Abstract
Long non-coding RNAs (lncRNAs) play critical regulatory roles in various cellular and metabolic processes in mosquitoes and all other organisms studied thus far. In particular, their involvement in essential processes such as reproduction makes them potential targets for the development of novel pest control approaches. However, their function in mosquito biology remains largely unexplored. To elucidate the role of lncRNAs in mosquitoes’ reproduction and vector competence for arboviruses, we have implemented a computational and experimental pipeline to mine, screen, and characterize lncRNAs related to these two biological processes. Through analysis of publicly available Zika virus (ZIKV) infection-regulated Aedes aegypti transcriptomes, at least six lncRNAs were identified as being significantly upregulated in response to infection in various mosquito tissues. The roles of these ZIKV-regulated lncRNAs (designated Zinc1, Zinc2, Zinc3, Zinc9, Zinc10 and Zinc22), were further investigated by dsRNA-mediated silencing studies. Our results show that silencing of Zinc1, Zinc2, and Zinc22 renders mosquitoes significantly less permissive to ZIKV infection, while silencing of Zinc22 also reduces fecundity, indicating a potential role for Zinc22 in trade-offs between vector competence and reproduction. We also found that silencing of Zinc9 significantly increases fecundity but has no effect on ZIKV infection, suggesting that Zinc9 may be a negative regulator of oviposition. Our work demonstrates that some lncRNAs play host factor roles by facilitating viral infection in mosquitoes. We also show that lncRNAs can influence both mosquito reproduction and permissiveness to virus infection, two biological systems with important roles in mosquito vectorial capacity.
Author summary
Long non-coding RNAs (lncRNAs) emerged during the last decades as important regulators of diverse biological processes. Despite their significance in various organisms, their role is still largely unexplored in mosquitoes that transmit human pathogens. Some of their functions in processes related to vectorial capacity could be exploited for the development of novel disease control strategies. Here we provide a comprehensive analysis of lncRNAs that regulate vector competence and reproduction of the notorious mosquito disease vector, Ae. aegypti. We show that the expression of several lncRNAs is stimulated by infection with Zika virus and that silencing three of them render mosquitoes more resistant to the virus infection, possibly through an immunity-related function. Moreover, we provide evidence regarding their impact on mosquitoes’ reproduction, such as fecundity and fertility. We also highlight the influence of Zinc22, an ovary-enriched lncRNA, that affects both reproduction and virus restriction, indicating its potential role in metabolic trade-offs between these two systems. This is the first study to analyze the influence of lncRNAs in vectorial capacity and reproduction of disease vectors, and thereby breaks new ground for their potential use in novel mosquito control and pathogen transmission-blocking strategies.
Introduction
Aedes aegypti is a major mosquito vector of many arthropod-borne viruses (arboviruses), including dengue virus (DENV), Zika virus (ZIKV), and chikungunya virus (CHIKV). Arboviruses are mainly spread through a horizontal transmission cycle between an arthropod vector and a vertebrate host. Arboviruses that are transmitted by mosquito vectors enter the mosquito’s body after a viremic blood meal has been taken from a vertebrate host. First, they infect midgut epithelial cells, where they replicate and produce viral particles. They then escape the midgut basal lamina and disseminate to the hemocoel and finally infect salivary glands, from where they can be transmitted to another vertebrate host through saliva during the next blood meal [1]. The average extrinsic incubation period (EIP) of the major flaviviruses, such as DENV and ZIKV, ranges from 7 to 14 days.
Beyond the physical barriers that protect against pathogens, mosquitoes mainly rely on their innate immune system to defend against arboviral infections [2]. Anti-viral defenses can be regulated by at least five distinct signaling pathways: the RNA interference (RNAi), Toll, immune deficiency (IMD), Janus kinase-signal transducer and activator of transcription (JAK-STAT), and mitogen-activated protein kinase (MAPK) pathways [3–6]. RNAi is one of the best described antiviral mechanisms relying on Dcr2, R2D2 and Ago2, the components of the RNA-induced silencing complex (RISC) [7] that mediate cleavage of the viral RNA [8]. Some factors of the other antiviral pathways have also been characterized, such as DENV restriction factors 1 & 2 (DVRF1, DVRF2) of the JAK/STAT pathway [9] and Rel1 and MyD88 of the Toll pathway [5,10]. However, many aspects of insect immune response against arboviruses are still not well understood. Moreover, viruses rely on numerous mosquito-encoded host factors to establish their replication and infection. Even though several host factors, including a vATPase and IMPDH, have been described [4,11], this field is still largely unexplored.
Long non-coding RNAs (lncRNAs), non-coding transcripts with sequences longer than 200 nucleotides [12], have been associated with multiple cellular functions. These functions include, but are not limited to: (i) guiding transcription factors or other enzymes, (ii) decoying proteins, (iii) sponging up microRNAs (miRNAs), (iv) acting as precursors of miRNAs, and (v) functioning as scaffolds for ribonucleoprotein complexes (reviewed in [13]). There is also evidence that lncRNAs are involved in insect innate immunity. In Drosophila melanogaster, a lncRNA named viral suppressor of RNAi (VSR)-interacting RNA (VINR), regulates the innate immune response upon infection with the Drosophila C virus (DCV) [14]. The VSRs are virulence factors produced by viruses to evade RNAi defense mechanisms. In the absence of VINR, viral titers of DCV increase dramatically. Interestingly, once a dsRNA-binding VSR is detected, VINR activity leads to the upregulation of antimicrobial peptide (AMP) genes. Induced by Infection (IBIN) is a D. melanogaster lncRNA that has been found to be highly expressed after infection with Micrococcus luteus, a Gram-positive bacterium [15]. Its upregulation results in high expression of genes involved in the Toll pathway, such as the AMP Drosomycin and the Toll pathway target IM1. Its overexpression also results in the upregulation of Npc2e, a gene that is involved in antimicrobial responses, through the IMD signaling pathway [16].
While our understanding of lncRNA biology has advanced significantly through studies in D. melanogaster, their role in Ae. aegypti biology is still limited. A study that focused on the role of lncRNAs that are regulated in the Ae. aegypti Aag2 cells upon infection with dengue virus [17] showed that 421 lncRNAs were differentially expressed (DE), and RNAi-mediated silencing of lincRNA-1317 led to a slight enhancement of viral replication. Another study using the same cell line identified 3,035 differentially expressed lncRNAs in response to infection with Wolbachia. Silencing of two of them (aae-lnc-7598 and aae-lnc-0165) revealed their influence on mosquito permissiveness to dengue virus. Specifically, aae-lnc-7598 expression modulated the antioxidant catalase 1B (CAT1B), which enhances production of reactive oxygen species (ROS), and aae-lnc-0165 activated REL1, a key regulator of the Toll pathway [18]. It is also likely that lncRNAs can influence infection by regulating the expression of virus host factors.
Beyond their role in immunity, lncRNAs have also been shown to regulate reproduction and may mediate trade-offs between reproduction and both vector competence and immunity. In many cases, increased reproductive effort results in reduced immunity and, reciprocally, infection and activation of the immune system reduce reproductive output. Several such findings have been reported in dipteran species, especially D. melanogaster [19–25] and the mosquitoes Ae. aegypti [26], An. gambiae [27,28] and An. stephensi [29]. Trade-offs are manifested by signaling mechanisms that are mainly regulated by a balance of the hormones 20-hydroxyecdysone (20E) and Juvenile hormone (JH) [30,31]. JH and 20E regulate the development of oogenesis and have been connected to ovarian follicle maturation and vitellogenesis [32]. JH and 20E also influence, in opposite ways, insect immunity. In D. melanogaster, JH has a negative effect, as it reduces the activation of AMPs [33], while 20E enhances the transcription of AMPs [33–36] and drives the expression of the PGRP-LC protein, which is an activator of the IMD immune pathway [37]. In Ae. aegypti, studies have revealed that JH and 20E modulate vitellogenesis and have a negative impact on both the expression of AMPs [38] and stimulation of the IMD pathway [39], respectively.
It is possible that lncRNAs involved in regulating vector competence of mosquitoes to arbovirus infection could be used to develop transmission-blocking strategies. Available transcriptomic data indicate that several lncRNAs are differentially expressed after infection with various arboviruses. However, published work that has aimed to characterize the role of specific lncRNAs in modulating virus infection has focused exclusively on the effect of infection in cell lines, and published in vivo data have been lacking regarding the influence of lncRNAs on the circulation and the infectivity of the virus in mosquitoes. Thus, the present study has been designed to identify specific lncRNAs that can regulate mosquito permissiveness to virus infection, possibly through antiviral immunity or other mechanisms, and assess the influence of such lncRNAs on female reproduction. These lncRNAs are likely to play roles in reproduction-vector competence and immunity trade-offs, and thereby represent potential disease control targets.
