Abstract
Age-dependent loss of skeletal muscle mass and function is a feature of sarcopenia, and increases the risk of many aging-related metabolic diseases. Here, we report phenotypic and single-nucleus transcriptomic analyses of non-human primate skeletal muscle aging. A higher transcriptional fluctuation was observed in myonuclei relative to other interstitial cell types, indicating a higher susceptibility of skeletal muscle fiber to aging. We found a downregulation of FOXO3 in aged primate skeletal muscle, and identified FOXO3 as a hub transcription factor maintaining skeletal muscle homeostasis. Through the establishment of a complementary experimental pipeline based on a human pluripotent stem cell-derived myotube model, we revealed that silence of FOXO3 accelerates human myotube senescence, whereas genetic activation of endogenous FOXO3 alleviates human myotube aging. Altogether, based on a combination of monkey skeletal muscle and human myotube aging research models, we unraveled the pivotal role of the FOXO3 in safeguarding primate skeletal muscle from aging, providing a comprehensive resource for the development of clinical diagnosis and targeted therapeutic interventions against human skeletal muscle aging and the onset of sarcopenia along with aging-related disorders.
Keywords: single-nucleus RNA sequencing, primate, aging, skeletal muscle, FOXO3
Introduction
Skeletal muscle is among the largest organs of the body, comprising roughly 40% of total body mass (Frontera and Ochala, 2015). With age, sarcopenia, manifested as loss of skeletal muscle mass and strength, leads to a decline in physical performance and impairs quality of life (Wilkinson et al., 2018; Larsson et al., 2019; Cai et al., 2022). In addition, as an essential endocrine and metabolic organ (Iizuka et al., 2014; Baskin et al., 2015), mounting evidence shows that skeletal muscle aging also contributes to perturbations in metabolic homeostasis probably by changing muscle-derived growth factors (myokines) or metabolites (Tieland et al., 2018; Gomarasca et al., 2020; Wang and Ng, 2022), ultimately increasing the risk of age-associated metabolic disorders such as diabetes and obesity (Kalinkovich and Livshits, 2017; Lipina and Hundal, 2017). Although resistance-type exercise training and diet can help maintain muscle mass (Stokes et al., 2018), effective treatments to delay or prevent skeletal muscle aging and sarcopenia progression in the elderly are still lacking. Therefore, an in-depth understanding of the mechanisms underlying skeletal muscle aging is of scientific and clinical importance.
Skeletal muscle is composed of multinucleated myofibers, embedded mononuclear muscle stem cells (MuSCs), and multiple interstitial cells (Wagers and Conboy, 2005; Frontera and Ochala, 2015). In the shared cytoplasm of muscle fibers, the distribution of diverse myonuclei with heterogeneous gene expression is known to meet functional requirements in distinct regions (Cadot et al., 2015; Orchard et al., 2021). In addition, homeostatic maintenance in skeletal muscle is also regulated by the niche compartment, which includes an array of interstitial cells, such as endothelial cells, smooth muscle cells, fibro-adipogenic progenitors, tendon fibroblasts, and different immune cells (macrophages, etc.) (Giordani et al., 2019; Dos Santos et al., 2020; Kim et al., 2020). The structure and physiology of skeletal muscle become progressively compromised during aging, which is reflected by declined muscle mass and strength at the tissue level (Wilkinson et al., 2018; Simon et al., 2019). At the cellular level, decreases in the number of MuSCs and extent of capillarization, concomitant with defects in neuromuscular junctions (NMJ), as well as a massive accumulation of interstitial adipocytes and infiltration of immune cells were observed with advanced age (Demontis et al., 2013; Livshits and Kalinkovich, 2019; Fuchs and Blau, 2020; Maeda et al., 2021), indicative of both impaired muscle fiber function and dysregulated muscle microenvironment. However, how myonuclei-specific transcriptomic programs shape the architectural and functional changes of multinucleated syncytial muscle cells, and how the interstitial microenvironment orchestrates myofiber deterioration during skeletal muscle aging remain largely unknown.
To develop approaches for treating human skeletal muscle aging and related diseases, it is imperative to gain a comprehensive understanding of human skeletal muscle aging. Despite its importance, ethical restrictions limit the availability of an adequate amount of age-matched skeletal muscle biopsies from healthy donors needed for systematic and in-depth surveys of skeletal muscle aging. As expected from the evolutionary conservation (Anderson and Colman, 2011), non-human primate (NHP) skeletal muscle is highly similar to that of humans in many aspects, including muscle fiber types, neuromuscular junctions, metabolic pathways, and inflammation regulation (Jones et al., 2017; Mercken et al., 2017). Therefore, a study in NHP skeletal muscle aging promises to put forward a better understanding of human skeletal muscle aging, which in turn can help identify biological processes or factors that can be targeted therapeutically.
Here, we generated the single-nucleus transcriptomic atlas of NHP skeletal muscle aging, and identified a panel of nuclear types with distinct and unique gene expression signatures. We also identified and experimentally validated that the downregulation of FOXO3 causally triggered human skeletal muscle aging. Our study provides a valuable resource for understanding the mechanistic underpinning of primate skeletal muscle aging, and facilitates the development of novel therapeutical intervention strategies to combat age-associated muscle degenerative disorders.
Results
Characterization of the histological alterations of NHP skeletal muscle aging
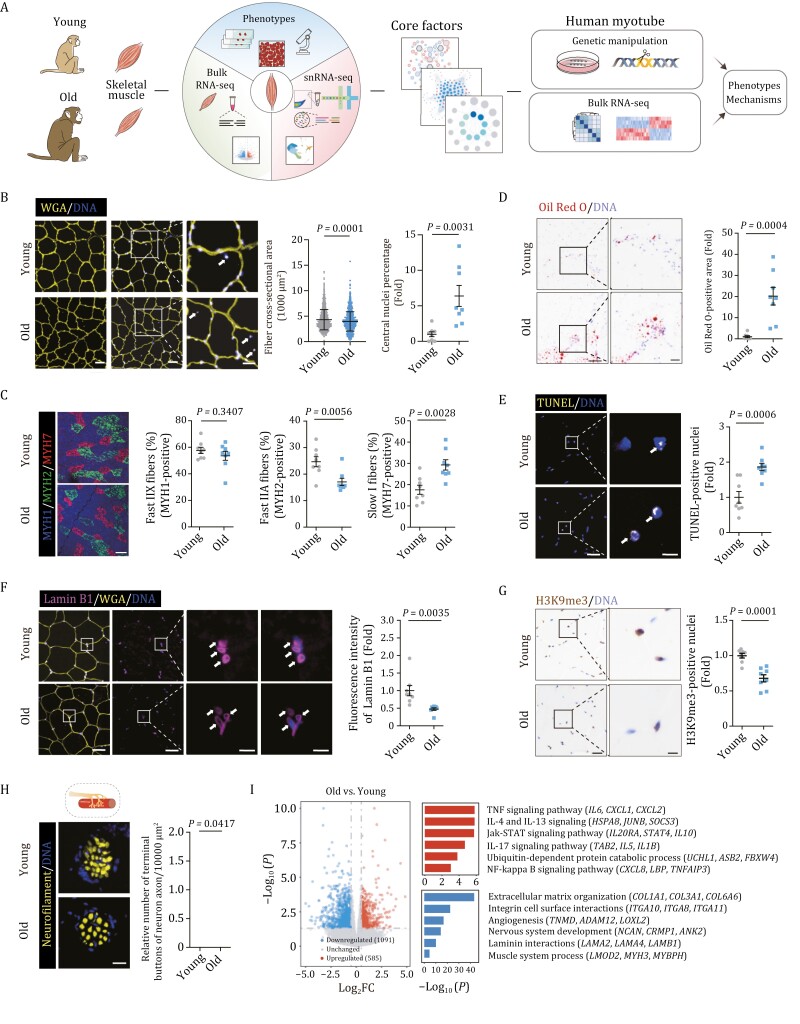
To dissect the phenotypic and molecular characteristics of NHP skeletal muscle aging, we obtained skeletal muscle from eight young (4–6 years old) and eight old (18–21 years old) cynomolgus monkeys, which are approximately ~ 16 and ~ 60 years old in human age, respectively (Figs. 1A and S1A) (Wang et al., 2020, 2021b; Li et al., 2021; Ma et al., 2021; Zhang et al., 2021, 2022; Zou et al., 2022). First, we found a decrease in the overall fiber cross-sectional area in aged NHP muscle (Fig. 1B), one of the classic indicators of skeletal muscle aging (Demontis et al., 2013). Additionally, we observed an increment in MYH7-positive type I slow-twitch fiber, while a decline in MYH2-labeled type IIA fast-twitch fiber (Fig. 1C), consistent with the aging-accompanying transition of fiber type (fast-twitch fiber to slow-twitch fiber) (Demontis et al., 2013). Notably, we also observed a panel of classic hallmarks for skeletal muscle aging and degeneration in aged NHP muscle (Dirks et al., 2006; Lipina and Hundal, 2017; Zhao and Chen, 2022; Zhang et al., 2015; 2020b), including an increased percentage of central nuclei in myofibers, accumulated intramuscular lipid deposition, elevated apoptosis, as well as accelerated loss of Lamin B1 and heterochromatin erosion (reflected by decreased H3K9me3 levels) (Fig. 1B and 1D–G). Concurrently, a decreased terminal button in the NMJ was also observed in the skeletal muscle of aged monkey, suggesting defects in neuromuscular junctions in skeletal muscle with advanced age (Fig. 1H). Collectively, histological analysis revealed multifaceted aging-associated damages in aged NHP skeletal muscle.
Figure 1.
Characterization of aging-related phenotypes in cynomolgus monkey skeletal muscle. (A) Workflow showing the procedure of phenotypic analysis, snRNA-seq, bulk RNA-seq of young and old monkey skeletal muscles and functional verification of core factors during aging. (B) Cross-sectional area of muscle fibers and relative central nuclei percentage from young and old monkeys. Representative images are shown on the left. Scale bars, 50 and 25 μm (zoomed-in image). Middle, the quantitative data for the cross-sectional area of myofibers are presented as mean ± SD. n = 8 monkeys for each group. Two hundred fibers were calculated for each individual. Right, the percentages of central nuclei were quantified as fold changes in old skeletal muscles vs. in young counterparts and are presented as mean ± SEMs. n = 8 monkeys for each group. Arrows indicate central nuclei in skeletal muscle. (C) Cross sections of skeletal muscle stained with antibodies specific for Fast IIX (marked by MYH1), Fast IIA (marked by MYH2), and Slow I fibers (marked by MYH7). Representative images are shown on the left. Scale bar, 100 μm. The percentages of each fiber type were quantified separately as fold changes in old skeletal muscles vs. in young counterparts and are presented as mean ± SEMs on the right. n = 8 monkeys for each group. (D) Oil Red O staining of muscle cross sections from young and old monkeys. Scale bars, 200 and 50 μm (zoomed-in image). Oil Red O-positive areas were quantified as fold changes in old skeletal muscles vs. in young counterparts and are presented as mean ± SEMs on the right. n = 8 monkeys for each group. (E) TUNEL staining in the skeletal muscles from young and old monkeys. Representative images are shown on the left. Scale bars, 50 and 10 μm (zoomed-in image). The numbers of TUNEL-positive nuclei were quantified as fold changes in old skeletal muscles vs. in young counterparts and are presented as mean ± SEMs on the right. n = 8 monkeys for each group. Arrows indicate TUNEL-positive nuclei in skeletal muscle. (F) Lamin B1 immunofluorescence staining in the skeletal muscles from young and old monkeys. Representative images are shown on the left. Scale bars, 50 and 10 μm (zoomed-in image). The fluorescence intensity of Lamin B1 was quantified as fold changes in old skeletal muscles vs. in young counterparts and are presented as mean ± SEMs on the right. n = 8 monkeys for each group. Arrows indicate Lamin B1-positive nuclei in skeletal muscle. (G) H3K9me3 immunohistochemical staining in skeletal muscles from young and old monkeys. Representative images are shown on the left. Scale bars, 50 and 25 μm (zoomed-in image). The numbers of H3K9me3-positive nuclei were quantified as fold changes in old skeletal muscles vs. in young counterparts and are presented as mean ± SEMs on the right. n = 8 monkeys for each group. (H) Analysis of terminal buttons in the NMJ by immunofluorescence staining of neurofilament in young and old monkeys. Representative images are shown on the left. Scale bar, 25 μm. The numbers of terminal buttons in the NMJ in fixed regions were quantified as fold changes in old skeletal muscles vs. in young counterparts and are presented as mean ± SEMs on the right. n = 8 monkeys for each group. (I) Left, volcano plot showing the relative expression levels of DEGs between old and young (O/Y) monkey skeletal muscles. Right, representative Gene Ontology (GO) terms and corresponding DEGs.
