Abstract
PANX2 forms large-pore channels mediating ATP release in response to physiological and pathological stimuli. Although PANX2 shows involvements in glioma genesis, the underlying mechanism remains unclear. PANX2 mRNA expression was analyzed via Oncomine and was confirmed via Gene Expression Profiling Interactive Analysis (GEPIA). The influence of PANX2 on overall survival (OS) of glioma was evaluated using LinkedOmics and further assessed through Cox regression analysis. The correlated genes with PANX2 acquired from LinkedOmics were validated through GEPIA and cBioPortal. Protein-protein interaction (PPI) of these genes was then obtained using Search Tool for the Retrieval of Interacting Genes (STRING) and Cytoscape with MCODE plug-in. All the PANX2-related genes underwent Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. The correlation between PANX2 and cancer immune infiltrates was evaluated via Tumor Immune Estimation Resource (TIMER). A higher expression of PANX2 only revealed a better OS in brain low grade glioma (LGG). PANX2-related genes in LGG functionally enriched in neuroactive ligand-receptor interaction, synaptic vesicle cycle, and calcium signaling. The hub genes from highest module of PPI were mainly linked to chemical synaptic transmission, plasma membrane, neuropeptide, and the pathway of neuroactive ligand-receptor interaction. Besides, PANX2 expression was negatively associated with infiltrating levels of macrophage, dendritic cells, and CD4+ T cells. This study demonstrated that PANX2 likely participated in LGG pathogenesis by affecting multiple molecular pathways and immune-related processes. PANX2 was associated with LGG prognosis and might become a promising therapeutic target of LGG.
Keywords: PANX2, Brain Lower Grade Gliomas, Immune Infiltrates, Microenvironment, Tumorigenesis
Introduction
Pannexins (Panxs) have similar topological structure and share common gating mechanisms with classical gap junctions: connexins (Cxs).1–3 Panxs form large-pore channels which mediate ATP release in response to a variety of physiological and pathological stimuli.4,5 Panx channels also permit the passage of small molecules or metabolites up to <1 kDa, delivering signals in a paracrine or endocrine fashion through targeting cell surface receptors.6,7 So far, there are three subtypes in human Panx family, namely PANX1, PANX2, and PANX3. PANX1 is ubiquitously expressed in most mammalian cells and many organs of human body, including brain, heart, and skeletal muscle. 8 PANX2 is a brain-specific channel, which is mainly enriched in neurons of the central nervous system with different expression in postnatal neural progenitor cells and mature neurons.9,10 PANX3 is located in osteoblasts, chondrocytes, and keratinocyte, serving an important role in cell proliferation and differentiation.11,12 All members of Panx family have been tightly linked to multiple diseases, particularly in cancers. 13
A growing body of publications elucidate that Panxs are involved in cancer progression. The up-regulated PANX1 promoted migration and invasion in I-10 testicular cancer cells. 14 Blocking channel function with PANX1 inhibitors remarkedly decreased cell growth and migration in melanoma cell lines. 15 Besides, chemotherapy using a pan-caspase inhibitor: benzyloxycarbonyl-VAD, might activate PANX1 channel to regulate interaction between dying tumor cells and leukocytes. 16 On the other hand, PANX2 is proposed to function as a potential tumor suppressor, especially in brain and hepatocellular carcinomas.17,18 Down-regulated PANX2 has been linked to human gliomas. In clear cell renal cell carcinomas (CCRCCs), PANX2 and FAM83H might be participate in tumor development in a co-operative manner. 19 In terms of PANX3, a significantly reduced protein level was reported in human basal and squamous cell carcinomas compared with normal human facial skin, indicating its potential role in skin cancer and osteosarcomas. 20
Despite numerous evidences demonstrating the involvements of Panxs in various tumors, the underlying mechanism remains unknown, especially in gliomas. In present study, we are trying to address this issue through bioinformatic approach. In recent years, the public cancer datasets provide favorable tools in oncological researches, allowing to conduct integrated bioinformatic analyses to expand our knowledge of Panxs in tumorigenesis and uncover the functional cellular networks. Hence, we first employed several databases, such as Oncomine to gain a general idea regarding the expression pattern of different Panx isoforms in multiple cancers. Then PANX2 which was closely associated with brain low grade gliomas (LGG) became our focus. Subsequent analyses were carried out to reveal its role in glioma genesis and clinical prognosis. Finally, considering Panxs have been well reported to be critical in inflammation,21,22 we investigated the potential relationship between PANX2 and tumor-infiltrating immune cells in LGG microenvironment as well. Our findings represent a novel area for bridging our current gaps in knowledge with respect to the function of PANX2 in LGG.
Methods
Oncomine database
Oncomine datasets (https://www.oncomine.org/) provides robust, peer-reviewed analysis methods, and a powerful set of analysis functions that compute gene expression signatures, clusters, and gene-set modules, automatically extracting biological insights from the data. We used Oncomine datasets to analyze the mRNA expression of PANX family (PANX1, PANX2, and PANX3) in different cancers. Clinical tumor samples were compared with that in normal controls. The thresholds of p value and fold change were defined as 0.05 and 1.5, respectively.
CCLE database
Cancer Cell Line Encyclopedia (CCLE, https://www.broadinstitute.org/ccle) provides public access to genomic data, analysis, and visualization for more than 1000 cell lines. 23 Herein, only PANX2 in glioma cell lines was assessed by the CCLE dataset and visualized through GENE-E (https://software.broadinstitute.org/GENE-E/download.html) in order to gain the knowledge regarding expression of this molecule.
GEPIA
The expression of PANX2 in glioblastomas (GBM) and LGG compared to normal tissue was tested by Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn), which is an interactive web tool for analyzing the RNA sequencing expression data of 9736 tumors and 8587 normal samples from the Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) projects. 24
LinkedOmics database
LinkedOmics (http://www.linkedomics.orglogin.php) is publicly available portal that includes multi-omics data from all 32 TCGA cancer types. 25 This online tool provides a unique platform for biologists and clinicians to access, analyze and compare cancer multi-omics data within and across tumor types. PANX2-related genes in LGG were acquired from this database with the selection of Pearson correlation test and the top three positive and negative PANX2-related genes were both confirmed by GEPIA. The survival analyses of these two brain tumors correlated to PANX2 expression were primarily estimated by this tool as well.