Results
Classification of lncRNAs in Ae. aegypti
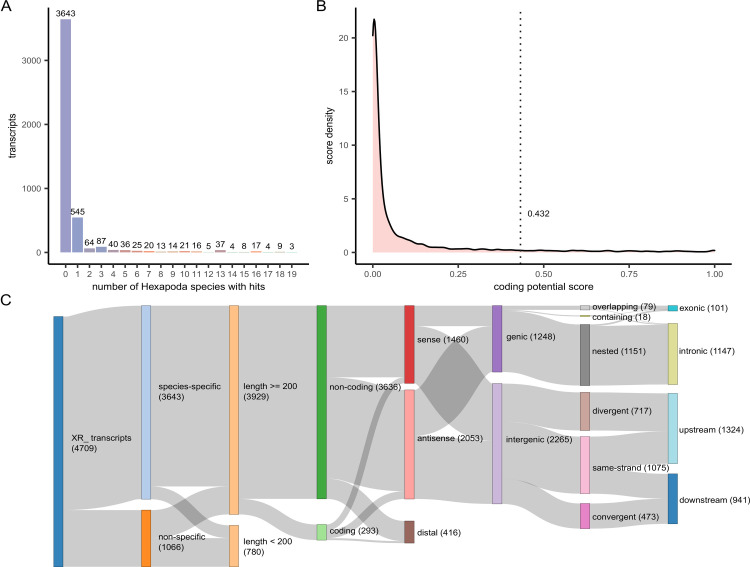
To identify Ae. aegypti-specific lncRNAs, we compared automatically annotated ncRNA transcript sequences of Ae. aegypti against all Hexapoda RefSeq RNAs. The number of species with hits were tallied for each transcript (Fig 1A). Out of 4,709 sequences, 3,643 (77.4%) had no significant Blast hits and were denoted as species-specific. To exclude lncRNAs bearing spurious coding potential from downstream experiments, we trained and utilized a coding potential classifier using the annotated protein-coding transcripts [40]. Based on the model’s optimal cutoff, 3,636 transcripts were classified as non-coding (Fig 1B). Additional information regarding the localization features of lncRNAs was extracted through the analysis of their overlapping with annotated protein-coding elements of their genomic region (Fig 1C and S3 Table).
Fig 1. Conservation, coding probability, and genomic localization information for NCBI automatically annotated lncRNAs in Aedes aegypti.
(A) The number of lncRNA sequences vs. the number of species against which they were found to exhibit Blast hits (top 20 hits shown). (B) Density plot of the coding potential model scores for all lncRNAs. The vertical dotted line indicates the cutoff point (0.432), below which transcripts were classified as non-coding. (C) Sankey plot displaying the overall annotation process and metrics for all RefSeq ncRNA transcripts (denoted by the “XR” prefix).
ZIKV infection modulates the expression of lncRNAs
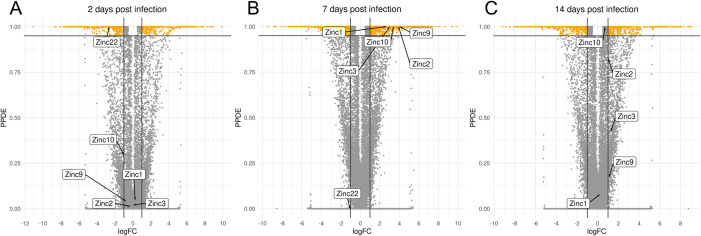
A publicly available RNA-Seq dataset [41] derived from whole-body Ae. aegypti specimens, comparing Zika virus infected to uninfected controls at 2, 7, and 14 days post-infection (dpi) [41], was analyzed to identify infection-responsive lncRNAs. Given our significant differential expression (DE) criteria of an absolute log2-fold change of at least 1 (equivalent to a two-fold change) and a minimum 95% posterior probability of belonging to the DE class, 823 (348 up- and 475 down-regulated upon infection), 1947 (1417 up- and 530 down-regulated) and 1303 (665 up- and 638 down-regulated) DE transcripts were found in the 2-, 7-, and 14-day comparisons, respectively (Fig 2A–2C). The lncRNA transcripts that displayed differential expression were termed Zika-modulated non-coding RNAs (Zinc). Five Zincs (Zinc1, Zinc2, Zinc3, Zinc9, and Zinc10) were selected for further functional studies due to their statistically significant upregulation, ranging between 6.6- to 17.1-fold, upon ZIKV infection at 7dpi (Fig 2B) and their species-specific nucleotide composition. In addition, the Zinc22 lncRNA was selected because of its significant downregulation at 2 dpi (Fig 2A) and its ovary-specific expression. Not wanting to limit our analysis to one category of lncRNAs with regard to their genomic localization, we chose three gene intron locating lncRNAs and three intergenic lncRNAs upstream/downstream of protein-coding genes, in sense/antisense orientation (S1 Table).
Fig 2. Volcano plot depicting differentially expressed transcripts in different time-points post Zika virus infection.
(A) 2 days post-infection (dpi) with ZIKV, (B) at 7dpi, and (C) at 14dpi. The y-axis shows the posterior probability that each transcript was differentially expressed. Vertical lines indicate the applied log2FC threshold (|log2(FC)| ≥ 1), and the horizontal line indicates the statistical significance threshold, set at probability ≥95% (corresponding to a 5% false-discovery rate).
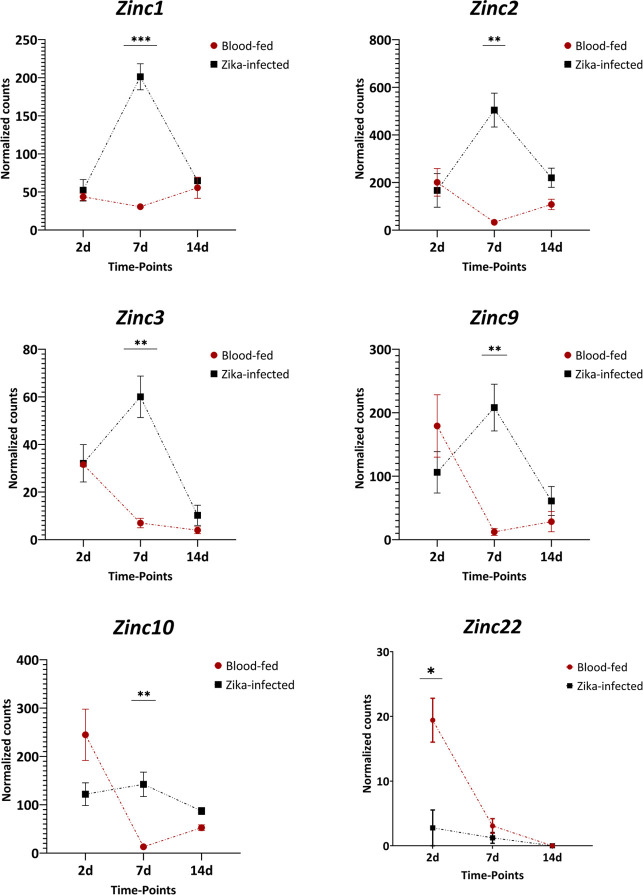
In Fig 2A–2C, volcano plots present the state of differential abundance for each comparison; the six lncRNAs of interest are also annotated (Fig 2A–2C and S1 Data). According to the transcriptomic data that were analyzed, five of the six shortlisted DE lncRNAs were upregulated in infected mosquitoes at 7 dpi. Specifically, Zinc1 displayed a 6.6-fold upregulation (P<0.001), Zinc2 a 15.4-fold upregulation (P<0.01), Zinc3 a 8.6-fold upregulation (P<0.01), Zinc9 a 17.1-fold upregulation (P<0.01), and Zinc10 an 10.9-fold upregulation (P<0.01). Zinc22 showed a 7.0-fold downregulation in the infected mosquitoes at 2 dpi (P<0.05) (Fig 3). When we compared the expression levels of Zinc22 in a detailed developmental dataset [42], this lncRNA emerged as ovary-specific and enriched in the insect’s post-vitellogenic stages. Validation of Zinc22 expression by qPCR revealed a 10-fold upregulation in mock infected ovaries at 2 days post-blood meal (PBM), whereas it exhibited a 3-fold down-regulation upon ZIKV-infection (S1 Fig). The available transcriptomic data did not reveal tissue-specificity for the other Zinc genes.
Fig 3. Expression of six Zinc lncRNAs in Ae. aegypti mosquitoes after a Zika virus-infected vs. non-infected blood meal.
