ABSTRACT
Lactococcus lactis and Lactococcus cremoris are Gram-positive lactic acid bacteria widely used as starter in milk fermentations. Lactococcal cells are covered with a polysaccharide pellicle (PSP) that was previously shown to act as the receptor for numerous bacteriophages of the Caudoviricetes class. Thus, mutant strains lacking PSP are phage resistant. However, because PSP is a key cell wall component, PSP-negative mutants exhibit dramatic alterations of cell shape and severe growth defects, which limit their technological value. In the present study, we isolated spontaneous mutants with improved growth, from L. cremoris PSP-negative mutants. These mutants grow at rates similar to the wild-type strain, and based on transmission electron microscopy analysis, they exhibit improved cell morphology compared to their parental PSP-negative mutants. In addition, the selected mutants maintain their phage resistance. Whole-genome sequencing of several such mutants showed that they carried a mutation in pbp2b, a gene encoding a penicillin-binding protein involved in peptidoglycan biosynthesis. Our results indicate that lowering or turning off PBP2b activity suppresses the requirement for PSP and ameliorates substantially bacterial fitness and morphology.
IMPORTANCE Lactococcus lactis and Lactococcus cremoris are widely used in the dairy industry as a starter culture. As such, they are consistently challenged by bacteriophage infections which may result in reduced or failed milk acidification with associated economic losses. Bacteriophage infection starts with the recognition of a receptor at the cell surface, which was shown to be a cell wall polysaccharide (the polysaccharide pellicle [PSP]) for the majority of lactococcal phages. Lactococcal mutants devoid of PSP exhibit phage resistance but also reduced fitness, since their morphology and division are severely impaired. Here, we isolated spontaneous, food-grade non-PSP-producing L. cremoris mutants resistant to bacteriophage infection with a restored fitness. This study provides an approach to isolate non-GMO phage-resistant L. cremoris and L. lactis strains, which can be applied to strains with technological functionalities. Also, our results highlight for the first time the link between peptidoglycan and cell wall polysaccharide biosynthesis.
KEYWORDS: Lactococcus, bacteriophage resistance, cell wall, lactic acid bacteria, penicillin-binding proteins, polysaccharide pellicle
INTRODUCTION
Lactococcus lactis and Lactococcus cremoris are members of the lactic acid bacteria group that are widely used in industrial starter cultures for the production of cheese and other fermented milk products. These starter cultures regularly suffer from bacteriophage infections which may result in reduced or failed milk acidification with associated economic losses (1, 2). Numerous lactococcal phages have been isolated and classified in 10 groups on the basis of morphological criteria and genetic analysis (3). They belong predominantly to the Caudoviricetes class with groups 936 (renamed as the Skunavirus genus), P335 and c2 (renamed as the Ceduovirus genus) accounting for around 80, 10, and 5% of the isolates of this family, respectively (4). The phage infection cycle starts with phage adsorption to its host, through the specific recognition of receptor molecules present at the bacterial surface. Whereas phages from the c2 group recognizes a protein receptor, phages from the 936 and P335 groups recognize a saccharidic receptor (5, 6).
L. lactis and L. cremoris are Gram-positive bacteria with a typical cell wall consisting of a peptidoglycan (PG) sacculus functionalized with other glycopolymers and proteins (7, 8). In recent years, these species have been shown to produce, like other Gram-positive ovoid-shaped cocci or ovococci, a large amount of a rhamnose-rich cell wall polysaccharide (Rha-CWPS), that appears to represent a functional analog of wall teichoic acid (9). In a given L. lactis or L. cremoris strain, the Rha-CWPS possesses a complex structure consisting of a poly-rhamnose linear chain (the rhamnan) decorated by oligo- or polymeric substituents containing for most of them phosphate groups (9). Rhamnan was found to be embedded inside the PG layer and covalently bound to it, whereas polysaccharide substituents (previously termed polysaccharide pellicle [PSP]) were detected at the bacterial surface (10, 11). Previous studies have established that the surface-exposed PSP is the receptor of bacteriophages of the Skunavirus and P335 phage families (10, 12–14). Moreover, PSP structure was shown to be highly variable between strains (15). This variability parallels the diversity found at the genetic level, which facilitated the identification of four L. lactis and L. cremoris genotypes (A, B, C, and D), with five C-subgroups (C1 to C5) (16). Differences in the associated PSP structures was found to be the major determinant of bacteriophage sensitivity and explains, at least partially, the narrow host range of lactococcal phages (12). Interestingly, PSP biosynthesis is independent of that of rhamnan, as mutants producing undecorated rhamnan were obtained, either spontaneously (10) or by genetic engineering (17). These mutants devoid of PSP are phage-resistant. However, since PSP is a cell wall component essential for normal growth and division, PSP-negative mutants exhibit dramatic alterations of cell shape, impairment of cell division leading to cell aggregates and strong growth defects (10, 17), which limits their technological value.
The PSP of L. cremoris MG1363 (and its NZ9000 derivative [18]) is composed of hexasaccharide subunits linked by phosphodiester bonds, and was shown to be the receptor of phages sk1 and p2 of the Skunavirus genus (10, 17). We previously proposed a model for PSP subunit biosynthesis requiring the activity of six glycosyltransferases, encoded by genes wpsACDEF (17) and csdD involved in side chain glucose addition (19). Genes wpsH and wpsI were proposed to encode proteins involved in PSP subunit polymerization (17). Consequently, wpsA and wpsH mutants are devoid of surface-exposed PSP and are resistant to sk1 and p2 phages (10, 17). In the present study, we successfully isolated suppressor mutations that improve the fitness of the two L. cremoris PSP-negative wpsA and wpsH mutants. Seven independently selected suppressor mutants contained a mutation mapped in the pbp2b gene encoding a class B penicillin-binding protein (PBP) involved in PG biosynthesis. We show that secondary pbp2b mutations increase the growth rate and alleviate the morphology and division defects of wpsA and wpsH mutants, while retaining their resistance to phages sk1 and p2. Finally, we evaluated the impact of the pbp2b mutations on PG structure in several selected mutants. Our results highlight the coordination between Rha-CWPS biosynthesis and PG assembly in lactococci.
RESULTS
Isolation of L. cremoris suppressor mutants with improved fitness in the absence of PSP.
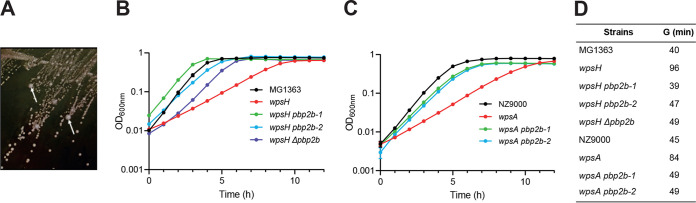
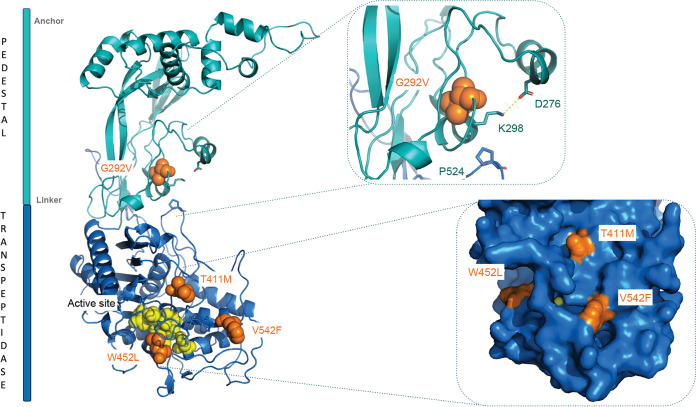
In a previous study, we isolated VES5748 as a spontaneous PSP-deficient strain derived from L. cremoris MG1363 (10). It carries a point mutation in the wpsH gene (locus tag llmg_0226), whose product WpsH is involved in PSP biosynthesis, presumably as a regulator of the putative polymerase WpsI (17). Since PSP serves as bacteriophage sk1 receptor, VES5748 is resistant to sk1; however, it exhibits slow growth, altered cell morphology and cell division impairment (10). As a result, VES5748 grows more slowly than WT strain and forms very small colonies on GM17 agar plates. As the slow growth creates selective pressure, larger colonies easily appear on the plates (Fig. 1A). To investigate which genetic changes result in appearance of these larger colonies, we first tested nine of them for their resistance to sk1, and we found that five of them were bacteriophage sensitive. Therefore, we concluded that these mutants were true revertants, restoring PSP biosynthesis, and excluded them from further study. In contrast, the four remaining mutants were bacteriophage resistant. Whole-genome sequencing of three of these mutants revealed a point mutation in gene pbp2b (locus tag llmg_0358), leading to a missense mutation in the PBP2b protein in strains VES7483 (G292V), VES7484 (T411M), and VES7485 (V542F) (Table 1). The mutations were confirmed by PCR amplification and sequencing of the pbp2b gene. In VES7483, we did not find any additional mutation, whereas VES7484 had also a point mutation in llmg_2412 encoding prolyl-tRNA synthase, and VES7485 had additional mutations in the genes lmg_pseudo_18 encoding mobile element transposase, and in llmg_0729 encoding PTS system protein PtnAB. A point mutation in pbp2b (leading to the change W452L in PBP2b) was also found in the fourth mutant (VES7482) through PCR amplification and sequencing of the gene. Thus, four independently isolated larger colony-forming mutants all carried different mutations in pbp2b. For this reason, we concentrated our studies on the role of the pbp2b mutation. This gene encodes a class B monofunctional PBP with transpeptidase activity, involved in PG synthesis (20, 21). To get insight in the possible impact of the obtained mutations on PBP2b activity, we projected the obtained mutations on a three-dimensional (3D) homology model of L. cremoris PBP2b (Fig. 2). We observed that the mutations in the strains VES7482, VES7484, and VES7485 are in the vicinity of the protein active site, indicating a possible loss of function, whereas the mutation in strain VES7483 (G292V) is located in the N-terminal pedestal domain of PBP2b, suggesting possible retention of the transpeptidase activity. We retained two of the pbp2b mutant suppressor strains, the ones which carry mutations near the active site, here named wpsH pbp2b-1 (VES7484) and wpsH pbp2b-2 (VES7485) (Table 1), for further characterization. Also, to avoid the influence of other mutations, a pbp2b deletion was introduced in VES5748, giving rise to strain VES7554 (wpsH Δpbp2b) that was used as a control in the rest of the study.