COX regression analysis and receiver operating characteristic (ROC) curve
RNA expression data and clinical information of 509 LGG patients were download from the TCGA database (https://portal.gdc.cancer.gov/). The clinical features used in the analyses included gender, age (>60/≤60), tumor type, tumor grade, and radiation treatment. After data preprocessing, univariate and multivariate Cox regression were performed through package “survival” of “R” to further demonstrate the association between PANX2 expression and patient survival. p < 0.05 was considered as significance. The sensitivity and specificity of the PANX2 prognostic model to predict clinical outcome were evaluated by calculating the area under curve (AUC) of the ROC curve in the R package of “pROC.”
CBioPortal
The cBioPortal for Cancer Genomics (http://cbioportal.org) provides an open web resource for exploring, visualizing, and analyzing multidimensional cancer genomics and clinical data. The public instance of this platform hosts more than 200 cancer genomics studies, including all of the data from TCGA. 26 Through this web tool, we verified the correlations between PANX2 and PANX2-related genes in LGG.
GO enrichment and KEGG pathway analyses of PANX2-related genes in LGG
To gain more insights concerning PANX2 function in the context of LGG, some of the PANX2-related genes obtained from LinkedOmics were chosen as key genes and further carried out GO enrichment and KEGG pathway analyses by using an online tool: Database for Annotation, Visualization and Integration Discovery (DAVID, https://david.ncifcrf.gov). |Pearson correlation| ≥ 0.45 and p-value ≤ 0.01 were set as the cut-off criteria. The top 15 records of GO analysis and KEGG pathways for hub genes were visualized by “Ggplot2” package and “ClusterProfiler” package, respectively.
Construction of protein-protein interaction (PPI) network
PPI network of PANX2-related key genes was evaluated through the online search tool-STRING website (Search Tool for the Retrieval of Interacting Genes, https://string-db.org). The plug-in Molecular Complex Detection (MCODE) of Cytoscape was employed to check the modules and to visualize the protein interactions. The criteria of selection were as follows: degree cutoff = 2, max. depth = 100, k-core = 2, and node score cutoff = 0.2. The hub genes of highest module were re-analyzed with GO enrichment and KEGG pathway.
Timer
Tumor Immune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/) is a comprehensive resource for systematical analysis of immune infiltrates across diverse cancer types, which allows users to input function-specific parameters, with resulting figures dynamically displayed to conveniently access the tumor immunological, clinical, and genomic features.27,28 Gene module was employed to analyze the correlation of PANX2 expression with the abundance of immune infiltrates in LGG, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells. Moreover, correlations between PANX2 expression and gene markers of various tumor-infiltrating immune cells were test via Correlation modules. These markers included: CD19, CD79A (B cells); CD8A, CD8B (CD8+ T cells); CD3D, CD3E (T cells general); NOS2, IRF5, PTGS2 (Macrophages M1); CD163, VSIG4, MS4A4A (Macrophages M2); EACAM8, ITGAM, CCR7 (neutrophils) and HLA-DBP1, HLA-DQB1, HLA-DRA (dendritic cells). The Spearman’s correlation and the estimated statistical significance were presented in the diagram and all the gene expression levels were displayed with log2 RSEM.
Statistical analysis
Correlation between two groups was determined using Pearson correlation coefficient analysis. p < 0.01 was considered to indicate a statistically significant value except for special instruction.
Results
The mRNA expression of Panx family in diverse types of tumors
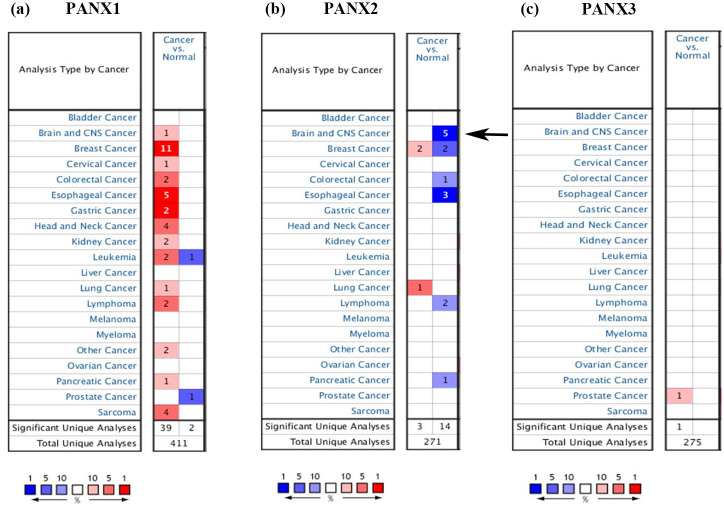
We first employed Oncomine databases to gain an overall understanding of Panx family in multiple kinds of cancers. Figure 1(a) to (c) showed that PANX1 was up-regulated in breast cancer, esophageal cancer, head and neck cancer, and sarcoma; Less PANX2 was observed in brain and CNS cancer, esophageal cancer, breast cancer, and lymphoma; The altered expression of PANX3 only presented in prostate cancer. As listed in Table 1, in terms of brain and CNS cancer, increased PANX1 expression was discovered in dataset of Liang Brain, 29 in which PANX1 was overexpressed in oligoastrocytoma patients with a fold change of 1.682. In Bredel Brain 2 dataset, 30 PANX1 had a fold change of 2.443 in anaplastic oligodendroglioma patients, a fold change of 2.020 in oligodendroglioma patients and a fold change of 2.405 in glioblastoma patients. In French Brain dataset, 31 enhanced PANX1 was also discovered with a fold change of 2.069 in anaplastic oligodendroglioma patients and a fold change of 1.887 in anaplastic oligoastrocytoma patients. PANX2 levels were down-regulated in datasets below: Bredel Brain 2 dataset showed that PANX2 had a fold change of −6.018 in anaplastic oligodendroglioma patients, a fold change of −6.021 in glioblastoma patients, a fold change of −4.866 in anaplastic oligoastrocytoma patients, and a fold change of −5.237 in oligodendroglioma patients. Data from Sun Brain 32 indicated PANX2 was low-expressed with a fold change of −3.240 in patients of diffuse astrocytoma. However, there was no obvious change of PANX3 based on Oncomine database. Given that the altered PANX2 was involved in brain and CNS cancer with more significance, this molecule turned into our focus for the subsequent analyses.
Figure 1.
The transcriptional expression of Pannexin family in different types of tumors from Oncomine database. The arrow indicates that decreased PANX2 are mainly discovered in tissues of brain and CNS cancers. (a) PANX1 expression in various cancers; (b) PANX2 expression in various cancers; (c) PANX3 expression in various cancers.
Table 1.