Data were collected at three different time points (2, 7, and 14 days post-blood-feeding). Comparisons were made between Zika virus-infected and non-infected samples at the same time point. On average, in infected samples, 7dpi Zinc1 was upregulated 6.6-fold, Zinc2 was up-regulated 15.4-fold, Zinc3 8.6-fold, Zinc9 17.1-fold, and Zinc10 10.9-fold, whereas Zinc22 was down-regulated 7.0-fold at 2dpi. Each sample contained three biological replicates. Expression values are transcript-level read counts, normalized for library size using the median-by-ratio method. Bars indicate mean value ± SEM. The significance was determined by Student’s t-test. *:P<0.05, **:P<0.01, ***:P<0.001
Silencing of Zinc lncRNAs influences mosquito permissiveness to virus infection
To elucidate the influence of Zinc lncRNAs on the mosquito’s susceptibility to virus infection, we used a standard RNAi approach based on dsRNA injection into the mosquito thorax. Target-specific dsRNA was generated against each of the Zincs and administered by microinjection into inseminated adult females at 7 days post-eclosion; control mosquitoes were injected with dsRNA against GFP (dsGFP). Silencing efficiency of Zincs was measured at three different time-points (3-, 7- and 10-days) post dsRNA injection. All Zincs showed silencing efficiencies from 51,7–83.9% for Zinc1, 89.1–96.6% for Zinc2, 86.5–97.3% for Zinc3, 35.7–56.8% for Zinc9, 62.3–93.8% for Zinc10 and 52.1–96.5% for Zinc22 (Fig 4). After confirming effective silencing, an infectious blood meal that contained a suspension of ZIKV mixed with human blood was provided to the injected mosquitoes at 2 days post-injection. Then, individual midguts and carcasses (i.e., the whole body, excluding the midgut) were dissected at 7 dpi and 10 dpi, respectively. The viral particles were extracted from the tissues, and the viral titer was measured through standard plaque assays in BHK-21 cells.
Fig 4. Zinc gene silencing efficiency.
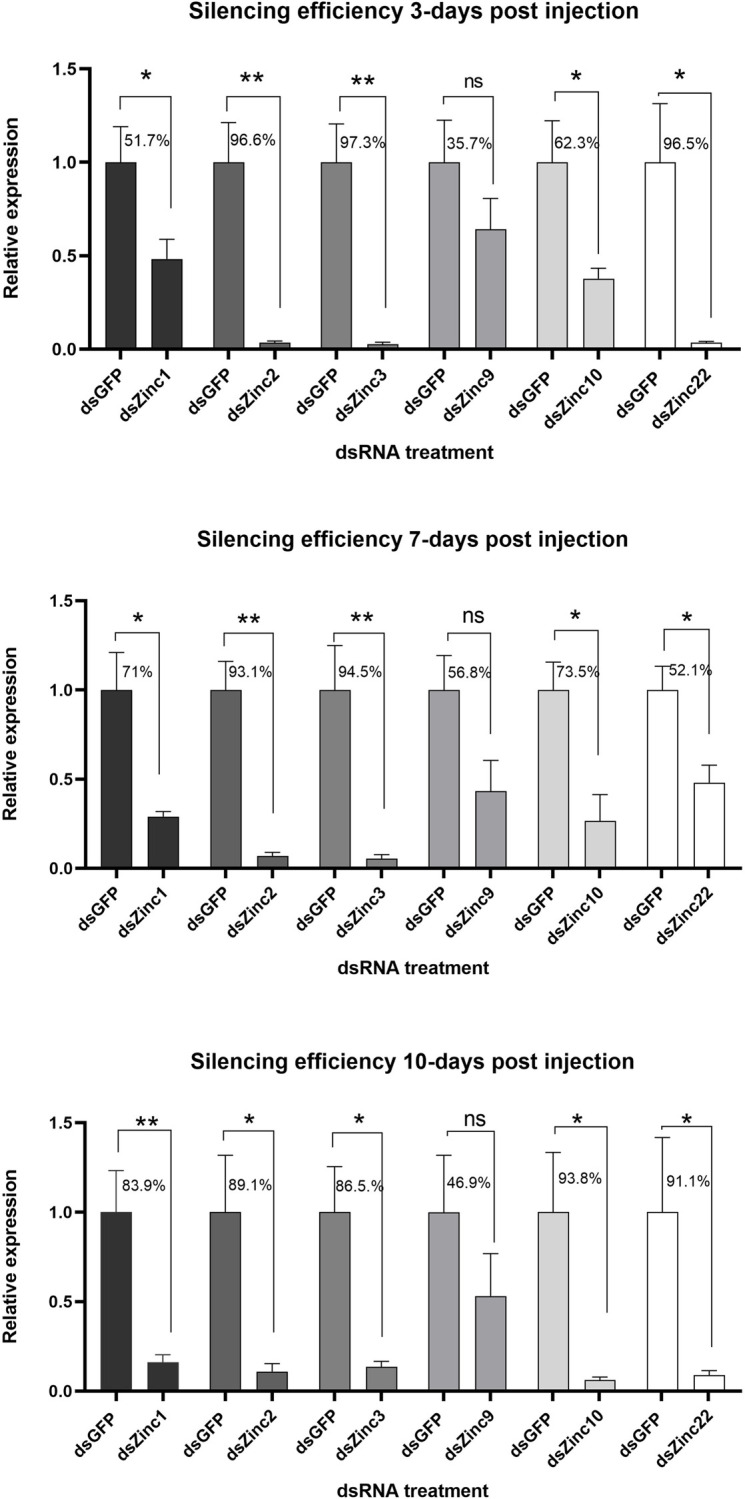
Relative quantification of Zinc1, Zinc2, Zinc3, Zinc9, Zinc10 and Zinc22 RNA abundance in Zinc-silenced vs GFP dsRNA-treated control mosquitoes. Whole mosquitoes were collected and pooled at three time-points (3-, 7- and 10-days) post-dsRNA injection. 3–5 biological replicates were performed for each Zinc and the average expression of GFP dsRNA treated replicates was set as 1 and the decreased dsZinc replicates, compared to GFP dsRNA, was measured. Samples were normalized using RNA abundance of the ribosomal RpS7 gene and are presented as the mean ± SEM. The significance was determined by Student’s t-test. *: P<0.05, **:P<0.01.
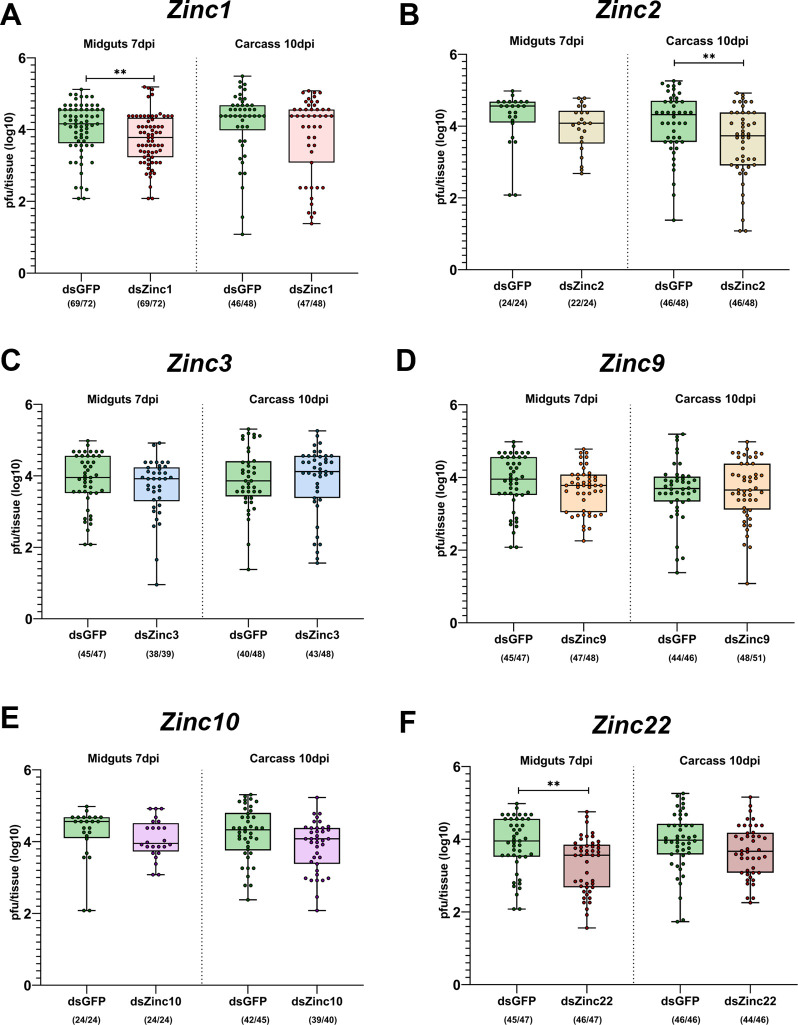
Our results revealed that dsRNA-mediated silencing of Zinc1, Zinc2, and Zinc22 resulted in a statistically significant decrease in the viral titers of Zika, whereas silencing of Zinc3, Zinc9, and Zinc10 did not produce any significant changes in viral titer. Specifically, Zinc1-silenced midguts dissected at 7 dpi had a median titer of 103.7 pfu, as compared to a median titer of 104.2 (P<0.01) for the GFP dsRNA-injected control (Fig 5A); Zinc2 silenced mosquito carcasses dissected at 10 dpi had a median titer of 103.7 pfu, as compared to 104.3 (P<0.01) for the control (Fig 5B); and Zinc22-silenced mosquito midguts dissected at 7 dpi had a median of 103.5 pfu, as compared to 104 (P<0.01) for the control (Fig 5F).
