Abstract
In Europe, very few studies are available regarding the diversity of Listeria monocytogenes (L. monocytogenes) clonal complexes (CCs) and sequence types (ST) in poultry and on the related typing of isolates using whole genome sequencing (WGS). In this study, we used a WGS approach to type 122 L. monocytogenes strains isolated from chicken neck skin samples collected in two different slaughterhouses of an integrated Italian poultry company. The studied strains were classified into five CCs: CC1-ST1 (21.3%), CC6-ST6 (22.9%), CC9-ST9 (44.2%), CC121-ST121 (10.6%) and CC193-ST193 (0.8%). CC1 and CC6 strains presented a virulence gene profile composed of 60 virulence genes and including the Listeria Pathogenicity Island 3, aut_IVb, gltA and gltB. According to cgMLST and SNPs analysis, long-term persistent clusters belonging to CC1 and CC6 were found in one of the two slaughterhouses. The reasons mediating the persistence of these CCs (up to 20 months) remain to be elucidated, and may involve the presence and the expression of stress response and environmental adaptation genes including heavy metals resistance genes (cadAC, arsBC, CsoR-copA-copZ), multidrug efflux pumps (mrpABCEF, EmrB, mepA, bmrA, bmr3, norm), cold-shock tolerance (cspD) and biofilm-formation determinants (lmo0673, lmo2504, luxS, recO). These findings indicated a serious risk of poultry finished products contamination with hypervirulent L. monocytogenes clones and raised concern for the consumer health. In addition to the AMR genes norB, mprF, lin and fosX, ubiquitous in L. monocytogenes strains, we also identified parC for quinolones, msrA for macrolides and tetA for tetracyclines. Although the phenotypical expression of these AMR genes was not tested, none of them is known to confer resistance to the primary antibiotics used to treat listeriosis The obtained results increase the data on the L. monocytogenes clones circulating in Italy and in particular in the poultry chain.
Keywords: Listeria monocytogenes, whole genome sequencing, poultry, hypervirulent profiles, persistence, slaughterhouse
1. Introduction
The European Union (EU) is one of the world’s largest poultry meat producers and a net exporter of poultry products with annual production of around 13.4 million tons [1]. The EU agricultural outlook for 2021–2031 estimated a pro capite increase from 23.5 kg in 2021 to 24.8 kg in 2031 for the poultry sector. This should be managed through a healthier image of poultry relative to other meats, greater convenience in its preparation and the absence of religious constraints in its consumption.
L. monocytogenes is a ubiquitous bacterium causing human listeriosis, the most serious foodborne disease under EU surveillance with the highest proportion of hospitalized cases (96.5% in 2021) and fatality rate (13.7% in 2021). Invasive forms of the disease mainly affect people at risk causing abortion and stillbirth in pregnant women and meningitis septicemia and death in the elderly, immunocompromised people, and new-borns [2].
Listeriosis associated with meat products is a significant food safety concern and slaughterhouses and market sites are ideal environments for the proliferation of L. monocytogenes [3]. Although the occurrence of L. monocytogenes in meat products is the highest in products of bovine or porcine origin [2,4], it is also present in chicken and other poultry meats, including packaged and non-packaged products, whole chickens and various types of sliced meat. Contamination of chicken meat and foods prepared using chicken meat can occur at various stages before marketing, including the primary production stage, abattoir, processing plant, and retail stores [5]. In recent years, numerous studies on the presence of L. monocytogenes in these matrices have been conducted worldwide [5,6,7,8,9]. The consumption of cooked chicken products was previously associated with the transmission of listeriosis and caused several listeriosis outbreaks in different countries [10,11,12,13].
Some L. monocytogenes strains are able to survive and persist in food-producing environments (FPE) even for years, due to the adaptation to different environmental stresses, resistance to disinfectants and biofilm formation. In a FPE, hard to reach surfaces during clean-up and sanitation could be ideal niches for L. monocytogenes persistence and act as persistent source of contamination [14,15,16,17,18].
Currently, the whole genome sequencing (WGS) offers the highest level of discrimination for surveillance of pathogens and for epidemiological investigations of foodborne outbreaks, also allowing the characterization of pathogenic microorganisms. WGS is considered to be the best approach to obtain the most detailed results on the nature and localization of genes associated with virulence, biological fitness and antimicrobial resistance [19].
WGS data on L. monocytogenes in poultry samples are reported worldwide, in particular are available data from poultry meat in China, from chicken in South Korea and from turkey in USA [9,20,21]. On the contrary, at European level, particularly in Italy, very few studies regarding the spread of L. monocytogenes clonal complexes (CCs) in poultry and the related genomic typing of isolates using WGS are available. More data on the genetic virulence profile is needed in order to precisely determine the pathogenic potential of L. monocytogenes isolates and to acquire a better comprehension of the risk exposure for the consumers. Moreover, the investigation of genetic traits for stress resistance and tolerance to disinfectants and heavy metals is important to understand the ability of the strains to adapt and persist in the production environment. The latter aspect is becoming relevant in intensive poultry production which increases the risk of microbial contamination and promotes the persistence of food-borne pathogens in environmental niches. It is well known that certain CCs, such as CC1, CC2, CC4, and CC6, are more frequently associated with clinical cases and are hypervirulent in a humanised mouse model, whereas others like CC9 and CC121 are mainly of foodborne origin and show hypovirulence in vivo [14,22,23,24,25]. Different virulence profiles have been observed between hyper- and hypovirulent clones of Listeria monocytogenes with the former carrying additional virulence determinants such as Listeria Pathogenicity Islands (LIPIs) in addition to the ubiquitous LIPI-1 [22,26,27]. Conversely, hypovirulent clones are better adapted to food processing environments, with a higher prevalence of stress resistance and benzalkonium chloride tolerance genes [22,27].
In this study, we used a WGS approach to type 122 L. monocytogenes strains isolated from poultry neck skin samples collected in two different slaughterhouses of an integrated Italian poultry company during a previous work performed by Iannetti et al. [7]. These authors used pulsed field gel electrophoresis (PFGE) to type strains and identify the persistent pulsotypes.
Specifically, in the present study we carried out WGS data analysis: (i) to identify the main L. monocytogenes CCs circulating in two poultry slaughterhouses; (ii) assess the clustering of the strains evaluating their persistence over time, (iii) determine the virulence genetic profiles of the isolates identifying hypervirulent clones and (iv) detect genes involved in stress response and tolerance to heavy metals and disinfectants.
2. Materials and Methods
2.1. Strain Collection
The strains collection derived from a previous study in which prevalence of L. monocytogenes in poultry along an Italian production chain was evaluated [7]. A total of 2080 samples (1560 faeces/caeca contents and 520 neck skin samples) from broiler chickens were analysed for L. monocytogenes detection. The broiler chickens were bred in farms of Northern and Central Italy and slaughtered in two different slaughterhouses of Central Italy, in particular M1 (first slaughterhouse) and M2 (second slaughterhouse).
The overall positivity of the collected samples was 6.63%, with 138 positive samples out of 2080. No positivity was recorded in samples of faeces, while an overall prevalence of 26.7% was detected in neck skin samples collected in the two slaughterhouses.
In this study 122 isolates (71 collected at M1 and 51 at M2), derived from positive neck skin samples, were typed using WGS and different bioinformatics analysis (Supplementary Table S1).
2.2. Whole Genome Sequencing (WGS) Analysis
DNA extraction was performed using QIAamp DNA Mini Kit (Qiagen Hilden, Germany) according to the manufacturer’s protocol with minor modifications according to Portmann et al. [28]. DNA quantity and quality were evaluated with Qubit fluorometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and Eppendorf BioSpectrometer fluorescence (Eppendorf s.r.l., Milano, Italy). DNA integrity was assessed with Agilent 4200 TapeStation system (Agilent, Santa Clara, CA, USA).
Starting from 1 ng of input DNA, the Nextera XT DNA chemistry (Illumina, San Diego, CA) for library preparation was used according to the manufacturer’s protocols. WGS was performed on the NextSeq 500 platform (Illumina, San Diego, CA, USA) with the NextSeq 500/550 mid output reagent cartridge v2 (300 cycles, standard 150-bp paired-end reads).