Single-nucleus transcriptome profiling of young and old NHP skeletal muscles
To unravel the transcriptomic alterations underlying age-associated changes in NHP skeletal muscle, we performed genome-wide RNA sequencing analysis (Fig. 1I and Table S1). We found that upregulated aging-associated differentially expressed genes (DEGs) were involved in the pro-inflammatory response, including TNF-signaling pathway (IL6, CXCL1, CXCL2) and NF-κB signaling pathway (CXCL8, LBP, TNFAIP3) (Fig. 1I). Importantly, pathway related to the ubiquitin-dependent protein catabolic processes (UCHL1, ASB2, FBXW4) was also activated in the aged skeletal muscle (Fig. 1I). Downregulated DEGs included genes broadly involved in the muscle structure (LMOD2, MYH3, MYBPH) and nervous system development (NCAN, CRMP1, ANK2), in line with the aging phenotypes observed in the aged NHP muscle (Fig. 1B and 1H). Altogether, these findings pinpointed molecular features underlying aging-associated defects in aged NHP skeletal muscle.
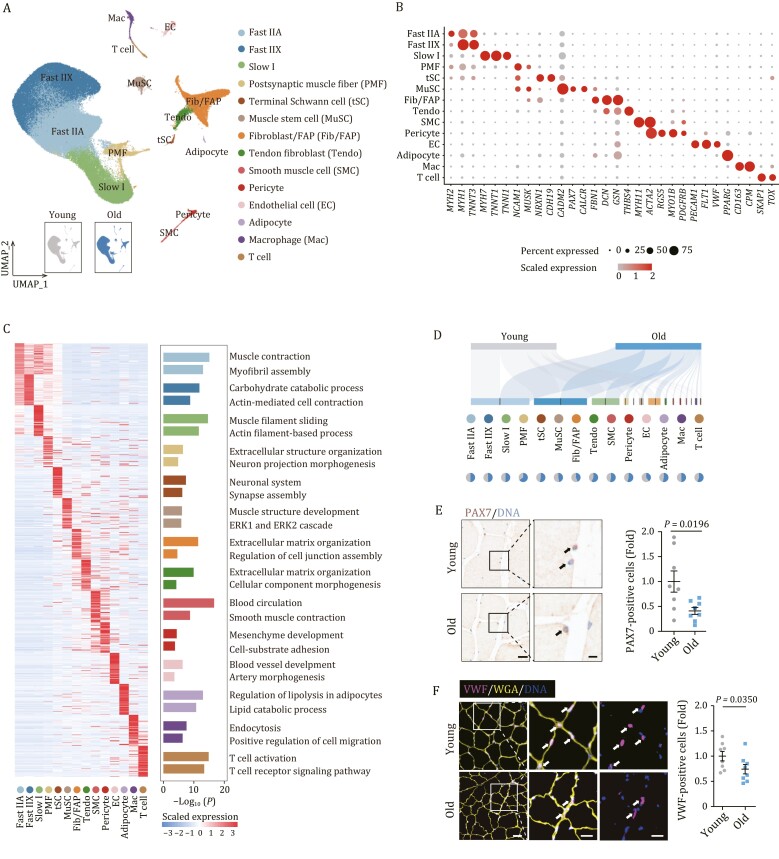
To resolve the cell type-specific transcriptional alterations of the aged NHP skeletal muscle, we performed single-nucleus RNA sequencing (snRNA-seq). After stringent filtration, we retained 112,493 qualified single-nucleus transcriptomes from 16 individuals (Fig. S1B–D). Using unbiased clustering and uniform manifold approximation and projection (UMAP) analysis, we identified 14 major populations in skeletal muscle with distinct transcriptomic signatures (Fig. 2A and Table S2). Among these, we identified four types of myonuclei from myofibers based on canonical markers, including Slow I, Fast IIA, Fast IIX, and postsynaptic endplates (postsynaptic muscle fiber, PMF), which is known for functional specialization at NMJ (Figs. 2A and S1E–G). In addition, based on the expression of classical marker genes, we annotated MuSC and a total of nine non-myonuclear cell types, including EC, SMC, pericyte, tendon fibroblast (Tendo), fibroblast/FAP (Fib/FAP), adipocyte, terminal Schwann cell (tSC), and two immune cell types (macrophage (Mac) and T cell) (Fig. 2A and 2B). In functional enrichment analysis of the top 50 marker genes of each cell type, we further revealed the transcriptional characteristics related to their unique physiological functions (Fig. 2C). For example, the marker genes of nuclei from Fast IIA, Fast IIX, and Slow I muscle fiber aligned with muscle functions, such as muscle contraction and muscle filament sliding, whereas those of PMF and tSC aligned with neuron projection morphogenesis and neuronal system (Fig. 2C). Consistent with the compromised muscle regenerative capacity in the elders (Larsson et al., 2019), we found the number of PAX7-positive MuSCs declined with age, indicative of attrition of the stem cell pool (Figs. 2D, 2E, and S2A). Concurrently, in the aged skeletal muscle, we noticed a prominent reduction in cell number of EC relative to young counterparts (Figs. 2D, 2F, and S2B), indicative of a dysregulated interstitial microenvironment in aged muscle.
Figure 2.
Single-nucleus transcriptomic atlas of cynomolgus monkey skeletal muscle aging. (A) Uniform manifold approximation and projection (UMAP) plot showing the 14 cell types of monkey skeletal muscle. Cells are annotated to the right. PMF, postsynaptic muscle fiber; tSC, terminal Schwann cell; MuSC, muscle stem cell; Fib/FAP, Fibroblast/fibro-adipogenic progenitor; Tendo, Tendon fibroblast; SMC, Smooth muscle cell; EC, Endothelial cell; Mac, Macrophage. UMAP plot in the lower left showing the distribution of cells from young and old groups. (B) Dot plot showing the expression signatures of representative marker genes for each cell type. (C) Left, heatmap showing the expression profiles of top 50 genes ranked by LogFC of each cell type. Right, enriched GO terms for marker genes of each cell type. (D) Sankey plot showing the distribution of young and aged cells across different cell types. Pie plots showing the relative cell proportion of between old and young groups across different cell types. (E) PAX7 immunohistochemical staining in skeletal muscles from young and old monkeys. Representative images are shown on the left. Scale bars, 20 and 5 μm (zoomed-in image). The numbers of PAX7-positive cells were quantified as fold changes in old skeletal muscles vs. in young counterparts and the quantitative data are presented as mean ± SEMs on the right. n = 8 monkeys for each group. Arrows indicate PAX7-positive cells in skeletal muscle. (F) VWF immunofluorescence staining in skeletal muscles from young and old monkeys. Representative images are shown on the left. Scale bars, 50 and 25 μm (zoomed-in image). The numbers of VWF-positive cells were quantified as fold changes in old skeletal muscles vs. in young counterparts and are presented as mean ± SEMs on the right. n = 8 monkeys for each group. Arrows indicate VWF-positive cells in skeletal muscle.
Cell type-specific alterations in gene expression programs of NHP skeletal muscle aging
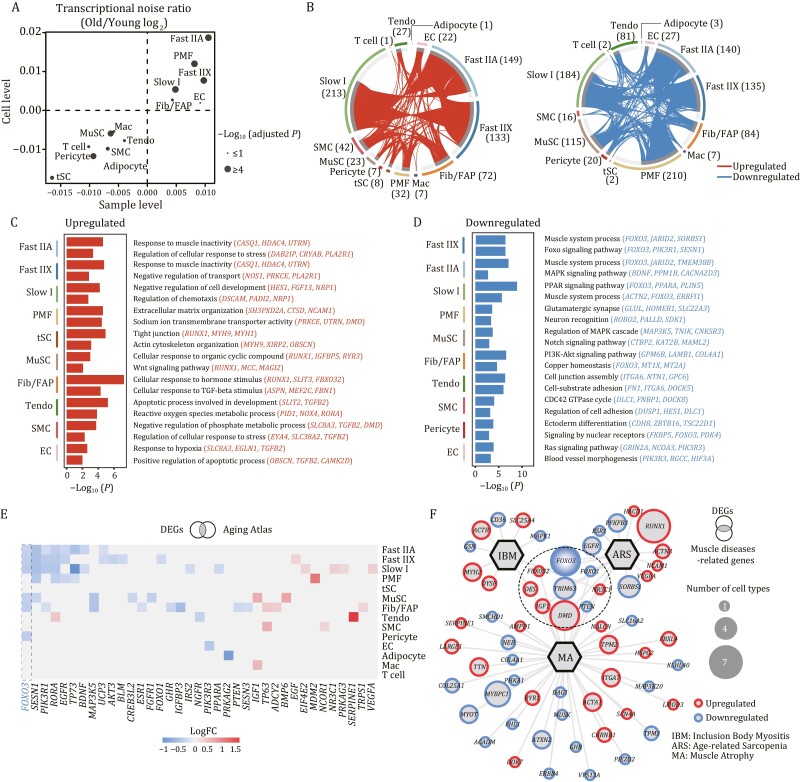
Given that increased transcriptional heterogeneity and perturbance are regarded as one of the features of mammalian aging (Wang et al., 2020; Li et al., 2021; Ma et al., 2021; Zhang et al., 2021; Leng and Pawelec, 2022; Zou et al., 2022), we sought to assess transcriptional noise during NHP skeletal muscle aging. A calculation of age-associated coefficient of variation demonstrated that aged Fast IIA, Fast IIX, Slow I, and PMF, the major components of skeletal muscle, exhibited a higher cell-to-cell transcriptional noise relative to other populations (Fig. 3A), suggesting a higher susceptibility of skeletal muscle fiber to aging.
Figure 3.
Cell type-specific transcriptional alterations during cynomolgus monkey skeletal muscle aging. (A) Scatter plot showing the log2 ratio of transcriptional noise between old and young samples as calculated using sample averages (n = 16) and single cells on the X and Y axes, respectively. (B) Circos plots showing the aging-related differentially expressed genes (O/Y) DEGs, adjusted P values < 0.05 and |LogFC| > 0.25 for each cell type between old and young groups. Each connecting curve represents a gene up- or downregulated in two cell types. The number of DEGs for each cell type is also annotated. (C and D) Bar charts showing the enriched GO terms for upregulated (C) or downregulated (D) DEGs between old and young groups across different cell types in monkey skeletal muscle. (E) Heatmap showing the DEGs included in Aging Atlas database. (F) Network plot showing the DEGs associated with aging-related muscle diseases. The node size of genes indicates the number of cell types in which this gene was differentially expressed with age. The genes in the circle with a dashed line are associated with at least two kinds of diseases.