Fig 5. Effects of Ae. aegypti Zinc lncRNA silencing on ZIKV infection.
Viral titers of Zika virus in midguts and carcasses obtained from dsRNA-treated mosquitoes at 7- or 10-days post-infection, respectively. Each panel corresponds to the comparison between Zinc-silenced mosquitoes and GFP dsRNA-treated controls: A) Zinc1-silenced, B) Zinc2-silenced, C) Zinc3-silenced, D) Zinc9-silenced, E) Zinc10-silenced, F) Zinc22-silenced. Each dot corresponds to the logarithmic number of plaque-forming units (pfu) that were counted per tissue and represents data from individual mosquitoes. The numbers in parentheses represent the number of virus-infected mosquitoes plotted against the number of mosquitoes checked. Data are presented as box plots in which the horizontal lines indicate the median value and whiskers show the minimum to maximum values. The significance was determined by Mann-Whitney U-test. **:P<0.01
Phenotypic impact of Zinc lncRNA silencing on fecundity and fertility
Given that the expression of the selected Zincs was modulated by infection with arboviruses, we next tested the hypothesis that their expression influenced reproduction in Ae. aegypti. Immunity and reproduction are two energy-demanding physiological processes that consume a significant proportion of an insect’s resources. To examine the possible involvement of selected Zincs in reproduction, we studied the impact of their RNAi-mediated silencing on three reproductive phenotypes: fecundity, fertility and progeny. dsRNA against either Zinc or GFP was administered by microinjection to inseminated adult female mosquitoes at 7 days post-eclosion, and 2 days later the injected mosquitoes were blood-fed on healthy mice. Their laid eggs were counted (fecundity assay) and the hatch rate was determined by comparing the percentage of successfully hatched larvae to the total number of eggs laid per mosquito (fertility assay). Progeny was defined as the number of live larvae produced by each mosquito.
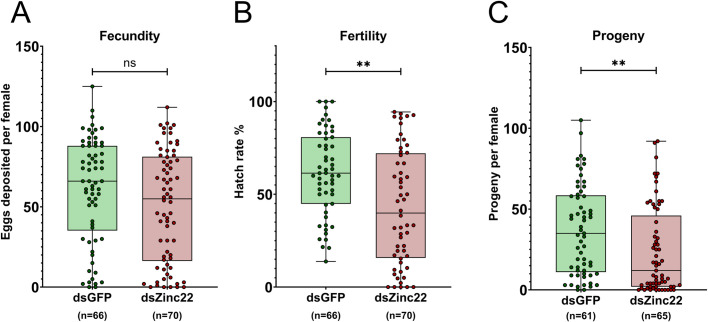
We first examined the effect of Zinc22-silencing on reproductive phenotypes. The Zinc22 is an ovary-enriched lncRNA that is upregulated by blood-feeding, but downregulated by Zika infection as shown by both RNA sequencing [41,42] and qPCR data (S1 Fig). Zinc22-silenced mosquitoes showed a marginally non-significant decrease in the number of eggs laid when compared to the GFP dsRNA-treated control mosquitoes. Specifically, Zinc22-silenced females laid a median of 55 eggs, as compared to a median of 66 eggs for the GFP dsRNA-treated controls (P = 0.08) (Fig 6A). However, there was a significant decrease in the hatch rate of the Zinc22 silenced females; showing a median egg hatch rate of 38.85%, whereas the GFP dsRNA-treated control mosquitoes produced eggs that had a median hatch rate of 61.40% (P<0.001) (Fig 6B). Finally, Zinc22-silenced females produced a significantly lower number of offspring than did the GFP dsRNA-treated control females; Zinc22-silenced mosquitoes had a median of 12 offspring each, whereas each of the GFP dsRNA-treated control mosquito cohorts had a median of 35 offspring (P<0.001) (Fig 6C).
Fig 6. Impact of silencing of Zinc22 lncRNAs on fecundity (oviposition), fertility (hatching) and progeny of Ae. aegypti mosquitoes.
(A) Each dot corresponds to the number of eggs that were laid by each individual Zinc22-silenced or GFP dsRNA-treated female mosquito. (B) The fertility was defined as the percentage (%) of hatched larvae in relation to the total number of laid eggs per mosquito. (C) The number of offspring produced by each Zinc-silenced or GFP dsRNA-treated mosquito. Data are presented as box plots in which the horizontal lines indicate the median value and whiskers the minimum to maximum values. The significance was determined by Mann-Whitney U-test. **P<0.001.
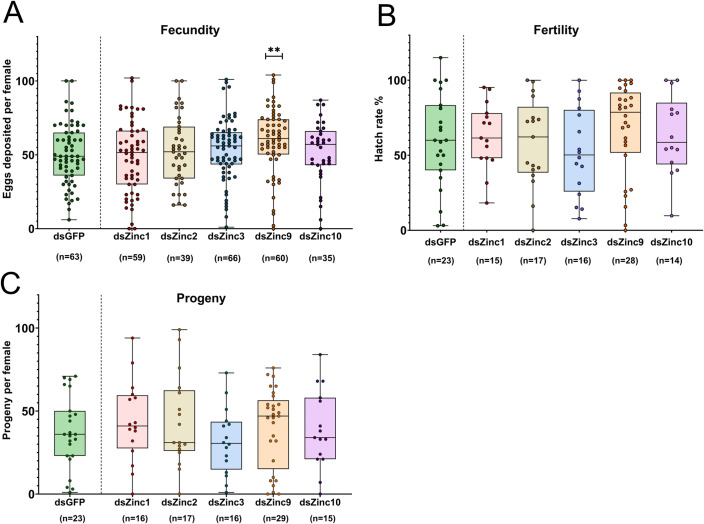
Next, we expanded our silencing studies to another five Zinc lncRNAs. Silencing of Zinc9 had a significant effect on fecundity (Fig 7A), while silencing of the other four Zincs had no effect, when compared to the GFP dsRNA-treated control mosquitoes, in either the fecundity, fertility or progeny assays (Fig 7A–7C). Silencing of dsZinc9 resulted in an increased oviposition rate, to a median number of 61 laid eggs, as compared to a median of 49 eggs for GFP dsRNA-treated mosquitoes (P<0.001) (Fig 7A). An increase in the hatch rate of eggs was also evident for the Zinc9-silenced females, although this increase was not statistically significant. Specifically, eggs from the Zinc9-silenced females showed a median hatch rate of 79%, as compared to 60% for the GFP dsRNA-treated controls (Fig 7B). Zinc9-silenced females also produced more progeny compared to the GFP dsRNA-treated controls. Specifically, Zinc9-silenced mosquitoes had a median of 47 offspring each, whereas each of the GFP dsRNA-treated control mosquitoes had a median of 36 offspring which was not a statistically significant difference.
Fig 7. Impact of Zinc1, Zinc2, Zinc3, Zinc9 and Zinc10 lncRNA silencing on fecundity, fertility and progeny.
(A) Each dot corresponds to the number of eggs that were laid by each individual Zinc-silenced or GFP dsRNA-treated female mosquito. (B) The fertility was defined as the percentage (%) of hatched larvae in relation to the total number of laid eggs per mosquito. (C) The number of offspring produced by each Zinc-silenced or GFP dsRNA-treated mosquito. Data are presented as box plots in which the horizontal lines indicate the median value and whiskers the minimum to maximum values. The significance was determined by Mann-Whitney U-test. **P<0.001.
Discussion
The role of lncRNAs in insect immunity and permissiveness to pathogens has been investigated in other species, particularly D. melanogaster, for which significant findings regarding its immune response against pathogenic viruses and bacteria have been reported [15,43,44]. Research has also been carried out in lepidopteran species such as Bombyx mori and Pectinophora gossypiella, in which specific lncRNAs have been implicated in the response to viral infection [45,46]. Despite the progress made in other species, lncRNA research in mosquito species has until now been limited and their implication in vector competence and immune responses to human pathogens has not been extensively addressed. Previous Ae. aegypti transcriptome studies have identified putative infection-regulated lncRNAs [41,47], and other studies have addressed specific Ae. aegypti lncRNAs that appear to interfere with arbovirus infection in cell lines [17,18,48]. Similarly, limited knowledge exists regarding the role of lncRNAs in Ae. aegypti reproduction. Transcriptomic analysis of Ae. aegypti ovaries has identified differentially expressed lncRNAs after a blood-meal [42,49], while studies in Ae. albopictus have revealed species-specific lncRNAs that influence fecundity and fertility [50].