For the analysis of WGS data, an in-house pipeline [29] was used which included steps for trimming (Trimmomatic v0.36) [30] and quality control check of the reads (FastQC v0.11.5) [31]. Genome de novo assembly of paired-end reads was performed using SPAdes v3.11.1 [32] with default parameters for the Illumina platform 2 × 150 chemistry (–only-assembler –careful –k21, 33, 55, 77). Subsequently, the genome assembly quality check was performed with QUAST v.4.3 [33].
All the genomes that met the quality parameters recommended by Timme et al. [34], such as average read quality Q score for R1 and R2 ≥ 30; Average coverage ≥ 20, number of contigs ≤ 300, and were used for the analysis.
The genome assemblies were deposited at DDBJ/ENA/GenBank under the BioProject PRJNA947067 (Supplementary Table S1).
2.3. Multilocus Sequence Typing, Core Genome Multilocus Sequence Typing and Single-Nucleotide Polymorphism Analysis
A multilocus sequence typing (MLST) analysis was performed by deducing the sequence type (ST) and the CC in silico, using the specific tool available from the BIGSdb-Lm database (https://bigsdb.pasteur.fr/listeria/; accessed on 20 February 2023) [35]. The MLST scheme used, included the seven housekeeping genes abcZ, bglA, cat, dapE, dat, ldh, and lhlA [35].
To verify the relatedness among the isolates, identifying genomic clusters, a core genome MLST (cgMLST) analysis was performed using the chewBBACA allele calling algorithm [36] and the Pasteur Institute cgMLST scheme of 1748 loci [37]. According to the guidelines for L. monocytogenes cgMLST typing [37], only the genomes with at least 1660 called loci (95% of the full scheme) were considered. GrapeTree [38] was used for the visualization of the minimum spanning tree (MSTreeV2 method).
A core single-nucleotide polymorphism (SNPs) analysis was performed using the CFSAN pipeline [39], and the sequence reads were mapped against an EGD-e reference genome (NC_003210.1). The resulting maximum likelihood (ML) tree was visualized using Interactive Tree of Life (iTOL) (https://itol.embl.de/; accessed on 20 February 2023).
The threshold values adopted in this study for cluster definition were an allelic distance (AD) of 7 for the cgMLST analysis [37] and 21 pairwise SNPs [40,41].
In order to find clones matching with strains belonging to the national collection of the Italian National Reference Laboratory for L. monocytogenes (NRL-Lm), the relative genome database (about 5000 L. monocytogenes genomes stored from food, environment and animals) was interrogated. In particular, a cgMLST analysis was performed comparing the genomes of the isolates of the study with all the genomes belonging to the same CC collected in the Italian database.
2.4. Genetic Determinants Involved in Virulence Potential, Antimicrobial Resistance, Stress Adaptation and Heavy Metal and Disinfectant Resistance
In silico analysis was performed using tools built in the BIGSdb-Lm platform for the detection of virulence genes, antimicrobial resistance (AMR) genes, heavy metal and disinfectants resistance determinants, stress survival islets (SSIs) and biofilm-associated genes.
The detection of additional determinants, not included in these schemes, was performed automatically using Prokka v.1.1211 [42].
3. Results
3.1. MLST Analysis
All the 122 genomes met the quality parameters recommended, such as average coverage ≥ 20X, de novo assembly length 2.7–3.2 Mbp and number of contings ≤ 300, and were used for the analysis.
The 122 isolates from M1 (n = 71) and M2 (M = 51) were classified into 5 STs and as many CCs (Table 1; Figure 1a; Supplementary Table S1): CC1-ST1 (26 strains; 21.3%), CC6-ST6 (28 strains; 22.9%), CC9-ST9 (54 strains; 44.2%), CC121-ST121 (13 strains; 10.6%) and CC193-ST193 (1 strain; 0.8%). Among the strains isolated from neck skin samples collected in M1, 28 belonged to CC6 (39.4%), 26 to CC1 (36.6%), 10 to CC121 (14.1%), 6 to CC9 (8.5%) and one strain belonged to CC193. Strains from M2 belonged to CC9 (48 strains; 94.1%) and CC121 (3 strains; 5.9%).
Table 1.
MLST analysis: number of strains belonging to different CCs and STs isolated in each slaughterhouse.
| Slaughterhouse | CC1-ST1 | CC6-ST6 | CC9-ST9 | CC121-ST121 | CC193-ST193 | Total |
|---|---|---|---|---|---|---|
| M1 | 26 | 28 | 6 | 10 | 1 | 71 |
| M2 | 0 | 0 | 48 | 3 | 0 | 51 |
| Total | 26 | 28 | 54 | 13 | 1 | 122 |
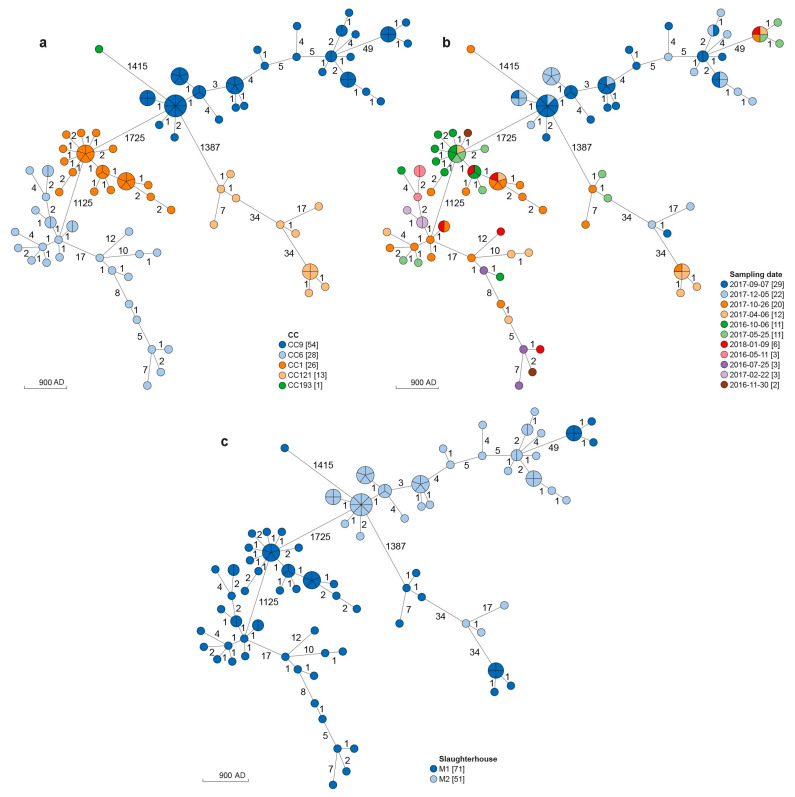
Figure 1.
Cluster analysis of L. monocytogenes strains based on cgMLST profiles. (a) In the minimum spanning tree (MSTv2), strains are colored according to the Clonal Complex (CC). (b) In the MSTv2, strains are colored according to the sampling date. (c) In the MSTv2 strains are colored according to the slaughterhouse from which they were isolated. The scale bar indicates the allelic distance (AD).
3.2. cgMLST and SNPs Analyses
The cluster analysis of the studied strains was performed using the results from both cgMLST and SNPs analysis (Figure 1 and Figure 2).