We next analyzed aging-associated DEGs (averaged Log|FoldChange| > 0.25 and adjusted P values < 0.05) between old (O) and young (Y) groups across different populations (Figs. 3B, S2C, and Table S1). We found that the majority of upregulated DEGs converged in Slow I, Fast IIA, and Fast IIX (213, 149, and 133 genes, respectively) (Fig. 3B), while the majority of downregulated DEGs were identified in PMF, Slow I, Fast IIA, and Fast IIX (210, 184, 140, and 135 genes, respectively) (Fig. 3B), again implying the sensitivity of myofibers to aging. We noticed a prominent upregulation of genes associated with response to muscle inactivity in fast-twitch myonuclei (Fig. 3C), in accordance with the loss of muscle mass and strength that is observed during aging (sarcopenia) and muscle disuse. This upregulation was presented in both Fast IIA and Fast IIX, but not in Slow I (Fig. 3C), consistent with the early onset of muscle degeneration in fast-twitch fiber (Demontis et al., 2013; Murgia et al., 2017). Interestingly, we detected upregulation of genes involved in Wnt signaling pathway in MuSC (Fig. 3C), which has been reported to correlate with the bias in cell fate determination, that is, the transition from a myogenic to a fibrogenic lineage (Domingues-Faria et al., 2016). These data were consistent with an age-dependent loss of the MuSC pool, concordant with the previously observed phenotype (Fig. 2E). Downregulated DEGs in Fast IIA, Fast IIX, and Slow I were consistently related to the maintenance of muscle structure and function (Fig. 3D), suggesting a coordinated muscle dysfunction at advanced age among different fibers. In addition, downregulated genes related to the Notch signaling pathway in MuSC further underlined the age-dependent depletion of MuSC (Fig. 3D) (Jiang et al., 2014; Verma et al., 2018). Further, we found downregulation of genes associated with glutamatergic synapse and neuron recognition in PMF (Fig. 3D), which was in concert with age-related defects in neuromuscular junctions as previously observed (Fig. 1H). Overall, these analyses deciphered the cellular and molecular programs underpinning NHP skeletal muscle aging.
Next, we identified a total of 39 DEGs that were shared in at least five cell types, of which 9 genes were consistently upregulated and 30 genes were consistently downregulated (Fig. S2C). Globally, multiple dysregulated genes were predominantly found in myofibers (Fig. S2C), strengthening the notion that muscle fibers were most responsive to aging. Amongst cell type-specific DEGs were genes that regulate corresponding cellular functions (Fig. S2D). Further, we conducted an integrated analysis of aging-associated DEGs and genes from the Aging Atlas database to identify the expression alterations of genes associated with aging (Guang-Hui Liu, 2021). Among these, FOXO3 exhibited the most prominent changes, whose downregulation was found in more than five cell types (Fig. 3E). FOXO3, encoding a member of the forkhead transcription factor (TF) family, is a well-known longevity gene (Morris et al., 2015; Zhang et al. 2020a; Yan et al. 2019). Its downregulation is likely correlated to the onset of skeletal muscle aging and degeneration. We then asked if these aging-associated DEGs are linked to degenerative diseases in skeletal muscle. To this end, we performed a joint comparative analysis of aging-associated DEGs and hotspot genes known to be involved in age-related muscle diseases (sarcopenia, muscle atrophy, and inclusion body myositis) (Navarro, 2001; Askanas and Engel, 2007; Yu et al., 2010; Askanas et al., 2012; González et al., 2015; Wilkinson et al., 2018) (Figs. 3F, S2E and Table S3). Interestingly, we noticed that FOXO3 and its homologous gene FOXO1, as the prominent hub genes, were closely linked to various kinds of age-related muscle disorders (Fig. 3F), suggesting a putative crucial role of FOXO family in the regulation of NHP skeletal muscle aging.
Transcriptional regulatory network pinpoints FOXO3 as a core transcription factor during NHP skeletal muscle aging
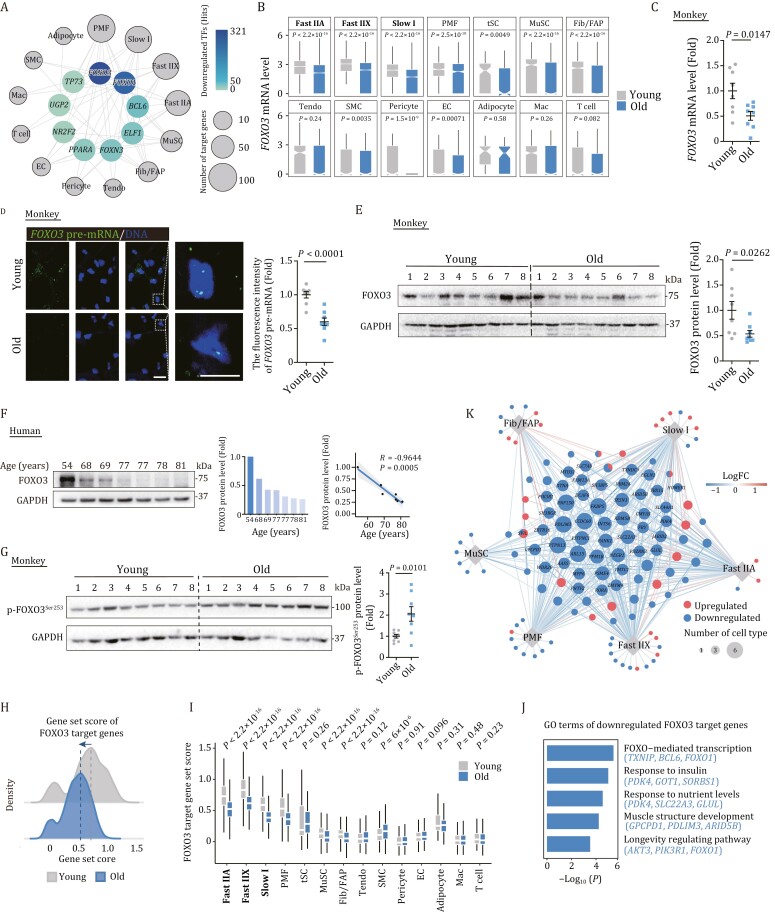
To further explore the core TFs governing aging-associated DEGs, we constructed a transcriptional regulatory network through Single-Cell Regulatory Network Inference and Clustering (SCENIC) analysis (Figs. 4A, S2F, and Table S4). Of note, we identified FOXO3 as one of the top downregulated transcriptional regulators in aged skeletal muscle (Fig. 4A). Indeed, FOXO3 transcripts were downregulated in the majority of cell types in aged skeletal muscle, along with a reduction in protein levels of FOXO3 in aged NHP muscle tissues as an entirety (Figs. 4B–E and S2G). Interestingly, we also found an age-dependent decline of FOXO3 protein in human skeletal muscle (Fig. 4F). More strikingly, phosphorylated FOXO3, an inactive form of this protein, that tends to be excluded from the nucleus and undergoes consequent ubiquitination and degradation (Sandri et al., 2004; Zhao et al., 2007; Yan et al., 2019), was elevated in the aged NHP muscle (Fig. 4G).
Figure 4.
Transcriptional network pinpoints FOXO3 as a crucial transcription factor in regulating primate skeletal muscle aging. (A) Network visualization of downregulated core transcriptional regulators in different cell types between old and young groups. Outer nodes display different cell types and the node size positively correlates with the number of target genes differentially expressed in corresponding cell types. (B) Box plots showing FOXO3 mRNA expression level across different cell types in monkey skeletal muscles from old and young groups. (C) RT-qPCR analysis showing the FOXO3 mRNA expression changes in young and old monkey skeletal muscles. FOXO3 mRNA levels were quantified as fold changes (old vs. young), and are presented as mean ± SEMs. n = 8 monkeys for each group. (D) RNA-FISH assay showing the FOXO3 expression changes in young and old monkey skeletal muscle tissues. Representative images are shown on the left. Scale bars, 20 and 10 μm (zoomed-in image). Right, fluorescence intensity of FOXO3 pre-mRNA level was quantified as fold changes (old vs. young) and are presented as mean ± SEMs. n = 8 monkeys for each group. (E) Western blot and band intensity quantification of FOXO3 protein levels in young and old monkey skeletal muscle samples. Data were quantified as fold changes in old skeletal muscles vs. in young counterparts and are presented as mean ± SEMs. n = 8 monkeys for each group. (F) Left, western blot and band intensity quantification of FOXO3 protein levels in human skeletal muscle samples. GAPDH was used as the loading control. Middle, bar plot showing the relative expression of FOXO3 protein levels for each individual. Right, the negative correlation of relative FOXO3 protein levels in skeletal muscle across different ages. The shadow indicates the 0.95 confidence interval around smooth. n = 7 donors. (G) Western blot and band intensity quantification of phospho-FOXO3 protein levels in young and old monkey skeletal muscle samples. Data were quantified as fold changes in old skeletal muscles vs. in young counterparts and are presented as mean ± SEMs. n = 8 monkeys for each group. (H) Ridge map showing the global distribution density of gene set score of FOXO3 target genes identified by SCENIC. The corresponding dashed line represents the peak position of each group. (I) Box plot showing the gene set score of FOXO3 target genes across each cell type of monkey muscles from old and young groups. (J) Bar plot showing representative GO terms of downregulated FOXO3 target genes. (K) Network visualization of FOXO3 target genes in Slow I, Fast IIA, Fast IIX, PMF, MuSC, and Fib/FAP cell types. Node size positively correlates with the number of cell types. Each connecting line represents gene differentially expressed in the corresponding cell type.
We next investigated the transcriptional activity of the FOXO3-modulated regulatory network (Calissi et al., 2021), and found that the scores of predicted FOXO3-target genes were lower in aged skeletal muscle and particularly in each fiber type relative to their young counterparts (Fig. 4H, 4I and Table S3). Furthermore, the downregulated FOXO3 target genes were relevant to longevity pathways (AKT3, PIK3R1, FOXO1) and muscle structure development or homeostasis maintenance (GPCPD1, PDLIM3, ARID5B) (Fig. 4J). For instance, PDLIM3 encodes cytoskeletal-related protein colocalizing with alpha-actinin-2 at the Z lines of skeletal muscle and is involved in cytoskeletal assembly (Ohsawa et al., 2011), whose downregulation may lead to a pronounced instability in skeletal muscle structure (Fig. 4K). The aforementioned results suggested that the inactivation of FOXO3 and its downstream cascade may underlie NHP skeletal muscle degeneration.