Here we provide in vivo evidence regarding the influence of specific lncRNAs on the vector competence for Zika virus which to a significant degree is regulated by the innate immune system. We focused on vector competence and reproduction because of their significance for disease transmission and survival and their potential for the development of novel mosquito control strategies. Our screening revealed that the vast majority of Ae. aegypti lncRNAs (77.4%) displayed a high species specificity, as judged by their nucleotide composition. High species specificity is a common feature of insect lncRNAs, as has been observed in other related species such as Ae. albopictus [50], because of the absence of functional domains and a faster evolutionary rate of non-coding genes [51].
Subsequent analysis of ZIKV-infected Ae. aegypti samples revealed an abundance of lncRNAs that are differentially expressed at various time-points after infection. Silencing of Zinc1, Zinc2 and Zinc22 rendered the mosquitoes less permissive to virus infection while differences in viral titers were also observed upon silencing of other Zinc genes, although without statistical significance. The impact of Zinc1, Zinc2 and Zinc22 silencing indicates that these lncRNAs may act as, or regulate, pro-viral host factors that facilitate ZIKV infection. The effects of Zinc1 and Zinc22 silencing was mainly observed in the mosquito midguts at 7dpi suggesting that these lncRNAs may be involved in the early stages of ZIKV infection. The effect of Zinc2-silencing on infection of the carcass tissues at 10dpi suggests that its role in infection may be after the midgut stage of infection. Host factors that have been implicated in these processes are the Inosine-5′-monophosphate dehydrogenase (IMPDH) that promotes RNA synthesis of ZIKV and DENV [4] and TER94/VCP that is involved in uncoating of the ZIKV genome into the cytoplasm [52]. Another putative host factor that has been suggested to function as a DENV receptor is prohibitin [53]. Further studies are required to clarify the specific host factor mechanisms that Zinc1, Zinc2 and Zinc22 are implicated in.
The effects of Zinc silencing may also indicate their potential roles as negative regulators of the mosquito anti-viral defense. The Toll and JAK/STAT immune signaling pathways, that have been shown to respond to ZIKV infection [4], comprise negative regulators such as Cactus Toll [54] and PIAS, respectively [9]. While no ncRNAs have been linked to the regulation of either pathway in Ae. aegypti, the D. melanogaster lncRNA CR11538 has been shown to suppress the Toll pathway [43], and miRNAs such as miR958, miR964 and miR317 are also a negatively regulating Toll signaling [55–57]. While these findings do not imply that the same interactions occur in Ae. aegypti, they do introduce a novel research target for investigation.
Given that vector competence and immunity have been connected to reproduction [24,25,27,28,39], as evidenced by the reproduction-immunity trade-off, a tempting question was whether the ZIKV-regulated Zinc lncRNAs had any impact on Ae. aegypti reproductive functions. Interestingly, Zinc22 silencing resulted in a significant reduction in egg hatching, as well as a decrease in the number of progeny. Considering that Zinc22 expression is downregulated upon ZIKV infection at 2 days PBM, while its silencing results in a suppression of infection in midguts at 7dpi it is likely that it may be involved in reproduction-immunity trade-offs. Based on the silencing phenotypes, one can hypothesize that a decrease in Zinc22 expression results in a stronger immune response against the virus and the trade-off of this energetically costly process is a reduction in reproduction. The trade-offs between the two systems have been shown to be regulated by several endocrine signals, and one of them is the production of 20-hydroxyecdysone (20E) which is essential for the production of yolk protein precursors (YPPs) involved in vitellogenesis and egg development. The 20E is produced in the fat body upon hydroxylation of ecdysone, a hormone that is produced by the female ovarian follicles in response to a blood-meal (reviewed in [32]. Upregulation of ecdysone increases mosquito reproductive ability, while its decrease its immune competence [58]. Zinc22 was identified as an ovary-specific lncRNA and may therefore be involved in the regulation of metabolic processes of ovaries. The apparent roles of Zinc22 as a positive regulator of reproduction and a negative regulator of immunity suggests it could be implicated in such processes.
It is noteworthy that silencing of Zinc9 produced an increase in mosquito fecundity, and to a lesser extent, fertility. Although Zinc9 was selected for our studies as a potential immune-related lncRNA, its silencing did not have any significant effect on infection with ZIKV. This could be a result of reduced silencing efficiency (35.7–56.8%), as compared to the silencing effects of dsRNA treatments for other Zincs. Additional experiments evaluating the influence of Zinc9 on the resistance to other pathogens, including other arboviruses and bacteria, should be conducted to further investigate its potential role in immunity. Considering its demonstrated effects on reproduction, any immune-related outcome could suggest its involvement in trade-offs.
In conclusion, our studies point at the potential influence of four mosquito lncRNAs on reproduction and vector competence. As regulators of these critical processes, these lncRNAs have the potential for development of novel vector and disease transmission control strategies. For example, reproductive enhancement through transgenic overexpression of a reproduction-regulating lncRNA could provide a fitness advantage to genetically modified pathogen-resistant mosquitoes [59,60]. Similarly, lncRNAs that either boost or disable antiviral immunity could be exploited to create transgenic virus-resistant mosquitoes. Refractory mosquitoes can be generated either by overexpressing antiviral effector genes or by inactivating pro-viral host factors [61,62].
Materials and methods
Ethics statement
This study was conducted in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, the Animal Care and Use Committee (ACUC) of Johns Hopkins University, and the institutional Ethics Committee. The Institutional Animal Care and Use Committee (IACUC) approved the protocol RA21H388. Mice were used for the mosquito rearing and reproductive assays. Anonymous, commercial blood from human donors was used for virus infection assays in mosquitoes.
lncRNA annotation
Non-coding RNA (ncRNA) sequences (XR accessions) were obtained from the Ae. aegypti GCF_002204515.2 gene models. Blastn [63] was executed locally against RefSeq RNAs of Hexapoda (TaxID 6960) for each transcript at default settings. Hits on opposite strands (+/-) as well as within Ae. aegypti (TaxID 7159) were filtered out.
FEELnc [40] was used to perform species-specific estimation of the coding potential of all non-coding RNAs. In brief, FEELnc_filter.pl was used to filter out ncRNAs overlapping with the exons of protein-coding transcripts (XM accessions) in the sense orientation as well as ncRNAs smaller than 200 nucleotides. The parameters for FEELnc_codpot.pl were established to build a coding potential estimation model using protein-coding transcripts (mode = shuffle). Using 10-fold cross validation, 0.432 was identified as the model’s optimal cutoff point to denote coding and non-coding transcripts (i.e., at this point the model’s sensitivity and specificity is maximized). The coding potential of the filtered ncRNAs (n = 3929) was estimated, classifying 3,636 (92.5%) of them as non-coding. In addition, FEELnc_classifier.pl was used to annotate ncRNA genes regarding their genomic localization relative to overlapping/closest protein-coding genes. Overlapping instances were annotated as sense/antisense, nested/overlapping and intronic/exonic, and intergenic ones were annotated as sense/antisense, same-strand/convergent/divergent and upstream/downstream. ncRNA genes exhibiting no neighboring genes within 100kb were annotated as distal (n = 416).
The R packages ggplot2 [64] and networkD3 [65] were used to render the plots shown in Fig 1. The annotation of all Ae. aegypti ncRNAs regarding their species-specificity, coding-potential, and genomic localization with respect to protein-coding transcripts is provided in S3 Table.
Transcriptome analysis
The RNA-Seq dataset produced and described by Etebari et al. [41] was retrieved from the Sequence Read Archive. FASTQC [66] and Cutadapt [67] were used for quality control and removal of adapters and low-quality bases, respectively. The Ae. aegypti GCF_002204515.2 AaegL.5.0 genome assembly and respective gene models were used to create indices for the STAR v2.7.9a aligner [68] (a splice-aware index, taking into account the dataset’s read length) and for the RSEM v1.3.1 quantification tool [69]. Genomic alignment of paired-end reads was conducted with STAR and parameterized according to the ENCODE options stated in its manual, and subsequently RSEM quantification was conducted. EBSeq v1.30.0 [70], which is tailored to isoform-level analyses, was used to identify DE transcripts. In brief, transcript-level raw counts were used to obtain library normalization factors with the default median-by-ratio method [71]; they were then subjected to empirical Bayes testing for 15 iterations (maxround argument), ensuring that the model’s hyper-parameter estimations converged. Differential expression was called at a 5% target false-discovery rate, setting FDRMethod argument at “hard” and disabling the Threshold_FCRatio filter. Transcripts exhibiting a two-fold expression change and at least a 95% posterior probability of being DE (PPDE) were considered to be DE.