FIG 1.
(A) Isolation of suppressor mutants from a L. cremoris PSP-negative strain on solid GM17 medium. Larger colonies (indicated by white arrows) arise spontaneously among the small colonies formed by VES5748 (wpsH) PSP-negative strain. (B and C) Growth in liquid GM17 medium of MG1363, wpsH mutant, and its pbp2b derivatives (B) and of NZ9000, wpsA mutant, and its pbp2b derivatives (C). Each point represents the mean value of three biological replicates. The standard deviations for each value were very low and could not be clearly represented on the corresponding curves. (D) Generation time (G) determined in the exponential growth phase for each strain.
TABLE 1.
Strains, plasmids, and bacteriophages used in this study
| Strain, plasmid, or phage | Relevant genotype and/or characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| L. cremoris | ||
| MG1363 | Plasmid-free strain | 43 |
| NZ9000 | MG1363 pepN::nisRK | 44 |
| VES7552 | NZ9000 Δpbp2b, NZ9000 derivative carrying deletion of pbp2b | This study |
| VES5748 | MG1363 wpsH, spontaneous MG1363 derivative, C-to-T transition in wpsH (llmg_0226) gene | 10 |
| VES7482 | Spontaneous VES5748 derivative, G1355-to-T transversion in pbp2b (llmg_0358), corresponding to W452L mutation in PBP2b | This study |
| VES7483 | Spontaneous VES5748 derivative, G881-to-T transversion in pbp2b, corresponding to G292V mutation in PBP2b | This study |
| VES7484 | wpsH pbp2b-1, spontaneous VES5748 derivative, C1232-to-T transition in pbp2b, corresponding to T411M mutation in PBP2b | This study |
| VES7485 | wpsH pbp2b-2, spontaneous VES5748 derivative, G1624-to-T transversion in pbp2b, corresponding to V542F mutation in PBP2b | This study |
| VES7554 | wpsH Δpbp2b, VES5748 derivative carrying a deletion of pbp2b | This study |
| NZ9000 wpsA | NZ9000 with an in-frame stop codon inserted in wpsA (llnz_0135) | 17 |
| VES7806 | wpsA pbp2b-1, spontaneous NZ9000 wpsA derivative, deletion of A in position 430 in pbp2b (llnz_01875) | This study |
| VES7807 | Spontaneous NZ9000 wpsA derivative, deletion of C in position 1727 in pbp2b | This study |
| VES7808 | wpsH pbp2b-2, spontaneous NZ9000 wpsA derivative, deletion of 33 nucleotides from position 1335 to 1367 in pbp2b | This study |
| E. coli | ||
| JIM4646 | TG1 strain with a chromosomal copy of repA gene, allowing replication of L. lactis pGhost9 plasmid | P. Renault, INRAE, France |
| Plasmids | ||
| pGhost9 | Thermosensitive plasmid for gene replacement by double-crossover integration; Eryr | 45 |
| pGhost9-Δpbp2b | pbp2b integration vector for construction of pbp2b deletion mutants | This study |
| Bacteriophages | ||
| sk1 | Skunavirus, propagated on NZ9000 | 46 |
| p2 | Skunavirus, propagated on NZ9000 | 47 |
| MCC1 | Skunavirus, derivative of sk1, propagated on NZ9000 | 17 |
| MCC17 | Skunavirus, derivative of sk1, propagated on NZ9000 | 17 |
FIG 2.
3D homology model of PBP2b. The pedestal domain is shown in teal, and transpeptidase domain is shown in blue. Amino acid substitutions found in the four wpsH pbp2b mutants are displayed as orange spheres. The catalytic amino acid residues are highlighted as yellow spheres. Inserted at the upper right is a closeup view of the G292V mutation and its interaction sphere, with a highlight on the salt bridge between D276 and K298 and on the interaction between K298 and P524. Inserted at the lower right is a closer view of the accessible surface of the active site that contains the three other substitutions.
L. cremoris NZ9000 wpsA mutant was previously shown to be devoid of PSP and also exhibited the associated defects. The wpsA gene encodes a glycosyltransferase proposed to initiate the synthesis of the PSP subunit by transfer of a GlcNAc residue on the undecaprenyl-phosphate lipidic carrier (17). To demonstrate that the involvement of pbp2b in the fitness restoration of PSP-negative mutants is not restricted to wpsH mutants, spontaneous large-colony morphotype derivatives were also isolated in the wpsA mutant background. With the procedure described above, three better-growing derivatives were isolated from the NZ9000-wpsA strain. pbp2b gene was PCR-amplified and its DNA sequence determined. As in the case of VES5748 (wpsH) derivatives, the three suppressor strains also carried mutations in pbp2b gene (locus tag llnz_01875): the deletion of A in position 430 (VES7806) and the deletion of C in position 1727 (VES7807), both resulting in frameshifts in pbp2b translation and in a premature stop codon, and the deletion of 33 nucleotides at positions 1335 to 1367 (VES7808), resulting in a 11-amino-acid deletion in PBP2b protein. We retained two of the pbp2b mutant suppressor strains, here named wpsA pbp2b-1 (VES7806) and wpsA pbp2b-2 (VES7808) (Table 1), for further characterization. Although all isolated derivatives were mutated in the same pbp2b gene, it is noteworthy that the four derivatives of VES5748 (MG1363 wpsH) carried point mutations, leading to a single amino acid substitution in PBP2b, whereas all three NZ9000-wpsA derivatives carried frameshift or deletion mutations resulting in the absence of full-length PBP2b.
The growth of the double mutants is improved.
The growth rate in GM17 medium of the selected mutants was compared to their wpsH or wpsA parent strains. The PSP-negative mutant wpsH (VES5748) has a low growth rate compared to MG1363 (Fig. 1B). The double mutants wpsH pbp2b-1 and wpsH pbp2b-2 exhibited improved growth (with a generation time [G] during exponential growth phases of 39 and 47 min, respectively), compared to their parent strain wpsH (G of 96 min) (Fig. 1B and D). In comparison, the generation time of wild-type (WT) MG1363 was 40 min. Interestingly, the same observation was made with wpsH Δpbp2b mutant (G of 49 min), confirming the role of pbp2b mutation in improving the growth rate of wpsH mutants. A similar growth improvement was observed for wpsA pbp2b-1 and wpsA pbp2b-2 mutants (Fig. 1C and D), with calculated generation times of 49 min for both, close to WT NZ9000 (45 min), versus 84 min for the NZ9000-wpsA mutant.