FOXO3 acts as a major effector gene to protect skeletal muscle from aging
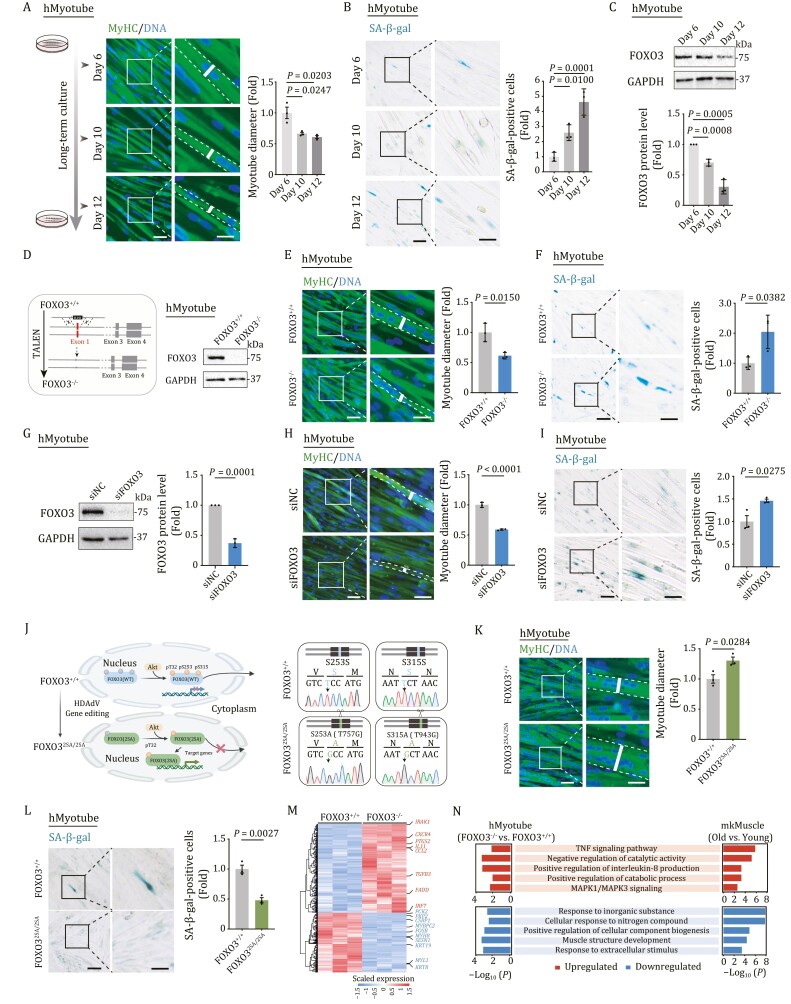
To investigate the function and molecular mechanism of FOXO3 in human skeletal muscle aging, we first generated human myotube (hMyotube) via directed differentiation from human embryonic stem cell (hESC) in a culture dish (Maffioletti et al., 2015). Prolonged culturing of human myotubes in vitro can result in a range of aging-related phenotypes, including decreased myotube diameter and elevated senescence-associated β-galactosidase (SA-β-gal) activity (Fig. 5A and 5B). Furthermore, the expression level of FOXO3 in hMyotubes was downregulated with prolonged in vitro culture (Fig. 5C), consistent with what we observed in the aged primate skeletal muscle (Fig. 4E and 4F). To investigate the effects of FOXO3 deficiency on hMyotube homeostasis, we generated FOXO3 knockout (FOXO3−/−) hMyotubes using TALEN-mediated gene editing technology (Fig. 5D) (see Methods section) (Zhang et al., 2020a). FOXO3−/− hMyotubes underwent accelerated aging, mainly characterized by reduced myotube diameter and increased SA-β-gal activity (Figs. 5E, 5F, and S3A). Similarly, siRNA-mediated knockdown of FOXO3 mimicked the human myotube aging phenotypes we observed in the FOXO3-deficient hMyotubes (Figs. 5G–I and S3B). Most importantly, we found that the aging phenotypes in hMyotubes were alleviated by gene editing-based activation of endogenous FOXO3 via alanine substitution on two of the three classical phosphorylation sites and generation of a constitutively active version of FOXO3 [FOXO3(2SA)] (Yan et al., 2019), as evidenced by an increment in human myotube diameter and concomitant reduction of SA-β-gal activity (Figs. 5J–L and S3C). When we performed genome-wide RNA sequencing, we found that FOXO3 deficiency in hMyotubes led to gene expression changes resembling those we had observed in skeletal muscle from aged monkeys (Figs. 5M, 5N, S3D, S3E, and Table S1). These included upregulated genes involved in the TNF-signaling pathway and interleukin-8 production, indicative of augmented age-associated inflammation. Upregulated DEGs also commonly converged on positive regulation of catabolic processes (Fig. 5N), suggesting that catabolism is favored upon FOXO3 ablation. Dampened transcriptional programs were commonly linked with muscle structure (Fig. 5N), supporting that FOXO3 downregulation underlies dysregulations of primate skeletal muscle homeostasis and degeneration. Taken together, these data suggest that FOXO3 plays a pivotal role in safeguarding primate muscle integrity against aging.
Figure 5.
FOXO3 protects human myotubes from senescence. (A) Left, representative immunofluorescence images of MyHC-positive hMyotube after long-term culture for 6, 10, and 12 days. Scale bars, 50 and 25 μm (zoomed-in image). Right, the diameters of the hMyotubes were quantified as fold changes in hMyotubes at Day 10 or Day 12 vs. at Day 6, and are shown as mean ± SEMs. n = 3 biological replicates. (B) Left, representative images of SA-β-gal-positive cells of WT hMyotubes after a long-term culture for 6, 10, and 12 days. Scale bars, 100 and 50 μm (zoomed-in image). Right, SA-β-gal-positive cells of the myotubes were quantified as fold changes in hMyotubes at Day 10 or Day 12 vs. at Day 6, and are presented as mean ± SEMs. n = 3 biological replicates. (C) Western blot analysis showing the protein levels of FOXO3 in WT hMyotubes after a long-term culture for 6, 10, and 12 days. GAPDH was used as the loading control. Band intensity were quantified as fold changes in hMyotubes at Day 10 or Day 12 vs. at Day 6 and are presented as mean ± SEMs. n = 3 independent experiments. (D) Left, schematic showing siRNA-mediated knockdown of FOXO3 in WT hMyotubes. Right, Western blot analysis of FOXO3 protein expression in FOXO3+/+ and FOXO3−/− hMyotubes. GAPDH was used as the loading control. (E) MyHC immunofluorescence staining in FOXO3+/+ and FOXO3−/− hMyotubes. Representative images are shown on the left. Scale bars, 50 and 25 μm (zoomed-in image). Right, the diameters of the hMyotubes were quantified as fold changes (FOXO3−/− vs. FOXO3+/+) and are presented as mean ± SEMs. n = 3 biological replicates. (F) Left, representative SA-β-gal staining images of FOXO3+/+ and FOXO3−/− hMyotubes. Scale bars, 100 and 50 μm (zoomed-in image). Right, SA-β-gal-positive cells of the myotubes were quantified as fold changes (FOXO3−/− vs. FOXO3+/+) and are presented as mean ± SEMs. n = 3 biological replicates. (G) Western blot showing the protein levels of FOXO3 in hMyotubes upon the knockdown of FOXO3. GAPDH was used as the loading control. Band intensity were quantified as fold changes (si-FOXO3 vs. si-NC) and are presented as mean ± SEMs. n = 3 independent experiments. (H) MyHC immunofluorescence staining of the hMyotubes transfected with si-NC or si-FOXO3. Representative images are shown on the left. Scale bars, 50 and 25 μm (zoomed-in image). The diameters of hMyotubes were quantified as fold changes (si-FOXO3 vs. si-NC) and are presented as mean ± SEMs on the right. n = 3 biological replicates. (I) SA-β-gal-positive cells of the hMyotubes transfected with si-NC or si-FOXO3. Representative images are shown on the left. Scale bars, 100 and 50 μm (zoomed-in image). Data were quantified as fold changes (si-FOXO3 vs. si-NC) and are presented as mean ± SEMs on the right. n = 3 biological replicates. (J) Left, the schematic shows that edited FOXO3 [FOXO3(2SA)] cannot be phosphorylated by AKT and is constitutively activated in the nucleus. Right, DNA sequencing demonstrates base conversion in FOXO32SA/2SA hMyotubes. The base conversion of T757G and T943G in genomic DNA results in the change of S253A and S315A in the protein sequence, respectively. (K) MyHC immunofluorescence staining in FOXO3+/+ and FOXO32SA/2SA hMyotubes. Representative images are shown on the left. Scale bars, 50 and 25 μm (zoomed-in image). The diameters of hMyotubes were quantified as fold changes (FOXO32SA/2SA vs. FOXO3+/+) and are presented as mean ± SEMs on the right. n = 3 biological replicates. (L) Left, representative SA-β-gal staining images of FOXO3+/+ and FOXO32SA/2SA hMyotubes. Scale bars, 100 and 50 μm (zoomed-in image). Right, SA-β-gal-positive myotubes were quantified as fold changes (FOXO32SA/2SA vs. FOXO3+/+) and are presented as mean ± SEMs. n = 3 biological replicates. (M)Heatmap showing the expression levels of DEGs between FOXO3−/− and FOXO3+/+ hMyotubes based on RNA-seq analysis. (N) GO terms shared by aging-associated DEGs in monkey skeletal muscle (mkMuscle) and DEGs between FOXO3−/− and FOXO3+/+ hMyotubes.
Discussion
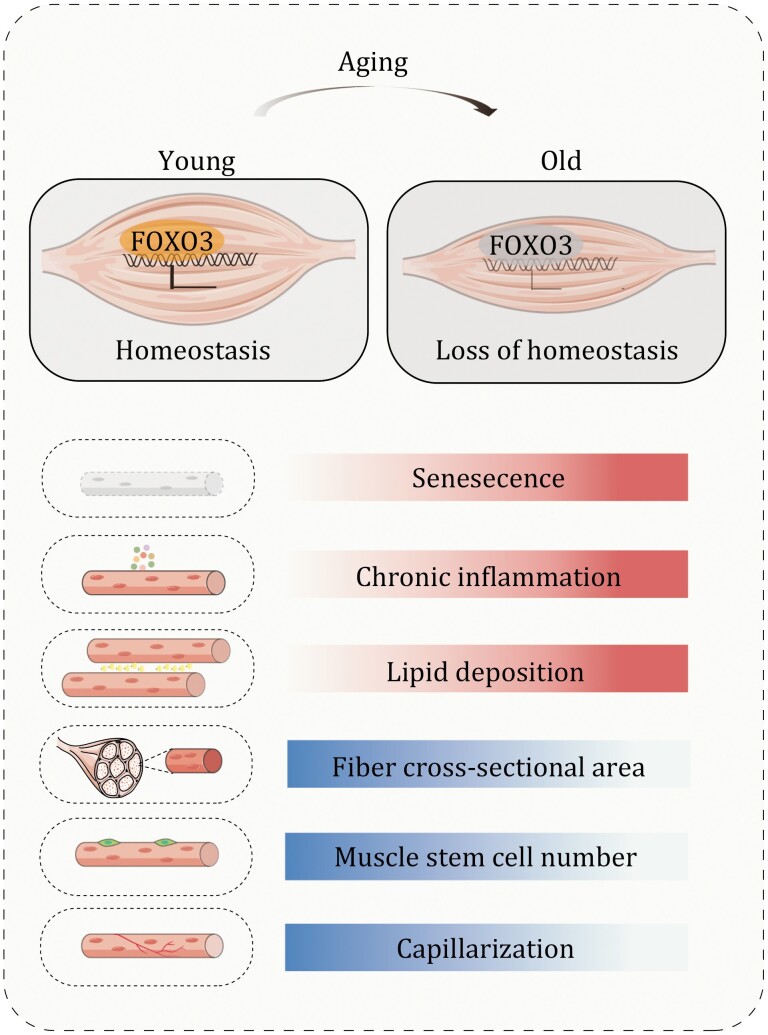
Age-related decline in skeletal muscle mass and strength is a hallmark feature of sarcopenia (McKellar et al., 2021). As such, this decline compromises physical performance and augments the risk of metabolic disease in the elderly. Here, we systemically analyzed aging-related phenotypes and established a single-nucleus transcriptome atlas of primate skeletal muscle aging. These efforts led to the identification of FOXO3 as a protective gatekeeper against primate skeletal muscle aging, and therefore pave the way for the development of novel diagnostics and intervention therapies for human skeletal muscle aging (Fig. 6).