Median-by-ratio normalized read counts were utilized to render plots of differentially expressed transcripts were produced using the ggplot2 R package. S1 Data provides the entire DE analysis results.
Mosquito rearing
Aedes aegypti mosquitoes (Liverpool strain) were maintained on a 10% sucrose solution in the insectary of the Johns Hopkins Malaria Research Institute at 27°C under 85% relative humidity and a 12h light/12h dark cycle.
Cell cultures
An Ae. albopictus C6/36 cell line was used for the ZIKV (Cambodia, FSS13025) propagation. Cells were grown in minimal essential medium (MEM, Gibco, Carlsbad, CA, USA) with 10% heat inactivated FBS, 1% L-glutamine, 1% penicillin-streptomycin, and 1% non-essential amino acids at 32°C with 5% CO2. A cell line of baby hamster kidney BHK-21 cells was used for the plaque assays. They were maintained on Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Carlsbad, CA) supplemented with 10% FBS, 1% L-glutamine, 1% penicillin-streptomycin, and 5 μg/ml plasmocin (Invitrogen, Carlsbad, CA) at 37°C with 5% CO2.
dsRNA synthesis and gene silencing
Total RNA was extracted from ten 7-day old Ae. aegypti females using Trizol (Invitrogen, Carlsbad, CA). cDNA was synthesized from total RNA using an oligo-dT primer and random hexamers with the MMLV reverse transcriptase kit (Invitrogen, Carlsbad, CA). Total cDNA was used to amplify Zinc lncRNAs by PCR, using gene-specific primer sets with the T7 RNA polymerase promoter (TAATACGACTCACTATAGGG) attached to their 5’ ends. GFP gene was similarly amplified using a GFP plasmid as a template (S2 Table). Amplified PCR products were purified with a Zymo DNA cleanup kit (Zymo Research, Irvine, CA). A total of 1 μg of PCR product was used to synthesize dsRNA using a NEB T7 HiScribe kit (NEB, Ipswich, MA) according to the manufacturer’s instructions. dsRNA was purified using isopropanol-sodium acetate precipitation. dsRNA was dissolved in sterile distilled water, and the concentration was measured with a Nanodrop spectrophotometer (ThermoFischer Scientific, Waltham, MA). The final concentration of the dsRNA was adjusted to 3 μg/μL.
One-week-old inseminated female mosquitoes were anesthetized on a cold block and intrathoracically injected with 69 nL of 3 μg/μL dsRNA against each of the Zinc genes or GFP as a control. Mosquitoes were reared for 2 days in the insectary to recover the injection before any downstream experiment. For the purpose of analyzing silencing efficiency whole mosquitoes were collected at three time-points (3-,7-,10-days) post-dsRNA injection and 5 females were pooled for each biological replicate. 3–5 biological replicates and two technical replicates per lncRNA were performed. PCR conditions are described in the Real-Time qPCR section.
Zika infections and plaque assays
C6/36 cells in T25 flasks were infected with ZIKV at a multiplicity of infection (MOI) of 0.5. After 4–5 days of incubation at 32°C with 5% CO2, infected cell culture medium was harvested and mixed with human red blood cells (RBC) supplemented with human serum containing 10 mM ATP (50% RBC, 40% ZIKV medium, 10% serum, 1% ATP). Injected females at 2 days post-injection were starved overnight and transferred to small (16 oz) paper cups. They were allowed to feed for 30 min on a blood-virus mixture at 37°C on a single glass artificial feeder. Fully engorged females were recovered by sorting and maintained on 10% sterile sucrose solution. Midguts from individual females at 7 dpi and carcasses from individual females at 10 dpi were dissected on ice and kept at −80°C. Each experiment had at least two biological replicates, and each replicate included at least 18 individual females.
Mosquito midguts and carcasses were homogenized in 300 μl DMEM with a Bullet Blender (Next Advance, Inc., Averill Park, NY) with glass beads. They were then centrifuged at 10,000 × g for 3 min, and 50 μl of the filtered supernatants was used for plaque assays with BHK 21 cells in 24-well plates. After the plates were incubated for 4 days for ZIKV, the plaques were visualized by staining with 1% crystal violet and counted under a microscope. Differences in viral intensity and prevalence of infection in samples dissected from Zinc-silenced and GFP dsRNA-treated females were compared by using a nonparametric Mann-Whitney U test and Fisher’s exact test, respectively.
Reproductive assays
Oviposition
Four days after blood-feeding, individual female mosquitoes were placed in 50-mL Falcon tubes that contained a moist filter paper at the bottom. A range of 35 to 70 mosquitoes were included in each Zinc-silenced/GFP dsRNA treated sample. The mosquitoes were left in the Falcon tubes in the insectary for 2 days to allow egg laying to occur. Moisture was regularly added to the tubes to keep the filter paper wet. Then mosquitoes were removed from the Falcon tubes, and the total number of eggs deposited by each individual was photographed and counted using ImageJ software [72]. Mosquitoes that deceased during the oviposition process were excluded from the analysis.
Hatching assay
The filter papers with eggs were collected, and the eggs were dried and then stored in sealed Petri dishes that contained a wet cotton ball as a source of humidity. The eggs laid by 14 to 56 individual mosquitoes were included in each Zinc-silenced/GFP dsRNA-treated sample. They were stored in separate Petri dishes and three days (72h) after egg-laying, 30ml of hatching broth was added to each dish. Hatching broth consisted of a fish food pellet diluted in 1L of autoclaved water. The eggs were then incubated in the hatching broth for up to 14 days. Hatching broth was replaced with fresh broth every 4 days. Larvae that hatched from each egg batch were counted daily and removed from the Petri dishes. The hatch rate was estimated as the percentage of emerged larvae divided by the total number of eggs laid:
Real-time qPCR
Ovaries were dissected from mosquitoes that were either: (i) non-blood fed (NBF), (ii) blood-fed with a blood mixture from healthy humans (50% RBC, 40% MEM, 10% serum, 1% ATP) at 48h PBM or (iii) blood-fed with human blood infected with Zika virus (50% RBC, 40% Zika medium, 10% serum, 1% ATP) at 48h PBM. The dissected ovaries were pooled into three groups of six ovaries for each condition and stored in 100μL of TRIzol reagent (Invitrogen, Carlsbad, CA). Total RNA was extracted according to the TRIzol manufacturer’s instructions. The integrity of the RNA was assessed by agarose gel electrophoresis. Total RNA was treated with DNase I (ThermoFisher Scientific, Waltham, MA), and 1μg of RNA was reverse-transcribed to cDNA by using oligo-dT primers and MMLV-RT (Invitrogen, Carlsbad, CA).
Gene-specific primers (S2 Table) were used for qPCR amplification. Amplification and analysis were carried out using the StepOnePlus real-time PCR system (Applied Biosystems, Warrington, UK). The final reaction volume was 15μl when using the ABI SYBR Green supermix. The PCR program was as follows: hold at 95°C for 10 min, 95°C for 15 s, and 60°C for 1 min, repeated for 40 cycles. The specificity of the SYBR Green PCR signal was further confirmed by melting curve analysis. The relative abundance of the gene transcripts was normalized and calculated by comparison to the ribosomal protein S7 gene (AAEL009496) as an endogenous reference using the Livak (2-ΔΔCt) method.