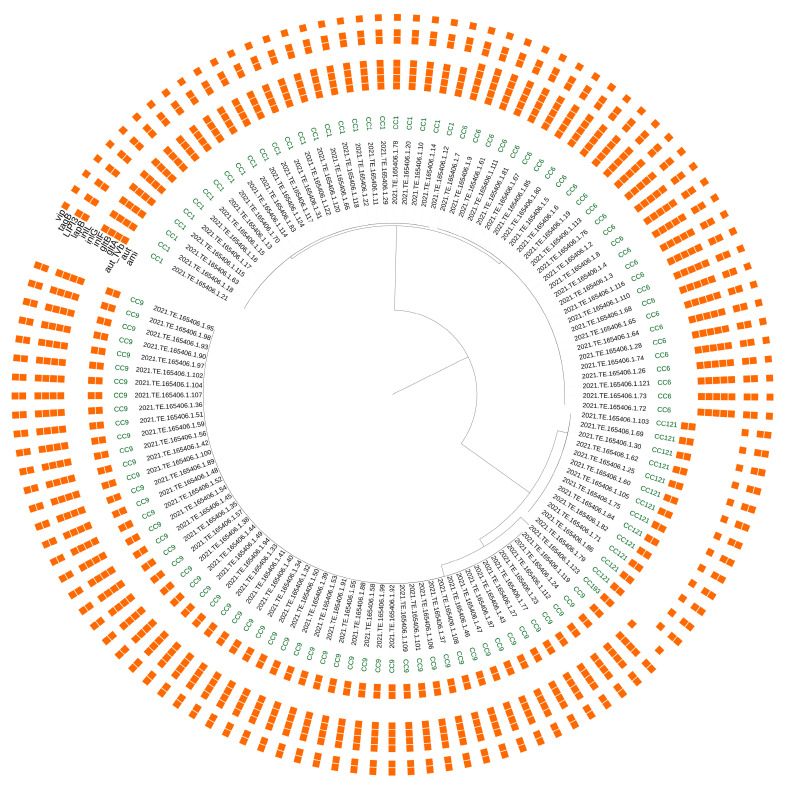
Figure 2.
SNPs analysis of L. monocytogenes strains. The first circle indicates the CC; the presence/absence matrix represents virulence genes that vary between CCs.
According to the minimum spanning tree (MST) obtained from cgMLST analysis (Figure 1a–c) and the SNPs distance matrix (Supplementary Table S2), all the CC1 strains grouped into a single cluster isolated in the M1 slaughterhouse from October 2016 to January 2018 (AD ranging from 0 to 7; SNPs of difference ranging from 4 to 10) (Table 2; Figure 1b,c).
Table 2.
Main clusters detected, CC, number of isolates, slaughterhouse, timeframe, ranges of allelic distances and SNPs.
| CC | Cluster | N° of Strains | Slaughterhouse | Timeframe | AD | SNPs |
|---|---|---|---|---|---|---|
| CC1 | I | 26 | M1 | October 2016–January 2018 | 0–7 | 4–10 |
| CC6 | I | 16 | M1 | May 2016–January 2018 | 0–7 | 1–17 |
| II | 4 | M1 | July 2016–January 2018 | 1–7 | 6–12 | |
| III | 3 | M1 | July 2016–January 2018 | 0–1 | 1–3 | |
| IV | 2 | M1 | April–October 2017 | 1 | 0 | |
| V | 2 | M1 | April 2017 | 1 | 2 | |
| CC9 | I | 30 | M2 | September–December 2017 | 0–7 | 0–16 |
| II | 15 | M2 | September–December 2017 | 0–5 | 1–20 | |
| III | 2 | M2 | September 2017 | 1 | 0 | |
| IV | 6 | M1 | April 2017–January 2018 | 0–1 | 1–6 | |
| CC121 | I | 4 | M1 | May–October 2017 | 0–7 | 1–19 |
| II | 6 | M1 | April–October 2017 | 0–1 | 0–5 | |
| III | 2 | M2 | September–December 2017 | 1 | 1 |
CC—Clonal Complex; AD—allelic distance.
Strains belonging to CC6 grouped into five main clusters. The largest CC6 cluster (16 strains; 0–7 AD; 1–17 SNPs) was isolated from May 2016 to January 2018, the second largest one (4 strains; 7 AD; 6–12 SNPs) from July 2016 to January 2018 and the third one (3 strains; 1 AD; 1–3 SNPs) from July 2016 to January 2018. One of the two smallest clusters was isolated from April to October 2017 (2 strains; 1 AD; 0 SNPs) and the other one included two strains both isolated at April 2017 (1 AD; 2 SNPs) (Table 2; Figure 1b,c).
Strains belonging to CC9 showed four main clusters. Three of them, all isolated in M2, were highly related and the remaining one, isolated in M1, was more distant. Among the M2 clusters, the largest one (30 strains; AD ranging from 0 to 7; 0–16 SNPs) and the second largest one (15 strains; AD ranging from 0 to 5; 1–20 SNPs) were both isolated from September to December 2017, while the third one (2 strains; a single AD and 0 SNPs of difference) was isolated in September 2017. The remaining CC9 cluster (6 strains; 0–1 AD; 1–6 SNPs) was isolated from samples collected from April 2017 to January 2018 (Table 2; Figure 1b,c).
The CC121 strains grouped into three main clusters. Two clusters were isolated in M1 from May to October 2017 (4 strains; 0–7 AD; 1–19 SNPs) and from April to October 2017 (6 strains; 0–1 AD; 0–5 SNPs) respectively. The remaining one (2 strains; a single AD and 1 SNP of difference) was isolated in M2 from September to December 2017 (Table 2; Figure 1b,c).
None of the 122 strains had a genetic correlation with those present in the NRL-Lm genome database. The cgMLST analysis showed that no distances ≤ 7 AD were found comparing the strains of this study with all the isolates of the same CC stored in the database.
3.3. Genetic Determinants Involved in Virulence Potential, Antimicrobial Resistance, Stress Adaptation and Heavy Metal and Disinfectant Resistance
Using the BIGSdb-Lm platform, 71 virulence genes were detected on a scheme of 93 targets. Fifty-two genes were carried by all the strains. Among these ubiquitous targets there were the conventional Listeria Pathogenicity Island (LIPI) 1, 9 internalins genes including inlA, inlB, inlC, inlC2, inlD, inlE, inlH, inlJ and inlK and the virR/virS virulence regulatory system. The variable genes showing differences in their presence/absence among different CCs were 19 (Figure 2). Overall, the CC1 and CC6 strains presented 66 virulence genes, while CC9, CC121 and CC193 carried 60, 57 and 56 virulence determinants, respectively.
More in detail, all the CC1 and the CC6 strains presented the additional LIPI-3, the teichoic acid biosynthesis genes gltA and gltB, and the invasion gene aut_IVb. The lapB gene was present in all the CCs except the CC193 strain. The virulence genes ami and tagB were carried by all the isolates belonging to CC9, CC121 and CC193. All the CC9 strains carried the Internalins genes inlF, also detected in the only CC193 strain, inlG and inlL.
The CC9, CC121 and CC193 strains showed different mutations in the inlA gene leading to a premature stop codon (PMSC). In particular, five PMSC mutation types were detected: PMSC_6 (all CC121 strains), PMSC_8 (48 CC9 strains), PMSC_25 (CC193) and PMSC_29 (6 CC9 strains). All the CC1 and CC6 strains carried a full length inlA.
All the tested strains presented the same AMR gene pattern including fos (fosfomycin), lmo0919 (lincosamide), lmo1695 (cationic antimicrobials), norB (fluoroquinolones) and sul (sulfanilamides). The additional AMR genes, parC for quinolones; msrA for macrolides and tetA for tetracyclines were also detected in all the strains. In addition to these AMR genes, different multidrug efflux pumps (emrB, bmrA, bmr3, norM) were detected in all the isolates.
The genetic determinants for different heavy metal resistance, multidrug efflux pumps and response to environmental stresses were reported in Table 3.
Table 3.
Relevant features for virulence, stress response and environmental adaptation.
| Main function | Gene/Island | Clonal Complex | |
|---|---|---|---|
| Virulence | LIPI-3 | CC1, CC6 | |
| gltA-gltB | CC1, CC6 | ||
| aut_IVb | CC1, CC6 | ||
| lapB | CC1, CC6, CC9, CC121 | ||
| ami | CC9, CC121, CC193 | ||
| tagB | CC9, CC121, CC193 | ||
| inlF | CC9, CC193 | ||
| inlG | CC9 | ||
| inlL | CC9 | ||
| Metal resistance |
Cadmium | cadA, cadC | CC1 |
| Arsenic | arsB, arsC | CC1, CC6, CC9, CC121, CC193 | |
| arsA, arsD | CC9 | ||
| Copper | CsoR-copA-copZ | CC1, CC6, CC9, CC121, CC193 | |
| copB | CC9, CC193 | ||
| Stress response |
Acid Tolerance | gadB, gadC | CC1, CC6, CC9, CC121, CC193 |
| SSI-1 | CC9 | ||
| Alkali response | mrpA, mrpB, mrpC, mrpE, mrpF | CC1, CC6, CC9, CC121, CC193 | |
| SSI-2 | CC121 | ||
| Low pH, high salt concentration, refrigeration | SSI-1 | CC9 | |
| Oxidative stress | SSI-2 | CC121 | |
| Cold-shock | cspD | CC1, CC6, CC9, CC121, CC193 | |
| Biocides resistance |
EmrB, mepA, bmrA, bmr3, norM | CC1, CC6, CC9, CC121, CC193 | |
| QACs | Tn6188_qacH | CC121 | |
| Biofilm production |
Lmo0673, lmo2504, luxS, recO | CC1, CC6, CC9, CC121, CC193 | |
| inlL | CC9 | ||
| PMSC inlA | CC9, CC121, CC193 | ||
QACs—quaternary ammonium compounds.