Figure 6.
Working model. A schematic illustration showing the phenotypic and transcriptomic signatures of primate skeletal muscle aging.
While a few studies captured non-myofiber mononuclear cells by single-cell RNA sequencing (scRNA-seq) (Dell’Orso et al., 2019; Giordani et al., 2019; De Micheli et al., 2020; He et al., 2020; Rubenstein et al., 2020; Limbad et al., 2022), the myofiber syncytium does not easily lend itself to single-cell analyses using conventional dissociation approaches. To overcome this obstacle, we employed snRNA-seq analysis of primate skeletal muscle nuclei in suspension to generate, for the first time, a comprehensive transcriptomic roadmap of physiological private muscle aging. Furthermore, to deepen explorations of molecular events, we complemented our single-nucleus transcriptome profiles with a human pluripotent stem cell-derived myotube model that permits genetic manipulations. Using this combined research platform, we demonstrated a geroprotective role of FOXO3 in primate skeletal muscle aging.
As one of the most conserved and widely recognized human longevity genes, the cytoprotective effect of FOXO3 on various tissues has been widely reported, and FOXO3 is therefore a key target for the prevention and treatment of aging-related diseases (Salih and Brunet, 2008; Yan et al., 2019; Zhang et al., 2020a; Calissi et al., 2021; Lei et al., 2021). Indeed, supporting our observations in aged primate skeletal muscle and prolonged cultured senescent human myotubes, several previous findings demonstrated decreased FOXO3 mRNA and total or nuclear FOXO3 (active form) protein levels in aged human skeletal muscle (Leger et al., 2008; Dalle and Koppo, 2021). Although mouse-based pioneering studies suggested a deleterious role for FOXO3 in regulating skeletal muscle homeostasis (Sandri et al., 2004; Mammucari et al., 2007; Zhao et al., 2007), the function of FOXO3 during primate skeletal muscle aging remains unexplored. In particular, identifying the causal mechanism underlying primate skeletal muscle aging is challenging, given the lack of a suitable research system. It is for this reason that, in the current study, leveraging extensive mechanistic studies using engineered human myotubes, we for the first time reveal that activation of FOXO3 alleviates its senescence, suggesting an unappreciated protective role of FOXO3 against progressive primate skeletal muscle degeneration.
In conclusion, we here established a comprehensive single-nucleus transcriptomic landscape of primate skeletal muscle aging, which helped identify the inactivation of FOXO3 as a novel biomarker and a potential driver for primate skeletal muscle aging. The single-cell atlas of primate skeletal muscle and the human myotube aging research platform produced by this study promisingly support further explorations of additional diagnostic biomarkers of human skeletal muscle aging and the development of novel therapeutic interventions to treat aging-associated muscle diseases.
Materials and methods
Ethical statement
This study was conducted following the guidelines for the Ethical Treatment of nonhuman primates and was approved by the Institutional Animal Care and Use Committee of the Institute of Zoology (Chinese Academy of Sciences). All animal experiments in this study have been approved by the Ethics Review Committee of the Institute of Zoology of the Chinese Academy of Sciences. The collection and use of human skeletal muscle in this study were performed under the approval of the Research Ethics committee of the Peking University First Hospital.
Experimental animals
Eight young monkeys (4–6 years old) and eight old monkeys (18–21 years old) originated from Southeast Asia were raised at 25°C, 12 h light and dark cycle, and fed a normal diet at a certified Primate Research Center in Beijing (Xieerxin Biology Resource) (Wang et al., 2020; Zhang et al., 2020a, 2021; 2022; Li et al., 2021; Ma et al., 2021). All animals have no clinical or experimental history potentially affecting the physiological aging process.
Cell culture
Human embryonic stem cells (hESCs, Line H9, from WiCell Research) were maintained on mitomycin C-inactivated Mouse Embryonic Fibroblasts in hESC culture medium (Hu et al., 2020) containing 80% DMEM/F12 (Gibco), 20% Knockout Serum Replacement (Gibco), 2 mmol/L GlutaMAX (Gibco), 0.1 mmol/L nonessential amino acids (NEAA, Gibco), 55 μmol/L β-mercaptoethanol (Invitrogen), and 10 ng/mL bFGF (Joint Protein Central). hESCs were also cultured on Matrigel (BD Biosciences) with mTeSR medium (STEMCELL Technologies) at 37°C with 5% CO2. Human myotubes were cultured in high-glucose DMEM medium (Hyclone) containing 2% horse serum (Gibco) and 2 mmol/L GlutaMAX (Gibco) at 37°C with 5% CO2 and 3%–5% O2 (Maffioletti et al., 2015). HEK293T cells were cultured in high-glucose DMEM (Hyclone) supplemented with 10% fetal bovine serum (FBS, Gibco) at 37°C with 5% CO2. There was no mycoplasma contamination observed during cell culture.
Tissue sampling
Monkey quadriceps muscle tissues were harvested as previously described (Zhang et al., 2018; Liu et al., 2022). Afterwards, the attached fat or fascia tissues were carefully removed. The tissues were then fixed in 4% paraformaldehyde (PFA) at 4°C and embedded in paraffin or directly embedded in Tissue-Tek O.C.T compound (Sakura Finetek) and frozen in liquid nitrogen for histological analysis. The remaining tissues were stored in liquid nitrogen until the nuclei were extracted for sequencing analysis as well as for other RNA and protein analyses. Human quadriceps muscle samples were obtained and washed twice in pre-cold PBS and immediately frozen and stored in liquid nitrogen for further analyses.
Oil Red O staining
Oil Red O staining was performed as previously described (Ma et al., 2020). In brief, a Leica CM3050S cryostat was used to perform 10-μm-thick cryosections of monkey skeletal muscle embedded in Tissue-Tek O.C.T compound. After air-drying, the sections were fixed in a solution containing 4% PFA at room temperature (RT) for 10 min, and then stained in a freshly prepared Oil Red O staining solution (Sigma-Aldrich) for 30 min, rinsed with tap water and counterstained with hematoxylin. Images were taken with PerkinElmer Vectra Polaris. Image J was used to quantify the percentage of Oil Red O-positive area.
Immunofluorescence staining
Immunofluorescence staining was performed according to the previous method with minor modifications (Wang et al., 2020). For tissues, muscle tissue was embedded in Tissue-Tek O.C.T compound by the Leica CM3050S cryostat. The 10-μm cryosections were air-dried for 15 min, and then washed three times in PBS. Next, the sections were fixed with 4% PFA for another 20 min, and permeabilized with 0.4% Triton X-100 in PBS for 1 h, and again rinsed in PBS three times. Human myotubes were fixed with 4% PFA in PBS for 20 min and rinsed with PBS twice, and permeabilized with 0.4% Triton X-100 (Sigma-Aldrich) for 1 h at RT. The sections and cells were incubated with blocking buffer (10% donkey serum in PBS) for 1 h at RT and then with primary antibodies overnight at 4°C. After washing several times with PBS, the samples were incubated with fluorescently labeled secondary antibodies for 1 h at RT. The nuclei were stained with Hoechst 33342 (Thermo Fisher Scientific), and the sections were washed three times in PBS and then mounted in VECTERSHIELD anti-fading mounting medium (Vector Laboratories). Images were acquired using a confocal laser-scanning microscope (Leica TCS SP5 II). MyHC (myosin heavy chain)-positive cell containing at least three nuclei and displaying a tube-like structure was defined asa myotube. The diameters of 30–50 myotubes from randomly selected fields in each well were measured using the Image J software. The average diameter of randomly selected myotubes in each well was used for statistical analysis, and there are in total three wells as biological replicates (n = 3) (Liu et al., 2018). The antibodies used for immunofluorescence analyses are listed in Table S5.
Immunohistochemistry staining
Immunohistochemistry staining was performed as previously described with minor modifications (Zou et al., 2021). Briefly, the paraffin-embedded sections were deparaffinized and rehydrated using xylene and graded ethanol. Antigen retrieval was performed by steaming in citrate buffer four times for 2 min each. Sections were rinsed three times in PBS after being slowly cooled down to RT, and then permeabilized with 0.4% Triton X-100 in PBS for 1 h. And then sections were incubated with 3% H2O2 for 10 min to inactivate endogenous peroxidase and blocked with 10% donkey serum in PBS for 1 h. Sections were then incubated with primary antibodies at 4°C overnight and HRP-conjugated secondary antibodies at RT for 1 h the next day. Sections were performed using the DAB Staining Kit (ZSGB-BIO) according to the manufacturer’s instructions. Finally, sections were dehydrated in a series of graded alcohols (50%, 70%, 80%, 90%, 100%, and 100%) and xylene before being mounted in the neutral resinous mounting medium. Images were taken with PerkinElmer Vectra Polaris. The antibodies used in this study are listed in Table S5.
Lentivirus packaging
For lentivirus packaging, HEK293T cells were co-transfected with lentiviral vectors, psPAX2 (Addgene) and pMD2G (Addgene) using Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific). The supernatants containing viral particles were collected at 48 and 72 h after transfection for ultracentrifugation at 19,400 ×g at 4°C for 2.5 h.
Generation of FOXO3-knockout and FOXO3-enhanced human myotube (hMyotube)
Knockout of FOXO3 in human embryonic stem cells (hESCs) was performed by TALEN-based homologous recombination as previously reported (Zhang et al., 2020a). Genetic activation of FOXO3 in hESCs was performed by helper-dependent adenoviral vector (HDAdV) for FOXO32SA/2SA (Ser253Ala and Ser315Ala) knock-in as previously described (Yan et al., 2019). Human myotubes were then derived from the FOXO3-deficient and FOXO3-activated hESCs using a previously published protocol with some modifications (Maffioletti et al., 2015). In brief, hESCs were cultured in differentiation medium [MEMα (Thermo Fisher Scientific) containing 10% FBS (Thermo Fisher Scientific), 1% penicillin/streptomycin (Thermo Fisher Scientific), 10 ng/mL bFGF (Joint Protein Central) and 5 ng/mL TGFβ (StemImmune)] for approximately 10 days to generate human myotube progenitor cells. And human myotube progenitor cells of early passages were then transduced with MyoD-ER(T) lentiviral vector and transferred to Matrigel-coated culture dishes. At 37°C, 1 μmol/L 4-OH-Tamoxifen (Sigma-Aldrich) was added into the myotube progenitor cells culture medium with 5% CO2 and 3%–5% O2 when the cells reached 70%–80% confluence. After 24 h, myotube progenitor cell culture medium was replaced with fresh hMyotube differentiation medium (high-glucose DMEM medium containing 2% horse serum) supplemented with 1 μmol/L 4-OH-Tamoxifen and then replaced with fresh hMyotube differentiation medium every 2 days. The day for the first administration of 4-OH-Tamoxifen was defined as Day 1. The mature myotubes form on Day 5 or Day 6.