Supporting information
Expression data were generated by Real-Time qPCR. Values were normalized with the ribosomal gene Rps7. Each sample included three biological replicates. Bars indicate mean value ± SEM. The significance was determined by Student’s t-test. *:P<0.05
(TIF)
Data include coding and non-coding transcripts that were either up- or down- regulated according to RNA seq data.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We thank the Johns Hopkins Malaria Research Institute Insectary for providing the mosquito-rearing facility and the Parasitology Core facilities for providing the naive human blood. We thank Dr. Deborah McClellan for editorial assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
AB-T was supported by the Fulbright U.S. Student Program, which is sponsored by the U.S. Department of State and The Fulbright Foundation in Greece. This work was supported by NIH/NIAID grant R01AI141532 and the Bloomberg Philanthropies. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bartholomay LC, Michel K. Mosquito immunobiology: the intersection of vector health and vector competence. Annual review of entomology. 2018;63:145–67. [DOI] [PubMed] [Google Scholar]
- 2.Sim S, Jupatanakul N, Ramirez JL, Kang S, Romero-Vivas CM, Mohammed H, et al. Transcriptomic profiling of diverse Aedes aegypti strains reveals increased basal-level immune activation in dengue virus-refractory populations and identifies novel virus-vector molecular interactions. PLoS neglected tropical diseases. 2013;7(7):e2295. doi: 10.1371/journal.pntd.0002295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sim S, Jupatanakul N, Dimopoulos G. Mosquito immunity against arboviruses. Viruses. 2014;6(11):4479–504. doi: 10.3390/v6114479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angleró-Rodríguez YI, MacLeod HJ, Kang S, Carlson JS, Jupatanakul N, Dimopoulos G. Aedes aegypti molecular responses to Zika virus: modulation of infection by the toll and jak/stat immune pathways and virus host factors. Frontiers in microbiology. 2017;8:2050. doi: 10.3389/fmicb.2017.02050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xi Z, Ramirez JL, Dimopoulos G. The Aedes aegypti toll pathway controls dengue virus infection. PLoS pathogens. 2008;4(7):e1000098. doi: 10.1371/journal.ppat.1000098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tikhe CV, Dimopoulos G. Mosquito antiviral immune pathways. Developmental & Comparative Immunology. 2021;116:103964. doi: 10.1016/j.dci.2020.103964 [DOI] [PubMed] [Google Scholar]
- 7.Liu X, Jiang F, Kalidas S, Smith D, Liu Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. Rna. 2006;12(8):1514–20. doi: 10.1261/rna.101606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123(4):607–20. doi: 10.1016/j.cell.2005.08.044 [DOI] [PubMed] [Google Scholar]
- 9.Souza-Neto JA, Sim S, Dimopoulos G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proceedings of the National Academy of Sciences. 2009;106(42):17841–6. doi: 10.1073/pnas.0905006106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramirez JL, Dimopoulos G. The Toll immune signaling pathway control conserved anti-dengue defenses across diverse Ae. aegypti strains and against multiple dengue virus serotypes. Developmental & Comparative Immunology. 2010;34(6):625–9. doi: 10.1016/j.dci.2010.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang S, Shields AR, Jupatanakul N, Dimopoulos G. Suppressing dengue-2 infection by chemical inhibition of Aedes aegypti host factors. PLoS neglected tropical diseases. 2014;8(8):e3084. doi: 10.1371/journal.pntd.0003084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–8. doi: 10.1126/science.1138341 [DOI] [PubMed] [Google Scholar]
- 13.Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome biology. 2017;18(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Xu W, Gao X, Li W, Qi S, Guo D, et al. lncRNA sensing of a viral suppressor of RNAi activates non-canonical innate immune signaling in Drosophila. Cell Host & Microbe. 2020;27(1):115–28. e8. doi: 10.1016/j.chom.2019.12.006 [DOI] [PubMed] [Google Scholar]
- 15.Valanne S, Salminen TS, Järvelä-Stölting M, Vesala L, Rämet M. Immune-inducible non-coding RNA molecule lincRNA-IBIN connects immunity and metabolism in Drosophila melanogaster. PLoS pathogens. 2019;15(1):e1007504. doi: 10.1371/journal.ppat.1007504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi X-Z, Zhong X, Yu X-Q. Drosophila melanogaster NPC2 proteins bind bacterial cell wall components and may function in immune signal pathways. Insect biochemistry and molecular biology. 2012;42(8):545–56. doi: 10.1016/j.ibmb.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etebari K, Asad S, Zhang G, Asgari S. Identification of Aedes aegypti long intergenic non-coding RNAs and their association with Wolbachia and dengue virus infection. PLoS neglected tropical diseases. 2016;10(10):e0005069. doi: 10.1371/journal.pntd.0005069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mao W, Zeng Q, She L, Yuan H, Luo Y, Wang R, et al. Wolbachia Utilizes lncRNAs to Activate the Anti-Dengue Toll Pathway and Balance Reactive Oxygen Species Stress in Aedes aegypti Through a Competitive Endogenous RNA Network. Frontiers in cellular and infection microbiology. 2022:1464. doi: 10.3389/fcimb.2021.823403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bashir-Tanoli S, Tinsley MC. Immune response costs are associated with changes in resource acquisition and not resource reallocation. Functional Ecology. 2014;28(4):1011–9. [Google Scholar]
- 20.Fellowes M, Kraaijeveld A, Godfray H. The relative fitness of Drosophila melanogaster (Diptera, Drosophilidae) that have successfully defended themselves against the parasitoid Asobara tabida (Hymenoptera, Braconidae). Journal of Evolutionary Biology. 1999;12(1):123–8. [Google Scholar]
- 21.Nystrand M, Dowling DK. Dose-dependent effects of an immune challenge at both ultimate and proximate levels in D rosophila melanogaster. Journal of evolutionary biology. 2014;27(5):876–88. doi: 10.1111/jeb.12364 [DOI] [PubMed] [Google Scholar]
- 22.Zerofsky M, Harel E, Silverman N, Tatar M. Aging of the innate immune response in Drosophila melanogaster. Aging cell. 2005;4(2):103–8. doi: 10.1111/j.1474-9728.2005.00147.x [DOI] [PubMed] [Google Scholar]
- 23.Fedorka KM, Linder JE, Winterhalter W, Promislow D. Post-mating disparity between potential and realized immune response in Drosophila melanogaster. Proceedings of the Royal Society B: Biological Sciences. 2007;274(1614):1211–7. doi: 10.1098/rspb.2006.0394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Short SM, Lazzaro BP. Female and male genetic contributions to post-mating immune defence in female Drosophila melanogaster. Proceedings of the Royal Society B: Biological Sciences. 2010;277(1700):3649–57. doi: 10.1098/rspb.2010.0937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Short SM, Wolfner MF, Lazzaro BP. Female Drosophila melanogaster suffer reduced defense against infection due to seminal fluid components. Journal of insect physiology. 2012;58(9):1192–201. doi: 10.1016/j.jinsphys.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan G, Severson DW, Christensen BM. Costs and benefits of mosquito refractoriness to malaria parasites: implications for genetic variability of mosquitoes and genetic control of malaria. Evolution. 1997;51(2):441–50. doi: 10.1111/j.1558-5646.1997.tb02431.x [DOI] [PubMed] [Google Scholar]
- 27.Ahmed A, Baggott S, Maingon R, Hurd H. The costs of mounting an immune response are reflected in the reproductive fitness of the mosquito Anopheles gambiae. Oikos. 2002;97(3):371–7. [Google Scholar]
- 28.Ahmed AM, Hurd H. Immune stimulation and malaria infection impose reproductive costs in Anopheles gambiae via follicular apoptosis. Microbes and Infection. 2006;8(2):308–15. doi: 10.1016/j.micinf.2005.06.026 [DOI] [PubMed] [Google Scholar]
- 29.Hogg J, Hurd H. Plasmodium yoelii nigeriensis: the effect of high and low intensity of infection upon the egg production and bloodmeal size of Anopheles stephensi during three gonotrophic cycles. Parasitology. 1995;111(5):555–62. doi: 10.1017/s0031182000077027 [DOI] [PubMed] [Google Scholar]
- 30.Harshman LG, Zera AJ. The cost of reproduction: the devil in the details. Trends in ecology & evolution. 2007;22(2):80–6. doi: 10.1016/j.tree.2006.10.008 [DOI] [PubMed] [Google Scholar]
- 31.Flatt T, Tu MP, Tatar M. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. Bioessays. 2005;27(10):999–1010. doi: 10.1002/bies.20290 [DOI] [PubMed] [Google Scholar]
- 32.Hansen IA, Attardo GM, Rodriguez SD, Drake LL. Four-way regulation of mosquito yolk protein precursor genes by juvenile hormone-, ecdysone-, nutrient-, and insulin-like peptide signaling pathways. Frontiers in physiology. 2014;5:103. doi: 10.3389/fphys.2014.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flatt T, Heyland A, Rus F, Porpiglia E, Sherlock C, Yamamoto R, et al. Hormonal regulation of the humoral innate immune response in Drosophila melanogaster. Journal of Experimental Biology. 2008;211(16):2712–24. doi: 10.1242/jeb.014878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dimarcq J-L, Imler J-L, Lanot R, Ezekowitz RAB, Hoffmann JA, Janeway CA, et al. Treatment of l (2) mbn Drosophila tumorous blood cells with the steroid hormone ecdysone amplifies the inducibility of antimicrobial peptide gene expression. Insect biochemistry and molecular biology. 1997;27(10):877–86. doi: 10.1016/s0965-1748(97)00072-6 [DOI] [PubMed] [Google Scholar]