Analysis of the CWPS content of the double mutants.
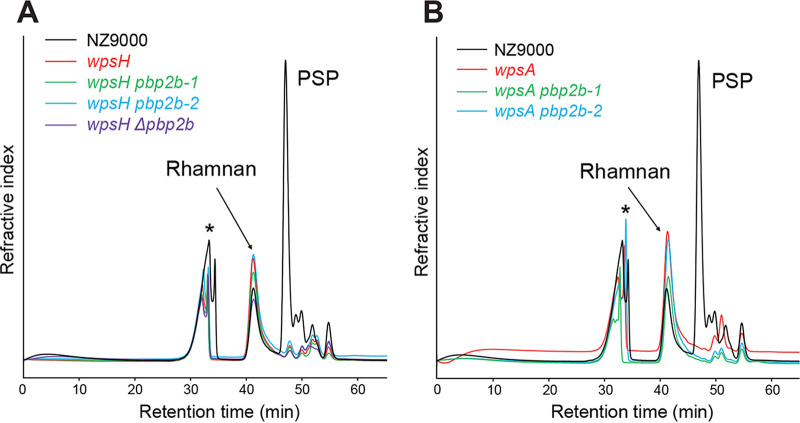
We verified that the mutations in pbp2b did not affect the CWPS content. For that, cell wall polysaccharides (CWPS) were extracted by hydrofluoric acid (HF) treatment of cell walls of WT and mutant strains. HF allows to release CWPS by cleaving the pyrophosphate bond between rhamnan and PG and by depolymerizing PSP by cleavage of the phosphodiester bonds between PSP subunits (11). Solubilized rhamnan and PSP oligosaccharides were then separated by size exclusion high pressure liquid chromatography (SEC-HPLC) (Fig. 3). The rhamnan peak was present in all tested strains whereas the PSP oligosaccharide peak was absent in wpsH and wpsA mutants, as well as in all of the double mutants. These results confirm the absence of PSP as expected in the four double mutants.
FIG 3.
SEC-HPLC analysis of CWPS extracted from L. cremoris NZ9000, wpsH and wpsA mutants, and their respective pbp2b mutant derivatives. Peaks containing rhamnan and PSP oligosaccharides are indicated. *, Nonpolysaccharidic compounds.
pbp2b mutations alleviate cell morphology defects of PSP-negative strains.
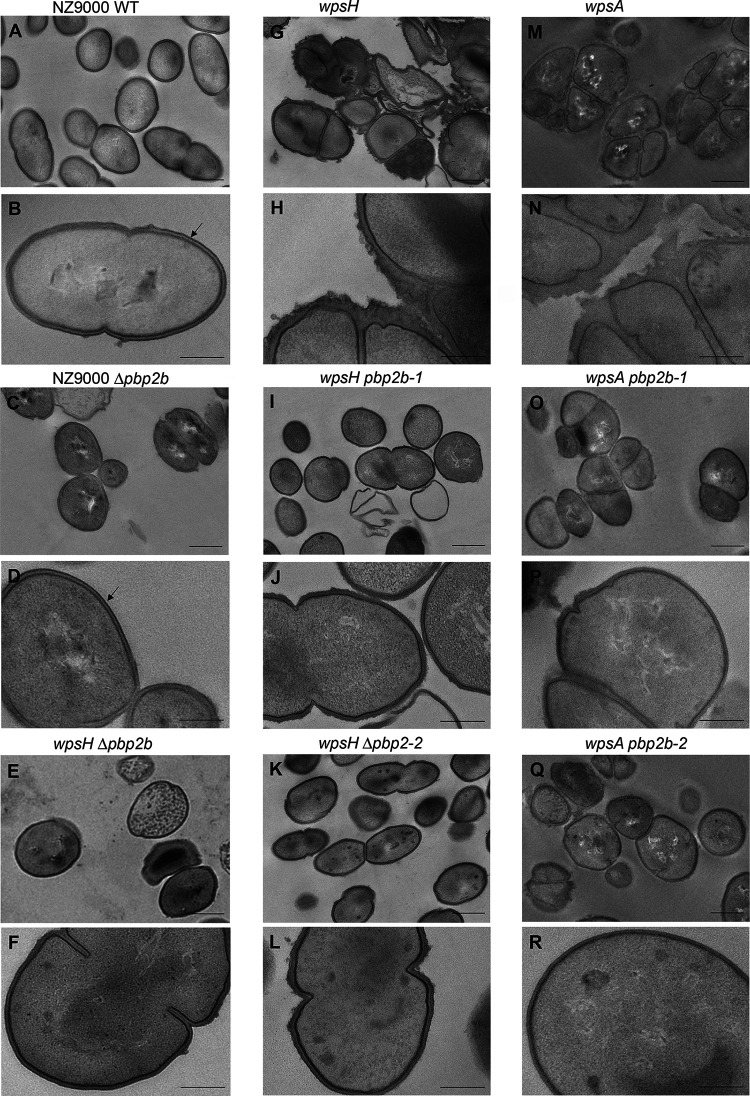
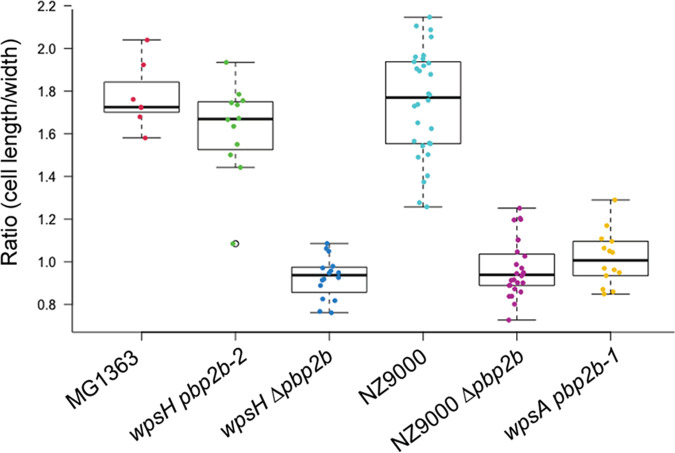
L. cremoris strains devoid of PSP exhibit severe cell morphology alterations when observed by transmission electron microscopy (TEM) including abnormal cell shape, misplaced septa, and poor separation of daughter cells leading to aggregates (10, 17) (Fig. 4G, H, M, and N). Moreover, the outer electron-dense layer detected by TEM and attributed to PSP in WT NZ9000 (Fig. 4B) is absent in wpsA and wpsH mutants, and irregularities and thickening of their cell walls are visible. TEM observations of the wpsH pbp2b-1 mutant (Fig. 4I and J) showed an improved morphology, characterized by cells with a more regular shape and a more symmetrical division process accompanied with a decrease of aggregates compared to its parent wpsH (Fig. 4G and H). Similar features were observed in the wpsH pbp2b-2 mutant (Fig. 4K and L). Of note, wpsH pbp2b-2 cells recovered ovoid shape as estimated by the ratio of the cell length to width (Fig. 5). Deletion of pbp2b gene in the wpsH mutant (wpsH Δpbp2b mutant) (Fig. 4E and F) resulted also in improved morphology compared to the wpsH mutant (Fig. 4G and H). In this case, the double mutant showed cells that were rounder than the WT (Fig. 5), as observed previously for L. cremoris pbp2b mutants (21) and for the NZ9000 Δpbp2b deletion mutant constructed here (Fig. 4C and D and Fig. 5). Regarding wpsA pbp2b-1 (Fig. 4O and P) and wpsA pbp2b-2 (Fig. 4Q and R), fewer cell aggregates were observed and the cell shape was partially restored compared to the wpsA mutant (Fig. 4M and N). The double mutants exhibited round cells resembling these of the wpsH Δpbp2b mutant (Fig. 5). Unlike in NZ9000, no PSP layer was visible at the surface of any of the wps and wps pbp2b mutant cells.
FIG 4.
TEM images of L. cremoris NZ9000, PSP-negative mutants, and their pbp2b derivatives. Black arrows indicate the outer electron dense layer attributed to PSP. Scale bars, 500 nm (A, C, E, G, I, K, M, O, and Q) and 200 nm (B, D, F, H, J, L, N, P, and R). (A and B) NZ9000 WT strain, showing typical ovoid shape, septation at midcell and PSP layer at the cell surface. (C and D) NZ9000 Δpbp2b (VES7552). (E and F) wpsH Δpbp2b (VES7552). (G and H) wpsH (VES5748). (I and J) wpsH pbp2b-1 (VES7484). (K and L) wpsH pbp2b-2 (VES7485). (M and N) wpsA (VES7810). (O and P) wpsA pbp2b-1 (VES7806). (Q and R) wpsA pbp2b-2 (VES7808).