The changes of PANX expression in brain and CNS cancer in ONCOMINE database.
| Gene | Type of tumor vs control sample | Fold change | p Value | t-test | References |
|---|---|---|---|---|---|
| PANX1 | Oligoastrocytoma (n = 3) vs postmortem specimens of normal brains from Stanford University Hospital (n = 2) | 1.682 | 0.021 | 3.589 | Liang Brain |
| Anaplastic oligoastrocytoma (n = 6) vs normal brain from Stratagene (n = 4) | 2.443 | 0.019 | 3.345 | Bredel Brain 2 | |
| Oligodendroglioma (n = 5) vs normal brain from Stratagene (n = 4) | 2.020 | 0.048 | 1.922 | Bredel Brain 2 | |
| Glioblastoma (n = 27) vs normal brain from Stratagene (n = 4) | 2.405 | 0.017 | 3.150 | Bredel Brain 2 | |
| Anaplastic oligodendroglioma (n = 23) vs control brain samples, 4 from whole cortex and 2 from white matter only (n = 6) | 2.069 | 0.002 | 3.993 | French Brain | |
| Anaplastic oligoastrocytoma (n = 4) vs. control brain samples, 4 from whole cortex and 2 from white matter only (n = 6) | 1.887 | 0.024 | 2.444 | French Brain | |
| PANX2 | Anaplastic oligodendroglioma (n = 3) vs normal brain from Stratagene (n = 4) | −6.018 | 2.55e−5 | −15.087 | Bredel Brain 2 |
| Glioblastoma (n = 27) vs normal brain from Stratagene (n = 4) | −6.021 | 3.18e−11 | −13.031 | Bredel Brain 2 | |
| Anaplastic oligoastrocytoma (n = 6) vs normal brain from Stratagene (n = 4) | −4.866 | 1.71e−4 | −7.018 | Bredel Brain 2 | |
| Oligodendroglioma (n = 5) vs normal brain from Stratagene (n = 4) | −5.237 | 0.001 | −6.144 | Bredel Brain 2 | |
| Diffuse astrocytoma (n = 7) vs nontumor brain tissues from epilepsy surgery (n = 23) | −3.240 | 0.003 | −3.499 | Sun Brain |
The confirmation of decreased PANX2 in gliomas
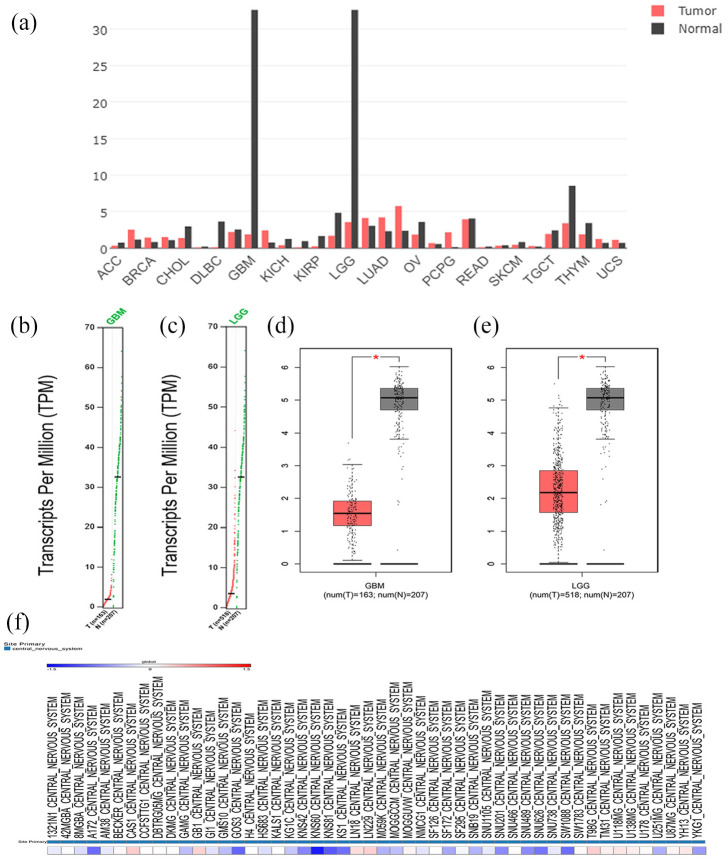
We next confirmed the mRNA expression of PANX2 in GBM and LGG through GEPIA. PANX2 was both significantly decreased in these two brain tumors compared to normal tissues (Figure 2(a)–(e)). Additionally, via capitalizing CCLE, we found that PANX2 was down-regulated in several cell lines of glioma, especially in KNS60, KNS81, and KNS42 (Figure 2(f)).
Figure 2.
The expression of PANX2 in gliomas tissues and cell lines: (a) the expression of PANX2 in pan-cancer. Down-regulated PANX2 is discovered in GBM and LGG compared to normal tissues, (b) PANX2 expression in GBM, (c) PANX2 expression in LGG, (d) the boxplot of PANX2 expression in GBM, (e) the boxplot of PANX2 expression in LGG, and (f) the expression of PANX2 in glioma cell lines assessed by CCLE.
The prognostic value of PANX2 in GBM and LGG
To assess the influence of PANX2 expression on clinical prognosis in GBM and LGG, we preliminarily investigated the value of overall survival using LinkedOmics. It is noteworthy that higher PANX2 was associated with better OS in LGG only (Figure 3(b)). As for GBM, lower PANX2 was likely to be related to better OS, but with no significance according to our threshold (Figure 3(a)). Univariate COX regression analysis also showed that PANX2 expression pattern had a significant association with LGG OS, whereas multivariate Cox analysis identified that PANX2 could become an independent prognosis factor for LGG (Table 2). However, ROC curves revealed the prognosis significance of PANX2 with AUC of 0.605, suggesting that the prognostic value of PANX2 might be limited (Figure 3(c)).
Figure 3.
The prognostic values of PANX2 in gliomas: (a) the prognostic value of mRNA level of PANX2 in GBM, analyzed by LinkedOmic, (b) the prognostic value of mRNA level of PANX2 in LGG, analyzed by LinkedOmic, and (c) the ROC curve of PANX2.
Table 2.