Knockdown of FOXO3 using small interfering RNA (siRNA)
siRNA-mediated knockdown of FOXO3 in human myotubes was performed as previously described (Wang et al., 2021a). Briefly, 25 μmol/L of negative control duplex or siRNAs against FOXO3 was mixed with 100 μL of Opti-MEM (Gibco) and 2μL Lipofectamine™RNAiMAX Transfection Reagent (Thermo Fisher Scientific) and then added to 1-well of 12-well plates. After an 8-h incubation, the culture medium was replaced with fresh medium. The cells were collected for RT-qPCR and Western blot 72 h after transfection. FOXO3 siRNAs were derived from a previous study and synthesized by RiboBio (China) (Wang et al., 2021a).
SA-β-gal staining
SA-β-gal staining of hMyotube was conducted as previously described (Wang et al., 2018). Briefly, cells were fixed in 2% formaldehyde and 0.2% glutaraldehyde at RT for 5 min and stained with fresh staining solution containing X-gal at 37°C overnight after washing twice with PBS. SA-β-gal-positive cells were counted in at least three fields selected randomly by Image J software.
TUNEL staining
TUNEL staining was performed using a TUNEL apoptosis detection Kit (Beyotime, C1088) according to the manufacturer’s instructions (Ma et al., 2022). Images were acquired using a Leica SP5 laser-scanning confocal microscope, and the percentages of positive nucleus were quantified using Image J software.
RNA isolation and quantitative reverse transcription PCR (RT-qPCR)
Total RNA was extracted using TRIzol (Life Technologies) according to the manufacturer’s protocol. Then the GoScript™ Reverse Transcription System (Promega) was used to reverse transcribe the cDNA from 2 μg of total RNA as a template. At least three independent samples were used for RT-qPCR assay with THUNDERBIRD SYBR qPCR Mix (Toyobo) on a CFX384 Real-Time PCR system (Bio-Rad). Primer sequences used in this study are listed in Table S6.
Western blot analysis
Western blot was performed as previously described (Wang et al., 2022). Tissues or cells were lysed with SDS lysis buffer [containing 4% SDS and 100 mmol/L Tris-HCl (pH = 6.8)] and incubated at 100°C for 10 min. To detect phosphorylated protein, RIPA lysate buffer (Beyotime, P0013B) containing a phosphatase inhibitor (Roche,4906837001) was used. A BCA Kit was used to perform protein quantification. The protein lysates were subjected to SDS-PAGE and subsequently transferred to a PVDF (polyvinylidene fluoride) membrane (Millipore). The membranes were blocked in 5% skimmed milk powder (BBI Life Sciences) in 1× TBST, incubated with the primary antibodies overnight at 4°C and with the HRP-conjugated secondary antibodies for 1 h at RT (ZSGB-BIO), followed by visualization using the ChemiDoc XRS system (Bio-Rad). The quantification was performed with Image J software. The antibodies used are listed in Table S5.
RNA-fluorescence in situ hybridization (RNA-FISH)
RNA-FISH was performed as reported previously (Kishi et al., 2019). Briefly, 8-μm sections of the monkey muscle tissues embedded in the Tissue-Tek O.C.T compound were cut using a cryotome and immediately placed on the polylysine-coated coverslips. One hundred and nineteen probes used for visualization of monkey FOXO3 pre-mRNA were designed by PaintSHOP (Hershberg et al., 2021). Briefly, sections were fixed in 4% PFA for 15 min and then treated by 0.5% Triton X-100 in 1× PBS (PBST) for 30 min. After 10 min of 10 μg/mL proteinase K (in PBST) treatment and 30 min of prehybridization at 43°C, the sections were then incubated with a hybridization mixture containing probes targeting monkey FOXO3 pre-mRNA for at least 16 h at 43°C. The slides were washed several times and incubated for 1 h with a hybridization mixture containing Alexa488-labeled fluorescent probes at 37°C. After 10 min of Hoechst 33342 (Thermo Fisher Scientific, H3570) (in 1× PBS) incubation and several washes, the sections were mounted with VECTASHIELD Antifade Mounting Medium (Vector Laboratories, H-1000), and images were captured with a confocal laser-scanning microscope (Zeiss 900 confocal system) and analyzed by Image J software.
Nuclei isolation and snRNA-seq on the 10× Genomics platform
Nuclei isolation was performed following a previously published protocol with some modifications (Krishnaswami et al., 2016). In brief, a piece of frozen monkey gastrocnemius muscle tissues (n = 16) was ground into powder separately with liquid nitrogen. This power was homogenized in 1.0 mL homogenization buffer [250 mmol/L sucrose, 25 mmol/L KCl, 5 mmol/L MgCl2, 10 mmol/L Tris buffer, 1 μmol/L DTT, 1× protease inhibitor, 0.4 U/μL RNaseIn, 0.2 U/μL Superasin, 0.1% Triton X-100, 1 μmol/L propidium iodide (PI), and 10 ng/mL Hoechst 33342 in Nuclease-Free water] using a freezing multisample tissue grinding system (60 Hz and 30 s per time, four times in total). Samples were filtered through a 40-micron cell strainer (BD Falcon), centrifuged at 3,000 ×g for 8 min at 4°C, and resuspended in PBS supplemented with 0.3% BSA, 0.4 U/μL RNaseIn and 0.2 U/μL Superasin. Hoechst 33342 and PI double-positive nuclei were sorted using FACS (BD Influx) and counted with a dual-fluorescence cell counter (Luna-FLTM, Logos Biosystems). Approximately 7,000 nuclei were captured for each sample with 10× Genomics Chromium Single Cell Kit (10× Genomics) version 3 following the standard protocol and then sequenced in a NovaSeq 6000 sequencing system (Illumina, 20012866).
Bulk RNA-seq library construction and sequencing
Total RNA of monkey skeletal muscles and hMyotubes were extracted using TRIzol reagent (Thermo Fisher Scientific). RNA quality control, library construction, and high-throughput sequencing were performed for each sample as previously described (Podnar et al., 2014). Briefly, sequencing libraries were prepared using NEBNext® UltraTM RNA Library Prep Kit for Illumina® (NEB, USA) and individually indexed. The resultant libraries were sequenced on an Illumina paired-end sequencing platform by 150-bp read length by Novogene Bioinformatics Technology Co. Ltd.
Processing and quality control of snRNA-seq data
Raw sequencing reads of monkey skeletal muscle were aligned to the pre-mRNA reference (Ensemble, Macaca_fascicularis_5.0) and counted using Cell Ranger (version 2.2.0) with the default parameters. The raw count matrix was filtered using CellBender (version 0.2.0) software in order to eliminate the contamination of background mRNA (Fleming et al., 2019). Seurat (version 3.2.2) object of each sample was constructed from the decontaminated matrix and nuclei with genes fewer than 200 or mitochondrial ratio more than 1% were discarded (Butler et al., 2018). Doublet removal was performed with DoubletFinder (version 2.0.3) (McGinnis et al., 2019). Afterwards, the clusters lacking specific marker genes and with relatively low gene content were also discarded.
Integration, clustering, and identification of cell types
The following steps were processed for monkey skeletal muscle dataset as the recommended pipeline of Seurat package. First, the count matrix of each sample was normalized using the “SCTransform” function. Features and anchors for downstream integration were selected with the corresponding pipeline using the “FindIntegrationAnchors” and “IntegrateData” functions, ensuring that the calculation was based on all necessary Pearson residuals. After data integration and scaling, principal component analysis (PCA) was performed with the “RunPCA” function, and the clustering analysis was conducted with the “FindNeighbors” and “FindClusters” functions. Dimensionality was reduced with the “RunUMAP” function. Cell types were identified according to the expression levels of the classic marker genes. The marker genes of each cell type were calculated using the “FindAllMarkers” function with the cutoff of |LogFC| > 0.5 and adjusted P values < 0.05 using t-test. Marker genes for each cell type are shown in Table S2.
Analysis of differentially expressed genes from snRNA-seq data
DEGs between different cell types of old and young groups in monkey skeletal muscle were analyzed by the function of “FindMarkers” in Seurat using Wilcoxon Rank Sum test, and were identified with the cutoff of |LogFC| > 0.25 and adjusted P values < 0.05 DEG lists for each cell type are shown in Table S1.
Gene expression noise analysis
Analysis of age-relevant transcriptional noise was used to analyze the aging effects on different cell types following previous work (Angelidis et al., 2019). For each cell type, with at least 10 young and old cells, the transcriptional noise was quantified in the following manner. In brief, to account for differences in total UMI counts, all cells were down-sampled so that all cells had equal library size. To account for differences in cell-type frequency, cell numbers were down-sampled to obtain the same number of young and old cells. Next, the Euclidean distance between each cell and the corresponding cell-type mean within each age group was calculated. Transcriptional noise of each cell was then measured by this Euclidean distance. Furthermore, the Euclidean distances for each monkey and the transcriptional noise ratio between old and young groups were also calculated to indicate the change of transcriptional noise across samples during aging.
Analysis of transcription factor regulatory network
Transcriptional regulatory network was analyzed by SCENIC (version 1.1.2.2) (Aibar et al., 2017) with default parameters. The reference of TFs was downloaded from RcisTarget (version 1.6.0) with hg19. The inference of regulatory network among the 1,824 aging-associated DEGs of monkey skeletal muscle was carried out by GENIE3 and GRNBoost in python 3.0 (Irrthum et al., 2010). Then, the enriched binding motifs and the target genes of each TF were analyzed by RcisTarget. Only target genes with high-confidence annotation were selected for the downstream analysis. The transcriptional regulatory network was visualized in Cytoscape (version 3.8.0) (Shannon et al., 2003). Core regulatory TFs of DEGs in monkey skeletal muscle during aging are shown in Table S4.
Gene Ontology enrichment analysis
Gene Ontology (GO) enrichment analysis was performed with Metascape (Zhou et al., 2019). Representative terms were selected with the cutoff of P values < 0.01 and visualized with ggplot2 R package (version 3.3.2) (Wickham, 2016).
Gene set score analysis
Public gene sets were acquired from KEGG (Kanehisa et al., 2021), MSigDB database (Liberzon et al., 2015), Aging Atlas (Guang-Hui Liu, 2021), and Regeneration Roadmap (Kang et al., 2022). FOXO3 target gene set was obtained with the SCENIC analysis described in the previous section. Gene sets were used for scoring each input cell with the Seurat function “AddModuleScore.” Differences in the scores between young and old samples were analyzed using ggpubr R package (version 0.2.4) via the Wilcoxon test. Gene sets used in this study are shown in Table S3.
Bulk RNA-seq data processing
RNA-seq data were processed as previously described (Ma et al., 2021). In brief, to trim adapter sequence and remove low-quality reads, the raw sequencing reads were first processed with Trim Galore (version 0.6.6). Then, the cleaned reads were mapped against monkey genome reference macFas5 or human genome reference hg19 downloaded from Ensembl (Yates et al., 2020) with HISAT2 (version 2.2.1) (Kim et al., 2015), and then the mapped reads were counted with HTSeq (version 0.13.5) (Anders et al., 2015). DEG analysis was conducted with DESeq2 (version 1.28.1) in R (Love et al., 2014), and DEGs were identified as the genes with |Log2FC| > 0.5 and adjusted P values < 0.05 or P values < 0.05 (for monkey skeletal muscle samples). DEG lists are shown in Table S1.
Identification of the binding motif of FOXO3 target genes
To investigate the transcriptional changes of FOXO3 target genes in skeletal muscle aging, the target genes and the binding motif sequences of FOXO3 were retrieved from the SCENIC analysis result.