FIG 5.
Cell length to width ratios measured on SEM images of each indicated strain. A whiskers plot and string chart are shown. Each dot indicates a measured cell.
Test of bacteriophage sensitivity of the double mutants.
Bacteriophages sk1 and p2 of the Skunavirus genus infecting L. cremoris NZ9000 recognize PSP as a receptor for their adsorption at the bacterial surface of their host. Consequently, wpsH and wpsA mutants are resistant to sk1 and p2 (10, 17). Nevertheless, we have shown previously that phage escape mutants derived from sk1 can be isolated on the wpsH mutant (17). These phages carried mutations in their baseplate involved in adsorption to host cells and were hypothesized to bind remnant PSP subunit detected in the wpsH mutant by mass spectrometry analysis. In contrast, the wpsA mutant was totally resistant to these phage escape mutants, as a result of the absence of any PSP fragments in this mutant, in agreement with WpsA being the initiating glycosyltransferase of the PSP subunit synthesis (17).
We next wanted to establish if the double mutants with improved growth and morphology had retained the phage resistance phenotype of their parental wpsH or wpsA strain. The sensitivity to phages sk1 and p2, as well as to two phage escape mutants, named MCC1 and MCC17, derived from phage sk1 and isolated against L. lactis wpsH, was assessed in plaque assays (Table 2). Both wpsH pbp2b-1 and wpsH pbp2b-2 mutants were found to remain highly resistant to phages sk1 and p2. Noteworthy, the efficiency of plaquing (EOP) values were slightly higher on the two wpsH pbp2b double mutants than on the wpsH mutant. We hypothesize that this is due to the improved growth rate of the double mutants. We then evaluated the sensitivity of wpsH pbp2b mutants to the previously isolated phage escape mutants, MCC1 and MCC17. Both MCC1 and MCC17 are able to infect WT and PSP-negative wpsH strains. The EOP values of both phages were very similar between wpsH pbp2b and wpsH mutants (Table 2). This result suggests that the MCC1 and MCC17 phage receptor on the bacterial cell surface is not further modified in wpsH pbp2b-1 and wpsH pbp2b-2 compared to wpsH.
TABLE 2.
Sensitivity of L. cremoris NZ9000, MG1363 wpsH, and NZ9000 wpsA strains and their respective pbp2b derivatives to phages sk1, p2, MCC1, and MCC17
| Strain or strain derivative | Mean EOP ± SEMa |
|||
|---|---|---|---|---|
| sk1 | p2 | MCC1 | MCC17 | |
| NZ9000 | 1 | 1 | 1 | 1 |
| wpsH | ≤3 × 10−9 | (1 ± 1.68) × 10−8 | 0.16 ± 0.07 | (1 ± 0.21) × 10−3 |
| wpsH pbp2b-1 | (2.67 ± 0.95) × 10−6 | (1.95 ± 1.30) × 10−8 | 0.23 ± 0.15 | (7.94 ± 0.97) × 10−3 |
| wpsH pbp2b-2 | (2.87 ± 1.34) × 10−7 | (1.61 ± 1.45) × 10−8 | 0.21 ± 0.06 | (5.76 ± 1.16) × 10−3 |
| wpsA | ≤3 × 10−9 | ≤6 × 10−9 | ≤4 × 10−9 | ≤2 × 10−9 |
| wpsA pbp2b-1 | ≤3 × 10−9 | ≤6 × 10−9 | ≤4 × 10−9 | ≤2 × 10−9 |
| wpsA pbp2b-2 | ≤3 × 10−9 | ≤6 × 10−9 | ≤4 × 10−9 | ≤2 × 10−9 |
EOP values were calculated as the ratio of the phage titer on the tested strain by the phage titer on NZ9000. Values are means of three independent experiments.
Similarly, phage sensitivity was assessed on the wpsA mutant and its pbp2b derivatives. No infection was observed on those strains by either of the tested phages (Table 2). In particular, the wpsA pbp2b mutants remained completely resistant to the phage escape mutants MCC1 and MCC17 like the wpsA mutant.
All these results show that the wps pbp2b double mutants retain the phage resistance trait of their parent PSP-negative strain while exhibiting an improved fitness.
Peptidoglycan analysis of the double mutants.
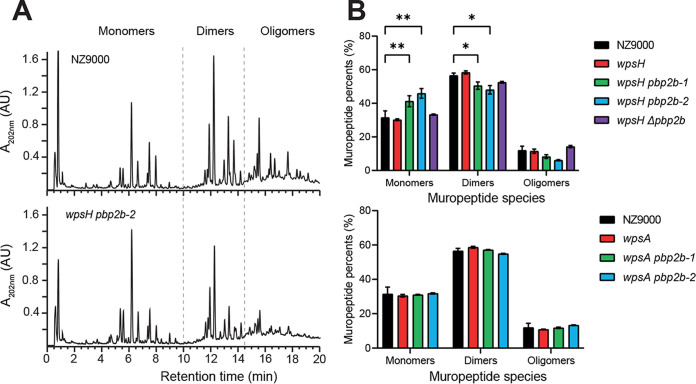
PBP2b is a class B monofunctional PBP with transpeptidase activity (22, 23). It catalyzes the formation of peptide bonds between the carbonyl group of the fourth d-Ala of the donor stem peptide chain with the amino group of d-Asp/d-Asn bound to Lys in the third position of an adjacent receptor chain. Thus, PBP2b contributes to PG reticulation. We evaluated the potential impact of pbp2b mutations on PG structure. To this end, we extracted PG from WT and the mutant strains, digested PG by mutanolysin and analyzed the resulting muropeptides by RP-UHPLC (Fig. 6A). Quantification of the muropeptides showed that the proportion of monomers was significantly increased in wpsH pbp2b-1 and wpsH pbp2b-2 mutants compared to WT (Fig. 6B). Concomitant significant decrease of dimers and slight decrease of oligomers were observed. These results indicate a reduced level of PG reticulation in the two wpsH pbp2b-1 and wpsH pbp2b-2 mutants compared to WT NZ9000. Of note, PG from the wpsH single mutant did not exhibit any difference in reticulation compared to NZ9000, confirming that modifications in wpsH pbp2b-1 and wpsH pbp2b-2 PG are not due to the initial mutation in wpsH, but rather to the pbp2b mutations. This strongly suggests that PBP2b with mutation T411M or V542F found in the two respective double mutants exhibits decreased transpeptidase activity. In line with that, as shown on the 3D homology model of L. cremoris PBP2b (Fig. 2), the two mutated residues are located in the vicinity of the PG binding site and could thus modify binding or access of the substrate to the active site. This substitution into larger residues could result in decreased activity or inactivation of the mutated PBP2b and could account for the lower PG reticulation.
FIG 6.
PG analysis of L. cremoris NZ9000, wpsH and wpsA mutants, and their respective pbp2b derivatives. (A) Representative muropeptide profiles obtained after PG digestion with mutanolysin and separation by RP-UHPLC for NZ9000 and wpsH pbp2b-2 strains. Identification of peaks containing monomers, dimers, or oligomers is based on mass spectrometry analysis and previous muropeptide identification (39). Monomers correspond to disaccharide peptides, dimers to two cross-linked monomers, and oligomers to three or more cross-linked monomers. (B) Relative abundances (%) of different muropeptide species for NZ9000, the wpsH mutant and its pbp2b derivatives (upper panel), and the wpsA mutant and its pbp2b derivatives (lower panel). Each value is the mean of three biological replicates. Errors bars represent ± the standard deviations of the mean. Abundance of each muropeptide species in mutant strains was compared to that of WT using a two-way analysis of variance, followed by Dunnett’s multiple comparison with a 95% confidence interval. The P values of significant differences are indicated by asterisks (*, P < 0.05; **, P < 0.01).