Univariate and multivariate Cox regression analyses of LGG prognostic factors.
| Characteristic | Univariate Cox |
Multivariate Cox |
||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95% CI | p-Value | Hazard ratio | 95% CI | p-Value | |
| PANX2 | 2.32 | 1.50–3.58 | 1.6e−4 | 2.43 | 1.17–5.06 | 0.017 |
| Gender | 1.09 | 0.73–1.65 | 0.67 | – | – | – |
| Age | 5.78 | 3.47–9.61 | 1.57e−11 | 5.30 | 2.36–11.94 | 5.45e−5 |
| Tumor type | 40.32. | 5.61–289.8 | 2.4e−4 | 12.54 | 1.70–92.33 | 0.013 |
| Tumor grade | 3.47 | 2.20–5.48 | 8.35e−8 | 3.55 | 1.59–9.51 | 0.003 |
| Radiation treatment | 2.07 | 1.07–4.01 | 0.03 | 0.87 | 0.40–1.92 | 0.74 |
PANX2-related genes in LGG
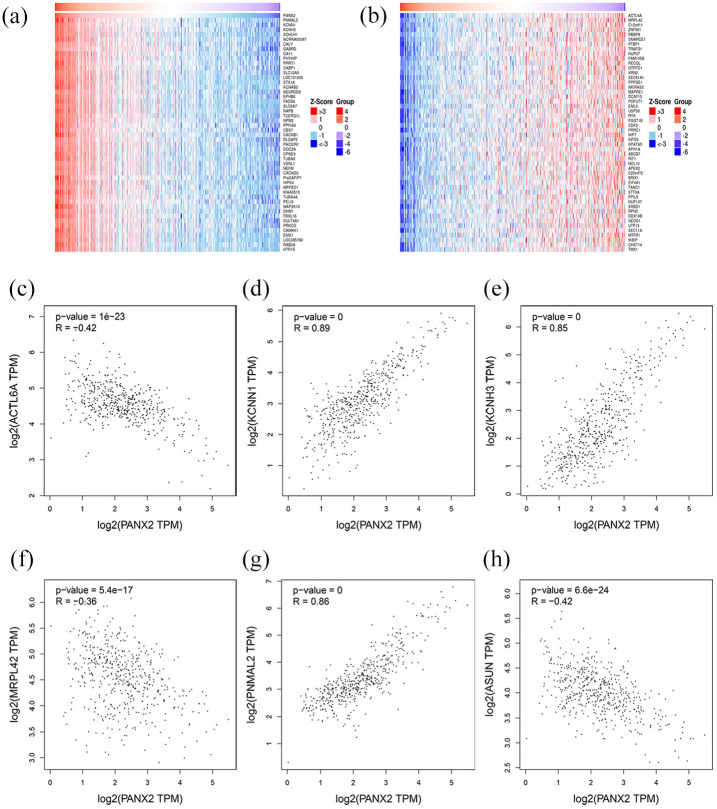
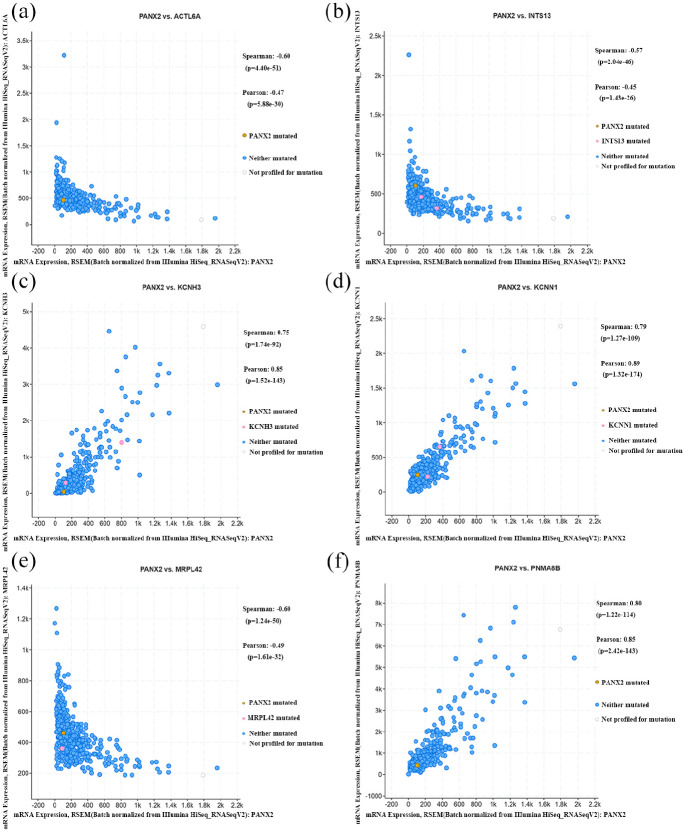
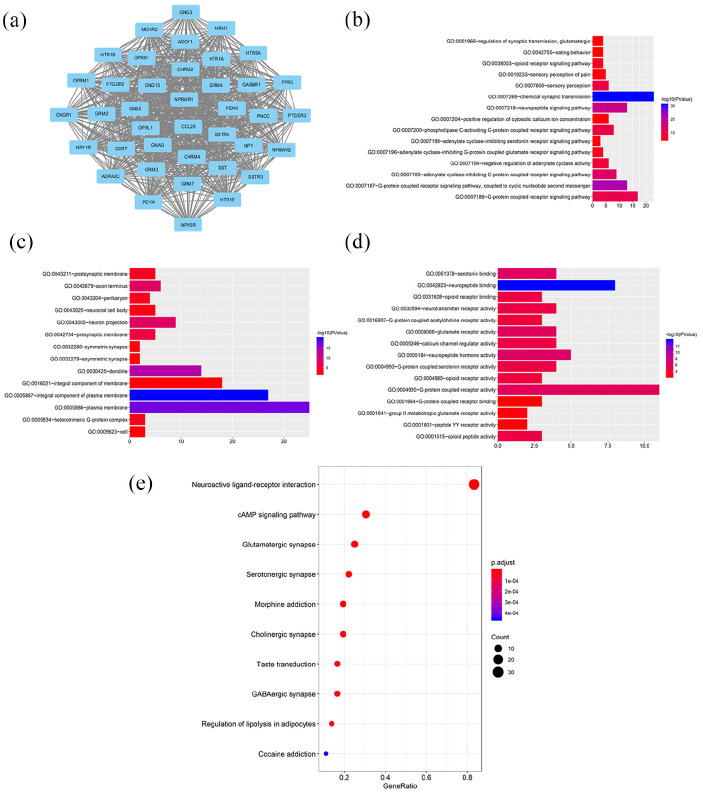
Then, the analysis concentrating on PANX2-related genes was carried out through LinkedOmics. A total of 1619 genes were identified as PANX2-related genes based on our criteria mentioned above, including 1357 positively correlated genes and 262 negatively correlated genes. Partial genes were presented in Figure 4(a) and (b) and the entire list was in Supplemental Table 1. Panx2 was positively associated with PNMAL2 (PNMA8B), KCNN1, KCNH3, SOHLH1, NCRNA00087, etc. and was negatively associated with ACTL6A, MRPL42, C12orf11 (INTS13, ASUN), ZNF561, RBBP8, etc. We also verified top three genes with most significance using GEPIA dataset (Figure 4(c)–(h)) and cBIOportal database (Figure 5(a)–(f)). Then, a functional enrichment analysis of these PANX2-related genes revealed that they were mainly involved in signal transduction, plasma membrane, and ATP binding as well as pathways of neuroactive ligand-receptor interaction, calcium signaling, and cAMP signal (Supplemental Tables 2 and 3). In the PPI network of PANX2-related genes shown in Figure 6(a), the highest module was composed of 39 nodes and 741 interactive edges and the hub genes from this module were enriched in chemical synaptic transmission, G-protein coupled receptor activity and plasma membrane as well as pathways of neuroactive ligand-receptor interaction, glutamatergic synapse, and cAMP signaling pathway (Figure 6(b)–(e), Supplemental Tables 4 and 5).