In particular, all the CC9 strains carried a complete SSI-1, while in all the CC121 a complete SSI-2 was found. Moreover, all the CC121 strains presented the Tn6188 transposon carrying the qacH gene which can confer reduced susceptibility to both quaternary ammonium compounds (QACs) and ethidium bromide. Only CC1 strains carried cadA and cadC for cadmium resistance.
Among the genetic determinants for arsenic resistance, arsB and arsC were found in all the studied strains while arsA and arsD were detected only in CC9 isolates. The csoR-copA-copZ copper resistance operon was present in all the strains while copB was only carried by 35 strains (34 CC9 and the CC193 isolate). All the isolates carried the cold-shock protein gene cspD and the genetic markers for biofilm production lmo0673, lmo2504, luxS and recO.
4. Discussion
Very few data are available at the European level on L. monocytogenes CCs and WGS analysis in poultry. Most of the existing studies have used old typing techniques such as serotyping and PFGE [5,6,43,44].
In this study 122 L. monocytogenes strains, isolated from two slaughterhouses of one of the main integrated Italian poultry companies during a previous study by Iannetti et al. (2020) [7], were typed using a WGS approach.
Ianneti et al., [7] used serotyping and PFGE to type the L. monocytogenes isolates identifying 18 different pulsotypes grouping into 8 main clusters. They also assumed the persistence of the pathogen since they isolated undistinguishable pulsotypes in carcasses slaughtered in the same slaughterhouse after periods up to 18 months long.
The WGS approach used in this study, allowed to better discriminate the strains and to obtain information on their genomes such as their virulence profile, the presence of AMR genes and of several determinants involved in environmental stress adaptation. Specifically, the MSTree, obtained from the cgMLST analysis, showed 78 different allelic profiles, grouping into 12 clusters and 6 singleton strains. The results obtained confirmed the long-term persistence of the pathogen in one of the slaughterhouse identifying clusters persisting up to 20 months.
As concluded by Iannetti et al. (2020) [7], considering the high number of carcasses processed in these slaughterhouses, it is probable that the environmental contamination derived from the sporadic introduction of L. monocytogenes strains through faeces of healthy carrier chickens and that once introduced, L. monocytogenes was able to persist in favourable niches thanks to its adaptation and surviving skills. Therefore, the slaughterhouse could be considered as an important hotspot of microbial contamination giving information on L. monocytogenes strains circulating at different point of the poultry chain.
In this study, we reported CCs already found by other authors worldwide in poultry and in particular CC1, CC6, CC9, CC121 and CC193. However, our results increased the data currently available at European and Italian level. Previous studies, mostly performed in China, reported the isolation of the following CCs from poultry meat: CC2, CC3, CC5, CC8, CC9, CC11, CC14, CC19, CC87, CC101, CC121, CC155, CC193 and CC307 [3,9,45,46]. Brown et al. [21] reported the isolation of a wide CCs diversity from the environment of turkey processing plants in the United States (CC1, CC2, CC3, CC5, CC6, CC7, CC8, CC9, CC11, CC29, CC87, CC89, CC155, CC224, CC321, CC506, CC554, CC570, CC1084) and the identification of persistent clones. Lee and Ruy reported the isolation of a CC1 strain from a chicken in South Korea [20].
Among the CCs found in this study, CC9 and CC121 have been previously defined hypovirulent clones, with a better adaptation to food processing environments. On the contrary, previous studies defined CC1 and CC6 as hypervirulent clones since they resulted most commonly associated with human listeriosis [14,22,23,24,25,47,48,49,50,51].
The microbial population associated with M1 was more heterogeneous than that isolated in M2. The results of cgMLST and SNPs analysis showed the presence of several clusters and none of them included strains isolated from both M1 and M2, indicating that each slaughterhouse presented an associated and unique microbial population. In M1, longer persistent clusters were detected (up to 20 months). Very interestingly, in this slaughter environment, CC1 and CC6 persisted for longer than CC9 and CC121, although the latter are commonly considered better adapted to food processing environments with a higher prevalence of stress resistance genes and a higher survival and biofilm formation [14,27].
All the genetic clusters detected in M2, belonging to CC9 and CC121, included strains isolated from September to December 2017 and so persisted for a shorter period.
Other authors also reported similar results [21,27,52,53] with hypervirulent clones such as CC1, CC2 and CC6 persisting for long time. These findings could indicate that the fitness of a strain is relative to the environment with which it is interacting. Therefore, within M1 may have been a selection pressure that favored CC1 and CC6 clones rather than the others. Moreover, besides the specific characteristics of the FPE (presence of ecological niches, non-compliant structures and equipment) and the survival abilities of the strains, other factors can influence L. monocytogenes persistence such as reintroduction of contaminated raw materials, inappropriate processing and ineffective cleaning and sanitizing protocols [27]. A limitation of this study was the lack of an intensive environmental sampling with multiple sampling sessions in the two slaughterhouses, in order to detect other persistent clones and to identify the surfaces acting as niches of persistence for L. monocytogenes. This should be useful to apply effective mitigation strategies. Another limitation was the lack of a genome comparison between the hypervirulent clones isolated in this study and those collected in the Italian genome database of L. monocytogenes clinical strains.
From the evaluation of the virulence profiles, it was quite evident that CC1 and CC6 isolates presented a higher number of virulence genes if compared with other CCs. In particular, all the CC1 and CC6 isolates presented both LIPI-1 and also the additional LIPI-3 known to confer a greater virulence to L. monocytogenes [54,55]. LIPI-1 carries the major virulence genes, such as prfA, the genes needed to escape from vacuoles (hly and plcA), genes for actin-based motility (actA) and genes needed for cell-to-cell spread (mpl and plcB). The LIPI-3 encodes the bacteriocin LLS, highly expressed in the intestine to alter host intestinal microbiota and allowing L. monocytogenes colonization of the intestine [56,57,58].
The aut_IVb, gltA, and gltB genes were also important virulence markers involved in invasion and teichoic acid biosynthesis respectively. These genes are typically carried by hypervirulent isolates and are absent in other L. monocytogenes leading a higher virulence potential [14,22,37].
All CC9, CC121 and CC193 strains did not present any LIPI other than the ubiquitous LIPI-1, and they harboured different types of PMSCs within the inlA gene [23,59,60] which are associated with attenuated virulence in mammal hosts [24,61,62].
All the strains isolated from M1 and M2 carried several determinants for stress adaptation, heavy metal resistance and tolerance to biocides. The prevalence of environmental adaptation determinants is commonly higher in hypovirulent CCs such as CC9 and CC121 [14,27,63]. However, in this study, many genes were also found in hypervirulent strains with some even exclusive to them, as in the case of cadA and cadC, only carried by CC1 strains. Genetic elements for arsenic and copper resistance [64,65], acid tolerance [66,67,68] and alkali response [66,69,70,71] were found in all the CCs as well as several multidrug efflux pumps, known to be involved in non-specific tolerance to disinfectants [26,72,73,74,75] (Table 1). The same result was obtained for the biofilm production markers lmo0673, lmo2504, luxS and recO [76] and for cspD, known to be essential for adaptation against various food-relevant stress conditions including cold growth [77]. Certain genetic determinants for environmental adaptation such as arsAD, copB, inlL and SSI-1 were only found in CC9 strains. The SSI-2 [78], and the Tn6188_qac gene were detected only in CC121 strains [79,80]. The presence of PMSCs mutations in the inlA gene, was previously found to be associated with increased biofilm production and was detected in CC9, CC121 and CC193 isolates [62]. However, further studies in this field are needed to identify factors that predispose some strains of L. monocytogenes towards increased biofilm formation [62].