Statistical analyses
All experimental data were statistically analyzed using PRISM software (GraphPad 8 Software). Comparisons were conducted using the two-tailed Student’s t-test or Wilcoxon Rank Sum test. P values are presented in indicated figures.
Supplementary information
The online version contains supplementary material available at https://doi.org/10.1093/procel/pwac061.
Acknowledgements
We thank Fang Cheng and Wei Wang for their help in harvesting monkey skeletal muscle tissues; Wenlei Bao for the technical support in human myotube differentiation; and Siyu Chen for her assistance in bioinformatic analysis. We thank Junying Jia from the Institute of Biophysics, Chinese Academy of Sciences, for his help in fluorescence-activated cell sorting (FACS), and Shiwen Li from the Institute of Zoology, Chinese Academy of Sciences, for her help in image scanning of immunohistochemical staining. We are also grateful to Lei Bai, Qun Chu, Jing Lu, Ying Yang, Ruijun Bai, Luyang Tian, Xiuping Li, and Jing Chen for administrative assistance. This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDA16000000), the National Natural Science Foundation of China (Nos. 82071588, 81921006, 82125011, 92149301, 92168201, 92049116, 32121001, 82192863, 91949209, 92049304, 82122024, 82001477, 31900523, 81861168034, 32000500, 82271600, 82201714), the National Key Research and Development Program of China (Nos. 2018YFC2000100, 2020YFA0804000, 2018YFA0107203, 2020YFA0112200, 2021YFF1201005, 2021ZD0202401, 2018YFC2000400, 2020YFA0113400, 2021YFE0111800, 2022YFA1103700), the Program of the Beijing Natural Science Foundation (No. Z190019), K. C. Wong Education Foundation (Nos. GJTD-2019-06, GJTD-2019-08), Beijing Medical Research (2021-8), the Pilot Project for Public Welfare Development and Reform of Beijing-affiliated Medical Research Institutes (No. 11000022T000000461062), Young Elite Scientists Sponsorship Program by CAST (Nos. YESS20200012, YESS20210002), CAS Project for Young Scientists in Basic Research (No. YSBR-076, YSBR-012, YSBR-036), Youth Innovation Promotion Association of CAS (Nos. E1CAZW0401, 2020085, 2022083). the Informatization Plan of Chinese Academy of Sciences (Nos. CAS-WX2022SDC-XK14, CAS-WX2021SF-0301, CAS-WX2021SF-0101), the Tencent Foundation (No. 2021-1045), CAMS Innovation Fund for Medical Sciences (No. 2021-I2M-1-050), and the Fellowship of China Postdoctoral Science Foundation (2022M712216).
Glossary
Abbreviations
- snRNA-seq
single-nucleus RNA sequencing
- PMF
postsynaptic muscle fiber
- tSC
terminal Schwann cell
- MuSC
muscle stem cell
- Fib/FAP
fibroblast/fibro-adipogenic progenitor
- Tendo
Tendon fibroblast
- SMC
smooth muscle cell
- EC
endothelial cell
- Mac
macrophage
- DEGs
differentially expressed genes
- FOXO3
Forkhead box O3
- NMJ
neuromuscular junctions
- NHP
non-human primate
- t-SNE
t-distributed stochastic neighbor embedding
- UMAP
uniform manifold approximation and projection
- CV
coefficient of variation
- siRNA
small interfering RNA.
Contributor Information
Ying Jing, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yuesheng Zuo, University of Chinese Academy of Sciences, Beijing 100049, China; CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Yang Yu, Department of Obstetrics and Gynecology, Center for Reproductive Medicine, Peking University Third Hospital, Beijing 100191, China; Clinical Stem Cell Research Center, Peking University Third Hospital, Beijing 100191, China.
Liang Sun, The Key Laboratory of Geriatrics, Beijing Institute of Geriatrics, Institute of Geriatric Medicine, Chinese Academy of Medical Sciences, Beijing Hospital/National Center of Gerontology of National Health Commission, Beijing 100730, China.
Zhengrong Yu, Department of Orthopaedics, Peking University First Hospital, Beijing 100034, China.
Shuai Ma, State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China.
Qian Zhao, Advanced Innovation Center for Human Brain Protection, National Clinical Research Center for Geriatric Disorders, Xuanwu Hospital Capital Medical University, Beijing 100053, China; Aging Translational Medicine Center, International Center for Aging and Cancer, Beijing Municipal Geriatric Medical Research Center, Xuanwu Hospital, Capital Medical University, Beijing 100053, China.
Guoqiang Sun, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Huifang Hu, State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China.
Jingyi Li, State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China.
Daoyuan Huang, Advanced Innovation Center for Human Brain Protection, National Clinical Research Center for Geriatric Disorders, Xuanwu Hospital Capital Medical University, Beijing 100053, China; Aging Translational Medicine Center, International Center for Aging and Cancer, Beijing Municipal Geriatric Medical Research Center, Xuanwu Hospital, Capital Medical University, Beijing 100053, China.
Lixiao Liu, University of Chinese Academy of Sciences, Beijing 100049, China; CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Jiaming Li, University of Chinese Academy of Sciences, Beijing 100049, China; CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; Sino-Danish College, University of Chinese Academy of Sciences, Beijing 101408, China.
Zijuan Xin, State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China.
Haoyan Huang, Advanced Innovation Center for Human Brain Protection, National Clinical Research Center for Geriatric Disorders, Xuanwu Hospital Capital Medical University, Beijing 100053, China; Aging Translational Medicine Center, International Center for Aging and Cancer, Beijing Municipal Geriatric Medical Research Center, Xuanwu Hospital, Capital Medical University, Beijing 100053, China.
Juan Carlos Izpisua Belmonte, Altos Labs, Inc., San Diego, CA 94022, USA.
Weiqi Zhang, University of Chinese Academy of Sciences, Beijing 100049, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; Sino-Danish College, University of Chinese Academy of Sciences, Beijing 101408, China; Sino-Danish Center for Education and Research, Beijing 101408, China.
Si Wang, Advanced Innovation Center for Human Brain Protection, National Clinical Research Center for Geriatric Disorders, Xuanwu Hospital Capital Medical University, Beijing 100053, China; Aging Translational Medicine Center, International Center for Aging and Cancer, Beijing Municipal Geriatric Medical Research Center, Xuanwu Hospital, Capital Medical University, Beijing 100053, China; The Fifth People’s Hospital of Chongqing, Chongqing 400062, China.
Jing Qu, State Key Laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China.
Guang-Hui Liu, State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China; Advanced Innovation Center for Human Brain Protection, National Clinical Research Center for Geriatric Disorders, Xuanwu Hospital Capital Medical University, Beijing 100053, China; University of Chinese Academy of Sciences, Beijing 100049, China; Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, Beijing 100101, China; Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China.
Conflict of interest
The authors declare no competing interests.
Ethics approval
All animals used in this study have been approved by the Ethics Review Committee of the Institute of Zoology of the Chinese Academy of Sciences. The collection and use of human skeletal muscle in this study were performed under the approval of the Research Ethics committee of the Peking University First Hospital.
Consent to participate
The authors declare their agreement to participate.
Consent for publication
The authors declare their agreement to publish.
Author contributions
G.-H.L., J.Q., S.W., and W.Z. conceptualized this project and supervised the overall experiments. S.W. and Y.J. performed isolation of monkey skeletal muscle snRNA-seq library construction and sequencing. Y.J. performed the phenotypic, mechanistic analyses. Y.Z. performed bioinformatics analysis of the snRNA-seq and bulk RNA-seq. Q.Z. performed some immunofluorescence staining. G.S. performed the RNA-FISH of monkey skeletal muscle. G.-H.L., S.W., J.Q., W.Z., Y.J., Y.Z., S.M., Q.Z., G.S., Jingyi Li., D.H., Jiaming Li., L.L., L.S, Y.Y., Z.Y., Z.X., H.H., and J.C.I.B performed manuscript writing, review, and editing. All authors reviewed the manuscript.
Data availability
The accession numbers for the raw snRNA-seq data and bulk RNA-seq data reported in this paper are GSA: CRA006505 and HRA002196.
Code availability
No specific custom code was used in this manuscript. All the codes are publicly available and the source is annotated in the text and/or in the Methods. Scripts are available upon reasonable request.