Figure 4.
PANX2-related genes in LGG: (a) genes which are positively related with PANX2, (b) genes which are negatively related with PANX2, and (c–h) the confirmations of top PANX2-related genes by GEPIA.
Figure 5.
The confirmations of top PANX2-related genes: (a–f) the genes which had positive and negative associations with PANX2 were further validated by cBioPortal.
Figure 6.
GO, KEGG, and PPI analyses of hub genes: (a) the highest module from PPI analysis composed by hub genes, (b–d) the top 15 terms of GO categories of biological process (BP), cellular component (CC) and molecular function MF, respectively, and (e) KEGG pathway analysis of hub genes.
The size of each spot represents the gene number, while the color represents the p-value. p-Value <0.01 was considered significantly.
PANX2 may correlated with immune infiltration in LGG
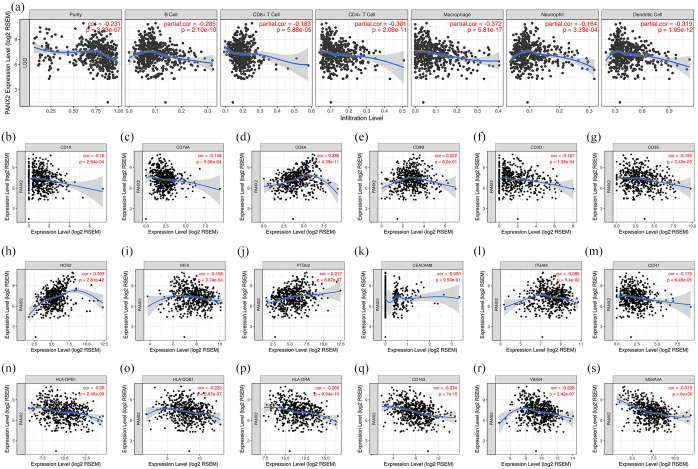
Given the well-documented roles of Panx family in inflammation, we wondered whether PANX2 would be related to immune infiltration levels in LGG. The findings from TIMER demonstrated that PANX2 expression had significant negative correlations with various immune cells, especially with macrophage (partial. Cor = −0.372, p = 5.81e−17), dendritic cell (partial. Cor = −0.319, p = 1.05e−12), and CD4+ T cell (partial. Cor = −0.301, p = 2.09e−11) (Figure 7(a)). As shown in Figure 7(b) to (s), the assessment of correlations between gene markers of different immune cells and PANX2 expression discovered that PANX2 was negatively associated with CD19 and CD79A (gene markers of B cells), CD8A (gene marker of CD8+ T cells), CD3D and CD3E (gene markers of general T cells), NOS2, IRF5, and PTGS2 (gene markers of macrophage M1), CD163, VSIG4, and MS4A4A (gene markers of macrophage M2), CCR7 (gene marker of neutrophil), HLA-DPB1, HLA-DQB1, and HLA-DRA (gene markers of dendritic cells). 33 Therefore, these information implied that PANX2 might play a fundamental role in the LGG immune microenvironment.
Figure 7.
PANX2 is associated with immune infiltrates in the context of LGG: (a) the relationship between PANX2 and different types of immune cells; (b–s) the relationship between PANX2 and gene markers of immune cells.
Discussion
The ectopic expressions of Panxs have been reported in many human diseases, ranging from infectious disorders to cancers. Panxs are vital to cell behaviors regulation, including cell proliferation, differentiation and migration. Hence, the dysfunction of Panxs probably contributes to the loss of control in cell growth, which facilitates carcinogenesis. However, there is still a huge knowledge gap as to the role of Panx in tumor. As far as we know, the present study is the first report focusing on how Panxs member (PANX2) might be involved in glioma occurrence through bioinformatics analysis using public databases. Herein, our findings indicated the transcriptional expression pattern of Panx isoforms in multiple types of neoplasms. The over-expression of PANX1 was prevalent in tumors compare to normal tissues with the exception of leukemia and prostate cancer. Most reports of increased PANX1 were from breast cancers, in which this change was frequently considered as a tumor-promoting effect. Bioinformatic and lab work both confirmed excess PANX1 was associated with metastasis via influencing the shift toward the phenotype of epithelial-to-mesenchymal in breast cancer patients. 34 The mutation of PANX1 discovered in metastatic breast cancer cell lines likely favored tumor cell survival during extravasation. 35 Intriguingly, the absence of PANX1 in rat C6 gliomas led to tumorigenesis, implying its potential anti-tumor activity. 36 On the other hand, the decreased expression of PANX2 was observed in tumors compare to normal tissues with the exception of breast cancer and lung cancer. A down-regulation of human PANX2 transcripts was discovered via Affymetrix chip analysis, particularly in GBM. 17 The trend in C6 glioma cells was consistent with decreased PANX2 in other human glioma cell lines, whereas over-expressed PANX2 tagged green fluorescent protein (GFP) significantly inhibited tumor growth in nude mice compared to controls. 37 The observation in hepatocellular carcinomas (HCC) also supported PANX2 as a tumor suppressor. 18 Moreover, the study concerning PANX3 in oncogenesis is still limited. Less PANX3 was previously discovered in human keratinocyte-derived basal cell carcinomas and squamous cell carcinomas, 20 which was not in line with what we found in prostate cancer from Oncomine datasets. 38 All these findings suggested complicated roles of Panx isoforms in different types of cancers, which is surely worth to gain more attention.
