sequence. Appl Environ Microbiol 71:8472–8480. doi: 10.1128/AEM.71.12.8472-8480.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Albert H, Dale EC, Lee E, Ow DW. 1995. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J 7:649–659. doi: 10.1046/j.1365-313x.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- 122.Lenz O, Schwartz E, Dernedde J, Eitinger T, Friedrich B. 1994. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J Bacteriol 176:4385–4393. doi: 10.1128/jb.176.14.4385-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Grosse C, Grass G, Anton A, Franke S, Santos AN, Lawley B, Brown NL, Nies DH. 1999. Transcriptional organization of the czc heavy metal homoeostasis determinant from Alcaligenes eutrophus. J Bacteriol 181:2385–2393. doi: 10.1128/JB.181.8.2385-2393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wagegg W, Braun V. 1981. Ferric citrate transport in Escherichia coli requires outer membrane receptor protein FecA. J Bacteriol 145:156–163. doi: 10.1128/jb.145.1.156-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kirsten A, Herzberg M, Voigt A, Seravalli J, Grass G, Scherer J, Nies DH. 2011. Contributions of five secondary metal uptake systems to metal homeostasis of Cupriavidus metallidurans CH34. J Bacteriol 193:4652–4663. doi: 10.1128/JB.05293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lee SM, Grass G, Rensing C, Barrett SR, Yates CJD, Stoyanov JV, Brown NL. 2002. The Pco proteins are involved in periplasmic copper handling in Escherichia coli. Biochem Biophys Res Commun 295:616–620. doi: 10.1016/s0006-291x(02)00726-x. [DOI] [PubMed] [Google Scholar]
- 127.Roberts SA, Weichsel A, Grass G, Thakali K, Hazzard JT, Tollin G, Rensing C, Montfort WR. 2002. Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc Natl Acad Sci USA 99:2766–2771. doi: 10.1073/pnas.052710499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gonzalez-Guerrero M, Argüello JM. 2008. Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc Natl Acad Sci USA 105:5992–5997. doi: 10.1073/pnas.0711446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rensing C, Fan B, Sharma R, Mitra B, Rosen BP. 2000. CopA: an Escherichia coli Cu(I)-translocating P-type ATPase. Proc Natl Acad Sci USA 97:652–656. doi: 10.1073/pnas.97.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Odermatt A, Krapf R, Solioz M. 1994. Induction of the putative copper ATPases, CopA and CopB, of Enterococcus hirae by Ag+ and Cu2+, and Ag+ extrusion by CopB. Biochem Biophys Res Commun 202:44–48. doi: 10.1006/bbrc.1994.1891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.00567-23-s0001.pdf, PDF file, 5.6 MB (5.6MB, pdf)