References
- Aibar S, González-Blas CB, Moerman Tet al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017;14:1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W.. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Colman RJ.. Prospects and perspectives in primate aging research. Antioxid Redox Signal 2011;14:203–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelidis I, Simon LM, Fernandez IEet al. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nat Commun 2019;10:963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askanas V, Engel WK.. Inclusion-body myositis, a multifactorial muscle disease associated with aging: current concepts of pathogenesis. Curr Opin Rheumatol 2007;19:550–559. [DOI] [PubMed] [Google Scholar]
- Askanas V, Engel WK, Nogalska A.. Pathogenic considerations in sporadic inclusion-body myositis, a degenerative muscle disease associated with aging and abnormalities of myoproteostasis. J Neuropathol Exp Neurol 2012;71:680–693. [DOI] [PubMed] [Google Scholar]
- Baskin KK, Winders BR, Olson EN.. Muscle as a “mediator” of systemic metabolism. Cell Metab 2015;21:237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert Pet al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadot B, Gache V, Gomes ER.. Moving and positioning the nucleus in skeletal muscle – one step at a time. Nucleus 2015;6:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Song W, Li Jet al. The landscape of aging. Sci China Life Sci 2022; 65(12), 2354–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calissi G, Lam EW, Link W.. Therapeutic strategies targeting FOXO transcription factors. Nat Rev Drug Discov 2021;20:21–38. [DOI] [PubMed] [Google Scholar]
- Dalle S, Koppo K.. Cannabinoid receptor 1 expression is higher in muscle of old vs. young males, and increases upon resistance exercise in older adults. Sci Rep 2021;11:18349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Micheli AJ, Spector JA, Elemento Oet al. A reference single-cell transcriptomic atlas of human skeletal muscle tissue reveals bifurcated muscle stem cell populations. Skelet Muscle 2020;10:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Orso S, Juan AH, Ko KDet al. Single cell analysis of adult mouse skeletal muscle stem cells in homeostatic and regenerative conditions. Development 2019;146:dev174177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F, Piccirillo R, Goldberg ALet al. Mechanisms of skeletal muscle aging: insights from Drosophila and mammalian models. Dis Model Mech 2013;6:1339–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks AJ, Hofer T, Marzetti Eet al. Mitochondrial DNA mutations, energy metabolism and apoptosis in aging muscle. Ageing Res Rev 2006;5:179–195. [DOI] [PubMed] [Google Scholar]
- Domingues-Faria C, Vasson MP, Goncalves-Mendes Net al. Skeletal muscle regeneration and impact of aging and nutrition. Ageing Res Rev 2016;26:22–36. [DOI] [PubMed] [Google Scholar]
- Dos Santos M, Backer S, Saintpierre Bet al. Single-nucleus RNA-seq and FISH identify coordinated transcriptional activity in mammalian myofibers. Nat Commun 2020;11:5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming SJ, Marioni JC, Babadi M.. CellBender remove-background: a deep generative model for unsupervised removal of background noise from scRNA-seq datasets. bioRxiv 2019. doi:10.1101/791699. [Google Scholar]
- Frontera WR, Ochala J.. Skeletal muscle: a brief review of structure and function. Calcif Tissue Int 2015;96:183–195. [DOI] [PubMed] [Google Scholar]
- Fuchs E, Blau HM.. Tissue stem cells: architects of their niches. Cell Stem Cell 2020;27:532–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordani L, He GJ, Negroni Eet al. High-dimensional single-cell cartography reveals novel skeletal muscle-resident cell populations. Mol Cell 2019;74:609–621.e6. [DOI] [PubMed] [Google Scholar]
- Gomarasca M, Banfi G, Lombardi G.. Myokines: the endocrine coupling of skeletal muscle and bone. Adv Clin Chem 2020;94:155–218. [DOI] [PubMed] [Google Scholar]
- González JP, Queralt-Rosinach N, Bravo Aet al. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database 2015;2015:bav028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang-Hui Liu YB, Qu J, Zhang Wet al. Aging Atlas: a multi-omics database for aging biology. Nucleic Acids Res 2021;49:D825–d830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Memczak S, Qu Jet al. Single-cell omics in ageing: a young and growing field. Nat Metab 2020;2:293–302. [DOI] [PubMed] [Google Scholar]
- Hershberg EA, Camplisson CK, Close JLet al. PaintSHOP enables the interactive design of transcriptome- and genome-scale oligonucleotide FISH experiments. Nat Methods 2021;18:937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Ji Q, Song Met al. ZKSCAN3 counteracts cellular senescence by stabilizing heterochromatin. Nucleic Acids Res 2020;48:6001–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka K, Machida T, Hirafuji M.. Skeletal muscle is an endocrine organ. J Pharmacol Sci 2014;125:125–131. [DOI] [PubMed] [Google Scholar]
- Irrthum A, Wehenkel L, Geurts P.. Inferring regulatory networks from expression data using tree-based methods. PLoS One 2010;5:e12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Wen Y, Kuroda Ket al. Notch signaling deficiency underlies age-dependent depletion of satellite cells in muscular dystrophy. Dis Model Mech 2014;7:997–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RA, Harrison C, Eaton SLet al. Cellular and molecular anatomy of the human neuromuscular junction. Cell Rep 2017;21:2348–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinkovich A, Livshits G.. Sarcopenic obesity or obese sarcopenia: a cross talk between age-associated adipose tissue and skeletal muscle inflammation as a main mechanism of the pathogenesis. Ageing Res Rev 2017;35:200–221. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Furumichi M, Sato Yet al. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res 2021;49:D545–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang W, Jin T, Zhang Tet al. Regeneration Roadmap: database resources for regenerative biology. Nucleic Acids Res 2022;50:D1085–D1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL.. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Franke V, Brandt Bet al. Single-nucleus transcriptomics reveals functional compartmentalization in syncytial skeletal muscle cells. Nat Commun 2020;11:6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishi JY, Lapan SW, Beliveau BJet al. SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat Methods 2019;16:533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswami SR, Grindberg RV, Novotny Met al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc 2016;11:499–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson L, Degens H, Li Met al. Sarcopenia: aging-related loss of muscle mass and function. Physiol Rev 2019;99:427–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger B, Derave W, De Bock Ket al. Human sarcopenia reveals an increase in SOCS-3 and myostatin and a reduced efficiency of Akt phosphorylation. Rejuvenation Res 2008;11:163–175B. [DOI] [PubMed] [Google Scholar]
- Lei J, Wang S, Kang Wet al. FOXO3-engineered human mesenchymal progenitor cells efficiently promote cardiac repair after myocardial infarction. Protein Cell 2021;12:145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng SX, Pawelec G.. Single-cell immune atlas for human aging and frailty. Life Med 2022:lnac013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zheng Y, Yan Pet al. A single-cell transcriptomic atlas of primate pancreatic islet aging. Natl Sci Rev 2021;8:nwaa127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir Het al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbad C, Doi R, McGirr Jet al. Senolysis induced by 25-hydroxycholesterol targets CRYAB in multiple cell types. iScience 2022;25:103848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipina C, Hundal HS.. Lipid modulation of skeletal muscle mass and function. J Cachexia Sarcopenia Muscle 2017;8:190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Liu X, Bai Yet al. Neuregulin-1β modulates myogenesis in septic mouse serum-treated C2C12 myotubes in vitro through PPARγ/NF-κB signaling. Mol Biol Rep 2018;45:1611–1619. [DOI] [PubMed] [Google Scholar]
- Liu Z, Li W, Geng Let al. Cross-species metabolomic analysis identifies uridine as a potent regeneration promoting factor. Cell Discov 2022;8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livshits G, Kalinkovich A.. Inflammaging as a common ground for the development and maintenance of sarcopenia, obesity, cardiomyopathy and dysbiosis. Ageing Res Rev 2019;56:100980. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Sun S, Geng Let al. Caloric restriction reprograms the single-cell transcriptional landscape of Rattus Norvegicus aging. Cell 2020;180:1001.e22. [DOI] [PubMed] [Google Scholar]
- Ma S, Sun S, Li Jet al. Single-cell transcriptomic atlas of primate cardiopulmonary aging. Cell Res 2021;31:415–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Wang S, Ye Yet al. Heterochronic parabiosis induces stem cell revitalization and systemic rejuvenation across aged tissues. Cell Stem Cell 2022;29:1005.e10. [DOI] [PubMed] [Google Scholar]
- Maeda S, Miyagawa S, Kawamura Tet al. Notch signaling-modified mesenchymal stem cells improve tissue perfusion by induction of arteriogenesis in a rat hindlimb ischemia model. Sci Rep 2021;11:2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffioletti SM, Gerli MF, Ragazzi Met al. Efficient derivation and inducible differentiation of expandable skeletal myogenic cells from human ES and patient-specific iPS cells. Nat Protoc 2015;10:941–958. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello Vet al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 2007;6:458–471. [DOI] [PubMed] [Google Scholar]
- McGinnis CS, Murrow LM, Gartner ZJ.. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst 2019;8:329–337.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKellar DW, Walter LD, Song LTet al. Large-scale integration of single-cell transcriptomic data captures transitional progenitor states in mouse skeletal muscle regeneration. Commun Biol 2021;4:1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercken EM, Capri M, Carboneau BAet al. Conserved and species-specific molecular denominators in mammalian skeletal muscle aging. NPJ Aging Mech Dis 2017;3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris BJ, Willcox DC, Donlon TAet al. FOXO3: A major gene for human longevity -- a mini-review. Gerontology 2015;61:515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia M, Toniolo L, Nagaraj Net al. Single muscle fiber proteomics reveals fiber-type-specific features of human muscle aging. Cell Rep 2017;19:2396–2409. [DOI] [PubMed] [Google Scholar]
- Navarro G. A guided tour to approximate string matching. ACM Comput Surv 2001;33:31–88. [Google Scholar]
- Ohsawa N, Koebis M, Suo Set al. Alternative splicing of PDLIM3/ALP, for α-actinin-associated LIM protein 3, is aberrant in persons with myotonic dystrophy. Biochem Biophys Res Commun 2011;409:64–69. [DOI] [PubMed] [Google Scholar]
- Orchard P, Manickam N, Ventresca Cet al. Human and rat skeletal muscle single-nuclei multi-omic integrative analyses nominate causal cell types, regulatory elements, and SNPs for complex traits. Genome Res 2021;31:2258–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podnar J, Deiderick H, Hunicke-Smith S.. Next-generation sequencing fragment library construction. Curr Protoc Mol Biol 2014;107:7.17.11–7.17.16. [DOI] [PubMed] [Google Scholar]
- Rubenstein AB, Smith GR, Raue Uet al. Single-cell transcriptional profiles in human skeletal muscle. Sci Rep 2020;10:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salih DA, Brunet A.. FoxO transcription factors in the maintenance of cellular homeostasis during aging. Curr Opin Cell Biol 2008;20:126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert Aet al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 2004;117:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier Oet al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Van Meter M, Ablaeva Jet al. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab 2019;29:871–885.e5 e875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes T, Hector AJ, Morton RWet al. Recent perspectives regarding the role of dietary protein for the promotion of muscle hypertrophy with resistance exercise training. Nutrients 2018;10:180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieland M, Trouwborst I, Clark BC.. Skeletal muscle performance and ageing. J Cachexia Sarcopenia Muscle 2018;9:3–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma M, Asakura Y, Murakonda BSRet al. Muscle satellite cell cross-talk with a vascular niche maintains quiescence via VEGF and notch signaling. Cell Stem Cell 2018;23:530–543.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagers AJ, Conboy IM.. Cellular and molecular signatures of muscle regeneration: current concepts and controversies in adult myogenesis. Cell 2005;122:659–667. [DOI] [PubMed] [Google Scholar]
- Wang S, Cheng F, Ji Qet al. Hyperthermia differentially affects specific human stem cells and their differentiated derivatives. Protein Cell 2022;13:615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Hu B, Ding Zet al. ATF6 safeguards organelle homeostasis and cellular aging in human mesenchymal stem cells. Cell Discov 2018;4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Yao X, Ma Set al. A single-cell transcriptomic landscape of the lungs of patients with COVID-19. Nat Cell Biol 2021a;23:1314–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zheng Y, Li Jet al. Single-cell transcriptomic atlas of primate ovarian aging. Cell 2020;180:585–600.e19. [DOI] [PubMed] [Google Scholar]
- Wang S, Zheng Y, Li Qet al. Deciphering primate retinal aging at single-cell resolution. Protein Cell 2021b;12:889–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Ng S-C.. Sphingolipids mediate lipotoxicity in muscular dystrophies. Life Med 2022:lnac015. [Google Scholar]
- Wickham, H. 2016. ggplot2: Elegant Graphics for Data Analysis. Springer [Google Scholar]
- Wilkinson DJ, Piasecki M, Atherton PJ.. The age-related loss of skeletal muscle mass and function: measurement and physiology of muscle fibre atrophy and muscle fibre loss in humans. Ageing Res Rev 2018;47:123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan P, Li Q, Wang Let al. FOXO3-engineered human ESC-derived vascular cells promote vascular protection and regeneration. Cell Stem Cell 2019;24:447–461.e8. [DOI] [PubMed] [Google Scholar]
- Yates AD, Achuthan P, Akanni Wet al. Ensembl 2020. Nucleic Acids Res 2020;48:D682–D688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Clyne M, Khoury MJet al. Phenopedia and genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics 2010;26:145–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Li J, Suzuki K, et al. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Sci 2015;348:1160–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li J, Ren Jet al. Single-nucleus transcriptomic landscape of primate hippocampal aging. Protein Cell 2021;12:695–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wan H, Feng Get al. SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature 2018;560:661–665. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhang S, Yan Pet al. A single-cell transcriptomic landscape of primate arterial aging. Nat Commun 2020a;11:2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qu J, Liu G-H, Belmonte JCI.. The ageing epigenome and its rejuvenation. Nat Rev Mol Cell Biol 2020b;21:137–150. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zheng Y, Wang Set al. Single-nucleus transcriptomics reveals a gatekeeper role for FOXP1 in primate cardiac aging. Protein & Cell 2022:pwac038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Chen S.. Failures at every level: breakdown of the epigenetic machinery of aging. Life Med 2022:lnac016. [Google Scholar]
- Zhao J, Brault JJ, Schild Aet al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 2007;6:472–483. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache Let al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, X., Dai, X., Mentis, A.F.A., Esteban, M.A., Liu, L., and Han, L.. From monkey single-cell atlases into a broader biomedical perspective. Life Med. 2022:lnac028. [Google Scholar]
- Zou Z, Long X, Zhao Qet al. A single-cell transcriptomic atlas of human skin aging. Dev Cell 2021;56:383–397.e8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the raw snRNA-seq data and bulk RNA-seq data reported in this paper are GSA: CRA006505 and HRA002196.