We next focused on PANX2 and gliomas in the subsequent analyses. Based on criteria from World Health Organization (WHO), gliomas are classified on scale of I to IV, including LGG (WHO grade II), such as astrocytoma and oligodendroglioma, and high-grade gliomas (HGG, WHO grade III and IV), such as GBM.39,40 In present study, we assessed the prognostic values of PANX2 in LGG and GBM through GEPIA, and further discovered that PANX2 could become a relative independent factor for clinical outcomes predictions in LGG. It has been demonstrated that PANX2 level was associated with post diagnosis survival for patients of glial tumors based on another brain cancer gene expression database REMBRANDT. 17 However, our analysis discovered that higher PANX2 could only predict better OS in LGG, but not in GBM. Considered that less PANX2 was found in HGG than in LGG, 41 we speculated that the roles of PANX2 might be various in different grades of gliomas. To clarify the mechanism of PANX2 in LGG, we further mined the correlated genes of PANX2 and carried out GO functional analysis. The abnormal PANX2 might cause dysfunctions of these genes and trigger cascades with respect to signal transduction, plasma membrane and ATP binding, promoting glioma progression or metastasis. Albeit lack of evidence regarding the direct interplay of PANX2 and PANX1 in oncogenesis, PANX2 was demonstrated to form membrane-bound vesicles and to be recruited by PANX1 for eliciting its activity in a plasma membrane channel-independent fashion.42,43 We also noticed that multiple PANX2-related genes highly enriched in “calcium signaling” in KEGG pathway analysis. Acting as a critical second messenger, calcium is involved in quiescence, maintenance, proliferation, and migration in glioma cells, while the ionotropic ATP-gated P2X7 receptor, a well-known PANX1 interactor, plays a fundamental role in this process.44,45 Therefore, whether PANX2 is involved in lower grade glioma genesis through the crosstalk with PANX1 and P2X7 receptor should be taken into account for at least a component of the mechanism. Moreover, PPI network and module analysis found that the hub genes were chiefly associated with chemical synaptic transmissions and neuropeptide binding. Serving as a regulator of chemical synaptic transmission, genes encoding metabotropic glutamate receptors (mGluRs) became novel oncogenes of gliomas. 46 Additionally, neuropeptide has been recognized as emerging molecular targets for glioma therapy. 47 Taken together, keep monitoring these molecular pathways may be of great importance in clarifying PANX2’s role in LGG occurrence and progression.
The microenvironment orchestrated by carcinomas and adjacent cells is essential to tumor formation and development, with at least 25% of cancers associated with it. 48 Infiltrated inflammatory immune cells are part of this complex ecosystems, whereas chronic inflammation is frequently considered as one of the critical hallmarks of cancer. 49 As an ATP release channels, the involvement of Panxs in regulating inflammation has been identified in multiple literatures.50–52 In multiple cell types of neurological system, including macrophages, microglia, neurons and astrocytes, PANX1 was well reported to activate inflammasomes.53–56 Hence, we hypothesized that as a Panx family member, PANX2 might be associated with tumor-infiltrated cells in LGG. Based on our analysis, PANX2 expression had significant negative correlation with various immune cells, particularly macrophage. Gene markers of pro-inflammatory macrophages M1, such as NOS2, PTGS2 and IRF5 showed relatively weaker correlations with PANX2 expression compared to alternatively-activated immunoregulatory M2 macrophage markers, such as CD163, VSIG4, and MS4A4A, while M2 macrophages are frequently predicted to be pro-tumoral instead of anti-tumoral. 57 These results disclosed the potential regulating role of PANX2 in polarization of tumor-associated macrophages (TAM). Moreover, the decreased PANX2 might led to the enhanced number of several types of immune cells in the preliminary phase of LGG to eliminate cancer cells. Nevertheless, the survival cells which were able to escape the immunological recognitions would inversely favor tumor progression, namely “immune evasion.” Apart from the findings mentioned above, the limitation of the current project is that all data analyzed were retrieved from online databases. Thereby experimental validations are necessary to further investigate the potential mechanisms of PANX2 in LGG occurrence in the future researches using animal models or cell lines.
Conclusions
The present study provided a thorough understanding of the Panx family expression in distinct cancers and shed light on the importance of PANX2 in LGG. We discovered that PANX2-related genes in the context of LGG were highly enriched in several pathways, such as calcium signaling. Synaptic transmission and neuropeptide signaling were both highlighted in PANX2 function. Besides, PANX2 likely played a critical role in immune cell infiltration in LGG microenvironments, implying new perspectives for exploring the underlying mechanism of LGG formation. In summary, this work makes important contributions to the understanding of the cellular roles of PANX2 in LGG and suggests that PANX2 might be a potential therapeutic target of LGG.
Supplemental Material
Supplemental material, sj-docx-1-sci-10.1177_00368504211011836 for PANX2 and brain lower grade glioma genesis: A bioinformatic analysis by XiaoXue Xu, YueHan Hao, Shuang Xiong and ZhiYi He in Science Progress
Supplemental material, sj-docx-2-sci-10.1177_00368504211011836 for PANX2 and brain lower grade glioma genesis: A bioinformatic analysis by XiaoXue Xu, YueHan Hao, Shuang Xiong and ZhiYi He in Science Progress
Supplemental material, sj-docx-3-sci-10.1177_00368504211011836 for PANX2 and brain lower grade glioma genesis: A bioinformatic analysis by XiaoXue Xu, YueHan Hao, Shuang Xiong and ZhiYi He in Science Progress
Supplemental material, sj-docx-4-sci-10.1177_00368504211011836 for PANX2 and brain lower grade glioma genesis: A bioinformatic analysis by XiaoXue Xu, YueHan Hao, Shuang Xiong and ZhiYi He in Science Progress
Supplemental material, sj-docx-5-sci-10.1177_00368504211011836 for PANX2 and brain lower grade glioma genesis: A bioinformatic analysis by XiaoXue Xu, YueHan Hao, Shuang Xiong and ZhiYi He in Science Progress
Acknowledgments
We deeply appreciate Professor Qiwen Xie for his assistance of study design, data analysis, and manuscript revision.
Author biographies
XiaoXue Xu, PhD, is an Associate professor. Research interests include Neuropeptide in epileptogenesis, the role of Pannexin 1 (Panx1) channel in the physiological and pathological processes, and non-coding RNA in neurological diseases.