- 35.Meister M, Lagueux M. Drosophila blood cells. Cellular microbiology. 2003;5(9):573–80. doi: 10.1046/j.1462-5822.2003.00302.x [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z, Palli S. Identification of a cis-regulatory element required for 20-hydroxyecdysone enhancement of antimicrobial peptide gene expression in Drosophila melanogaster. Insect molecular biology. 2009;18(5):595–605. doi: 10.1111/j.1365-2583.2009.00901.x [DOI] [PubMed] [Google Scholar]
- 37.Rus F, Flatt T, Tong M, Aggarwal K, Okuda K, Kleino A, et al. Ecdysone triggered PGRP-LC expression controls Drosophila innate immunity. The EMBO journal. 2013;32(11):1626–38. doi: 10.1038/emboj.2013.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang M-M, Wang Y-H, Yang Q-T, Wang X-L, Wang M, Raikhel AS, et al. Regulation of antimicrobial peptides by juvenile hormone and its receptor, Methoprene-tolerant, in the mosquito Aedes aegypti. Insect biochemistry and molecular biology. 2021;128:103509. doi: 10.1016/j.ibmb.2020.103509 [DOI] [PubMed] [Google Scholar]
- 39.Wang M, Wang Y, Chang M, Wang X, Shi Z, Raikhel AS, et al. Ecdysone signaling mediates the trade-off between immunity and reproduction via suppression of amyloids in the mosquito Aedes aegypti. PLoS Pathogens. 2022;18(9):e1010837. doi: 10.1371/journal.ppat.1010837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wucher V, Legeai F, Hedan B, Rizk G, Lagoutte L, Leeb T, et al. FEELnc: a tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic acids research. 2017;45(8):e57–e. doi: 10.1093/nar/gkw1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Etebari K, Hegde S, Saldaña MA, Widen SG, Wood TG, Asgari S, et al. Global transcriptome analysis of Aedes aegypti mosquitoes in response to Zika virus infection. MSphere. 2017;2(6):e00456–17. doi: 10.1128/mSphere.00456-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akbari OS, Antoshechkin I, Amrhein H, Williams B, Diloreto R, Sandler J, et al. The developmental transcriptome of the mosquito Aedes aegypti, an invasive species and major arbovirus vector. G3: Genes, Genomes, Genetics. 2013;3(9):1493–509. doi: 10.1534/g3.113.006742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou H, Li S, Wu S, Jin P, Ma F. LncRNA-CR11538 decoys Dif/Dorsal to reduce antimicrobial peptide products for restoring Drosophila Toll immunity homeostasis. International Journal of Molecular Sciences. 2021;22(18):10117. doi: 10.3390/ijms221810117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou H, Ni J, Wu S, Ma F, Jin P, Li S. lncRNA-CR46018 positively regulates the Drosophila Toll immune response by interacting with Dif/Dorsal. Developmental & Comparative Immunology. 2021;124:104183. doi: 10.1016/j.dci.2021.104183 [DOI] [PubMed] [Google Scholar]
- 45.Lin S, Zhang SL, Yin HT, Zhao ZM, Chen ZK, Shen MM, et al. Cellular Lnc_209997 suppresses Bombyx mori nucleopolyhedrovirus replication by targeting miR-275-5p in B. mori. Insect Molecular Biology. 2022;31(3):308–16. doi: 10.1111/imb.12760 [DOI] [PubMed] [Google Scholar]
- 46.Li S, Hussain F, Unnithan GC, Dong S, UlAbdin Z, Gu S, et al. A long non-coding RNA regulates cadherin transcription and susceptibility to Bt toxin Cry1Ac in pink bollworm, Pectinophora gossypiella. Pesticide biochemistry and physiology. 2019;158:54–60. doi: 10.1016/j.pestbp.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 47.Vedururu RK, Neave MJ, Tachedjian M, Klein MJ, Gorry PR, Duchemin J-B, et al. Rnaseq analysis of aedes albopictus mosquito midguts after chikungunya virus infection. Viruses. 2019;11(6):513. doi: 10.3390/v11060513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azlan A, Obeidat SM, Theva Das K, Yunus MA, Azzam G. Genome-wide identification of Aedes albopictus long noncoding RNAs and their association with dengue and Zika virus infection. PLoS neglected tropical diseases. 2021;15(1):e0008351. doi: 10.1371/journal.pntd.0008351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nag DK, Dieme C, Lapierre P, Lasek-Nesselquist E, Kramer LD. RNA-Seq analysis of blood meal induced gene-expression changes in Aedes aegypti ovaries. BMC genomics. 2021;22(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Belavilas-Trovas A, Gregoriou M, Tastsoglou S, Soukia O, Giakountis A, Mathiopoulos K. A species-specific lncRNA modulates the reproductive ability of the asian tiger mosquito. Frontiers in Bioengineering and Biotechnology. 2022;10:885767-. doi: 10.3389/fbioe.2022.885767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell reports. 2015;11(7):1110–22. doi: 10.1016/j.celrep.2015.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gestuveo RJ, Royle J, Donald CL, Lamont DJ, Hutchinson EC, Merits A, et al. Analysis of Zika virus capsid-Aedes aegypti mosquito interactome reveals pro-viral host factors critical for establishing infection. Nature Communications. 2021;12(1):2766. doi: 10.1038/s41467-021-22966-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuadkitkan A, Wikan N, Fongsaran C, Smith DR. Identification and characterization of prohibitin as a receptor protein mediating DENV-2 entry into insect cells. Virology. 2010;406(1):149–61. doi: 10.1016/j.virol.2010.07.015 [DOI] [PubMed] [Google Scholar]
- 54.Hoffmann JA. The immune response of Drosophila. Nature. 2003;426(6962):33–8. doi: 10.1038/nature02021 [DOI] [PubMed] [Google Scholar]
- 55.Li S, Li Y, Shen L, Jin P, Chen L, Ma F. miR-958 inhibits Toll signaling and Drosomycin expression via direct targeting of Toll and Dif in Drosophila melanogaster. American Journal of Physiology-Cell Physiology. 2017;312(2):C103–C10. doi: 10.1152/ajpcell.00251.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li S, Xu J, Sun L, Li R, Jin P, Ma F. Drosophila miR-964 modulates Toll signaling pathway in response to bacterial infection. Developmental & Comparative Immunology. 2017;77:252–8. doi: 10.1016/j.dci.2017.08.008 [DOI] [PubMed] [Google Scholar]
- 57.Li R, Huang Y, Zhang Q, Zhou H, Jin P, Ma F. The miR-317 functions as a negative regulator of Toll immune response and influences Drosophila survival. Developmental & Comparative Immunology. 2019;95:19–27. doi: 10.1016/j.dci.2019.01.012 [DOI] [PubMed] [Google Scholar]
- 58.Schwenke RA, Lazzaro BP, Wolfner MF. Reproduction–immunity trade-offs in insects. Annual review of entomology. 2016;61:239–56. doi: 10.1146/annurev-ento-010715-023924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Scolari F, Siciliano P, Gabrieli P, Gomulski L, Bonomi A, Gasperi G, et al. Safe and fit genetically modified insects for pest control: from lab to field applications. Genetica. 2011;139(1):41–52. doi: 10.1007/s10709-010-9483-7 [DOI] [PubMed] [Google Scholar]
- 60.Marrelli MT, Moreira CK, Kelly D, Alphey L, Jacobs-Lorena M. Mosquito transgenesis: what is the fitness cost? Trends in parasitology. 2006;22(5):197–202. doi: 10.1016/j.pt.2006.03.004 [DOI] [PubMed] [Google Scholar]
- 61.Dong S, Dong Y, Simões ML, Dimopoulos G. Mosquito transgenesis for malaria control. Trends in parasitology. 2021. doi: 10.1016/j.pt.2021.08.001 [DOI] [PubMed] [Google Scholar]
- 62.Powell JR. Modifying mosquitoes to suppress disease transmission: Is the long wait over? Genetics. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, et al. BLAST+: architecture and applications. BMC bioinformatics. 2009;10(1):1–9. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wickham H, Chang W, Wickham MH. Package ‘ggplot2’. Create elegant data visualisations using the grammar of graphics Version. 2016;2(1):1–189.
- 65.Allaire J, Ellis P, Gandrud C, Kuo K, Lewis B, Owen J, et al. Package ‘networkD3’. D3 JavaScript network graphs from R. 2017. [Google Scholar]
- 66.Andrews S. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom; 2010. [Google Scholar]
- 67.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17(1):10–2. [Google Scholar]
- 68.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29(8):1035–43. doi: 10.1093/bioinformatics/btt087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Anders S, Huber W. Differential expression analysis for sequence count data. Nature Precedings. 2010:1-. doi: 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9(7):676–82. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression data were generated by Real-Time qPCR. Values were normalized with the ribosomal gene Rps7. Each sample included three biological replicates. Bars indicate mean value ± SEM. The significance was determined by Student’s t-test. *:P<0.05
(TIF)
Data include coding and non-coding transcripts that were either up- or down- regulated according to RNA seq data.
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.