In CC1 and CC6 strains, the presence of genetic determinants involved in stress response, such as heavy metal resistance, alkali and acid response and cold tolerance, may have contributed to the long-term persistence of these hypervirulent CCs in the M1 slaughterhouse. In the future, it might be interesting to evaluate the ability of these hypervirulent clones to produce biofilm in vitro.
The ability of L. monocytogenes strains to persist in food processing environments once introduced throughout the raw materials, together with the tendency of poultry slaughterhouses to act as contamination hotspots, could contribute to these food-associated environments becoming reservoirs of different L. monocytogenes genetic variants, including hypervirulent clones with adaptation skills [21].
The AMR genetic profile was the same in all the strains studied and included genes with mechanisms of antibiotic efflux (norB), antibiotic target alteration (mprF), and antibiotic inactivation (lin, fosX) [81,82,83,84]. Previous studies showed fosX and lin to be present in nearly all L. monocytogenes isolates [82,85]. This can be explained by native resistance to fosfomycin and lincosamides reported in L. monocytogenes strains. In addition to these ubiquitous AMR genes, we identified, in all tested strains, parC for quinolones, msrA for macrolides and tetA for tetracyclines, which were recently reported in L. monocytogenes by other authors [81,86]. Although the phenotypical expression of these AMR genes was not tested, none of them is known to confer resistance to the primary antibiotics used to treat listeriosis [85].
5. Conclusions
This study deepens the current knowledge on L. monocytogenes CCs circulating in the Italian poultry chain and on their genetic features, using WGS data analysis. Useful information was provided to integrate the limited data available in this regard at European level and in particular in Italy.
Among the detected CCs, CC1 and CC6, known to be hypervirulence clones, were found to persist for long time in the slaughterhouse.
These findings indicate a serious risk of finished food contamination with hypervirulent L. monocytogenes clones and raise concern for the consumer health.
As a future perspective, it would be interesting to evaluate the genomic correlation between the hypervirulent strains of this study and those isolated from clinical cases of listeriosis occurred in Italy.
Further WGS-based studies are needed, including intensive environmental sampling for L. monocytogenes at the slaughterhouse and analysis of finished products are needed, in order to deepen our knowledge on the genetic population of L. monocytogenes in the poultry chain, identify persistence niches and understand the dynamics of food contamination. Such data would support Food Business Operators to implement effective mitigation strategies to prevent and/or minimize poultry meat contamination.
Acknowledgments
The authors would like to thank the team of the Italian National Reference Laboratory for Listeria monocytogenes and the Italian National Reference Centre for Whole Genome Sequencing of microbial pathogens: database and bioinformatic analysis. The authors also would like to thank Paola Di Giuseppe from the Library Services and Communication unit of Istituto Zoopofilattico Sperimentale dell’Abruzzo e del Molise “G. Caporale” for her graphic support.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11061543/s1, Table S1: Metadata of all the 122 L. monocytogenes used in this study; Table S2: SNPs distance matrix.
Author Contributions
Conceptualization, M.T., F.G., G.C. and A.C. (Alexandra Chiaverini); formal analysis, M.T., G.C., F.G., S.S., A.C. (Alessandra Cornacchia) and A.C. (Alexandra Chiaverini); investigation, M.T., G.C., F.G., S.S., A.C. (Alessandra Cornacchia) and A.C. (Alexandra Chiaverini); resources, L.I.; data curation, L.I., F.P., M.T., G.C., F.G. and A.C. (Alexandra Chiaverini); writing—original draft preparation, M.T., F.G., G.C. and A.C. (Alexandra Chiaverini); writing—review and editing, M.T., F.G., G.C., A.C. (Alexandra Chiaverini), A.C. (Alessandra Cornacchia), S.S., M.S., P.V., L.I., N.D. and F.P.; visualization, F.G.; supervision, M.T., L.I., F.P. and N.D.; project administration, L.I., M.T. and F.P.; funding acquisition, L.I., F.P. and N.D. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Sequencing data are available within the NCBI BioProject under accession PRJNA947067.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was funded by the Italian Ministry of Health in the framework of the Ricerca Finalizzata Project n. GR-2011-02349917 entitled “Farm Animal Welfare and Food Safety: Effect of on-farm and pre-slaughtering stress factors on microbiological contamination of poultry meat”.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.European Commission. [(accessed on 15 February 2023)]. Available online: https://agriculture.ec.europa.eu/farming/animal-products/poultry_en.
- 2.European Food Safety Authority European Centre for Disease Prevention and Control the European Union One Health 2021 Zoonoses Report. EFSa J. 2022;20:07666. doi: 10.2903/j.efsa.2022.7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X., Shi X., Song Y., Yao S., Li K., Shi B., Sun J., Liu Z., Zhao W., Zhao C., et al. Genetic Diversity, Antibiotic Resistance, and Virulence Profiles of Listeria Monocytogenes from Retail Meat and Meat Processing. Food Res. Int. 2022;162:112040. doi: 10.1016/j.foodres.2022.112040. [DOI] [PubMed] [Google Scholar]
- 4.Tsai Y.-H., Moura A., Gu Z.-Q., Chang J.-H., Liao Y.-S., Teng R.-H., Tseng K.-Y., Chang D.-L., Liu W.-R., Huang Y.-T., et al. Genomic Surveillance of Listeria Monocytogenes in Taiwan, 2014 to 2019. Microbiol. Spectr. 2022;10:e01825-22. doi: 10.1128/spectrum.01825-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.López-Alonso V., Ortiz S., Corujo A., Martínez-Suárez J.V. Analysis of Benzalkonium Chloride Resistance and Potential Virulence of Listeria Monocytogenes Isolates Obtained from Different Stages of a Poultry Production Chain in Spain. J. Food Prot. 2020;83:443–451. doi: 10.4315/0362-028X.JFP-19-289. [DOI] [PubMed] [Google Scholar]
- 6.Coban A., Pennone V., Sudagidan M., Molva C., Jordan K., Aydin A. Prevalence, Virulence Characterization, and Genetic Relatedness of Listeria Monocytogenes Isolated from Chicken Retail Points and Poultry Slaughterhouses in Turkey. Braz. J. Microbiol. 2019;50:1063–1073. doi: 10.1007/s42770-019-00133-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iannetti L., Schirone M., Neri D., Visciano P., Acciari V.A., Centorotola G., Mangieri M.S., Torresi M., Santarelli G.A., Di Marzio V., et al. Listeria Monocytogenes in Poultry: Detection and Strain Characterization along an Integrated Production Chain in Italy. Food Microbiol. 2020;91:103533. doi: 10.1016/j.fm.2020.103533. [DOI] [PubMed] [Google Scholar]
- 8.Agostinho Davanzo E.F., dos Santos R.L., Castro V.H.d.L., Palma J.M., Pribul B.R., Dallago B.S.L., Fuga B., Medeiros M., Titze de Almeida S.S., da Costa H.M.B., et al. Molecular Characterization of Salmonella Spp. and Listeria Monocytogenes Strains from Biofilms in Cattle and Poultry Slaughterhouses Located in the Federal District and State of Goiás, Brazil. PLoS ONE. 2021;16:e0259687. doi: 10.1371/journal.pone.0259687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y., Zhang J., Chang X., Qin S., Song Y., Tian J., Ma A. Analysis of 90 Listeria Monocytogenes Contaminated in Poultry and Livestock Meat through Whole-Genome Sequencing. Food Res. Int. 2022;159:111641. doi: 10.1016/j.foodres.2022.111641. [DOI] [PubMed] [Google Scholar]