YueHan Hao, MD of Neurology. Research interests include the neuropathology of autoimmune disorders, such as multiple sclerosis and mechanism of low grade glioma.
Shuang Xiong’s major research interest is pharmaceutical synthesis of novel medicines for neurological diseases and brain tumors.
ZhiYi He, MD, PhD, Professor Director of Department of Neurology, Research fields include cerebrovascular disease, neurodegenerative disease, and brain tumor control and treatment, etc.
Footnotes
The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was kindly supported by National Natural Science Foundation of China (81601129) and Science Foundation of Shenyang (F16-205-1-48).
ORCID iD: XiaoXue Xu  https://orcid.org/0000-0003-1831-3657
https://orcid.org/0000-0003-1831-3657
Supplemental material: Supplemental material for this article is available online.
References
- 1.Esseltine JL, Laird DW. Next-Generation Connexin and Pannexin Cell Biology. Trends Cell Biol 2016; 26(12): 944–955. [DOI] [PubMed] [Google Scholar]
- 2.Penuela S, Gehi R, Laird DW. The biochemistry and function of pannexin channels. Biochim Biophys Acta 2013; 1828(1): 15–22. [DOI] [PubMed] [Google Scholar]
- 3.Graham S, Jiang J, Mesnil M. Connexins and pannexins: important players in tumorigenesis, metastasis and potential therapeutics. Int J Mol Sci 2018; 19(6): 1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyce AKJ, Epp AL, Nagarajan A, et al. Transcriptional and post-translational regulation of pannexins. Biochim Biophys Acta 2018; 1860(1): 72–82. [DOI] [PubMed] [Google Scholar]
- 5.Xu X, Wicki-Stordeur LE, Sanchez-Arias JC, et al. Probenecid Disrupts a Novel Pannexin 1-Collapsin Response Mediator Protein 2 Interaction and Increases Microtubule Stability. Front Cell Neurosci. 2018; 12: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scemes E, Spray DC, Meda P. Connexins, pannexins, innexins: novel roles of “hemi-channels”. Pflugers Archiv Eur J Physiol 2009; 457(6): 1207–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sosinsky GE, Boassa D, Dermietzel R, et al. Pannexin channels are not gap junction hemichannels. Channels 2011; 5(3): 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baranova A, Ivanov D, Petrash N, et al. The mammalian pannexin family is homologous to the invertebrate innexin gap junction proteins. Genomics 2004; 83(4): 706–716. [DOI] [PubMed] [Google Scholar]
- 9.Le Vasseur M, Lelowski J, Bechberger JF, et al. Pannexin 2 protein expression is not restricted to the CNS. Front Cell Neurosci 2014; 8: 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swayne LA, Sorbara CD, Bennett SAL. Pannexin 2 is expressed by postnatal hippocampal neural progenitors and modulates neuronal commitment. J Biol Chem 2010; 285(32): 24977–24986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bond SR, Lau A, Penuela S, et al. Pannexin 3 is a novel target for Runx2, expressed by osteoblasts and mature growth plate chondrocytes. J Bone Miner Res 2011; 26(12): 2911–2922. [DOI] [PubMed] [Google Scholar]
- 12.Penuela S, Bhalla R, Gong XQ, et al. Pannexin 1 and pannexin 3 are glycoproteins that exhibit many distinct characteristics from the connexin family of gap junction proteins. J Cell Sci 2007; 120(21): 3772–3783. [DOI] [PubMed] [Google Scholar]
- 13.Jiang JX, Penuela S. Connexin and pannexin channels in cancer. BMC Cell Biol 2016; 17(S1): 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, Yuan M, Yao Y, et al. In vitro effect of pannexin 1 channel on the invasion and migration of I-10 testicular cancer cells via ERK1/2 signaling pathway. Biomed Pharmacother 2019; 117: 109090. [DOI] [PubMed] [Google Scholar]
- 15.Freeman T, Sayedyahossein S, Johnston D, et al. Inhibition of pannexin 1 reduces the tumorigenic properties of human melanoma cells. Cancers 2019; 11(1): 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyd-Tressler A, Penuela S, Laird DW, et al. Chemotherapeutic drugs induce ATP release via caspase-gated pannexin-1 channels and a caspase/pannexin-1-independent mechanism. J Biol Chem 2014; 289(39): 27246–27263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Litvin O, Tiunova A, Connell-Alberts Y, et al. What is hidden in the pannexin treasure trove: the sneak peek and the guesswork. J Cell Mol Med 2006; 10(3): 613–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie C-R, Sun H, Wang F-Q, et al. Integrated analysis of gene expression and DNA methylation changes induced by hepatocyte growth factor in human hepatocytes. Mol Med Rep 2015; 12(3): 4250–4258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim KM, Hussein UK, Bae JS, et al. The expression patterns of FAM83H and PANX2 are associated with shorter survival of clear cell renal cell carcinoma patients. Front Oncol 2019; 9: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowan KN, Langlois S, Penuela S, et al. Pannexin1 and Pannexin3 exhibit distinct localization patterns in human skin appendages and are regulated during keratinocyte differentiation and carcinogenesis. Cell Commun Adhes 2012; 19(3–4): 45–53. [DOI] [PubMed] [Google Scholar]
- 21.Makarenkova H, Shah SB, Shestopalov VI. The two faces of pannexins: new roles in inflammation and repair. J Inflamm Res 2018; 11: 273–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makarenkova HP, Shestopalov VI. The role of pannexin hemichannels in inflammation and regeneration. Front Physiol 2014; 5: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barretina J, Caponigro G, Stransky N, et al. 22 The cancer cell line encyclopedia – using preclinical models to predict anticancer drug sensitivity. Eur J Cancer 2012; 48: S5–S7. [Google Scholar]
- 24.Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017; 45(W1): W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018; 46(D1): D956–D963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu P, Heins ZJ, Muller JT, et al. Integration and analysis of CPTAC proteomics data in the context of cancer genomics in the cBioPortal. Mol Cell Proteomics 2019; 18(9): 1893–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017; 77(21): e108–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, Severson E, Pignon J-C, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 2016; 17(1): 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang Y, Diehn M, Watson N, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci USA 2005; 102(16): 5814–5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bredel M, Bredel C, Juric D, et al. Functional network analysis reveals extended gliomagenesis pathway maps and three novel MYC-interacting genes in human gliomas. Cancer Res 2005; 65(19): 8679–8689. [DOI] [PubMed] [Google Scholar]