Surprisingly, PG reticulation was not significantly affected in the wpsH Δpbp2b mutant (Fig. 6B), although it is not able to synthesize PBP2b. Also, in wpsA pbp2b-1 and wpsA pbp2b-2 mutants, PG reticulation was shown to be very close to that of the WT strain NZ9000 (Fig. 6B). The wpsA pbp2b-1 mutant carries a single nucleotide deletion in position 430 of the pbp2b gene, leading to a frameshift and a premature stop codon, causing the predicted synthesis of a truncated protein. This protein corresponds to just a portion (160 amino acids out of the 721 amino acids of the full-length protein) located in the N-terminal pedestal domain of PBP2b, which is thus lacking enzymatic activity (Fig. 2). Regarding the wpsA pbp2b-2 mutant, it carries a 33-nucleotide deletion in pbp2b leading to a mutated PBP2b with an 11-amino-acid residue deletion in its sequence (positions 448 to 458). This deletion is located between the two conserved motifs of the transpeptidase domain (S414-X-X-K417 containing the catalytic serine and S470-X-N472) (23), which may result in a misfolded domain devoid of enzymatic activity. In conclusion, in all three mutants—wpsH Δpbp2b, wpsA pbp2b-1, and wpsA pbp2b-2—PBP2b is partially or completely deleted though this does not appear to cause modification of PG reticulation. In contrast, in both wpsH pbp2b-1 and wpsH pbp2b-2 mutants with reduced PG reticulation, full-length PBP2b with a unique missense mutation is present.
DISCUSSION
Isolation of spontaneous bacteriophage insensitive variants constitutes one of the strategies to design starter culture to counter bacteriophage infection. However, in the case of Caudoviricetes phages recognizing PSP as a receptor at the bacterial surfaces of L. lactis and L. cremoris for adsorption, phage resistance of the isolated lactococcal variants lacking PSP, is obtained at the cost of bacterial fitness. This study outlines an approach to isolate phage-resistant strains with improved fitness. Moreover, our results have more fundamental implications regarding the coordination of CWPS and PG biosynthesis in Gram-positive ovococci.
We took advantage of the poor growth phenotype of the PSP-mutants derived from L. cremoris MG1363 and NZ9000 to isolate spontaneous suppressor mutants able to form larger colonies on agar plates. In this way, we successfully isolated and characterized two sets of spontaneous suppressor mutants, in two different PSP-negative backgrounds, which were caused by wpsA or wpsH mutations. These mutants displayed improved general fitness (exponential growth rate, cell morphology) while retaining the phage resistance inherited from their respective parents. Notably, the wpsH mutant (VES5748) was a spontaneous long-chain forming mutant previously isolated without the use of genetic tools (10). Also, the derived suppressor mutants were obtained as spontaneous mutants without genetic manipulation and can be classified as food-grade. Thus, our results constitute a proof of concept of the possibility to isolate non-GMO, phage-resistant L. lactis and L. cremoris strains with a fitness close to the WT parental strain. PSP-negative mutants may also be isolated as spontaneous bacteriophage-insensitive mutants obtained upon exposure to virulent bacteriophages (1, 24). Therefore, despite the previously described structural diversity of PSP across the species (15), this method may be applicable to a wide range of lactococcal strains with technological functionalities.
From a fundamental perspective, our results for the first time highlight the link between Rha-CWPS, more specifically the PSP in lactococci, and the PG synthesis and cell division machineries. Rhamnan, the conserved component of Rha-CWPS in L. lactis and L. cremoris, as well as other streptococcal Rha-CWPS, was previously found to play a key role in cell morphology and division (11, 25, 26). Impairment of PSP biosynthesis also leads to alterations of cell shape and septum positioning in L. lactis mutants, in particular in NZ9000-wpsA mutant that is mutated in the initial step of PSP biosynthesis (17). In the latter mutant, we can rule out that the observed alterations result from sequestration of undecaprenyl-phosphate lipid carrier into biosynthetic intermediates, which would hamper PG biosynthesis. Therefore, synthesis of PSP, the variable component of lactococcal Rha-CWPS, also appears to be required for the proper functioning of the cell division machinery. In this study, we have isolated suppressor mutants with (partially) restored growth and shape, exhibiting mutations in pbp2b gene, encoding PBP2b shown previously to ensure peripheral growth and septum positioning in Lactococcus (20, 21). Our results further substantiate the interactions between Rha-CWPS and proteins of the cell division machinery. We hypothesize that Rha-CWPS (PSP or rhamnan) in lactococcal strains directly interact with proteins participating in cell division and in PG synthesis, and in such a way, mediate their correct positioning, conformation and/or function. When PSP is absent, the PBP2b activity may be too high and/or may not occur at the correct location in the cell wall, potentially leading to uncontrolled PG synthesis and cell division and to the morphological and growth defects depicted above. Hence, the suppressor effect of the pbp2b mutations in the absence of PSP would result in an improvement of cell wall assembly by lowering or shutting off PBP2b activity. As observed in the TEM micrographs, a leakage of cell wall material normally localized between the cytoplasmic membrane and PSP is clearly visible in the wpsA and wpsH mutant strains, whereas the cell wall appears thinner and more organized in their respective pbp2b derivatives.
Interestingly, a recent study has reported on the role of Rha-CWPS in cell division in Streptococcus mutans, another ovoid-shaped bacterium (27). In S. mutans, Rha-CWPS consists of a poly-rhamnose backbone substituted by Glc side chains further modified by glycerol phosphate (GroP). More specifically, it was shown that GroP modifications are required for proper localization of the PG hydrolase AtlA involved in cell separation and of MapZ, a protein involved in FtsZ ring positioning during cell division (28). In the proposed model, rhamnan deprived of negatively charged GroP modification is localized at the cell poles and equators and directs the correct positioning of MapZ and AtlA. In a similar way, we propose that PSP, which may be considered a negatively charged extracellular modification of rhamnan (9) and which covers the bacterial surface (10, 29), guides certain proteins of the cell division machinery, such as PBP2b, to their correct location. Whether this interaction is direct or not requires further investigations.
Although suppressor mutations were obtained in the pbp2b gene in the two wpsH and wpsA genetic backgrounds, TEM observations and PG analysis suggest somewhat different compensatory mechanisms. In the wpsH pbp2b suppressor mutants, cells exhibited ovoid shape and decreased PG reticulation, whereas wpsA pbp2b mutant cells were shown to be rounder and PG reticulation was similar to the WT. To explain these differences, we propose that the point mutations leading to single amino acid substitutions in PBP2b (obtained in the wpsH background) cause this protein be less active and retain its tertiary structure. This implies that the divisome complex is not disrupted and that PBP2b may still synthesize peripheral PG in complex with its cognate SEDS protein, RodA with transglycosylase activity (30), although at a reduced efficiency. The lower activity of PBP2b would then result in lower PG cross-linking detected by PG global analysis with cells keeping an ovoid shape. In contrast, in wpsH Δpbp2b mutant and the wpsA pbp2b suppressor mutants, which all are deletions of different sizes, PBP2b would be absent or completely inactive, and no PG peripheral synthesis could occur, leading to rounder cells. Regarding PG reticulation similar to the WT, this could result from compensation by other PG transpeptidase as, for, example by class A bifunctional PBPs, proposed to have a repair and maintenance activity of the PG meshwork in Gram-positive bacteria (31).
In conclusion, our results validate the isolation of PSP deficiency suppressor mutations in L. lactis and L. cremoris as a powerful approach with both applied and fundamental implications.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains used in this study are listed in Table 1. L. cremoris was grown at 30°C in M17 broth (Difco) supplemented with 0.5% (wt/vol) glucose (GM17). Solid GM17 for plate culture contained 1.5% (wt/vol) agar. Escherichia coli was grown in Luria-Bertani (LB) broth (Difco) with shaking at 37°C or on LB plates containing 1.5% (wt/vol) agar. The antibiotics erythromycin (Sigma-Aldrich) and chloramphenicol (Sigma-Aldrich) were used at concentrations of 3 and 5 μg/mL for L. lactis and 100 and 10 μg/mL for E. coli, respectively.
Whole-genome DNA sequence determination and analysis.
L. cremoris chromosomal DNA was extracted using the Wizard Genomic DNA purification kit (Promega Corporation). DNA sequencing was carried out on an Illumina HiSeq at GATC-Biotech (Constance, Germany) to generate about 5 to 6 million paired-end reads (150 bases in length). Assembly and mutant analysis were performed on services implemented on the web server BV-BRC (https://www.bv-brc.org/). The paired-end reads were merged, and de novo assembly was performed using SPAdes 3.9. (32).