- 10.Marcus R., Hurd S., Mank L., Mshar P., Phan Q., Jackson K., Watarida K., Salfinger Y., Kim S., Ishida M.L., et al. Chicken Salad as the Source of a Case of Listeria Monocytogenes Infection in Connecticut. J. Food Prot. 2009;72:2602–2606. doi: 10.4315/0362-028X-72.12.2602. [DOI] [PubMed] [Google Scholar]
- 11.Little C.L., Amar C.F.L., Awofisayo A., Grant K.A. Hospital-Acquired Listeriosis Associated with Sandwiches in the UK: A Cause for Concern. J. Hosp. Infect. 2012;82:13–18. doi: 10.1016/j.jhin.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Zwizwai R. Infectious Disease Surveillance Update. Lancet Infect. Dis. 2019;19:699. doi: 10.1016/S1473-3099(19)30320-2. [DOI] [PubMed] [Google Scholar]
- 13.McLauchlin J., Aird H., Amar C., Barker C., Dallman T., Elviss N., Jørgensen F., Willis C. Listeria Monocytogenes in Cooked Chicken: Detection of an Outbreak in the United Kingdom (2016 to 2017) and Analysis of L. Monocytogenes from Unrelated Monitoring of Foods (2013 to 2017) J. Food Prot. 2020;83:2041–2052. doi: 10.4315/JFP-20-188. [DOI] [PubMed] [Google Scholar]
- 14.Maury M.M., Bracq-Dieye H., Huang L., Vales G., Lavina M., Thouvenot P., Disson O., Leclercq A., Brisse S., Lecuit M. Hypervirulent Listeria Monocytogenes Clones’ Adaption to Mammalian Gut Accounts for Their Association with Dairy Products. Nat. Commun. 2019;10:2488. doi: 10.1038/s41467-019-10380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoller A., Stevens M., Stephan R., Guldimann C. Characteristics of Listeria Monocytogenes Strains Persisting in a Meat Processing Facility over a 4-Year Period. Pathogens. 2019;8:32. doi: 10.3390/pathogens8010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons C., Lee S., Kathariou S. Dissemination and Conservation of Cadmium and Arsenic Resistance Determinants in Listeria and Other Gram-positive Bacteria. Mol. Microbiol. 2020;113:560–569. doi: 10.1111/mmi.14470. [DOI] [PubMed] [Google Scholar]
- 17.Palaiodimou L., Fanning S., Fox E.M. Genomic Insights into Persistence of Listeria Species in the Food Processing Environment. J. Appl. Microbiol. 2021;131:2082–2094. doi: 10.1111/jam.15089. [DOI] [PubMed] [Google Scholar]
- 18.Gray J.A., Chandry P.S., Kaur M., Kocharunchitt C., Bowman J.P., Fox E.M. Characterisation of Listeria Monocytogenes Food-Associated Isolates to Assess Environmental Fitness and Virulence Potential. Int. J. Food Microbiol. 2021;350:109247. doi: 10.1016/j.ijfoodmicro.2021.109247. [DOI] [PubMed] [Google Scholar]
- 19.EFSA Panel on Biological Hazards (EFSA BIOHAZ Panel) Koutsoumanis K., Allende A., Alvarez-Ordóñez A., Bolton D., Bover-Cid S., Chemaly M., Davies R., De Cesare A., Hilbert F., et al. Whole Genome Sequencing and Metagenomics for Outbreak Investigation, Source Attribution and Risk Assessment of Food-borne Microorganisms. EFSa J. 2019;17:05898. doi: 10.2903/j.efsa.2019.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S., Ryu S. Draft Genome Sequence of Listeria Monocytogenes Clonal Complex 1 Strain SNU3 from South Korea. Microbiol. Resour. Announc. 2023;12:e01226-22. doi: 10.1128/mra.01226-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown P., Chen Y., Siletzky R., Parsons C., Jaykus L.-A., Eifert J., Ryser E., Logue C.M., Stam C., Brown E., et al. Harnessing Whole Genome Sequence Data for Facility-Specific Signatures for Listeria Monocytogenes: A Case Study with Turkey Processing Plants in the United States. Front. Sustain. Food Syst. 2021;5:742353. doi: 10.3389/fsufs.2021.742353. [DOI] [Google Scholar]
- 22.Maury M.M., Tsai Y.-H., Charlier C., Touchon M., Chenal-Francisque V., Leclercq A., Criscuolo A., Gaultier C., Roussel S., Brisabois A., et al. Uncovering Listeria Monocytogenes Hypervirulence by Harnessing Its Biodiversity. Nat. Genet. 2016;48:308–313. doi: 10.1038/ng.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moura A., Criscuolo A., Pouseele H., Maury M.M., Leclercq A., Tarr C., Björkman J.T., Dallman T., Reimer A., Enouf V., et al. Whole Genome-Based Population Biology and Epidemiological Surveillance of Listeria Monocytogenes. Nat. Microbiol. 2016;2:16185. doi: 10.1038/nmicrobiol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vázquez-Boland J.A., Wagner M., Scortti M. Why Are Some Listeria Monocytogenes Genotypes More Likely to Cause Invasive (Brain, Placental) Infection? mBio. 2020;11:e03126-20. doi: 10.1128/mBio.03126-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi D., Anwar T.M., Pan H., Chai W., Xu S., Yue M. Genomic Determinants of Pathogenicity and Antimicrobial Resistance for 60 Global Listeria Monocytogenes Isolates Responsible for Invasive Infections. Front. Cell. Infect. Microbiol. 2021;11:718840. doi: 10.3389/fcimb.2021.718840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H., Chen W., Wang J., Xu B., Liu H., Dong Q., Zhang X. 10-Year Molecular Surveillance of Listeria Monocytogenes Using Whole-Genome Sequencing in Shanghai, China, 2009–2019. Front. Microbiol. 2020;11:551020. doi: 10.3389/fmicb.2020.551020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guidi F., Orsini M., Chiaverini A., Torresi M., Centorame P., Acciari V.A., Salini R., Palombo B., Brandi G., Amagliani G., et al. Hypo- and Hyper-Virulent Listeria Monocytogenes Clones Persisting in Two Different Food Processing Plants of Central Italy. Microorganisms. 2021;9:376. doi: 10.3390/microorganisms9020376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Portmann A.-C., Fournier C., Gimonet J., Ngom-Bru C., Barretto C., Baert L. A Validation Approach of an End-to-End Whole Genome Sequencing Workflow for Source Tracking of Listeria Monocytogenes and Salmonella Enterica. Front. Microbiol. 2018;9:446. doi: 10.3389/fmicb.2018.00446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cito F., Di Pasquale A., Cammà C., Cito P. The Italian Information System for the Collection and Analysis of Complete Genome Sequence of Pathogens Isolated from Animal, Food and Environment. Int. J. Infect. Dis. 2018;73:296–297. doi: 10.1016/j.ijid.2018.04.4090. [DOI] [Google Scholar]
- 30.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wingett S.W., Andrews S. FastQ Screen: A Tool for Multi-Genome Mapping and Quality Control. F1000Research. 2018;7:1338. doi: 10.12688/f1000research.15931.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timme R.E., Wolfgang W.J., Balkey M., Venkata S.L.G., Randolph R., Allard M., Strain E. Optimizing Open Data to Support One Health: Best Practices to Ensure Interoperability of Genomic Data from Bacterial Pathogens. One Health Outlook. 2020;2:20. doi: 10.1186/s42522-020-00026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ragon M., Wirth T., Hollandt F., Lavenir R., Lecuit M., Le Monnier A., Brisse S. A New Perspective on Listeria Monocytogenes Evolution. PLoS Pathog. 2008;4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva M., Machado M.P., Silva D.N., Rossi M., Moran-Gilad J., Santos S., Ramirez M., Carriço J.A. ChewBBACA: A Complete Suite for Gene-by-Gene Schema Creation and Strain Identification. Microb. Genom. 2018;4:000166. doi: 10.1099/mgen.0.000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moura A., Tourdjman M., Leclercq A., Hamelin E., Laurent E., Fredriksen N., Van Cauteren D., Bracq-Dieye H., Thouvenot P., Vales G., et al. Real-Time Whole-Genome Sequencing for Surveillance of Listeria Monocytogenes, France. Emerg. Infect. Dis. 2017;23:1462–1470. doi: 10.3201/eid2309.170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Z., Alikhan N.-F., Sergeant M.J., Luhmann N., Vaz C., Francisco A.P., Carriço J.A., Achtman M. GrapeTree: Visualization of Core Genomic Relationships among 100,000 Bacterial Pathogens. Genome Res. 2018;28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis S., Pettengill J.B., Luo Y., Payne J., Shpuntoff A., Rand H., Strain E. CFSAN SNP Pipeline: An Automated Method for Constructing SNP Matrices from next-Generation Sequence Data. PeerJ Comput. Sci. 2015;1:e20. doi: 10.7717/peerj-cs.20. [DOI] [Google Scholar]