- 31.French PJ, Swagemakers SMA, Nagel JHA, et al. Gene expression profiles associated with treatment response in oligodendrogliomas. Cancer Res 2005; 65(24): 11335–11344. [DOI] [PubMed] [Google Scholar]
- 32.Sun L, Hui AM, Su Q, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 2006; 9(4): 287–300. [DOI] [PubMed] [Google Scholar]
- 33.Pan JH, Zhou H, Cooper L, et al. LAYN is a prognostic biomarker and correlated with immune infiltrates in gastric and colon cancers. Front Immunol 2019; 10: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jalaleddine N, El-Hajjar L, Dakik H, et al. Pannexin1 is associated with enhanced epithelial-to-mesenchymal transition in human patient breast cancer tissues and in breast cancer cell lines. Cancers 2019; 11(12): 1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Furlow PW, Zhang S, Soong TD, et al. Mechanosensitive pannexin-1 channels mediate microvascular metastatic cell survival. Nat Cell Biol 2015; 17(7): 943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai CPK, Bechberger JF, Thompson RJ, et al. Tumor-suppressive effects of pannexin 1 in C6 glioma cells. Cancer Res 2007; 67(4): 1545–1554. [DOI] [PubMed] [Google Scholar]
- 37.Lai CPK, Bechberger JF, Naus CC. Pannexin2 as a novel growth regulator in C6 glioma cells. Oncogene 2009; 28(49): 4402–4408. [DOI] [PubMed] [Google Scholar]
- 38.Arredouani MS, Lu B, Bhasin M, et al. Identification of the transcription factor single-minded homologue 2 as a potential biomarker and immunotherapy target in prostate cancer. Clin Cancer Res 2009; 15(18): 5794–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braunstein S, Raleigh D, Bindra R, et al. Pediatric high-grade glioma: current molecular landscape and therapeutic approaches. J Neurooncol 2017; 134(3): 541–549. [DOI] [PubMed] [Google Scholar]
- 40.Morshed RA, Young JS, Hervey-Jumper SL, et al. The management of low-grade gliomas in adults. J Neurosurg Sci 2019; 63(4): 450–457. [DOI] [PubMed] [Google Scholar]
- 41.Xu J, He J, Huang H, et al. MicroRNA-423-3p promotes glioma growth by targeting PANX2. Oncol Lett 2018; 16: 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Bruzzone R, Hormuzdi SG, Barbe MT, et al. Pannexins, a family of gap junction proteins expressed in brain. Proc Natl Acad Sci USA 2003; 100(23): 13644–13649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Penuela S, Harland L, Simek J, et al. Pannexin channels and their links to human disease. Biochem J 2014; 461(3): 371–381. [DOI] [PubMed] [Google Scholar]
- 44.Maklad A, Sharma A, Azimi I. Calcium signaling in brain cancers: roles and therapeutic targeting. Cancers 2019; 11(2): 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrone FB, Gehring MP, Nicoletti NF. Calcium channels and associated receptors in malignant brain tumor therapy. Mol Pharmacol 2016; 90(3): 403–409. [DOI] [PubMed] [Google Scholar]
- 46.Yu LJ, Wall BA, Wangari-Talbot J, et al. Metabotropic glutamate receptors in cancer. Neuropharmacol 2017; 115: 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roesler R, Brunetto AT, Abujamra AL, et al. Current and emerging molecular targets in glioma. Expert Rev Anticancer Ther 2010; 10(11): 1735–1751. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez H, Hagerling C, Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev 2018; 32(19–20): 1267–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coussens LM, Werb Z. Inflammation and cancer. Nature 2002; 420: 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gulbransen BD, Bashashati M, Hirota SA, et al. Activation of neuronal P2X7 receptor-pannexin-1 mediates death of enteric neurons during colitis. Nat Med 2012; 18: 600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pelegrin P. Targeting interleukin-1 signaling in chronic inflammation: focus on P2X(7) receptor and Pannexin-1. Drug News Perspect 2008; 21: 424–433. [DOI] [PubMed] [Google Scholar]
- 52.Kanneganti T-D, Lamkanfi M, Kim Y-G, et al. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of Toll-like receptor signaling. Immunity 2007; 26: 433–443. [DOI] [PubMed] [Google Scholar]
- 53.Pelegrin P, Surprenant A. Pannexin-1 couples to maitotoxin- and nigericin-induced interleukin-1beta release through a dye uptake-independent pathway. J Biol Chem 2007; 282: 2386–2394. [DOI] [PubMed] [Google Scholar]
- 54.Pelegrin P, Surprenant A. Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. EMBO J 2006; 25: 5071–5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albalawi F, Lu W, Beckel JM, et al. The P2X7 receptor primes IL-1β and the NLRP3 inflammasome in astrocytes exposed to mechanical strain. Front Cell Neurosci 2017; 11: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silverman WR, de Rivero Vaccari JP, Locovei S, et al. The pannexin 1 channel activates the inflammasome in neurons and astrocytes. J Biol Chem 2009; 284: 18143–18151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Domingues P, González-Tablas M, Otero Á, et al. Tumor infiltrating immune cells in gliomas and meningiomas. Brain Behav Immun. 2016; 53: 1–15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-sci-10.1177_00368504211011836 for PANX2 and brain lower grade glioma genesis: A bioinformatic analysis by XiaoXue Xu, YueHan Hao, Shuang Xiong and ZhiYi He in Science Progress
Supplemental material, sj-docx-2-sci-10.1177_00368504211011836 for PANX2 and brain lower grade glioma genesis: A bioinformatic analysis by XiaoXue Xu, YueHan Hao, Shuang Xiong and ZhiYi He in Science Progress
Supplemental material, sj-docx-3-sci-10.1177_00368504211011836 for PANX2 and brain lower grade glioma genesis: A bioinformatic analysis by XiaoXue Xu, YueHan Hao, Shuang Xiong and ZhiYi He in Science Progress
Supplemental material, sj-docx-4-sci-10.1177_00368504211011836 for PANX2 and brain lower grade glioma genesis: A bioinformatic analysis by XiaoXue Xu, YueHan Hao, Shuang Xiong and ZhiYi He in Science Progress
Supplemental material, sj-docx-5-sci-10.1177_00368504211011836 for PANX2 and brain lower grade glioma genesis: A bioinformatic analysis by XiaoXue Xu, YueHan Hao, Shuang Xiong and ZhiYi He in Science Progress