Annotations were performed with Rapid Annotation using the Subsystem Technology server (33) Mutations were searched by alignment of the reads with BWA-mem (34), followed by SNP calling with FreeBayes (35).
PCR amplification and sequencing of pbp2b gene.
pbp2b gene was amplified from chromosomal DNA by PCR using Phusion High-Fidelity DNA polymerase (Biolabs) and primers alpF1 and recR2 (Table 3). Gene sequencing was performed with the primers pbp2b3s, pbp2b5, pbp2b6, and pbp2b7 (Table 3) by Eurofins Genomics.
TABLE 3.
Primers used in this study
| Primer | Sequence (5′–3′)a |
|---|---|
| alpF1 | ATTGTGTCAATGGGAAGTGG |
| recR2 | GAGCAAAAGCGTAAATCTCG |
| pbp2b3s | TCAGGTAGCGAATAAGCTAG |
| pbp2b5 | AAAAGGAAAACGTGGGGATG |
| pbp2b6 | CAATTGGCAGTTTATGCTGC |
| pbp2b7 | TTTGTTCCGCCGTAAAGTTC |
| Pbp2B 1 pGh9 L | AGCTTGATATCGAATTCCTGCAGCCCTTGTCCTCATTGTGATGGTC |
| Pbp2B 2 | ACATTTGGACTCCAAGAATGAG |
| Pbp2B 3 | CTCATTCTTGGAGTCCAAATGTAAAAGCCCTCTTGAATGAGG |
| Pbp2B 4 pGh9 R | CGGCCGCTCTAGAACTAGTGGATCCCCCTCTCCACGGCTTTTGATAAG |
| pGH9L | ATGTTACAGTCTATCCCTGG |
| pGH9R | GTATACTACTGACAGCTTCC |
| Pbp2B del L | TTGGCGTGATTTAGGAATCC |
| Pbp2B del R | TTTACTGACTGCCTTGTCAG |
The overlaps between sequences to assemble pbp2b gene are shaded gray; the overlap between sequences to assemble amplified pbp2b gene into pGh9 are underlined.
Construction of PBP2b-deficient strains.
L. cremoris pbp2b deletion mutants were constructed by double crossing over, using plasmid pGhost9-Δpbp2b containing DNA fragments surrounding pbp2b. To construct this plasmid, the fragments surrounding pbp2b gene were amplified from genomic DNA using Phusion High-Fidelity DNA polymerase (Biolabs) and the primer pairs Pbp2B 1 pGh9 L and Pbp2B 2 (Table 3) (to generate a 703-bp fragment upstream pbp2b), and Pbp2B 3 and Pbp2B 4 pGh9 R (Table 3) (to generate a 720-bp fragment downstream pbp2b). The PCR products were fused to the plasmid pGhost9, previously linearized with SmaI (NEB), by the strand overlap extension method (36), using Gibson Assembly Master Mix (NEB). The reaction mixture was transformed to E. coli strain TG1repA, with the selection of erythromycin (100 μg/mL)-resistant clones on LB plates. The DNA sequence of the cloned fragment in the resulting plasmid pGhost9-Δpbp2b was confirmed by PCR amplification and sequencing, using the primers pGH9L and pGH9R (Table 3). Plasmid pGhost9-Δpbp2b was introduced into NZ9000 or VES5748 (wpsH mutant) by electroporation, with the selection of erythromycin (3 μg/mL)-resistant clones on GM17 plates at 30°C. To select single crossing-over events, an exponential culture grown with erythromycin (3 μg/mL) at 30°C was plated on GM17 plates containing erythromycin, followed by incubation at a nonpermissive temperature (42°C). The second recombination event leading to plasmid excision was selected by inoculating obtained colonies in liquid GM17 medium without antibiotic at 42°C, shifting exponentially grown culture to 30°C for 2 h and then growing the culture overnight at 42°C. The culture was then plated on GM17 agar plates without antibiotic at 30°C. A strain harboring a pbp2b deletion was selected as an erythromycin-sensitive clone, and the presence of the deletion was confirmed by PCR amplification and nucleotide sequence determination, using the primers Pbp2B del L and Pbp2B del R (Table 3). The resulting mutant harboring pbp2b deletion derived from NZ9000 was named VES7552 (called NZ9000 Δpbp2B here), and the one derived from VES5748 was named VES7554 (called wpsH Δpbp2b here).
Determination of bacterial growth rate.
Bacterial strains were inoculated in GM17 broth. Exponential-growth-phase cultures were diluted at an optical density at 600 nm (OD600) of 0.01 in GM17 and distributed (200 μL per well) into a 96-well plate (Cellstar; Greiner Bio-One). Growth was monitored in a Tekan Infinite 200 plate-reader for 16 h at 30°C. Growth of each strain was measured in triplicate. The generation time (G) was calculated during the exponential growth phase as follows: [t2 − t1/log2(OD2)] − log2(OD1).
TEM.
Samples were fixed with 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) for 1 h at room temperature. Samples were contrasted with 0.5% Oolong tea extract in cacodylate buffer and postfixed with 1% osmium tetroxide containing 1.5% potassium cyanoferrate. The samples were dehydrated and embedded in Epon (Delta Microscopies, France), as described previously (17). Thin sections (70 nm) were collected onto 200-mesh copper grids and counterstained with lead citrate. Grids were examined with a Hitachi HT7700 electron microscope operated at 80 kV, and images were acquired with a charge-coupled device camera (Advanced Microcopy Techniques; facilities were located on the MIMA2 platform [INRAE, Jouy-en-Josas, France; https://doi.org/10.15454/1.5572348210007727E12]).
SEM.
Bacterial cells in exponential-phase cultures were directly fixed in Glutaraldehyde 2% buffered by sodium cacodylate 0.2 M, 1 h at room temperature then overnight at 4°C on glass slides. Samples were rinsed 10 min in 0.2 M sodium cacodylate buffer, dehydrated in successive baths of ethanol (50, 70, 90, 100, and anhydrous 100%), and then dried using Leica EM300 critical point apparatus with slow 20 exchange cycles, with a 2-min delay. Samples were mounted on aluminum stubs with adhesive carbon and coated with 6 nm of Au/Pd using a Quorum SC7620, 50 Pa of Ar, 180 s of sputtering at 3.5 mA. Samples were observed using the SE detector of a FEG SEM Hitachi SU5000, 2 KeV, 30 spot size, 5 mm working distance (facilities located on the MIMA2 platform, INRAE, Jouy-en-Josas, France; https://doi.org/10.15454/1.5572348210007727E12). Scanning electron microscopy (SEM) images were analyzed by using FIJI software and calibrated using the printed scale bar on the image during the acquisitions, and then we manually applied the measuring tool to determine the lengths and widths of cells that are at the same division stage.
Phage sensitivity assays.
Phages used in this study are listed in Table 1. Propagation of phages on their host strain was performed as previously described (37). The E.O.P. of a particular phage on a specific host was determined on solid and semisolid agar prepared using GM17 broth containing 1.0% or 0.4% agar, respectively, and supplemented by 10 mM CaCl2 as described previously (38). Serial 10-fold dilutions of phage suspensions were prepared using SM buffer (100 mM NaCl, 20 mM CaCl2, 10 mM MgSO4, 10 mM Tris-HCl [pH 7.0]). They were mixed with an overnight culture of the tested strain and semisolid GM17 and poured on top of solid GM17 medium. The EOP was calculated as the ratio of the titer obtained on the tested mutant strain to the one on the WT strain.
Cell wall preparation.
L. cremoris strains were grown in GM17 medium and harvested in exponential phase (OD600 of 0.6 to 0.8). After a wash with MilliQ H2O, the cells were boiled for 10 min and centrifuged at 5,000 × g at 4°C. Cell walls were prepared as described previously (39, 40), with some modifications. Heat-inactivated cells were resuspended in 1 mL of 5% sodium dodecyl sulfate (SDS) preheated at 60°C and boiled for 25 min. After centrifugation at 20,000 × g, the pellets were resuspended in 1 mL of 5% SDS preheated at 60°C, boiled for 15 min, and then washed with MilliQ water several times to remove SDS traces. Next, the pellets were sequentially treated under shaking (in an Eppendorf Thermomixer) with 2 mg mL−1 of Pronase (Roche) in 50 mM Tris-HCl (pH 7.0) for 90 min at 60°C, 200 μg mL−1 of trypsin (Sigma-Aldrich) in 20 mM Tris-HCl (pH 8.0) for 16 h at 37°C, and DNase (Sigma-Aldrich) and RNase (Sigma-Aldrich) (50 μg mL−1) in 20 mM Tris-HCl (pH 7.0) for 4 h at 37°C. After centrifugation at 20,000 × g for 10 min, the pellets were washed three times with MilliQ water. Purified cell walls were lyophilized and stored at −80°C until use.