- 40.Yang H., Hoffmann M., Allard M.W., Brown E.W., Chen Y. Microevolution and Gain or Loss of Mobile Genetic Elements of Outbreak-Related Listeria Monocytogenes in Food Processing Environments Identified by Whole Genome Sequencing Analysis. Front. Microbiol. 2020;11:866. doi: 10.3389/fmicb.2020.00866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pightling A.W., Pettengill J.B., Luo Y., Baugher J.D., Rand H., Strain E. Interpreting Whole-Genome Sequence Analyses of Foodborne Bacteria for Regulatory Applications and Outbreak Investigations. Front. Microbiol. 2018;9:1482. doi: 10.3389/fmicb.2018.01482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seemann T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 43.Alonso-Calleja C., Gómez-Fernández S., Carballo J., Capita R. Prevalence, Molecular Typing, and Determination of the Biofilm-Forming Ability of Listeria Monocytogenes Serotypes from Poultry Meat and Poultry Preparations in Spain. Microorganisms. 2019;7:529. doi: 10.3390/microorganisms7110529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jamshidi A., Zeinali T. Significance and Characteristics of Listeria Monocytogenes in Poultry Products. Int. J. Food Sci. 2019;2019:7835253. doi: 10.1155/2019/7835253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fagerlund A., Langsrud S., Schirmer B.C.T., Møretrø T., Heir E. Genome Analysis of Listeria Monocytogenes Sequence Type 8 Strains Persisting in Salmon and Poultry Processing Environments and Comparison with Related Strains. PLoS ONE. 2016;11:e0151117. doi: 10.1371/journal.pone.0151117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji S., Song Z., Luo L., Wang Y., Li L., Mao P., Ye C., Wang Y. Whole-Genome Sequencing Reveals Genomic Characterization of Listeria Monocytogenes from Food in China. Front. Microbiol. 2023;13:1049843. doi: 10.3389/fmicb.2022.1049843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin Y., Tan W., Wang G., Kong S., Zhou X., Zhao D., Jia Y., Pan Z., Jiao X. Geographical and Longitudinal Analysis of Listeria Monocytogenes Genetic Diversity Reveals Its Correlation with Virulence and Unique Evolution. Microbiol. Res. 2015;175:84–92. doi: 10.1016/j.micres.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 48.Hilliard A., Leong D., O’Callaghan A., Culligan E., Morgan C., DeLappe N., Hill C., Jordan K., Cormican M., Gahan C. Genomic Characterization of Listeria Monocytogenes Isolates Associated with Clinical Listeriosis and the Food Production Environment in Ireland. Genes. 2018;9:171. doi: 10.3390/genes9030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee S., Chen Y., Gorski L., Ward T.J., Osborne J., Kathariou S. Listeria Monocytogenes Source Distribution Analysis Indicates Regional Heterogeneity and Ecological Niche Preference among Serotype 4b Clones. mBio. 2018;9:e00396-18. doi: 10.1128/mBio.00396-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scaltriti E., Bolzoni L., Vocale C., Morganti M., Menozzi I., Re M.C., Pongolini S. Population Structure of Listeria Monocytogenes in Emilia-Romagna (Italy) and Implications on Whole Genome Sequencing Surveillance of Listeriosis. Front. Public Health. 2020;8:519293. doi: 10.3389/fpubh.2020.519293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lepe J.A. Current Aspects of Listeriosis. Med. Clínica. 2020;154:453–458. doi: 10.1016/j.medcli.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 52.Burnett E., Kucerova Z., Freeman M., Kathariou S., Chen J., Smikle M. Whole-Genome Sequencing Reveals Multiple Subpopulations of Dominant and Persistent Lineage I Isolates of Listeria Monocytogenes in Two Meat Processing Facilities during 2011–2015. Microorganisms. 2022;10:1070. doi: 10.3390/microorganisms10051070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nüesch-Inderbinen M., Bloemberg G.V., Müller A., Stevens M.J.A., Cernela N., Kollöffel B., Stephan R. Listeriosis Caused by Persistence of Listeria Monocytogenes Serotype 4b Sequence Type 6 in Cheese Production Environment. Emerg. Infect. Dis. 2021;27:284–288. doi: 10.3201/eid2701.203266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su X., Cao G., Zhang J., Pan H., Zhang D., Kuang D., Yang X., Xu X., Shi X., Meng J. Characterization of Internalin Genes in Listeria Monocytogenes from Food and Humans, and Their Association with the Invasion of Caco-2 Cells. Gut Pathog. 2019;11:30. doi: 10.1186/s13099-019-0307-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tavares R.d.M., da Silva D.A.L., Camargo A.C., Yamatogi R.S., Nero L.A. Interference of the Acid Stress on the Expression of LlsX by Listeria Monocytogenes Pathogenic Island 3 (LIPI-3) Variants. Food Res. Int. 2020;132:109063. doi: 10.1016/j.foodres.2020.109063. [DOI] [PubMed] [Google Scholar]
- 56.Cotter P.D., Draper L.A., Lawton E.M., Daly K.M., Groeger D.S., Casey P.G., Ross R.P., Hill C. Listeriolysin S, a Novel Peptide Haemolysin Associated with a Subset of Lineage I Listeria Monocytogenes. PLoS Pathog. 2008;4:e1000144. doi: 10.1371/journal.ppat.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quereda J.J., Morón-García A., Palacios-Gorba C., Dessaux C., García-del Portillo F., Pucciarelli M.G., Ortega A.D. Pathogenicity and Virulence of Listeria monocytogenes: A Trip from Environmental to Medical Microbiology. Virulence. 2021;12:2509–2545. doi: 10.1080/21505594.2021.1975526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Quereda J.J., Meza-Torres J., Cossart P., Pizarro-Cerdá J. Listeriolysin S: A Bacteriocin from Epidemic Listeria monocytogenes Strains That Targets the Gut Microbiota. Gut Microbes. 2017;8:384–391. doi: 10.1080/19490976.2017.1290759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gelbícová T., Kolácková I., Pantu R. A Novel Mutation Leading to a Premature Stop Codon in InlA of Listeria Monocytogenes Isolated from Neonatal Listeriosis. New Microbiol. 2015;38:293–296. [PubMed] [Google Scholar]
- 60.Van Stelten A., Simpson J.M., Ward T.J., Nightingale K.K. Revelation by Single-Nucleotide Polymorphism Genotyping That Mutations Leading to a Premature Stop Codon in InlA Are Common among Listeria Monocytogenes Isolates from Ready-To-Eat Foods but Not Human Listeriosis Cases. Appl. Environ. Microbiol. 2010;76:2783–2790. doi: 10.1128/AEM.02651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nightingale K.K., Ivy R.A., Ho A.J., Fortes E.D., Njaa B.L., Peters R.M., Wiedmann M. InlA Premature Stop Codons Are Common among Listeria Monocytogenes Isolates from Foods and Yield Virulence-Attenuated Strains That Confer Protection against Fully Virulent Strains. Appl. Environ. Microbiol. 2008;74:6570–6583. doi: 10.1128/AEM.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Di Ciccio P., Rubiola S., Panebianco F., Lomonaco S., Allard M., Bianchi D.M., Civera T., Chiesa F. Biofilm Formation and Genomic Features of Listeria Monocytogenes Strains Isolated from Meat and Dairy Industries Located in Piedmont (Italy) Int. J. Food Microbiol. 2022;378:109784. doi: 10.1016/j.ijfoodmicro.2022.109784. [DOI] [PubMed] [Google Scholar]