CWPS extraction and SEC-HPLC analysis.
CWPS were extracted by HF treatment of purified cell walls as described previously (11). Cell walls (containing PG and CWPS) were treated with 48% HF for 48 h at 4°C to extract CWPS. The samples were centrifuged at 20,000 × g, and the supernatant containing CWPS was dried under a stream of nitrogen. The residue was solubilized in MilliQ H2O and lyophilized. Rhamnan and PSP oligosaccharides present in the sample were separated by SEC-HPLC with two columns in tandem (Shodex Sugar KS-804 and KS-803 columns; Showa Denko, Japan) as described previously (11). Elution was performed with MilliQ H2O, and detection of eluted compounds was performed with a refractometer (2414 refractive index detector; Waters).
PG extraction and RP-UHPLC analysis.
PG was extracted from purified cell walls (containing PG and CWPS) as described previously (39, 40). Lyophilized cell walls were treated with hydrofluoric acid (48%) for 19h at 4°C to remove covalently bound CWPS. The remaining insoluble PG was washed twice with 250 mM Tris-HCl (pH 8.0) and four times with MilliQ H2O and then lyophilized. PG was digested with mutanolysin (Sigma-Aldrich), and the resulting soluble muropeptides were reduced by NaBH4, as described previously (39). The reduced muropeptides were separated by reversed-phase ultrahigh-pressure liquid chromatography (RP-UHPLC) with a 1290 chromatography system (Agilent Technologies) and a Zorbax RRHD Eclipse Plus C18 column (100 by 2.1 mm; particle size, 1.8 μm; Agilent Technologies) at 50°C at 0.4-mL min−1 flow rate with a 1-min isocratic step of 10 mM ammonium phosphate buffer, followed by a methanol linear gradient to 20% in 19 min. The eluted muropeptides were detected by UV absorbance at 202 nm. Muropeptides in NZ9000 were identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry analysis, as described previously (39), with a Voyager DE STR mass spectrometer (Applied Biosystems) and according to the structures published previously (39). For the mutant strains, muropeptides were identified by their retention times by comparison to the NZ9000 muropeptide reference chromatogram. The different muropeptides were quantified by integration of the peak areas, and the percentage of each peak was calculated as the ratio of its area over the sum of all peak areas.
Protein 3D modeling.
At the time of computation, homology modeling of the PBP2b from L. lactis NZ9000 WT and mutants strains was performed using the building software Modeller (v9.18) (41) and the structure of PBP2b from Streptococcus pneumoniae strain 5204 as a 3D template (PDB ID 2wad) (23). For each PBP2b, WT and mutant proteins, a hundred models were computed and ranked according to the Modeller function and Dope scores. Eventually, a final model was selected for the WT and each of the mutants, upon these combined lowest scores, as they merged the highest probability to satisfy the spatial constraints. Last, the stereochemistry of each selected model was checked using Molprobity (42) and assisted by visual inspection using PyMOL (Schrödinger).
ACKNOWLEDGMENTS
We are very grateful to Pierre Renault (Micalis, INRAE) for his help for whole-genome sequence analysis. We thank Brian McDonnell (UCC) for his valuable technical advice for phage assays.
H.G. received a Ph.D. fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche (MESR, France) and was awarded a FEMS Research and Training Grant.
Contributor Information
Saulius Kulakauskas, Email: saulius.kulakauskas@inrae.fr.
Marie-Pierre Chapot-Chartier, Email: marie-pierre.chapot-chartier@inrae.fr.
Charles M. Dozois, INRS Armand-Frappier Sante Biotechnologie Research Centre
REFERENCES
- 1.Ortiz Charneco G, de Waal PP, van Rijswijck IMH, van Peij N, van Sinderen D, Mahony J. 2023. Bacteriophages in the dairy industry: a problem solved? Annu Rev Food Sci Technol 14:367–385. doi: 10.1146/annurev-food-060721-015928. [DOI] [PubMed] [Google Scholar]
- 2.Garneau JE, Moineau S. 2011. Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb Cell Fact 10(Suppl 1):S20. doi: 10.1186/1475-2859-10-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deveau H, Labrie SJ, Chopin MC, Moineau S. 2006. Biodiversity and classification of lactococcal phages. Appl Environ Microbiol 72:4338–4346. doi: 10.1128/AEM.02517-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spinelli S, Veesler D, Bebeacua C, Cambillau C. 2014. Structures and host-adhesion mechanisms of lactococcal siphophages. Front Microbiol 5:3. doi: 10.3389/fmicb.2014.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahony J, Cambillau C, van Sinderen D. 2017. Host recognition by lactic acid bacterial phages. FEMS Microbiol Rev 41:S16–S26. doi: 10.1093/femsre/fux019. [DOI] [PubMed] [Google Scholar]
- 6.Romero DA, Magill D, Millen A, Horvath P, Fremaux C. 2020. Dairy lactococcal and streptococcal phage-host interactions: an industrial perspective in an evolving phage landscape. FEMS Microbiol Rev 44:909–932. doi: 10.1093/femsre/fuaa048. [DOI] [PubMed] [Google Scholar]
- 7.Chapot-Chartier MP, Kulakauskas S. 2014. Cell wall structure and function in lactic acid bacteria. Microb Cell Fact 13(Suppl 1):S9. doi: 10.1186/1475-2859-13-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez B, Rodriguez A, Kulakauskas S, Chapot-Chartier MP. 2020. Cell wall homeostasis in lactic acid bacteria: threats and defences. FEMS Microbiol Rev 44:538–564. doi: 10.1093/femsre/fuaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guerin H, Kulakauskas S, Chapot-Chartier MP. 2022. Structural variations and roles of rhamnose-rich cell wall polysaccharides in Gram-positive bacteria. J Biol Chem 298:102488. doi: 10.1016/j.jbc.2022.102488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapot-Chartier MP, Vinogradov E, Sadovskaya I, Andre G, Mistou MY, Trieu-Cuot P, Furlan S, Bidnenko E, Courtin P, Pechoux C, Hols P, Dufrene YF, Kulakauskas S. 2010. The cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J Biol Chem 285:10464–10471. doi: 10.1074/jbc.M109.082958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadovskaya I, Vinogradov E, Courtin P, Armalyte J, Meyrand M, Giaouris E, Palussiere S, Furlan S, Pechoux C, Ainsworth S, Mahony J, van Sinderen D, Kulakauskas S, Guerardel Y, Chapot-Chartier MP. 2017. Another brick in the wall: a rhamnan polysaccharide trapped inside peptidoglycan of Lactococcus lactis. mBio 8:e01303-17. doi: 10.1128/mBio.01303-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ainsworth S, Sadovskaya I, Vinogradov E, Courtin P, Guerardel Y, Mahony J, Grard T, Cambillau C, Chapot-Chartier MP, van Sinderen D. 2014. Differences in lactococcal cell wall polysaccharide structure are major determining factors in bacteriophage sensitivity. mBio 5:e00880-14. doi: 10.1128/mBio.00880-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bebeacua C, Tremblay D, Farenc C, Chapot-Chartier MP, Sadovskaya I, van Heel M, Veesler D, Moineau S, Cambillau C. 2013. Structure, adsorption to host, and infection mechanism of virulent lactococcal phage p2. J Virol 87:12302–12312. doi: 10.1128/JVI.02033-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCabe O, Spinelli S, Farenc C, Labbe M, Tremblay D, Blangy S, Oscarson S, Moineau S, Cambillau C. 2015. The targeted recognition of Lactococcus lactis phages to their polysaccharide receptors. Mol Microbiol 96:875–886. doi: 10.1111/mmi.12978. [DOI] [PubMed] [Google Scholar]
- 15.Mahony J, Frantzen C, Vinogradov E, Sadovskaya I, Theodorou I, Kelleher P, Chapot-Chartier MP, Cambillau C, Holo H, van Sinderen D. 2020. The CWPS Rubik’s cube: linking diversity of cell wall polysaccharide structures with the encoded biosynthetic machinery of selected Lactococcus lactis strains. Mol Microbiol 114:582–596. doi: 10.1111/mmi.14561. [DOI] [PubMed] [Google Scholar]