- 63.Hurley D., Luque-Sastre L., Parker C.T., Huynh S., Eshwar A.K., Nguyen S.V., Andrews N., Moura A., Fox E.M., Jordan K., et al. Whole-Genome Sequencing-Based Characterization of 100 Listeria Monocytogenes Isolates Collected from Food Processing Environments over a Four-Year Period. mSphere. 2019;4:e00252-19. doi: 10.1128/mSphere.00252-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parsons C., Lee S., Kathariou S. Heavy Metal Resistance Determinants of the Foodborne Pathogen Listeria Monocytogenes. Genes. 2018;10:11. doi: 10.3390/genes10010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corbett D., Schuler S., Glenn S., Andrew P.W., Cavet J.S., Roberts I.S. The Combined Actions of the Copper-Responsive Repressor CsoR and Copper-Metallochaperone CopZ Modulate CopA-Mediated Copper Efflux in the Intracellular Pathogen Listeria Monocytogenes. Mol. Microbiol. 2011;81:457–472. doi: 10.1111/j.1365-2958.2011.07705.x. [DOI] [PubMed] [Google Scholar]
- 66.Cotter P.D., Ryan S., Gahan C.G.M., Hill C. Presence of GadD1 Glutamate Decarboxylase in Selected Listeria Monocytogenes Strains Is Associated with an Ability to Grow at Low PH. Appl. Environ. Microbiol. 2005;71:2832–2839. doi: 10.1128/AEM.71.6.2832-2839.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ryan S., Begley M., Hill C., Gahan C.G.M. A Five-Gene Stress Survival Islet (SSI-1) That Contributes to the Growth of Listeria Monocytogenes in Suboptimal Conditions: Stress Survival Islet in L. Monocytogenes. J. Appl. Microbiol. 2010;109:984–995. doi: 10.1111/j.1365-2672.2010.04726.x. [DOI] [PubMed] [Google Scholar]
- 68.Liu Y., Orsi R.H., Gaballa A., Wiedmann M., Boor K.J., Guariglia-Oropeza V. Systematic Review of the Listeria Monocytogenes σ B Regulon Supports a Role in Stress Response, Virulence and Metabolism. Future Microbiol. 2019;14:801–828. doi: 10.2217/fmb-2019-0072. [DOI] [PubMed] [Google Scholar]
- 69.Abdel-Motaal H., Meng L., Zhang Z., Abdelazez A.H., Shao L., Xu T., Meng F., Abozaed S., Zhang R., Jiang J. An Uncharacterized Major Facilitator Superfamily Transporter From Planococcus Maritimus Exhibits Dual Functions as a Na+(Li+, K+)/H+ Antiporter and a Multidrug Efflux Pump. Front. Microbiol. 2018;9:1601. doi: 10.3389/fmicb.2018.01601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu N., Zheng Y., Wang X., Krulwich T.A., Ma Y., Liu J. The Lysine 299 Residue Endows the Multisubunit Mrp1 Antiporter with Dominant Roles in Na + Resistance and PH Homeostasis in Corynebacterium Glutamicum. Appl. Environ. Microbiol. 2018;84:e00110-18. doi: 10.1128/AEM.00110-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan L., Wang C., Jiang J., Liu S., Zheng Y., Yang M., Zhang Y. Nitrate Removal by Alkali-Resistant Pseudomonas Sp. XS-18 under Aerobic Conditions: Performance and Mechanism. Bioresour. Technol. 2022;344:126175. doi: 10.1016/j.biortech.2021.126175. [DOI] [PubMed] [Google Scholar]
- 72.Slipski C.J., Zhanel G.G., Bay D.C. Biocide Selective TolC-Independent Efflux Pumps in Enterobacteriaceae. J. Membr. Biol. 2018;251:15–33. doi: 10.1007/s00232-017-9992-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sharma A., Gupta V., Pathania R. Efflux Pump Inhibitors for Bacterial Pathogens: From Bench to Bedside. Indian J. Med. Res. 2019;149:129. doi: 10.4103/ijmr.IJMR_2079_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cherifi T., Arsenault J., Pagotto F., Quessy S., Côté J.-C., Neira K., Fournaise S., Bekal S., Fravalo P. Distribution, Diversity and Persistence of Listeria Monocytogenes in Swine Slaughterhouses and Their Association with Food and Human Listeriosis Strains. PLoS ONE. 2020;15:e0236807. doi: 10.1371/journal.pone.0236807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matereke L.T., Okoh A.I. Listeria Monocytogenes Virulence, Antimicrobial Resistance and Environmental Persistence: A Review. Pathogens. 2020;9:528. doi: 10.3390/pathogens9070528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gorski L., Cooley M.B., Oryang D., Carychao D., Nguyen K., Luo Y., Weinstein L., Brown E., Allard M., Mandrell R.E., et al. Prevalence and Clonal Diversity of over 1200 Listeria Monocytogenes Isolates Collected from Public Access Waters near Produce Production Areas on the Central California Coast during 2011 to 2016. Appl Environ. Microbiol. 2022;88:e00357-22. doi: 10.1128/aem.00357-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muchaamba F., Stephan R., Tasara T. Listeria Monocytogenes Cold Shock Proteins: Small Proteins with A Huge Impact. Microorganisms. 2021;9:1061. doi: 10.3390/microorganisms9051061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Harter E., Wagner E.M., Zaiser A., Halecker S., Wagner M., Rychli K. Stress Survival Islet 2, Predominantly Present in Listeria Monocytogenes Strains of Sequence Type 121, Is Involved in the Alkaline and Oxidative Stress Responses. Appl. Environ. Microbiol. 2017;83:e00827-17. doi: 10.1128/AEM.00827-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Müller A., Rychli K., Muhterem-Uyar M., Zaiser A., Stessl B., Guinane C.M., Cotter P.D., Wagner M., Schmitz-Esser S. Tn6188—A Novel Transposon in Listeria Monocytogenes Responsible for Tolerance to Benzalkonium Chloride. PLoS ONE. 2013;8:e76835. doi: 10.1371/journal.pone.0076835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cherifi T., Carrillo C., Lambert D., Miniaï I., Quessy S., Larivière-Gauthier G., Blais B., Fravalo P. Genomic Characterization of Listeria Monocytogenes Isolates Reveals That Their Persistence in a Pig Slaughterhouse Is Linked to the Presence of Benzalkonium Chloride Resistance Genes. BMC Microbiol. 2018;18:220. doi: 10.1186/s12866-018-1363-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parra-Flores J., Holý O., Bustamante F., Lepuschitz S., Pietzka A., Contreras-Fernández A., Castillo C., Ovalle C., Alarcón-Lavín M.P., Cruz-Córdova A., et al. Virulence and Antibiotic Resistance Genes in Listeria Monocytogenes Strains Isolated From Ready-to-Eat Foods in Chile. Front. Microbiol. 2022;12:796040. doi: 10.3389/fmicb.2021.796040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fredriksson-Ahomaa M., Sauvala M., Kurittu P., Heljanko V., Heikinheimo A., Paulsen P. Characterisation of Listeria Monocytogenes Isolates from Hunted Game and Game Meat from Finland. Foods. 2022;11:3679. doi: 10.3390/foods11223679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mafuna T., Matle I., Magwedere K., Pierneef R.E., Reva O.N. Whole Genome-Based Characterization of Listeria Monocytogenes Isolates Recovered from the Food Chain in South Africa. Front. Microbiol. 2021;12:669287. doi: 10.3389/fmicb.2021.669287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guidi F., Chiaverini A., Repetto A., Lorenzetti C., Centorotola G., Bazzucchi V., Palombo B., Gattuso A., Pomilio F., Blasi G. Hyper-Virulent Listeria Monocytogenes Strains Associated With Respiratory Infections in Central Italy. Front. Cell. Infect. Microbiol. 2021;11:765540. doi: 10.3389/fcimb.2021.765540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hanes R.M., Huang Z. Investigation of Antimicrobial Resistance Genes in Listeria Monocytogenes from 2010 through to 2021. Int. J. Environ. Res. Public Health. 2022;19:5506. doi: 10.3390/ijerph19095506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mpondo L., Ebomah K.E., Okoh A.I. Multidrug-Resistant Listeria Species Shows Abundance in Environmental Waters of a Key District Municipality in South Africa. Int. J. Environ. Res. Public Health. 2021;18:481. doi: 10.3390/ijerph18020481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available within the NCBI BioProject under accession PRJNA947067.