- 16.Mahony J, Kot W, Murphy J, Ainsworth S, Neve H, Hansen LH, Heller KJ, Sorensen SJ, Hammer K, Cambillau C, Vogensen FK, van Sinderen D. 2013. Investigation of the relationship between lactococcal host cell wall polysaccharide genotype and 936 phage receptor binding protein phylogeny. Appl Environ Microbiol 79:4385–4392. doi: 10.1128/AEM.00653-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theodorou I, Courtin P, Palussiere S, Kulakauskas S, Bidnenko E, Pechoux C, Fenaille F, Penno C, Mahony J, van Sinderen D, Chapot-Chartier MP. 2019. A dual-chain assembly pathway generates the high structural diversity of cell-wall polysaccharides in Lactococcus lactis. J Biol Chem 294:17612–17625. doi: 10.1074/jbc.RA119.009957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linares DM, Kok J, Poolman B. 2010. Genome sequences of Lactococcus lactis MG1363 (revised) and NZ9000 and comparative physiological studies. J Bacteriol 192:5806–5812. doi: 10.1128/JB.00533-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Theodorou I, Courtin P, Sadovskaya I, Palussiere S, Fenaille F, Mahony J, Chapot-Chartier MP, van Sinderen D. 2020. Three distinct glycosylation pathways are involved in the decoration of Lactococcus lactis cell wall glycopolymers. J Biol Chem 295:5519–5532. doi: 10.1074/jbc.RA119.010844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Nunez D, Briandet R, David B, Gautier C, Renault P, Hallet B, Hols P, Carballido-Lopez R, Guedon E. 2011. A new morphogenesis pathway in bacteria: unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci. Mol Microbiol 79:759–771. doi: 10.1111/j.1365-2958.2010.07483.x. [DOI] [PubMed] [Google Scholar]
- 21.David B, Duchene MC, Haustenne GL, Perez-Nunez D, Chapot-Chartier MP, De Bolle X, Guedon E, Hols P, Hallet B. 2018. PBP2b plays a key role in both peripheral growth and septum positioning in Lactococcus lactis. PLoS One 13:e0198014. doi: 10.1371/journal.pone.0198014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinho MG, Kjos M, Veening JW. 2013. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat Rev Microbiol 11:601–614. doi: 10.1038/nrmicro3088. [DOI] [PubMed] [Google Scholar]
- 23.Contreras-Martel C, Dahout-Gonzalez C, Martins ADS, Kotnik M, Dessen A. 2009. PBP active site flexibility as the key mechanism for beta-lactam resistance in pneumococci. J Mol Biol 387:899–909. doi: 10.1016/j.jmb.2009.02.024. [DOI] [PubMed] [Google Scholar]
- 24.Dupont K, Janzen T, Vogensen FK, Josephsen J, Stuer-Lauridsen B. 2004. Identification of Lactococcus lactis genes required for bacteriophage adsorption. Appl Environ Microbiol 70:5825–5832. doi: 10.1128/AEM.70.10.5825-5832.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caliot E, Dramsi S, Chapot-Chartier MP, Courtin P, Kulakauskas S, Pechoux C, Trieu-Cuot P, Mistou MY. 2012. Role of the group B antigen of Streptococcus agalactiae: a peptidoglycan-anchored polysaccharide involved in cell wall biogenesis. PLoS Pathog 8:e1002756. doi: 10.1371/journal.ppat.1002756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Sorge NM, Cole JN, Kuipers K, Henningham A, Aziz RK, Kasirer-Friede A, Lin L, Berends ET, Davies MR, Dougan G, Zhang F, Dahesh S, Shaw L, Gin J, Cunningham M, Merriman JA, Hutter J, Lepenies B, Rooijakkers SH, Malley R, Walker MJ, Shattil SJ, Schlievert PM, Choudhury B, Nizet V. 2014. The classical Lancefield antigen of group A Streptococcus is a virulence determinant with implications for vaccine design. Cell Host Microbe 15:729–740. doi: 10.1016/j.chom.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zamakhaeva S, Chaton CT, Rush JS, Ajay Castro S, Kenner CW, Yarawsky AE, Herr AB, van Sorge NM, Dorfmueller HC, Frolenkov GI, Korotkov KV, Korotkova N. 2021. Modification of cell wall polysaccharide guides cell division in Streptococcus mutans. Nat Chem Biol 17:878–887. doi: 10.1038/s41589-021-00803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fleurie A, Lesterlin C, Manuse S, Zhao C, Cluzel C, Lavergne JP, Franz-Wachtel M, Macek B, Combet C, Kuru E, VanNieuwenhze MS, Brun YV, Sherratt D, Grangeasse C. 2014. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature 516:259–262. doi: 10.1038/nature13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andre G, Kulakauskas S, Chapot-Chartier MP, Navet B, Deghorain M, Bernard E, Hols P, Dufrene YF. 2010. Imaging the nanoscale organization of peptidoglycan in living Lactococcus lactis cells. Nat Commun 1:1–8. doi: 10.1038/ncomms1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sjodt M, Rohs PDA, Gilman MSA, Erlandson SC, Zheng S, Green AG, Brock KP, Taguchi A, Kahne D, Walker S, Marks DS, Rudner DZ, Bernhardt TG, Kruse AC. 2020. Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex. Nat Microbiol 5:813–820. doi: 10.1038/s41564-020-0687-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Straume D, Piechowiak KW, Kjos M, Havarstein LS. 2021. Class A PBPs: it is time to rethink traditional paradigms. Mol Microbiol 116:41–52. doi: 10.1111/mmi.14714. [DOI] [PubMed] [Google Scholar]
- 32.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aziz RK, Devoid S, Disz T, Edwards RA, Henry CS, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Stevens RL, Vonstein V, Xia F. 2012. SEED servers: high-performance access to the SEED genomes, annotations, and metabolic models. PLoS One 7:e48053. doi: 10.1371/journal.pone.0048053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv https://arxiv.org/abs/1207.3907.
- 36.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 37.Mahony J, McGrath S, Fitzgerald GF, van Sinderen D. 2008. Identification and characterization of lactococcal-prophage-carried superinfection exclusion genes. Appl Environ Microbiol 74:6206–6215. doi: 10.1128/AEM.01053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lillehaug D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J Appl Microbiol 83:85–90. doi: 10.1046/j.1365-2672.1997.00193.x. [DOI] [PubMed] [Google Scholar]
- 39.Courtin P, Miranda G, Guillot A, Wessner F, Mezange C, Domakova E, Kulakauskas S, Chapot-Chartier MP. 2006. Peptidoglycan structure analysis of Lactococcus lactis reveals the presence of an l,d-carboxypeptidase involved in peptidoglycan maturation. J Bacteriol 188:5293–5298. doi: 10.1128/JB.00285-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyrand M, Boughammoura A, Courtin P, Mezange C, Guillot A, Chapot-Chartier M-P. 2007. Peptidoglycan N-acetylglucosamine deacetylation decreases autolysis in Lactococcus lactis. Microbiology (Reading) 153:3275–3285. doi: 10.1099/mic.0.2007/005835-0. [DOI] [PubMed] [Google Scholar]
- 41.Sali A, Blundell TL. 1993. Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol 234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 42.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gasson MJ. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol 154:1–9. doi: 10.1128/jb.154.1.1-9.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuipers OP, de Ruyter PGGA, Kleerebezem M, de Vos WM. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol 64:15–21. doi: 10.1016/S0168-1656(98)00100-X. [DOI] [Google Scholar]
- 45.Maguin E, Prevost H, Ehrlich SD, Gruss A. 1996. Efficient insertional mutagenesis in lactococci and other Gram-positive bacteria. J Bacteriol 178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chandry PS, Moore SC, Boyce JD, Davidson BE, Hillier AJ. 1997. Analysis of the DNA sequence, gene expression, origin of replication and modular structure of the Lactococcus lactis lytic bacteriophage sk1. Mol Microbiol 26:49–64. doi: 10.1046/j.1365-2958.1997.5491926.x. [DOI] [PubMed] [Google Scholar]
- 47.Higgins DL, Sanozky-Dawes RB, Klaenhammer TR. 1988. Restriction and modification activities from Streptococcus lactis ME2 are encoded by a self-transmissible plasmid, pTN20, that forms cointegrates during mobilization of lactose-fermenting ability. J Bacteriol 170:3435–3442. doi: 10.1128/jb.170.8.3435-3442.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]